Abstract
Exosomes are a subgroup of nanosized extracellular vesicles enclosed by a lipid bilayer membrane and secreted by most eukaryotic cells. They represent a route of intercellular communication and participate in a wide variety of physiological and pathological processes. The biological roles of exosomes rely on their bioactive cargos, including proteins, nucleic acids, and lipids, which are delivered to target cells. Their distinctive properties—innate stability, low immunogenicity, biocompatibility, and good biomembrane penetration capacity—allow them to function as superior natural nanocarriers for efficient drug delivery. Another notably favorable clinical application of exosomes is in diagnostics. They hold various biomolecules from host cells, which are indicative of pathophysiological conditions; therefore, they are considered vital for biomarker discovery in clinical diagnostics. Here, we use data from the CAS Content Collection and provide a landscape overview of the current state and delineate trends in research advancement on exosome applications in therapeutics and diagnostics across time, geography, composition, cargo loading, and development pipelines. We discuss exosome composition and pathway, from their biogenesis and secretion from host cells to recipient cell uptake. We assess methods for exosome isolation and purification, their clinical applications in therapy and diagnostics, their development pipelines, the exploration goals of the companies, the assortment of diseases they aim to treat, development stages of their research, and publication trends. We hope this review will be useful for understanding the current knowledge in the field of medical applications of exosomes, in an effort to further solve the remaining challenges in fulfilling their potential.
Keywords: exosome, extracellular vesicle, drug delivery, diagnostics, biomarker, nanoparticle, nanocarrier, blood−brain barrier, therapeutics
Nearly 20 years after the discovery of liposomes,1 it was found out that similar lipid vesicles form naturally in living organisms.2,3 These include membrane-contained nanosized extracellular vesicles (EVs), secreted from cells as part of their normal process or certain pathologies. Based on the origin and size of the EVs, as well as on the current understanding of their biogenesis, they are grouped as follows: exosomes (diameter ∼30–150 nm); microvesicles or ectosomes (100 nm–1 μm); and apoptotic bodies (50 nm–5 μm).4,5
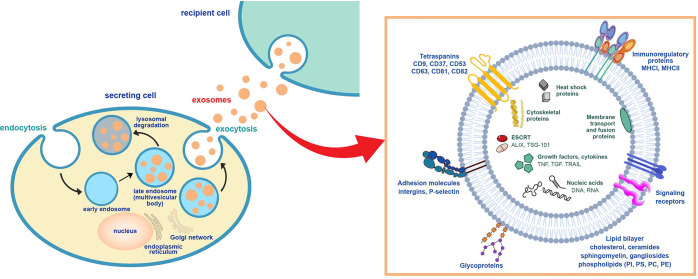
Exosomes are produced in the endosomes of most eukaryotic cells and subsequently released in the extracellular space by fusion with the cellular biomembrane (Figure 1). Their functions are still largely unknown but a subject of a recent burst of interest as their important roles in physiological and pathophysiological processes are steadily revealed. They have been shown to provide means of efficient intercellular communication and signaling, including transport of bioactive molecules such as proteins, lipids, and nucleic acids, between cells and across biological barriers.6,7 These results and the physicochemical properties of exosomes are reasons that they are viewed as the rising star in drug delivery and diagnostics.5,8,9 However, there is still insufficient knowledge regarding exosome physiology. In order to make use of the clinical potential of exosomes, it is necessary to better understand the cellular processes that govern their biology and membrane trafficking.
Figure 1.
Scheme of exosome biogenesis and secretion. The inset exemplifies the molecular constituents of the exosomes.
For a long time, synthetic drug nanocarriers have been developed to improve the efficacy of therapeutics, to refine their pharmacokinetics and pharmacodynamics, while lessening the toxicity and side effects.10,11 Many smart artificial delivery systems such as various functionalized, stimuli-responsive, targeted lipidic or polymeric nanocarriers have been invented to improve key features of the delivery systems such as circulation time in the bloodstream, biodistribution, cellular interactions, and drug loading and release. However, synthetic drug delivery systems still come across many setbacks, such as non-specific drug targeting and toxicity of the carriers, immunogenicity, and unsatisfactory efficacy.12 Specifically, lipid nanoparticles (LNPs) have been recognized as favorable vehicles to protect, transport, and deliver a wide variety of drugs and vaccines to cells.10 Liposomes, an early kind of lipid nanoparticles, are a flexible and resourceful nanomedicine delivery system. They can significantly enhance drug pharmacokinetics. By encapsulating drugs in liposomes, they are protected against dilution and degradation or inactivation in the blood.10,13 Lipid nanoparticle technologies together with other nanotechnological platforms for drug delivery have improved the efficiency, selectivity, residence time, and biodistribution of traditional drug carrier systems while reducing their drawbacks. However, the clinical application of the lipid nanocarriers has experienced substantial difficulties such as low bioavailability, toxicity, removal from the bloodstream, or stimulation of innate immune reactions.
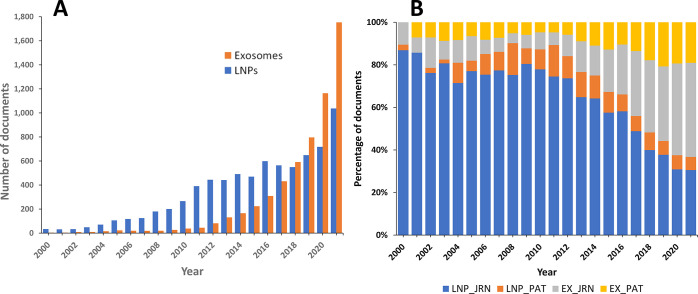
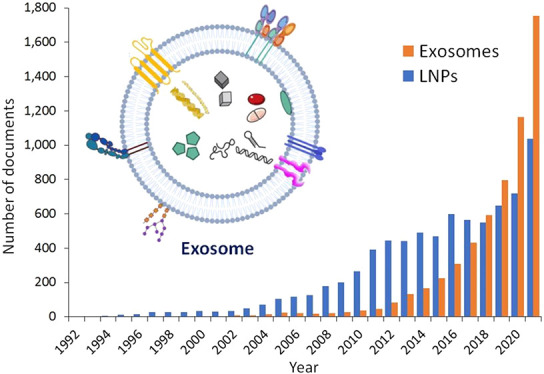
After the discovery of exosomes, it was realized that they are quite similar to liposomes, in fact a more complex version of liposomes, but originating from biological systems. Despite the evident similarities, exosomes exhibit certain advantages, which make them a preferable drug delivery vehicle. Their lipid composition is rich in non-lamellar forming lipids, which may give rise to favorable curvatures in their lipid bilayer, which has been proven beneficial in drug delivery.14 Furthermore, the exosome lipid bilayer is highly asymmetrical, which could be particularly advantageous for their interaction with the plasma membrane and especially with their target cells. While liposomes generally do not contain proteins, a large variety of integral and peripheral membrane proteins are found in exosomes, another favorable feature in their application in drug delivery. As a result, in the last 3–4 years, exosomes have become preferable over lipid nanoparticles as prospective drug carriers. The number of documents, both patents and journal articles, related to exosomes applied in drug delivery has significantly surpassed that of lipid nanoparticles, as revealed by a search in the CAS Content Collection15 (Figure 2).
Figure 2.
Publication trends of exosomes and lipid nanoparticles applied to drug delivery. (A) Comparison of the trends in the number of publications related to exosomes and lipid nanoparticles. The number of publications has been estimated by combining drug-delivery-related search terms such as “drug delivery”, “pharmaceutic”, and “carrier” with the terms “lipid nanoparticle” vs “exosome” or “extracellular vesicle”. (B) Corresponding yearly percentages of publications related to exosomes (EX) and lipid nanoparticles (LNP) in journal articles (JRN) and patents (PAT) calculated for each specific year are compared.
In enhancing exosome efficiency, valuable lessons learned from liposome development have been employed. Various techniques found useful and significantly refined in liposome/lipid nanoparticle production and drug loading, such as sonication, extrusion, freeze–thaw cycles, microfluidics, and others, have been successfully applied in exosomes. Functional modifications that have significantly improved liposome efficiency have been found useful in exosomes as well. The most noteworthy of these include targeting by surface-attached ligands for specific receptors on cells and coating with biocompatible inert polymers, typically polyethylene glycol (PEG), making the carriers invisible to phagocytes (PEGylation), considerably extending their circulatory half-life.10
The applications of exosomes as a natural carrier platform to deliver drugs have been regarded as a hope and promise to overcome the limitations associated with many previously studied drug delivery systems. For instance, exosomes are originated from biological systems and their components can be readily metabolized and excreted at the end of the delivery journey. In addition, exosomes produce a minimal immune response related to cell therapies, which might be rejected by the recipient.16 Furthermore, exosomes are believed to exhibit minimum tumorigenicity,17 as they could be readily absorbed and excreted via the blood and urine.18 Various studies have shown the capacity of exosomes for promoting angiogenesis, providing cytoprotection, and controlling apoptosis.17 The exciting observations on the delivery potential of exosomes such as their ability to overcome barriers for conventional colloidal delivery systems, in particular the blood–brain barrier (BBB), and effectiveness for hard to deliver molecules such as proteins and RNAs have inspired intense research on their application as drug delivery vehicles.
Another especially promising clinical application of exosomes is in diagnostics. They transport biomolecules from their cells of origin, which may contain signs of pathophysiological conditions; therefore, they are widely considered to be essential for biomarker discovery in clinical diagnostics. Recent studies have shown that exosomes contain proteins and nucleic acids implicated in cancer and numerous other diseases, such as neurodegenerative, metabolic, infectious, inflammatory, and others. Moreover, exosomes can be obtained from easily achievable body fluids such as blood and urine and are thus appropriate targets for diagnostic application.19,20
Since the EV terminology is often confusing and has not been standardized due to the current limitations in isolating a particular type of EVs, the International Society for Extracellular Vesicles21 on the Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV 2018) guidelines suggested the use of alternative terms such as “small EVs” (<200 nm) or “large EVs” (>200 nm).22 However, the term “exosome” is still largely used and dominates in the literature for vesicles of diameter ∼30–150 nm. The term “exosome” should also not be mixed up with “exosome complex”, a multiprotein membraneless intracellular complex.23
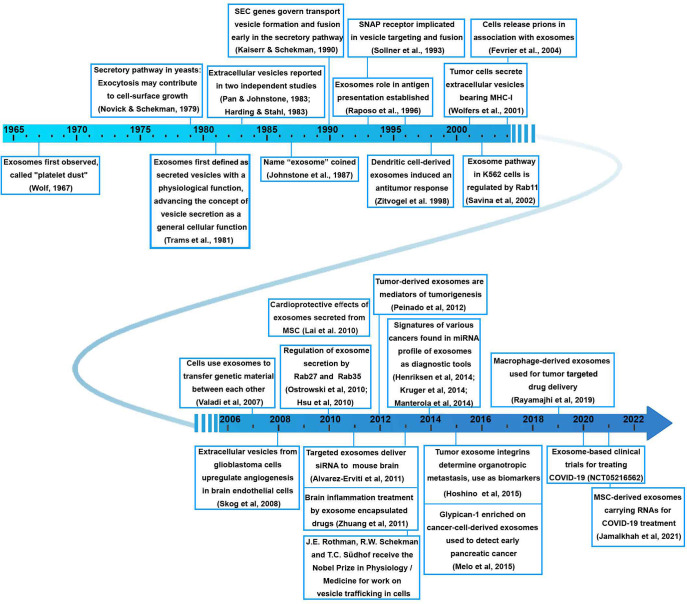
Observation of small particles in plasma referred to as “platelet dust” was reported over 50 years ago.24 The discovery of exosomes is related to two independent studies from 1983 focused on the transferrin receptor externalization.2,3 It was subsequently realized that most viable cell types, such as B and T lymphocytes, dendritic cells, mast cells, intestinal epithelial cells, neurons, tumor cells, and various kinds of stem cells, release exosomes. It has become well-established that exosomes play an important role as messengers of intercellular communication. The interest in them was strongly enhanced after the power of antigen-loaded exosomes to eliminate tumors in mice was demonstrated25 and phase I clinical trials in metastatic melanoma patients vaccinated with autologous dendritic-cell-derived exosomes were completed,26 so exosomes emerged as a promising tool for autologous treatments in cancer. A timeline exemplifying some of the significant breakthroughs in the field of exosome research2,3,24−51 is shown in Figure 3.
Figure 3.
Timeline of major research and development milestones related to exosomes and their medical applications.2,3,24−51
In this paper, we review the advances in the exosome applications in drug delivery and diagnostics. We examine data from the CAS Content Collection,15 the largest human-curated collection of published scientific knowledge, and analyze the publication landscape of recent research on exosome applications in therapeutics and diagnostics to provide insights into the research advances in the area. We also discuss the exosome composition and pathway, from their biogenesis and secretion from the host cells to the recipient cellular uptake. Subsequently, we assess the methods for isolation and purification of exosomes, their clinical applications in therapy and diagnostics, their development pipelines with company research focuses, disease categories, development stages, and publication trends. We hope this review can serve as a useful resource in understanding the current state of knowledge in the field of clinical applications of exosomes, in an effort to further solve the remaining challenges for fulfilling their potential.
Landscape of Exosome Research—Insights from the CAS Content Collection
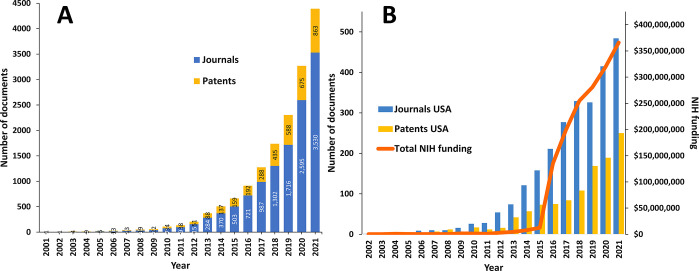
The CAS Content Collection15 is the largest human-curated collection of published scientific knowledge, representing a comprehensive resource to access and keep up to date on the world’s published scientific literature across disciplines including chemistry, biomedical sciences, engineering, materials science, agricultural science, and many more, thus empowering quantitative analysis of global research publications against parameters such as time, scientific area, medical application, disease, and chemical composition. Currently, there are over 40,000 scientific publications (mainly journal articles and patents) in the CAS Content Collection related to exosomes/extracellular vesicles. Over 25,000 of them are related to the application of exosomes in drug delivery and diagnostics. There is a steady, exponential growth of these documents over time (Figure 4A). On Figure 4B, the number of documents (journal articles and patents) originating from organizations in the USA have been correlated with the funding from the National Institutes of Health (NIH),52 increasing sharply after 2015.53
Figure 4.
Journal and patent publication trends of exosome research in drug delivery and diagnostics and the association with research funding. (A) Trends in the number of publications related to exosomes in drug delivery and diagnostics, including journal articles and patents. (B) Number of documents originating from organizations in the USA as correlated with the annual NIH funding.
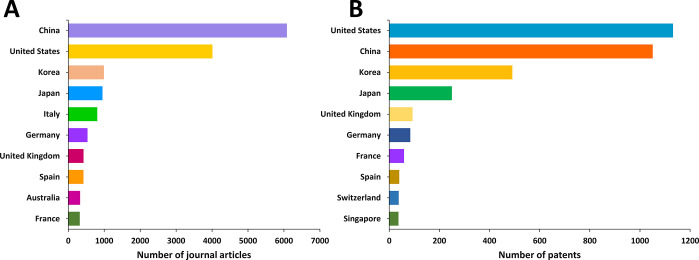
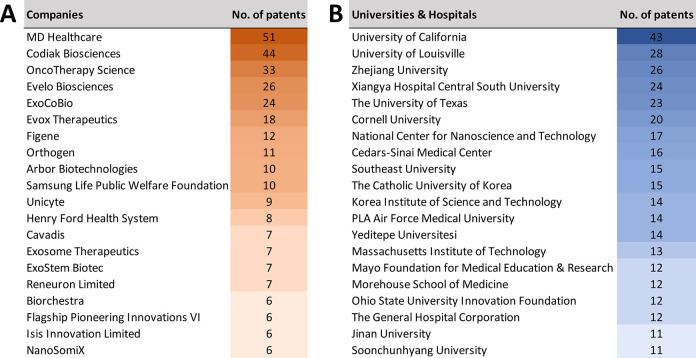
United States, China, Korea, and Japan are the leaders in the number of published journal articles (Figure 5A) and patents (Figure 5B) related to exosomes in therapeutics and diagnostics. Patenting activity related to exosomes is nearly equally shared between corporate and academic players (Figure 6). MD Healthcare, Codiak Biosciences, and OncoTherapy Science have the largest number of patents among the companies (Figure 6A), while University of California, University of Louisville, and Zhejiang University are the leaders among the universities and hospitals (Figure 6B).
Figure 5.
Top countries publishing journal articles (A) and patents (B) related to exosomes in drug delivery and diagnostics.
Figure 6.
Top patent assignees from companies (A) and universities and hospitals (B) for patents related to exosome applications in drug delivery and diagnostics.
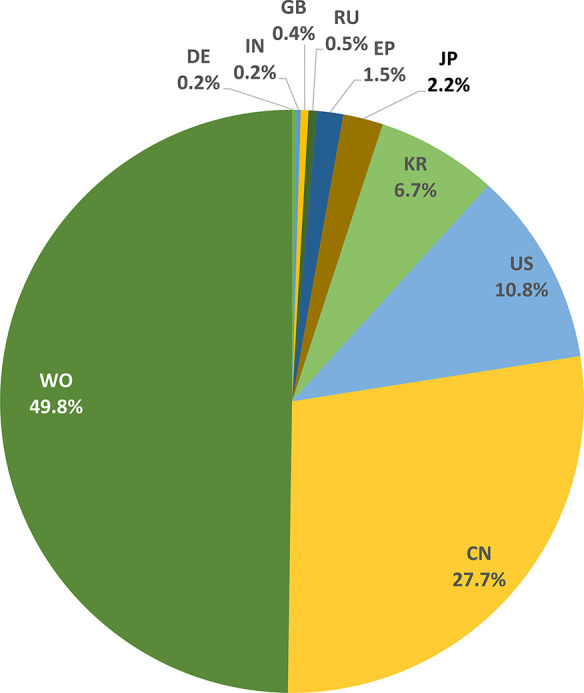
Figure 7 presents the distribution of patents related to the application of exosomes in drug delivery and diagnostics with respect to the patent office. The World Intellectual Property Organization (WIPO) received the most patent applications, followed by the US and China patent offices, the European Patent Office (EPO), and the Korean and Japan patent offices. The percentage of Chinese patents, 27.2%, is well below the average number (63%) of chemistry-related Chinese patents in the CAS Content Collection from the last 10 years. This shows that exosome applications are emerging areas, and it may take some time to establish the technologies. At the same time, the percentage (49.8%) of patents filed through WIPO is significantly higher than the average number (18%) of chemistry-related WIPO patents in the CAS Content Collection, which indicates a strong desire of patenting exosome-related technologies internationally.
Figure 7.
Top patent offices receiving patent applications for exosomes in drug delivery and diagnostics.
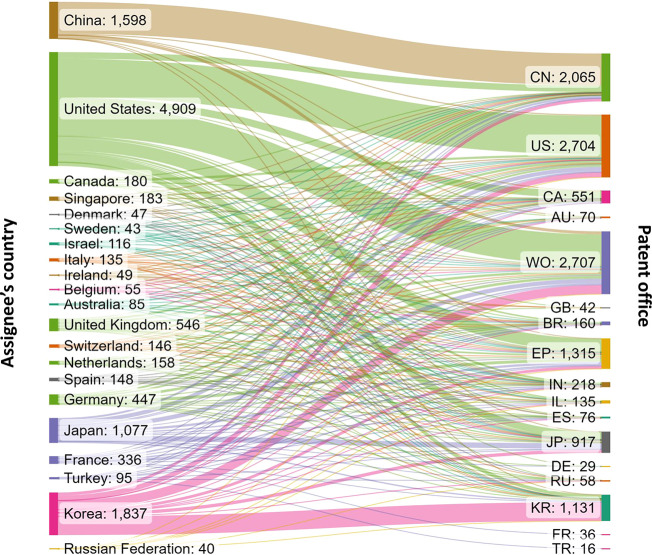
Patent protection is territorial, and thus, the same invention may be filed for patent protection in two or more jurisdictions. Therefore, we looked at all related filings on exosome applications in drug delivery and diagnostics. One patent family may be counted multiple times when it is applied in multiple patent offices. Figure 8 presents the flow of patent filings from different applicant locations to various patent offices of filing. There are diverse patent filing strategies: some patent assignees, such as those from China, file foremost in their home country patent office (CN), with a smaller proportion filing through the World International Patent Office WIPO (WO), or other jurisdictions. Others, for instance United States-based applicants, have a nearly equal number of US and WO filings and a considerable number of filings at other patent offices such as the European Patent Office (EP).
Figure 8.
Flow of patent filings related to exosome applications in therapy and diagnostics from different patent assignee locations (left) to various patent offices of filing (right). The abbreviations on the right indicate the patent offices of China (CN), United States (US), Canada (CA), Australia (AU), World Intellectual Property Organization (WO), Great Britain (GB), Brazil (BR), European Patent Office (EP), India (IN), Israel (IL), Spain (ES), Japan (JP), Germany (DE), Russian Federation (RU), Korea (KR), France (FR), and Turkey (TR).
