Abstract
Caffeine, a methylxanthine analog of purine bases, is a compound that is largely consumed in beverages and medications for psychoactive and diuretic effects and plays many beneficial roles in neuronal stimulation and enhancement of anti-tumor immune responses by blocking adenosine receptors in higher organisms. In single-cell eukaryotes, however, caffeine somehow impairs cellular fitness by compromising cell wall integrity, inhibiting target of rapamycin (TOR) signaling and growth, and overriding cell cycle arrest caused by DNA damage. Among its multiple inhibitory targets, caffeine specifically interacts with phosphatidylinositol 3-kinase (PI3K)-related kinases causing radiosensitization and cytotoxicity via specialized intermediate molecules. Caffeine potentiates the lethality of cells in conjunction with several other stressors such as oxidants, irradiation, and various toxic compounds through largely unknown mechanisms. In this review, recent findings on caffeine effects and cellular detoxification schemes are highlighted and discussed with an emphasis on the inhibitory interactions between caffeine and its multiple targets in eukaryotic microorganisms such as budding and fission yeasts.
Keywords: Caffeine, growth inhibition, radiosensitization, DNA damage checkpoint, unicellular eukaryotes
Introduction
Caffeine (1,3,7-trimethylpurine-2,6-dione) is a naturally occurring purine derivative found in beverages such as coffee, tea, energy drinks, and several medications, as well as a myriad of dietary sources [1]. In higher animals, caffeine acts as a non-selective antagonist for A1 and A2A adenosine receptors in both heart and brain and has antidepressant and diuretic effects [2, 3]. Low doses of caffeine (< 65 mg) have been known to improve cognitive performance, working memory, and mood, whereas approximately 200 mg increases alertness, arousal, task accuracy, and energy in normal human populations [4, 5].
Caffeine is known to affect cell growth, proliferation, and energy metabolism by inhibiting the mammalian target of rapamycin (mTOR) signaling pathway [6]. Caffeine elicits pleiotropic physiological responses by triggering intracellular Ca2+ mobilization in various cell types [7]. Caffeine also inhibits DNA damage repair and perturbs cell cycle checkpoint function, which might lead to mutagenesis, apoptosis, and carcinogenesis [8]. Many earlier studies indicated that caffeine impairs cell cycle progression delays induced by chemicals or radiation, and enhances the toxicity of anti-cancer agents [9, 10]. Controversially, however, an accumulating body of evidence suggests that caffeine may both enhance and antagonize carcinogenic effects [11].
Caffeine elicits chemopreventive effects in mouse skin tumorigenesis models by inducing apoptosis [12]. Caffeine specifically suppresses epidermal growth factor (EGF)-induced malignant cell transformation and promotes human keratinocyte apoptosis with unrepaired DNA damage by blocking ultraviolet B (UVB)-induced phosphorylation of Chk1 and AKT, thereby preventing tumorigenesis. However, the mechanisms by which these effects are triggered remain largely unknown [13, 14]. Recently, it has been reported that caffeine enhances anti-tumor immune responses in mice by blocking the A2A receptor [15].
Several meta-analyses have reported that there is no significant association between caffeine intake and ovarian cancer risk [16]. Nonetheless, caffeine consumption is somehow inversely associated with the incidence of melanoma, liver and endometrial cancer, and type 2 diabetes in a dose-dependent manner [17-19]. Therefore, despite extensive analyses, the effects of caffeine on cell cycle progression and proliferation remain ambiguous.
Caffeine has also been reported to inhibit bacterial growth. Particularly, earlier studies demonstrated that caffeine impairs thymidine metabolism, thereby inhibiting DNA synthesis in Escherichia coli [20, 21]. At high doses, caffeine effectively kills E. coli and Salmonella enterica, albeit not as effectively as standard antibiotic ampicillin, and is more effective against Staphylococcus aureus and Enterobacter aerogenes [22-24]. Caffeine is a secondary metabolite produced by over one hundred plant species and possesses antibacterial activity against several plant pathogenic bacteria such as Ralstonia solanacearum, Clavibacter michiganensis, Xanthomonas campestris, and others. [25]. Caffeine alters cell morphology, substantially delays cell division time, and suppresses RNA biosynthesis [25]. Intriguingly, however, Pseudomonas aeruginosa and Pseudomonas putida can reportedly degrade caffeine to use it as a nutrient and carbon source [26].
Yeast, a relatively simple unicellular eukaryote, is also responsive to caffeine. Specifically, caffeine affects yeast cell growth, morphology, and a variety of cellular metabolic pathways to maintain homeostasis [27]. High concentrations of caffeine act as a cell wall antagonist triggering the mitogen-activated protein (MAP) kinase cascade for cell wall integrity signaling, and exert mutagenic effects leading to cell cycle alterations through the suppression of Tel1 and Mec1, two yeast homologs of mammalian ataxia-telangiectasia mutated (ATM) and ATM-related (ATR) kinases [28, 29].
Despite its use as a genotoxic agent over 40 years, the molecular mechanisms underlying the adverse effects of caffeine on cell proliferation and maintenance have not been elucidated. This review on the pleiotropic effects of caffeine on unicellular eukaryotes provides a comprehensive overview of how cells respond to stressful environmental conditions and toxic substances similar to caffeine via complex cellular processes, including cell signaling, cell cycle regulation, and damage checkpoint activation.
Caffeine Has Multiple Targets that Impair Diverse Cellular Mechanisms
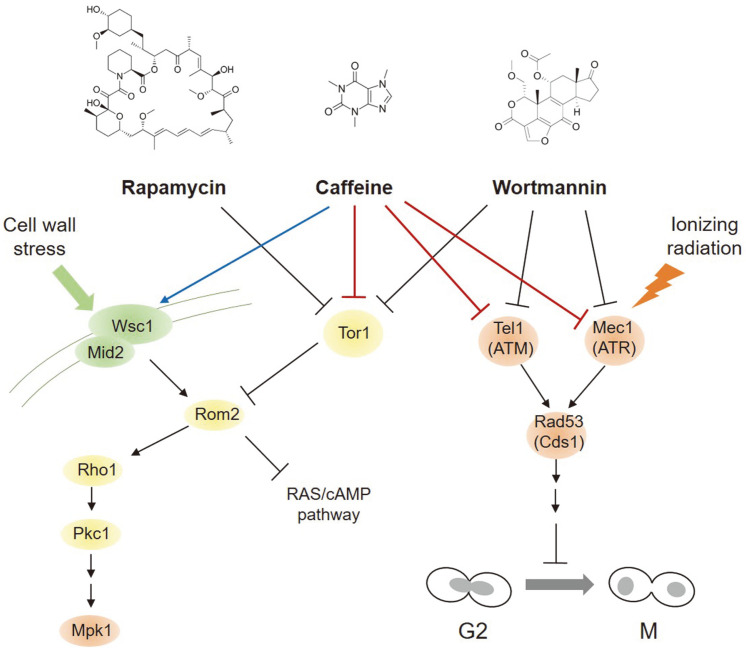
Caffeine is a relatively non-selective agent and affects multiple cellular processes related to cell growth and metabolism, mostly by acting as a low-affinity adenosine analog [30]. The physiological activity of caffeine has been reported to inhibit alkaline phosphatase, cAMP phosphodiesterase, and the nucleotide exchange activity of RCC1 (regulator of chromosome condensation) [31-34]. In yeasts and fungi, caffeine is categorized as a cell wall perturbing agent, such as Congo red and Calcofluor-white, as mutants that lack cell surface sensors or components of the MAP kinase cascade of the cell wall integrity pathway are sensitive to caffeine [35, 36]. Caffeine induces rapid phosphorylation of Mpk1, the downstream MAP kinase of the Pkc1-mediated cell wall integrity pathway in yeast, and Mpk1 phosphorylation by caffeine is abolished in Tor1 kinase-defective mutant cells (Fig. 1). Genetic and biochemical data from genome-wide transcriptome analyses have shown that caffeine activates a subset of cell wall-related genes through the Pkc1-Mpk1 cascade and inhibits the Ras/cAMP pathway through Tor1-mediated signaling [37, 38].
