Summary
The emergence of Omicron sublineages impacts the therapeutic efficacy of anti-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) monoclonal antibodies (mAbs). Here, we evaluate neutralization and antibody-dependent cellular cytotoxicity (ADCC) activities of 6 therapeutic mAbs against Delta, BA.2, BA.4, and BA.5. The Omicron subvariants escape most antibodies but remain sensitive to bebtelovimab and cilgavimab. Consistent with their shared spike sequence, BA.4 and BA.5 display identical neutralization profiles. Sotrovimab is the most efficient at eliciting ADCC. We also analyze 121 sera from 40 immunocompromised individuals up to 6 months after infusion of Ronapreve (imdevimab + casirivimab) or Evusheld (cilgavimab + tixagevimab). Sera from Ronapreve-treated individuals do not neutralize Omicron subvariants. Evusheld-treated individuals neutralize BA.2 and BA.5, but titers are reduced. A longitudinal evaluation of sera from Evusheld-treated patients reveals a slow decay of mAb levels and neutralization, which is faster against BA.5. Our data shed light on antiviral activities of therapeutic mAbs and the duration of effectiveness of Evusheld pre-exposure prophylaxis.
Keywords: SARS-CoV-2, antibodies, Omicron, neutralization, ADCC
Graphical abstract
Highlights
-
•
Cilgavimab and bebtelovimab neutralize Delta, BA.2, BA.4, and BA.5 with similar potency
-
•
Neutralization does not correlate to antibody-dependent cellular cytotoxicity
-
•
Serum neutralization of BA.5 is detectable up to 6 months after Evusheld administration
Bruel et al. show that BA.4 and BA.5 Omicron sublineages escape most therapeutic antibodies, but cilgavimab and bebtelovimab remain fully active. Sotrovimab is the most efficient at eliciting antibody-dependent cellular cytotoxicity. Sera from individuals receiving Evusheld (cilgavimab/tixagevimab) pre-exposure prophylaxis neutralize Delta, BA.2, and BA.5 up to 6 months after injection.
Introduction
Nine months after its emergence, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron lineage outcompeted previous variants of concern (VOCs). Sublineages with improved transmissibility have replaced the initial Omicron BA.1 strain. As of September 2022, Omicron was composed of 5 main lineages, BA.1, BA.2, BA.3, BA.4, and BA.51,2, which further diversify into sublineages, such as BA.2.75, BA.2.75.2, BA.4.6, or BQ.1.1.3 The BA.1 strain has 34 mutations in its spike, which are associated with antibody escape,4,5,6,7,8,9,10,11,12,13,14,15 CD8 T cell evasion,16 and modified tropism.17,18,19 BA.2 harbors 30 mutations, 21 of which are not present in BA.1.2 The BA.4 and BA.5 sublineages share the same spike sequence, which differs from BA.2 by three mutations (including one reversion) in the receptor-binding domain (RBD) and one deletion in the N-terminal domain (NTD).1 The BA.5 sublineage was dominant in many countries in September 2022.20
Several monoclonal antibodies (mAbs) targeting the spike protein of SARS-CoV-2 are used in therapeutic, pre-exposure prophylaxis (PrEP) and post-exposure prophylaxis (PEP) settings.21 Therapeutic administration of mAbs is highly effective, reaching 85% efficacy in preventing coronavirus disease 19 (COVID-19)-related hospitalization or death.22,23,24 Antibody-based prophylaxis also achieves high levels of protection. Ronapreve (imdevimab + casirivimab) and Evusheld (cilgavimab + tixagevimab) cocktails provide 81% and 77% protection against symptomatic infection, respectively.25,26 These successes are mitigated by viral evolution. Omicron variants display considerable escape from mAbs.4,5,7,8,9,10,13,15,27,28,29 The use of Ronapreve (imdevimab + casirivimab) and Sotrovimab was discouraged after BA.1 and BA.2 emergence.30 It is recommended to inject a double dose of Evusheld instead, as serum neutralization is decreased against BA.1 and BA.2 in individuals receiving these antibodies as PrEP.29,30,31,32 Bebtelovimab is similarly effective against ancestral strains and Omicron BA.1 and BA.2,33 but its access is so far restricted to the United States.34 Thus, a continuous evaluation of mAb efficacy against new variants is needed to optimize their utilization.
As other Omicron sublineages, BA.4 and BA.5 escape most neutralizing mAbs.35,36,37,38,39,40,41,42 The neutralization profile of BA.4 and BA.5 is similar to that of BA.2, with only cilgavimab and bebtelovimab being efficient against these strains with high potency.35,36,37,38,39 Animal models revealed that some mAbs do not only rely on neutralization for therapeutic efficacy.43,44 Antibodies can trigger effector mechanisms through their fragment crystallizable (Fc) region. These Fc-effector functions mediate killing of infected cells through activation of antibody-dependent cellular cytotoxicity (ADCC) by natural killer (NK) cells and antibody-dependent complement-mediated lysis (ADCML) by complement, or clearance of viral particles, through macrophage-mediated antibody-dependent cellular phagocytosis (ADCP).45 Interaction between the Fc region and cognate Fc receptors (FcRs) may also promote inflammation and antibody-dependent enhancement (ADE) of infection.46 Hence, some therapeutic mAbs were mutated in the Fc region to abrogate FcR recognition and eliminate a putative risk of ADE.21 This is the case for cilgavimab and tixagevimab, which bear a triple mutation (TM) motif (L234F, L235E, and P331S). Fc engineering may also modulate neonatal Fc receptor (FcRn) affinity and extend antibody half-life.46 Such modifications were introduced in sotrovimab (M428L and N434S, called LS) and cilgavimab and tixagevimab (M252Y, S254Y, and T256E, called YTE). In contrast, imdevimab, casirivimab, and bebtelovimab have an unmutated Fc domain.21 Overall, the therapeutic activity of antibodies is the sum of neutralization potency, Fc-effector functions, and bio-disponibility.
Here, we evaluated the neutralization and ADCC activity of 6 therapeutic mAbs against BA.4 and BA.5 isolates. To consider variations in pharmacokinetic, dosage, or drug interactions, we also analyzed serum neutralization from 40 immunocompromised individuals up to 6 months post-infusion of Ronapreve (imdevimab + casirivimab) and Evusheld (cilgavimab + tixagevimab).
Results
In vitro neutralization of BA.4 and BA.5
We first investigated the sensitivity of two isolates of BA.4 and BA.5 to neutralization by mAbs. We selected 6 antibodies that are either used in patients (cilgavimab, tixagevimab, and bebtelovimab) or that were withdrawn because of Omicron escape (sotrovimab, casirivimab, and imdevimab). We used the commercial formulation, except for eebtlovimab, which is not available in France. We also tested Ronapreve (imdevimab + casirivimab) and Evusheld (cilgavimab + tixagevimab) cocktails. As controls, we included Delta and BA.2 strains.27,29 We used our S-Fuse neutralization assay,26,28,47,48 based on syncytia formation, to quantify infection via a green fluorescent protein (GFP) split system. As control, we included isolates of Delta and Omicron BA.2. Figure S1 summarizes the mutational landscape of VOCs included in the study compared with the ancestral Wuhan strain.
