Abstract
Despite the obligatory role of ethylene in climacteric fruit ripening and the identification of 77 ethylene response factors (ERFs) in the tomato (Solanum lycopersicum) genome, the role of few ERFs has been validated in the ripening process. Here, using a comprehensive morpho-physiological, molecular, and biochemical approach, we demonstrate the regulatory role of ERF D7 (SlERF.D7) in tomato fruit ripening. SlERF.D7 expression positively responded to exogenous ethylene and auxin treatments, most likely in a ripening inhibitor-independent manner. SlERF.D7 overexpression (OE) promoted ripening, and its silencing had the opposite effect. Alterations in its expression modulated ethylene production, pigment accumulation, and fruit firmness. Consistently, genes involved in ethylene biosynthesis and signaling, lycopene biosynthesis, and cell wall loosening were upregulated in the OE lines and downregulated in RNAi lines. These transgenic lines also accumulated altered levels of indole-3-acetic acid at late-breaker stages. A positive association between auxin response factor 2 (ARF2) paralog’s transcripts and SlERF.D7 mRNA levels and that SlARF2A and SlARF2B are direct targets of SlERF.D7 underpinned the perturbed auxin–ethylene crosstalk for the altered ripening program observed in the transgenic fruits. Overall, this study uncovers that SlERF.D7 positively regulates SlARF2A/B abundance to amalgamate auxin and ethylene signaling pathways for controlling tomato fruit ripening.
An ethylene response factor regulates tomato fruit ripening by activating auxin response factors involved in ripening.
Introduction
Owing to the agronomical position tomato (Solanum lycopersicum) upholds, a comprehensive dissection of its intrinsic ripening program is imperative to unravel the molecular mechanisms governing vital qualitative and quantitative fruit-related traits. The original concept of the fruit maturation process in climacteric fruits is based on a classic linear prototype guided by the precise spatio-temporal expression of the genes of ethylene biosynthesis and signaling (Alba et al., 2005; Kumar et al., 2016; Sravankumar et al., 2018). However, zooming in on the transcription networks activated during ripening in fleshy fruits has revealed a well-defined information system in which a multitude of hidden layers tightly control the ethylene biosynthesis and signaling to produce ripened fruits as an output. One such regulatory circuit involves crosstalk between ethylene and auxin, in which the latter antagonistically targets major facets of ethylene-regulated fruit ripening (Böttcher et al., 2010; Schaffer et al., 2013; Sravankumar et al., 2018). Auxin and ethylene are two cornerstones of overall fruit development and are assumed to be involved in inevitable trade-offs, that is, the ability of ethylene to trigger fruit ripening occurs at the expense of disruption of auxin biosynthetic and signaling machinery (Given et al., 1988; Zaharah et al., 2012; Kumar et al., 2014; Sravankumar et al., 2018). Experimental validation of this trade-off was reported when the exogeneous application of auxin inhibitor, p-chlorophenoxyisobutyric acid, to tomato fruits mimicked 1-aminocyclopropane-1-carboxylic acid (ACC) treatment and displayed early signs of fruit ripening (Su et al., 2015).
Deepening insights into the connection between signal transduction components of auxin and ethylene roots back to the era of identification of ethylene insensitive mutants with defects in auxin transporters, aux1 and ethylene insensitive root1/pinformed 2 (eir1/pin2) (Pickett et al., 1990; Luschnig et al., 1998). These mutants exhibited root growth inhibition synergistic with the effect of auxin on this process (Rahman et al., 2001; Swarup et al., 2002). Apart from being collaborators, ethylene and auxin have exemplified competitiveness in lateral root initiation in thale cress (Arabidopsis thaliana) whereby auxin have been shown to promote lateral root formation, and elongation with ethylene negatively regulated the primary as well as lateral root elongation (Ivanchenko et al., 2008; Negi et al., 2008, 2010; Muday et al., 2012). The centrality of auxins in defining regions of meristem growth in roots and shoots has long been recognized (Benková et al., 2003; Blilou et al., 2005). What is fascinating is that the pattern of auxin transport in these regions is strongly influenced by ethylene (Růžička et al., 2007). Once accumulated, auxin then initiates the repression of ethylene mediated root growth phenotype (Lewis et al., 2011). Additionally, the points of convergence between auxin and ethylene at the transcriptional level have been extensively studied due to documentation of their receptor and signaling mutants in the public domain. Ethylene-insensitive root growth phenotypes were observed in auxin receptor mutant Transport inhibitor response 1 (TIR1), auxin transport mutants (AUX1 and EIR/AGR/PIN2) as well as in mutants deficient in auxin response (AXR2/IAA7 and AXR3/IAA17) (Pickett et al., 1990; Luschnig et al., 1998; Stepanova et al., 2005; Muday et al., 2012). In parallel, ethylene receptors are up-regulated in fruits by auxins (Gillaspy et al., 1993; Jones et al., 2002; Trainotti et al., 2007). These points of intersection serve two separate and important roles: regulating global plant architecture and conferring local robustness on development.
In climacteric fruits, an absolute requirement of increased ethylene production via the upregulation of 1-aminocyclopropane-1-carboxylate synthase 2 (ACS2) and 1-aminocyclopropane-1-carboxylate oxidase 1 (ACO1) transcripts coordinated with low auxin levels sets the stage for ripening (Alba et al., 2005; Sravankumar et al., 2018). Interestingly, mRNA levels of ACS2, ACS4, and ACO1 are upregulated by auxin in tomato and peach (Jones et al., 2002; Trainotti et al., 2007). The physiological effects of both the hormones, at the genetic level, are brought about by their main transcriptional regulators, such as EIN3-like proteins (EILs) and ethylene response factors (ERFs) in the case of ethylene and auxin response factors (ARFs), AUX/indole-3-acetic acid (IAA), and Topless proteins in the case of auxin. Genome-wide identification studies have revealed that the tomato genome harbors 77 ERF (Sharma et al., 2010; Pirrello et al., 2012) and 22 ARF members. Accumulated evidence shows that ethylene controls the accumulation of some ARF transcripts during tomato fruit development signifying the possibility of involvement of auxin in climacteric fruit ripening (Jones et al., 2002). Likewise, several ERF genes are regulated by auxin (Trainotti et al., 2007; Pirrello et al., 2012). This two-way communication channel is feasible due to the multi-member gene families of ERFs and ARFs and the presence of both auxin and ethylene cis-regulatory elements in the promoter regions of these two sets of transcription factors (Muday et al., 2012; Zouine et al., 2014). Consistent with antagonism to ethylene, tomato fruit firmness and sugar metabolism have been reported to be partly regulated by SlARF4 (Jones et al., 2002; Sagar et al., 2013). More recently, another ARF, SlARF2 has been proven to be a quintessential component of the regulatory network controlling fruit ripening. Silencing of SlARF2 brought about dramatic ripening defects with a concomitant reduction in ethylene production and down-regulation of key ripening regulators RIN, NOR, and CNR in tomato (Hao et al., 2015; Breitel et al., 2016).
Several ripening-induced ERF genes have been implicated in the regulation of fruit ripening in tomato (Sharma et al., 2010; Liu et al., 2014, 2016). Overexpression (OE) of one such gene, LeERF1, resulted in constitutive ethylene response with accelerated fruit ripening and enhanced fruit softening (Li et al., 2007; Liu et al., 2015). Another key piece of evidence further substantiating the importance of ERFs in ripening has come from SlERF6, which integrates ethylene and carotenoid pathways in tomato (Lee et al., 2012; Liu et al., 2016). Due to the issue of proposed overlapping functions among ERF members, evidence for the involvement of another ERF, SlERF.B3, in controlling carotenoid accumulation and ethylene production was demonstrated using a dominant repressor strategy (Liu et al., 2014). Although the contribution of a few of the ripening-induced ERFs in tomato fruit ripening has been elucidated, the function of the majority of these genes largely remains undocumented. Moreover, due to the constitutive OE or silencing of these genes in earlier studies, pleiotropic phenotypes unrelated to fruit ripening have also been reported (Li et al., 2007; Liu et al., 2018). Previously, we have identified and reported several ripening-induced ERFs (Sharma et al., 2010; Kumar et al., 2012a; Srivastava and Kumar, 2019). We have also characterized a fruit ripening-specific RIP1 promoter (Agarwal et al., 2017). In the present study, we report the functional validation of a yet to be described ripening-induced ERF gene, SlERF.D7, for its roles in the regulation of fruit ripening traits in tomato by silencing it under RIP1 promoter. First, we report that SlERF.D7 transcripts are inhibited during fruit ripening in rin and nor mutants and in-house RIN-silenced lines (Kumar et al., 2012a). Transcript profiling using reverse transcription-quantitative PCR (RT-qPCR) revealed strong induction of SlERF.D7 mRNA levels upon exogenous ethylene and auxin applications. Using a reverse genetic approach, we demonstrate that SlERF.D7 plays a pivotal role in fruit ripening via directly modulating the expression of SlARF2, thereby serving as a critical point of intersection between ethylene and auxin signaling pathways. Fruit-specific silencing of SlERF.D7 under RIP1 promoter results in a severe reduction in ethylene production, fruit firmness, and pigment accumulation of the transgenic fruit, a phenotype that resembles the SlARF2 down-regulation line fruits (Hao et al., 2015; Breitel et al., 2016). In contrast, RIP1 driven ripening-specific OE of SlERF.D7 fastened the ripening progression and enhanced fruit lycopene levels. Thus, using a suite of morpho-physiological, biochemical, pharmacological, and molecular tools, we identify a regulator of tomato fruit ripening and provide critical insight into the auxin–ethylene controlled molecular circuitry that controls the ripening traits in tomato.
Results
Transcript profiling of SlERF.D7 reveals a potential role in fruit ripening
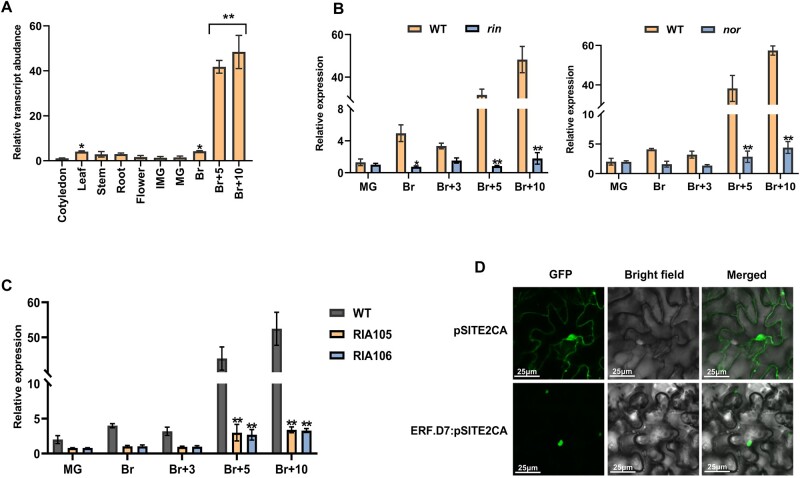
First, to predict the function of SlERF.D7, its transcript abundance was assessed by RT-qPCR in different plant tissues/organs/stages, including cotyledons, leaf, stem, root, flower, and at different stages of tomato fruit development and ripening. The transcript levels of SlERF.D7 were found to be low in vegetative organs. In contrast, it displayed a drastic up-regulation at the 5 days post breaker (Br) stage and reached its maximum levels at 10 days post Br, indicating its prospective requirement in the ripening process (Figure 1A). Next, we evaluated its mRNA abundance at different ripening stages, namely mature green (MG), Br, 3 days post Br (Br + 3), 5 days post Br (Br + 5), and 10 days post Br (Br + 10) in wild-type (WT), RIAI05, and RIAI06 transgenic lines and same-age fruits of two ripening mutants, ripening-inhibitor (rin) and non-ripening (nor) (Figure 1, B and C). Contrary to the WT fruits, the RT-qPCR analysis revealed no ripening-associated induction of SlERF.D7 transcripts at B + 5 and B + 10 stages in rin mutant fruits (Figure 1B). However, fruits from RIN suppressed transgenic lines (RIA105 and RIAI06), and nor mutant fruits exhibited a slight increase in the expression level of SlERF.D7 at late-Br stages, but the enhancement was significantly lower than the WT fruits. These results indicated the functional relevance of the SlERF.D7 gene during tomato fruit ripening.
Figure 1.
Transcript profiling of SlERF.D7 (Solyc03g118190) and its subcellular localization. A, Expression of SlERF.D7 in various tissues, including cotyledon, roots, stems, leaves, flowers, and fruit at different developmental stages: immature green (IMG), MG, Br, 5-day after Br (Br+5), and 10-day after Br (Br+10) in WT (Pusa Ruby). Values are means ± sd of three independent replicates. Asterisks indicate the statistical significance using ANOVA: *, 0.01 < P-value < 0.05; **, 0.001 < P-value < 0.01. B and C, Relative expression levels (in fold change) of SlERF.D7 in WT (Ailsa Craig), ripening-inhibitor (rin; accession no. LA1795, in the unknown background) mutant and nor mutant (in Ailsa Craig background), and 3S:RIN knockdown lines (RIA105 and RIA106; in Pusa Ruby background) at various stages of fruit ripening. Expression profiles were studied at different stages of fruit ripening by employing the RT-qPCR technique. The mRNA levels of SlERF.D7 at the MG stage in WT were used as the reference for all stages. Values are means ± sd of three independent replicates. Asterisks indicate the statistical significance using Student’s t test: *, 0.01 < P-value < 0.05; **, 0.001 < P-value < 0.01. D, Subcellular localization of SlERF-D7 in the nucleus of N. benthamiana epidermal cells.
