Abstract
The gene expression analysis of formalin-fixed paraffin-embedded (FFPE) tissues is often hampered by poor RNA quality, which results from the oxidation, cross-linking and other chemical modifications induced by the inclusion in paraffin. Yet, FFPE samples are a valuable source for molecular studies and can provide great insights into disease progression and prognosis. With the advancement of genomic technologies, new methods have been established that offer reliable and accurate gene expression workflows on samples of poor quality. NanoString is a probe-based technology that allows the direct counting of the mRNA transcripts and can be applied to degraded samples. Here, we have tested 2 RNA extraction methods for FFPE samples, and we have performed a titration experiment to evaluate the impact of RNA degradation and RNA input on the gene expression profiles assessed using the NanoString IO360 panel. We have selected FFPE samples of different DV200 values and assessed them on the nCounter platform with 2 different amounts of input RNA. This study concludes that the nCounter is a robust and reliable platform to assess the gene expression of RNA samples with DV200 > 30%; its robustness and ease of use could be of particular benefit to clinical settings.
Keywords: titration, RNA, gene expression, nCounter, FFPE
Introduction
The rapid advancement of genomic technologies over the past decades has led to individualized diagnostic and therapeutic approaches, allowing the implementation of precision medicine.1–4 Yet, the histological assessment remains the “gold standard” for pathology diagnostics,5 despite the lack of reproducibility due to the subjective interpretation of the histopathologists. Molecular technologies can help overcome arbitrary histological interpretation; however, their application has been so far limited by the cost and by the technical challenges that can arise when working with archival samples, such as formalin-fixed paraffin-embedded (FFPE) samples.5,6 FFPE samples are a valuable source for molecular studies and can provide great insights into disease progression and prognosis.6,7 Nevertheless, the quality of the RNA isolated from FFPE samples is generally poor due to oxidation, cross-linking, and other chemical modifications, which are also enhanced by prolonged air exposure.8,9 In fact, formalin induces the cross-linking between nucleic acids and proteins, which limits the conversion of mRNA to cDNA.10 Reliable and reproducible RNA extraction methods are warranted to ensure the isolation of sufficient amounts of good quality nucleic acids that can be processed for downstream molecular analyses. Some of the common methods used for gene expression analysis, including quantitative real-time PCR (qPCR) and microarrays, have been shown to lack reproducibility when applied to RNA of poor quality.7,10–12 The NanoString technology offers many advantages for gene expression profiling of RNA isolated from FFPE samples.13 NanoString is a probe-based technology that allows the direct counting of the mRNA transcripts. The technology employs molecular barcodes to count unique transcripts in a given reaction using single molecule imaging. The isolated RNA can be used directly for gene expression analysis without the need for cDNA conversion. Thus, the technology is particularly advantageous for gene expression analysis of specimens in which the nucleic acid cannot directly be amplified by PCR.12,13 NanoString showed higher sensitivity when compared to microarray technologies, and it has also been shown to work well with RNA of low input.13 Another study found that NanoString showed superior gene expression quantification results when compared to qPCR in archived FFPE samples.7 However, NanoString needs specific RNA requirements to be met for successful results with regards to the DV200, input amount and purity. Those requirements are explained below. NanoString recommends using the percentage of RNA fragments > 200 nt (DV200) as a reliable measure of RNA quality for the gene expression assays. In fact, although the RNA integrity number (RIN) is a reliable parameter to assess the integrity of RNA isolated from fresh, frozen tissues and cell cultures, it is not a definite quality metric for RNA isolated from FFPE samples. Thus, NanoString recommends measuring the DV200 for RNA isolated from FFPE samples. The DV200 is also used to calculate RNA input adjustments prior to sample processing. The NanoString IO360 panel, which we employed here, requires ∼100 ng of total RNA as starting material, nevertheless input corrections should be calculated by taking the DV200 values into account, for example, doubling the RNA input for samples that display a DV200 of 50%. In addition, purity is also an important factor to consider. For NanoString gene expression assays, the A260/A280 and A260/A230 ratios should be as close as possible to 2 and certainly above 1. In this study, we have compared 2 RNA extraction methods for FFPE samples, and we have performed a titration experiment to evaluate the impact of mRNA degradation and mRNA input on the gene expression profiles using the NanoString IO360 panel. We have selected 5 FFPE samples of different DV200 values and performed NanoString IO360 profiling with 2 different input RNA amounts per sample. We then evaluated the robustness and feasibility of the NanoString nCounter platform on RNA samples displaying different DV200 values.
