Abstract
Sclerotinia sclerotiorum causes substantial damage and loss of yield in oilseed rape (Brassica napus). The molecular mechanisms of oilseed rape defense against Sclerotinia remain elusive. In this study, we found that in the early stages of B. napus infection a conserved mitogen-activated protein kinase (MAPK) cascade mediated by BnaA03.MKK5-BnaA06.MPK3/BnaC03.MPK3 module phosphorylates the substrate BnWRKY33, enhancing its transcriptional activity. The activated BnWRKY33 binds to its own promoter and triggers a transcriptional burst of BnWRKY33, thus helping plants effectively resist the pathogenic fungi by enhancing the expression of phytoalexin synthesis-related genes. The expression of BnWRKY33 is fine-tuned during defense. Ongoing Sclerotinia infection induces BnaA03.WRKY28 and BnaA09.VQ12 expression. BnaA09.VQ12 interacts physically with BnaA03.WRKY28 to form a protein complex, causing BnaA03.WRKY28 to outcompete BnWRKY33 and bind to the BnWRKY33 promoter. BnaA03.WRKY28 induction suppresses BnWRKY33 expression in the later stages of infection but promotes branch formation in the leaf axils by regulating the expression of branching-related genes such as BnBRC1. BnaA03.WRKY28 participates in the trade-off between defense and growth. These findings suggest that oilseed rape plants may modulate defense-response strength and develop alternative reproduction and survival strategies in the face of lethal pathogens.
BnaA03.WRKY28 interacts with BnaA09.VQ12 and takes precedence over phosphorylated BnWRKY33 to bind to the BnWRKY33 promoter, thereby weakening Sclerotinia resistance but promoting branching.
Introduction
In nature, plants are challenged frequently by pathogens, activating their innate immune system (Jones and Dangl, 2006). Brassica napus (oilseed rape), derived from natural hybridization between Brassica rapa and Brassica oleracea, is one of the most important oil crops worldwide (Chalhoub et al., 2014). Sclerotinia sclerotiorum, a necrotrophic fungal pathogen, is considered one of the most economically damaging diseases of oilseed rape (Derbyshire and Denton-Giles, 2016).
The WRKY superfamily of transcription factors (TFs), which contain a conserved WRKYGQK sequence and an atypical zinc-finger structure, can bind specifically to T/CTGACC/T (W-box) cis-acting elements of target genes in response to diverse abiotic and biotic stresses (Eulgem et al., 2000; Ülker and Somssich, 2004; Rushton et al., 2010). AtWRKY33 plays a critical role in stress response. The wrky33 mutant is sensitive to NaCl stress (Jiang and Deyholos, 2009), and WRKY33 can bind to RAP2.2, regulating its expression during submergence-induced hypoxia response (Tang et al., 2021). AtWRKY33 overexpression (OE) enhances resistance to the necrotrophic fungal pathogen Botrytis cinerea (Birkenbihl et al., 2012; Liu et al., 2017). WRKY33 regulates the expression of CYP71B15 (PAD3) and CYP71A13, which encode cytochrome P450 enzymes that are required for the synthesis of the antimicrobial phytoalexin camalexin, and WRKY33 can directly target the W-box in the PAD3 promoter (Qiu et al., 2008; Mao et al., 2011). WRKY33 regulates its own expression by binding to the WRKY33 promoter (Mao et al., 2011). In oilseed rape, BnWRKY33 has been reported to play a positive regulatory role in Sclerotinia resistance. BnWRKY33-overexpressing lines are more resistant than wild-type (WT) plants to S. sclerotiorum (Wang et al., 2014b; Liu et al., 2018). In addition, activated AtMPK3/6 can directly phosphorylate downstream substrates including AtWRKY33, to induce defense-related genes in response to pathogens (Mao et al., 2011).
WRKY superfamily TFs participate both in pathogen-induced defense responses and in plant growth and development (Eulgem et al., 2000; Rushton et al., 2010). AtWRKY28 is specifically expressed in the hypodermal somatic cells surrounding the megaspore mother cell (MMC), inhibiting them from attaining MMC-like fate. The cytochrome P450 gene KLU promotes WRKY28 expression via H2A.Z deposition at WRKY28, mediated by chromatin remodeling complex SWR1 (Zhao et al., 2018). Moreover, WRKY28 positively regulates salicylic acid (SA) biosynthesis by promoting SA biosynthesis-related gene expression (van Verk et al., 2011). A light-signaling protein, FAR-RED ELONGATED HYPOCOTYL3, binds to the WRKY28 promoter and inhibits WRKY28 expression, thereby negatively regulating SA biosynthesis and leaf senescence (Tian et al., 2020). BnWRKY28 (a putative ortholog of AtWRKY28) is induced by S. sclerotiorum in B. napus (Yang et al., 2009).
VQ proteins, which contain a conserved FxxxVQxLTG motif, are involved in developmental processes (Li et al., 2014; Lei et al., 2017), biotic stress responses (Wang et al., 2015; Chen et al., 2018), and abiotic stress responses (Perruc et al., 2004; Hu et al., 2013b). AtVQ12-overexpressing plants show susceptibility to B. cinerea (Wang et al., 2015). Most VQ proteins interact with WRKY TFs as co-factors (Cheng et al., 2012; Jing and Lin, 2015). For example, VQ9 and WRKY8 form a complex that contributes to salinity tolerance (Hu et al., 2013b). By contrast, VQ10 interacts with WRKY8 and participates in plant defense against the fungal pathogen B. cinerea (Chen et al., 2018).
In nature, oilseed rape plants are frequently attacked by S. sclerotiorum, but the immune mechanisms of oilseed rape response to Sclerotinia remain obscure; particularly, our knowledge about function modes of interacting factors during infection is limited. In this study, we found that BnaA03.WRKY28 and BnWRKY33 play opposing roles in Sclerotinia defense by competitively binding to the BnWRKY33 promoter. In the late stage of infection, BnaA03.WRKY28 dampens the immune response and promotes plant growth. The BnaA03.WRKY28-BnWRKY33 module fine-tunes the timing of the defense response against Sclerotinia. This work improves our understanding of plant immunity.
Results
BnaA03.WRKY28 negatively regulates S. sclerotiorum resistance in B. napus
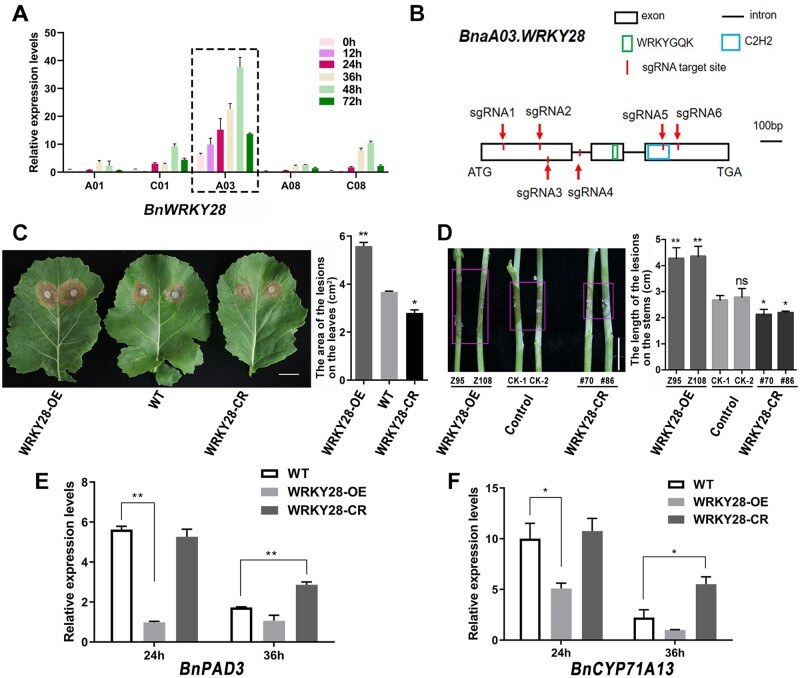
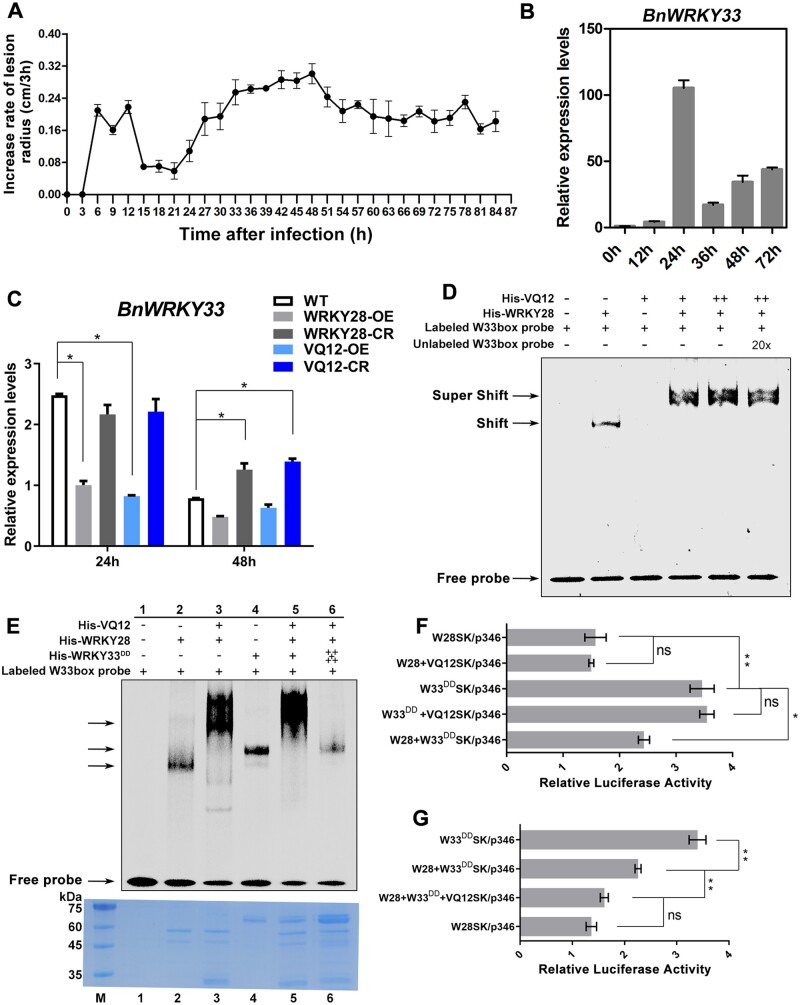
Many WRKY TFs respond to S. sclerotiorum and hormone treatments in B. napus, including WRKY28, which could be induced after the inoculation of Sclerotinia (Yang et al., 2009). In oilseed rape, we identified five copies of BnWRKY28 on chromosomes A01, C01, A03, A08, and C08, namely BnaA01.WRKY28, BnaC01.WRKY28, BnaA03.WRKY28, BnaA08.WRKY28, and BnaC08.WRKY28, all of which were orthologous to AtWRKY28 (Supplemental Figure S1). Of these, only BnaA03.WRKY28 was strongly and dramatically induced when WT plants were challenged with S. sclerotiorum (Figure 1A and Supplemental Table S1). BnaA03.WRKY28 was gradually induced during infection; expression level peaked 48 h after inoculation, then decreased. To elucidate the biological function of BnaA03.WRKY28 in response to Sclerotinia, both OE and CRISPR/Cas9-mediated knockout (CR) lines (Figure 1B) were generated. We investigated the BnaA03.WRKY28 expression in OE lines and the edited form in CR lines, and compared the lesion areas after 48 h inoculation in WT, OE, and homozygous out-of-frame insertion/deletion CR lines (Supplemental Figure S2). Compared with WT plants, WRKY28-OE plants showed more severe symptoms and fungal invasion, whereas WRKY28-CR plants exhibited better resistance (Figure 1, C and D and Supplemental Figure S2). At 24 h after inoculation with Sclerotinia, the transcript levels of two pathogen-induced marker genes, BnPAD3 and BnCYP71A13, were similar in WRKY28-CR and in the WT, but were significantly lower in WRKY28-OE lines; while at 36 h after inoculation, BnPAD3 and BnCYP71A13 showed higher expression in WRKY28-CR lines than in the WT (Figure 1, E and F). Taken together, these data suggest that BnaA03.WRKY28 negatively regulates Sclerotinia resistance in B. napus.