Fig. 1. Schematic diagram showing how caffeine and its related drugs affect Pkc1-Mpk1 kinase pathway, Tor1-mediated signaling, and Tel1/Mec1 (ATM/ATR in mammals)-mediated damage checkpoint responses by inhibiting PI3K-related protein kinases in S. cerevisiae cells.
Proteins mainly localized to cell wall and outer membrane are colored in green; cytoplasm in yellow; and nucleus in orange. G2; cell cycle phase Gap 2, M; mitosis.
TOR signaling is a well-known pathway for cellular homeostasis and growth, and caffeine exhibits a remarkably similar effect to that of rapamycin on the inhibition of TOR complex 1 (TORC1) and the ensuing alteration of global gene expression patterns in yeast [39]. Mutant cells lacking the genes encoding Tor1, Kog1, or Tco89, three non-essential TORC1-specific components, exhibit hypersensitivity to caffeine, suggesting that TORC1 is a specific caffeine target [39, 40]. Caffeine, wortmannin, and many other compounds with similar structures markedly inhibit the phosphorylation of mammalian TOR (mTOR)-dependent substrates both in vivo and in vitro [41]. Wanke et al. [42] proposed that caffeine extends the life span of yeast cells by targeting TORC1 and its downstream kinase cascade. TOR kinase belongs to the phosphatidylinositol 3-kinase-related kinase (PIKK) family, which commonly contains a c-terminus serine/threonine protein kinase domain similar to the catalytic domain of phosphatidylinositol 3-kinases (PI3Ks) [43]. This family includes ATM, ATR, and DNA-dependent protein kinase (DNA-PK), the catalytic activity of which can be inhibited to varying degrees by various xanthine alkaloids, including caffeine [44, 45].
Caffeine inhibits sugar transport by binding at the nucleotide-binding site of GLUT1, the primary facilitative glucose transporter in mammals [46]. GLUT1 is also allosterically inhibited by ATP, and AMP acts as a competitive antagonist of ATP-mediated glucose uptake inhibition. Interestingly, kinetic analyses have revealed that ATP can antagonize caffeine-mediated uncompetitive inhibition of glucose uptake, suggesting that caffeine and adenosine share structural similarities [47].
Caffeine also regulates calcium mobilization by inhibiting extracellular Ca2+ uptake in S. cerevisiae [48]. Caffeine directly binds to and inhibits voltage-gated Ca2+ channels, and may have indirect effects by inhibiting cAMP phosphodiesterase, resulting in PKA-dependent inhibition of Ca2+ channels [48, 49]. Lack of components in the calcineurin pathway also led to caffeine sensitivity in fission yeast, further suggesting that Ca2+ levels are regulated by caffeine [50].
Fcy2, a purine-cytosine permease, is thought to act as a non-specific transporter for caffeine uptake in S. cerevisiae, as intracellular caffeine accumulation is not observed in fcy2 mutant cells even when treated with high doses [51]. Due to its structural similarity to adenine, guanine, hypoxanthine, and cytosine, all of which are readily uptaken by cells, caffeine could affect several nucleic acid metabolic pathways including both DNA synthesis and degradation mechanisms involved in DNA replication and damage repair. In fact, caffeine reportedly alters cell cycle control by inducing mitosis even before DNA replication is completed in mammalian cells. Moreover, caffeine also potentiates the lethal effects of several genotoxic agents primarily due to inhibition of timely DNA repair [52]. For instance, synchronized BHK and CHO hamster fibroblasts arrested in the early S phase with hydroxyurea (HU) underwent premature mitotic events when exposed to caffeine [53, 54]. Additionally, caffeine was shown to override S-M checkpoint induction by inhibiting Cds1 activation and Chk1 phosphorylation in the fission yeast Schizosaccharomyces pombe [55].
Caffeine Acts as a DNA Damage-Sensitizing Agent
It has been reported in earlier studies that caffeine acts as a DNA damage repair inhibitor and reduces the duration of radiation-induced cell cycle arrest in the G2 phase [10, 56]. Although caffeine alone did not affect timely cell cycle progression, caffeine led to radiosensitization to X-rays and induced G2/M override in several mammalian cell lines, which was more pronounced in p53 null cells than their wild-type counterparts [57-59]. As p53-deficient cells fail to arrest at G1, irradiation-induced cell cycle control becomes completely dependent on G2 arrest, which is abrogated by caffeine-mediated activation of Cdk1 (also known as Cdc2), leading to sensitization to apoptosis [60]. Given that p53-deficient primary and tumor cells can be preferentially vulnerable to DNA damage-inducing reagents with caffeine exposure, caffeine could serve as a useful anticancer genotoxic adjunct in radiation therapy and chemotherapy [61].
Caffeine is believed to have multiple molecular targets and may possess an especially high affinity to protein kinases due to its chemical properties [58]. Predictably, caffeine treatment before irradiation, but not after irradiation, inhibited the radiation-mediated activation of Cds1 and its upstream kinase ATM in HeLa cells [62]. In vitro kinase assays also revealed that caffeine directly inhibits ATM kinase, not Cds1, which was consistent with the effects of wortmannin, a selective phosphoinositide 3-kinase (PI3K) inhibitor [62, 63]. In turn, inhibition of Cds1 phosphorylation by caffeine prevents the inactivation of Cdc25, leading to activation of Cdk1 and premature G2/M transition (Fig. 1).
This DNA damage-sensitizing effect of caffeine is associated with the inhibition of multiple components of the damage checkpoint signaling machinery. The activity of another major PI3K-related kinase, ATR, is also suppressed by caffeine in vivo as well as in vitro even in the absence of its substrate DNA molecules [53, 44, 64]. In contrast, in vitro kinase assays determined that DNA-PK and hChk1 are relatively resistant to caffeine-induced radiosensitivity [44]. Several other methylxanthine-derived drugs such as theobromine, theophylline, paraxanthine, and pentoxifylline also lead to cell radiosensitization at low concentrations [44].
Caffeine-Mediated Inhibition of PI3K-Related Protein Kinases
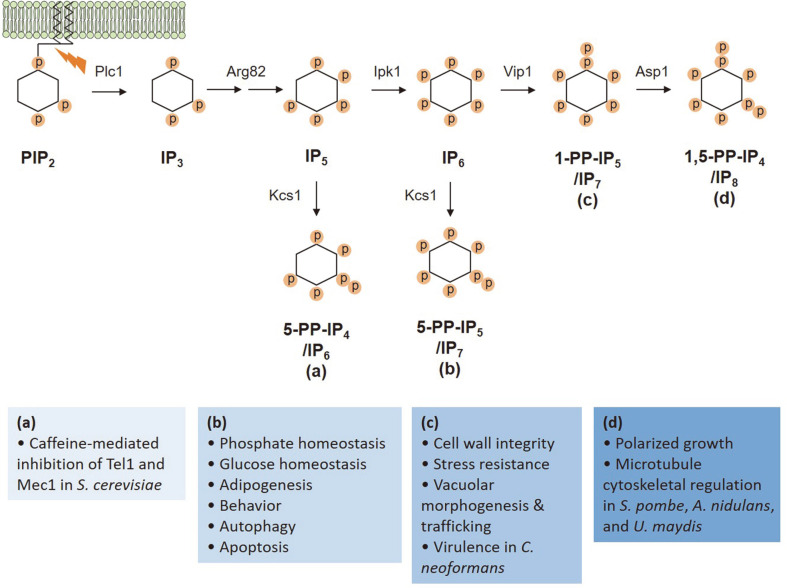
The lethal effects of caffeine and wortmannin via the inhibition of Mec1 and Tel1 kinase activity in S. cerevisiae are selectively blocked in mutants lacking inositol pyrophosphate (PP-IP) synthesis [29]. Inositol pyrophosphates are a specialized group of signaling molecules that are highly conserved from yeast to higher eukaryotes and regulate numerous energetic biological processes such as cell growth, environmental stress responses, pathogenicity, and autophagy (Fig. 2) [65-68]. The budding yeast kcs1 mutant, which is unable to produce high energy inositol pyrophosphates, displays slow growth, defective endocytic trafficking, and resistance to caffeine and wortmannin [69]. Moreover, inositol polyphosphate multikinase (encoded by ARG82)- or phospholipase C (PLC1)-deficient mutant strains are also resistant to the harmful effects of caffeine, whereas the ipk1 mutant, which lacks inositol polyphosphate kinase 1, is not caffeine resistant [29]. These observations suggest that the lethal effects of caffeine are mediated by a specific form of inositol pyrophosphate such as PP-IP4 (Fig. 2).