SlERF.D7 is a nuclear-localized gene that responds positively to exogenous auxin and ethylene treatment
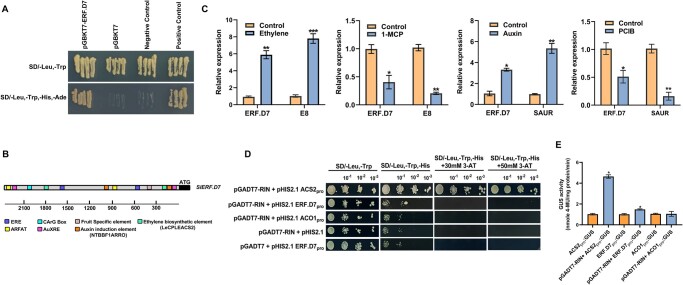
Next, we studied the subcellular localization of SlERF.D7 by transiently expressing SlERF.D7::GFP construct in Nicotiana benthamiana leaves. The CaMV35S-driven SlERF.D7::GFP fusion protein was found to be exclusively localized to the nucleus, consistent with its putative role in transcriptional regulation (Figure 1D andSupplemental Figure S1). To further characterize the SlERF.D7 protein, we determined its transactivation potential via transient expression assay using a GAL4-responsive reporter system. As hypothesized, SlERF.D7 displayed a strong transcriptional activation potential in the yeast system (Figure 2A).
Figure 2.
RIN-independent and ethylene- and auxin-dependent mode of action of SlERF.D7. A, Analysis of transactivation potential of SlERF.D7 in yeast by growing transformants on synthetic dextrose (SD) media-lacking leucine (Leu), tryptophan (Trp), histidine (His), and adenine (Ade). B, The presence of CArG Box, a putative fruit-specific element in addition to putative ethylene and AuxREs in the promoter of SlERF.D7 gene. The cis-acting regulatory elements identified are represented by different color boxes. C, RT-qPCR analysis of SlERF.D7 transcripts in total RNA samples extracted from WT MG fruit samples treated with 100 µM ethrel, 100 µM 1-MCP, 100 μM IAA, and 100 µM PCIB. Error bars, mean ± sd of three biological replicates. Asterisks indicate the statistical significance using Student’s t test: *, 0.01 < P-value < 0.05; **, 0.001 < P-value < 0.01, *** P-value < 0.001. E8, an ethylene response gene; SAUR68, an auxin response gene. D and E, Identification of DNA binding activity of RIN to the promoter of SlERF.D7 with (D) growth performance of transformants on SD/−Leu−/Trp/−His medium containing 50 mM 3-AT. Binding of RIN to the promoter of tomato ACC synthase2 (ACS2) gene-: positive control, binding of RIN to the promoter of tomato ACC oxidase1 (ACO1) gene-: negative control (E, in vivo interaction study of RIN to promoters of SlERF.D7, SlACS2, and SlACO1 via GUS reporter assays in N. benthamiana leaves.
We have previously observed the activation of ethylene-related ripening-associated genes by exogenous auxin treatment and vice versa (Kumar et al., 2012a; Sravankumar et al., 2018). Mining of the 2.5-kb upstream sequence of SlERF.D7 for cis-acting regulatory elements using the PLACE/signal search tool (http://www.dna.affrc.go.jp/PLACE/signalscan.html) revealed the presence of two putative ethylene response elements (ERE-CCGAC) and two auxin response elements (AuxRE-TGTCTC) in its promoter region (Figure 2B). Furthermore, a putative fruit-specific element variant with a sequence motif TCTTCACA was also identified in the promoter region. We next investigated the influence of these two hormones on the transcriptional regulation of SlERF.D7. Exogenous treatment of both ethylene and auxin led to induction of SlERF.D7 mRNA levels (Figure 2C). Furthermore, a decline in its mRNA levels upon treatment with either 1-methylcyclopropane (1-MCP) (100 µM), an inhibitor of ethylene perception, or p-chlorophenoxyisobutyric acid (PCIB) (100 µM), an antagonist of auxin function, validated the positive effect of both ethylene and auxin on SlERF.D7 transcription (Figure 2C). We estimated the efficacy of hormonal treatments by evaluating the transcript abundance of known ethylene (E8) and auxin (SAUR) responsive genes.
SlERF.D7 is not transcriptionally activated by RIN
MADS-box proteins such as RIN, TAGL1, FUL1, and FUL2 have been previously reported to bind to CArG box elements in the promoters of ripening-related genes such as SlACS2 and SlPSY1 (Fujisawa et al., 2011; Li et al., 2019). The presence of a CArG box element in the upstream region of SlERF.D7 (Figure 2B) instigated us to examine its direct regulation by the ripening master regulator RIN using yeast one-hybrid (Y1H) assay. Failure of survival of yeast cells containing both RIN protein and SlERF.D7 promoter on selection media suggested no direct binding of RIN to the promoter. In contrast, yeast cells with RIN-activated SlACS2 promoter, the positive control taken in the study, survived on the selection media, thereby conferring resistance in the cells to successfully grow on the selection medium (Figure 2D). To further corroborate this result in vivo, we conducted a transient transactivation assay of SlERF.D7 promoter by RIN in N. benthamiana leaves using GUS and GFP reporter systems. For this purpose, 4-week-old Nicotiana leaves were co-infiltrated with the effector construct carrying the RIN coding sequence (CDS) driven by the CAMV35S promoter. The reporter constructs were GUS or GFP CDSs driven by SlERF.D7 promoter. As noticed in the Y1H assay, there was no significant alteration in SlERF.D7-driven GUS activity or GFP fluorescence, in contrast to the positive control SlACS2 driven enhanced GUS activity and GFP fluorescence transactivation assays confirming that RIN is incapable of binding to the SlERF.D7 promoter (Figure 2E). These observations indicated that SlERF.D7 induction during fruit ripening is independent of RIN.
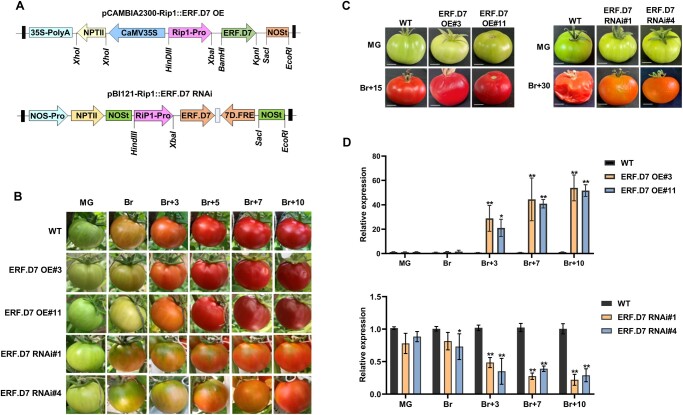
Ripening-specific over-expression and silencing of SlERF.D7 display dramatic but contrasting ripening-related changes in the transgenic fruits
To further elucidate the role of SlERF.D7 in tomato fruit ripening, we generated SlERF.D7 over-expression and RNAi knock-down transgenic lines in tomato cultivar “Pusa Ruby” under a ripening-specific RIP1 promoter, characterized in our laboratory earlier (Figure 3A; Agarwal et al., 2017). RIP1 gene is activated at post Br stage of ripening. A total of 12 independent over-expression and eight independent silencing transgenic lines were obtained (Supplemental Table S2 and Supplemental Figure S2). Based on fruits exhibiting the highest and the lowest transcript levels of SlERF.D7, we selected two homozygous T2 generation representative transgenic lines for OE and silencing (RNAi) for further characterization. At the molecular level, the selected two OE lines, SlERF.D7-OE#3 and SlERF.D7-OE#11 exhibited the significantly elevated transcript accumulation at Br + 3 and Br + 10 stages compared with the tissue culture grown WT control fruits (Figure 3D). Similarly, two RNAi lines, SlERF.D7-RNAi#1 and SlERF.D7-RNAi#4, displayed the strongest suppression of its transcripts, showing only 15%–20% accumulation of its WT fruits transcripts at the Br + 10 stage (Figure 3B). No visible phenotypic differences in plant height, flower phenotype, fruit morphology, fruit set, and fruit development in both OE and RNAi lines compared with the WT control plants were observed by us. Considering the ripening-related expression pattern of SlERF.D7, we next examined the phenotype of OE and RNAi transgenic fruits in detail. SlERF.D7-OE#3 and SlERF.D7-OE#11 OE genotypes developed an intense red color at the Br + 10 stage compared with the WT fruits, whereas SlERF.D7-RNAi#1 and SlERF.D7-RNAi#4 fruits exhibited inhibited red color development and failed to turn fully red. The RNAi fruits developed a mottled ripening pattern with partial degradation of chlorophyll at the final ripe stage (Figure 3C). However, we noticed discernable phenotypic alterations pertaining to fruit softening in the fruits of SlERF.D7 OE lines as both SlERF.D7-OE#3 and SlERF.D7-OE#11 fruits displayed wrinkling of the outer pericarp tissue, possibly accounting for early signs of cell wall loosening than their WT controls (Figure 3C). Off-wine analysis of SlERF.D7 OE and RNAi fruits exhibited the same ripening pattern, with OE lines fruits displaying signs of premature ripening compared with the WT control fruits. Contrastingly, SlERF.D7 RNAi-silenced fruits showed less pigment accumulation coupled with increased firmness than their same-age WT control fruits (Figure 3D). Altogether, we observed contrasting fruit phenotypes regarding fruit color and chlorophyll degradation in OE and RNAi lines. These observations further indicated that the degradation of chlorophyll, synchronous with the synthesis of carotenoids and cell wall degradation during the ripening process, is directly targeted by SlERF.D7.
Figure 3.
Morphological and molecular characterization of SlERF.D7 OE and RNAi lines. A, Vector diagram of pCAMBIA2300-Rip1::SlERF.D7 OE (over-expression) and pBI121-Rip1::SlERF.D7 RNAi (silencing) construct. B, Phenotypic analysis of WT, SlERF.D7 OE, and RNAi line fruits at MG, Br, 3-day after Br (Br+3), 5-day after Br (Br+5), and 7-day after Br (Br+7). C, Off-vine phenotypic assessment of WT, SlERF.D7 OE, and RNAi line fruits harvested at the MG stage and photographed till the first sign of shriveling appears, 15-day post Br (Br+15) in SlERF.D7 OE lines, and 30-day post Br (Br+30) in SlERF.D7 RNAi line. D, Transcript levels of SlERF.D7 in WT, OE, and RNAi transgenic fruit analyzed at MG, Br, 3-day after Br (Br+3), 7-day after Br (Br+7), and 10-day after Br (Br+10) by RT-qPCR with Actin gene as an internal control. Error bars mean±sd of three biological replicates. Asterisks indicate the statistical significance using Student’s t test: *, 0.01 < P-value < 0.05; **, 0.001 < P-value < 0.01.
Down-regulation of SlERF.D7 leads to reduced pigment accumulation and enhanced fruit firmness
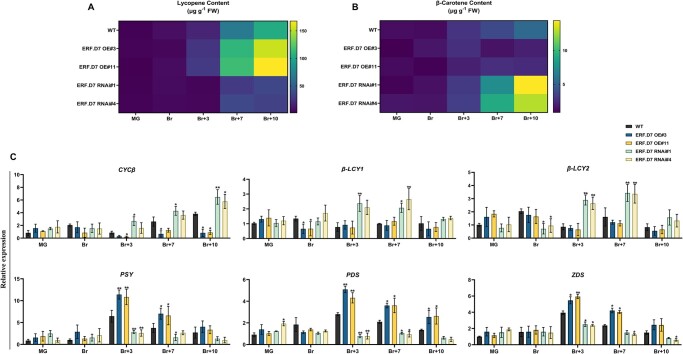
To investigate the possible cause of altered pigmentation in SlERF.D7 OE and RNAi fruits, we performed HPLC-based carotenoid profiling in WT and transgenic line pericarp tissue at MG, Br + 3, Br + 7, and Br + 10 stages. In terms of lycopene accumulation, a 55%–60% reduction was observed in SlERF.D7-RNAi#1 and SlERF.D7-RNAi#4 fruits compared with WT fruits at the Br + 10 stage. In contrast, the fruits of SlERF.D7-OE#3 and SlERF.D7-OE#11 lines displayed a 40%–45% enhancement in lycopene levels at the Br + 10 stage compared with WT fruits, consequently imparting them an intense red color phenotype (Figure 4A). Concomitantly, a sharp increase in β-carotene content was detected in SlERF.D7 RNAi fruits at the Br + 10 stage, keeping with an orange fruit phenotype (Figure 4B). Similar to an opposite trend observed for lycopene content in OE and RNAi lines, β-carotene content also exhibited contrasting profiles in the fruits of the two sets of transgenic plants. To uncover the molecular basis of this modulation in carotenoid composition, we analyzed the transcript level of genes involved in the carotenoid biosynthetic pathway of pericarp tissue at different stages during fruit ripening by RT-qPCR. Transcript abundance of phytoene synthase, PSY1, a key regulator of the carotenoid pathway, was severely repressed in SlERF.D7-RNAi#1 and SlERF.D7-RNAi#4 fruits at B + 3 and later ripening stages, concomitant to the silencing pattern of this gene in the RNAi lines (Figure 4C). A similar reduction in mRNA levels of phytoene desaturase (SlPDS) was also observed in SlERF.D7 RNAi fruits (Figure 4C). In contrast, transcripts accumulation of all the three lycopene-β-cyclases (β-LYC1, β-LYC2, and CYC-β) were significantly upregulated in SlERF.D7 RNAi fruits when compared with their WT counterparts, probably responsible for the elevated β-carotene accumulation in these transgenic fruits (Figure 4C). Likewise, a substantial increase in the transcript levels of SlPSY1 and SlPDS with a coordinated decline in expression levels of β-LYC1, β-LYC2, and CYC-β in SlERF.D7-OE#3 and SlERF.D7-OE#11 fruits, at Br + 3 and later stages, accounted for the enhanced carotenoid accumulation in these fruits (Figure 4C). The data indicate that repression of SlERF.D7 culminates in modified lycopene to β-carotene ratio via the alteration in expression levels of key carotenoid pathway genes such as PSY1, PDS, and lycopene β-cyclases.
