Abstract
High ambient temperature suppresses Arabidopsis (Arabidopsis thaliana) rosette leaf area and elongates the stem and petiole. While the mechanism underlying the temperature-induced elongation response has been extensively studied, the genetic basis of temperature regulation of leaf size is largely unknown. Here, we show that warm temperature inhibits cell proliferation in Arabidopsis leaves, resulting in fewer cells compared to the control condition. Cellular phenotyping and genetic and biochemical analyses established the key roles of PHYTOCHROME-INTERACTING FACTOR4 (PIF4) and TEOSINTE BRANCHED1/CYCLOIDEA/PCF4 (TCP4) transcription factors in the suppression of Arabidopsis leaf area under high temperature by a reduction in cell number. We show that temperature-mediated suppression of cell proliferation requires PIF4, which interacts with TCP4 and regulates the expression of the cell cycle inhibitor KIP-RELATED PROTEIN1 (KRP1) to control leaf size under high temperature. Warm temperature induces binding of both PIF4 and TCP4 to the KRP1 promoter. PIF4 binding to KRP1 under high temperature is TCP4 dependent as TCP4 regulates PIF4 transcript levels under high temperature. We propose a model where a warm temperature-mediated accumulation of PIF4 in leaf cells promotes its binding to the KRP1 promoter in a TCP4-dependent way to regulate cell production and leaf size. Our finding of high temperature-mediated transcriptional upregulation of KRP1 integrates a developmental signal with an environmental signal that converges on a basal cell regulatory process.
High ambient temperature suppresses Arabidopsis leaf size by reducing cell proliferation via coordination of two diverse transcription factors integrating environmental and developmental signals.
Introduction
A suite of morphological adaptations that occurs in response to high ambient temperature is termed thermomorphogenesis, which includes hypocotyl and petiole elongation, accelerated flowering, etc. (Quint et al., 2016). While hypocotyl elongation is predicted to play a role in protecting meristematic tissue from the heat-absorbing soil, hyponastic petiole movement and open rosette structure help in increasing the leaf cooling capacity (Crawford et al., 2012; Kim et al., 2019). Leaves, on the other hand, show a reduction in blade area and thickness, reduced epidermal cell and stomatal density, early senescence, and reduced net carbon assimilation (Jin et al., 2011; Vile et al., 2012; Ibañez et al., 2017; Kim et al., 2020a). Leaves perceive and respond to various environmental signals, and are major above-ground mediators between a plant and its environment (Efroni et al., 2010; Lippmann et al., 2019). In the current warming climate, studying the impact of environmental changes on leaf growth and development and understanding the genetic mechanism for leaf phenotypic plasticity under varied environmental stimuli has become particularly important. While the mechanism underlying temperature-induced hypocotyl elongation is extensively studied, the genetic basis of temperature regulation of leaf features is largely unknown (Gray et al., 1998; Koini et al., 2009; Quint et al., 2016). Leaves are lateral organs that arise as outgrowths from L1 to L3 layers of the shoot apical meristem. Recent reports show that the leaf is highly thermoresponsive and facilitates temperature-mediated growth in other aerial tissues (Kim et al., 2020b).
PHYTOCHROME-INTERACTING FACTOR4 (PIF4) is a key transcription factor for thermomorphogenesis that integrates high-temperature cues to downstream phytohormone signaling and cellular response to manifest hypocotyl and petiole elongation (Koini et al., 2009; Quint et al., 2016). PIF4 is expressed in all aerial tissues but its leaf epidermis-specific expression drives hypocotyl elongation and leaf hyponastic movements under high temperature (Kim et al., 2020b). In warm conditions, PIF4-mediated induction of auxin biosynthesis in the leaves and ASYMMETRIC LEAVES1 (AS1)- and PINOID-dependent directional auxin flow toward the abaxial side of the petiole determines the leaf thermonastic movements that help in leaf cooling (Park et al., 2019). PIF4-mediated transcriptional activation of growth-promoting genes, on the other hand, is antagonized by AT-hook transcription factors to suppress petiole elongation, suggesting a general involvement of PIF4 in growth regulatory responses (Favero et al., 2020). Recently, a role of PIF7 in temperature signaling was also reported in the early seedling growth and particularly for daytime regulation of high-temperature response, suggesting that different PIFs play specific roles in temperature-mediated growth adaptations (Chung et al., 2020; Fiorucci et al., 2020).
Several reports have suggested that PIF4 functions in concert with other growth regulators to control a variety of growth responses in a context-dependent fashion. For example, PIF4 regulates hypocotyl elongation in response to light and temperature via induction of auxin and brassinosteroids biosynthetic and response genes that involve integration with other transcription regulators, such as AUXIN RESPONSE FACTORs (ARFs), BRASSINAZOLE-RESISTANT1 (BZR1), SEUSS, HOOKLESS1, etc. (Huai et al., 2018; Ibañez et al., 2018; Reed et al., 2018; Jin et al., 2020). Apart from these transcription regulators, more recently, a small aromatic compound, Heatin, was shown to promote hypocotyl elongation via the PIF4-auxin module in response to temperature (van der Woude et al., 2021). Contrary to the elongation response in the hypocotyl and petioles, PIF4 is shown to suppress the formation of axillary meristems in the dark along with BZR1 via inhibition of ARGONAUTE10 and by antagonizing with ARF5 (Zhang et al., 2020). Temperature-mediated inhibition of meristemoid division via suppression of SPEECHLESS in the stomatal lineage cells is further reported to operate in a PIF4-dependent manner that leads to the production of fewer stomata in the cotyledons of Arabidopsis (Arabidopsis thaliana) seedlings grown under high temperature (Lau et al., 2018). These reports suggest that PIF4 functions with other transcription factors in a context-dependent manner to play a pivotal role in linking environmental responses to the core developmental pathways. Thus, PIF4 crosstalk with other growth regulators needs to be explored for a better mechanistic understanding of these pathways and for their exploitation to optimize growth under a variable environment.
Five members of class II TEOSINTE BRANCHED1/CYCLOIDEA/PCFs (TCPs) regulated by miR319, control multiple developmental pathways including leaf growth, shape, and senescence (Palatnik et al., 2003; Schommer et al., 2012; Koyama et al., 2017). TCP4, one of the most characterized class II CIN-TCP transcription factors, suppresses cell proliferation by activating the transcription of a cell cycle inhibitor gene KIP-RELATED PROTEIN1 (KRP1/ICK1) and indirectly by repressing GROWTH-REGULATING FACTORs (GRFs) via miR396 (Schommer et al., 2014). TCP4 was shown to promote cotyledon opening during the de-etiolation process by antagonizing the dark-induced PIF3, providing a molecular link between environmental signal and developmental response (Dong et al., 2019).
Plants display architectural plasticity in response to their surrounding environment via changes in organ shape or size by regulating basal cellular processes like cell division and expansion. Several reports have highlighted the importance of cell proliferation in mediating plant growth plasticity under various environmental stimuli (Tardieu and Granier, 2000; Rymen et al., 2007; Casadevall et al., 2013; Okello et al., 2016; Romanowski et al., 2021). This is achieved by tight regulation of cell cycle progression and arrest by various genes including several transcription factors, such as the regulation of cell cycle arrest under heat by NAC (NAM, ATAF1/2 and CUC2)-type transcription factors ANAC044 and ANAC085, regulation of cell cycle under high temperature by ICARUS1, and miR396-mediated repression of GRFs to regulate cell proliferation under UV radiations (Casadevall et al., 2013; Zhu et al., 2015; Takahashi et al., 2019). A recent study showed an antagonistic regulation of cell cycle regulators by PIF7 and ANGUSTIFOLIA (AN3) to constrain leaf blade cell division by end-of-day far-red (Hussain et al., 2022).
High temperature elicits different growth responses in different organs. Growth promotion by cell elongation is a well-documented response in plants under various environmental cues including high temperature. As opposed to hypocotyl and petiole elongation, high temperature regulation of leaf size has not been studied in detail and the underlying genetic regulation is largely unknown. Here, we show that high temperature suppresses leaf area by inhibiting cell division in the leaves. We, further, show that temperature-mediated induction of PIF4 and TCP4 expression regulates KRP1 transcription, leading to inhibition of cell proliferation in leaves of the plants grown under high temperature.
Results
High-temperature-mediated suppression of leaf size involves inhibition of cell division
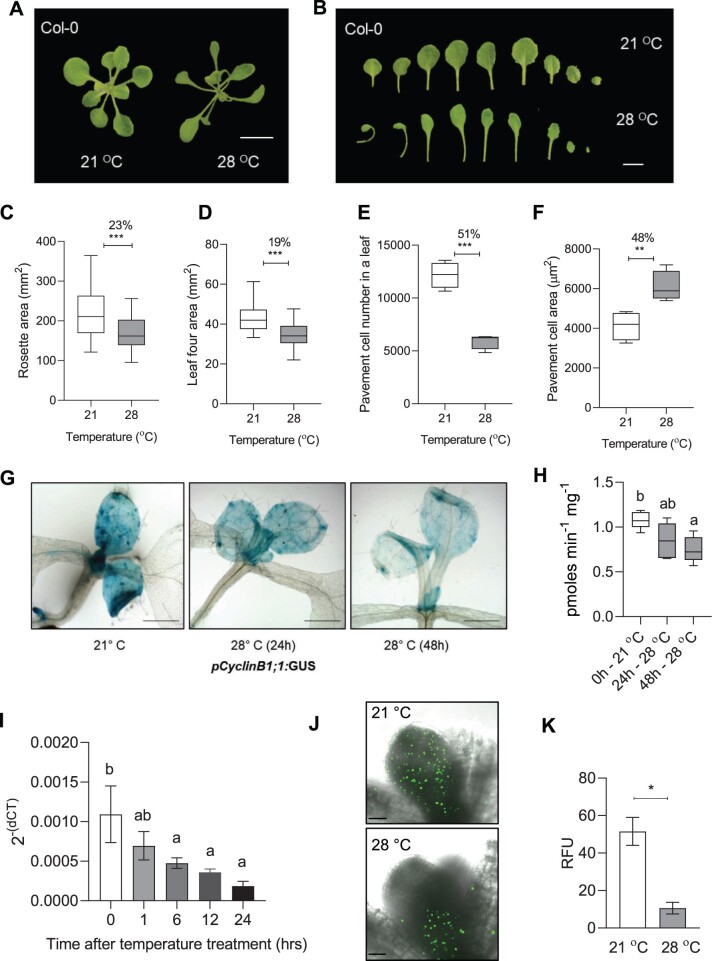
To investigate the effects of high ambient temperature on leaf morphological features, we sequentially examined the rosette area, individual rosette leaf area, epidermal cells, and palisade mesophyll cells of leaf four of the wild-type A. thaliana ecotype, Col-0, under control temperature (21°C) and high ambient temperature (28°C). The Col-0 rosette showed an open structure (i.e. increased convex hull) with a reduced total area under high temperature compared to the control (Figure 1, A and C). Individual leaves also showed a reduction in their respective sizes at high temperature compared to the control (Figure 1B; Supplemental Figure S1A). Leaf four exhibited the most significant difference between the two temperature conditions among all the rosette leaves and, therefore, was used in this study for cellular phenotyping (Figure 1D; Supplemental Figure S1A). As expected, temperature-mediated changes in the lamina and petiole resulted in a decreased ratio of lamina and petiole length under high temperature (Supplemental Figure S1B). To find the cellular basis of leaf size reduction, we performed cellular profiling of leaf four by quantifying cell number and area in abaxial epidermal pavement cells and palisade mesophyll cells. We found a drastic 50% reduction in the number of epidermal cells per leaf and a 66% reduction in palisade cells under high temperature compared to the control (Figure 1E; Supplemental Figure S1, C–E). In contrast, the average area of epidermal and palisade cells showed an increase under high temperature (Figure 1F; Supplemental Figure S1F). To confirm the results, we also quantified the epidermal cell number and area of the first pair of Col-0 leaves under control and high temperature. The first pair of leaves also showed results similar to that of leaf four (Supplemental Figure S2, A–C). These results suggested that prolonged exposure to high temperature reduced leaf area by negatively impacting cell number that could likely result due to temperature-triggered inhibition of the cell division process.
