Abstract
The diversity and plant growth-promoting ability of fungal endophytes that are associated with five halophytic plant species (Phragmites australis, Suaeda australis, Limonium tetragonum, Suaeda glauca Bunge, and Suaeda maritima) growing in the Buan salt marsh on the west coast of South Korea have been explored. About 188 fungal strains were isolated from these plant samples’ roots and were then studied with the use of the internal transcribed spacer (ITS) region (ITS1-5.8S-ITS2). The endophytic fungal strains belonged to 33 genera. Alternaria (18%) and Fusarium (12.8%), of the classes Dothideomycetes and Sordariomycetes, were most rampant in the coastal salt marsh plants. There was a higher diversity in fungal endophytes that are isolated from S. glauca Bunge than in isolates from other coastal salt marsh plants. Plant growth-promoting experiments with the use of Waito-C rice seedlings show that some of the fungal strains could encourage a more efficient growth than others. Furthermore, gibberellins (GAs) GA1, GA3, and GA9 were seen in the Sa-1-4-3 isolate (Acrostalagmus luteoalbus) culture filtrate with a gas chromatography/mass spectrometry.
Keywords: Coastal salt marsh plants, fungal endophytes, Buan salt marsh, growth promotion, gibberellin
Introduction
The world’s population is estimated to rise to 9 billion by 2050 [1]. There is a need to increase food production under less than optimal conditions due to population growth and climate change. In the mid- to late twentieth century, the cultivation efforts’ purpose is to improve crop varieties, introduce hybrids, and increase agricultural productions in terms of fertilizer, crop management practices, herbicides, water delivery systems, and pesticides resulted in the “Green Revolution” [2, 3]. Worldwide, there is a similar global challenge, leading to a need of breed-improved varieties as well as agricultural production practices [4]. The expansion of agricultural production to marginal lands and the effects of global climate change also need an increased biotic and abiotic stress tolerance and efficient nutrient utilization in crop plants for the future global food requirements to be met.
Endophytic fungi are organisms that are found anywhere (intercellularly or intracellularly in plants from nearly all genera of Kingdom Plantae). These fungi live in host plants for at least a portion of their lives without generating any immediate overt disease symptoms. These associations can encourage tissue differentiation and plant growth and can help in managing abiotic and biotic stresses to which the host plants are subjected [5, 6]. In addition, endophytic fungi may prevent pathogenic organisms and provide nutrients, benefiting the plant host [7]. These well-distributed fungi form diverse plant associations and thus constitute outstanding sources of new bioactive secondary metabolites [8]. Accordingly, there are several new bioactive compounds with insecticidal, antimicrobial, cytotoxic, and anticancer activities that have been separated from endophytic fungi recently [9, 10].
Plant signaling compounds, also called phytohormones, regulate plant responses to environmental change as well as control plant growth and development [11]. Notably, recent studies have reported that certain endophytes encourage host plant growth through the synthesis of phytohormones, for example, gibberellins (GAs), cytokinins, and indole-3-acetic acid (IAA) [12-14]. Indeed, endophytic fungi promote plant growth by secreting gibberellins in the rhizosphere of their hosts, which leads to an increase in plant biomass production as well as disease resistance. Apart from this, some endophytic fungi secrete both IAA and GAs into culture media [15].
This research surveys both distribution and diversity of fungal endophytes in a certain coastal region of Korea. We have investigated fungal isolates’ capacity to encourage growth of Waito-C rice seedlings and have identified whether secondary metabolites like gibberellins were seen in fungal culture filtrates.
Materials and Methods
Plant Materials and Sampling Sites
Healthy plants and roots of Phragmites australis, Suaeda australis, Limonium tetragonum, Suaeda glauca Bunge, and Suaeda maritima were gathered from different places in the Buan salt marsh in South Korea. The samples were carefully sealed in sterile plastic bags and were then processed in the laboratory within 24 hours of collection. The scientific names, codes, and local sites of the five plant species are listed in Table 1.
Table 1.
Geographic coordinates and scientific names of plants native to the Buan salt marsh.
| No | Scientific name | Code | Site of collection | Habitat |
|---|---|---|---|---|
| 1 | Phragmites australis | Pa | 35°35'34.69"N / 126°36'2.42"E | Halophytic |
| 2 | Suaeda australis | Sa | 35°35'10.99"N / 126°32'44.70"E | Halophytic |
| 3 | Limonium tetragonum | Lt | 35°35'10.73"N / 126°32'51.40"E | Halophytic |
| 4 | Suaeda glauca Bunge | Su | 35°35'10.80"N / 126°32'51.90"E | Halophytic |
| 5 | Suaeda maritima | Sm | 35°35'34.92"N / 126°36'3.95"E | Halophytic |
Sterilization and Isolation of Endophytes from the Roots of Halophytes
Fungal endophytes were isolated from healthy roots of halophytic plants that were gathered from the salt marsh. Each plant’s root samples were washed with tap water, cut into 2–2.5-cm-long segments, and treated with Tween 80 solution for 10 min on a shaker with 160 rotations per minute (rpm). Afterward, root segments were incubated in a solution of 1% (w/v) perchloric acid for 10 min and then rinsed with double-distilled water [16, 17]. Then, they were dehydrated for 5–6 min at a temperature of 22°C on a clean bench, and two to three root segments were placed in a 90-mm Petri plate with Hagem minimal media that contain 80 ppm streptomycin. Samples were incubated at 25°C in dark conditions until there is growth of fungi from the root segments seen [18, 19]. Lastly, pure fungal strains were separated from the root segments and were kept on potato dextrose agar at 25°C [20].
DNA Extraction, PCR Amplification, and the Identification of Fungal Strains
Fungal endophyte cultures were grown in an Erlenmeyer flask that contains 50 ml potato dextrose broth for 7–10 days at a temperature of 26°C on a shaker at 120 rpm. All the 188 lyophilized endophyte samples were known. There was a fungal genomic DNA extraction using a DNeasy Plant Mini Kit (Qiagen, USA). There was identification of fungi performed by sequencing the internal transcribed spacer (ITS) region with the universal primers ITS-1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS-4 (5′-TCCTCCGCTTATTGATATGC-3′). Reaction cycling comprised of an initial denaturation step at 95°C for 2 min, which is then followed by 35 cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 1 min, and extension at 72°C for 1 min. The final extension was done at 72°C for 7 min. PCR products were electrophoresed on agarose gels with an ethidium bromide stain and purified using the QIAquick PCR purification kit (Qiagen). The products were then sequenced with the use of the ABI PRISM BigDye terminator cycle sequencing kit (PE Biosystems, USA) on an ABI 310 DNA automated sequencer (Perkin, USA). The sequences were identified with the use of the BLAST (Basic Local Alignment Search Tool) tool of the National Center for Biotechnology Information (NCBI).
Statistical Analyses
The fungal endophytes’ diversity at the genus level was indicated by the Shannon diversity index (H’), Fisher’s alpha index (α), and Simpson’s index of diversity. Richness was assessed by Menhinick’s richness index (Dmn) and Margalef ’s index (Dmg) [21, 22]. The Menhinick’s index was calculated through the following formula: ; Dmg=(S−1)/ln(N), where S is the number of genera in a sample, and N is the total number of individuals in a community. Both indices ranged from 0 to ∞.