Figure 4.
Pigment accumulation assessment in SlERF.D7 OE and RNAi fruits. A and B, Estimation of (A) lycopene and (B) β-carotene in WT and SlERF.D7 OE and RNAi lines at different stages of fruit ripening. C, RT-qPCR analysis of carotenoid biosynthetic pathway genes in WT and SlERF.D7 OE and RNAi tomato lines at MG, Br, 3-day after Br (Br+3), 7-day after Br (Br+7), and 10-day after Br (Br+10) with Actin gene as an internal control. Error bars are means ±sd of three biological replicates. Asterisks indicate the statistical significance using Student’s t test: *, 0.01 < P-value < 0.05; **, 0.001 < P-value < 0.01. β-LCY1, β-LCY2, CYC-β lycopene b-cyclases; PSY1 phytoene synthase; PDS phytoene desaturase; and ZDS, carotenoid desaturase.
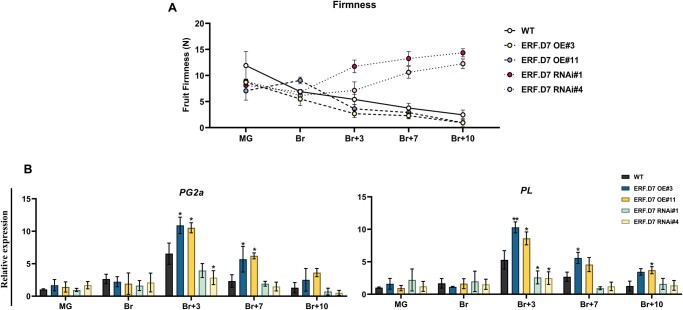
Given that fruit softening is another critical parameter of fruit ripening, we assessed the progression of firmness in SlERF.D7 OE and RNAi lines fruits from MG to Br + 10 stage. Acceleration of 35% in fruit softening was detected in SlERF.D7-OE#3 and SlERF.D7-OE#11 fruits at the Br + 3 stage compared with their WT control (Figure 5A). On the other hand, SlERF.D7 RNAi-silenced fruits were associated with a noticeable increase in fruit firmness, which reached a maximum of two to three times higher than that in WT fruit at the Br + 7 stage (Figure 5A). To further substantiate these findings at the genetic level, we examined the mRNA abundance of cell wall modifying enzymes such as polygalacturonase-2a (PG2A) and pectate lyase (PL) using RT-qPCR. SlERF.D7-OE#3 and SlERF.D7-OE#11 lines fruits accumulated higher PG2A and PL transcripts than the WT fruits at the Br + 3 and Br + 7 stages, consistent with the shriveled appearance of these fruits (Figure 5B). Contrastingly, transcripts levels of these two genes were significantly inhibited at the B + 3 stage onward in the fruits of SlERF.D7 RNAi lines compared with their WT controls. Altogether, data indicate that once the ripening program initiates, it proceeds at a much more intense rate in SlERF.D7 OE transgenic fruits and milder in the SlERF.D7 RNAi fruits in comparison to their WT control counterparts.
Figure 5.
Modulations in levels of fruit firmness in SlERF.D7 OE and RNAi fruits. A, Fruit firmness analysis in WT, SlERF.D7 OE, and RNAi line fruits at different stages of ripening, with fruits being harvested at MG stage. A total of 15 fruits were used for each measurement, and the values shown are the means ±sd. B, RT-qPCR analysis of polygalacturonase gene (SlPG2A) and PL (SlPL) at MG, Br, 3-day after Br (Br+3), 7-day after Br (Br+7), and 10-day after Br (Br+10) in SlERF.D7 OE, RNAi, and WT fruits. GAPDH and Actin were used as the internal controls. The error bars represent ±sd of three biological replicates. Asterisks indicate the statistical significance using Student’s t test: *, 0.01 < P-value < 0.05; **, 0.001 < P-value < 0.01.
Ethylene emission and perception are altered in SlERF.D7 OE fruits
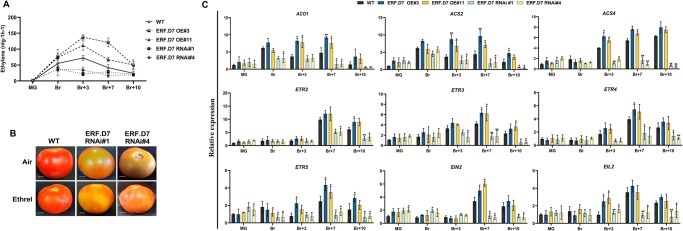
Since increased ethylene output is instrumental in determining the speed of ripening, carotenoid accumulation, and fruit firmness, we next assessed ethylene production in SlERF.D7 OE and RNAi fruits from MG to B + 10 stage. A 40%–45% rise in ethylene emission was recorded in SlERF.D7-OE#3 and SlERF.D7-OE#11 transgenic fruits compared with the same-stage WT control fruits. In contrast, substantially lower ethylene production was observed in SlERF.D7 RNAi-silenced fruits at the B + 3 stage. The atypical ethylene maxima observed at the Br + 3 stage in the WT fruits were missing in the RNAi lines fruits (Figure 6A). Assessment of the expression of key ethylene biosynthetic genes by RT-qPCR revealed elevated SlACO1, SlACS2, and SlACS4 transcripts in SlERF.D7 OE fruits at B + 3 or later ripening stages (Figure 6C). On the other hand, SlERF.D7 RNAi fruits displayed reduced mRNA levels at Br + 3, Br + 7, and Br + 10 stages compared with their same-stage WT counterparts (Figure 6C). Interestingly, to compensate for low ethylene production, exogenous ethylene application to SlERF.D7 RNAi fruits at the MG stage failed to reverse the inhibited ripening phenotype (Figure 6B and Supplemental Figure S3). To corroborate this, we conducted an expression profiling of genes involved in the ethylene signaling pathway in WT, SlERF.D7 RNAi, and OE fruits at different stages of ripening. We noticed a drastic reduction in the transcripts accumulation of ethylene receptors such as ETR2, ETR3 (NR), ETR4, and ETR5 in post Br stages of SlERF.D7 RNAi fruits when compared with the WT fruits (Figure 6C). Among the other important ethylene signaling genes, ethylene-insensitive 2 (EIN2) and EIN2-like (EIL2) were also down-regulated during ripening in SlERF.D7 RNAi line fruits (Figure 6C). To further narrow down to ERFs, altered but mostly opposite expression patterns of numerous ERFs were observed in SlERF.D7 OE and RNAi fruits to each other. Five ERF genes (ERF.A3, ERF.C1, ERF.E1, ERF.E2, and ERF.E4) displayed up-regulation at the onset of ripening starting from the Br stage in SlERF.D7 OE lines (Supplemental Figure S4). In contrast, a concomitant decrease in transcripts of these five ERFs was observed in SlERF.D7 RNAi fruits at post-Br stages. By contrast, the mRNA abundance of three ERFs (SlERF.B1, SlERF.B2, and SlERF.B3) exhibited enhanced mRNA abundance in SlERF.D7 RNAi line fruits (Supplemental Figure S4). To rule out any possibility of off-target effects in SlERF.D7 transgenic line, we further investigated the expression profiles of all SlERF.D clade members. We found no significant changes in any SlERF.D clade member’s transcript abundance in transgenic fruits when investigated using Student’s t test with P-value ranging from 0.05 < P-value < 0.001 (Supplemental Figure S5). In conclusion, these results signify that ethylene biosynthesis coupled with ethylene perception and signaling contributes to the altered ripening phenotypes observed in the OE and RNAi lines.
Figure 6.
Alterations in ethylene biosynthesis and perception in SlERF.D7 transgenic fruits. A, Ethylene production of WT and SlERF.D7 OE and RNAi fruits assessed at MG, Br, 3-day after Br (Br+3), 7-day after Br (Br+7), and 10-day after Br (Br+10) stages. Values represent the means of at least five individual fruits. The error bars represent ±sd of three biological replicates. B, Exogenous ethylene treatment on WT and SlERF.D7 RNAi fruit. MG fruits from WT and SlERF.D7 RNAi lines were injected with a buffer solution containing 10 mM MES, pH 5.6, sorbitol (3% w/v), and 100 µM ethrel (2-chloroethylphosphonic acid, 40% solution, SRL Diagnostics). After the treatment, fruits were incubated in a culture room at 26°C, under a 16-h light/8-h dark cycle with a light intensity of 100 μmol m−2 s−1 and photographed after 7 days. C, RT-qPCR analysis of ethylene biosynthesis and perception pathway genes at MG, Br, 3-day after Br (Br+3), 7-day after Br (Br+7), and 10-day after Br (Br+10) in SlERF.D7 OE, SlERF.D7 RNAi, and WT fruits with Actin gene as an internal control. Error bars represent ±sd of three biological replicates. Asterisks indicate the statistical significance using Student’s t test: *, 0.01 < P-value < 0.05; **, 0.001 < P-value < 0.01. ACO1, aminocyclopropane-1-carboxylic acid oxidase 1; ACS2 and ACS4, aminocyclopropane-1-carboxylic acid synthases; ETR2, ETR3, ETR4, and ETR5, ethylene receptors; EIN2, ethylene signaling protein; and EIL2, protein.
Transcription of key ripening regulators is modulated in SlERF.D7 transgenic line fruits
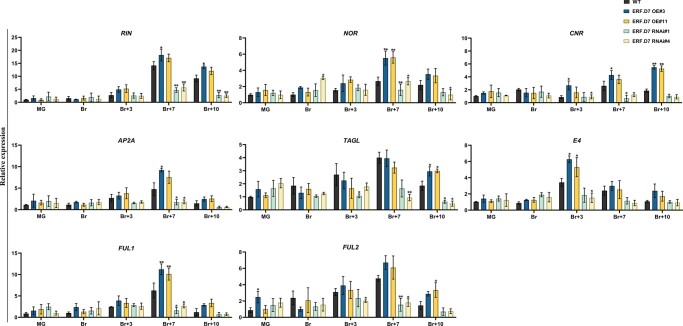
To unveil the possible molecular mechanism responsible for this altered ripening phenotypes of SlERF.D7 transgenic lines, we assessed the transcript profiles of major ripening regulator genes at different stages of ripening. Compared with WT fruits, RIN and CNR mRNA levels were significantly reduced at Br + 7 and Br + 10 stages in the RNAi fruits (Figure 7). Likewise, 50%–60% reduction in the transcripts of FRUITFUL1 (FUL1) and FRUITFUL2 (FUL2) were observed in SlERF.D7 RNAi fruits at post Br stages (Figure 7). Similar inhibition was also observed in mRNA levels of NOR, SlAP2a, and TAGL1 genes in the RNAi lines fruits (Figure 7). The altered expression pattern of most of these genes in SlERF.D7 transgenic fruits is consistent with the delayed onset of ripening observed in the RNAi lines. Correspondingly, SlERF.D7 OE fruits accumulated elevated transcripts of RIN, CNR, NOR, AP2A, and TAGL1, compared with the WT control fruits, at post Br stages.
Figure 7.
Transcript profiling of key ripening regulator genes in WT and SlERF.D7 OE and RNAi tomato lines during fruit ripening. Total RNA was extracted from MG, Br, 3-day after Br (Br + 3), 7-day after Br (Br + 7), and 10-day after Br (Br + 10) stages of SlERF.D7 OE, SlERF.D7 RNAi, and WT fruits. The relative mRNA levels of each gene were normalized using the Actin gene as an internal control. Error bars represent ±sd of three biological replicates. Asterisks indicate the statistical significance using Student’s t test: *, 0.01 < P-value < 0.05; **, 0.001 < P-value < 0.01. NOR, non-ripening; CNR, colorless non-ripening; AP2a, APETALA2/ERF gene; TAGL1, tomato AGAMOUS-LIKE 1; E4, ethylene-responsive and ripening regulated genes; FUL1 and FUL2, fruitful MADS-box transcription factor homologs.
SlERF.D7 alters fruit ripening by influencing auxin sensitivity
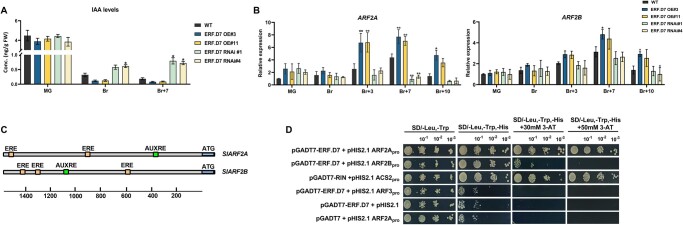
Because SlERF.D7 positively responded to IAA treatment, and the fact that SlERF.D7 RNAi fruits closely mimicked the ripening phenotype of SlARF2A and SlARF2B RNAi (or OE) fruits reported earlier (Hao et al., 2015; Breitel et al., 2016), we next assessed the levels of IAA in transgenic fruits at MG, Br, and Br + 7 stages. Remarkably, a significant increase in IAA concentration was observed in SlERF.D7 RNAi fruits at the Br + 7 stage (Figure 8A). In contrast, the OE lines showed slightly decreased IAA levels. Considering the modulated IAA levels in transgenic fruits and the known roles of tomato ARFs in ripening, we then examined the expression profile of all tomato ARF gene family members by RT-qPCR (Kumar et al., 2015; Supplemental Figure S6). Strikingly, the transcript accumulation of SlARF2 paralogs was significantly affected in the transgenic fruits. Transcription of no other ARF gene family members was affected in SlERF.D7 transgenic fruits (Supplemental Figure S6). In particular, SlARF2A displayed a dramatic down-regulation in SlERF.D7-RNAi#1 and SlERF.D7-RNAi#4 fruits at the Br + 3 stage, whereas SlARF2B exhibited a slight reduction in its mRNA levels at this stage (Figure 8B). However, transcript accumulation of SlARF2B was reduced to 30% in SlERF.D7 RNAi fruits at the Br + 7 stage (Figure 8A). On a similar level, SlERF.D7 OE line fruits showed a significant upregulation in the expression of SlARF2A at post Br stages. These results signified a possible role of SlERF.D7 in controlling ripening traits by moderating auxin responses, plausibly through SlARF2 paralogs, in tomato fruits.
