Abstract
Regulation of seed germination is important for plant survival and propagation. ABSCISIC ACID (ABA) INSENSITIVE5 (ABI5), the central transcription factor in the ABA signaling pathway, plays a fundamental role in the regulation of ABA-responsive gene expression during seed germination; however, how ABI5 transcriptional activation activity is regulated remains to be elucidated. Here, we report that C-type Cyclin1;1 (CycC1;1) is an ABI5-interacting partner affecting the ABA response and seed germination in Arabidopsis (Arabidopsis thaliana). The CycC1;1 loss-of-function mutant is hypersensitive to ABA, and this phenotype was rescued by mutation of ABI5. Moreover, CycC1;1 suppresses ABI5 transcriptional activation activity for ABI5-targeted genes including ABI5 itself by occupying their promoters and disrupting RNA polymerase II recruitment; thus the cycc1;1 mutant shows increased expression of ABI5 and genes downstream of ABI5. Furthermore, ABA reduces the interaction between CycC1;1 and ABI5, while phospho-mimic but not phospho-dead mutation of serine-42 in ABI5 abolishes CycC1;1 interaction with ABI5 and relieves CycC1;1 inhibition of ABI5-mediated transcriptional activation of downstream target genes. Together, our study illustrates that CycC1;1 negatively modulates the ABA response by interacting with and inhibiting ABI5, while ABA relieves the CycC1;1 interaction with and inhibition of ABI5 to activate ABI5 activity for the ABA response, thereby inhibiting seed germination.
The C-type cyclin protein CycC1;1 interacts with ABSCISIC ACID INSENSITIVE5 (ABI5) to downregulate the expression of ABA-responsive genes and ABI5 itself, thereby inhibiting ABA signaling during seed germination.
Introduction
Seed dormancy is a crucial process for angiosperms to adapt to changing environments especially stressful conditions (Finch-Savage and Leubner-Metzger, 2006). When sensing the favorable circumstance for growth, plants start with the release of seeds dormancy and launch of germination for seedling establishment (Liu et al., 2016; Nonogaki, 2017). Thus, the balance between seed dormancy and germination is precisely controlled in plants, which plays a vital role in plant survival and propagation as well as agricultural production (Shu et al., 2018). Various endogenous phytohormones affecting seed germination have been widely studied, and among them, abscisic acid (ABA) plays a pivotal role in promoting seed dormancy, inhibiting seed germination, and arresting seedling growth (Zhao et al., 2018). During the seed maturation and dormancy induction processes, the endogenous ABA level is substantially increased and ABA signaling pathway is strongly activated; on the contrary, ABA accumulation is decreased and its signaling is suppressed during seed germination and postgerminative seedling growth (Shu et al., 2017). Under adverse conditions or ABA treatment, increased ABA level in seeds is perceived by its receptors PYRABACTIN RESISTANCE PROTEINS/PYR-LIKE PROTEINS/REGULATORY COMPONENTS OF ABA RECEPTOR, relieving PHOSPHATASE2C-mediated repression of SNF1-RELATED PROTEIN KINASE2 (SnRK2) kinases (Ma et al., 2009; Park et al.,2009; Zhao et al., 2018). Many ABA-responsive element-binding factors (ABFs) implicated in plant ABA response are phosphorylated by the activated SnRK2 kinases, resulting in changes of gene expression (Fujii et al., 2009; Yoshida et al., 2014; Vishwakarma et al., 2017).
The basic leucine zipper (bZIP) transcription factor, ABA INSENSITIVE5 (ABI5) is a central player in ABA-mediated inhibition of seed germination (Zhao et al., 2020). ABI5 recognizes and binds to the ABA response elements (ABREs), a G-box of numerous ABA-responsive genes, such as EARLY METHIONINE-LABELED1 (EM1), EM6, and RESPONSIVE TO DESICCATION29A (RD29A), which encode products important for plant ABA response during seed germination, and transcriptionally activates the expression of these genes (Yamaguchi-Shinozaki et al., 1992; Nakashima et al., 2009). Both EM1 and EM6 are belonging to the late embryogenesis abundant proteins, and have been used as molecular markers of late embryogenesis (Gaubier et al., 1993; Yang et al., 2021). Although EM1 and EM6 have different expression pattern, but both of them are strongly activated by ABA and thus involved in the acquisition of desiccation response and establishment of embryo dormancy (Wise, 2003; del Viso et al., 2007). ABI5 expression and activity are tightly regulated in plants to ensure optimal responses to internal and external cues. SnRK2-mediated phosphorylation of ABI5 results in its stabilization and activation in plant response to ABA. Serine (Ser)-42, Ser-145, and Threonine (Thr)-201 are the major phosphorylation sites of ABI5, and the phospho-dead forms of ABI5 at these sites severely block its transactivation of the targeted genes while the phospho-mimic forms promote such activity (Wang et al., 2013; Zhou et al., 2015). On the contrary, ABI5 activity can be inactivated by phytochrome-associated Ser/Thr protein phosphatase/Ser/Thr-specific phosphoprotein phosphatase 6-mediated de-phosphorylation (Dai et al., 2013). In addition, ABI5 is also posttranslationally regulated by DWD HYPERSENSITIVE TO ABA1-mediated ubiquitination (Lee et al., 2010), nitric oxide-mediated S-nitrosylation (Albertos et al., 2015), and SAP AND MIZ1 DOMAIN-CONTAINING LIGASE1-mediated sumoylation (Zheng et al., 2012), which disrupts plant ABA response through promoting ABI5 degradation.
In addition to the posttranslational regulation, ABI5 is also regulated at the transcriptional level. Several transcription factors that positively or negatively regulate ABI5 expression by binding to ABI5 promoter have been identified, including ANAC060, ABI3, WRKY40, HY5, a B box-containing protein BBX19, MCM1/AGAMOUS/DEFICIENS/SRF-box transcription factors AGL21 and RELATED TO ABI3/VP1 (Xu et al., 2014; Yu et al., 2017, 2020; Bai et al., 2019; Wang et al., 2020). In addition, ABI5 can upregulate ABI5 transcription by binding to its own promoter (Xu et al., 2014; Bai et al., 2019). Notably, a REGULATOR OF CHROMATIN CONDENSATION1 (RCC1) family protein SENSITIVE TO ABA1 (SAB1) and the transcription factor BRINSENSITIVE1-EMS-SUPPRESSOR1 suppress the expression of ABI5 and ABI5-targed genes through physical interaction, thereby interfering plant responses to ABA (Ji et al., 2019; Zhao et al., 2019).
The regulatory information of most targeted gene transcription conveyed by transcription factors generally needs to be delivered to RNA polymerase II (RNAP II), and the Mediator complex as a signal processor plays a necessary role in this process by establishing the linkage (Asturias et al., 1999; Agrawal et al., 2021). Mediator complex is conserved in eukaryotes, and comprises four modules, including head, middle, tail, and kinase modules. The head, middle, and tail modules form the core part of Mediator complex, while the kinase module is a separable part of the complex consisting of CYCLIN-DEPENDENT KINASE8 (CDK8), C-type cyclin (CycC), MEDIATOR COMPLEX SUBUNIT12 (MED12), and MED13 (Wang and Chen, 2004; Mathur et al., 2011; MaJi et al., 2019). The head and the tail modules serve as the physical interaction interfaces for RNAP II and sequence-specific transcription factors, respectively, and the kinase module is able to alter the function of Mediator through association or de-association with the core part, thereby negatively or positively regulating gene transcription (Poss et al., 2013). Emerging evidence has revealed the involvement of the Mediator complex in plant ABA responses. For example, MED25, a subunit of Mediator tail module, physically interacts with ABI5 and exerts a negative effect on the expression of ABI5-targeted genes (Chen et al., 2012). MED16 can compete with MED25 to interact with ABI5, thus positively regulating plant ABA signaling (Guo et al., 2021). The biological role of CDK8, a subunit of the kinase module, has also been studied in plant ABA response, where it is associated with ERF/AP2 transcription factor RAP2.6 and SnRK2.6 to positively modulate ABA signaling and drought response in Arabidopsis (Arabidopsis thaliana) (Zhu et al., 2020). In contrast, CycC in Arabidopsis is a small family consisting with two members, CycC1;1 and CycC1;2, and in addition to their role in resistance to necrotrophic pathogens in Arabidopsis (Zhu et al., 2020), functions of CycC in plant stress responses remains elusive.
In this study, we report that CycC1;1 is an ABI5-interacting protein, and negatively modulates ABA-inhibited seed germination by inhibiting the transcriptional activation activity of ABI5 for the downstream targeted genes including ABI5 itself. Mutation of CycC1;1 resulted in increased sensitivity in plant response to ABA whereas its overexpression led to decreased sensitivity. CycC1;1 physically interacts with ABI5 to occupy the promoters of ABI5-targeted genes and ABI5 itself, and thus interfering its transcriptional activation activity for its target genes as well as ABI5; while ABA-induced ABI5 phosphorylation at Ser-42 relieves its repression of ABI5 by impairing their physical interaction. Thus, our study unravels the role of CycC1;1 in ABA signaling transduction by interacting with and inhibiting ABI5 during seed germination.