Figure 1.
Negative effect of BnaA03.WRKY28 on S. sclerotiorum resistance in B. napus. A, Induced expression of five copies of BnWRKY28, identified 0, 12, 24, 36, 48, and 72 h after inoculation with S. sclerotiorum. The latest fully unfolded leaves of oilseed rape (Jia 9709, ca. six-week-old) were used for infection. Data are shown as means ± sd (n = 3). B, Schematic diagram to show the characterization of the BnaA03.WRKY28 sequence and the target sites of sgRNAs using the CRISPR/Cas9 system. sgRNA1/2, sgRNA3/4, and sgRNA5/6 were included in one construct respectively. C, The lesions of detached leaves from BnaA03.WRKY28 transgenic T3 homozygous lines and WT plants (Jia 9709, ca. six-week-old) were imaged. Lesion areas was calculated after inoculation using a 7-mm agar block containing S. sclerotiorum hyphae grown at 22°C for 48 h. Scale bar, 2 cm. D, Lesions on the stems of transgenic lines and transformation controls at the flowering stage were imaged, and the lesion lengths were calculated after 3 days of inoculation with S. sclerotiorum. Scale bar, 2 cm. E and F, Relative expression levels of two camalexin biosynthesis-related genes, BnPAD3 and BnCYP71A13, in BnaA03.WRKY28 transgenic lines and WT plants after Sclerotinia inoculation for 24 and 36 h. In (C–F), WRKY28-OE, BnaA03.WRKY28-OE line; WRKY28-CR, homozygous out-of-frame BnaA03.WRKY28 editing lines mediated by the CRISPR/Cas9 system. Data are shown as means ± sd (n = 3). Asterisks indicate significant differences compared with the WT/Control (t test; *P < 0.05, **P < 0.01; ns, P > 0.05); and in (D), that CK-2 as the reference shows the same trend.
BnaA03.WRKY28 directly targets BnWRKY33
To examine negative regulation by BnaA03.WRKY28 in Sclerotinia resistance, we performed RNA-seq and chromatin immunoprecipitation (ChIP)-seq analyses, using BnaA03.WRKY28-fused FLAG-tag OE lines. The fused target protein was stably expressed in line Z95 (Supplemental Figure S3A). In ChIP-seq, most of the binding sites of BnaA03.WRKY28-bound peaks were located in promoter regions (Supplemental Figure S3B), with strong enrichment of the TTGACT/C motif (Supplemental Figure S3C). In RNA-seq, 3,972 upregulated and 3,322 downregulated genes were identified (Supplemental Table S2). Gene ontology (GO) analysis of up- and downregulated gene clusters indicated that the differentially expressed genes (DEGs) were significantly enriched in response to stimulus/stress (Supplemental Figure S3, D and E). Of the 7,294 DEGs identified by RNA-seq, the promoters of 3,980 genes were shown to be BnaA03.WRKY28 bound in the ChIP-seq (Supplemental Figure S3F; genes were listed in Supplemental Table S3). Among the overlapping genes, most were involved in stress response (Supplemental Figure S3G), and there were about 360 TF-encoding genes (Supplemental Figure S3H and Supplemental Table S3). Bound-peaks were found in the promoters of the four BnWRKY33 copies among the DEGs (Figure 2A).
Figure 2.
BnaA03.WRKY28 binds to the BnWRKY33 promoter in vivo and in vitro. A, ChIP-seq showing specifically enrichment in the promoter regions of four copies of BnWRKY33. B, Characterization of the BnWRKY33 promoter sequence and the positions of W-boxes. The core sequence of the W-box is marked. C, A diagram of the BnWRKY33 locus showing the positions of amplicons (P1–P5) used for ChIP-qPCR. W-box elements are marked with circles. Amplicons: P1 (–370 to –166 bp), P2 (–330 to –181 bp), P3 (–328 to –233 bp), P4 (–1085 to –894 bp), and P5 (located in gene body). D, ChIP-qPCR to verify the binding of BnaA03.WRKY28 to the W33box region of BnWRKY33. The input and precipitated DNA were analyzed by qPCR primers. BnACTIN2 (P0), the promoter region distant from the start codon (P4), and the exon region of BnWRKY33 (P5) were used as negative controls. % input method was employed to determine the enrichment of BnaA03.WRKY28 at designated loci. Asterisks indicate significant differences compared with BnACTIN2 (t test; **P < 0.01). E and F, Y1H assays to explore the binding capacity of BnaA03.WRKY28 to the BnWRKY33 promoter. DNA fragments (W33box, –326 to –246 bp; W1/W2/W3, −326 to −313 bp, −309 to −296 bp, and −259 to −246 bp for three times tandem duplication, respectively) from the BnWRKY33 promoter were cloned into the pAbAi vector, and the CDS of BnaA03.WRKY28 was cloned into the pGADT7 vector (pGADT7-WRKY28). SD/-Leu, selective dropout medium without leucine. AbA300, medium containing 300 ng/mL AbA; AbA0, medium without AbA. p53 was used as a positive control. Oblique triangles above the images indicate a 10°, 10−1, and 10−2 yeast-cell concentration gradient. G and H, EMSA to test the interaction between BnaA03.WRKY28 and the BnWRKY33 promoter in vitro. Probes labeled with Cy5 were used. The mutated probe replaced the core W-box sequence (W1, W2, and W3) with GAGAGA. I, Schematic of effector and reporter constructs using the pGreenII vector set. The CDSs of the TFs were introduced into the pGreenII 62-SK (SK) vector as effectors. Truncated promoter regions of BnWRKY33 were inserted in front of the Firefly LUC as reporters. Renilla LUC was used as an internal control. J, Dual-LUC assays were conducted to detect relative LUC activity (the ratio of Firefly to Renilla LUC activity) when co-transformed with effector and reporter. Co-transformed empty pGreenII 62-SK plasmids and reporter constructs were used as negative controls. Data are shown as means ± sd (n = 3). Asterisks indicate significant differences compared with the negative controls (t test; ns, P > 0.05, *P < 0.05, **P < 0.01).
WRKY TFs can bind to W-box cis-elements with the core sequence 5′-T/CTGACC/T-3′ (Eulgem et al., 2000). The BnWRKY33 promoter contains three W-box elements in the –346 to –1 bp region (Figure 2B), namely W1, W2, and W3. Additionally, a region from −326 to −246 bp containing three W-boxes was defined as W33box. To confirm whether BnaA03.WRKY28 binds to the BnWRKY33 promoter in vivo, ChIP-qPCR experiments were performed. Amplicons P1, P2, and P3, all covering the W33box, were significantly enriched compared with amplicons P4 (distant from W33box), P5 (located in gene body), or to the negative control BnACTIN2 (P0) (Figure 2, C and D). In Y1H assay, the bait strain pW33-box-AbAi was transformed with the prey construct pGADT7-BnaA03.WRKY28 and grew on SD/-Leu medium containing 300 ng/mL AbA (Figure 2E), indicating that BnaA03.WRKY28 can bind to the W33box in the BnWRKY33 promoter region. Moreover, truncating and mutating the W33box revealed that W1 and W3, but not W2, were the target sites of BnaA03.WRKY28 (Figure 2F and Supplemental Figure S4).
To further confirm that BnaA03.WRKY28 could directly target the cis-acting elements of BnWRKY33, electrophoretic mobility shift assay (EMSA) was carried out. When we added BnWRKY28 protein, a specific shifted band was produced; this shifted band was completely competitive when the unlabeled cold probe was added (Figure 2, G and H). We also detected the effect of BnaA03.WRKY28 on BnWRKY33 transcription using dual-luciferase (LUC) transient transcriptional activity assays. The Firefly LUC reporter gene was activated when Pro35S:BnaA03.WRKY28 was co-introduced into Arabidopsis (Arabidopsis thaliana) protoplasts with p346:LUC, pΔW1:LUC, pΔW2:LUC, or pΔW1W2:LUC (Figure 2, I and J). These results indicate that BnaA03.WRKY28 directly targets W1 and W3 in the promoter region of BnWRKY33 and activates BnWRKY33 expression.
Conserved MAPK cascade phosphorylates and activates BnWRKY33
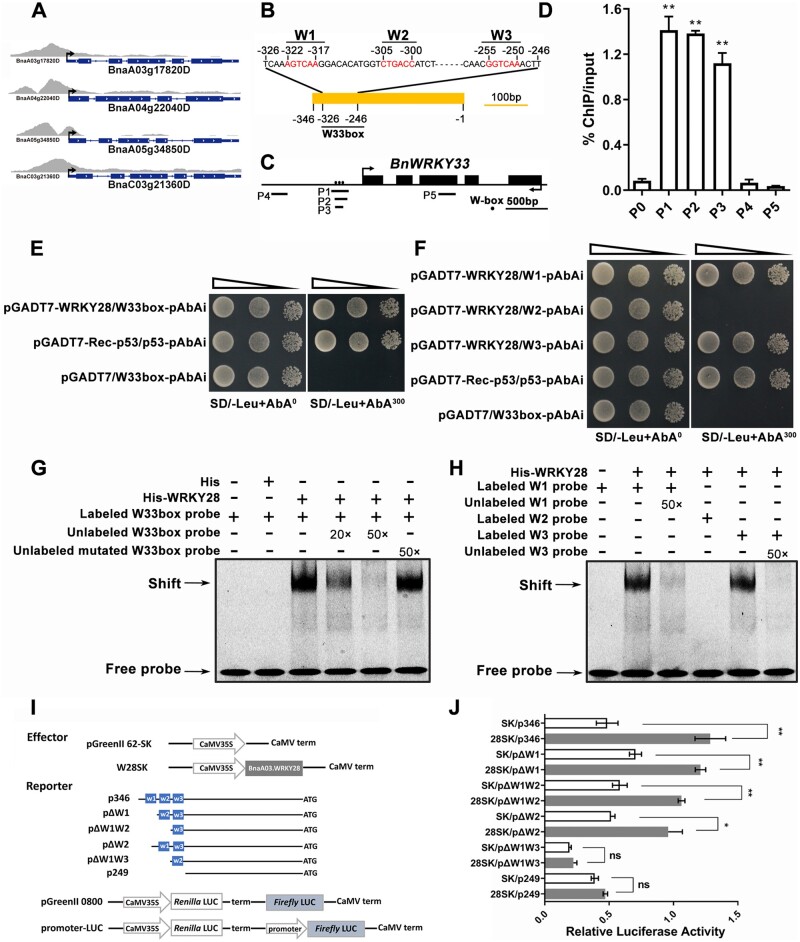
We demonstrated that BnaA03.WRKY28 was able to bind to BnWRKY33 promoter and promoted BnWRKY33 expression, negatively regulating Sclerotinia resistance. The expression level of BnWRKY33 in BnaA03.WRKY28 OE lines was higher than that in the WT (Supplemental Figure S5). Similarly, WRKY33 upregulates its expression through direct association with the WRKY33 promoter to amplify its positive effects on pathogen resistance (Mao et al., 2011; Liu et al., 2018). These indicate that both BnaA03.WRKY28 and BnWRKY33 can activate the expression of BnWRKY33, but lead to different resistance phenotypes, which was speculated to be the result of the weaker activation ability of BnaA03.WRKY28 on BnWRKY33 than BnWRKY33. However, BnWRKY33 does not have a stronger activation effect on BnWRKY33 than BnaA03.WRKY28 (Figure 3A). The transcription activity of BnaA03.WRKY28 was not lower than that of BnWRKY33 (Figure 3B). In Arabidopsis, WRKY33 can be phosphorylated by MPK3/6 in B. cinerea resistance (Mao et al., 2011). In this study, BnWRKY33 expression in BnaA03.WRKY28 OE lines and LUC activity was measured in the static state (without inoculation). Therefore, we considered that BnWRKY33 may be modified after inoculation with Sclerotinia. A cluster of five Ser residues in the N-terminus of WRKY33 was conserved (Figure 3C), which are the phosphorylation sites of AtWRKY33 mediated by AtMPK3. By site-directed mutagenesis, all five Ser residues were mutated to Asp (BnWRKY33DD), generating constitutively active BnWRKY33. The relative LUC activity of the co-transformed Pro35S:BnWRKY33DD with p346:LUC (W33DDSK/p346) was much higher than that of W33SK/p346 or W28SK/p346 (Figure 3A). Similar results were found when comparing W33DDGAL4BD and W33GAL4BD or W28GAL4BD (Figure 3B), suggesting that phosphorylated BnWRKY33 had greater activation capacity than BnaA03.WRKY28.