Figure 1.
High temperature reduces leaf area by controlling cell number in the Arabidopsis leaves. A, Rosette picture and (B) leaf series of 21°C and 28°C grown Arabidopsis Col-0 wild-type plants at 20 DAS. The images were digitally extracted for size comparison. Scale = 1 cm. C–F, Quantification of rosette area (n = 80–100, C), leaf four area (n = 19–23, D), average pavement cell number on the abaxial side of leaf four (n = 4 leaves, E), and average pavement cell area (n = 4 leaves, F) in Col-0 plants grown at 21°C and 28°C at 20 DAS. Box plot extends from 25th to 75th percentiles, where the center line represents the median and whiskers minimum and maximum values. Percent change is indicated above the graphs. G and H, Images showing GUS staining (G) and its quantification (n = 6, H) in pCyclinB1;1:GUS reporter line at 21°C and 28°C. Scale = 1 cm. Box plot extends from 25th to 75th percentiles, where the center line represents the median and whiskers minimum and maximum values. I, Transcript levels of CyclinB1;1 in the proliferating leaves in Col-0 at different time points after high-temperature (28°C) treatment presented as 2-(dCt) values. Values represent mean ± se (n = 3). J, EdU fluorescence showing the distribution of mitotic cells in 5-day-old Col-0 leaf grown at 21°C or 28°C. Scale = 50 µm. K, Quantification of fluorescent intensity in EdU-treated Col-0 leaves. RFU = relative fluorescent unit (arbitrary). Values represent mean ± se (n = 3). Stars in C–F and K indicate Student’s t test ***P< 0.001, **P < 0.01, *P< 0.05. Letters indicate statistically significant difference between treatments (H) or the time points (I) based on one-way ANOVA and Tukey’s test, P < 0.05.
To test the effect of temperature on cell division, we first used a cell cycle marker line pCyclinB1;1: β-glucuronidase (GUS) (Donnelly et al., 1999). The marker line showed reduced staining in the GUS assay at 28°C, thus indicating suppression of cell division with increasing exposure to high temperature (Figure 1, G and H). Consistent with the pCyclinB1;1:GUS marker line, we detected reduced expression of CyclinB1;1 in the proliferating leaves under high temperature (Figure 1I). We also detected reduced cell cycle progression using 5-ethynyl-2′-deoxyuridine (EdU)-based S-phase assay in high temperature-grown seedlings compared to the control (Figure 1, J and K). Further, we used 4′,6-diamidino-2-phenylindole (DAPI) to visualize mitotic figures in the basal region of young proliferating leaves as a readout for dividing cells and found a greater number of cells undergoing division under the control condition than in high temperature (Supplemental Figure S3, A and B). Taken together, these results showed that the high ambient temperature regulates the leaf size by restricting cell division.
Temperature regulation of leaf size via cell division control is dependent on PIF4
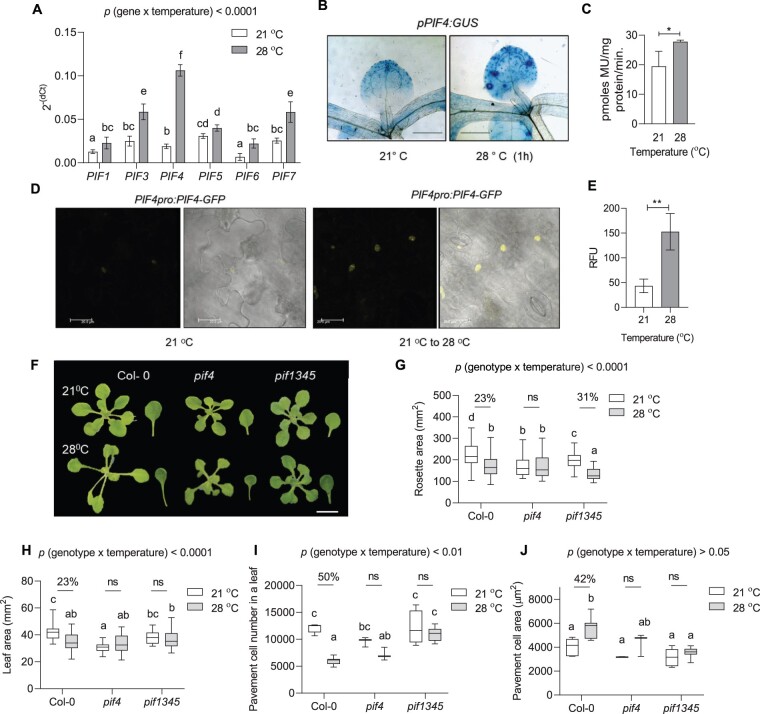
The roles of PIF4 and PIF7 transcription factors in temperature-induced hypocotyl elongation response are well characterized (Koini et al., 2009; Chung et al., 2020; Fiorucci et al., 2020). However, the role of PIF transcription factors in temperature control of leaf growth is unexplored. Therefore, we first examined the expression levels of various PIFs in the leaves under high temperature. All the tested PIFs were upregulated shortly after high-temperature treatment, where PIF4 showed the highest induction in its expression (Figure 2A). The induction in the expression, however, subsided at 8 h of treatment for all the PIFs except PIF4 (Supplemental Figure S4). Consistent with this, the pPIF4:GUS line showed an increased spatio-temporal expression level after high-temperature treatment (Figure 2, B and C). Interestingly, unlike the control leaves, high-temperature treated leaves showed increased GUS staining in the basal part of the leaf that coincides with the site of cell proliferation (Andriankaja et al., 2012). Using a translational reporter line, pPIF4:PIF4-GFP, we further showed comparatively more accumulation of PIF4 protein in the leaf epidermal cells under high temperature than the control (Figure 2, D and E).
Figure 2.
High temperature-mediated suppression of cell number in the leaves involves PIF4. A, Expression level of PIFs relative to an endogenous control, ACTIN2, in young proliferating leaves of Col-0 at 21°C and 1 h of 28°C temperature treatment plotted as 2−(dCt). Values represent mean ± se (n = 7–9), P < 0.0001 (temperature), P < 0.0001 (gene). B, GUS staining in the leaves of PIF4:GUS reporter lines at 21°C and 28°C, Scale = 1 cm. C, Quantification of the GUS staining in PIF4:GUS reporter lines. Values represent mean ± se (n = 3). Star indicate Student’s t test *P-value < 0.05. D, Translation fusion reporter line pPIF4:PIF4-GFP showing protein localization in epidermal cells of 7-day-old seedling at 21°C and 21°C–28°C shift for 4 h. Scale = 20 µm. E, Quantification of the fluorescence in the epidermal cells of the translation fusion reporter line pPIF4:PIF4-GFP. Values represent mean ± se (n = 4). Stars indicate Student’s t test **P< 0.001. F, Rosette and leaf pictures of different genotypes grown at 21°C or 28°C at 20 DAS. The images were digitally extracted for size comparison. Scale = 1 cm. G, Quantification of rosette area (n = 87–100), P < 0.0001 (temperature), P < 0.0001 (genotype). H, Quantification of leaf four area (n = 30–38), P < 0.0001 (temperature), P < 0.05 (genotype). I, Average pavement cell number in the abaxial side of leaf four (n = 7–8 leaves), P < 0.001 (temperature), P < 0.0001 (genotype). J, Average pavement cell area in the abaxial side of leaf four (n = 7–8 leaves), P < 0.0001 (temperature), P< 0.001 (genotype). Box plot extends from 25th to 75th percentiles, where the center line represents the median and whiskers minimum and maximum values. The letters in (A) and (G–J) indicate significant differences based on two-way ANOVA and Tukey’s test (P < 0.05). Percent change between 21°C and 28°C is indicated above the graphs. The P-value for the interaction (genotype × temperature) is shown at the top of each plot. ns, not significant.
Next, we investigated the leaf phenotypes of pif4, pif7, and pif1345 mutants under control and at high temperature. The rosette and leaf phenotype of pif4 remained unchanged under high temperature compared to control condition, unlike Col-0 (Figure 2, F–H; Supplemental Figures S2, A and S5, A). Quadruple mutant pif1345 showed a decrease in rosette area; however, the area of leaf four remained unchanged under high temperature (Figure 2, G and H). Notably, unlike Col-0, the pavement cell number in pif4 and pif1345 mutants remained unchanged between two temperature treatments, suggesting that PIFs substantially impact leaf size under warm conditions by affecting the number of cells in the leaves (Figure 2I; Supplemental Figure S2B). Next, we analyzed the changes in cellular expansion under high temperature by quantifying cell area. Col-0 showed increased cell area of pavement cells under high temperature, while pif4 and pif1345 mutants showed no changes between the contrasting temperature conditions (Figure 2J; Supplemental Figure S2C). However, the statistical interaction of genotype and temperature was not significant for pavement cell area in contrast to pavement cell number, suggesting a regulatory role of PIF4 in leaf size determination under high temperature via control of cell number (Figure 2, I and J). We found similar results for palisade cells that further proved PIF4 to be important for the regulation of cell proliferation in different layers of the leaf under high temperature (Supplemental Figure S5, B and C). Similar to leaf four, the average pavement cell number and cell area in the first pair of leaves of the pif4 mutant remained unchanged under high temperature (Supplemental Figure S2). Interestingly, pif7, unlike pif4, showed reduced rosette and leaf area and reduced cell number; however, cell area remained unchanged (Supplemental Figure S6). Overall, these results suggested a direct and more important role of PIF4 in cell division control under high temperature possibly via changes in its spatio-temporal expression. While others PIFs might still play a role, a clear phenotype of pif4 suggests that the other PIFs are dispensable. The significantly different rosette and leaf area of pif4 than Col-0 under normal growth temperature suggests a pleiotropic effect of PIF4 on Arabidopsis leaf growth (Figure 2, F–H). Moreover, the similar cell number in the leaves of Col-0 and pif4 under normal growth temperature (21°C) indicates that PIF4 may regulate cell number specifically under warm (28°C) growth conditions (Figure 2I).
