Abstract
A plant’s oxygen supply can vary from normal (normoxia) to total depletion (anoxia). Tolerance to anoxia is relevant to wetland species, rice (Oryza sativa) cultivation, and submergence tolerance of crops. Decoding and transmitting calcium (Ca) signals may be an important component to anoxia tolerance; however, the contribution of intracellular Ca transporters to this process is poorly understood. Four functional cation/proton exchangers (CAX1–4) in Arabidopsis (Arabidopsis thaliana) help regulate Ca homeostasis around the vacuole. Our results demonstrate that cax1 mutants are more tolerant to both anoxic conditions and submergence. Using phenotypic measurements, RNA-sequencing, and proteomic approaches, we identified cax1-mediated anoxia changes that phenocopy changes present in anoxia-tolerant crops: altered metabolic processes, diminished reactive oxygen species production post anoxia, and altered hormone signaling. Comparing wild-type and cax1 expressing genetically encoded Ca indicators demonstrated altered cytosolic Ca signals in cax1 during reoxygenation. Anoxia-induced Ca signals around the plant vacuole are involved in the control of numerous signaling events related to adaptation to low oxygen stress. This work suggests that cax1 anoxia response pathway could be engineered to circumvent the adverse effects of flooding that impair production agriculture.
Plants lacking a vacuolar H+/Ca transporter are more tolerant to anoxia and submergence.
Introduction
Plants are frequently exposed to limited oxygen availability, influencing plant growth and, in an agricultural context, plant productivity (Loreti et al., 2016; Pedersen et al., 2017; Schmidt et al., 2018). The lack of oxygen (anoxia) reduces plant energy production (Rawyler et al., 2002). To survive and limit post-anoxic injuries, plants often regulate the flow of the little energy produced under stress conditions to essential processes (Blokhina et al., 2001; Virolainen et al., 2002; Huang et al., 2008; Atwell et al., 2015). In a natural context, submergence leads to multiple abiotic stress-related constraints, including limited light availability and the occurrence of oxidative stress either during or after the period of hypoxic conditions (Sasidharan et al., 2018). Plants respond to submergence by responding to ethylene, which accumulates in plant tissues because of the lower underwater gas diffusion rate. The perception and response to cellular oxygen (O2) levels per se is mediated by a subset of ethylene response factors (ERFs) belonging to the group VII of this large class of transcription factors (ERF-VII; Gibbs et al., 2015). ERF-VIIs induce a set of genes which are required to adapt plant metabolism and growth during low O2 conditions (Paul et al., 2016). Calcium (Ca) signaling is another, less studied, component of plant responses to hypoxia (Igamberdiev and Hill, 2018; Huang et al., 2022). Ca signaling was proposed to control the induction of hypoxia-responsive genes (HRGs) and this would imply a cross-talk of this signaling pathway with the ERF-VII-dependent regulation of HRGs (Subbaiah et al., 1994), although this is not yet been demonstrated. In response to changes in O2 levels, Ca signaling differs among plants, with marked differences between Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) in the spatiotemporal parameters of the observed Ca signatures (Bailey-Serres and Chang, 2005; Behera et al., 2015). Ca signaling differences appear to precede gene expression differences and metabolic fluxes, although precisely how Ca is triggering downstream events remains unclear (Schmidt et al., 2018). Conceptualizing signaling events involved in anoxia perception and tolerance might be useful to improve plant performance both during and after anoxic conditions.
Ca is at the nexus between many facets of plant biology (Ali et al., 2007; Ma and Berkowitz, 2007; Choi et al., 2017; Costa et al., 2018; Demidchik et al., 2018). Ca transporters on various membranes help to orchestrate responses to stresses such as changes in temperature and anoxia (Catalá et al., 2003; Wang et al., 2016; Choi et al., 2017; Costa et al., 2018; Weigand et al., 2021). Ca pumps and exchangers are found in multiple membrane systems, including the endoplasmic reticulum (ER), vacuole, and plasma membrane (PM; Blumwald and Poole, 1986). Ca pumps are considered low-capacity, high-affinity efflux systems, but within this broad group of transporters, H+/Ca eXchangers (CAXs) are distinct because they have high capacity and low affinity (Kong et al., 2020; Tian et al., 2020). While both Ca pumps and CAXs remove Ca from the cytoplasm, their individual contributions to the control of the magnitude or duration of different stimulus-specific Ca signals remains enigmatic.
There are six CAXs in Arabidopsis, but only CAX1–4 are expressed and appear to be predominately localized to the vacuole (Manohar et al., 2011). CAX2 and CAX4 are expressed at lower levels in plants, and most of the research on Ca signaling and homeostasis in plants has focused on the more highly expressed CAX1 and CAX3 (29). CAX1 is expressed in aerial tissue and is the dominant mechanism for Ca accumulation in the leaf (Conn et al., 2011; Whitt et al., 2020); CAX3 is primarily expressed in the roots (Hocking et al., 2017). The absence of CAX1 induces ectopic expression of other CAXs, which is often sufficient to maintain mesophyll cellular uptake of Ca from the apoplast (Cheng et al., 2005). The numerous cax1 lines characterized have subtle phenotypes throughout their life cycle and are virtually indistinguishable from controls in normal growth conditions (Manohar et al., 2011). These subtle phenotypes could be due to the interplay between CAX transporters and other transporters; deletions in CAX1 reduce vacuolar H+/Ca antiport activity by 50%, decrease vacuolar V-type proton ATPase activity by 40% and increase vacuolar Ca-ATPase activity by 36% (Cheng et al., 2003).
Plant responses to hypoxia and anoxia have been extensively studied (Atwell et al., 2015; Voesenek and Bailey-Serres, 2013; Voesenek et al., 2016); however, less is known about the contribution of Ca signaling processes governing a plant’s tolerance to low oxygen. Traditional approaches to study these events include differential gene expression studies and comparative genomic analysis (Yeung et al., 2018). Here, our analysis focuses on CAX1, a gene that is neither highly expressed during oxygen stress conditions nor thought to be modified in plants adapted to oxygen limitations. We demonstrate that cax1 plants are extremely tolerant to anoxia conditions and submergence. We established that during post anoxia, cax1 showed reduced reactive oxygen species (ROS) production; additionally, transcriptomic and proteomic analysis revealed cax1 plants displayed apparent changes in transcripts and proteins for Ca transport and numerous signaling pathways. We also investigated how cax1 changed the amplitude and duration of Ca signals during reoxygenation. Our work highlights a central role of tonoplast localized H+/Ca2+ transport in anoxia responses while further defining the molecular choreography of anoxia signaling.
Results
CAX1 mediates tolerance to anoxia and submergence
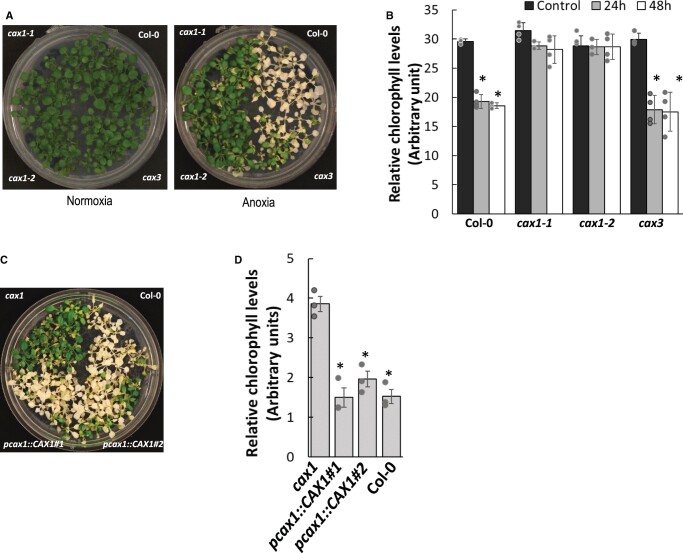
Multiple studies have shown that Ca-regulated transporters and proteins like calmodulin demonstrate modified expression during conditions of low O2 (Otsuka et al., 2010; Lokdarshi et al., 2016; Wang et al., 2016). We sought to clarify the role of CAX genes in anoxia tolerance using a seedling survival assay where plants are exposed to anoxic conditions for a period of time and survival monitored upon return to normoxic conditions (ambient air, i.e. ∼21% O2). In our assay, 21-day-old plants (nine rosette leaf development stage (Boyes et al., 2001) were placed in a GasPack anaerobic system that reduced O2 levels in the experimental chamber to ∼0% (Supplemental Figure S1A). After anoxia, phenotypes of the cax lines relative to Col-0 plants were well-defined: the cax1 lines (both cax1-1 and cax1-2) were more tolerant to anoxic conditions and this phenotype was clearly visible 4 days after being removed from the anoxic chamber (Figure 1A). Chlorophyll loss measurements were made to compare the damage during reoxygenation among the tolerant and susceptible lines. At 24-h and 48-h post-anoxia, Col-0 and cax3 displayed more chlorophyll loss, while both cax1 alleles maintained their leaf chlorophyll content (Figure 1B; Liang et al., 2017). After these anoxic conditions, >80% of the cax1 plants remained alive, whereas <15% of the cax3 and Col-0 grew to maturity (Supplemental Figure S1, B and S1C). This tolerance was lost when CAX1 driven by the CAX1 promoter region was expressed in cax1-1 (Figure 1C; Supplemental Figure S1D). Again, this susceptibility was verified by the loss of chlorophyll (Figure 1D). cax2, cax3, and cax4 had instead similar anoxia phenotypes as Col-0 (Supplemental Figure S1E).
Figure 1.
Arabidopsis cax1 lines are anoxia tolerant. A, Enhanced anoxia tolerance in cax1-1 and cax1-2 compared to Col-0 and cax3. Twenty-one-day-old plants (nine-rosette leaf stage) were placed in a GasPack Anaerobic system. After 8 h, plants were returned to normoxia conditions and photographed after 4 days. Photographs represent more than 15 biological replicates in each condition. B, Post anoxia cax1 exhibited less chlorophyll loss than Col-0 and cax3. Plants belonging to nine-rosette leaf stage were treated as described in Figure 1B and aerial portions of the plants were sampled 24 and 48 h after returning to normoxia conditions. Plants sampled prior to the anoxic conditions were used as controls. All results are means ± sem, n = 3 and error bars indicate standard error of the mean. Individual data points represented with a gray circle and asterisk indicate significant difference from the pre-treatment conditions at P =0.01, as calculated from Student’s t test in Excel. C, Expressing CAX1 under the CAX1 promoter (pcax1::CAX1) in cax1-1 caused sensitivity to anoxia. Assay conditions similar to those described in (B). The data are a representative of 14 different transgenic lines analyzed. D, Chlorophyll loss following anoxia in two cax1-1 lines expressing CAX1 under its native promoter (pcax1::CAX1#1, pcax1::CAX1#2). The data are a representative of 14 different transgenic lines analyzed. Individual data values from three biological replicates are represented with a grey circle and error bars indicate standard error of the mean. Student’s t test was used to calculate the statistical significance and an asterisk indicates a P ≤0.001. The increase in CAX1 expression in the transgenic lines has been confirmed by RT–qPCR as shown in Supplemental Figure S1E. Assay conditions similar to those described in (B) and (C).
