Abstract
PspC is one of three designations for a pneumococcal surface protein whose gene is present in approximately 75% of all Streptococcus pneumoniae strains. Under the name SpsA, the protein has been shown to bind secretory immunoglobulin A (S. Hammerschmidt, S. R. Talay, P. Brandtzaeg, and G. S. Chhatwal, Mol. Microbiol. 25:1113–1124, 1997). Under the name CbpA, the protein has been shown to interact with human epithelial and endothelial cells (C. Rosenow et al., Mol. Microbiol. 25:819–829, 1997). The gene is paralogous to the pspA gene in S. pneumoniae and was thus called pspC (A. Brooks-Walter, R. C. Tart, D. E. Briles, and S. K. Hollingshead, Abstracts of the 97th General Meeting of the American Society for Microbiology 1997). Sequence comparisons of five published and seven new alleles reveal that this gene has a mosaic structure, and modular domains have contributed to gene diversity during evolution. Two major clades exist: clade A alleles are larger and contain an extra module that is shared with many pspA alleles; clade B alleles are smaller and lack this pspA-like domain. All alleles have a proline-rich domain and a choline-binding repeat domain that show 0% divergence from similar domains in the PspA protein. Immunization of a rabbit with a recombinant clade B PspC molecule produced antiserum that cross-reacted with both PspC and PspA from 15 pneumococcal isolates. The cross-reactive antibodies afforded cross-protection in a mouse model system. Mice immunized with PspC were protected against challenge with a strain that expressed PspA but not PspC. The PspA- and PspC-cross-reactive antibodies were directed to the proline-rich domain present in both molecules.
Streptococcus pneumoniae is a major cause of bacterial pneumonia, otitis media, bacterial meningitis, and bacteremia. Despite the availability of a licensed vaccine and effective antibiotic treatments, the morbidity and mortality attributed to S. pneumoniae remain significant in both developed and developing countries. Licensed pneumococcal vaccines and many vaccines currently under development stimulate immunity to the pneumococcus by eliciting antibodies that recognize many of the different capsular polysaccharides. Pneumococcal proteins can also elicit protective immunity, and an enhanced understanding of these proteins should lead to the development of improved vaccines and treatments.
S. pneumoniae possesses a family of proteins that bind the phosphocholine (4, 22) present in the teichoic acid and the lipoteichoic acid of the cell membrane and the cell wall (25). The choline-binding proteins of pneumococci and other gram-positive organisms all contain structurally similar choline-binding domains, which are composed of multiple tandem amino acid repeats. Autolysin, PspA (pneumococcal surface protein A), and PcpA (pneumococcal choline-binding protein A) of S. pneumoniae, toxins A and B of Clostridium difficile, glucosyltransferases from Streptococcus downei and Streptococcus mutans, CspA of Clostridium acetobutylicum, and PspA of Clostridium perfringens all contain similar regions (2, 3, 9, 11, 23, 24).
In PspA from S. pneumoniae, these choline-binding repeats are responsible for the attachment of PspA to the surface of the pneumococcus (30). PspA molecules interfere with complement activation (6, 26), slow the clearance of pneumococci from the blood of infected mice (21, 26), and elicit protection against pneumococcal sepsis and nasal carriage (19, 27). A single non-pspA locus whose product has greater similarity to the choline-binding and proline-rich regions of PspA than to any of the other choline-binding proteins has been identified (20). We designated the molecule PspC because of its strong molecular and serologic similarities to PspA (7).
Two other laboratories have independently sequenced alleles at this same locus. Hammerschmidt et al. identified a protein, SpsA, which is reported to bind secretory immunoglobulin A (IgA) (13). Rosenow et al. isolated from a pspA mutant strain a choline-binding protein, CbpA, which appears to be responsible for binding a moiety on eukaryotic surfaces (22). Immunization with a crude extract of pooled non-PspA choline-binding proteins containing CbpA elicited protection to a lethal challenge of pneumococci introduced intraperitoneally into mice (22).
In the present studies, we have demonstrated that immunization with purified PspC is able to elicit protection against sepsis and that this protection is apparently mediated by antibodies cross-reactive with PspA. We have also examined the genetic diversity present within the genetic locus, herein called pspC, by the examination of 12 sequenced alleles. These include the previously sequenced alleles of cbpA and spsA, an allele from The Institute for Genomic Research (TIGR) genomic sequencing project, and seven newly sequenced pspC genes presented here for the first time. We have also included the sequence of PbcA, a C3-binding protein that has high sequence identity to PspC (15).
The previously published sequences of cbpA and spsA both included sequences of D39 or its derivatives. Rosenow et al. sequenced cbpA from LM91, a pspA mutant of D39 (22). Hammerschmidt et al. sequenced spsA from an encapsulated derivative of R36A (ATCC 11733) (13). From a comparison of these two sequences, it was apparent that the spsA sequence contained a 480-bp deletion. Because of this discrepancy, we also report here a sequence of pspC from a cloned HindIII-EcoRI chromosomal fragment of D39 that was determined contemporaneously with the cbpA and spsA sequences (7). Other sequences that were used for sequence alignment comparisons included two spsA sequences from capsular serotype 1 and 47 serotype strains (13) and the pspC sequence from the capsular serotype 4 strain sequenced in TIGR genome project (30a).
MATERIALS AND METHODS
Bacterial strains, plasmids, and recombinant DNA techniques.
Chromosomal DNA from S. pneumoniae EF6796, a serotype 6A clinical isolate (5), and D39, a serotype 2 isolate, was isolated by a cesium chloride gradient procedure (1). The HindIII-EcoRI fragment of EF6796 and D39 was cloned in a modified pZero vector (Invitrogen, San Diego, Calif.), in which the Zeocin resistance cassette was replaced by a kanamycin resistance cassette kindly provided by Randall Harris. Recombinant plasmids were electroporated into Escherichia coli TOP10F′ cells [F′ (lacIq Tetr) mcrA Δ(mrr-hsdRMS-mcrBC) f80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG] (Invitrogen). DNA was purified from agarose with Gene Clean (Bio 101, Inc., Vista, Calif.).