TCP4 regulates leaf growth under high temperature
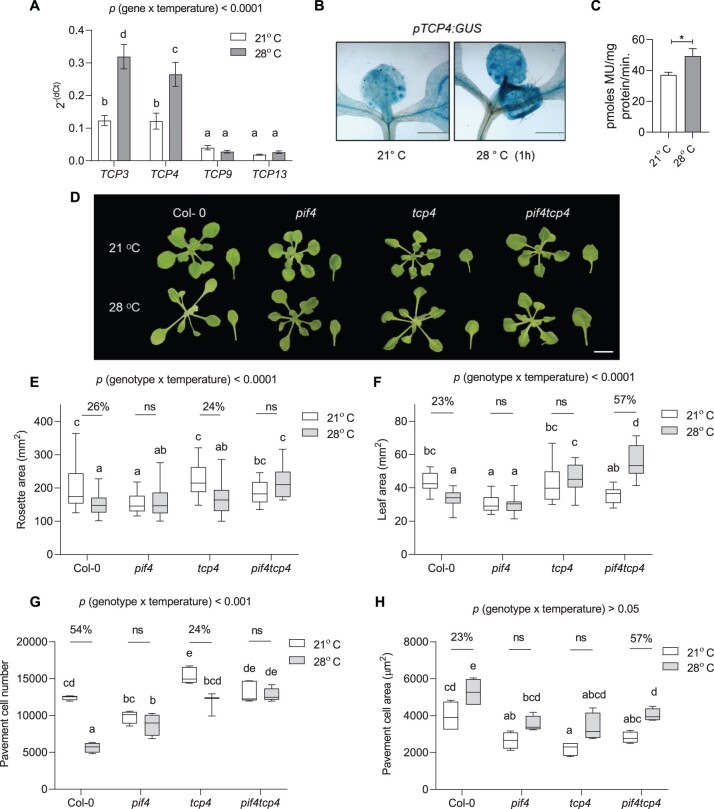
To get further insight on leaf size regulation under high temperature, we examined the publicly available transcriptome data to look for genes that expressed in the leaves and were also induced by high temperature (Stavang et al., 2009; Sidaway-lee et al., 2014). We then compared the list of temperature-induced genes with the list of literature curated leaf development-related genes (Supplemental Table S1). Several genes encoding transcription factors, including several TCPs and GRFs, were found to be expressed in leaves as well as responsive to high temperature. We checked the transcript levels of temperature-responsive TCPs in leaves and found class II TCPs, TCP3 and TCP4, to be highly upregulated by high temperature (Figure 3A). Consistent with this, a strong induction in β-Glucuronidase (GUS) staining was visible in pTCP4:GUS reporter line within 1 h of high-temperature treatment suggesting a potential involvement of TCP4 in temperature signaling in the leaves (Figure 3, B and C).
Figure 3.
PIF4 and TCP4 function together to regulate high temperature-induced cell number suppression. A, Expression level of TCPs relative to an endogenous control, ACTIN2, at 21°C and 1 h of 28°C temperature treatment in young proliferating leaves of Col-0 plotted as 2−(dCt). Values represent mean ± se (n = 6–8). B and C, GUS staining (scale = 1 cm, B) and its quantification (C) in leaves of TCP4:GUS reporter lines at 21°C or 28°C. Values represent mean ± se (n = 4). Stars indicate Student’s t test *P < 0.05. D, Rosette and leaf pictures of different genotypes grown at 21°C or 28°C at 20 DAS. The images were digitally extracted for size comparison. Scale = 1 cm. E, Quantification of rosette area in PIF4- and TCP4-related mutants grown at 21°C or 28°C (n = 45–55), P < 0.0001 (temperature), P < 0.001 (genotype). F, Quantification of leaf four area (n = 25), P < 0.001 (temperature), P < 0.05 (genotype). G, Average pavement cell number in the abaxial side of leaf four (n = 4–5 leaves), P < 0.0001 (temperature), P < 0.0001 (genotype). H, Average pavement cell area in the abaxial side of leaf four (n = 4–5 leaves), P < 0.001 (temperature), P < 0.001 (genotype). Box plot extends from 25th to 75th percentiles, where the center line represents the median and whiskers minimum and maximum values. The letters in (A) and (E–H) indicate significant differences based on two-way ANOVA and Tukey’s test (P < 0.05). Percent change between 21°C and 28°C is indicated above the graphs. The P-value for the interaction (genotype × temperature) is shown at the top of each plot. ns, not significant.
TCP4 regulates leaf size by repression of cell proliferation (Schommer et al., 2014). A significant cell number difference between Col-0 and tcp4 verified the role of TCP4 in regulation of cell number under normal growth conditions (Figure 3G). To investigate the role of TCP4 in temperature-regulated leaf size control, we further investigated the phenotype of the tcp4 mutant under warm temperatures. The leaf area remained unchanged in tcp4; however, the mutant showed an open rosette structure like Col-0 (Figure 3, D–F). Unlike the complete attenuation of the cell number phenotype in pif4, tcp4 showed a significant partial reduction in cell number under high temperature (Figure 3G). However, the extent of cell number reduction in tcp4 was lesser than Col-0, suggesting a role for TCP4 in temperature control of cell division in the leaves (Figure 3G). The interaction of genotype and temperature was significant for pavement cell number and was not significant for pavement cell area (Figure 3, G and H). To check functional redundancy with other members of miR319-regulated class II TCPs in temperature-mediated leaf size regulation, we used miR319 gain-of-function line JAGGED AND WAVY-1D (jaw-1D). jaw-1D line shows downregulation of multiple CIN-related class II TCPs, including TCP2, 3, 4, 10, and 24, and has a phenotype of highly serrated and cranky leaves (Palatnik et al., 2003). jaw-1D rosette area showed reduction under high temperature due to excessive leaf curling (Supplemental Figure S6, A and B). jaw-1D line displayed no change in cell number, cell area, or final leaf size between control and high temperature (Supplemental Figure S6). Complete abolishment of cell number reduction response under high temperature in jaw-1D compared to the single tcp4 mutant suggested the involvement of multiple Class II TCPs in temperature-controlled leaf growth and cell number regulation (Supplemental Figure S6D). Overall, these results established a role of Class II TCPs, including the TCP4, in temperature-regulated cell division control to determine leaf size.
Both PIF4 and TCP4 are required for temperature-mediated regulation of cell number in leaves
The expression levels of both PIF4 and TCP4 were induced under high temperature, and pif4 and tcp4 mutants showed unchanged leaf area with attenuation of cell number reduction under high temperature compared to Col-0 (Figure 3, F and G). Based on these results, we hypothesized that the two transcription factors may function in a common signaling pathway, perhaps at the same time, to regulate leaf size under high temperature. To test this genetic interaction, we generated a pif4tcp4 mutant line and it showed no change in rosette area but increased leaf four area under high temperature (Figure 3, E and F). Interestingly, pif4tcp4 did not show any significant change in their cell number under high temperature and was irresponsive to the temperature-induced cell number suppression compared to Col-0 (Figure 3G). Partial attenuation of temperature-induced cell number reduction response in tcp4 and complete attenuation of the same in pif4tcp4, similar to pif4, suggested that PIF4 and TCP4 act in a same pathway to regulate leaf thermomorphogenesis. Unlike cell number changes, the cell expansion response in pif4tcp4 was similar to Col-0 (Figure 3H).
PIF4- and TCP4-mediated cell cycle inhibition under high temperature occurs via KRP1
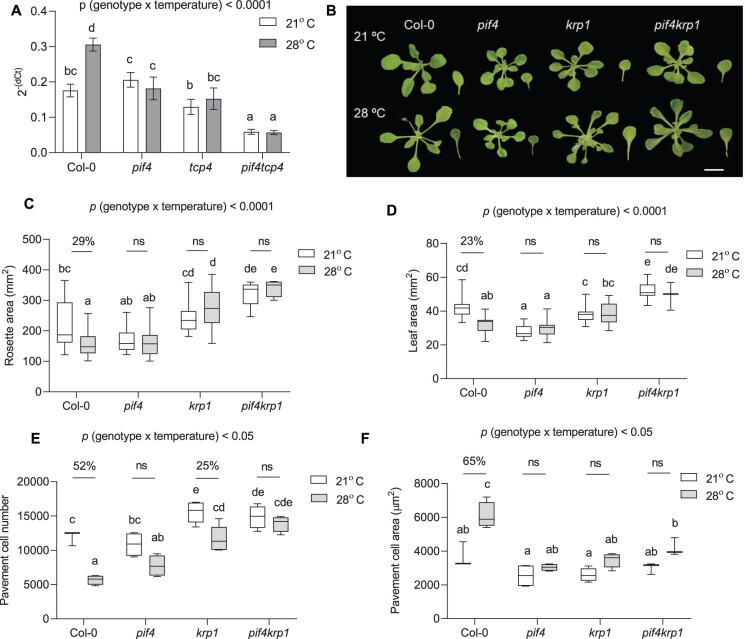
Since we hypothesized that PIF4 and TCP4 function in the same signaling pathway to control the temperature-mediated leaf size regulation, we intrigued about the signaling component downstream to PIF4 and TCP4. TCP4 is known to inhibit cell division via the upregulation of KRP1 (Schommer et al., 2014). Hence, we first asked whether KRP1 could be involved in temperature-mediated cell cycle inhibition. The expression of KRP1 was induced within 1 h of high-temperature treatment in Col-0. In contrast, the temperature-induced expression of KRP1 was significantly affected in pif4, tcp4, and pif4tcp4 (Figure 4A). Interestingly, KRP1 transcript levels were significantly lower in pif4tcp4 compared to Col-0 or either of the single mutants. Together, these results showed that temperature regulates KRP1 expression via PIF4 and TCP4 and further raised the possibility of KRP1 functioning downstream to PIF4 and TCP4 to inhibit cell division.
Figure 4.