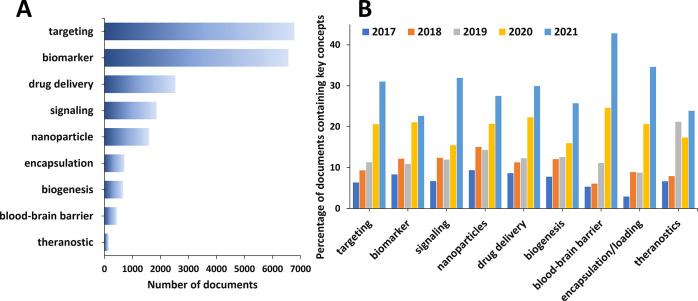
We explored the presence and trends of selected essential concepts relevant to the exosome applications in drug delivery and diagnostics as they appear in the scientific publications (Figure 9). With respect to the cumulative number of documents, “targeting” and “biomarker” appear as top concepts in the area (Figure 9A), reflecting the rising interest in the application of exosomes in therapeutics with specificity and diagnostics. It is noteworthy that the “blood–brain barrier” concept, although with a relatively low cumulative number of publications, exhibits the greatest growth rate in the past 2 years (Figure 9B), characterizing it as the trendiest concept in the field.
Figure 9.
Key concepts in the scientific publications relevant to the exosome applications in drug delivery and diagnostics. (A) Number of publications exploring key concepts related to exosome applications in therapy and diagnostics. (B) Trends in key concepts presented in the articles related to exosome applications in therapy and diagnostics during the years 2017–2021.
The landscape of exosome research as revealed from the CAS Content Collection is further explored in the later sections of this paper with respect to the exosome components and their roles.
Characterization of Exosomes
Exosome Pathway—Biogenesis, Secretion, Transport, Uptake
Exosomes are a population of extracellular vesicles. They are being secreted by many cell types using the endocytic pathway.54 The formation of exosomes includes three steps: (i) the endocytic vesicles form from the plasma membrane; these early endosomes mature into late endosomes; (ii) the endosomal membrane experiences inward budding, forming multiple intraluminal vesicles (ILVs) encapsulated within multivesicular bodies (MVB); (iii) the latter either fuse with the lysosome and bring the ILVs to degradation or access the cell membrane and discharge the ILVs in the form of exosomes (Figure 1).28,55 Thus, MVBs and late endosomes comprise ILVs, capturing certain proteins, lipids, and substances from the cytosol. The cytoskeleton and the microtubule network are the routes by which MVBs are transported to the cell membrane where they fuse with the cell membrane and undergo exocytosis. This way, the ILVs are being secreted as exosomes.56,57 Other MVBs exhibit degradation through lysosomes.
Indications exist that the endosomal mode of exosome formation—by endosomal budding—is not the only way of exosome biogenesis. Evidence has been accumulated indicating that exosomes may also bud from the plasma membrane directly.4,58−60 Altogether, the exosome biogenesis is a complex process with multiple participants involved in essential cellular functions.
The extracellular circulation half-life of exosomes has been estimated to be approximately 2–30 min according to reported pharmacokinetic profiles.61 Currently, there is certain knowledge regarding the exosome biogenesis and secretion, but there is still insufficient data regarding the uptake of exosomes by various cells and their signaling pathways. Internalization of the exosomes by the recipient cells follows the common endocytic pathways; e.g., it might be mediated by clathrin, lipid rafts, caveolins, through phagocytosis, or through micropinocytosis.57 Likewise, after internalization, exosomes follow the usual endosomal routes.62
Exosomes are membrane-bound carriers. Like other EVs, they are surrounded by a lipid membrane, which encloses their cargo. The typical exosome cargo includes mainly peptides, small proteins, and nucleic acids, such as mRNA, microRNA (miRNA), and non-coding RNA (ncRNA).63 These are used by the cell for signaling, to manage biological functions and to preserve homeostasis.64
Physiological Functions of Exosomes in Health and Disease
The intercellular traffic of exosomes plays a significant role in many physiological and pathological processes, including immune response, tissue homeostasis and regeneration, as well as in development of diseases such as cancer, neurodegenerative, cardiovascular, and other disorders. They are key players in cell–cell communication, signal transduction, extracellular matrix support and remodeling, and various other important physiological activities (Table 1). Furthermore, exosomes play significant roles in viral infections.65
Table 1. Roles of Exosomes in Health and Disease.
| Exosome role | Details and references |
|---|---|
| cell–cell communication | Exosomes can participate in an autocrine, paracrine, or endocrine communication reaching their target cells via the systemic or local circulation. They are important participants in cell communication including cell migration, proliferation, and senescence.66,67 |
| immune response | The cells of the immune system are known to release exosomes.29 Exosomes mediate immune modulation, both immunosuppression and immunostimulation.68 |
| signal transduction | Exosomes enable intercellular communication between various types of cells, regulating gene expressions and cellular signaling pathways of recipient cells by delivering their components, such as specific lipids, proteins, and RNAs. Certain lipid components including sphingomyelin, cholesterol, and ceramides have been involved in signaling;69,70 phosphatidylinositol-3-phosphate (PI3P) is also known to participate in regulating cell signaling.71 The presence of multiple kinds of signaling molecules—lipids, proteins, and RNAs—results in rapid signal changes in the target cell. |
| material (cargo) transport | Exosomes transport their constituents involving proteins, nucleic acids, lipids, and metabolites between cells, both in the close vicinity of the parent cell and at distant sites in the body carried by biofluids. It has been reported that RNA cargo of exosomes can modify gene expression in recipient cells.72,73 |
| pathogenesis | Viruses are known to make use of exosome biogenesis pathways to release a variety of pathogenic factors. Thus, a number of pathogen-derived components have been detected on exosomes after infection. These include, e.g., human immunodeficiency virus, Epstein–Barr virus, cytomegalovirus, hepatitis C virus, and herpes simplex virus.74 Exosomes play multiple roles in the progression of cancer via various communication pathways.75 Exosomes are more often released by tumor cells than by healthy ones and facilitate communication within the tumor microenvironment.76 |
| blood–brain communication | Exosomes are able to cross the BBB in both directions—from the brain to the bloodstream and from the blood to the CNS. Moreover, exosomes can interact with the BBB, leading to changes in the barrier’s properties.77 |
| target cell delivery | The delivery of cargos such as bioactive RNAs, proteins, metabolites, and/or lipid makes the capture of exosomes by target cells of vital importance in a variety of key biological processes such as angiogenesis,78 bone development,79 and cell migration.80 |
Exosome Composition
Nearly 100,000 proteins and over 1,000 lipids are found related to exosomes, along with a multitude of mRNAs and miRNAs, according to various available database collections such as ExoCarta,81 a web-based compendium of exosome proteins, RNA, and lipid database information;82 Vesiclepedia,83 a community compendium for extracellular vesicles; and EVpedia,84 a web-based resource providing bioinformatics tools for extracellular vesicles research.85 The contents enclosed into exosomes vary depending on the cell types and cellular conditions. Exosomes include proteins originating from the intracellular endosomal component. They include heat shock proteins, membrane transport proteins and fusion proteins, as well as a multitude of tetraspanins, a transmembrane protein family.86,87 With respect to lipid constituents, the exosomal content of cholesterol, sphingomyelin, saturated phosphatidylcholines, and phosphatidylethanolamines is higher than that of the plasma membrane.88
With respect to substance classes represented in the publications related to the exosome applications in drug delivery and diagnostics in the CAS Content Collection, nucleic acids have the highest share (Supporting Information Figure S1). Indeed, the capability of exosomes to carry nucleic acids from cell to cell is one of their major features attracting attention nowadays. As natural intercellular shuttles of RNAs, they affect many physiological and pathological processes and are the appropriate nanocarriers for targeted delivery of nucleic acids.89 Moreover, they have been identified as biomarkers for diagnosing of diseases, particularly various cancers. The RNAs, which have been examined include ncRNAs: microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) carried by exosomes.90
Lipids
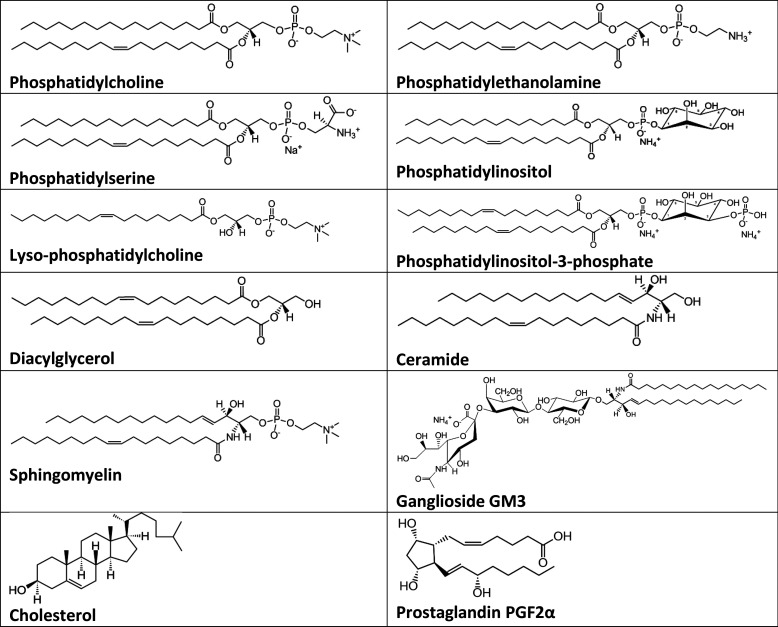
Lipids are essential constituents of biological membranes. As such, they are also abundant in exosomes. The major membrane lipid classes include phosphoglycerolipids, sphingolipids, and sterols. Certain lipid kinds are enriched in exosomes compared to their parent cells, suggesting that some membrane reorganization occurs upon exosome biogenesis.91 Lipid species enriched in exosomes include ceramides (Cer), sphingomyelins (SM), and gangliosides GM3 from the class of sphingolipids; phosphatidylserines (PS), phosphatidylethanolamines (PE), phosphatidylcholines (PC), lyso-phosphatidylcholines, and phosphatidylinositols (PI) from the class of phospholipids; diacylglycerols (DAG); as well as cholesterol (Figure 10).92,93
Figure 10.
Representative molecular structures of the major lipid classes in exosomes.
The parental cell types and their physiological status are determinants of the proportion of the lipid content in exosomes.94−96 Lipids are critical players in exosome biogenesis. As a result of their various molecular shapes—cone or inverse cone—they tend to generate negative or positive membrane curvatures. Lipids with large headgroups such as PIs and gangliosides or single-chain lipids such as lyso-PCs induce positive membrane curvature, while smaller headgroup lipids such as PEs or lipids lacking a hydrophilic headgroup such as ceramides and DAGs induce negative membrane curvature.97,98 Since these lipid classes are enriched in exosomes, they can significantly modify their membrane curvature. As a rule, membrane curvature is very important for cellular functions and trafficking.99
It has been reported that exosomes adjust their lipid composition to adjust to their biological function. Such enrichment of specific lipid classes with respect to the parental cells has been commonly observed.94,100,101 Thus, exosomes are typically enriched in cholesterol102 which supposedly accumulates in MVBs. It is important for the generation of intraluminal vesicles, the precursors of exosomes.100 Plasma membrane lipid rafts—ordered and tightly packed membrane microdomains organizing the assembly of signaling molecules for promoting signal transduction—are supposedly the origin of the high sphingomyelin content in exosomal membranes.100,103
The distribution of lipids in the two layers of the exosomal membrane lipid bilayer has been reported to be asymmetrical.93 Generally, asymmetric arrangement of lipids in the two membrane leaflets is a fundamental feature of the biological membranes. It is a consequence of various factors, including the biophysical properties of lipids, including their miscibility, the ionic composition of the media on both sides of the membrane, as well as the presence of transporter enzymes that actively support and maintain the lipid distribution across the bilayer, including flippases, floppases, and scramblases.104−106 Membrane asymmetry is believed to be associated with important biological functions such as apoptosis, cell fusion, and signaling.107 In the exosomal membranes, the sphingomyelin is typically found mostly in the outer leaflet and phosphatidylserines in the inner leaflet, while phosphatidylethanolamines seem to be randomly distributed across the bilayer.108 However, phosphatidylserine is externalized in apoptotic and malignant cells, attracting macrophages from the immune system.109 This finding is also useful from the viewpoint of possible use as exosomal lipid biomarkers for cancer diagnosis.110 Certain lipids are predominantly allocated in certain types of exosomes.111 The process of biogenesis of exosomes and cargo packaging appears as a well-controlled process with lipids playing an important role.112
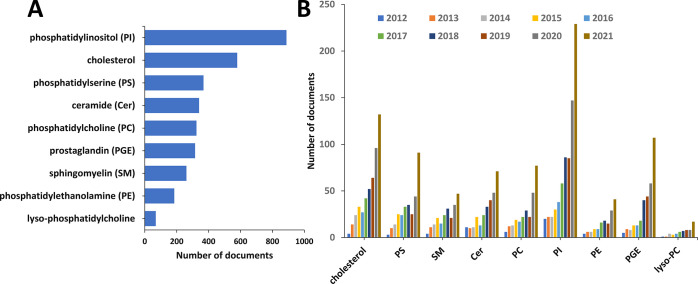
Figure 11 illustrates the results from a search on various lipid classes in the CAS Content Collection in documents related to the medical application of exosomes. Phosphatidylinositol and its derivatives appear to attract significant attention in the field. Seven phosphorylated phosphatidylinositols (PIPs) have been identified in membranes. They can be transformed into each other by phosphorylation or dephosphorylation of the 3-, 4-, and 5-hydroxyl groups of the inositol headgroup (see the example of PIP3 in Figure 10). PIPs are known as precursors for certain second messengers involved in signal transduction and, noteworthy, regulate membrane dynamics and vesicular transport. They have been reported to significantly affect exosome secretion69 and macrophage targeting,113 which may have provoked the strong attention in the published literature (Figure 11).
Figure 11.
Number of documents mentioning specific types of lipids related to exosome applications in therapeutics and diagnostics. (A) A top list of classes of lipids with the numbers of associated documents. (B) Annual growth of document numbers.
Cholesterol is another lipid that attracts strong attention in the recent exosome publications (Figure 11). Indeed, cholesterol has been reported to be essential for the entire development of exosomes, for their biogenesis and release, for their membrane stability, and for entrance into the target cells.114 Furthermore, reports show that exosomes constitute part of the cellular machinery taking care of the cholesterol balance and that they can assist in detecting and combating cholesterol-related pathologies.114
Exosomal enzymes are responsible for the production of bioactive lipids in exosomes. For example, exosomes contain A2 phospholipases, which hydrolyze glycerophospholipids to generate arachidonic acid and other free fatty acids.115 Arachidonic acid can be processed to release leukotrienes such as LTB4, involved in the inflammation, and LTC4/LTD4 which promote angiogenesis. Exosomal cyclooxygenases COX1/2 and the PGE synthase promote the transformation of arachidonic acid to the pro-inflammatory prostaglandin E2 (PGE2) (structure included in Figure 10) or to the anti-inflammatory and tumor-suppressing 15-deoxy-prostaglandin J2.63,115 The role of exosomes in mediating lipid metabolism during cancer progression is attracting much attention recently. Bioactive exosomal lipids, e.g., the prostaglandins PGE2α, PGE1, and PGE2, are known to be released from macrophages into the cancer microenvironment.116 Such bioactive lipids are a subject of increasing interest in the recent exosome publications (Figure 11).
Proteins
Exosomes comprise a broad collection of proteins including transmembrane proteins, lipid-anchored membrane proteins, peripheral membrane proteins, as well as soluble proteins inside the exosome core (Table 2).4
Table 2. Exosomal Proteins with respect to Their Location and Role.
| Proteins | Examples |
|---|---|
| Exosomal Proteins with respect to Their Location | |
| integral transmembrane proteins117−119 | tetraspanins (CD81, CD82, CD37, CD63) |
| lipid-anchored outer membrane proteins43,120−123 | ectonucleotidases (CD39, CD73); sperm receptor Juno; complement-inhibiting proteins (CD55, CD59); glypican-1; prion proteins (PrPC, PrPSC) |
| lipid-anchored inner membrane proteins34,35,58,124−127 | prenylated GTPases (Rabs, Ras, Rho), myristoylated signaling kinases (Src), palmitoylated membrane proteins, acylated Gag proteins |
| peripheral outer surface proteins128−135 | wingless (Wnt) proteins; BMPs; TGF β; tumor necrosis factor (TNF); cytokines; extracellular matrix (ECM) proteins (fibronectin, tenascin C, ECM1) |
| peripheral inner membrane proteins4,102,136−142 | scaffolding factors including the ezrin-radixin-moesin (ERM) proteins and ERM ligands (EBP50, CD43, CD44, IGSF8, PTGFRN); syntenin; ALIX; heat shock proteins (HSP70, Hsp40/DnaJ proteins, Hsp90, Hsp20, Hsp27, α/β-crystallins) |
| enzymes27,120,143,144 | ATPase, pyruvate kinase, fatty acid synthases, phosphatases, pyrophosphatases, calcium-binding annexins, phosphate transporters; RNA editing enzymes, lipases, proteases, glycosyl transferases, glycosidases, metabolic enzymes |
| cytosolic proteins117,145 | clathrin, HSC70, HSP70, HSP60, HSP90, ALIX, YWHAE, ubiquitin, TSG101, ESCRT |
| Exosomal Proteins with respect to Their Role | |
| membrane transport and fusion-related proteins | annexin, Rab-GTPase, heat shock proteins (HSPs), e.g., Hsp60, Hsp70, Hsp90 |
| membrane organization and trafficking | tetraspanins: CD9, CD63, CD81, CD82, CD106, Tspan8, ICAM-1 |
| multivesicular-body (MVB)-related proteins | ALIX, TSG101 |
| cell-adhesion-related proteins | integrins |
| cytoskeletal proteins | actin, myosin |
Exosomes are rich in certain tetraspanins such as CD81, CD82, CD37, and CD63, representatives of the class of the integral membrane proteins.117 They are membrane proteins including four transmembrane α-helices. Tetraspanins are believed to have an important role as organizers of transmembrane and cytosolic proteins, as well as lipids (e.g., cholesterol) into a membrane network, the tetraspanin web. Tetraspanins do not exhibit catalytic activities; their function is to facilitate the trafficking, functioning, and stability of the other membrane proteins.118 CD81 and CD63 are frequently used exosomal marker proteins, as well as CD9, another exosomal tetraspanin, which mediates the membrane metalloendopeptidase CD10 loading into exosomes.118,119 Certain surface proteins alter the exosome circulation time. For example, the occurrence of CD47 or CD55/CD59 on the exosomal membrane can extend the blood circulation time by avoiding phagocytic clearance.12 CD47 has been found on the surface of EVs secreted by fibroblasts, T-cells, and MSCs,146 and EVs secreted by antigen-presenting cells and retinal pigmented epithelium express CD55 and CD59.147
Lipid-anchored proteins including some glycosyl-phosphatidylinositol-anchored proteins are present on the exosome surface. Between them are the ectonucleotidases CD39 and CD73,120 the complement-inhibiting proteins CD55 and CD59,121 glypican-1,43 prion proteins,122 and the Hedgehog morphogens attached to the outer layer by their cholesterol portion.123
Exosomes also involve peripheral surface proteins playing key roles in signaling. Representatives of this class are the wingless (Wnt) proteins,128 surface-bound bone morphogenetic proteins,129 transforming growth factor β,130 tumor necrosis factor,131 cytokines,132 and a large collection of other surface signaling molecules. Extracellular matrix proteins including fibronectin, tenascin C, and ECM1 are also at the exosome surface.133,134 Some peripheral proteins are attached to the exosomal phosphatidylserine.135
The exosome inner membrane leaflet carries acylated, lipid-anchored proteins, including prenylated GTPases, myristoylated signaling kinases, and palmitoylated membrane proteins.34,35,124 A large part of the composition of exosomes released by infected cells include acylated Gag proteins.58,125 Furthermore, some Gag proteins, such as the activity-regulated cytoskeletal (ARC) protein, play a critical role in cognition.126,127
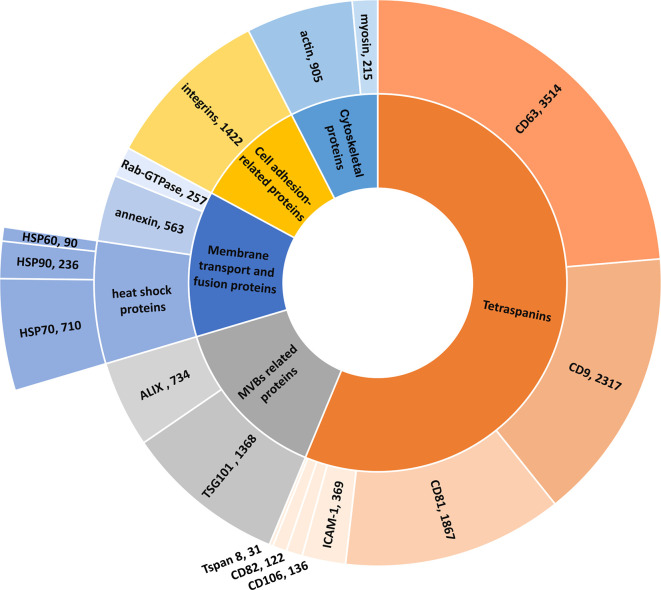
The distribution of publications in the CAS Content Collection among exosomal proteins is presented in Figure 12. Because of the high content of tetraspanins in exosomes, they are most often used as specific exosome markers,117 which explains the abundance of tetraspanin-related publications in those concerning exosome applications in therapy and diagnostics (Figure 12). The distribution of documents concerning the major tetraspanin classes as well as their roles, in particular in therapy (THU) vs diagnostics (DGN), are shown in Supporting Information Figure S2.