The genus diversity was evaluated using the Shannon diversity index (H’), Fisher’s alpha index (α), and Simpson’s index of diversity [23]. Fisher’s alpha index (α) was calculated as follow; S=α·ln(1+N/α), where S is the number of genera, and N is the total number of individuals. The formula for Shannon’s diversity index is , where pi is the proportion of individuals found in genera i in a sample. The values of the Shannon diversity index generally range from 1.5 to 3.5. Simpson’s index of diversity (1-D) was calculated as follow; , where N is the total number of individuals in a sample, and ni is the number of individuals found in genera i in a sample. The magnitude of this index ranges from 0 to 1; the greater the magnitude, the greater the sample diversity.
Screening of Fungal Cultures on Waito-C rice Seedlings
To test whether the fungi have growth-promoting capacities, Waito-C rice sprouts were exposed to fungal culture filtrates. The fungal strains were grown in the Czapek Dox broth medium on a shaking incubator for 7 days at 25°C and 180 rpm and were harvested using filtration. The harvested fungal culture filtrates were immediately stored at a temperature of −70°C and then lyophilized. The lyophilized culture filtrates were then mixed together with 1 ml of distilled water. Waito-C rice grains were treated with uniconazole for 24 h to lessen gibberellins’ activity in the seed coat. The treated rice seeds were washed and soaked in distilled water until radical emergence occurs, and then the young seedlings were placed in glass tubes with 0.6% water agar medium to grow in a growth chamber [24]. Concentrated, lyophilized culture filtrates (10 microliters) from each fungal isolate were applied to apical meristems after the rice seedlings reached the two-leaf stage. Both the plant and shoot lengths of rice were seen after 1 week application and compared against rice seedlings treated with wild-type Gibberella fujikuroi, which was the positive control in this study.
Analysis of Gibberellins
Gibberellins were extracted from culture filtrates after the incubation of fungal isolates for 7 days in Czapek Dox broth medium that contains 1% (w/v) glucose and peptone. Reverse-phase C18 high-performance liquid chromatography followed by gas chromatography/mass spectrometry (GC/MS) with selected ion monitoring (SIM) investigated extracted gibberellins. Three major ions of the supplemented [2H2] gibberellin internal standards as well gibberellins were simultaneously monitored. The GC/MS data were eventually collected and investigated. The retention time was known with the use of hydrocarbon standards in calculating the Kovats retention index (KRI) value, and peak area ratios between non-deuterated and deuterated gibberellins were used for the quantification of the gibberellins [25].
Results
Identification of Endophytic Fungi
There is a total of 188 endophytic fungal strains separated from the roots of five species of plants collected from the Buan salt marsh on the west coast of South Korea. Fifty-two fungal strains from P. australis, 40 strains from S. australis, 25 strains from L. tetragonum, 41 strains from S. glauca Bunge, and 30 strains from S. maritima were isolated.
The endophytic fungi nucleotide sequences from each plant sample were deposited in the NCBI GenBank database under the accession numbers KP018214–KP018265 from P. australis, KP018266–KP018305 from S. australis, KP018306–KP018330 from L. tetragonum, KP018331–KP018371 from S. glauca Bunge, and KP018372–KP018401 from S. maritima (Table 2).
Table 2.
Identification of endophytic fungal isolates from roots of plants.
| Sample No. | Closely related fungal sequences | Similarity (%) | Accession No. |
|---|---|---|---|
| Pa-1-2-1 | Pestalotiopsis sp. 1 MJ-2014 (KJ572189) | 100 | KP018214 |
| Pa-1-3-1 | Alternaria sp. BOP212b (KC771455) | 100 | KP018215 |
| Pa-1-3-3 | Fusarium incarnatum strain LS 03 (KJ721990) | 99 | KP018216 |
| Pa-1-4-3 | Trichoderma aureoviride strain SL (KJ610807) | 99 | KP018217 |
| Pa-1-6-1 | Alternaria rosae strain CM24T-EY-E (KF815569) | 99 | KP018218 |
| Pa-1-6-2 | Lewia sp. OUCMBI101191 (HQ914885) | 100 | KP018219 |
| Pa-1-8-1 | Alternaria alternata strain HMA1D (KJ677246) | 100 | KP018220 |
| Pa-1-8-2 | Penicillium oxalicum strain SY20-5 (KJ619622) | 99 | KP018221 |
| Pa-1-8-3 | Fusarium incarnatum strain FI-00602 (KJ572780) | 100 | KP018222 |
| Pa-1-8-4 | Talaromyces verruculosus (JN676121) | 99 | KP018223 |
| Pa-1-9-1 | Fusarium incarnatum strain FI-00602 (KJ572780) | 100 | KP018224 |
| Pa-1-9-2 | Alternaria alternata strain HMA1D (KJ677246) | 100 | KP018225 |
| Pa-1-9-3 | Alternaria sp. HT-M18-LS (KJ527010) | 100 | KP018226 |
| Pa-2-2-2 | Lewia sp. OUCMBI101191 (HQ914885) | 99 | KP018227 |
| Pa-2-2-3 | Simplicillium obclavatum (AB604000) | 100 | KP018228 |
| Pa-2-2-4 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018229 |
| Pa-2-3-1 | Alternaria sp. DX-FOF7 (KF558883) | 100 | KP018230 |
| Pa-2-3-4 | Alternaria alternata strain HMA1D (KJ677246) | 100 | KP018231 |