Results
CycC1;1 physically interacts with ABI5
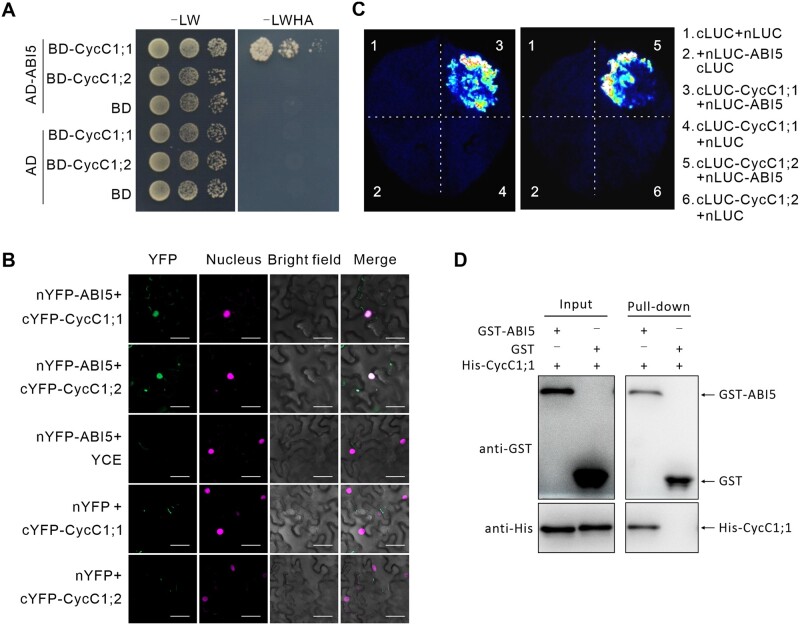
ABI5, as a central transcription factor in ABA signaling pathway, plays a fundamental role in seed dormancy and germination by regulating ABA-triggered gene expression. We assumed that identification of ABI5-interacting proteins would gain more insights into our understanding for the precise regulation of ABI5 in seed germination and ABA response. Using ABI5 as a bait, a C-type Cyclin family member CycC1;1 (AT5G48630) was identified as a putative partner that could interact with ABI5 in yeast two-hybrid (Y2H) system. To verify this interaction, we cloned full-length coding sequence (CDS) of CycC1;1 and re-examined its interaction between ABI5 in yeast cells, our results showed that CycC1;1 and ABI5 indeed had interaction in yeast cells (Figure 1A). The interaction was also confirmed by bimolecular fluorescence complementation (BiFC) assay, in which ABI5 fused to the N-terminal half of yellow fluorescent protein (YFP), CycC1;1 fused to the C-terminal half of YFP and nuclear marker H2B-mCherry (Rosa et al., 2014) were co-expressed in Nicotiana benthamiana leaves. Reconstituted fluorescence was observed in the nucleus while no fluorescence was observed in negative controls (Figure 1B), indicating that CycC1;1 interacts with ABI5 in planta. Similarly, our luciferase (LUC) complementation imaging (LCI) experiments further confirmed the interaction between CycC1;1 and ABI5, as LUC activity was only detected in N. benthamiana leaves where CycC1;1 fused with the N-terminal of LUC and ABI5 fused with the C-terminal of LUC were co-expressed (Figure 1C). To examine whether CycC1;1 has physical interaction with ABI5 in vitro, we purified 6×histidine (His)-tagged CycC1;1 and glutathione S-transferase (GST)-tagged ABI5 proteins expressed in Escherichia coli, and performed GST pull-down assay using glutathione-sepharose beads. Our results showed that His-CycC1;1 could be specifically pulled down by GST-ABI5 but not by GST tag alone (Figure 1D), indicating that CycC1;1 physically interacts with ABI5 in vitro. These results reveal that CycC1;1 has interaction with ABI5 in vitro and in vivo.
Figure 1.
CycC1;1 and CycC1;2 interact with ABI5. A, Y2H assay shows the interaction between ABI5 and CycC1;1 or CycC1;2. CycC1;1 or CycC1;2 and ABI5 were fused to the transcriptional activation domain (AD) or DNA-BD of GAL4. Protein interactions were examined based on the growth of yeast cells on selective media. -L/W indicates Leu and Trp, -L/W/H/A indicates Trp, Leu, His, and Ade drop-out plates. B, BiFC analysis. ABI5-nYFP and cYFP-CycC1;1 or cYFP-CycC1;2 were transiently co-expressed in N. benthamiana leaves. H2B-mCherry (Rosa et al., 2014) was used as a nuclear marker. Scale bars = 20 μm. C, Split LUC assay. nLUC-ABI5 and cLUC-CycC1;1 or cLUC-CycC1;2 constructs were transiently expressed in N. benthamiana leaves. cLUC, C-terminus of LUC; nLUC, N-terminus of LUC. D, GST pull-down analysis. 6×His-CycC1;1 were mixed with GST-ABI5 or GST and immobilized on glutathione sepharose beads. After washing, the eluted proteins were subjected to immunoblot analysis with anti-GST and anti-His antibodies, respectively.
The C-type Cyclin family in Arabidopsis (A. thaliana) has another member, CycC1;2 (AT5G48630) in addition to CycC1;1, thus we further investigated whether CycC1;2 has interaction with ABI5 in plants. For this end, we performed Y2H experiment, and found that the yeast cells co-expressing both CycC1;2 and ABI5 could grow on triple dropout medium but not quadruple dropout medium (Figure 1A andSupplemental Figure S1A), revealing weaker interaction between CycC1;2 and ABI5 than that between CycC1;1 and ABI5 in yeast cells. We also carried out BiFC and LCI assays, showing that CycC1;2 could interact with ABI5 in N. benthamiana leaves (Figure 1, B and C). These results revealed that similar to CycC1;1, CycC1;2 also has interaction with ABI5 in planta. Considering that C-type cyclin and CDK8 are members of the kinase module in the Mediator complex, we wondered whether CDK8 could also interact with ABI5. CDK8 could indeed interact with CycC1;2; however, no interaction between CDK8 and ABI5 was observed in yeast cells (Supplemental Figure S1B).
CycC1;1 is epistatic to ABI5 to negatively regulate ABA-inhibited seed germination
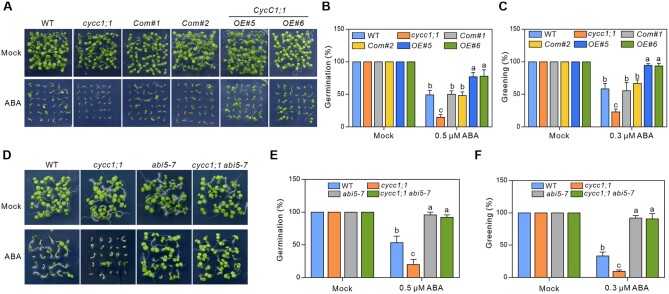
The interaction of CycC1;1/CycC1;2 with ABI5 prompted us to test whether they functions in ABA-regulated seed germination. We first obtained a previously reported T-DNA insertion line of CycC1;1, SALK_053291 (Zhu et al., 2014), where the T-DNA is inserted in the second intron of CycC1;1, resulting in reduced expression of CycC1;1 but not CycC1;2 (Supplemental Figure S2, A–C). Our results showed that both the wild-type (WT) and cycc1;1 mutant exhibited similar germination and cotyledon greening rates in the absence of ABA, whereas when subjected to ABA treatment, the mutant displayed significantly higher sensitivity to ABA than the WT as evidenced by less germination and cotyledon greening rates (Figure 2, A–C), revealing that CycC1;1 negatively regulates ABA-mediated seed germination. To confirm that the increased ABA sensitivity of the cycc1;1 mutant is due to defective CycC1;1 expression caused by T-DNA insertion in the mutant, a binary vector harboring CycC1;1::CycC1;1 was transformed into cycc1;1 mutant for the complementation analysis. Decreased CycC1;1 expression level in the cycc1;1 mutant was fully restored in the transgenic complementation lines (Supplemental Figure S2B). Consistently, increased ABA sensitivity of the mutant in terms of the seed germination and cotyledon greening rates was also rescued by the expression of CycC1;1 under the control of its native promoter (Figure 2, A–C), further supporting the negative effects of CycC1;1 on ABA-inhibited seed germination. We also tested ABA sensitivity of the CycC1;1-overexpressing transgenic lines where the expression of CycC1;1-GFP was driven by the 35S promoter (Figure 2B). In contrast to the cycc1;1 mutant, CycC1;1-overexpressing lines exhibited remarkably insensitive phenotype to ABA as they had higher seed germination and cotyledon greening rates than the WT in the presence of ABA (Figure 2, A–C), further validating the negative regulation of CycC1;1 in ABA-inhibited seed germination.
Figure 2.
CycC1;1 suppresses ABA-inhibited seed germination through ABI5. A, Seedlings of 7-day-old various genotypes germinated on MS medium supplemented without or with 0.3 μM ABA. B, Germination percentages of the various genotypes in response to ABA. Seed germination on MS medium without or with 0.5 μM ABA was recorded after 3 days of stratification. C, Percentages of greening cotyledon of various genotypes. Cotyledon greening was recorded 7 days after stratification on MS medium supplemented without or with 0.3 μM ABA. D, Seedlings of 7-day-old WT, cycc1;1, abi5-7, and cycc1;1 abi5-7 germinated on MS medium supplemented without or with 0.3 μM ABA. E, Germination percentages of the WT, cycc1;1, abi5-7, and cycc1;1 abi5-7 in response to ABA. Seed germination on MS medium without or with 0.5 μM ABA was recorded after 3 days of stratification. F, Percentages of greening cotyledon of the WT, cycc1;1, abi5-7, and cycc1;1 abi5-7. Cotyledon greening was recorded 7 days after stratification on MS medium supplemented without or with 0.3 μM ABA. Data indicate mean ± sd (n = 3). Bars with different letters indicate significant differences at P < 0.05, revealed using ANOVA analysis.
The genes of CycC1;1 and CycC1;2 are linked in the Arabidopsis genome (Supplemental Figure S2A). We could not obtain the single mutant of CycC1;2, but another T-DNA insertion line SALK_039400, where the T-DNA is inserted in the intergenic region between CycC1;1 and CycC1;2, was identified (Supplemental Figure S2A). Consistent with the previously reports (Zhu et al., 2014; Chen et al., 2019), the SALK_039400 line indeed had reduced expression of both CycC1;1 and CycC1;2, thus it was named cycc1;1/1;2 (Supplemental Figure S3A). Similar to cycc1;1 mutant, the cycc1;1/1;2 mutant also showed higher ABA sensitivity than the WT in terms of seed germination and cotyledon greening rate (Supplemental Figure S3, B–D), suggesting a similar negative role of CycC1;2 to CycC1;1 in regulating ABA-inhibited seed germination. To further confirm the role of CycC1;2 in ABA response, we generated CycC1;2-overexpressing lines and tested their ABA sensitivity (Supplemental Figure S3A). As expected, the CycC1;2-overexpressing lines had higher seed germination and cotyledon greening rate than the WT in the presence of ABA (Supplemental Figure S3, B–D). These results further support that both CycC1;1 and CycC1;2 negatively regulate ABA-inhibited seed germination.