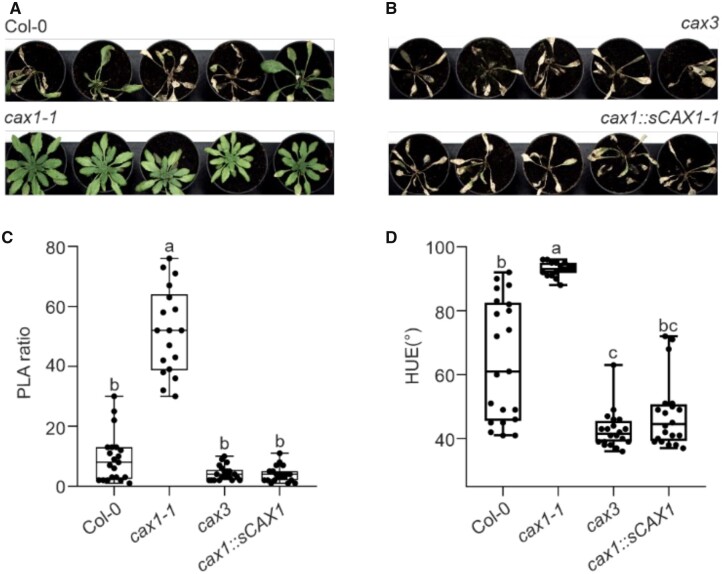
We investigated if knockingout CAX1 resulted in enhanced tolerance to submergence and waterlogging in adult Arabidopsis plants grown in soil. Although cax1 growth rate was initially reduced compared to the Col-0, it reached a similar size when grown in air. When plants were submerged for 48 h, followed by 7 days of aerobic recovery, Col-0 plants displayed stunted growth (Figure 2A; Supplemental Figure S2), indicating that the length of oxygen deprivation was long enough to severely affect the ability of the plants to recover from the stress. Remarkably, cax1 was extremely tolerant to submergence, showing none of the symptoms of prolonged hypoxia, namely inability to recover from the submergence treatment and yellowing. Both these parameters were quantified utilizing a phenotyping setup (Ventura et al., 2020), as described in Figure 2, B and C. Similar to the Col-0, the cax3 mutant was also intolerant, as was a CAX1 complementation line (cax1:sCAX1 in cax1-1).
Figure 2.
Effect of mutations in CAX genes on submergence tolerance. A, Photographs of representative plants (≥18 plants) after 48 h of submergence followed by 7 days of aerobic recovery. B, Rosette area shown as PLA ratio of plants after 48 h of submergence followed by 7 days of aerobic recovery. The ratio has been obtained between the PLA values of individual plants subjected to submergence and the average area value of aerobic plants. C, Color (HUE) of plants after 48 h of submergence followed by 7 days of aerobic recovery. In the box-blots, the dots represent the single data points, whiskers denote the min/max values, the box defines the interquartile range, center represents the median, box bounds represent the lower and upper quartiles (at least 15 biological replicates). Different letters indicate significant difference (P < 0.05) after Kruskal–Wallis test with Dunn’s post hoc test. Outliers have been identified using GraphPad 8.0.1 software (Q = 0.1%) and removed from the plots.
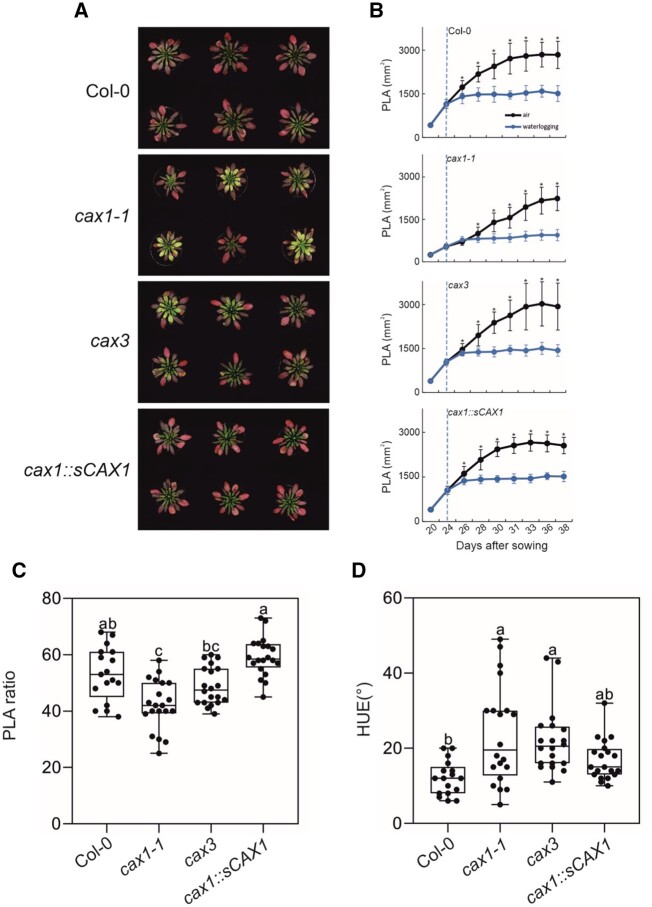
Next, we explored the possibility that mutations in CAX1 would also result in a tolerant phenotype to waterlogging, whereby only the root system is submerged. This is of interest, given that CAX1 is predominantly expressed in leaves and CAX3 in roots. The phenotype of all genotypes under waterlogging were similar, with enhanced anthocyanin synthesis in the leaves (Figure 3A). The growth of all genotypes stopped under waterlogging (Figure 3B). The size of cax1-1 plants was slightly smaller than the other genotypes under waterlogging (Figure 3C), while the leaf color of cax1-1 turned from red to yellowish (Figure 3D). Overall, while tolerance to submergence was greatly enhanced in cax1-1, CAX1 seems to have a limited role in oxygen deprivation when it is restricted to the root system.
Figure 3.
Effect of mutations in CAX genes on waterlogging tolerance. A, Photographs of representative plants (≥17 plants) after 2 weeks of waterlogging. B, Rosette area shown as PLA of control and waterlogged plants for each genotype. Student’s t test: *P ≤ 0.05; error bars: standard deviation from mean. C, PLA ratio of plants after 2 weeks of waterlogging. The ratio has been obtained between the PLA values of individual plants subjected to waterlogging and the average area value of aerobic plants. D, Color (HUE) of plants after 2 weeks of waterlogging. In the box-blots, the dots represent the single data points, whiskers denote the min/max values, the box defines the interquartile range, center represents the median, box bounds represent the lower and upper quartiles (at least 20 biological replicates). Different letters indicate significant difference (P < 0.05) after Kruskal–Wallis test with Dunn’s post hoc test. GraphPad 8.0.1 software (Q = 0.1%) did not identify any outliers and hence all the data points are all represented in the plot.
CAX1 controls post-anoxia production of reactive oxygen species
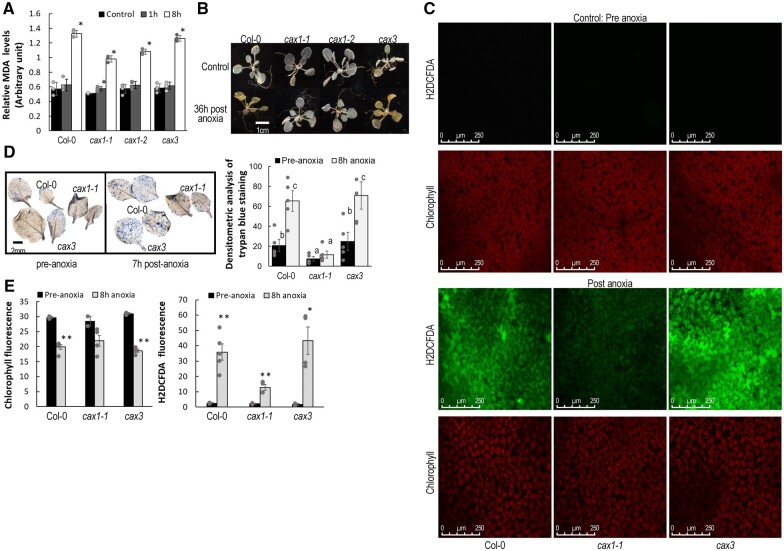
Given that CAX transporters have been shown to play various roles in altering plant signaling (Shigaki and Hirschi, 2006), we sought to assess its impact on anoxic responses. Oxidative-stress-induced lipid peroxidation (by malondialdehyde measurement in leaves) was elevated in Col-0 and cax3 at 8-h post-anoxia, but both cax1 lines displayed lower lipid peroxidation (Figure 4A; Sattler et al., 2006). Since ROS are important signal molecules mediating plant stress responses and have been closely linked to hypoxia response (Chapman et al., 2019; Wu et al., 2021), staining with 3,3′-diaminobenzidine (DAB) was performed (Daudi and O'Brien, 2012). Although the leaves often varied in their responses, the cax1 lines consistently showed lower H2O2 accumulation (Figure 4B). We also measured hydrogen peroxide levels using the cell permeable dye H2DCFDA in plants exposed to anoxic conditions or to normoxia (Fichman et al., 2019). Although there was also variability among the leaves, Col-0 and cax3 plants generally displayed elevated H2O2 post anoxia while cax1 displayed lower levels (Figure 4C). Trypan blue staining of seedlings was performed to investigate cell death during the anoxia stress response. We observed a mild degree of cell death in standard aerobic conditions. Post anoxia, Col-0 and cax3 mutants displayed significantly greater cell death throughout the leaves, compared to cax1 (Figure 4D), which also showed greater survival at the whole plant level.
Figure 4.
Effect of mutations in CAX genes on post-anoxia production of reactive oxygen species. A, Col-0 and cax3 displayed higher induction of lipid peroxidation as measured by MDA. Plants belonging to nine-rosette leaf stage were treated as described in Figure 1B and aerial portions of the plants were sampled 1 h and 8 after returning to normoxia conditions. Plants sampled prior to the anoxic conditions were used as controls. All results are means ± sem, n = 3. Asterisk indicate significant differences compared to pre-anoxia conditions at P < 0.01, as calculated from Student’s t test in Excel. B, Reduced H2O2 in cax1 compared to Col-0 and cax3 when recovering from anoxia as measured via DAB staining. A dark brown precipitate indicates the presence of H2O2. Whole rosettes from 3-week-old plants were exposed to anoxic conditions for 7 h. Rosettes were sampled 36 h after the plants were brought back to normoxia growth conditions. Unstressed plants were used as the control. Although there is variability among plants of the same genotype, the results were consistent. These data are representative of more than three independent experiments. C, Col-0 and cax3 plants demonstrated elevated H2O2 fluorescence using the cell permeable dye H2DCFDA in rosette leaves post anoxia compared to cax1. In the presence of ROS, H2DCF is oxidized to DCF, giving the fluorescence in Col-0 and cax3. Three-week-old plants were exposed to anoxic conditions for 8 h in dark. Following anoxia stress, mature whole rosette leaves were detached from the plants and were used for staining, Scale bar = 250 µM. Fluorescence signals quantified by ImageJ are given in the lower panel. Asterisks indicate significant differences compared to pre-anoxic conditions using Student’s t test in Excel (*P < 0.05 and **P < 0.005). Although there is variability among plants, the trends were consistent. Data are a representative three independent experiments and error bars indicate standard error of the mean. D, Cell death in leaves as measured by trypan blue staining was higher in Col-0 and cax3 post anoxia compared to cax1. Plants belonging to nine-rosette leaf stage were subject to anoxia as described in Figure 1A and leaves were sampled 7 h after the plants were returned to normoxia. Nine leaves were used to quantify the cell death intensity. The letters indicate that there is a statistically significant difference between two columns (level of significance 95%) as calculated from Student’s t test in Excel. Error bars indicate standard error from mean.
RNA-sequencing reveals cax1 changes in transcripts during anoxia and post-anoxia
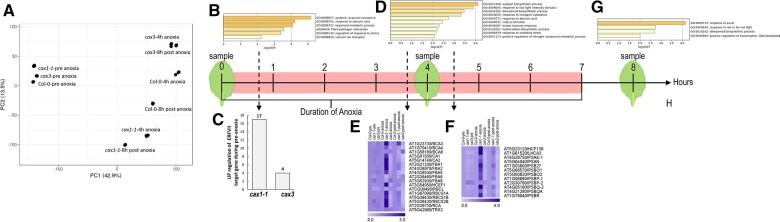
To identify molecular processes contributing to the observed difference between the sensitive lines (Col-0, cax3) and the tolerant line (cax1-1), the leaves of 21-day-old plants (nine-rosette leaf stage) were subject to RNA-sequencing (RNA-seq) analysis before, during, and after anoxic conditions (Safavi-Rizi et al., XXXX). Leaves were harvested from plants at the start of the treatment (0-h control-pre), after being in the anoxia chamber for 4 h (anoxia) and 1-h post 7-h anoxia treatment (post-anoxia; Figure 5). Each library consisted of at least 38 million reads mapped to the Col-0 genome. A large number of genes responded significantly to both treatments (anoxia and post-anoxia; Supplemental Figure S3). In Col-0, 5,292 genes were upregulated during anoxia, while 6,216 genes were downregulated. Post anoxia, 5,025 genes were upregulated and 5,050 genes downregulated (Supplemental Figure S3A). Enriched gene ontology (GO) categories for the genes up- and downregulated demonstrates that the anoxic conditions resulted in responses similar to those from previous hypoxia studies (van Veen et al., 2016; Yeung et al., 2018; Supplemental Tables S1–S12 and Supplemental Figure S4). Treatments and mutants clearly clustered separately in a principal component analysis (PCA) plot showing the first and second PCs which together explained 56.4% of variances, suggesting the RNA-seq was likely revealing differences between responses in the various genotypes (Figure 5A). We validated the RNA-seq data for several of the genes using reverse transcription–quantitative polymerase chain reaction (RT–qPCR; Supplemental Figure S5).
