Abstract
pANCA is a marker antibody associated with inflammatory bowel disease (IBD), including most patients with ulcerative colitis and a subset with Crohn's disease. This study addressed the hypothesis that pANCA reacts with an antigen(s) of microbial agents potentially relevant to IBD pathogenesis. Using a pANCA monoclonal antibody, we have previously identified the C-terminal basic random-coil domain of histone H1 as a pANCA autoantigen. BLAST analysis of the peptide databases revealed H1 epitope homologues in open reading frames of the Mycobacterium tuberculosis genome. Western analysis of extracts from six mycobacterial species directly demonstrated reactivity to a single, conserved ∼32-kDa protein. Direct protein sequencing, followed by gene cloning, revealed a novel 214-amino-acid protein, an iron-regulated protein recently termed HupB. Sequence analysis demonstrated its homology with the mammalian histone H1 gene family, and recombinant protein expression confirmed its reactivity with the 5-3 pANCA monoclonal antibody. Binding activity of patient serum immunoglobulin G (IgG) to HupB did not correlate with reactivity to histone H1 or pANCA, indicating the complex character of the pANCA antigen. However, anti-HupB IgA was strongly associated with Crohn's disease (P < 0.001). These findings indicate that the 5-3 pANCA monoclonal antibody detects a structural domain recurrent among mycobacteria and cross-reactive with a DNA-binding domain of histone H1. The association of HupB-binding serum IgA with IBD provides new evidence for the association of a mycobacterial species with Crohn's disease.
Inflammatory bowel disease (IBD) encompasses several closely related chronic inflammatory diseases involving T-cell-mediated destruction of the intestinal mucosa (1, 45, 50). Familial disease pattern and genetic susceptibility loci reflect a hereditary component of disease pathogenesis. (9, 15, 20, 34–36, 42, 44, 52, 63, 69). These findings have often been interpreted as evidence of an autoimmune basis. However, variation in penetrance and demographic and epidemiologic features indicate an important role for environmental factors in the inflammatory process.
Intestinal bacteria have been increasingly implicated as an environmental factor in IBD, due to their mucosal localization and antigenic and immunomodulatory components. This concept is supported by correlative clinical evidence and by direct validation in several rodent IBD model systems (6, 7, 18, 32, 33, 43, 47, 64, 67). Notably, Elson and colleagues have demonstrated that colonic bacteria are antigenic targets of disease-associated T- and B-cell immune responses in the C3H/HeJBir mouse (6, 16). Immunoregulation mediated by gut flora is directly relevant to disease pathology, since CD4+ T cells transfer disease in mouse model systems (39, 48).
These observations imply that the human disease-specific immune response might be useful in the identification of microorganisms which contribute to human disease. High serum levels of anti-neutrophil cytoplasmic antibodies (pANCA) are a marker immune response in IBD associated with 60 to 70% of patients with ulcerative colitis (UC) and a subset of Crohn's disease (CD) patients. These findings are interpreted as evidence that pANCA expression is an immunologic trait related to disease susceptibility (21, 51, 55, 68). Notably, pANCA and IBD-associated antibacterial serum antibodies were recently reported to cross-compete for bacterial and pANCA antigen binding (54). However, the bacterial species and proteins involved in this cross-reaction have not been defined.
Our laboratory has isolated human pANCA monoclonal antibodies and characterized their autoantigen and epitope specificity (a COOH-terminal recurrent peptide motif in histone H1) (21a, 22). The present study employed these antibodies and sequence information to search for a cross-reactive microbial antigen, resulting in the identification and cloning of HupB, a new protein of mycobacterial origin.
MATERIALS AND METHODS
Antibodies and detection reagents.
Fab 5-2, Fab 5-3, and P313 anti-tetanus toxoid rFabs were produced and purified as previously described (22). P313 producing vector was a generous gift of Carlos Barbas III (4). Alkaline phosphatase-conjugated goat anti-human Fab, immunoglobulin G (IgG), and IgA antibodies were purchased from Pierce (Rockford, Ill.); goat anti-mouse IgG was from Sigma Chemical Co. (St. Louis, Mo.).
Human sera.
Sera from 70 UC and CD patients and healthy controls were obtained from the serum archive of the Cedars-Sinai IBD Research Center. Sera were produced from standard phlebotomy blood specimens, anonymously number coded, aliquoted, and stored at −80°C until use. The methodology for nonbiased specimen selection from this archive has been previously described (55). Quantitation of UC pANCA binding activity was previously performed on all archival specimens (53). Procedures for subject recruitment, informed consent, and specimen procurement were in accordance with protocols approved by the Institutional Human Subject Protection Committees of the University of California at Los Angeles (UCLA) and the Cedars-Sinai Medical Center.
Mycobacterial culture.
Mycobacterium tuberculosis Erdman (ATCC 35801), M. avium (ATCC 25291), M. avium subsp. paratuberculosis (ATCC 19698), M. avium subsp. paratuberculosis “Linda” (ATCC 43015, isolated from a CD patient), M. smegmatis (ATCC 14468), M. bovis (ATCC 19210), and M. bovis BCG (bacille Calmette-Guérin; ATCC 19274) were grown in unshaken 300-cm2 Falcon tissue culture flasks (Becton Dickinson, Oxnard, Calif.) for 3 weeks (7 to 10 days for the M. avium and M. smegmatis strains) in 7H9 (Difco Laboratorie, Detroit, Mich.) or Sauton's medium with glycerol but without bovine albumin and Tween 80 at pH 6.7 and 37°C in a 5% CO2–95% air atmosphere. M. smegmatis ATCC 14468 was grown in shaken Erlenmeyer flasks for 3 days in 7H9 (Difco) or Sauton's medium with glycerol but without bovine albumin and Tween 80 at pH 6.7 and 37°C in an environmental incubator. During the entire growth phase, all mycobacterial cultures were subjected weekly or daily (M. avium and M. smegmatis) to gentle sonication three pulses of 1 min each at 50 or 60 Hz) to maintain the cultures as single-cell suspensions, to counter their strong tendency to grow in clumps that continuously increase in size. Typically, mycobacteria were cultured from an initial cell density of 1 × 105 to 5 × 105/ml to a final density of 1 × 105 to 5 × 108/ml.
Preparation of subcellular fractions.
Mycobacterial cultures were separated into cell pellets and culture supernatants by centrifugation at 3,000 × g for 30 min at 4°C. Cell pellets were taken up in a small volume of phosphate-buffered saline (PBS; 50 mM sodium phosphate, 150 mM sodium chloride, pH 7.2) and lysed by vigorous vortexing with 60-mesh crystalline alumina beads (Fisher, Pittsburgh, Pa.) for 5 min at room temperature and boiling for 10 min in polyacrylamide gel loading buffer (125 mM Tris/Cl [pH 6.8], 30% glycerol, 4% sodium dodecyl sulfate [SDS], 500 mM 2-mercaptoethanol, 0.02% Coomassie brilliant blue R). Insoluble cellular material was collected by centrifugation at 10,000 × g for 10 min, and solubilized cellular proteins were adjusted to a final volume such that 1 μl contained the proteins of ∼108 lysed cells. Culture supernatants were first filtered through Tuffryn 0.45- and 0.22-μm-pore-size filters (Gelman Sciences, Ann Arbor, Mich.) and then concentrated by tangential flow through a Filtron polyethersulfone membrane with a 3-kDa cutoff (Gelman Sciences). Proteins in these concentrates were precipitated with ammonium sulfate at 100% saturation, pelleted by centrifugation at 10,000 × g for 20 min, dialyzed against PBS at 4°C, and finally brought up in polyacrylamide gel loading buffer such that 1 μl contained the proteins of ∼108 cell equivalents. Proteins in the cell pellets and culture supernatants were analyzed for integrity by electrophoresis on standard 10% denaturing polyacrylamide gels and then stained with Coomassie brilliant blue R. Protein concentrations in the cell pellets and culture supernatants were determined by the bicinchoninic acid reagent (Pierce).