Figure 8.
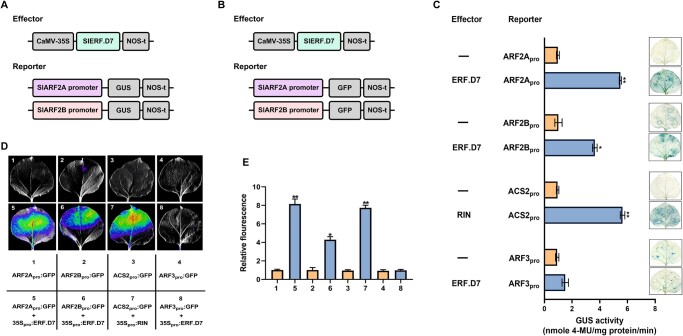
Altered auxin levels and ARF2 orthologs transcription in SlERF.D7 transgenic fruits. A, Determination of IAA levels during fruit ripening in WT, SlERF.D7 OE, and RNAi line fruits at MG, Br, and 7-day post Br (Br + 10) stages. Error bars represent ±sd of three biological replicates. Asterisks indicate the statistical significance using Student’s t test: *, 0.01 < P-value < 0.05; **, 0.001 < P-value < 0.01. B, Expression profiling of SlARF2 orthologs, SlARF2A and SlARF2B in transgenic fruits of SlERF.D7 OE and RNAi lines during different fruit maturation stages, MG, Br, 3-day after Br (Br+3), 7-day after Br (Br+7), and 10-day after Br (Br+10). The relative mRNA levels of both the genes were normalized using Actin and GAPDH genes as an internal control. Error bars represent ±sd of three biological replicates. Asterisks indicate the statistical significance using Student’s t test: *, 0.01 < P-value < 0.05; **, 0.001 < P-value < 0.01. C, Analysis of putative ethylene and auxin-responsive cis-elements in the 2.5-kb promoter region of SlARF2A and SlARF2B genes. D, Y1H analysis of binding of SlERF.D7 protein to putative ethylene-responsive elements in the promoter regions of SlARF2A and SlARF2B genes displaying growth performance of transformants on SD/−Leu−/Trp/−His medium containing 50 mM 3-AT. Binding of RIN to the promoter of SlACS2 gene-: positive control, binding of SlERF.D7 to the promoter of SlARF3 gene-: negative control.
SlERF.D7 binds to and activates the promoters of SlARF2A/B
Because of the observed alterations in the transcript level of SlARF2A and SlARF2B in SlERF.D7 transgenic lines fruits, we hypothesized that SlERF.D7 might bind to the promoters of the two ARF2 paralogs. In silico analysis of 2.5-kb promoter sequences of SlARF2A and SlARF2B revealed the presence of two and three conserved ethylene responsive elements (ERE), respectively (Figure 8C). These elements are the putative targets of ERF-type transcription factors in plants. To validate this assumption, we performed a Y1H assay. A pGAD::SlERF.D7 plasmid (containing the SlERF.D7 putative DNA domain fused to the GAL4 active domain) and a pHIS-cis-acting reporter construct (2-kb PCR amplified promoter regions of SlARF2A and SlARF2B) were co-transformed into yeast strain Y187. The results displayed a strong binding of SlERF.D7 to the SlARF2A promoter, whereas a weak interaction between this protein and SlARF2B promoter was noticed (Figure 8D). As indicated by the modulations in the transcript levels of these reporter genes, SlERF.D7 could directly bind to the promoters of SlARF2A and SlARF2B and regulate the expression of these target genes.
To further determine SlERF.D7-mediated direct activation of SlARF2 genes in planta, we evaluated GUS and GFP activity driven by SlARF2A and SlARF2B promoter fusion reporter constructs in N. benthamiana leaves that also transiently expressed the SlERF.D7 gene under the control of CaMV35S promoter (effector construct) (Figure 9, A and B). Transactivation of the SlARF2A promoter by SlERF.D7 significantly enhanced the GUS reporter activity. However, the transient co-expression of SlARF2B promoter and SlERF.D7 displayed less GUS activity elevation than SlARF2A and the positive controls (Figure 9C). Similar observations were made with GFP fluorescence driven by transient co-expression of SlARF2A and SlARF2B reporter constructs and SlERF.D7 effector construct in N. benthamiana leaves (Figure 9, B and D). Upon quantification of the relative fluorescence using flow cytometry, transactivation of SlARF2A reporter construct by SlERF.D7 exhibited an approximate seven-fold increase in fluorescence when compared with an approximate four-fold elevation observed in transactivation of SlARF2B reporter construct (Figure 9E).
Figure 9.
In vivo transactivation of SlARF2 promoters by SlERF.D7. A and B, Schematic diagrams of the reporter and effector constructs. The GUS and GFP reporter plasmids contain the promoters of SlARF2A and SlARF2B. The effector plasmids contain the SlERF.D7 CDS under the control of the CaMV35S promoter. C, Confirmation of the activation of SlARF2A and SlARF2B promoters by SlERF.D7 via the GUS complementation assay in N. benthamiana leaves. GUS assay was performed after 48 h of infiltration. GUS activity is expressed in nmole 4-methylumelliferone mg−1 protein min−1. Activation of the SlACS2 promoter by RIN-: positive control, Activation of the SlARF3 promoter by SlERF.D7-: negative control. Error bars represent ±sd of three biological replicates. Asterisks indicate the statistical significance using Student’s t test: *, 0.01 < P-value < 0.05; **, 0.001 < P-value < 0.01. D and E, Transient expression assay shows that SlERF.D7 activates the expression of the GFP reporter gene. The bottom panel indicates the combination of reporter and effector plasmids infiltrated. The GFP reporter gene is driven by ARF2A and ARF2B promoters. Representative images of N. benthamiana leaves 48 h after infiltration captured via NightSHADE Plant Imaging System are shown. Activation of the SlACS2 promoter by RIN-: positive control, activation of the SlARF3 promoter by SlERF.D7-: negative control. E, Quantitative analysis of fluorescence intensity in three independent determinations was assessed. Error bars represent ±sd of three biological replicates. Asterisks indicate the statistical significance using Student’s t test: *, 0.01 < P-value < 0.05; **, 0.001 < P-value < 0.01.
The data indicate that SlERF.D7 regulates the expression of SlARF2 paralogs by directly binding, most likely to the typical ERE elements, in their promoter regions.
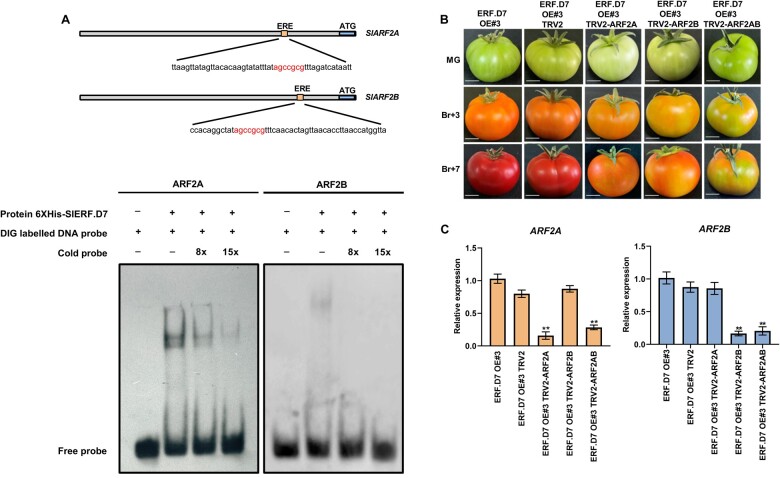
Although the transient GUS and GFP transactivation assays suggested a direct regulation of SlARF2 promoters by SlERF.D7 protein, we further conducted an electrophoretic mobility shift assay (EMSA) to assess the ability of SlERF.D7 to directly bind to SlARF2A and SlARF2B promoters. Indeed SlERF.D7 exhibited direct binding to the DNA probe containing the ERE element present in the SlARF2A promoter, whereas the unlabeled promoter fragment displaced the binding of the labeled probe in a dose-dependent manner (Figure 10A). However, SlERF.D7 displayed weak binding to ERE motif in the SlARF2B promoter (Figure 10A). These results revealed the ability of SlERF.D7 to specifically bind to ERE motif in the SlARF2A promoter. Combined together, the data confirm that SlARF2 promoters are direct targets of SlERF.D7 in planta.
Figure 10.
SlARF2 promoters are a direct target of SlERF.D7. A, SlERF.D7 binding to the promoters of SlARF2 harboring the ERE element. The WT probe containing the ERE motif was digoxigenin-labeled. Competition for SlERF.D7 binding was performed with 8× and 15× cold probes. The symbols − and + represent the absence or presence of the probes and 6× Histidine-tagged SlERF.D7 protein. B, Phenotypes of SlARF2A, SlARF2B, and SlARF2AB silenced fruits in SlERF.D7 OE background. Un-infiltrated and vector-only (pTRV) fruits were used as the control. Photographs were taken at MG, 3-day post Br (Br+3), and 7-day post Br (Br+7) stages. C, The silencing efficiency of the SlARF2 orthologs in SlARF2A, SlARF2B, and SlARF2AB infiltrated fruits. The relative mRNA levels of both the genes were normalized using GAPDH and Actin gene as an internal control. Error bars represent ±sd of three biological replicates. Asterisks indicate the statistical significance using Student’s t test: *, 0.01 < P-value < 0.05; **, 0.001 < P-value < 0.01.
Silencing of SlARF2 paralogs in SlERF.D7 OE lines fruits partially reverse the fastened ripening phenotype
To confirm whether the elevated transcript levels of SlARF2 paralogs were responsible for the enhanced carotenoid synthesis in SlERF.D7 OE line fruits, we performed virus-induced gene silencing (VIGS) assays of SlARF2A, SlARF2B, and double knock-down of both SlARF2A/B in SlERF.D7-OE#3 fruits at the MG stage. A spotted ripening pattern was observed in SlARF2A single knock-down VIGS fruit, with the fruits remaining orange at the final maturation stage (Figure 10B). Similarly, SlARF2B single knock-down VIGS fruits exhibited more distinct variegation of yellow and orange color on the outer pericarp and never achieved the intense red color of SlERF.D7 OE fruits (Figure 10B). The simultaneous double knock-down SlARF2A/B VIGS fruits displayed more severe ripening defects. These fruits showed mottled green and orange sectors, separated by distinct borders, contrasting with the uniform red color observed in SlERF.D7 OE and SlERF.D7 OE tobacco rattle virus (TRV) control fruits, suggesting a redundant function of SlARF2A and SlARF2B in ripening (Figure 10B). Previously, RNAi-mediated stable transgenic silencing lines of SlARF2A, SlARF2B along with a double knock-down SlARF2A/B have already been investigated for fruit ripening alterations by two groups (Hao et al., 2015; Breitel et al., 2016). The authors have described a similar delayed ripening phenotype in RNAi lines as we noticed with a VIGS construct in SlERF.D7-OE#3 fruits. Also, SlERF.D7 RNAi-silenced fruits in this study yield a similar but milder phenotype, which can be attributed to using a weaker but more ripening-specific RIP1 promoter, instead of a constitutive CaMV35S, in VIGS experiments. To verify that the phenotype obtained was associated with the silencing of ARF2A and ARF2B in single and double VIGS-silenced fruits, we analyzed the expression of these genes at the molecular level. A 70%–75% reduction in transcript levels of SlARF2 genes was observed in single and double knock-down VIGS fruits compared with SlERF.D7-OE#3 fruits (Figure 10C). Additionally, the mRNA abundance of SlARF2B in SlARF2B single and double knock-down VIGS fruits decreased to about 15%–20% of their control OE fruits (Figure 10C).
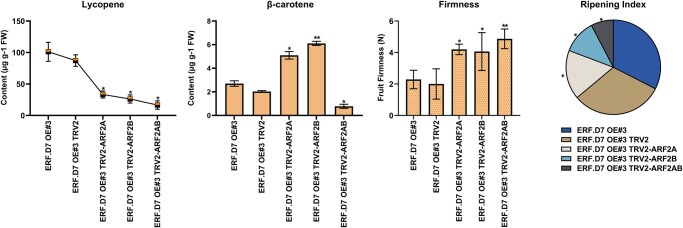
HPLC-based assessment of color change in VIGS fruit for lycopene and β-carotene further emphasized the difference in carotenoid production in SlERF.D7-OE#3 and its SlARF2 paralogs VIGS fruits. The lycopene levels were significantly compromised in all the VIGS fruits, with SlARF2A/B double knock-down fruits displaying the maximum reduction (Figure 11). In the context of β-carotene, an increase of 40%–45% was observed in SlARF2B lines compared with SlERF.D7-OE#3 line control fruits (Figure 11). Therefore, it can be interpreted that this decreased lycopene and increased β-carotene levels were responsible for the yellow–orange phenotype observed in ARF2 VIGS SlERF.D7-OE#3 line fruits. Furthermore, SlARF2A/B VIGS fruits exhibited maximum fruit firmness followed by SlARF2B and SlARF2A, thereby indicating that a noticeable delay in ripening of VIGS fruits was due to reduced transcript accumulation of SlARF2 paralogs (Figure 11). Further, ARF2A/B VIGS fruits displayed the lowest ripening index, followed by the SlARF2B- and SlARF2A-silenced fruits, respectively (Figure 11). These results indicate that SlERF.D7 acts upstream of SlARF2 paralogs and performs its ripening-related function by directly regulating them.