Figure 3.
BnMKK5–BnMPK3-mediated phosphorylation of BnWRKY33 is necessary to increase BnWRKY33 transcription. A, Relative LUC activity of BnaA03.WRKY28, BnWRKY33, and BnWRKY33DD when binding to the BnWRKY33 promoter. DD, conserved Ser residues mutated to Asp. B, Comparisons of the transcriptional activities of BnaA03.WRKY28, BnWRKY33, and BnWRKY33DD. The TF fused with the yeast GAL4 binding domain (GAL4BD) was used as an effector; GAL4-fLUC was used as a reporter and AtUbi:rLUC was used as an internal control. C, A diagram of the N-terminus of BnWRKY33 showing the positions of five conserved Ser residues according to AtWRKY33. Asterisks indicate the positions of conserved residue; marked S represents serine residue, and marked D represents aspartic acid residue. D, Interaction detection between BnWRKY33 and BnaA06.MPK3/BnaC03.MPK3 in Y2H. E, Co-IP to explore the interactions of BnWRKY33 with BnaA06.MPK3/BnaC03.MPK3 in vivo. Fusion proteins were co-expressed in N. benthamiana leaves. Total proteins were extracted (Input) and immunoprecipitated (IP) with GFP beads. F, Phosphorylation detection of BnWRKY33 by activated BnaA06.MPK3 (MPK3-A6) and BnaC03.MPK3 (MPK3-C3) in vitro. MPK3-A6 and MPK3-C3 were activated with constitutively active BnaA03.MKK5DD (MKK5DD). WRKY33-N, the N-terminus of BnWRKY33 containing the five conserved Ser residues; WRKY33AA-N, five conserved Ser residues in the BnWRKY33 N-terminus all mutated to Ala. Phosphorylation reactions were incubated in protein kinase buffer containing [γ-32P] ATP. Phosphorylation signals were detected via autoradiography (top panels). Protein inputs were assessed using Coomassie Brilliant Blue (CBB) staining (bottom panels). G, Dual-LUC transient transcriptional activity assays showing the effect of MAPK cascade on BnWRKY33-BnWRKY33 module. BnWRKY33 (W33), BnaA06.MPK3 (MPK3-A6), BnaC03.MPK3 (MPK3-C3), and BnaA03.MKK5DD (MKK5DD) were introduced into pGreenII 62-SK (SK) as effectors. The BnWRKY33 promoter (p346, –326 to –246 bp) was inserted into pGreenII-0800 as reporter. H, In vitro EMSA to explore the binding of phosphorylated BnWRKY33 (WRKY33DD) to the BnWRKY33 promoter. W33box, W1, W2, and W3 were used as probes. In (A), (B), and (G), data are shown as means ± sd (n = 3). Asterisks indicate significant differences (t test; ns, P > 0.05, *P < 0.05).
The study of WRKY33 phosphorylation by MPK3/6 in Arabidopsis inspired us to conjecture that Sclerotinia infection induces BnWRKY33 to be phosphorylated and activated by MAPK cascade in oilseed rape. Y2H assays revealed that BnWRKY33 interacted with BnaA06.MPK3 and BnaC03.MPK3 (Figure 3D). These interactions were further confirmed via co-immunoprecipitation (Co-IP) assays (Figure 3E). Therefore, in vitro phosphorylation assays were performed to determine whether BnWRKY33 could be phosphorylated by the MAPK cascade. We previously demonstrated that the BnaA03.MKK5-BnaA06.MPK3/BnaC03.MPK3 module plays a conserved and positive role in S. sclerotiorum resistance of B. napus (Zhang et al., 2022), in which BnaA03.MKK5DD (an active BnaA03.MKK5 mutant) was used. The autoradiogram revealed that the recombinant GST-BnaA06.MPK3 and GST-BnaC03.MPK3 strongly phosphorylated BnWRKY33 and the truncated BnWRKY33 (N-terminal containing the conserved Ser residues) with constitutively active BnaA03.MKK5DD (Figure 3F). However, when the five Ser residues were mutated into Ala (generating WRKY33AA-N), BnWRKY33 was not phosphorylated by BnMPK3 (Figure 3F). In the dual-LUC transient transcriptional activity assays, we found that without activation by the constitutively active BnaA03.MKK5DD, neither BnaA06.MPK3 nor BnaC03.MPK3 enhanced BnWRKY33 activity. However, relative LUC activity was significantly higher when the MAPK cascade was activated (Figure 3G and Supplemental Figure S6). Overall, these results demonstrate that the conserved MAPK cascade module mediated by BnaA03.MKK5-BnaA06.MPK3/BnaC03.MPK3 phosphorylates BnWRKY33 and activates the transcriptional activity of BnWRKY33. In addition, EMSA indicates that the phosphorylated BnWRKY33 is able to bind to W33box, W1, W2, and W3 (Figure 3H).
As a co-factor, BnaA09.VQ12 interacts with BnaA03.WRKY28, forming a protein complex that negatively regulates Sclerotinia resistance
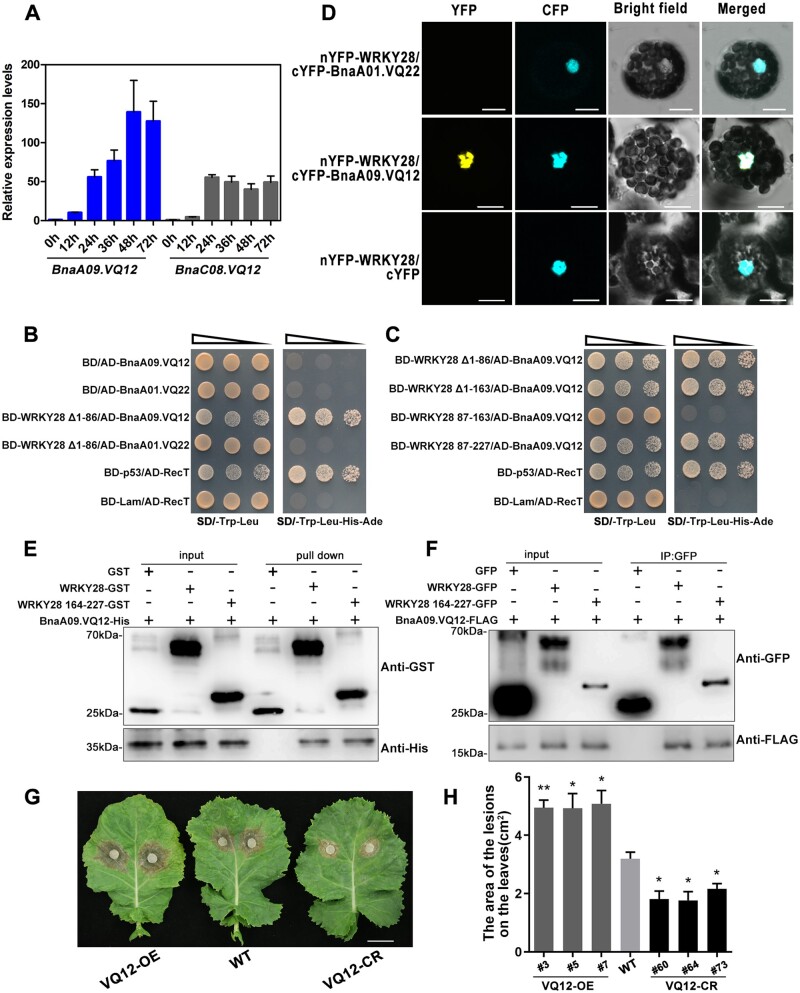
VQ proteins are a class of plant-specific proteins with the conserved FxxxVQxLTG amino acid sequence motif. VQ proteins have been reported to regulate gene expression by interacting with WRKY TFs (Cheng et al., 2012). AtVQ12 plays a negative role in resistance to B. cinerea (Wang et al., 2015). There were two homoeologous copies of BnVQ12 (BnaA09.VQ12 and BnaC08.VQ12) in B. napus. BnaA09.VQ12 was markedly induced by S. sclerotiorum; and its induction trend was similar to that of BnaA03.WRKY28 (Figure 4A). We first tested the interaction between BnaA03.WRKY28 and BnaA09.VQ12 using Y2H assays. In yeast, full-length BnaA03.WRKY28 was able to activate reporter genes, thus truncated domains of BnaA03.WRKY28 were used (Supplemental Figure S7). BnaA03.WRKY28, with a deleted domain (WRKY28 Δ1-86), interacted with BnaA09.VQ12 but not BnA01.VQ22 (Figure 4B), indicating the specificity of the BnaA03.WRKY28–BnaA09.VQ12 interaction. Furthermore, BnaA09.VQ12 interacted with BnaA03.WRKY28 via the 164–227 amino acid domain of BnaA03.WRKY28, which is the DNA-binding domain containing the WRKY motif and C2H2 zinc finger (Figure 4C). The bimolecular fluorescence complementation (BiFC) assays showed the proximity of BnaA03.WRKY28 and BnaA09.VQ12 in the nucleus (Figure 4D). Subcellular localization analysis showed that BnaA03.WRKY28 was localized in the nucleus, whereas BnaA09.VQ12 was present in the cytoplasm and nucleus (Supplemental Figure S8). Glutathione S-transferase (GST) pull-down assays were conducted to confirm the interactions in vitro. GST-BnaA03.WRKY28 and GST-BnaA03.WRKY28 164–227, but not the GST control, pulled down His-BnaA09.VQ12 (Figure 4E). Co-IP assays further showed that BnaA09.VQ12-FlAG interacted with BnaA03.WRKY28-GFP and even with BnaA03.WRKY28 164–227 (Figure 4F). Thus, the co-factor BnaA09.VQ12 forms a complex with BnaA03.WRKY28 in the nucleus by binding to the DNA-binding domain of BnaA03.WRKY28.
Figure 4.
