Abstract
Dental pathogens lead to chronic diseases like periodontitis, which causes loss of teeth. Here, we examined the plausible antibacterial efficacy of copper nanoparticles (CuNPs) synthesized using Cupressus macrocarpa extract (CME) against periodontitis-causing bacteria. The antimicrobial properties of CME-CuNPs were then assessed against oral microbes (M. luteus. B. subtilis, P. aerioginosa) that cause periodontal disease and were identified using morphological/ biochemical analysis, and 16S-rRNA techniques. The CME-CuNPs were characterized, and accordingly, the peak found at 577 nm using UV–Vis spectrometer showed the formation of stable CME-CuNPs. Also, the results revealed the formation of spherical and oblong monodispersed CME-CuNPs with sizes ranged from 11.3 to 22.4 nm. The FTIR analysis suggested that the CME contains reducing agents that consequently had a role in Cu reduction and CME-CuNP formation. Furthermore, the CME-CuNPs exhibited potent antimicrobial efficacy against different isolates which was superior to the reported values in literature. The antibacterial efficacy of CME-CuNPs on oral bacteria was compared to the synergistic solution of clindamycin with CME-CuNPs. The solution exhibited a superior capacity to prevent bacterial growth. Minimum inhibitory concentration (MIC), minimum bactericidal concentration (MBC), and fractional inhibitory concentration (FIC) of CME-CuNPs with clindamycin recorded against the selected periodontal disease-causing microorganisms were observed between the range of 2.6–3.6 μg/ml, 4-5 μg/ml and 0.312-0.5, respectively. Finally, the synergistic antimicrobial efficacy exhibited by CME-CuNPs with clindamycin against the tested strains could be useful for the future development of more effective treatments to control dental diseases.
Keywords: Antibiotic, antimicrobial activity, copper nanoparticles, periodontal disease
Introduction
As of 2017, it was estimated that about 3.5 billion people have dental diseases such as tooth caries and periodontitis infections in their permanent teeth [1]. Periodontal disease is one of the major public health problems in many countries [2, 3]. It presents as a chronic, inflammatory, and infectious disease that affects the teeth and surrounding structures [4, 5], causing in the most severe cases, excessive gingival bleeding and inflammation, increased dental mobility, and finally, tooth loss [6]. Studies have shown that disturbance of microbiota may activate P. aeruginosa, E. coli, S. pyogenes, and B. cereus [7-9]. In the periodontal site of chronic patients, high distribution of B. cereus, S. pyogenes, P. aeruginosa, and E. coli commonly occurs [10]. Research has also suggested periodontal disease as the most common oral infectious disease that is predominated by microorganisms [11]. Also, bacterial resistance in biofilms to anti-microbial treatment can be up to 1,000 times greater than that of planktonic organisms [12] which presents a major global health problem [13] and causes tooth loss [14, 15]. Clindamycin is an antibiotic that is usually advised as effective therapy because of its excellent coverage of typical oral flora [16]. Currently, microbial resistance to conventional antibiotics is considered an important health problem and has raised the demand for more effective solutions [17]. The discovery of novel, strategic and effective antimicrobial agents to prevent microbial infections is needed [18] to prevent plaque formation and maturation as the first line of therapy [19]. The study of synergistic effects of metal nanoparticles with antibiotics and their antimicrobial effect on periodontal disease-causing microorganisms is an important aim.
Copper nanoparticles (CuNPs) exhibit high antimicrobial activity against different species of microorganisms including fungi, gram-negative and gram-positive bacteria; however, there have been no reports on their activity against dental pathogens. In antimicrobial therapies, the use of CuNPs would require and depend on therapeutic time and concentration [20]. Eco-friendly materials such as microbes, plants, and enzymes are used for the biosynthesis of CuNPs because they are nontoxic materials as well as being simple to use and low cost [21-23].
In this study, we adopted dental prophylaxis as our objective and sought to synthesize antibacterial nano copper using an herbal formulation of CME as a reducing agent against periodontal infectious disease-causing microorganisms such as gram-positive B. subtilis, M. luteus, in addition to gram-negative P. aeruginosa. We also evaluated the antimicrobial efficiency of combining antibiotics and biosynthesized CuNPs to overcome antibiotic resistance.
Materials and Methods
Preparation of Cupressus macrocarpa Aqueous Extract
Cupressus macrocarpa plant was identified by Prof. Ahmed A. El-Settawy (Head of Forestry and Wood Technology Department at the Faculty of Agriculture, Alexandria University) and given the voucher number Zidan313 [24]. Aqueous extract of Cupressus macrocarpa leaves was prepared by taking 25 g of leaves, rinsing it in a glass beaker containing deionized water (100 ml), and heating at 85°C for 30 min and then filtering and storing it for subsequent use [25].
Biosynthesis of CuNPs
Forty milliliters of copper acetate (Sigma-Aldrich, USA) and then 10 ml of CME were added in an Erlenmeyer flask. Then, 10 ml of the CME was added. The mixture was irradiated in a microwave oven (Matsushita Electric Industrial Co., Ltd. Panasonic, Japan) at 800 W for 1 h until the formation of copper nanoparticles (CME-CuNPs), and allowed to cool at room temperature [26].
Characterization of CME-CuNPs
Characterization of CME-CuNPs was done using the following methods:
UV-Visible spectrum: UV–vis spectrometer (Shimadzu UV-1650) was used to record absorbance in the range of 300–700 nm and monitor the rate of CME-CuNP formation [27].
Transmission electron microscopy (TEM): TEM analysis was done using the TEM JEOL 100 kV (Assiut Electron Microscope Unit). The CME-CuNPs were prepared by placing a drop of the suspension on carbon-coated copper grids and allowing it to dry on the grid for 4 min. The shape and size of CME-CuNPs were determined from the TEM micrograph [28].
X-Ray Diffraction Analysis
The nature and size of the CME-CuNPs were determined using Shimadzu XRD (Shimadzu XD-3A, Japan). The nanoparticle size was calculated using the formula of Debye–Scherrer [29].