KRP1 functions downstream of PIF4 to regulate cell number in leaves under high temperature. A, Expression level of KRP1 relative to an endogenous control, ACTIN2, in young proliferating leaves at 21°C and 1 h of 28°C temperature treatment in different genotypes plotted as 2− (dCt). Values represent mean ± se (n = 6), P < 0.0001 (temperature), P < 0.0001 (gene). B, Representative rosette and leaf four images of 21°C and 28°C grown plants of Col-0, pif4, krp1, and pif4krp1 at 20 DAS. The images were digitally extracted for size comparison. Scale = 1 cm. C, Quantification of rosette area in PIF4- and KRP1-related mutants grown at 21°C or 28°C (n = 51–53), P < 0.0001 (temperature), P < 0.05 (genotype). D, Quantification of leaf four area at 20 DAS (n = 19–22), P < 0.0001 (temperature), P < 0.05 (genotype). E, Average pavement cell number in the abaxial side of leaf four (n = 5 leaves), P < 0.0001 (temperature), P < 0.0001 (genotype). F, Average pavement cell area in the abaxial side of leaf four (n = 5 leaves), P < 0.0001 (temperature), P < 0.0001 (genotype). Box plot extends from 25th to 75th percentiles, where the center line represents the median and whiskers minimum and maximum values. The letters in (A) and (C–F) indicate significant differences based on two-way ANOVA and Tukey’s test (P < 0.05). Percent change between 21°C and 28°C is indicated above the graphs. The P-value for the interaction (genotype × temperature) is shown at the top of each plot. ns, not significant.
Significant cell number differences between Col-0 and krp1 verified the role of KRP1 in regulation of cell number under normal growth conditions (Jakoby et al., 2006; Figure 4E). To investigate the involvement of KRP1 in cell number regulation under warm temperature, we performed cellular phenotyping in krp1 under high temperature. Rosette and leaf area, and cell area showed no significant change between the two temperature treatments (Figure 4, B–D and F). Similar to tcp4, krp1 showed attenuated response for cell number reduction compared to Col-0 under high temperature (Figure 4E). To test for functional redundancy among the KRPs, we checked the phenotype of the triple mutant krp4/6/7. The triple mutant showed a smaller rosette and leaf with a significant reduction in their cell number under high temperature, similar to Col-0, excluding the possibility of their involvement in temperature-regulated leaf size control (Supplemental Figure S6). To test whether PIF4 and KRP1 work in the same pathway to control leaf size under high temperature we examined the phenotype of the pif4krp1 double mutant. Rosette and leaf area were unchanged between the control and high-temperature grown plants (Figure 4, B and C). As hypothesized, cell numbers were completely unaffected in pif4krp1 under high-temperature treatment, unlike the krp1, when compared to Col-0 (Figure 4E). A significant loss of response in cell area increase in pif4krp1 could mean that the two have a pleiotropic effect on growth (Figure 4F). Overall, these results supported our hypothesis that PIF4 and KRP1 may act in the same pathway to control cell number in high-temperature-grown leaves.
PIF4 and TCP4 coordinate to regulate KRP1 expression
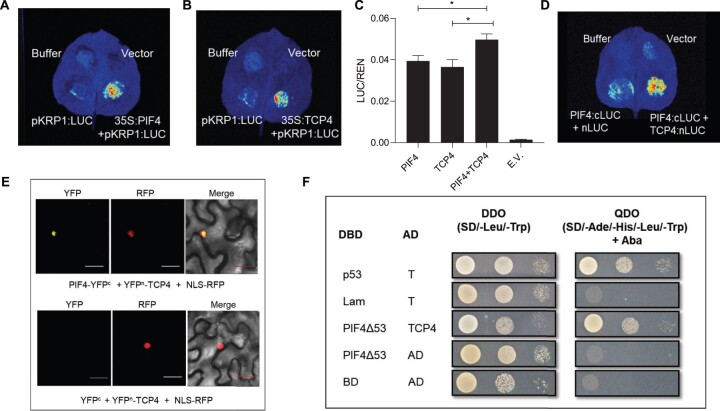
TCP4 is known to directly activate the expression of KRP1, we checked whether PIF4 regulates KRP1 expression (Schommer et al., 2014). Using luciferase transactivation assay, we showed that PIF4 induced KRP1 expression (Figure 5, A and C). We also validated the binding of TCP4 to the KRP1 promoter to induce the expression (Figure 5, B and C). In addition, we quantified the relative activity of the KRP1 promoter using dual luciferase assay. We found more KRP1 promoter activity in the presence of both PIF4 and TCP4 compared to the individual transcription factors (Figure 5C). This ruled out competitive binding between the two transcription factors on the KRP1 promoter and suggested that both could potentially co-regulate KRP1 transcription. This further raised the possibility of physical interaction between PIF4 and TCP4 to co-regulate the downstream target(s). We, therefore, performed protein–protein interaction assays using split luciferase assay and bimolecular fluorescence complementation (BiFC) in Nicotiana benthamiana leaves and showed a positive interaction between PIF4 and TCP4 (Figure 5, D and E). We further proved this interaction in a heterologous system using a yeast two-hybrid assay (Figure 5F). Since the temperature-mediated reduction in cell number was completely abolished in the jaw-1D mutant, it is plausible that PIF4 might interact with other members of Class II TCPs. Protein–protein interaction study using BiFC showed that indeed PIF4 interacted with TCP2, TCP3, TCP10, and TCP24 apart from TCP4 in that class (Supplemental Figure S7).
Figure 5.
PIF4 interacts with TCP4 and regulates KRP1 expression. A and B, Luciferase transactivation assay showing binding of PIF4 (A) and TCP4 (B) to the KRP1 promoter in N. benthamiana leaves. C, Dual-luciferase assay in N. benthamiana leaves co-infiltrated with different transcription factors. LUC/REN ratio represents the relative activity of the KRP1 promoter, where REN was used as an internal control. Values represent mean ± se (n = 5) E.V., empty vector. Stars indicate Student’s t test P < 0.05. D, Split Luciferase assay showing protein–protein interaction between PIF4 (cloned with cLUC) and TCP4 (cloned with nLUC). E, BiFC assays in N. benthamiana showing an interaction between PIF4 and TCP4. NLS-RFP is used as a nuclear marker. Scale = 30 µm. F, Yeast two-hybrid assays showing an interaction between PIF4Δ53 and TCP4. The DBD was fused to truncated PIF4 named as PIF4Δ53, and the activation domain was fused to TCP4.
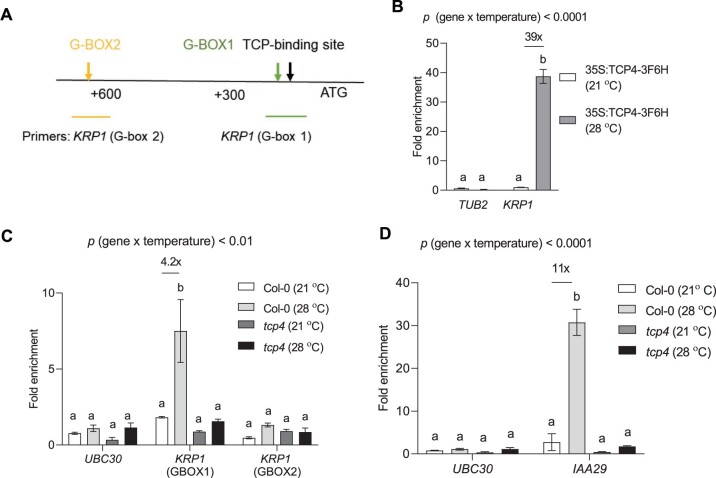
To get further insight into the co-regulatory mechanism of KRP1 by PIF4 and TCP4, we scanned the promoter of KRP1 for putative PIF4-binding sites. Within 1-kb upstream of the start codon, we found two putative PIF4-binding G-boxes close to the known TCP4-binding site on the KRP1 promoter (Figure 6A). Using chromatin immunoprecipitation (ChIP)-PCR, we first confirmed the binding of TCP4 on the KRP1 promoter and interestingly found it to increase by nearly 40-fold by exposure to high temperature (Figure 6B). Next, we did ChIP-qPCR using anti-PIF4 antibodies under control and high temperature-treated samples in different genotypes. We observed a four-fold enrichment of putative G-box1 at KRP1 promoter using anti-PIF4 antibody after high-temperature treatment in Col-0 (Figure 6C). Interestingly, we did not find increased enrichment of G-boxes on the promoter of KRP1 in the tcp4 mutant under high temperature, suggesting a requirement of TCP4 for PIF4 to regulate KRP1. We confirmed the TCP4-dependent PIF4 gene regulatory functions under high temperature using IAA29, which is an auxin-responsive gene known to be thermo-responsive in a PIF4-dependent manner (Figure 6D).
Figure 6.
PIF4 regulates KRP1 in a TCP4-dependent manner. A, Schematic diagram of KRP1 promoter with ATG start site showing the presence of TCP4 binding site and putative G-boxes. B, ChIP assay showing enrichment of TCP-binding element in the promoter of KRP1 region in 35S:TCP4-3F6H line using anti-FLAG antibody under control and high temperature, P < 0.0001 (temperature), P < 0.0001 (gene). C, ChIP assay showing enrichment of G-box region using the anti-PIF4 antibody in the promoter of KRP1 in Col-0 and tcp4 under control (21°C) and high temperature (28°C) along with negative control UBC30, P < 0.001 (temperature), P < 0.01 (gene). D, Enrichment of G-box region using the anti-PIF4 antibody in the promoter of IAA29 under control and high temperature in Col-0 and tcp4 along with negative control UBC30, P < 0.0001 (temperature), P < 0.0001 (gene). Values represent mean ± se (n = 3–4). The letters indicate significant differences based on two-way ANOVA and Tukey’s test (P < 0.05) with fold change for significant difference between 21°C and 28°C presented above the graphs. The P-value for the interaction (genotype × temperature) is shown at the top of each plot.
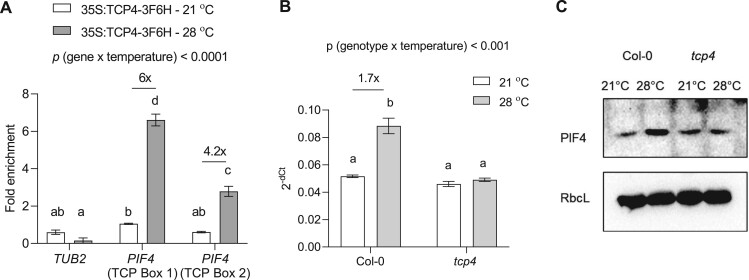
Since TCP4 is a transcription factor, the regulation of PIF4 expression by TCP4 would be the most plausible mechanism for this. We detected increased enrichment of the two putative TCP binding elements on the promoter of PIF4 in high temperature (Figure 7A). In support, PIF4 expression failed to elicit in response to high temperature in tcp4, contrary to Col-0, suggesting transcriptional-level regulation that could further affect PIF4 protein abundance and thus PIF4 availability for binding to its targets (Figure 7B). Indeed, western blot results confirmed the TCP4-dependent regulation of PIF4 level under high temperature, as the tcp4 mutant failed to show increased PIF4 levels under high temperature as seen in Col-0 (Figure 7C). Taken together, we showed that TCP4 acts upstream to PIF4, and PIF4-mediated regulation of KRP1 under high temperature requires TCP4 to regulate leaf size in warmth.
Figure 7.