Sequence and database analyses.
Homologues of the histone H1-1 (H1d) amino acid (aa) 108 to 212 sequence were identified in the National Institutes of Health (NIH) nonredundant database with the National Center for Biotechnology Information BLASTP program (version 1.4.6.MP, June 1994) and a BLOSUM 62 scoring matrix. Homologues of the N-terminal (aa 1 to 107) and C-terminal (aa 108 to 214) segments of HupB were similarly identified by a search of the database with BLASTP (version 1.4.11, November 1997) (3). Alignments were performed by using the CLUSTAL W Multiple Sequence Alignment Program (version 1.7, June 1997) (61), and CLUSTAL W absolute scores were used as a measure of protein identity. The number of histone H1 peptide motifs (PAKKAA, SPKKAKK, PKKAKK, and PKKA) in each homologue was determined by manual sequence inspection.
Western immunoblot analysis.
Mycobacterial cell lysates (10 μg/well) were separated on 13% polyacrylamide gels under reducing conditions in Laemmli buffer. Proteins were transferred overnight to nitrocellulose membranes (Amersham Life Sciences, Buckinghamshire, England) in Tris glycine buffer (National Diagnostics, Atlanta, Ga.) and verified by Ponceau S red staining (Sigma). Membranes were blocked in 5% nonfat milk (Carnation, Glendale, Calif.) in PBS with 0.1% Tween 20 for 1 h. Primary and secondary antibodies diluted in 1% milk in PBS-Tween 20 were incubated with membranes for 1 h. Fab 5-3 and P313 anti-tetanus toxoid were used at 1 μg/ml and detected with goat anti-human Fab-alkaline phosphatase and 5-bromo-4-chloro-3-indolylphosphate (BCIP)–nitroblue tetrazolium (Sigma).
Preparative gel electrophoresis and peptide sequencing.
Samples were electrophoresed on a full-size 13% polyacrylamide gel (Bio-Rad, Richmond, Calif.). Proteins were transferred overnight onto polyvinylidene difluoride membranes (Bio-Rad) in 10 mM 4-chloro-1-aminoethylphenylsulfate (CAPS)–20% methanol buffer at pH 11.0. Membranes were Coomassie stained, and the reactive protein band was identified by immunoblotting performed on one half of a lane. Identified bands were excised from the membrane and subjected to solid-phase NH2-terminal microsequencing using a Beckman-Porton 2090E sequencer (Beckman Instruments, Anaheim, Calif.) at the UCLA protein microsequencing core facility.
Construction of HupB-GST and histone H1(69-171) fusion proteins.
M. tuberculosis Erdman was cultured as already described, and genomic DNA to be used as a template was extracted by phenol-chloroform. Two sets of nested oligonucleotide primers were designed to amplify the complete 214-aa HupB open reading frame (ORF) (accession no. Z83018; see Fig. 2) and to add EcoRI and HindIII sites (5′ and 3′ primers, respectively). PCR products were ligated by in-frame fusion to the glutathione S-transferase (GST) gene of pGEX-KG (29). HupB-pGEX fusion plasmids were transformed into Escherichia coli XL-1 Blue (Stratagene, La Jolla, Calif.), and clones were validated by PCR amplification and sequencing using pGEX sequencing primers (Pharmacia, Piscataway, N.J.). A 102-aa peptide of histone H1 (aa positions 69 to 171) was similarly constructed in pGEX-KG to yield a GST-H1(69-171) fusion 21a). The predicted sizes of the fusion proteins are 29, 41, and 54 kDa for GST, GST-H1(69-171), and GST-HupB, respectively.
FIG. 2.
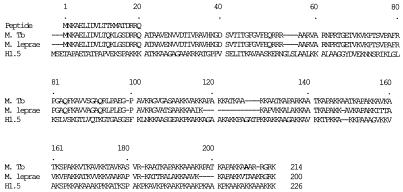
Direct protein sequencing identifies the reactive protein as a mycobacterial homologue of histone H1. The N-terminal protein sequence from the biochemically isolated immunoreactive mycobacterial protein (Peptide) is aligned with two homologue mycobacterial proteins identified in the NIH nonredundant sequence database, Z83018 from M. tuberculosis (M. Tb.) and Z99263 from M. leprae, and the H1.5 isoform of human histone H1. Strain H37Rv (shown) and strain Erdman differed at aa 208 by a single missense polymorphism resulting in alanine and threonine, respectively (bold). Dashes are spaces for alignment of sequences defined by the CLUSTAL program.
Recombinant GST fusion protein production.
HupB-pGEX and empty pGEX vectors were transformed into XL-1 Blue, and H1/69-171-pGEX was transformed into XL-21. For expression, 10 ml of a 24-h bacterial culture was inoculated into 0.5 liter of Luria-Bertani broth with ampicillin (0.1 mg/ml), cultured at 37°C in a shaker running at 200 rpm to mid-log phase (optical density at 600 nm [OD600], 0.6), and then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h. Cultures were harvested by centrifugation and resuspended in lysis buffer (50 mM Tris-Cl [pH 7.5], 300 mM NaCL, 10 mM EDTA, 0.1% SDS, and protease inhibitors). Cells were lysed by two periods of 1 min of sonication at 50% intensity using a Misonix Ultrasonic Processor sonicator (Misonix, Farmingdale, N.Y.). The soluble fraction of each lysate was isolated by centrifugation (12,000 × g for 18 min). Purified recombinant proteins were quantified by Bradford assay and analyzed by enzyme-linked immunosorbent assay (ELISA) or gel electrophoresis, followed by silver staining or immunoblotting.
ELISA analysis.
Costar 3069 microtiter plates (Costar, Cambridge, Mass.) were coated with GST fusion proteins (1 μg/well in 50 μl of Dulbecco's PBS) for 15 h at 4°C. Wells were washed with PBS–0.05% Tween 20, blocked with 1% bovine serum albumin in PBS–0.05% Tween 20 for 1 h, and washed again prior to incubation with sera. Fab monoclonal antibody, human sera, or mouse anti-GST monoclonal antibody were tested in duplicate at various dilutions for 2 h at room temperature. Primary antibodies were washed four times with Tween 20-PBS and then reacted for 1 h with a 1:1,000 dilution of alkaline phosphatase-labeled goat anti-human IgG or IgA [F(ab′)2] or goat anti-mouse IgG. Plates were washed three times in Tween 20-PBS and twice with Tris-buffered saline and then developed for 15 min with Sigma 104 Phosphatase Substrate. A405 was measured with a Bio-Rad ELISA reader and Macintosh analytic software. OD values of nonspecific binding of sera to GST alone were subtracted from values for HupB fusion protein to obtain specific absorbances.