Figure 11.
VIGS of SlARF2 orthologs in SlERF.D7 OE transgenic fruits. Assessment of lycopene, β-carotene, fruit firmness levels and ripening index in SlERF.D7 OE, SlERF.D7 OE TRV (control), and the ARF2A, ARF2B, and ARF2AB VIGS-silenced fruits. Error bars represent ±sd of three biological replicates. Asterisks indicate the statistical significance using Student’s t-test: *, 0.01 < P-value < 0.05; **, 0.001 < P-value < 0.01.
Discussion
SlERF.D7, a ripening-associated gene, encodes a nuclear-localized transcriptional activator
Tomato fruit ripening is a multilayer perception process in which different hormonal inputs operate in parallel or at various levels to define an output transcriptome. For years, the center stage of this ripening process has been occupied by ethylene (Giovannoni, 2004). However, given a large number of ERFs are present in fruit species and the fact that ripening-related roles of many of these are yet to be elucidated, the complete molecular circuitry by which ethylene regulates the ripening-associated genes is not entirely understood (Solano et al., 1998; Pirrello et al., 2012; Liu et al., 2016; Srivastava and Kumar, 2019). For the last two decades, patterns of mRNA accumulation of tomato ERF gene family members during ripening have been under scrutiny. Among the 77 ERFs characterized in tomato, the expression dynamics of several ERFs have demonstrated a classic ripening-associated behavior with an increase in their transcript levels at the onset of ripening, peaking at 5 days post Br, followed by a subsequent decline at late ripening stages (Sharma et al., 2010; Liu et al., 2016). So far, a limited number of ERFs have been functionally validated for their role in regulating tomato fruit ripening (Liu et al., 2016). In this regard, the current study presents several lines of evidence substantiating the physiological importance of SlERF.D7 as an active ripening modulator which corroborates with the well-documented involvement of ERFs in tomato fruit ripening (Srivastava and Kumar, 2019). The presence of ethylene-responsive elements in the promoter region of SlERF.D7 and its direct regulation by ethylene forms the first line of evidence on this subject. The comprehensive expression profiling of SlERF.D7 during different stages of fruits in WT, RIN-silenced genotypes, and ripening impaired rin and nor mutants further establish a strong positive link between SlERF.D7 and tomato fruit ripening in multiple genetic backgrounds. A strong transactivation potential in yeast coupled with exclusive nuclear localization, SlERF.D7 protein displays consistency in being designated as a transcriptional factor, as shown previously for the key ripening regulator RIN protein (Ito et al., 2008). Although the mRNA abundance of SlERF.D7 is dramatically repressed in rin mutant fruits in parallel with the presence of CArG box in its promoter region, the present research indicates that SlERF.D7 and RIN do not seem to operate in the same regulatory network and its ripening-associate transcription is possibly controlled by some other ripening regulator.
SlERF.D7 influences all the major facets of fruit ripening
To functionally validate the importance of SlERF.D7 in the ripening process, we generated stable OE and RNAi transgenic lines of SlERF.D7 using a fruit ripening-specific RIP1 promoter (Agarwal et al., 2017). RIP1 promoter was used in this study to avoid any pleiotropic non-ripening effects in the SlERF.D7 transgenic lines. The altered phenotypes associated with over and underexpression of SlERF.D7, such as ethylene production and fruit pigmentation, signify that this transcription factor broadly impacts the fruit ripening process. As per the fact that a respiratory burst coupled with an increase in ethylene production at the onset of ripening is a hallmark of the climacteric fruits, such as tomato, measurements of ethylene emission in OE and RNAi fruits were in synchronization with the observed delay in ripening of SlERF.D7 RNAi fruits and an early ripening in the fruits of SlERF.D7 OE lines (Alexander and Grierson, 2002; Giovannoni, 2004). The paramount role of ACC synthase and ACC oxidase genes in mediating the fruit transition from System 1 (auto-inhibitory) to System 2 (auto-catalytic) ethylene production is well known. Tomato ACS1 and ACS6 have shown to mediate System 1 ethylene production in immature fruit, whereas the induction of ACS2, ACS4, ACO1, and ACO4 genes brings about the characteristic sharp increase in ethylene biosynthesis in System 2 (Nakatsuka et al., 1998; Barry et al., 2000; Barry and Giovannoni, 2007). The suppression of these genes has been found to inhibit tomato fruit ripening (Hamilton et al., 1990; Oeller et al., 1991; Lincoln et al., 1993; Nakatsuka et al., 1998; Barry et al., 2000). A disturbed expression pattern of SlACO1, SlACS2, and SlACS4 genes in SlERF.D7 transgenic fruits is seemingly responsible for the altered ethylene levels observed in these fruits. Strikingly, exogenous ethylene treatment to compensate for low ethylene levels observed in SlERF.D7 RNAi fruits failed to restore the normal ripening process. This impairment of SlERF.D7 RNAi fruits to activate autocatalytic ethylene production could result from the inhibited transcript levels of ethylene receptor genes, SlETR2, SlETR3, SlETR4, and SlETR5. A critical evaluation of the contribution of the SlETR3 (NR) receptor in regulating ethylene biosynthesis and carotenoid accumulation has already been well-documented (Tieman et al., 2000). However, targeting a single ethylene receptor to achieve a delay in fruit ripening has been reported to result in compensatory over-expression dynamics of other ethylene receptor genes (Tieman et al., 2000). Thus, the inhibition of four ethylene receptors could account for a considerable loss of ability to perceive and channel the ethylene responses to downstream targets in the RNAi fruits. The inhibited ethylene signaling was validated by the downregulation of EIN2 and EIL3 transcripts, the positive regulators of ethylene responses, in SlERF.D7 RNAi fruits, which may explain the defective ripening phenotype. Downstream of EILs, the signal branches to ERFs, manipulation of which can result in alterations specific to color, flavor, and texture during ripening (Liu et al., 2015, 2016; Xie et al., 2016). Notably, a report on the functional relevance of SlERF6 in mediating carotenoid accumulation during ripening has already been published (Lee et al., 2012). Another APETALA2/ERF gene family member, SlAP2a, negatively regulates tomato fruit ripening (Chung et al., 2010; Karlova et al., 2011). Also, downregulation of another ERF, SlERF.B3, via the dominant repressor technology has been shown to cause a delay in tomato ripening (Liu et al., 2014). On analyzing the expression profiles of major ERFs, SlERF.D7 transgenic fruits displayed dramatic modulations in transcript accumulation of a high number of ERFs consistent with alterations in ethylene production and observed ripening phenotypes. A disparity in mRNA levels of SlERF.E1 in SlERF.D7#OE and SlERF.D7#RNAi lines is interesting and needs further investigation to validate if it is upregulated due to fluctuations in the transcript abundance of SlARF2A and SlARF2B (Hao et al., 2015).
The enhanced lycopene accumulation during tomato fruit ripening is promoted by an up-regulation of PSY1 and PDS and concomitant repression of lycopene cyclases transcripts. Contrastingly, inhibition in the expression of PSY1 and PDS and the enhanced expression of β-LYC and CYC-β cyclases promote the conversion of lycopene to β-carotene in tomato fruits (Giuliano et al., 1993; Fray and Grierson, 1993; Pecker et al., 1996; Ronen et al., 2000; Fantini et al., 2013; Liu et al., 2014). Compared with the WT, lycopene to β-carotene ratio in transgenic fruits is substantially modified, conferring a deep red color to SlERF.D7 OE fruits whereas imparting an orange coloration to SlERF.D7 RNAi fruits at Br + 10 stage. This off-balance is probably a result of modulations in transcript levels of SlPDS, SlPSY1, and SlCYCB genes. The incapacity of SlERF.D7 RNAi fruits to reach a red color could also be attributed to low levels of ethylene production as the rate-limiting enzyme of carotenoid biosynthesis, SlPSY1, is known to be ethylene inducible (Fraser et al., 1994). Apart from severe impairment of ethylene biosynthetic genes, the effect of SlERF.D7 RNAi on the transcript levels of carotenoid pathway genes may also result from the down-regulation of the ripening master regulators, such as RIN, NOR, and CNR. These TFs are known to impact carotenoid accumulation in tomato fruits (Alba et al., 2005; Manning et al., 2006; Vrebalov et al., 2009; Osorio et al., 2011). In addition, the enhanced expression of SlLCYB and SlCYCB in SlERF.D7 RNAi fruits perfectly explains the changes in carotenoid content and pigmentation observed in these fruits.
The impact of altered transcript levels of SlERF.D7 on fruit firmness was evident from the texture analysis of SlERF.D7 OE and RNAi transgenic lines. Numerous studies have elucidated the importance of early cell wall disintegration as one of the biggest challenges responsible for the post-harvest deterioration of tomato fruit (Meli et al., 2010; Yang et al., 2017). Although PG2a, PL, and PME enzymes are considered the major contributors to the cell wall degradation process, no significant fluctuations were observed in fruit texture in knockout fruits of PG2a (Grierson et al., 1993; Hall et al., 1993; Yang et al., 2017). However, CRISPR-generated mutation in PL resulted in firmer fruits with increased shelf life (Li et al., 2019). The enhanced softening phenotype of SlERF.D7 OE fruits is in line with the elevated transcript levels of both SlPG2a and SlPL at late-Br stages. It is widely accepted that RIN has hundreds of target genes, including those involved in pathways regulating cell wall loosening, such as PG2a (Fujisawa et al., 2011, 2013; Zhong et al., 2013). We speculate that the OE of RIN in SlERF.D7 OE fruits may also contribute to the early fruit softening phenotype observed in these fruits. Likewise, a significant reduction in mRNA accumulation of SlPL and SlPG2a combined with the decreased levels of RIN in SlERF.D7 RNAi fruits impeccably synchronizes with fruit firmness studies and the phenotype associated with down-regulation of SlERF.D7.
SlERF.D7 regulates the expression dynamics of major ripening regulators
Tomato genetics and genomics resources are very well developed due to detailed and informative research on pleiotropic mutants such as rin, nor, and cnr (colorless non-ripening) (Vrebalov et al., 2002; Giovannoni, 2004; Manning et al., 2006; Barry and Giovannoni, 2007). Inhibition of these genes dramatically and irreversibly impairs the ripening signaling cascade and therefore is termed as master regulators of fruit ripening. Interestingly, SlERF.D7 RNAi lines fruits also display analogous through milder ripening defects as that of rin and nor mutant fruits, such as low ethylene emission, disturbed carotenoid accumulation, with altered fruit levels firmness. At the genetic level, ripening-specific silencing of SlERF.D7 exhibits strong repression of RIN, NOR, and CNR genes at late-Br stages, plausibly contributing to the incompetency of the RNAi fruits to ripen normally. Apart from RIN, other MADS-domain containing transcription factors such as FUL1, FUL2, and AGAMOUS-like1 (TAGL1) play critical roles in fruit ripening regulation by forming multimeric protein complexes with RIN. Suppression of these genes substantially inhibits ripening by blocking ethylene biosynthesis and decreases carotenoid accumulation (Leseberg et al., 2008; Vrebalov et al., 2009; Itkin et al., 2009; Martel et al., 2011; Bemer et al., 2012). Previous reports on the down-regulation of TAGL1 have shown to yield a yellow–orange pigmented fruit phenotype accompanied by a drastic decrease in levels of ethylene (Itkin et al., 2009; Vrebalov et al., 2009). In addition, FUL1 and FUL2 are functionally redundant in controlling the expression module of pigment-producing genes during ripening (Bemer et al., 2012). Interestingly, the fruit phenotypes observed in FUL1, FUL2, and TAGL1 suppression lines are similar to the orange-colored SlERF.D7 RNAi-silenced fruits. Correspondingly, the transcript abundance of these genes in SlERF.D7 RNAi fruits displays a downward expression profile, keeping in line with the observed phenotype of these fruits. Taken together, SlERF.D7 emerges as a critical networking component that is transcriptionally wired into a complex regulatory interplay involving several MADS-box proteins essential for ripening to occur.