Chromosomal DNA used for PCR was isolated by a chloroform-isoamyl alcohol procedure. Oligonucleotide primers ABW13 (5′CGACGAATAGCTGAAGAGG3′) and SKH2 (5′CATACCGTTTTCTTGTTTCCAGCC3′) were used to amplify the DNA encoding the α-helical region through the proline-rich region of pspC in 100 additional S. pneumoniae strains. These primers correspond to nucleotides 215 to 235 and nucleotides 1810 to 1834, respectively, of the EF6796 pspC gene. PCR products from L81905 (serotype 4), BG9163 (serotype 6B), DBL6A (serotype 6A), BG8090 (serotype 19), and E134 (serotype 23) were cloned in a pGem vector (Promega, Madison, Wis.) or a Topo TA vector (Invitrogen), each of which utilizes the A overhangs generated by Taq polymerase.
Cloning and expression of recombinant truncated PspC molecules.
Oligonucleotides were used to amplify a 1.2-kb fragment of strain L81905 which encodes amino acids (aa) 263 to 482 of the α-helical region and the proline-rich region of PspC. The amplified PCR fragment was cloned into pQE40 (Qiagen, Chatsworth, Calif.) to create a construct containing a fusion product with a polyhistidine tag at the amino-terminal end, dihyrofolate reductase, and the fragment of L81905 PspC described above. Expression of the fusion protein in E. coli BL21(DE3) was induced during growth at room temperature by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The overexpressed fusion protein was purified by affinity chromatography under nondenaturing conditions over a nickel resin according to the manufacturer's protocols. The purified fusion protein was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and quantitated by a Bio-Rad (Hercules, Calif.) protein assay. Two fragments of D39 PspC (aa 1 to 445 and aa 255 to 445) and three fragments of Rx1 PspA (aa 1 to 301, aa 1 to 314, and aa 1 to 370) were expressed as fusion proteins with a six-histidine tag in the pET20b expression system (Novagen, Madison, Wis.). In this case, the overexpressed fusion proteins contain a PelB leader peptide, the PspC or PspA fragments, and the His tag at the carboxy terminus. Expression of the pET20b-based constructs in the expression strain BL21(DE3) was induced with 0.4 mM IPTG, and the constructs were purified according to the manufacturer's protocols.
Production of a polyclonal antiserum, SDS-PAGE, and immunoblotting.
A truncated PspC molecule (aa 263 to 482) from S. pneumoniae L81905 was overexpressed in E. coli, purified by metal affinity chromatography, and used to immunize a rabbit. Approximately 4 μg of purified PspC was injected subcutaneously into a rabbit twice in 2 consecutive weeks, and blood was collected 10 days after the last injection. The primary immunization was given with Freund's complete adjuvant, and the booster immunization was given with saline. Polyclonal rabbit antiserum was diluted 1:50 and used to analyze pneumococcal lysates by SDS–7.5% PAGE (Bio-Rad). Pneumococcal lysates and immunoblots were prepared as described by Yother and White (30).
Immunization and challenge studies.
CBA/N mice were immunized with purified recombinant PspC proteins originating from strain L81905 (aa 263 to 482), the full α-helical region of PspC from strain D39 (aa 1 to 445), or a truncated portion of the PspC protein from strain D39 (aa 255 to 445). Each mouse received only one of the above recombinant proteins, and groups of five or six mice were immunized in each experiment. The mice were immunized subcutaneously with approximately 1 μg of purified protein emulsified in 0.1 ml of complete Freund's adjuvant and 0.1 ml of Ringer's saline. Three weeks later, they were boosted with 1 μg of purified protein in Ringer's saline. Three weeks after the boost, the mice were challenged with approximately 700 CFU of pneumococcal strain WU2 or 2000 CFU of BG7322 injected intravenously in 0.2 ml of Ringer's injection solution. Control mice were immunized in the same manner with buffer and complete Freund's adjuvant.
Analysis of immune sera.
Mice were bled retro-orbitally 24 h before challenge. Each 75-μl blood sample was collected into 0.5 ml of 1% bovine serum albumin–phosphate-buffered saline (PBS). Samples were centrifuged for 1 min (1,000 × g), and the supernatants were collected and stored at −20°C until used in direct enzyme-linked immunosorbent assays (ELISA). Microtiter 96-well plates (Nunc, Weisbaden, Germany) were coated overnight at 4°C with 5 μg of recombinant protein per ml. Separate ELISA plates were used to measure reactivity to either recombinant PspC protein from D39 (aa 255 to 445) or one of three recombinant PspA proteins from D39 (UAB055 [aa 1 to 302], UAB15 [aa 1 to 314], and UAB103 [aa 1 to 370]). Plates were blocked with 1% bovine serum albumin–PBS, followed by incubation with immune sera for 3 h at 37°C. Plates were washed with PBS–0.15% Tween–100 mM NaCl–0.5 mM NaH2PO4–1.5 mM Na2HPO4 and incubated with biotin-conjugated goat anti-mouse immunoglobulin antiserum and streptavidin-alkaline phosphatase (Southern Biotechnology Associates, Birmingham, Ala.). They were developed with p-nitrophenyl phosphate (Sigma, St. Louis, Mo.). Antibody reactivity to PspC protein or cross-reactivity to different PspA proteins was determined and depicted as the titer giving 33% maximum binding in each assay. Data is presented as the log reciprocal titer of this 33% maximal titer.
Sequencing and DNA analysis.
Sequencing of pspC was completed by automated DNA sequencing (ABI 377; Applied Biosystems, Inc., Foster City, Calif.). Sequence analyses were performed with University of Wisconsin Genetics Computer Group programs (8), Mac Vector 6.5 (Oxford Molecular), and Sequencer 3.0 (GeneCodes, Inc.). Sequence similarities of pspC were determined with NCBI BLAST. The coiled-coil structure predicted by the pspC sequence was analyzed with the Matcher program (10).
Nucleotide sequence accession numbers.
The GenBank/EMBL accession numbers for the nucleotide sequence of pspC are as follows: EF6796, U72655; DBL6A, AF068645; D39, AF068646; E134, AF068647; BG8090, AF068648; L81905, AF068649; and BG9163, AF068650. Preliminary sequence data was obtained from TIGR Website (30a).
RESULTS
Sequence analysis of the pspC gene—aspects relating to domain structure and function.