TCP4 regulates PIF4 expression under high temperature. A, ChIP assay showing enrichment of TCP binding sites on the promoter of PIF4 under 21°C and 28°C in 35S:TCP4-3F6H line using anti-FLAG antibody, P < 0.0001 (temperature), P < 0.0001 (gene). B, Quantification of PIF4 transcript level in proliferating leaves of Col-0 and tcp4 under 21°C and 3-h 28°C treatment represented as 2−(dCt). Values represent mean ± se (n = 4), P < 0.0001 (temperature), P < 0.0001 (genotype). The letters indicate significant differences based on two-way ANOVA and Tukey’s test (P < 0.05) with fold change for significant difference between 21°C and 28°C presented above the graphs. The P-value for the interaction (genotype × temperature) is shown at the top of each plot. C, Western blot comparison of PIF4 protein levels in Col-0 and tcp4 background at 21°C and after 4 h of 28°C treatment between Zt1 to Zt5. RbcL was used as the loading control.
Discussion
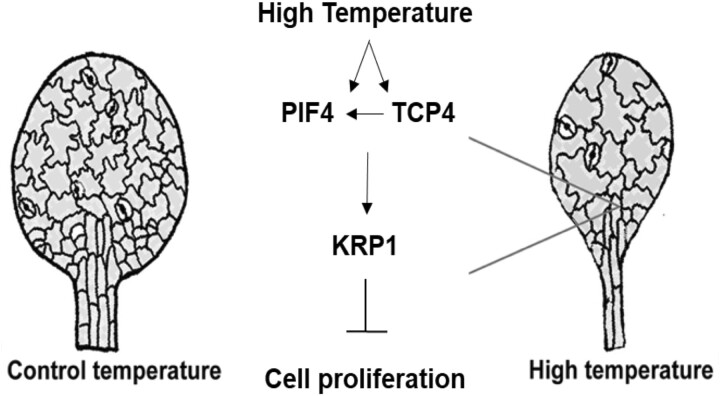
Plants show remarkable plasticity in their growth and development in response to their surroundings by tightly regulating the final organ size and shape. Shade avoidance syndrome is one of the well-studied classical examples of such growth plasticity where plants exhibit various architectural adaptions including the elongation response to compete with neighboring vegetation for light (Pierik and De Wit, 2014). Recently, a role of cell division in regulating leaf size under shade was highlighted, where shade-grown leaves showed fewer cells and thus small size as opposed to longer hypocotyl with elongated epidermal cells (Romanowski et al., 2021). Cell division and cell expansion are the key processes to mediate plant growth and developmental plasticity at the cellular level (Hepworth and Lenhard, 2014). Temperature-induced growth plasticity varies in different organs, and these organ-specific responses become evident at the early seedling development itself. While studies report a slight increase in the number of epidermal cells in the hypocotyl, the organ elongation response under various environmental cues including high temperature is largely attributed to the cell expansion process (Gray et al., 1998; Ibañez et al., 2018; Bellstaedt et al., 2019). However, at least one study reported a reduction in stomatal density in the high-temperature-grown cotyledons (Lau et al., 2018). Here we show a central role of cell division for leaf growth plasticity in response to temperature. We report TCP4 induces PIF4 expression in high temperature and that PIF4 and TCP4 play critical and co-dependent roles to regulate cell number in high-temperature-grown leaves by suppressing the expression of KRP1 (Figure 8). Together, we showed that coordinated interplay between PIF4, an environmental regulator, and TCP4, a developmental regulator, determines the fate of final leaf size under high temperature.
Figure 8.
A schematic diagram representing high-temperature-mediated suppression of leaf size. Warm temperature positively regulates expression of PIF4 and TCP4 and accumulation of PIF4 in leaf cells. High temperature further promotes binding of PIF4 and TCP4 to the promoter of cell cycle inhibitor KRP1 to induce KRP1 expression, which in turn suppresses cell proliferation and thus leaf size. Our results show that TCP4 is required for transcriptional induction of PIF4 expression (left facing arrow) that subsequently affects accumulation of PIF4 protein and thus its enrichment on KRP1 promoter under warm temperature. The physical interaction between TCP4 and PIF4 may further add to their co-regulatory role for induction of KRP1 expression under high temperature.
Temperature regulates leaf size via cell division control
Temperature-induced cell elongation response in the petiole and hypocotyl is shown to drive leaf thermonastic movement that assists in plant cooling via transpiration (van Zanten et al., 2010; Crawford et al., 2012). Smaller and thinner leaves, on the other hand, help in heat dissipation via evaporation and convection (Franklin and Wigge, 2014; Quint et al., 2016). While a 1°C–3°C rise in temperature was found to be advantageous for Arabidopsis, a 5°C or more rise is known to adversely affect leaf functional traits, including photosynthesis. Thus, the reallocation of reserved carbon in driving elongation growth could be a mean to increase cooling via transpiration or to reach a light source to increase photosynthetic capacity (Jin et al., 2011; Romero-Montepaone et al., 2021). In agreement with this, Vasseur et al. (2011) have shown that plants only exhibit temperature-induced elongation response under low to medium light. The effect of temperature on plant phenotype subsides when light is not limiting. This suggests that elongation response seen under high temperature could be primarily to increase light capture and photosynthetic capacity. However, as high temperature affects a wide array of changes in both vegetative and yield-related traits, this remains to be seen how those will vary under varying light and temperature conditions (Ibañez et al., 2017).
Previously, temperature-mediated hypocotyl elongation was shown to involve the expansion of epidermal cells (Gray et al., 1998). We studied the cellular and molecular basis of smaller leaf size under high temperature compared to the control temperature. Cellular profiling of leaves in combination with the expression pattern of cell cycle markers in response to high temperature confirmed that temperature-mediated restriction of leaf size involves suppression of cell division (Figure 1). Interestingly, a previous report showed enrichment of several cyclins, KRPs, and cyclin-dependent kinases (CDKs) involved in cell cycle regulation among the genes differentially expressed in Arabidopsis plants grown under high temperature (Sidaway-lee et al., 2014). Our results suggested possibilities of overall reduced cell division and/or changes in the duration of the cell division phase in the leaves under high temperature. Prolonged exposure to shade, showing plant responses similar to high temperature, has been reported to cause an early exit from cell proliferation, suggesting that high temperature and shade likely control the leaf size via control of cell division (Carabelli et al., 2018). Consistent with this, a recent report has suggested a Phytochrome B (PhyB)-dependent suppression of CYCB1;1 expression and cell proliferation in the leaves under shade (Romanowski et al., 2021). PhyB is reported to be a thermosensor in Arabidopsis, suggesting PhyB likely integrates high-temperature signal to repression of cell division response via downstream transcription factors, such as PIFs (Jung et al., 2016; Legris et al., 2016).
PIF4 as a mediator of plant’s environment to growth plasticity
PIF4 expression domain is widespread throughout the leaf development as it accumulates in the epidermis of young leaves, at the shoot apical meristem (SAM)/Leaf boundary, in the middle zone of leaf coinciding with leaf vasculature framework, and later at the axils of the young leaves (Zhang et al., 2020). The spatio-temporal expression pattern of PIF4 suggests a potential role of the gene at various stages of leaf and meristem patterning. Our results showed that PIF4 accumulates in the leaf epidermal cells under warm temperature and suppresses cell number, as was evident from the pif4 mutant showing attenuation of cell number reduction response under high temperature compared to Col-0 (Figure 2). The role of PIF4 in elongation response is extensively studied in the past decade (Koini et al., 2009; Choi and Oh, 2016; Ibañez et al., 2018). More recently, evidence for the involvement of PIF4 in the regulation of cell division is reported in the axillary meristem and stomatal lineage cells (Lau et al., 2018; Zhang et al., 2020). This suggests that PIF4 could potentially control the fate of the cells leaving the meristems for division or differentiation in the leaves. Whether it is the thermonastic movements to induce leaf cooling or reduction in leaf size to help in heat dissipation, both the responses involve differential regulation of PIF4 for cell elongation versus cell division. Together, these emphasize the pivotal importance of PIF4 in complex growth regulatory decisions that plants need to make under high temperature. Hussain et al. (2022) reported a role of PIF7 in limiting cell division in shade-grown plants via competition with AN3, known to promote cell proliferation along with GRFs. Interestingly, based on the cellular phenotype we did not find major involvement of PIF7 in cell number suppression rather in cell elongation response under high temperature (Supplemental Figure S6). Several reports have shown roles of various PIFs in signal integration to regulate plant growth and development in response to different intrinsic and extrinsic cues (reviewed in Paik et al., 2017). Our results point to additional mechanisms whereby different PIFs selectively transduce signals in a context-dependent manner to optimize growth in response to temperature.
TCP4 as a regulator of leaf growth plasticity in warm conditions
Classes I and II of TCP transcription factors antagonistically control growth. Class I members, in general, promote cell proliferation, while Class II members promote cell differentiation and progression of cell cycle arrest front (Gutie et al., 2005; Efroni et al., 2008; Kieffer et al., 2011). Recent reports have shown the involvement of three Class I TCPs in temperature signaling, where TCP 5, 13, and 17 were shown to regulate as well as interact with PIF4 in temperature-induced hypocotyl elongation response (Han et al., 2019; Zhou et al., 2019). We observed a similar regulatory response between TCP4 and PIF4 in leaf size regulation in warm temperature. TCP4 was shown to promote CONSTANS levels to induce photoperiodic flowering (Kubota et al., 2017). However, the role of Class II TCPs in the regulation of temperature response is unexplored. A lack of cell number suppression response in jaw-1D compared to Col-0 points to a role for Class II TCPs in temperature signaling (Supplemental Figure S6D). Acknowledging that TCP4 is one of the five TCPs downregulated in jaw-1D line and based on the temperature-mediated induction of TCP4 expression and the reduced response to changes in cell number in tcp4 under warm conditions, we propose a role for TCP4 in temperature-induced leaf size reduction (Figure 3, A and G; Palatnik et al., 2003). TCP4 is already known to inhibit cell division and promote cell differentiation in leaves, our study adds to our understanding of TCP4 function in restriction of leaf size via cell division inhibition under high temperature (Schommer et al., 2014).
TCP4 regulates PIF4, and together they function as co-regulators
Similar to PIF4, TCP4 is known to promote cell elongation via activation of genes involved in auxin and brassinosteroids biosynthesis and signaling (Challa et al., 2016). This suggests that TCP4, along with PIF4, could be instrumental in integrating environmental signals and developmental responses. Our cellular, genetic, and biochemical analyses proved that TCP4 works alongside PIF4. Based on attenuated response to cell number reduction under high temperature in pif4, tcp4, and pif4tcp4, we propose a critical and co-regulatory role of PIF4 and TCP4 in the cell division process (Figure 3). Cell expansion changes in pif4 or tcp4 leaves were comparable to Col-0 making them dispensable for the process. However, the response was reduced in jaw1-D mutants, suggesting redundancy in the gene families for cell expansion response (Figure 3H; Supplemental Figure S6E). A TCP4-dependent regulation of PIF4 transcript levels and, in turn, PIF4 protein levels and its availability on its target-binding sites, along with evidence of a physical protein–protein interaction between the two further highlights the complexity and degree of regulation of the growth pathways. To summarize, our genetic analyses, as well as DNA–protein interactions, showed that TCP4 regulates PIF4 and PIF4 and TCP4 work in a common pathway to control cell proliferation, and possibly other aspects of leaf growth and development. We speculate that the physical protein–protein interaction between PIF4 and TCP4 may aid in PIF4 accumulation under high temperature along with their regulation of target genes.