Considering that both CycC1;1 and CycC1;2 could interact with ABI5, we hypothesized that CycC1;1 and CycC1;2 may affect ABA-mediated seed germination through ABI5. If this was the case, mutation of ABI5 in the cycc1;1 mutant should revert its ABA hypersensitivity as the ABI5 loss-of-function mutants are insensitive to ABA (Zhao et al., 2020). Therefore, to genetically analyze the relationship between CycC1;1 and ABI5, we crossed cycc1;1 with abi5-7 (Zhao et al., 2020), resulting in cycc1;1 abi5-7 double mutant. The abi5-7 mutant was indeed very insensitive to ABA (Figure 2, D–F), which is consistent with previous reports (Zhao et al., 2020), the cycc1;1 mutant was hypersensitive to ABA while the cycc1;1 abi5-7 double mutant showed identical ABA sensitivity to abi5-7 in terms of seed germination and cotyledon greening rates upon ABA treatment (Figure 2, D–F). These results demonstrate that CycC1;1 acts epistatically to ABI5 in the negative regulation of ABA-mediated seed germination.
CycC1;1 interferes ABI5 transcriptional activation of the downstream targets
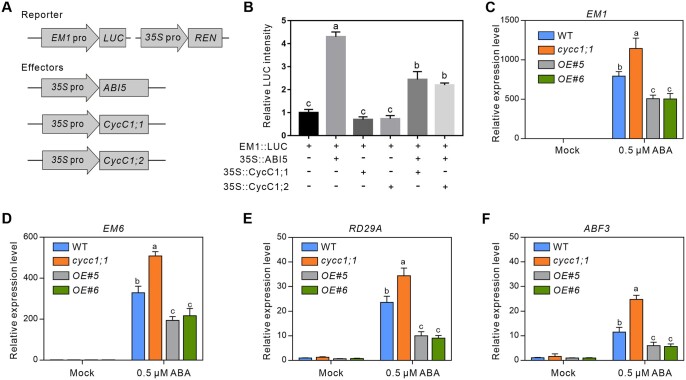
Our above results reveal that CycC1;1 negatively regulates ABA-inhibited seed germination, while ABI5 is a positive factor favoring plant ABA responses including inhibiting seed germination. We speculated that CycC1;1 may affect the transcriptional activation activity of ABI5 through their physical interaction. Because EM1 is reported as a direct target gene of ABI5 during seed germination, we performed dual-LUC reporter gene assays in N. benthamiana leaves, where LUC was used as a reporter under the control of EM1 promoter and Renilla LUC (REN) under the control of the constitutive 35S promoter was used an internal control (Figure 3A). Our results showed that LUC activity was extensively stimulated when 35S::ABI5 as an effector was expressed but remained unchanged when 35S::CycC1;1 alone was expressed, while this ABI5-induced LUC activity was significantly compromised by the co-expression of ABI5 and CycC1;1 (Figure 3B), revealing that CycC1;1 interferes the activation of EM1 expression by ABI5. Similarly, ABI5-induced LUC activity could also be suppressed by the expression of CycC1;2 (Figure 3B). Consistent with this, our reverse-transcription quantitative PCR (RT-qPCR) data showed that ABA-induced EM1 transcripts in the WT were significantly enhanced in the cycc1;1 mutant but repressed in the CycC1;1-overexpressing plants (Figure 3C), further supporting the negative effects of CycC1;1 on ABI5-induced EM1 expression in plant response to ABA. In addition to EM1, we also examined the expression levels of other ABI5 target genes that are implicated in seed germination, including EM6, RD29A, and ABF3. The RT-qPCR results showed that the expression levels of these ABI5-targeted genes in the WT were greatly induced by ABA, while such changes were further promoted in the cycc1;1 mutant and suppressed in the CycC1;1-overexpressing plants, respectively (Figure 3, D–F). Together, these data demonstrated that CycC1;1 interferes ABI5-mediated transcriptional activation of the downstream target genes during ABA-inhibited seed germination.
Figure 3.
CycC1;1 and CycC1;2 suppress the transcriptional activation activity of ABI5 for the downstream genes. A and B, Dual-LUC reporter gene assay to examine the role of CycC1;1 and CycC1;2 in ABI5-activated EM1 expression. The schematic diagram (A) shows the reporters and effectors used in the assay. The effector and reporter genes were co-expressed in N. benthamiana leaves, and the activities of LUC and REN were detected. The relative LUC intensity (B) represents the EM1::LUC activity relative to the internal control (REN driven by the 35S promoter). The activity of EM1::LUC alone was set to 1. Error bars indicate mean ± sd (n = 3). C–F, Expression of EM1, EM6, RD29A, and ABF3 in germinating seeds of the WT, cycc1;1 and CycC1;1-overexpression lines. Seeds were treated with MS medium or MS medium containing 10 μM ABA for 24 h. Error bars indicate mean ± sd (n = 3). Different letters indicate significant differences as determined using one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
CycC1;1 represses ABI5 expression by associating with its genomic DNA
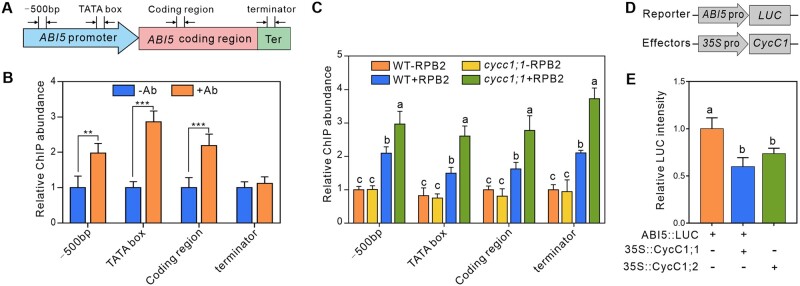
Considering that ABI5 can bind to the promoter of its own gene and activate gene expression, we therefore further explored whether CycC1;1 affects ABI5 expression at the transcriptional level. To achieve this, we first assayed whether CycC1;1 is associated with ABI5 genomic DNA by chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR). 35S::GFP-CycC1;1 transgenic plants and anti-GFP antibody were used for the ChIP, and quantitative PCR primers designed at different positions, including 500-bp upstream of translation start site, TATA box (the RNAP II binding site), coding and terminator regions of ABI5 were used for the subsequent qPCR (Figure 4A). Our results showed that DNA fragments in −500 bp upstream, TATA box, and coding region but not terminator were significantly enriched by anti-GFP antibody compared with the control without antibody (Figure 4B), revealing that CycC1;1 is associated with ABI5 promoter DNA. The core part of Mediator regulates gene transcription by acting as a bridge that connects RNAP II and transcription factors, and this connection can be affected positively or negatively by the kinase module (Agrawal et al., 2021). We then performed ChIP-qPCR to investigate the recruitment of RNAP II to ABI5 in the WT and cycc1;1 mutant plant using a specific antibody raised against the C-terminal domain of RNAP II. Our results showed that RNAP II was associated with the ABI5 genomic DNA in the WT while this association was significantly enhanced in the cycc1;1 mutant (Figure 4C), revealing that CycC1;1 disrupted RNAP II recruitment to ABI5 in planta. We speculated that this enhanced association of RNAP II with ABI5 promoter in cycc1;1 mutant likely leads to increased expression of ABI5. To test this hypothesis, we carried out dual-reporter gene assay in N. benthamiana leaves by expressing LUC driven by ABI5 promoter in the presence or absence of CycC1;1 or CycC1;2 (Figure 4D). The result showed that LUC expression under the control of ABI5 promoter was significantly repressed by the co-expression of either CycC1;1 or CycC1;2 (Figure 4E). These data indicate that CycC1;1 and CycC1;2 can associate with ABI5 promoter to reduce RNAP II recruitment, thereby repressing ABI5 transcription.
Figure 4.
CycC1;1 decreases ABI5 transcription by interfering RNAP II association with ABI5 promoter. A, A diagram showing the positions of ABI5 gene primers used for ChIP-qPCR. B, CycC1;1 associates with the ABI5 promoter. Chromatin was extracted from 7-day-old CycC1;1-overexpression (35S::GFP-CycC1;1) lines and then precipitated with anti-GFP antibody (+Ab) or only IgG (−Ab). Error bars indicate mean ± sd (n = 3). Asterisks indicate significant differences determined by Student’s t test (**P < 0.01 and ***P < 0.001). C, CycC1;1 reduces RNAP II association to the ABI5 promoter. Chromatin was extracted from 7-day-old WT and cycc1;1 mutant seedlings and precipitated with anti-RPB2 antibody raised against the C-terminal domain of RNAP II (+RPB2) or only IgG (−RPB2). Error bars indicate mean ± sd (n = 3). D and E, Dual-LUC reporter gene assay to examine the effects of CycC1;1 and CycC1;2 on ABI5 expression. LUC under the control of ABI5 promoter was co-expressed with CycC1;1 or CycC1;2 in N. benthamiana leaves. The activity of ABI5::LUC alone was set to 1. Error bars indicate mean ± sd (n = 3). Different letters indicate significant differences as determined using one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
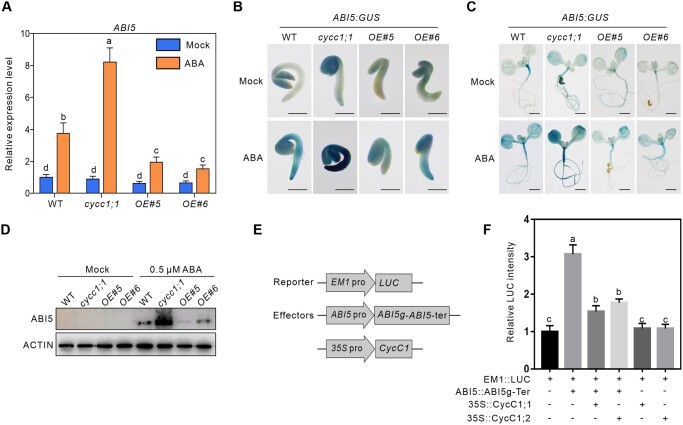
Consistent with the above results, the RT-qPCR data showed that ABI5 transcript levels in ABA-treated cycc1;1 mutant and CycC1;1-overexpressing plants were significantly higher and lower than that in the treated WT, respectively (Figure 5A). In addition, GUS reporter activity under the control of ABI5 promoter was also monitored in the WT, cycc1;1 and CycC1;1-overexpressing background plants. We found that ABA-induced GUS activity in the germinating seeds and seedlings of the WT was strongly increased in the cycc1;1 mutant but extensively decreased in the CycC1;1-overexpressing plants as evidenced by the GUS staining experiment (Figure 5, B and C). Furthermore, ABI5 protein abundance was increased in the cycc1;1 mutant but decreased in the CycC1;1-overexpressing plants, respectively, compared with the WT when treated with ABA (Figure 5D).