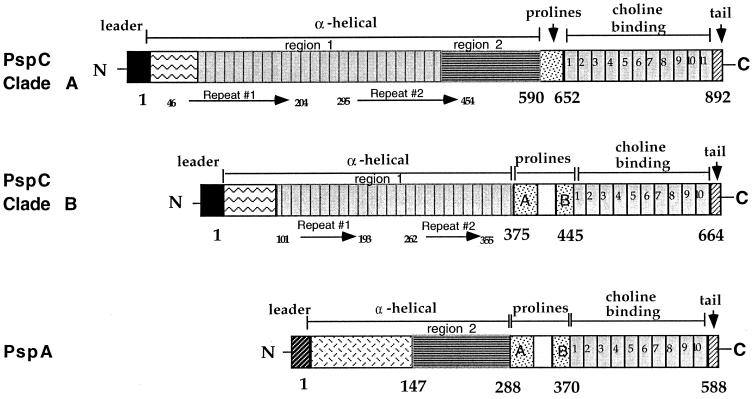
The sequences of the pspC, spsA, cbpA, and pbcA gene products were aligned with Mac Vector 6.5 (Fig. 1 and 2) (13, 15, 22). The predicted amino acid sequences encode proteins ranging in size from 59 to 105 kDa. The signal sequences of 37 aa are highly conserved (84 to 100% identity). The major part of each protein is composed of a large α-helical domain (Fig. 1 and 2). The N-terminal 100 to 150 aa of this α-helical domain are hypervariable in both size and sequence and are unique for each sequenced PspC of unrelated parentage (Fig. 2; the D39 PspC protein, SpsA2, CbpA, and PbcA are all from a related lineage). In the hypervariable regions of capsular serotype 1 and 4 strains, there is a unique 23-aa serine-rich sequence (aa 112 to 135).
FIG. 1.
Cartoon of the PspC clades compared to a representative PspA molecule. Long arrows represent the direct repeats found within the α helix. The hypervariable region is indicated by the box containing zigzag lines. The region showing homology to the α helix is indicated by the box containing horizontal lines.
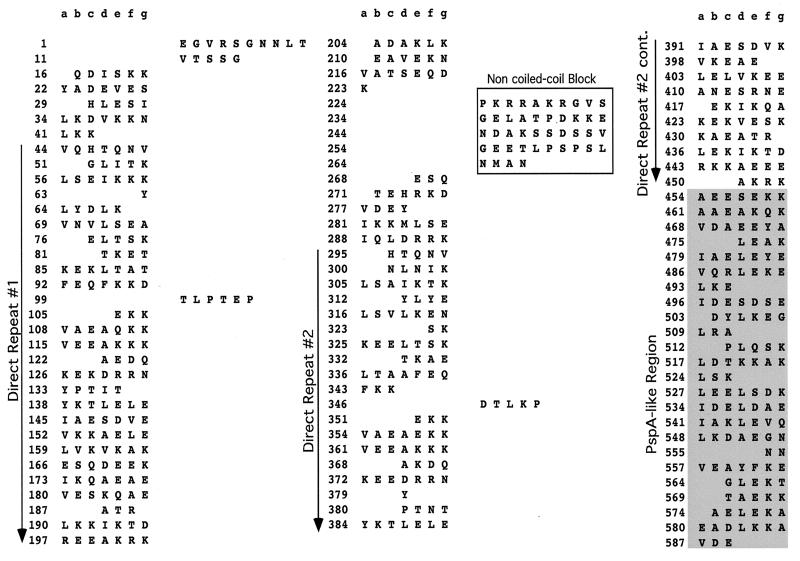
FIG. 2.
Alignment of PspC. The amino acid sequences which included the α-helical region and the proline-rich region of PspC were aligned with Mac Vector 6.5. The direct repeats within the α helix, the non-coiled-coil block, and the proline-rich region are indicated with arrows. Conserved regions are shaded, and gaps are shown with a dash. Entries are named for the strain from which the gene was cloned, with the exception of GenBank entries: SpsA1 (Y10818) from strain ATCC 33400 (serotype 1), SpsA2 (AJ002054) from strain ATCC 11733 (serotype 2), SpsA47 (AJ002055) from strain NCTC10319 (serotype 47), CbpA (AF019904) from strain LM91 (serotype 2), PbcA (a C3-binding protein) (AF067128), and TIGR sequence for a serotype 4 clinical isolate (30a). The capsular serotypes of the other strains are as follows: EF6796, 6A; BG8090, 19; L81905, 4; DBL6A, 6A; BG9163, 6B; D39, 2; and E134, 23.
Downstream of the hypervariable region and central to the α-helical domain is the first of two direct repeats. The amino acid repeats (Fig. 2 and 3) vary in size in individual PspC proteins from 101 to 205 aa and are approximately 79 to 89% identical at the amino acid level. Smaller amino acid repeats in some strains differ from the larger repeats in other strains only by the lack of a sequence at the NH2-terminal end, which accounts for their smaller size. The first repeat in each strain is more like the corresponding first repeat in other strains than it is like the second repeat in the same strain. This pattern suggests that a duplication formed this repeat in an ancestral gene, prior to the diversification of pspC into the numerous divergent alleles seen today. These repeats are highly charged, with approximately 45% of their sequence being either lysine or glutamic acid residues. These α-helical repeats were present in all alleles examined, except for the spsA alleles from serotype 1 and serotype 2 strains (13) (Fig. 2).
FIG. 3.
Coiled-coil motif of the α helix of EF6796 PspC. Amino acids that are not in the coiled-coil motif are shifted to the right column. The direct repeats of the α helix and the non-coiled-coil block are indicated. The region with homology to PspA is shaded. This is the output from the Matcher program (10, 24a).
Between the amino acid repeats of the α-helical domain is a highly conserved 40-aa sequence break in the coiled-coil motif which was identified by use of the Matcher program (10) (Fig. 2 and 3). Matcher examines the characteristic seven-residue periodicity of coiled-coil proteins arising largely from the predominance of hydrophobic residues in the first and fourth positions (a and d) and nonhydrophobic residues in the remaining positions (10). The coiled-coil region of the α helix of EF6796 PspC has three breaks in the heptad repeat motif (Fig. 3). These interruptions of the heptad motif in the seven-residue periodicity were 6, 44, and 5 aa long. Similar breaks at corresponding sequence positions were found in all PspC molecules.
In some molecules of PspC, the proline-rich region followed the second amino acid repeat of the α helix (Fig. 1 and 2). However, in the three larger PspC molecules, a region very similar to a corresponding region of the gene product from pspA genetic locus was present. Based on whether this PspA-like region was present or absent and on a distance-based cluster analysis, PspC molecules were classified into two clades (Fig. 4). Clade A molecules contained the PspA-like element and were larger. PspC clade B molecules were smaller and lacked the PspA-like region. This PspA-like region (α-helical region 2) was present in PspC proteins from BG9163, EF6796, and BG7322 (Fig. 1 and 2 and Table 1) as well as in the gene products from many pspA genes (14).