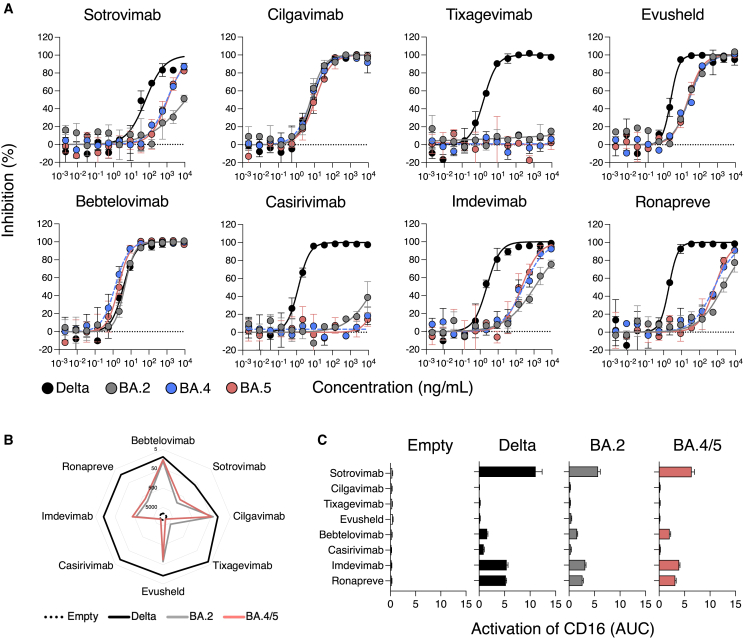
The IC50 of 4 out of the 6 mAbs (sotrovimab, tixagevimab, casirivimab, and imdevimab) were higher for BA.4 and BA.5 than for Delta (Figure 1A; Table 1). Tixagevimab and casirivimab lacked neutralization in the range of concentrations tested (Figure 1A; Table 1). Sotrovimab and imdevimab remained active but lost potency. Compared with Delta, sotrovimab was 15- and 17-fold less potent against BA.4 and BA.5, respectively. The increase in IC50s was higher for imdevimab: 110- and 86-fold against BA.4 and BA.5, respectively. Imdevimab remained more potent than sotrovimab against both strains (IC50 of 265 and 996 ng/mL for BA.4 and 208 and 1088 ng/mL for BA.5, respectively) (Figure 1A; Table 1). Importantly, cilgavimab and bebtelovimab displayed no or only minimal changes compared with Delta and remained highly potent against BA.4 and BA.5. When compared with BA.2, BA.4 and BA.5 display slightly improved neutralization by imdevimab (4.2- and 5.3-fold) and sotrovimab (9- and 8.3-fold) (Figure 1A; Table 1). We also analyzed the combination of cilgavimab and tixagevimab (Evusheld by Astrazeneca) and casirivimab and imdevimab (Ronapreve by Regeneron). Both displayed a drop in potency compared with Delta, which was less marked for Evusheld (BA.4: 10.4-fold and BA.5: 9-fold) than Ronapreve (BA.4: 330-fold and BA.5: 350-fold) (Figure 1A; Table 1).
Figure 1.
Neutralization and antibody-dependent cellular cytotoxicity of Omicron BA.4 and BA.5 by therapeutic mAbs
(A) Neutralization curves of mAbs using the S-Fuse system. Dose-response analysis of the neutralization by the indicated antibodies and by Evusheld, a combination of cilgavimab and tixagevimab, and Ronapreve, a combination of casirivimab and imdevimab. Data are mean ± SD of 2 independent experiments. The IC50 values for each antibody are presented in Table 1. The dashed line indicates the limit of detection.
(B) mAbs binding at the surface of Raji cells stably expressing the indicated spikes. Raji cells transduced with a control empty vector not coding for any spike (Empty). Depicted are EC50, calculated with a curve fitting the percentage of mAb-positive cells measured by flow cytometry against antibody concentration in limiting dilutions. Data are mean of 2 independent experiments. The EC50 values for each antibody are also presented in Table 1.
(C) Activation of the CD16 pathway as a surrogate of the capacity of each mAb to elicit antibody-dependent cellular cytotoxicity (ADCC). The area under curve of a dose-response analysis of CD16 activation by each mAb against each SARS-CoV-2 variant is depicted. Data are mean ± SD of 2 independent experiments.
Table 1.
Neutralization, binding, and ADCC of therapeutic mAbs against Delta, BA.2 BA.4, and BA.5
| Neutralization, IC50 (ng/ML) |
Binding, EC50 (ng/mL) |
ADCC, AUC |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Delta | BA.2 | BA.4 | BA.5 | Delta | BA.2 | BA.4/5 | Delta | BA.2 | BA.4/5 | |
| Sotrovimab | 64.18 | >9,000 | 996 | 1,088 | 72.3 | 1,429 | 848.2 | 11.1 | 5.7 | 6.4 |
| Cilgavimab | 7.9 | 6.1 | 6.5 | 11 | 22 | 39 | 49.3 | 0.1 | 0.2 | 0.2 |
| Tixagevimab | 1.6 | >9,000 | >9,000 | >9,000 | 7.8 | 4,226 | >9,000 | 0.2 | 0.2 | 0.1 |
| Evusheld (cilgavimab/tixagevimab) | 2.5 | 24.7 | 26.1 | 22.6 | 13.5 | 72.9 | 92.3 | 0.2 | 0.4 | 0.1 |
| Bebtelovimab | 3.8 | 4.5 | 1.3 | 2 | 12.2 | 20.2 | 17.4 | 1.6 | 1.6 | 2.1 |
| Casirivimab | 1.3 | >9,000 | >9,000 | >9,000 | 11.1 | >9,000 | >9,000 | 0.9 | 0.3 | 0.2 |
| Imdevimab | 2.4 | 1,120 | 265 | 208 | 12.8 | 610 | 391 | 5.4 | 3.2 | 3.9 |
| Ronapreve (imdevimab/casirivimab) | 2 | 1,985 | 660 | 700 | 12.8 | 1,106 | 739 | 5.2 | 2.6 | 3.2 |
Overall, our data reveal a large escape of therapeutic mAbs by Omicron BA.4 and BA.5. BA.2., BA.4, and BA.5 have a similar profile of neutralization by these mAbs. Cilagavimab and bebtelovimab remain fully active against these variants.
Antibody binding to BA.4/5 spike and induction of ADCC
Next, we evaluated the capacity of these mAbs to bind to the BA.4 and BA.5 spike (referred to as BA.4/5 spike) and trigger ADCC. We assessed antibody binding by flow cytometry using Raji cells stably expressing the BA4/5 spike. As controls, we included Delta, BA.2 spikes, and cells transduced with an empty vector (Empty). To confirm spike expression and compare the various cells lines, we stained the cells with bebtlovimab, which neutralized Delta, BA.2, BA.4, and BA.5 with a similar potency, and analyzed them by flow cytometry (Figures S2A and S2B). All cell lines showed high levels of spike expression with a similar median fluorescence intensity (MFI) across variants (MFIs of 97,853, 71,735, and 68,635 for Delta, BA.2, and BA.4/5, respectively) (Figure S2B). No binding was observed with Raji-Empty cells (MFI of 784) (Figure S2B).
We analyzed the binding of the therapeutic mAbs and their combinations against these cells. We performed limiting dilution tests to calculate exact effective concentrations 50% (EC50) (Figures 1B and S2C; Table 1). All antibodies bound the Delta spike, with EC50 <100 ng/mL. This was expected given their neutralizing potency against this strain. Binding profiles were generally similar between BA.2 and BA.4/5, with both spikes displaying a high level of escape compared with Delta (Figure 1B). Bebtelovimab and cilgavimab displayed similar binding levels. Tixagevimab and casirivimab did not recognize the BA.4/5 spike, even at a high concentration (10 μg/mL). Sotrovimab and imdevimab recognized the BA.4/5 spike, with a loss of potency compared with delta (11.7- and 30.5-fold, respectively) (Figures 1B and S2C; Table 1).
Then, we investigated the capacity of these mAbs to trigger ADCC. We used a surrogate assay that measures the activation of the CD16 pathway. We previously demonstrated that this assay correlates to killing of infected cells by primary NK cells.47 We tested the antibodies by limiting dilution. We measured the area under the curve (AUC) to depict the ADCC capacity of the mAbs against each viral spike (Figures 1C and S2D; Table 1). As expected, none of the mAbs elicited CD16 activation against the Raji-Empty cells. CD16 activation was detectable against Raji-Delta, -BA.2, and -BA.4/5 cells. Sotrovimab was the most efficient mAb regardless of the viral strain, albeit the AUC was reduced against BA.2 and BA.4/5 compared with Delta (Figures 1C and S2D; Table 1). Cilgavimab and tixagevimab alone or in the Evusheld cocktail did not activate ADCC, in line with the mutations in their Fc domain that decrease binding to FcR (Figures 1C and S2D; Table 1). Bebtelovimab induced similar levels of ADCC activation against all strains, yet at low levels (Figures 1C and S2D; Table 1). Imdevimab and casirivimab, alone or in the Ronapreve cocktail, displayed intermediate levels of activation (Figures 1C and S2D; Table 1).