Methods
Tissue Samples and RNA Isolation
A total of 46 samples were included in this study. Of those 46 samples, 11 were FFPE samples obtained from gut tissues surgically resected from patients with diagnosis of colorectal dysplasia or neoplasia and a previous history of inflammatory bowel disease (IBD) admitted at Hamad Medical Corporation (HMC, Doha, Qatar) (Table 1; Supplementary Table 1) and 35 were FFPE biopsies from either IBD, inflamed, or normal tissues obtained from pediatric patients admitted at Sidra Medicine (Supplementary Table 1). This study was approved by Sidra Medicine IRB (IRB# 1804022817; July 2019) and by HMC IRB (IRB# MRC-02-18-096; October 2019). Subjects submitted their written informed consent prior to participation in the study. Two consecutive 10-µm sections were obtained from each FFPE block, while the microtome blade was replaced, and the equipment sterilized after processing different blocks. The sections were transferred into microcentrifuge tubes and stored at room temperature. Two RNA extraction methods were tested in this study, the AllPrep DNA/RNA FFPE kit (Qiagen, Hilden, Germany) and the RNAstorm FFPE RNA extraction kit (Cell Data Sciences, Fremont, CA, USA). Samples were processed according to the manufacturer's recommendations.
Table 1.
Samples Employed in the Study.
| Sample ID | Concentration (ng/μL) | A260/A280 | A260/A230 | DV200 | Recommended input (ng) | Volume required (µL) | Input amount to test (ng) |
|---|---|---|---|---|---|---|---|
| HMC-FFPE-1 | 119.50 | 1.75 | 0.62 | 30–50% | 200 | 1.67 | 200; 400 |
| HMC-FFPE-2 | 49.77 | 2.00 | 1.08 | NA | NA | NA | - |
| HMC-FFPE-3 | 124.30 | 2.05 | 1.49 | >70% | 100 | 0.80 | - |
| HMC-FFPE-4 | 208.80 | 2.06 | 1.59 | >70% | 100 | 0.48 | - |
| HMC-FFPE-5 | 94.80 | 1.89 | 0.22 | 30–50% | 200 | 2.11 | 200; 400 |
| HMC-FFPE-6.1 | 165.10 | 2.20 | 1.61 | 50–70% | 150 | 0.91 | 150; 300 |
| HMC-FFPE-6.2 | 19.55 | 2.17 | 0.20 | 30–50% | 200 | 10.23 | - |
| HMC-FFPE-7 | 234.70 | 2.05 | 1.52 | >70% | 100 | 0.43 | 100; 200 |
| HMC-FFPE-8 | 794.90 | 2.11 | 1.73 | <30% | 300 | 0.38 | 300; 600 |
| HMC-FFPE-9 | 159.30 | 2.09 | 0.46 | <30% | 300 | 1.88 | - |
| HMC-FFPE-10 | 12.48 | 1.79 | 0.24 | 50–70% | 150 | 12.02 | - |
Note. In bold are the samples that have been processed for NanoString IO360 gene expression.
Abbreviations: FFPE: formalin-fixed paraffin-embedded; HMC: Hamad Medical Corporation.
The isolated RNA was assessed for quantity using Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). We report average RNA yield and concentration for the RNA isolation methods tested. As the RNA amount was of sufficient quantity only for the tissues collected from the adult patients, we have selected only those 11 samples for further analysis. The RNA quality was assessed using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The samples were run on Bioanalyzer using the Agilent RNA 6000 Pico Kit. Their concentration, absorbance ratios, and DV200 values are reported in Table 1. For each sample, we have calculated the “recommended input” that takes into account the DV200 correction and the “volume required” for NanoString assay based on the sample concentration and assuming 100 ng as the ideal input RNA amount (Table 1). Five samples displaying different DV200 values were further selected for the titration experiment, each sample was processed with 2 different RNA input amounts, for a total of 10 samples. The samples obtained from the same FFPE block were considered belonging to the same “sample pair”.
The nonparametric Mann–Whitney U test was applied to check significant differences in the concentration, yield, and absorbance ratios between the 2 RNA extraction methods tested in this study.
Gene Expression Using NanoString Technology
The PanCancer IO360 panel was used to assess gene expression. The panel includes 750 genes that cover the key pathways at the interface of the tumor, tumor microenvironment, and immune response and 20 internal reference genes for data normalization. The gene list is available at https://www.nanostring.com/wp-content/uploads/2021/01/LBL-10498-02_IO_360_Gene_List.xlsx.