Fourier-transform infrared analysis (FTIR): The spectra of CME and CME-CuNPs were determined using a Shimadzu IR-470 Spectrometer (Shimadzu) in the range of 500–4,000 cm-1[30].
Isolation and Identification of Bacterial Isolates
Ethical approval for the current study (Ref. No. 41- 1107-00152) was provided by the Ethics Committee of Research, Taif University, Taif, KSA. Bacterial isolates were obtained from the Faculty of Dentistry Medical Diagnosis and Infection Control Unit, Taif University, Taif, KSA. Isolation of bacteria collected from the mouths of patients (dental pulp, and dental plaque) was conducted by provisional researchers from patients infected with periodontal disease-causing microorganisms using sterile root canal instruments and the isolates were stored in sterile saline in a 10 ml saline kit. One hundred isolates were isolated on Mueller-Hinton agar (Thermo Scientific Oxoid), for 24 h at 37°C under aerobic condition and suspected single colonies were then identified by morphological/biochemical tests according to the criteria [31, 32].
Molecular Characterization Using 16S rRNA Gene
Bacterial cells were used for collecting genomic DNA with the Wizard Genomic DNA Purification Kit (Promega, USA) and used as a PCR template. In 20 μl of the polymerase chain reaction (PCR) reaction solution containing forward primer 27F and reverse primer 1492R, one microliter of DNA template was added. Then, 35 amplification cycles of 16S rRNA genes were performed at 94°C for 45 s in the denaturation step, at 55°C for 60 s in the annealing step, and then at 72°C for 60 s in the extension step. DNA fragments were amplified up to 1,400 bp. The products of 16S rRNA of approximately 1,400 bp were sequenced using 518F/800R primer set. The sequencing of 16S rRNA has been analyzed on an automated Applied BioSystems, DNA sequencing system,(model 3730XL, USA). The similarity of bacterial strains was aligned using CLUSTAL W (1.81) phylogenetic tree obtained from the nucleotide sequence databases.
Antibacterial Activity of CME-CuNPs
Micro-Dilution Test for Determination of MIC and MBC
The MBC of CME-CuNPs, clindamycin, or both is defined as the lowest concentration of an antimicrobial agent killing the majority (99.99%) of bacterial inoculums. Since the MIC of CME-CuNPs, clindamycin, or both relates to their inhibitory ability, it is possible that if the antimicrobial agent were removed, the bacteria would begin to grow again [33]. The test was performed with serial dilutions of clindamycin (from 64 μg/ml to 8 μg/ml), CME-CuNPs (from 27 μg/ml to 9 μg/ml) or clindamycin with CME-CuNPs (from 5 μg/ml to 2.6 μg/ml) arranged across the rows in Mueller-Hinton broth and inoculation of the wells of a micro-dilution plate with the bacterial culture. The MIC and MBC of CME-CuNPs, clindamycin, or both were determined against the tested isolates [34]. All samples of clindamycin, CME-CuNPs, or CME-CuNPs with clindamycin (1 μg/ml:1 μg/ml) were tested in triplicate, and the test was repeated five separate times.
Antibacterial Synergy Test
The antibacterial synergy in vitro test was carried out using a checkerboard synergy method based on the value of the FIC (fractional inhibitory concentration). The FIC value was calculated by comparing the MIC of each agent CME-CuNP and clindamycin with the MIC of the CME-CuNP and clindamycin combination. Clindamycin was added in concentrations between 64 and 8 μg/ml and CME-CuNPs in a range of 10–30 μg/ml. These effects can be quantified by the application of mathematical expressions: the fractional inhibitory concentration (FIC) of combinations among CME-CuNPs and clindamycin against tested strains. For two antibacterial agents, CME-CuNPs (A) and clindamycin (B) acting individually or in combination:
FICA=MIC (A in the combination of B)/MIC (A alone)
FICB=MIC (B in the combination of A)/MIC (B alone)
FIC= FICA + FICB
An FIC index of < 0.5 indicates synergism, > 0.5–1 indicates additive effects, > 1 to < 2 indifference, and ≥ 2 is considered to be antagonism [34].
Statistical Analysis
Descriptive summary statistics were obtained for all independent and outcome variables. The mean difference was tested using analysis of variance (ANOVA) followed by Tukey's post hoc and t-test. The analysis was obtained using the Statistical Package for Social Science version 17 (SPSS INC Chicago link). All statistical tests were two-sided, and the significance level was set at p < 0.05.
Results
Biosynthesis of CME-CuNPs
In the current research, CME-CuNPs were biosynthesized by reducing copper acetate to CME-CuNPs using CME. The formation of CME-CuNPs in the reaction mixture is indicated by a change in the color from blue to brown.
Characterization of CME-CuNPs
UV–Vis spectroscopy of CME-CuNPs: UV–Vis absorption was studied after the formation of CME-CuNPs was dispersed and absorbance was measured by using a UV–visible spectrophotometer between 450 and 800 nm. CME-CuNPs showed peaks at 577 nm as shown in Fig. 1. The UV–visible peak at 577 nm was for pure CME-CuNPs.
Fig. 1. (A) Illustration showing biosynthesis of CME-CuNPs. Notes: Photograph showing the formation of CME-CuNPs using Cupressus macrocarpa extract. (B) UV–visible spectrum of CME-CuNPs.
Transmission Electron Microscope Analysis
The CME-CuNPs were characterized by TEM analysis (Fig. 2). The average size of the CME-CuNPs was estimated to be at 11.3-22.4 nm, via TEM analysis. TEM analysis of the CME-CuNPs showed spherical to oblong, polydispersed shapes (Fig. 2).
Fig. 2. TEM images of the synthesized CME-CuNPs.

X-Ray Diffraction (XRD)
X-ray diffraction (XRD) pattern showed four major peaks at 2θ values of 47°, 54°, and 72°. These characteristic peaks could be attributed to reflection planes (111), (200), and (220) of the face-centered cubic crystalline (FCC) structure of pure metallic copper (Fig. 3).
Fig. 3. XRD patterns of the synthesized CME-CuNPs with aqueous extract of Cupressus macrocarpa.