| Pa-2-4-1-2 | Penicillium sp. CCF3828 (FJ430753) | 99 | KP018232 |
| Pa-2-4-2 | Trichoderma ovalisporum strain (KC847168) | 100 | KP018233 |
| Pa-2-5-2 | Penicillium sp. CCF3828 (FJ430753) | 99 | KP018234 |
| Pa-2-6-2 | Alternaria alternata strain HMA1D (KJ677246) | 100 | KP018235 |
| Pa-2-6-5 | Cladosporium sp. HT-Z1-V (KJ527013) | 100 | KP018236 |
| Pa-2-7-1 | Phoma sp. P17E3 (JN207293) | 99 | KP018237 |
| Pa-2-7-3 | Alternaria alternata strain HMA1D (KJ677246) | 100 | KP018238 |
| Pa-2-8-1-2 | Fusarium caeruleum (KF887087) | 100 | KP018239 |
| Pa-2-8-2 | Fusarium oxysporum strain P43 (JX045812) | 100 | KP018240 |
| Pa-2-8-3-1 | Fusarium incarnatum strain LS 03 (KJ721990) | 100 | KP018241 |
| Pa-2-9-1 | Verticillium saksenae (KF472156) | 99 | KP018242 |
| Pa-2-9-2 | Lewia sp. OUCMBI101191 (HQ914885) | 100 | KP018243 |
| Pa-2-9-3 | Sclerostagonospora phragmiticola strain (KF251230) | 100 | KP018244 |
| Pa-2-9-4 | Cladosporium cladosporioides strain (KJ589558) | 100 | KP018245 |
| Pa-2-9-5 | Verticillium saksenae (KF472156) | 99 | KP018246 |
| Pa-2-9-7 | Lewia sp. OUCMBI101191 (HQ914885) | 100 | KP018247 |
| Pa-2-9-8 | Lewia sp. OUCMBI101191 (HQ914885) | 100 | KP018248 |
| Pa-2-10-2 | Fusarium longipes (HG423537) | 99 | KP018249 |
| Pa-2-10-6 | Lewia sp. OUCMBI101191 (HQ914885) | 100 | KP018250 |
| Pa-3-1-1 | Alternaria alternata strain HMA1D (KJ677246) | 100 | KP018251 |
| Pa-3-1-3 | Fusarium longipes (HG423537) | 99 | KP018252 |
| Pa-3-2-3 | Fusarium longipes (HG423537) | 99 | KP018253 |
| Pa-3-2-4 | Talaromyces verruculosus (JN676121) | 100 | KP018254 |
| Pa-3-3-1 | Phoma sp. P17E3 (JN207293) | 99 | KP018255 |
| Pa-3-4-1 | Fusarium incarnatum strain LS 03 (KJ721990) | 100 | KP018256 |
| Pa-3-6-2 | Fusarium incarnatum strain LS 03 (KJ721990) | 100 | KP018257 |
| Pa-3-7-2 | Fusarium commune strain dH 23113 (JX162390) | 100 | KP018258 |
| Pa-3-7-3 | Penicillium spinulosum isolate FFJC 16 (KF876837) | 100 | KP018259 |
| Pa-3-7-4-1 | Cladosporium cladosporioides isolate (KJ572146) | 100 | KP018260 |
| Pa-3-7-4-2 | Phoma sp. P17E3 (JN207293) | 99 | KP018261 |
| Pa-3-9-1 | Fusarium commune strain dH 23113 (JX162390) | 100 | KP018262 |
| Pa-3-10-1 | Lecanicillium sp. O_3_BESC_246b (KC007329) | 100 | KP018263 |
| Pa-3-10-2 | Fusarium longipes (HG423537) | 99 | KP018264 |
| Pa-3-10-3 | Lewia sp. OUCMBI101191 (HQ914885) | 100 | KP018265 |
| Sa-1-3-1 | Fusarium oxysporum strain HPA2 (KJ677253) | 100 | KP018266 |
| Sa-1-3-2 | Aspergillus brasiliensis strain HPA8 (KJ677257) | 100 | KP018267 |
| Sa-1-4-3 | Acrostalagmus luteoalbus isolate AcLu2 (JQ387575) | 100 | KP018268 |
| Sa-1-5-1 | Phoma sp. JX1203 (KC203049) | 100 | KP018269 |
| Sa-1-5-2-2 | Phoma sp. JX1203 (KC203049) | 100 | KP018270 |
| Sa-1-6-2 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018271 |
| Sa-1-7-2 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018272 |
| Sa-1-9-1 | Fusarium incarnatum strain FI-00602 (KJ572780) | 100 | KP018273 |
| Sa-1-9-2 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018274 |
| Sa-2-1-2 | Alternaria alternata strain HMA1D (KJ677246) | 100 | KP018275 |
| Sa-2-2-1 | Exophiala oligosperma (AB480204) | 100 | KP018276 |
| Sa-2-2-1-1 | Paraconiothyrium cyclothyrioides strain (KC215138) | 100 | KP018277 |
| Sa-2-2-3 | Exophiala oligosperma (AB777520) | 100 | KP018278 |
| Sa-2-3-1 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018279 |
| Sa-2-3-2 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018280 |
| Sa-2-4-1 | Pleospora bjoerlingii (JX045842) | 100 | KP018281 |
| Sa-2-6-2 | Penicillium canescens strain CV0198 (JX140832) | 100 | KP018282 |
| Sa-2-7-1 | Plectosphaerella sp. MF-1 (AB520859) | 100 | KP018283 |
| Sa-2-7-2 | Aspergillus terreus isolate D34 (KF971363) | 100 | KP018284 |
| Sa-2-8-1 | Penicillium lapidosum (KJ676451) | 100 | KP018285 |
| Sa-2-9-1 | Sclerostagonospora phragmiticola strain (KF251230) | 99 | KP018286 |
| Sa-2-9-4 | Paraphoma sp. BJ18 (KJ702586) | 100 | KP018287 |
| Sa-2-10-1 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018288 |
| Sa-2-10-2 | Alternaria sp. BJ35 (KJ702610) | 100 | KP018289 |
| Sa-3-1-1 | Hypocrea sp. SFCF20120803-50 (KF313111) | 100 | KP018290 |
| Sa-3-1-2 | Aspergillus aff. fumigatus A28 (JN246065) | 100 | KP018291 |
| Sa-3-2-2 | Cladosporium sp. XJ18 (KF143793) | 100 | KP018292 |
| Sa-3-3-1 | Acrostalagmus luteoalbus strain PTV-1 (GU813970) | 100 | KP018293 |
| Sa-3-4-2 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018294 |
| Sa-3-4-3 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018295 |
| Sa-3-6-1 | Penicillium citrinum strain NF7 (KJ653821) | 100 | KP018296 |
| Sa-3-6-3 | Fusarium oxysporum isolate F102 (KJ512160) | 100 | KP018297 |
| Sa-3-8-1 | Penicillium simplicissimum strain (KF815055) | 99 | KP018298 |
| Sa-3-8-1-1 | Alternaria sp. BJ35 (KJ702610) | 100 | KP018299 |
| Sa-3-8-2-1 | Colletotrichum gloeosporioides strain (KJ632430) | 100 | KP018300 |
| Sa-3-8-3 | Aspergillus sp. BJ39 (KJ702608) | 100 | KP018301 |
| Sa-3-9-1 | Fusarium oxysporum isolate F102 (KJ512160) | 100 | KP018302 |
| Sa-3-9-2 | Fusarium oxysporum strain HPA2 (KJ677253) | 99 | KP018303 |