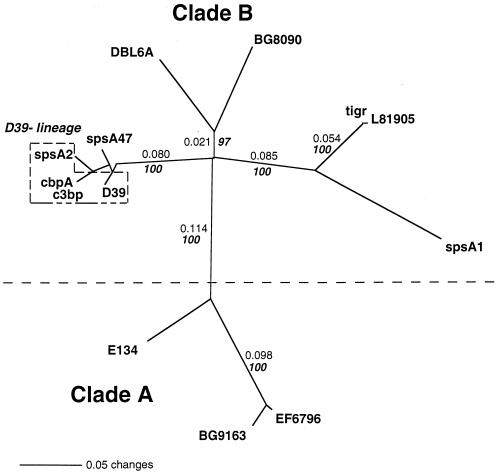
FIG. 4.
Tree of the PspC proteins from this study and related proteins SpsA and CbpA from GenBank. PspC proteins were truncated after the proline-rich region (Fig. 1) before being aligned with the Clustal W algorithm and the Blosum30 amino-acid-scoring matrix in Mac Vector. The tree is an unrooted phylogram generated by the neighbor-joining method with mean character distances in the program PAUP 4.0B. Nonitalic numbers on the tree indicate distances along the branch lengths as calculated by PAUP. Italic bold numbers indicate the percentage of time that the branches were joined together under bootstrap analysis (1,000 replicates were performed). Clade A and clade B are monophyletic groups which were separated by a distance of greater than 0.1 and which clustered together 100% of the time. Clade A PspC proteins share a 120-aa domain with many PspA proteins (Fig. 2). Clade B PspC proteins lack the 120-aa domain, but the other PspC, SpsA, or CbpA proteins share the proline-rich domain with the PspA proteins. The boxed D39 lineage indicates different sequences originating from strains that are laboratory descendents of strain D39. The entries used were the same as those described in the legend to Fig. 2.
TABLE 1.
Conservation of PspC domains
| PspC domain | Range of pairwise % amino acid
identity fora:
|
|||
|---|---|---|---|---|
| PspC vs PspC (orthologous) | Clade A PspC vs PspA (paralogous) | Clade B PspC vs PspA (paralogous) | PspA vs PspA (orthologous) | |
| Upstream through signal peptide | >97 | No alignment possible | No alignment possible | >95 |
| Whole gene | 68–99 | 14–29 | 14–21 | 22–79 |
| α-Helical region 1 | 67–99 | 12–22 | 15–23 | Not present |
| α-Helical region 2 | 100 | 13–89 | Not present | 14–99 |
| Proline richb | High | High | High | High |
| Choline binding | 87 | 77 | 79–99 | 77–98 |
| 17-aa Tail | 100 | 89 | 89–94 | 98–100 |
| 3′ Downstream | 99 | No alignment possible | No alignment possible | ND |
Pairwise percent amino acid identities were calculated with a distance matrix from PAUP 4.0B. ND, not determined.
All PspA and PspC molecules have in this region a repetitive segment of protein with the motif PEPK or PAPAP. Clade B PspC molecules have a conserved nonrepetitive break in the proline-rich region. Distance ranges are uninformative because it is not possible to align these sequences in a meaningful way.
Although there is some variation within the proline-rich region of the sequenced PspC proteins (Fig. 1 and 2), the region is not distinguishable from the proline-rich region of PspA molecules. Within PspA molecules, two types of proline-rich region have been identified. One type, which corresponds to about 60% of PspA proteins (14), contains a central region of 27 nonproline aa which is highly conserved. The other type of proline-rich region in PspA proteins lacks this conserved non-proline-rich region. In the case of PspC, clade A strains lacked the 27-aa non-proline-rich block, whereas the four clade B PspC molecules had this conserved block. When present, the sequence of the 27-aa non-proline-rich region is highly conserved between PspC and PspA molecules. No correlation was observed for the expression of this conserved region within PspA and PspC molecules produced by the same strain (data not shown). The proline-rich region of SpsA from serotype 1 strains was different from those of the PspC molecules. This SpsA molecule has a truncated proline-rich region which contains the 27-aa nonproline break but lacks the NH2 end of the proline-rich region.
The choline-binding repeat domains of the PspC, CbpA, and SpsA proteins each contained between 4 and 11 repeats of about 20 aa (Fig. 5). The repeats found in the center of the choline-binding domain were closest to the consensus sequence, while repeats on the NH2-terminal and COOH-terminal ends of the block were more distant from the consensus sequence. The arrangements of similar repeats within the choline-binding regions of five PspC and three PspA molecules for which the entire choline-binding domain was sequenced were examined (17, 28).
FIG. 5.
Consensus sequences for choline-binding regions of PspC and PspA. The repeat regions of eight proteins, which included three PspA and five PspC molecules, were aligned with the Clustal W algorithm. The alignment was adjusted to maintain divergent repeats together to account for the various sizes of the choline-binding domains. The consensus for repeat number 1 through repeat number 9 of the PspA proteins is in the upper half of figure and that of the PspC proteins is in the bottom half of the figure. The consensus for all nine repeats (1–9) representing at least 60% of the individual amino acids in each position is shown at the top of the diagram. The amino acids that diverge from the consensus sequence are indicated in light gray, and the C-terminal tail is indicated in dark gray. Five amino acids that vary in a gene-specific manner are indicated in white letters on a black background.
The following findings all suggested a very close relationship between PspA and PspC in the choline-binding regions of the molecules. (i) The NH2-terminal divergent repeats are identical between the paralogous proteins (PspA and PspC). (ii) Similarly, the COOH-terminal divergent repeats are very similar between PspC and PspA (see repeats 10 and 11 of the PspC consensus sequence and repeats 9 and 10 of the PspA consensus sequence; Fig. 5), yet these repeats are highly diverged from the rest of the repeat block. (iii) The conserved central repeats of the choline-binding domain in each case have a single amino acid at position 6 which is frequently asparagine in PspC proteins but is usually tyrosine in PspA proteins. Other than position 6, the consensus central repeats for both genes are identical. (iv) The areas of divergence of individual amino acids within the 20-aa repeat from the repeat consensus sequence are identical between PspA and PspC (positions 4, 6, 9, 12, 13, 15, 16, and 18). (v) The repeat block is followed by a 17-aa partially hydrophobic “tail” that is nearly identical in PspC and PspA except for an additional asparagine present at the end of the PspC proteins but absent from the PspA proteins. Overall, the choline-binding domains of PspA and PspC are so similar that it would not be possible to determine with certainty whether any particular choline-binding domain from either of these two proteins belongs to PspA or PspC without knowledge of its flanking DNA.