RESULTS
Database screen.
The available sequence databases were searched for histone H1 homologous sequences by BLAST analysis using as the query a 105-aa sequence corresponding to the pANCA-reactive histone H1 COOH terminus. The only high-probability matches were two mycobacterial ORFs (14). These two sequences, accession no. Z83018 and Z99263, were also notable for high overall sequence identity (48.3%) with human histone H1.
Fab 5-3 immunoreactivity of various mycobacterial strains.
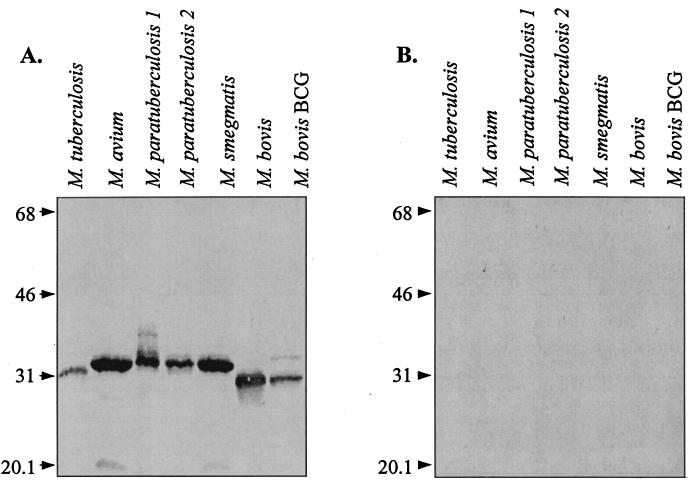
Since the mycobacterial genome revealed putative proteins with primary structure similarity to histone H1, we tested experimentally for immunoreactive pANCA antigens in this microorganism by immunoblot analysis with the Fab 5-3 pANCA monoclonal antibody. We cultured a panel of mycobacterial strains and prepared whole-cell lysates. Equivalent amounts of protein from each sample were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose filters for immunoblotting. Filters were probed with Fab 5-3 or anti-tetanus toxoid Fab P313 as a negative control (Fig. 1). While no reactivity was observed for the negative control Fab P313, a single strongly reactive ∼32-kDa protein was detected by Fab 5-3 in all of the strains tested.
FIG. 1.
Immunoblot analysis of mycobacterial cell lysates. Equivalent amounts of cellular proteins (10 μg/lane) were separated on a 13% polyacrylamide gel, transferred to nitrocellulose filters, and probed with a monoclonal antibody. Panels: A, pANCA Fab 5-3; B, anti-tetanus toxoid Fab P313. The values to the left of each panel are molecular masses in kilodaltons.
Among the tested strains, M. avium and M. paratuberculosis consistently expressed the highest reactivity. Variation in band intensity among the other strains might represent differences in protein expression levels or disruptive sequence variations adjacent to the Fab 5-3 epitope. The reactive proteins expressed by the different strains showed slight variation in apparent molecular mass: ∼31 kDa for M. tuberculosis, M. bovis, and M. bovis BCG and ∼32 kDa for M. avium, M. paratuberculosis, and M. smegmatis. These minor differences might reflect variations in amino acid sequence length, posttranslational modifications, or sequence differences affecting the net pI.
Mycobacterial antigen identification.
We characterized the reactive protein band further by direct NH2-terminal amino acid sequencing. Mycobacterial proteins were resolved by SDS-PAGE and transferred to polyvinylidene difluoride filters, and the reactive protein band was excised for solid-phase Edman degradation sequencing. Sequences of 20 aa were obtained for M. avium and two M. paratuberculosis strains. Sequence analysis confirmed that the proteins were identical in this region, and the obtained amino acid sequence was used to search protein databases for homologues in mycobacterial sequences. Residues 1 to 20 were 85% identical to a 214-aa putative ORF in the H37Rv M. tuberculosis genome (accession no. Z83018) and a 200-aa putative ORF in the M. leprae genome (accession no. Z99263) (14). The sequence identity, similar apparent molecular weights, and consistent monoclonal immunoreactivity indicated that the reactive proteins expressed by the different mycobacterial species and strains represent a single species-conserved protein.
Gene cloning.
It was possible that the authentic pANCA-reactive antigen was not the protein identified but, instead, comigrated with this major protein. Therefore, we validated the reactivity to this protein by gene cloning and expression. Two sets of nested primers were designed that corresponded to the M. tuberculosis H37Rv 214-aa ORF DNA sequence. Primers were used for sequential amplification of the gene by using M. tuberculosis Erdman chromosomal DNA as a template. An ORF of 642 nucleotides was obtained of a sequence nearly identical to a strain H37Rv ORF sequence, with the only difference being an A-to-G change specifying a change at position 208 from threonine to alanine (Fig. 2). Because of this change, the sequence from the more pathogenic Erdman strain had higher identity to histone H1 than did the H37Rv strain. The ORF has recently been named HupB by the research consortium for the M. tuberculosis genome (14). Interestingly, this protein was identified as a major iron-regulated protein of M. tuberculosis, with two forms differing slightly in apparent mass—one form (referred to as Irp28) upregulated by low iron concentrations and the other form (Irp29) upregulated by high iron high iron concentrations (8).
HupB gene expression.
HupB was subcloned into vector pGEX-KG as a GST fusion under β-galactosidase promoter control and expressed in E. coli (29). Production of the recombinant protein proved difficult, since its expression was toxic for E. coli, resulting in cell death in some strains and slow growth of others. XL-1 Blue cells were least affected by the gene product, with a doubling time of ∼1.2 h (versus a doubling time of ∼25 min for XL-1 Blue cells expressing GST alone). In addition, efficient protein purification was hindered by the fusion protein's limited solubility and high susceptibility to proteolysis. However, high levels of expression allowed purification of the recombinant protein to 50% of the total protein, as assessed by SDS-PAGE and protein staining.
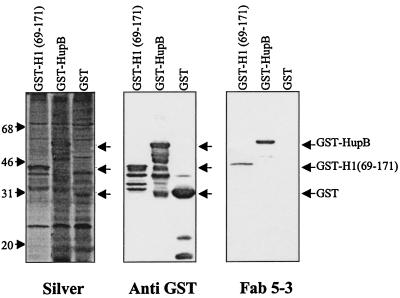
The recombinant GST-HupB fusion protein migrated at the predicted molecular mass of ∼54 kDa (Fig. 3, right panel), and its identity was validated by immunoreactivity with a mouse anti-GST monoclonal antibody (Fig. 3, middle panel). This panel shows that Fab 5-3 binding was similar for the GST-HupB and GST-H1(69-171) fusion proteins, the latter expressing a distinct peptide with the Fab 5-3 epitope (21a). Fab 5-3 binding was specific, since no binding was observed with the GST protein alone (Fig. 3, right panel) and the negative control Fab p313 did not react with any of the recombinant proteins (data not shown). Immunoblots with anti-GST and Fab 5-3 revealed that the GST-HupB and GST-H1(69-171) fusion proteins underwent substantial proteolysis, reflected by laddering of smaller immunoreactive peptides (Fig. 3, middle and right panels). Analysis of the HupB proteolytic ladder indicated significant loss of reactivity for species of 125 aa or less, thus localizing the Fab 5-3-binding epitope to the 90 aa of the HupB COOH terminus (Fig. 3, middle and right panels).