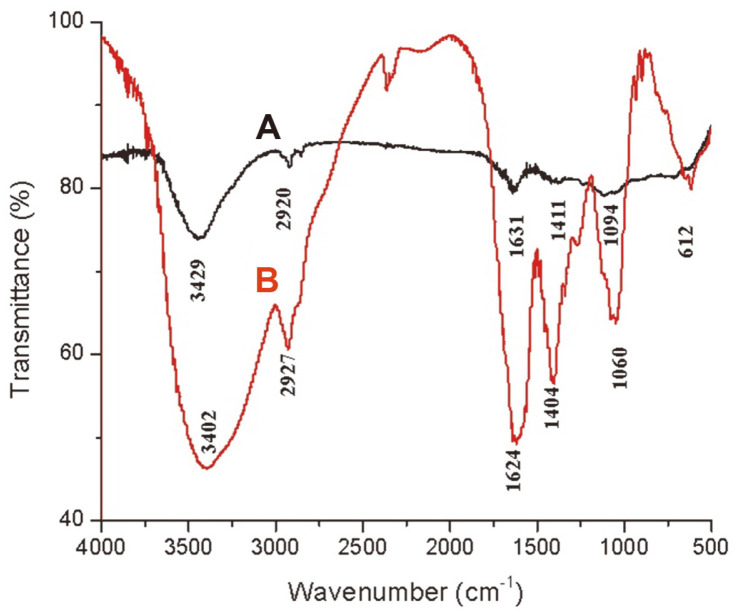
Comparison of FTIR Spectra of Cupressus macrocarpa Extract, CME-CuNPs
Fourier-transform infrared analysis (FTIR) was used to characterize the CME and the resulting CME-CuNPs. Absorbance bands were observed in the region 500 to 4,000 cm-1, (Figs. 4A and 4B). Fig. 4A showed the absorption bands of plant extract. There is a broad and strong band in the region 3,429 cm−1 ascribed to O–H stretching vibrations. According to a known standard, a band in the 2,920 cm−1 region originates from C–H (hydrocarbon) stretching vibration, 1,631 cm-1, denoting an amine group related to proteins, 1,411 cm-1, representing methylene -CH bending bond, and 1,094 cm-1 of primary alcohol/C-O- stretching. The CME caused a reduction of copper ions. Fig. 4B showed the absorption bands of CME-CuNPs. Peaks at 3,402, 2,927, 1,631, 1,404, and 1,060 cm-1 represent the OH functional groups, an amine group (NH), stretching -CH, and C-O stretching vibrations, respectively. Polyphenolics, proteins, amino acids, and carbohydrate compounds were adsorbed on the surface of the CME-CuNPs.
Fig. 4. FTIR spectra of Cupressus macrocarpa extract (A) and the synthesized CME-CuNPs (B).
Isolation and Identification of Drug-Resistant Bacteria
One hundred isolates were collected on Mueller-Hinton agar at 37°C. Then, the suspected single colonies were identified as M. luteus (50%), B. subtilis (12.5%), and P. aeruginosa (12.5%) by morphological/biochemical tests according to the criteria.
The strains were morphologically and biochemically characterized as M. luteus, B. subtilis, and P. aeruginosa and designated as M. luteus MIC1, B. subtilis BAC1, and P. aeruginosa PSE1, respectively. The aerobic strains MIC1, ABC1, and PSE5 were chosen for the current study (Table 1).
Table 1.
Morphological and biochemical characteristics of bacteria strains (MIC1, BAC1 and PSE5).
(-): Negative, (+): Positive
16S rRNA Analysis
Partial 16S rRNA gene sequences were used to identify multidrug-resistant bacterial isolates. MIC1, BAC1, and PSE5 were carried and sequenced. The 16S rRNA gene sequences of the bacterial isolates from the dental pulp and plaque were deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases with the following accession numbers (Table 2): MIC1 strain (LC628029), BAC1 strain (LC628030), and PSE5 strain (LC628031). The 16S rRNA gene sequences of strains were found to be 1310, 1240, and 1312 nt in length, for strain MIC1, BAC1, and PSE5, respectively.
Table 2.
The bacterial isolates (MIC1, BAC1 and PSE5) of oral infections.
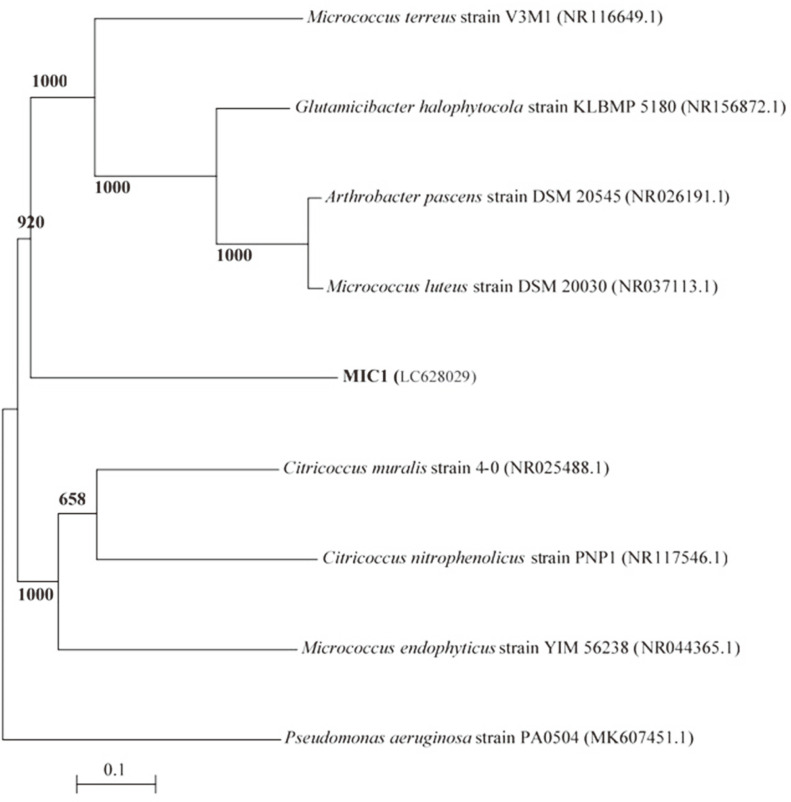
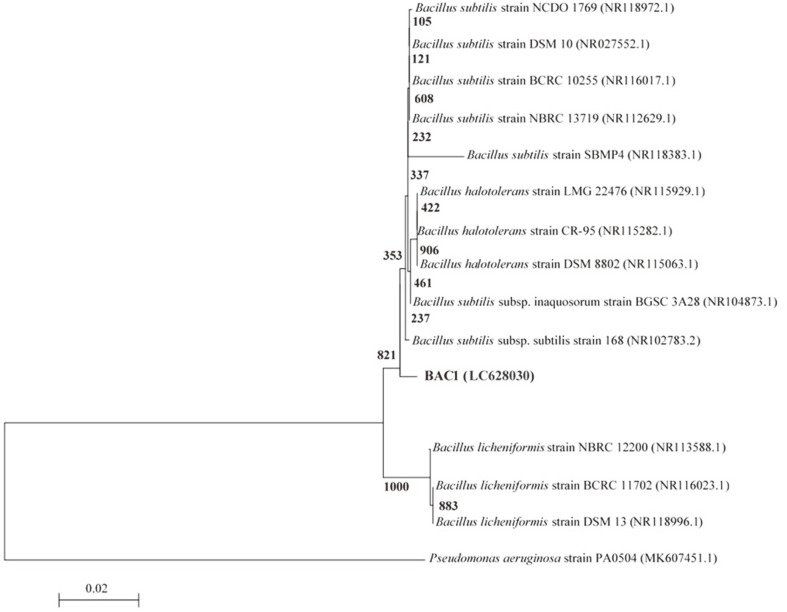
A dendrogram demonstrating the results of the 16S rRNA of M. luteus (MIC1), B. subtilis (BAC1), and P. aeruginosa (PSE5) analysis is displayed in Figs. 5A, 5B, 5C. The results show the highest similarity of isolates MIC1, BAC1, and PSE5 to members of the Micrococcus, Bacillus, and Pseudomonas group, respectively. Moreover, the 16S rRNA sequences of the isolates MIC1, BAC1, and PSE5 are the most closely associated with M. luteus, B. subtilis, and P. aeruginosa (Table 2). The 16S rRNA gene of MIC1, BAC1, and PSE5 displayed 98.31%, 96.43%, and 97.22 similarities to Micrococcus luteus DSM 20030 (NR037113.1), Bacillus subtilis subsp. subtilis 168 (NR102783.2), and Pseudomonas aeruginosa ATCC 10145 (NR114471.1), respectively.