BnaA03.WRKY28 interacts with BnaA09.VQ12 and negatively contributes to Sclerotinia resistance. A, Induced expression profiles of two copies of BnVQ12 were identified at 0, 12, 24, 36, 48, and 72 h after inoculation with S. sclerotiorum. The latest fully unfolded leaves of oilseed rape (Jia 9709, ca. six-week-old) were used for infection. Data are shown as means ± sd (n = 3). B and C, Y2H assays to identify the interactions between BnaA03.WRKY28 and BnaA09.VQ12 (B), and the specific domain of BnaA03.WRKY28 that interacts with BnaA09.VQ12 (C). The yeast cells were cultured on SD/-Trp-Leu (selective dropout medium without tryptophan and leucine) or SD/-Trp-Leu-His-Ade (selective dropout medium without tryptophan, leucine, histidine, and adenine). AD, pGADT7/activation domain; BD, pGBKT7/binding domain. BD-p53/AD-RecT, positive control; BD-Lam/AD-RecT, negative control. Oblique triangles above the images indicate a 10°, 10−1, and 10−2 yeast-cell concentration gradient. D, BiFC to confirm the interaction between BnaA03.WRKY28 and BnaA09.VQ12 in Arabidopsis protoplasts. BnaA01.VQ22 and cYFP served as negative controls. Scale bars, 10 μm. E, In vitro pull-down assays to test the direct interactions between BnaA09.VQ12 and BnaA03.WRKY28, especially and the DNA-binding domain of BnaA03.WRKY28 (comprising 164–227 amino acids). GST, GST-WRKY28, or GST-WRKY28 164–227 was immobilized on GST beads and incubated with His-BnaA09.VQ12 protein. F, In vivo Co-IP assays with BnaA03.WRKY28-GFP and BnaA09.VQ12-FLAG co-expressed in N. benthamiana leaves. Proteins were extracted (Input) and immunoprecipitated (IP) with GFP beads. G and H, Phenotype of BnVQ12 transgenic homozygous lines and the WT (Jia 9709) after 48 h inoculation with S. sclerotiorum; lesion areas were analyzed statistically. Scale bar, 2 cm. Data are shown as means ± sd (n = 3). Asterisks indicate significant differences compared with the WT (t test; *P < 0.05, **P < 0.01).
To characterize the biological function of BnaA09.VQ12 in response to Sclerotinia infection, three independent BnaA09.VQ12 OE lines and three independent loss-of-function homozygous mutants with two VQ12 copies were obtained (Supplemental Figure S9, A and B). At 48 h post-inoculation, the necroses in the WT plants were more severe than those in the VQ12 knockout lines, but less severe than those in the BnaA09.VQ12 OE lines (Figure 4, G and H and Supplemental Figure S9C). After S. sclerotiorum inoculation, BnPAD3 and BnCYP71A13 transcription was significantly lower in BnaA09.VQ12 OE lines, but greater in VQ12 knockout lines, relative to levels in the WT. In WT plants, expression of camalexin synthesis-related genes was significantly upregulated 24 h after incubation, but was significantly lower at 36 h. By contrast, in the BnaA09.VQ12 OE lines, their expression was not induced noticeably at 24 h, and remained low at 36 h. Relative to the levels in the WT, camalexin synthesis-related gene expression in VQ12 knockout lines was similar at 24 h, but was significantly higher at 36 h (Supplemental Figure S9, D and E). These results suggest that BnaA09.VQ12, similar to its partner BnaA03.WRKY28, contributes negatively to Sclerotinia resistance.
BnaA03.WRKY28–BnaA09.VQ12-complex formation causes BnaA03.WRKY28 to have stronger BnWRKY33-promoter-binding capacity than BnWRKY33
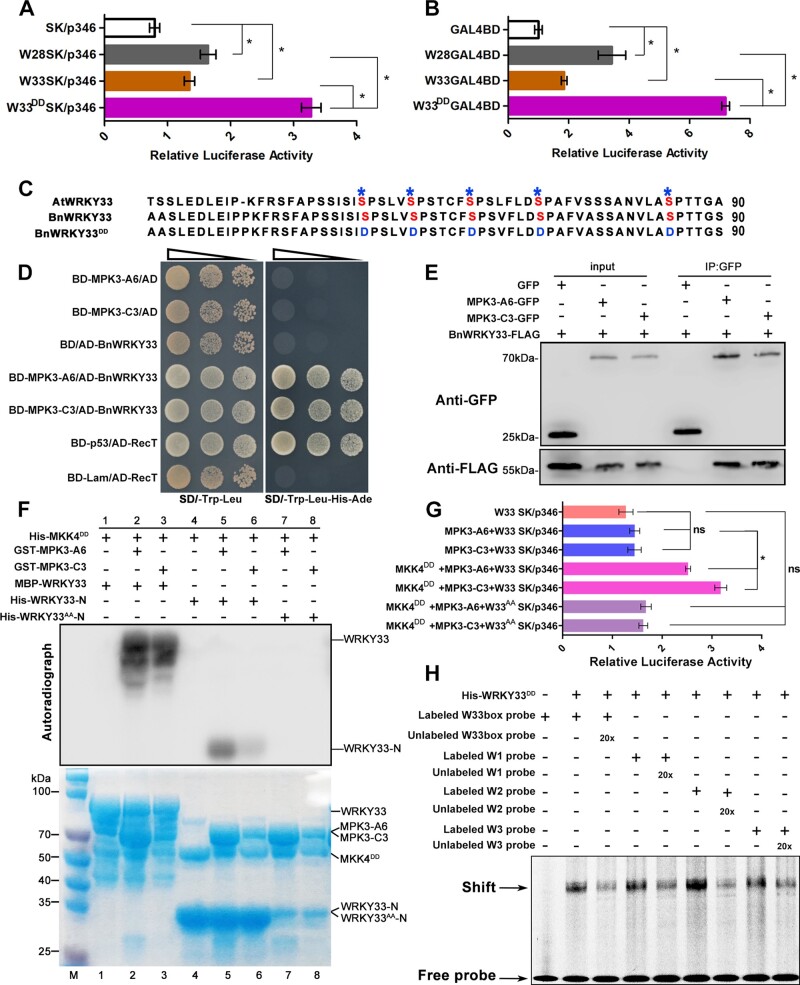
We monitored lesion development on oilseed rape leaves infected with S. sclerotiorum within 84 h, and plotted the increased rate of lesions every 3 h (Figure 5A). The increased rate of lesions was at the lowest level within 15–24 h after inoculation, the period of greatest BnWRKY33 induction (Figure 5B). However, from 36 h after inoculation, lesion area increased rapidly, peaking at 42–48 h, the period when BnaA03.WRKY28 and BnaA09.VQ12 were substantially induced (Figures 1A and 4A). The expression of BnWRKY33 decreased sharply 36 h after inoculation (Figure 5B). In addition, we detected the expression levels of WRKY28, WRKY33, and VQ12 at 0, 1, 2, 3, 4, and 5 days after inoculation of S. sclerotiorum in oilseed rape stems, and found that these genes were all induced during infection time (Supplemental Figure S10). The induction trends were almost consistent with those in infected leaves, indicating that the function mode of these genes in leaf and stem may be similar.
Figure 5.
The BnaA09.VQ12–BnaA03.WRKY28 protein complex inhibits BnWRKY33 transcription by competitively binding to the BnWRKY33 promoter. A, Increase in lesion radius on oilseed rape leaves infected by Sclerotinia over 84 h. Lesions were measured every 3 h. B, Induced expression of BnWRKY33 in the WT, 0, 12, 24, 36, 48, and 72 h after inoculation with S. sclerotiorum. C, Relative expression levels of BnWRKY33 in BnaA03.WRKY28/BnaA09.VQ12 transgenic lines and WT plants after Sclerotinia inoculation for 24 and 48 h. Asterisks indicate significant differences compared with the WT at the corresponding time points (t test; *P < 0.05). D, EMSA showing the effect of the BnaA09.VQ12 on the binding capacity of BnaA03.WRKY28 to the BnWRKY33 promoter. Shift, indicating the binding of BnaA03.WRKY28; Super shift, indicating the binding of BnaA09.VQ12–BnaA03.WRKY28 complex. E, In vitro binding assays to compare the binding capacity of BnaA09.VQ12–BnaA03.WRKY28 complex and phosphorylated BnWRKY33 to the BnWRKY33 promoter. Equal addition of BnaA03.WRKY28 protein in Lanes 2, 3, 5, and 6; equal addition of BnWRKY33 protein in Lanes 4 and 5; equal addition of BnWRKY33DD and BnaA03.WRKY28 was mixed in Lane 5; 5:1 ratio of BnWRKY33DD:BnaA03.WRKY28 was mixed in Lane 6. Protein inputs were assessed using CBB staining (bottom panels). F and G, In vivo dual-LUC transient transcriptional activity assays to explore competitive binding of BnaA09.VQ12, BnaA03.WRKY28, and BnWRKY33 to the BnWRKY33 promoter. In (F) and (G), data are shown as means ± sd (n = 3); asterisks indicate significant differences (t test; ns, P > 0.05, *P < 0.05, **P < 0.01).
BnWRKY33 expression was significantly lower in BnaA03.WRKY28 or BnaA09.VQ12 OE lines than in the WT, especially at 24 h after inoculation, when the expression of BnWRKY33 reached its peak in the WT. However, 24 h after inoculation, BnWRKY33 expression in BnaA03.WRKY28 or BnaA09.VQ12 knockout lines was similar to that in the WT. By contrast, at 48 h, BnWRKY33 expression was significantly higher in BnaA03.WRKY28 or BnaA09.VQ12 knockout plants than in the WT (Figure 5C). These results indicate that the BnaA03.WRKY28–BnaA09.VQ12 complex may inhibit the expression of BnWRKY33. Since both BnaA03.WRKY28 and BnWRKY33 were able to bind to the BnWRKY33 promoter, and BnaA03.WRKY28 had lower transcriptional activity than activated BnWRKY33, we considered that BnaA03.WRKY28 may inhibit BnWRKY33 binding to the BnWRKY33 promoter.
To evaluate this, we first examined the effects of the interaction between BnaA09.VQ12 and BnaA03.WRKY28 on the binding of BnaA03.WRKY28 to the BnWRKY33 promoter (Figure 5D). In the absence of BnaA09.VQ12, BnaA03.WRKY28 bound to the BnWRKY33 promoter and formed a specific shifted band, whereas BnaA09.VQ12 alone failed to target the BnWRKY33 promoter. Adding BnaA09.VQ12 to the mixture of BnaA03.WRKY28 and a DNA probe produced a specific super-shifted band, whose intensity was enhanced when more BnaA09.VQ12 was added. These results suggest that BnaA09.VQ12 enhances the binding capacity of BnaA03.WRKY28 to BnWRKY33 by forming a complex with BnaA03.WRKY28. In vitro EMSA was subsequently conducted to evaluate the competitive binding abilities of BnaA03.WRKY28 and BnWRKY33 to the BnWRKY33 promoter (Figure 5E). BnaA03.WRKY28, BnaA03.WRKY28–BnaA09.VQ12 complex and BnWRKY33DD could target the BnWRKY33 promoter, respectively, and produced corresponding shifted bands. However, when equal addition of BnaA03.WRKY28 and BnWRKY33DD protein was incubated together with the DNA probe, BnaA03.WRKY28–BnaA09.VQ12 complex outcompeted phosphorylated BnWRKY33 for binding to the BnWRKY33 promoter; when a 5:1 ratio of BnWRKY33DD:BnaA03.WRKY28 was used, the shifted band produced by phosphorylated BnWRKY33 binding was observed. In addition, dual-LUC transient transcriptional activity assays revealed that the relative LUC activity displayed marked trends when W28SK, W33DDSK, and VQ12SK were used as effector combinations. Similar relative LUC activity was observed when using W28SK or W33DDSK as independent effectors, and using VQ12SK and W28SK, VQ12SK and W33DDSK as simultaneous effectors (Figure 5F), indicating that BnaA09.VQ12 did not change the transcriptional activity of BnaA03.WRKY28 and BnWRKY33. Using W28SK and W33DDSK simultaneously, the relative LUC activity was between that achieved when they were used as independent effectors (Figure 5F). However, when VQ12SK was reintroduced into the mixture of W28SK and W33DDSK, the relative LUC activity tended to be similar to that of W28SK as an independent effector (Figure 5G). Based on these data, we believe that BnaA03.WRKY28 and BnWRKY33 compete to bind to the BnWRKY33 promoter when present simultaneously, but that the presence of BnaA09.VQ12 causes BnaA03.WRKY28 to have stronger BnWRKY33-promoter-binding capacity than BnWRKY33.