The samples’ preparation, hybridization, detection, and scanning were performed as per manufacturer's instructions. Briefly, the hybridization master mix was created by adding 70 µL of hybridization buffer to 42 µL of the reporter code sets. Eight µL of the hybridization master mix were combined with 5 µL of sample containing 100 to 600 ng of total RNA. Then, 2 µL of Capture ProbeSet was added to each tube and mixed by inverting and flicking the tubes. The hybridization was carried out at 65 °C for 16 h. After hybridization, samples were immobilized and washed using the automated nCounter Prep Station (NanoString Technologies) and then imaged on the nCounter Digital Analyzer (NanoString Technologies) at 555 fields of view (FOV). We have tested 5 pairs of samples with different DV200 values. For each sample pair, we have assessed 2 different RNA inputs (Table 1). Along with the 5 sample pairs, we have included the PanCancer IO360 panel standard and an internal pooled control RNA obtained from blood samples of 10 healthy donors, 100 ng were used as total RNA input. The panel standard contains a pool of synthetic DNA oligonucleotides that correspond to the target sequence of each of the 770 unique probe targets in the panel. The nSolver data analysis software version 4.0 (NanoString Technologies) was used for QC and for the normalization of the raw gene expression counts. We used the recommended default parameters for QC flagging; briefly, flags were generated if samples did not meet the following QC criteria: imaging threshold with FOV registration of at least 75%, binding density between 0.1 and 2.25, positive control linearity with R2>0.95, positive control limit of detection of 0.5fM, positive control > or = 2 standard deviations above the mean of the negative controls. Background subtraction was performed by subtracting the mean of the negative controls from all data points. All the samples were normalized to the geometric mean of the positive control probes and to the geometric mean of the housekeeping (HK) genes.
The Advanced Analysis module of nSolver data analysis software version 4.0 was used to generate heatmaps and principal component analysis plots where relevant.
Results
FFPE RNA Isolation and QC
To assess yield, concentration, and absorbance ratios, we have divided the samples into 3 groups, namely: (i) samples obtained from adults and isolated using the AllPrep DNA/RNA FFPE kit (n = 11); (ii) samples obtained from pediatric biopsies and isolated using the AllPrep DNA/RNA FFPE kit (n = 20); and (iii) samples obtained from pediatric biopsies and isolated using the RNAstorm FFPE RNA extraction kit (n = 15). For the 11 samples obtained from adults and isolated using the AllPrep DNA/RNA FFPE kit, the average RNA yield was 5.41 μg (range: 0.38-23.85 μg) with an average concentration of 180.29 ng/μL (range: 12.48-795.0 ng/μL), an average A260/A280 ratio of 2.01 (range: 1.75-2.2), and an average A260/A230 ratio of 0.97 (range: 0.22-1.73). For the 20 samples obtained from pediatric biopsies and isolated using the AllPrep DNA/RNA FFPE kit, the average RNA yield was 392.02 ng (range: 39.0-2039.0 ng) with an average concentration of 13.07 ng/μL (range: 1.31-68.0 ng/μL), an average A260/A280 ratio of 2.11 (range: 1.12-9.67), and an average A260/A230 ratio of 0.15 (range: 0.01-1.22). For the 15 samples obtained from pediatric biopsies and isolated using the RNAstorm FFPE RNA extraction kit, the average RNA yield was 999.0 ng (range: 33.0-2534.0 ng) with an average concentration of 19.98 ng/μL (range: 0.67-50.7 ng/μL), an average A260/A280 ratio of 1.8 (range: 0.68-2.53), and an average A260/A230 ratio of 0.89 (range: 0.35-2.09). It should be noted that the high variability in the concentration, yield, and absorbance ratios might depend on the quality of the tissue samples and the time the inclusion in paraffin occurred. Additionally, Nanodrop sensitivity decreases proportionally to the decrease in sample concentration and can lead to off-scale values.
We sought to determine whether the 2 different isolation methods led to different concentration, yield, and absorbance ratios. As the RNAstorm FFPE RNA extraction kit was tested only on the pediatric biopsies, we compared the 2 extraction methods for the samples obtained from the pediatric biopsies only. The nonparametric Mann–Whitney U test was used to assess the statistical significance of the comparisons performed. No statistical difference was observed between the concentration and A260/A280 values. However, the A260/A230 values were statistically different between the RNAstorm FFPE RNA kit and the AllPrep DNA/RNA FFPE kit (Mann–Whitney U test; Z = −4.5586; P < .0001), suggesting that the RNAstorm FFPE RNA kit might perform better in removing contaminants. Additionally, the RNAstorm FFPE RNA kit resulted in a higher yield when compared to the AllPrep DNA/RNA FFPE kit (Mann–Whitney U test; Z = −2.2168; P = .0266).
As only the 11 samples obtained from adult tissues had an acceptable amount of RNA for NanoString profiling, we have selected only those 11 samples for further analyses. These samples were run on the Bioanalyzer for the assessment of the DV200. The DV200 values obtained from the samples assessed are reported in Table 1. Five samples displaying different DV200 values and absorbance ratios were selected for the titration experiment (Table 1).