Next, we investigated the association between neutralization, binding, and ADCC capacity. Neutralization is positively correlated to binding (r = 0.97; p < 0.0001) but not to ADCC (r = 0.0053; p = 0.98) (Figure S3).
Overall, our results indicate that BA.4/5 avoid antibody recognition and ADCC activation by most of therapeutic mAbs tested. Sotrovimab is the most efficient ADCC inducer, and cilgavimab and tixagevimab lack ADCC activity.
Serum neutralization of BA.5 in individuals receiving mAbs
Next, we investigated antibody levels and neutralization in sera from 40 immunocompromised individuals who received, by intra-muscular injection, either 300 (n = 29) or 600 mg (n = 11) Evusheld as PrEP. Patients were sampled prior to and at a median of 26 (range 10–40) or 37 (range 14–48) days after the single or double dose, respectively. Among the 29 individuals who received 300 mg Evusheld, 17 previously received Ronapreve as PrEP. The last injection of Ronapreve occurred at a median of 35 days (range 29–49) before the first Evusheld injection. Two out of the 11 individuals who received 600 mg Evusheld also received Ronapreve. The injections were spaced by >160 days, which is ∼5 times above the half-life of Ronapreve.48 Participants were included in the study in two places, the Centre Hospitalier Regional (CHR) in Orléans (France; n = 8) or the Hôpital Cochin in Paris (France; n = 32). Most of the patients were female (n = 28), were diagnosed with anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis (n = 26) and treated with rituximab as immunosuppressive therapy (n = 31). A complete description of the patients’ characteristics is provided in Table 2. The 8 individuals recruited at the CHR Orléans were longitudinally sampled every month as part of an ongoing prospective cohort.
Table 2.
Characteristics of patients
| Orléans cohort | Cochin group | Total (%) | |
|---|---|---|---|
| Patient characteristics | |||
| N | 8 | 32 | 40 (100) |
| Gender, female (♀) | 6 | 22 | 28 (70) |
| Gender, male (♂) | 2 | 10 | 12 (30) |
| Obesity | 3 | 1 | 4 (10) |
| Diseases | |||
| Rheumatoid arthritis | 5 | 2 | 7 (17.5) |
| Kidney graft | 2 | 0 | 2 (5) |
| Myelodysplasia | 1 | 0 | 1 (2.5) |
| ANCA-associtaed vasculitis | 0 | 26 | 26 (65) |
| Polychondritis | 0 | 1 | 1 (2.5) |
| Lupus | 0 | 1 | 1 (2.5) |
| Systemic sclerosis | 0 | 1 | 1 (2.5) |
| Cryoglobuminemic vasculitis | 0 | 1 | 1 (2.5) |
| Medications | |||
| Rituximab (anti-CD20) | 5 | 26 | 31 (77.5) |
| Infliximab (anti-tumor necrosis factor [TNF]) | 0 | 1 | 1 (2.5) |
| Mepolizumab (anti-interleukin-5 [IL-5]) | 0 | 1 | 1 (2.5) |
| Prednisone | 4 | 10 | 14 (35) |
| Mycofenolate mofetil | 2 | 1 | 3 (7.5) |
| Methotrexate | 0 | 3 | 3 (7.5) |
| 5-Azacytidine | 1 | 0 | 1 (2.5) |
| Tacrolimus | 1 | 0 | 1 (2.5) |
| Cyclosporin | 1 | 0 | 1 (2.5) |
| Cyclophosphamide | 0 | 1 | 1 (2.5) |
| Vaccines doses | |||
| 2 | 0 | 1 | 1 (2.5) |
| 3 | 5 | 25 | 30 (75) |
| 4 | 3 | 6 | 9 (22.5) |
| Previous COVID-19 | 0 | 8 | 8 (20) |
| PrEP | |||
| Ronapreve | 3 | 17 | 20 (50) |
| Evusheld 300 mg | 8 | 21 | 29 (72.5) |
| Evusheld 600 mg | 0 | 11 | 11 (27.5) |
We analyzed the 40 individuals before and after infusion of Evusheld. We categorized the patients into 5 groups: naive of any antibody administration (n = 11; before treatment); Ronapreve (n = 18; who received 1200 mg Ronapreve); Ronapreve + Evusheld (n = 18; who received 1,200 mg Ronapreve followed by 300 mg Evulsheld); Evusheld (n = 11; 300 mg); and Evusheld×2 (n = 11; 600 mg in a single injection). All individuals received at least two doses of vaccine but failed to mount an antibody response >260 binding antibody unit (BAU)/mL, making them eligible for Evusheld according to French recommendations. Eight out the 40 individuals included in the cohort had COVID-19 but remained >260 BAU/mL. Three patients had COVID-19 after Evusheld administration. These breakthrough infections were previously described.29
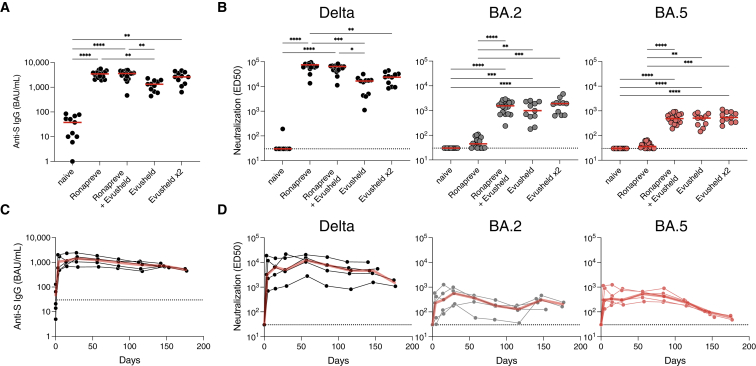
We first measured the levels of anti-spike immunoglobulin Gs (IgGs) in BAU (BAU/mL) using the S-Flow assay.49 Compared with the naive group, sera containing mAbs display a sharp increase in anti-spike (S) IgG (median of 38 versus 3,449, 3,591, 1,323, and 2,623 BAU/mL for Ronapreve, Ronapreve + Evusheld, Evusheld, and Evusheld×2, respectively) (Figure 2A). We then investigated serum neutralization against Delta, BA.2, and BA.5 with the S-Fuse assay. We tested sera in limiting dilutions to calculate titers as effective dilution 50% (ED50). We did not include BA.4 as its profile of neutralization is identical to BA.5. Untreated individuals (naive group) did not neutralize the three strains, except for one patient who slightly neutralized Delta. Infusion of mAbs dramatically increased Delta neutralization, with increases from 552- to 2,484-fold compared with the naive group. Individuals who received Ronapreve neutralized BA.2 and BA.5 at low levels (non-significant compared with naive individuals; two-sided Kruskall-Wallis test with Dunn’s multiple comparison correction). The groups that included Evusheld neutralized BA.2 and BA.5 at levels significantly higher than the naive and Ronapreve groups (Figure 2B). Of note, neutralization titers were higher in the Evusheld×2 group with all variants tested (16,585 versus 23,772, 992 versus 1,908, and 511 versus 539 against Delta, BA.2, and BA.5, respectively), without reaching statistical significance. In this group, the two individuals who received Ronapreve >160 days prior to Evusheld did not harbor significantly higher levels of neutralization (Mann-Whitney test; p = 0.1455, p = 0.1455, and p = 0.2182, for Delta, BA.2, and BA.5, respectively) (Figure S4A). Neutralization titers tended to be lower against BA.5 than BA.2, but this difference was significant only for individuals who received 300 mg Evusheld (1,549 versus 489; p = 0.0228) (Figure S4B).
Figure 2.
Antibody levels and neutralization of Delta, BA.2, and BA.5 in sera of immunocompromised individuals receiving mAbs
(A) Anti-S IgGs were measured using the flow cytometry-based S-Flow assay in sera of individuals before PrEP (naive; n = 11), treated with Ronapreve (n = 18), treated with 300 (n = 11) or 600 mg (n = 11) Evusheld, or treated with both Ronapreve and 300 mg Evusheld (n = 18). Indicated are the binding antibody units (BAUs) per mL (BAU/mL) of anti-S IgGs. Two-sided Kruskall-Wallis test with Dunn’s multiple comparison correction. Each dot is an individual. Red bars indicate medians.