Fig. 5A. A phylogenetic tree of bacterial isolates relied on the nucleotide sequences of 16S rRNA genes, constructed by neighbor-joining method.
The scale bar displays the genetic distance. The number presented next to each node displays the percentage bootstrap value of 1000 replicates. The Pseudomonas aeruginosa strain PA0504 was treated as the out-group. The GenBank accession numbers of the bacteria are presented in parentheses.
Fig. 5B. A phylogenetic tree of bacterial isolates relied on the nucleotide sequences of 16S rRNA genes, constructed by neighbor-joining method.
The scale bar displays the genetic distance. The number presented next to each node displays the percentage bootstrap value of 1000 replicates. The Pseudomonas aeruginosa strain PA0504 was treated as the out-group. The GenBank accession numbers of the bacteria are presented in parentheses.
Fig. 5C. A phylogenetic tree of bacterial isolates relied on the nucleotide sequences of 16S rRNA genes, constructed by neighbor-joining method.
The scale bar displays the genetic distance. The number presented next to each node displays the percentage bootstrap value of 1000 replicates. The Bacillus subtilis strain BCRC 10255 was treated as the out-group. The GenBank accession numbers of the bacteria are presented in parentheses.
Antimicrobial Activity
Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) Assays
In this study, three strains resistant to clindamycin were used. The MIC of CME-CuNPs was found to be 13.5, 18, and 18 μg/ml against M. luteus, B. subtilis, and P. aeruginosa, respectively (Table 3), while the MIC of CME-CuNPs with clindamycin was found to be 2.6, 3.6, and 3.6 μg/ml, respectively. Moreover, the MBC of CME-CuNPs was determined to be 20, 27, and 27 μg/ml for M. luteus, B. subtilis, and P. aeruginosa, respectively, while the MBC of CME-CuNPs with clindamycin was determined to be 4, 5, and 5 μg/ml, respectively. The MICs of CME-CuNPs with clindamycin significantly decreased by more than 5 folds compared to CME-CuNPs alone while it decreased by more than 3-10 folds compared to clindamycin alone. Our data showed that CME-CuNPs have a small size with high antimicrobial activity. As shown in Table 4, MICs of CME-CuNPs were the lowest concentration (13.5–18 μg/ml) compared to MICs of CuNPs in earlier studies (40 mg/ml, 75 μg/ml, 100 μg/ml, and 150-225 μg/ml) on antibacterial activity against bacteria.
Table 3.
MIC, MBC and of CME-CuNPs and CME-CuNPs with clindamycin (μg/ml) against oral bacteria in Mueller Hinton broth.
| Isolate | Clindamycin (μg/ml) | CME-CuNPs (μg/ml) | CME-CuNPs with clindamycin (μg/ml) | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| MIC | MBC | MBC/MIC | MIC | MBC | MBC/MIC | MIC | MBC | MBC/MIC | |
| M. luteus MIC1 | 8 | 16 | 2 | 13.5 | 20 | 1.5 | 2.6 | 4 | 1.5 |
| B. subtilis BAC1 | 32 | 64 | 2 | 18 | 27 | 1.5 | 3.6 | 5 | 1.4 |
| P. aeruginosa PSE1 | 32 | 64 | 2 | 18 | 27 | 1.5 | 3.6 | 5 | 1.4 |
MIC: Minimum inhibitory concentration, MBC: Minimum bactericidal concentration.
Table 4.