BnaA03.WRKY28 promotes axillary bud activity and shoot branching
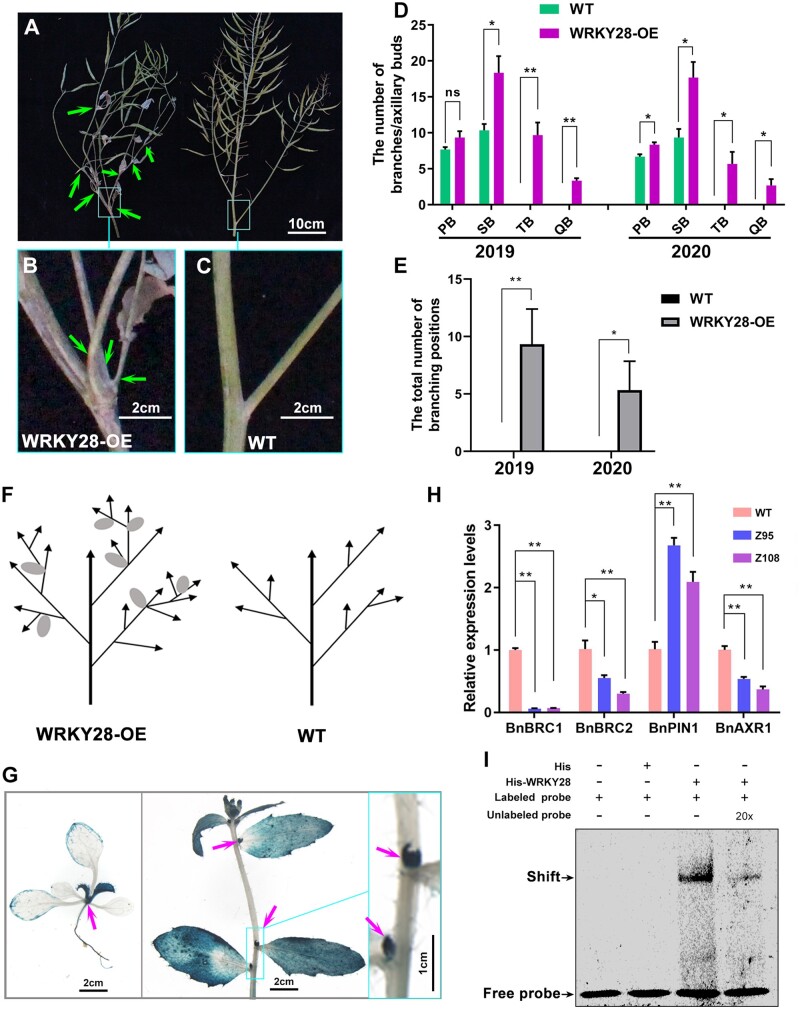
Notably, BnaA03.WRKY28 OE lines produced more branches than the WT, especially many higher-order branches and many new axillary buds in late-stage oilseed rape growth (Figure 6A). In BnaA03.WRKY28 OE lines, two or more branches were produced in one leaf axil (Figure 6, B and C). We quantified the number of branches and buds developing in the leaf axils of WRKY28-OE lines and WT plants in the field for two years (2019 and 2020). In the WT plants, at most one branch formed in each leaf axil, and secondary branching was the highest order branching. By contrast, in the BnaA03.WRKY28 OE lines, many more branches were formed; two or more branches formed in each leaf axil and quaternary branches were very common (Figure 6, D and E; detailed data can be found in Supplemental Figure S11), suggesting that BnaA03.WRKY28 promotes the initiation of axillary bud activity and shoot branching (Figure 6F).
Figure 6.
BnaA03.WRKY28 promotes axillary branch formation. A, Phenotype of branching (indicated by arrows) in BnaA03.WRKY28 OE lines (left) and WT (right). Scale bar, 10 cm. B and C, Close-up view of the representative leaf axil in the square in (A); scale bars, 2 cm. D and E, Statistical description of lateral branches or buds in every order branching (D) and branching positions that produce two or more branches in one axil (E) in BnaA03.WRKY28 OE lines and late-growth WT plants (n = 3) for two years (2019 and 2020). F, Schematic of branches in WRKY28-OE lines and WT plants. Arrows, branches; ellipses, cauline leaves. G, GUS staining showing the expression of BnaA03.WRKY28. Left, two-week-old plants; scale bar, 2 cm. Middle, six-week-old plants; scale bar, 2 cm. Right, close-up view of the leaf axils in the square in the middle image; scale bar, 1 cm. H, RT-qPCR to examine transcription levels of branching-related genes in BnaA03.WRKY28 OE lines and in the WT. Data are shown as means ± sd (n = 3). Asterisks indicate significant differences (t test; *P < 0.05, **P < 0.01). I, EMSA to detect BnaA03.WRKY28 binding to the BnaC03.BRC1 promoter. The sequence containing two W-box motifs in the BnaC03.BRC1 promoter was used as a DNA probe.
β-galactosidase (GUS) staining revealed that BnaA03.WRKY28 was expressed in the latest true leaves and axils at the seedling stage, and in leaf axils at the flowering stage (Figure 6G). Moreover, among the DEGs identified via RNA-seq, expression of genes involved in meristem growth and auxin polar transport was significantly higher in the BnaA03.WRKY28 OE lines than in the WT, whereas that of genes involved in auxin influx, cellular response to auxin stimulus, and auxin biosynthetic process was significantly lower (Supplemental Figure S12 and Supplemental Table S4). Among the overlapping genes of RNA-seq and ChIP-seq, Teosinte branched1/Cycloidea/Proliferating cell factor (TCP) TFs, including TCP18 (BRC1), were identified (Supplemental Table S3). TCP TFs have profound effects on meristem and lateral organ growth; BRC1 plays a central role in shoot branching, with BRC2 having a slightly weaker role (Aguilar-Martínez et al., 2007; Nicolas and Cubas, 2016). Axillary meristem initiation and axillary bud activation require minimization of auxin content in the leaf axil, which largely depends on PIN1-mediated auxin efflux (Wang et al., 2014a). AXR1 is required for auxin to inhibit axillary bud growth (Stirnberg et al., 1999). RT-qPCR revealed that BnBRC1, BnBRC2, and BnAXR1 were downregulated in BnaA03.WRKY28 OE lines, whereas BnPIN1 was upregulated, relative to the WT (Figure 6H). Two W-box elements were found approximately 1,000 bp upstream of the BnaC03.BRC1 start codon. A BnaC03.BRC1-promoter sequence containing two W-box elements was synthesized as a probe. EMSA revealed that BnaA03.WRKY28 bound to the DNA probe to produce a specific shifted band, which was diluted when an unlabeled probe was added (Figure 6I). These results support the involvement of BnaA03.WRKY28 in growth and development, and particularly in axillary bud outgrowth and branch formation.
Discussion
Although defense against pathogens is essential to ensure plant survival, it is costly: the immune response leads to the inhibition of plant growth and development (Gómez‐Gómez et al., 1999; Tian et al., 2003). In particular, constitutive defense reactions involving OE of resistance-related signaling molecules can lead to lesion-mimic phenotypes, reduced height, and early senescence (Delteil et al., 2010). A trade-off between plant growth and disease resistance has become a generally accepted concept (Karasov et al., 2017). Differential allocation of limited resources to life activities may be responsible for trade-offs between growth and defense (Heil and Baldwin, 2002; Matyssek et al., 2005).
Phytoalexins, important components of plant defense, are low molecular-mass antimicrobial compounds synthesized de novo and accumulated after a plant is infected by a pathogen (VanEtten et al., 1994; Ahuja et al., 2012). Camalexin, the major phytoalexin in Arabidopsis and some Brassicaceae species, participates in defense against S. sclerotiorum and B. cinerea (Mao et al., 2011; Stotz et al., 2011; Ahuja et al., 2012). Camalexin is synthesized from tryptophan, which is also precursor to indole-3-acetic acid (IAA) (Cohen et al., 2003; Glawischnig et al., 2004), potentially contributing to the antagonism between growth and defense. Two P450 enzymes, CYP71A13 and PAD3, are involved in camalexin biosynthesis. In the absence of stress, camalexin biosynthesis-related genes, including AtCYP71A13 and AtPAD3, are expressed at very low levels. Under pathogen infection, however, WRKY33 targets and induces these genes strongly, activating camalexin biosynthesis (Nafisi et al., 2007; Qiu et al., 2008; Zhou et al., 2020). Relative to WT plants, BnCYP71A13 and BnPAD3 expression was higher in BnMPK3/BnaA03.MKK5 OE and BnaA03.WRKY28/BnaA09.VQ12 knockout plants; this was associated with better disease resistance.
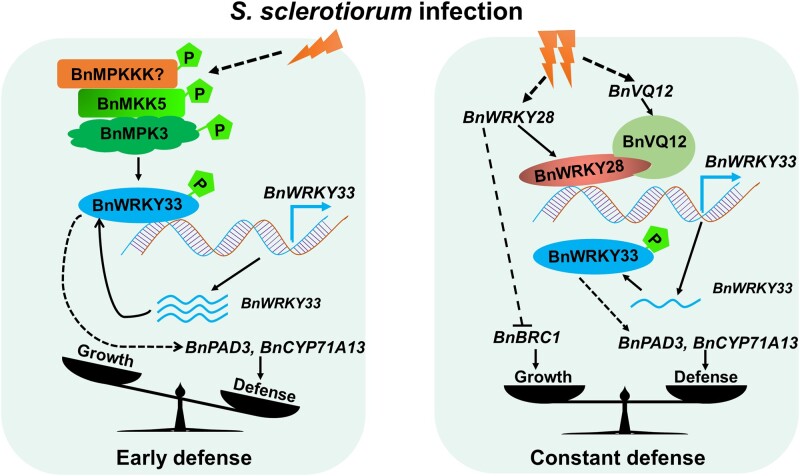
We previously characterized the positive regulatory role of BnWRKY33 in Sclerotinia resistance (Liu et al., 2018). In the present study, we found that two BnWRKY TFs (BnaA03.WRKY28 and BnWRKY33) dynamically regulate plant immunity against Sclerotinia by competitive binding to BnWRKY33 promoter. Based on these results, we propose a model that accounts for the actions of BnWRKY33 and BnaA03.WRKY28 in response to S. sclerotiorum in oilseed rape (Figure 7). When threatened by Sclerotinia, the plant innate immune system mediated by MAPK cascade is engaged. BnaA06.MPK3 and BnaC03.MPK3 are activated following phosphorylation by BnaA03.MKK5; the activated BnMPK3 phosphorylates BnWRKY33 and enhances its transactivation activity. BnWRKY33 binds to its own promoter to maintain high BnWRKY33 expression. BnWRKY33 regulates the high expression of disease resistance-related genes such as BnPAD3 and BnCYP71A13, which promote phytoalexin biosynthesis and defend against the fungal pathogen invasion. Defense is dominant during this early stage. With ongoing infection, the plant is at a potential risk of resistance breakdown, and continued resistance becomes highly costly. At this point, BnaA03.WRKY28 and BnaA09.VQ12 are strongly induced. BnaA09.VQ12 interacts with BnaA03.WRKY28, helping it to compete with BnWRKY33 in binding to the BnWRKY33 promoter. Because the transcriptional activity of BnaA03.WRKY28 is weaker than that of BnWRKY33, the expression of BnWRKY33 is lower than that mediated by BnWRKY33 during the early infection stage. This in turn slows BnWRKY33-regulated resistance-related gene expression, thereby reducing disease resistance. During the later stages of infection, this reduction in defense response is accompanied by recommencement of active growth: BnaA03.WRKY28, which is expressed in leaf axils, stimulates axillary meristem initiation and the development of axillary buds that grow into complete branches. This inhibition of disease resistance contributes to the trade-off between defense and growth.