PIF4 and TCP4 integrate high-temperature signal to cell cycle inhibitor KRP1
Previous studies have shown leaf area reduction via a reduction in cell number under mild osmotic stress, UV irradiation, and simulated shade suggesting that inhibition of cell cycle machinery could be a general response by which plants modulate their growth under diverse environmental conditions (González et al., 1998; Wargent et al., 2009; Skirycz et al., 2011). Leaf growth and development is a complex growth process, for simplicity and dissection of interlinked pathways under high temperature, we largely focused on temperature-mediated cell number suppression and leaf size regulation in plants grown under continuous high-temperature conditions postgermination. We provided a molecular link connecting the core leaf development pathway and high-temperature signaling via KRP1. KRP1 showed upregulation under high temperature in a PIF4- and TCP4-dependent manner to regulate cell number and leaf size (Figure 4). Increased expression of KRP1 negatively impacts cell division and plant growth by inhibiting the activity of CDKs (Wang et al., 2000). Attenuated cell number suppression phenotype in the mutants and TCP4-dependent abundance of PIF4 on the KRP1 promoter suggest that the three represent the core temperature signaling components that regulate leaf size (Figure 8). In addition, the large-sized leaf phenotype in pif4tcp4 suggests that the two transcription factors may be involved in pleiotropic leaf growth effects in addition to the proposed cell cycle pathway. Since PIF4 and TCP4 physically interact with each other, additional signaling components downstream to PIF4 and TCP4 together remain to be explored in response to environmental cues.
Conclusions
The interplay between environmental signaling and leaf developmental pathway is understudied at the genetic level than either of the processes itself. Here, we showed the convergence of a developmental pathway and environmental signaling on the core cell cycle machinery. We report the high-temperature-mediated transcriptional induction of KRP1, TCP4-dependent regulation of PIF4, and its involvement in leaf size regulation. Our study added to the diverse functions of PIF4 not only through its direct transcriptional regulatory roles but also by integrating with other growth regulatory pathway via TCP4 in optimizing leaf growth plasticity under warm temperature conditions. This mechanism has a potential to be explored further to understand leaf growth plasticity in response to the environmental cues that can be targeted to optimize leaf growth.
Materials and methods
Plant material and growth conditions
Arabidopsis (A. thaliana) plants (Columbia ecotype) were grown on half-strength Murashige and Skoog (1/2 MS; Duchefa) plates at 21°C or 28°C with 100 μmolm−2s−1 light intensity under 16-h/8- h light/dark conditions. The seeds of pCyclinB1:GUS (CS68143), pPIF4:GUS (CS69169), pif4-2 (CS66043), and pif7-1 (CS68809) were obtained from the Arabidopsis Biological Resource Center at Ohio State University. The mutants tcp4-2 (GABI_363H08), jaw1-D, krp1, krp4/6/7, and the transgenic lines, 35S::TCP-3F6H and pTCP4:GUS, were described previously (Jakoby et al., 2006; Sarvepalli and Nath, 2011; Challa et al., 2016; Kubota et al.,2017; Sizani et al., 2019). The homozygous seeds of mutants pif4tcp4 and pif4krp1 were obtained by crossing. Primer details for genotyping of the mutants are available in Supplemental Table S2.
Microscopic morphological analysis and cellular analysis
Rosette pictures from the top were taken from three replicate media plates using a Canon EOS 700D camera. Images were converted to black and white images after color threshold changes and the rosette area was measured (as a sum of the area of all canopy leaves and their petioles) using ImageJ version 1.48 (http://rsbweb.nih.gov/ij/). Leaves were fixed overnight with 3:1 ethanol:acetic acid (v/v), followed by overnight clearing in 100% lactic acid. Cleared leaves were then pictured under a Nikon AZ-100 microscope equipped with a Nikon DS-Ri1 digital camera. Leaves with an area close to the average were chosen for drawing cells in ImageJ using a drawing tablet (Wacom) and analyzed as per Andriankaja et al. (2012). Each genotype used in phenotyping experiments was tested in minimum of two to maximum of five independent experiments. Cellular analyses were performed on fully grown fourth leaf of each plant. From each leaf roughly 400–500 cells were used from different sectors in the middle of the leaf blade avoiding the ones close to mid vein to plot and interpret the cellular data.
GUS assay
Whole plants were harvested at 10–12 days after stratification (DAS) in GUS solution (Phosphate buffer (pH 7), 0.1% (v/v) Triton X-100, 0.5-mM K4[Fe(CN)6].3H2O, 0.5-mM K3[Fe(CN)6]) with X-gluc (958-µM X-gluc dissolved in 1% DMSO w/v) and kept at 37°C for required time. Samples were fixed with ethanol:acetic acid (3:1, v/v) and cleared with 100% lactic acid before mounting on slides. The quantification of GUS staining was done using 4-methylumbelliferyl β-d-glucuronide (Sigma) as a substrate with excitation at 365 nm and emission at 455 nm read using a plate reader POLARstar Omega (BMG Labtech, Ortenberg, Germany) as previously described (Blazquez, 2007).
Expression profiling
RNA was extracted from proliferating leaves at 10–12 DAS using QIAzol lysis reagent (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. One microgram of total RNA was quantified using a nanodrop spectrophotometer Nanovue (GE Healthcare, Chicago, IL, USA) and used for making cDNA using Verso cDNA synthesis kit (Thermo Fischer Scientific, Waltham, MA, USA). Reverse transcription-quantitative PCR was performed using gene-specific primers using PowerUp SYBR Green master mix (Applied Biosystems, Waltham, MA, USA) and Bio-Rad CFX9 connect thermocycler (Bio-Rad, Hercules, CA, USA). Primers’ details can be found in Supplemental Table S2. This was done as three biological and technical repetitions using ACT2 as the reference gene. The transcript levels were estimated using 2−(dCt) method (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008).
Confocal microscopy
Seven-day-old seedlings of pPIF4:PIF4-GFP were grown at 21°C were shifted to 21°C in dark or 28°C in dark for 4 h before taking images using Leica TCS SP5 confocal laser scanning microscope (Leica Microsystems, Wetzlar, Germany) using a 63× oil objective, white light laser (WLL) at 40% intensity, and PMT2 detector at 877% gain with excitation wavelength of 488 nm and emission at 493–531 nm for GFP. DAPI (Invitrogen, Carlsbad, CA, USA) images were taken using 6-day-old young seedling using Leica TCS SP8 confocal laser scanning microscope (Leica Microsystems) using a 63× water immersion objective, Diode 405 laser at 14% intensity, and HyD detector at 195% gain with excitation at 405 nm and emission at 420–480 nm.
EdU fluorescence
Control and high temperature-grown 5-day-old seedlings were harvested and incubated in 1/2 MS containing 10-nM EdU (Click-iT EdU Cell Proliferation Kit) for 30 min in dark at room temperature. Seedlings were further incubated and washed with 1/2 MS followed by washing in PBS buffer. As per the manufacturer’s protocol, seedlings were then incubated in a Click-iT reaction cocktail for 30 min in dark followed by washes in phosphate buffered saline (PBS) buffer. Fluorescence was detected using Alexa Fluor 488 settings with Leica TCS SP8 confocal laser scanning microscope (Leica Microsystems) using a 40× (water) objective, Argon laser at 5.5% intensity, and PMT detector at 839% gain with 488 nm excitation and 500–540 nm emission range.
Yeast two-hybrid assays
The yeast two-hybrid assay was performed using the GAL4-based two-hybrid system (Clonetech, Mountain View, CA, USA). To check the interaction of PIF4 with TCP4, the full-length coding sequence of TCP4 with stop codon was amplified and cloned in pENTR-D-TOPO vector (Invitrogen). The prey construct that is, TCP4-AD was generated via LR reaction between TCP4-TOPO and pGADT7g. Since we found that PIF4 can activate the reporter in yeasts, we excised 53 amino acids from the N-terminus of PIF4 to generate bait constructs. PIF4Δ53 was amplified from the cDNA using primers PIF4Δ53-CDS-F and PIF4-CDS-SC-R and cloned into the vector pENTR-D-TOPO to generate PIF4Δ53-TOPO. Further, the bait construct DNA-binding domain (DBD)-PIF4Δ53 was generated using LR reaction between PIF4Δ53-TOPO and pGBKT7g. Interaction between p53 (pGBKT7-p53) and large T-antigen (pGADT7-T) was used as a positive control and negative control was performed using pGBKT7-Lam and pGADT7-T. The interaction was checked between the bait construct DBD-PIF4Δ53 with each prey construct (TCP4-AD) or empty pGADT7g after co-transforming in yeast Y2H gold strain by using the EZ-Yeast Transformation Kit (MP Biomedicals) according to the manufacturer’s protocol.
BiFC assays
For the BiFC assay, the full-length coding sequence of PIF4 (without stop codon) and TCP class II proteins (TCP2, TCP3, TCP4, TCP10, and TCP24) with stop codon were amplified and cloned into the vector pENTR-D-TOPO (Invitrogen). Further, using GatewayLR Clonase II Enzyme mix (Invitrogen), PIF4 CDS was cloned into pSITEcEYFP-N1 (CD3-1651) to generate PIF4-cYFP and TCPs CDS were cloned into pSITE-nEYFP-C1 (CD3-1648) to generate nYFP-TCPs constructs (Primer for each construct can be found in Supplemental Table S2). These constructs were then transformed into Agrobacterium strain, EHA105. Agrobacterium cells harboring our constructs (PIF4-cYFP, nYFP-TCPs) and empty vectors were incubated in infiltration buffer (10-mM MES pH 5.6, 10-mM MgCl2, 200-μM acetosyringone) for 4 h at 28°C. For co-infiltration, an equal volume of appropriate Agrobacterium cells were mixed and were co-infiltrated into the leaves of N. benthamiana. The infiltrated N. benthamiana plants were kept for overnight incubation in dark followed by light for 48 h. YFP fluorescence signified a positive interaction. NLS-RFP was used as a nuclear marker. The fluorescence was detected using Leica SP5 confocal laser scanning microscope (Leica Microsystems) under 20× water objective. YFP was detected using WLL at 40% intensity, and PMT detector at 754% gain with excitation wavelength of 514 nm and emission at 524–570 nm, whereas RFP was detected using white light laser (WLL) at 22% intensity and PMT detector at 166% gain with excitation wavelength of 558 nm and emission at 570–647 nm.