(B) Serum neutralization of Delta and Omicron BA.2 and BA.5 in the same individuals as in (A). Indicated are effective dilution 50% (ED50; titers) as calculated with the S-Fuse assay. Two-sided Kruskall-Wallis test with Dunn’s multiple comparison correction. Each dot is an individual. Red bars indicate median. The dashed line indicates the limit of detection.
(C) Longitudinal measurement of anti-S levels in 5 immunocompromised individuals who initiated an Evusheld PrEP with no history of Ronapreve. All individuals and sampling points are depicted (black lines and dots). The red lines indicate medians. Indicated are the BAU/mL of anti-S IgGs. The dashed line indicates the limit of detection.
(D) Sero-neutralization of Delta and Omicron BA.2 and BA.5 in the same individuals as in (C). Indicated are ED50 (titers) as calculated with the S-Fuse assay. Two-sided Kruskall-Wallis test with Dunn’s multiple comparison correction. Each dot is an individual. Red bars indicate medians. The dashed line indicates the limit of detection.
Overall, these data show that the serum neutralization activity of individuals receiving Ronapreve and Evusheld as PrEP is decreased against BA.2 and BA.5. The diminution is less marked with Evusheld- than Ronapreve-treated individuals.
Kinetics of serum neutralization up to 6 months after infusion of Evusheld
Longitudinal sampling was performed for 8 out of the 40 individuals (for patients’ characteristics, see Table 2). The serum samples were available up to 186 days post-administration of Evusheld (300 mg). We investigated anti-S IgG levels and neutralization (Figures 2C, 2D, and S5). We first analyzed 5 patients who were naive at the time of Evusheld injection (the 3 others were previously under Ronapreve PrEP) (Figures 2C and 2D). Antibody levels peaked at 28 (range 28–30) days after Evusheld administration, with a median of 1,400 (range 646–4,014) BAU/mL. Anti-S IgG then slowly decreased, reaching 500 (range 452–548) BAU/mL at 176 (range 175–177) days post-administration (Figure 2C). Neutralization of Delta, BA.2, and BA.5 mirrored anti-S levels, showing a sharp increase upon administration and a steady decrease until month 6 (Figure 2D). Neutralization of Delta remains consistently higher than that of BA.2 and BA.5, in line with our other observations (Figures 1A, 2B, and S4B). After almost 6 months of follow up, the five patients who received Evusheld harbored detectable levels of neutralization against the tested strains. The neutralization levels against the two Omicron subvariants were low at 6 months (ED50 of 1,503, 202, and 59, for Delta, BA.2, and BA.5, respectively) (Figure 2D). We also analyzed the 3 individuals who received Ronapreve prior to Evusheld (Figure S5). Together, their profiles were similar to those who only received Evusheld. However, they consistently harbored higher levels of anti-S and neutralization titers in the first 2 months, suggesting a disappearance of Ronapreve, but a maintenance of Evusheld, as expected given the longer half-life of Evusheld than Ronapreve. We also analyzed ADCC activity in these sera by evaluating their capacity to activate the CD16 pathway. Some individuals displayed slight activation against cells expressing the delta S, but generally ADCC activation was undetectable (Figures S6A and S6B). This is consistent with the mutated Fc of cilgavimab and tixagevimab.
Overall, these results show that a single administration of Evusheld allowed serum neutralization of BA.5 for 6 months, with reduced titers compared with Delta.
Discussion
We show here that BA.4 and BA.5 escape neutralization by most therapeutic mAbs, in line with previous reports.35,36,37,38,39,40,41,42 Some antibodies remain effective. Bebtelovimab is the most potent, followed by cilgavimab. Tixagevimab and casirivimab lost any neutralizing activity, and imdevimab and sotrovimab were poorly active against BA.4/BA.5. We observed a slightly higher neutralization of BA.5 by sotrovimab compared with BA.2. Similar findings were reported,38,39,42 but others have found decreased or identical neutralization of BA.5 compared with BA.235,36,37,40,41. There is also some discrepancy in the literature regarding cilgavimab. We observed a similar neutralization against BA.2 and BA.5 for this antibody, whereas some reports showed a slight decrease against BA.5 compared with BA.236,37,39. Further investigations are needed to determine why some mAbs more susceptible to experimental variations. It may be due to the use of different target cells (which vary in their ACE2 levels), viral isolates, or pseudotypes.50 Overall, the data presented here and the literature indicate that BA.2 and BA.5 have a very close spectrum of neutralization by clinically available mAbs.
In addition to our in vitro evaluation of mAbs neutralization, we analyzed the sera of immunocompromised individuals receiving Ronapreve or Evusheld as PrEP. In line with our in vitro observation, Ronapreve-treated individuals barely neutralized Omicron sublineages. Evusheld-treated individuals had detectable neutralization against BA.2 and BA.5, albeit decreased compared with Delta. We observed a trend for a higher neutralization of BA.2 than BA.5 in individuals receiving Evusheld. Longitudinal evaluation in 8 patients showed a slightly faster decay of antibody responses against BA.5. This difference between BA.2 and BA.5 may be explained by the loss of BA.5 binding and neutralization by tixagevimab. This decrease may be negligible when the Evusheld antibodies are tested alone but is more visible in the serum. How serum components might affect Evusheld potency against BA.5 deserve further investigations. Cilgavimab may also be slightly less potent against BA.4/5 than BA.2, as reported by others.36,37,39 The difference of BA.2 and BA.5 serum neutralization in Evusheld-treated individuals and the observation of a faster antibody decay stress the need for a booster dose of mAbs after 6 months, as is currently recommended. It may be of great interest to evaluate the impact of an earlier booster dose of Evusheld to compensate for Omicron escape.
How vaccination and antibody-based PrEP may be combined is an interesting question. Infusion of the HIV-1 broadly neutralizing antibody 3BNC117 increases humoral immunity and cross-neutralization in patients infected with HIV-1.51 The mechanisms underlying this improvement of autologous immune response remain ill defined but likely include the formation of immune complexes and their processing by antigen-presenting cells.52 Besides this “vaccinal effect,” mAbs may mask epitopes and alter immune responses, as suggested by an in-depth analysis of the B cell compartment in a cohort of individuals receiving COVID-19 vaccination after infusion of two anti-S mAbs.53 Thus, the interplay between mAbs and the immune system is complex, with outcomes that remain difficult to predict. Whether these antibody feedbacks loops may be manipulated to improve immunotherapies and prophylaxis of immunocompromised individuals deserves further investigation.
There was large inter-individual variability in neutralization and antibody levels after mAb administration. Recent work demonstrated an impact of the body mass index (BMI) on antibody levels after Evusheld injection, with a high BMI associated with low titers.31 This is consistent with the unique recommended dosage of Evusheld (initially 300 and then 600 mg). In our study, we observed a non-significant trend for higher titers in individuals receiving 600 mg and no association with BMI. This lack of significance is likely due to the small number of individuals tested and to additional factors accounting for the inter-individual’s variability. An investigation of a larger cohort previously demonstrated a significant increase in antibody levels in individuals received 600 compared with 300 mg.32 It will be interesting to evaluate whether adapting the dose to BMI may homogenize the response to Evusheld PrEP and improve its efficacy.
We also tested the binding and ADCC capacity of these mAbs. Binding correlated to neutralization but not to ADCC. The most potent antibody to activate ADCC against Omicron sublineages was sotrovimab, even if its neutralization IC50 was relatively high compared with other antibodies. This is likely the consequence of a distinct binding mode of sotrovimab, with an orientation and/or a positioning of its Fc that may facilitate the interaction with FcRs.54 This ADCC activity may help understand why sotrovimab remains clinically active against BA.2 despite its very limited neutralization.55 Similarly, it has been reported that non-neutralizing antibodies capable of mediating Fc-effector functions display some efficacy in animal models.56 It may be worth examining whether a combination of sotrovimab and Evusheld or bebtelovimab may improve therapeutic efficacy of the mAbs.
