Abstract
Cytotoxic necrotizing factor types 1 and 2 (CNF1 and -2) produced by pathogenic Escherichia coli strains have 90% conserved residues over 1,014-amino-acid sequences. Both CNFs are able to provoke a remarkable increase in F-actin structures in cultured cells and covalently modify the RhoA small GTPases. In this study, we demonstrated that CNF2 reduced RhoA GTPase activity in the presence and absence of P122RhoGAP. Subsequently, peptide mapping and amino acid sequencing of CNF2-modified FLAG-RhoA produced in E. coli revealed that CNF2 deamidates Q63 of RhoA-like CNF1. In vitro incubation of the C-terminal domain of CNF2 with FLAG-RhoA resulted also in deamidation of the FLAG-RhoA, suggesting that this region contains the enzymatic domain of CNF2. An oligopeptide antibody (anti-E63) which specifically recognized the altered G-3 domain of the Rho family reacted with glutathione S-transferase (GST)-RhoA and GST-Rac1 but not with GST-Cdc42 when coexpressed with CNF2. In addition, CNF2 selectively induced accumulation of GTP form of FLAG-RhoA and FLAG-Rac1 but not of FLAG-Cdc42 in Cos-7 cells. Taken together, these results indicate that CNF2 preferentially deamidates RhoA Q63 and Rac1 Q61 and constitutively activates these small GTPases in cultured cells. In contrast, anti-E63 reacted with GST-RhoA and GST-Cdc42 but not with GST-Rac1 when coexpressed with CNF1. These results indicate that CNF2 and CNF1 share the same catalytic activity but have distinct substrate specificities, which may reflect their differences in toxic activity in vivo.
Several bacterial toxins and enzymes have been shown to modify eukaryotic small GTPases. Most of them were found to target members of the Rho GTPase family. The Rho GTPase family consists of the Rho, Rac, Cdc42, and G25K subfamilies in humans (3, 35). Among them, the Rho GTPase subfamily, including RhoA, RhoB, and RhoC, controls the formation of focal adhesions and actin stress fibers in cells (42, 43). These GTPases are either in a GDP-bound inactive form or in a GTP-bound active form (2, 35). The GDP-bound form is converted to the GTP-bound form by a GDP-GTP exchange reaction, which is regulated by GDP-GTP exchange protein (GEP) (19, 21, 26, 27, 33). The GTP-bound form is converted to the GDP-bound form by a GTPase reaction, which is regulated by GTPase-activating protein (GAP) (23, 30, 48).
The biochemistry involved in the toxin-mediated modification of Rho GTPases includes ADP-ribosylation, glucosylation, and N-acetylglucosaminylation, all of which result in deactivation of Rho GTPases. By contrast, the dermonecrotic toxin (DNT) of Bordetella bronchiseptica (24) and cytotoxic necrotizing factor types 1 and 2 (CNF1 and -2), produced by certain pathogenic Escherichia coli strains, activate Rho GTPase (16, 17, 38).
CNF1 and CNF2 are 110- to 115-kDa monomeric toxins (6, 10, 36) sharing 90% conserved residues over 1,014 amino acids (14, 38). CNF1 is encoded by the chromosome (14), whereas a transmissible plasmid called pVir codes for CNF2 (37). CNF1 and CNF2 are highly lethal in mice and share the ability to induce multinucleation in different cell lines and necrosis in the rabbit skin (7, 8). However, CNF1 is more potent than CNF2 in inducing multinucleation but less necrotic in the rabbit skin test and mouse footpad (8). In addition, CNF2 was found to migrate faster than CNF1 in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (36). The involvement of the Rho signaling pathway in CNF toxicity was first suggested by the remarkable reorganization of the F-actin cytoskeleton into stress fibers after treatment of cultivated cells with CNF1 or CNF2 (15, 38). The Rho GTPase isolated from CNF-treated cells was then found to migrate more slowly in SDS-PAGE, suggesting that Rho is covalently modified by CNF (16, 38). Indeed, CNF1 was shown to directly deamidate Rho at Q63, an amino acid present in the G-3 domain conserved in small GTPases and known to be essential for GTPase activity (18, 45). The deamidated Rho showed a marked decrease in intrinsic GTPase activity, and thus the Rho stayed in a constitutively active form. In addition, CNF1 was shown to decrease intrinsic GTPase activity of another member of the Rho GTPase family, Cdc42, suggesting that CNF1 modifies other members of the Rho family (45).
The apparent differences between CNF1 and CNF2 in toxic activities and electrophoretic properties led us to investigate the nature of biochemical modification of RhoA after treatment by CNF2. We report here that CNF2 also deamidates Rho at Q63 and abolishes GTPase activity in the presence or absence of Rho GAP. Moreover, Western blotting analysis with anti-E63, an oligopeptide antibody specifically recognizing DTAGEEDYDRLRPLS, the G-3 domain of Rho family with altered E63 (underlined), revealed that, in vitro, CNF2 preferentially deamidates RhoA and Rac1 and has low to no enzymatic activity on Cdc42. This was confirmed in Cos-7 cells, where CNF2 selectively induced accumulation of the GTP-bound form of RhoA and Rac1.
Taken together, our results clearly indicate that CNF2 and CNF1 share the same catalytic activity and deamidation of members of Rho family but that their substrate specificities are distinct.
MATERIALS AND METHODS
Materials.
CNF2 was purified to homogeneity from an E. coli C600 (pEOSW30) (38) homogenate by three column chromatography steps with DEAE Toyopearl 650M, TSK-gel Phenyl 5PW, and TSK-gel HA1000 columns (Tosoh, Tokyo, Japan). The CNF2 activity was assayed as described earlier (9). The detailed purification procedure will be described elsewhere. Briefly, the supernatant of an E. coli cell homogenate prepared by use of a French press was dialyzed against 20 mM Tris-Cl (pH 8.5). The dialyzed sample was loaded into DEAE Toyopearl 650M equilibrated with the same buffer and was eluted with a linear gradient from 0 to 0.5 M NaCl in 20 mM Tris-Cl (pH 8.5). The active fractions were pooled and dialyzed against 50 mM phosphate buffer containing 1.5 M NaCl (pH 7.0) and then loaded into TSK-gel Phenyl 5PW (7.5 by 75 mm). The sample was eluted with a linear gradient from 1.5 to 0 M NaCl in 50 mM phosphate buffer (pH 7.0). The active fractions were pooled and dialyzed against 10 mM phosphate buffer (pH 7.0). The dialyzed sample was passed through a hydroxyapatite column, TSK-gel HA1000 (7.5 by 75 mm) equilibrated with 10 mM phosphate buffer (pH 7.0). The unbound fraction was collected and used as purified CNF2. The purity of the CNF2 was checked by SDS-PAGE, followed by Coomassie brilliant blue staining. [α-32P]GTP, [γ-32P]GTP, [α-32P]NAD, and 32Pi were obtained from DuPont NEN. Guanine nucleotides were from Sigma Chemical Co., St. Louis, Mo. The plasmids used in this study are listed in Table 1. pGEX-5X-3-Cdc42Hs, pGEX2T-P122RhoGAP, and pGST-RhoA were gifts from Akira Kikuchi (Department of Biochemistry, Hiroshima University School of Medicine), Yoshimi Homma (Institute for Signal Transduction, Fukushima Medical College), and Hiroshi Maruta (Ludwig Institute for Cancer Research), respectively. Rabbit immunoglobulin G generated against synthetic peptide DTAGEEDYDRLRPLS, which corresponds to the conserved region of the GTPase domain (the underlined amino acid [E] was altered from Q) (anti-E63), was prepared as described earlier (25). Swiss 3T3 and Cos-7 cells were cultured as described previously (38). Swiss 3T3 cells were washed three times with cold phosphate-buffered saline (PBS), pooled by scraping, pelleted by centrifugation, and homogenized as described by Oswald et al. (37).
TABLE 1.