Figure 5.
CAX mutants transcriptome-wide responses to anoxia. A, PCA representation of Col-0, cax1, and cax3 transcriptomes before, during, and after anoxic treatment. B, GO groups identified in cax1 pre-anoxia that overlap with genes differentially expressed in col-0 and cax3 during anoxia (4-h) and post anoxia (1 h post 7-h anoxia). C, Bar graph denotes the upregulated HRGs downstream to ERFVII in pre-anoxic conditions. D, Substantially differential expression GO groups specifically found in cax1 after 4 h of anoxia. E, Preferential upregulation of genes involved in photosystem maintenance and cax1 following anoxia. F, Preferential upregulation of genes involved in carbohydrate metabolism in cax1 following anoxia. Fold change expression of the genes were used for constructing heat map using MeV_4_9_0 in both (c) and (F). G, Substantially differential expression GO groups specifically found in cax1 during post-anoxic conditions. H, Scheme and timing of leaf tissue harvesting for RNA-seq analysis. Plants belonging to nine rosette leaf stage were taken for RNA-seq analysis before anoxia (pre-conditions), after 4 h of anoxia (anoxia), and 1 h after a 7-h anoxia treatment (post anoxia).
We posited that enhanced expression or repression of specific genes prior to the stress could prime cax1 for tolerance (cax1 1,423 genes upregulated–146 genes downregulated; cax3 526 genes upregulated–167 downregulated; Figure 5; Supplemental Figure S3B and Supplemental Tables S13–S15). We thus asked if any of the genes highly differentially expressed in cax1 in conditions (Figure 5B) were genes highly differentially expressed in Col-0 during anoxic or post-anoxic conditions (Supplemental Figure S6 and Supplemental Tables S15–S21). This proved to be true. Of the 1,423 genes upregulated in cax1 pre-anoxia, ∼50% of these genes (717) were upregulated in Col-0 during or post anoxia (Supplemental Figure S6A and Supplemental Table S16). The gene concept network indicates that specifically up- and downregulated genes could be assigned to different clusters. The upregulated genes appear to be involved in root morphogenesis and cellular response to hypoxia (Supplemental Figure S6B). Only 34 genes were downregulated in cax1 with 80% (Kong et al., 2020) of these downregulated in Col-0 during or post anoxia. These overlapping genes appeared to be involved in cold acclimation and photosynthesis (Supplemental Figure S6, C and S6D and Supplemental Table S17). Meanwhile, the sensitive cax3 line displayed 141 genes upregulated during normoxia and 33% (41 genes) were upregulated in Col-0 with many of these overlapping genes involved in phenylpropanoid biosynthesis and root morphogenesis (Supplemental Figure S6, I and S6J and Supplemental Table S18). In terms of downregulated genes, 32 genes in cax3 were downregulated during normoxia, and 66% (Wang et al., 2016) were also downregulated in Col-0 during or post anoxia. These overlapping genes were involved in cold acclimation, defense responses, and leaf senescence (Supplemental Figure S6, K and S6L and Supplemental Table S19).
Some of the ERF-VIIs target genes were expressed in cax1 prior to the stress (Figure 5, B and C). These ERF-VIIs are plant-specific transcription factors that are important regulators of low oxygen tolerance (Gibbs et al., 2015). Previously, a core hypoxia-specific gene set (n ≈ 49) have been identified that are primary targets of ERF-VIIs (Mustroph et al., 2009; Gasch et al., 2016). In cax1, 17 of these genes were substantially expressed at a higher level in cax1 even under aerobic conditions, indicating that the altered Ca homeostasis in cax1 influences the ERF-VII-dependent gene expression, even prior to anoxia stress (Figure 5C; Supplemental Table S20); in contrast, cax3 expressed only four of these genes at high levels prior to the stress (Figure 5C; Supplemental Table S20). This observation is consistent with the concept that cax1 is primed for low-oxygen environments during normal growth conditions. However, analysis of the expression of all the 49 core anaerobic genes revealed a marked dampening of the expression of these HRGs during anoxia (Figure 5D; Supplemental Figure S7). Indeed, cax1-1 only showed a modest induction of most of the 49 core HRGs, indicating that CAX1 is required to ensure a normal expression of the ERF-VII-dependent HRGs. This suggests that the tolerance to anoxia and submergence observed in cax1-1 may not be mediated by the ERF-VII-dependent pathway.
During the anoxia phase, cax1-1 also displayed heightened expression of metabolic and photosystem genes (cax1 2,000 genes upregulated–2,225 downregulated; Figure 5, D–F; Supplemental Figure S3B). Apparently, the loss of CAX1 caused substantial changes in the way the plant manages depleted light and energy levels.
To investigate cax1-1 anoxia tolerance more thoroughly during the stress, 141 and 586 unique (not found in either cax3 or Col-0) up- and downregulated were submitted for GO enrichment analysis (Supplemental Tables S21 and S22 and Supplemental Figure S8). The genes clustered with oxidative stress, defense responses, and nitrogen and hydrocarbon metabolic processes (Figure 5D). In contrast, genes involved in cell signaling, response to photooxidative stress, generation of precursor metabolites and energy, were downregulated (Supplemental Figure S8B). These may be related to downregulated genes for plastid organization and chloroplast organization, which likely results in suppressed carbohydrate metabolism (Supplemental Figures S9 and S10). The cluster response to oxidative stress were connected to a number of genes that showed both up- and downregulation during anoxia.
To further conceptualize the cax1 anoxia tolerance recovery phase (cax1 1,432 genes upregulated–2,464 downregulated), 131 and 751 unique (not found in either cax3 or Col-0) up- and downregulated genes were also submitted for GO enrichment analysis (Supplemental Figure S3B and Supplemental Tables S23 and S24). The enriched GO category for these genes post-anoxia were predominately auxin-responsive genes, response to red light, and diterpenoid metabolism (Figure 5G). Meanwhile, the enriched GO category post-anoxia downregulated genes were cell wall biogenesis, anion transport, development, and cell growth related (Supplemental Figure S8C).
Protein changes in cax1 post anoxia
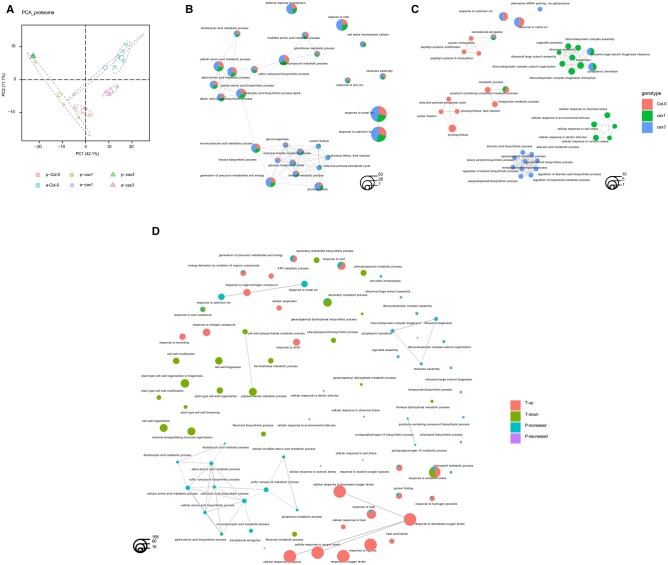
To provide additional insight into how the CAX proteins are involved in anoxia responses, global changes in protein abundance between Col-0, cax1, and cax3, pre- and post-anoxia were analyzed. In total, among the three genotypes, 463 unique proteins were identified from a total of 17,339 spectra. Normalized spectral abundance factor (NSAF) (Zybailov et al., 2006) of each protein served as the basis for PCA. Clear separations were shown for clusters in accordance with genotypes and treatments represented by 95% confidence ellipses from three biological replicates, with only slight overlap between the clusters of Col-0 and cax1 proteome post anoxia (Figure 6A).
Figure 6.
Proteomic analysis of cax1 post-anoxia treatment. A, PCA of the proteome of Col-0, cax1, and cax3 pre (p) and post anoxia (a) conditions. Sample scores for the first and second principal components were plotted, the explained percentage of variance of PC1 and PC2 are indicated along the x and y axes. Clusters corresponding to the groups are represented by 95% confidence ellipses and include data from three biological replicates. The enlarged dots represent the means of the groups. Enrichment network depicting the top 20 most significantly enriched GO terms (hypergeometric test with Bonferroni correction, P < 0.05) for abundance increased (B) and decreased (C) proteins in Col-0, cax1, and cax3 post anoxia. D, A combined GO enrichment study of transcriptomic and proteomic regulations in cax1 post-anoxia treatment. Each GO term is represented by a circle, and different groups are shown. The size of the GO term circle reflects the number of genes/proteins enriched in the corresponding item. T-up/down, enriched GO terms for significantly up/downregulated transcripts; P-increased/decreased, enriched GO terms for significantly increased/decreased proteins.
A total of 242, 218, and 356 differentially abundant proteins (DAPs) were identified for Col-0, cax1, and cax3 post-anoxia treatment, respectively, based on their relative abundance, with details of increased and decreased proteins in each of the genotypes listed in Supplemental Table S25. DAPs in all three lines were subjected to enrichment analysis based on the functional annotation of GO terms. The top 20 significantly enriched GO terms (padj < 0.05, Bonferroni method) for increased and decreased proteins in all three lines (represented by different colors) are presented in Figure 6, B and C. The results suggested that increased proteins in all three lines were enriched in GO terms, including response to metal ion, photosynthesis, carbohydrate, and small molecule metabolic process, organic acid metabolic process, and sulfur compound metabolic process, while increased proteins from only cax1 and cax3 were enriched in carbon fixation, reductive pentose-phosphate cycle, and photosynthesis dark reaction (Figure 6B). In contrast, these GO terms were specifically enriched in Col-0 for proteins that had decreased abundance (Figure 6C). This is likely related to the decrease in proteins related to the GO term, chlorophyll metabolic processes, in this line which was also detected in the analysis as shown in Figure 6C.
In addition, although decreased proteins in all three lines were enriched in GO terms for translational elongation, only Col-0 showed enrichment in decreased proteins for GO terms related to protein modification, such as peptidyl-amino acid modifications and protein nitrosylation (Figure 6C). GO enrichment analyses of significantly decreased proteins in cax1, and cax3 post anoxia also showed different clustering. For example, decreased proteins in cax1 were enriched for processes including response to stimulus and ribosome biogenesis, while decreased proteins in cax3 were enriched for metabolic processes (Figure 6C).
A combined GO enrichment analysis of significantly altered transcripts and proteins provided insight into coordinated regulation occurring in cax1 post-anoxia treatment and is demonstrated by overlapping of clusters of significantly enriched GO terms (Figure 6D; Supplemental Figure S11). This highlighted processes such as increased energy metabolism, represented by generation of precursor metabolites, energy derivation by the oxidation of organic compounds, as well as an increase in oxidative scavenging shown as response to oxidative stress, response to reactive oxygen species, and response to hydrogen peroxide in cax1 post anoxia (Figure 6D; Supplemental Figure S11). Worth mentioning is that although proteomic analysis captured a decreased response to environmental stimulus such as chemical stress and salt stress (Figure 6, C and D), both transcriptomic and proteomic studies demonstrated an enrichment in GO terms for response to temperature (heat and cold) in cax1 post anoxia.