In conclusion, we provide here an in-depth evaluation of the efficacy of therapeutic mAb and serum from mAb-treated patients against Omicron sublineages. The BA.5 variant remains sensitive to Evusheld, but the decay of the serum neutralizing activity in treated individuals is accelerated, compared with previously circulating variants.
Limitations of the study
Our study has limitations. First, our sample size is small, precluding the analysis of patient characteristics associated with high serum neutralization titers. Whether gender, age, ongoing medication, or underlying conditions modulate bio-disponibility of mAbs remain open questions. Second, we did not have access to mucosal samples. Systemic levels of antibodies are known to be key to prevent severe COVID-19, whereas mucosal mAb levels may correlate with protection from infection. Third, we restricted our investigation of Fc-effector functions to ADCC. Thus, it will be worth determining how ADCP and ADCML contribute to the antiviral activity of therapeutic anti-S mAbs. Our study was also limited to BA.4 and BA.5, and we did not analyze the sensitivity of other Omicron subvariants, such as BA.2.75, BA.2.75.2, BA.4.6, or BQ1.13,57,58,59. We did not have access to the medical formulation of bebtelovimab.34 Fc-effector functions are influenced by the method of antibody preparation, the isotype, Fc glycosylation, and mutations. Our observation that bebtelovimab is a poor ADCC inducer deserves to be confirmed using the medical formulation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bamlanivimab | Kind gift of Dr Thierry Prazuck (CHR d’Orléans, France) | N/A |
| Casirivimab | Kind gift of Dr Thierry Prazuck (CHR d’Orléans, France) | N/A |
| Etesevimab | Kind gift of Dr Thierry Prazuck (CHR d’Orléans, France) | N/A |
| Imdevimab | Kind gift of Dr Thierry Prazuck (CHR d’Orléans, France) | N/A |
| Cilgavimab | Kind gift of Dr Thierry Prazuck (CHR d’Orléans, France) | N/A |
| Tixagevimab | Kind gift of Dr Thierry Prazuck (CHR d’Orléans, France) | N/A |
| Sotrovimab | Kind gift of Dr Thierry Prazuck (CHR d’Orléans, France) | N/A |
| Bebtelovimab | Kind gift of Dr Thierry Prazuck (CHR d’Orléans, France) | N/A |
| anti-IgG AlexaFluor647 | Jackson ImmunoResearch | Cat#A-21445 |
| Bacterial and virus strains | ||
| D614G(hCoV-19/France/GE1973/2020) | National Reference Centre for Respiratory Viruses (Institut Pasteur, Paris, France) | EPI_ISL_41463 |
| Delta | Laboratory of Virology of Hopital Européen Georges Pompidou (Assistance Publique – Hopitaux de Paris) | EPI_ISL_2029113 |
| Omicron BA.2 | NRC UZ/KU Leuven, Belgium | EPI_ISL_10654979 |
| Omicron BA.4 | NRC UZ/KU Leuven, Belgium | EPI_ISL_15728568 |
| Omicron BA.5 | CHU de Tours, France | EPI_ISL_13660702 |
| Chemicals, peptides, and recombinant proteins | ||
| Hoechst 33342 | Invitrogen | Cat#H3570 |
| Paraformaldehyde 4% | Alfa Aesar | Cat#J19943.K2 |
| Critical commercial assays | ||
| ADCC Reporter Bioassay | Promega | Cat#G7010 |
| Bright-Glo Luciferase Assay System | Promega | Cat#E2620 |
| Deposited data | ||
| D614G(hCoV-19/France/GE1973/2020) | GISAID | EPI_ISL_41463 |
| Delta | GISAID | EPI_ISL_2029113 |
| Omicron BA.2 | GISAID | EPI_ISL_10654979 |
| Omicron BA.4 | GISAID | EPI_ISL_15728568 |
| Omicron BA.5 | GISAID | EPI_ISL_13660702 |
| Experimental models: Cell lines | ||
| 293T | ATCC | Cat#CRL-3216 |
| U2OS cells | ATCC | Cat#HTB-96 |
| Raji | ATCC | Cat#CCL-86 |
| Software and algorithms | ||
| Harmony High-Content Imaging and Analysis Software | PerkinElmer | Cat#HH17000012 |
| Excel 365 | Microsoft | https://www.microsoft.com/en-ca/microsoft-365/excel |
| Prism 8 | Graphpad | https://www.graphpad.com/ |
| FlowJo v10 | Tree Star | https://www.flowjo.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Timothée Bruel (timothee.bruel@pasteur.fr).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Human subjects
Individuals under Evusheld PreP were recruited in the French cities of Orléans and Paris (CHR d’Orléans and Hôpital Cochin). The Neutralizing Power of Anti-SARS-CoV-2 Serum Antibodies (PNAS) cohort is an ongoing prospective, monocentric, longitudinal, observational cohort clinical study aiming to describe the kinetics of neutralizing antibodies after SARS-CoV-2 infection or vaccination (ClinicalTrials.gov identifier: NCT05315583). The cohort takes place in Orélans, France and enrolled immunocompromised individuals receiving Evusheld PreP. This study was approved by the Est II (Besançon) ethical committee. At enrollment, written informed consent was collected, and participants completed a questionnaire that covered sociodemographic characteristics, clinical information and data related to anti-SARS-CoV-2 vaccination. Blood sampling was performed on the day of Evusheld infusion and after 3 days, 15 days and then every months. None of the patients self-reported a COVID-19 during the study period. The ‘Cochin’ cohort is a prospective, monocentric, longitudinal, observational clinical study (NCT04870411) enrolling immunocompromised individuals with rheumatic diseases, aiming at describing immunological responses to COVID-19 vaccine in patients with autoimmune and inflammatory diseases treated with immunosuppressants and/or biologics. Ethics approval was obtained from the Comité de Protection des Personnes Nord-Ouest II. Leftover sera from usual care were used from these individuals in the setting of the local biological samples collection (RAPIDEM). A written informed consent was collected for all participants. None of the study participants received compensation.
Viral strains
All strain were isolated from a nasopharyngeal swab using Vero E6 cells (ADCC: CRL-1586™) tested negative for mycoplasma. The Delta and Omicron BA.2 strains were previously described.27,29 BA.4 and BA.5 strains were isolated from Belgian and French patients, respectively. BA.4 was isolated and sequenced by the NRC UZ/KU Leuven (Belgium). BA.5 was isolated from a 67-year-old female patient. On May 15, she experienced mild and unspecific symptoms, she tested positive for COVID-19 using a lateral flow assay. Due to pre-existing conditions (polymyalgia rheumatica), she presented at the hospital on 17/05, where a nasal swab was collected. A PCR testing (kit: Eurobioplex Fast-SVD-EBX-047 from Eurobio scientific) identified BA.5, which was confirmed by sequencing using the NEBNext ARTIC SARS-COV-2 Companion Kit for Oxford Nanopore (New England BioLabs) with Varskip Short v2 and BA.2 Spike-in supplemental primers (https://github.com/nebiolabs/VarSkip/commit/3fd0283adb878fe24e16b161c4e5c1c4364cd4c0) as per the manufacturer’s instruction. Her COVID-19 symptoms remain mild (arthralgia and cough). Both patients provided informed consent for the use of the biological materials. The sequences of the isolates were deposited on GISAID immediately after their generation, with the following Delta ID: EPI_ISL_2029113; Omicron BA.2 GISAID ID: EPI_ISL_10654979; Omicron BA.4 GISAID ID: EPI_ISL_15728568; Omicron BA.5: EPI_ISL_13660702. viral stocks were titrated in limiting dilution on Vero E6 cells and on S-Fuse cells.
mAbs
Bamlanivimab, Casirivimab, Etesevimab, Imdevimab, Cilgavimab, Tixagevimab and Sotrovimab were provided by CHR Orleans. Bebtelovimab was produced as previously described.29
Cell lines
Raji cells (ATCC CCL-86) were grown in complete RPMI medium (10% Fetal Calf Serum (FCS), 1% Penicillin/Streptomycin (PS)). 293T cells (ATCC CRL-3216) and U2OS cells (ATCCa HTB-96) were grown in complete DMEM medium (10% FCS, 1% PS). U2OS stably expressing ACE2 and the GFPsplit system (GFP1-10 and GFP11; S-Fuse cells) were previously described.60 Blasticidin (10 mg/mL) and puromycin (1 mg/mL) were used to select for ACE2 and GFPsplit transgenes expression, respectively. Raji cells stably expressing the SARS-CoV-2 Spike protein of Delta, BA.2 and BA.4/5 (GenBank: QHD43416.1, UJP23605.1 and UPN16705.1) were generated by lentiviral transduction and selection with puromycin (1 mg/mL). Absence of mycoplasma contamination was confirmed in all cell lines with the Mycoalert Mycoplasma Detection Kit (Lonza). All cell lines were cultured at 37°C and 5% CO2.