Plasmids used
| Plasmid | Vector | Relevant properties | Source or reference |
|---|---|---|---|
| pGEX-5X-3 | E. coli expression vector | Pharmacia Biotech | |
| pK184 | E. coli expression vector | 38 | |
| pET21d | E. coli expression vector | Novagen | |
| pFLAG-MAC | E. coli expression vector | Sigma | |
| pROEX-1 | E. coli expression vector | Life Technologies | |
| pEF-BOS | Mammalian expression vector | Nagata | |
| pEOSW30 | pK184 | 3,332-bp BanI-HoeII fragment carrying cnf2 | 38 |
| pSPEO1/2 | pK184 | 3,000-bp SmaI fragment carrying cnf1 | 37 |
| pGST-RhoA | pGEX-5X-3 | H. Maruta | |
| pGST-Rac1 | pGEX-5X-3 | H. Maruta | |
| pGST-Cdc42 | pGEX-5X-3 | A. Kikuchi | |
| pGST-P122RhoGAP | pGEX-2T | Y. Homma | |
| pFLAG-RhoA | Y. Horiguchi | ||
| pFLAG-RhoA63E | Y. Horiguchi | ||
| pFLAG-Rac1 | pFLAG-MAC | This study | |
| pFLAG-Cdc42 | pFLAG-MAC | This study | |
| pSPEO6/33 | pROEX-1 | 1,014-bp fragment of cnf2 (entire cnf2 structural gene) | This study |
| pSPEO3/3 | pROEX-1 | 497-bp fragment from nucleotides 1 to 497 of cnf2 (N-terminal region of cnf2) | This study |
| pEOFH3 | pROEX-1 | 872-bp fragment from nucleotides 442 to 1014 of cnf2 (C-terminal region of cnf2) | This study |
| pMH-RhoA | pEF-BOS | EcoRI fragment of pFLAG-RhoA | This study |
| pMH-RhoA63E | pEF-BOS | EcoRI fragment of pFLAG-RhoA63E | This study |
| pMH-Rac1 | pEF-BOS | EcoRI fragment of pFLAG-Rac1 | This study |
| pMH-Cdc42 | pEF-BOS | EcoRI fragment of pFLAG-Cdc42 | This study |
Preparation of small GTPase, P122RhoGAP, and CNF.
E. coli C600(pGST-RhoA) and E. coli BL21(DE3)(pFLAG-RhoA) or BL21(D3)(pFLAG-RhoA63E) were prepared as described earlier (25). E. coli harboring a vector containing the small GTPase gene was transformed with either pEOSW30, which encodes CNF2; pSPEO1/2 (37), which encodes CNF1; or vector alone, pK184 (38). To purify glutathione S-transferase (GST) fused to small G protein from E. coli, transformed E. coli was grown in Luria-Bertani broth at 37°C to an absorbance of 0.8 (optical density at 660 nm). Then isopropyl-β-d-thiogalactopyranoside was added at a final concentration of 40 μM, and further incubation was carried out for 3 h. GST fused to P122RhoGAP was produced in E. coli as described above except that E. coli was grown at 25°C. The recombinant RhoA, Cdc42, Rac1, and P122RhoGAP expressed as GST fusion proteins were purified by using glutathione-Sepharose beads according to the manufacturer's instructions. The recombinant RhoA and RhoA E63 expressed as N-terminal FLAG-tagged proteins were purified by using anti-FLAG M2 affinity gel (Sigma) according to the manufacturer's instructions. Some experiments used histidine-tagged (His-tag) fusion proteins containing full-length CNF2, N-terminal CNF2 fragments, or C-terminal CNF2 fragments. The pROEX-1 vector (Life Technologies, Inc.) was used to construct and express these fusion proteins. When necessary, an NdeI restriction site was introduced upstream the ATG by PCR. The constructs were pSPEO3, encoding His-tag fused with the N-terminal region of CNF2 (amino acids 1 to 522) (His-tag–CNF2 [N-term]); pSPE01, encoding His-tag fused with the C-terminal region of CNF2 (amino acids 450 to 1014) (His-tag-CNF2 [C-term]); and pSPE6, encoding His-tag fused with full-length CNF2 (amino acids 1 to 1014) (His-tag–CNF2 [full]). The predicted molecular masses of the fusion proteins were 59.8, 66.3, and 117.5 kDa, respectively. The recombinant His-tagged proteins were purified by using TSK-gel AF-chelating Toyopearl 650M (Tosoh) and Ni2+ as the metal ion as described previously (51). All purification procedures were performed at 4°C.
Enzymatic digestions and peptide mapping.
The purified FLAG-RhoA (1 mg) was digested with endopeptidase Lys-C from Achromobacter lyticus (Wako Pure Chemical Industries, Ltd., Osaka, Japan). The proteolytic digestion was carried out as described elsewhere. Products from Lys-C were separated by reversed-phase high-performance liquid chromatography (HPLC) by using a C18 column (2.0 by 150 mm; Nomura Chemical Co., Ltd., Seto, Japan) attached to a Waters 600E HPLC system. The peptides were eluted at a flow rate of 0.15 ml/min with a multistep, linear gradient of 70% solvent A (0.06% trifluoroacetic acid in water) and 30% solvent B (0.054% trifluoroacetic acid in acetonitrile-water [4:1, vol/vol]) to 40% solvent A–60% solvent B over 85 min, followed by a linear gradient of 40% solvent A–60% solvent B to 20% solvent A–60% solvent B over 10 min. The A210 was monitored with a Waters 486 UV detector. Some isolated peptides were further digested with trypsin in 0.1 M NH4HCO3–2 M urea at 37°C for 24 h, and the products were separated by the reversed-phase HPLC system. The peptides were eluted at a flow rate of 0.15 ml/min with a multistep linear gradient from 98% solvent A and 2% solvent B to 100% solvent B by using the following program: 2 to 37% solvent B for 75 min, 37 to 75% solvent B for 10 min, and 75 to 100% solvent B for 5 min.
N-terminal amino acid sequencing of peptides.
Peptides isolated by reversed-phase HPLC were spotted directly on a polyvinylidene difluoride membrane (ABI ProBlot). Immobilized peptides were subjected to automated Edman degradation on an ABI 476A protein sequencer with standard blot sequencing cycles.
MS analysis of peptides.
Peptide fractions resulting from the reversed-phase HPLC were analyzed by electrospray ionization-mass spectrometry (ESI-MS) on a TSQ-700 triple-sector quadrupole mass spectrometer (Finnigan-MAT, San Jose, Calif.) equipped with an ESI source operating at atmospheric pressure. The molecular weights of peptides were determined by mathematically transforming the raw ESI-MS spectrum of each peptide with the BIOMASS Deconvolution program (Finnigan-MAT).
[35S]GTPγS-binding assay.
Binding of 35S-labeled guanosine 5′-3(O-thio)triphospate with sulfur linked to the phosphate group at the third position (GTPγS) to GST-RhoA was determined by using the nitrocellulose method (49). Purified GST-RhoA was incubated at 30°C in 100 μl of a reaction mixture containing 50 mM Tris-Cl at pH 7.5, 1 mM dithiothreitol (DTT), 5 mM MgCl2, and various concentration of [35S]GTPγS (1,000 to 2,000 cpm/pmol). The reaction was stopped by the addition of 4.75 ml of ice-cold 20 mM Tris-Cl (pH 7.5), 1 mM DTT, and 5 mM MgCl2, followed by rapid filtration on nitrocellulose filters. Filters were washed three times with the same ice-cold buffer. Thereafter, the filters were dissolved in 4 ml of Scintisol EX-H (Dojin Chemical Institute, Kumamoto, Japan), and the radioactivity was measured. The experiments were carried out in triplicate. The Kd values were determined as described elsewhere (22).
GDP dissociation assay.