Figure 5.
CycC1;1 represses ABI5 expression during seed germination. A, The expression of ABI5 in germinating seeds of the WT, cycc1;1, and CycC1;1-OE lines. Seeds were treated with MS medium or MS medium containing 10 μM ABA for 24 h. Error bars indicate mean ± sd (n = 3). B and C, GUS staining of germinating seeds (B) and seedlings (C) of ABI5::GUS transgenic line in the WT, cycc1;1 and CycC1;1-OE background plants. One-day-old and 7-day-old seedlings grown on MS medium were transferred to MS medium containing 10 μM ABA for 24 h, and then subjected to GUS staining. Bar, 0.2 cm. D, ABI5 protein abundance in the seeds of WT, cycc1;1 and CycC1;1-OE lines. Seeds were treated with MS medium without or with 10 μM ABA for 24 h. The anti-ABI5 antibody was used in the immune analysis. E and F, Dual-LUC reporter gene assay to examine the effects of ABI5 and CycC1;1 on EM1 expression. The intact ABI5 genomic DNA containing its promoter, coding region, and terminator was cloned into a binary vector to transiently express ABI5 protein in N. benthamiana leaves. The activity of EM1::LUC alone was set to 1. Error bars indicate mean ± sd (n = 3). Different letters indicate significant differences as determined using one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
To examine the effect of CycC1;1-repressed ABI5 transcription on the activation of ABI5-targeted genes, intact ABI5 genomic DNA ABI5::ABI5g-Ter including promoter, coding region, and terminator was constructed and employed as an effector, and EM1::LUC was used as a reporter (Figure 5, E and F). The dual-reporter gene assay results showed that expression of ABI5 but not CycC1;1 alone could significantly stimulate LUC activity while when ABI5 and CycC1;1 are co-expressed, ABI5-increased LUC activity was remarkably suppressed (Figure 5F). Such effects of CycC1;1 on inhibiting ABI5-promoted EM1 expression were also observed when CycC1;2 was co-expressed (Figure 5F). These results indicate that CycC1;1 and CycC1;2 negatively regulate ABI5 activity at the transcription level, thus down-regulating the expression of ABI5-targeted genes.
ABA-induced phosphorylation of ABI5 at Ser42 dampens its interaction with CycC1;1
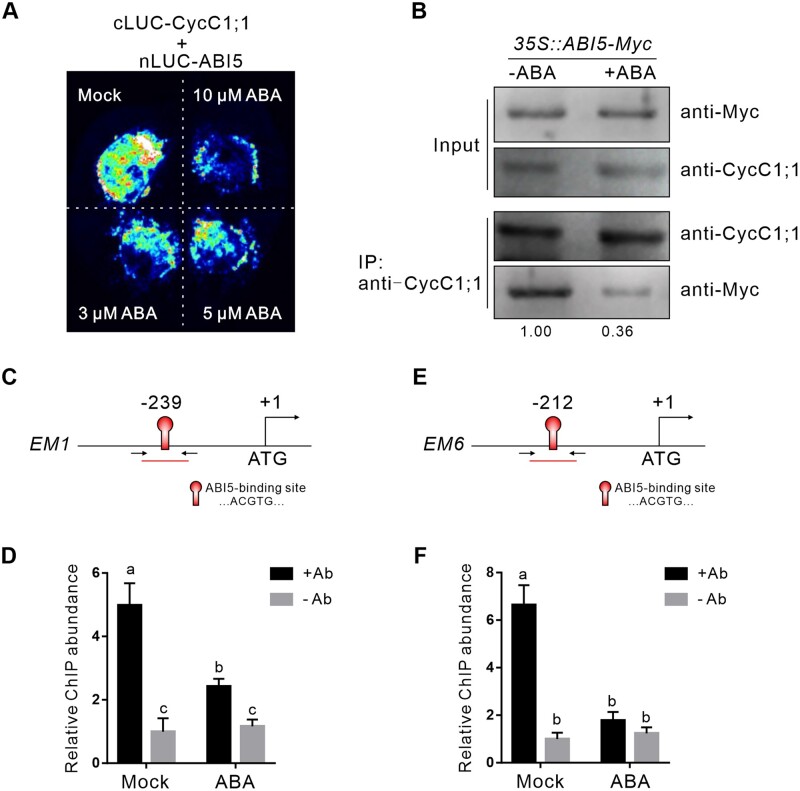
We have demonstrated that CycC1;1 negatively regulates ABI5 activity through physical interaction, whereas whether and how ABA affects their interaction remains elusive. We carried out split-LUC experiments by expressing both cLUC-CycC1;1 and nLUC-ABI5 in N. benthamiana leaves. Following treatment with different concentrations of ABA, LUC activity was monitored, and we found that the LUC signal intensity was extensively suppressed by the ABA treatments compared with the untreated control in the N. benthamiana leaves (Figure 6A), revealing ABA reduced the interaction between CycC1;1 and ABI5. This was confirmed by the co-immunoprecipitation (Co-IP) assay where untreated and ABA-treated 35S::ABI5-Myc seedlings were employed to the isolate the CycC1;1-interacting proteins using anti-CycC1;1 antibody (Supplemental Figure S2D) and then Myc-tagged ABI5 protein was analyzed by anti-Myc antibody in the precipitants, and our result showed that the precipitated ABI5-Myc protein was less in the ABA-treated seedlings than the untreated control (Figure 6B). These results imply that ABA could reduce CycC1;1 interaction with ABI5 in planta. If this is really the case in plants, association of CycC1;1 on ABI5-targeted genes could be also suppressed by ABA. Previous reports documented that ABI5 binds to the ABRE motif containing ACGTG in the EM1 or EM6 promoter. To test this hypothesis, we performed ChIP-qPCR by analyzing the enrichment of CycC1;1 on these ABRE sites in EM1 and EM6 promoters. Without ABA treatment, CycC1;1 was indeed significantly associated with EM1 or EM6 promoter DNA, while ABA treatment remarkably inhibited such association (Figure 6, C–F), further supporting that ABA could dampen CycC1;1 interaction with ABI5 in plants.
Figure 6.
ABA reduces CycC1;1 interaction with ABI5 and association with the promoters of EM1 and EM6. A, Split LUC assay to examine the interaction between CycC1;1 and ABI5 in the absence or presence of different concentrations of ABA. nLUC-ABI5 and cLUC-CycC1;1 constructs were transiently expressed in N. benthamiana leaves for 2 days, and then different concentrations of ABA solution were injected into the leaves. Following the injection for 1 day, the LUC images were captured. B, Co-IP to assay the effect of ABA on the interaction between CycC1;1 and ABI5 in Arabidopsis. Seven-day-old 35S::ABI5-Myc transgenic seedlings treated with or without 10 μM ABA for 12 h were employed to isolate the total proteins, and then proteins were immunoprecipitated by the anti-CycC1;1 antibody. After immunoprecipitation, anti-Myc antibody was used to examine the ABI5-Myc protein in the precipitate. C–F, ChIP-qPCR to assay the association of CycC1;1 with the ABI5-binding sites on the EM1 or EM6 promoters. Diagrams showing the positions of ABI5-binding sites on the EM1 and EM6 promoters and the primers used for the ChIP-qPCR (C and E). Chromatin was extracted from 7-day-old 35S::GFP-CycC1;1 seedlings treated with or without 10 μM ABA for 12 h, and then precipitated with anti-GFP antibody (+Ab) or only IgG (−Ab). Error bars indicate mean ± sd (n = 3). Different letters indicate significant differences as determined using two-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
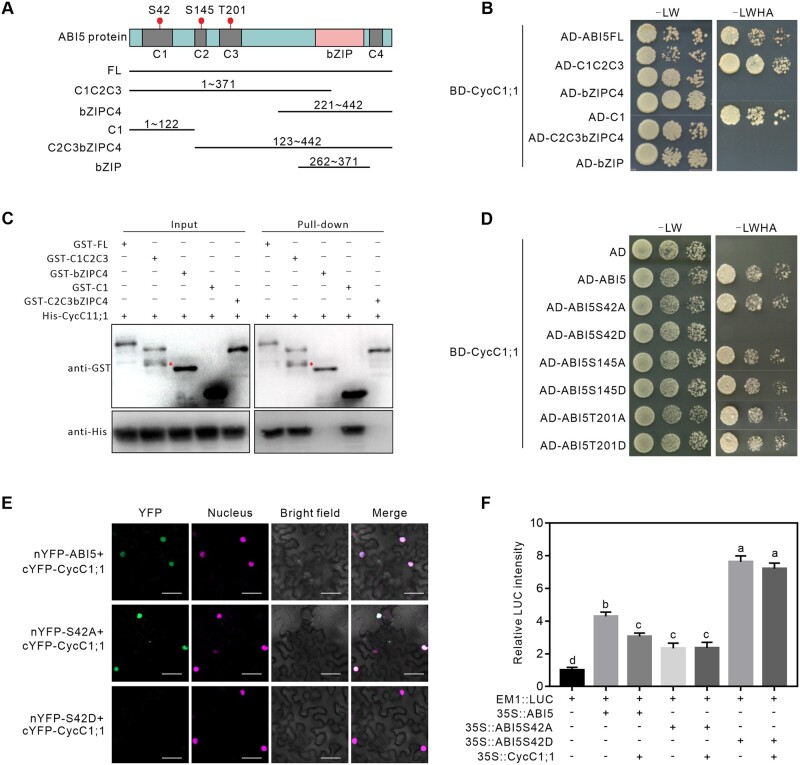
To explore how ABA affects CycC1;1 interaction with ABI5, we mapped CycC1;1-interacting domain of ABI5 protein by Y2H. Based on the previous functional analysis of ABI5 protein structure, ABI5 has four conserved regions (C1, C2, C3, and C4) and a bZIP DNA binding domain (BD) (Bensmihen et al., 2002; Chen et al., 2012), thus we generated several truncated forms of ABI5 (Figure 7A). The Y2H results showed that only the C1 domain or the truncated C1C2C3 fragment covering the C1 region had interaction with CycC1;1, while the truncated fragments without C1 region including bZIPC4 and C2C3bZIPC4 did not (Figure 7B), suggesting that the C1 region of ABI5 is the major site for CycC1;1 interaction. To verify this, we purified GST-tagged full-length and truncated ABI5 proteins expressed in E. coli, and incubated them with 6×His-tagged CycC1;1 protein for in vitro GST pull-down assay. Our result showed that His-CycC1;1 could be specifically pulled down by intact or truncated ABI5 proteins that contains the C1 region including C1 and C1C2C3, but not by the truncated forms without C1 region including bZIPC4 or C2C3bZIPC4 (Figure 7C), further supporting that CycC1;1 interacts with the C1 region of ABI5.