Figure 12.
Number of documents concerning major exosome proteins in the documents related to exosome applications in therapeutics and diagnostics.
Nucleic Acids
Exosomes comprise RNAs and can transfer them to other cells and tissues. The exosome-mediated RNA transfer has been initially reported for mRNAs and miRNAs.32,33,148,1494,149,150 Exosomal RNA pools are enriched in small non-coding RNAs (sncRNAs) and differ from the cellular RNA profile.72,151,152 A wide variety of RNA species are embedded in the extracellular vesicles. The extracellular vesicles compendium Vesiclepedia lists over 10,000 entries of EV miRNAs and over 27,000 entries of EV mRNAs.83 Upon internalization of exosomes by the recipient cells, the variety of cargo RNA species can be released. The subsequent translation of mRNA into active proteins may result in phenotypical changes.32
miRNA is the dominating RNA found in exosomes. It has been reported that exosomal miRNAs play an important role in intercellular communication. Multiple examples of EV-mediated transfer of miRNAs have been established for a variety of physiological and pathological events. Reports show that it is highly abundant in exosomes, with the five most common miRNAs being miR-99a-5p, miR-128, miR-124-3p, miR-22-3p, and miR-99b-5p.153 mRNAs carried in the extracellular vesicles can serve as a source of proteins in recipient cells. Active translation of exosomal mRNAs into recipient cells was reported, such as expression of reporter proteins from mRNA transferred by extracellular vesicles between mast cells and from glioblastoma to endothelial cells.32,33
Exosomes also contain multiple kinds of DNAs, including single-stranded, double-stranded, genomic, and mitochondrial DNAs.154−157 Certain inflammatory processes and cell aging are thought to rationalize the presence of DNA in exosomes.64 There is more DNA in exosomes derived from cancer cells than from healthy cells. It has been suggested that DNA secretion originating form exosomes may affect the inflammation regulation. It is viewed as a potential marker of cancer, viral infection, or chemotherapeutic resistance.4,155,158
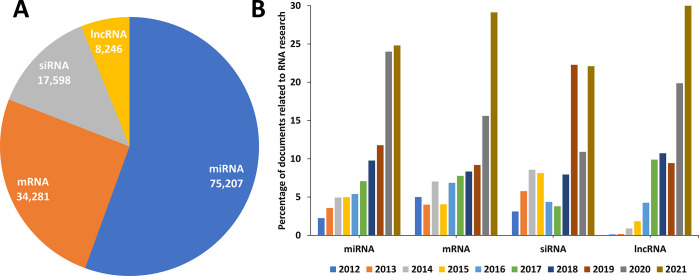
Analysis of the data available in the CAS Content Collection confirmed that exosomal RNAs are dominated by miRNAs (Figure 13A). Annual distribution of the RNA-related documents within the pool of publications concerning exosome applications in therapy and diagnostics shows an explosive growth in the last 2–3 years for all RNA types (Figure 13B), which is impressive, even considering the overall rapid growth in interest in all RNA medicines.150
Figure 13.
Types of RNA molecules in exosome applications and their document counts. (A) Distribution of the number of documents related to exosome applications in therapy and diagnostics concerning various RNAs during the years 2012–2021. (B) Annual trends in the number of the same documents. (Percentages are calculated with yearly publication numbers for each RNA, normalized by the total number of publications for the same RNA in the same time period.)
Glycoconjugates
Polysaccharides and glycans are other exosomal constituents located on their outer surface.159,160 They are predominantly enriched in
mannose
α-2,3- and α-2,6-sialic acids
complex N-linked glycans
high-mannose N-glycans
heparan sulfate
polylactosamine
The role of glycans in exosome biology is less well understood than that of proteins, lipids, and nucleic acids, yet there is evidence that surface glycoconjugates play important roles in exosome biogenesis, release, targeting, and uptake by cells.161 Glycoconjugates appear to be an additional source of exosome biomarkers as well (Table 3), since variation in glycosylation is characteristic of different types of cancers.162,163
Table 3. Examples of Glycoconjugate Tumor Markers in Exosomes.
| Tumor markera | Cancer | Body fluid/cells |
|---|---|---|
| α-fetoprotein | liver and germ cell tumors | blood |
| β-human chorionic gonadotropin | choriocarcinoma, germ cell tumors | urine, blood |
| C-kit/CD117 | gastrointestinal tumor, melanoma | tumor cells |
| CA15-3/CA27.29 (MUC1) | breast cancer | blood |
| CA19-9 | pancreatic, gallbladder, bile duct, stomach cancers | blood |
| CA-125 (MUC16) | ovary cancer | blood |
| carcinoembryonic antigen (CEA) | colorectal cancer | blood |
| estrogen receptor (ER) | breast cancer | tumor cells |
| HE4 | ovary cancer | blood |
| HER2/neu | breast, stomach, gastroesophageal adenocarcinoma | tumor cells |
| prostate-specific antigen (PSA) | prostate cancer | blood |
Tumor markers selected from https://www.cancer.gov/about-cancer/diagnosis-staging/diagnosis/tumor-markers-fact-sheet.
Applications of Exosomes
Methods for Exosome Isolation/Purification
Speedy, straightforward isolation methods offering high purity and recovery are a requirement for large-scale applications of extracellular vesicles in clinics. Each of the available methods discussed below brings about certain advantages and disadvantages to exosome isolation and purification. Based on the application purpose, different methods can be applied for exosome separation and analysis.
Ultracentrifugation-Based Isolation Techniques
Ultracentrifugation is capable of generating very high centrifugal forces, up to 1,000,000g, and is currently one of the most frequently used methods for exosome isolation,164 considered as a gold standard before 2015 (Figure 14). This approach does not require much expertise and is affordable over time. Furthermore, it is not rather time-consuming, without considerable sample pretreatment. Differential ultracentrifugation (DUC) and density gradient ultracentrifugation (DGUC) are the two popular kinds of preparative ultracentrifugation.
Figure 14.
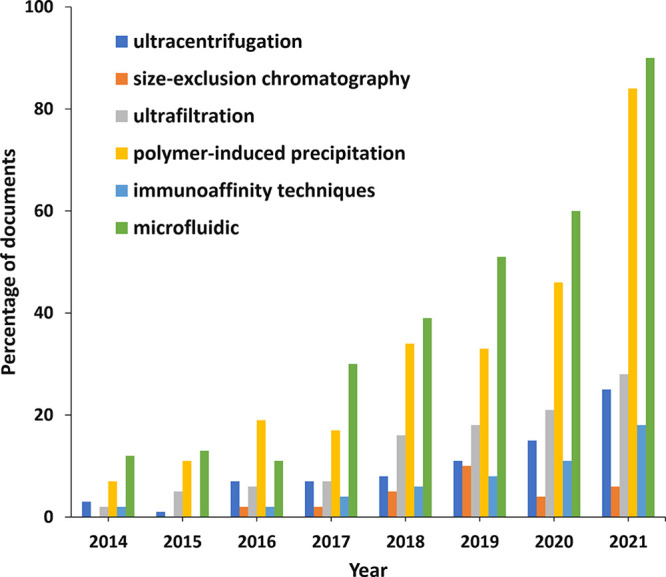
Trends in the number of documents related to exosome applications in therapeutics and diagnostics concerning various exosome isolation methods during the years 2014–2021. (Percentages have been calculated with yearly publication numbers for each isolation method, normalized by the total number of publications for the same isolation method in the same time period.)
DUC includes several steps with continuously increasing centrifugation forces and durations, with the purpose of sequentially isolating smaller particles from large ones, such as whole cells, cellular debris, and macromolecular proteins. Finally, exosomes are separated by ultracentrifugation at 100,000–150,000g. The technique is time- and effort-consuming compared with DGUC, because of the multiple steps. In addition, heterogeneity of exosomes and overlap in the size of extracellular vesicles lead to contaminations.165−167
The separation of particles by DGUC is based on their size, shape, and density by utilizing an inert medium of graded densities.168 Under a given centrifugal force, components of a sample will reside in the zone corresponding to their density. This technique has a relatively higher separation efficacy and thus provides higher purity. It is noteworthy that exosomes are not likely to be crushed during the separation.166 However, this method also has the issue of unwanted aggregation of particles as well as the contamination of proteins and nucleic acids.
Ultrafiltration
Ultrafiltration is a size-based technique frequently used for exosome isolation. Exosomes are being isolated using membrane filters with a specific pore size defining their molecular weight or size-exclusion limits. A microfluidic device consisting of ciliated micropillars has been fabricated to isolate exosomes.169 Commercial exosome isolation kits have been designed for exosome isolation from serum, cell culture medium, and other body fluid.170 The kits apply rapid fractionation including a syringe filter with two membranes to capture the exosomes and the larger extracellular vesicles. It is noteworthy that a combination method of ultrafiltration with size-exclusion chromatography (SEC) has been successfully applied in the isolation of exosomes from adipose tissue.171
Immunoaffinity Techniques
As more cell-type and disease-specific protein receptors in the exosomal membrane are being identified, opportunities are created to develop highly efficient techniques for exosome isolation. Immunoaffinity techniques have been developed employing the affinity interactions between surface proteins and corresponding antibodies. Preferably, markers for exosome immunoisolation are membrane-attached without soluble parts and are only located on the surface of exosomes. Thus, the widely popular enzyme-linked immunosorbent assay (ELISA) has been established for isolating and quantifying exosomes from sources such as blood serum, plasma, and urine.172−176 ELISA results are typically expressed as absorbance measures to enable a quick comparison with standards of known exosome counts, thus enabling absorbance measures to be calibrated to quantify the resultant exosomes. By means of the microplate immunoaffinity approach, the distinction and yield of exosomes can be assessed with respect to ultracentrifugation. This method is highly specific, resulting in extremely pure exosome populations. A corresponding kit was developed for exosome isolation based on this theory and enables fast isolation of high-purity and high-yield exosomes.177
Exosome Precipitation—Polymer-Based Precipitation Techniques
By modifying their solubility or ability to disperse, exosomes can be precipitated from biological bodily fluids. Polymers that exclude water such as PEG are utilized for this purpose. Such polymers bind water and components of lower solubility out of the solution. Samples are incubated with a PEG (MW ∼ 8000 Da) precipitation solution. PEG binds water molecules and thus expels less soluble components out of the solution.178 Subsequently, the sediment containing exosomes is settled out by centrifugation or filtration. This approach is easy to conduct and scalable for large sample size, which allows easy transition to clinical applications. To date, several commercial kits utilizing PEG for the isolation of exosomes have been developed. One of the most widely used kits is ExoQuick (System Biosciences, Mountain View, CA, USA).171 Some of these kits are developed to be compatible with body fluids, as well as culture medium. Selected kits are summarized and discussed in the following section. Samples usually require precleaning from cells and cellular debris before carrying out the precipitation. Urinary exosome precipitation by these kits has been shown to achieve the highest yield compared to other techniques. The disadvantage of the method is that the presence of the polymer may affect downstream analysis due to the positive charged molecules.179
Microfluidics-Based Isolation Techniques
These techniques utilize high-throughput microfluidic tools to isolate exosomes based on concepts including size, density, and immunoaffinity. The immuno-microfluidic method is developed based on the immunoaffinity capture technique. Exosomes are isolated by the specific binding of antibodies immobilized on the microfluidic chips and bind specifically to exosome markers (antigens).180−182 The advantages of this technique include efficient, speedy processing and high grade of purity. Modifications of the microfluidic method such as size-based microfluidic isolation using the exosome total isolation chip (ExoTIC),183 acoustofluidic,184 and dielectrophoretic185 techniques have been successfully applied. The tools are very complicated and expensive with requirement of specific fabrication skills. In summary, microfluidics is an advanced and promising technology, but it still needs certain regularization in order to be considered as a standard exosome isolation method.
Size-Exclusion Chromatography
According to the accumulating evidence, size-exclusion chromatography (SEC) has been considered as the most preferred method for isolation and purification of exosomes.186 Exosome isolation using SEC has a low level of contaminants, resulting in a homogeneous isolation of exosomes. This circumstance has promoted the use of SEC among its competitor techniques for body fluid exosome-related biomarker identification. SEC has been used successfully for isolation, purification, and enrichment of exosomes from an assortment of biological fluids including plasma, serum, urine, cerebrospinal fluid, saliva, milk, and tears. SEC is advantageous because it does not require a large sample volume and the shearing force generated in SEC does not likely damage the original structure of the vesicles. These distinctive properties make this technique preferable compared to centrifugation.187 Presently, SEC is a commonly used technique for isolation of exosomes from both blood and urine samples.188,189
A summary of the most widely used exosome isolation techniques is highlighted in Table 4, including the isolation mechanisms, advantages, and disadvantages.168,186,190−192 Other isolation methods and method modifications have also been applied, e.g., asymmetric flow field-flow fractionation (AF4),193 aptamer-based isolation,194 and others.168 Even though various exosome purification approaches have been developed, it is difficult for one method to solve all the associated challenges such as low yield, contaminations, and variations between batches. The combination of several methods to isolate and purify exosomes would be needed to characterize exosomes effectively and comprehensively. And it has been suggested as a promising strategy for improvement of the isolation outcome, in order to provide exosome subsets with high purity, in particular with respect to size, morphology, density, number, presence of exosome-enriched markers, and lack of contaminants.168
Table 4. Major Methods of Exosome Isolation/Purification.
| Method | Principle | Advantages | Disadvantages |
|---|---|---|---|
| ultracentrifugation | density- and size-based sequential separations | • appropriate for large-volume samples | • high equipment cost |
| • markers not introduced | • labor-intensive | ||
| • cost-effective | • potential damage of exosomes | ||
| • low yield | |||
| ultrafiltration | using a membrane filter with a defined size-exclusion limit or molecular weight cutoff | • low cost | • potential damage of exosomes |
| • time efficient | • membrane clogging and blockage | ||
| • simple | |||
| immunoaffinity | exosome capture based on antigen–antibody-specific recognition and binding | • high specificity | • potential damage of exosome integrity |
| • simple | • expensive reagents | ||
| • scalability | • non-specific binding | ||
| polymer precipitation | hydrophilic water-excluding polymer adhering and precipitating exosomes | • broad applicability | • lack of specificity and selectivity |
| • simple and rapid | • low purity | ||
| • no exosome deformation | • contamination with polymers | ||
| microfluidics | immunoaffinity, size, density | • high efficiency | • large volumes of starting materials |
| • fast processing | • low sample capacity | ||
| • good portability | |||
| • easy automation and integration | |||
| size-exclusion chromatography | exosome separation based on hydrodynamic radii | • preserve biological activity | • potential contamination |
| • no preprocessing | • high equipment cost | ||
| • high yield |
The annual trends in the number of documents related to exosome applications in therapeutics and diagnostics concerning various exosome isolation methods during the years 2014–2021 are shown in Figure 14. The precipitation and microfluidic methods are dominating the field, because of their broad applicability and high efficiency.
Exosomes as Drug Delivery Vehicles
Exosomes provide distinct benefits as highly efficient drug carriers. They have been recognized as a successful platform for delivery of various drugs because of their ability to mediate cellular communications.195 Exosomes can be modified by means of their parental cells to exhibit the desired targeting capability and being loaded with therapeutic agents with anticipated biological activity. Exosomal drug formulations are applicable to many diseases including cancers and infectious, cardiovascular, and neurodegenerative disorders. Generally, exosomes exhibit a combination of advantages characteristic of both synthetic drug carriers and cell-mediated delivery methods, at the same time preventing their drawbacks.
Multiple encapsulation approaches for exosomes utilizing physical/chemical/biological techniques have been developed for stocking therapeutic agents into exosomes, to achieve diverse therapeutic effects and optimum efficiency.
Cargo Loading
Therapeutic agents can be introduced into exosomes either before or after exosome isolation.5,195−197 Pre-isolation loading methods introduce the therapeutic molecules into the parental cells before the EV production, so that they are encapsulated before exosome biogenesis.
Cell transfection of RNA, peptides, and proteins has been used.198,199 This is the most commonly used approach for loading therapeutic molecules into exosomes. Another way of pre-isolation cargo loading comprises simple incubation of the parental cells with the drug, allowing passive diffusion of the drugs into cells or exosomes during their biogenesis.200
Advantages: Appropriate for loading nucleic acids and proteins; large cargos
Disadvantages: Cytotoxicity, difficult purification
Post-isolation loading methods introduce the therapeutic agent after the exosome being collected applying techniques such as co-incubation, sonication, electroporation, freeze–thaw cycles, and extrusion.197,201 Most of these methods have been acquired from their application in the liposome-based drug delivery.
In the direct co-incubation method, the therapeutic agent and the exosomes are mixed and incubated for a certain time period at room temperature. It is driven by the passive transport mechanism exploiting the concentration gradient. As a result, therapeutic small molecule drugs enter through the membrane into the exosomes or cells, with subsequent secretion of drug-loaded exosomes.202 Loading is highly dependent on the drug hydrophobicity, with hydrophobic molecules being loaded more efficiently into exosomes.203 An incubation time of ∼90 min has been reported to result in the most efficient loading of exosomes with synthetic oligonucleotides.204 The size of the drug molecule is a substantial loading controlling factor.205 Loading capacity can be strongly modulated by tuning the cells/exosomes–drug ratio.
Advantages: Simple, convenient, mild
Disadvantages: Low loading efficiency
Sonication is a technique using mechanical energy to produce temporary pores in the exosomal membranes allowing the cargo to be encapsulated, with subsequent reorganization and recovery of the lipid bilayer.197,206 Sonication exhibits higher loading efficiency, but it could cause deformation of exosomes with subsequent compromising of their integrity. Also, sonication may lead to heating and damage of the active agents; therefore, careful temperature and process controls are critical.197,205
Advantages: High loading efficiency
Disadvantages: Heat generation, possible active agent damage, aggregation
Electroporation makes possible the entry of the therapeutic cargo by applying electrical pulses to modify the dielectric properties of the membrane, thereby opening recoverable pores and enhancing its permeability.205 This way of permeabilization of exosome membranes is one of the most common techniques applied for exosome loading. The applied potential can vary significantly in different cases, from 0.1 to 1000 kV. Disruption of the membrane lipid bilayer allows hydrophilic compounds such as small DNAs,207 miRNAs,208,209 and siRNAs210 to diffuse into exosomes. The method is simple to operate, has a high loading efficiency, and has been widely applied to encapsulate siRNAs or miRNAs. However, possible aggregation of therapeutic nucleic acids during loading caused by metal ions originating from the electrodes is a likely disadvantage.211,212 Aggregation can be prevented by using protectors such as the trehalose, citric acid, and EDTA.212
Advantages: High loading efficiency, controllable
Disadvantages: Cargo aggregation
Freeze–thaw cycles are also successfully used for drug loading after exosome isolation. The exosomes are being frozen together with the drug in liquid nitrogen at −80 °C with subsequent thawing at room temperature for several cycles.205 A minimum of three freezing–thawing cycles is needed, and 5–10 cycles are recommended.213 It is a relatively mild method appropriate for miRNA and protein loading.210 Drug penetrates through exosomal membrane as a result of minor disordering of the lipid bilayer during the procedure. Moderate loading efficiency is characteristic for this method.210 It can be combined with the co-incubation and/or sonication techniques for enhancing efficiency.214 The freeze–thaw technique has been successfully used to fuse exosomes and liposomes, thus producing exosome-mimetic particles.213
Advantages: Mild and simple, appropriate for RNA and protein loading
Disadvantages: Uncertain efficiency, aggregation
The extrusion technique includes forcing the exosomes to mechanical destruction by using an extruder device. The device has a heating block and polycarbonate filters with specific pore sizes (usually ∼100–400 nm). The exosome components are subsequently reconstructed into a population of nanovesicles incorporating the intended drug.215 The method exhibits good loading efficiency, but the applied excessive shear stress can damage the vesicles and their protein components.205 The extrusion method has been found to be appropriate in constructing exosome-mimetics.5
Advantages: Good loading efficiency, uniform size
Disadvantages: Possible damage of the exosome membrane
The distribution of documents in the CAS Content Collection related to exosome applications in therapy and diagnostics with respect to the applied exosome loading methods is illustrated in Figure 15. Dominating are physical methods—electroporation, freeze–thaw, sonication, and extrusion—while chemical and biological methods, such as transfection and incubation, are less popular. In fact, various methods turn out to be appropriate for different cargo loadings.
Figure 15.
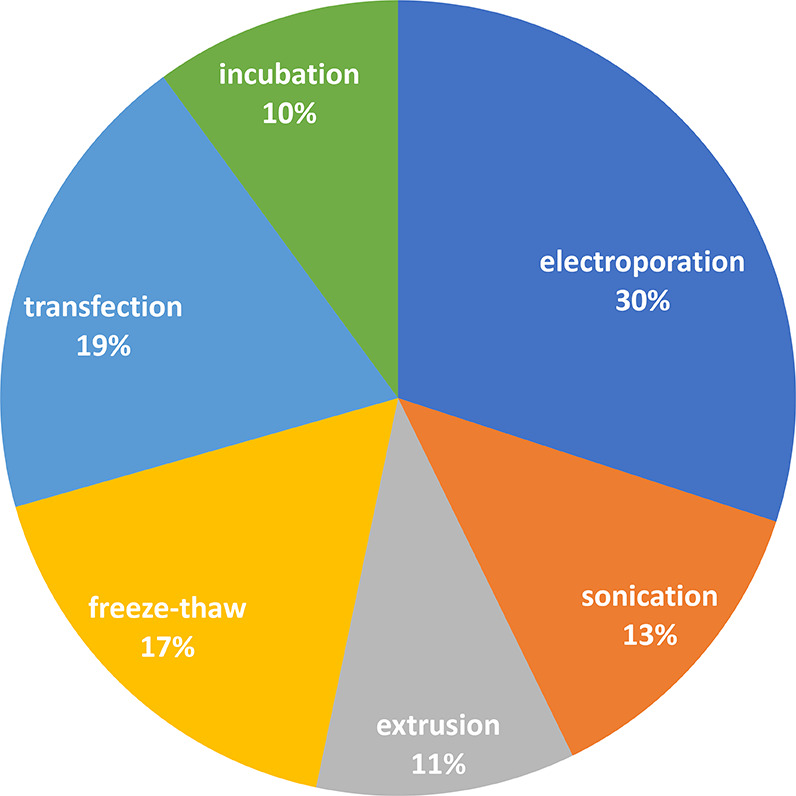
Percentages of documents related to exosome applications in therapy and diagnostics concerning various exosome loading methods.