QC Metrics Overview
We have chosen 555 FOV for the NanoString imaging setting. The imaging was satisfactory across the samples assessed; the average counted FOV was 551 (range: 547-554 FOV). The binding density was within the recommended range (average 0.24, range: 0.1–0.46); we expected to see an increase in the binding density proportionate to the increase of input RNA; however, this was not the case for all the pairs assessed, except for the sample HMC_FFPE6.1 (DV200 = 50%-70%), whose binding density increased from 0.23 to 0.46 proportionally to the increase of RNA input from 150 to 300 ng. The positive controls displayed linearity as expected.
All the samples were normalized to the geometric mean of the positive control probes and to the geometric mean of the HK genes. Upon normalization the 2 replicates with DV200 < 30% displayed a QC flag, indicating that they had a content normalization factor at least 10-fold different from the average sample in the same experiment. This was likely caused by an insufficient number of RNA targets counted. Since the QC analysis did not return any other QC flags, the assay worked well; however, the flagged samples contained low RNA molecules to count, as expected from degraded samples with low DV200.
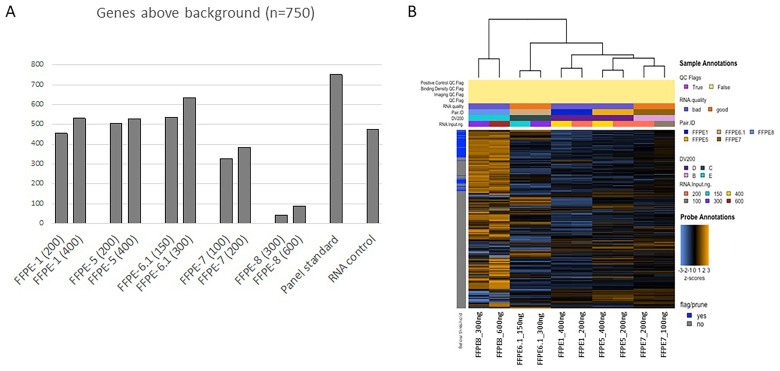
For each sample assessed, we have calculated the background, which is represented by the mean of the negative controls plus 2 standard deviations. We have then used 30 counts as the cut-off to calculate the number of targets that are robustly above the background. As expected, the number of genes above the background was higher when a higher amount of RNA was used as input for each of the 5 sample pairs assessed (Figure 1A). Nevertheless, the sample pair with DV200 <30% displayed a number of genes above background lower than 100 even when 600 ng of RNA were used as input, suggesting that samples with a DV200 value lower than 30% might be too degraded and could lead to unsuccessful results even when RNA input corrections are taken into account (Figure 1A). It should also be noted that the number of genes above background for the RNA control sample was lower when compared to the other samples, because the RNA was isolated from the blood of 10 healthy individuals, hence, it should not express cancer-related genes.
Figure 1.
(A) Bar plot of the genes detected above the background; for each sample, the RNA input in ng is reported in brackets. (B) Unsupervised heatmap of the 5 sample pairs assessed in the study. Samples are categorized into A, with DV200 = 100%; B, with DV200 > 70%; C, with DV200 = 50% to 70%; D, with DV200 = 30% to 50%; and E, with DV200 < 30%. None of the samples displayed in the figure had a DV200 = 100%; hence no sample is represented into category “A”. The “good” category refers to samples with DV200 > 50% while the “bad” category refers to samples with DV200 < 50%. Orange indicates high expression; blue indicates low expression.