To determine the GDP dissociation constant (K−1), GST-RhoA in [3H]GDP-bound form (100 nM) was incubated in the buffer containing 50 mM Tris-Cl (pH 7.5), 1 mM DTT, 5 mM EDTA-Na (pH 7.5), 10 mM MgCl2, 0.1% bovine serum albumin (BSA), and 100 μM cold GTP for various periods of time at 30°C. The experiments were carried out in triplicate. The K−1 values were determined as described earlier (49).
GTPase assay.
The steady-state rate of GTPase activity was determined by incubating 5 pmol of GTPase with 1 μM [γ-32P]GTP in a reaction mixture containing 50 mM Tris-Cl (pH 7.5), 5 mM MgCl2, EDTA-Na (pH 7.5), 1 mM DTT, and 0.1% BSA for various periods of time at 30°C. An aliquot (60 μl) was taken out and mixed with 390 μl of ice-cold charcoal suspension (5% Norit SX-2 [wt/vol]). After a vortexing for 10 s, the supernatant was obtained by centrifugation at 8,000 × g for 10 min, and the residual radioactivity was measured. The steady-state rate (Kss) was expressed as the turnover number as described earlier (29). The actual catalytic rate (Kcat) of GTPase activity was determined in the presence or absence of P122RhoGAP (50 nM). Five picomoles of GTPase was incubated with 2 μM [γ-32P]GTP in a reaction mixture containing 20 mM Tris-Cl (pH 7.5), 1.3 mM MgCl2, 2 mM EDTA-Na (pH 7.5), 1 mM DTT, and 40 mg of BSA per ml for 10 min at 30°C. The reaction was then stopped by the addition of stop buffer containing 100 mM Tris-Cl (pH 7.5), 100 mM MgCl2, and 10 mM DTT. Next, the mixture was incubated with cold reaction buffer containing 1 mM GTP with or without 50 nM P122RhoGAP for various periods of time at 30°C. An aliquot was filtered through a prewetted nitrocellulose filter. Thereafter, the filters were dissolved in 4 ml of distilled water, and the radioactivity was measured. The experiments were carried out in triplicate. The steady-state rate of GTPase activity was determined as described elsewhere (22).
Transient transfection.
Mammalian FLAG-tagged small GTPase expression plasmid was constructed by the following procedures. The coding sequences for the small GTPase in pGST-RhoA, pGST-Rac1, and pGST-Cdc42 with HindIII/EcoRI sites in both ends were amplified by PCR. The DNA fragment was cut at the HindIII and EcoRI sites and ligated to pFLAG-MAC (Sigma). The full length FLAG-tagged small GTPase coding sequence with the EcoRI site upstream of the initiation methionine codon and downstream of the termination codon was synthesized by PCR. The fragment was digested with EcoRI and ligated into the EcoRI site of the mammalian expression vector pEF-BOS. Similarly, pFLAG-RhoA and pFLAG-RhoA63E were digested with EcoRI, and the DNA fragments were ligated into the EcoRI site of pEF-BOS. Cos-7 cells were transfected with the FLAG-tagged small GTPase expression plasmid by using DEAE-dextran as described earlier (44).
Analysis of GDP and GTP bound to FLAG-tagged small GTPase expressed in Cos-7 cells.
Nucleotides bound to FLAG-tagged small GTPases were measured essentially as described previously (34). Cos-7 cells transfected with certain DNA were cultured for 2 days and treated with either purified CNF2 or saline. After incubation for 12 h, the cells were washed twice with 3 ml of ice-cold Tris-buffered saline (TBS) and once with 0.5 ml of phosphate-free RPMI 1640. Cos-7 cells were labeled for 12 h with 32PPi (NEN) at 0.1 mCi/ml in phosphate-free RPMI 1640. The cells were washed once with ice-cold TBS, and then a lysis buffer (10 mM Tris-Cl [pH 7.5], 150 mM NaCl, 20 mM MgCl2, 0.5% Triton X-100, 20 μg of leupeptin per ml, 20 μg of aprotinin per ml, 1 mM phenylmethylsulfonyl fluoride) was added to the dishes. The cells were scraped off with a rubber policeman and collected into a 1.5-ml plastic tube; this was followed by centrifugation at 10,000 × g for 5 min. The supernatant was gently mixed with anti-FLAG M2 affinity gel (Sigma) for 2 h at 4°C. The precipitate was then washed four times with the lysis buffer and suspended in the sample buffer (20 mM Tris-Cl [pH 7.5], 20 mM EDTA, 2% SDS, 1 mM GDP, 1 mM GTP), followed by incubation at 65°C for 5 min. The supernatant was spotted onto a polyethyleneimine-cellulose thin-layer sheet (Marchery-Nägel Catalog number 801063) and developed with 1 M KH2PO4-H3PO4 (pH 3.4). The radioactivity was quantitated with a BAS-2000 bioimaging analyzer (Fujix, Tokyo, Japan). The molar ratio of GDP to GTP bound to FLAG-tagged small GTPase was determined by multiplying the ratio of their radioactivities by 1.5.
Other procedures.
SDS-PAGE and immunoblotting were carried out as described earlier (50). Immunodetection was carried out by using an ECL immunodetection kit (Amersham) according to the kit's manual. The anti-63E was diluted 1,000-fold for immunodetection. Protein concentrations were determined by the method of Bradford (4), with BSA as the standard.
RESULTS
CNF2 inhibits intrinsic and RhoGAP-dependent GTPase activities of RhoA.
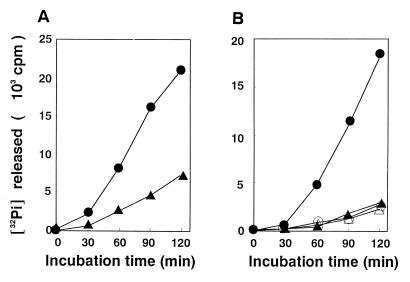
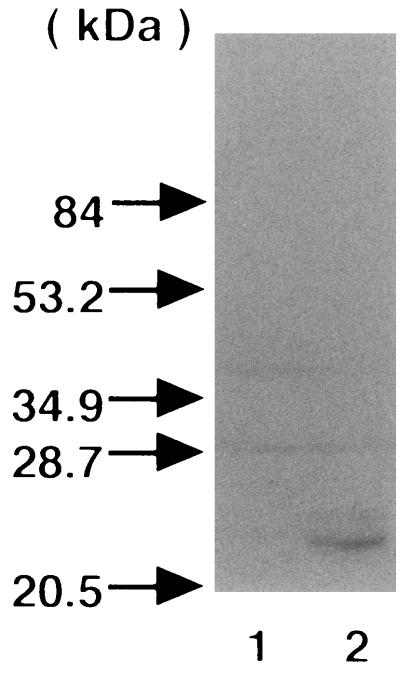
We used the coexpression system of CNF2 and RhoA in E. coli described previously (38). When the GST-RhoA purified from E. coli C600(pK184, pGST-RhoA) and from C600(pEOSW30, pGST-RhoA) were analyzed by SDS-PAGE, we observed that the GST-RhoA coexpressed with CNF2 had an apparently slower electrophoretic mobility, although the difference was only slight (Fig. 1). This finding indicated that GST-RhoA is as susceptible as untagged RhoA (38) to chemical modification by coexpressed CNF2. We then analyzed the guanine nucleotide binding activities of the purified CNF2-modified and -unmodified GST-RhoA. For this purpose, GST-RhoA was incubated with a nonhydrolyzable analog of guanine nucleotide, [35S]GTPγS, and the reaction was terminated at various time points. The amount of [35S]GTPγS bound was quantified by filtration of the terminated reactions through nitrocellulose filters. The Kd values of CNF2-unmodified and -modified GST-RhoA for GTPγS were 11 ± 1 and 16 ± 3 nM, respectively, which were markedly lower levels than that of posttranslationally modified RhoA but were consistent with those expressed in bacteria (47) (Table 2). This suggested that the affinities of CNF2-modified and CNF2-unmodified GST-RhoA were similar. The GDP dissociation constants of CNF2-modified and -unmodified GST-RhoA were measured. The K−1 values of CNF2-unmodified and -modified GST-RhoA were 270 ± 70 and 160 ± 40, respectively (Table 2). Next, the intrinsic GTPase activities of the GST-RhoA were determined. GST-RhoA was incubated with [γ-32P]GTP, and aliquots were taken at various time points. The amount of 32Pi released from [γ-32P]GTP was quantified. As shown in Fig. 2A, CNF2-modified GST-RhoA showed decreased GTPase activity compared to unmodified GST-RhoA, which hydrolyzed [γ-32P]GTP and released 32Pi in a time-dependent fashion. The steady-state rates (Kss) of GTPase activity of CNF2-unmodified and -modified GST-RhoA were (33 ± 6) × 10−3 and (11 ± 1) × 10−3 min−1, respectively (Table 2). P122RhoGAP stimulated the actual catalytic rate of GTPase activity of GST-RhoA 8.4-fold, while that of CNF-modified GST-RhoA was stimulated only 2.2-fold (Table 2). These results suggested that CNF2-induced modification inhibited intrinsic and RhoGAP-stimulated GTPase activities. We also purified GST-Rac1 and GST-Cdc42 expressed in the presence or absence of CNF2 in E. coli, respectively, and assessed the effect of coexpression on the GTPase activity of these small GTPases. Both the steady-state rate and the actual catalytic rate of GTPase activity of GST-Rac1 were lowered when GST-Rac1 was coexpressed with CNF2. On the other hand, coexpression of CNF2 hardly affected GTPase activities of GST-Cdc42.