Fig. 2. Schematic model showing major steps in inositol pyrophosphate (PP-IP) metabolism and enzymes catalyzing each converting step in S. cerevisiae.
PP-IPs and their metabolizing enzymes regulate a variety of biological processes as shown in (a)~(d). Caffeine inhibition of Tel1 and Mec1 activity seems to be mediated by PP-IP4.
In order to respond to a wide variety of stimuli, the high-energy pyrophosphate group of inositol pyrophosphates might conceivably drive or inhibit phosphotransfer reactions even in a kinase-independent manner, thus affecting signaling via the PI3K-related protein kinase family [70]. Inositol pyrophosphates produced by mammalian IP6 kinase 1 impact insulin sensitivity and weight gain by inhibiting Akt, a serine/threonine-specific protein kinase [71]. The kcs1 yeast mutant displays longer telomeres, which is mainly regulated by PI3K-related protein kinase, Tel1 [29, 72]. Altogether, these results indicate that methylxanthine-induced inhibition of Mec1 and Tel1 might be involved in the suppression of their phosphotransferase activity mediated by the specific group of inositol pyrophosphates [44]. Intriguingly, however, Cryptococcus neoformans mutant strains lacking Ipk1 and/or Kcs1 show a significant growth defect in the presence of caffeine, which is inconsistent with the resistant phenotypes observed in S. cerevisiae [73].
In contrast, Cortez [74] reported that neither ATM nor ATR activity in vivo is inhibited by caffeine. Caffeine treatment abrogated ionizing radiation (IR)- or hydroxyurea (HU)-initiated G2/M checkpoint activation without any decrease in ATM- or ATR-dependent phosphorylation of CHK1 and CHK2 in human cell lines. In line with these observations, caffeine has also been found to intercalate into DNA molecules and prevent the binding of damage repair proteins, thereby interfering with DNA repair activities and promoting hyperactivation of ATM and ATR due to feedback mechanisms [52, 75].
Although checkpoint abrogation could provide a mechanistic explanation for caffeine-induced radiosensitization, relevant studies have only identified a relatively weak correlation between checkpoint disruption and caffeine-induced radiosensitization levels [76, 77]. Moreover, ataxia-telangiectasia (A-T) cells defective in ATM are still significantly radiosensitized by caffeine treatment, suggesting that other factors might mediate caffeine-induced cytotoxicity, including the inhibition of DNA repair systems [78].
Caffeine Inhibits DNA Damage Repair Pathways
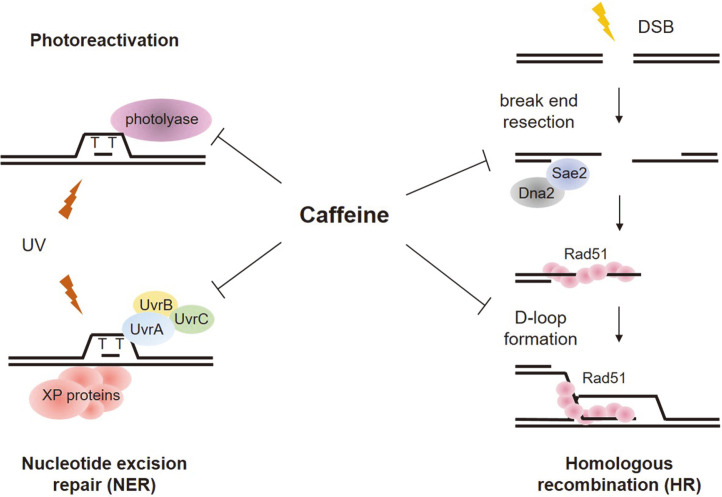
Among several DNA damage repair mechanisms, photoreactivation and nucleotide excision repair (NER) were reportedly inhibited by caffeine, whereas none of the tested major repair enzymes for base excision repair (BER) were affected [52, 79]. Caffeine inhibits photoreactivation by interfering with the binding of DNA photolyase to damaged DNA lesions (Fig. 3). Interestingly, however, NER-mediated repair is inhibited by caffeine because it promotes nonspecific binding of UvrA, the damage recognition subunit of ABC excinuclease in E. coli, to undamaged sites, thereby inhibiting proper nicking of damaged DNA (Fig. 3) [52]. The inhibitory effect of caffeine is also prominent in xeroderma pigmentosum (XP)-variant cells in humans, which can be reversed by the expression of DNA polymerase η, a crucial component for accurate translesion DNA synthesis through bulky DNA damage caused by UV radiation [80, 81]. Other DNA intercalating agents such as ethidium bromide and acridine orange also inhibit excinuclease activity through a similar mechanism, suggesting that caffeine inhibits DNA repair by intercalating into DNA and distorting its native helical structure [52]. This conclusion is consistent with the observation that BER enzymes, which are involved in small-scale base damage repair, are not largely affected by caffeine treatment.
Fig. 3. Inhibitory effects of caffeine on various DNA damage repair proteins.
Caffeine inhibits repair of pyrimidine dimers interfering with the binding of DNA photolyase and UvrA to damaged DNA lesions. Caffeine also inhibits several pivotal steps in HR pathway by removing Sae2 and Dna2 nucleases, and by promoting non-productive Rad51 nucleofilament formation.
The inhibitory effects of caffeine on homology-dependent damage repair have been previously documented in many organisms [82-84]. Several pivotal steps in homologous recombination (HR) for DSB repair are inhibited by caffeine. It has been observed in budding yeast and HeLa cells that caffeine impairs DSB-induced DNA end resection by rapid loss of Sae2 and Dna2, two nucleases that play important roles in early stages in the homologous recombination pathway (Fig. 3) [85]. The amount of functional Sae2 and Dna2 is reduced by caffeine-induced proteasomal degradation even in the absence of DNA damage, and this effect is independent of DNA damage checkpoint inhibition. It is also worth noting that autophagy can be stimulated in yeasts and mammalian cells by caffeine, as well as rapamycin and valproic acid (VPA), which are well-known autophagy-inducing agents [86-88].