| Sa-3-10-1 | Penicillium citrinum strain NF7 (KJ653821) | 100 | KP018304 |
| Sa-3-10-2 | Trichoderma harzianum (HG940484) | 100 | KP018305 |
| Lt-1-1-1 | Pestalotiopsis sp. 1 AE-2013 strain F4872 (KF746123) | 100 | KP018306 |
| Lt-1-2-2 | Pleospora bjoerlingii (JX045842) | 100 | KP018307 |
| Lt-1-3-2 | Penicillium lapidosum (KJ676451) | 100 | KP018308 |
| Lt-1-4-1 | Fusarium longipes (KJ412506) | 100 | KP018309 |
| Lt-1-5-1 | Paraconiothyrium cyclothyrioides strain (KC215138) | 100 | KP018310 |
| Lt-1-6-1 | Pleospora bjoerlingii (JX045842) | 100 | KP018311 |
| Lt-1-8-2 | Alternaria alternata strain SR/I/90 (KJ767532) | 100 | KP018312 |
| Lt-1-9-1 | Fusarium oxysporum f. sp. conglutinans (KF381081) | 100 | KP018313 |
| Lt-1-10-1 | Pestalotiopsis clavispora strain P44 (JX045813) | 100 | KP018314 |
| Lt-1-10-2 | Cochliobolus kusanoi isolate SH8 (KJ572135) | 100 | KP018315 |
| Lt-2-1-1 | Colletotrichum acutatum strain 11E031 (KF717039) | 100 | KP018316 |
| Lt-2-4-1 | Penicillium paneum strain M-18 (JQ422610) | 100 | KP018317 |
| Lt-2-5-1-1 | Phoma sp. EIODSF018 (KJ173541) | 100 | KP018318 |
| Lt-2-5-1-2 | Trichoderma sp. BCC 3579 (AY550911) | 99 | KP018319 |
| Lt-2-6-2 | Penicillium sp. SK14JW2P (KC545799) | 100 | KP018320 |
| Lt-2-7-1 | Alternaria sp. BJ35 (KJ702610) | 100 | KP018321 |
| Lt-2-8-2 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018322 |
| Lt-3-2-2 | Trichoderma harzianum strain ML16-1 (KJ619615) | 100 | KP018323 |
| Lt-3-3-1 | Lewia sp. OUCMBI101191 (HQ914885) | 100 | KP018324 |
| Lt-3-3-2 | Meira sp. JCM 18504 (AB778892) | 99 | KP018325 |
| Lt-3-6-1 | Penicillium sp. 12140 (JX657339) | 100 | KP018326 |
| Lt-3-7-1 | Alternaria sp. BJ35 (KJ702610) | 100 | KP018327 |
| Lt-3-9-1 | Alternaria sp. BJ35 (KJ702610) | 99 | KP018328 |
| Lt-3-9-2 | Aspergillus brasiliensis strain HPA8 (KJ677257) | 100 | KP018329 |
| Lt-3-9-4 | Aspergillus clavatus strain USMO08 (KF669482) | 99 | KP018330 |
| Su-1-1-1 | Talaromyces pinophilus isolate SCLB5 (KF913534) | 100 | KP018331 |
| Su-1-2-1 | Talaromyces pinophilus isolate SCLB5 (KF913534) | 100 | KP018332 |
| Su-1-2-2 | Penicillium sp. OY18307 (FJ571475) | 100 | KP018333 |
| Su-1-4-1 | Hypocrea sp. SFCF20120803-50 (KF313111) | 100 | KP018334 |
| Su-1-6-2 | Fusarium sp. BJ9 (KJ702598) | 100 | KP018335 |
| Su-1-6-2-1 | Alternaria sp. DX-FOF7 (KF558883) | 100 | KP018336 |
| Su-1-7-2 | Phomopsis sp. H4243 (GU595056) | 99 | KP018337 |
| Su-1-10-2 | Alternaria sp. BJ35 (KJ702610) | 100 | KP018338 |
| Su-2-1-1 | Pleospora bjoerlingii (JX045842) | 100 | KP018339 |
| Su-2-2-3 | Purpureocillium lilacinum strain (KC157754) | 100 | KP018340 |
| Su-2-3-1 | Phoma sp. Y19 (KJ572232) | 100 | KP018341 |
| Su-2-3-2 | Pleospora bjoerlingii (JX045842) | 100 | KP018342 |
| Su-2-4-3 | Purpureocillium lilacinum strain (KC157756) | 100 | KP018343 |
| Su-2-5-3 | Aspergillus brasiliensis (KJ445022) | 100 | KP018344 |
| Su-2-6-2 | Pleospora bjoerlingii (JX045842) | 100 | KP018345 |
| Su-2-7-1 | Gibberella fujikuroi (KC752592) | 100 | KP018346 |
| Su-2-7-2 | Penicillium sp. OY18307 (FJ571475) | 100 | KP018347 |
| Su-2-8-1 | Alternaria sp. BJ35 (KJ702610) | 100 | KP018348 |
| Su-2-8-2 | Metacordyceps chlamydosporia (FN598950) | 99 | KP018349 |
| Su-2-9-1 | Penicillium sp. Cs/13/2 (JN585948) | 100 | KP018350 |
| Su-2-9-2 | Alternaria sp. BJ35 (KJ702610) | 100 | KP018351 |
| Su-2-9-3 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018352 |
| Su-2-10-1 | Alternaria sp. 174wat (KF811432) | 100 | KP018353 |
| Su-2-10-2 | Penicillium sp. KJ-2012 strain GZU(JQ965022) | 100 | KP018354 |
| Su-3-1-1 | Davidiella macrospora (KJ529009) | 100 | KP018355 |
| Su-3-1-1-1 | Davidiella macrospora (KJ529009) | 100 | KP018356 |
| Su-3-1-2 | Pleospora bjoerlingii (JX045842) | 99 | KP018357 |
| Su-3-3-1 | Fusarium oxysporum strain P43 (JX045812) | 100 | KP018358 |
| Su-3-4-1 | Pestalotiopsis clavispora strain P44 (JX045813) | 100 | KP018359 |
| Su-3-4-3 | Pleospora bjoerlingii (JX045842) | 100 | KP018360 |
| Su-3-5-1 | Phomopsis sp. H4243 (GU595056) | 99 | KP018361 |
| Su-3-5-2 | Aspergillus terreus isolate D34 (KF971363) | 100 | KP018362 |
| Su-3-6-1 | Colletotrichum gloeosporioides strain (KJ632430) | 100 | KP018363 |
| Su-3-6-3 | Aspergillus allahabadii strain CBS (GQ342626) | 100 | KP018364 |
| Su-3-6-5 | Pestalotiopsis clavispora strain P44 (JX045813) | 100 | KP018365 |
| Su-3-7-2 | Purpureocillium lilacinum strain E303 (KJ540087) | 100 | KP018366 |
| Su-3-7-3 | Cladosporium sp. BJ45 (KJ702611) | 100 | KP018367 |
| Su-3-9-1 | Clonostachys rosea strain F-3-51 (KF887020) | 100 | KP018368 |
| Su-3-9-2 | Gliomastix murorum (AB540558) | 100 | KP018369 |
| Su-3-10-1 | Alternaria sp. BJ35 (KJ702610) | 100 | KP018370 |
| Su-3-10-2-1 | Colletotrichum gloeosporioides strain (KJ632430) | 100 | KP018371 |
| Sm-1-1-2 | Aspergillus sp. SL-F20 (KJ528990) | 99 | KP018372 |
| Sm-1-3-1 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018373 |
| Sm-1-3-2 | Tetracladium setigerum isolate (HQ647302) | 99 | KP018374 |