Changes in cytosolic Ca levels at the onset of anoxia and during recovery from anoxia in cax1
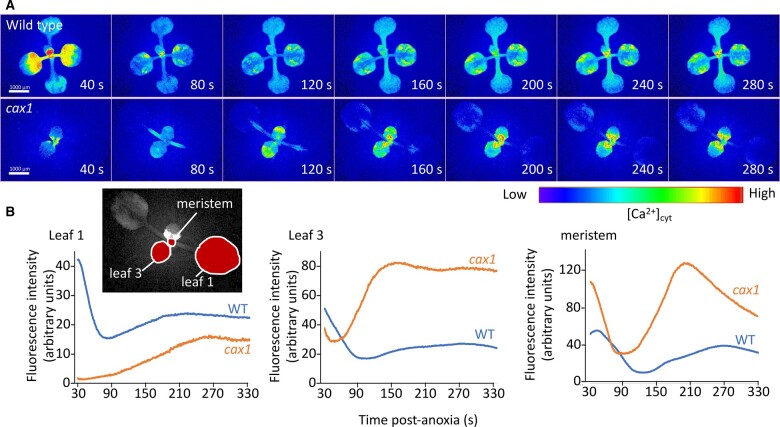
Ca plays an important role in plant sensing of external signals (Sanders et al., 2002). CAX mutants may alter the distribution of Ca and thus influence anoxia responses. In order to investigate the relationship between CAX1 function and changes in cytosolic Ca and anoxia tolerance, we generated stable Col-0, cax3, and cax1 lines expressing the Ca biosensor GCaMP3, a GFP-based fluorescent protein made Ca2+-sensitive by the insertion of a calmodulin domain (Toyota et al., 2018). Under normal growth conditions, without anoxia stress, the Ca signals in Col-0 and cax1 were similar (Supplemental Figure S12). After 4 h of anoxia treatment, these lines were reoxygenated and imaged for their cytosolic Ca signature. In this assay, it took ∼15–30 s to unseal the container and begin the imaging, once the imaging progressed, Col-0 lines showed a prominent initial Ca signal which dissipated within 90 s (Figure 7; Supplemental Movie 1); a second less prominent Ca signal peaked around 200- 300-s post reoxygenation depending on the organ monitored. In cax1, the initial cytosolic Ca signal seen to decline over the initial 90 s in the Col-0 was absent. Meanwhile, a stronger Ca signal peaked at around 100–200 s, which was especially evident in the younger leaves and meristematic region (Figure 7; Supplemental Movie 1). The spatial distribution of the signal also changed: the second Ca signal appeared to have a more pronounced temporal and spatial progression in cax1 moving from the base of the meristem of the rosette along the petiole and into the leaf blade of the leaves. Although it is tempting to speculate that this represents a wave of response propagating outwards through the leaves, it is important to note that the reoxygenation post anoxia occurs plant wide and so these dynamics could equally report temporal differences in Ca signal generation in different parts of the plant.
Figure 7.
Calcium changes in Col-0 and cax1 post anoxia. Plants were removed from their anoxic environment to normoxic conditions and then imaged using a fluorescence microscope. A, Images of plants expressing the gCaMP3 Ca2+ biosensor have had background subtracted and then fluorescence intensity pseudo-color coded according to the inset scale. Time is post transfer to normoxic conditions. B, Quantification of fluorescence of plants shown in (A). Fluorescence intensity was monitored in the regions of interest outlined in the inset. Representative of ≥10 independent plants.
Initial assessment for the roles of Ca signaling, ROS, and vacuolar function on anoxia tolerance
Previous studies on anoxia stress responses (Loreti and Perata, 2020) and our RNA-seq, proteomics, and Ca imaging suggest anoxia tolerance may involve vacuolar function, Ca signaling, pH homeostasis, and signaling pathways similar to those involved in abiotic and biotic stresses (Voesenek et al., 2016; Schmidt et al., 2018). To further investigate the roles of these processes, we assessed various T-DNA insertion lines with alterations in vacuolar function, pH homeostasis, Ca transport, stress, and ROS signaling for anoxia tolerance (Table 1; Supplemental Figure S13). Using a simple visual screen, lines with altered expression of a tonoplast localized Ca-ATPase (Geisler et al., 2000), Na/H transporters (Reguera et al., 2015), and V-ATPase (Kriegel et al., 2015) displayed no changes in anoxia tolerance (Table 1; Supplemental Figure S13). Additionally, mutations in genes related to Ca signaling, ROS, and defense signaling, despite showing changes in gene expression in cax1, had loss-of-function phenotypes that closely resembled Col-0 (Supplemental Table 26).
Table 1.
Response of various lines previously identified as involved in altering Ca transport; production of ROS and regulation of vacuolar pH
| SI. No. | Germplasm_ID | Mutant_Name | Gene_ID | Gene_Name | Specific function of the gene in pathway | Response to anoxia | |
|---|---|---|---|---|---|---|---|
| Ca signaling mutants | |||||||
| 1 | SALK_078400 | cml39 | AT1G76640 | CALMODULIN LIKE 39 | Ca-binding EF-hand family protein | S | |
| 2 | CS872685 | aca11 | AT3G57330 | AUTOINHIBITED Ca2+-ATPase 11 | Ca2+-ATPase | S | |
| 3 | SALK_029620C | aca4 | AT2G41560 | AUTOINHIBITED Ca2+-ATPase 4 | Ca2+-ATPase | S | |
| ROS signaling mutants | |||||||
| 4 | CS9555 | atrbohD | AT5G47910 | RESPIRATORY BURST OXIDASE HOMOLOGUE D | Regulates ROI production | S | |
| 5 | SALK_057686C | apx2 | AT3G09640 | ASCORBATE PEROXIDASE 2 | Scavenge hydrogen peroxide | S | |
| 6 | CS6962C | rev-6 | AT5G60690 | REVOLUTA | ROS-sensitive TF involved in senescence | S | |
| Mutants associated with regulation of vacuolar pH | |||||||
| 7 | SALK_034001c | nhx1 | AT5G27150 | Na+/H+ EXCHANGER 1 | Vacuolar Na+/H+ antiporter | S | |
| 8 | SALK_084844c | nhx2-3 | AT3G05030 | Na+/H+ EXCHANGER 2 | vacuolar K+/H+ exchanger | S | |
| 9 | SALK_142642c | vha-a2 | AT2G21410 | VACUOLAR PROTON ATPase A2 | Vacuolar proton ATPase subunit VHA-a isoform 2 | S | |
| 10 | SALK_029786c | vha-a3 | AT4G39080 | VACUOLAR PROTON ATPase A3 | Vacuolar proton ATPase subunit VHA-a isoform 3 | S | |
| Other mutants | |||||||
| 11 | SALK_142672C | hai1-1 | AT5G59220 | HIGHLY ABA-INDUCED PP2C GENE 1 | Negative regulator of ABA signaling | S | |
| 12 | CS874182 | pr1 | AT2G14610 | PATHOGENESIS-RELATED GENE 1 | Induced in SAR | S | |
| 13 | SALK_055063C | pr5 | AT1G75040 | PATHOGENESIS-RELATED GENE 5 | Involved in response to pathogens | S | |
| 14 | CS16353 | rgl1-2 | AT1G66350 | RGA-LIKE 1 | Negative regulator of GA response | S | |
| 15 | SALK_131604C | crk37 | AT4G04500 | CYSTEINE-RICH RLK 37 | Protein kinase | S | |
Anoxia sensitivity is indicated by the letter “S”; wherein the mutant plants showed a phenotype similar to col-0 plants; no lines analyzed showed a tolerance to anoxia similar to cax1. Each of these mutants have been tested thrice; with a minimum of 15 seeds planted for each experiment.
Discussion
Hypoxia exerts a negative impact on energy production (Atwell et al., 2015). Hypoxic events can be viewed as a sequential, compound stress, where both the lack of oxygen and the reoxygenation periods pose distinct stressors; lack of light exerts another stress, eventually leading to carbon starvation (Loreti et al., 2018). Tolerance is determined by the plant’s ability to acclimate to all these stresses (Yeung et al., 2018). Plants may use spatial and temporal dynamics in Ca to convey information about anoxic conditions (Farnese et al., 2016; Wang et al., 2016). Here, we identified CAX1 as a Ca transporter whose expression is detrimental to anoxic conditions and submergence (Figures 1 and 2). Using this mutant, we have sought to identify various molecular and physiological processes influencing anoxia tolerance.
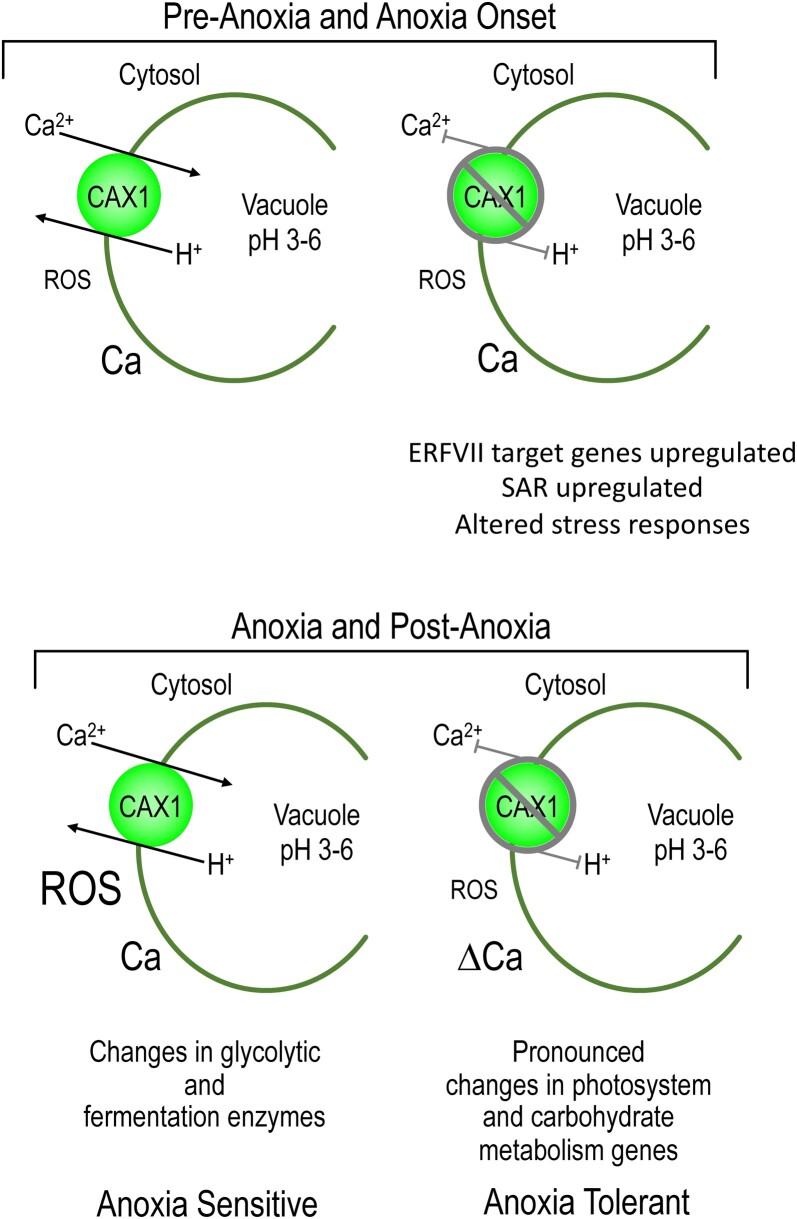
Our work suggests that cax1 has numerous adaptations to endure anoxia stress (Figure 8): (1) The plant is primed for anoxia through altered expression of ROS, HRGs, and temperature perception genes (Figures 4 and 5; Supplemental Figures S6 and S13 and Supplemental Table S20); (2) During anoxia, changes in light and carbohydrate metabolism may mitigate the stress (Figures 5 and 6); (3) Post anoxia, altered Ca signaling could help dissipate the reoxygenation stress (Figure 7). These findings suggest that altered tonoplast H+/Ca transport impacts an array of biological processes associated with anoxia (Figures 4, 5, 6 and 8); however, mutants in tonoplast localized Ca transporters with different transport kinetics did not impact anoxia tolerance (Table 1). The loss of high-capacity low-affinity Ca transport caused by disruption of CAX1 led to numerous changes in gene expression and protein abundance; the breadth of these changes makes the identification of a single causative pathway for this submergence/anoxia tolerance a challenging endeavor.