FIG. 3.
Fab 5-3 binding to HupB and histone H1 GST fusion proteins. Equivalent numbers of bacteria expressing recombinant GST fusions with histone H1 (aa 69 to 171), HupB, or GST alone were subjected to SDS–13% PAGE and electrotransferred to nitrocellulose membranes. Membranes were analyzed by silver staining for protein composition (left panel), immunoblotted with anti-GST to detect GST fusion protein expression (middle panel), or immunoblotted with Fab 5-3 to detect expression of the Fab 5-3 pANCA epitope (right panel). Arrows to right of each panel indicate sizes of full-length recombinant products for GST, GST-H1(69-171), and GST-HupB (29, 41, and 54 kDa, respectively). Smaller products detected by immunoblotting are proteolytic fragments of the full-length products. The values to the left are molecular masses in kilodaltons.
HupB sequence analysis.
The HupB protein sequence was analyzed in comparison with other DNA-binding proteins in the available databases (Tables 1 and 2). Close identity was observed to bacterial HU DNA-binding proteins and mammalian histone H1. HU similarity was localized to the NH2-terminal half of HupB, and histone H1 similarity was localized to the COOH-terminal half of the protein. Alignment of the HupB aa 1 to 107 sequence with similar sequences indicated closer identity to a yeast HupB-like protein than to HU proteins expressed by other Bacillus species (Table 1). Alignment of HupB aa 108 to 214 indicated a closer similarity to prokaryote, plant, and insect proteins than to mammalian histone H1 (Table 2). In addition, a repeating prokaryotic DNA-binding (PAKKAA) motif was prevalent, whereas histone H1 COOH terminus-specific (SPKKAK) motifs were absent (2, 31, 46).
TABLE 1.
Sequence similarities of bacterial histone homologues to the HupB N terminusa
| Organism | Protein | Size (aa) | HupB aa 1–107 | Histone H1 | P(AKKA)A | SPKKAK | PKKAKK | PKKA |
|---|---|---|---|---|---|---|---|---|
| M. tuberculosis | HupB | 214 | 10−64 | 10−09 | 5 (9) | 0 | 0 | 0 |
| M. leprae | Histone like | 200 | 10−62 | 10−01 | 4 (9) | 0 | 0 | 0 |
| S. coelicolor | Histone like | 218 | 10−27 | 10−04 | 1 (6) | 0 | 0 | 0 |
| Bacillus caldotenax | HB HU | 90 | 10−21 | >0.4 | 0 | 0 | 0 | 0 |
| Bacillus subtilis | HBsu | 98 | 10−21 | >0.4 | 0 | 0 | 0 | 0 |
| B. caldolyticus | HU | 90 | 10−20 | >0.4 | 0 | 0 | 0 | 0 |
| Bacillus globigii | HB | 92 | 10−20 | >0.4 | 0 | 0 | 0 | 0 |
| Anabaena sp. | HU | 94 | 10−20 | >0.4 | 0 | 0 | 0 | 0 |
| Clostridium pasteurianum | HU | 91 | 10−19 | >0.4 | 0 | 0 | 0 | 0 |
| B. stearothermophilus | HU | 90 | 10−19 | >0.4 | 0 | 0 | 0 | 0 |
| E. coli | HU-α (NS2) | 90 | 10−18 | >0.4 | 0 | 0 | 0 | 0 |
| Campylobacter jejuni | HupB | 98 | 10−17 | >0.4 | 0 | 0 | 0 | 0 |
| Homo sapiens | Histone H1.3 | 221 | 1 | 10−132 | 0 (4) | 0 | 1 | 2 |
The N terminal segment of HupB (aa 1 to 107) was used to probe for homologues in the NIH nonredundant sequence database by using the BLASTP program with a BLOSUM 62 matrix. CLUSTAL W absolute scores were calculated for each homologue with BLASTP for the N-terminal HupB sequence (HupB aa 1–107) and the C-terminal sequence (aa 69 to 225) of human histone H1.5. The numbers of occurrences of four H1 peptide motifs (PAKKAA, SPKKAKK, PKKAKK, and PKKA) are shown. The number of occurrences of the peptide motif are denoted in parentheses.
TABLE 2.
Sequence similarities of bacterial histone homologues to the HupB C terminusa
| Organism | Protein | Size (aa) | HupB aa 108–214 | Histone H1 | P(AKKA)A | SPKKAK | PKKAKK | PKKA |
|---|---|---|---|---|---|---|---|---|
| M. tuberculosis | HupB | 214 | 10−57 | 10−09 | 5 (9) | 0 | 0 | 0 |
| M. leprae | Histone like | 200 | 10−15 | 10−01 | 4 (9) | 0 | 0 | 0 |
| Pseudomonas putida | pprB | 298 | 10−12 | 10−07 | 0 | 0 | 0 | 0 |
| Triticum aestivum | Wheat H1 | 288 | 10−11 | 10−10 | 3 (4) | 0 | 0 | 1 |
| Bordetella pertussis | bpH1 | 182 | 10−11 | 10−07 | 3 (22) | 0 | 0 | 1 |
| Lytechinus pictus | Urchin H1 | 210 | 10−11 | 10−11 | 3 (10) | 0 | 0 | 0 |
| P. aeruginosa | ORF | 217 | 10−11 | 10−07 | 0 | 0 | 0 | 0 |
| Apium graveolens | Celery H1 | 302 | 10−10 | 10−14 | 1 (2) | 0 | 0 | 0 |
| Chironomus thummi subsp. thummi | Orphon H1 | 244 | 10−10 | 10−15 | 0 (4) | 0 | 0 | 1 |
| Lycopersicon esculentum | Tomato H1 | 287 | 10−09 | 10−08 | 2 (2) | 0 | 0 | 0 |
| Chaetopterus variopedatus | Polichaete H1 | 202 | 10−09 | 10−16 | 0 | 0 | 2 | 2 |
| Caenorhabditis elegans | Histone H1.4 | 253 | 10−09 | 10−16 | 1 (4) | 0 | 0 | 1 |
| Glyptotendipes barbipes | H1-I | 233 | 10−08 | 10−09 | 1 (3) | 1 | 0 | 0 |
| Oncorhynchus mykiss | Trout H1 | 206 | 10−08 | 10−52 | 1 (5) | 2 | 0 | 2 |
| Volvox carteri | H1-II | 241 | 10−08 | 10−14 | 0 (2) | 4 | 4 | |
| Gallus gallus | Chicken H1 | 220 | 10−08 | 10−62 | 0 (1) | 2 | 0 | 3 |
| Mytilus trossulus | PHI-2B | 203 | 10−08 | 10−10 | 0 (1) | 5 | 0 | 0 |
| Chironomus thummi | Midge H1-I-1 | 241 | 10−08 | 10−16 | 0 (4) | 0 | 0 | 1 |
| Mus musculus | Mouse H1D | 221 | 10−08 | 10−116 | 0 (3) | 1 | 1 | 2 |
| Homo sapiens | Histone H1C | 221 | 10−08 | 10−132 | 0 (4) | 0 | 1 | 2 |
| Tigriopus californicus | Copepod H1 | 181 | 10−08 | 10−24 | 0 | 0 | 2 |
Same analysis as for Table 1 but with the C-terminal segment of HupB (aa 108 to 214).
HupB serum immunoreactivity.