SlERF.D7 integrates ethylene and auxin signal transduction pathways via the activation of SlARF2 paralogs during ripening
Ethylene and auxin signaling components have previously been reported to be arranged and integrated into ways that can modulate the state and output of numerous plant developmental processes. However, precise signal integrators responsible for these interactions remain poorly understood despite a well-defined networking system. There is overwhelming evidence that reinforces ethylene–auxin complex interactions at the molecular level. It has been shown that mutations in auxin signaling display abnormal ethylene responses (Pickett et al., 1990). In addition, a mutual regulation, by auxin and ethylene, of genes involved in the biosynthetic machinery of both the hormones has also been reported (Muday et al., 2012). In this regard, ERF1 has been designated as a mediator between ethylene and auxin biosynthesis via regulating ASA1 expression in Arabidopsis during root elongation (Mao et al., 2016). Likewise, auxin biosynthesis during lateral root formation is directly impacted by AtERF109 (Cai et al., 2014). Recently, SlERF.B3 has been reported to directly target SlIAA27 in mediating root length in tomato (Liu et al., 2018). Previously, we have also demonstrated the role of SlGH3-2, a ripening-induced IAA-amido synthetase, in regulating fruit ripening aspects by controlling ethylene biosynthesis and auxin homeostasis (Sravankumar et al., 2018). This study’s detailed functional analysis combined with various bioinformatic approaches has rendered SlERF.D7 an active ripening regulator. To add to the complexity, we found that SlERF.D7 functions as an integrator in the interplay between ethylene and auxin through the regulation of ARF2paralogs, members of the tomato ARF family of transcriptional regulators. The transcriptional regulation of SlARF2 by both auxin and ethylene has already been reported (Hao et al., 2015; Breitel et al., 2016). Furthermore, SlERF.D7 positively mediates ethylene and auxin sensitivity, and its silencing results in modifications in ethylene levels, alterations in pigment accumulation, and a decrease in fruit firmness, reminiscent of the phenotypes of the tomato fruits under-expressing ARF2 paralogs (Hao et al., 2015; Breitel et al., 2016). Apart from the resemblance in the fruit phenotypes, a slight increase in accumulation of IAA levels in SlERF.D7 RNAi fruits, in conjunction with significant alterations in the expression of SlARF2A and SlARF2B in the silenced lines, indicates these genes to be the putative target of SlERF.D7. The presence of ERE elements in the promoters of SlARF2A and SlARF2B further supported our notion. The heterologous and in planta confirmation of SlERF.D7 binding to SlARF2A and SlARF2B promoters in yeast and N. benthamiana, respectively, validated this hypothesis. Consistent with the idea of SlARF2 being a direct target of SlERF.D7 in Y1H and electrophoretic mobility shift experiments, subsequent transactivation assays using GUS and GFP reporter systems in N. benthamiana evidently demonstrated a direct association between the two classes of transcription factors. Another key piece of evidence further substantiating the activation of ARF2 promoters by SlERF.D7 is obtained from the VIGS assay of single and chimeric knock-down of ARF2A/B in SlERF.D7 OE lines. The ripening phenotypes exhibited by SlARF2A, SlARF2B, and SlARF2A/B VIGS fruits are in agreement with the similarity of phenotypes observed in SlERF.D7 RNAi and SlARF2-silenced fruits (Hao et al., 2015; Breitel et al., 2016). Altogether, the data reveal that SlERF.D7 acts upstream of SlARF2 transcription factors and impacts fruit ripening via their transcriptional activation.
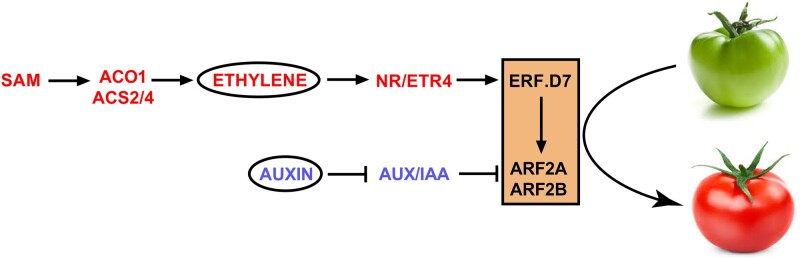
In summary, our results underpin a prototype in which the ripening-induced ERF, SlERF.D7, emerges as a critical positive regulator of fruit ripening. SlERF.D7 amalgamates auxin and ethylene signaling pathways via regulating the transcript accumulation of SlARF2 paralogs in tomato (Figure 12). Silencing of SlERF.D7 culminates in down-regulation of SlARF2A and SlARF2B genes, negatively impacting tomato fruit ripening. The SlERF.D7-ARF2A/B association seems critical for fine-tuning ethylene and auxins aspects, such as pigment accumulation and shelf-life during fruit ripening. Nonetheless, further research is warranted to delineate the molecular mechanisms underlying SlERF.D7 controlled ripening regulation and validate if SlERF.D7 directly activates the transcription of ripening master regulators such as RIN, NOR, and CNR by binding to their promoters. Besides identifying the transcription factor responsible for its activation during ripening, SlERF.D7 physical binding to ERE elements of the promoters of ARF2 paralogs and other transcriptionally affected genes in this study also remains to be ascertained.
Figure 12.
A general regulatory model depicting the role of SlERF.D7 in controlling tomato fruit ripening. SlERF.D7 functions in ethylene and auxin-dependent manner and directly activates the transcription of SlARF2A and SlARF2B. Thus, down-regulation of SlERF.D7 leads to severe impairment of ARF2 orthologs and causes a delayed fruit ripening phenotype. On the contrary, OE of SlERF.D7 promotes ripening traits by directly activating ARF2 orthologs. Further functional and physiological dissection reveals a positive connection between the mode of action of ARF2 orthologs and fruit ripening process. Taken together, SlERF.D7 emerges as an integrator of ethylene and auxin biosynthesis and signaling pathways, which work synergistically with SlARF2A or with both SlARF2A and SlARF2B to achieve the competency of the fruits to ripen. SAM, S-adenosyl methionine; NR, never-ripe; ETR4, ethylene receptor4; and AUX/IAA, auxin/indole-3-acetic acid.
Materials and methods
Plant materials and growth conditions
Tomato (S. lycopersicum L.cv Pusa Ruby) WT, 35S:RIN-RNAi transgenic lines in Pusa Ruby background generated in our lab (RIAI05 and RIAI06) (http://hdl.handle.net/10603/389794), Ailsa Crag WT and rin (accession no. LA1795, in an unknown background) and nor (Ailsa Craig background) fruit ripening defective mutants were grown under standard greenhouse conditions: 14-h-day/10-h-night cycle, 25°C/20°C day/night temperature, 60% relative air humidity, and 250 µmol m−2 s−1 intense luminosity. Fruit pericarp tissue samples were collected from different fruit development and ripening stages, as described earlier (Kumar et al., 2012a,b). For the measurement of time to ripen, flowers were tagged at anthesis. The number of days counted from anthesis to the appearance of the first symptoms of ripening was designated as the number of days taken to reach the Br stage. The MG stage was fixed one day before the Br stage. All fruit stages were harvested in three biological replicates (each replicate was represented by a pool of several fruits from different plants for each genotype used in the present study). Upon harvest, fruits were snap-frozen in liquid nitrogen to avoid injury.
Plasmid construction and VIGS assay
Tobacco rattle virus (TRV)-based pTRV1 and pTRV2 vectors were used for VIGS experiments (Liu et al., 2002). A 300-bp fragment of the coding region corresponding to SlARF2A and SlARF2B was retrieved by us using the VIGS tool of the SOL Genomics Network Database (https://vigs.solgenomics.net/). Each 300-bp fragment was then PCR amplified from S. lycopersicum cv Pusa Ruby cDNA and inserted in the pTRV2 vector using Xba1 and EcoR1 restriction sites for SlARF2A and Xho1 and Sac1 for SlARF2B. For double knock-down of SlARF2A and SlARF2B, a chimeric construct with fragments corresponding to both the genes was cloned and ligated into the same pTRV2 vector. The primers used for VIGS assay cloning are listed in the Supplemental Table S1. The empty pTRV vector was used as a control in the VIGS assays. The pTRV::ARF2A, pTRV::ARF2B, and pTRV::ARF2AB plasmids were verified by sequencing and mobilized into Agrobacterium tumefaciens strain GV3101. As described previously, VIGS was carried out on MG fruits (35DPA) (Fu et al., 2005).
Subcellular localization of SlERF.D7
The CDS of SlERF.D7 was cloned in frame with GFP-reported gene into pSITE-2CA vector (primers are listed in Supplemental Table S1). The empty pSITE-2CA vector was used as a control. SlERF.D7-pSITE-2CA and the control vectors were transferred to A. tumefaciens strain GV3101 and injected into 4-week-old N. benthamiana leaves as described previously (Martin et al., 2009). GFP fluorescence was observed and captured by a laser confocal microscope (Leica TCS SP8, Germany) after 48 h of infiltration, as described previously by us (Kumar et al., 2015).
In silico analysis of SlERF.D7, SlARF2A, and SlARF2B promoters
To identify putative cis-acting elements in the promoter sequences of SlERF.D7, SlARF2A, and SlARF2B, 2.5-kb upstream regions from the transcription start site of each gene were retrieved from the SOL Genomics Network database. After that, the sequences were scanned using the PLACE/signal search tool (http://www.dna.affrc.go.jp/PLACE/signalscan.html) to identify ethylene/auxin/fruit-specific/ripening-related cis-acting regulatory elements.
Phytohormones treatment
For ethylene treatment to fruits, tomato fruits were harvested at the MG stage of development and injected with a buffer solution containing 10 mM MES, pH 5.6, sorbitol (3% w/v), and 100 μM of Ethrel (2-chloroethylphosphonic acid, 40% solution, Sisco Research Laboratories Pvt. Ltd., India). Similarly, auxin treatment to the MG fruits was given by injecting a buffer solution with 10 mM MES, pH 5.6, and sorbitol (3% w/v) having 100 µM IAA. For inhibitors treatment, tomato fruits harvested at Br fruits were infiltrated with the abovementioned buffer containing 100 µM 1-MCP and 100 µM PCIB, respectively. Briefly, similar-sized tomato fruits were injected using a 1-mL syringe with a 0.5-mm needle and inserted 3–4 mm into the fruit tissue through the stylar apex. The infiltration solution was gently injected into the fruit until the solution ran off the stylar apex and the hydathodes at the tip of the sepals. Only completely infiltrated fruits were used in the experiments. Control fruits were treated with the corresponding buffers only. After the treatment, fruits were incubated in a culture room at 26°C, under a 16-h light/8-h dark cycle with a light intensity of 100 μmol m−2 s−1. After 24 h, the fruit pericarp was collected and frozen at −80°C until further use.
RNA isolation and RT-qPCR
Total RNA from the different tissues/organs/stages (cotyledon, root, leaf, stem, flower, and pericarp of fruits at IMG, MG, Br, Br + 3, Br + 5, and Br + 10) of tomato plants was isolated using RNeasy Plant Mini Kit (Qiagen, Germany) by following manufacturer’s instructions. A provision for on-column DNase treatment was provided in the kit. After the RNA isolation, 1-µg of total RNA for each sample was subjected to cDNA synthesis with high-capacity cDNA reverse transcription kit (Applied Biosystems, USA), according to the instructions provided in the manual. Gene-specific primers for RT-qPCR were designed using PRIMER EXPRESS version 2.0 (PE Applied Biosystems, USA) with default parameters. Further, 2× Brilliant III SYBR Green QPCR master mix (Agilent Technologies, USA) was used for a RT-qPCR reaction carried out in a Stratagene Mx3005P qPCR machine (Agilent Technologies, USA). Three independent RNA isolations along with three technical replicates were used for mRNA quantification. The expression values of genes were normalized using GAPDH and ACTIN gene expression values. Relative expression values were calculated by employing the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Construction of SlERF.D7 OE and silencing vectors and tomato transformation
To generate the RIP1::SlERF.D7 OE transgenic plants, we amplified the full-length CDS and first cloned in pGEM-T Easy vector (Promega, USA) and finally mobilized into a binary vector, pCAMBIA2300-RIP1-NOSt, where we have already cloned the RIP1 promoter. Similarly, to generate the RIP1::SlERF.D7 RNAi transgenic plants, a 380-bp unique CDS of SlERF.D7 was PCR amplified and cloned first in pUC19. The same fragment in sense and antisense orientation was cloned in juxtaposition, where an intronic sequence separated the two fragments. The assembled construct was finally mobilized into another binary vector pBI121-RIP1-NOSt. The cloning was confirmed by PCR, restriction digestion, and Sanger’s sequencing. The sequence-confirmed OE and silencing (RNAi) plasmids were mobilized from E scherichia coli into A. tumefaciens strain AGL1. After confirmation of this mobilization, the transformed Agrobacterium cultures were used to generate stable transgenic RIP1::SlERF.D7 OE and RNAi lines in Pusa Ruby, as described previously (Maligeppagol et al., 2011). Primers used in constructing RIP1::SlERF.D7 OE and RNAi constructs are listed in Supplemental Table S1. The OE and RNAi lines were grown and analyzed for their morphological, biochemical, molecular, and physiological characterization in the T2 generation. The segregation analysis of kanamycin resistance in T2 progeny of SlERF.D7 OE and SlERF.D7 RNAi transgenic tomato plants was done to obtain homozygous lines. Final experiments were carried out using homozygous lines from T2 or later generations.
Fruit firmness measurement
The assessment of fruit firmness was carried out using TA.XT Plus Texture Analyzer (Stable Micro Systems, UK). Fruits that had undergone VIGS at the red ripe (Br + 7) stage were subjected to a puncture test with a 2-mm needle probe, and the force and distance measurements were recorded. Fifteen fruits from each transgenic and tissue culture generated WT genotype were used in the study. Fruit firmness calculated is equivalent to the amount of force applied to penetrate the surface of the fruit. For fruit shelf-life measurement, 10 fruits from the WT and transgenic lines were harvested at the MG stage and kept in the dark at room temperature, and photographed at regular intervals during the experiment.
Ethylene measurement
Fruits at each developmental stage were harvested and placed in open 120-mL jars for 2 h to minimize the effect of wound ethylene caused by picking. Jars were then sealed and incubated at room temperature for 35 min, and 1 mL of headspace gas was injected into an Agilent 7820A gas chromatograph equipped with a flame ionization detector (Agilent, Santa Clara, California, USA). Samples were compared with reagent-grade ethylene standards of known concentration and normalized for fruit weight. Fruits at each developmental stage were harvested and placed in open 120-mL jars for 2 h to minimize the effect of wound ethylene caused by picking. Jars were then sealed and incubated at room temperature for 35 min, and 1 mL of headspace gas was injected into an Agilent 7820A gas chromatograph equipped with a flame ionization detector (Agilent). Samples were compared with reagent-grade ethylene standards of known concentration and normalized for fruit weight. Fruits at each developmental stage were harvested and placed in open 120-mL jars for 2 h to minimize the effect of wound ethylene caused by picking. Jars were then sealed and incubated at room temperature for 35 min, and 1 mL of headspace gas was injected into an Agilent 7820A gas chromatograph equipped with a flame ionization detector (Agilent). Samples were compared with reagent-grade ethylene standards of known concentration and normalized for fruit weight.