Comparison of antibacterial activity for CME-CuNPs with earlier studies.
| Materials | Size (nm) | Concentrations (μg/ml) | Bacteria | References |
|---|---|---|---|---|
| CuNPs | 55 - 350 nm | 40 mg/disc | Staphylococcus aureus (S. aureus), Escherichia coli (E. coli) | [50] |
| CuNPs | 4.7 to 17.4 nm | 100 μg/ml | Bacillus, S. aureus, E. coli, and P. aeruginosa | [51] |
| CuNPs | 60–90 nm | 75 μg/ml | Streptococcus sp. E. coli | [52] |
| CuNPs | 131 nm | 40 μg/ml | Streptococcus mutans | [53] |
| CuNPs | 80 nm | 150 -225 μg/ml | Enterococcus faecalis | [54] |
| CME-CuNPs | 11.3-22.4 nm | 13.5-18 μg/ml | B. subtilis, M. luteus, P. aeruginosa | Present study |
Antibacterial Synergy Test
The FIC of CME-CuNPs and the clindamycin combination were investigated and are summarized in Table 5. This experiment showed that CME-CuNPs acted synergistically (p < 0.5) with clindamycin against B. subtilis, M. luteus, in addition to P. aeruginosa.
Table 5.
FIC index of combinations among CME-CuNPs and clindamycin against tested strains.
| Bacterial isolates | FIC index | |
|---|---|---|
| MIC1 | 0.500 | (S) |
| BAC1 | 0.312 | (S) |
| PSE5 | 0.312 | (S) |
An FIC index of < 0.5 indicates synergism (S), > 0.5–1 indicates additive effects (AD), > 1 to < 2 indifference (ID), and ≥ 2 is considered to be antagonism (AN).
Mechanism of Action
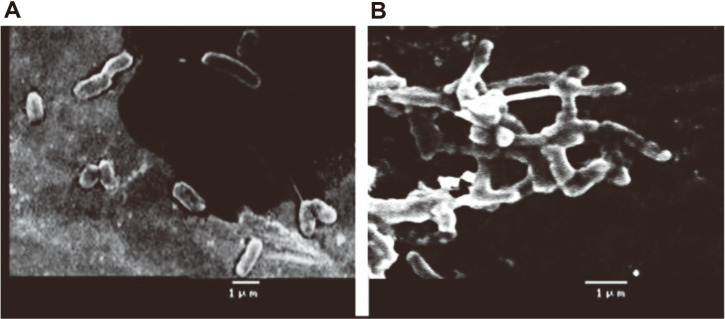
The CME-CuNP treated B. subtilis was observed with distorted membrane morphology, and cell elongation,(Fig. 6B) while the untreated control B. subtilis cells were found to be compact, tightly packed cells (Fig. 6A). In our results, the bacteria cells were distorted and CME-CuNPs act as an inhibitor for selected bacteria.
Fig. 6.
SEM observation of control B. subtilis (a) and images shows the CME-CuNP treated B. subtilis (b) showing membrane damage, and cell elongation in treated cells.
Discussion
In this study, we applied a combination of CME-CuNPs and clindamycin. When CME-CuNPs with clindamycin antibacterial properties were compared with CME-CuNPs alone against tested bacteria, CME-CuNPs with clindamycin proved to be stronger. Based on our results, the present study confirmed the success of the biosynthesis of CME-CuNPs from CME. The CME-CuNPs biosynthesis reaction progress was monitored for color change and UV-Vis spectrometric analysis. The change of color from blue to brown in the reaction mixture forming CME-CuNPs agreed with Wu et al. (2020) [35] who similarly observed the color change during the synthesis of CuNPs. The absorbance peaks recorded at 577 nm are specific to CME-CuNPs and were attributed to the surface plasmon resonance phenomenon which indicates the formation of CME-CuNPs. Various studies have also recorded similar CuNPs SPR peaks [26, 36, 37]. TEM images of nano copper: the appearance is spherical to oblong with a size of about 11.3-22.4 nm [38]. There are three lattice planes that are (111), (200), and (220) of the face-centered cubic (fcc) structure that was observed for CME-CuNPs at De Bragg’s reflection angles of 47°, 54°, and 72°, respectively. No other impurity peak was detected in the CME-CuNP sample. The XRD results reflected the purity of CME-CuNPs in the present study and this is in accordance with previous studies [39, 40].
The organic compounds in the CME reduced and stabilized the CME-CuNPs, thereby preventing agglomeration. The CME contains FTIR peaks at 3,402, 2,927, 1,631, 1,404, and 1,060 cm-1 representing the OH functional groups, an amine group (NH), stretching -CH, and C-O stretching vibrations, respectively. These functional groups of the CME related to polyphenolics, proteins, amino acids, and carbohydrate compounds were adsorbed on the surface of CME-CuNPs [41], and these components are functionalized in reducing and stabilizing the CME-CuNPs. Moreover, CME contains secondary metabolite components such as phenolics, flavonoids, saponins, tannins, and terpenes. For this reason, we used CME in the biosynthesis of CME-CuNPs. Also, CME has wound-healing, antibacterial, and anti-inflammatory properties [42].
Our results show that MIC1, BAC1, and PSE5 are compatible with the conclusions of the morphological and biochemical characterization. Various studies have demonstrated that M. luteus, B. subtilis, and P. aeruginosa can be isolated from dental plaque and dental pulp [43, 44].
The present study's antimicrobial results document the high potential of CME-CuNPs with clindamycin against periodontal disease-causing microorganisms (M. luteus, B subtilis, and P. aeruginosa) compared to CME-CuNPs or clindamycin alone. Our results demonstrated the greater synergistic effect of the solution containing CME-CuNPs and clindamycin and anti-periodontal disease agents against tested clinical strains, because of its low MIC values. The antibacterial efficacy of some nanoparticles such as CME-CuNPs have been evaluated in previous studies and different mechanisms have been proposed for their effects [45]. The antibacterial efficacy of CME-CuNPs was related to its capability to distort the bacterial cell and cause multiple effects of reactive oxygen species (ROS) formation, and liberation of Cu ions, which lead to lipid peroxidation, protein oxidation, and DNA degradation in bacterial cells [46]. The inhibition of growth in selected bacteria and ROS formation was caused by electrostatic attraction between negatively charged bacterial cells and positively charged nanoparticles [47].