Figure 7.
Model showing the actions of BnWRKY33 and BnaA03.WRKY28 in response to S. sclerotiorum in B. napus. A hypothetical model of the BnaA03.WRKY28-BnWRKY33 module fine-tuning the timing of the defense response against Sclerotinia. Early defense, dominant defense mediated by BnWRKY33; constant defense, a balance between growth and defense mediated by BnaA03.WRKY28.  indicates phosphorylation;
indicates phosphorylation;  represents transcription of BnWRKY33;
represents transcription of BnWRKY33;  indicates promotion; and
indicates promotion; and  indicates inhibition. The solid line indicates confirmation and the dotted line indicates potential.
indicates inhibition. The solid line indicates confirmation and the dotted line indicates potential.
We focused on the negative regulation mechanism of BnaA03.WRKY28 in plant immunity and demonstrated its function in promoting branch formation. BnaA03.WRKY28 OE lines showed more active axillary buds and produced more leaf-axil branches than the WT. AtWRKY71, which is in the same clade as AtWRKY28, promotes axillary meristem initiation by positively regulating key meristem formation-related TFs (including AtRAX1, AtRAX2, and AtRAX3) in branches (Guo et al., 2015). Guo et al. (2015) reported that AtWRKY28 OE also causes excessive branching; particularly, five closely related WRKY members (WRKY8, WRKY28, WRKY48, WRKY57, and WRKY71) are functionally redundant in shoot branching, and the phenotype of branch formation defect can be only identified in the quintuple mutants. Our RNA-seq and ChIP-seq analyses revealed the involvement of lateral-branch formation-related genes, such as TCP TFs (including BnBRC1) and auxin polar transporters (including BnPIN1); changes in their expression caused excessive branching of the BnaA03.WRKY28 OE lines.
VQ proteins are thought to regulate gene expression by interacting with WRKY TFs involved in plant growth, development, and stress responses (Cheng et al., 2012; Jing and Lin, 2015). In Arabidopsis, WRKY superfamily TFs are classified into three groups (group I, group II, and group III) (Eulgem et al., 2000), and group II is further divided into IIa, IIb, IIc, IId, and IIe (Rushton et al., 2010). VQ proteins interact physically with group I or group IIc WRKY proteins via their WRKY domains (Cheng et al., 2012; Jing and Lin, 2015). AtWRKY33 (group I) interacts with VQ23 (SIB1) or VQ16 (SIB2) in resistance to B. cinerea (Lai et al., 2011). The interaction between VQ and WRKY can affect the DNA-binding activity of WRKY TFs (Jing and Lin, 2015). Here, we identified another group IIc member, BnaA03.WRKY28, which interacted with BnaA09.VQ12 via its DNA-binding domain. Formation of the BnaA09.VQ12–BnaA03.WRKY28 complex enhances the DNA-binding activity of BnaA03.WRKY28. Further, although AtQ22 is speculated to interact with AtWRKY28 (Hu et al., 2013a), we found that, in oilseed rape Sclerotinia response, BnaA09.VQ12 but not BnA01.VQ22 was the co-factor of BnaA03.WRKY28.
MAPK cascade phosphorylation mediated by MEKK–MKK–MAPK is an essential event in plant defense. In the study of MAPK cascades, MPK3/6 is well elucidated and is conservatively involved in response to biotic and abiotic stresses. The MKK4/5–MPK3/6 module contributes to plant immunity (Ichimura et al., 2002; Meng and Zhang, 2013; Zhang et al., 2022). Kinase phosphorylates TF and alters its transactivation activity or DNA-binding activity (Mao et al., 2011; Zhou et al., 2020). In this study, we verified that phosphorylation of BnWRKY33 by the BnMKK5–BnMPK3 module is conserved in Sclerotinia resistance in oilseed rape, and is necessary to enhance the transactivation activity of BnWRKY33. When plants were incubated with Sclerotinia, no phosphorylation signal was detected on BnaA03.WRKY28 OE protein, but the phosphorylation signal was detected on BnWRKY33 protein (Supplemental Figure S13), suggesting that BnWRKY33 is phosphorylated in response to S. sclerotiorum, but BnaA03.WRKY28 is probably not phosphorylated after infection, indicating that the difference in the transcriptional activity of these two WRKY TFs leads to differences in resistance response severity to S. sclerotiorum.
Plants have evolved intricate and precise gene regulatory networks to modulate their growth and cope with the changing environment. In the early stage of defense, BnWRKY33 is activated by MAPK cascade, and the activated BnWRKY33 directly binds to its own promoter, contributing to the transcriptional burst of BnWRKY33. The promoters of both AtWRKY33 and BnWRKY33 contain a cluster of W-boxes, and WRKY33 can bind directly to its own promoter (Mao et al., 2011; Liu et al., 2018). In this study, we identified that BnaA03.WRKY28 was also able to bind to the BnWRKY33 promoter. Specifically, BnWRKY33 binds to W1, W2, and W3 of the BnWRKY33 promoter, whereas BnaA03.WRKY28 targets W1 and W3. Additionally, BnWRKY33 and BnaA03.WRKY28 exhibited different induced trends after infection. BnWRKY33 expression was significantly induced earlier, peaked at 24 h, and then declined, whereas that of BnaA03.WRKY28 was induced later and peaked at 48 h. The later-stage decline in BnWRKY33 expression and simultaneous increase in BnaA03.WRKY28 expression were due by the fact that BnaA03.WRKY28, which has lower transcriptional activity, replaced BnWRKY33 in binding to the BnWRKY33 promoter. It is suggested that BnaA03.WRKY28 has a higher affinity for BnWRKY33 promoter than BnWRKY33 with the help of BnaA09.VQ12. This counts much for fine-tuning of the late-stage defense mechanism. Further, BnaA03.WRKY28, which was induced during late-stage infection, promoted branch formation by regulating the expression of branching-related genes. BnaC03.BRC1 is the likely target gene of BnaA03.WRKY28. In response to lethal pathogens, BnaA03.WRKY28 acted as a brake factor of defense and an activated factor of growth, contributing to the strategies of survival and reproduction.
Materials and methods
Plant materials, growth conditions, and plasmid construction
Transgenic lines were established from spring oilseed rape (B. napus) varieties Jia9709 or Westar. OE and CRISPR/Cas9 (CR) lines were generated, including BnaA03.WRKY28-OE/Jia9709, BnaA03.WRKY28-OE/Westar, BnaA03.WRKY28-CR/Jia9709, BnaA09.VQ12-OE/Jia9709, and BnVQ12-CR/Jia9709. Plants were grown at an experimental station in Wuhan under natural conditions for stem inoculation and seed propagation. For oilseed rape leaf inoculation and growth of A. thaliana and Nicotiana benthamiana, seedlings were grown in a greenhouse at 22°C under a long-day condition (16-h day/8-h night). The target fragments were cloned based on B. napus reference genome Darmor-bzh V5 and BnTIR (http://yanglab.hzau.edu.cn/BnTIR) database, and were amplified from Jia9709/Westar cDNA. OE constructs were driven by CaMV35S (modified from pCAMBIA2300 vector); 35S:Kpn1-BnaA03.WRKY28-FLAG-Xba1 was transformed into Jia9709, 35S:Sal1-BnaA03.WRKY28-BamH1 was transformed into Jia9709 and Westar, and 35S:Sal1-BnaA09.VQ12-BamH1 was transformed into Jia9709. CRISPR/Cas9-mediated gene-editing constructs were designed using the binary vector pKSE401 (Xing et al., 2014). The sgRNA was designed using CRISPR-P (http://cbi.hzau.edu.cn/cgi-bin/CRISPR). The constructed plasmids were confirmed by restriction digestion analysis and sequencing. The transgenic plants of empty-loading (empty OE vector backbone) transformation were used as negative test lines. Construct primers are listed in Supplemental Table S5.
Pathogen inoculation and disease resistance assay
Sclerotinia sclerotiorum strain A367 was cultured on potato dextrose agar (PDA) medium (20% [w/v] potato, 2% [w/v] dextrose, and 1.5% [w/v] agar) for inoculation. The leaf inoculation assay was performed as previous described (Liu et al., 2018; Wang et al., 2018), using the latest fully unfolded leaves of ca. 6-week-old plants. At 48 h post-inoculation, images of the inoculated leaves were taken, and lesion area was measured using ImageJ software. For stem inoculation, the assay was performed as previously described (Wei et al., 2016). At the flowering period, the mycelial side of the fungus agar block was attached to the stem ca. 30 cm above the soil, using plastic wrap. The stems were sprayed with water mist. At 72 h after inoculation, images of inoculated stems were taken, and lesion length was measured using ImageJ software.
RNA extraction and RT-qPCR
Total RNA was extracted from 100 mg oilseed rape leaves (collected from infected plants, uninfected plants, or OE lines) using the RNAprep plant kit (TIANGEN, DP441, China). The RNA-seq samples were from BnaA03.WRKY28 OE lines and WT plants for three biological replicates. Twio micrograms of total RNA was used for cDNA synthesis using the RevertAid First Strand cDNA kit (Fermentas, #K1622, USA). RT-qPCR analysis was performed with the CFX96 Real-Time system (Bio-Rad, USA) using the SYBR Green Realtime PCR Master Mix (TOYOBO, QPK-201, Japan). BnACTIN2 was used as a control to normalize expression levels according to the 2−ΔΔCT method (Livak and Schmittgen, 2001). The RT-qPCR primers are given in Supplemental Table S5.
Yeast one-hybrid assay
Yeast one-hybrid (Y1H) assays were performed as the manual of Matchmaker Gold Y1H System (Clontech). The amplified coding sequence (CDS) of BnaA03.WRKY28 was cloned into the pGADT7 vector. WT and mutated BnWRKY33 promoter fragments (−326 to −246 bp), and three W-box elements of BnWRKY33 promoter (−326 to −313, −309 to −296, and −259 to −246 bp for three times tandem duplication, respectively) were cloned into the pAbAi vector, and then were transformed into the Y1HGold yeast strain. The prey plasmid was introduced into the bait strains. Clones were cultured on selective dropout medium SD/-Leu with or without 300 ng/mL Aureobasidin A (AbA). Construct primers are listed in Supplemental Table S5.
Yeast two-hybrid assay
The full-length CDS of BnWRKY33, BnaA09.VQ12, and BnaA01.VQ22 was cloned into the pGADT7 vector. The CDS of BnaA03.WRKY28, truncated BnaA03.WRKY28, BnaA06.MPK3, and BnaC03.MPK3 was cloned into the pGBKT7 vector. The recombinant pGADT7 and pGBKT7 constructs were co-transformed into AH109 yeast strain. The yeast two-hybrid (Y2H) assays were performed following the manufacturer’s protocol using the Matchmaker GAL4 Two-Hybrid System (Clontech). Clones were grown on selective dropout medium (lacking either Trp and Leu or Trp, Leu, His, and Ade). Construct primers are listed in Supplemental Table S5.