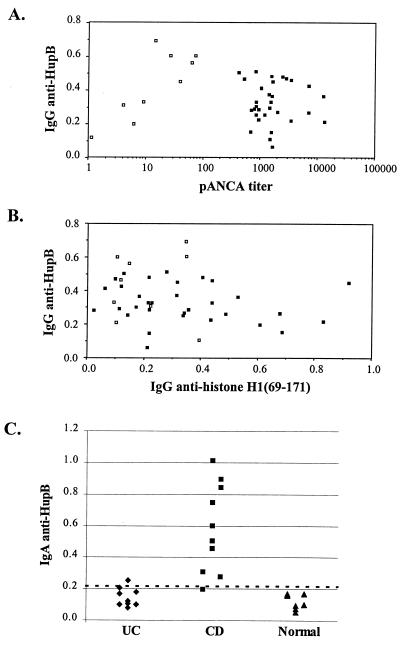
Recombinant HupB was tested by direct ELISA for immunoreactivity with IgG antibodies in sera from 31 UC patients with a wide range of pANCA titers and nine healthy pANCA-negative controls. Under these ELISA conditions, Fab 5-3 displayed specific and strong binding to HupB-GST (OD >0.8 and <0.05 for GST; data not shown). Significant binding to HupB-GST was detected among certain sera in this study set, but there was no correlation between the frequency or signal level of HupB-GST binding when the data were stratified by disease state (UC patients versus healthy subjects), pANCA titer, or histone H1(69-171)-binding activity (Fig. 4A and B).
FIG. 4.
Serum IgG and IgA binding to HupB. (A and B) ELISA wells coated with GST-HupB or GST-histone H1(69-171) were reacted with sera from 31 UC patients (closed symbols) and nine healthy controls (open symbols) and detected with anti-human IgG. Specific absorbances were calculated by subtracting nonspecific binding to GST alone. pANCA titers were previously determined for this serum panel. (A) Comparison of IgG anti-HupB binding and pANCA titer. (B) Comparison of IgG anti-HupB and anti-histone H1(69-171). Data are shown for a 1:1,000 dilution of primary sera; qualitatively similar results were observed for 1:200 to 1:2,000 dilutions. (C) Sera from 30 patients (10 each for UC and CD patients and healthy controls [Normal]) were reacted with GST-HupB and detected with anti-human IgA. Specific absorbances (after subtraction of binding to GST alone) are shown for a 1:200 serum dilution; qualitatively similar results were obtained with dilutions between 1:100 and 1:1,000. Positive and negative values are defined as those above or below the mean plus 2 standard deviations for the healthy control group.
An additional set of 30 sera (10 each for CD and UC patients and healthy controls) were tested for serum IgA binding activity (Fig. 4C). This revealed strong HupB binding in 9 of 10 CD patients but no samples with binding above the cutoff level (mean plus 2 standard deviations of the normal group) in UC patients or healthy controls. The mean absorbance for the CD group was significantly higher compared to the UC or healthy control group (P < 0.001; Student's t test). This binding activity was not related to UC pANCA, since none of the 10 CD sera had UC pANCA absorbances above the cutoff level.
DISCUSSION
UC pANCA has been a research focus in IBD pathogenesis based on the premise that disease-specific antibodies identify disease-specific antigens. The present study addressed this observation by employing pANCA monoclonal antibodies to identify a microbial UC pANCA target antigen. Our findings identify a new species-conserved mycobacterial protein, HupB, as one such pANCA antigen and demonstrate that anti-HupB IgA is associated with CD.
Characterization of HupB.
We initially identified HupB through a database screen for pANCA-reactive sequences of histone H1, revealing significant homologies only among putative ORFs in the M. tuberculosis and M. leprae genomes. Western analysis and N-terminal peptide sequencing demonstrated the expression of an ∼32-kDa protein (consistent with the predicted size of the HupB ORF and bearing the HupB N-terminal amino acid sequence) by a diverse set of pathogenic and nonpathogenic mycobacterial strains. Molecular cloning and recombinant expression of HupB directly confirmed its immunoreactivity with the Fab 5-3 pANCA monoclonal antibody.
The cross-reactivity of HupB and histone H1 raises the issue of the evolutionary origin of HupB. Sequence analysis indicated that HupB is globally similar in primary amino acid sequence to human histone H1 and bears typical histone H1 structural features, including a prominent alanine-lysine-rich COOH-terminal random coil. HupB is also a close homologue to bacterial HU type DNA-binding proteins with a histone H1-like COOH-terminal extension (27). Histone H1, like the COOH terminus of HupB, shows significant sequence variation from mammalian histone H1 and is more closely related to histone H1-like proteins of lower taxa. In addition, bacterial DNA-binding (PAKKAA) motifs are expressed extensively at the COOH terminus whereas histone H1 COOH-terminal (SPKKA) motifs were not found (2, 31, 46). Histone H1 (SPKKAK) motifs were implicated in linker DNA binding and the posttranslational regulation of histone H1 activity in the formation and stabilization of packed chromatin. Such motifs are highly conserved in higher organisms (57–59). These observations suggest that HupB originated earlier in evolution and do not favor gene capture as a mechanism of acquisition. The specific immunoreactivity thus reflects a convergent evolutionary process rather than a restricted protein genealogy (19, 28, 37). Moreover, we emphasize that the present study did not distinguish whether the cross-reactive Fab 5-3 epitopes detected in histone H1 and HupB are conferred by linear peptide homologies or conformational epitopes shared by these positively charged random-coil molecules.
Relationship of HupB with the pANCA antigen identified by serum antibodies.
The foregoing indicated that HupB is a bacterial antigen recognized by a pANCA monoclonal antibody, Fab 5-3. However, this study also shows that in patient sera, anti-HupB IgG activity did not correlate with serum pANCA activity. Specifically, anti-HupB IgG activity was discordant for UC disease status, serum pANCA IgG titer, or anti-histone H1(69-171) IgG activity. Histone H1 is a large protein with diverse linear and conformational peptide epitopes (38, 56). Moreover, histone H1 is only a minor specificity of serum pANCA antibodies (21a). Thus, unlike Fab 5-3, it appears that the major antibodies responsible for serum pANCA and serum anti-histone H1 IgG are specific for epitopes of these antigens which are not cross-reactive with HupB.
Anti-HupB IgA provides new evidence associating mycobacteria with CD.
In contrast to serum IgG, anti-HupB serum IgA was associated with CD. The disease association of IgA versus anti-HupB IgG may relate to the divergent antigenic repertoires of these isotypes and the origin of a significant proportion of IgA (but not IgG) in the mucosal immune system (40, 62). Other studies have reported an association of antimycobacterial antibodies with CD (5, 23, 24, 65, 66). It is notable that as in the present findings, several of these studies related the CD-specific antibodies to the IgA component. The present study is distinguished from these preceding ones because it identified a specific mycobacterial antigen for the antibody response, HupB.
It is possible that anti-HupB IgA is only indicative of mycobacterial presence rather than pathogenesis. Mycobacteria are common inhabitants of the healthy gut, and an antimycobacterial antibody response may simply be secondary to CD- or UC-associated mucosal disruption and local immune activity (26). Several groups have attempted to validate the relationship of mycobacterial infection (particularly M. paratuberculosis) with CD based on the important similarities of human CD with bovine Johne's disease (13). Some studies have associated CD lesions with M. paratuberculosis by using species-specific PCR, although the frequency and specificity of this association are controversial (10–12, 17, 25, 41, 49). Antimycobacterial therapy has also been evaluated in CD but has thus far been ineffective (30, 60).
