Abstract
The Wildsoydb DataHub is an integrated interface for biologists and breeders to access soybean genomic resources easily, allowing them to fully utilize the results of genomic research.
Dear Editor,
The rapid development of sequencing technology in the last decade has ushered soybean (Glycine max and Glycine soja) research into the genomic age. Mining information out of these datasets is challenging for biologists without intensive bioinformatic training, but web applications with intuitive data retrieval and visualization functions can empower general users to access both the genome sequences and genomics resources. Although there are some online platforms for soybean (Machado et al., 2020; Yu et al., 2022), they are primarily focused on RNA-Seq data and lack the association to gene regulation information, like chromatin accessibility. While the leading platform Soybase (https://soybase.org/) has deposited multiomic data, most of their data are based on prior versions of the soybean reference genome (Wm82 a1v1.1, Wm82 a2v1) (Brown et al., 2021). Furthermore, none of the aforementioned platforms allow biologists to easily retrieve intergenic sequences (e.g. promoters) or design primers. With the increasing amount of sequencing data generated from various cultivars, there is a growing demand for a curated web portal hosting multiple reference genomes with associated multiomic data. To address this gap, we created an integrated online platform Wildsoydb DataHub (https://datahub.wildsoydb.org/) hosting four high-quality genome assemblies, including two G. max cultivars: Williams 82 (Wm82 a2v1 and Wm82 a4v1) (Schmutz et al., 2010; Valliyodan et al., 2019) and Zhonghuang 13 (v2) (Liu et al., 2020) and a G. soja cultivar: W05 (v1) (Xie et al., 2019). We aim to provide an easy-to-use web interface for biologists to fully benefit from the genomic resources. Aside from comprehensive functional annotations of all the genes, the sequencing data from 21 soybean genomics studies and 11 public SRA BioProjects of various data types were integrated. Furthermore, a variety of functional modules were developed to provide an intuitive user-centric interface. Modules currently available on this platform include Gene Search, BLAST, Jbrowse, Synteny, SeqExtractor, and Primer3. All functions are aggregated to the Gene Search result and can also be invoked from their respective pages, allowing users to (1) retrieve functional annotations, genome sequences, and gene expressions; (2) cross-compare sequences of interest with BLAST and synteny analysis; (3) visualize expression and methylation levels as well as other genomics information from a well-organized browser; (4) design primers for selected genes; and (5) retrieve sequence from unannotated regions.
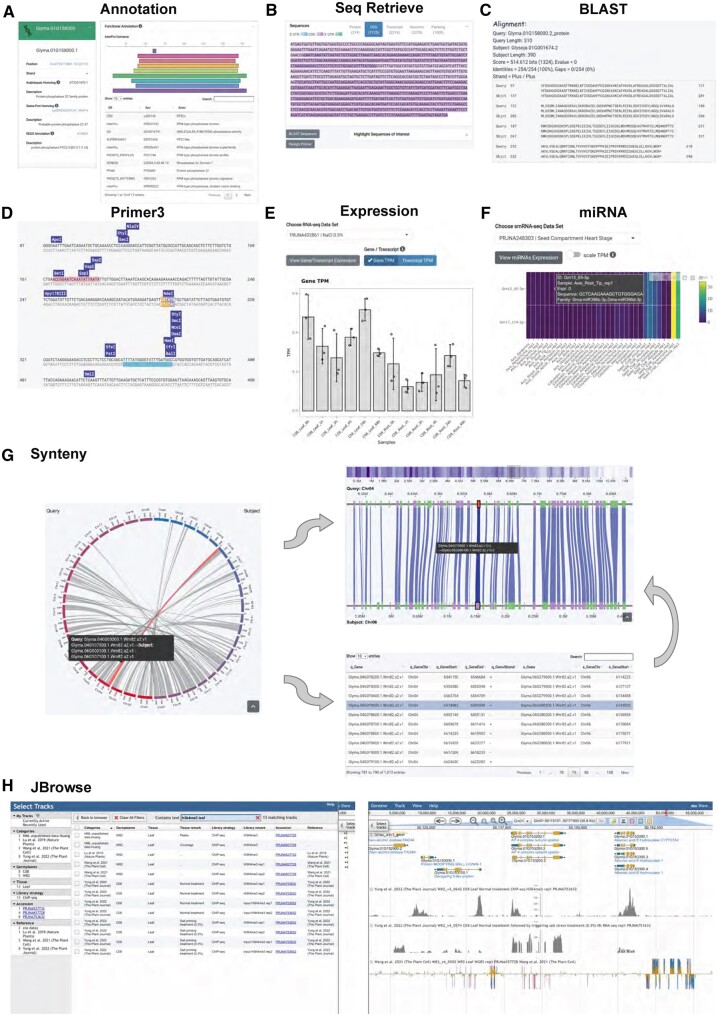
Gene Search is designed to be versatile. Apart from the soybean gene ID, a search can be made using the Arabidopsis (Arabidopsis thaliana) gene ID, annotation database identifiers (GO, KO, PFAM, PATHER, IPR, Swiss-Prot ID, EC number, etc.), gene functions, and genomics coordinates. The search result page incorporates detailed information on the query gene, including a brief description, the Arabidopsis homologs (TAIR10), Swiss-Prot ID (2021_03), and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation (Figure 1A). The protein motifs discovered by InterProScan (5.48–83.0) are displayed in a table with their locations in an interactive plot (Figure 1A). Besides annotations, the genomic sequence, transcripts, coding regions, proteins, and flanking sequences are returned in a tab box container. The output sequence is shaded according to the genomic contexts (Figure 1B). The sequence can be sent for BLAST (Figure 1C) or primer design by a simple click (Figure 1D). Fully functional BLAST programs were integrated to adapt the usage to different scenarios. The Primer3 module generates an interactive plot showing the positions of candidate primers as well as the predicted restriction enzyme digestion sites on the templates (Figure 1D). Moreover, if any expression dataset of the selected reference genome is available, the expression of the gene or transcripts can be displayed as transcript per million values in a bar chart (Figure 1E). Meanwhile, if the query gene is predicted to be the target of any miRNA from the selected smRNA-seq dataset, the expression of all miRNA candidates will be depicted as a heatmap, and the miRNA families and the mature sequences will be displayed in tooltips (Figure 1F).