Gene Expression Analysis
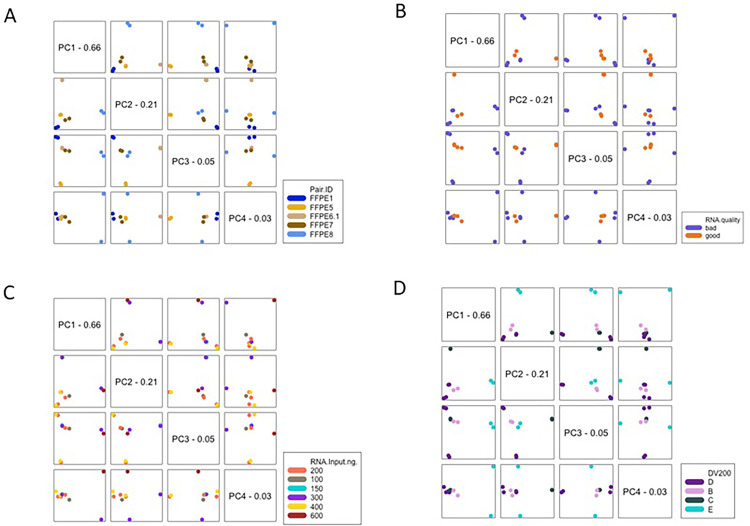
To gain a high-level exploratory view of the data, we have generated an unsupervised heatmap of the normalized counts, scaled to give all the genes equal variance (Figure 1B). Samples were categorized into A, with DV200 = 100%; B, with DV200 > 70%; C, with DV200 = 50% to 70%; D, with DV200 = 30% to 50%; and E, with DV200 < 30%. None of the samples displayed in Figure 1B had a DV200 = 100%; hence no sample is represented into category “A”. The “good” category referred to samples with DV200 > 50% while the “bad” category referred to samples with DV200 < 50% (Figure 1B). The self-organizing heatmap was highly predictive of the sample pairs, while the RNA control sample clustered independently. We next generated principal component plots according to the different covariates assessed in this study. Principal component analysis maps high-dimensional datasets into a smaller number of informative dimensions. We have plotted the first 4 principal components of the gene expression data against each other and colored the samples according to the different covariates. Specifically, we have generated correlation matrices of the first 4 principal components for the following covariates: pair ID (Figure 2A), RNA quality (Figure 2B), RNA input (Figure 2C), and DV200 categories (Figure 2D). We have excluded the panel standard and the RNA control sample from this analysis to avoid any confounding effect.
Figure 2.
Correlation matrices of the first 4 principal components according to the (A) Pair ID, (B) RNA quality, (C) RNA input, and (D) DV200.
Sample belonging to the same pair were found closer to each other while no significant effect was found for the other covariates, suggesting that the biological variability across sample pairs was higher than the technical variability due to the different RNA input used or the different RNA quality (Figure 2A–2D).
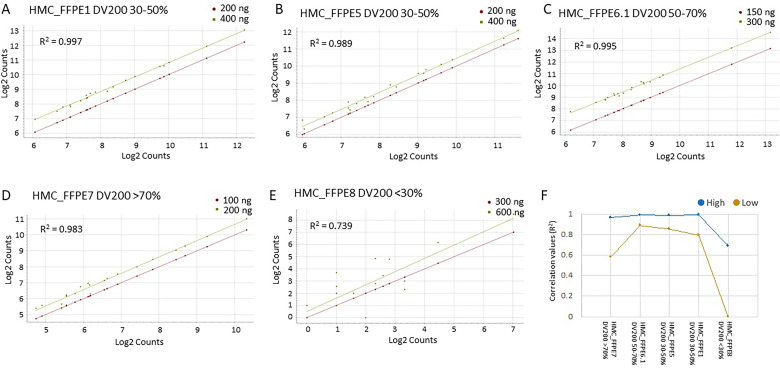
To gain a better understanding of the effect of the DV200 on gene expression, we have used the 20 HK genes to generate correlation plots for each sample pair assessed. As expected, the expression values of the HK genes, represented as Log2 counts, were higher for the sample with higher RNA input for each pair.
For all the sample pairs with DV200 > 30%, the R2 of the Log2 counts was higher than 0.9, while the sample pair with DV200 <30% displayed an R2 value of 0.739, suggesting that samples with DV200 <30% might be too degraded to generate data with high correlation (Figure 3A–3E). The sample pair with DV200 <30% added variability to the HK gene expression. In fact, the expression of the HK genes was less variable when the sample pair with DV200 <30% was removed from the box and whisker plot (Supplementary Figure 1).
Figure 3.
Correlation analysis of the Log2 Counts of the HK genes (N = 20) for the following sample pair: (A) FFPE1, (B) FFPE5, (C) FFPE6.1, (D) FFPE7, and (E) FFPE8. (F) Line graph of the R2 values for the high-expressed (blue) and low-expressed (yellow) genes for the 5 sample pairs assessed in this study.
Abbreviations: FFPE: formalin-fixed paraffin-embedded; HK: housekeeping.
We next sought to determine whether the degradation of the RNA, hence the DV200 values, affected the low-abundance genes more than the high-abundance genes. Toward this aim, the total 750 genes (excluding the 20 HK genes) were ranked based on the average counts and divided into 2 groups of equal numerosity (N = 375) of low and high expression in each of the sample pairs assessed. The correlation analysis returned an R2 value > 0.9 for the high-abundance genes in all the sample pairs except for the sample pair with DV200 < 30%, for which the R2 = 0.739 (Figure 3F). The low-abundance genes showed a poorer correlation across the sample pairs, although the R2 value was still above 0.8 for the sample pairs with DV200 > 30%, except for the sample pair HMC_FFPE7 with DV200 >70%. This was an interesting finding as we expected a higher correlation for samples displaying a high DV200. When looking back at the gel image from the Bioanalyzer QC, we realized that the sample FFPE7 had an abnormal smear, suggesting the presence of contaminants that might have interfered with the hybridization especially for the low-expressed genes. We concluded that a thorough evaluation of the Bioanalyzer smear is required to make sure that the DV200 estimate is accurate (Supplementary Figure 2). The absorbance ratios did not affect the success of the analysis. For instance, sample HMC_FFPE5 produced good quality data even if the A260/A230 was 0.22. Similarly, samples HMC_FFPE5 and HMC_FFPE1 had A260/A280 values lower than 2.0; yet, their NanoString analysis returned good quality data.