The present findings introduce independent immunologic evidence for the association of CD and mycobacterial infection. HupB may be a useful antigen for evaluation of antimycobacterial immunity and serodiagnosis of CD, a clinical issue which merits validation in a population-based study.
ACKNOWLEDGMENTS
This research was supported by NIH grant DK46763, CA12800, DK43026, AI07126, the Crohn's and Colitis Foundation of America, the UCLA Jonnson Comprehensive Cancer Center, and the Feintech Family Chair of Inflammatory Bowel Disease.
We acknowledge Karin Reimann for expert and dedicated laboratory experimentation, Audrey Fowler for advice and direction of protein microsequencing, and Dominica Salvatore for administrative support. We thank Loren C. Karp for critical review of the manuscript.
REFERENCES
- 1.Abreu-Martin M T, Targan S R. Regulation of immune responses of the intestinal mucosa. Crit Rev Immunol. 1996;16:277–309. doi: 10.1615/critrevimmunol.v16.i3.30. [DOI] [PubMed] [Google Scholar]
- 2.Allan J, Hartman P G, Crane-Robinson C, Aviles F X. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Barbas C F, Kang A S, Lerner R A, Benkovic S J. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaser M J, Miller R A, Lacher J, Singleton J W. Patients with active Crohn's disease have elevated serum antibodies to antigens of seven enteric bacterial pathogens. Gastroenterology. 1984;87:888–894. [PubMed] [Google Scholar]
- 6.Brandwein S L, McCabe R P, Cong Y, Waites K B, Ridwan B U, Dean P A, Ohkusa T, Birkenmeier E H, Sundberg J P, Elson C O. Spontaneously colitic C3H/HeJBir mice demonstrate selective antibody reactivity to antigens of the enteric bacterial flora. J Immunol. 1997;159:44–52. [PubMed] [Google Scholar]
- 7.Cahill R J, Foltz C J, Fox J G, Dangler C A, Powrie F, Schauer D B. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun. 1997;65:3126–3131. doi: 10.1128/iai.65.8.3126-3131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calder K M, Horwitz M A. Identification of iron-regulated proteins of Mycobacterium tuberculosis and cloning of tandem genes encoding a low iron-induced protein and a metal transporting ATPase with similarities to two-component metal transport systems. Microb Pathog. 1998;24:133–143. doi: 10.1006/mpat.1997.9999. [DOI] [PubMed] [Google Scholar]
- 9.Cariappa A, Sands B, Forcione D, Finkelstein D, Podolsky D K, Pillai S. Analysis of MHC class II DP, DQ and DR alleles in Crohn's disease. Gut. 1998;43:210–215. doi: 10.1136/gut.43.2.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cellier C, De Beenhouwer H, Berger A, Penna C, Carbonnel F, Parc R, Cugnenc P H, Le Quintrec Y, Gendre J P, Barbier J P, Portaels F. Mycobacterium paratuberculosis and Mycobacterium avium subsp. silvaticum DNA cannot be detected by PCR in Crohn's disease tissue. Gastroenterol Clin Biol. 1998;22:675–678. [PubMed] [Google Scholar]
- 11.Chiba M, Fukushima T, Horie Y, Iizuka M, Masamune O. No Mycobacterium paratuberculosis detected in intestinal tissue, including Peyer's patches and lymph follicles, of Crohn's disease. J Gastroenterol. 1998;33:482–487. doi: 10.1007/s005350050119. [DOI] [PubMed] [Google Scholar]
- 12.Clarkston W K, Presti M E, Petersen P F, Zachary P E J, Fan W X, Leonardi C L, Vernava A M, Longo W E, Kreeger J M. Role of Mycobacterium paratuberculosis in Crohn's disease: a prospective, controlled study using polymerase chain reaction. Dis Colon Rectum. 1998;41:195–199. doi: 10.1007/BF02238248. [DOI] [PubMed] [Google Scholar]
- 13.Cocito C, Gilot P, Coene M, De Kesel M, Poupart P, Vannuffel P. Paratuberculosis. Clin Microbiol Rev. 1994;7:328–345. doi: 10.1128/cmr.7.3.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 15.Compton R F, Sandborn W J, Yang H-Y, Lindor N M, Tremaine W J, Davis M D, Khalil A A, Tountas N A, Tyan D B, Landers C J, et al. A new syndrome of Crohn's disease and pachydermoperiostosis in a family. Gastroenterology. 1997;112:241–249. doi: 10.1016/s0016-5085(97)70241-5. [DOI] [PubMed] [Google Scholar]
- 16.Cong Y, Brandwein S L, McCabe R P, Lazenby A, Birkenmeier E H, Sundberg J P, Elson C O. CD4+T cells reactive to enteric bacterial antigens in spontaneously colitic C3H/HeJBir mice: increased T helper cell type 1 response and ability to transfer disease. J Exp Med. 1998;187:855–864. doi: 10.1084/jem.187.6.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Prete R M, Quaranta, Lippolis A, Giannuzzi V, Mosca A, Jirillo E, Miragliotta G. Detection of Mycobacterium paratuberculosis in stool samples of patients with inflammatory bowel disease by IS900-based PCR and colorimetric detection of amplified DNA. J Microbiol Methods. 1998;33:105–114. [Google Scholar]
- 18.Dianda L, Hanby A M, Wright N A, Sebesteny A, Hayday A C, Owen M J. T cell receptor-alpha, beta-deficient mice fail to develop colitis in the absence of a microbial environment. Am J Pathol. 1997;150:91–97. [PMC free article] [PubMed] [Google Scholar]
- 19.Doolittle R F. Convergent evolution: the need to be explicit. Trends Biochem Sci. 1994;19:15–18. doi: 10.1016/0968-0004(94)90167-8. [DOI] [PubMed] [Google Scholar]
- 20.Duerr R H, Barmada M M, Zhang L, Davis S, Preston R A, Chensny L J, Brown J L, Ehrlich G D, Weeks D E, Aston C E. Linkage and association between inflammatory bowel disease and a locus on chromosome 12. Am J Hum Genet. 1998;63:95–100. doi: 10.1086/301929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duerr R H, Targan S R, Landers C J, Sutherland L R, Shanahan F. Anti-neutrophil cytoplasmic antibodies in ulcerative colitis. Comparison with other colitides/diarrheal illnesses. Gastroenterology. 1991;100:1590–1596. doi: 10.1016/0016-5085(91)90657-7. [DOI] [PubMed] [Google Scholar]
- 21a.Eggena, M., O. Cohavy, M. H. Parseghian, B. A. Hamkalo, D. Clemens, S. R. Targan, L. K. Gordon, and J. Braun. Identification of histone H1 as a cognate antigen of the ulcerative colitis-associated marker antibody pANCA. J. Autoimmun., in press. [DOI] [PubMed]
- 22.Eggena M, Targan S R, Iwanczyk L, Vidrich A, Gordon L K, Braun J. Phage display cloning and characterization of an immunogenetic marker (perinuclear anti-neutrophil cytoplasmic antibody) in ulcerative colitis. J Immunol. 1996;156:4005–4011. [PubMed] [Google Scholar]