Figure 8.
Model comparing pre-anoxia and anoxia responses in CAX1 and cax1 lines. Before anoxia, plants appear to have altered regulation of ERF-VII-targeted transcripts and stress response genes such as those involved in systemic-acquired resistance (SAR). During anoxia and post anoxia, cax1 appears to have dampened ROS production, altered (Δ)Ca signaling, and differences in the expression of carbohydrate and metabolism genes.
The fact that a subset of HRGs is upregulated in normoxia in cax1, while most of the HRGs are downregulated during hypoxia is intriguing. This suggests that altered Ca homeostasis caused by perturbed CAX1 activates some HRGs, most likely though an ERF-VII-independent pathway, given that this class of transcription factors are highly unstable and thus inactive under aerobic conditions (Weits et al., 2014). Under hypoxia, instead, ERF-VII are stable because of the inability of plant cysteine oxidases to utilize molecular oxygen (Weits et al., 2014). Under these conditions, which promote a vigorous expression of ERF-VII-dependent HRGs, we observed a dampening of the response, which could be beneficial to preserve the carbon reserves required during the recovery phase. Monitoring the protein stability of ERF-VII (e.g. by western blot) as a consequence of CAX1-dependent alterations in Ca fluxes could be difficult but could shed light on the role of Ca on the stability of ERF-VII. Furthermore, future experiments with erfVII/cax1 will be informative to define whether the tolerance in cax1 requires the activity of ERF-VIIs or it is an ERF-VII-independent mechanism of tolerance.
As the lack of a waterlogging phenotypes demonstrates, plant roots and shoots have different mechanisms for hypoxia/anoxia stress tolerance (Ellis et al., 1999). Previous work in Arabidopsis has used tissue-specific root ion profiling to show roles of putative CAXs and Ca-ATPase transporters in hypoxia (Wang et al., 2016). These studies demonstrated a gene termed CCX5 (misidentified as CAX11) was involved in hypoxia responses; 3 weeks of waterlogging resulted in dramatic loss of shoot biomass in ccx5 (cax11) plants. It is important to clarify here that CCX5 is not a CAX (Shigaki et al., 2006) and is more closely related to mammalian K+-dependent Na+/Ca2+ antiporters (Pittman and Hirschi, 2016). Our results showed that the leaf-expressed CAX1 is detrimental for both anoxia and submergence tolerance (Figures 1 and 2). Furthermore, the expression of CAX1 in cax1-1 suppressed these tolerance phenotypes (Figures 1 and 2). Waterlogging, which results in hypoxia restricted to the root system, provided support to the role of CAX1 limited to the aerial part of the plant (Figure 3). Additionally, CAX3 does not appear to be required for hypoxia signaling given that cax3 remained sensitive to both submergence and waterlogging.
In cax1, signaling and metabolic mechanisms for anoxia tolerance are activated, suggesting that CAX1 plays a negative role in low oxygen tolerance (Figure 8). However, the mechanism by which the absence of this transporter positively impacts multiple biochemical mechanisms toward tolerance requires further research. In yeast, the tonoplast localized Ca/H+ antiporter, VCX1, functions to rapidly sequester cytosolic Ca and attenuate the activation of Ca signaling pathways (Miseta et al., 1999). However, the relationship between Ca signaling and CAX1 during limited oxygen conditions is less clear. Defining this relationship is confounded by the variable Ca responses among leaves within the same plant. This work focused on the two older leaves in the four-leaf rosette stage in young plants, where the changes between cax1 and Col-0 were most obvious. In cax1, during the first minute after exposure to oxygen, plants appeared to accumulate less cytosolic Ca in these leaves (Figure 7). One interpretation of this observation is that CAX1 has a minimal role in sequestering cytosolic Ca during this response as its loss would be expected to cause a larger Ca increase if it was removing signaling-related Ca from the cytosol. Indeed, such an increase in the magnitude of the pathogen elicitor-triggered Ca signal is seen when the two tonoplasts localized Ca2+-ATPases ACA4 and ACA 11 are knocked out, providing evidence that for these two pumps, they do normally act to attenuate the Ca signal (Hilleary et al., 2020). From our results, it seems likely that CAX1 is instead functioning to create an exchangeable Ca store that can be drawn upon to generate signals, and this is lost in the mutant. Alternatively, cax1 may cause compensatory upregulation of numerous endomembrane Ca transporters to diminish cytosolic Ca levels (Cheng et al., 2003, 2005). In yeast, the absence of Vcx1p results in heightened Ca-ATPase expression (Callewaert et al., 2020) and may result in the induction or activation of a set of transporters capable of coupling vacuolar Ca uptake to the vacuolar H+ gradient (Miseta et al., 1999). Indeed, our analyses reveal significant (P < 0.01) constitutive transcriptional upregulation to more than two-fold of some classic Ca transporters including ACAs 1 and 9–13 in the cax1 background and as noted above, ACA11 is thought to act to attenuate Ca signaling (Hilleary et al., 2020). We also cannot discount activation of another transport system, or post-translational regulation of, for example the ACAs or CAXs playing an important role in these processes. Caution regarding ascribing any direct mechanistic relationship between cax1-mediated Ca levels and anoxia tolerance is also warranted. For example, the phenotype of anoxia tolerance displayed in the mutant could be caused by heightened H+ pumping. For adequate energy production, residual H+ pumping is critical to cope with anoxia by importing energy-rich compounds (Felle, 2005). The perturbation of Arabidopsis H+-ATPase activities at both the plasma membrane and tonoplast in mutants lacking tonoplast Ca/H+ antiporters (Pittman et al., 2005; Barkla et al., 2008) suggests future studies that address how cax1 impacts transmembrane H+ gradients during oxygen limiting conditions (Reguera et al., 2015). This information may provide important insight into the molecular events underlying cax1 tolerance.
A limitation of our model (Figure 8) is its inability to address the known variability in anoxic responses among different tissues and leaves (Banti et al., 2013; Klecker et al., 2014; Ventura et al., 2020). These differences were seen here in both the ROS and Ca imaging experiments (Figures 4 and 7). While the anoxic response is variable, the role of CAX1 in different tissues and leaves also fluctuates. In dicots, CAX1 is found predominately in the tonoplast of mesophyll cells (Conn et al., 2011). Each tissue will have a different proportion of mesophyll cells, and individual cells will have variations in vacuole size and shape (Tan et al., 2019). Thus, there will be developmental and spatial differences in the contribution of CAX1 to Ca and H+ fluxes. For example, in contrast to the major mesophyll expression observed in dicots, in monocots CAX1 is localized to epidermal cells (Conn et al., 2011), we posit monocot loss-of-function mutants will show very different responses to anoxic conditions.
Loss-of-function mutations in Arabidopsis H+/Ca transporter CAX1 display a robust conditional phenotype, they gain tolerance to anoxic conditions (Figure 1). The disruption of CAX transport during this stress impacts a myriad of cellular processes: cell wall biosynthesis, metabolism, ROS, hormone, and Ca signaling (Figure 5). Furthermore, the mutants appear to be primed for the stress by having heightened expression of ROS-related transcripts and diminished expression of cold tolerance genes during normal growth conditions (Figure 4; Supplemental Figure S6). The cax1 lines also appear to be “warmed up” as they display heightened expression of heat-responsive transcripts during the anoxia recovery phase (Supplemental Figure S14). Heat shock response involves the induction of a wide range of molecular chaperones to aid in protecting and refolding damaged proteins and so this constitutive production of, for example heat shock proteins may equip cax1 with readymade machinery to cope with some of the cellular damage that anoxia and reoxygenation imposes. Previous work has also shown that cax1 has altered cold-acclimation responses (Catalá et al., 2003) and temperature perception may be an important factor in our anoxia tolerance phenotype.
Conclusions
This study adds granularity to the role of tonoplast localized Ca transport in plant stress responses. The cax1-mediated anoxia tolerance is caused by extensive changes in both metabolism and signaling. Indeed, one of the most important findings of this study is the breath and amplitude of changes caused by the loss of a single tonoplast H+/Ca transporter (Figures 5 and 6). To harness this knowledge for translational applications, judicious application is advised: CAX transporters impact a variety of plant signal transduction pathways (Pittman and Hirschi, 2016; Bradshaw, 2005; Baliardini et al., 2015, 2016). However, the CAX transporters appear to be autoinhibited, requiring a protein partner for activation (Shigaki and Hirschi, 2006). During anoxic conditions, CAX1 may be activated by a specific regulatory protein and downregulation of this activator during anoxic conditions may be a means of transiently impairing CAX1 function and improving anoxia tolerance.
Materials and methods
Plant materials
The Arabidopsis (A. thaliana) Col-0 and CAX mutant seeds (cax1-1, cax1-2, cax3-1) and various CAX1 construct used in the study have been previously characterized (Catalá et al., 2003; Cheng et al., 2005). In normal growth conditions, the cax1 lines are virtually indistinguishable from Col-0 in terms of size and fertility (Cheng et al., 2003). The CAX mutants were genotyped to confirm the presence of the T-DNA insertions. The plants expressing the binary construct harboring p35S:sCAX1 or pCAX1::CAX1 have both been previously described and growth and stress response phenotypes are similar to Col-0 (Cheng et al., 2003; Conn et al., 2011). Col-0 harboring a stable p35S::GCaMP3 Ca biosensor construct (Toyota et al., 2018; GCamP3/Col-0) was crossed with cax1 to obtain GCamP3/cax1. F3 plants that were verified as cax1-1 were then assayed for expression of GCamP3.
Plant growth, anoxia, and flooding treatment
Arabidopsis seeds were sterilized in 20% bleach (v/v) for 15 min before they were planted on 1/2X Murashige and Skoog (MS) media containing 0.8% agar (w/v) and 0.5% sucrose (w/v), and were grown under equal day/night light conditions (12-h light, 22°C, 180 μmol·m−2·s −1, and 12-h night 20°C). For anoxia treatments, 21-day-old plants (nine-rosette leaf developmental stage) were foil covered at 2:00 pm and then placed in a GasPak using anaerobic atmosphere generation bags (Sigma) for 7 h or an anoxia chamber (Anaerobe Systems, AS-580, Morgan Hill, CA, USA). After the treatment the plants are returned to the growth chamber. Images were taken 3 or 4 days after the anoxia treatment to assess plant tolerance.
Plants for the submergence and waterlogging experiments were grown as described (Ventura et al., 2020). Briefly, were grown in pots for 3–4 weeks at 23°C with a 12-/12-h photoperiod at 120 μmol photons m−2 s−1 before being used in our experiments. Waterlogging treatments were performed on 3-week-old plants, during which all the root system was immersed in water, but the leaves and petioles were above the water level. For the submergence experiments, the plants were submerged in tanks with a water level of 10-cm above leaf level. The submergence treatment was carried out in the dark (also the controls in air were in the dark), while waterlogged plants were kept under the normal day/night cycle.
Plant phenotyping
Plant trays were placed in a LabScanalyzer (LemnaTec, GmbH, Aachen, Germany) imaging box equipped with a Manta G-1236 camera (Allied Vision Technologies GmbH, Stadtroda, Germany) and a Kowa LM12XC lens (Kowa Optimed Deutschland GmbH, Düsseldorf, Germany). Trays were illuminated by two cool white LED panels (polyscale GmbH & Co. KG, Aachen, Germany) mounted beside the camera at an angle of 30° to prevent direct reflection from the imaging area. The raw images acquired as previously described (Ventura et al., 2020) and analyzed with LemnaGrid software (LemnaTec, GmbH, Aachen). The parameters measured were the projected leaf area (PLA, area of the rosette) and color (HUE) as described in Ventura et al. (2020). In order to allow comparison of plants with different size in air, we used the ratio between the PLA values of individual plants subjected to either submergence or waterlogging and the average PLA value of aerobic plants of the same genotype (PLA ratio).