| Sm-1-5-3 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018375 |
| Sm-1-6-2 | Alternaria sp. BJ35 (KJ702610) | 100 | KP018376 |
| Sm-1-9-2-2 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018377 |
| Sm-1-10-1 | Fusarium andiyazi strain CBS 134430 (KC954400) | 100 | KP018378 |
| Sm-1-10-2 | Alternaria sp. BJ35 (KJ702610) | 100 | KP018379 |
| Sm-2-1-3 | Alternaria sp. BJ35 (KJ702610) | 100 | KP018380 |
| Sm-2-6-1 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018381 |
| Sm-2-6-2 | Alternaria sp. BJ35 (KJ702610) | 100 | KP018382 |
| Sm-2-7-2 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018383 |
| Sm-2-8-1 | Talaromyces trachyspermus strain (KF147920) | 99 | KP018384 |
| Sm-2-8-2-1 | Exophiala oligosperma (AB777520) | 100 | KP018385 |
| Sm-2-8-2-2 | Meira sp. JCM 18504 (AB778892) | 99 | KP018386 |
| Sm-2-9-1 | Alternaria sp. DX-FOF7 (KF558883) | 100 | KP018387 |
| Sm-2-9-2 | Alternaria sp. BJ35 (KJ702610) | 100 | KP018388 |
| Sm-2-10-1 | Alternaria sp. HT-M18-LS (KJ527010) | 100 | KP018389 |
| Sm-2-10-2 | Cladosporium cladosporioides strain (KJ589558) | 100 | KP018390 |
| Sm-3-1-1 | Macrophoma sp. TXc4-6 (HQ262514) | 99 | KP018391 |
| Sm-3-1-3 | Cladosporium oxysporum strain B2F2 (KJ589590) | 100 | KP018392 |
| Sm-3-2-1 | Alternaria sp. BJ35 (KJ702610) | 100 | KP018393 |
| Sm-3-3-3 | Penicillium sp. JMG302 (KJ598874) | 98 | KP018394 |
| Sm-3-4-1 | Cochliobolus kusanoi isolate SH8 (KJ572135) | 100 | KP018395 |
| Sm-3-5-1 | Alternaria sp. DX-FOF7 (KF558883) | 100 | KP018396 |
| Sm-3-8-1 | Alternaria sp. DX-FOF7 (KF558883) | 100 | KP018397 |
| Sm-3-8-2 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018398 |
| Sm-3-9-2 | Macrophoma sp. TXc4-6 (HQ262514) | 100 | KP018399 |
| Sm-3-10-2 | Alternaria sp. HT-M18-LS (KJ527010) | 100 | KP018400 |
| Sm-3-10-3 | Cladosporium cladosporioides strain (KJ589558) | 100 | KP018401 |
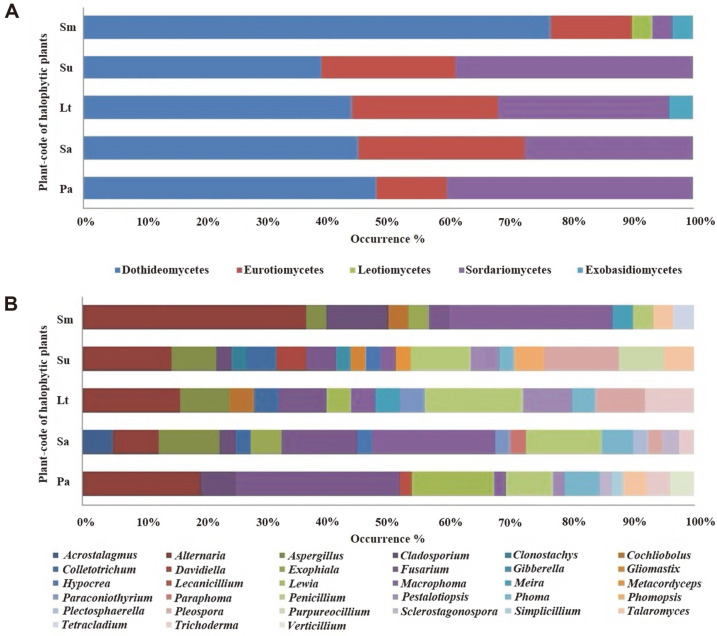
There was a total of 188 strains categorized into the phyla Ascomycota (186 strains) and Basidiomycota (two strains). The class Dothideomycetes (93 strains) has the largest number of strains, followed by the following classes: Sordariomycetes (56 strains), Eurotiomycetes (36 strains), Exobasidiomycetes (two strains), and Leotiomycetes (one strain). At the genus level, Alternaria has the largest proportion of isolates (34 strains) followed by Fusarium (24 strains).
The genus of each strain was observed, and each group’s proportion at the class and genus levels was assessed (Fig. 1). Dothideomycetes has the highest percentage of isolates at the class level. With the exception of isolates from S. glauca Bunge, Dothideomycetes is the majority of fungi in every plant sample. At the genus level, Alternaria was the most prevalent (18%) fungal isolate, which is followed by Fusarium (12.7%).
Fig. 1. Distribution of fungal isolates in different plant samples at the class (A) and genus (B) levels.
Pa, Phragmites australis; Sa, Suaeda australis; Lt, Limonium tetragonum; Su, Suaeda glauca Bunge; and Sm, Suaeda maritima.
Diversity of Endophytic Fungi isolates
About 188 culturable fungal strains were isolated from the roots of five halophytes based on colony morphologies. All endophytic fungi from halophytes belonged to 33 genera in accordance with molecular identification. Fungal isolates were then classified into 14 genera, 16 species, and 19 unclassified strains from P. australis; 17 genera, 17 species, and 17 unclassified strains from S. australis; 14 genera, 13 species, and 11 unclassified strains from L. tetragonum; 19 genera, 14 species, and 17 unclassified strains from S. glauca Bunge; and 11 genera, 7 species, and 22 unclassified strains from S. maritima (Table 3).
Table 3.
Endophytic fungi (188 strains) isolated from five plants with scientific names, plant codes, taxa of fungal strain, and number of fungal isolates.
| Scientific name of plant sample | Abbreviated plant name | Taxon of fungal strains | No. of isolates |
|---|---|---|---|
| Phragmites australis | Pa | 14 genera, 16 species | 52 |
| Suaeda australis | Sa | 17 genera, 17 species | 40 |
| Limonium tetragonum | Lt | 14 genera, 13 species | 25 |
| Suaeda glauca Bunge | Su | 19 genera, 14 species | 41 |
| Suaeda maritima | Sm | 11 genera, 7 species | 30 |
Pa, Phragmites australis; Sa, Suaeda australis; Lt, Limonium tetragonum; Su, Suaeda glauca Bunge; and Sm, Suaeda maritima.
As regards generic diversity, S. glauca Bunge had the highest score in Shannon’s index (2.76), Fisher’s (α) (13.75), and Simpson’s index of diversity (0.970). S. maritima had the lowest scores in Shannon’s index and Fisher’s (α) (1.86 and 6.26, respectively), and P. australis had the lowest Simpson’s index of diversity (0.904). As regards generic richness, S. glauca Bunge displayed the highest scores in Menhinick’s index (2.97) and Margalef ’s index (4.85). The lowest scores in Menhinick’s and Margalef ’s indices were in P. australis (1.94) and S. maritima (2.94), respectively (Table 4).
Table 4.
Diversity indices and distribution of endophytic fungi isolated from plants native to the Buan salt marsh.