Discussion
Diagnostic workflows require reproducible methods that can be paired to the current standard of care.5 Understanding the transcriptomic makeup within tissue samples is of paramount relevance to the goal of personalized medicine. The gene expression analysis of FFPE samples can be challenging due to a generally more degraded RNA when compared to RNA samples isolated from other sources.9 In fact, the sample preservation by formalin inclusion hinders several molecular applications.10 FFPE samples are challenging when employing standard RNA extraction workflows and often lead to RNA of poor quality and quantity.14,15 In fact, formaldehyde induces the formation of adducts and crosslinks and interferes with the analysis of RNA itself.14 Additionally, the RNA isolated from FFPE samples is typically fragmented and its utility might also be hampered by the presence of contaminants and inhibitors.16,17 Thus, it is of paramount importance to establish a robust and reproducible method to obtain RNA of sufficient quality and quantity for downstream analyses. This study compares 2 different extraction methods, the AllPrep DNA/RNA FFPE RNA extraction kit (Qiagen) and the RNAstorm FFPE RNA extraction kit (Cell Data Sciences) for downstream NanoString-based gene expression. The AllPrep DNA/RNA FFPE RNA kit is a dual extraction kit that provides the advantage of the concurrent isolation of DNA and RNA from the same tissue. The RNAstorm FFPE RNA extraction kit employs chemical catalysts that accelerates the destruction of adducts induced by the formaldehyde, including alkylated and crosslinked bases.18 In this study we show that the RNAstorm FFPE RNA extraction kit performed slightly better than the AllPrep DNA/RNA FFPE kit, as it led to significantly higher yield and purity. It is known that by spectrophotometric assessment, RNA extraction kits can lead to low A260/A230 ratios, indicating the presence of potential organic contaminants in the eluent.15,17,19–21 Contaminants can inhibit downstream applications,15,22 thus extraction kits leading to higher A260/A230 ratios are preferred.
The RNA quantity obtained from the pediatric biopsies was insufficient to proceed with the NanoString workflow, thus only the samples obtained from the adult cohort were used for the further steps. It should be noted that the grade of inflammation and infiltration as well as the stromal component might affect the RNA yield. The lack of such correlation represents a limitation of the current study.
Another limitation is the sample size. No power analysis was performed for the current study. We are planning to validate our findings in a bigger cohort to assure adequate power to detect statistical significance, and to test samples coming from different disease models.
NanoString showed higher sensitivity when compared to microarray technologies, and it has also been shown to work well with RNA of low input.13
Another study found that NanoString showed superior gene expression quantification results when compared to qPCR in archived FFPE samples.7 Here, we have assessed not only the effect of RNA input but also the DV200 (i.e., RNA quality) on NanoString gene expression profiling. We demonstrate that samples with DV200 > 30% produce consistent results. When taking RNA input corrections into account we did not find any significant difference in the number of genes detected above background. Correlative analyses between the 2 different RNA input amounts were also strong for all the samples with DV200 >30%, pointing to NanoString as a reliable and methodologically robust platform. In light of these findings, we plan to test the NanoString gene expression profiles on FFPE samples of low quantity/quality for different disease models.
The nCounter system offers many advantages as it is technically simple, requiring overall ∼15 min of hands-on time and being methodologically robust even when working with samples of poor quality. NanoString also offers the possibility to run as little as 10 ng of RNA isolated from FFPE samples using the low RNA input kit. This kit is specifically optimized for FFPE samples and crude cell lysates and overcomes limitations related to sample quantity. All these advantages make NanoString a great platform to implement into clinical routine settings as it delivers sensitive and reproducible results without requiring advanced wet and dry lab skills.23 The other 2 major gene expression technologies include microarrays and next-generation sequencing. Nevertheless, they have several disadvantages for their implementation in the clinical settings. Microarrays are in fact limited by the low dynamic range of detection and by their inability to process nucleic acids of poor quality and of limited amount.23–26 In contrast, RNA sequencing is highly sensitive and can offer precise measurement of transcript level. However, its cost and its relatively complex manipulation during library preparation and data analysis might limit its application in clinical settings.27,28
Conclusions
We believe that the data provided in this paper supports the use of NanoString technology in the clinical settings for the development of predictive and prognostic signatures toward the implementation of personalized medicine. Further studies employing bigger cohorts are warranted to confirm our results.