Figure 7.
ABI5 Ser-42 phosphoamino acid affects ABI5 interaction with CycC1;1. A and B, Identification of the CycC1;1-interacting domain of ABI5 via Y2H assay. The diagram in the box (A) indicates the conserved domains in ABI5 and the truncated fragments tested in the assays. The full-length and truncated ABI5 proteins were fused to the AD of GAL4, and CycC1;1 was fused to the BD of GAL4. Protein interactions were examined based on the growth of yeast cells on selective media (B). C, GST pull-down assay to analyze the CycC1;1-interacting domain of ABI5. 6×His-CycC1;1 were mixed with GST-tagged full-length or truncated ABI5 proteins and then immobilized on glutathione sepharose beads. After washing, the eluted proteins were subjected to immunoblot analysis with anti-GST and anti-His antibodies, respectively. D, Y2H assay to analyze the interaction between CycC1;1 and phospho-mimic or phospho-dead forms of ABI5 at the Ser42, Ser145, and Thr201 sites. E, BiFC analysis to examine the interaction between CycC1;1 and ABI5, ABI5S42A or ABI5S42D in N. benthamiana leaves. Scale bars = 20 μm. F, Dual-LUC reporter gene assay to examine the effects of ABI5, ABI5S42A, or ABI5S42D on EM1 expression in the presence or absence of CycC1;1. The activity of EM1::LUC alone was set to 1. Error bars indicate mean ± sd (n = 3). Different letters indicate significant differences as determined using one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
Previous reports demonstrate that phosphorylation of ABI5 plays a crucial role in plant response to ABA, and Ser-42, Ser-145, and Thr-201 are the major phosphorylation sites substantially affecting ABI5 protein activity (Wang et al., 2013; Zhou et al., 2015). Considering that ABI5 C1 region that interacts with CycC1;1 contains the Ser-42, we herein asked if phosphorylation of Ser-42 influences its interaction with CycC1;1. For this end, we mutated Ser-42 of ABI5 as alanine (Ala, A) or aspartic acid (Asp, D) to mimic the de-phosphorylated or phosphorylated form, respectively, and tested their interaction with CycC1;1. Our Y2H experiment showed that, similarly to ABI5, ABI5-S42A, the phospho-dead form of ABI5 at Ser-42, could still interact with CycC1;1 (Figure 7D). Strikingly, ABI5-S42D, the phospho-mimic form of ABI5 at Ser-42, lost its interaction with CycC1;1 (Figure 7D), suggesting that phosphorylation of ABI5 at Ser-42 deprives its interaction with CycC1;1. This note was reinforced by the BiFC experiment, where ABI5 and ABI5-S42A but not ABI5-S42D could interact with CycC1;1 in N. benthamiana leaves (Figure 7E). In addition, phospho-mimic and phospho-dead forms of ABI5 at Ser-145 and Thr-201 were also produced to test the changes of their interaction with CycC1;1. Consistent with the interaction analysis, mutations of Ser-145 and Thr-201 in ABI5 as Ala or Asp did not affect ABI5 interaction with CycC1;1 (Figure 7D) as the two sites are localized in the C2 and C3 domains, respectively.
Further, we performed dual-reporter gene assay in N. benthamiana leaves to examine the effect of phospho-dead ABI5-S42A and phospho-mimic ABI5-S42D on the transactivation of EM1 in the presence or absence of CycC1;1. Our results showed that when EM1::LUC was used as a reporter, CycC1;1 significantly suppressed ABI5- and ABI5-S42A-promoted LUC activity, but ABI5-S42D-activated LUC was not significantly impacted by CycC1;1 (Figure 7F), suggesting that CycC1;1 interferes ABI5 transcriptional activity through interacting with its un-phosphorylated form, whereas phosphorylation of Ser-42 blocks CycC1;1 interaction with and inhibition of ABI5. These results imply that ABI5 activity is interfered by CycC1;1 through their physical interaction while ABA-induced Ser-42 phosphorylation relieves their interaction, thereby promoting ABI5 activity and the expression of ABI5-targeted genes. In addition to the changes of CycC1;1 interaction with ABI5 caused by ABA-mediated ABI5 phosphorylation, whether ABA modulates CycC1;1 expression remains unclear. Therefore, we performed RT-qPCR experiment to determine the expression of CycC1;1 and CycC1;2 in ABA-treated WT plant seedlings, and our result showed that the transcripts of CycC1;1 and CycC1;2 in ABA-treated plants were less than that in the untreated control (Supplemental Figure S4), indicating that ABA suppresses the expression of both CycC1;1 and CycC1;2 in plants for the further activation of ABI5 activity.
Discussion
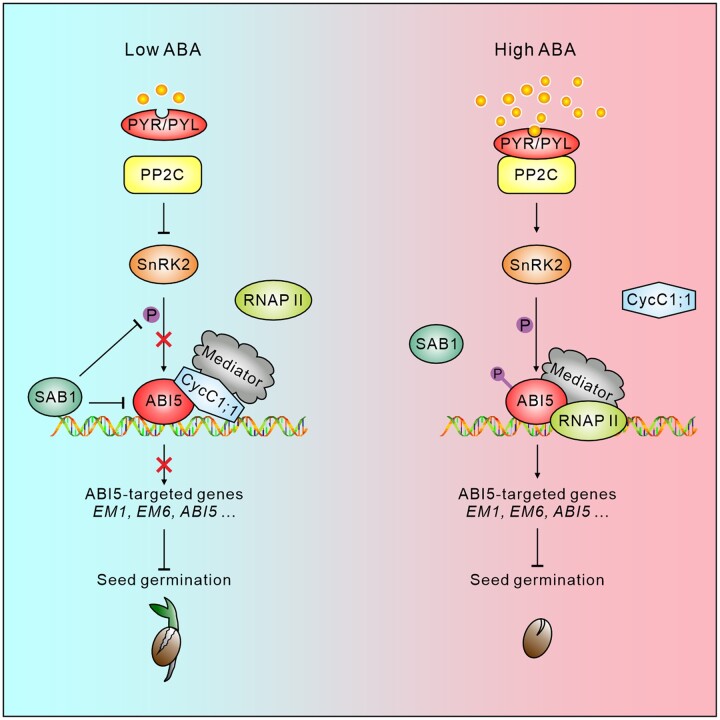
Seed dormancy and germination are important physiological processes for angiosperms species, and crucial to plant survival and propagation. Long-term evolution enables plants to precisely determine whether seed germination starts or not by sensing the changes of external environments. ABA is one of the most important phytohormones inhibiting seed germination. ABI5 functions as a central transcription factor in ABA signaling, facilitates plant versatile ABA responses, including inhibition of seed germination and repression of postgerminative seedling growth. It has now been widely accepted that ABI5 is crucial to regulate the expression of ABA-responsive genes, but how plants precisely regulate the transcriptional activation activity of ABI5 for the downstream genes especially during seed germination remains to be further elucidated. Here, we provide extensive evidence demonstrating that CycC1;1 functions in ABA-inhibited seed germination and cotyledon greening processes by interacting with and inhibiting ABI5. CycC1;1 interacts with ABI5 in a de-phosphorylated form of Ser-42 to occupy the promoters of ABI5-targeted genes, thus interfering ABI5 transcriptional activation effects for the downstream target genes including ABI5 itself under low ABA conditions; when ABA level is increased in plants, phosphorylation of ABI5 at Ser-42 relieves CycC1;1 repression of ABI5 by dampening their physical interaction, thereby promoting the expression of ABA response and inhibiting seed germination. These results illustrate a CycC1;1-involved regulatory network that precisely regulates plant ABI5 activity and expression at multiple layers, and also provide insights into the understanding of ABA–ABI5 module-mediated regulation of seed germination (Figure 8).
Figure 8.