Figure 1.
Overview of the Wildsoydb DataHub web interface, using Glyma.01G158000 queried against the published soybean genome, Wm82 a4v1, as an example. A, Comprehensive annotation of the query gene, including the homolog in the Arabidopsis genome and Swiss-Prot, the annotation in the KEGG database (left), and functional annotations of the protein motif discovered by Interproscan (right). B, The DNA sequence of the query gene. C, Protein BLAST results. D, Primer3 result of the query gene. E, Expression levels of the query gene in a salt treatment RNA-seq dataset. F, Expressions of the miRNAs are predicted to target the query gene from a seed development dataset. G, Intra-genome synteny analysis of Wm82 a2v1. The macro-synteny blocks were illustrated with a circular layout (left), while the gene density and local micro-synteny regions were represented as a heatmap and in a parallel layout (right top). All the genes within the macro-synteny block were shown in a searchable table (right bottom). H, Jbrowse faceted track selector (left) and a demonstration view of the genomic regions around Glyma.01G153200 with an H3K4me3 ChIP-seq track, an RNA-seq track, as well as a BS-seq track on display (right; top to bottom).
The synteny inference functional module in Wildsoydb DataHub could associate the target gene to neighboring genes in the same region, providing a clearer sense of the gene–gene relationship in the evolutionary sense. We performed synteny analyses with primary transcripts to generate both intra- and inter-genome synteny blocks using the MCscan pipeline (Tang et al., 2008). The macro-synteny result is illustrated on the whole-chromosome scale using a circular layout. By clicking on the syntenic region of interest, an interactive micro-synteny plot will be shown for local gene analyses. Meanwhile, gene pairs discovered from the chosen macro-synteny block will be listed in a table. Users can search the table for their gene of interest, and re-center and highlight the selected query gene in the micro-synteny view by clicking on the record (Figure 1G). The synteny module uses the same core as the standard alone version ShinySyn developed by us (Xiao and Lam, 2022).
To better elucidate soybean genomics data, we incorporated JBrowse (Buels et al., 2016) into the platform as a module, which provides a faster and more fluent user experience, and integrated the RNA-seq, bisulfite sequencing (BS)-seq, smRNA-seq, ATAC-seq, and ChIP-seq data, yielding 664 data tracks for Williams 82 a4v1, 110 data tracks for ZH13 v2, 115 data tracks for W05 v1, and 4 data tracks for Williams 82 a2v1. A full list of the studies included can be found at https://docs.datahub.wildsoydb.org/jbrowse/genomics_data/. A faceted track selector was implemented, making all the properties of the metadata searchable for users. One can search via publication, SRA accession ID, library type, germplasm, tissue, or treatment (salt, auxin, etc.), enabling easy access to specific groups of data (e.g. H3K4me3 leaf) and cross-referencing of results from different studies (Figure 1H).
In summary, we added more extensive multiomic datasets than the current soybean websites and developed unique functionalities like primer design and universal sequence retrieval to greatly minimize biologists’ efforts while studying gene regulation. All of these features work together to make Wildsoydb DataHub a user-centric web interface for accessing soybean genomes and genomic resources from multiple high-quality references. We believe it will be an efficient platform for biologists and breeders and accelerate their studies.