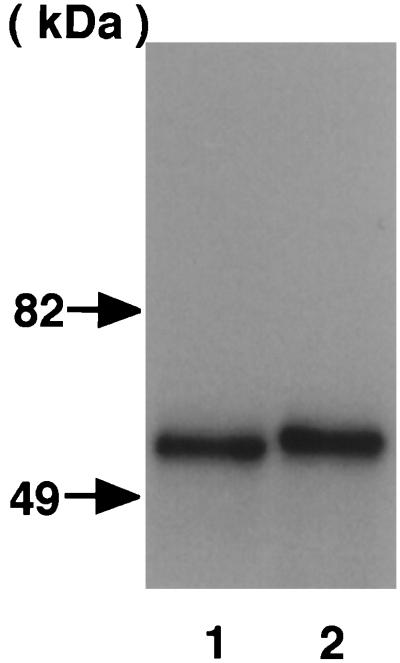
FIG. 1.
Effect of CNF2 on the electrophoretic mobility of GST-RhoA. GST-RhoA was coexpressed with (lane 2) or without (lane 1) CNF2 in E. coli. GST fusion protein was purified from the E. coli homogenate by using glutathione-Sepharose beads according to the manufacturer's instructions. Proteins were analyzed by SDS-PAGE in a 10% gel and by subsequent staining with Coomassie brilliant blue. The molecular masses of the standard are indicated by arrows.
TABLE 2.
Characterization of small GTPases coexpressed with CNF2a
| GTPase | Kd (GTPγS) (nM) | GDP dissociation constant (K−1 [103, min]) | GTPase (steady-state rate) (Kss [103, min]) | GTPase (actual catalytic rate)
(Kcat [103,
min−1])
|
GAP sensitivity (fold) | |
|---|---|---|---|---|---|---|
| GAP (−) | GAP (+) | |||||
| RhoA | 11 ± 1 | 270 ± 70 | 33 ± 6 | 61 ± 10 | 510 ± 100 | 8.4 |
| RhoACNF | 16 ± 3 | 160 ± 40 | 11 ± 1* | 20 ± 4* | 45 ± 9* | 2.2 |
| Rac1 | ND | ND | 12.9 ± 0.6 | 95 ± 20 | ND | ND |
| Rac1CNF | ND | ND | 7 ± 1** | 22 ± 1* | ND | ND |
| Cdc42 | ND | ND | 11.3 ± 0.9 | 175 ± 1 | ND | ND |
| Cdc42CNF | ND | ND | 13 ± 1 | 169 ± 20 | ND | ND |
The results shown are means ± the standard errors of the means of three to five independent experiments. Significant differences between Rho and RhoCNF or Rac and RacCNF are indicated by asterisks. ∗, P < 0.05; ∗∗, P < 0.01 (t test). ND, not determined.
FIG. 2.
Effect of the CNF2-induced modification on GTPase activity of Rho. (A) Effect of CNF2-induced modification on GTPase activity of GST-RhoA. GST-RhoA coexpressed with (▴) or without (●) CNF2 in E. coli was purified and assayed for GTPase activity as described in Materials and Methods. (B) Effect of CNF2-induced modification on GTPase activity of FLAG-RhoA Q63 and FLAG-RhoA E63. FLAG-RhoA Q63 (wild type) coexpressed with (▴) or without (●) CNF2 and FLAG-RhoA E63 coexpressed with (▵) or without (○) CNF2 in E. coli were purified and assayed for GTPase activity as described in Materials and Methods. The results shown are representative of three independent experiments.
CNF2 deamidates Q63 of RhoA coexpressed in E. coli.
Next, we tried to identify the structural change induced by CNF2. Since the GST-RhoA we used did not contain a protease cleavage site to separate GST from fused protein, we switched to a FLAG-RhoA system for the preparation of FLAG peptide-tagged RhoA (FLAG-RhoA). FLAG-RhoA was also susceptible to chemical modification by CNF2 in the E. coli coexpression system (data not shown). When purified CNF2-modified and -unmodified FLAG-RhoA were analyzed by ESI-MS performed on a TSQ-700 triple-sector quadrupole mass spectrometer, no differences were observed between Mrs of the proteins (not shown). This suggested that the nature of chemical modification by CNF2 was too subtle to detect by MS. Therefore, we generated peptide fragments by proteolytic digestion of CNF2-modified and -unmodified FLAG-RhoA and analyzed peptides by N-terminal sequencing, together with MS. Table 3 shows peptide mapping of the CNF2-modified FLAG-RhoA digested with endopeptidase Lys-C. When compared to the results with the coding amino acid sequence of FLAG-RhoA, only Q71 (Q63 in RhoA) was changed to E71 in the 30-L7 fragment. Amino acid sequencing of the corresponding fragment, 184-L7, revealed that the 71st amino acid was Q in unmodified FLAG-RhoA ESI-MS, and N-terminal amino acid sequence analysis of peptides generated by endoproteinase Lys-C digestion of FLAG-RhoA revealed that fragment 184-L7 (amino acids 60 to 106; QVELALWDTAGQEDYDRLRPLSYPDTDVILMCFSIDSPDSLENIPEK) had a predicted mass of 5,399.9 Da and an observed mass of 5,969.2 Da. The amino acid alteration at the 71st amino acid was confirmed with peptides generated by trypsin digestion of 30-L7 fragment as shown in Table 4. Of note, the observed mass corresponding to the L7 fragment in both CNF2-modified and -unmodified FLAG-RhoA was larger than that predicted from deduced amino acid sequences (see above and Table 3). However, mass data of trypsin-digested fragments, which cover most of the L7 fragment, revealed no difference between the observed and the predicted masses (Table 4). The reason for the discrepancy between observed and predicted masses of L7 remains, however, unknown.
TABLE 3.