Consistent with earlier studies on caffeine toxicity in recombination-dependent DNA repair, genome-wide screening of caffeine-sensitive mutants in the fission yeast has shown that rad3, ssb3, rad51, and rad54 mutants defective in homologous recombination are all sensitive to caffeine [50]. In fact, caffeine disturbs gene targeting by promoting non-productive Rad51 nucleofilament formation with random genomic regions (Fig. 3). As a result, caffeine repels the previously assembled Rad51 foci [89]. Caffeine interferes with Rad51-mediated strand exchange during DNA repair by homologous recombination (HR), whereas γH2AX and Exo1 remain activated at DNA breaks, which renders cells more sensitive to genome instability [90, 91]. Homologous recombination interference by low caffeine concentrations (2~4 mM) appears to be independent of its inhibition of Tel1- or Mec1-mediated damage checkpoint responses, suggesting that caffeine does not affect gene targeting by specific checkpoint inhibition but rather by different mechanisms. Moreover, DNA replication inhibition, another previously reported caffeine effect, was not observed upon caffeine treatment at similar concentrations as those mentioned above [90].
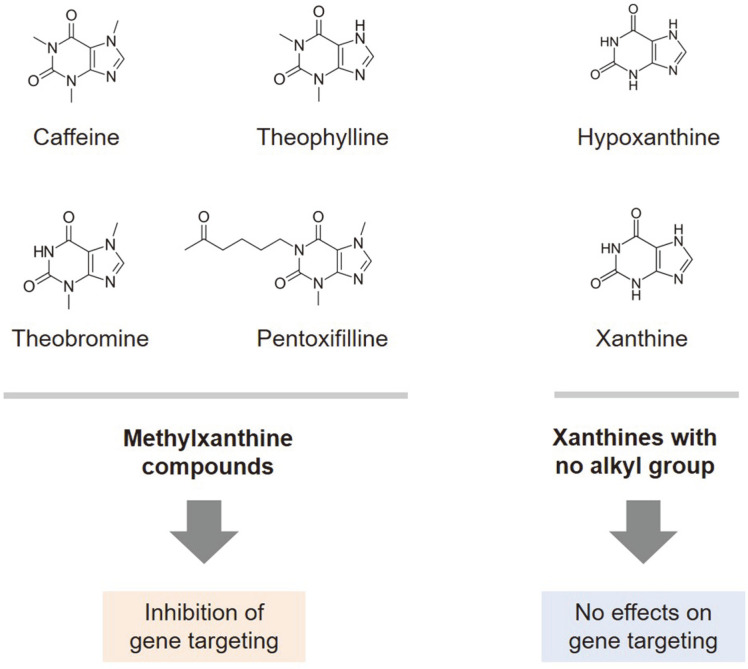
The aforementioned observations have led to speculation regarding the specificity of caffeine-mediated inhibition. Interestingly, some related methylxanthine compounds lacking only one methyl group compared to caffeine, such as theophylline, theobromine, and pentoxifylline, exhibit similar or even more severe inhibitory effects on homologous recombination than caffeine, which are consistent with the results of checkpoint activation inhibition experiments, whereas hypoxanthine and xanthine with no alkyl groups have almost no negative effects on gene targeting efficiency (Fig. 4) [90, 92]. Suppression of homologous recombination is caused by alkylxanthine-specific inhibition of D-loop formation, which has been demonstrated as not being due to intercalation-induced topological DNA structure change but to the direct formation of non-productive and homology-independent Rad51 nucleoprotein filaments [90].
Fig. 4. Effects of caffeine and its structurally-related compounds on gene targeting efficiency via Rad51-mediated homologous recombination.
Methylxanthines, but not xanthines with no alkyl groups, suppress Rad51-mediated D-loop formation and strand exchange.
Caffeine-induced radiosensitizing effects are significantly diminished in mutant cells deficient in XRCC2 and XRCC3, as well as in two RAD51 paralogs in mammals [78, 93]. Additionally, the strong inhibition of homology-directed repair by caffeine observed in wild-type cells was substantially reduced in mutant cells lacking XRCC2 and XRCC3. Moreover, these mutants show nearly intact damage checkpoint responses, i.e., timely G2 arrest and delayed S phase. Intriguingly, no significant effect of caffeine has been experimentally measured on non-homologous end joining (NHEJ) [78]. Together, these observations suggest that checkpoint disruption by itself is not sufficient for radiosensitization and the inhibition of homologous recombination proteins is likely an additional component of the caffeine radiosensitization mechanisms.
Integrated Intracellular Mechanisms Conferring Caffeine Tolerance
Screening for caffeine-resistance genes using a S. cerevisiae multicopy vector library revealed that two multidrug resistance (MDR) transporters, Snq2 and Pdr5, mediate caffeine efflux in yeast [94]. Snq2 is functionally homologous to Pdr5, which is one of the most abundant ATP-binding cassette (ABC) transporters involved in cellular detoxification in yeast; overexpression of either one of them is sufficient to confer resistance to caffeine although Snq2 is slightly more effective [94, 95]. Another set of studies showed that combinatorial deletions of the HSE1, RTS3, SDS23, and/or SDS24 genes, none of which are involved in caffeine efflux, render budding yeast cells hypersensitive to caffeine, indicating that caffeine tolerance is not solely attributed to its efflux from the cell [96]. In mammals, intracellular caffeine is metabolized and detoxified in the liver by the cytochrome P450 oxidase enzyme system; however, no specific caffeine transporter has been identified [97].
Several caffeine-resistant S. pombe mutant strains were isolated in previous studies via mutant screenings [98-100]. Interestingly, all of the identified mutants reacted to caffeine through Pap1, an AP1-like transcription factor in S. pombe. Hydrogen peroxide triggers nuclear accumulation of Pap1 for the maintenance of redox homeostasis [101]. Caffeine tolerance was observed in mutants lacking Crm1, the nuclear exporter of Pap1, as well as in mutants defective in Hba1, a cofactor of Crm1-mediated Pap1 export [98, 99, 102]. A loss-of-function mutation in trr1, which encodes thioredoxin reductase, induces constitutive oxidation and the constant activation of Pap1 leads to caffeine tolerance. Overexpression of Pap1 itself confers caffeine resistance, but interestingly, caffeine treatment does not enhance Pap1-mediated transcription [98]. These results suggest that Pap1 is responsible for transcriptional regulation of downstream target genes specifically involved in caffeine tolerance or detoxification. In fact, multicopy plasmid-based overproduction of Caf5, which encodes an ABC transporter, causes caffeine resistance and the expression of Caf5 is dependent on Pap1 [103].
Several ABC transporters that mediate multiple drug resistance are known to belong to oxidative stress-responsive regulons. Conversely, many toxic drugs could trigger the production of reactive oxygen species (ROS) in the cell. A genome-wide screening of caffeine-sensitive mutants in S. pombe revealed that Pap1 and another H2O2-induced MAP kinase, Sty1, were both required for caffeine tolerance. Moreover, Pap1-mediated caffeine tolerance is largely due to Hba2, a Pap1-dependent drug efflux pump [50]. However, only the overexpression of Pap1, but not Sty1, can allow a cell to overcome the toxic effects of caffeine, and intriguingly, only the Sty1 pathway, but not Pap1, is activated by caffeine treatment [50, 98]. It is therefore likely that these complex mechanistic changes are not specific to caffeine but might confer protection against a broad range of stressors.
Several caffeine-sensitive mutants are also defective in oxidative stress response mechanisms, and several mutants lacking important components for general stress response, cell wall integrity, vesicle-mediated protein trafficking, and genome stability are both sensitive to H2O2 and caffeine. This highlights the importance of characterizing the genetic and biochemical crosstalk between the molecular targets of caffeine and oxidative stress signaling pathways to understand the integration of cellular detoxification mechanisms [50]. Overexpression of Yap1, the Pap1 homolog in S. cerevisiae, also confers cell resistance to several toxic drugs such as diazaborine and cadmium, and this tolerance is dependent on Ycf1 and Flr1, two multidrug transporters, the expression of which is mainly controlled by Yap1 [104, 105].