A selection of small molecule drugs, which have been frequently used as exosome cargo in drug delivery, as seen from the CAS Content Collection, have been exemplified in Supporting Information Table S1.
Cell Sources for Derived Exosomes
As a form of cell–cell messenger, exosomes play a crucial role in different physiological processes. Exosomes secreted by different tissues and cells exhibit specific properties. Moreover, understanding the properties of different cell-derived exosomes can also help us understand the pathogenesis mechanism of various diseases.
Exosomes Derived from Tumor Cells
Tumor-derived exosomes are able to modify tumor progression, including growth, angiogenesis, invasion, and metastasis. They promote cell development, adhesion, and cell polarity.216,217 Exosomes derived from tumors may be involved in various immunomodulatory outcomes, since they carry both immunosuppressive and immunostimulatory mediators. Furthermore, tumor-derived exosomes may be used as immunoadjuvants and antigens in cancer vaccines.218 They secrete cytokines and growth factors and can thus protect T-cells from cancer-cell-mediated apoptosis.219 Also, tumor-derived exosomes exhibit a composition related to that of their cells of origin. When administered, they prefer to fuse with their parent cancer cells, indicating that exosomes might be distinctively suitable, as Trojan horses, to deliver anticancer therapeutics.220
Exosomes secreted by cancer stem cells mediate cell–cell communication and substance exchange, thus regulating processes of tumor growth metastasis, epithelial–mesenchymal transition, and angiogenesis by transporting tumor-related mRNA, non-coding RNA, surface proteins, and encapsulated proteins.221 In colorectal cancer, exosomes derived from fibroblasts activate the Wnt signaling pathway, rendering cancer cells to exhibit stem cell properties, including spherocytosis and tumorigenicity, and increase the number of cancer stem cells in colorectal cancer cells.222 Also, exosomes derived from mesenchymal stem cells can boost breast cancer cell proliferation by activating the Wnt signaling pathway.223 Growing evidence indicates that targeting signal pathways regulated by exosomes could act on CSCs to inhibit the incidence and development of tumors, which has become a trending topic in recent years.
Mesenchymal-Stem-Cell-Derived Exosomes
Mesenchymal stem cells (MCS) are pluripotent stem cells and can be derived from certain adult tissues and organs. MSCs are an ideal exosome source. They inhibit the proliferation of immune cells. MSC-derived exosomes inherit the immunomodulatory properties.224 In addition, MSCs have exhibited the highest amount of CD81 expressed exosomes.225,226 It has been shown that upregulated miRNAs, especially miR-320C, from MSC-derived exosomes promote osteoarthritis chondrocyte proliferation. In a myocardial I/R injury study, MSC-derived exosomes carrying miR-182-5p showed a cardioprotective effect with improving cardiac function and reducing myocardial infarction, accompanied with reduced inflammation in vivo.227 Exosomes from mesenchymal stem cells play an important role in many diseases and can be used as an adjuvant in supporting and complementing other therapeutic modalities. Bone marrow MSC-derived exosomes are being utilized by Direct Biologics, a regenerative biologic company, in many different clinical trials.228 Their therapeutic product ExoFlo is currently available under FDA expanded use protocol for the treatment of COVID-19 acute respiratory distress syndrome (ARDS) (NCT04657458).229,230 It is also under clinical trial for ulcerative colitis (NCT05176366),231 Crohn’s disease and irritable bowel disease (NCT05130983),232 solid organ transplant rejection (NCT05215288),233 and mild/moderate COVID-19 (NCT05125562).234
Macrophage-Derived Exosomes
Macrophages are known to exhibit phagocytic ability in the immune system.235 They are able to identify and eliminate pathogenic microbial products and tumor cells and are thus important for the prevention of diseases.236 Studies have reported that they are an essential regulator in injury and repair. After chemotherapy, macrophage-derived exosomes stimulate breast cancer proliferation and metastasis. Thus, inhibition of exosome secretion is identified as beneficial for breast tumor prevention.237 M2-macrophage-derived exosomes could promote cardiac repair in a mouse model of acute myocardial infarction. miR-1271-5p-enriched macrophage-derived exosomes suppressed cell apoptosis and enhanced the viability of hypoxia-induced cardiomyocytes. By downregulating SOX6, miR-1271-5p decreased cardiomyocyte apoptosis induced by hypoxia and alleviated cardiac injury.238
Exosomes Derived from T-Cells
Exosomes derived from T-cells are a subject of growing interest because of their potential role in controlling innate immune responses. Similarly to the exosomes from other sources,32,239 these exosomes carry bioactive miRNA.240 Exosomal carriers can transport miRNA from T-cells to antigen-presenting cells.240
In addition to modulating the immune response, T-cell-derived exosomes participate in tumor inhibition. T-cell secreted exosomes containing Fas ligand promote tumor infiltration in lungs by enhancing the expression of matrix metalloproteinase 9,241 and exosomes released from CD8+CD45+ regulatory T-cells inhibit the response of the CD8+ cytotoxic T-lymphocyte and the antitumor activity.242 Exosomes contribute to the tolerance to transplantation as well.243 Through a clinical trial (NCT04389385), TC Erciyes University in Turkey is researching the use of COVID-19-specific T-cell-derived exosomes.244 This clinical trial is testing the safety and efficacy of the agent following a metered inhalation for targeted delivery (Turk-Patent Application Number: PCT/TR2020/050302).244
Exosomes Derived from Other Cells
A lot of studies have been dedicated to identifying the roles of other living-cell-derived exosomes. Exosomes obtained from fibroblasts rich in miR-21-3p could induce cardiomyocyte hypertrophy by targeting SORBS2 and PDLIM5. Inhibition of miR-21-3p diminished cardiac hypertrophy in animals treated with Ang II. Exosomes extracted from endothelial cells expressing KLF2 can attenuate the formation of atherosclerosis. Exosomes derived from neural stem cells are being researched by Aruna Biomedical for the treatment of stroke along with other neurological and neurodegenerative diseases. Their candidate AB126 shows the ability to cross the BBB and demonstrates central nervous system specificity.245 Their preclinical data supports that neural stem-cell-derived exosomes were more effective than MSC-derived exosomes in improving cellular, tissue, and functional outcomes in the tested mouse thromboembolic stroke model.245
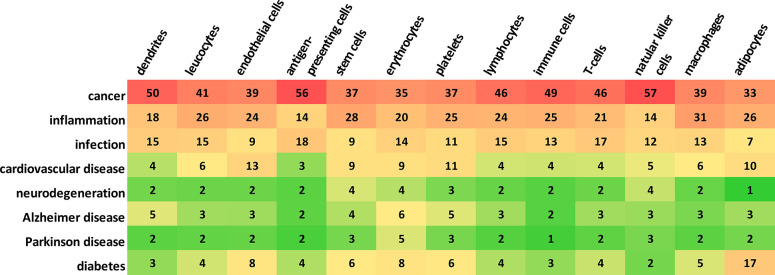
The frequency of using various kinds of exosome donor cells in the studies related to exosome applications in therapy and diagnostics, as presented by the number of documents in the CAS Content Collection, is illustrated in Supporting Information Figure S3. Tumor cells and stem cells (specifically, mesenchymal stem cells, MSC) are the most frequently used exosome sources. Figure 16 illustrates the correlation between the exosome donor cells and the diseases to which the exosomes have been applied to in studies related to exosome application in therapeutics and diagnostics, as represented by the number of documents in the CAS Content Collection. Cancer studies clearly dominate, followed by inflammation and infection studies. Furthermore, in cancer studies, antigen-presenting cells and natural killer cells have been frequently used. Macrophages and stem cells are the most frequently used in inflammation, while antigen-presenting cells and T-cells are frequently used in infection.
Figure 16.
Correlation between exosome donor cells and the diseases to which the exosomes have been applied to in the studies related to exosomes in therapy and diagnostics, as represented by the number of documents in the CAS Content Collection.
Delivery of Small Molecules
Exosomes have been recognized as prospective vehicles for therapeutic small molecules. Generally, exosomal delivery vehicles exhibit higher biocompatibility due to their endogenous origin, tissue-specific targeted delivery, drug deposition in target cells, and favorable drug stability and blood circulation time, thus improving the effectiveness and pharmacokinetics of the small molecule drugs, such as curcumin, paclitaxel, doxorubicin, and withaferin. Exosome-encapsulated curcumin has been reported as able to reduce inflammation.246 Exosomes derived from macrophages and packed with the antitumor drug paclitaxel produced a strong antitumor effect.247 Paclitaxel, doxorubicin, and withaferin were encapsulated in exosomes isolated from bovine milk and exhibited better antiproliferative activities against A549 lung cancer cells than the free drugs.
Delivery of Proteins
Exosomes have been examined and found particularly promising as delivery vehicles for macromolecular proteins. The routes of inserting proteins into exosomes include either genetic engineering—by transfecting the donor with a plasmid carrying the gene of interest—or direct loading into the exosomes. A delivery construct for the potent antioxidant catalase has been developed for treating inflammatory and neurodegenerative disorders, in particular Parkinson’s disease.248 SIRPα has been loaded into exosomes for antitumor therapy by blocking the CD47 receptor on tumor cells.249 Hyaluronan degradation has been applied to stimulate tumor penetration by using exosomes holding PH20 hyaluronidase.250 Moreover, it was reported that the exosome codelivery of PH20 hyaluronidase and doxorubicin inhibit tumors.250
Delivery of Nucleic Acids
Because of their ability to protect nucleic acids from degradation, exosomes have been identified also as superior carriers for nucleic acids for gene therapy. Thus, B-cell-derived exosomes has been employed for delivery of a miRNA-155 inhibitor in order to decrease the lipopolysaccharide-stimulated TNFα production in macrophages.251 The tumor-suppressing agent miR-199a-3p encapsulated into exosomes from fibroblasts of ovarian cancer successfully suppressed c-Met production and inhibited cancer cell proliferation and invasion.208 Substantial inhibition of postoperative breast cancer metastases was attained by an exosome-based siRNA delivery system comprising biomimetic nanoparticles including albumin and siS100A4 with an exosome membrane coating.208,251−253 CRISPR/Cas9 genome editing technology has recently become a preferred tool due to the high precision and efficiency modifying, deleting, or replacing specific genes.254 Exosomal nanocarriers were reported to have achieved efficient delivery of CRISPR/Cas9 plasmids with cancer cell tropism and produced advanced antitumor effects.255
Table 5 exemplifies some exosome drug delivery systems with relation to diseases and exosome sources.
Table 5. Exemplary Exosome-Based Drug Delivery Systems.
| Exosome source | Disease | Drug/therapeutic agent | Study type/disease model or cell line |
|---|---|---|---|
| non-small-cell lung cancer H1299 cells and MRC9 lung fibroblasts256 | lung cancer | doxorubicin/gold nanoparticles | in vitro/human cell |
| Raw264.7 macrophages257 | pulmonary metastases | paclitaxel | in vivo/mouse model |
| pancreatic adenocarcinoma PANC-1 or MIA PaCa-2 cells258 | pancreatic cancer | curcumin | in vitro/human cell |
| mesenchymal stromal cells (SR4987)200 | pancreatic adenocarcinoma | paclitaxel | in vitro/mouse cell |
| human brain glioblastoma–astrocytoma U-87 cells and endothelial bEND.3 cells259 | brain cancer | doxorubicin and paclitaxel | in vivo/zebrafish model |
| mouse macrophages Raw264.7260 | glioma | curcumin/SPIONsa | in vitro/mouse and human cell |
| human brain glioblastoma–astrocytoma U-87 cells261 | glioblastoma | paclitaxel | in vitro/human cell |
| human endometrial stem cells (hEnSCs)262 | glioblastoma | atorvastatin | in vitro/human cell |
| immature mouse dendritic cell transfected by vector expressing iRGD-Lamp2b fusion protein263 | breast cancer | doxorubicin | in vivo/mouse model |
| bone marrow mesenchymal stem cells264 | neuroinflammation | miR-193b-3p | in vivo/mouse model |
| mesenchymal stem cells265 | traumatic brain injury | MSC generated exosomes | in vivo/rat model |
| macrophages266 | Alzheimer’s disease | curcumin | in vivo/mouse model |
| blood plasma267 | Alzheimer’s disease | quercetin | in vivo/mouse model |
| adipose-derived stem cells268 | Alzheimer’s disease | neprilysin | in vivo/mouse model |
| blood plasma269 | Parkinson disease | dopamine | in vivo/mouse model |
| human mesenchymal stem cells270 | Parkinson disease | catalase mRNA | in vivo/mouse model |
| murine dendritic cells271 | Parkinson disease | shRNA | in vivo/mouse model |
| Raw264.7 macrophages248 | Parkinson disease | catalase | in vivo/mouse model |
| HEK 293 cells272 | Huntington disease | miR-124 | in vivo/mouse model |
| Schwann cells273 | Huntington disease | siRNA | in vivo/mouse model |
| mesenchymal stem cells274 | bacterial infection | antimicrobial peptides: cathelicidin LL-37, β-defensin-2, hepcidin, lipocalin-2 | in vitro/mouse and human cells and in vivo/mouse model |
| human amniotic fluid275 | COVID-19 | zofin | clinical trial identifier NCT04657406 for expanded access use/in vivo/human |
SPIONs, superparamagnetic iron oxide nanoparticles.
Exosomes as Therapeutics
Exosomes are considered a promising drug delivery system due to their specific structure and composition allowing them to be used as efficient natural nanocarriers, as well as their impressive preclinical success. Yet another rapidly expanding and noteworthy application of exosomes is their use as therapeutic agents.276−281
Exosomes can modify tumor growth because of some proteins and RNAs which they deliver to the tumor cells.282 Reports show that tumorigenesis is being controlled, specifically downregulated, by the transport of miR-139-5p encapsulated into exosomes in bladder cancer cells.283 A similar effect has been observed when miR-381 packed in exosomes is transfected into triple negative breast cancer cells. miR-140-3p in exosomes isolated from human colorectal cancer blood samples inhibits cancer cell proliferation. miR-5100 in exosomes derived from mouse breast cancer xenograft model-associated macrophages hinders the CXCL12/CXCR4 spreading tumor cells to regional nodes of the primary tumor.284,285 Other miRNAs secreted by colorectal cancer exosomes hinder angiogenesis in colorectal cancer.286
With respect to neurodegenerative diseases, the therapeutic power of exosomes is augmented thanks to the capability of exosomes to cross the BBB. For example, the enzymes neprilysin and insulin degrading enzyme (IDE), which degrade amyloid β peptide, can be found in exosomes. Uptake of these enzymes results in reduction of amyloid β levels.287 Exosomal miRNAs have been found helpful in neurological diseases including Alzheimer’s disease. For example, analysis of exosomal miRNAs isolated from mesenchymal stem cells has been shown to improve various brain disorder pathologies, including Alzheimer’s disease, Parkinson’s disease, subarachnoid hemorrhage, and traumatic brain injury.288,289
Exosomes are reported to be helpful for treatment of cardiovascular diseases as well. An example includes cellular conditioning after acute myocardial infarction. Stem cell exosomes have been reported to promote angiogenesis, impart cytoprotection, and control apoptosis.17,279 Progenitor cell exosomes supplemented with certain cardioprotective miRNAs reduce infarct size in an animal model of ischemia-reperfusion injury.290 Such exosomes also protect from ischemia-reperfusion injury, advancing cardiac performance.291
In infectious diseases, exosomes have been shown to incorporate pathogen-originated molecules or immunomodulators favoring eradication of the microorganism and immune balance.280,281 Hence, exosomes are considered as appropriate carriers of substances to prevent or manage infection, e.g., to control bacterial infections, sepsis, and COVID-19.281 MSC-derived exosomes have been shown to be able to treat infections by expression of microbicidal peptides cathelicidin LL-37, human β-defensin-2, hepcidin, and lipocalin-2 and/or by immunomodulation.274,292 Compared to antibiotics, antimicrobial peptides exhibit certain advantages, such as lower toxicity and immunomodulatory activities, and are thus preferable.292 Exosomes have also become a valuable tool for treatment of sepsis.293 Thus, miR-27b carried by MSC-derived exosomes induces decline of the production of pro-inflammatory cytokines.294 miR-21 carried by exosomes gives rise to substantial renoprotection imparted by remote ischemic preconditioning, proposed as an efficient therapeutic approach for renal damage caused by sepsis.295 Patients with severe COVID-19 disease have been reported to develop a “cytokine storm syndrome”, leading to acute lung injury, acute respiratory distress syndrome, organ failure, and ultimately death.296 Using the model developed for sepsis, exosomes might perform as a therapeutic strategy for the immunomodulatory cure of COVID-19.297 The safety and therapeutic efficacy of exosomes overexpressing CD24 have been assessed, as they are able to directly suppress a cytokine storm.294,298,299 CD24 is an important factor in many human cancers. It is also a significant participant in controlling the T-cell proliferation and as such may suppress inflammation. T-cell-derived exosomes have also been suggested as a useful medication for pneumonia in patients with early stage COVID-19 infection. A clinical study assessing the safety and efficacy of such exosomes has been delivered for inhalation by aerosol.244 Research on treating severe COVID-19 pneumonia is carried out based on exosome inhalation as well.300,301
The variety of diseases to which exosome systems have been applied as therapeutic or diagnostic tools, as demonstrated by our exploration of the publications in the CAS Content Collection, is shown in Figure 17. The largest part (68%) of the publications are associated with cancer, and neurodegenerative, inflammatory, and cardiovascular diseases are also highly represented (Figure 17).
Figure 17.
Distribution of the publications in the CAS Content Collection related to exosome applications in therapy and diagnostics with respect to the target diseases.
Advantages and Disadvantages of Exosomes in Drug Delivery versus Lipid Nanoparticles
Exosomes are small and flexible and exhibit adhesive proteins on their surface, so they can cross the BBB. At the same time, they are endogenous; their membrane is composed of cellular lipids, which imparts them with negligible immunogenicity and toxicity. Exosomes are rich in proteins and genetic material and are thus useful for early and accurate diagnosis. More studies on exosome in vivo biodistribution are required to establish biodistribution mechanisms and their important features, such as the route of administration, disease progression, their cells of origin, and the recipient cell types available to uptake circulating exosomes.302
The endogenous origin of exosomes may also have disadvantages in the clinical practice. The yield of exosomes is considerably lower than that of liposomes. The yield of exosomes is strictly limited by the secretion abilities of cells, the complexity and expenses for large-scale cell culturing, and the time- and effort-consuming, low-efficiency procedures for exosome production, making industrial scale-up manufacture of exosomes a hard to ignore obstacle.303
Additionally, the cargo carrying efficiency of exosomes is restricted. They intrinsically carry a large load of natural components, which significantly complicates and restrains the anticipated cargo loading.304 Although approaches to engineer exosomes with enhanced loading capacity are being elaborated, they are still less efficient than the synthetic liposomes.
As an additional drawback, the quality control of exosomes is harder than that of liposomes. Exosomes are highly heterogenic, even when generated by a single cell type. Due to the lack of sensitive high-throughput analysis methods, it is hard to separate the heterogeneous exosome population into homogeneous ones.4 Furthermore, since one of the essential functions of exosomes is to remove the harmful substances from cells, they may be left with undesired and unsafe macromolecules from their parent cells.305 Strategies to precisely control the contents of exosomes are currently still insufficient.