Moreover, clindamycin has a synergy effect on the antibacterial efficacy of the CME-CuNPs on tested bacteria. The reaction between clindamycin and CME-CuNPs led to synergism. The clindamycin molecule contains active groups like hydroxyl and amide, which react easily with CME-CuNPs by chelation [48]. The rationale for using drug combinations (CME-CuNPs with clindamycin) is the expectation that effective combinations might lower the incidence of bacterial resistance, reduce the host toxicity of the antimicrobial agents or enhance bactericidal activity [44, 49].
In conclusion, the CME-CuNPs prepared by biological synthesis using CME are more secure, cost-effective, and eco-friendly. The biological synthesis approach for CME-CuNPs has many advantages, such as being simple to use with commercial viability. The synergistic solution of clindamycin with CME-CuNPs displayed more significant antimicrobial activity compared to clindamycin or CME-CuNPs alone against selected bacteria, such as M. luteus, B. subtilis, and P. aeruginosa. Therefore, the obtained eco-friendly biosynthesized CME-CuNPs can be used for their enhanced efficacy in bio-dental fields for exploiting the existing antibiotics and other drugs and will result in the development of cost-effective treatments for infections and periodontal diseases in the future.
Acknowledgments
We would like to express our gratitude to Deanship of Scientific Research, Taif University, Taif, Saudi Arabia for financial support under the research project number (1/439/6084). This study was approved by the Research Ethics Committee of Taif University, Taif, Saudi Arabia (No. 41- 1107-00152). The authors declare that the funding bodies had no role in the design of the study, the collection, analysis, and interpretation of data, or in writing the manuscript.
Footnotes
Funding
This work was funded by Deanship of Scientific Research, Taif University (research project No. 1/439/6084), Taif, Saudi Arabia.
Conflict of Interest
The authors have no financial conflicts of interest to declare.
REFERENCES
- 1.Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 392:1789–8583. doi: 10.1016/S0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dias HB, Carrera ET, Bortolatto JF, De Andrade MF, De Souza Rastelli AN. LED and low-level laser therapy association in tooth bleaching using a novel low concentration H2O2/N-doped TiO2 bleaching agent. Laser Phys. 2016;26:015602. doi: 10.1088/1054-660X/26/1/015602. [DOI] [Google Scholar]
- 3.Petersen PE, Ogawa H. The global burden of periodontal disease: Towards integration with chronic disease prevention and control. Periodontol. 2000. 2012;60:15–39. doi: 10.1111/j.1600-0757.2011.00425.x. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Wu G, Feng Z, Dong Y, Zhou W, Li B, et al. Advanced biomaterials and their potential applications in the treatment of periodontal disease. Crit. Rev. Biotechnol. 2016;36:760–775. doi: 10.3109/07388551.2015.1035693. [DOI] [PubMed] [Google Scholar]
- 5.Osorio R, Alfonso-Rodríguez CA, Medina-Castillo AL, Alaminos M, Toledano M. Bioactive polymeric nanoparticles for periodontal therapy. PLoS One. 2016;11:e0166217. doi: 10.1371/journal.pone.0166217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knight ET, Liu J, Seymour GJ, Faggion CM, Jr, Cullinan MP. Risk factors that may modify the innate and adaptive immune responses in periodontal diseases. Periodontol. 2000. 2016;71:22–51. doi: 10.1111/prd.12110. [DOI] [PubMed] [Google Scholar]
- 7.Souto R, Silva-Boghossian CM, Colombo APV. Prevalence of Pseudomonas aeruginosa and Acinetobacter spp. in subgingival biofilm and saliva of subjects with chronic periodontal infection. Braz. J. Microbiol. 2014;45:495–501. doi: 10.1590/S1517-83822014000200017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viciani E, Montagnani F, Tordini G, Romano A, Salerni L, De Luca A, et al. Prevalence of M75 Streptococcus pyogenes strains harboring slaA gene in patients affected by pediatric obstructive sleep apnea syndrome in central Italy. Front. Microbiol. 2017;8:294. doi: 10.3389/fmicb.2017.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Culotti A, Packman AI. Pseudomonas aeruginosa promotes Escherichia coli biofilm formation in nutrient-limited medium. PLoS One. 2014;9:e107186. doi: 10.1371/journal.pone.0107186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colombo AP, Magalhaes CB, Hartenbach FA, do Souto RM, da Silva-Boghossian CM. Periodontal-disease-associated biofilm: a reservoir for pathogens of medical importance. Microb. Pathog. 2016;94:27–34. doi: 10.1016/j.micpath.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Sudiono J, Sandra F, Halim NS, Kadrianto TA, Melinia M. Bactericidal and cytotoxic effects of Erythrina fusca leaves aquadest extract. Dent. J. Majal. Kedokt. Gigi. 2013;46:9–13. doi: 10.20473/j.djmkg.v46.i1.p9-13. [DOI] [Google Scholar]
- 12.Mah TF. Biofilm-specific antibiotic resistance. Future Microbiol. 2012;7:1061–1072. doi: 10.2217/fmb.12.76. [DOI] [PubMed] [Google Scholar]