A simplified model showing the role of CycC1;1 in ABA-inhibited seed germination by interacting with and modulating ABI5. Under low ABA conditions, CycC1;1, as a subunit of the Mediator complex interacts with de-phosphorylated ABI5 to form a transcriptional repressor complex and occupy the promoters of ABI5-targeted downstream genes including ABI5 itself, therefore interfering the RNAP II recruitment and suppressing the transcriptional activation activity of ABI5 for the expression of ABI5 and ABI5 downstream genes; Under high ABA conditions, ABA-induced phosphorylation of ABI5 at Ser-42 reduces CycC1;1 interaction with ABI5 to relieve its repression of ABI5 transcriptional activation activity, and thus the expression of ABI5 and ABI5 downstream genes was promoted and seed germination was inhibited.
In this study, we identified and employed two previously-reported T-DNA insertion lines, SALK_053291 and SALK_039400 (Zhu et al., 2014; Chen et al., 2019), where the T-DNA is inserted in the second intron of CycC1;1 and the intergenic region between CycC1;2 and CycC1;1, resulting in reduced expression of CycC1;1 alone or both genes, respectively (Figure 2, A, B, and F), and our results showed that both the mutants are hypersensitive to ABA in terms of seed germination and cotyledon greening rate. Although we could not obtain the T-DNA insertion mutant that only affects the expression of CycC1;2, the ABA insensitive phenotype of the CycC1;2-overexpressing lines (Figure 2, G–I) and the inhibitory effects of CycC1;2 on ABI5 transactivation of targeted genes (Figures 3, A and B, 4F, and 6, D and E) also clearly support that CycC1;2 negatively regulates ABA-inhibited seed germination through interfering ABI5 activity via protein physical interaction in a similar way to CycC1;1.
Mediator is a large protein complex comprised with multiple subunits, and CycC1;1/CycC1;2 together with MED12, MED13, and CDK8 belong to the kinase module that can be associated with or separated away from the core part of Mediator. Our study showed that CycC1;1 can physically interact with ABI5 whereas CDK8 has no interaction with ABI5, suggesting that CycC1;1 possibly modulates ABI5 activity for ABA response in a CDK8-independent manner. This hypothesis is at least partially supported by the findings that cdk8 mutant is insensitive to ABA as reported by a previous study (Zhu et al., 2020) while cycc1;1 mutant is hypersensitive to ABA as shown in our study (Figure 2). In addition to the regulation of plant ABA response, CycC1;1 and CDK8 also have complicated relationships in different biological processes, including growth, development, and stress responses. For example, it has been reported that the ckd8 and cycc1;1/1;2 mutants exhibits similar susceptibility to necrotrophic pathogen Alternaria brassicicola but different resistance against another necrotrophic pathogen Botrytis cinerea (Zhu et al., 2014), suggesting that CDK8 and CycC have both convergent and divergent functions in plant response to different pathogens infection. Interestingly, CDK8 and CycC also elicit similar roles in the regulation of plant flowering as a recent study reported that both the mutants of CDK8 and CycC showed delayed flowering and prolonged reproductive phase in pea (Pisum sativum L.) (Hasan et al., 2020). Based on these reports and our findings, we infer that CycC regulates plant growth, development, and environmental responses in a CDK8-dependent and CDK8-independent manners. Identification of more CDK8- and CycC-implicated biological processes and unraveling the underlying mechanisms will shed more light on how CDK8 and CycC work together or separately in plants.
In addition to the kinase module, the core module of Mediator complex also functions in plant response to ABA. For instance, MED25, a subunit of Mediator tail module, can also interact with ABI5 and negatively regulate the transcriptional activation activity of ABI5 for the downstream target genes and ABI5 itself (Chen et al., 2012), which is very similar to the role of CycC1;1 in the regulation of ABI5 for plant ABA response. However, we have noticed that MED25 interacts with the C3 domain of ABI5 (Chen et al., 2012) but CycC1;1 interacts with its C1 domain (Figure 7, A–C). Also, phospho-mimic mutation of Ser-42 with C1 domain of ABI5 blocks its interaction with CycC1;1, whereas phospho-mimic mutation of Thr-201 within C3 domain of ABI5 did not alter its interaction with CycC1;1 (Figure 7, D–E), further suggesting distinct mechanisms of CycC1;1 and MED25 in the regulation of ABI5 activity through their interaction during plant response to different doses of ABA. From this point of view, CycC1;1 likely plays its role in the regulation of ABI5 activity for ABA response independently of MED25.
Interestingly, ABI5 can stimulate expression of its own gene through binding to the ABRE sites on its own gene promoter, forming a positive feedback regulatory loop in ABA-ABI5 module-regulated processes (Xu et al., 2014; Bai et al., 2019). It is noteworthy that the expression of LUC driven by ABI5 native promoter in N. benthamiana leaves could be suppressed by CycC1;1 (Figure 4, D and E), and the ChIP-qPCR experiments demonstrated that CycC1;1 could mainly associate with ABI5 promoter regions to restrict recruitment of RNAP II (Figure 4, A–C). Therefore, we infer that CycC1;1 as a subunit of Mediator complex interacts with ABI5 to form a repressor complex to repress ABI5 expression by associating with ABRE site on ABI5 promoter. The observation of higher ABI5 expression in the cycc1;1 mutant than the WT by the RT-qPCR, GUS staining, and immunoblotting analysis (Figure 5, A–D) can also be ascribed to more RANP II recruitment to ABI5 promoter caused by the absence of CycC1;1 in the cycc1;1 mutant. Similarly, CycC1;1 can also associate to the ABRE sites of other ABI5-targeted genes promoter including EM1 and EM6 through interacting with ABI5 (Figure 6, C–F), which might disrupt the RNAP II recruitment to these genes and thus repressing their transcription. In addition, ABA treatment strongly repressed CycC1;1 interaction with ABI5 and CycC1;1 association with EM1 and EM6 promoters at the ABRE sites (Figure 6, C–F). Different from those previously reported transcription factors regulating ABI5 expression or interacting partners affecting ABI5 activity, our results reveal a previously unidentified mechanism of ABA-mediated ABI5-activated expression of EM1 and EM6, where CycC1;1 interacts with ABI5 to form a transcriptional repressor to modulate ABA signaling during seed germination while the effects of such complex are dynamically modulated by ABA, conferring plants flexible adaptive responses to the internal and external cues. From another point of view, it is worthy of further explorations to study whether and how CycC1;1 modulates plant growth and development because the trade-off between plant growth and stress response is tightly modulated in plants under changing environments (Liu et al., 2022a).
Structural analysis of ABI5 reveals that ABI5 contains four conserved regions and a bZIP domain for DNA binding. We identified that the C1 region of ABI5 provides the interface for CycC1;1 physical interaction (Figure 4, A–C). Further study found that CycC1;1 could interact with de-phosphorylated or phospho-dead ABI5 at Ser-42, but phospho-mimicking Asp-42 destroyed its interaction with CycC1;1 (Figure 4, D and E). Considering that Ser-42 is one of the major phosphorylation sites mediated by SnRK2s and that SnRK2 kinase activity is activated by increased ABA accumulation in plants (Wang et al., 2013; Zhou et al., 2015), we speculated that changes of phosphorylation status of Ser-42 in ABI5 caused by ABA/SnRK2s act as a molecular switch for its interaction with CycC1;1 and subsequent ABI5 activation. In addition to Ser-42, Ser-145, and Thr-201 of ABI5 can also be phosphorylated by SnRK2s (Lopez-Molina et al., 2002; Wang et al., 2013; Zhou et al., 2015), whereas neither phospho-dead nor phospho-mimic mutations of these two amino acids affect ABI5 interaction with CycC1;1 (Figure 4D), which is due to that the C2C3 regions of ABI5 covering Ser-145 and Thr-201 had no interaction with CycC1;1 (Figure 4, A–C). We have noticed that Ser-145 is the major site for its interaction with SAB1, a previously reported negative regulator of ABI5 stability and transcriptional binding activity (Ji et al., 2019). The different interaction sites of ABI5 for CycC1;1 an SAB1 suggest distinct regulatory mechanisms of CycC1;1 and SAB1 on ABI5 activity in ABA-mediated seed germination. Apart from posttranslational regulation by CycC1;1 and SAB1 on ABI5 activity, they also negatively regulate ABI5 expression at the transcription level with contrasting mechanisms: SAB1 is an RCC1 family protein and downregulates ABI5 expression by increasing the level of histone H3K27me2 in the ABI5 promoter (Ji et al., 2019), while CycC1;1 as a subunit of the Mediator complex reduces ABI5 transcription by disrupting RNAP II association with its genomic DNA. These multiple layers of regulation on ABI5 mediated by CycC1;1 as well as other factors should enable plants sophisticated and accurate responses to the external surroundings during seed germination and seedling establishment.
Materials and methods
Plant materials and growth conditions
Arabidopsis (A. thaliana) ecotype Columbia-0 was used as the WT. The cycc1;1 (SALK_053291) and cycc1;1/1;2 (SALK_039400) mutants were obtained from the Arabidopsis Biological Resource Center. The abi5-7 mutant (Zhao et al., 2020) and ABI5::GUS transgenic line were used in our study. The cycc1;1 abi5-7 double mutant and ABI5::GUS line in the cycc1;1 mutant and CycC1;1-overexpressing background plants were obtained by genetic cross. Plants were grown at 22°C under a 16-/8-h light/dark photoperiod at 100 µmol m−2 s−1 on a half-strength Murashige and Skoog (0.5 MS) medium containing 0.8% (w/v) agar.
To generate transgenic overexpression plants, the full-length CDSs of CycC1;1 and CycC1;2 cloned into the pEGAD and pCambia1300 vectors at the BamHI and SalI sites, respectively, under the control of the cauliflower mosaic virus 35S promoter. The constructs were used to transform the WT Arabidopsis plants by Agrobacterium tumefaciens-mediated transformation. The expression of CycC1;1 and CycC1;2 of the transgenic plants was examined by RT-qPCR. T4 homozygous transgenic lines were used for subsequent analysis. The primers used are listed in Supplemental Table S1.