Chlorophyll fluorescence measurement
Chlorophyll content was measured via spectroscopic absorbance of chlorophyll a and b, as previously described (Yeung et al., 2018). Briefly chlorophyll was extracted from 3-week-old whole rosettes (nine-rosette leaf stage) using 96% (v/v) dimethyl sulfoxide (DMSO). Approximately 100 mg of fresh leaf sample was ground in 96% DMSO and incubated at 65°C water bath for 4 h. After cooling, the supernatant was collected and absorbance at 664, 647, and 750 nm was measured with a spectrophotometer (Cary 50) in 1-mL cuvettes. Chlorophyll a and b concentrations, and total chlorophyll content were calculated following the equations of reference and were normalized to tissue fresh weight.
RNA-seq transcriptomic analysis
Approximately 100 mg of full rosettes from anoxia-treated or untreated plants were frozen and ground in liquid nitrogen, and were subjected to Trizol total RNA extraction. Total RNA samples were sent to BGI Genomics (BGI Americas Corporation, Cambridge, MA, USA) for transcriptome sequencing on their DNBseq platform using 150-bp pair-end sequencing chemistry. Data processing and differentially expressed gene (DEG) analysis were also conducted by Beijing Genomics Institute (BGI). Briefly, the reads obtained after sequencing were filtered to remove any low-quality, adapter-polluted, and high content of unknown base (N) reads. The clean reads thus obtained were mapped to the Arabidopsis reference genome (TAIR10_Araport11_www.arabidopsis.org_20190423) using HISAT2 (Kim et al., 2015). New transcripts were identified by StringTie (Pertea et al., 2015), Cuffcompare (Trapnell et al., 2012), and CP5 (Kong et al., 2007); and SNP and INDEl variants by GATK (McKenna et al., 2010). Total clean reads were again mapped to reference using Bowtie2 (Langmead and Salzberg, 2012), and then calculated the gene expression level with RSEM (Li and Dewey, 2011). DEGs were detected with DEseq2, based on the negative binomial distribution (Love et al., 2014).
Leaf tissue from 21-day-old plants were used for transcriptome analysis. RNA-seq data were obtained from three independent biological replicates for each of the conditions analyzed. On average, sequencing from the DNBseq platform generated 6.77 Gb bases per sample. All the samples had clean raw reads higher than 44 Mb. The average mapping ratio with the reference genome was 90.03% and the average mapping ratio was 78.29. Complete RNA-seq data have been deposited in the SRA database with the accession no. PRJNA666693.
RNA extraction and RT–qPCR
Total RNA was extracted using trizol from 50-mg full rosette leaves. For RT–qPCR, single-stranded cDNA was synthesized from 1-μg RNA using oligo dT18 primers (Invitrogen). RT–qPCR was performed on a Biorad CFX96 Real-Time PCR System (Biorad) with iTAQ SYBR Green Master Mix (Bio-Rad). Primers used are listed in Supplemental Table S27. Relative transcript abundance was calculated using the comparative 2−ΔΔCT method normalized to UBQ10 (Czechowski et al., 2005). The absence of a complete functional transcript in the mutants were confirmed by semi-quantitative PCR using the primers spanning either sides of T-DNA insertion (Supplemental Figure S15).
Mass spectrometry proteomic analysis
Arabidopsis whole rosette leaves were harvested from 2.5-week-old plants (six-rosette leaf stage) and proteins were extracted from leaves (Barkla et al., 2009). Protein identification was carried out at the Institute for Molecular Biosciences proteomics facility at the University of Queensland, Brisbane. Proteins were analyzed using an Eksigent, Ekspert nano LC400 uHPLC coupled to a TripleTOF 6600+ System (SCIEX, Canada) equipped with a PicoView nanoflow ion source (New Objective, USA). For protein isolation, 3 g of tissue were ground in liquid N2 and were then subjected to extraction using prechilled buffer containing: 100-mm Tris-MES, pH 8.0, 1-mm EGTA, 5-mm dithiothreitol, 4-mm MgSO4, 5% [w/v] insoluble POLYVINYLPYRROLIDONE (PVP), and plant protease inhibitor cocktails (Sigma Aldrich, St Louis, MO, USA) to the recommended concentration by manufacturer. The homogenate was then filtrated through Miracloth (Calbiochem, La Jolla, CA, USA), and subsequently clarified by centrifugation to remove cellular debris. The total proteins in supernatant (500 μL) were then precipitated using 200-μL 10X TE, 200 μL of 0.3% sodium deoxycholate, and 200 μL of cold 72% TCA. The protein pellets were washed in 90% methanol at room temperature and subsequently lyophilized.
Proteins were analyzed using an Eksigent, Ekspert nano LC400 uHPLC coupled to a TripleTOF 6600+ System (SCIEX, Canada) equipped with a PicoView nanoflow ion source (New Objective, USA). Protein extract (up to 5 μL) was injected onto a ChromXP C18-CL column (3 μm, 75 μm × 150 mm; SCIEX, Canada). Mobile phase solvents consisted of solvent A: 0.1% formic acid in water and solvent B: 0.1% formic acid in acetonitrile. Linear gradients of 5%–30% solvent B were run over 120 min at 400-nL min−1 flow rate, followed by 30%–90% solvent B for 3 min, then 90% solvent B for 17 min, for peptide elution. The gradient was then returned to 5% solvent B for equilibration prior to the next sample injection. Column temperature was maintained at 45°C throughout. The ion spray voltage was set to 2,600 V, declustering potential at 80V, curtain gas flow 25 psi, nebulizer gas 30 psi, and interface heater at 150°C. The mass spectrometer was set to acquire 100 ms of full scan time-of-flight mass spectrometry (TOF-MS) data over the mass range 350–1,500 m/z, followed by up to fifty 50-ms full scan product ion data in IDA mode over the mass range 100–1,500 m/z. Ions observed in the TOF-MS scan exceeding a threshold of 100 counts and a charge state of +2 to +5 were set to trigger the acquisition of product ion tandem mass spectrometry (MS/MS) spectra of the resultant 50 most intense ions.
Protein identification and quantification
Protein Pilot 5.0.2 (SCIEX, Canada) was used to search spectra against the Uniprot Arabidopsis database (129,652 proteins, 12 May 2020) and encode the output mzIdentML file for the downstream analysis. Scaffold 4.8.6 (Proteome Software, Portland, OR, USA) was used to validate MS/MS-based protein identifications and quantification. Protein identifications were accepted if they could be established at >99% probability and contained at least two unique peptides. NSAF was used for protein quantification (Gulcicek et al., 2005; Searle, 2010).
Bioinformatics analysis for GO enrichment analysis
Proteins identified in at least two out of three biological replicates of leaf tissue from Col-0, cax1, and cax3 post anoxia (8 h) were considered as present in the corresponding genotypes. To evaluate the significance of comparative quantification by different genotypes, Student’s t test was performed on the data, and the differences were assigned to be significant at a P <0.05. DAPs (including exclusively present proteins) were submitted to GO enrichment using ClusterProfiler package in R (Wu et al., 2021).
Malondialdehyde measurements
Malondialdehyde (MDA) was quantified using a colorimetric method (Stewart and Bewley, 1980). Briefly, full rosette leaves of 3-week-old plants (nine-rosette leaf stage) were pulverized in liquid nitrogen and resuspended in 80% (v/v) ethanol. The supernatant was collected and mixed with a reactant mixture of 0.65% (w/v) thiobarbituric acid and 20% (w/v) trichloroacetic acid. Absorbance of the supernatant was measured at 532 nm with a spectrophotometer (Cary 50) in 1-mL cuvettes. To reduce the nonspecific absorption from other organic compounds, absorbance of the supernatant was measured at 600 nm and subtracted from the absorbance at 532 nm.
In situ staining of hydrogen peroxide and cell death in Arabidopsis leaves
Production of hydrogen peroxide in the cell was visualized by DAB staining and quantitated by measuring the oxidation of 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) (Daudi and O'Brien, 2012). Cell death was assayed by trypan blue staining (Bhadauria et al., 2010).
In situ staining of hydrogen peroxide and cell death assays in Arabidopsis leaves
DAB staining
The in situ staining of hydrogen peroxide in Arabidopsis leaves was performed following established protocols (Daudi and O'Brien, 2012). Whole rosettes from 3-week-old plants were stained with 1 mg/mL 3,3′-freshly prepared DAB dissolved in 20-mM Na2HPO4 (pH 5.6) and 0.05% Tween-20. The leaves were stained at room temperature with shaking at 80–100 rpm under dark conditions for 6h. Following the incubation, the DAB staining solution was replaced with the destaining solution (80% ethanol) and incubated at 80°C for 15–20 min to remove the chlorophyll. This process was repeated until the chlorophyll was completely removed.
H2DCFDA staining
Cellular levels of H2O2 were studied by H2DCFDA staining (Pedersen et al., 2017). The mature outer rosette leaves from 3-week-old plants were detached and stained with 10-µM H2DCFDA prepared in phosphate buffer (pH 7.4) for 30 min. After incubation, the leaves were washed twice with potassium phosphate buffer to remove the extra staining solution. The leaves were examined under a Leica TCS SPE Microscope (Model: DMi8 automated) using an excitation wavelength of 488 nm, under 10× magnification.
Trypan blue staining
Arabidopsis leaves were assayed for cell death using trypan blue (Rawyler et al., 2002). Whole rosettes from 3-week-old plants were covered with lactophenol–trypan blue solution (10 mL of lactic acid, 10 mL of glycerol, 10 g of phenol, 10 mg of trypan blue, dissolved in 10 mL of distilled water). Leaves were stained at room temperature without vacuum infiltration in the solution for 1 h. Following incubation, the staining solution was replaced with 100% ethanol solution and incubated at room temperature overnight. The leaves were washed repeatedly in absolute ethanol until the green tissue became colorless. Leaves were then mounted onto glass microscopic slide with 60% glycerol (v/v) and were observed under Nikon SMZ1500 Microscope (WD136-Nikon). The blue precipitation indicating location of cell death was quantified using ImageJ software.
Real-time measurements of cytosolic Ca in plant leaves post anoxia
Plants were transformed with p35S:GCaMP3 using Agrobacterium tumefaciens GV3101 and floral dip according to (Atwell et al., 2015). Transformants were selected on hygromycin and T2 (or greater) lines used for analysis. Arabidopsis plants expressing the GFP-based Ca indicator GCaMP3 were imaged with a motorized fluorescence stereomicroscope (Zeiss Axio Zoom V16; Model, EMS3/SyCoP3) equipped with a PlanNeoFluar Z 1.0× objective lens and a Zeiss AxioCam HRm sCMOS camera. GCaMP3 was excited using a mercury lamp (with an intensity of 100% power of 150W Metal Halide Zeiss HXP 200c Illuminator), with a 470-/40-nm excitation filter, and a 500-nm dichroic mirror. The green fluorescent signal (Zeiss Filter Set 10 – GF) passing through a 535-/50-nm emission filter was acquired every 1 s for a total of 6 min with 210 Frames (360.55 s) using Zeiss Zen pro imaging software. Col-0 or cax1-1 harboring the GCaMP3 sensor constructs were grown on half-strength MS media supplemented with 0.5% (w/v) sucrose. For experiments that monitored the onset of anoxia, plants were grown in small Petri dishes then placed in a plastic bag with 1 AnaeroPack (Thermo Fisher, Waltham, MA, USA). The bag was modified to contain a glass-slide as a viewing window. Upon the onset of anoxia, the plant was immediately viewed through the window under the microscope as anoxic conditions formed within the bag. To minimize condensation, and not obstruct the imaging, a small ice pack was placed away from the plant inside prior to sealing the bag. Numerous no anoxia controls were performed (n > 20) to establish that the ice did not impact Ca signaling. Additionally, numerous (n > 30) experiments were performed for 30 min or more to establish that no signaling occurred after the initial Ca changes at the onset of anoxia. For post-anoxia measurements, the surgical tape around the plates was removed to allow air exchange in and out of the plates. The plates were then foil covered and placed in heat-sealed AnaeroPouch with one bag of AnaeroPack (Thermo Fisher, Waltham, MA, USA). GCamP3 fluorescence was measured to assay cytosolic Ca concentrations immediately after the plants were re-exposed to atmospheric oxygen. It took 15–30 s to reproducibly transfer the plants from the anoxic chamber to the microscope, precluding precise measurement during this initial period. Fluorescence was analyzed using ImageJ. Mean fluorescence intensity over time was then measured in the background and subtracted from the plant measurement. It is important to note that GCamP3 shows a very low signal under low Ca2+ and then up to a six-fold increase when Ca2+ levels rise. Therefore, the background represents a substantial proportion of the unstimulated signal (Supplemental Figure S16). Background was Col-0: 11.7 ± 3.4; cax1: 16.8 ± 5.7, mean ± sd. Experiments were run in duplicate each day and experiments repeated on different days over the course of multiple months until data from at least 10 replicates was recorded. See supplementary data for full description.