- 13.Victor T Noronha, Amauri J Paula, Gabriela Durán, Andre Galembeck, Karina Cogo-Müller, Michelle Franz-Montan, et al. Silver nanoparticles in dentistry. Dent. Mater. 2017;33:1110–1126. doi: 10.1016/j.dental.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Roshna T, Nandakumar K. Generalized aggressive periodontitis and its treatment options: case reports and review of the literature. Case Rep. Med. 2012;2012:535321. doi: 10.1155/2012/535321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michaud DS, Fu Z, Shi J, Chung M. Periodontal disease, tooth loss, and cancer risk. Epidemiol. Rev. 2017;39:49–58. doi: 10.1093/epirev/mxx006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rieuwpassa IE, Achmad H, Rahmasari R. Effectiveness of clindamycin in treatment of Periodontitis. Indian J. Public Health Res. Dev. 2019;10:1223. doi: 10.5958/0976-5506.2019.02062.X. [DOI] [Google Scholar]
- 17.Wyszogrodzka G, Marszałek B, Gil B, Doro˙zy'nski P. Metalorganic frameworks: mechanisms of antibacterial action and potential applications. Drug Discov. Today. 2016;21:1009–1018. doi: 10.1016/j.drudis.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 18.Dφaz-Visurraga J, Daza C, Pozo C, Becerra A, von Plessing C, Garcφa A study on antibacterial alginate-stabilized copper nanoparticles by FT-IR and 2D-IR correlation spectroscopy. Int. J. Nanomed. 2012;7:3597. doi: 10.2147/IJN.S32648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holla G, Yeluri R, Munshi AK. Evaluation of minimum inhibitory and minimum bactericidal concentration of nano-silver base inorganic anti-microbial agent (Novaron) against Streptococcus mutans. Contemp. Clin. Dent. 2012;3:288–293. doi: 10.4103/0976-237X.103620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenstein G, Tonetti M. The role of controlled drug delivery for periodontitis. The Research, Science and Therapy Committee of the American Academy of Periodontology. J. Periodontol. 2000;71:125–140. doi: 10.1902/jop.2000.71.1.125. [DOI] [PubMed] [Google Scholar]
- 21.Rajeshkumar S. Anticancer activity of eco-friendly gold nanoparticles against lung and liver cancer cells. J. Genet. Eng. Biotechnol. 2016;14:195–202. doi: 10.1016/j.jgeb.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Podstawczyk D, Pawłowska A, Bastrzyk A, Czeryba M, Oszmiánski J. Reactivity of (+)-catechin with copper (II) ions: the green synthesis of size-controlled Sub-10 nm copper nanoparticles. ACS Sustain. Chem. Eng. 2019;7:17535–17543. doi: 10.1021/acssuschemeng.9b05078. [DOI] [Google Scholar]
- 23.Molnár Z, Bódai V, Szakacs G, Erdélyi B, Fogarassy Z, Sáfrán G, Varga T, et al. Green synthesis of gold nanoparticles by thermophilic filamentous fungi. Sci. Rep. 2018;8:3943. doi: 10.1038/s41598-018-22112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salem MZM, Elansary HO, Ali HM, El-Settawy AA, Elshikh MS, Abdel-Salam EM, et al. Bioactivity of essential oils extracted from Cupressus macrocarpa branchlets and Corymbia citriodora leaves grown in Egypt. BMC Complement Altern. Med. 2018;18:23. doi: 10.1186/s12906-018-2085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enan ET, Ashour AA, Basha S, Felemban NH, Gad El-Rab SMF. Antimicrobial activity of biosynthesized silver nanoparticles, Amoxicillin and glass-ionomer cement against Streptococcus mutans and Staphylococcus aureus. Nanotechnology. 2021;32:215101 (11pp) doi: 10.1088/1361-6528/abe577. [DOI] [PubMed] [Google Scholar]
- 26.Yaqub A, Malkani N, Shabbir A, Ditta S, Tanvir F, Ali S, et al. Novel biosynthesis of copper nanoparticles using Zingiber and Allium sp. with synergic effect of doxycycline for anticancer and bactericidal activity. Curr. Microbiol. 2020;77:2287–2299. doi: 10.1007/s00284-020-02058-4. [DOI] [PubMed] [Google Scholar]
- 27.Gad El-Rab SMF, Abo-Amer AE, Asiri AM. Biogenic synthesis of ZnO nanoparticles and its potential use as antimicrobial agent against multidrug-resistant pathogens. Curr. Microbiol. 2020;77:1767–1779. doi: 10.1007/s00284-020-01991-8. [DOI] [PubMed] [Google Scholar]
- 28.Gad El-Rab SMF, Halawani EM, Hassan AM. Formulation of ceftriaxone conjugated gold nanoparticles and their medical applications against extended-spectrum β-Lactamase producing bacteria and breast cancer. World J. Microbiol. Biotechnol. 2018;28:1563–1572. doi: 10.4014/jmb.1711.11037. [DOI] [PubMed] [Google Scholar]
- 29.Dubey SP, Lahtinen M, Sillanpaa M. Tansy fruit mediated greener synthesis of silver and gold nanoparticles. Process Biochem. 2010;45:1065–1071. doi: 10.1016/j.procbio.2010.03.024. [DOI] [Google Scholar]
- 30.Halawani EM, Hassan AM, Gad El-Rab SMF. Nanoformulation of biogenic cefotaxime-conjugated-silver nanoparticles for enhanced antibacterial efficacy against multidrug-resistant bacteria and anticancer studies. Int. J. Nanomed. 2020;5:1889–1901. doi: 10.2147/IJN.S236182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.John G., Holt PhD . Bergey's manual of determinative bacteriology 9th edn (Baltimore, Maryland: Williams & Wilkins) 1994. pp. 20: 527–558. [Google Scholar]
- 32.MaccFadin JK. Biochemical test for identification of medical bacteria 3rd edn (New York: Lippincott Williams and Winkins, AwolterKlumer Company. Philadelphia Baltimore) 2000. [Google Scholar]
- 33.Kanmani P, Lim ST. Synthesis and characterization of pullulan-mediated silver nanoparticles and its antimicrobial activities. Carbohydr. Polym. 2013;12:421–428. doi: 10.1016/j.carbpol.2013.04.048. [DOI] [PubMed] [Google Scholar]
- 34.Iskandarsyah NH, Rosana Y. Sinergicity test of silver nanoparticles and clindamycin against Staphylococcus aureus. Int. J. Res. Pharm. Sci. 2020;11:1192–1198. doi: 10.26452/ijrps.v11i1.1957. [DOI] [Google Scholar]