Method details
Anti-spike antibody binding and serology
Circulating levels of anti-S antibodies were measured with the S-Flow assay. This assay uses 293T cells stably expressing the spike protein (293T spike cells) and 293T control cells as control to detect anti-spike antibodies by flow cytometry.49 In brief, the cells were incubated at 4 °C for 30 minutes with sera (1:300 dilution) in PBS containing 0.5% BSA and 2 mM EDTA. Cells were then washed with PBS and stained with an anti-human IgG Fc Alexa Fluor 647 antibody (109-605-170, Jackson Immuno Research). After 30 minutes at 4 °C, cells were washed with PBS and fixed for 10 minutes using 4% PFA. A standard curve with serial dilutions of a human anti-spike monoclonal antibody (mAb48) was acquired in each assay to standardize the results as a binding Unit (BU). Data were acquired on an Attune NxT instrument using Attune NxT software version 3.2.2 (Life Technologies) and analyzed with FlowJo version 10.7.1 software (see Extended Data Figure 4 for gating strategy). The sensitivity is 99.2% with a 95% confidence interval of 97.69–99.78%, and the specificity is 100% (98.5–100%)40. To determine BAU/mL (Binding Antibody Units per mL), we analyzed a series of vaccinated (n = 144), convalescent (n = 59) samples and World Health Organization international reference sera (20/136 and 20/130) on S-Flow and on two commercially available ELISAs (Abbott 147 and Beckmann 56). Using this dataset, we performed a Passing–Pablok regression, which shows that the relationship between BU the S-Flow (see above) and BAU/mL is linear, allowing calculation of BAU/mL using S-Flow data.61 The binding mAbs to Delta, BA.2 and BA.4/5 spikes was assessed using Raji cells stably expressing these spikes. Stainings were performed at the indicated concentration of mAbs and following the S-Flow protocol, except that antibodies were biotinylated and revealed with a streptavidin conjugated to AlexaFluor647 (Life Technologies; dilution 1:400).
S-Fuse neutralization assay
U2OS-ACE2 GFP1-10 or GFP11 cells, also termed S-Fuse cells, become GFP + when they are productively infected by SARS-CoV-2. Cells tested negative for mycoplasma. Cells were mixed (ratio 1:1) and plated at 8 × 103 per well in a μClear 96-well plate (Greiner Bio-One). The indicated SARS-CoV-2 strains were incubated with serially diluted mAb or sera for 15 minutes at room temperature and added to S-Fuse cells. The sera were heat-inactivated for 30 minutes at 56 °C before use. Eighteen hours later, cells were fixed with 2% paraformaldehyde (PFA), washed and stained with Hoechst (dilution 1:1,000, Invitrogen). Images were acquired with an Opera Phenix high-content confocal microscope (PerkinElmer). The GFP area and the number of nuclei were quantified using Harmony software version 4.9 (PerkinElmer). The percentage of neutralization was calculated using the number of syncytia as value with the following formula: 100 × (1 − (value with serum −value in ‘non-infected’)/(value in ‘no serum’ − value in ‘non-infected’)). Neutralizing activity of each serum was expressed as the ED50. ED50 values (in μg ml−1 for mAbs and in dilution values for sera) were calculated with a reconstructed curve using the percentage of the neutralization at the different concentrations. We previously reported correlations between neutralization titers obtained with the S-Fuse assay and both pseudovirus neutralization and microneutralization assays.62,63 Of note, we previously reported that the neutralization assay with the S-Fuse system is not affected by differences in fusogenicity between variants.27
Antibody-dependent cellular cytotoxicity reporter assay
ADCC was quantified using the ADCC Reporter Bioassay (Promega) as previously described.47 Briefly, 5 × 104 Raji stably expressing the indicated spikes were co-cultured with 5 × 104 Jurkat-CD16-NFAT-rLuc cells in presence or absence of mAbs at the indicated concentration. Luciferase was measured after 18 h of incubation using an EnSpire plate reader (PerkinElmer). ADCC was measured as the fold induction of Luciferase activity compared to the ‘‘no serum’’ condition. Sera were tested at a 1:100 dilution and normalized to the control condition to account for inter-individual variations of the background.
Quantification and statistical analysis
Statistical analysis
No statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded. Flow cytometry data were analyzed with FlowJo version 10 software. Calculations were performed using Excel 365 (Microsoft). Figures were drawn on Prism 9 (GraphPad Software). Statistical analysis was conducted using GraphPad Prism 9. Statistical significance between different groups was calculated using Kruskall–Wallis test with Dunn’s multiple comparisons, Friedman tests with Dunn’s multiple comparison correction and Spearman non-parametric correlation test. All tests were two-sided.
Acknowledgments
We thank the European Health Emergency Preparedness and Response Authority (HERA) for supporting the work being done at Institut Pasteur and UK Leuven. We thank the patients who participated in this study. We thank members of the Virus and Immunity Unit for discussions and help. We thank Ludivine Grzelak for her help in creating an illustration of SARS-CoV-2 variant mutations. We thank N. Aulner and the UtechS Photonic BioImaging (UPBI) core facility (Institut Pasteur), a member of the France BioImaging network, for image acquisition and analysis. The Opera system was co-funded by Institut Pasteur and the Région Ȋle-de-France (DIM1Health). We thank F. Peira, V. Legros, and L. Courtellemont for their help with the cohorts. UZ Leuven, as a national reference center for respiratory pathogens, is supported by Sciensano, which is gratefully acknowledged. We thank Hélène Péré and David Veyer for their help in sequencing viral strains and helpful discussions. Work in the O.S. lab is funded by Institut Pasteur; Urgence COVID-19 Fundraising Campaign of Institut Pasteur; Fondation pour la Recherche Médicale (FRM); ANRS; the Vaccine Research Institute (ANR-10-LABX-77); Labex IBEID (ANR-10-LABX-62-IBEID); ANR/FRM Flash Covid PROTEO-SARS-CoV-2; and ANR Coronamito and IDISCOVR. Work in the UPBI facility is funded by grant ANR-10-INSB-04-01 and the Région Ȋle-de-France program DIM1Health. D.P. is supported by the Vaccine Research Institute. P.M. acknowledges the support of a COVID-19 research grant from “Fonds Wetenschappelijk Onderzoek” /Research Foundation Flanders (grant G0H4420N) and “Internal Funds KU Leuven” (grant 3M170314). E.S.-L. acknowledges funding from the INCEPTION program (Investissements d’Avenir grant ANR-16-CONV-0005). The funders of this study had no role in study design, data collection, analysis, and interpretation or writing of the article.
Author contributions
Experimental strategy and design, T.B. and O.S.; laboratory experiments, T.B., D.T., I.S., F.P., F.G.-B., W.-H.B., D.P., M.P., E.S.-L., and J.B.; cohort management and clinical research, Y.N., J.H., L.M., A.S., T.P., B.T., and L.H.; viral strains and antibodies, K.S., L.H., C.P., E.A., G.B., L.C., and H.M.; manuscript writing, T.B. and O.S.; manuscript editing, T.B., Y.N., W.-H.B., B.T., L.H., and O.S.
Declaration of interests
T.B., C.P., H.M., and O.S. have a pending patent application for an anti-RBD mAb not used in this study (PCT/FR2021/070522).
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100850.
Contributor Information
Timothée Bruel, Email: timothee.bruel@pasteur.fr.
Olivier Schwartz, Email: olivier.schwatrtz@pasteur.fr.