Luciferase transactivation assay and dual-luciferase assay
For luciferase assay, the 1,467-bp promoter region of KIP-RELATED PROTEIN1 (KRP1) was amplified and cloned in the pENTR-D-TOPO vector (pKRP1-TOPO). Using Gateway LR Clonase II Enzyme mix (Invitrogen) and pKRP1-TOPO construct, pKRP1:LUC was generated using Gateway binary vector pGWB435:LUC. Full-length CDS of transcription factors, PIF4 and TCP4 were cloned under the control of constitutive 35S promoter into plant Gateway binary vector pGWB402, to generate 35S:PIF4 and 35S:TCP4, respectively. All these constructs and empty vectors were introduced into A. tumefaciens strain, EHA105. Agrobacterium cells containing 35S:PIF4+pKRP1:LUC and 35S:TCP4 + pKRP1:LUC constructs were co-injected into N. benthamiana leaves. After 48 h, the transformed leaves were sprayed with 100-mM luciferin, kept in dark for 3–5 min, and luminescence was detected using low light cooled CCD imaging apparatus (Bio-Rad). Further, to quantify luciferase activity under pKRP1, the same promoter region was used to generate pKRP1:LUC construct using gateway vector p635nRRF containing 35S:REN. The expression of REN was used as an internal control. LUC/REN ratio represented the relative activity of the KRP1 promoter. All the mixed cultures (1) 35S:PIF4 + pKRP1:LUC; (2) 35S:TCP4 + pKRP1:LUC; (3) 35S:PIF4 + 35S:TCP4 + pKRP1:LUC)) were co-infiltrated into N. benthamiana leaves. Firefly luciferase and Renilla luciferase activities were assayed in extracts from leaf disc (1 cm in diameter) using Dual-Luciferase Assay System E1910 (Promega, Madison, WI, USA) according to manufacturer’s protocol. The quantification was done using luminometer POLARstar Omega (BMG Labtech).
Split luciferase assay
PIF4 and TCP4 CDS were amplified using CDS primers with kpn1 and Sal1 restriction site and cloned into pJET1.2 vector (Thermo Fischer Scientific). Following restriction digestion, the products into the destination vectors pCAMBIA-cLUC (CD3-1700) and pCAMBIA-nLUC (CD3-1699), respectively, and co-infiltrated in N. benthamiana before imaging using Luciferase assay system E1500 (Promega) as mentioned above.
ChIP
Arabidopsis seedlings were collected in dark at 12 DAS grown at 21°C or shifted to 28°C for 8 h before harvesting them in crosslinking buffer containing formaldehyde. ChIP was performed using antiPIF4 antibody (AS16 3157; Agrisera) or Anti-FLAG(R) M2-Antibody (F1804; Sigma) as described in Saleh et al. (2008). An aliquot of 2 µL chromatin was used for amplification using 40 cycles for qPCR analysis PowerUp SYBR Green master mix (Applied Biosystems) and CFXconnect thermocycler (Bio-Rad). Details for primers used in ChIP can be found in Supplemental Table S2.
Western blot
Eight-day-old seedlings were harvested at Zt 5 (Zeitgeber time 5) for control and high-temperature samples (treated between Zt1 and Zt5). Protein from 200-mg tissue was isolated using YODA-buffer (50-mM Tris pH 7.5; 150-mM NaCl; 1-mM EDTA; 10% glycerol v/v; 5-mM DTT; 1% protease inhibitor cocktail v/v (Sigma); 10-µM MG132; 0.1% Triton X-100 v/v). Protein electrophoresis and western blot were performed (Bolt mini blot module, Invitrogen) using antiPIF4 antibody (AS16 3157; Agrisera) and Anti-RbcL antibody (AS03 037A; Agrisera), and detected using ECL chemiluminescence kit (BioRad). Imaging was done using Hi-sensitivity chemiluminescence settings under ChemiDoc XRS+ (BioRad).
Quantification and statistical analysis
Quantitative data from all experiments were compiled and analyzed in GraphPad Prism version 8.3.2 (https://www.graphpad.com/scientific-software/prism/) and interpreted based on one-way or two-way analysis of variance (ANOVA) and appropriate post hoc test (Tukey/Sidak) as mentioned with the relevant figure legends. Student’s t tests were done using GraphPad prism wherever necessary. Conditions of normality of distribution and homogeneity of variance were checked and met. Details of the statistical tests applied, including the statistical methods, the total number of replicates from all independent experiments, mean, and error bar details, and significance are indicated in the relevant figure legends. Fully grown fourth leaf of each plant was used for cellular analyses with number of leaves (n) indicated in figure legends. Phenotyping experiments were performed from a minimum of two to maximum 5 times for individual genotypes.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers PIF1 (AT2G20180), PIF3 (AT1G09530), PIF4 (AT2G43010), PIF5 (AT3G59060), PIF6 (AT3G62090), PIF7 (AT5G61270), TCP2 (AT4G18390), TCP3 (AT1G53230), TCP4 (AT3G15030), TCP9 (AT2G45680), TCP13 (AT3G02150), TCP10 (AT2G31070), TCP24 (AT1G30210), KRP1 (AT2G23430), KRP4 (AT2G32710), KRP6 (AT3G19150), and KRP7 (AT1G49620).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Leaf phenotype in control and high-temperature grown Arabidopsis plants.
Supplemental Figure S2. Phenotyping of the first pair of leaves under high temperature.
Supplemental Figure S3. Proliferating leaves of Col-0 seedlings stained with DAPI.
Supplemental Figure S4. Expression level of PIFs at 21°C and 8 h of 28°C temperature.
Supplemental Figure S5. PIFs control temperature-mediated leaf architectural changes.
Supplemental Figure S6. Leaf features of different Arabidopsis mutants along with wild-type Col-0 under control and high temperature conditions.
Supplemental Figure S7. BiFC assays in N. benthamiana showing protein–protein interaction between PIF4 and class II TCP members.
Supplemental Table S1. Literature curated leaf development-related genes that were among DEGs under high-temperature conditions in Arabidopsis.
Supplemental Table S2. List of primers used in the study.
Supplementary Material
Acknowledgments
We are grateful to Gerrit Beemster for sharing with us the seeds of krp1, and krp4/6/7, Christian Fankhauser for pif1345, Utpal Nath for tcp4-2 and pTCP4:GUS lines, On Sun Lau for pPIF4:PIF4-GFP, and to Takato Imaizumi for the 35S::TCP-3F6H line. We thank Praveen Verma for providing the p635nRRF vector for dual luciferase assay. K.S. acknowledges her DBT-RA and SERB-NPDF fellowship and A.D. acknowledges her UGC-JRF fellowship. We thank the Confocal facility and other Central Instrumentation Facility (CIF) at NIPGR for their support.
Funding
This work was supported by Ramalingaswamy Re-entry Fellowship (BT/RLF/Re-entry/05/2013) from the Department of Biotechnology, Ministry of Science and Technology, India, Science & Engineering Research Board (SERB)-Core Research Grant (CRG/2020/002179) and Science & Engineering Research Board-National Post Doctoral Fellowship (SERB-NPDF) scheme (PDF/2018/002387) from Science and Engineering Research Board, India.
Conflict of interest statement. The authors declare that there is no conflict of interest.
Contributor Information
Kumud Saini, National Institute of Plant Genome Research, New Delhi 110067, India.
Aditi Dwivedi, National Institute of Plant Genome Research, New Delhi 110067, India.
Aashish Ranjan, National Institute of Plant Genome Research, New Delhi 110067, India.
K.S. and A.R. conceived the project and designed the experiments. K.S. and A.D. performed the experiments. K.S. analyzed the data and wrote the manuscript with contributions from A.R. and A.D. A.R. supervised the project, provided scientific discussions, and agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Aashish Ranjan (aranjan@nipgr.ac.in).