- 35.Wu S, Rajeshkumar S, Madasamy M, Mahendran V. Green synthesis of copper nanoparticles using Cissus vitiginea and its antioxidant and antibacterial activity against urinary tract infection pathogens. Artif. Cells Nanomed. Biotechnol. 2020;48:1153–1158. doi: 10.1080/21691401.2020.1817053. [DOI] [PubMed] [Google Scholar]
- 36.Joseph AT, Prakash P, Narvi S. Phytofabrication and Characterization of copper nanoparticles using Allium sativum and its antibacterial activity. Int. J. Sci. Eng. Technol. 2016;4:463–472. [Google Scholar]
- 37.Sengan M, Veerappan A. N-myristoyltaurine capped copper nanoparticles for selective colorimetric detection of Hg2+ in wastewater and as effective chemocatalyst for organic dye degradation. Microchem. J. 2019;148:1–9. doi: 10.1016/j.microc.2019.04.049. [DOI] [Google Scholar]
- 38.Suarez-Cerda J, Espinoza-Gómez H, Alonso-Núñez G, et al. A green synthesis of copper nanoparticles using native cyclodextrins as stabilizing agents. J. Saudi Chem. Soc. 2017;21:341–348. doi: 10.1016/j.jscs.2016.10.005. [DOI] [Google Scholar]
- 39.Fan D, Zhou Q, Lv X, Jing J, Ye Z, Shao S, Xie J. Synthesis, thermal conductivity and anti-oxidation properties of copper nanoparticles encapsulated within few-layer h-BN. Ceram. Int. 2018;44:1205–1208. doi: 10.1016/j.ceramint.2017.10.018. [DOI] [Google Scholar]
- 40.Dutta D, Phukan A, Dutta DK. Nanoporous montmorillonite clay stabilized copper nanoparticles: efficient and reusable catalyst for oxidation of alcohols. Mol. Catal. 2018;451:178–185. doi: 10.1016/j.mcat.2017.12.032. [DOI] [Google Scholar]
- 41.Gurunathan S, Kalishwaralal K, Vaidyanathan R, Venkataraman D, Pandian SR, Muniyandi J, et al. Biosynthesis, purification and characterization of silver nanoparticles using Escherichia coli. Colloids Surf. B. 2009;74:328–335. doi: 10.1016/j.colsurfb.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 42.Harraz FM, Hammoda, El-Hawiet A, Radwan MM, Wanas, Eid AME, lSohly MA. From natural product research chemical constituents, Antibacterial and Acetylcholine esterase inhibitory activity of Cupressus macrocarpa leaves. Nat. Prod. Res. 2018;34:816–822. doi: 10.1080/14786419.2018.1508140. [DOI] [PubMed] [Google Scholar]
- 43.Fuloria NK, Fuloria S, Chia KY, Karupiah S, Sathasivam K. Response of green synthesized drug blended silver nanoparticles against periodontal disease triggering pathogenic microbiota. J. Appl. Biol. Biotechnol. 2019;7:46–56. doi: 10.7324/JABB.2019.70408. [DOI] [Google Scholar]
- 44.Emmanuel R, Palanisamy S, Chen S, Chelladurai K, Padmavathy S, Saravanan M, et al. Antimicrobial efficacy of green synthesized drug blended silver nanoparticles against dental caries and periodontal disease-causing microorganisms. Mater. Sci. Eng. C. 2015;56:374–379. doi: 10.1016/j.msec.2015.06.033. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Zhang W, Niu J, Chen Y. Mechanism of photogenerated reactive oxygen species and correlation with the antibacterial properties of engineered metal-oxide nanoparticles. ACS Nano. 2012;6:5164–5173. doi: 10.1021/nn300934k. [DOI] [PubMed] [Google Scholar]
- 46.Chatterjee AK, Chakraborty R, Basu T. Mechanism of antibacterial activity of copper nanoparticles. Nanotechnology. 2014;25:135101. doi: 10.1088/0957-4484/25/13/135101. [DOI] [PubMed] [Google Scholar]
- 47.Arumugam A, Karthikeyan C, Haja Hameed AS, Gopinath K, Gowri S, Karthika V. Synthesis of cerium oxide nanoparticles using Gloriosa superba L. leaf extract and their structural, optical and antibacterial properties. Mater. Sci. Eng. C. 2015;49:408–415. doi: 10.1016/j.msec.2015.01.042. [DOI] [PubMed] [Google Scholar]
- 48.Mandava K, Kadimcharla K, Keesara NR, Sumayya NF, Prathyusha B, Batchu UR. Green synthesis of stable copper nanoparticles and synergistic activity with antibiotics. Indian J. Pharm. Sci. 2017;79:695–700. doi: 10.4172/pharmaceutical-sciences.1000281. [DOI] [Google Scholar]
- 49.Gad El-RabSMF, Halawani EM, Alzahrani SSS. Biosynthesis of silver nano-drug using Juniperus excelsa and its synergistic antibacterial activity against multidrug-resistant bacteria for wound dressing applications. 3 Biotech. 2021;11:255. doi: 10.1007/s13205-021-02782-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel BH, Channiwala MZ, Chaudhari SB, Mandot AA. Biosynthesis of copper nanoparticles; its characterization and efficacy against human pathogenic bacterium. J. Environ. Chem. Eng. 2016;4:2163–2169. doi: 10.1016/j.jece.2016.03.046. [DOI] [Google Scholar]
- 51.Hassanien R, Husein DZ, Al-Hakkani MF. Biosynthesis of copper nanoparticles using aqueous Tilia extract: antimicrobial and anticancer activities. Heliyon. 2018;4:e01077. doi: 10.1016/j.heliyon.2018.e01077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajeshkumar S, Menon S, Kumar SV, Tambuwala MM, Bakshi H A, Mehta M, et al. Antibacterial and antioxidant potential of biosynthesized copper nanoparticles mediated through Cissus arnotiana plant extract. J. Photochem. Photobiol. B. 2019;197:111531. doi: 10.1016/j.jphotobiol.2019.111531. [DOI] [PubMed] [Google Scholar]
- 53.Covarrubias C, Trepiana D, Corral C. Synthesis of hybrid copper-chitosan nanoparticles with antibacterial activity against cariogenic Streptococcus mutans. Dent. Mater. J. 2018;37:379–384. doi: 10.4012/dmj.2017-195. [DOI] [PubMed] [Google Scholar]
- 54.Mardones J, Gómez ML, Díaz C, Galleguillos C, Covarrubias C. In vitro antibacterial properties of copper nanoparticles as endodontic medicament against Enterococcus faecalis. J. Dent. Oral Disord. 2018;4:1107. [Google Scholar]