| Fungal taxon | Pa | Sa | Lt | Su | Sm |
|---|---|---|---|---|---|
| Acrostalagmus | 2 | ||||
| Alternaria | 10 | 3 | 4 | 6 | 11 |
| Aspergillus | 4 | 2 | 3 | 1 | |
| Cladosporium | 3 | 1 | 1 | 3 | |
| Clonostachys | 1 | ||||
| Cochliobolus | 1 | 1 | |||
| Colletotrichum | 1 | 1 | 2 | ||
| Davidiella | 2 | ||||
| Exophiala | 2 | 1 | |||
| Fusarium | 14 | 5 | 2 | 2 | 1 |
| Gibberella | 1 | ||||
| Gliomastix | 1 | ||||
| Hypocrea | 1 | 1 | |||
| Lecanicillium | 1 | ||||
| Lewia | 7 | 1 | |||
| Macrophoma | 1 | 8 | 1 | 1 | 8 |
| Meira | 1 | 1 | |||
| Metacordyceps | 1 | ||||
| Paraconiothyrium | 1 | 1 | |||
| Paraphoma | 1 | ||||
| Penicillium | 4 | 5 | 4 | 4 | 1 |
| Pestalotiopsis | 1 | 2 | 2 | ||
| Phoma | 3 | 2 | 1 | 1 | |
| Phomopsis | 2 | ||||
| Plectosphaerella | 1 | ||||
| Pleospora | 1 | 2 | 5 | ||
| Purpureocillium | 3 | ||||
| Sclerostagonospora | 1 | 1 | |||
| Simplicillium | 1 | ||||
| Talaromyces | 2 | 2 | 1 | ||
| Tetracladium | 1 | ||||
| Trichoderma | 2 | 1 | 2 | ||
| Verticillium | 2 | ||||
| N | 52 | 40 | 25 | 41 | 30 |
| S | 14 | 17 | 14 | 19 | 11 |
| Shannon diversity index (H') | 2.22 | 2.55 | 2.50 | 2.76 | 1.86 |
| Simpson’s index of diversity (1-D) | 0.904 | 0.936 | 0.967 | 0.970 | 0.929 |
| Menhinick's index (Dmn) | 1.94 | 2.69 | 2.80 | 2.97 | 2.01 |
| Margalef's index (Dmg) | 3.29 | 4.34 | 4.04 | 4.85 | 2.94 |
| Fisher's diversity (α) | 6.29 | 11.17 | 13.14 | 13.75 | 6.26 |
Pa, Phragmites australis; Sa, Suaeda australis; Lt, Limonium tetragonum; Su, Suaeda glauca Bunge; and Sm, Suaeda maritima.
Screening for Fungal Metabolites Promoting Plant Growth in Waito-C rice Seedlings
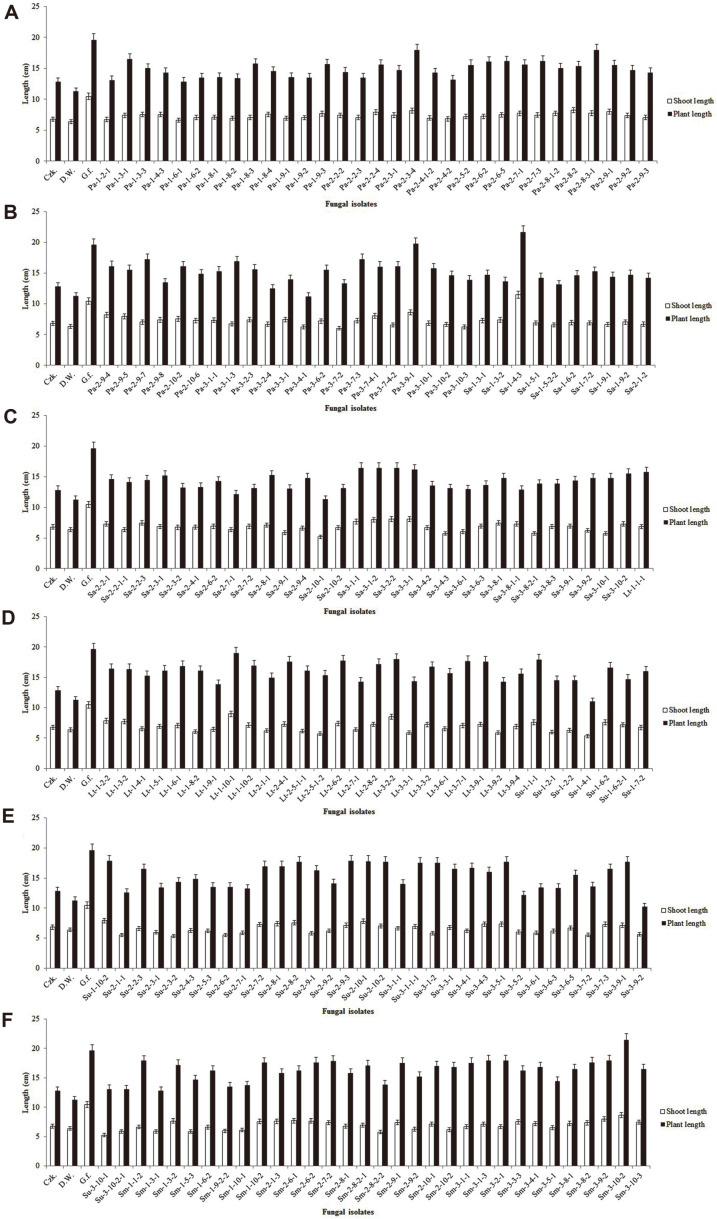
The fungal culture filtrates were applied on Waito-C rice seedlings, and the seedling lengths were measured after 1 week of fungal culture filtrate application. Out of 188 isolated fungi, 4 fungal isolates considerably encouraged shoot lengths of rice seedlings. The fungal isolate Sa-1-4-3 promoted maximum plant and shoot lengths of 21.6 cm and 11.5 cm, respectively, while culture filtrates Pa-3-9-1, Lt-1-10-1, and Sm-3-10-2 produced plant and shoot lengths of 19.7 and 8.6 cm, 19 and 9 cm, and 21.4 and 8.6 cm, respectively. There was a much lower plant and shoot growth promotion by Gibberella fujikuroi than by Sa-1-4-3 (Fig. 2); consequently, Sa-1-4-3 was chosen for further analysis.
Fig. 2. Screening for plant growth promoting of Waito-C rice seedlings with culture filtrates of fungal endophytes isolated from plant samples A-F.
Ten microliters of lyophilized culture filtrates was treated to Waito-C rice seedlings. The shoot length and plant length of the Waito-C rice seedlings were measured after 7 days of treatment. The standard deviation from means was calculated using Microsoft Excel.
Extraction and Quantification of Gibberellins
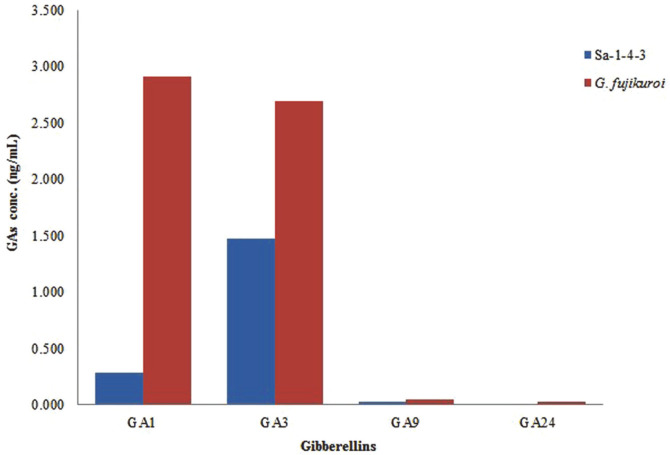
After the growth of Sa-1-4-3 fungal strain for 7 days in Czapek Dox broth medium (at 25°C; 180 rpm), the culture was then filtered with the use of a filter paper to obtain a clear supernatant (150 ml). The supernatant was extracted and chromatographed for the detection of gibberellins. The GC/MS-SIM analysis has discovered that different physiologically active and inactive gibberellins have different quantities. The physiologically bioactive gibberellins were GA1 (0.285 ng/ml) and GA3 (1.479 ng/ml), while the physiologically inactive gibberellins were GA9 (0.029 ng/ml) and GA24. GA3 was considered to be more abundant than other gibberellins (Fig. 3).