To generate the complementation (Com) lines of the cycc1;1 mutant, the genomic sequence of 1,030 bp upstream of CycC1;1 translation start codon (ATG) and the full length CDS of CycC1;1 were amplified, and cloned into pCambia1381 vector at the PstI and HindIII sites, respectively. The resulting plasmid was introduced into the cycc1;1 mutant via A. tumefaciens-mediated floral transformation. Com T4 transgenic plants were used for phenotypic analysis.
Seed germination assays
Mature Arabidopsis seeds were harvested and dried at room temperature for 3 weeks, and then used for the germination assays. Sterilized seeds were plated on 0.5 MS medium without or with the indicated concentration of ABA (Sigma, St. Louis, Missouri, USA). Following stratification at 4°C for 3 days in the dark, the plates were transferred to a growth chamber with a 16-h light/8-h dark cycle at 23°C. Seeds were identified as germinated when a radicle had emerged from the seed coat (Nie et al., 2022).
RT-qPCR analysis
Treated or untreated germinating seeds or seedlings were collected for total RNA isolation, first-strand cDNA synthesis, and RT-qPCR as we described previously (Wang et al., 2021). The constitutively expressed ACTIN2 gene was used as an internal control. The primers used for RT-qPCR are listed in Supplemental Table S1.
Y2H assay
The full-length CDS of CycC1;1 or CycC1;2 was cloned into pGBKT7 vector. The full-length and truncated forms of ABI5 were cloned into pGBKT7 vector. The yeast transformation and growth were carried out with the Matchmaker system (Clontech, Mountain View, California, USA). Yeast cells were selected on double dropout medium lacking Leu and Trp (-LW) to select transformants, triple dropout medium lacking Leu, Trp, and His (-LWH) or quadruple dropout medium lacking Leu, Trp, His, and Ade (-LWHA) to analyze protein interactions. Primer sequences are listed in Supplemental Table S1.
LCI assay
The CDS of ABI5 and CycC1;1 or CycC1;2 were cloned into the JW772 or JW771 vector containing the N-terminal of LUC (nLUC) and or the C-terminal of LUC (cLUC) at the BamHI and SalI sites. The resultant plasmids were transformed into A. tumefaciens and used to infiltrate N. benthamiana leaves. Three days after infiltration, the leaves were incubated in PBS solution containing 150 µg/mL D-Luciferin potassium salt in the dark for 10 min before luminescence assay. The LUC image was captured by NightSHADE LB 985 according to the manufactory’s instructions. Primer sequences are listed in Supplemental Table S1.
BiFC assays
The CDS of ABI5 and CycC1;1 or CycC1;2 were cloned into the pSPYNE or pSPYCE vector containing the N-terminal of YFP (nYFP) and or the C-terminal of YFP (cYFP), respectively. nYFP-ABI5 and cYFP-CycC1;1 or cYFP-CycC1;2 were co-expressed in N. benthamiana leaves. YFP fluorescence (excitation 488 nm, emission 543 nm) was detected under a Zeiss LSM980 laser scanning confocal microscope. H2B-mCherry (Rosa et al., 2014) was used as a nuclear marker, and the red fluorescence (excitation 561 nm, emission 600–630 nm) was also detected by Zeiss LSM980 laser scanning confocal microscope. Primer sequences are listed in Supplemental Table S1.
Dual-LUC reporter gene assay
The dual-LUC reporter gene assay was performed according to the previous reports (Liu et al., 2022b). The plasmid containing the EM1 promoter upstream of the LUC gene was used as the reporter, and the REN under the control of 35S promoter was used as the internal control. The reporter and internal control were transiently co-expressed with pCambia1307-ABI5, pEGAD-CycC1;1, or pCambia1300-CycC1;2 in N. benthamiana leaves, and then the activities of LUC and REN were detected.
To assay the role of CycC1;1 or CycC1;2 in the regulation of ABI5 expression, the genomic DNA sequence of 2,024 bp upstream of ABI5 translation start codon (ATG) was cloned into pGreenII0800-LUC vector at the BamHI site, resulting in the reporter ABI5::LUC. The reporter and internal control were transiently co-expressed with pEGAD-CycC1;1 or pCambia1300-CycC1;2 in N. benthamiana leaves, and the activities of LUC and REN were then detected. Primer sequences are listed in Supplemental Table S1.
To test the effects of CycC1;1 and CycC1;2 on ABI5-activated EM1 expression using ABI5 genomic DNA sequence, the genomic sequence of 2,024 bp upstream of ABI5 translation start codon (ATG) was cloned into pCambia1381 vector at the EcoRI site, and then the DNA sequence from the translation start codon (ATG) to the end of terminator of ABI5 was cloned into the above plasmid at the HindIII site, resulting in ABI5pro::ABI5g-Ter. The reporter and internal control were co-expressed with ABI5pro::ABI5g-Ter, pEGAD-CycC1;1 or pCambia1300-CycC1;2 in N. benthamiana leaves, and then the activities of LUC and REN were detected. The primers used for these plasmids construction are listed in Supplemental Table S1.
Co-IP assays
The Co-IP experiment was performed according to our previously reported methods with some modifications (Liu et al., 2022a; Zhang et al., 2022a). Total proteins were isolated from 7-day-old 35S::ABI5-Myc transgenic seedlings (Zhang et al., 2022b) treated with or without 10 μM ABA for 12 h, and then proteins were immunoprecipitated by the anti-CycC1;1 antibody. After immunoprecipitation, the precipitated proteins were separated on SDS-PAGE gel, and ABI5-Myc protein was immunoblotted by anti-Myc antibody (#AE060, ABclonal, China). The anti-CycC1;1 polyclonal antibody was generated in GenScript (Nanjing, China) using the peptide (VDVVHDLDKERGISC) specific for CycC1;1 protein, and verified by immunoblot analysis using the proteins isolated from the WT and cycc1;1 mutant seedlings.
ChIP-qPCR analysis
To assay whether CycC1;1 was associated with ABI5 genomic DNA, 35S::GFP-CycC1;1 transgenic lines was were used for ChIP analysis according to previously reported method (Zhu et al., 2014). In brief, chromatin was isolated from 7-day-old 35S::GFP-CycC1;1 transgenic plant seedlings. Monoclonal anti-GFP antibody (#AE064; ABclonal, Wuhan, China) was used for protein immunoprecipitation. DNA fragments in both input and immunoprecipitated samples were quantified by qPCR.
To examine the effect of CycC1;1 on RNAP II recruitment to the ABI5 promoter, Chromatin was extracted from 7-day-old WT and cycc1;1 mutant seedlings and precipitated with anti-RPB2 antibody (ABclonal) raised against the C-terminal domain of RNAP II. DNA fragments in both input and immunoprecipitated samples were quantified by qPCR. The ACTIN7 was used as a reference genes. At least three independent experiments were performed. The primers used for ChIP-qPCR are listed in Supplemental Table S1.
GST pull-down assay
The CDS of full-length or truncated ABI5 and CycC1;1 or CycC1;2 was in cloned into the pGEX4t-1 and pET28a vectors at the SmaI and BamHI sites, respectively, and the GST-tagged ABI5 or truncated ABI5 and 6×His-CycC1;1 were purified from E. coli. 6×His-CycC1;1 was mixed with GST alone, GST-tagged ABI5, or truncated ABI5 on ice for 1 h. The protein mixture was then incubated with glutathione sepharose beads at 4°C for 3 h. After washing, the eluted proteins with Elution Buffer (10 mM GSH in 50 mM Tris–HCl, pH 8.0) were detected with anti-His (#E12-004-3; EnoGene) and anti-GST (#E12-007; EnoGene) antibodies, respectively. Primer sequences are listed in Supplemental Table S1.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers_CycC1;1 (AT5G48640), CycC1;2 (AT5G48630), ABI5 (AT2G36270), EM1 (AT3G51810), EM6 (AT2G40170), RD29A (AT5G52310), ABF3 (AT4G34000), and CDK8 (AT5G63610).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. The interaction of CycC1;2 with ABI5 and CDK8 with ABI5 in yeast cells.
Supplemental Figure S2. CycC1;1 suppresses ABA-inhibited seed germination through ABI5.
Supplemental Figure S3. Analysis of the ABA sensitivity of the cycc1;1/1;2 mutant and CycC1;2-overexpression lines.
Supplemental Figure S4. ABA represses the expression of CycC1;1 and CycC1;2.
Supplemental Table S1. List of the primers used in this study.
Supplementary Material
Acknowledgments
We thank Prof. Ying-Tang Lu (Wuhan University, China) for providing guidance and valuable advice. We also thank Prof. Xingliang Hou (South China Botanical Garden, Chinese Academy of Sciences, China) and Prof. Yuan Zheng (Henan University, China) for the seeds of ABI5::GUS line and abi5-7 mutant, respectively. Finally, we thank Prof. Yingfang Zhu (Henan University, China) for sharing the pGBKT7-CDK8 plasmid.
Funding
This work was supported by the National Natural Science Foundation of China (#32000150) and the Natural Science Foundation of Henan Province (#222300420401).
Conflict of interest statement. The authors declare no conflict of interest.
Contributor Information
Jia-Xing Guo, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
Ru-Feng Song, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
Kai-Kai Lu, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
Yu Zhang, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
Hui-Hui Chen, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
Jia-Xin Zuo, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
Ting-Ting Li, Jiangsu Key Laboratory of Marine Pharmaceutical Compound Screening, Jiangsu Ocean University, Lianyungang 222005, China.
Xue-Feng Li, Anyang Wenfeng District Natural Resources Bureau, Anyang 455000, China.
Wen-Cheng Liu, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, College of Life Sciences, Henan University, Kaifeng 475004, China.
These authors contributed equally (J.-X.G., R.-F.S., and K.-K.L.)
W.-C.L. conceived and designed the project, and wrote the article. J.-X.G., R.-F.S., K.-K.L., Y.Z., H.-H.C., J.-X.Z., T.-T.L., and X.-F.L. performed the experiments. R.-F.S., J.-X.G., and K.-K.L. analyzed the data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Wen-Cheng Liu (liuwencheng@henu.edu.cn).