Supplemental information
Data and code availability
SARS-CoV-2 variants genomes have been deposited at GISAID and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This study did not generate any new codes. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Tegally H., Moir M., Everatt J., Giovanetti M., Scheepers C., Wilkinson E., Subramoney K., Makatini Z., Moyo S., Amoako D.G., et al. Emergence of SARS-CoV-2 omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022;28:1785–1790. doi: 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao Y., Jian F., Wang J., Yu Y., Song W., Yisimayi A., Wang J., An R., Zhang N., Wang Y., et al. Imprinted SARS-CoV-2 humoral immunity induces converging Omicron RBD evolution. bioRxiv. 2022 doi: 10.1101/2022.09.15.507787. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iketani S., Liu L., Guo Y., Liu L., Chan J.F.-W., Huang Y., Wang M., Luo Y., Yu J., Chu H., et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.VanBlargan L.A., Errico J.M., Halfmann P.J., Zost S.J., Crowe J.E., Purcell L.A., Kawaoka Y., Corti D., Fremont D.H., Diamond M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022;28:490–495. doi: 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carreño J.M., Alshammary H., Tcheou J., Singh G., Raskin A.J., Kawabata H., Sominsky L.A., Clark J.J., Adelsberg D.C., Bielak D.A., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 7.Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takashita E., Kinoshita N., Yamayoshi S., Sakai-Tagawa Y., Fujisaki S., Ito M., Iwatsuki-Horimoto K., Chiba S., Halfmann P., Nagai H., et al. Efficacy of antibodies and antiviral drugs against covid-19 omicron variant. N. Engl. J. Med. 2022;386:995–998. doi: 10.1056/nejmc2119407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Touret F., Baronti C., Bouzidi H.S., de Lamballerie X. In vitro evaluation of therapeutic antibodies against a SARS-CoV-2 Omicron B.1.1.529 isolate. Sci. Rep. 2022;12:4683. doi: 10.1038/s41598-022-08559-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Chivukula V., Herati R.S., Hubbard S.R., Mulligan M.J., Landau N.R. Increased resistance of SARS-CoV-2 Omicron variant to neutralization by vaccine-elicited and therapeutic antibodies. EBioMedicine. 2022;78 doi: 10.1016/j.ebiom.2022.103944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457–466.e4. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérez-Then E., Lucas C., Monteiro V.S., Miric M., Brache V., Cochon L., Vogels C.B.F., Malik A.A., De la Cruz E., Jorge A., et al. Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat. Med. 2022;28:481–485. doi: 10.1038/s41591-022-01705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L., Iketani S., Guo Y., Chan J.F.-W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 16.Dolton G., Rius C., Hasan M.S., Wall A., Szomolay B., Behiry E., Whalley T., Southgate J., Fuller A., Morin T., et al. COVID-19 Genomics UK COG-UK consortium Emergence of immune escape at dominant SARS-CoV-2 killer T-cell epitope. Cell. 2022;185:2936–2951.e19. doi: 10.1016/j.cell.2022.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willett B.J., Grove J., MacLean O.A., Wilkie C., De Lorenzo G., Furnon W., Cantoni D., Scott S., Logan N., Ashraf S., et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat. Microbiol. 2022;7:1161–1179. doi: 10.1038/s41564-022-01143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hui K.P.Y., Ho J.C.W., Cheung M.C., Ng K.C., Ching R.H.H., Lai K.L., Kam T.T., Gu H., Sit K.-Y., Hsin M.K.Y., et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature. 2022;603:715–720. doi: 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- 19.Meng B., Abdullahi A., Ferreira I.A.T.M., Goonawardane N., Saito A., Kimura I., Yamasoba D., Gerber P.P., Fatihi S., Rathore S., et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ECDC . 2022. Country Overview Report: Week 27 2022.https://www.ecdc.europa.eu/en/covid-19/country-overviews [Google Scholar]
- 21.Gruell H., Vanshylla K., Weber T., Barnes C.O., Kreer C., Klein F. Antibody-mediated neutralization of SARS-CoV-2. Immunity. 2022;55:925–944. doi: 10.1016/j.immuni.2022.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dougan M., Nirula A., Azizad M., Mocherla B., Gottlieb R.L., Chen P., Hebert C., Perry R., Boscia J., Heller B., et al. Bamlanivimab plus Etesevimab in mild or moderate covid-19. N. Engl. J. Med. 2021;385:1382–1392. doi: 10.1056/nejmoa2102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta A., Gonzalez-Rojas Y., Juarez E., Crespo Casal M., Moya J., Falci D.R., Sarkis E., Solis J., Zheng H., Scott N., et al. Early treatment for covid-19 with SARS-CoV-2 neutralizing antibody Sotrovimab. N. Engl. J. Med. 2021;385:1941–1950. doi: 10.1056/nejmoa2107934. [DOI] [PubMed] [Google Scholar]
- 24.Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Xiao J., Hooper A.T., Hamilton J.D., Musser B.J., et al. REGEN-COV antibody combination and outcomes in outpatients with covid-19. N. Engl. J. Med. 2021;385:e81. doi: 10.1056/nejmoa2108163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levin M.J., Ustianowski A., De Wit S., Launay O., Avila M., Templeton A., Yuan Y., Seegobin S., Ellery A., Levinson D.J., et al. Intramuscular AZD7442 (Tixagevimab–Cilgavimab) for prevention of covid-19. N. Engl. J. Med. 2022;386:2188–2200. doi: 10.1056/nejmoa2116620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Brien M.P., Forleo-Neto E., Musser B.J., Isa F., Chan K.-C., Sarkar N., Bar K.J., Barnabas R.V., Barouch D.H., Cohen M.S., et al. Subcutaneous REGEN-COV antibody combination to prevent covid-19. N. Engl. J. Med. 2021;385:1184–1195. doi: 10.1056/nejmoa2109682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.-H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 28.Yu J., Collier A.R.Y., Rowe M., Mardas F., Ventura J.D., Wan H., Miller J., Powers O., Chung B., Siamatu M., et al. Neutralization of the SARS-CoV-2 omicron BA.1 and BA.2 variants. N. Engl. J. Med. 2022;386:1579–1580. doi: 10.1056/nejmc2201849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruel T., Hadjadj J., Maes P., Planas D., Seve A., Staropoli I., Guivel-Benhassine F., Porrot F., Bolland W.-H., Nguyen Y., et al. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat. Med. 2022;28:1297–1302. doi: 10.1038/s41591-022-01792-5. [DOI] [PubMed] [Google Scholar]
- 30.NIH NIH COVID-19 treatment guidelines, anti-SARS-CoV-2 antibody products, summary recommendations. 2022. https://www.covid19treatmentguidelines.nih.gov/therapies/anti-sars-cov-2-antibody-products/summary-recommendations/ Anti-SARS-CoV-2 Antibody Products.