Exosomes in Diagnostics
To be practicable in clinical use, a blood biomarker needs to be easy to assess, cost-effective, specific for the targeted disease, highly sensitive, and easily and reliably measured. Exosomes are favorable with respect to conventional biomarkers especially in their higher diagnostic sensitivity and accuracy. Thus, exosomes are an appropriate tool for clinical diagnostics for the following reasons:
The disease progression strongly modulates the content of exosomes; exosomal bioactive substances have been shown to be altered and are thus highly informative regarding the pathological status.306,307
Exosomes can be obtained non-invasively from easily available biological fluids including urine, blood, saliva, and even tears for early and fast diagnosis of diseases such as cancers, cardiovascular diseases, and neurodegenerative diseases such as Alzheimer’s disease.308,309
Exosomes are highly stable due to their lipid bilayer membrane. They can thus circulate even in a harsh tumor microenvironment. Moreover, the biomembrane protects the exosomal content from degradation by extracellular proteases.309,310
Exosomes express surface markers characteristic to their cells of origin, so their source can be identified.311
Exosomes can be stored by freezing, freeze-drying, or spray-drying and are highly stable, which is of significant importance for their clinical application.312
Exosomes can permeate through the BBB in both directions; they thus afford collecting information about brain cells non-invasively.310,313
Exosomes exhibit advantages compared to conventional biomarkers in their higher diagnostic sensitivity and accuracy.314−316
The significant potential of exosomes in diagnostics is already widely appreciated, and exosomes are drawing intense attention as evidence is being accumulated that exosomes contain biological molecules characteristic of cancer, neurodegenerative, infectious, and metabolic diseases and can be possibly used as diagnostic biomarkers.19,41,276,317−320
Exosomal Proteins as Diagnostic Biomarkers
Tetraspanins, a group of membrane scaffolding proteins, are abundant in exosomes. One of the members of this family is the exosomal marker CD63. It has been reported that there is a much higher amount of plasma exosomes comprising the CD63 marker in patients with melanoma as compared to healthy ones.321 Furthermore, CD63 has been found to be elevated in exosomes from various types of human cancer cells. Thus, exosomal CD63 is suggested as an appropriate protein marker for cancer.322 Another tetraspanin, CD81, is found essential in hepatitis C pathology, seemingly associated with inflammation and fibrosis. It has thus been identified as a marker for hepatitis C diagnosis.323 A higher expression of CD151, CD171, and tetraspanin 8 (TSPAN8) is reported in blood serum exosomes collected from lung cancer patients.324 These findings suggest exosomal proteins are appropriate biomarkers for cancer diagnosis. Other members of the tetraspanin family such as CD91, CD82, CD147, CD9, and TSPAN8 have also been explored as cancer biomarkers.319
Numerous exosomal protein biomarkers have been identified that can be used to diagnose diseases of the central nervous system. Glioblastoma-specific receptor EGFRvIII has been detected in glioblastoma-patient-derived exosomes, suggesting that exosomal EGFRvIII is an appropriate source of glioblastoma diagnostic information.33 Exosomes from brain tumor patients were found to comprise EGFR, EGFRvIII, and TGF-β.325 Exosomal amyloid peptides are found in brain plaques indicative for Alzheimer’s disease.326 Tau protein phosphorylated at Thr-181, which is an established biomarker for Alzheimer’s disease, was detected at elevated levels in exosomes isolated from cerebrospinal fluid of Alzheimer’s disease patients.307 Thus, exosomes are possibly valuable for early diagnosis of Alzheimer’s disease. Exosome Sciences327 partnered with Boston University researched the use of their TauSome biomarker (exosomal tau) for diagnosis and monitoring of chronic traumatic encephalopathy (CTE) in living individuals.328,329 With Boston University’s DIAGNOSE CTE study (clinical trial NCT02798185),330 they have enrolled 120 former National Football League Players, 60 former college football players, and 60 healthy controls to develop methods to diagnosis CTE and to examine potential risk factors.330 Another biomarker, α-synuclein, the aggregation of which is considered to play a key role in Parkinson’s disease pathology, has been reported to be released from exosomes in a Parkinson’s disease model system.331 The study showed that lysosomal dysfunction typical for Parkinson’s disease increases exosomal α-synuclein release.
The easily and non-invasively attainable proteins in urinary exosomes have also been examined as diagnostic biomarkers, especially for urinary tract diseases. Urinary exosomal fetuin-A has been found to be elevated in acute kidney injury occurrences.332 Exosomal proteins in urine have also been examined as potential biomarkers for bladder cancer and prostate cancer. Eight urinary exosomal proteins have been identified as possible biomarkers for bladder cancer.333 Two identified prostate cancer biomarker proteins were found in urine exosomes from prostate cancer patients.334 Twenty-four urinary exosomal proteins notably differ between bladder cancer and control patients.335
Table 6 exemplifies some candidate exosomal protein biomarkers reported to date for diagnostic applications.
Table 6. Examples of Exosomal Proteins for Clinical Diagnostic Applications.
| Protein(s) | Disease | Body fluid |
|---|---|---|
| CD81323 | chronic hepatitis C | blood plasma |
| CD63, caveolin-1, TYRP2, VLA-4, HSP70, HSP9037,321 | melanoma | blood plasma |
| epidermal growth factor receptor VIII33 | glioblastoma | blood plasma |
| survivin336 | prostate cancer | blood plasma |
| c-src337 | plasma cell dyscrasias | blood plasma |
| NY-ESO-1338 | lung cancer | blood plasma |
| PKG1, RALGAPA2, NFX1, TJP2339 | breast cancer | blood plasma |
| Her2340 | breast cancer | blood plasma |
| glypican-143 | breast cancer | blood serum |
| glypican-143 | pancreatic cancer | blood serum |
| glypican-1341 | colorectal cancer | blood plasma |
| CEA342 | colorectal cancer | blood serum |
| AMPN VNN1, PIGR343 | cholangiocarcinoma | blood serum |
| PSA344 | prostate cancer | blood plasma |
| GGT1345 | prostate cancer | blood serum |
| CD24, EpCAM, CA-125346 | ovarian cancer | blood plasma |
| CD91347 | lung cancer | blood serum |
| TSPAN8, CD151348 | lung cancer | blood plasma |
| CD82349 | breast cancer | blood serum |
| CD9, CD147350 | colorectal cancer | blood serum |
| TSPAN8351 | pancreatic cancer | blood serum |
| fetuin-A, ATF 3332,352 | acute kidney injury | urine |
| CD26, CD81, S1c3A1, CD10353 | liver injury | urine |
| NKCC2354 | Bartter syndrome type 1 | urine |
| EGF, α subunit of Gs, resisitin, retinoic acid-induced protein 3333 | bladder cancer | urine |
| PSA, PCA3, ERG, SPDEF334,355 | prostate cancer | urine |
| L1CAM, CD24, ADAM10, EMMPRIN, claudin356,357 | ovarian cancer | blood plasma, cell culture medium, ascites |
| A2M, HPA, MUC5B, LGALS3BP, IGHA1, PIP, PKM1/M2, GAPDH358 | squamous cell carcinoma | saliva |
| Annexin Al, A2, A3, A5, A6, All, NPRL2, CEACAM1, HIST1H4A, MUC1, PROM1, TNFAIP3359 | lung cancer | saliva |
| LMP1, galectin-9, BARF-1360 | nasopharyngeal cancer | blood, saliva |
| CALML5, KRT6A, and S100P361 | dry eye disease | tears |
Exosomal Nucleic Acids as Diagnostic Biomarkers
Exploration of exosomal RNAs as diagnostic biomarkers has been triggered by the finding that exosomes contain RNAs.32,362 Indeed, exosomal RNAs are protected from RNase degradation by the lipid bilayer membrane and thus can be steadily detected in blood, making them perfect diagnostic biomarkers. Among all exosomal cargo substances, miRNA has drawn attention because of its complex roles in regulating the cancer microenvironment involving angiogenesis, cell proliferation, and metastasis. Its roles in regulating cellular behaviors in situ or in the remote recipient cells are under intensive investigation.363−365
Exosomal miRNAs are being most commonly utilized as cancer biomarkers. Thus, eight miRNAs were identified in serum exosomes from ovarian cancer patients which are missing in healthy controls, suggesting that easily attainable exosomal miRNAs are appropriate diagnostic markers.363 Exosomal miRNAs from lung adenocarcinoma were significantly different from the control patients. Thus, exosomal miRNAs are a possible tool for screening for lung adenocarcinoma.366 Similarly, the miR-141 level is supposedly a forceful diagnostic marker for prostate cancer.364 Researchers from Hackensack University Medical Center are also currently recruiting for a clinical trial (NCT03694483) that will purify prostate-cancer-derived exosomes and characterize their miRNA for the potential development of a prostate cancer liquid biopsy assay.367
A simple, urine-based liquid biopsy test has been developed by Exosome Diagnostics called ExoDx368 and is commercially available to provide risk probabilities of aggressive prostate cancer in patients. The ExoDx test was granted FDA Breakthrough Device Designation in 2019369 and uses RNA copy numbers of ERG, PCA3, and SPDEF to develop a predictive count to correlate the probability that a patient may develop prostate cancer.355
Exosomal miRNAs are reported as hopeful biomarkers for esophageal squamous cell cancer. Exosomal miR-21 has been found to be high in esophageal squamous cell cancer patients’ serum.370 Serum miRNA-1246 exhibits a sensitivity of 71.3% and a specificity of 73.9% for esophageal squamous cell cancer diagnosis. Serum miRNA-1246 has been found to also be significantly correlated with the tumor, lymph node, and metastasis stage and is a strong risk factor for poor survival.371
Exosomal miRNAs have been identified as possible biomarkers for diagnosing cardiovascular diseases and renal fibrosis as well.372−375 Recently, a study of tear exosomes concluded that miR-145-5p, miR-214-3p, miR-218-5p, and miR-9-5p are dysregulated during diabetic retinopathy development.361 Furthermore, tears were established as another easily accessible body fluid expected to improve molecular diagnostics to diagnose ocular, neurodegenerative, and systemic diseases, as well as cancer. Thus, the study of miRNAs in tear exosomes has shown that miR-145-5p, miR-214-3p, miR-218-5p, and miR-9-5p are dysregulated during diabetic retinopathy development361
Another possible diagnostic biomarker besides miRNAs are the exosomal mRNAs.375,376 For example, specific features for diagnosing prostate cancer have been identified in circulating exosomal mRNA.377 Urinary exosome mRNA has been suggested as a tool for non-invasive detection of kidney disease.375
Searching for biomarkers among RNAs to be used in non-invasive diagnostics has been booming in recent years. Representative examples of exosomal miRNAs reported as cancer therapeutic and diagnostic agents are shown in Table 7.
Table 7. Exosomal miRNAs as Cancer Therapeutic and Diagnostic Agents.
| miRNAs | Cancer types | Applications |
|---|---|---|
| miR-378378 | non-small-cell lung cancer | prognostic |
| miR-323-3p, miR-1468-3p, miR-5189-5p, and miR-651359379 | non-small-cell lung cancer | prognostic; osimertinib therapy management |
| miR-486-5p and miR-146a-5p380 | non-small-cell lung cancer | early diagnosis |
| miR-375-3p381 | non-small-cell lung cancer | therapeutic |
| miR-433382 | non-small-cell lung cancer | therapeutic |
| miR-148a383 | breast cancer | prognostic |
| miR-423, miR-424, let7-i, and miR-660384 | breast cancer | diagnostic |
| miR-567385 | breast cancer | therapeutic; reversing trastuzumab resistance |
| miR-9 and miR-181a386 | breast cancer | therapeutic; expanding early myeloid-derived suppressor cells (MDSCs) |
| miR-423-3p387 | prostate cancer | prognostic; castration resistance |
| miR-16-5p, miR-451a, miR-142-3p, miR-21-5p, and miR-636388 | prostate cancer | prognostic; metastasis |
| miR-125a-5p and miR-141-5p389 | prostate cancer | diagnostic |
| miR-375 and miR-451a390 | prostate cancer | diagnostic |
| miR-143 (from cancer tissue)391 | prostate cancer | therapeutic |
| miRNA-92a-1-5p392 | prostate cancer | therapeutic |
| miR-24-3p393 | oral squamous cell carcinoma | diagnostic |
| miR-130a394 | oral squamous cell carcinoma | diagnostic and prognostic |
| miR-30a395 | oral squamous cell carcinoma | therapeutic; cisplatin sensitivity |
| miR-130b-3p396 | oral squamous cell carcinoma | therapeutic |
| miR-139-3p383 | colorectal cancer | diagnostic |
| miR-126, miR-1290, miR-23a, and miR940397 | colorectal cancer | diagnostic |
| miR-106b-3p398 | colorectal cancer | therapeutic |
| miR-221/222399 | colorectal cancer | therapeutic |
A search in the CAS Content Collection15 found an extensive increase of the number of documents related to exosome applications in diagnostics (Figure 18A). A comparison with the therapy-related exosome documents demonstrates that, although at present they outnumber the diagnostic-related documents (Figure 18), the annual growth of the diagnostic exosome documents has begun to dominate (Figure 18A, inset).
Figure 18.
Diagnostic vs therapeutic applications of exosomes. (A) Comparison of the number of documents related to exosome applications in therapy vs diagnostics. Inset: Annual growth of the number of documents related to exosome applications in therapy vs diagnostics. (B) Comparison of the number of documents related to exosome applications in therapy vs diagnostics with respect to their role indicators (THU, therapeutic; DGN, diagnostic).
Exosomes as Therapeutic Targets
Exosomes are known to be related to the pathogenesis of various illnesses such as cancer, neurodegenerative, cardiovascular, and others. Provided that exosome amounts are frequently enhanced and related to the severity of the diseases, in particular for cancers, a successful therapeutic strategy may involve reducing exosome production and circulation to normal levels to prevent disease progression.400−403 With this perception, numerous studies are intended to modify the exosome pathway at its various steps, including production, release, and uptake.404
A number of approaches have been explored for inhibiting exosome formation. The endosomal sorting complexes required for transport (ESCRT) are known to be involved in multivesicular body biogenesis112 Several reports have correlated the exosome secretion to the ESCRT-0 protein hepatocyte growth factor-regulated tyrosine kinase substrate (HGS, HRS), by reporting decreased exosome release in HRS depleted dendritic cells and tumor cells.405,406 Mechanisms of exosome formation which do not depend on ESCRT are known too. They include ceramide or the tetraspanins. The small-molecule inhibitors of sphingomyelinase, the enzyme generating ceramide from sphingomyelin, are able to reduce endosomal sorting and production, causing a reduction in tumor growth.407,408 Otherwise, the formation of exosomes may be controlled by certain signaling pathways triggered by Ras homologue family member A or ADP-ribosylation factor 6 (ARF6).409,410 Targeting these pathways may produce a distinct therapeutic effect on tumor progression.
Other strategies that block exosome secretion have been developed as well. The sphingomyelinase inhibitor drug GW4869 causes inhibition of intraluminal vesicle formation and release of exosomes.239,411 Inhibition of exosome production has been accomplished by attenuation of sphingomyelinase 2, which manipulates the synthesis of ceramide and restrains angiogenesis and metastasis in breast cancer.412 Numerous modulators of exosome fabrication from prostate cancer cells have also been reported recently.413
Another way of modulating extracellular exosome levels is by inhibiting exosome release. Certain proteins, such as small GTPases of the Rab family, are associated with the discharge of exosomes. Thus, Rab27a and Rab27b are significant regulators of exosome release and the same is true for their effector proteins.34,414 Silencing Rab27a by RNA interference can reduce tumor growth.415 Lipids are also shown to be involved in the exosome secretion regulation. Diacylglycerol kinase downregulation results in the suppression of the secretion of exosomes containing the Fas ligand.416 Exosome discharge includes fusion of MVBs with the cell membrane as a final step. This process is mediated by the SNARE complex machinery, with the SNARE protein Ykt6 involved.417 Lastly, cellular pH also modulates exosome secretion, via modulation of proton pump inhibitors.418 Furthermore, studying the exosome roles has revealed that suppression of melanoma progression is correlated with exosomes released by natural killer cells.419
Exosome uptake inhibition is another way to modulate exosome activity. Cells uptake exosomes using various endocytic pathways, such as clathrin-dependent endocytosis as well as clathrin-independent routes, e.g., macropinocytosis and phagocytosis.420−422 Exosome treatment with proteinase K has been reported to significantly reduce uptake by ovarian cancer cells, which is an indication that proteins located on the exosomal surface may operate as uptake receptors.423 The uptake of tumor exosomes is supposedly mediated by the membrane phosphatidylserine that is possibly inhibited by diannexin.424 Heparan sulfate proteoglycans allegedly operate as internalization receptors of cancer cell exosomes. Such an uptake route appears to be significant, since heparin treatment considerably inhibits the cancer cell migration stimulation mediated by exosomes.425 Besides, exosome uptake is inhibited by dynamin2 knockdown, required for clathrin and caveolin endocytosis pathways.421
Another successful strategy of treating cancer has been exploited by physical elimination of exosomes secreted by cancer cells. Communication among cells in tumors is mostly via chemokines, cytokines, or growth factors.426,427 Exosomes from tumor cells are noted to facilitate these kinds of communications, thus playing a role in tumor progression.428 Therefore, the removal of exosomes secreted by cancer cells is one of the exosome-targeting therapeutic approaches. A hemofiltration system capable of targeting cancer cell exosomes by specifically targeting at human epidermal growth factor receptor 2 (HER2) on the exosome surface was utilized.429 That caused selective elimination of cancer-derived exosomes, which proved to be very valuable for cancer treatment.219
Collectively, these data reinforce the hypothesis that elimination of exosomes or inhibition of their secretion, release, or internalization mechanisms may have favorable effects in cancer therapy. Thus, a good understanding of the disease-specific mechanism of exosome pathways is needed in finding specific therapies intervened by targeting exosomes.427
Other Applications
Exosomes in Food and Cosmetics
Prospective applications of exosomes are also in cosmetics and food.430 It has been reported that stem-cell-derived exosomes are able to perform significantly in skin cosmetology, specifically in promoting wound healing, alleviating skin aging, and preventing scar formation.431,432 For example, exosomes derived from induced pluripotent stem cells are able to modulate the expression of MMP-1/3 and enhance the expression of type I collagen in senescence skin fibroblasts.433 Exosomes from adipose stem cells were reported as able to promote wound healing through the PI3K/Akt signaling route and to increase the amount of collagen type I and type III in fibroblasts.434 A search in the CAS Content Collection revealed a sharp growth in the number of documents related to applications of exosomes in cosmetics in the last 3 years (Supporting Information Figure S4A).
Bioactive compounds—polyphenols, vitamins, polyunsaturated fatty acids, and others—are common food supplements aiming to elevate nutritional value. However, their effect can be compromised by their poor bioavailability, limited water solubility, and metabolic alterations; thus, they require carriers. While extracellular vesicles and specifically their exosome subclass have emerged demonstrating an impressive potential to realize efficient delivery of bioactive compounds, they can successfully serve as carriers of such food-related bioactive compounds, as well. Indeed, the interest in applications of exosomes in food has rapidly grown in the recent years (Supporting Information Figure S4B).
Recent studies verified isolation of exosomes from food stuff such as lemon, ginger, and milk.435 Such food-derived exosomes can be uptaken in the intestine to act locally and can allegedly play roles in alleviating diseases and especially in modulating gut microbiota, yet the underlying mechanism is still unclear.
Exosomes from Plant Cells
The existence of EVs in plants has been long debated because of the existence of the cell wall. Growing evidence implies however that plants also secrete EVs performing various functions such as unconventional protein secretion, RNA transport, and pathogen defense.436
It has been hypothesized that edible structures within cells of plants such as ginger, aloe, and others might have clinically valuable anti-inflammatory effects on the intestinal lining of patients with inflammatory bowel disease (clinical trial NCT04879810).437 Exosomes from ginger or aloe are being tested for the treatment of polycystic ovary syndrome (NCT03493984).438 Grape exosomes are in a clinical trial as an anti-inflammatory agent to decrease the frequency of oral mucositis following radiation and chemotherapy treatment of head and neck tumors (NCT01668849).439
Exosomal Drug/Biomarker in the Development Pipeline
Companies are working to progress exosome research from conception to commercialization. To start, many companies are offering services and products for exosomal research. Many other companies, medical centers, universities, and research organizations are looking to utilize exosomes for therapy and diagnostics to target diseases with high unmet needs. Promising preclinical therapeutic and diagnostic exosome research is explored in this section. Selected clinical trials utilizing exosomes as therapeutics and diagnostics are also highlighted. Lastly, clinical trials that research exosomes as the disease target are examined.
Companies Offering Services and Products for Exosome Isolation, Purification, Characterization, and Engineering
As exosome research has grown dramatically within the past decade (Figure 4), so have the number of companies offering services and products for exosome isolation, purification, characterization, and engineering for both therapy and diagnosis. A selection of these companies is discussed along with their services and products within Table 8.