- 23.Elsaghier A, Prantera C, Moreno C, Ivanyi J. Antibodies to Mycobacterium paratuberculosis-specific protein antigens in Crohn's disease. Clin Exp Immunol. 1992;90:503–508. doi: 10.1111/j.1365-2249.1992.tb05874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Zaatari F A, Naser S A, Graham D Y. Characterization of a specific Mycobacterium paratuberculosisrecombinant clone expressing 35,000-molecular-weight antigen and reactivity with sera from animals with clinical and subclinical Johne's disease. J Clin Microbiol. 1997;35:1794–1799. doi: 10.1128/jcm.35.7.1794-1799.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fidler H M, Thurrell W, Johnson N M, Rook G A, McFadden J J. Specific detection of Mycobacterium paratuberculosis DNA associated with granulomatous tissue in Crohn's disease. Gut. 1994;35:506–510. doi: 10.1136/gut.35.4.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finegold S M, Sutter V L, Sugihara P T, Elder H A, Lehmann S M, Phillips R L. Fecal microbial flora in Seventh Day Adventist populations and control subjects. Am J Clin Nutr. 1977;1:1781–1792. doi: 10.1093/ajcn/30.11.1781. [DOI] [PubMed] [Google Scholar]
- 27.Goyard S. Identification and characterization of BpH2, a novel histone H1 homolog in Bordetella pertussis. J Bacteriol. 1996;178:3066–3071. doi: 10.1128/jb.178.11.3066-3071.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graumann P, Marahiel M A. A case of convergent evolution of nucleic acid binding modules. Bioessays. 1996;18:309–315. doi: 10.1002/bies.950180409. [DOI] [PubMed] [Google Scholar]
- 29.Guan K L, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 30.Gui G PH, Thomas P RS, Tizard M LV, Lake J, Sanderson J D, Hermon-Taylor J. Two-year-outcomes analysis of Crohn's disease treated with rifabutin and macrolide antibiotics. J Antimicrob Chemother. 1997;39:393–400. doi: 10.1093/jac/39.3.393. [DOI] [PubMed] [Google Scholar]
- 31.Hayes J J, Kaplan R, Ura K, Pruss D, Wolffe A. A putative DNA binding surface in the globular domain of a linker histone is not essential for specific binding to the nucleosome. J Biol Chem. 1996;271:25817–25822. doi: 10.1074/jbc.271.42.25817. [DOI] [PubMed] [Google Scholar]
- 32.Herfarth H H, Mohanty S P, Rath H C, Tonkonogy S, Sartor R B. Interleukin 10 suppresses experimental chronic, granulomatous inflammation induced by bacterial cell wall polymers. Gut. 1996;39:836–845. doi: 10.1136/gut.39.6.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hermiston M L, Gordon J I. Inflammatory bowel disease and adenomas in mice expressing a dominant negative N-cadherin. Science. 1995;270:1203–1207. doi: 10.1126/science.270.5239.1203. [DOI] [PubMed] [Google Scholar]
- 34.Hesresbach D, Alizadeh M, Bretagne J F, Gautier A, Quillivic F, Lemarchand B, Gosselin M, Genetet B, Semana G. Investigation of the association of major histocompatibility complex genes, including HLA class I, class II and TAP genes, with clinical forms of Crohn's disease. Eur J Immunogenet. 1996;23:141–151. doi: 10.1111/j.1744-313x.1996.tb00275.x. [DOI] [PubMed] [Google Scholar]
- 35.Hofmeister A, Neibergs H L, Pokorny R M, Galandiuk S. The natural resistance-associated macrophage protein gene is associated with Crohn's disease. Surgery. 1997;122:173–178. doi: 10.1016/s0039-6060(97)90006-4. [DOI] [PubMed] [Google Scholar]
- 36.Hugot J-P, Laurent-Puig P, Gower-Rousseau C, Olson J M, Lee J C, Beaugerie L, Naom I, Dupas J-L, Van Gossum A, Orholm M, Bonaiti-Pellie C, Weissenbach J, Mathew C G, Lennard-Jones J E, Cortot A, Colombel J-F, Thomas G Groupe d'Etude Therapeutique des Affections Inflammatoires Digestives. Mapping of a susceptibility locus for Crohn's disease on chromosome 16. Nature. 1996;379:821–823. doi: 10.1038/379821a0. [DOI] [PubMed] [Google Scholar]
- 37.Mattevi A, Vanoni M A, Todone F, Rizzi M, Teplyakov A, Coda A, Bolognesi M, Curti B. Crystal structure of d-amino acid oxidase: a case of active site mirror-image convergent evolution with flavocytochrome b2. Proc Natl Acad Sci USA. 1996;93:7496–7501. doi: 10.1073/pnas.93.15.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morino N, Sakurai H, Yamada A, Yazaki Y, Minota S. Rabbit anti-chromatin antibodies recognize similar epitopes on a histone H1 molecule as lupus autoantibodies. Clin Immunol Immunopathol. 1995;77:52–58. doi: 10.1016/0090-1229(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 39.Morrissey P J, Charrier K, Braddy S, Liggitt D, Watson J D. CD4+ T cells that express high levels of CD45RB induce wasting disease when transferred into congenic severe combined immunodeficient mice. Disease development is prevented by cotransfer of purified CD4+T cells. J Exp Med. 1993;178:237–244. doi: 10.1084/jem.178.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mortari F, Wang J Y, Schroeder H W., Jr Human cord blood antibody repertoire: mixed population of VH gene segments and CDR3 distribution in the expressed Calpha and Cgamma repertoires. J Immunol. 1993;150:1348–1357. [PubMed] [Google Scholar]
- 41.Moss M T, Sanderson J D, Tizard M L, Hermon-Taylor J, el-Zaatari F A, Markesich D C, Graham D Y. Polymerase chain reaction detection of Mycobacterium paratuberculosis and Mycobacterium avium subsp silvaticum in long term cultures from Crohn's disease and control tissues. Gut. 1992;33:1209–1213. doi: 10.1136/gut.33.9.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohmen J D, Yang H-Y, Yamamoto K K, Zhao H Y, Ma Y, Bentley L G, Huang Z, Gerwehr S, Pressman S, McElree C, et al. Susceptibility locus for inflammatory bowel disease on chromosome 16 has a role in Crohn's disease, but not in ulcerative colitis. Hum Mol Genet. 1996;5:1679–1683. doi: 10.1093/hmg/5.10.1679. [DOI] [PubMed] [Google Scholar]
- 43.Peeters M, Geypens B, Claus D, Nevens H, Ghoos Y, Verbeke G, Baert F, Vermeire S, Vlietinck R, Rutgeerts P. Clustering of increased small intestinal permeability in families with Crohn's disease. Gastroenterology. 1997;113:802–807. doi: 10.1016/s0016-5085(97)70174-4. [DOI] [PubMed] [Google Scholar]
- 44.Plevy S E, Targan S R, Yang H-Y, Fernandez D, Rotter J I, Toyoda H. Tumor necrosis factor microsatellites define a Crohn's disease-associated haplotype on chromosome 6. Gastroenterology. 1996;110:1053–1060. doi: 10.1053/gast.1996.v110.pm8612993. [DOI] [PubMed] [Google Scholar]