Fruits at each developmental stage were harvested and placed in open 250-mL jars for 2 h to minimize the effect of wound-induced ethylene caused by the harvesting of the fruit. Jars were then sealed and incubated at room temperature for 4 h, and 1 mL of headspace gas was injected into Shimadzu QP-2010 Plus with Thermal Desorption System TD 20 (Shimadzu, Japan). Samples were compared with reagent-grade ethylene standards of known concentration and normalized for fruit weight. Ethylene in the headspace gas was measured thrice for each sample, with at least three biological samples for each ripening stage.
Estimation of fruit pigments
Lycopene and β-carotenoid extractions for HPLC experiments were performed as described previously (Fantini et al., 2013). Briefly, 150 mg of ground lyophilized tomato fruit powder was extracted with chloroform and methanol (2:1 v/v); subsequently, 1 volume of 50 mM Tris buffer (pH 7.5, containing 1 M NaCl) was added, and followed by the incubation of samples on ice for 20 min. After centrifugation (15,000 g for 10 min at 4°C), the organic phase was collected and the aqueous phase was re-extracted with the same amount of chloroform. The combined organic phases were then dried by centrifugal evaporation and resuspended in 100 µL of ethyl acetate. A final volume of 20 µL was injected into a C-18 column in HPLC (Shimadzu, Japan) analysis. For each genotype, at least five independent extractions were performed.
Estimation of titrable acids
The amount of titrable acids (TA) was determined by titrating the fruit homogenates against 0.1 N NaOH solution using phenolphthalein as an indicator to the endpoint at pH 8.1 (Singh and Pal, 2008). The TAs were measured by using the following equation:
Determination of total soluble solids
The quantification of total soluble acids (TSSs) was done by measuring the refractive index of tomato juice through a portable refractometer (Erma Handheld Refractometer, India). The refractive index of water was initially calculated with the device to serve as zero error. A drop of the sample (tomato juice) of all the test samples was placed individually on the measuring surface beneath the Viewpoint Illuminator. Through the eyepiece, the readings were recorded at the point where the contrast line crossed the scale. The results have been expressed in degrees Brix (percentage of TSS in solution at 20°C).
The ripening index for all the samples was calculated using the following formula:
Measurement of IAA in fruit pericarp tissue
Extraction and purification of IAA were carried out as previously reported with slight modifications (Edlund et al., 1995). Frozen samples were ground in liquid nitrogen, 50 mg fresh weight of the powdered sample was mixed with 1 mL of 80% methanol containing 1% acetic acid (v/v), and 2 ng of [13C6]-IAA was added as an internal standard (Cambridge Isotope Laboratories, Andover, Massachusetts, USA). The sample was extracted for 2 h at 4°C under continuous shaking. After centrifugation (10,000 g, 5 min), the supernatant was collected and concentrated in a vacuum. The sample was resuspended in 1 mL 0.01 M HCl slurried for 10 min at 4°C under continuous shaking with 15 mg AmberLite XAD-7HP (Organo, Tokyo, Japan). After removing the supernatant, the XAD-7HP was washed twice with 1% acetic acid. Samples were then extracted with CH2Cl2 (once with 400 µL and twice with 200 µL), and the combined CH2Cl2 fraction was passed through a 0.2-µm filter. After concentration in a vacuum, the sample was analyzed by GC-MS. Five independent samples were extracted and analyzed.
Y1H assay
For Y1H experiments, 1.5 kb long promoter sequences of SlARF2A and SlARF2B were amplified using tomato (S. lycopersicum L.cv Pusa Ruby) genomic DNA. The amplified products were cloned into the yeast expression vector pHIS2.1 and co-transformed with the pGAD-T7-SlERF.D7 into yeast strain Y187. The binding of RIN to the promoter of LeACS2 was taken as a positive control for this experiment. For negative control, the interaction of SlARF3 promoter with SlERF.D7 was used. The DNA–protein interaction was validated by transformant growth assays on SD/-Leu/-Trp/-His plates supplemented with 50 mM of 3-AT. Primers used in this section are listed in the Supplemental Table S1.
Transactivation of SlARF2A and SlARF2B promoters in N. benthamiana
To verify the DNA–protein interaction in planta, 1.5 kb upstream regions of SlARF2A and SlARF2B were used to drive the expression of GUS and GFP, and were designated as reporter vectors. The effector vector was constructed by cloning the full open reading frame (ORF) of SlERF.D7 in the binary vector pBI121 driven by CaMV35S promoter. As previously described, both reporter and effector constructs were co-transformed into 4-week-old N. benthamiana leaves. GUS histochemical staining assay and GFP fluorescence measurement assay using flow cytometry (FACS calibur microflow cytometer) (Becton Dickinson, Franklin Lakes, New Jersey, USA) done after 48 h of infiltrations. The excitation for GFP was carried out by argon laser and the fluorescence was detected using a bandpass filter (530/30 nm) in the FL1 channel. The NightSHADE LB 985 (Berthold Technologies, USA) in vivo plant imaging system was employed to detect fluorescent signals with 5 s exposure time. Data were analyzed by the IndiGO software. The average fluorescence signal for each sample was collected (cps, count per second). It was normalized using the N. benthamiana leaves transformed with a reporter vector combined with the vector used as effector but lacking the ERF or ARF CDSs. Three independent replicates were used for each analysis. Positive and negative controls were the same as reported earlier for the Y1H assay. Primers used in this experiment are listed in the Supplemental Table S1.
Electrophoretic mobility shift assay
The full-length SlERF.D7 CDS was cloned in frame into pET-28a (to fuse in frame with 6× Histidine tag) for heterologous protein expression. The fusion protein construct was expressed in BL21 strain of E. coli. Recombinant SlERF.D7-6XHIS protein was purified by Ni2+ gravity flow chromatography according to the manufacturer’s protocol (Ni-NTA Agarose, Qiagen) as described. For EMSA, the 50-bp probe covering the ERE element (AGCCGCC) derived from SlARF2A and SlARF2B promoter was labeled with digoxigenin as per the manufacturer’s protocol using DIG Oligonucleotide 3′-End Labeling Kit (Roche Diagnostics). The same unlabeled DNA fragment was used as a competitor. The binding reactions were performed at room temperature in binding buffer (10 mM Tris [pH7.5], 50 mM KCl, 1 mM DTT, 2.5% glycerol, 5 mM MgCl2, 0.5 mM EDTA, and 50 ng/ml poly [dI-dC]) containing 1 µg purified SlERF.D7-6XHIS fusion protein and 5 ng probes. The reaction products were analyzed on 6% (w/v) native polyacrylamide gel electrophoresis. The products then transferred from the gel to Hybond N+ Nylon membrane (Amersham Biosciences) and detected using DIG Nucleic Acid Detection Kit (Roche Diagnostics) according to manufacturer’s protocol.
GUS histochemical and activity assays
Histochemical staining of GUS in N. benthamiana leaves was performed as described previously. Briefly, N. benthamiana infiltrated leaves were harvested and incubated in a substrate solution (50 mM sodium phosphate, pH 7.5, 0.5 mM K3Fe(CN)6, 0.5 mMK4Fe(CN)6, 1 mM X-gluc [5-bromo-4-chloro-3-indolyl-b-d-glucuronide]) at 37°C overnight. Tissues were then cleared in 70% ethanol overnight. For the fluorometric GUS assay, the infiltrated N. benthamiana leaf discs were homogenized in 1 mL of extraction buffer (50 mM NaH2PO4, pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 0.1% [w/v] sarcosyl [w/v], and 10 mM β-mercaptoethanol). The homogenate was centrifuged for 10 min at 12,000 g at 4°C, and 100 µL of the supernatant was mixed with 900 µL of assay buffer (1 mM 4-methylumbelliferyl-d-glucuronide in the extraction buffer). The reaction mixture was incubated at 37°C for 1 h and eventually stopped by adding a stop buffer (0.2 M sodium acetate). GUS activity was normalized to protein concentration in each crude extract and was calculated as pmole or nmole of 4-methylumbelliferone (4-MU) min−1 mg−1 protein. The Bradford method assessed the protein content using bovine serum albumin as a standard.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: Sl-ERF.A3 (Solyc06g063070), SlERF.B1 (Solyc05g052040), SlERF.B2 (Solyc02g077360), SlERF.B3 (Solyc05g052030), SlERF.C1 (Solyc05g051200), SlERF.D1 (Solyc04g051360), SlERF.D2 (Solyc12g056590), SlERF.D3 (Solyc01g108240), SlERF.D4 (Solyc10g050970), SlERF.D5 (Solyc04g012050), SlERF.D6 (Solyc04g071770), SlERF.D7 (Solyc03g118190), SlERF.D8 (Solyc12g042210), SlERF.D9 (Solyc06g068830), Sl-ERF.E1 (Solyc09g075420), Sl-ERF.E2 (Solyc09g089930), SlERF.E2 (Solyc06g063070), SlERF.E4 (Solyc01g065980), PSY1 (Solyc03g031860), PDS (Solyc03g123760), ZDS (Solyc01g097810), βLCY1 (Solyc04g040190), βLCY2 (Solyc10g079480), CYCβ (Solyc06g074240), ACS2 (Solyc01g095080), ACS4 (Solyc05g050010), ACO1 (Solyc07g049530), E4 (Solyc03g111720), E8 (Solyc09g089580), PG2a (Solyc10g080210), RIN (Solyc05g012020), CNR (Solyc02g077920), NOR (Solyc10g006880), TAGL1 (Solyc07g055920), AP2a (Solyc03g044300), EIN2 (Solyc09g007870), EIL2 (Solyc01g009170), EIL3 (Solyc01g096810), ETR2 (Solyc07g056580), ETR3 (Solyc09g075440), ETR4 (Solyc06g053710), ETR5 (Solyc11g006180), FUL1 (Solyc06g069430), FUL2 (Solyc03g114830), ACO2 (Solyc12g005940), ACO3 (Solyc07g049550), ACO4 (Solyc02g081190), SAUR (Solyc09g007970.1.1), ARF2A (Solyc03g118290), and ARF2B (Solyc12g042070).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Table S1. List of primers used in this study.
Supplemental Table S2. Segregation analysis of kanamycin resistance in T2 progeny of SlERF.D7 OE and SlERF.D7 RNAi transgenic tomato plants.
Supplemental Figure S1. DAPI staining of SlERF.D7 in N. benthamiana.
Supplemental Figure S2. Kanamycin-based PCR screening of SlERF.D7 OE and silencing lines.
Supplemental Figure S3. Phenotypic time-line of ethrel-treated and non-treated control fruits from MG to red ripe stage.
Supplemental Figure S4. The expression of ERF genes in WT and SlERF.D7 OE and SlERF.D7 RNAi plants.
Supplemental Figure S5. The expression of ERF.D clade genes in WT and SlERF.D7 OE and SlERF.D7 RNAi plants.
Supplemental Figure S6. The expression of SlARF in WT and SlERF.D7 OE and SlERF.D7 RNAi plants.
Supplementary Material
Acknowledgements
The authors acknowledge the Department of Science and Technology, India, for the Purse Grant. The SAP Grant of the University Grants Commission and FIST grant of DST, India, to the Department of Plant and Molecular Biology for infrastructure support are also acknowledged.
Funding
This work is financially supported by grants received from the Department of Biotechnology (DBT), Government of India. R.K. acknowledges the Department of Science and Technology, India, for the INSPIRE-Faculty (IF-LSPA-15), Department of Biotechnology, Government of India (BT/PR31630/AGIII/103/1119/2019) and MHRD-IoE ([RC1-20-018] and [F11/9/2019-U3(A)]) grants. A.K.S. acknowledges DBT grant BT/PR6983/PBD/16/1007/2012.
Conflict of interest statement. There are no conflicts of interest to be declared.
Contributor Information
Priya Gambhir, Department of Plant Molecular Biology, University of Delhi South Campus, New Delhi 110021, India.
Vijendra Singh, Department of Plant Molecular Biology, University of Delhi South Campus, New Delhi 110021, India.
Adwaita Parida, Department of Plant Molecular Biology, University of Delhi South Campus, New Delhi 110021, India.
Utkarsh Raghuvanshi, Department of Plant Molecular Biology, University of Delhi South Campus, New Delhi 110021, India.
Rahul Kumar, Department of Plant Sciences, School of Life Sciences, University of Hyderabad, Hyderabad 500046, India.
Arun Kumar Sharma, Department of Plant Molecular Biology, University of Delhi South Campus, New Delhi 110021, India.
These authors contributed equally (V.S. and A.P.)
A.K.S. and R.K.: conceptualization. P.G., A.P., U.R., R.K., and V.S.: investigation, validation, methodology, and formal analysis. P.G.: writing original draft. A.K.S. and R.K.: reviewing, editing, and supervision.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Arun K. Sharma (arun.sharma@south.du.ac.in).