Conclusion
Caffeine is a natural purine analog that has been widely used for studies of cellular response to external toxic materials due to its wide-ranging pleiotropic effects on cells including cell growth, DNA damage repair, and cell cycle regulation, as well as on changes in cell morphology and energy metabolism. Caffeine and other methylxanthine-derived drugs with structural similarities have multiple molecular targets in the cell but possess a particularly high affinity to PI3K-related kinases such as Mec1, Tel1, and Tor1. These inhibitory interactions leading to radiosensitization and cell toxicity presumably require the presence of a specialized group of signaling molecules including PP-IPs and/or properly organized interactions with critical factors involved in homology-directed repair such as Rad51-related nucleoproteins.
Growing evidence suggests that caffeine toxicity could be relieved mainly by several ABC transporters associated with multiple drug resistance, and interestingly, many of them are involved in oxidative stress response as well. Moreover, ROS-responsive transcription factors and a few relevant MAP kinases are genetically- and functionally-linked to caffeine tolerance. In line with these observations, it is strongly suggested that caffeine potentiates cell lethality in conjunction with several other exogenous or endogenous stimuli such as irradiation, toxic chemicals, ROS, and genomic infidelity. Therefore, cells need to be equipped with integrated detoxification mechanisms to maintain physiological homeostasis.
Taken together, the points discussed in this review present a novel framework for future studies to elaborate on the cytotoxic effects of caffeine and its mechanisms of action through its interactions with cell integrity pathways, and could also provide a better understanding of how cells react to caffeine in the context of holistic and pleiotropic stress responses.
Acknowledgments
The work for this review was supported by the Duksung Women’s University Research Grants 2018.
Footnotes
Conflict of Interests
The author has no financial conflicts of interest to declare.
REFERENCES
- 1.Wikoff D, Welsh BT, Henderson R, Brorby GP, Britt J, Myers E, et al. Systematic review of the potential adverse effects of caffeine consumption in healthy adults, pregnant women, adolescents, and children. Food Chem. Toxicol. 2017;109(Pt 1):585–648. doi: 10.1016/j.fct.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Fredholm BB, Bättig K, Holmén J, Nehlig A, Zvartau EE. Actions of caffeine in the brain with special reference to factors that contribute to its widespread use. Pharmacol. Rev. 1999;51:83–133. [PubMed] [Google Scholar]
- 3.López-Cruz L, Salamone JD, Correa M. Caffeine and selective adenosine receptor antagonists as new therapeutic tools for the motivational symptoms of depression. Front. Pharmacol. 2018;9:526. doi: 10.3389/fphar.2018.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malinauskas BM, Aeby VG, Overton RF, Carpenter-Aeby T, Barber-Heidal K. A survey of energy drink consumption patterns among college students. Nutr. J. 2007;6:35. doi: 10.1186/1475-2891-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith AP. Caffeine, extraversion and working memory. J. Psychopharmacol. 2013;27:71–76. doi: 10.1177/0269881112460111. [DOI] [PubMed] [Google Scholar]
- 6.Zhou H, Luo Y, Huang S. Updates of mTOR inhibitors. Anticancer Agents Med. Chem. 2010;10:571–581. doi: 10.2174/187152010793498663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Combettes L, Berthon B, Claret M. Caffeine inhibits cytosolic calcium oscillations induced by noradrenaline and vasopressin in rat hepatocytes. Biochem. J. 1994;301:737–744. doi: 10.1042/bj3010737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson LR, Philpott M. Nutrition and mutagenesis. Annu. Rev. Nutr. 2008;28:313–329. doi: 10.1146/annurev.nutr.28.061807.155449. [DOI] [PubMed] [Google Scholar]
- 9.Tolmach LJ, Jones RW, Busse PM. The action of caffeine on x-irradiated Hela cells. I. Delayed inhibition of DNA synthesis. Radiat. Res. 1977;71:653–665. doi: 10.2307/3574633. [DOI] [PubMed] [Google Scholar]
- 10.Busse PM, Bose SK, Jones RW, Tolmach LJ. The action of caffeine on X-irradiated HeLa cells III. Enhancement of X-rayinduced killing during G2 arrest. Radiat. Res. 1978;76:292–307. doi: 10.2307/3574780. [DOI] [PubMed] [Google Scholar]
- 11.Porta M, Vioque J, Ayude D, Alguacil J, Jariod M, Ruiz L, et al. Coffee drinking: the rationale for treating it as a potential effect modifier of carcinogenic exposures. Eur. J. Epidemiol. 2003;18:289–298. doi: 10.1023/A:1023700216945. [DOI] [PubMed] [Google Scholar]
- 12.Lu YP, Lou YR, Xie JG, Peng QY, Liao J, Yang CS, et al. Topical applications of caffeine or (-)-epigallocatechin gallate (EGCG) inhibit carcinogenesis and selectively increase apoptosis in UVB-induced skin tumors in mice. Proc. Natl. Acad. Sci. USA. 2002;99:12455–12460. doi: 10.1073/pnas.182429899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nomura M, Ichimatsu D, Moritani S, Koyama I, Dong Z, Yokogawa K, et al. Inhibition of epidermal growth factor-induced cell transformation and akt activation by caffeine. Mol. Carcinog. 2005;44:67–76. doi: 10.1002/mc.20120. [DOI] [PubMed] [Google Scholar]
- 14.Han W, Ming M, He YY. Caffeine promotes ultraviolet B-induced apoptosis in human keratinocytes without complete DNA repair. J. Biol. Chem. 2011;286:22825–22832. doi: 10.1074/jbc.M111.222349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkata Charan Tej GN, Neogi K, Verma SS, Chandra Gupta S, Nayak PK. Caffeine-enhanced anti-tumor immune response through decreased expression of PD1 on infiltrated cytotoxic T lymphocytes. Eur. J. Pharmacol. 2019;859:172538. doi: 10.1016/j.ejphar.2019.172538. [DOI] [PubMed] [Google Scholar]
- 16.Shafiei F, Salari-Moghaddam A, Milajerdi A, Larijani B, Esmaillzadeh A. Coffee and caffeine intake and risk of ovarian cancer: a systematic review and meta-analysis. Int. J. Gynecol. Cancer. 2019;29:579–584. doi: 10.1136/ijgc-2018-000102. [DOI] [PubMed] [Google Scholar]
- 17.Ding M, Bhupathiraju SN, Chen M, van Dam RM, Hu FB. Caffeinated and decaffeinated coffee consumption and risk of type 2 diabetes: a systematic review and a dose-response meta-analysis. Diabetes Care. 2014;37:569–586. doi: 10.2337/dc13-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Micek A, Godos J, Lafranconi A, Marranzano M, Pajak A. Caffeinated and decaffeinated coffee consumption and melanoma risk: a dose-response meta-analysis of prospective cohort studies. Int. J. Food. Sci. Nutr. 2018;69:417–426. doi: 10.1080/09637486.2017.1373752. [DOI] [PubMed] [Google Scholar]