Phylogenetic analysis.
The pspA and pspC genes are paralogous because they are both present in the genomes of most pneumococci and because they have high identity in the sequences encoding their COOH-terminal halves (Table 1). An alignment of 12 PspC, CbpA, and SpsA sequences was constructed by use of the Clustal W algorithm (Fig. 2). An unrooted phylogram was produced with PAUP 4.0B and the neighbor-joining method from the mean amino acid distances calculated over this alignment (Fig. 4). Figure 4 incorporates both distance measurements along the branch lengths and bootstrap analysis of 1,000 repetitions. The branch length between molecules is proportional to the similarity of the sequences. The tree represents the evolutionary hypothesis that PspC molecules arose in two main clusters representing clades A and B. One clade, A, consisted of the larger PspC molecules and contained strong identity in α-helical region 2 with some PspA molecules. The second clade, B, did not contain this region of identity with the PspA α-helical region.
Analysis of pspC by PCR.
PCR was used to amplify pspC from different strains of S. pneumoniae to permit studies of the variability of PspC. Two oligonucleotides which recognized the common sequence regions of pspC but which did not amplify the pspA genes were designed in an effort to permit the specific amplification of pspC alleles from all pneumococcal strains. Oligonucleotide ABW13 is specific to DNA upstream of the promoter sequence of the pspC gene locus. Oligonucleotide SKH2 is specific to the DNA encoding the C-terminal end of the proline-rich region of both the pspA and the pspC gene loci. These oligonucleotides were used to amplify fragments of pspC from 100 S. pneumoniae strains. Seventy-eight of the 100 strains produced PCR-generated fragments, which varied from 1.5 to 2.2 kb in size. The remaining 22 strains failed to produce a PCR product. Based on the strains with known sequences, it was observed that the sizes of the amplified products correlated with whether they were clade A or clade B. Because of the absence of the pspA conserved region, the clade B pspC sequences were smaller than the clade A pspC sequences. The product amplified from clade A molecules with oligonucleotides ABW13 and SKH2 was 2.0 kb or larger. The fragment amplified from clade B molecules was approximately 1.6 kb. Approximately 4% of the 75 strains from which a pspC gene was amplified were clade A by this criterion, and 96% were clade B.
Cross-reactivity of antiserum made to L81905 PspC with PspA and other PspC molecules.
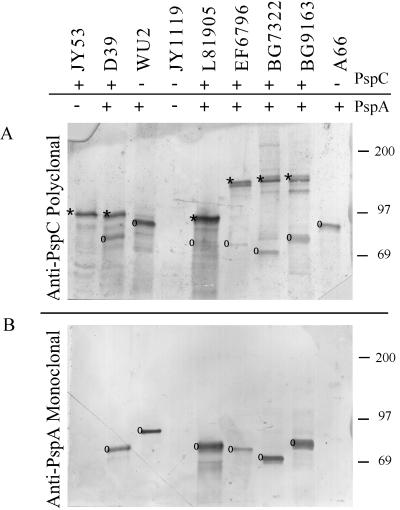
A truncated product (aa 263 to 482) of the L81905 clade B PspC protein was expressed in E. coli by use of the Qiagen expression system (see Materials and Methods). It should be noted that L81905 PspC is clade B and lacks the PspA-like region in its α helix. The truncated (aa 263 to 482) clade B PspC protein was purified by metal affinity chromatography and used to immunize a rabbit to generate a polyclonal antiserum to PspC. Pneumococcal lysates were separated on SDS-polyacrylamide gels and blotted to nitrocellulose. The blots were developed either with Xi126, a monoclonal antibody (MAb) to PspA, or with the anti-PspC rabbit polyclonal antiserum. The reactivity of the antiserum to PspC with selected pneumococcal lysates in a Western immunoblot is shown in Fig. 6.
FIG. 6.
Western immunoblot of pneumococcal lysates. Panel A was developed with anti-PspC polyclonal serum, and panel B was developed with anti-PspA MAb Xi126. An asterisk indicates PspC proteins, and a circle indicates PspA proteins. Molecular mass markers (in kilodaltons) are indicated on the right. Cross-reaction of the polyclonal serum to PspC was observed with all strains tested, but the polyclonal serum reacted weakly to the PspA molecule from L81905.
The reactivity pattern of the antiserum to PspC was deciphered in part by use of lysates from S. pneumoniae JY1119 and JY53. These strains are derivatives of pneumococcal strains WU2 and D39, respectively, in which the pspA genes have been insertionally inactivated (29). From the Western blot, it is apparent that the polyclonal antiserum reacts with a 90-kDa band in JY53, even though the pspA gene has been inactivated in this strain. This band is assumed to represent PspC. Both JY1119 and its parent, WU2, lack the pspC gene (20). An 85-kDa band from WU2 reacts with the anti-PspC antiserum and with the anti-PspA MAb. This band is not present in JY1119, which contains an insertionally inactivated pspA gene.
The rabbit antiserum was reactive with proteins in lysates from all pneumococcal strains tested. The relative molecular weights of the proteins detected also made it apparent that the antiserum was reacting with both PspA and PspC molecules. To distinguish cross-reactivity with the PspA molecule from direct reactivity with the PspC molecule in untested strain lysates, a second, identical Western blot was developed with a MAb specific to PspA molecules (Fig. 6B). PspC bands could be identified through a comparison of banding patterns in Fig. 6. Bands reactive with the anti-PspC rabbit antiserum but not with the anti-PspA MAb were identified as PspC. Bands that were stained by the rabbit antiserum and that comigrated with those also stained by the MAb were PspA molecules that cross-reacted with the antiserum to PspC. Besides failing to react with the MAb, the PspC bands were of a higher molecular weight than the PspA bands. By these criteria, the anti-PspC antiserum cross-reacted with PspA in all strains tested except A66. For A66, a single band was detected. Further testing determined this band to be PspA derived rather than PspC derived. In this case, A66 lacked a pspC gene and the PspA of A66 was not reactive with the MAb used (Xi126), even though anti-PspA immune serum does detect PspA in this strain (data not shown). From the above patterns of reactivity, it was concluded that the anti-PspC polyclonal antiserum does cross-react specifically with the PspA molecule.
Ability of PspC to elicit protective immunity in mice.