References
- Agrawal R, Jiří F, Thakur JK (2021) Kinase module of Mediator complex: important signalling processor for development and survival of plants. J Exp Bot 72: 224–240 [DOI] [PubMed] [Google Scholar]
- Albertos P, Romero-Puertas MC, Tatematsu K, Mateos I, Sanchez-Vicente I, Nambara E, Lorenzo O (2015) S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat Commun 6: 8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD (1999) Conserved structures of mediator and RNA polymerase II holoenzyme. Science 283: 985–987 [DOI] [PubMed] [Google Scholar]
- Bai M, Sun J, Liu J, Ren H, Dehesh K (2019) B-box protein bbx 19 suppresses seed germination via induction of ABI5. Plant J 99: 1192–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, Giraudat J, Parcy F (2002) The homologous ABI5 and EEL transcription factors function antagonistically to finetune gene expression during late embryogenesis. Plant Cell 14: 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Mohan R, Zhang Y, Li M, Fu ZQ (2019) NPR1 promotes its own and target gene expression in plant defense by recruiting CDK8. Plant Physiol 181: 289–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, Liu X, Li H, Zheng W, Sun J, Li C (2012) The Arabidopsis Mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Xue Q, McCray T, Margavage K, Chen F, Lee JH, Nezames CD, Guo L, Terzaghi W, Wan J, et al. (2013) The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 25: 517–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Viso F, Casaretto JA, Quatrano RS (2007) 14-3-3 Proteins are components of the transcription complex of the ATEM1 promoter in Arabidopsis. Planta 227: 167–175 [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubier P, Raynal M, Hull G, Huestis GM, Grellet F, Arenas C, Pages M, Delseny M (1993) Two different Em-like genes are expressed in Arabidopsis thaliana seeds during maturation. Mol Gen Genet 238: 409–418 [DOI] [PubMed] [Google Scholar]
- Guo P, Chong L, Wu F, Hsu CC, Li C, Zhu JK, Zhu Y (2021) Mediator tail module subunits MED16 and MED25 differentially regulate abscisic acid signaling in Arabidopsis. J Integr Plant Biol 63: 802–815 [DOI] [PubMed] [Google Scholar]
- Hasan A, Schoor J, Hecht VF, Weller JL (2020) The cyclin-dependent kinase module of the Mediator complex promotes flowering and reproductive development in pea. Plant Physiol 182: 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Wang S, Cheng C, Li R, Wang Z, Jenkins GI, Kong FJ, Li X (2019) The RCC1 family protein SAB1 negatively regulates ABI5 through multidimensional mechanisms during postgermination in Arabidopsis. New Phytol 222: 907–922 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoon HJ, Terzaghi W, Martinez C, Dai M, Li J, Byun MO, Deng XW (2010) DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 22: 1716–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hu P, Huang M, Tang Y, Li Y, Li L, Hou X (2016) The NF-YC–RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat Commun 7: 12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WC, Song RF, Zheng SQ, Li TT, Zhang BL, Gao X, Lu YT (2022a) Coordination of plant growth and abiotic stress responses by Tryptophan Synthase β Subunit1 through modulating tryptophan and ABA homeostasis in Arabidopsis. Mol Plant 15: 1–18 [DOI] [PubMed] [Google Scholar]
- Liu WC, Song RF, Qiu YM, Zheng SQ, Li TT, Wu Y, Song CP, Lu YT, Yuan HM (2022b) Sulfenylation of ENOLASE2 facilitates H2O2-conferred freezing tolerance in Arabidopsis. Dev Cell 57: 1883–1898 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Maji S, Dahiya P, Waseem M, Dwivedi N, Bhat DS, Dar TH, Thakur JK (2019) Interaction map of Arabidopsis Mediator complex expounding its topology. Nucleic Acids Res 47: 3904–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur S, Vyas S, Kapoor S, Tyagi AK (2011) The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol 157: 1609–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, et al. (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Nie K, Zhao H, Wang X, Niu Y, Zhou H, Zheng Y (2022) The MIEL1-ABI5/MYB30 regulatory module fine tunes abscisic acid signaling during seed germination. J Integr Plant Biol 64: 930–941. [DOI] [PubMed] [Google Scholar]
- Nonogaki H (2017) Seed biology updates-highlights and new discoveries in seed dormancy and germination research. Front Plant Sci 8: 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss ZC, Ebmeier CC, Taatjes DJ (2013) The Mediator complex and transcription regulation. Critic Rev Biochem Mol Biol 48: 575–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa S, Ntoukakis V, Ohmido N, Pendle A, Abranches R, Shaw P (2014) Cell differentiation and development in Arabidopsis are associated with changes in histone dynamics at the single-cell level. Plant Cell 26: 4821–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Liu XD, Xie Q, He ZH (2017) Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9: 34–45 [DOI] [PubMed] [Google Scholar]
- Shu K, Zhou W, Yang W (2018) Apetala 2-domain-containing transcription factors: focusing on abscisic acid and gibberellins antagonism. Phytologist 217: 977–983 [DOI] [PubMed] [Google Scholar]
- Vishwakarma K, Upadhyay N, Kumar N, Yadav G, Singh J, Mishra RK, Kumar V, Verma R, Upadhyay RG, Pandey M, et al. (2017) Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front Plant Sci 8: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Chen X (2004) HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131: 3147–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li L, Ye T, Lu Y, Chen X, Wu Y (2013) The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J Exp Bot 64: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Huang S, Zhang A, Guo P, Liu Y, Xu C, Cong W, Liu B, Xu ZY (2020) JMJ17–WRKY40 and HY5–ABI5 modules regulate the expression of ABA-responsive genes in Arabidopsis. New Phytol 230: 567–584 [DOI] [PubMed] [Google Scholar]
- Wang LF, Lu KK, Li TT, Zhang Y, Guo JX, Song RF, Liu WC (2021) Maize PHYTOMELATONIN RECEPTOR1 functions in plant osmotic and drought stress tolerance. J Exp Bot 10.1093/jxb/erab553 [DOI] [PubMed] [Google Scholar]
- Wise MJ (2003) LEAping to conclusions: a computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinformatics 4: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Li J, Gangappa SN, Hettiarachchi C, Lin F, Andersson MX, Jiang Y, Deng XW, Holm M (2014) Convergence of light and ABA signaling on the ABI5 promoter. PLoS Genetics 10: e1004197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Han X, Yang J, Jiang Y, Hu Y (2021) The Arabidopsis circadian clock protein PRR5 interacts with and stimulates ABI5 to modulate abscisic acid signaling during seed germination. Plant Cell 33: 3022–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Koisumi M, Urao S, Shinozaki K (1992) Molecular cloning and characterization of 9 cDNAs genes that are responsive to desiccation in Arabidopsis thaliana: sequence analysis of one cDNA clone that encodes a putative transmembrane channel protein. Plant Cell Physiol 33: 217–224 [Google Scholar]
- Yu LH, Wu J, Zhang ZS, Miao ZQ, Zhao PX, Wang Z, Xiang CB (2017) Arabidopsis MADS-Box transcription factor AGL21 acts as environmental surveillance of seed germination by regulating ABI5 expression. Mol Plant 10: 834–845 [DOI] [PubMed] [Google Scholar]
- Yu B, Wang Y, Zhou H, Li P, Liu C, Chen S, Peng Y, Zhang Y, Teng S (2020) Genome-wide binding analysis reveals that ANAC060 directly represses sugar-induced transcription of ABI5 in Arabidopsis. Plant J 103: 965–979 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Mogami J, Yamaguchi-Shinozaki K (2014) ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol 21: 133–139 [DOI] [PubMed] [Google Scholar]
- Zhao H, Nie K, Zhou H, Yan X, Zhan Q, Zheng Y, Song CP (2020) ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytol 228: 596–608 [DOI] [PubMed] [Google Scholar]
- Zhao X, Dou L, Gong Z, Wang X, Mao T (2019) BES1 hinders ABSCISIC ACID INSENSITIVE5 and promotes seed germination in Arabidopsis. New Phytol 221: 908–918 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang Z, Gao J, Wang P, Hu T, Wang Z, Hou YJ, Wan Y, Liu W, Xie S, et al. (2018) Arabidopsis duodecuple mutant of PYL ABA receptors reveals PYL repression of ABA-independent SnRK2 activity. Cell Rep 23: 3340–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Schumaker KS, Guo Y (2012) Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc Natl Acad Sci USA 109: 12822–12827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang LF, Han SY, Ren F, Liu WC (2022a) Sorting Nexin1 negatively modulates phosphate uptake by facilitating phosphate transporter1;1 degradation in Arabidopsis. Plant J 111: 72–84 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li TT, Wang LF, Guo JX, Lu KK, Song RF, Zuo JX, Chen HH, Liu WC (2022b) Abscisic acid facilitates phosphate acquisition through the transcription factor ABA INSENSITIVE5 in Arabidopsis. Plant J 111: 269–281 [DOI] [PubMed] [Google Scholar]
- Zhou X, Hao H, Zhang Y, Bai Y, Zhu W, Qin Y, Yuan F, Zhao F, Wang M, Hu J, et al. (2015) SOS2-LIKE PROTEIN KINASE5, an SNF1-RELATED PROTEIN KINASE3-type protein kinase, is important for abscisic acid responses in Arabidopsis through phosphorylation of ABSCISIC ACID-INSENSITIVE5. Plant Physiol 168: 659–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Huang P, Guo P, Chong L, Yu G, Sun X, Hu T, Li Y, Hsu CC, Tang K, et al. (2020) CDK8 is associated with RAP2.6 and SnRK2.6 and positively modulates abscisic acid signaling and drought response in Arabidopsis. New Phytol 228: 1573–1590 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Schluttenhoffer CM, Wang P, Fu F, Thimmapuram J, Zhu JK, Lee SY, Yun DJ, Mengiste T (2014) CYCLIN-DEPENDENT KINASE8 differentially regulates plant immunity to fungal pathogens through kinase-dependent and -independent functions in Arabidopsis. Plant Cell 26: 4149–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.