ESI-MS and N-terminal amino acid sequence analysis of peptides generated by endoproteinase Lys-C digestion of CNF2-modified FLAG-RhoA
| Fragment | Position | Amino acid sequence | Predicted mass (Da) | Observed mass (Da) |
|---|---|---|---|---|
| 30-L1 | 1–4 | MDYK | 555.7 | 555.2 |
| 30-L2 | 5–9 | DDDDK | 606.6 | 606.5 |
| 30-L3 | 10–14 | AAIRK | 557.7 | 557.2 |
| 30-L4 | 16–26 | LVIVGDGACGK | 1,031.2 | 1,031.4 |
| 30-L5 | 27–35 | TCLLIVFSK | 1,023.3 | 1,022.9 |
| 30-L6 | 36–59 | DQFPEVYVPTVFENYVADIEVDGK | 2,774.0 | 2,774.4 |
| 30-L7 | 60–106 | QVELALWDTAGEEDYDRLRPLSYPDTDVILMCFSIDSPDSLENIPEK | 5,400.9 | 5,970.9 |
| 30-L8 | 107–112 | WTPEVK | 758.8 | 758.3 |
| 30-L9 | 113–126 | HFCPNVPIILVGNK | 1,550.9 | 1,551.6 |
| 30-L10 | 113–141 | HFCPNVPIILVGNKKDLRNDEHTRRELAK | 3,413.9 | 3,409.4 |
| 30-L11 | 128–141 | DLRNDEHTRRELAKMK | 1,752.9 | 1,752.5 |
| 30-L12 | 144–170 | QEPVKPEEGRDMANRIGAFGYMECSAK | 3,014.4 | 3,012.5 |
| 30-L13 | 144–193 | QEPVKPEEGRDMANRIGAFGYMECSAKTKDGVREVFEMATRAALQARRGK | 5,587.3 | 5,588.3 |
| 30-L14 | 173–193 | DGVREVFEMATRAALQARRGK | 2,361.7 | 2,360.9 |
TABLE 4.
ESI-MS and N-terminal amino acid sequence analysis of peptides generated by trypsin digestion of CNF2-modified FLAG-RhoA L7 fragment
| Fragment | Position | Amino acid sequence | Predicted mass (Da) | Observed mass (Da) |
|---|---|---|---|---|
| L7-T1 | 65–76 | ALWDTAGEEDYDR | 1,540.5 | 1,541.6 |
| L7-T1 | 77–92 | LRPLSYPDTDVILMCF | 1,883.2 | 1,881.3 |
| L7-T1 | 93–106 | SIDSPDSLENIPEK | 1,543.6 | 1,543.9 |
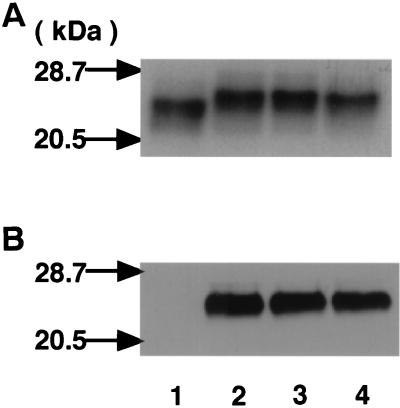
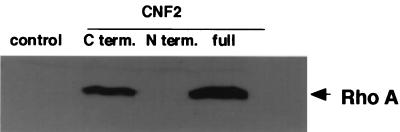
Q63 belongs to one of the conserved regions among small GTPases, G-3, which is critical in GDP-GTP exchange, GTP-induced conformational change, and GTP hydrolysis (3). Mutation of Q63 to L63 of Rho inhibited GTPase and GAP stimulation and induced the dominant positive phenotype of Rho GTPase (11, 40, 41). We measured intrinsic GTPase activities of FLAG-RhoA Q63 (wild type) and FLAG-RhoA E63 either expressed alone or coexpressed with CNF2. As shown in Fig. 2B, FLAG-RhoA E63 as well as FLAG-RhoA coexpressed with CNF2 revealed decreased intrinsic GTPase activity. We compared the electrophoretic mobilities of CNF2-modified FLAG-RhoA and FLAG-RhoA E63. As shown in Fig. 3A, FLAG-RhoA coexpressed with CNF2 revealed an electrophoretic mobility similar to that of FLAG-RhoA E63. FLAG-RhoA E63 coexpressed with CNF2 did not alter the electrophoretic mobility in SDS-PAGE either. These results further support the idea that RhoA with Q63 deamidated to E63 by CNF2 has the lowered GTPase activity. We analyzed the structural alteration of FLAG-RhoA coexpressed with CNF2 by using anti-oligopeptide antibody generated against DTAGEEDYDRLRPLS (anti-E63), the G-3 domain, with altered E63 (underlined) conserved in small GTPases. As shown in Fig. 3B, anti-E63 reacted with FLAG-RhoA coexpressed with CNF2 and with FLAG-RhoA E63 coexpressed with or without CNF2 but not with FLAG-RhoA alone. We then undertook to the express C-terminal and N-terminal portions of CNF2 as fusion proteins with a six-histidine tag. The fusion proteins were incubated with purified FLAG-RhoA to see whether deamidation takes place with one of the fragments of CNF2 in vitro. Treatment of CNF2 alone resulted in deamidation of FLAG-RhoA, and the C-terminal portion of CNF2 was also able to deamidate FLAG-RhoA but the N-terminal portion was not (Fig. 4). This was not the case when heat-inactivated CNF2 or CNF2 fragment was used (not shown).
FIG. 3.
SDS-PAGE and Western blotting analyses of FLAG-RhoA. FLAG-RhoA Q63 (wild type) coexpressed with (lane 2) or without (lane 1) CNF2 and FLAG-RhoA E63 coexpressed with (lane 4) or without (lane 3) CNF2 in E. coli were purified as described in Materials and Methods. Purified proteins were analyzed by SDS-PAGE with Coomassie brilliant blue staining (A) or by Western blotting with anti-E63 antibody (dilution, 1,000-fold) (B).
FIG. 4.
In vitro modification of FLAG-RhoA by CNF2. Results of Western blotting analysis of FLAG-RhoA with anti-E63 antibody are shown. Purified FLAG-RhoA was coincubated with nothing (control), purified His-tag–CNF2 (C term.), His-tag–CNF2 (N term.), or His-tag–CNF2 (full) at a molar ratio of 2:3 in PBS at 37°C for 30 min. The treated samples were subjected to SDS-PAGE and probed with anti-E63 antibody (dilution, 1,000-fold).
CNF2 selectively deamidates RhoA and Rac1 coexpressed in E. coli and 21-kDa protein in Swiss 3T3 cells.
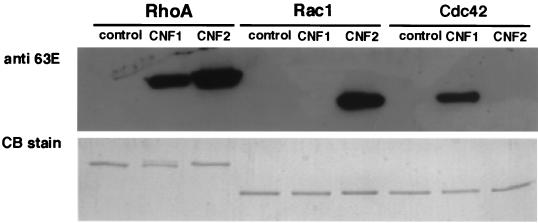
We further analyzed the structural alterations of small GTPases of the Rho subfamily coexpressed with CNF2 by SDS-PAGE and Western blotting by using anti-E63 antibody. Anti-E63 reacted with GST-RhoA and GST-Rac1 coexpressed with CNF2, but it did not react with GST-Cdc42 coexpressed with CNF2 (Fig. 5). On the other hand, anti-E63 reacted with GST-RhoA and GST-Cdc42 coexpressed with CNF1 but did not react with GST-Rac1 coexpressed with CNF1 (Fig. 5). These results clearly indicated that CNF2 and CNF1 have different substrate specificities: CNF2 preferentially deamidates RhoA and Rac1, and CNF1 preferentially deamidates RhoA and Cdc42. The data for CNF1 further supported the observation described by Schmidt et al. (45). To know whether the deamidation event by CNF2 takes place in Swiss 3T3 cells, homogenates of CNF2-treated and -untreated Swiss 3T3 cells were immunodetected by using anti-E63. As shown in Fig. 6, an immunoreactive band of 21 kDa was observed in the homogenate of CNF2-treated cells but not in that of CNF2-untreated Swiss 3T3 cells, suggesting that protein(s) belonging to the Rho family was deamidated by CNF2 treatment in vivo.
FIG. 5.
SDS-PAGE and Western blotting analyses of GST-fused small GTPases. GST-RhoA (RhoA), GST-Rac1 (Rac1), and GST-Cdc42 (Cdc42) coexpressed without (control) or with CNF1 or CNF2 in E. coli were purified. Purified proteins were analyzed by SDS-PAGE with Coomassie brilliant blue staining (lower panel) or by Western blotting with anti-E63 antibody (dilution, 1,000-fold) (upper panel).