Electrophoretic mobility shift assay
Two complementary oligonucleotide strands isolated from the BnWRKY33 promoter were labeled with Cy5 and annealed to generate probes. The recombinant His-WRKY28, His-VQ12, and His-WRKY33 proteins were expressed in Escherichia coli BL21 or Rosetta (DE3) with 0.2 mM isopropyl-beta-d-thiogalactopyranoside (IPTG) at 16°C for 16 h. The cells were lysed using a high-pressure cell disrupter (JNBIO, China), and the supernatant was added onto a column equipped with Ni2+ affinity resin (BBI, C600033, China) after centrifugation at 12,000 g at 4°C for 30 min. The DNA–protein binding activities were incubated in the reaction system containing EMSA/Gel-shift Binding Buffer (Beyotime, GS005) and 25 nM Cy5-labeled probe at 23°C for 30 min. For the competition, unlabeled probes were added to the reactions. The reaction mixture was loaded on a 6% native polyacrylamide gel and electrophoresed in 0.5× TBE (45 mM Tris-base, 45 mM boric acid, 0.5 mM EDTA, pH 8.3) at 4°C for 1 h at 80 V in the dark. Fluorescence-labeled DNA on the gel was directly detected using Fujifilm FLA-9000 (FujiFilm, Japan). Probe and primer sequences are given in Supplemental Table S5.
ChIP assay
ChIP was performed as previously described (Zhang et al., 2021). Briefly, oilseed rape leaves were crosslinked in 1% (v/v) formaldehyde for 15 min and quenched with 0.2 M glycine. Samples were lysed at 4°C and the chromatin was fragmented by sonication using a Bioruptor (Diagenode, Belgium). The chromatin solution was incubated with 10 μL anti-FLAG (antibody of the tag of a short peptide composed of DYKDDDDK amino acids) (Sigma) for 6 h at 4°C. The immunoprecipitated chromatin was washed with low-salt buffer, high-salt buffer, wash buffer, and TE buffer. ChIP DNA was used for ChIP-seq and ChIP-qPCR. The ChIP-seq samples were from BnaA03.WRKY28 OE lines for two biological replicates, and ChIP-qPCR was analyzed according to percentage input method (Haring et al., 2007). The raw data of ChIP-seq and RNA-seq have been deposited in GenBank (accession number, PRJNA855127) The ChIP-qPCR primers are listed in Supplemental Table S5.
Dual-LUC transient transcriptional activity assay
Genes were cloned into pGreenII 62-SK and pGAL4BD vectors as effectors (Hellens et al., 2005; Zong et al., 2016). BnWRKY33 promoter sequences (p346, –346 to –1 bp; pΔW1, –313 to –1 bp; pΔW1W2, –313 to –1 bp; p249, and –249 to –1 bp) were inserted into the pGreenII 0800-LUC vector to construct reporters. The CaMV35S-driven Renilla LUC was used as an internal control. The effector, reporter, and internal control were co-transformed into Arabidopsis protoplasts as Yoo et al. (2007). LUC activity was measured according to the manufacturer’s instructions (Promega, E1910, USA). Construct primers are listed in Supplemental Table S5.
In vitro pull-down assay
His-tagged BnaA09.VQ12, GST-tagged BnaA03.WRKY28, and BnaA03.WRKY28 164–227 (164–227 amino acid domain) proteins were used. GST or GST-tagged protein samples were incubated with GST beads (BBI, C600031, China) at 4°C for 1 h, washed five times, then incubated with His-tagged protein at 4°C for 2 h. The beads were then washed five times. The isolated precipitate was boiled with loading buffer at 100°C for 6 min, and detected by western blotting using anti-His (Abclonal, AE003, China, 1:10,000) or anti-GST (Abclonal, AE001, China, 1:10,000) antibodies. The ECL western-specific luminescence detection kit (BIO-RAD, #1705060, USA) was used to collect the signal, and the images were scanned using the Image Quant luminescence LAS 4000. Construct primers are listed in Supplemental Table S5.
BiFC assay
The BiFC vectors were based on those used in Waadt et al. (2008). BnaA03.WRKY28 and BnaA03.WRKY28 164–227 were amplified and cloned into the pSPYCE vector, and the full-length CDS of BnaA09.VQ12 and BnaA01.VQ22 was inserted into the pSPYNE vector. The constructs were transformed into Arabidopsis protoplasts in different combinations. After 16 h of incubation in the dark, the fluorescence signals were visualized using a laser scanning confocal microscope (TCS SP2; Leica). For excitation of fluorescent proteins, the following lines of an argon ion laser were used: 405 nm for cyan fluorescent protein (CFP) and 488 nm for yellow fluorescent protein (YFP). Emission at 420–485 nm for CFP and 500–542 nm for YFP. Construct primers are listed in Supplemental Table S5.
Co-IP assay
BnaA06.MPK3, BnaC03.MPK3, BnaA03.WRKY28, and BnaA03.WRKY28 164-227 fused with GFP tag were cloned into pH7LIC6.0, and BnWRKY33 and BnaA09.VQ12 fused with FLAG tag were cloned into pH7LIC4.1. The fused proteins were expressed in four-week-old N. benthamiana leaves. After incubation for 3 days, the total protein was extracted from 5 g of infiltrated leaves with 5 mL extraction buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% [v/v] Triton X-100, 5 mM EDTA, 10% [v/v] glycerol, and 1× protease inhibitor [Roche]) and incubated with 25 μL GFP-Trap-MA (Chromotek, gtma-20, Germany) at 4°C for 1 h. Next, the beads were washed five times with wash buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1% [v/v] Triton X-100, 5 mM EDTA, and 10% [v/v] glycerol) and eluted with 50 μL SDS loading buffer. Immunoblot assays were conducted using anti-FLAG (Abclonal, AE005, China, 1:10,000) and anti-GFP (Abclonal, AE012, China, 1:10,000) antibodies. Construct primers are listed in Supplemental Table S5.
In vitro phosphorylation assay
Phosphorylation assays were performed as per Mao et al. (2011). MBP-tagged BnWRKY33 was purified with amylose resin (Sangon, C500096, China). Briefly, 0.5 μg GST-tagged BnaA06.MPK3/BnaC03.MPK3 was activated by 0.1 μg His-tagged BnaA03.MKK5DD in the reaction buffer (50 mM Tris–HCl pH 7.5, 50 μM ATP, 10 mM MgCl2, and 1 mM dithiothreitol) at 25°C for 1 h. The superscript DD indicates that the Ser residues were all mutated to Asp. Activated BnaA06.MPK3/BnaC03.MPK3 was used to phosphorylate BnWRKY33 and BnWRKY33-N (5:1 substrate/enzyme ratio) in the reaction buffer (50 mM Tris–HCl pH 7.5, 10 μM ATP, 10 mM MgCl2, 1 μCi [γ-32P] ATP, and 1 mM dithiothreitol) at 25°C for 1 h. The reactions were stopped with SDS loading buffer. Phosphorylation signals for BnWRKY33 and BnWRKY33-N were visualized via autoradiography after being resolved in a 10% SDS-PAGE gel. The BnaA03.WRKY28 protein samples were collected from BnaA03.WRKY28 OE line Z95 when infected with Sclerotinia for 24 and 48 h. The BnWKRY33-FLAG protein was transiently expressed in N. benthamiana leaves; total protein samples were collected when infected with Sclerotinia for 24 and 48 h. Total proteins were immunoprecipitated with FLAG beads (MBL, M185-10, Japan). Phosphorylation signal was detected using phosphoserine antibody (Abclonal, AP0893, China, 1:2,000).
Subcellular localization
To determine the subcellular localization of BnaA03.WRKY28 and BnaA09.VQ12, full-length CDS without stop codon was cloned into pM999 vector to construct a recombinant of fused green fluorescent protein driven by CaMV35S. The OsGHD7 fusion CFP or red fluorescent protein (RFP) located in the nucleus was used as the control. The fusion constructs were transformed into Arabidopsis protoplasts. The fluorescence signals were visualized using a laser scanning confocal microscope (TCS SP2; Leica) after 16 h of incubation in the dark. For excitation of fluorescent proteins, the following lines of an argon ion laser were used: 405 nm for CFP, 488 nm for GFP, and 594 nm for RFP. Emission at 420–485 nm for CFP, 495–542 nm for GFP, and 600–650 nm for RFP.
Histochemical GUS staining
The samples of ProBnaA03.WRKY28:GUS (the β-glucuronidase gene driven by native BnaA03.WRKY28 promoter) transgenic plants were immersed in 90% (v/v) acetone on ice for 20–30 min, then washed twice with 50 mM phosphate buffer (mixing 39 mL 0.2 M NaH2PO4·2H2O and 61 mL 0.2 M Na2HPO4·12H2O in 500 mL solution, pH 7.2). Thereafter, the samples were stained in 1 mg/mL X-Gluc solution overnight at 37°C, and 70% (v/v) ethanol was used to remove the chlorophyll. Images were captured using a Leica stereoscope. The sequence ca. 1,000 bp upstream of the start codon of BnaA03.WRKY28 is used as the promoter.
Statistical analysis
Statistical analyses for the significance differences among the mean values between independent samples were conducted in Microsoft Office Excel using Student’s t test (P, 0.05). Samples included in the analysis were arranged based on at least three biological replicates.
Accession numbers
The accession numbers from https://www.genoscope.cns.fr/brassicanapus/ are as follows: BnaA03.WRKY28 (BnaA03g43640D), BnWRKY33 (BnaA05g34850D), BnaA09.VQ12 (BnaA09g42280D), BnaA06.MPK3 (BnaA06g18440D), BnaC03.MPK3 (BnaC03g55440D), BnaA03.MKK5 (BnaA03g36120D), and BnaC03.BRC1 (BnaCnng23770D).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequences analysis of BnWRKY28.
Supplemental Figure S2. Expression level and Sclerotinia resistance detection of BnaA03.WRKY28 transgenic lines.
Supplemental Figure S3. ChIP-seq and RNA-seq analysis of BnaA03.WRKY28 OE lines.
Supplemental Figure S4. BnaA03.WRKY28 binds to W1/W3 of BnWRKY33 promoter in yeast.
Supplemental Figure S5. BnWRKY33 expression level in BnaA03.WRKY28 OE lines.
Supplemental Figure S6. Dual-LUC transient transcriptional activity assays showing the effect of MAPK cascade on BnWRKY33 transcription.
Supplemental Figure S7. Self-activation test of full-length BnaA03.WRKY28 in yeast.
Supplemental Figure S8. Subcellular localization of BnaA03.WRKY28 and BnaA09.VQ12 in Arabidopsis protoplasts.
Supplemental Figure S9. Analysis of BnaA09.VQ12 transgenic lines.
Supplemental Figure S10. Induced expression profiles in oilseed rape stem.
Supplemental Figure S11. Statistical analysis branches in WRKY28-OE lines and WT plants.
Supplemental Figure S12. Analysis of branching-related genes in RNA-seq data.
Supplemental Figure S13. Phosphorylation detection of BnWRKY33 and BnaA03.WRKY28 when incubation with Sclerotinia for 0, 24, and 48 h.
Supplemental Table S1. Significance analysis of induced expression levels of WRKY28, VQ12, and WRKY33 in B. napus.
Supplemental Table S2. List of BnaA03.WRKY28-regulated genes in B. napus.
Supplemental Table S3. List of overlapping genes bound and regulated by BnaA03.WRKY28.
Supplemental Table S4. Genes used for heatmapping.
Supplemental Table S5. All primers used in this study.
Supplementary Material
Acknowledgments
We thank Prof. Daohong Jiang (Huazhong Agriculture University) for providing S. sclerotiorum strains, Prof. Xingwang Li (Huazhong Agriculture University) for ChIP assays assistance, Prof. Feng Li (Huazhong Agriculture University) for providing CO-IP vectors, Dr. Yongliang Wang for phosphorylation assays assistance, Mr. Pugang Yu for ChIP-seq and RNA-seq data analysis, and Prof. Shunping Yan (Huazhong Agriculture University) and Prof. Guoyong Xu (Wuhan University) for manuscript modifying.
Funding
This work was financed by the funding from the National Key Research and Development Program of China (2016YFD0100305) and the National Natural Science Foundation of China (31376120).
Conflict of interest statement. The authors have no conflicts of interest to declare.