Table 8. Highlighted Companies Offering Services and/or Products for Exosome Isolation, Purification, Characterization, and Engineering for Research and/or Commercialization.
| Company (location) | Summary |
|---|---|
| Ciloa (France) | Ciloa is an exosome spin-off company from the French National Center for Scientific Research and the University of Montepellier. Ciloa is dedicated to in vivo development of recombinant exosomes for therapeutic and preventative applications. Their recombinant exosomes allow for loading of two types of protein cargo: the first one, at the surface for disease targeting; the second one, as the cargo inside the exosome to deliver a signal for modification, multiplication, or death.440 |
| Clara Biotech (USA) | Clara provides exosome isolation using their developed ExoRelease Isolation Platform and characterization as a service for researchers. They have developed a starter kit version of their ExoRelease Isolation Platform for researchers to perform isolation of exosomes in their own lab, as well. They also offer the services of nanoparticle tracking analysis for exosome characterization, exosome proteomic analysis, exosome nucleic acid analysis, and exosome imaging.441 |
| Creative Biolabs (USA) | Creative Biolabs offers a wide range of exosome-related research services. These services include exosome isolation, purification, characterization, quantification, profiling, proteomics, lipidomic and metabolomics assays, RNA sequencing, exosome engineering and manufacturing, and exosome antibody development and display and have in vitro and in vivo model platforms.442 |
| EverZom (France) | EverZom is an exosome service company who provides a large panel of services for exosome development including exosome production, characterization, isolation/purification, and engineering services.443 |
| Exosome Plus (Republic of Korea) | Exosome Plus manufactures MSC-derived exosomes, plant-derived exosomes, human-derived exosomes, and animal-derived exosomes. Their therapy platform is called ExoThera, and they are hoping to develop their liquid biopsy platform to diagnose 11 major cancers using body fluid exosomes. They also sell an exosome isolation kit called Exo2D and an EV characterization system called ExoCope which is a single exosome multicolor fluorescence colocalization and particle tracking analysis system.444 |
| Exosomics (Italy) | Exosomics offers the services of exosome isolation and characterization, nucleic acid extraction, protein separation, nucleic acid analytical assays, and protein analytical assays.445 They also offer kits for researchers to use in their lab including exosome purification kits and exosome-based reference standards.446 |
| FUJIFILM Wako Chemicals USA Corporation (USA) | FUJIFILM offers many different exosome kits and products including exosome isolation kits, ELISA kits, and flow cytometry kits. They also offer exosome marker antibodies, blocking reagents, purified exosomes, exosome cell cultures, and labware for researchers to utilize in their own laboratories.447 |
| HansaBioMed Life Sciences (Estonia) | HansaBioMed is entirely dedicated to research and development in the exosome sciences field. Their services include purification of exosomes from condition media, biofluids, or plant extracts, exosome characterization, biomarker assessment by mass spectrometry, and RNA sequencing. They also sell a broad range of purified exosomes, tools for purification, enrichment, and characterization.448 |
| Lonza (USA) | Lonza acquired HansaBioMed Life Sciences in 2017. More recently in 2021, Lonza acquired Codiak’s (Therapeutic exosome company) manufacturing facility and is the strategic manufacturing partner for Codiak’s pipeline. Additionally, in 2021, they announced they acquired Exosomics.449 |
| NanoFCM (UK) | NanoFCM has a commercial product available called the flow nanoanalyzer which is a high-sensitivity flow cytometry for exosome analysis. They also offer exosome sample analysis service.450 |
| NanoView Biosciences (USA) | Nanoview Biosciences creates exosome products to help with exosome characterization. Their ExoView R200 product allows for automated exosome measurement. Their ExoView kits allow for standard or customizable assays for purification of free exosomes. The ExoView chip washer offers reliable hands-free sample preparation, along with their ExoView software suite that offers reporting of exosome size, counts, and biomarker colocalization.451 |
| ReNeuron (UK) | Exosomes produced by the ReNeuron’s stem cell lines or via its induced pluripotent stem cell platform have the possibility to be manufactured through a scalable process and loaded with a broad range of payloads, such as nucleic acids, proteins, as well as small molecules. They are in collaboration with universities, global pharma, and biotech companies in various stages from discovery to in vivo late-stage studies.452 |
| RoosterBio (USA) | RoosterBio is dedicated to accelerating exosome product and process development for exosome therapeutics. They have developed an extensive panel of exosome analytical methods to support this including exosome NTA for characterization, purification, protein analysis, surface marker expression, cytosolic marker expression, miRNA quantitation and analysis, lipid content, albumin contamination, CD63 quantitation, and scratch assays for wound healing.453 They also produce exosome production media for both research and manufacturing.454 |
| Systems Biosciences (USA) | Exosome research products and services are offered by Systems Biosciences to help advance exosome research studies. They offer exosome isolation, detection, quantification, labeling, biomarker discovery, engineering, and design kits and products.455 |
| ThermoFisher Scientific (USA) | ThermoFisher Scientific is a world leader in serving science, staying one step ahead for advancing science, and their products for exosomes are no different. They offer a wide range of exosome products for isolation, analysis, and cargo isolation. They also offer exosome depleted fetal bovine serum along with reagents for automated preparation products for exosome analysis.456 |
Therapeutic Exosome Companies
The number of companies that are utilizing exosomes for therapy is also expanding. Both preclinical and clinical works are progressing exosome therapeutics through companies’ pipelines. A thorough review of exosome therapeutic companies reveals that the most highly represented diseases are cancer, neurological and neurodegenerative diseases, lung diseases, and wound healing (Figure 19 and Supporting Information Table S2).
Figure 19.
Promising exosome therapeutic companies and targeted diseases.
Preclinical Therapeutic Exosomes
A growing number of companies are researching exosomes in hopes of advancing their therapeutic discoveries to the clinic. While historically MSC-derived exosomes were researched for therapy, a shift is taking place and companies are starting to focus research effort on organ-specific exosomes such as cardiac-derived exosomes or neural-derived exosomes for more targeted specificity in treating diseases. Table 9 displays selected preclinical companies focusing their research efforts to the highly represented targeted diseases from Figure 19.
Table 9. Highlighted Companies Working on Preclinical Therapeutic and Cosmetic Exosomes along with Their Summaries.
| Company (location) | Summary |
|---|---|
| Anjarium Biosciences (Switzerland) | Anjarium is researching and developing precision exosome therapeutics. Their Hybridosome platform utilized nanotechnology and biochemistry to increase the efficiency of exosome loading with therapeutic cargo.457 Anjarium is looking to use its exosome-based therapy platform to treat cancers and rare genetic diseases. |
| Aruna Bio (USA) | Aruna Bio is transforming treatment for neurological and neurodegenerative diseases. They utilize neural exosomes derived from neural stem cells that have CNS specificity and the ability to cross the BBB. Their candidate AB126 shows high uptake in the cerebellum and basal ganglia showing treatment potential for diseases such as stroke and neurodegenerative diseases.245 Their pipeline shows that AB126 can be loaded with different cargos including siRNA, ASO, progranulin, and tripeptidyl-peptidase 1.458 |
| Capricor (USA) | Capricor is developing multiple exosome platforms including cardiosphere-derived cell exosomes (CDC exosomes), engineered exosomes, and an exosome-based vaccine. They are currently researching the use of CDC exosomes for the treatment of Duchenne muscular dystrophy and engineered exosomes for RNA and protein delivery in trauma-related injuries and conditions in collaboration with the U.S. Army Institute of Surgical Research. They are also in preclinical trials for an exosome-based multivalent vaccine for COVID-19 and other infectious diseases.459 |
| Carmine Therapeutics (USA) | Carmine Therapeutics utilizes red blood cell exosomes.460 Their Red Cell EV Gene Therapy (REGENT) platform will be used to generate a pipeline of therapies for treatment of a wide range of diseases.461 |
| EV Therapeutics (USA) | EV Therapeutics is developing modified exosomes (mEVs) (miR-424i and miR-424 KO) in combination with an immune checkpoint inhibitor for treatment of metastatic colorectal cancer and other GI cancers.462 mTEV is a CD-28–CD80/86 costimulatory pathway technology platform that functions in combination with checkpoint inhibitors to enhance T-cell immunomodulation to prevent solid tumor cancer recurrence.463 |
| Evora BioSciences (France) | The EVOGEX therapeutic platform was developed by Evora. Their lead product EVOGEX-Digest aims to treat digestive fistula and improve patient outcomes.464 |
| Evox (UK) | Evox is an exosomal therapeutic company using its DeliverEX platform to deliver proteins and nucleic acids to treat a variety of rare diseases. Their internal program is researching rare metabolic disorders. They have partnered with Takeda to treat lysosomal storage disease and other undisclosed rare diseases. Evox has also recently partnered with Lilly to research neurological treatment.465 |
| Exocel Bio (USA) | Exocel utilizes placental MSC-derived exosomes for both skincare and hair care. Their products include the Evovex line called Evovex Restore, Evovex Revive, Evovex Renew, and Evovex Reveal.466 These products are used in conjunction with facial and scalp microneedling and energy-based aesthetic device treatments to enhance results and improve recovery time.467 |
| ExoCoBio (Republic of Korea) | ExoCoBio is focusing its research on stem-cell-derived exosomes to create both therapeutic and cosmetic products. They have developed ExoSCRT Exosome for the treatment of atopic dermatitis,468 irritable bowel syndrome, acute kidney injury, and alopecia.469 An immune-oncology drug based on exosomes derived from immune cells is also in their pipeline. |
| Exogenus Therapeutics (Portugal) | Exogenus’s lead candidate Exo-101 is produced from umbilical cord blood mononuclear cells. It has been shown to have regenerative, anti-inflammatory, and immunomodulatory properties. Exo-101 is being investigated for treatment in inflammatory skin diseases such as psoriasis, inflammatory lung disorders such as COVID-19 ARDS,470 and chronic wound healing.471 |
| Florica Therapeutics (USA) | Florica Therapeutics aims to use hypothalamus stem-cell-derived exosome therapeutics to increase lifespan and deter neurological diseases of aging.472 |
| Ilias Biologics (South Korea) | Ilias developed the platform EXPLOR that allows the loading of proteins into exosomes in a more controlled manner than conventional passive loading.473 Ilias’ lead compound ILB-202 consists of an exosome loaded with an anti-inflammatory protein super-repressor IkB targeting both acute and chronic inflammatory diseases. This lowers the risk of an off-target effect by targeting core inflammation signals.474 |
| Innocan Pharma (Israel) | Innocan is a pharmaceutical company researching cannabidiol (CBD) drugs and enhancing their targeting due to its low bioavailability. Innocan is researching with Tel Aviv University the development of CBD-loaded exosomes to target inflammatory diseases and central nervous system diseases.475 |
| Kimera Laboratories (USA) | Kimera specializes in the use of perinatal MSC-derived exosome products for both cosmetics and scientific research.476 Their cosmetic products are XoGlo, XOGloPro, and Vive. They also produce a veterinarian wound healing agent called Equisome HC.477 |
| MDimune (Republic of Korea) | MDimmune developed a platform technology called BioDrone that uses cell-derived vesicles for targeted drug delivery.478 Their internal pipeline includes treatment for chronic obstructive pulmonary disease (COPD) and an undisclosed rare disease with therapeutics BDR-231 and BDR-331, respectively. They have partnered with Ildong, Kainos Medicine, and NeoCura for the treatment of cancer using various mRNAs and small molecules for cargo for therapeutic products BDR-165, BDR-166, and BDR-167. They are also partnered with Reyon for a vaccine with therapeutic BDR-761 and treatment of an undisclosed rare disease with therapeutic BDR-762 using mRNA as cargo.479 |
| OmniSpirant (Ireland) | OmniSpirant’s platform technology is based on inhalation and is very efficient at delivering cargos to treat respiratory diseases. The mucus penetrating exosomes will be used to develop a regenerative gene therapy for cystic fibrosis and other respiratory diseases.480 |
| Regen Suppliers (USA) | Regen Suppliers developed an exosome product called ReBellaXO, derived from umbilical stem cell tissue and Wharton’s jelly used for regenerative cosmetic procedures involving facial, hair, and sexual rejuvenation.481 |
| Xollent (USA) | Xollent is advancing a diversified pipeline of therapeutics including exosome therapeutics treating myocardial infarction through an intravenous patch, alopecia through a spray, and skin aging through a needle-free injection.482 |
Preclinical Diagnostic Exosomes
Companies, medical centers, and universities are also focusing their research efforts on discovering exosome biomarkers and representative tests for diagnosis of hard-to-treat diseases earlier, to help aid in the treatment success and patient survival. While cancer is the most highly represented disease for diagnosis through exosomes (Supporting Information Table S2), many other diseases can be diagnosed with exosome detection and organizations are working to develop these appropriate assays (Table 10). Many universities are also hard at work researching exosome disease diagnosis. Table 10 explores promising preclinical companies, medical centers, and universities researching exosome disease diagnosis. The current field of exosome diagnostics and developed assays is still relatively small with room to grow as more promising early disease biomarkers are researched and discovered.
Table 10. Highlighted Companies and Universities on Preclinical Research of Exosomes as Biomarkers for Diagnosis of Various Diseases and Their Summaries.
| Companies/medical centers/universities (location) | Summary |
|---|---|
| Aarhus University Hospital (Denmark) | Researchers discovered that the biomarkers CD151, CD171, and tetraspanin 8 were the main dividing factors for patients with non-small-cell lung cancer of all types versus patients without cancer.348 |
| Craif (Japan) | Craif developed a medical device consisting of a zinc oxide nanowire embedded in a microfluidic channel that collects urinary miRNA for exosome-based liquid biopsy. They are using machine learning technology to analyze miRNA profiles with their original miRNA database to identify biomarkers for early cancer detection.483 |
| Frankfurt University Hospital (Germany) | Researchers studied how CD81 is increased in the exosomal serum of patients with chronic hepatitis C and appears to be associated with inflammatory activity and severity of liver fibrosis.323 |
| Harvard Medical School (USA)/Wenzhou Medical University (China) | Researchers have developed an incorporated tear-exosome analysis via rapid-isolation system (iTEARS) via nanotechnology to discover if exosomes from tears can diagnose ocular disorders and systemic diseases. Data show that iTEARS might be used to improve the molecular diagnostics of dry eye disease, along with diabetic retinopathy.361 There is also a possibility that iTEARS could be used to detect other neurodegenerative diseases and cancer. |
| Mercy Bioanalytics (USA) | Mercy developed the Halo test for early cancer detection test with initial focus on hard-to-treat cancers such as ovarian and lung cancers.484 Preliminary results from studies researching Halo detection of both early stage ovarian and lung cancers were positive.485,486 |
| Osako University (Japan) | Researchers discovered that three p53-responsive microRNAs, miR-194, miR-34a, and miR-192 are elevated in exosomes of patients with acute myocardial infarction, suggesting that these microRNAs function as circulating regulators of heart failure. They feel that these three microRNAs are worth further exploration as biomarkers for ischemic heart failure after acute myocardial infarction.487 |
| UCSF Medical Center (USA) | Researchers discovered that levels of P-S396-tau, P-T181-tau, and Aβ1–42 from neural-derived blood exosomes can predict the development of Alzheimer’s disease up to 10 years before clinical onset of symptoms.306 |
| University of Texas MD Anderson Cancer Center (USA) | Researchers identified a cell surface proteoglycan, glypican-1 (GPC1), specifically enriched on cancer-cell-derived exosomes. GPC1(+) circulating exosomes may serve as a potential diagnostic and screening biomarker for assays to detect early stages of pancreatic cancer.43 |
Conclusions and Perspectives
As demonstrated by data analysis of the CAS Content Collection, the interest in exosome exploration has grown significantly in the recent years. A growing number of studies provide valuable knowledge regarding this notable subtype of EVs. Indeed, exosomes exhibit distinctive functions as intercellular messengers, potential to modulate cellular bioactivities, as well as substantial therapeutic capacity, in disease diagnostics and targeted drug delivery. Their advantages over traditional pharmaceutical nanocarriers distinguish them as a rising star in both therapeutics and diagnostics. In this review, we provide a landscape of the global research effort for exosome development for medical applications, along with the challenges and growth opportunities for fulfilling their potential.
Exosomes are released from most cell types into the extracellular space following fusion of multivesicular bodies with the cellular membrane.4,7,54,488 During the process of exosome secretion, parent cell information is stored in the exosomes, in their constituent lipids, proteins, and nucleic acids, which are then able to manipulate the functions of recipient cells on arrival. The content of the exosomes is therefore characteristic to the cell of origin, permitting parent cell signals to be communicated to neighboring cells without direct cell-to-cell contact. A foremost advantage over signaling molecule secretions is that exosomes are able to deliver signals at large distances without any dilution or degradation, because the biomolecules are being safely transported within their lipid bilayer capsule.
With significant research being devoted to exosome medical applications—in drug delivery, in diagnostics, as therapeutic targets, or as therapeutics themselves—it is vital to review and recapitulate the progress made, along with the persisting challenges.9,19,20 Although exosome analyses have intensely evolved in the recent years, their exact mechanisms of biogenesis and uptake are still largely unknown. Furthermore, the challenges in efficient and successful exosome isolation are still persisting, primarily due to the complexity of bodily fluids, the extensive overlap of the physicochemical and biochemical characteristics among the exosomes, lipoproteins, viruses, and other extracellular vesicles, as well as the heterogeneity of exosomes. Thus, developing efficient and reliable isolation and characterization techniques is critical to further advance in this area, in order to examine the cargo contents and functions, which would shed light on the biogenesis and uptake in return. Furthermore, fundamental questions in the field such as the secretory regulation mechanism of exosomes, the exosomal content sorting mechanism, and their intercellular transduction pathway are still to be answered too. To fully utilize the exosome potential, basic research and emerging advanced technologies need to be combined, which will set forth their therapeutic applications.
Clinical applications of exosomes, although highly promising, are hindered by the lack of standardization in exosome isolation and analysis, which has become a major challenge in the field.489 The use of inconsistent protocols for sample handling, analysis, and data control leads to discrepancy that significantly affects analysis, makes interstudy comparisons difficult, and overall complicates the knowledge development. Thus, standardization in exosome preparation such as specimen handling, isolation, and quantification still has to be established.205,206
Thus, some appealing challenges in exosome knowledge include the following:
Potent isolation methods that do not compromise on the purity of the isolated specimens are required in order to exploit exosomes in biomedical research and therapeutics—such methods are the primary prerequisite for exosomal large-scale application in medical practice. Additionally, recent studies have shown that an appropriate combination of several methods to extract and purify exosomes can effectively contribute to solving this problem.168,171
The exact mechanisms involved in the biogenesis, secretion, and fusion of exosomes have not yet been fully elucidated and require further research. It is also mostly unknown whether incorporating cargo into exosomes is a selective or a random process, although data is accumulating that suggests a certain degree of cellular control.
The underlying mechanism of how exosomes communicate with the target cells and how selectivity is achieved is not yet well understood. Advanced knowledge on these processes is a prerequisite to develop effective therapeutics that target exosome communication and for the development of engineered exosome-derived therapeutic vehicles.
Exosome loading capacity and methods for enhancing their targeting need to be optimized and improved for their large-scale application in clinic.
Studies have demonstrated that exosomes are able to permeate the BBB from the brain to the bloodstream as well as from the blood to the CNS; however, only limited knowledge exists about the mechanisms exosomes use to cross the BBB.77,245 Understanding of the surface markers required to cross the barriers protecting the brain and the ones needed to target the cells or tissues responsible for a pathology is needed in order to make use of this notable ability of exosomes.77
Standardization of exosome preparation, including source selection, isolation, characterization, drug loading, stability, targeting, and quality control, in compliance with good manufacturing practice, is an important aspect in the clinical application of exosomes and needs to be advanced. There is thus an urgent need to develop guidelines for manufacturing, storage, and administration of therapeutically relevant exosomes, with respect to safety and quality GMP standards to be followed.
The prospective use of exosomes as a delivery vector needs further deep assessment. The tractability of the exosomes needs to be improved, and the possibility to package multiple drugs for combination (immuno)therapy needs to be explored. With personalized medicine models emerging and being advanced, it is important to assess the potential for developing personalized approaches for delivering therapeutically relevant exosomes.
Advanced knowledge on the pharmacokinetic profile and biodistribution of exosomes is still particularly insufficient and is a required step toward their practical utility in clinics.
The nature of the cargo in exosomes soundly depends on the origin of the cells where the exosomes are released. It is thus important to know how the cargo is packed in exosomes, since cancer cells are known for their heterogeneity and the nature of cargo from each cancer cell will be distinctive. Such knowledge would advance designing strategies for early diagnosis and monitoring treatment response by using exosomes.490
Cells modulate the composition of exosomes in response to exogenous stress. Understanding the mechanisms involved might result in the development of therapeutics that take advantage of this property.
An emerging area of exosome research currently gaining considerable attention is their potential application in cancer immunotherapy, in particular developing anticancer vaccines. Various cells such as B-cells, dendritic cells, macrophages, cancer cells, and normal cells have been employed for isolating exosomes as possible agents in cancer immunotherapy. These cells all exhibit characteristic composition profiles directly involved in anticancer immunotherapy.
Since exosomes represent an advanced potential treatment strategy in a wide range of therapeutic areas, possibly as cell-free regenerative medicines, as treatments for cardiovascular, CNS, and oncological disorders, as vectors for gene therapy, as immune modulators, and as drug delivery vehicles, their innovation and range of uses means that there will be specific regulatory classification and jurisdiction issues to be clarified to enable development plans to be established. In recent years, concurrent advances have been witnessed in the exosome expertise for next-generation diagnostics, disease supervision, and individualized diagnosis and therapy. These are widely applied for early diagnosis and delivery systems with high efficacy. Further advances in the drug loading strategies and modification methods will enable clinical translation in the future, with a tangible patient benefit.
Glossary
Vocabulary
- extracellular vesicles
lipid-bilayer-surrounded particles, which are secreted by cells into the extracellular space; they represent a route of intercellular communication and contribute to a wide range of physiological and pathological processes
- exosomes
a nanosized subset of extracellular vesicles (diameter ∼30–150 nm) comprising bioactive cargos, including proteins, nucleic acids, lipids, and metabolites
- biomarker
indicator of normal or pathogenic biological processes, or pharmacological responses to a therapeutic intervention, which can be measured objectively, accurately, and reproducibly
- drug targeting
delivering medication to a patient in a manner that results in predominant drug accumulation in a specific body area (organ, cellular, and subcellular level of specific tissue) in order to overcome the toxic effect of conventional drug delivery
- blood–brain barrier
highly selective boundary of endothelial cells that prevents solutes in the circulating blood from non-selectively crossing into the extracellular fluid of the central nervous system
Appendix: Exosomes in Clinical Trials
Exosome Therapeutics in Clinical Trials
Currently, the total number of clinical trials registered at https://clinicaltrials.gov for exosomal therapeutics is 59 clinical trials (Supporting Information Table S2). The most highly researched targeted diseases for exosome therapeutics include lung disease (11 clinical trials), SARS-CoV-2 infections (9 clinical trials), and cancer, heart disease, and neurological diseases (all with 4 clinical trials). Highlighted clinical trials with respect to these diseases are listed in Table 11.