Mice were immunized with one of three purified fragments of clade B PspC from L81905 (aa 263 to 482), D39 (aa 1 to 445), and D39 (aa 255 to 445). None of these immunogens contained PspA-like α-helical region 2, but all of them contained the proline-rich region. Mice immunized with PspC or control mice immunized with adjuvant only were challenged with WU2 or BG7322. WU2 is a capsular serotype 3 strain that produces no detectable PspC and does not contain the structural gene for pspC (Fig. 6). BG7322 is a capsular serotype 6B strain that contains a clade A PspC molecule. Significant protection against death was seen with both challenge strains in mice immunized with the three different PspC clade B molecules (Table 2). Protective immunity in mice challenged with WU2 was presumably mediated by antibodies that cross-react with the PspA molecule present on the surface of strain WU2. The ability of PspC to elicit immunity directed against PspA was expected, since PspC had been shown to elicit antibodies cross-reactive with PspA (Fig. 6). Protection of the mice challenged with BG7322 was statistically significant, even though only 62% of the mice were protected, as opposed to 96% of mice challenged with WU2.
TABLE 2.
Cross-protection of CBA/N mice immunized with recombinant PspCa
| Immunogen (PspC fragment) | Challenge strain | No. of live mice/no. of dead mice
|
||
|---|---|---|---|---|
| Immunized | Nonimmunized | Pb | ||
| L81905 (aa 263 to 482) | WU2 (PspA+ PspC−) | 13/0 | 1/12 | <0.0001 |
| D39 (aa 1 to 445) | WU2 (PspA+ PspC−) | 5/0 | 0/5 | 0.008 |
| D39 (aa 255 to 445) | WU2 (PspA+ PspC−) | 4/1 | 0/5 | 0.048 |
| D39 (aa 255 to 445) | BG7322 (PspA+ PspC+) | 13/8 | 1/19 | 0.0002 |
Immunized mice received PspC with adjuvant; nonimmunized mice received adjuvant and buffer. Mice were challenged 21 days postimmunization with 700 CFU of WU2 or 2,000 CFU of BG7322 injected intravenously in 0.2 ml of Ringer's injection solution.
The statistical difference between immunized and nonimmunized mice was calculated with the Fisher exact test.
Antibody elicited to recombinant PspC.
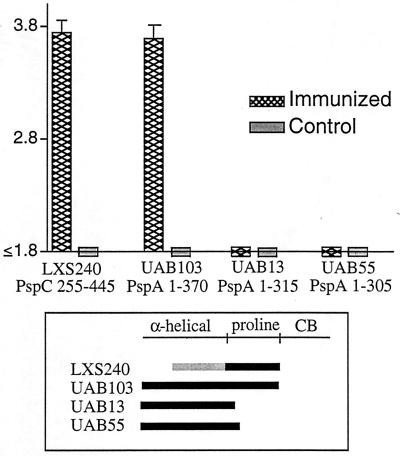
For this study, sera were obtained from mice immunized with a PspC protein fragment (LXS240), which encodes aa 255 to 445 of clade B PspC from D39. This sequence contains the entire proline-rich region of PspC from D39. Direct binding ELISA were conducted to localize the epitope yielding the cross-reactivity with PspA. Microtiter 96-well plates were coated with fragments of PspC from D39 and PspA from Rx1. Each of the cloned PspA molecules from Rx1 used in these assays expressed the PspA α-helical region and differed from the others only in the number of amino acids it contained in the proline-rich region. UAB55 contained 15 aa in the proline-rich region, UAB13 contained 26 aa in the proline-rich region, and UAB103 contained the entire proline-rich region. The results from the ELISA are depicted in Fig. 7. Mouse antiserum reacted only with the PspA molecules containing the entire proline-rich region. The antiserum did not react with PspA molecules UAB55 and UAB13, which contained truncated proline-rich regions. These results strongly suggest that the antibodies that are elicited by PspC and that cross-protect against PspA are probably directed at the proline-rich regions of these molecules.
FIG. 7.
Antibody response patterns in cross-protection. The antibody responses to recombinant PspC and PspA in sera of mice immunized with PspC were measured by an ELISA, and log reciprocal titers were determined. Each bar on the graph represents the mean of the log reciprocal titer and the upper boundary of the standard error for sera from five mice. The limit of detection for the log reciprocal antibody titers in these assays was 1.8. CB, choline-binding region.
DISCUSSION
PspC is a chimeric protein which has acquired domains from both interspecies and intraspecies genetic exchanges. The protein contains a signal sequence that has 75% nucleotide identity to the bac gene from group B streptococci (accession numbers X59771 and X58470) (13). The bac gene encodes the β antigen of group B streptococci, a cell surface receptor that binds the constant region of human IgA. The similar sequence in the signal peptide region suggests potential interspecies genetic exchange between group B streptococci and S. pneumoniae. This exchange event would have formed a chimeric locus including the bac regulatory region and a partial pspA gene or a pspA-like locus to create an ancestral gene for pspC. The origin of the central region specific to the current pspC genes is unknown. The direct amino acid repeats of the α helix suggest that this region of PspC has evolved by a domain duplication event which has led to gene elongation. The region of the α helix is presumably the functional region of the molecule and reportedly binds secretory IgA (13). Further intraspecies variation events are hinted at in the finding that 4% of PspC proteins are of clade A. This clade appears to have been derived from a recombination event with PspA (or visa versa), providing further evidence of the chimeric structure of PspC and possibly PspA molecules.
Several functions have been attributed to the PspC molecule (also called CbpA, SpsA, or PbcA). In addition to binding secretory IgA and a moiety on the surface of epithelial cells, it has been reported to bind the complement component C3 (15). Recent studies have shown that PspA inhibits complement activation by inhibiting the formation of the C3 convertase (26). With the similar structural domains of PspA and PspC, it is conceivable that the virulence properties of the two proteins may complement each other in the host. WU2 is a strain of S. pneumoniae that does not contain a structural gene for PspC. PspA mutants of WU2 show a 10,000-fold decrease in virulence (6). When PspA is mutated in D39, a strain that contains both PspA and PspC, there is only a 10-fold decrease in virulence (6). These data, combined with the preliminary data of Hostetter et al. (15), suggest that PspA and PspC may complement each other in their abilities to block the clearance of pneumococci by interfering with the complement pathway.