- 45.Podolsky D K. Inflammatory bowel disease. N Engl J Med. 1991;325:928–937. doi: 10.1056/NEJM199109263251306. [DOI] [PubMed] [Google Scholar]
- 46.Pruss D, Bartholomew B, Persinger J, Hayes J, Arents G, Moudrianakis E N, Wolffe A P. An asymmetric model for the nucleosome: a binding site for linker histones inside the DNA gyres. Science. 1996;274:614–617. doi: 10.1126/science.274.5287.614. [DOI] [PubMed] [Google Scholar]
- 47.Rath H C, Herfarth H H, Ikeda J S, Grenther W B, Hamm T E, Jr, Balish E, Taurog J D, Hammer R E, Wilson K H, Sartor R B. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J Clin Investig. 1996;98:945–953. doi: 10.1172/JCI118878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rudolphi A, Bonhagen K, Reimann J. Polyclonal expansion of adoptively transferred CD4+alpha beta T cells in the colonic lamina propria of scid mice with colitis. Eur J Immunol. 1996;26:1156–1163. doi: 10.1002/eji.1830260529. [DOI] [PubMed] [Google Scholar]
- 49.Sanderson J D, Moss M T, Tizard M L, Hermon-Taylor J. Mycobacterium paratuberculosis DNA in Crohn's disease tissue. Gut. 1992;33:890–896. doi: 10.1136/gut.33.7.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sartor R B. Current concepts of the etiology and pathogenesis of ulcerative colitis and Crohn's disease. Gastroenterol Clin N Am. 1995;24:475–507. [PubMed] [Google Scholar]
- 51.Satsangi J, Landers C J, Welsh K I, Koss K, Targan S R, Jewell D P. The presence of anti-neutrophil antibodies reflects clinical and genetic heterogeneity within inflammatory bowel disease. Inflamm Bowel Dis. 1998;4:18–26. doi: 10.1097/00054725-199802000-00004. [DOI] [PubMed] [Google Scholar]
- 52.Satsangi J, Parkes M, Louis E, Hashimoto L, Kato N, Welsh K, Terwilliger J D, Lathrop G M, Bell J I, Jewell D P. Two stage genome-wide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12. Nat Genet. 1996;14:199–202. doi: 10.1038/ng1096-199. [DOI] [PubMed] [Google Scholar]
- 53.Saxon A, Shanahan F, Landers C, Ganz T, Targan S. A distinct subset of antineutrophil cytoplasmic antibodies is associated with inflammatory bowel disease. J Allergy Clin Immunol. 1990;86:202–210. doi: 10.1016/s0091-6749(05)80067-3. [DOI] [PubMed] [Google Scholar]
- 54.Seibold F, Brandwein S, Simpson S, Terhorst C, Elson C O. pANCA represents a cross-reactivity to enteric bacterial antigens. J Clin Immunol. 1998;18:153–160. doi: 10.1023/a:1023203118100. [DOI] [PubMed] [Google Scholar]
- 55.Shanahan F, Duerr R H, Rotter J I, Yang H-Y, Sutherland L R, McElree C, Landers C J, Targan S R. Neutrophil autoantibodies in ulcerative colitis: familial aggregation and genetic heterogeneity. Gastroenterology. 1992;103:456–461. doi: 10.1016/0016-5085(92)90834-l. [DOI] [PubMed] [Google Scholar]
- 56.Stemmer C, Briand J-P, Muller S. Mapping of linear epitopes of human histone H1 recognized by rabbit anti-H1/H5 antisera and antibodies from autoimmune patients. Mol Immunol. 1994;31:1037–1046. doi: 10.1016/0161-5890(94)90099-x. [DOI] [PubMed] [Google Scholar]
- 57.Suzuki M. SPKK, a new nucleic acid-binding unit of protein found in histone. EMBO J. 1989;8:797–804. doi: 10.1002/j.1460-2075.1989.tb03440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki M, Sohma H, Yazawa M, Yagi K, Ebashi S. Histone H1 kinase specific to the SPKK motif. J Biochem (Tokyo) 1990;108:356–364. doi: 10.1093/oxfordjournals.jbchem.a123206. [DOI] [PubMed] [Google Scholar]
- 59.Suzuki M, Sugiura M, Ebashi S. Sea urchin protease specific to the SPKK motif in histone. J Biochem (Tokyo) 1990;108:347–355. doi: 10.1093/oxfordjournals.jbchem.a123205. [DOI] [PubMed] [Google Scholar]
- 60.Thomas G A O, Swift G L, Green J T, Newcombe R G, Braniff-Mathews C, Rhodes J, Wilkinson S, Strohmeyer G, Kreuzpainter G. Controlled trial of antituberculosis chemotherapy in Crohn's disease: a five year follow up study. Gut. 1998;42:497–500. doi: 10.1136/gut.42.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomasi T B. Regulation of the mucosal IgA response—an overview. Immunol Investig. 1989;18:1–15. doi: 10.3109/08820138909112223. [DOI] [PubMed] [Google Scholar]
- 63.Toyoda H, Wang S-J, Yang H-Y, Redford A, Magalong D, Tyan D, McElree C, Pressman S, Shanahan F, Targan S R, Rotter J I. Distinct associations of HLA class II genes with inflammatory bowel disease. Gastroenterology. 1993;104:741–748. doi: 10.1016/0016-5085(93)91009-7. [DOI] [PubMed] [Google Scholar]
- 64.Van Kruiningen H J, Colombel J F, Cartun R W, Whitlock R H, Koopmans M, Kangro H O, Hoogkamp-Korstanje J A A, Lecomte-Houcke M, Devred M, Paris J C, Cortot A. An in-depth study of Crohn's disease in two French families. Gastroenterology. 1993;104:351–360. doi: 10.1016/0016-5085(93)90401-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vannuffel P, Dieterich C, Naerhuyzen B, Gilot P, Coene M, Fiasse R, Cocito C. Occurrence, in Crohn's disease, of antibodies directed against a species-specific recombinant polypeptide of Mycobacterium paratuberculosis. Clin Diagn Lab Immunol. 1994;1:241–243. doi: 10.1128/cdli.1.2.241-243.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wayne L G, Hollander D, Anderson B, Sramek H A, Vadheim C M, Rotter J I. Immunoglobulin A (IgA) and IgG serum antibodies to mycobacterial antigens in Crohn's disease patients and their relatives. J Clin Microbiol. 1992;30:2013–2018. doi: 10.1128/jcm.30.8.2013-2018.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang H-Y, McElree C, Shanahan F, Targan S R, Rotter J I. Familial empiric risks for inflammatory bowel disease. Differences between Jews and non-Jews. Gut. 1993;34:517–522. doi: 10.1136/gut.34.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang H-Y, Rotter J I, Toyoda H, Landers C, Tyan D, McElree C K, Targan S R. Ulcerative colitis: a genetically heterogeneous disorder defined by genetic (HLA class II) and subclinical (antineutrophil cytoplasmic antibodies) markers. J Clin Investig. 1993;92:1080–1084. doi: 10.1172/JCI116613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang H-Y, Vora K A, Targan S R, Toyoda H, Beaudet A, Rotter J I. Intercellular adhesion molecule 1 gene association with immunologic subsets of inflammatory bowel disease. Gastroenterology. 1995;109:440–446. doi: 10.1016/0016-5085(95)90331-3. [DOI] [PubMed] [Google Scholar]