Table 11. Highlighted Exosome Therapeutic Clinical Trialsa.
| Companies/medical centers/universities (location) | Exosome | Disease treated | Clinical trial number | Clinical trial stage or status (date initiated) | Summary |
|---|---|---|---|---|---|
| M.D. Anderson Cancer Center (USA) | MSC-derived exosomes with KrasG12D siRNA (iExosomes) | metastatic pancreatic cancer with KrasG12D mutation | NCT03608631 | phase I (2018) | This study researches the optimal dose and the drug toxicity of using iExosomes in treating metastatic pancreatic cancer patients.491 |
| Neurological Associates of West Los Angeles (USA) | exosomes | cranial facial neuralgia | NCT04202783 | suspended (due to COVID-19 pandemic) (2019) | This study will evaluate the safety and efficacy of exosome treatment in patients with craniofacial neuralgia.492 |
| Organicell Regenerative Medicine (USA) | amniotic-fluid-derived exosomes/Zofin (organicell flow) | mild/moderate COVID-19 | NCT04657406 | expanded access status available (2020) | The therapeutic Zofin is currently undergoing clinical trials for COVID-19, COPD, and osteoarthritis.493 |
| Direct Biologics (USA) | bone marrow MSC-derived exosomes/DB-001/ExoFlo | COVID-19 ARDS | NCT04657458 | expanded access status available (2020) | Their therapeutic ExoFlo is currently undergoing clinical trials for COVID-19-associated moderate to severe ARDS, ulcerative colitis, Crohn’s disease/irritable bowel syndrome, and organ transplant rejection.494 |
| Rion (USA) | purified exosome product (PEP) | skin graft | NCT04664738 | phase I (2020) | Rion is researching with this clinical trial the application of their PEP therapeutic (a leukocyte depleted blood preparation derived from apheresed platelets) in patients with a skin graft for wounds to determine if it offers improvement in healing properties over the standard wound dressing treatment.495 |
| Ruijin Hospital (China) | adipose mesenchymal stem-cell-derived exosomes (MSCs-Exos) | Alzheimer’s-disease-induced dementia | NCT04388982 | phase I/II (2020) | The purpose of this study is to explore the safety and efficacy of MSCs-Exos in the treatment of mild to moderate dementia due to Alzheimer’s disease.496 |
| Rion (USA) | purified exosome product (PEP) | acute myocardial infarction | NCT04327635 | phase II (2020) | Rion’s PEP exosome therapeutic is currently in preclinical and clinical studies for multiple indications497 including acute myocardial infarction, wound healing,498 pelvic floor disorders,499 hair loss treatment, and degenerative joint disease500,501 with many encouraging results. |
| University of Louisville (USA) | ginger exosomes with/without curcumin | irritable bowel disease | NCT04879810 | recruiting (2021) | The purpose of this clinical trial is to test if edible ginger exosomes will have clinically relevant anti-inflammatory action on the gut lining of patients with inflammable bowel disease.437 The University of Louisville also has an active clinical trial exploring edible plant exosomes conjugated to curcumin for the treatment of colon cancer.502 They also conducted a completed clinical trial researching grape exosomes dosed as grape powder to reduce the incidence of oral mucositis during radiation and chemotherapy cancer treatments.439 |
| OBCTCD24 (Israel) | exosomes overexpressing CD24/CovenD24/EXO-CD24 | moderate or severe COVID-19 | NCT04902183 | phase I (2021) | OBCTCD24 has their product EXO-CD24/CovenD24 in one other clinical trial. CovenD24 is an exosome overexpressing CD24 administered through inhalation dosing for the treatment of moderate or severe COVID-19.298 |
| Aegle Therapeutics (USA) | bone marrow MSC-derived exosomes/AGLE-102 | burns | NCT05078385 | phase I (2021) | Preclinical data reveals that exosomes isolated by Aegle accelerated healing, minimized scars, and promoted blood vessel and nerve regeneration, as well as hair follicle growth.503 In addition to burns, AGLE-102 is also in a clinical trial for the treatment of dystrophic epidermolysis bullosa, a group of rare genetic disorders that presents with blistering or erosion of the skin in response to little or no trauma.504 |
| Maimónides Biomedical Research Institute of Córdoba (Spain) | MSC-derived exosomes | wound healing/skin ulcers in diabetic patients | NCT05243368 | not yet recruiting (2022) | The focus of this clinical trial is to develop a therapeutic process to accelerate the healing of diabetic chronic skin ulcers, based on nutritional intervention and the application of MSC-derived exosomes to the wound, to improve skin regeneration.505 |
| Codiak BioSciences (USA) | exosomes loaded with a synthetic lipid-tagged oligonucleotide/CDK-004/exoASO-STAT6 | advanced hepatocellular carcinoma (HCC)/liver metastasis | NCT05375604 | phase I (2022) | In addition to HCC,506 Codiak also has clinical trials for treatment of cutaneous T-cell lymphoma, solid tumors, and non-small-cell lung cancer. They have also developed an exosome-based vaccine on their engEx platform for the treatment of beta coronavirus, Epstein–Barr virus, and HIV.507 Recent preclinical data resulted in positive results for their pan-beta coronavirus vaccine, ecoVACC showing the probable protection from additional beta-coronaviruses and emerging variants.508 |
| Vitti Laboratories (USA) | umbilical-cord-derived exosomes/EV-Pure in combination with Wharton’s jelly MSCs (WJ-Pure) | moderate to severe ARDS associated with COVID-19 | NCT05387278 | phase I (2022) | While Vitti Laboratories has two current clinical trials,509 they have many more disease indications in their pipeline for their EV-Pure exosomal product along with exosomal-based topical and serum applications for wound healing and age-related macular degeneration, respectively. Their EV-Pure product is currently researched preclinically for treatment of COPD, osteoarthritis, traumatic brain injury, Crohn’s disease, polycystic ovarian syndrome, and Alzheimer’s disease.510 |
Details obtained from https://clinicaltrials.gov/.
Exosome Diagnostics in Clinical Trials
Currently on https://clinicaltrials.gov, there is a total of 208 clinical trials with exosomes being used for diagnosis (Supporting Information Table S2). Over half of these clinical trials (108 clinical trials) are related to cancer diagnosis utilizing exosomes. Other highly represented diseases include neurological diseases (15 clinical trials), cardiovascular diseases (13 clinical trials), and lung diseases (6 clinical trials). Early diagnosis of these diseases is crucial for better prognosis. The large number of clinical trials of exosomes in diagnosis highlighted the value and advantage of using exosomes in early disease diagnosis. Table 12 highlights the companies, medical centers, and universities related to exosome diagnosis of these diseases.
Table 12. Highlighted Exosome Diagnostic Clinical Trialsa.
| Companies/medical centers/universities (location) | Exosome (disease target) | Disease diagnosed | Clinical trial number | Clinical trial status (date initiated) | Summary |
|---|---|---|---|---|---|
| University of Alabama at Birmingham (USA) | blood- or urine-derived exosomes (LRRK2) | Parkinson’s disease | NCT04350177 | completed (2013) | Researchers used this study to determine exosome biomarkers for Parkinson’s disease and to determine if LRKK2 expression within exosomes from LRRK2 kinase inhibitor sunitinub treated patients decreased after treatment. They hope to use this information to build an assay for on-target effects for future LRRK2 inhibitor clinical trials.511 |
| Boston University (USA) | plasma-derived exosome (tau) | chronic traumatic encephalopathy | NCT02798185 | active (2016) | Boston University researchers collaborated with Exosome Sciences and Aethlon Medical for the DETECT CTE research project, which aims to validate exosomal tau as a non-invasive CTE biomarker. Preliminary findings look promising that plasma exosomal tau may be an accurate, non-invasive biomarker for CTE.328 Researchers are using this clinical trial as an advancement to the previous study with the goal of diagnosing CTE during life for the prevention and treatment of the disease. |
| Exosome Diagnostics (USA) | urine-derived exosome (ERG, PCA3, and SPDEF) | prostate cancer | NCT02702856 | completed (2016) | Exosome Diagnostics developed the ExoDx test that utilizes circulating cancer exosomes from urine-derived exosomes and is commercially available. The ExoDx test was granted FDA Breakthrough Device Designation in 2019.512 They also have current clinical trials for the use of exosomes in diagnosis of non-small-cell lung cancer513 and kidney transplant rejection.514 Preliminary data from their breast cancer trial reveals specific gene signatures could be isolated from plasma-derived exosomes,515 and their kidney transplant trial showed the discovery of two separate gene signatures for the monitoring of kidney transplant rejections.516 They have also had success preclinically with identifying plasma biomarkers for glioblastoma517 and a saliva exosomal RNA signature for Sjogren’s syndrome.518 |
| miR Scientific (USA) | urine-derived exosome (442 sncRNA) | bladder cancer | NCT04155359 | recruiting (2019) | miR Scientific developed the miR Sentinel test currently commercially available for prostate cancer detection with extracted sncRNA in urine-derived exosomes.519 They are investigating through this clinical trial if there is evidence that they can also diagnose bladder cancer with the miR Sentinel test. The miR Sentinel test received FDA Breakthrough Device Designation in 2020.520 |
| University of Utah Center for Clinical and Translational Science (USA) | urine-derived exosomes (sodium transporters) | heart failure with preserved ejection fraction (HFpEF) | NCT03837470 | completed (2019) | This trial examines sodium transporters in the exosomes from patients with HFpEF for characterization to aid in diagnosis and treatment of these patients.521 |
| Aarhus University Hospital (Denmark) | plasma-derived exosomes | acute ischemic stroke | NCT04266639 | completed (2020) | This trial was performed to determine if exosome isolation with characterization of the nucleic acid (DNA and RNA, including miRNA) content will show any decrease in stroke complications and any advantage of remote ischemic conditioning.522 |
| Lithuanian University of Health Sciences (Lithuania) | eosinophil-derived exosome | asthma | NCT04542902 | recruiting (2020) | This clinical trial’s investigation of ncRNA in eosinophil-derived exosomes will provide insights on eosinophils subtypes in airway remodeling. ncRNAs are key regulators for gene transcription, and researchers predict that altered blood levels of ncRNAs could be a diagnostic biomarker in asthma.523 |
| Peking Union Medical College Hospital (China) | blood and bronchoalveolar lavage fluid (BALF)-derived exosomes | ARDS | NCT05451342 | recruiting (2022) | This clinical trial will characterize exosomes from blood and BALF with transcriptome and metabolomic analysis to aid in the diagnosis of ARDS.524 |
Details obtained from https://clinicaltrials.gov/.
Exosomes as the Disease Target in Clinical Trials
Using exosomes as targets is another avenue that is being explored for disease treatment. Aetholon Medical is a California-based clinical company that has designed an investigational medical device called the Hemopurifier. Targeting circulating exosomes, the Hemopurifier captures viral and bacterial toxins and cancer exosomes to treat disease. To date, Aetholon has used the Hemopurifer to treat patients with ebola, hepatitis C, HIV, and COVID-19.525 Their two current clinical trials are explored in Table 13.
Table 13. Highlighted Clinical Trials That Target Exosomes (Physical Elimination) for Disease Treatmenta.
| Company (location) | Exosome | Disease treated | Clinical trial number | Clinical trial status (date initiated) | Summary |
|---|---|---|---|---|---|
| Aethlon Medical (PA, USA) | circulating exosomes | squamous cell carcinoma of the head and neck | NCT04453046 | recruiting (2020) | Aethon is conducting an early feasibility study for the treatment of head and neck cancer with their Hemopurifier in combination with the antibody drug pembrolizumab.526 |
| Aethlon Medical (PA, USA) | circulating exosomes | COVID-19 | NCT04595903 | recruiting (2021) | Preclinical results had promising results showing Galanthus nivalis agglutinin affinity resin of Aethlon’s Hemopurifier captures seven clinically relevant variants of SARS-CoV-2.527 They are currently in an early feasibility study for the treatment of COVID-19, where their first patient successfully completed treatment.528 |
Details obtained from https://clinicaltrials.gov/.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.2c08774.
Table S1, exemplary small molecule drugs delivered by exosomes; Figure S1, classes of substances represented in the documents related to exosome applications in therapy and diagnostics as found in the CAS Content Collection; Figure S2, percentages of documents concerning the major tetraspanin classes in the documents related to exosome applications in therapy and diagnostics, along with role indicators for the top three tetraspanin classes; Figure S3, number of documents related to exosome applications in therapy and diagnostics, in which various kinds of cells have been used as exosome donors; Figure S4, annual trend of the number of documents in the CAS Content Collection related to the exosome applications in cosmetics and food (PDF)
Table S2, therapeutic and diagnostic exosome clinical trials (XLSX)
The authors declare no competing financial interest.
Supplementary Material
References
- Bangham A. D.; Horne R. W. Negative staining of phospholipids and their structural modification by surface-active agents as observed in the electron microscope. J. Mol. Biol. 1964, 8, 660–668. 10.1016/S0022-2836(64)80115-7. [DOI] [PubMed] [Google Scholar]
- Harding C.; Stahl P. Transferrin recycling in reticulocytes: pH and iron are important determinants of ligand binding and processing. Biochem. Biophys. Res. Commun. 1983, 113, 650–658. 10.1016/0006-291X(83)91776-X. [DOI] [PubMed] [Google Scholar]
- Pan B. T.; Johnstone R. M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell 1983, 33, 967–978. 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- Pegtel D. M.; Gould S. J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. 10.1146/annurev-biochem-013118-111902. [DOI] [PubMed] [Google Scholar]
- Chen H.; Wang L.; Zeng X.; Schwarz H.; Nanda H. S.; Peng X.; Zhou Y. Exosomes, a New Star for Targeted Delivery. Frontiers in Cell and Developmental Biology 2021, 9, 751079. 10.3389/fcell.2021.751079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S.-A.; Xie Y.; Fu Z.; Wang Y.; Wang J.-A.; Xiang M. Emerging role of exosome-mediated intercellular communication in vascular remodeling. Oncotarget 2017, 8, 25700–25712. 10.18632/oncotarget.14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol. Rep. 2011, 3, 15. 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross R.Meet the exosome, the rising star in drug delivery. Chem. Eng. News 2018. https://cen.acs.org/business/start-ups/Meet-exosome-rising-star-drug/96/i31. [Google Scholar]
- Kumar D. N.; Chaudhuri A.; Aqil F.; Dehari D.; Munagala R.; Singh S.; Gupta R. C.; Agrawal A. K. Exosomes as Emerging Drug Delivery and Diagnostic Modality for Breast Cancer: Recent Advances in Isolation and Application. Cancers (Basel) 2022, 14, 1435. 10.3390/cancers14061435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenchov R.; Bird R.; Curtze A. E.; Zhou Q. Lipid Nanoparticles—From Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano 2021, 15, 16982–17015. 10.1021/acsnano.1c04996. [DOI] [PubMed] [Google Scholar]
- Begines B.; Ortiz T.; Pérez-Aranda M.; Martínez G.; Merinero M.; Argüelles-Arias F.; Alcudia A. Polymeric Nanoparticles for Drug Delivery: Recent Developments and Future Prospects. Nanomaterials (Basel) 2020, 10, 1403. 10.3390/nano10071403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meel R.; Fens M. H.; Vader P.; van Solinge W. W.; Eniola-Adefeso O.; Schiffelers R. M. Extracellular vesicles as drug delivery systems: lessons from the liposome field. J. Controlled Release 2014, 195, 72–85. 10.1016/j.jconrel.2014.07.049. [DOI] [PubMed] [Google Scholar]
- Gregoriadis G. Liposomes in Drug Delivery: How It All Happened. Pharmaceutics 2016, 8, 19–19. 10.3390/pharmaceutics8020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koynova R.; Tenchov B.; MacDonald R. C. Nonlamellar Phases in Cationic Phospholipids, Relevance to Drug and Gene Delivery. ACS Biomaterials Science & Engineering 2015, 1, 130–138. 10.1021/ab500142w. [DOI] [PubMed] [Google Scholar]
- CAS Content Collection. https://www.cas.org/about/cas-content (accessed January 5, 2022).
- Bradley J. A.; Bolton E. M.; Pedersen R. A. Stem cell medicine encounters the immune system. Nat. Rev. Immunol. 2002, 2, 859–871. 10.1038/nri934. [DOI] [PubMed] [Google Scholar]
- Dougherty J. A.; Kumar N.; Noor M.; Angelos M. G.; Khan M.; Chen C. A.; Khan M. Extracellular Vesicles Released by Human Induced-Pluripotent Stem Cell-Derived Cardiomyocytes Promote Angiogenesis. Front. Physiol. 2018, 9, 1794. 10.3389/fphys.2018.01794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. P.; Mardini O.; Ericsson M.; Prabhakar S.; Maguire C.; Chen J. W.; Tannous B. A.; Breakefield X. O. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano 2014, 8, 483–494. 10.1021/nn404945r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.; Li J.; Huang B.; Liu J.; Chen X.; Chen X.-M.; Xu Y.-M.; Huang L.-F.; Wang X.-Z. Exosomes: Novel Biomarkers for Clinical Diagnosis. Scientific World Journal 2015, 2015, 657086. 10.1155/2015/657086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huda M. N.; Nafiujjaman M.; Deaguero I. G.; Okonkwo J.; Hill M. L.; Kim T.; Nurunnabi M. Potential Use of Exosomes as Diagnostic Biomarkers and in Targeted Drug Delivery: Progress in Clinical and Preclinical Applications. ACS Biomaterials Science & Engineering 2021, 7, 2106–2149. 10.1021/acsbiomaterials.1c00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Society for Extracellular Vesicles. https://www.isev.org/ (accessed August 3, 2022).
- Théry C.; Witwer K. W.; Aikawa E.; Alcaraz M. J.; Anderson J. D.; Andriantsitohaina R.; Antoniou A.; Arab T.; Archer F.; Atkin-Smith G. K.; Ayre D. C.; Bach J.-M.; Bachurski D.; Baharvand H.; Balaj L.; Baldacchino S.; Bauer N. N.; Baxter A. A.; Bebawy M.; Beckham C.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles 2018, 7, 1535750. 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P.; Petfalski E.; Shevchenko A.; Mann M.; Tollervey D. The Exosome: A Conserved Eukaryotic RNA Processing Complex Containing Multiple 3′→5′ Exoribonucleases. Cell 1997, 91, 457–466. 10.1016/S0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Wolf P. The Nature and Significance of Platelet Products in Human Plasma. Br. J. Hamaetol. 1967, 13, 269–288. 10.1111/j.1365-2141.1967.tb08741.x. [DOI] [PubMed] [Google Scholar]
- Zitvogel L.; Regnault A.; Lozier A.; Wolfers J.; Flament C.; Tenza D.; Ricciardi-Castagnoli P.; Raposo G.; Amigorena S. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell derived exosomes. Nat. Med. 1998, 4, 594–600. 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- Escudier B.; Dorval T.; Chaput N.; André F.; Caby M. P.; Novault S.; Flament C.; Leboulaire C.; Borg C.; Amigorena S.; Boccaccio C.; Bonnerot C.; Dhellin O.; Movassagh M.; Piperno S.; Robert C.; Serra V.; Valente N.; Le Pecq J. B.; Spatz A.; et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of thefirst phase I clinical trial. J. Transl. Med. 2005, 3, 10. 10.1186/1479-5876-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trams E. G.; Lauter C. J.; Salem N. Jr.; Heine U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta 1981, 645, 63–70. 10.1016/0005-2736(81)90512-5. [DOI] [PubMed] [Google Scholar]
- Johnstone R. M.; Adam M.; Hammond J. R.; Orr L.; Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. 10.1016/S0021-9258(18)48095-7. [DOI] [PubMed] [Google Scholar]
- Raposo G.; Nijman H. W.; Stoorvogel W.; Liejendekker R.; Harding C. V.; Melief C. J.; Geuze H. J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfers J.; Lozier A.; Raposo G.; Regnault A.; Théry C.; Masurier C.; Flament C.; Pouzieux S.; Faure F.; Tursz T.; Angevin E.; Amigorena S.; Zitvogel L. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med. 2001, 7, 297–303. 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- Savina A.; Vidal M.; Colombo M. I. The exosome pathway in K562 cells is regulated by Rab11. J. Cell Sci. 2002, 115, 2505–2515. 10.1242/jcs.115.12.2505. [DOI] [PubMed] [Google Scholar]
- Valadi H.; Ekström K.; Bossios A.; Sjöstrand M.; Lee J. J.; Lötvall J. O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Skog J.; Würdinger T.; van Rijn S.; Meijer D. H.; Gainche L.; Curry W. T.; Carter B. S.; Krichevsky A. M.; Breakefield X. O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski M.; Carmo N. B.; Krumeich S.; Fanget I.; Raposo G.; Savina A.; Moita C. F.; Schauer K.; Hume A. N.; Freitas R. P.; Goud B.; Benaroch P.; Hacohen N.; Fukuda M.; Desnos C.; Seabra M. C.; Darchen F.; Amigorena S.; Moita L. F.; Thery C. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- Hsu C.; Morohashi Y.; Yoshimura S.; Manrique-Hoyos N.; Jung S.; Lauterbach M. A.; Bakhti M.; Grønborg M.; Möbius W.; Rhee J.; Barr F. A.; Simons M. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J. Cell Biol. 2010, 189, 223–232. 10.1083/jcb.200911018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L.; Seow Y.; Yin H.; Betts C.; Lakhal S.; Wood M. J. A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- Peinado H.; Alečković M.; Lavotshkin S.; Matei I.; Costa-Silva B.; Moreno-Bueno G.; Hergueta-Redondo M.; Williams C.; García-Santos G.; Ghajar C. M.; Nitadori-Hoshino A.; Hoffman C.; Badal K.; Garcia B. A.; Callahan M. K.; Yuan J.; Martins V. R.; Skog J.; Kaplan R. N.; Brady M. S.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 2013 Nobel Prize in Physiology or Medicine. https://www.nobelprize.org/prizes/medicine/2013/press-release/ (accessed July 15, 2022). [DOI] [PubMed]
- Henriksen M.; Johnsen K. B.; Olesen P.; Pilgaard L.; Duroux M. MicroRNA Expression Signatures and Their Correlation with Clinicopathological Features in Glioblastoma Multiforme. Neuromolecular Med. 2014, 16, 565–577. 10.1007/s12017-014-8309-7. [DOI] [PubMed] [Google Scholar]
- Kruger S.; Elmageed Z. Y. A.; Hawke D. H.; Wörner P. M.; Jansen D. A.; Abdel-Mageed A. B.; Alt E. U.; Izadpanah R. Molecular characterization of exosome-like vesicles from breast cancer cells. BMC Cancer 2014, 14, 44. 10.1186/1471-2407-14-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manterola L.; Guruceaga E.; Pérez-Larraya J. G.; González-Huarriz M.; Jauregui P.; Tejada S.; Diez-Valle R.; Segura V.; Samprón N.; Barrena C.; Ruiz I.; Agirre A.; Ayuso Á.; Rodríguez J.; González Á.; Xipell E.; Matheu A.; López de Munain A.; Tuñón T.; Zazpe I.; et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol. 2014, 16, 520–527. 10.1093/neuonc/not218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A.; Costa-Silva B.; Shen T. L.; Rodrigues G.; Hashimoto A.; Tesic Mark M.; Molina H.; Kohsaka S.; Di Giannatale A.; Ceder S.; Singh S.; Williams C.; Soplop N.; Uryu K.; Pharmer L.; King T.; Bojmar L.; Davies A. E.; Ararso Y.; Zhang T.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo S. A.; Luecke L. B.; Kahlert C.; Fernandez A. F.; Gammon S. T.; Kaye J.; LeBleu V. S.; Mittendorf E. A.; Weitz J.; Rahbari N.; Reissfelder C.; Pilarsky C.; Fraga M. F.; Piwnica-Worms D.; Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi S.; Nguyen T. D. T.; Marasini R.; Aryal S. Macrophage-derived exosome-mimetic hybrid vesicles for tumor targeted drug delivery. Acta Biomater. 2019, 94, 482–494. 10.1016/j.actbio.2019.05.054. [DOI] [PubMed] [Google Scholar]
- Efficacy and Safety of EXOSOME-MSC Therapy to Reduce Hyper-inflammation In Moderate COVID-19 Patients (EXOMSC-COV19). https://www.clinicaltrials.gov/ct2/show/NCT05216562?term=exosome&cond=COVID-19&draw=2&rank=2 (accessed July 15, 2022).