Fig. 3. Gibberellins content of fungal culture filtrates of the Sa-1-4-3 strain and wild type Gibberella fujikuroi.
GC/MS-SIM analysis of culture filtrate extracts from the Sa-1-4-3 fungal isolate detected two bioactive GAs. Sa-1- 4-3 showed the presence of bioactivity of GA1, GA3, and other inactive GA.
Discussion
Endophytes reside inside healthy plant tissues, providing shelter and nutrition. In return, these endophytes act as an excellent source of bioactive compounds and functional metabolites that impact both plant health and growth [26-28]. Some of these metabolites prompt resistance mechanisms that protect the plants against different biotic and abiotic stresses; therefore, plant fitness and productivity are increased [29].
In our study, 188 endophytic fungi were isolated from 5 plants growing in the Buan salt marsh and were identified by ITS1, 5.8S, and ITS2 sequencing. Thirty-three genera were recognized among the isolated fungal samples. Alternaria and Fusarium were the most commonly found fungi. The majority of endophytic fungi belong to the phylum Ascomycota [30, 31]. Recent studies have reported that the genus Alternaria was the most prominent fungus found in root, stem, and leaf tissues of Gossypium hirsutum [32]. Additionally, both Alternaria and Fusarium are the most frequently encountered endophytes in different types of plants [32].
Fungal endophytes are well known for their production of a wide range of secondary metabolites and enhancement of plant resistance to environmental stress. The plant growth-enhancing effects of these fungal culture filtrates were confirmed with the use of Waito-C rice seedling bioassays [33, 34]. Although other plants are also valuable for analyze the plant growth-enhancing effects, Waito-C rice has more benefits. Because of small size of plant and lack of gibberellin, uniconazole treated Waito-C rice reacts sensitively to gibberellin from foreign substances and shows rapidly the difference in growth following injection. The use of rice mutant Waito-C, which reduces gibberellin biosynthesis, was very efficient. The Sa-1-4-3 fungal strain has promoted a better plant growth in the Waito-C rice seedlings. These results are the same as those of a previous study wherein Talaromyces pinophilus Su-3-4-3, which is isolated from the roots of S. glauca Bunge, encouraged the growth in various rice plants [35].
The host plant benefits from the hormones produced by endophytic fungi. Several studies have shown plant growth-promoting characteristics and secretion of secondary metabolites, such as gibberellins, of endophytic fungi, most of which have a relationship with the roots [34, 36]. In growth of plant, many other growth promoting compounds including cytokinins and indole-3-acetic acid are also existing. But since gibberellin is the most representative compound, it was selected as a target material for growth promotion in this study. We have acquired a strain that was initially identified as Acrostalagmus luteoalbus (Sa-1-4-3) and detected gibberellin in this culture’s filtrate. According to the result of the Waito-C rice cultivation, the A. luteoalbus strain (Sa-1-4-3) showed the better growth contribution of plant length than the G. fujikuroi. Based on the period of plant cultivation in this study, it is expected that A. luteoalbus will promote early growth of Waito-C rice in particular. The GC/MS-SIM method, an established technique to identify secondary metabolites, analyzed the gibberellins in the culture filtrate of Sa-1-4-3.
In a nutshell, a total of 188 fungi were isolated from roots of five plants that are located in the Buan salt marsh. These fungi were classified into 2 phyla, 5 classes, 10 orders, 19 families, and 33 genera. Alternaria and Fusarium accounted for more than 30% of all isolates. Endophytic fungi that are isolated from S. glauca Bunge are the most diverse. This study informs us on the capacity of A. luteoalbus (Sa-1-4-3) to produce gibberellins. These results are expected to promote plant growth in areas with high salt concentrations, which are considered to be of great benefit to crop production and agriculture.
Acknowledgments
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), Ministry of Education (2016R1A6A1A05011910), Research Institute for Dok-do and Ulleung-do Island of Kyungpook National University, and Korea Ministry of Environment as “The Eco-Innovation Project”.
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
REFERENCES
- 1.Bologna M, Aquino G. Deforestation and world population sustainability: a quantitative analysis. Sci. Rep. 2020;10:7631. doi: 10.1038/s41598-020-63657-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Würschum T, Langer SM, Longin CFH, Tucker MR, Leiser WL. A modern Green Revolution gene for reduced height in wheat. Plant J. 2017;92:892–903. doi: 10.1111/tpj.13726. [DOI] [PubMed] [Google Scholar]
- 3.Wu K, Wang S, Song W, Zhang J, Wang Y, Liu Q, et al. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science. 2020;367:eaaz2046. doi: 10.1126/science.aaz2046. [DOI] [PubMed] [Google Scholar]
- 4.Martin RC, Dombrowski JE. Isolation and identification of fungal endophytes from grasses along the Oregon coast. Am. J. Plant Sci. 2015;6:3216. doi: 10.4236/ajps.2015.619313. [DOI] [Google Scholar]
- 5.Saikkonen K, Faeth SH, Helander M, Sullivan TJ. Fungal endophytes: a continuum of interactions with host plants. Annu. Rev. Ecol. Evol. Syst. 1998:319–343. doi: 10.1146/annurev.ecolsys.29.1.319. [DOI] [Google Scholar]
- 6.Schulz B, Boyle C. The endophytic continuum. Mycol. Res. 2005;109:661–686. doi: 10.1017/S095375620500273X. [DOI] [PubMed] [Google Scholar]
- 7.Singh LP, Gill SS, Tuteja N. Unraveling the role of fungal symbionts in plant abiotic stress tolerance. Plant Signal Behav. 2011;6:175–191. doi: 10.4161/psb.6.2.14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strobel GA. Endophytes as sources of bioactive products. Microbes Infect. 2003;5:535–544. doi: 10.1016/S1286-4579(03)00073-X. [DOI] [PubMed] [Google Scholar]
- 9.Nisa H, Kamili AN, Nawchoo IA, Shafi S, Shameem N, Bandh SA. Fungal endophytes as prolific source of phytochemicals and other bioactive natural products: A review. Microb. Pathog. 2015;82:50–59. doi: 10.1016/j.micpath.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Spaepen S, Vanderleyden J, Remans R. Indole‐3‐acetic acid in microbial and microorganism‐plant signaling. FEMS Microbiol. Rev. 2007;31:425–448. doi: 10.1111/j.1574-6976.2007.00072.x. [DOI] [PubMed] [Google Scholar]