- 19.Zhao LG, Li ZY, Feng GS, Ji XW, Tan YT, Li HL, et al. Coffee drinking and cancer risk: an umbrella review of meta-analyses of observational studies. BMC Cancer. 2020;20:101. doi: 10.1186/s12885-020-6561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandlie I, Solberg K, Kleppe K. The effect of caffeine on cell growth and metabolism of thymidine in Escherichia coli. Mutat. Res. 1980;73:29–41. doi: 10.1016/0027-5107(80)90133-5. [DOI] [PubMed] [Google Scholar]
- 21.Sandlie I, Lossius I, Sjåstad K, Kleppe K. Mechanism of caffeine-induced inhibition of DNA synthesis in Escherichia coli. FEBS Lett. 1983;151:237–242. doi: 10.1016/0014-5793(83)80077-5. [DOI] [PubMed] [Google Scholar]
- 22.Daglia M, Cuzzoni MT, Dacarro C. Antibacterial activity of coffees: Relationship between biological activity and chemical markers. J. Agric. Food Chem. 1994;42:2273–2277. doi: 10.1021/jf00046a036. [DOI] [Google Scholar]
- 23.Almeida AAP, Farah A, Silva DAM, Nunan EA, Glória MBA. Antibacterial activity of coffee extracts and selected coffee chemical compounds against enterobacteria. J. Agric. Food. Chem. 2006;54:8738–8743. doi: 10.1021/jf0617317. [DOI] [PubMed] [Google Scholar]
- 24.Al-Janabi AAHS. Potential activity of the purine compounds caffeine and aminophylline on bacteria. J. Glob. Infect. Dis. 2011;3:133–137. doi: 10.4103/0974-777X.81689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sledz W, Los E, Paczek A, Rischka J, Motyka A, Zoledowska S, et al. Antibacterial activity of caffeine against plant pathogenic bacteria. Acta Biochim. Pol. 2015;62:605–612. doi: 10.18388/abp.2015_1092. [DOI] [PubMed] [Google Scholar]
- 26.Dash SS, Gummadi, S Inhibitory effect of caffeine on growth of various bacterial strains. Res. J. Microbiol. 2008;3:457–465. doi: 10.3923/jm.2008.457.465. [DOI] [Google Scholar]
- 27.Ruta LL, Farcasanu IC. Saccharomyces cerevisiae and caffeine implications on the eukaryotic cell. Nutrients. 2020;12:2440. doi: 10.3390/nu12082440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubois E, Scherens B, Vierendeels F, Ho MM, Messenguy F, Shears SB. In Saccharomyces cerevisiae, the inositol polyphosphate kinase activity of Kcs1p is required for resistance to salt stress, cell wall integrity, and vacuolar morphogenesis. J. Biol. Chem. 2002;277:23755–23763. doi: 10.1074/jbc.M202206200. [DOI] [PubMed] [Google Scholar]
- 29.Saiardi A, Resnick AC, Snowman AM, Wendland B, Snyder SH. Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc. Natl. Acad. Sci. USA. 2005;102:1911–1914. doi: 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bode AM, Dong Z. The enigmatic effects of caffeine in cell cycle and cancer. Cancer Lett. 2007;247:26–39. doi: 10.1016/j.canlet.2006.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beavo JA, Rogers NL, Crofford OB, Hardman JG, Sutherland EW, Newman EV. Effects of xanthine derivatives on lipolysis and on adenosine 3',5'-monophosphate phosphodiesterase activity. Mol. Pharmacol. 1970;6:597–603. [PubMed] [Google Scholar]
- 32.Wharton W, Goz B. Induction of alkaline phosphatase activity in HeLa cells. Inhibition by xanthine derivatives and thermostability studies. Biochem. Pharmacol. 1979;28:763–768. doi: 10.1016/0006-2952(79)90356-3. [DOI] [PubMed] [Google Scholar]
- 33.Wells JN, Miller JR. Methylxanthine inhibitors of phosphodiesterases. Methods Enzymol. 1988;159:489–496. doi: 10.1016/0076-6879(88)59048-1. [DOI] [PubMed] [Google Scholar]
- 34.Nishijima H, Nishitani H, Saito N, Nishimoto T. Caffeine mimics adenine and 2'-deoxyadenosine, both of which inhibit the guanine-nucleotide exchange activity of RCC1 and the kinase activity of ATR. Genes Cells. 2003;8:423–435. doi: 10.1046/j.1365-2443.2003.00644.x. [DOI] [PubMed] [Google Scholar]
- 35.Jacoby JJ, Nilius SM, Heinisch JJ. A screen for upstream components of the yeast protein kinase C signal transduction pathway identifies the product of the SLG1 gene. Mol. Gen. Genet. 1998;258:148–155. doi: 10.1007/s004380050717. [DOI] [PubMed] [Google Scholar]
- 36.Martin H, Rodriguez-Pachon JM, Ruiz C, Nombela C, Molina M. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:1511–1519. doi: 10.1074/jbc.275.2.1511. [DOI] [PubMed] [Google Scholar]
- 37.Kuranda K, Leberre V, Sokol S, Palamarczyk G, Francois J. Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol. Microbiol. 2006;61:1147–1166. doi: 10.1111/j.1365-2958.2006.05300.x. [DOI] [PubMed] [Google Scholar]
- 38.Kim H, Thak EJ, Yeon JY, Sohn MJ, Choo JH, Kim JY, et al. Functional analysis of Mpk1-mediated cell wall integrity signaling pathway in the thermotolerant methylotrophic yeast Hansenula polymorpha. J. Microbiol. 2018;56:72–82. doi: 10.1007/s12275-018-7508-6. [DOI] [PubMed] [Google Scholar]
- 39.Reinke A, Chen JC, Aronova S, Powers T. Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J. Biol. Chem. 2006;281:31616–31626. doi: 10.1074/jbc.M603107200. [DOI] [PubMed] [Google Scholar]
- 40.Lum PY, Armour CD, Stepaniants SB, Cavet G, Wolf MK, Butler JS, et al. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell. 2004;116:121–137. doi: 10.1016/S0092-8674(03)01035-3. [DOI] [PubMed] [Google Scholar]
- 41.McMahon LP, Yue W, Santen RJ, Lawrence JC., Jr Farnesylthiosalicylic acid inhibits mammalian target of rapamycin (mTOR) activity both in cells and in vitro by promoting dissociation of the mTOR-raptor complex. Mol. Endocrinol. 2005;19:175–183. doi: 10.1210/me.2004-0305. [DOI] [PubMed] [Google Scholar]
- 42.Wanke V, Cameroni E, Uotila A, Piccolis M, Urban J, Loewith R, et al. Caffeine extends yeast lifespan by targeting TORC1. Mol. Microbiol. 2008;69:277–285. doi: 10.1111/j.1365-2958.2008.06292.x. [DOI] [PubMed] [Google Scholar]
- 43.Wullschleger S, Loewith R, Hall MN. TOR Signaling in growth and mtabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 44.Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, et al. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59:4375–4382. [PubMed] [Google Scholar]
- 45.Block WD, Merkle D, Meek K, Lees-Miller SP. Selective inhibition of the DNA-dependent protein kinase (DNA-PK) by the radiosensitizing agent caffeine. Nucleic Acids Res. 2004;32:1967–1972. doi: 10.1093/nar/gkh508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sage JM, Cura AJ, Lloyd KP, Carruthers A. Caffeine inhibits glucose transport by binding at the GLUT1 nucleotide-binding site. Am. J. Physiol. Cell Physiol. 2015;308:C827–834. doi: 10.1152/ajpcell.00001.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carruthers A, Helgerson AL. The human erythrocyte sugar transporter is also a nucleotide binding protein. Biochemistry. 1989;28:8337–8346. doi: 10.1021/bi00447a011. [DOI] [PubMed] [Google Scholar]
- 48.Courchesne WE, Ozturk S. Amiodarone induces a caffeine-inhibited, MID1-depedent rise in free cytoplasmic calcium in Saccharomyces cerevisiae. Mol. Microbiol. 2003;47:223–234. doi: 10.1046/j.1365-2958.2003.03291.x. [DOI] [PubMed] [Google Scholar]