Rosenow et al. demonstrated that CbpA is expressed more strongly by pneumococci in the nasopharynx than by pneumococci in the blood (22). Thus, it is feasible that the two forms of the molecule, PspC and PspA, serve the same general function, possibly in different host tissues and in different stages of infection. Furthermore, either molecule may be more critical to virulence in the absence of the other. This hypothesis is further strengthened by data from ongoing studies in our laboratory that indicate that mutants lacking both PspC and PspA show a decrease in virulence (6a).
In PspC immunization studies, we challenged mice with a strain expressing both PspC and PspA and a strain expressing PspA but not PspC. By including strains lacking the pspC gene, we could determine if protection elicited by PspC required the expression of PspC or might occur, at least in part, through cross-reactions with PspA. For the study presented, mice were immunized with clade B PspC. This molecule lacks the PspA-PspC region of homology near the C-terminal end of the α-helical region of PspC. Thus, this immunogen was expected to be one which would give less cross-reaction with PspA than would clade A PspC. Even so, immunization with PspC from D39 resulted in protection when mice were challenged with either strain BG7322, which expresses both PspA and PspC, or strain WU2, which expresses PspA but lacks PspC.
The protection-eliciting PspC immunogen contained the entire proline-rich region. The α-helical regions of PspA from WU2 and PspC from D39 have essentially no homology. However, the proline-rich region of PspC is repetitive and homologous with that of PspA. It was possible that antibody to this region was responsible for the cross-protection that we observed. This hypothesis was supported by the observation that antibodies elicited to PspC reacted with PspA fragments that contained the proline-rich region but not with those that lacked the proline-rich region in direct ELISA. Antibodies elicited by PspC also cross-reacted with PspA on Western blots. The likelihood that the protective cross-reaction of PspC immune sera was mediated through PspA was further strengthened by the sequence data released by TIGR (30a). Extensive searches of the largely completed genome failed to reveal other pneumococcal gene sequences with as high a similarity to the PspC sequence domains as the proline-rich region of PspA.
Electron microscopy surface labeling studies and epitope mapping studies have localized PspA on the surface of pneumococci with an exposed α-helical region (12, 16, 18). Studies by Yother and White have shown that PspA is attached by the C-terminal end to lipoteichoic acids (30). No information is available, however, about whether or not the proline-rich domain is surface exposed. Results from experiments indicating that antibodies to the proline-rich domain are protective suggest that this domain of PspA is probably accessible on the surface of pneumococci. This study also provides the first published evidence that antibodies reactive with the proline-rich region of PspA can be protective against pneumococcal infection.
PspA, PspC (also called CbpA or SpsA), LytA, and PcpA are proteins of S. pneumoniae that contain choline-binding domains. The consensus sequences of these domains of PspC and PspA are from 90 to 95% identical. The middle region of the choline-binding domains of PspA and PspC is conserved. The first and last two repeats of PspA and PspC differ substantially (by 40 to 65%) from the consensus sequence. The choline-binding domains of LytA and PcpA are quite different from that of PspA or PspC (42 to 62% identity) (11, 24). Whereas PspA and PspC have most likely evolved by gene duplication, PcpA, whose choline-binding domain is more like that of CspA of Clostridium beijerinckii, has probably arisen from horizontal gene transfer. The choline-binding regions of these proteins all support a modular form of evolution of this group of proteins.
This report provides a comprehensive study of the sequence of pspC and shows that PspC proteins can be divided into two clades based on the sequences in their α-helical and proline-rich domains. This study also demonstrates that immunity to the proline-rich domain of PspC can be protective through recognition of the proline-rich domain of PspA. The fact that the N-terminal α-helical domain of PspC is different from the α-helical domain of PspA suggests that PspC and PspA may serve somewhat distinct roles in virulence. However, the fact that the two molecules have a very similar domain structure and have similarity in much of their sequences raises the possibility that these two molecules may have similar functions. Although the sequences of a few pspC alleles have been previously reported, this is the first report that PspC contains two clades and that PspC shows homology to PspA within the cross-protective region of the α helix. The identification of two clades of PspC should be pertinent to future efforts to develop a PspC-containing vaccine. Moreover, the observation that antibodies to the proline-rich regions of PspA and PspC may be cross-protective may facilitate the design of a more efficacious vaccine.
ACKNOWLEDGMENTS
We thank Rebecca Tart, Larry McDaniel, Bill Benjamin, Tanya Kelly, Melissa Caimano, Edwin Swiatlo, and Kim Benton for interest in and advice during this project. We also thank Xinping Wu for technical assistance in the production of the cloned pspA fragments.
This work was supported in part by National Institutes of Health grants AI21548 and HL58418. Alexis Brooks-Walter was supported by a University of Alabama at Birmingham Comprehensive Minority Faculty Development Fellowship. Sequencing of S. pneumoniae at TIGR was accomplished with the support of the Merek Genome Research Institute and the National Institute of Allergy and Infectious Diseases (National Institutes of Health). The University of Wisconsin Genetics Computer Group programs were supported by the Center for AIDS Research (grant P30 AI27767). The University of Alabama at Birmingham (UAB) Sequencing Facility was supported in part by grants to the UAB Medical School from the Howard Hughes Medical Institute and the UAB Health Sciences Foundation.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 2.Banas J A, Russell R R B, Ferretti J J. Sequence analysis of the gene for the glucan-binding protein of Streptococcus mutansIngbritt. Infect Immun. 1990;58:667–673. doi: 10.1128/iai.58.3.667-673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barroso L A, Wang S-Z, Phelps C J, Johnson J L, Wilkins T D. Nucleotide sequence of Clostridium difficiletoxin B gene. Nucleic Acids Res. 1990;18:4004. doi: 10.1093/nar/18.13.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breise T, Hackenbeck R. Interactions of pneumococcal amidase with lipoteichoic acid and choline. Eur J Biochem. 1985;146:417–427. doi: 10.1111/j.1432-1033.1985.tb08668.x. [DOI] [PubMed] [Google Scholar]
- 5.Briles D E, Crain M J, Gray B M, Forman C, Yother J. A strong association between capsular type and mouse virulence among human isolates of Streptococcus pneumoniae. Infect Immun. 1992;60:111–116. doi: 10.1128/iai.60.1.111-116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briles D E, Hollingshead S K, Swiatlo E, Brooks-Walter A, Szalai A, Virolainen A, McDaniel L S, Benton K A, White P, Prellner K, Hermansson A, Aerts P C, Van Dijk H, Crain M J. PspA and PspC: their potential for use as pneumococcal vaccines. Microb Drug Resist. 1997;3:401–408. doi: 10.1089/mdr.1997.3.401. [DOI] [PubMed] [Google Scholar]
- 6a.Brooks-Walter, A., S. K. Hollingshead, and D. E. Briles. Unpublished data.