Contributor Information
Ka Zhang, National Key Laboratory of Crop Genetic Improvement, National Sub-Center of Rapeseed Improvement in Wuhan, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, China; Crop Research Institute, Sichuan Academy of Agricultural Sciences, Chengdu 610066, China.
Fei Liu, National Key Laboratory of Crop Genetic Improvement, National Sub-Center of Rapeseed Improvement in Wuhan, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, China.
Zhixin Wang, National Key Laboratory of Crop Genetic Improvement, National Sub-Center of Rapeseed Improvement in Wuhan, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, China.
Chenjian Zhuo, National Key Laboratory of Crop Genetic Improvement, National Sub-Center of Rapeseed Improvement in Wuhan, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, China.
Kaining Hu, National Key Laboratory of Crop Genetic Improvement, National Sub-Center of Rapeseed Improvement in Wuhan, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, China.
Xiaoxia Li, National Key Laboratory of Crop Genetic Improvement, National Sub-Center of Rapeseed Improvement in Wuhan, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, China.
Jing Wen, National Key Laboratory of Crop Genetic Improvement, National Sub-Center of Rapeseed Improvement in Wuhan, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, China.
Bin Yi, National Key Laboratory of Crop Genetic Improvement, National Sub-Center of Rapeseed Improvement in Wuhan, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, China.
Jinxiong Shen, National Key Laboratory of Crop Genetic Improvement, National Sub-Center of Rapeseed Improvement in Wuhan, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, China.
Chaozhi Ma, National Key Laboratory of Crop Genetic Improvement, National Sub-Center of Rapeseed Improvement in Wuhan, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, China.
Tingdong Fu, National Key Laboratory of Crop Genetic Improvement, National Sub-Center of Rapeseed Improvement in Wuhan, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, China.
Jinxing Tu, National Key Laboratory of Crop Genetic Improvement, National Sub-Center of Rapeseed Improvement in Wuhan, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, China.
J.T. directed the project. K.Z. and J.T. conceived and designed the research. K.Z., F.L., C.Z., and X.L. performed the experiments. K.Z., Z.W., and K.H. analyzed the data. All authors interpreted and discussed the results. K.Z. and Z.W. wrote the manuscript. K.Z., F.L., Z.W., and J.T. edited and modified the manuscript. J.W., B.Y., J.S., C.M., and T.F. supervised the research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Jinxing Tu (tujx@mail.hzau.edu.cn).
References
- Aguilar-Martínez J, Poza-Carrión C, Cubas P (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja I, Kissen R, Bones AM (2012) Phytoalexins in defense against pathogens. Trends Plant Sci 17: 73–90 [DOI] [PubMed] [Google Scholar]
- Birkenbihl R, Diezel C, Somssich I (2012) Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol 159: 266–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B (2014) Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345: 950–953 [DOI] [PubMed] [Google Scholar]
- Chen J, Wang H, Li Y, Pan J, Hu Y, Yu D (2018) Arabidopsis VQ10 interacts with WRKY8 to modulate basal defense against Botrytis cinerea. J Integr Plant Biol 60: 956–969 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Zhou Y, Yang Y, Chi Y-J, Zhou J, Chen J-Y, Wang F, Fan B, Shi K, Zhou Y-H (2012) Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol 159: 810–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JD, Slovin JP, Hendrickson AM (2003) Two genetically discrete pathways convert tryptophan to auxin: more redundancy in auxin biosynthesis. Trends Plant Sci 8: 197–199 [DOI] [PubMed] [Google Scholar]
- Delteil A, Zhang J, Lessard P, Morel J-B (2010) Potential candidate genes for improving rice disease resistance. Rice 3: 56–71 [Google Scholar]
- Derbyshire MC, Denton-Giles M (2016) The control of Sclerotinia stem rot on oilseed rape (Brassica napus): current practices and future opportunities. Plant Pathol 65: 859–877 [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Glawischnig E, Hansen BG, Olsen CE, Halkier BA (2004) Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc Natl Acad Sci USA 101: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Gómez L, Felix G, Boller T (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18: 277–284 [DOI] [PubMed] [Google Scholar]
- Guo D, Zhang J, Wang X, Han X, Wei B, Wang J, Li B, Yu H, Huang Q, Gu H, et al. (2015) The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in Arabidopsis. Plant Cell 27: 3112–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M (2007) Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M, Baldwin IT (2002) Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci 7: 61–67 [DOI] [PubMed] [Google Scholar]
- Hellens R, Allan A, Friel E, Bolitho K, Grafton K, Templeton M, Karunairetnam S, Gleave A, Laing W (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Zhou W, Cheng Z, Fan M, Wang L, Xie D (2013a) JAV1 controls jasmonate-regulated plant defense. Mol Cell 50: 504–515 [DOI] [PubMed] [Google Scholar]
- Hu Y, Chen L, Wang H, Zhang L, Wang F, Yu D (2013b) Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J 74: 730–745 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Shinozaki K, Tena G, Sheen J, Henry Y, Champion A, Kreis M, Zhang S, Hirt H, Wilson C (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Deyholos MK (2009) Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol 69: 91–105 [DOI] [PubMed] [Google Scholar]
- Jing Y, Lin R (2015) The VQ motif-containing protein family of plant-specific transcriptional regulators. Plant Physiol 169: 371–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Karasov TL, Chae E, Herman JJ, Bergelson J (2017) Mechanisms to mitigate the trade-off between growth and defense. Plant Cell 29: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu J, Chen Z (2011) Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23: 3824–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei R, Li X, Ma Z, Lv Y, Hu Y, Yu D (2017) Arabidopsis WRKY 2 and WRKY 34 transcription factors interact with VQ 20 protein to modulate pollen development and function. Plant J 91: 962–976 [DOI] [PubMed] [Google Scholar]
- Li Y, Jing Y, Li J, Xu G, Lin R (2014) Arabidopsis VQ MOTIF-CONTAINING PROTEIN29 represses seedling deetiolation by interacting with PHYTOCHROME-INTERACTING FACTOR1. Plant Physiol 164: 2068–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Li X, Wang M, Wen J, Yi B, Shen J, Ma C, Fu T, Tu J (2018) Interactions of WRKY15 and WRKY33 transcription factors and their roles in the resistance of oilseed rape to Sclerotinia infection. Plant Biotechnol J 16: 911–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Ziegler J, Zeier J, Birkenbihl R, Somssich I (2017) Botrytis cinerea B05.10 promotes disease development in Arabidopsis by suppressing WRKY33-mediated host immunity. Plant Cell Environ 40: 2189–2206 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyssek R, Agerer R, Ernst D, Munch JC, Osswald W, Pretzsch H, Priesack E, Schnyder H, Treutter D (2005) The plant’s capacity in regulating resource demand. Plant Biol 7: 560–580 [DOI] [PubMed] [Google Scholar]
- Meng X, Zhang S (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51: 245–266 [DOI] [PubMed] [Google Scholar]
- Nafisi M, Goregaoker S, Botanga CJ, Glawischnig E, Olsen CE, Halkier BA, Glazebrook J (2007) Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19: 2039–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M, Cubas P (2016) TCP factors: new kids on the signaling block. Curr Opin Plant Biol 33: 33–41 [DOI] [PubMed] [Google Scholar]
- Perruc E, Charpenteau M, Ramirez BC, Jauneau A, Galaud JP, Ranjeva R, Ranty B (2004) A novel calmodulin‐binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. Plant J 38: 410–420 [DOI] [PubMed] [Google Scholar]
- Qiu J, Fiil B, Petersen K, Nielsen H, Botanga C, Thorgrimsen S, Palma K, Suarez-Rodriguez M, Sandbech-Clausen S, Lichota J, et al. (2008) Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J 27: 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Stirnberg P, Chatfield S, Leyser H (1999) AXR1 acts after lateral bud formation to inhibit lateral bud growth in Arabidopsis. Plant Physiol 121: 839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz HU, Sawada Y, Shimada Y, Hirai MY, Sasaki E, Krischke M, Brown PD, Saito K, Kamiya Y (2011) Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate‐derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum. Plant J 67: 81–93 [DOI] [PubMed] [Google Scholar]
- Tang H, Bi H, Liu B, Lou S, Song Y, Tong S, Chen N, Jiang Y, Liu J, Liu H (2021) WRKY33 interacts with WRKY12 protein to up-regulate RAP2.2 during submergence induced hypoxia response in Arabidopsis thaliana. New Phytol 229: 106–125 [DOI] [PubMed] [Google Scholar]
- Tian D, Traw M, Chen J, Kreitman M, Bergelson J (2003) Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77 [DOI] [PubMed] [Google Scholar]
- Tian T, Ma L, Liu Y, Xu D, Chen Q, Li G (2020) Arabidopsis FAR-RED ELONGATED HYPOCOTYL3 integrates age and light signals to negatively regulate leaf senescence. Plant Cell 32: 1574–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7: 491–498 [DOI] [PubMed] [Google Scholar]
- van Verk M, Bol J, Linthorst H (2011) WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol 11: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanEtten HD, Mansfield JW, Bailey JA, Farmer EE (1994) Two classes of plant antibiotics: phytoalexins versus “phytoanticipins.” Plant Cell 6: 1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt L, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J Cell Mol Biol 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Wang H, Hu Y, Pan J, Yu D (2015) Arabidopsis VQ motif-containing proteins VQ12 and VQ29 negatively modulate basal defense against Botrytis cinerea. Sci Rep 5: 14185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang J, Shi B, Yu T, Qi J, Meyerowitz E, Jiao Y (2014a) The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26: 2055–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Fang H, Chen Y, Chen K, Li G, Gu S, Tan X (2014b) Overexpression of BnWRKY33 in oilseed rape enhances resistance to Sclerotinia sclerotiorum. Mol Plant Pathol 15: 677–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wan L, Xin Q, Chen Y, Zhang X, Dong F, Hong D, Yang G (2018) Overexpression of OsPGIP2 confers Sclerotinia sclerotiorum resistance in Brassica napus through increased activation of defense mechanisms. J Exp Bot 69: 3141–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Jian H, Lu K, Filardo F, Yin N, Liu L, Qu C, Li W, Du H, Li J (2016) Genome‐wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol J 14: 1368–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Dong L, Wang Z, Zhang H, Han C, Liu B, Wang X, Chen Q (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Jiang Y, Rahman MH, Deyholos MK, Kav NN (2009) Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol 9: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S, Cho Y, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang K, Zhuo C, Wang Z, Liu F, Wen J, Yi B, Shen J, Ma C, Fu T, Tu J (2022) BnaA03.MKK5-BnaA06.MPK3/BnaC03.MPK3 module positively contributes to Sclerotinia sclerotiorum resistance in Brassica napus. Plants 11: 609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Guan P, Zhao L, Ma M, Xie L, Li Y, Zheng R, Ouyang W, Wang S, Li H, et al (2021) Asymmetric epigenome maps of subgenomes reveal imbalanced transcription and distinct evolutionary trends in Brassica napus. Mol Plant 14: 604–619 [DOI] [PubMed] [Google Scholar]
- Zhao L, Cai H, Su Z, Wang L, Huang X, Zhang M, Chen P, Dai X, Zhao H, Palanivelu R, et al. (2018) KLU suppresses megasporocyte cell fate through SWR1-mediated activation of expression in. Proc Natl Acad Sci USA 115: E526–E535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang X, He Y, Sang T, Wang P, Dai S, Zhang S, Meng X (2020) Differential phosphorylation of the transcription factor WRKY33 by the protein kinases CPK5/CPK6 and MPK3/MPK6 cooperatively regulates camalexin biosynthesis in Arabidopsis. Plant Cell 32: 2621–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong W, Tang N, Yang J, Peng L, Ma S, Xu Y, Li G, Xiong L (2016) Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol 171: 2810–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.