FIG. 6.
Western blotting analysis of CNF2-treated Swiss 3T3 cell homogenate with anti-E63 antibody. Homogenates (10 μg) of Swiss 3T3 cells treated with (lane 2) or without (lane 1) CNF2 (50 ng/ml) for 30 min were subjected to SDS-PAGE and probed with anti-E63 antibody (dilution, 1,000-fold) as the secondary antibody.
CNF2 selectively induces accumulation of the GTP-bound form of the small GTPases in Cos-7 cells.
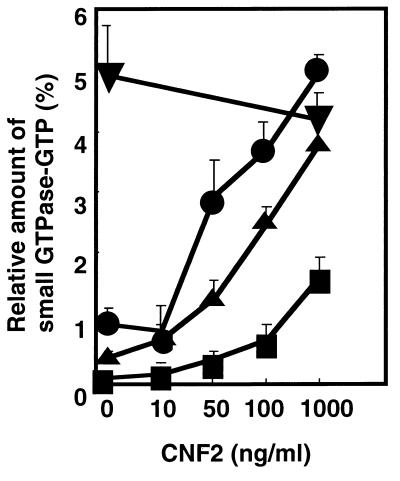
We addressed the question of whether CNF2-induced activation of small GTPases takes place in cultured cells. We used Cos-7 cells to transiently express FLAG-tagged small GTPase belonging to the Rho family, i.e., RhoA, Rac1, and Cdc42. The cells transfected with mammalian expression vector pEF-BOS harboring the FLAG-tagged small GTPase gene were treated with purified CNF2 and subsequently labeled with 32Pi for 12 h and harvested. Cells were lysed under mild conditions, and the small GTPase was recovered from the lysate with anti-FLAG monoclonal antibody-conjugated beads. Then the bound nucleotides were extracted from the immunoprecipitates and applied to a polyethyleneimine-cellulose thin-layer sheet for thin-layer chromatography (TLC), and the percentage of GTP to small GTPase-bound guanine nucleotide was determined. CNF2 dose dependently increased the proportion of FLAG-RhoA GTP and FLAG-Rac1 GTP (Fig. 7). On the other hand, CNF2 slightly increased the percentage of FLAG-Cdc42 GTP, but it was less than 2% even at the concentration of 1,000 ng of CNF2 per ml. These results strongly suggested that CNF2 selectively induces accumulation of the GTP form of RhoA and Rac1 but not of Cdc42.
FIG. 7.
Effect of CNF2 on accumulation of the GTP-bound form of Rho subfamily GTPases in Cos-7 cells. FLAG-RhoA (●), FLAG-RhoA E63 (▾), FLAG-Rac1 (▴), and FLAG-Cdc42 (■) were transiently expressed in Cos-7 cells. The cells were then treated with CNF2 at the indicated concentrations for 12 h and labeled with 32Pi. The cell lysates were subjected to immunoprecipitation with anti-FLAG monoclonal antibody. Immunoprecipitates were developed by TLC, and the radioactivity was analyzed with a BAS2000. The ratio of GTP to small GTPase-bound guanine nucleotides was indicated as a percentage.
DISCUSSION
Our findings indicate that CNF2 induces deamidation of RhoA Q63 to E63 in E. coli. The CNF2-modified RhoA exhibited lowered intrinsic GTPase activity. Furthermore, the modified RhoA showed decreased sensitivity to Rho GAP. Gln63 of Rho is conserved in small GTPases other than Rap and is essential for GTPase activity (3). Mutation of Q63 to L63 inhibited GTPase and GAP stimulation and induced the dominant positive phenotype of Rho GTPase (11, 40, 41). Somatic mutation of Ras Q61 equivalent to Rho Q63 was shown to reduce GTPase and was suggested to contribute to the generation of human tumors (1). Taken together, these results suggest that CNF2 deamidates Rho Q63, which inhibits the intrinsic GTPase activity of Rho, as well as GTPase activity stimulated by P122RhoGAP. This leads to the constitutive activation of Rho and induces reorganization of actin stress fibers (38). CNF1 was previously demonstrated to deamidate RhoA at Q63 and convert it to E63 and to constitutively activate the RhoA in an in vitro assay (18, 45). Furthermore, CNF1 (at a molar ratio of GTPase to toxin of 16:1) was shown to partly modify Cdc42, one of the members of the Rho subfamily (45). Rac1, however, was not modified with regard to the toxin/GTPase ratio. A higher concentration of CNF1 (at a molar ratio of GTPase to toxin of 5:1) was shown to inhibit the GTPase activities of Cdc42 and Rac1 in the presence or absence of p50GAP by approximately 60 and 80%, respectively, in vitro (32). However, it was not demonstrated whether these GTPases are substrates for CNF1 in vivo. By using an E. coli coexpression system, we found that CNF2 induced activation of not only RhoA but also Rac1. The GTPase activity of Cdc42 was not affected when coexpressed with CNF2. Furthermore, Western blotting analysis with anti-E63 antibody demonstrated that CNF2 deamidated RhoA Q63 and Rac1 Q61 but not Cdc42 Q61. On the other hand, CNF1 was shown to deamidate RhoA Q63 and Cdc42 Q61 but not Rac1 Q61. The data obtained with CNF1 are consistent with the previous observations of Schmidt et al. (45). Nevertheless, these results clearly indicate that CNF2 and CNF1 share the substrate, RhoA, but that their substrate specificities are distinct. In vivo experiments with Cos-7 cells transiently expressing FLAG-tagged small GTPase revealed that CNF2 preferentially deamidated RhoA and Rac1.
It is tempting to speculate that the substrate specificity of CNF toxins explains the differences between CNF1 and CNF2 toxic activities. For example, the more intense necrotic activity of CNF2 could be due to a stronger inflammatory effect of CNF2 associated with vascular endothelial cell damage. Indeed, CNF2 could be a more potent inducer of mast cell degranulation, since Rac activation and Rho activation are activated in the secretory response of mast cells to stimuli (39). By activating Rac1, CNF2 could also generate reactive oxygen species that are essential for NF-κB-dependent transcriptional regulation of interleukin-1α, which, in an autocrine manner, induce collagenase-1 gene expression (28). Remodeling of the extracellular matrix and consequent alterations of integrin-mediated adhesion and cytoarchitecture are central to wound healing and inflammation. Furthermore, CNF2 could also affect the host immune response, since Rac specifically regulates integrin-mediated spreading and increased adhesion of T lymphocytes (12). Finally, we also observed that CNF2 preferentially induces formation of membrane ruffling and reorganization of actin and vinculin in quiescent Swiss 3T3 cells (data not shown). Again, the differences in reorganization of the actin cytoskeleton and in the vinculin staining pattern may reflect differences in the substrate specificities of CNF2 and CNF1. However, in eukaryotic cells, there is a significant cross-talk among small GTPases belonging to the Rho family (20). Therefore, the functions of these GTPases are closely associated. For example, Rac activates Rho and Cdc42 activates Rac. A careful time course analysis of the effects of CNF1 and CNF2 may provide some clues as to the sequential regulation of the small GTPase cascades.