References
- Agarwal P, Kumar R, Pareek A, Sharma A (2017) Fruit preferential activity of a tomato RPP1 gene promoter in transgenic tomato and Arabidopsis. Mol Genet Genomics 292: 145–156 [DOI] [PubMed] [Google Scholar]
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ (2005) Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17: 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander L, Grierson D (2002) Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J Exp Bot 53: 2039–2055 [DOI] [PubMed] [Google Scholar]
- Barry CS, Giovannoni JJ (2007) Ethylene and fruit ripening. J Plant Growth Regul 26: 143 [Google Scholar]
- Barry CS, Llop-Tous MI, Grierson D (2000) The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol 123: 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M, Karlova R, Ballester AR, Tikunov YM, Bovy AG, Wolters-Arts M, Rossetto P de B, Angenent GC, de Maagd RA (2012) The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24: 4437–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Böttcher C, Keyzers RA, Boss PK, Davies C (2010) Sequestration of auxin by the indole-3-acetic acid-amido synthetase GH3-1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J Exp Bot 61: 3615–3625 [DOI] [PubMed] [Google Scholar]
- Breitel DA, Chappell-Maor L, Meir S, Panizel I, Puig CP, Hao Y, Yifhar T, Yasuor H, Zouine M, Bouzayen M. et al. (2016) AUXIN RESPONSE FACTOR 2 intersects hormonal signals in the regulation of tomato fruit ripening. PLoS Genet 12: e1005903–e1005903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X-T, Xu P, Zhao P-X, Liu R, Yu L-H, Xiang C-B (2014) Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat Commun 5: 5833. [DOI] [PubMed] [Google Scholar]
- Chung M-Y, Vrebalov J, Alba R, Lee J, McQuinn R, Chung J-D, Klein P, Giovannoni J (2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64: 936–947 [DOI] [PubMed] [Google Scholar]
- Edlund A, Eklof S, Sundberg B, Moritz T, Sandberg G (1995) A microscale technique for gas chromatography-mass spectrometry measurements of picogram amounts of indole-3-acetic acid in plant tissues. Plant Physiol 108: 1043–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini E, Falcone G, Frusciante S, Giliberto L, Giuliano G (2013) Dissection of tomato lycopene biosynthesis through virus-induced gene silencing. Plant Physiol 163: 986–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Truesdale MR, Bird CR, Schuch W, Bramley PM (1994) Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression). Plant Physiol 105: 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fray RG, Grierson D (1993) Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol Biol 22: 589–602 [DOI] [PubMed] [Google Scholar]
- Fu D-Q, Zhu B-Z, Zhu H-L, Jiang W-B, Luo Y-B (2005) Virus-induced gene silencing in tomato fruit. Plant J 43: 299–308 [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Ito Y (2011) Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biol 11: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Shima Y, Ito Y (2013) A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25: 371–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W (1993) Fruits: a developmental perspective. Plant Cell 5: 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ (2004) Genetic regulation of fruit development and ripening. Plant Cell 16(Suppl): S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Bartley GE, Scolnik PA (1993) Regulation of carotenoid biosynthesis during tomato development. Plant Cell 5: 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Given NK, Venis MA, Gierson D (1988) Hormonal regulation of ripening in the strawberry, a non-climacteric fruit. Planta 174: 402–406 [DOI] [PubMed] [Google Scholar]
- Grierson D, Schuch W, Bevan MW, Harrison BD, Leaver CJ (1993) Control of ripening. Phil Trans R Soc Lond Ser B Biol Sci 342: 241–250 [Google Scholar]
- Hall LN, Tucker GA, Smith CJS, Watson CF, Seymour GB, Bundick Y, Boniwell YM, Fletcher JD, Ray JA, Schuch W. et al. (1993) Antisense inhibition of pectin esterase gene expression in transgenic tomatoes. Plant J 3: 121–129 [Google Scholar]
- Hamilton AJ, Lycett GW, Grierson D (1990) Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plants. Nature 346: 284–287 [Google Scholar]
- Hao Y, Hu G, Breitel D, Liu M, Mila I, Frasse P, Fu Y, Aharoni A, Bouzayen M, Zouine M (2015) Auxin response factor SlARF2 is an essential component of the regulatory mechanism controlling fruit ripening in tomato. PLoS Genet 11: e1005649–e1005649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A (2009) TOMATO AGAMOUS-LIKE1 is a component of the fruit ripening regulatory network. Plant J 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T (2008) DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J 55: 212–223 [DOI] [PubMed] [Google Scholar]
- Ivanchenko MG, Muday GK, Dubrovsky JG (2008) Ethylene–auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana. Plant J 55: 335–347 [DOI] [PubMed] [Google Scholar]
- Jones B, Frasse P, Olmos E, Zegzouti H, Li ZG, Latché A, Pech JC, Bouzayen M (2002) Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant J 32: 603–613 [DOI] [PubMed] [Google Scholar]
- Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA (2011) Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23: 923–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Agarwal P, Pareek A, Tyagi AK, Sharma AK (2015) Genomic survey, gene expression, and interaction analysis suggest diverse roles of ARF and Aux/IAA proteins in Solanaceae. Plant Mol Biol Rep 33: 1552–1572 [Google Scholar]
- Kumar R, Agarwal P, Tyagi AK, Sharma AK (2012a) Genome-wide investigation and expression analysis suggest diverse roles of auxin-responsive GH3 genes during development and response to different stimuli in tomato (Solanum lycopersicum). Mol Genet Genomics 287: 221–235 [DOI] [PubMed] [Google Scholar]
- Kumar R, Sharma MK, Kapoor S, Tyagi AK, Sharma AK (2012b) Transcriptome analysis of rin mutant fruit and in silico analysis of promoters of differentially regulated genes provides insight into LeMADS-RIN-regulated ethylene-dependent as well as ethylene-independent aspects of ripening in tomato. Mol Genet Genomics 287: 189–203 [DOI] [PubMed] [Google Scholar]
- Kumar V, Irfan M, Ghosh S, Chakraborty N, Chakraborty S, Datta A (2016) Fruit ripening mutants reveal cell metabolism and redox state during ripening. Protoplasma 253: 581–594 [DOI] [PubMed] [Google Scholar]
- Kumar R, Khurana A, Sharma AK (2014) Role of plant hormones and their interplay in development and ripening of fleshy fruits. J Exp Bot 65: 4561–4575 [DOI] [PubMed] [Google Scholar]
- Lee JM, Joung J-G, McQuinn R, Chung M-Y, Fei Z, Tieman D, Klee H, Giovannoni J (2012) Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J 70: 191–204 [DOI] [PubMed] [Google Scholar]
- Leseberg CH, Eissler CL, Wang X, Johns MA, Duvall MR, Mao L (2008) Interaction study of MADS-domain proteins in tomato. J Exp Bot 59: 2253–2265 [DOI] [PubMed] [Google Scholar]
- Lewis DR, Negi S, Sukumar P, Muday GK (2011) Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development 138: 3485–3495 [DOI] [PubMed] [Google Scholar]
- Li S, Chen K, Grierson D (2019) A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling fleshy fruit ripening. New Phytol 221: 1724–1741 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhu B, Xu W, Zhu H, Chen A, Xie Y, Shao Y, Luo Y (2007) LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Rep 26: 1999–2008 [DOI] [PubMed] [Google Scholar]
- Lincoln JE, Campbell AD, Oetiker J, Rottmann WH, Oeller PW, Shen NF, Theologis A (1993) LE-ACS4, a fruit ripening and wound-induced 1-aminocyclopropane-1-carboxylate synthase gene of tomato (Lycopersicon esculentum). Expression in Escherichia coli, structural characterization, expression characteristics, and phylogenetic analysis. J Biol Chem 268: 19422–19430 [PubMed] [Google Scholar]
- Liu M, Chen Y, Chen Y, Shin J-H, Mila I, Audran C, Zouine M, Pirrello J, Bouzayen M (2018) The tomato ethylene response factor Sl-ERF.B3 integrates ethylene and auxin signaling via direct regulation of Sl-Aux/IAA27. New Phytol 219: 631–640 [DOI] [PubMed] [Google Scholar]
- Liu M, Diretto G, Pirrello J, Roustan J-P, Li Z, Giuliano G, Regad F, Bouzayen M (2014) The chimeric repressor version of an ethylene response factor (ERF) family member, Sl-ERF.B3, shows contrasting effects on tomato fruit ripening. New Phytol 203: 206–218 [DOI] [PubMed] [Google Scholar]
- Liu M, Gomes BL, Mila I, Purgatto E, Peres LEP, Frasse P, Maza E, Zouine M, Roustan J-P, Bouzayen M. et al. (2016) Comprehensive profiling of ethylene response factor expression identifies ripening-associated ERF genes and their link to key regulators of fruit ripening in tomato. Plant Physiol 170: 1732–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Pirrello J, Chervin C, Roustan J-P, Bouzayen M (2015) Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. Plant Physiol 169: 2380–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR (1998) EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev 12: 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maligeppagol M, Navale P, Chandra S, Asokan R, Nagesha SN (2011) An improved protocol for rapid and efficient Agrobacterium mediated transformation of tomato (Solanum Iycopersicum L.). J Appl Horticult 13: 3–7 [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Mao J-L, Miao Z-Q, Wang Z, Yu L-H, Cai X-T, Xiang C-B (2016) Arabidopsis ERF1 mediates cross-talk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PLoS Genet 12: e1005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ (2011) The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol 157: 1568–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162 [DOI] [PubMed] [Google Scholar]
- Meli VS, Ghosh S, Prabha TN, Chakraborty N, Chakraborty S, Datta A (2010) Enhancement of fruit shelf life by suppressing N-glycan processing enzymes. Proc Natl Acad Sci USA 107: 2413–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday GK, Rahman A, Binder BM (2012) Auxin and ethylene: collaborators or competitors? Trends Plant Sci 17: 181–195 [DOI] [PubMed] [Google Scholar]
- Nakatsuka A, Murachi S, Okunishi H, Shiomi S, Nakano R, Kubo Y, Inaba A (1998) Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening1. Plant Physiol 118: 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi S, Ivanchenko MG, Muday GK (2008) Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. Plant J Cell Mol Biol 55: 175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi S, Sukumar P, Liu X, Cohen JD, Muday GK (2010) Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. Plant J 61: 3–15. [DOI] [PubMed] [Google Scholar]
- Oeller PW, Lu MW, Taylor LP, Pike DA, Theologis A (1991) Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254: 437–439 [DOI] [PubMed] [Google Scholar]
- Osorio S, Alba R, Damasceno CMB, Lopez-Casado G, Lohse M, Zanor MI, Tohge T, Usadel B, Rose JK, Fei Z, et al. (2011) Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol 157: 405–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecker I, Gabbay R, Cunningham FX, Hirschberg J (1996) Cloning and characterization of the cDNA for lycopene β-cyclase from tomato reveals decrease in its expression during fruit ripening. Plant Mol Biol 30: 807–819 [DOI] [PubMed] [Google Scholar]
- Pickett FB, Wilson AK, Estelle M (1990) The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance 1 2. Plant Physiol 94: 1462–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrello J, Prasad BN, Zhang W, Chen K, Mila I, Zouine M, Latché A, Pech JC, Ohme-Takagi M, Regad F. et al. (2012) Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene. BMC Plant Biol 12: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Amakawa T, Goto N, Tsurumi S (2001) Auxin is a positive regulator for ethylene-mediated response in the growth of Arabidopsis roots. Plant Cell Physiol 42: 301–307 [DOI] [PubMed] [Google Scholar]
- Ronen G, Carmel-Goren L, Zamir D, Hirschberg J (2000) An alternative pathway to beta-carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. Proc Natl Acad Sci USA 97: 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E (2007) Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M, Chervin C, Mila I, Hao Y, Roustan JP, Benichou M, Gibon Y, Biais B, Maury P, Latché A. et al. (2013) SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol 161: 1362–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer RJ, Ireland HS, Ross JJ, Ling TJ, David KM (2013) SEPALLATA1/2-suppressed mature apples have low ethylene, high auxin and reduced transcription of ripening-related genes. AoB Plants 5: pls047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MK, Kumar R, Solanke AU, Sharma R, Tyagi AK, Sharma AK (2010) Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato. Mol Genet Genomics 284: 455–475 [DOI] [PubMed] [Google Scholar]
- Singh SP, Pal RK (2008) Controlled atmosphere storage of guava (Psidium guajava L.) fruit. Postharvest Biol Technol 47: 296–306 [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sravankumar T, Akash Naik N, Kumar R (2018) A ripening-induced SlGH3-2 gene regulates fruit ripening via adjusting auxin-ethylene levels in tomato (Solanum lycopersicum L.). Plant Mol Biol 98: 455–469 [DOI] [PubMed] [Google Scholar]
- Srivastava R, Kumar R (2019) The expanding roles of APETALA2/ethylene responsive factors and their potential applications in crop improvement. Brief Funct Genomics 18: 240–254 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM (2005) A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17: 2230–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Diretto G, Purgatto E, Danoun S, Zouine M, Li Z, Roustan J-P, Bouzayen M, Giuliano G, Chervin C (2015) Carotenoid accumulation during tomato fruit ripening is modulated by the auxin–ethylene balance. BMC Plant Biol 15: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Parry G, Graham N, Allen T, Bennett M (2002) Auxin cross-talk: integration of signalling pathways to control plant development. Plant Mol Biol 49: 409–424 [DOI] [PubMed] [Google Scholar]
- Tieman DM, Taylor MG, Ciardi JA, Klee HJ (2000) The tomato ethylene receptors NR and LeETR4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proc Natl Acad Sci USA 97: 5663–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainotti L, Tadiello A, Casadoro G (2007) The involvement of auxin in the ripening of climacteric fruits comes of age: the hormone plays a role of its own and has an intense interplay with ethylene in ripening peaches. J Exp Bot 58: 3299–3308 [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (Rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJM, McQuinn R, Chung M, Poole M, Rose J, Seymour G, Grandillo S, Giovannoni J. et al. (2009) Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Yin X, Chen K (2016) Roles of APETALA2/ethylene-response factors in regulation of fruit quality. Crit Rev Plant Sci 35: 120–130 [Google Scholar]
- Yang L, Huang W, Xiong F, Xian Z, Su D, Ren M, Li Z (2017) Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol J 15: 1544–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharah SS, Singh Z, Symons GM, Reid JB (2012) Role of brassinosteroids, ethylene, abscisic acid, and indole-3-acetic acid in mango fruit ripening. J Plant Growth Regul 31: 363–372 [Google Scholar]
- Zhong S, Fei Z, Chen Y-R, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J. et al. (2013) Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol 31: 154–159 [DOI] [PubMed] [Google Scholar]
- Zouine M, Fu Y, Chateigner-Boutin A-L, Mila I, Frasse P, Wang H, Audran C, Roustan J-P, Bouzayen M (2014) Characterization of the tomato ARF gene family uncovers a multi-levels post-transcriptional regulation including alternative splicing. PLoS One 9: e84203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.