Acknowledgments
We would like to thank all the soybean researchers who made their genomics data available to the public. We apologize for any omissions in the cited literature owing to space limitations. Wildsoydb DataHub is built on the Shiny/R framework; we would also like to thank all the developers, as well as Yihui Fan for sharing their experience in deploying Shiny apps. Jee Yan Chu copy-edited this manuscript. Any opinions, findings, conclusions, or recommendations expressed in this publication do not reflect the views of the Government of Hong Kong Special Administrative Region or the Innovation and Technology Commission.
Funding
This work is supported by the Hong Kong Research Grants Council Area of Excellence Scheme (AoE/M-403/16) and the Lo Kwee-Seong Biomedical Research Fund.
Conflict of interest statement. None declared.
Contributor Information
Zhixia Xiao, School of Life Sciences, Center for Soybean Research of the State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, Hong Kong.
Qianwen Wang, School of Life Sciences, Center for Soybean Research of the State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, Hong Kong; Department of Bioinformatics, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Man-Wah Li, School of Life Sciences, Center for Soybean Research of the State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, Hong Kong.
Mingkun Huang, School of Life Sciences, Center for Soybean Research of the State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, Hong Kong; Lushan Botanical Garden, Chinese Academy of Sciences, Jiujiang 332900, China.
Zhili Wang, School of Life Sciences, Center for Soybean Research of the State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, Hong Kong.
Min Xie, School of Life Sciences, Center for Soybean Research of the State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, Hong Kong; Guangdong Engineering Research Center of Plant and Animal Genomics, BGI Genomics, BGI-Shenzhen, Shenzhen 518083, China.
Rajeev K Varshney, State Agricultural Biotechnology Centre, Centre for Crop and Food Innovation, Food Futures Institute, Murdoch University, Murdoch, Western Australia 6150, Australia.
Henry T Nguyen, Division of Plant Sciences, National Center for Soybean Biotechnology, University of Missouri, Columbia, Missouri 65211, USA.
Ting-Fung Chan, School of Life Sciences, Center for Soybean Research of the State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, Hong Kong.
Hon-Ming Lam, School of Life Sciences, Center for Soybean Research of the State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, Hong Kong.
These authors contributed equally (Z.X. and Q.W.)
H.-M.L. and T.-F.C. coordinated this research and acquired funding and resources for the study. H.-M.L., T.-F.C., and Z.X. conceived the study. Z.X. constructed the website. Z.X., Q.W., M.H., and M.X. performed the data analysis. Z.X., Q.W., M.-W.L., Z.W., R.K.V., and H.T.N. interpreted the results. H.-M.L. and Z.X. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Hon-Ming Lam (honming@cuhk.edu.hk).
References
- Brown AV, Conners SI, Huang W, Wilkey AP, Grant D, Weeks NT, Cannon SB, Graham MA, Nelson RT (2021) A new decade and new data at SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res 49: D1496–D1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buels R, Yao E, Diesh CM, Hayes RD, Munoz-Torres M, Helt G, Goodstein DM, Elsik CG, Lewis SE, Stein L, et al. (2016) JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol 17: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Du H, Li P, Shen Y, Peng H, Liu S, Zhou G-A, Zhang H, Liu Z, Shi M, et al. (2020) Pan-genome of wild and cultivated soybeans. Cell 182: 162–176.e13 [DOI] [PubMed] [Google Scholar]
- Machado FB, Moharana KC, Almeida‐Silva F, Gazara RK, Pedrosa‐Silva F, Coelho FS, Grativol C, Venancio TM (2020) Systematic analysis of 1298 RNA‐Seq samples and construction of a comprehensive soybean (Glycine max) expression atlas. Plant J 103: 1894–1909 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH (2008) Synteny and collinearity in plant genomes. Science (80-) 320: 486–488 [DOI] [PubMed] [Google Scholar]
- Valliyodan B, Cannon SB, Bayer PE, Shu S, Brown AV, Ren L, Jenkins J, Chung CY‐L, Chan T, Daum CG, et al. (2019) Construction and comparison of three reference‐quality genome assemblies for soybean. Plant J 100: 1066–1082 [DOI] [PubMed] [Google Scholar]
- Xiao Z, Lam H-M (2022) ShinySyn: a Shiny/R application for the interactive visualization and integration of macro- and micro-synteny data. Bioinformatics btac503 [DOI] [PubMed] [Google Scholar]
- Xie M, Chung CY-L, Li M-W, Wong F-L, Wang X, Liu A, Wang Z, Leung AK-Y, Wong T-H, Tong S-W, et al. (2019) A reference-grade wild soybean genome. Nat Commun 10: 1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Zhang H, Long Y, Shu Y, Zhai J (2022) Plant public RNA‐seq database: a comprehensive online database for expression analysis of ∼45 000 plant public RNA‐seq libraries. Plant Biotechnol J (doi:10.1111/pbi.13798) [DOI] [PMC free article] [PubMed] [Google Scholar]