Bordetella species such as B. pertussis, B. parapertussis, and B. bronchiseptica produce DNTs (5, 13, 52). DNT and CNF possess similar biological properties. DNT induces dermonecrosis in a variety of animals when injected intradermally (5, 13, 24, 52). It inhibits cytokinesis, induces multinucleation of cultured cells, and generates the formation of thick actin stress fibers and focal adhesions (24). Horiguchi et al. demonstrated that purified DNT from B. bronchiseptica directly modifies RhoA in vitro, and the modified RhoA revealed a slower electrophoretic mobility in SDS-PAGE (24). Recently, these authors reported that DNT deamidates the Q63 residue of RhoA to E63 and alters RhoA to the constitutively active form (25). A comparison of amino acid sequences of both CNFs and DNT revealed that these dermonecrotizing cytotoxins are homologous in a localized C-terminal region, showing 35% identity between CNF amino acids 824 to 929 and DNT amino acids 1263 to 1368. The fact that CNF1, CNF2, and DNT possess similar activities suggests that they constitute a family of bacterial toxins which target eukaryotic Rho proteins and modify their function by deamidation of Q63. The C-terminal homologous regions of these toxins seems to be the active domain for deamidation activity. We have shown in the present study that the C-terminal portion of CNF2 was able to deamidate RhoA, and similar results were observed with CNF1 (31). In that study, the fact that the C-terminal domain lost its catalytic activity when deleted of various subdomains suggests a scattered distribution of catalytic-site amino acids. Nevertheless, Schmidt and coworkers have recently demonstrated that CNF1 has both deamidation and transglutamination activities and shares a catalytic dyad of cysteine and histidine residues with eukaryotic transglutaminases and cysteine proteases (46).
Despite similarities in biological activities, CNF and DNT toxins have several distinct characteristics. The detection of proteins [32P]ADP-ribosylated by clostridial C3 exoenzyme in DNT-treated cells revealed multiple bands with slower and faster electrophoretic mobilities than that of untreated control (24). On the other hand, protein [32P]ADP-ribosylated by EDIN or by C3 in CNF-treated cells revealed a single band in one-dimensional slab gel electrophoresis, and the band migrated more slowly than that of an untreated control (16, 17, 38). Swiss 3T3 cells were highly sensitive to intoxication by CNF2 but insensitive to that by DNT. These results suggest that the mode of action of these toxins differs and that CNF and DNT probably bind to different cell receptors. Comparative studies with CNF and DNT may provide insight into the mechanism of intoxication.
ACKNOWLEDGMENTS
We thank Akira Kikuchi and Shinya Koyama for GST-Cdc42 and helpful discussions on GTPase measurement, Yoshimi Homma for GST-P122RhoGAP, and Hiroshi Maruta for pGST-RhoA. We also thank Ryousuke Yamano and Junichi Yamagishi for their skillful assistance. We thank the Research Center for Molecular Medicine, Hiroshima University School of Medicine, The Research Facility, Hiroshima University School of Dentistry, and the Research Facility, Okayama University School of Dentistry, for the use of their facilities.
This work was supported in part by research grants from the Ministry of Education, Science, Sports, and Culture, Japan (1997), the European Community program FAIR (number 1335), and the Conseil Regional de la region Midi-Pyrenees (number 9507783). S.Y.P. was a recipient of a scholarship from the Ministère de l'Enseignement Supérieur et de la Recherche.
Footnotes
This study is dedicated to the memory of Henry C. Wu, who died 12 February 1996.
REFERENCES
- 1.Barbacid M A. rasgenes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 2.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–131. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 3.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–126. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Bruckner I E, Evans D G. The toxin of Br. parapertussis and the relationship of this organism to H. pertussis and Br. bronchiseptica. J Pathol Bacteriol. 1939;48:67–78. [Google Scholar]
- 6.Caprioli A, Donelli G, Falbo V, Possenti R, Roda L G, Roscetti G, Ruggeri F M. A cell division-active protein from Escherichia coli. Biochem Biophys Res Commun. 1984;118:587–593. doi: 10.1016/0006-291x(84)91343-3. [DOI] [PubMed] [Google Scholar]
- 7.Caprioli A, Falbo V, Roda L G, Ruggeri F M, Zona C. Partial purification and characterization of an Escherichia colitoxic factor that induces morphological alteration. Infect Immun. 1983;39:1300–1306. doi: 10.1128/iai.39.3.1300-1306.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Rycke J, Gonzalez E A, Blanco J, Oswald E, Blanco M, Boivin R. Evidence for two types of cytotoxic necrotizing factor in human and animal clinical isolates of Escherichia coli. J Clin Microbiol. 1990;28:694–699. doi: 10.1128/jcm.28.4.694-699.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Rycke J, Guillot J F, Boivin R. Cytotoxins in non-enterotoxigenic strains of Escherichia coliisolated from feces of diarrheic calves. Vet Microbiol. 1987;15:137–150. doi: 10.1016/0378-1135(87)90139-8. [DOI] [PubMed] [Google Scholar]
- 10.De Rycke J, Phan-Thanh L, Bernard B. Immunochemical identification and biological characterization of cytotoxic necrotizing factor from Escherichia coli. J Clin Microbiol. 1989;27:983–988. doi: 10.1128/jcm.27.5.983-988.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diekmann D, Hall A. In vitrobinding assay for interactions of Rho and Rac with GTPase-activating proteins and effectors. Methods Enzymol. 1995;256:207–215. doi: 10.1016/0076-6879(95)56025-4. [DOI] [PubMed] [Google Scholar]
- 12.D'Souza-Schorey C, Boettner B, Van Aelst L. Rac regulates integrin-mediated spreading and increased adhesion of T lymphocytes. Mol Cell Biol. 1998;18:3936–3946. doi: 10.1128/mcb.18.7.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans D G. The production of pertussis antitoxin in rabbits and the neutralization of pertussis, parapertussis and bronchisepticatoxins. J Pathol Bacteriol. 1940;51:49–58. [Google Scholar]
- 14.Falbo V, Pace T, Picci L, Pizzi E, Caprioli A. Isolation and nucleotide sequence of the gene encoding cytotoxic necrotizing factor 1 of Escherichia coli. Infect Immun. 1993;61:4909–4914. doi: 10.1128/iai.61.11.4909-4914.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falzano L, Fiorentini C, Donelli G, Michel E, Kocks C, Cossart P, Cabanie L, Oswald E, Boquet P. Induction of phagocytic behavior in human epithelial cells by Escherichia coli. Mol Microbiol. 1993;9:1247–1254. doi: 10.1111/j.1365-2958.1993.tb01254.x. [DOI] [PubMed] [Google Scholar]
- 16.Fiorentini C. Escherichia colicytotoxic necrotizing factor 1 increases actin assembly via the p21 Rho protein. Parasitenkd Zentbl Bakteriol. 1994;24(Suppl.):404–405. [Google Scholar]
- 17.Fiorentini C. Escherichia colicytotoxic necrotizing factor 1: evidence for induction of actin assembly by constitutive activation of the p21 Rho GTPase. Infect Immun. 1995;63:3936–3944. doi: 10.1128/iai.63.10.3936-3944.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flatau G, Lemichez E, Gauthier M, Chardin P, Paris S, Fiorentini C, Boquet P. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–733. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- 19.Fukumoto Y, Kaibuchi K, Hori Y, Fujioka H, Araki S, Ueda T, Kikuchi A, Takai Y. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for the rho proteins, rasp21-like small GTP-binding proteins. Oncogene. 1990;5:1321–1328. [PubMed] [Google Scholar]