Supplemental Material
Supplemental material, sj-pdf-1-tct-10.1177_15330338221129710 for Gene Expression Profiling of FFPE Samples: A Titration Test by Harshitha Shobha Manjunath, Moza Al Khulaifi, Heba Sidahmed, Adham Ammar, Jayakumar Vadakekolathu, Sergio Rutella, Muneera Jassim Al-Mohannadi, Mamoun Elawad, William Mifsud, Adrian Charles, Cristina Maccalli and Sara Tomei in Technology in Cancer Research & Treatment
Supplemental material, sj-xlsx-2-tct-10.1177_15330338221129710 for Gene Expression Profiling of FFPE Samples: A Titration Test by Harshitha Shobha Manjunath, Moza Al Khulaifi, Heba Sidahmed, Adham Ammar, Jayakumar Vadakekolathu, Sergio Rutella, Muneera Jassim Al-Mohannadi, Mamoun Elawad, William Mifsud, Adrian Charles, Cristina Maccalli and Sara Tomei in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-3-tct-10.1177_15330338221129710 for Gene Expression Profiling of FFPE Samples: A Titration Test by Harshitha Shobha Manjunath, Moza Al Khulaifi, Heba Sidahmed, Adham Ammar, Jayakumar Vadakekolathu, Sergio Rutella, Muneera Jassim Al-Mohannadi, Mamoun Elawad, William Mifsud, Adrian Charles, Cristina Maccalli and Sara Tomei in Technology in Cancer Research & Treatment
Abbreviations
- FOV
field of view
- FPPE
formalin-fixed paraffin-embedded
- HK
housekeeping
- HMC
Hamad Medical Corporation
- IBD
inflammatory bowel disease
- RIN
RNA integrity number
Footnotes
Author’s Contribution: ST conceived the study. HSM performed the RNA QC, the NanoString experiment, and the QC analysis of the NanoString data. MAK and HS performed the RNA isolation experiments. MJA and ME selected the patients. AA, WM, and AC retrieved and prepared the histological samples and provided their phenotypic information. JV and SR contributed to the manuscript editing. ST and CM wrote the manuscript and generated graphs and tables. All authors discussed the results and approved the final submitted version. The contents of the paper have not been published previously.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Ethical Approval: This study was approved by Sidra Medicine IRB (IRB# 1804022817; July 2019) and by HMC IRB (IRB# MRC-02-18-096; October 2019).
ORCID iDs: Harshitha Shobha Manjunath https://orcid.org/0000-0001-6626-9254
Cristina Maccalli https://orcid.org/0000-0002-6737-4715
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Mirnezami R, Nicholson J, Darzi A. Preparing for precision medicine. N Engl J Med. 2012;366:489-491. [DOI] [PubMed] [Google Scholar]
- 2.Ten Have H, Gordijn B. Precision in health care. Med Health Care Philos. 2018;21:441-442. [DOI] [PubMed] [Google Scholar]
- 3.Scatena C, Murtas D, Tomei S. Cutaneous melanoma classification: the importance of high-throughput genomic technologies. Front Oncol. 2021;11:635488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thareja G, Al-Sarraj Y, Belkadi Aet al. Whole genome sequencing in the Middle Eastern Qatari population identifies genetic associations with 45 clinically relevant traits. Nat Commun. 2021;12:1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adam B, Afzali B, Dominy KMet al. Multiplexed color-coded probe-based gene expression assessment for clinical molecular diagnostics in formalin-fixed paraffin-embedded human renal allograft tissue. Clin Transplant. 2016;30:295-305. [DOI] [PubMed] [Google Scholar]
- 6.Blow N. Tissue preparation: tissue issues. Nature. 2007;448:959-963. [DOI] [PubMed] [Google Scholar]
- 7.Reis PP, Waldron L, Goswami RSet al. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol . 2011;11:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27:4436-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kojima K, April C, Canasto-Chibuque Cet al. Transcriptome profiling of archived sectioned formalin-fixed paraffin-embedded (AS-FFPE) tissue for disease classification. PLoS One. 2014;9:e86961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norton N, Sun Z, Asmann YWet al. Gene expression, single nucleotide variant and fusion transcript discovery in archival material from breast tumors. PLoS One. 2013;8:e81925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott DW, Chan FC, Hong Fet al. Gene expression-based model using formalin-fixed paraffin-embedded biopsies predicts overall survival in advanced-stage classical Hodgkin lymphoma. J Clin Oncol. 2013;31:692-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veldman-Jones MH, Brant R, Rooney Cet al. Evaluating robustness and sensitivity of the NanoString technologies nCounter platform to enable multiplexed gene expression analysis of clinical samples. Cancer Res. 2015;75:2587-2593. [DOI] [PubMed] [Google Scholar]