- 49.Islam MS, Larsson O, Nilsson T, Berggren PO. Effects of caffeine on cytoplasmic free Ca2+ concentration in pancreatic betacells are mediated by interaction with ATP-sensitive K+ channels and L-type voltage-gated Ca2+ channels but not the ryanodine receptor. Biochem. J. 1995;306:679–686. doi: 10.1042/bj3060679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calvo IA, Gabrielli N, Iglesias-Baena I, García-Santamarina S, Hoe KL, Kim DU, et al. Genome-wide screen of genes required for caffeine tolerance in fission yeast. PLoS One. 4:e6619. doi: 10.1371/journal.pone.0006619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi Z, Xiong L. Characterization of a purine permease family gene OsPUP7 involved in growth and development control in rice. J. Integr. Plant. Biol. 2013;55:1119–1135. doi: 10.1111/jipb.12101. [DOI] [PubMed] [Google Scholar]
- 52.Selby CP, Sancar A. Molecular mechanisms of DNA repair inhibition by caffeine. Proc. Natl. Acad. Sci. USA. 1990;87:3522–3525. doi: 10.1073/pnas.87.9.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlegel R, Pardee AB. Caffeine-induced uncoupling of mitosis from the completion of DNA replication in mammalian cells. Science. 1986;232:1264–1266. doi: 10.1126/science.2422760. [DOI] [PubMed] [Google Scholar]
- 54.Downes CS, Musk SR, Watson JV, Johnson RT. Caffeine overcomes a restriction point associated with DNA replication, but does not accelerate mitosis. J. Cell Biol. 1990;110:1855–1859. doi: 10.1083/jcb.110.6.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moser BA, Brondello JM, Baber-Furnari B, Russell P. Mechanism of caffeine-induced checkpoint override in fission yeast. Mol. Cell. Biol. 2000;20:4288–4294. doi: 10.1128/MCB.20.12.4288-4294.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rowley R, Zorch M, Leeper DB. Effect of caffeine on radiation-induced mitotic delay: Delayed expression of G2 arrest. Radiat. Res. 1984;97:178–185. doi: 10.2307/3576199. [DOI] [PubMed] [Google Scholar]
- 57.Kirillova, TV, Rozanov, Iu M, Seregina, TB, Spivak, IM The Effect of caffeine on the duration of the mitotic phase cycle in CHO-K1 Chinese hamster cells, irradiated with X-rays. Tsitologiia. 1989;31:476–483. [PubMed] [Google Scholar]
- 58.Jung T, Streffer C. Effects of caffeine on protein phosphorylation and cell cycle progression in X-irradiated two-cell mouse embryos. Int. J. Radiat. Biol. 1992;62:161–168. doi: 10.1080/09553009214551971. [DOI] [PubMed] [Google Scholar]
- 59.Powell SN, DeFrank JS, Connell P, Eogan M, Preffer F, Dombkowski D, et a1. Differential sensitivity of p53(-) and p53(+) cells to caffeine-induced radiosensitization and override of G2 delay. Cancer Res. 1995;55:1643–1648. doi: 10.1016/0360-3016(95)97825-L. [DOI] [PubMed] [Google Scholar]
- 60.Yao SL, Akhtar AJ, McKenna KA, Bedi GC, Sidransky D, Mabry M, et al. Selective radiosensitization of p53-deficient cells by caffeine-mediated activation of p34cdc2 kinase. Nat. Med. 1996;2:1140–1143. doi: 10.1038/nm1096-1140. [DOI] [PubMed] [Google Scholar]
- 61.Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 62.Blasina A, Price BD, Turenne GA, McGowan CH. Caffeine inhibits the checkpoint kinase ATM. Curr. Biol. 1999;9:1135–1138. doi: 10.1016/S0960-9822(99)80486-2. [DOI] [PubMed] [Google Scholar]
- 63.Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 1998;58:4375–4382. [PubMed] [Google Scholar]
- 64.Hall-Jackson CA, Cross DA, Morrice N, Smythe C. ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene. 1999;18:6707–6713. doi: 10.1038/sj.onc.1203077. [DOI] [PubMed] [Google Scholar]
- 65.Taylor R Jr, Chen PH, Chou CC, Patel J, Jin SV. KCS1 deletion in Saccharomyces cerevisiae leads to a defect in translocation of autophagic proteins and reduces autophagosome formation. Autophagy. 2012;8:1300–1311. doi: 10.4161/auto.20681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Worley J, Luo X, Capaldi AP. Inositol pyrophosphates regulate cell growth and the environmental stress response by activating the HDAC Rpd3L. Cell Rep. 2013;3:1476–1482. doi: 10.1016/j.celrep.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shears SB. Intimate connections: Inositol pyrophosphates at the interface of metabolic regulation and cell signaling. J. Cell. Physiol. 2018;233:1897–1912. doi: 10.1002/jcp.26017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lev S, Li C, Desmarini D, Sorrell TC, Saiardi A, Djordjevic JT. Fungal kinases with a sweet tooth: Pleiotropic roles of their phosphorylated inositol sugar products in the pathogenicity of Cryptococcus neoformans present novel drug targeting opportunities. Front. Cell Infect. Microbiol. 2019;9:248. doi: 10.3389/fcimb.2019.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saiardi A, Sciambi C, McCaffery JM, Wendland B, Snyder SH. Inositol pyrophosphates regulate endocytic trafficking. Proc. Natl. Acad. Sci. USA. 2002;99:14206–14211. doi: 10.1073/pnas.212527899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burton A, Hu X, Saiardi A. Are inositol pyrophosphates signalling molecules? J. Cell. Physiol. 2009;220:8–15. doi: 10.1002/jcp.21763. [DOI] [PubMed] [Google Scholar]
- 71.Chakraborty A, Koldobskiy MA, Bello NT, Maxwell M, Potter JJ, Juluri KR, et al. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143:897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.York SJ, Armbruster BN, Greenwell P, Petes TD, York JD. Inositol diphosphate signaling regulates telomere length. J. Biol. Chem. 2005;280:4264–4269. doi: 10.1074/jbc.M412070200. [DOI] [PubMed] [Google Scholar]
- 73.Li C, Lev S, Saiardi A, Desmarini D, Sorrell TC, Djordjevic JT. Identification of a major IP5 kinase in Cryptococcus neoformans confirms that PP-IP5/IP7, not IP6, is essential for virulence. Sci. Rep. 2016;6:23927. doi: 10.1038/srep23927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cortez D. Caffeine inhibits checkpoint responses without inhibiting the ataxia-telangiectasia-mutated (ATM) and ATM- and Rad3-related (ATR) protein kinases. J. Biol. Chem. 2003;278:37139–37145. doi: 10.1074/jbc.M307088200. [DOI] [PubMed] [Google Scholar]
- 75.Tornaletti S, Russo P, Parodi S, Pedrini AM. Studies on DNA binding of caffeine and derivatives: evidence of intercalation by DNA-unwinding experiments. Biochim. Biophys. Acta. 1989;1007:112–115. doi: 10.1016/0167-4781(89)90138-3. [DOI] [PubMed] [Google Scholar]
- 76.Harvey AN, Savage JR. A case of caffeine-mediated cancellation of mitotic delay without enhanced breakage in V79 cells. Mutat. Res. 1994;304:203–209. doi: 10.1016/0027-5107(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 77.Ribeiro JC, Barnetson AR, Jackson P, Ow K, Links M, Russell PJ. Caffeine-increased radiosensitivity is not dependent on a loss of G2/M arrest or apoptosis in bladder cancer cell lines. Int. J. Radiat. Biol. 1999;75:481–492. doi: 10.1080/095530099140410. [DOI] [PubMed] [Google Scholar]