- 11.Tamaoki M. The role of phytohormone signaling in ozone-induced cell death in plants. Plant Signal. Behav. 2008;3:166–174. doi: 10.4161/psb.3.3.5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuswinanti T, Syam'un E, Masniawati A. The Potency of endophytic fungal isolates collected from local aromatic rice as indole acetic acid (IAA) producer. Procedia Food Sci. 2015;3:96–103. doi: 10.1016/j.profoo.2015.01.009. [DOI] [Google Scholar]
- 13.Morrison EN, Knowles S, Hayward A, Thorn RG, Saville BJ, Emery RJN. Detection of phytohormones in temperate forest fungi predicts consistent abscisic acid production and a common pathway for cytokinin biosynthesis. Mycologia. 2015;107:245–257. doi: 10.3852/14-157. [DOI] [PubMed] [Google Scholar]
- 14.You YH, Kwak TW, Kang SM, Lee MC, Kim JG. Aspergillus clavatus Y2H0002 as a new endophytic fungal strain producing gibberellins isolated from Nymphoides pe ltata in fresh water. Mycobiology. 2015;43:87–91. doi: 10.5941/MYCO.2015.43.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Redman RS, Kim YO, Woodward CJ, Greer C, Espino L, Doty SL, et al. Increased fitness of rice plants to abiotic stress via habitat adapted symbiosis: a strategy for mitigating impacts of climate change. PLoS One. 2011;6:e14823. doi: 10.1371/journal.pone.0014823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmad N, Hamayun M, Khan SA, Khan AL, Lee IJ, Shin DH. Gibberellin-producing endophytic fungi isolated from Monochoria vaginalis. J. Microbiol. Biotechnol. 2010;20:1744–1749. [PubMed] [Google Scholar]
- 17.Hamayun M, Khan SA, Khan AL, Rehman G, Kim YH, Iqbal I, et al. Gibberellin production and plant growth promotion from pure cultures of Cladosporium sp. MH-6 isolated from cucumber (Cucumis sativus L.) Mycologia. 2010;102:989–995. doi: 10.3852/09-261. [DOI] [PubMed] [Google Scholar]
- 18.Vázquez MM, César S, Azcón R, Barea JM. Interactions between arbuscular mycorrhizal fungi and other microbial inoculants (Azospirillum, Pseudomonas, Trichoderma) and their effects on microbial population and enzyme activities in the rhizosphere of maize plants. Appl. Soil Ecol. 2000;15:261–272. doi: 10.1016/S0929-1393(00)00075-5. [DOI] [Google Scholar]
- 19.Yamada A, Ogura T, Degawa Y, Ohmasa M. Isolation of Tricholoma matsutake and T. bakamatsutake cultures from fieldcollected ectomycorrhizas. Mycosci. 2001;42:43–50. doi: 10.1007/BF02463974. [DOI] [Google Scholar]
- 20.You YH, Kang SM, Choo YS, Lee JM. Fungal diversity and plant growth promotion of endophytic fungi from six halophytes in Suncheon Bay. J. Microbiol. Biotechnol. 2012;22:1549–1556. doi: 10.4014/jmb.1205.05010. [DOI] [PubMed] [Google Scholar]
- 21.Wearn JA, Sutton BC, Morley NJ, Gange AC. Species and organ specificity of fungal endophytes in herbaceous grassland plants. J. Ecol. 2012;100:1085–1092. doi: 10.1111/j.1365-2745.2012.01997.x. [DOI] [Google Scholar]
- 22.Mozaffari E, Saghafipour A, Arzamani K, Jesri N, Kababian M, Hashemi SA. Geographical distribution, biodiversity, and species richness of medically important necrophagous flies in central Iran. J. Med. Entomol. 2020;57:377–381. doi: 10.1093/jme/tjz203. [DOI] [PubMed] [Google Scholar]
- 23.Fisher RA, Corbet AS, Williams CB. The relation between the number of species and the number of individuals in a random sample of an animal population. J. Anim. Ecol. 1943;12:42–58. doi: 10.2307/1411. [DOI] [Google Scholar]
- 24.Hughes AR, Cebrian J, Heck K, Goff J, Hanley TC, Scheffel W, et al. Effects of oil exposure, plant species composition, and plant genotypic diversity on salt marsh and mangrove assemblages. Ecosphere. 2018;9:e02207. doi: 10.1002/ecs2.2207. [DOI] [Google Scholar]
- 25.Choi WY, Rim SO, Lee JH, Lee JM, Lee IJ, Cho KJ, et al. Isolation of gibberellins-producing fungi from the root of several Sesamum indicum plants. J. Microbiol. Biotechnol. 2005;15:22–28. [Google Scholar]
- 26.Lahrmann U, Ding Y, Banhara A, Rath M, Hajirezaei MR, Döhlemann S, et al. Host-related metabolic cues affect colonization strategies of a root endophyte. Proc. Natl. Acad. Sci. USA. 2013;110:13965–13970. doi: 10.1073/pnas.1301653110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitter B, Petric A, Chain P, Trognitz F, Nowak J, Compant S, Sessitsch A. Genome analysis, ecology, and plant growth promotion of the endophyte Burkholderia phytofirmans strain PsJN. Mol. Microb. Ecol. Rhizos. 2013;1:865–874. doi: 10.1002/9781118297674.ch81. [DOI] [Google Scholar]
- 28.Verma VC, Kharwar RN, Strobel GA. Chemical and functional diversity of natural products from plant associated endophytic fungi. Nat. Prod. Commun. 2009;4:1511–1532. doi: 10.1177/1934578X0900401114. [DOI] [PubMed] [Google Scholar]
- 29.Shoresh M, Harman GE, Mastouri F. Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 2010;48:21–43. doi: 10.1146/annurev-phyto-073009-114450. [DOI] [PubMed] [Google Scholar]
- 30.Min YJ, Park M, Fong J, Quan Y, Jung S, Lim, Y Diversity and saline resistance of endophytic fungi associated with Pinus thunbergii in coastal shelterbelts of Korea. J. Microbiol. Biotechnol. 2014;24:324–333. doi: 10.4014/jmb.1310.10041. [DOI] [PubMed] [Google Scholar]
- 31.Zhang T, Zhang YQ, Liu HY, Wei YZ, Li HL, et al. Diversity and cold adaptation of culturable endophytic fungi from bryophytes in the Fildes Region, King George Island, maritime Antarctica. FEMS Microbiol. Lett. 2013;341:52–61. doi: 10.1111/1574-6968.12090. [DOI] [PubMed] [Google Scholar]
- 32.Li ZF, Wang LF, Feng ZL, Zhao LH, Shi YQ, Zhu HQ. Diversity of endophytic fungi from different Verticillium-wilt-resistant Gossypium hirsutum and evaluation of antifungal activity against Verticillium dahliae in vitro. J. Microbiol. Biotechnol. 2014;24:1149–1161. doi: 10.4014/jmb.1402.02035. [DOI] [PubMed] [Google Scholar]
- 33.Khan SA, Hamayun M, Yoon H, Kim HY, Suh SJ, Hwang SK, et al. Plant growth promotion and Penicillium citrinum. BMC Microbiol. 2008;8:231. doi: 10.1186/1471-2180-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.You YH, Yoon H, Kang SM, Shin JH, Choo YS, Lee IJ, Kim JG. Fungal diversity and plant growth promotion of endophytic fungi from six halophytes in suncheon bay. J. Microbiol. Biotechnol. 2012;22:1549–1556. doi: 10.4014/jmb.1205.05010. [DOI] [PubMed] [Google Scholar]
- 35.Khalmuratova I, Kim H, Nam YJ, Oh Y, Jeong MJ, Choi HR, et al. Diversity and plant growth promoting capacity of endophytic fungi associated with halophytic plants from the west coast of Korea. Mycobiology. 2015;43:373–383. doi: 10.5941/MYCO.2015.43.4.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waqas M, Khan AL, Kamran M, Hamayun M, Kang SM, Kim YH, et al. Endophytic fungi produce gibberellins and indoleacetic acid and promotes host-plant growth during stress. Molecules. 2012;17:10754–10773. doi: 10.3390/molecules170910754. [DOI] [PMC free article] [PubMed] [Google Scholar]