Statistical analysis
Statistical analyses were performed with analysis of variance (ANOVA) and Tukey’s post hoc testing or the Student’s t test formula in Microsoft Excel. Significance was set at P < 0.05. Data were presented as means ± sems.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number PRJNA666693. cax1-A (ABRC: CS25435), cax1-D (SALK_021486C), cax3 (ABRC: CS25429).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Anoxia effects on CAX mutants.
Supplemental Figure S2. Phenotypic characterization of CAX mutants grown under aerobic conditions.
Supplemental Figure S3. Overview of transcript changes following anoxia.
Supplemental Figure S4. Enriched GO categories of DEGs in Col-0, cax1, and cax3 during and post anoxia.
Supplemental Figure S5. RT–qPCR to compare with RNA-seq expression data.
Supplemental Figure S6. Genes shared by Col-0, cax1, and cax3 following anoxia.
Supplemental Figure S7. Heatmap showing the expression of the target genes of ERF-VII during normoxia, anoxia, and post anoxia.
Supplemental Figure S8. GO categories identified in cax1.
Supplemental Figure S9. Gene-concept network of specifically up-/downregulated transcripts in cax1 during anoxia.
Supplemental Figure S10. Gene-concept network of specifically downregulated transcripts in cax1 post anoxia.
Supplemental Figure S11. Gene-concept network of specifically up-/downregulated transcripts and increased/decreased proteins in cax1 post anoxia.
Supplemental Figure S12. Ca changes in Col-0 and cax1 during normoxia.
Supplemental Figure S13. Phenotyping of lines previously identified as altering vacuolar function, pH homeostasis, Ca transport and ROS signaling.
Supplemental Figure S14. Increased expression of heat-responsive transcripts in cax1 post anoxia.
Supplemental Figure S15. Assessing the transcripts of cax1 and cax3.
Supplemental Figure S16. Range of Ca reporter fluorescence signal.
Supplemental Table S1. Genes highly upregulated in Col-0 during anoxia.
Supplemental Table S2. Genes highly upregulated in cax1 during anoxia.
Supplemental Table S3. Genes highly upregulated in cax3 during anoxia.
Supplemental Table S4. Genes highly upregulated in Col-0 post anoxia.
Supplemental Table S5. Genes highly upregulated in cax1 post anoxia.
Supplemental Table S6. Genes highly upregulated in cax3 post anoxia.
Supplemental Table S7. Genes highly downregulated in Col-0 during anoxia.
Supplemental Table S8. Genes highly downregulated in cax1 during anoxia.
Supplemental Table S9. Genes highly downregulated in cax3 during anoxia.
Supplemental Table S10. Genes highly downregulated in Col-0 post anoxia.
Supplemental Table S11. Genes highly downregulated in cax1 post anoxia.
Supplemental Table S12. Genes highly downregulated in cax3 post anoxia.
Supplemental Table S13. Genes specifically upregulated in cax1 during normoxia.
Supplemental Table S14. Genes specifically down regulated in cax1 during normoxia.
Supplemental Table S15. Genes specifically upregulated in cax1 during normoxia, and in cax3 and Col-0 during anoxia and post anoxia.
Supplemental Table S16. Genes (717) commonly upregulated in cax1 during normoxia and Col-0 during and/or post anoxia.
Supplemental Table S17. Genes (27) commonly downregulated in cax1 during normoxia and Col-0 during and/or post anoxia.
Supplemental Table S18. Genes (47) commonly upregulated in cax3 during normoxia and Col-0 during and/or post anoxia.
Supplemental Table S19. Genes (22) commonly downregulated in cax3 during normoxia and Col-0 during and/or post anoxia.
Supplemental Table S20. HRGs downstream to ERFVII that are upregulated in cax1 during normoxia.
Supplemental Table S21. Genes specifically upregulated in cax1 during anoxia.
Supplemental Table S22. Genes specifically downregulated in cax1 during anoxia.
Supplemental Table S23. Genes specifically upregulated in cax1 post anoxia.
Supplemental Table S24. Genes specifically downregulated in cax1 post anoxia.
Supplemental Table S25. Identified proteins and their regulations upon 8 h anoxia treatment.
Supplemental Table S26. Expression pattern of selected mutants involved in signaling pathways (Ca, ROS, and pH) following anoxia and post anoxia.
Supplemental Table S27. Primer sequences for genes selected for RT–qPCR validation of RNA-seq analysis.
Supplemental Movie S1. Ca changes in Col-0 and cax1 post anoxia.
Supplementary Material
Acknowledgments
We thank Ardawna Green and Alex Sosa for managing the growth facilities and Jon Pittman for reviewing a draft of this work.
Funding
This work was supported by grants (to K.D.H) from the National Science Foundation (1557890), USDA (3092-51000-061-00D), and National Institute of Health (R03 AI149201-02) and grants (to S.G) from NSF MCB2016177 and NASA 80NSSC19K0126.
Conflict of interest statement. None declared.
Contributor Information
Jian Yang, Pediatrics-Nutrition, Children’s Nutrition Research, Baylor College of Medicine, Houston, Texas 77030, USA.
Iny Elizebeth Mathew, Pediatrics-Nutrition, Children’s Nutrition Research, Baylor College of Medicine, Houston, Texas 77030, USA.
Hormat Rhein, Pediatrics-Nutrition, Children’s Nutrition Research, Baylor College of Medicine, Houston, Texas 77030, USA.
Richard Barker, Department of Botany, Birge Hall, University of Wisconsin, Wisconsin, USA.
Qi Guo, Southern Cross Plant Science, Southern Cross University, Lismore, New South Wales, Australia.
Luca Brunello, Plant Lab, Institute of Life Sciences, Scuola Superiore Sant'Anna, San Giuliano Terme, Pisa, Italy.
Elena Loreti, Institute of Agricultural Biology and Biotechnology, National Research Council, 56124 Pisa, Italy.
Bronwyn J Barkla, Southern Cross Plant Science, Southern Cross University, Lismore, New South Wales, Australia.
Simon Gilroy, Department of Botany, Birge Hall, University of Wisconsin, Wisconsin, USA.
Pierdomenico Perata, Plant Lab, Institute of Life Sciences, Scuola Superiore Sant'Anna, San Giuliano Terme, Pisa, Italy.
Kendal D Hirschi, Pediatrics-Nutrition, Children’s Nutrition Research, Baylor College of Medicine, Houston, Texas 77030, USA.
J.Y., B.J.B., S.G., E.L., P.P., and K.D.H. designed the research; J.Y., I.E.M., H.R., Q.G., E.L., L.B., and K.D.H. performed the research; J.Y., I.E.M., R.B., Q.G., B.J.B., S.G., P.P., and K.D.H. analyzed the data. J.Y., B.J.B., S.G., P.P., and K.D.H. wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Kendel Hirschi (kendalh@bcm.edu).
References
- Ali R, Ma W, Lemtiri-Chlieh F, Tsaltas D, Leng Q, von Bodman S, Berkowitz GA (2007) Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell BJ, Greenway H, Colmer TD (2015) Efficient use of energy in anoxia-tolerant plants with focus on germinating rice seedlings. New Phytol 206: 36–56. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Chang R (2005) Sensing and signalling in response to oxygen deprivation in plants and other organisms. Ann Bot 96: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliardini C, Corso M, Verbruggen N (2016) Transcriptomic analysis supports the role of CATION EXCHANGER 1 in cellular homeostasis and oxidative stress limitation during cadmium stress. Plant Signal Behav 11: e1183861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliardini C, Meyer CL, Salis P, Saumitou-Laprade P, Verbruggen N (2015) CATION EXCHANGER1 cosegregates with cadmium tolerance in the metal hyperaccumulator Arabidopsis halleri and plays a role in limiting oxidative stress in Arabidopsis Spp. Plant Physiol 169: 549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banti V, Giuntoli B, Gonzali S, Loreti E, Magneschi L, Novi G, Paparelli E, Parlanti S, Pucciariello C, et al. (2013) Low oxygen response mechanisms in green organisms. Int J Mol Sci 14: 4734–4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkla BJ, Hirschi KD, Pittman JK (2008) Exchangers man the pumps: functional interplay between proton pumps and proton-coupled Ca2+ exchangers (Article Addendum). Plant Signal Behav 3: 354–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkla BJ, Vera-Estrella R, Hernandez-Coronado M, Pantoja O (2009) Quantitative proteomics of the tonoplast reveals a role for glycolytic enzymes in salt tolerance. Plant Cell 21: 4044–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera S, Wang N, Zhang C, Schmitz-Thom I, Strohkamp S, Schultke S, Hashimoto K, Xiong L, Kudla J (2015) Analyses of Ca2+ dynamics using a ubiquitin-10 promoter-driven Yellow Cameleon 3.6 indicator reveal reliable transgene expression and differences in cytoplasmic Ca2+ responses in Arabidopsis and rice (Oryza sativa) roots. New Phytol 206: 751–760 [DOI] [PubMed] [Google Scholar]
- Bhadauria V, Miraz P, Kennedy R, Banniza S, Wei Y (2010) Dual trypan-aniline blue fluorescence staining methods for studying fungus-plant interactions. Biotech Histochem 85: 99–105 [DOI] [PubMed] [Google Scholar]
- Blokhina OB, Chirkova TV, Fagerstedt KV (2001) Anoxic stress leads to hydrogen peroxide formation in plant cells. J Exp Bot 52: 1179–1190 [PubMed] [Google Scholar]
- Blumwald E, Poole RJ (1986) Kinetics of Ca2+/H+ antiport in isolated tonoplast vesicles from storage tissue of beta vulgaris L. Plant Physiol 80: 727–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw HD Jr (2005) Mutations in CAX1 produce phenotypes characteristic of plants tolerant to serpentine soils. New Phytol 167: 81–88 [DOI] [PubMed] [Google Scholar]
- Callewaert G, D’Hooge P, Ma TY, Del Vecchio M, Van Eyck V, Franssens V, Winderickx J (2020) Decreased vacuolar Ca(2+) storage and disrupted vesicle trafficking underlie alpha-synuclein-induced Ca(2+) dysregulation in S. cerevisiae. Front Genet 11: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalá R, Santos E, Alonso JM, Ecker JR, Martinez-Zapater JM, Salinas J (2003) Mutations in the Ca2+/H+ transporter CAX1 increase CBF/DREB1 expression and the cold-acclimation response in Arabidopsis. Plant Cell 15: 2940–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JM, Muhlemann JK, Gayomba SR, Muday GK (2019) RBOH-dependent ROS synthesis and ROS scavenging by plant specialized metabolites to modulate plant development and stress responses. Chem Res Toxicol 32: 370–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N-H, Pittman JK, Barkla BJ, Shigaki T, Hirschi KD (2003) The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell 15: 347–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N-H, Pittman JK, Shigaki T, Lachmansingh J, LeClere S, Lahner B, Salt DE, Hirschi KD (2005) Functional association of Arabidopsis CAX1 and CAX3 Is required for normal growth and ion homeostasis. Plant Physiol 138: 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WG, Miller G, Wallace I, Harper J, Mittler R, Gilroy S (2017) Orchestrating rapid long-distance signaling in plants with Ca(2+), ROS and electrical signals. Plant J 90: 698–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn SJ, Gilliham M, Athman A, Schreiber AW, Baumann U, Moller I, Cheng N-H, Stancombe MA, Hirschi KD, et al. (2011) Cell-specific vacuolar calcium storage mediated by CAX1 regulates apoplastic calcium concentration, gas exchange, and plant productivity in Arabidopsis. Plant Cell 23: 240–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Navazio L, Szabo I (2018) The contribution of organelles to plant intracellular calcium signalling. J Exp Bot 69: 4175–4193 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudi A, O’Brien JA (2012) Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio Protoc 2: e263 [PMC free article] [PubMed] [Google Scholar]
- Demidchik V, Shabala S, Isayenkov S, Cuin TA, Pottosin I (2018) Calcium transport across plant membranes: mechanisms and functions. New Phytol 220: 49–69 [DOI] [PubMed] [Google Scholar]
- Ellis MH, Dennis ES, Peacock WJ (1999) Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol 119: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnese FS, Menezes-Silva PE, Gusman GS, Oliveira JA (2016) When bad guys become good ones: the key role of reactive oxygen species and nitric oxide in the plant responses to abiotic stress. Front Plant Sci 7: 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle HH (2005) pH regulation in anoxic plants. Ann Bot 96: 519–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichman Y, Miller G, Mittler R (2019) Whole-plant live imaging of reactive oxygen species. Mol Plant 12: 1203–1210 [DOI] [PubMed] [Google Scholar]