- 7.Brooks-Walter A, Tart R C, Briles D E, Hollingshead S K. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. 1997. The pspC gene encodes a second pneumococcal surface protein homologous to the gene encoding the protection-eliciting PspA protein of Streptococcus pneumoniae. [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dove C H, Wang S-Z, Price S B, Phelps C J, Lyerly D M, Wilkins T D, Johnson J L. Molecular characterization of the Clostridium difficiletoxin A gene. Infect Immun. 1990;58:480–488. doi: 10.1128/iai.58.2.480-488.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischetti V A, Landau G M, Sellers P H, Schmidt J P. Identifying periodic occurrences of a template with applications to protein structure. Inform Proc Lett. 1993;45:11–18. [Google Scholar]
- 11.Garcia P, Garcia J L, Garcia E, Lopez R. Nucleotide sequence and expression of the pneumococcal autolysin gene from its own promoter in Escherichia coli. Gene. 1986;43:265–272. doi: 10.1016/0378-1119(86)90215-5. [DOI] [PubMed] [Google Scholar]
- 12.Gray B M. Streptococcal infections. In: Brachman P E, Evans A S, editors. Bacterial infections of humans. 3rd ed. New York, N.Y: Plenum Publishing Corp.; 1998. pp. 673–711. [Google Scholar]
- 13.Hammerschmidt S, Talay S R, Brandtzaeg P, Chhatwal G S. SpsA, a novel pneumococcal surface protein with specific binding to secretory immunoglobulin A and secretory component. Mol Microbiol. 1997;25:1113–1124. doi: 10.1046/j.1365-2958.1997.5391899.x. [DOI] [PubMed] [Google Scholar]
- 14.Hollingshead, S. K., R. S. Becker, and D. E. Briles. Unpublished data.
- 15.Hostetter M K, Cheng Q, Finkel D A. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. 1997. C3-binding protein (pbcA) in Streptococcus pneumoniae: accession number AF067128, abstr. B-478. [Google Scholar]
- 16.McDaniel L S, Briles D E. Monoclonal antibodies against surface components of Streptococcus pneumoniae. In: Macario A J L, de Macario E C, editors. Monoclonal antibodies against bacteria. Vol. 3. Orlando, Fla: Academic Press, Inc.; 1986. pp. 143–164. [Google Scholar]
- 17.McDaniel L S, McDaniel D O, Hollingshead S K, Briles D E. Comparison of the PspA sequence from Streptococcus pneumoniaeto the previously identified PspA sequence from strain Rx1 and the ability of PspA from EF5668 to elicit protection against pneumococci of different capsular types. Infect Immun. 1998;66:4748–4754. doi: 10.1128/iai.66.10.4748-4754.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDaniel L S, Ralph B A, McDaniel D O, Briles D E. Localization of protection-eliciting epitopes on PspA of Streptococcus pneumoniaebetween amino acid residues 192 and 260. Microb Pathog. 1994;17:323–337. doi: 10.1006/mpat.1994.1078. [DOI] [PubMed] [Google Scholar]
- 19.McDaniel L S, Sheffield J S, Delucchi P, Briles D E. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect Immun. 1991;59:222–228. doi: 10.1128/iai.59.1.222-228.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDaniel L S, Sheffield J S, Swiatlo E, Yother J, Crain M J, Briles D E. Molecular localization of variable and conserved regions of pspA, and identification of additional pspA homologous sequences in Streptococcus pneumoniae. Microb Pathog. 1992;13:261–269. doi: 10.1016/0882-4010(92)90036-n. [DOI] [PubMed] [Google Scholar]
- 21.McDaniel L S, Yother J, Vijayakumar M, McGarry L, Guild W R, Briles D E. Use of insertional inactivation to facilitate studies of biological properties of pneumococcal surface protein A (PspA) J Exp Med. 1987;165:381–394. doi: 10.1084/jem.165.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenow C, Ryan P, Weiser J N, Johnson S, Fontan P, Ortqvist A, Masure H R. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol Microbiol. 1997;25:819–829. doi: 10.1111/j.1365-2958.1997.mmi494.x. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez-Beato A R, Garcia J L. Molecular characterization of a family of choline-binding proteins of Clostridium acetobutylicum NCIB 8052 accession number Z50008. 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez-Beato A R, Lopez R, Garcia J L. Molecular characterization of PcpA: a novel choline-binding protein of Streptococcus pneumoniae. FEMS Microbiol Lett. 1998;164:207–214. doi: 10.1111/j.1574-6968.1998.tb13087.x. [DOI] [PubMed] [Google Scholar]
- 24a.Schmidt, J. P. 1993. Matcher program. [Online.] http://catt.poly.edu/∼jps/. [22 October 1999, last date accessed.]
- 25.Tomasz A. Choline in the cell wall of a bacterium: novel type of polymer-linked choline in Pneumococcus. Science. 1967;157:694–697. doi: 10.1126/science.157.3789.694. [DOI] [PubMed] [Google Scholar]
- 26.Tu A-H, Fulgham R L, McCrory M A, Briles D E, Szalai A J. Pneumococcal surface protein A (PspA) inhibits complement activation by Streptococcus pneumoniae. Infect Immun. 1999;67:4720–4724. doi: 10.1128/iai.67.9.4720-4724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H-Y, Nahm M, Guo Y, Russell M, Briles D E. Intranasal immunization of mice with PspA (pneumococcal surface protein A) can prevent intranasal carriage and infection with Streptococcus pneumoniae. J Infect Dis. 1997;175:893–846. doi: 10.1086/513980. [DOI] [PubMed] [Google Scholar]
- 28.Yother J, Briles D E. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J Bacteriol. 1992;174:601–609. doi: 10.1128/jb.174.2.601-609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yother J, Handsome G L, Briles D E. Truncated forms of PspA that are secreted from Streptococcus pneumoniae and their use in functional studies and cloning of the pspAgene. J Bacteriol. 1992;174:610–618. doi: 10.1128/jb.174.2.610-618.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yother J, White J M. Novel surface attachment mechanism of the Streptococcus pneumoniaeprotein PspA. J Bacteriol. 1994;176:2976–2985. doi: 10.1128/jb.176.10.2976-2985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.The Institute for Genomic Research Website. Sequences [Online.] http://www.tigr.org.