References
- Andriankaja M, Dhondt S, DeBodt S, Vanhaeren H, Coppens F, DeMilde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GTSS, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 [DOI] [PubMed] [Google Scholar]
- Bellstaedt J, Trenner J, Lippmann R, Poeschl Y, Zhang X, Friml J, Quint M, Delkera C (2019) A mobile auxin signal connects temperature sensing in cotyledons with growth responses in hypocotyls. Plant Physiol 180: 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez M (2007) Quantitative GUS activity assay of plant extracts. Cold Spring Harb Protoc 2007: pdb.prot4690-pdb.prot4690. [DOI] [PubMed] [Google Scholar]
- Carabelli M, Possenti M, Sessa G, Ruzza V, Morelli G, Ruberti I (2018) Arabidopsis HD-Zip II proteins regulate the exit from proliferation during leaf development in canopy shade. J Exp Bot 69: 5419–5431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall R, Rodriguez RE, Debernardi JM, Palatnik JF, Casati P (2013) Repression of growth regulating factors by the MicroRNA396 inhibits cell proliferation by UV-B radiation in Arabidopsis leaves. Plant Cell 25: 3570–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challa KR, Aggarwal P, Nath U (2016) Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 28: 2117–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Oh E (2016) PIF4 integrates multiple environmental and hormonal signals for plant growth regulation in Arabidopsis. Mol Cells 39: 587–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BYW, Balcerowicz M, Di Antonio M, Jaeger KE, Geng F, Franaszek K, Marriott P, Brierley I, Firth AE, Wigge PA (2020) An RNA thermoswitch regulates daytime growth in Arabidopsis. Nat Plants 6: 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford AJ, McLachlan DH, Hetherington AM, Franklin KA (2012) High temperature exposure increases plant cooling capacity. Curr Biol 22: R396–R397 [DOI] [PubMed] [Google Scholar]
- Dong J, Sun N, Yang J, Deng Z, Lan J, Qin G, He H, Deng XW, Irish VF, Chen H, et al. (2019) The transcription factors tcp4 and pif3 antagonistically regulate organ-specific light induction of saur genes to modulate cotyledon opening during de-etiolation in arabidopsis. Plant Cell 31: 1155–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Efroni I, Blum E, Goldshmidt A, Eshed Y (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Eshed Y, Lifschitz E (2010) Morphogenesis of simple and compound leaves: a critical review. Plant Cell 22: 1019–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favero DS, Kawamura A, Shibata M, Takebayashi A, Jung JH, Suzuki T, Jaeger KE, Ishida T, Iwase A, Wigge PA, et al. (2020) AT-hook transcription factors restrict petiole growth by antagonizing PIFs. Curr Biol 30: 1454–1466.e6 [DOI] [PubMed] [Google Scholar]
- Fiorucci AS, Galvão VC, Ince YÇ, Boccaccini A, Goyal A, Allenbach Petrolati L, Trevisan M, Fankhauser C (2020) PHYTOCHROME INTERACTING FACTOR 7 is important for early responses to elevated temperature in Arabidopsis seedlings. New Phytol 226: 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Wigge P (2014) Temperature and Plant Development. Wiley-Blackwell, Hoboken, NJ [Google Scholar]
- González R, Mepsted R, Wellburn AR, Paul ND (1998) Non-photosynthetic mechanisms of growth reduction in pea (Pisum sativum L.) exposed to UV-B radiation. Plant Cell Environ 21: 23–32 [Google Scholar]
- Gray W, Ostin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutie RA, Doerner P, Li C, Colon-Carmona A, Potuschak T (2005) Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA 102: 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yu H, Yuan R, Yang Y, An F, Qin G (2019) Arabidopsis transcription factor TCP5 controls plant thermomorphogenesis by positively regulating PIF4 activity. iScience 15: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth J, Lenhard M (2014) Regulation of plant lateral-organ growth by modulating cell number and size. Curr Opin Plant Biol 17: 36–42 [DOI] [PubMed] [Google Scholar]
- Huai J, Zhang X, Li J, Ma T, Zha P, Jing Y, Lin R (2018) SEUSS and PIF4 coordinately regulate light and temperature signaling pathways to control plant growth. Mol Plant 11: 928–942 [DOI] [PubMed] [Google Scholar]
- Hussain E, Romanowski A, Halliday KJ (2022) PIF7 controls leaf cell proliferation through an AN3 substitution-repression mechanism. Proc Natl Acad Sci USA 119: e2115682119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañez C, Delker C, Martinez C, Bürstenbinder K, Janitza P, Lippmann R, Ludwig W, Sun H, James GV, Klecker M, et al. (2018) Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via BZR1. Curr Biol 28: 303–310 [DOI] [PubMed] [Google Scholar]
- Ibañez C, Poeschl Y, Peterson T, Bellstädt J, Denk K, Gogol-Döring A, Quint M, Delker C (2017) Ambient temperature and genotype differentially affect developmental and phenotypic plasticity in Arabidopsis thaliana. BMC Plant Biol 17: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby MJ, Weinl C, Pusch S, Kuijt SJH, Merkle T, Dissmeyer N, Schnittger A (2006) Analysis of the subcellular localization, function, and proteolytic control of the Arabidopsis cyclin-dependent kinase inhibitor ICK1/KRP1. Plant Physiol 141: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B, Wang L, Wang J, Jiang KZ, Wang YYL, Jiang XX, Ni CY, Wang YYL, Teng NJ (2011) The effect of experimental warming on leaf functional traits, leaf structure and leaf biochemistry in Arabidopsis thaliana. BMC Plant Biol 11: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Lin J, Zhu Z (2020) PIF4 and HOOKLESS1 impinge on common transcriptome and isoform regulation in thermomorphogenesis. Plant Commun 1: 100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S, et al. (2016) Phytochromes function as thermosensors in Arabidopsis. Science 354: 886–889 [DOI] [PubMed] [Google Scholar]
- Kieffer M, Master V, Waites R, Davies B (2011) TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J 68: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Kim SJ, Jeong J, Park E, Oh E, Park Y Il, Lim PO, Choi G (2020a) High ambient temperature accelerates leaf senescence via PHYTOCHROME-INTERACTING FACTOR 4 and 5 in Arabidopsis. Mol Cells 43: 645–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Park Y, Lee J, Park C, Young J, Park Y, Lee J, Park C (2019) Developmental polarity shapes thermo-induced nastic movements in plants. Plant Signal Behav 14: 1617609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Hwang G, Kim S, Thi TN, Kim H, Jeong J, Kim J, Kim J, Choi G, Oh E (2020b) The epidermis coordinates thermoresponsive growth through the phyB-PIF4-auxin pathway. Nat Commun 11: 1529–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA, Le L (2009) Report high temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Koyama T, Sato F, Ohme-Takagi M (2017) Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol 175: 874–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota A, Ito S, Shim JS, Johnson RS, Song YH, Breton G, Goralogia GS, Kwon MS, Laboy Cintrón D, Koyama T, et al. (2017) TCP4-dependent induction of CONSTANS transcription requires GIGANTEA in photoperiodic flowering in Arabidopsis. PLoS Genet 13: e1006856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Song Z, Zhou Z, Davies KA, Chang J, Yang X, Wang S, Lucyshyn D, Tay IHZ, Wigge PA, et al. (2018) Direct control of SPEECHLESS by PIF4 in the high-temperature response of stomatal development. Curr Biol 28: 1273–1280.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie ES, Rojas CC, Neme M, Hiltbrunner A, Wigge PA, Schäfer E, Vierstra RD, Casal JJ (2016) Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354: 897–900 [DOI] [PubMed] [Google Scholar]
- Lippmann R, Babben S, Menger A, Delker C, Quint M (2019) Development of wild and cultivated plants under global warming conditions. Curr Biol 29: R1326–R1338 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Okello RCO, de Visser PHB, Heuvelink E, Marcelis LFM, Struik PC (2016) Light mediated regulation of cell division, endoreduplication and cell expansion. Environ Exp Bot 121: 39–47 [Google Scholar]
- Paik I, Kathare PK, Kim J Il, Huq E (2017) Expanding roles of PIFs in signal integration from multiple processes. Mol Plant 10: 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Park YJ, Lee HJ, Gil KE, Kim JY, Lee JH, Lee H, Cho HT, Vu LD, De Smet I, Park CM (2019) Developmental programming of thermonastic leaf movement. Plant Physiol 180: 1185–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, De Wit M (2014) Shade avoidance: phytochrome signalling and other aboveground neighbour detection cues. J Exp Bot 65: 2815–2824 [DOI] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, Van Zanten M (2016) Molecular and genetic control of plant thermomorphogenesis. Nat Plants 2: 1–9 [DOI] [PubMed] [Google Scholar]
- Reed JW, Wu MF, Reeves PH, Hodgens C, Yadav V, Hayes S, Pierik R (2018) Three auxin response factors promote hypocotyl elongation1,2[open]. Plant Physiol 178: 864–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski A, Fursniss JJ, Hussain E, Halliday JK (2021) Phytochrome regulates cellular response plasticity and the basic molecular machinery of leaf development. Plant Physiol 55: 312–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Montepaone S, Sellaro R, Esteban Hernando C, Costigliolo-Rojas C, Bianchimano L, Ploschuk EL, Yanovsky MJ, Casal JJ (2021) Functional convergence of growth responses to shade and warmth in Arabidopsis. New Phytol 231: 1890–1905 [DOI] [PubMed] [Google Scholar]
- Rymen B, Fiorani F, Kartal F, Vandepoele K, Inzé D, Beemster GTS (2007) Cold nights impair leaf growth and cell cycle progression in maize through transcriptional changes of cell cycle genes. Plant Physiol 143: 1429–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A,, Alvarez-Venegas R,, Avramova Z. (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Sarvepalli K, Nath U (2011) Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J 67: 595–607 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schommer C, Bresso EG, Spinelli SV, Palatnik JF (2012) Role of MicroRNA miR319 in Plant Development. In R Sunkar, ed, Signal. Commun. Plants. Springer Berlin Heidelberg, pp 29–4722539031
- Schommer C, Debernardi JM, Bresso EG, Rodriguez RE, Palatnik JF (2014) Repression of cell proliferation by miR319-regulated TCP4. Mol Plant 7: 1533–1544 [DOI] [PubMed] [Google Scholar]
- Sidaway-lee K, Costa MJ, Rand DA, Finkenstadt B, Penfield S (2014) Direct measurement of transcription rates reveals multiple mechanisms for configuration of the Arabidopsis ambient temperature response. Genome Biol 15: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizani BL, Kalve S, Markakis MN, Domagalska MA, Stelmaszewska J, AbdElgawad H, Zhao X, De Veylder L, De Vos D, Broeckhove J, et al. (2019) Multiple mechanisms explain how reduced KRP expression increases leaf size of Arabidopsis thaliana. New Phytol 221: 1345–1358 [DOI] [PubMed] [Google Scholar]
- Skirycz A, Claeys H, de Bodt S, Oikawa A, Shinoda S, Andriankaja M, Maleux K, Eloy NB, Coppens F, Yoo SD, et al. (2011) Pause-and-stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell 23: 1876–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang JA, Gallego-Bartolomé J, Gómez MD, Yoshida S, Asami T, Olsen JE, García-Martínez JL, Alabadí D, Blázquez MA (2009) Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J 60: 589–601 [DOI] [PubMed] [Google Scholar]
- Takahashi N, Ogita N, Takahashi T, Taniguchi S, Tanaka M, Seki M, Umeda M (2019) A regulatory module controlling stress-induced cell cycle arrest in Arabidopsis. eLife 8: 2018–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu F, Granier C (2000) Quantitative analysis of cell division in leaves: methods, developmental patterns and effects of environmental conditions. Plant Mol Biol 43: 555–567 [DOI] [PubMed] [Google Scholar]
- Vasseur F, Pantin F, Vile D (2011) Changes in light intensity reveal a major role for carbon balance in Arabidopsis responses to high temperature. Plant, Cell Environ 34: 1563–1576 [DOI] [PubMed] [Google Scholar]
- Vile D,, Pervent M,, Belluau M,, Vasseur F,, Bresson J,, Muller B,, Granier C,, Simonneau T. (2012) Arabidopsis growth under prolonged high temperature and water deficit: independent or interactive effects? Plant Cell Environ 35: 702–718 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke LC (2000) Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J 24: 613–623 [DOI] [PubMed] [Google Scholar]
- Wargent JJ, Gegas VC, Jenkins GI, Doonan JH, Paul ND (2009) UVR8 in Arabidopsis thaliana regulates multiple aspects of cellular differentiation during leaf development in response to ultraviolet B radiation. New Phytol 183: 315–326 [DOI] [PubMed] [Google Scholar]
- van der Woude L, Piotrowski M, Klaasse G, Paulus JK, Krahn D, Ninck S, Kaschani F, Kaiser M, Novák O, Ljung K, et al. (2021) The chemical compound ‘Heatin’ stimulates hypocotyl elongation and interferes with the Arabidopsis NIT1‐subfamily of nitrilases. Plant J 106: 1523–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M, Pons TL, Janssen JAM, Voesenek LACJ, Peeters AJM (2010) On the relevance and control of leaf angle. CRC Crit Rev Plant Sci 29: 300–316 [Google Scholar]
- Zhang C, Fan L, Le BH, Ye P, Mo B, Chen X (2020) Regulation of ARGONAUTE10 expression enables temporal and spatial precision in axillary meristem initiation in Arabidopsis. Dev Cell 55: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Xun Q, Zhang D, Lv M, Ou Y, Li J (2019) TCP transcription factors associate with PHYTOCHROME INTERACTING FACTOR 4 and CRYPTOCHROME 1 to regulate thermomorphogenesis in Arabidopsis thaliana. iScience 15: 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Ausin I, Seleznev A, Méndez-Vigo B, Picó FX, Sureshkumar S, Sundaramoorthi V, Bulach D, Powell D, Seemann T, et al. (2015) Natural variation identifies ICARUS1, a universal gene required for cell proliferation and growth at high temperatures in Arabidopsis thaliana. PLoS Genet 11: 1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.