- Gasch P, Fundinger M, Muller JT, Lee T, Bailey-Serres J, Mustroph A (2016) Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 28: 160–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Frangne N, Gomes E, Martinoia E, Palmgren MG (2000) The ACA4 gene of Arabidopsis encodes a vacuolar membrane calcium pump that improves salt tolerance in yeast. Plant Physiol 124: 1814–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Conde JV, Berckhan S, Prasad G, Mendiondo GM, Holdsworth MJ (2015) Group VII ethylene response factors coordinate oxygen and nitric oxide signal transduction and stress responses in plants. Plant Physiol 169: 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulcicek EE, Colangelo CM, McMurray W, Stone K, Williams K, Wu T, Zhao H, Spratt H, Kurosky A, Wu B (2005) Proteomics and the analysis of proteomic data: an overview of current protein-profiling technologies. Curr Protoc Bioinformatics Chapter 13:Unit 13.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleary R, Paez-Valencia J, Vens C, Toyota M, Palmgren M, Gilroy S (2020) Tonoplast-localized Ca(2+) pumps regulate Ca(2+) signals during pattern-triggered immunity in Arabidopsis thaliana. Proc Natl Acad Sci USA 117: 18849–18857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking B, Conn SJ, Manohar M, Xu B, Athman A, Stancombe MA, Webb AR, Hirschi KD, Gilliham M (2017) Heterodimerization of Arabidopsis calcium/proton exchangers contributes to regulation of guard cell dynamics and plant defense responses. J Exp Bot 68: 4171–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Colmer TD, Millar AH (2008) Does anoxia tolerance involve altering the energy currency towards Ppi? Trends Plant Sci 13: 221–227 [DOI] [PubMed] [Google Scholar]
- Huang X, Shabala L, Zhang X, Zhou M, Voesenek L, Hartman S, Yu M, Shabala S.. Cation transporters in cell fate determination and plant adaptive responses to the low oxygen environment. J Exp Bot 2022; 73: 636–645 [DOI] [PubMed] [Google Scholar]
- Igamberdiev AU, Hill RD (2018) Elevation of cytosolic Ca(2+) in response to energy deficiency in plants: the general mechanism of adaptation to low oxygen stress. Biochem J 475: 1411–1425 [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klecker M, Gasch P, Peisker H, Dormann P, Schlicke H, Grimm B, Mustroph A (2014) A shoot-specific hypoxic response of Arabidopsis sheds light on the role of the phosphate-responsive transcription factor PHOSPHATE STARVATION RESPONSE1. Plant Physiol 165: 774–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35: W345–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Xu L, Jamieson P (2020) Plant sense: the rise of calcium channels. Trends Plant Sci 25: 838–841 [DOI] [PubMed] [Google Scholar]
- Kriegel A, Andres Z, Medzihradszky A, Kruger F, Scholl S, Delang S, Patir-Nebioglu MG, Gute G, Yang H, et al. (2015) Job sharing in the endomembrane system: vacuolar acidification requires the combined activity of V-ATPase and V-Ppase. Plant Cell 27: 3383–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Urano D, Liao KL, Hedrick TL, Gao Y, Jones AM (2017) A nondestructive method to estimate the chlorophyll content of Arabidopsis seedlings. Plant Methods 13: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokdarshi A, Conner WC, McClintock C, Li T, Roberts DM (2016) Arabidopsis CML38, a calcium sensor that localizes to ribonucleoprotein complexes under hypoxia stress. Plant Physiol 170: 1046–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Perata P (2020) The many facets of hypoxia in plants. Plants (Basel) 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Valeri MC, Novi G, Perata P (2018) Gene regulation and survival under hypoxia requires starch availability and metabolism. Plant Physiol 176: 1286–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, van Veen H, Perata P (2016) Plant responses to flooding stress. Curr Opin Plant Biol 33: 64–71 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Berkowitz GA (2007) The grateful dead: calcium and cell death in plant innate immunity. Cell Microbiol 9: 2571–2585 [DOI] [PubMed] [Google Scholar]
- Manohar M, Shigaki T, Hirschi KD (2011) Plant cation/H+ exchangers (CAXs): biological functions and genetic manipulations. Plant Biol (Stuttg) 13: 561–569 [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miseta A, Kellermayer R, Aiello DP, Fu L, Bedwell DM (1999) The vacuolar Ca2+/H+ exchanger Vcx1p/Hum1p tightly controls cytosolic Ca2+ levels in S. cerevisiae. FEBS Lett 451: 132–136 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka C, Minami I, Oda K (2010) Hypoxia-inducible genes encoding small EF-hand proteins in rice and tomato. Biosci Biotechnol Biochem 74: 2463–2469 [DOI] [PubMed] [Google Scholar]
- Paul MV, Iyer S, Amerhauser C, Lehmann M, van Dongen JT, Geigenberger P (2016) Oxygen sensing via the ethylene response transcription factor RAP2.12 affects plant metabolism and performance under both normoxia and hypoxia. Plant Physiol 172: 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen O, Perata P, Voesenek L (2017) Flooding and low oxygen responses in plants. Funct Plant Biol 44: iii–vi [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman JK, Hirschi KD (2016) CAX-ing a wide net: cation/H transporters in metal remediation and abiotic stress signalling. Plant Biol (Stuttg) 18: 741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman JK, Hirschi KD (2016) Phylogenetic analysis and protein structure modelling identifies distinct Ca(2+)/Cation antiporters and conservation of gene family structure within Arabidopsis and rice species. Rice (NY) 9: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman JK, Shigaki T, Hirschi KD (2005) Evidence of differential pH regulation of the Arabidopsis vacuolar Ca2+/H+ antiporters CAX1 and CAX2. FEBS Lett 579: 2648–2656 [DOI] [PubMed] [Google Scholar]
- Rawyler A, Arpagaus S, Braendle R (2002) Impact of oxygen stress and energy availability on membrane stability of plant cells. Ann Bot 90: 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera M, Bassil E, Tajima H, Wimmer M, Chanoca A, Otegui MS, Paris N, Blumwald E (2015) pH regulation by NHX-type antiporters is required for receptor-mediated protein trafficking to the vacuole in Arabidopsis. Plant Cell 27: 1200–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavi-Rizi V, Herde M, Stohr C (2020) RNA-Seq reveals novel genes and pathways associated with hypoxia duration and tolerance in tomato root. Sci Rep 10: 1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signaling. Plant Cell 14: S401–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R, Hartman S, Liu Z, Martopawiro S, Sajeev N, van Veen H, Yeung E, Voesenek L (2018) Signal dynamics and interactions during flooding stress. Plant Physiol 176: 1106–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Mene-Saffrane L, Farmer EE, Krischke M, Mueller MJ, DellaPenna D (2006) Nonenzymatic lipid peroxidation reprograms gene expression and activates defense markers in Arabidopsis tocopherol-deficient mutants. Plant Cell 18: 3706–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RR, Weits DA, Feulner CFJ, van Dongen JT (2018) Oxygen sensing and integrative stress signaling in plants. Plant Physiol 176: 1131–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle BC (2010) Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 10: 1265–1269 [DOI] [PubMed] [Google Scholar]
- Shigaki T, Hirschi KD (2006) Diverse functions and molecular properties emerging for CAX cation/H+ exchangers in plants. Plant Biol 8: 419–429 [DOI] [PubMed] [Google Scholar]
- Shigaki T, Rees I, Nakhleh L, Hirschi KD (2006) Identification of three distinct phylogenetic groups of CAX cation/proton antiporters. J Mol Evol 63: 815–825 [DOI] [PubMed] [Google Scholar]
- Stewart RR, Bewley JD (1980) Lipid peroxidation associated with accelerated aging of soybean axes. Plant Physiol 65: 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Zhang J, Sachs MM (1994) Involvement of intracellular calcium in anaerobic gene expression and survival of maize seedlings. Plant Physiol 105: 369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Li K, Wang Z, Zhu K, Tan X, Cao J (2019) A review of plant vacuoles: formation, located proteins, and functions. Plants (Basel) 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Wang C, Gao Q, Li L, Luan S (2020) Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat Plants 6: 750–759 [DOI] [PubMed] [Google Scholar]
- Toyota M, Spencer D, Sawai-Toyota S, Jiaqi W, Zhang T, Koo AJ, Howe GA, Gilroy S (2018) Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veen H, Vashisht D, Akman M, Girke T, Mustroph A, Reinen E, Hartman S, Kooiker M, van Tienderen P, et al. (2016) Transcriptomes of eight Arabidopsis thaliana accessions reveal core conserved, genotype- and organ-specific responses to flooding stress. Plant Physiol 172: 668–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura I, Brunello L, Iacopino S, Valeri MC, Novi G, Dornbusch T, Perata P, Loreti E (2020) Arabidopsis phenotyping reveals the importance of alcohol dehydrogenase and pyruvate decarboxylase for aerobic plant growth. Sci Rep 10: 16669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virolainen E, Blokhina O, Fagerstedt K (2002) Ca(2+)-induced high amplitude swelling and cytochrome c release from wheat (Triticum aestivum L.) mitochondria under anoxic stress. Ann Bot 90: 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voesenek LA, Bailey-Serres J (2013) Flooding tolerance: O2 sensing and survival strategies. Curr Opin Plant Biol 16: 647–653 [DOI] [PubMed] [Google Scholar]
- Voesenek LA, Sasidharan R, Visser EJ, Bailey-Serres J (2016) Flooding stress signaling through perturbations in oxygen, ethylene, nitric oxide and light. New Phytol 209: 39–43 [DOI] [PubMed] [Google Scholar]
- Wang F, Chen ZH, Liu X, Colmer TD, Zhou M, Shabala S (2016) Tissue-specific root ion profiling reveals essential roles of the CAX and ACA calcium transport systems in response to hypoxia in Arabidopsis. J Exp Bot 67: 3747–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigand C, Kim SH, Brown E, Medina E, Mares M 3rd, Miller G, Harper JF, Choi WG (2021) A ratiometric calcium reporter CGf reveals calcium dynamics both in the single cell and whole plant levels under heat stress. Front Plant Sci 12: 777975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weits DA, Giuntoli B, Kosmacz M, Parlanti S, Hubberten HM, Riegler H, Hoefgen R, Perata P, van Dongen JT, Licausi F (2014) Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nature Commun 5: 3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt L, Ricachenevsky FK, Ziegler GZ, Clemens S, Walker E, Maathuis FJM, Kear P, Baxter I (2020) A curated list of genes that affect the plant ionome. Plant Direct 4: e00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Su N, Huang X, Cui J, Shabala L, Zhou M, Yu M, Shabala S (2021) Hypoxia-induced increase in GABA content is essential for restoration of membrane potential and preventing ROS-induced disturbance to ion homeostasis. Plant Commun 2: 100188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, et al. (2021) clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (NY) 2: 100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung E, van Veen H, Vashisht D, Sobral Paiva AL, Hummel M, Rankenberg T, Steffens B, Steffen-Heins A, Sauter M, et al. (2018) A stress recovery signaling network for enhanced flooding tolerance in Arabidopsis thaliana. Proc Natl Acad Sci USA 115: E6085–E6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP (2006) Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res 5: 2339–2347 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.