- 78.Asaad NA, Zeng ZC, Guan J, Thacker J, Iliakis G. Homologous recombination as a potential target for caffeine radiosensitization in mammalian cells: reduced caffeine radiosensitization in XRCC2 and XRCC3 mutants. Oncogene. 2000;19:5788–5800. doi: 10.1038/sj.onc.1203953. [DOI] [PubMed] [Google Scholar]
- 79.Kane CM, Linn S. Purification and characterization of an apurinic/apyrimidinic endonuclease from HeLa cells. J. Biol. Chem. 1981;256:3405–3414. doi: 10.1016/S0021-9258(19)69623-7. [DOI] [PubMed] [Google Scholar]
- 80.Lehmann AR, Kirk-Bell S, Arlett CF, Harcourt SA, de Weerd-Kastelein EA, Keijzer W, Hall-Smith P. Repair of ultraviolet light damage in a variety of human fibroblast cell strains. Cancer Res. 1977;37:904–910. [PubMed] [Google Scholar]
- 81.Kaufmann WK, Heffernan TP, Beaulieu LM, Doherty S, Frank AR, Zhou Y, et al. Caffeine and human DNA metabolism: the magic and the mystery. Mutat. Res. 2003;532:85–102. doi: 10.1016/j.mrfmmm.2003.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grigg GW. Effects of coumarin, pyronin Y, 6,9-dimethyl 2-methylthiopurine and caffeine on excision repair and recombination repair in Escherichia coli. J. Gen. Microbiol. 1972;70:221–230. doi: 10.1099/00221287-70-2-221. [DOI] [PubMed] [Google Scholar]
- 83.Loprieno N, Barale R, Baroncelli S. Genetic effects of caffeine. Mutat. Res. 1974;26:83–87. doi: 10.1016/S0027-5107(74)80038-2. [DOI] [PubMed] [Google Scholar]
- 84.Yefremova GI, Filippova LM. Effect of caffeine on crossing-over in Drosophila melanogaster. Mutat. Res. 1974;23:347–352. doi: 10.1016/0027-5107(74)90108-0. [DOI] [PubMed] [Google Scholar]
- 85.Tsabar M, Eapen VV, Mason JM, Memisoglu G, Waterman DP, Long MJ, et al. Caffeine impairs resection during DNA break repair by reducing the levels of nucleases Sae2 and Dna2. Nucleic Acids Res. 2015;43:6889–6901. doi: 10.1093/nar/gkv520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Winter G, Hazan R, Bakalinsky AT, Abeliovich H. Caffeine induces macroautophagy and confers a cytocidal effect on food spoilage yeast in combination with benzoic acid. Autophagy. 2008;4:28–36. doi: 10.4161/auto.5127. [DOI] [PubMed] [Google Scholar]
- 87.Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011;471:74–79. doi: 10.1038/nature09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saiki S, Sasazawa Y, Imamichi Y, Kawajiri S, Fujimaki T, Tanida I, et al. Caffeine induces apoptosis by enhancement of autophagy via PI3K/Akt/mTOR/p70S6K inhibition. Autophagy. 2011;7:176–187. doi: 10.4161/auto.7.2.14074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsabar M, Mason JM, Chan YL, Bishop DK, Haber JE. Caffeine inhibits gene conversion by displacing Rad51 from ssDNA. Nucleic Acids Res. 2015;43:6902–6918. doi: 10.1093/nar/gkv525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zelensky AN, Sanchez H, Ristic D, Vidic I, van Rossum-Fikkert SE, Essers J, et al. Caffeine suppresses homologous recombination through interference with RAD51-mediated joint molecule formation. Nucleic Acids Res. 2013;41:6475–6489. doi: 10.1093/nar/gkt375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Choi EH, Yoon S, Park KS, Kim KP. The homologous recombination machinery orchestrates post-replication DNA repair during self-renewal of mouse embryonic stem cells. Sci. Rep. 2017;7:11610. doi: 10.1038/s41598-017-11951-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kimler BF, Leeper DB, Snyder MH, Rowley R, Schneiderman MH. Modification of radiation-induced division delay by caffeine analogues and dibutyryl cyclic AMP. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1982;41:47–58. doi: 10.1080/09553008214550041. [DOI] [PubMed] [Google Scholar]
- 93.Wang H, Boecker W, Wang H, Wang X, Guan J, Thompson LH, et al. Caffeine inhibits homology-directed repair of I-SceIinduced DNA double-strand breaks. Oncogene. 2004;23:824–834. doi: 10.1038/sj.onc.1207168. [DOI] [PubMed] [Google Scholar]
- 94.Tsujimoto Y, Shimizu Y, Otake K, Nakamura T, Okada R, Miyazaki T, et al. Multidrug resistance transporters Snq2p and Pdr5p mediate caffeine efflux in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 2015;79:1103–1110. doi: 10.1080/09168451.2015.1010476. [DOI] [PubMed] [Google Scholar]
- 95.Prasad R, Goffeau A. Yeast ATP-binding cassette transporters conferring multidrug resistance. Annu. Rev. Microbiol. 2012;66:39–63. doi: 10.1146/annurev-micro-092611-150111. [DOI] [PubMed] [Google Scholar]
- 96.Hood-DeGrenier JK. Identification of phosphatase 2A-like Sit4-mediated signalling and ubiquitin-dependent protein sorting as modulators of caffeine sensitivity in S. cerevisiae. Yeast. 2011;28:189–204. doi: 10.1002/yea.1830. [DOI] [PubMed] [Google Scholar]
- 97.Kot M, Daniel WA. Caffeine as a marker substrate for testing cytochrome P450 activity in human and rat. Pharmacol. Rep. 2008;60:789–797. [PubMed] [Google Scholar]
- 98.Kumada K, Yanagida M, Toda T. Caffeine-resistance in fission yeast is caused by mutations in a single essential gene, crm1+ Mol. Gen. Genet. 1996;250:59–68. doi: 10.1007/BF02191825. [DOI] [PubMed] [Google Scholar]
- 99.Benko Z, Miklos I, Carr AM, Sipiczki M. Caffeine-resistance in S. pombe: mutations in three novel caf genes increase caffeine tolerance and affect radiation sensitivity, fertility, and cell cycle. Curr. Genet. 1997;31:481–487. doi: 10.1007/s002940050233. [DOI] [PubMed] [Google Scholar]
- 100.Kuramae EE, Robert V, Snel B, Boekhout T. Conflicting phylogenetic position of Schizosaccharomyces pombe. Genomics. 2006;88:387–393. doi: 10.1016/j.ygeno.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 101.Vivancos AP, Castillo EA, Jones N, Ayté J, Hidalgo E. Activation of the redox sensor Pap1 by hydrogen peroxide requires modulation of the intracellular oxidant concentration. Mol. Microbiol. 2004;52:1427–1435. doi: 10.1111/j.1365-2958.2004.04065.x. [DOI] [PubMed] [Google Scholar]
- 102.Toda T, Shimanuki M, Saka Y, Yamano H, Adachi Y, Shirakawa M, et al. Fission yeast pap1-dependent transcription is negatively regulated by an essential nuclear protein, crm1. Mol. Cell. Biol. 1992;12:5474–5484. doi: 10.1128/MCB.12.12.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Benko Z, Fenyvesvolgyi C, Pesti M, Sipiczki M. The transcription factor Pap1/Caf3 plays a central role in the determination of caffeine resistance in Schizosaccharomyces pombe. Mol. Genet. Genomics. 2004;271:161–170. doi: 10.1007/s00438-003-0967-3. [DOI] [PubMed] [Google Scholar]
- 104.Wemmie JA, Szczypka MS, Thiele DJ, Moye-Rowley WS. Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J. Biol. Chem. 1994;269:32592–32597. doi: 10.1016/S0021-9258(18)31675-2. [DOI] [PubMed] [Google Scholar]
- 105.Jungwirth H, Wendler F, Platzer B, Bergler H, Högenauer G. Diazaborine resistance in yeast involves the efflux pumps Ycf1p and Flr1p and is enhanced by a gain-of-function allele of gene YAP1. Eur. J. Biochem. 2000;267:4809–4816. doi: 10.1046/j.1432-1327.2000.01537.x. [DOI] [PubMed] [Google Scholar]