- 20.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 21.Hart M J, Eva A, Evans T, Aaronson S A, Cerione R A. Catalysis of guanine nucleotide exchange on the CDC42Hs protein by the dbloncogene product. Nature. 1991;354:311–314. doi: 10.1038/354311a0. [DOI] [PubMed] [Google Scholar]
- 22.Higashijima T, Ferguson K M, Smigel M D, Gilman A D. The effect of GTP and Mg on the GTPase activity and the fluorescent properties of G0. J Biol Chem. 1987;262:757–761. [PubMed] [Google Scholar]
- 23.Homma Y, Emori Y. A dual functional signal mediator showing RhoGAP and phospholipase C-delta stimulating activities. EMBO J. 1995;14:286–291. doi: 10.1002/j.1460-2075.1995.tb07002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horiguchi Y, Senda T, Sugimoto N, Katahira J, Matsuda M. Bordetella bronchisepticadermonecrotizing toxin stimulates assembly of actin stress fibers and focal adhesions by modifying the small GTP-binding protein rho. J Cell Sci. 1995;108:3243–3251. doi: 10.1242/jcs.108.10.3243. [DOI] [PubMed] [Google Scholar]
- 25.Horiguchi Y, Inoue N, Masuda M, Kashimoto T, Katahira J, Sugimoto N, Matsuda M. Bordetella bronchiseptica dermonecrotizing toxin induces reorganization of actin stress fibers through deamidation of Gln-63 of the GTP-binding protein Rho. Proc Natl Acad Sci USA. 1997;94:11623–11626. doi: 10.1073/pnas.94.21.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horii Y, Beeler J F, Sakaguchi K, Tachibana M, Miki T. A novel oncogene, ost, encodes a guanine nucleotide exchange factor that potentially links Rho and Rac signaling pathways. EMBO J. 1994;13:4776–4786. doi: 10.1002/j.1460-2075.1994.tb06803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaibuchi K, Mizuno T, Fujioka H, Yamamoto T, Kishi K, Fukumoto Y, Hori Y, Takai Y. Molecular cloning of the cDNA for stimulatory GDP/GTP exchange protein for smg p21s (ras p21-like small GTP-binding proteins) and characterization of stimulatory GDP/GTP exchange protein. Mol Cell Biol. 1991;11:2873–2880. doi: 10.1128/mcb.11.5.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kheradmand F, Werner E, Tremble P, Symons M, Werb Z. Role of Rac1 and oxygen radicals in collagenase-1 expression induced by cell shape change. Science. 1998;280:898–902. doi: 10.1126/science.280.5365.898. [DOI] [PubMed] [Google Scholar]
- 29.Kikuchi A, Yamashita T, Kawata M, Yamamoto K, Ikeda K, Tanimoto T, Takai Y. Purification and characterization of a novel GTP-binding protein with a molecular weight of 24,000 from bovine brain membranes. J Biol Chem. 1988;263:2897–2904. [PubMed] [Google Scholar]
- 30.Lancaster C A, Taylor-Harris P M, Self A J, Brill S, van Erp H E, Hall A. Characterization of rhoGAP. A GTPase-activating protein for rho-related small GTPases. J Biol Chem. 1994;269:1137–1142. [PubMed] [Google Scholar]
- 31.Lemichez E, Flatau G, Bruzzone M, Boquet P, Gauthier M. Molecular localization of the Escherichia colicytotoxic necrotizing factor CNF1 cell-binding and catalytic domains. Mol Microbiol. 1997;24:1061–1070. doi: 10.1046/j.1365-2958.1997.4151781.x. [DOI] [PubMed] [Google Scholar]
- 32.Lerm M, Selzer J, Hoffmeyer A, Rapp U R, Aktories K, Schmidt G. Deamidation of Cdc42 and Rac by Escherichia colicytotoxic necrotizing factor 1: activation of c-Jun N-terminal kinase in HeLa cells. Infect Immun. 1999;67:496–503. doi: 10.1128/iai.67.2.496-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuno T, Kaibuchi K, Yamamoto T, Kawamura M, Sakoda T, Fujioka H, Matsuura Y, Takai Y. A stimulatory GDP/GTP exchange protein for smg p21 is active on the post-translationally processed form of c-Ki-ras and rhoAp21. Proc Natl Acad Sci USA. 1991;88:6442–6446. doi: 10.1073/pnas.88.15.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muroya K, Hattori S, Nakamura S. Nerve growth factor induces rapid accumulation of the GTP-bound form of p21ras in rat pheochromocytoma PC12 cells. Oncogene. 1992;7:277–281. [PubMed] [Google Scholar]
- 35.Nobes C, Hall A. Regulation and function of the Rho subfamily of small GTPases. Curr Opin Genet Dev. 1994;4:77–81. doi: 10.1016/0959-437x(94)90094-9. [DOI] [PubMed] [Google Scholar]
- 36.Oswald E, De Rycke J. A single protein of 110 kDa is associated with the multinucleating and necrotizing activity coded by the Vir plasmid of Escherichia coli. FEMS Microbiol Lett. 1990;68:279–284. [PubMed] [Google Scholar]
- 37.Oswald E, De Rycke J, Guillot J F, Boivin R. Cytotoxic effect of multinucleation in HeLa cell cultures associated with the presence of Vir plasmid in Escherichia coli. FEMS Microbiol Lett. 1989;58:95–100. doi: 10.1016/0378-1097(89)90349-2. [DOI] [PubMed] [Google Scholar]
- 38.Oswald E, Sugai M, Labigne A, Wu H C, Fiorentini C, Boquet P, O'Brien A D. Cytotoxic necrotizing factor type 2 produced by virulent Escherichia colimodifies the small GTP-binding proteins Rho involved in assembly of actin stress fibers. Proc Natl Acad Sci USA. 1994;91:3814–3818. doi: 10.1073/pnas.91.9.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price L S, Norman J C, Ridley A J, Koffer A. The small GTPases Rac and Rho as regulators of secretion in mast cells. Curr Biol. 1995;5:68–73. doi: 10.1016/s0960-9822(95)00018-2. [DOI] [PubMed] [Google Scholar]
- 40.Renshaw M W, Taksoz D, Schwartz M A. Involvement of the small GTPase Rho in integrin-mediated activation of mitogen-activated protein kinase. J Biol Chem. 1996;271:21691–21694. doi: 10.1074/jbc.271.36.21691. [DOI] [PubMed] [Google Scholar]
- 41.Ridley A J. Microinjection of Rho and Rac into quiescent Swiss 3T3 cells. Methods Enzymol. 1995;256:313–320. doi: 10.1016/0076-6879(95)56036-x. [DOI] [PubMed] [Google Scholar]
- 42.Ridley A J, Hall A. Signal transduction pathways regulating Rho-mediated stress fibre formation: requirement for a tyrosine kinase. EMBO J. 1994;13:2600–2610. doi: 10.1002/j.1460-2075.1994.tb06550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. New York, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K. Gln 63 Rho is deamidated by Escherichia colicytotoxic necrotizing factor-1. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt G, Selzer J, Lerm M, Aktories K. The Rho-deamidating cytotoxic necrotizing factor 1 from Escherichia colipossesses transglutaminase activity. Cysteine 866 and histidine 881 are essential for enzyme activity. J Biol Chem. 1998;273:13669–13674. doi: 10.1074/jbc.273.22.13669. [DOI] [PubMed] [Google Scholar]
- 47.Self A J, Hall A. Purification of recombinant Rho/Rac/G25K from Escherichia coli. Methods Enzymol. 1995;256:3–10. doi: 10.1016/0076-6879(95)56003-3. [DOI] [PubMed] [Google Scholar]
- 48.Settleman J, Albright C F, Foster L C, Weinberg R A. Association between GTPase activators for Rho and Ras families. Nature. 1992;359:153–154. doi: 10.1038/359153a0. [DOI] [PubMed] [Google Scholar]
- 49.Shoji I, Kikuchi A, Kuroda S, Takai Y. Kinetic analysis of the binding of guanine nucleotide to bovine brain smgp25A. Biochem Biophys Res Commun. 1989;162:273–281. doi: 10.1016/0006-291x(89)91992-x. [DOI] [PubMed] [Google Scholar]
- 50.Sugai M, Hashimoto K, Kikuchi A, Inoue S, Okumura H, Matsumoto K, Goto Y, Ohgai H, Moriishi K, Syuto B, et al. Epidermal cell differentiation inhibitor ADP-ribosylates small GTP-binding proteins and induces hyperplasia of epidermis. J Biol Chem. 1992;267:2600–2604. [PubMed] [Google Scholar]
- 51.Sugai M, Kawamoto T, Pérès S Y, Ueno Y, Komatsuzawa H, Fujiwara T, Kurihara H, Suginaka H, Oswald E. The cell cycle-specific growth-inhibitory factor produced by Actinobacillus actinomycetemcomitansis a cytolethal distending toxin. Infect Immun. 1998;66:5008–5019. doi: 10.1128/iai.66.10.5008-5019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wardlaw A C, Parton R. Bordetella pertussis toxins. Oxford, England: Pergamon Press, Ltd.; 1983. [Google Scholar]