- 13.Geiss GK, Bumgarner RE, Birditt Bet al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317-325. [DOI] [PubMed] [Google Scholar]
- 14.Groelz D, Sobin L, Branton P, Compton C, Wyrich R, Rainen L. Non-formalin fixative versus formalin-fixed tissue: a comparison of histology and RNA quality. Exp Mol Pathol. 2013;94:188-194. [DOI] [PubMed] [Google Scholar]
- 15.Patel PG, Selvarajah S, Guerard KPet al. et al. Reliability and performance of commercial RNA and DNA extraction kits for FFPE tissue cores. PLoS One. 2017;12:e0179732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okello JB, Rodriguez L, Poinar Det al. Quantitative assessment of the sensitivity of various commercial reverse transcriptases based on armored HIV RNA. PLoS One. 2010;5:e13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okello JB, Zurek J, Devault AMet al. Comparison of methods in the recovery of nucleic acids from archival formalin-fixed paraffin-embedded autopsy tissues. Anal Biochem. 2010;400:110-117. [DOI] [PubMed] [Google Scholar]
- 18.Karmakar S, Harcourt EM, Hewings DSet al. Organocatalytic removal of formaldehyde adducts from RNA and DNA bases. Nat Chem. 2015;7:752-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radstrom P, Knutsson R, Wolffs P, Lovenklev M, Lofstrom C. Pre-PCR processing: strategies to generate PCR-compatible samples. Mol Biotechnol. 2004;26:133-146. [DOI] [PubMed] [Google Scholar]
- 20.King C, Debruyne R, Kuch M, Schwarz C, Poinar H. A quantitative approach to detect and overcome PCR inhibition in ancient DNA extracts. Biotechniques. 2009;47:941-949. [DOI] [PubMed] [Google Scholar]
- 21.Schrader C, Schielke A, Ellerbroek L, Johne R. PCR inhibitors - occurrence, properties and removal. J Appl Microbiol. 2012;113:1014-1026. [DOI] [PubMed] [Google Scholar]
- 22.Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15:155-166. [PMC free article] [PubMed] [Google Scholar]
- 23.Tsang HF, Xue VW, Koh SP, Chiu YM, Ng LP, Wong SC. Nanostring, a novel digital color-coded barcode technology: current and future applications in molecular diagnostics. Expert Rev Mol Diagn. 2017;17:95-103. [DOI] [PubMed] [Google Scholar]
- 24.Heller MJ. DNA Microarray technology: devices, systems, and applications. Annu Rev Biomed Eng. 2002;4:129-153. [DOI] [PubMed] [Google Scholar]
- 25.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu X, Fu N, Guo Set al. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics. 2009;10:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S, Lin L, Jiang P, Wang D, Xing Y. A comparison of RNA-Seq and high-density exon array for detecting differential gene expression between closely related species. Nucleic Acids Res. 2011;39:578-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-tct-10.1177_15330338221129710 for Gene Expression Profiling of FFPE Samples: A Titration Test by Harshitha Shobha Manjunath, Moza Al Khulaifi, Heba Sidahmed, Adham Ammar, Jayakumar Vadakekolathu, Sergio Rutella, Muneera Jassim Al-Mohannadi, Mamoun Elawad, William Mifsud, Adrian Charles, Cristina Maccalli and Sara Tomei in Technology in Cancer Research & Treatment
Supplemental material, sj-xlsx-2-tct-10.1177_15330338221129710 for Gene Expression Profiling of FFPE Samples: A Titration Test by Harshitha Shobha Manjunath, Moza Al Khulaifi, Heba Sidahmed, Adham Ammar, Jayakumar Vadakekolathu, Sergio Rutella, Muneera Jassim Al-Mohannadi, Mamoun Elawad, William Mifsud, Adrian Charles, Cristina Maccalli and Sara Tomei in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-3-tct-10.1177_15330338221129710 for Gene Expression Profiling of FFPE Samples: A Titration Test by Harshitha Shobha Manjunath, Moza Al Khulaifi, Heba Sidahmed, Adham Ammar, Jayakumar Vadakekolathu, Sergio Rutella, Muneera Jassim Al-Mohannadi, Mamoun Elawad, William Mifsud, Adrian Charles, Cristina Maccalli and Sara Tomei in Technology in Cancer Research & Treatment