- 31.Benotmane I., Velay A., Gautier-Vargas G., Olagne J., Thaunat O., Fafi-Kremer S., Caillard S. Pre-exposure prophylaxis with 300 mg Evusheld elicits limited neutralizing activity against the Omicron variant. Kidney Int. 2022;102:442–444. doi: 10.1016/j.kint.2022.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stuver R., Shah G.L., Korde N.S., Roeker L.E., Mato A.R., Batlevi C.L., Chung D.J., Doddi S., Falchi L., Gyurkocza B., et al. Activity of AZD7442 (tixagevimab-cilgavimab) against Omicron SARS-CoV-2 in patients with hematologic malignancies. Cancer Cell. 2022;40:590–591. doi: 10.1016/j.ccell.2022.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westendorf K., Žentelis S., Wang L., Foster D., Vaillancourt P., Wiggin M., Lovett E., van der Lee R., Hendle J., Pustilnik A., et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hentzien M., Autran B., Piroth L., Yazdanpanah Y., Calmy A. A monoclonal antibody stands out against omicron subvariants: a call to action for a wider access to bebtelovimab. Lancet Infect. Dis. 2022;22:00495–00499. doi: 10.1016/s1473-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuekprakhon A., Nutalai R., Dijokaite-Guraliuc A., Zhou D., Ginn H.M., Selvaraj M., Liu C., Mentzer A.J., Supasa P., Duyvesteyn H.M.E., et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185:2422–2433.e13. doi: 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., Shah J.G., Nguyen N., Chen Z., Meyers K., Yin M.T., Sobieszczyk M.E., Sheng Z., Huang Y., Liu L., Ho D.D. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.608.1, BA.4, and BA.5. Nature. 2022:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arora P., Nehlmeier I., Kempf A., Cossmann A., Schulz S.R., Dopfer-Jablonka A., Baier E., Tampe B., Moerer O., Dickel S., et al. Augmented neutralisation resistance of emerging omicron subvariants BA.2.12.1, BA.4, and BA.5. Lancet Infect. Dis. 2022;22:1537–1538. doi: 10.1016/s1473-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamasoba D., Kosugi Y., Kimura I., Fujita S., Uriu K., Ito J., Sato K., Genotype to Phenotype Japan G2P-Japan Consortium Neutralisation sensitivity of SARS-CoV-2 omicron subvariants to therapeutic monoclonal antibodies. Lancet Infect. Dis. 2022;22:942–943. doi: 10.1016/s1473-3099(22)00365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takashita E., Yamayoshi S., Simon V., van Bakel H., Sordillo E.M., Pekosz A., Fukushi S., Suzuki T., Maeda K., Halfmann P., et al. Efficacy of antibodies and antiviral drugs against omicron BA.2.12.1, BA.4, and BA.5 subvariants. N. Engl. J. Med. 2022;387:468–470. doi: 10.1056/nejmc2207519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Touret F., Baronti C., Pastorino B., Villarroel P.M.S., Ninove L., Nougairède A., Lamballerie X. de. In vitro activity of therapeutic antibodies against SARS-CoV-2 Omicron BA.1, BA.2 and BA.5. Sci Rep-uk. 2022;12 doi: 10.1038/s41598-022-16964-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aggarwal A., Akerman A., Milogiannakis V., Silva M.R., Walker G., Kidinger A., Angelovich T., Waring E., Amatayakul-Chantler S., Roth N., et al. SARS-CoV-2 Omicron BA.5: evolving tropism and evasion of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern. medRxiv. 2022 doi: 10.1101/2022.07.07.22277128. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamin R., Jones A.T., Hoffmann H.-H., Schäfer A., Kao K.S., Francis R.L., Sheahan T.P., Baric R.S., Rice C.M., Ravetch J.V., Bournazos S. Fc-engineered antibody therapeutics with improved anti-SARS-CoV-2 efficacy. Nature. 2021;599:465–470. doi: 10.1038/s41586-021-04017-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Chong Z., Zost S.J., Jang H., Huang Y., Allen J.D., et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820.e16. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu L.L., Suscovich T.J., Fortune S.M., Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 2018;18:46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PubMed] [Google Scholar]
- 47.Dufloo J., Grzelak L., Staropoli I., Madec Y., Tondeur L., Anna F., Pelleau S., Wiedemann A., Planchais C., Buchrieser J., et al. Asymptomatic and symptomatic SARS-CoV-2 infections elicit polyfunctional antibodies. Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Regeneron . Regeneron; 2022. Fact sheet for health care providers emergency use authorization (EUA) of REGEN-Cov® (Casirivimab and Imdevimab).https://www.regeneron.com/downloads/treatment-covid19-eua-fact-sheet-for-hcp.pdf [Google Scholar]
- 49.Grzelak L., Velay A., Madec Y., Gallais F., Staropoli I., Schmidt-Mutter C., Wendling M.-J., Meyer N., Planchais C., Rey D., et al. Sex differences in the evolution of neutralizing antibodies to SARS-CoV-2. J. Infect. Dis. 2021;224:983–988. doi: 10.1093/infdis/jiab127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lempp F.A., Soriaga L.B., Montiel-Ruiz M., Benigni F., Noack J., Park Y.-J., Bianchi S., Walls A.C., Bowen J.E., Zhou J., et al. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature. 2021;598:342–347. doi: 10.1038/s41586-021-03925-1. [DOI] [PubMed] [Google Scholar]
- 51.Schoofs T., Klein F., Braunschweig M., Kreider E.F., Feldmann A., Nogueira L., Oliveira T., Lorenzi J.C.C., Parrish E.H., Learn G.H., et al. HIV-1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV-1. Science. 2016;352:997–1001. doi: 10.1126/science.aaf0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pelegrin M., Naranjo-Gomez M., Piechaczyk M. Antiviral monoclonal antibodies: can they Be more than simple neutralizing agents? Trends Microbiol. 2015;23:653–665. doi: 10.1016/j.tim.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaefer-Babajew D., Wang Z., Muecksch F., Cho A., Raspe R., Johnson B., Canis M., DaSilva J., Ramos V., Turroja M., et al. Antibody feedback regulation of memory B cell development in SARS-CoV-2 mRNA vaccination. medRxiv. 2022 doi: 10.1101/2022.08.05.22278483. Preprint at. [DOI] [Google Scholar]
- 54.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 55.Martin-Blondel G., Marcelin A.-G., Soulié C., Kaisaridi S., Lusivika-Nzinga C., Dorival C., Nailler L., Boston A., Melenotte C., Cabié A., et al. Sotrovimab to prevent severe COVID-19 in high-risk patients infected with Omicron BA.2. J. Infect. 2022;85:e104–e108. doi: 10.1016/j.jinf.2022.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beaudoin-Bussières G., Chen Y., Ullah I., Prévost J., Tolbert W.D., Symmes K., Ding S., Benlarbi M., Gong S.Y., Tauzin A., et al. A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection. Cell Rep. 2022;38:110368. doi: 10.1016/j.celrep.2022.110368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao Y., Yu Y., Song W., Jian F., Yisimayi A., Yue C., Feng R., Wang P., Yu L., Zhang N., et al. Neutralizing antibody evasion and receptor binding features of SARS-CoV-2 Omicron BA.2.75. bioRxiv. 2022 doi: 10.1101/2022.07.18.500332. Preprint at. [DOI] [Google Scholar]
- 58.Yamasoba D., Kimura I., Kosugi Y., Uriu K., Fujita S., Ito J., Sato K., Consortium, T.G. to P.J. (G2P-J) Neutralization sensitivity of Omicron BA.2.75 to therapeutic monoclonal antibodies. bioRxiv. 2022 doi: 10.1101/2022.07.14.500041. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheward D.J., Kim C., Fischbach J., Muschiol S., Ehling R.A., Björkström N.K., Hedestam G.B.K., Reddy S.T., Albert J., Peacock T.P., et al. Evasion of neutralizing antibodies by Omicron sublineage BA.2.75. bioRxiv. 2022 doi: 10.1101/2022.07.19.500716. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buchrieser J., Dufloo J., Hubert M., Monel B., Planas D., Rajah M.M., Planchais C., Porrot F., Guivel-Benhassine F., Van der Werf S., et al. Syncytia formation by SARS-CoV-2-infected cells. Embo J. 2020;39 doi: 10.15252/embj.2020106267. e2020106267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hadjadj J., Planas D., Ouedrani A., Buffier S., Delage L., Nguyen Y., Bruel T., Stolzenberg M.-C., Staropoli I., Ermak N., et al. Immunogenicity of BNT162b2 vaccine against the Alpha and Delta variants in immunocompromised patients with systemic inflammatory diseases. Ann. Rheum. Dis. 2022;81:720–728. doi: 10.1136/annrheumdis-2021-221508. annrheumdis-2021-221508. [DOI] [PubMed] [Google Scholar]
- 62.Pelleau S., Woudenberg T., Rosado J., Donnadieu F., Garcia L., Obadia T., Gardais S., Elgharbawy Y., Velay A., Gonzalez M., et al. Kinetics of the severe acute respiratory syndrome Coronavirus 2 antibody response and serological estimation of time since infection. J. Infect. Dis. 2021;224:1489–1499. doi: 10.1093/infdis/jiab375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sterlin D., Mathian A., Miyara M., Mohr A., Anna F., Claër L., Quentric P., Fadlallah J., Devilliers H., Ghillani P., et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abd2223. eabd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SARS-CoV-2 variants genomes have been deposited at GISAID and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This study did not generate any new codes. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.