- Jamalkhah M.; Asaadi Y.; Azangou-Khyavy M.; Khanali J.; Soleimani M.; Kiani J.; Arefian E. MSC-derived exosomes carrying a cocktail of exogenous interfering RNAs an unprecedented therapy in era of COVID-19 outbreak. J. Transl. Med. 2021, 19, 164. 10.1186/s12967-021-02840-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C. A.; Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 1990, 61, 723–733. 10.1016/0092-8674(90)90483-U. [DOI] [PubMed] [Google Scholar]
- Söllner T.; Whiteheart S. W.; Brunner M.; Erdjument-Bromage H.; Geromanos S.; Tempst P.; Rothman J. E. SNAP receptors implicated in vesicle targeting and fusion. Nature 1993, 362, 318–324. 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Fevrier B.; Vilette D.; Archer F.; Loew D.; Faigle W.; Vidal M.; Laude H.; Raposo G. Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 9683–9688. 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai R. C.; Arslan F.; Lee M. M.; Sze N. S. K.; Choo A.; Chen T. S.; Salto-Tellez M.; Timmers L.; Lee C. N.; El Oakley R. M.; Pasterkamp G.; de Kleijn D. P. V.; Lim S. K. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Research 2010, 4, 214–222. 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Zhuang X.; Xiang X.; Grizzle W.; Sun D.; Zhang S.; Axtell R. C.; Ju S.; Mu J.; Zhang L.; Steinman L.; Miller D.; Zhang H. G. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol. Ther. 2011, 19, 1769–1779. 10.1038/mt.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH RePORTER. https://reporter.nih.gov/ (accessed August 2, 2022).
- NIH Reporter - Projects Funding by Fiscal Year. https://reporter.nih.gov/search/x7G9Jfo4WU-Qfsl8HFH-_g/projects/charts?fy=2022;2021;2020;2019;2018;2017;2016;2015;2014;2013;2012;2011;2010;2009;2008;2007;2006;2005;2004;2003;2002 (accessed August 27, 2022).
- Delenclos M.; Trendafilova T.; Mahesh D.; Baine A. M.; Moussaud S.; Yan I. K.; Patel T.; McLean P. J. Investigation of Endocytic Pathways for the Internalization of Exosome-Associated Oligomeric Alpha-Synuclein. Front. Neurosci. 2017, 11, 172. 10.3389/fnins.2017.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanelli L.; Magini A.; Buratta S.; Brozzi A.; Sagini K.; Polchi A.; Tancini B.; Emiliani C. Signaling pathways in exosomes biogenesis, secretion and fate. Genes (Basel) 2013, 4, 152–170. 10.3390/genes4020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M.; Raposo G.; Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev Biol. 2014, 30, 255–289. 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- Gurung S.; Perocheau D.; Touramanidou L.; Baruteau J. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Communication and Signaling 2021, 19, 47. 10.1186/s12964-021-00730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth A. M.; Fang Y.; Fallon J. K.; Yang J. M.; Hildreth J. E.; Gould S. J. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J. Cell Biol. 2006, 172, 923–935. 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordjour F. K.; Daaboul G. G.; Gould S. J.. A shared pathway of exosome biogenesis operates at plasma and endosome membranes. 2019, 545228. bioRxiv. https://www.biorxiv.org/content/10.1101/545228v1 (accessed October 20, 2022). [DOI] [PMC free article] [PubMed]
- Casado S.; Lobo M. d. V. T.; Paíno C. L. Dynamics of plasma membrane surface related to the release of extracellular vesicles by mesenchymal stem cells in culture. Sci. Rep. 2017, 7, 6767. 10.1038/s41598-017-07265-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.; Shi J.; Xie J.; Wang Y.; Sun J.; Liu T.; Zhao Y.; Zhao X.; Wang X.; Ma Y.; Malkoc V.; Chiang C.; Deng W.; Chen Y.; Fu Y.; Kwak K. J.; Fan Y.; Kang C.; Yin C.; Rhee J.; et al. Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation. Nat. Biomed Eng. 2020, 4, 69–83. 10.1038/s41551-019-0485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J.; Schelhaas M.; Helenius A. Virus entry by endocytosis. Annu. Rev. Biochem. 2010, 79, 803–833. 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- Donoso-Quezada J.; Ayala-Mar S.; González-Valdez J. The role of lipids in exosome biology and intercellular communication: Function, analytics and applications. Traffic 2021, 22, 204–220. 10.1111/tra.12803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A.; Okada R.; Nagao K.; Kawamata Y.; Hanyu A.; Yoshimoto S.; Takasugi M.; Watanabe S.; Kanemaki M. T.; Obuse C.; Hara E. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 2017, 8, 15287. 10.1038/ncomms15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crenshaw B. J.; Gu L.; Sims B.; Matthews Q. L. Exosome Biogenesis and Biological Function in Response to Viral Infections. Open Virol. J. 2018, 12, 134–148. 10.2174/1874357901812010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C.; Ostrowski M.; Segura E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- Zduriencikova M.; Gronesova P.; Cholujova D.; Sedlak J. Potential biomarkers of exosomal cargo in endocrine signaling. Endocr. Regul. 2015, 49, 141–150. 10.4149/endo_2015_03_141. [DOI] [PubMed] [Google Scholar]
- Leone D. A.; Rees A. J.; Kain R. Dendritic cells and routing cargo into exosomes. Immunol Cell Biol. 2018, 96, 683. 10.1111/imcb.12170. [DOI] [PubMed] [Google Scholar]
- Skotland T.; Hessvik N. P.; Sandvig K.; Llorente A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 2019, 60, 9–18. 10.1194/jlr.R084343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryabin G. O.; Komelkov A. V.; Savelyeva E. E.; Tchevkina E. M. Lipid Rafts in Exosome Biogenesis. Biochemistry (Moscow) 2020, 85, 177–191. 10.1134/S0006297920020054. [DOI] [PubMed] [Google Scholar]
- Marat A. L.; Haucke V. Phosphatidylinositol 3-phosphates-at the interface between cell signalling and membrane traffic. EMBO J. 2016, 35, 561–579. 10.15252/embj.201593564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K.; Breyne K.; Ughetto S.; Laurent L. C.; Breakefield X. O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. 10.1038/s41580-020-0251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y.; Yamamoto Y.; Sato T. A.; Ochiya T. Extracellular vesicles as trans-genomic agents: Emerging roles in disease and evolution. Cancer Sci. 2017, 108, 824–830. 10.1111/cas.13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorey J. S.; Cheng Y.; Singh P. P.; Smith V. L. Exosomes and other extracellular vesicles in host–pathogen interactions. EMBO reports 2015, 16, 24–43. 10.15252/embr.201439363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood J. L.; San R. S.; Wickline S. A. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011, 71, 3792–3801. 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- Iero M.; Valenti R.; Huber V.; Filipazzi P.; Parmiani G.; Fais S.; Rivoltini L. Tumour-released exosomes and their implications in cancer immunity. Cell Death Differ. 2008, 15, 80–88. 10.1038/sj.cdd.4402237. [DOI] [PubMed] [Google Scholar]
- Saint-Pol J.; Gosselet F.; Duban-Deweer S.; Pottiez G.; Karamanos Y. Targeting and Crossing the Blood-Brain Barrier with Extracellular Vesicles. Cells 2020, 9, 851. 10.3390/cells9040851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro M. F.; Zhu H.; Millard R. W.; Fan G. C. Exosomes Function in Pro- and Anti-Angiogenesis. Curr. Angiogenes 2013, 2, 54–59. 10.2174/22115528113020020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y.; Luan J.; Li H.; Zhou X.; Han J. Exosomes derived from mineralizing osteoblasts promote ST2 cell osteogenic differentiation by alteration of microRNA expression. FEBS Lett. 2016, 590, 185–192. 10.1002/1873-3468.12024. [DOI] [PubMed] [Google Scholar]
- Sung B. H.; Ketova T.; Hoshino D.; Zijlstra A.; Weaver A. M. Directional cell movement through tissues is controlled by exosome secretion. Nat. Commun. 2015, 6, 7164. 10.1038/ncomms8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ExoCarta - Exosome Protein, RNA and Lipid Database. http://www.exocarta.org/ (accessed July 14, 2022).
- Mathivanan S.; Fahner C. J.; Reid G. E.; Simpson R. J. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012, 40, D1241–1244. 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesiclepedia. http://www.microvesicles.org/ (accessed July 28, 2022).
- EVpedia/Exosome RNA. https://exosome-rna.com/tag/evpedia/#:~:text=Tag%20Archives%3A%20EVpedia&text=Extracellular%20vesicles%20(EVs)%20are%20a,conserved%20across%20prokaryotes%20and%20eukaryoteshttps://exosome-rna.com/tag/evpedia/#:~:text=Tag%20Archives%3A%20EVpedia&text=Extracellular%20vesicles%20(EVs)%20are%20a,conserved%20across%20prokaryotes%20and%20eukaryotes. (accessed July 28, 2022).
- Kim D. K.; Kang B.; Kim O. Y.; Choi D. S.; Lee J.; Kim S. R.; Go G.; Yoon Y. J.; Kim J. H.; Jang S. C.; Park K. S.; Choi E. J.; Kim K. P.; Desiderio D. M.; Kim Y. K.; Lötvall J.; Hwang D.; Gho Y. S. EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J. Extracell Vesicles 2013, 2, 20384. 10.3402/jev.v2i0.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschuschke M.; Kocherova I.; Bryja A.; Mozdziak P.; Angelova Volponi A.; Janowicz K.; Sibiak R.; Piotrowska-Kempisty H.; Iżycki D.; Bukowska D.; Antosik P.; Shibli J. A.; Dyszkiewicz-Konwińska M.; Kempisty B. Inclusion Biogenesis, Methods of Isolation and Clinical Application of Human Cellular Exosomes. J. Clin Med. 2020, 9, 436. 10.3390/jcm9020436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A.; Bisht B.; Dutta S.; Paul M. K. Current advances in the use of exosomes, liposomes, and bioengineered hybrid nanovesicles in cancer detection and therapy. Acta Pharmacol. Sin. 2022, 43, 2759–2776. 10.1038/s41401-022-00902-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya-Beltri C.; Baixauli F.; Gutiérrez-Vázquez C.; Sánchez-Madrid F.; Mittelbrunn M. Sorting it out: regulation of exosome loading. Semin. Cancer Biol. 2014, 28, 3–13. 10.1016/j.semcancer.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boorn J. G.; Daßler J.; Coch C.; Schlee M.; Hartmann G. Exosomes as nucleic acid nanocarriers. Adv. Drug Delivery Rev. 2013, 65, 331–335. 10.1016/j.addr.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Ye M.; Wang J.; Pan S.; Zheng L.; Wang Z.-W.; Zhu X. Nucleic acids and proteins carried by exosomes of different origins as potential biomarkers for gynecologic cancers. Molecular Therapy - Oncolytics 2022, 24, 101–113. 10.1016/j.omto.2021.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentino T. R.; Rule B. D.; Mobley C. B.; Nikolova-Karakashian M.; Vechetti I. J. Skeletal Muscle Cell Growth Alters the Lipid Composition of Extracellular Vesicles. Membranes (Basel) 2021, 11, 619. 10.3390/membranes11080619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D.-S.; Kim D.-K.; Kim Y.-K.; Gho Y. S. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics 2013, 13, 1554–1571. 10.1002/pmic.201200329. [DOI] [PubMed] [Google Scholar]
- Skotland T.; Sandvig K.; Llorente A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41. 10.1016/j.plipres.2017.03.001. [DOI] [PubMed] [Google Scholar]
- Llorente A.; Skotland T.; Sylvänne T.; Kauhanen D.; Róg T.; Orłowski A.; Vattulainen I.; Ekroos K.; Sandvig K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta 2013, 1831, 1302–1309. 10.1016/j.bbalip.2013.04.011. [DOI] [PubMed] [Google Scholar]
- Carayon K.; Chaoui K.; Ronzier E.; Lazar I.; Bertrand-Michel J.; Roques V.; Balor S.; Terce F.; Lopez A.; Salomé L.; Joly E. Proteolipidic Composition of Exosomes Changes during Reticulocyte Maturation*. J. Biol. Chem. 2011, 286, 34426–34439. 10.1074/jbc.M111.257444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloribi S.; Ristorcelli E.; Breuzard G.; Silvy F.; Bertrand-Michel J.; Beraud E.; Verine A.; Lombardo D. Exosomal Lipids Impact Notch Signaling and Induce Death of Human Pancreatic Tumoral SOJ-6 Cells. PLoS One 2012, 7, e47480 10.1371/journal.pone.0047480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenegger M.; Kremser J.; Frewein M. P. K.; Ziherl P.; Bonthuis D. J.; Pabst G. Intrinsic lipid curvatures of mammalian plasma membrane outer leaflet lipids and ceramides. Biochim. Biophys. Acta 2021, 1863, 183709. 10.1016/j.bbamem.2021.183709. [DOI] [PubMed] [Google Scholar]
- Koynova R.; Tenchov B.. Phase Transitions and Phase Behavior of Lipids. In Encyclopedia of Biophysics; Roberts G. C. K., Ed.; Springer Verlag: Berlin, 2013; pp 1841–1854. [Google Scholar]
- McMahon H. T.; Boucrot E. Membrane curvature at a glance. J. Cell Sci. 2015, 128, 1065–1070. 10.1242/jcs.114454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subra C.; Laulagnier K.; Perret B.; Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 2007, 89, 205–212. 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Peterka O.; Jirásko R.; Chocholoušková M.; Kuchař L.; Wolrab D.; Hájek R.; Vrána D.; Strouhal O.; Melichar B.; Holčapek M. Lipidomic characterization of exosomes isolated from human plasma using various mass spectrometry techniques. Biochim. Biophys. Acta 2020, 1865, 158634. 10.1016/j.bbalip.2020.158634. [DOI] [PubMed] [Google Scholar]
- Wubbolts R.; Leckie R. S.; Veenhuizen P. T. M.; Schwarzmann G.; Möbius W.; Hoernschemeyer J.; Slot J.-W.; Geuze H. J.; Stoorvogel W. Proteomic and Biochemical Analyses of Human B Cell-derived Exosomes: Potential Implications for Their Function and Multivesicular Body Formation. J. Biol. Chem. 2003, 278, 10963–10972. 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- Simons K.; Sampaio J. L. Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 2011, 3, a004697. 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenchov B. G.; Koynova R. D. The effect of nonideal lateral mixing on the transmembrane lipid asymmetry. Biochim. Biophys. Acta 1985, 815, 380–391. 10.1016/0005-2736(85)90364-5. [DOI] [PubMed] [Google Scholar]
- Koynova R. D.; Tenchov B. G. Effect of ion concentration on phosphatidylethanolamine distribution in mixed vesicles. Biochim. Biophys. Acta 1983, 727, 351–356. 10.1016/0005-2736(83)90420-0. [DOI] [PubMed] [Google Scholar]
- van Meer G.; Voelker D. R.; Feigenson G. W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A.; Korte T.; Herrmann A.; Wohland T. Plasma membrane asymmetry of lipid organization: fluorescence lifetime microscopy and correlation spectroscopy analysis[S]. J. Lipid Res. 2020, 61, 252–266. 10.1194/jlr.D119000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa K.; Kurata S.; Yanagihashi Y.; Brummelkamp T. R.; Matsuda F.; Nagata S. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 2014, 344, 1164–1168. 10.1126/science.1252809. [DOI] [PubMed] [Google Scholar]
- Miyanishi M.; Tada K.; Koike M.; Uchiyama Y.; Kitamura T.; Nagata S. Identification of Tim4 as a phosphatidylserine receptor. Nature 2007, 450, 435–439. 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- Lea J.; Sharma R.; Yang F.; Zhu H.; Ward E. S.; Schroit A. J. Detection of phosphatidylserine-positive exosomes as a diagnostic marker for ovarian malignancies: a proof of concept study. Oncotarget 2017, 8, 14395. 10.18632/oncotarget.14795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P.; Mao K.; Xu J.; Wu F.; Wang X.; Wang S.; Zhou M.; Duan L.; Tan Q.; Ma G.; Yang G.; Du R.; Huang H.; Huang Q.; Li Y.; Guo M.; Jin Y. Metabolic characteristics of large and small extracellular vesicles from pleural effusion reveal biomarker candidates for the diagnosis of tuberculosis and malignancy. Journal of Extracellular Vesicles 2020, 9, 1790158. 10.1080/20013078.2020.1790158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M.; Moita C.; van Niel G.; Kowal J.; Vigneron J.; Benaroch P.; Manel N.; Moita L. F.; Théry C.; Raposo G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- Matsumoto A.; Takahashi Y.; Nishikawa M.; Sano K.; Morishita M.; Charoenviriyakul C.; Saji H.; Takakura Y. Role of Phosphatidylserine-Derived Negative Surface Charges in the Recognition and Uptake of Intravenously Injected B16BL6-Derived Exosomes by Macrophages. J. Pharm. Sci. 2017, 106, 168–175. 10.1016/j.xphs.2016.07.022. [DOI] [PubMed] [Google Scholar]
- Pfrieger F. W.; Vitale N. Thematic Review Series: Exosomes and Microvesicles: Lipids as Key Components of their Biogenesis and Functions, Cholesterol and the journey of extracellular vesicles. J. Lipid Res. 2018, 59, 2255–2261. 10.1194/jlr.R084210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subra C.; Grand D.; Laulagnier K.; Stella A.; Lambeau G.; Paillasse M.; De Medina P.; Monsarrat B.; Perret B.; Silvente-Poirot S.; Poirot M.; Record M. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J. Lipid Res. 2010, 51, 2105–2120. 10.1194/jlr.M003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.; Dubois R. N. Eicosanoids and cancer. Nat. Rev. Cancer 2010, 10, 181–193. 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escola J. M.; Kleijmeer M. J.; Stoorvogel W.; Griffith J. M.; Yoshie O.; Geuze H. J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998, 273, 20121–20127. 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- Hemler M. E. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev Biol. 2003, 19, 397–422. 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- Mazurov D.; Barbashova L.; Filatov A. Tetraspanin protein CD9 interacts with metalloprotease CD10 and enhances its release via exosomes. FEBS J. 2013, 280, 1200–1213. 10.1111/febs.12110. [DOI] [PubMed] [Google Scholar]
- Clayton A.; Al-Taei S.; Webber J.; Mason M. D.; Tabi Z. Cancer exosomes express CD39 and CD73, which suppress T cells through adenosine production. J. Immunol. 2011, 187, 676–683. 10.4049/jimmunol.1003884. [DOI] [PubMed] [Google Scholar]
- Rabesandratana H.; Toutant J. P.; Reggio H.; Vidal M. Decay-accelerating factor (CD55) and membrane inhibitor of reactive lysis (CD59) are released within exosomes during In vitro maturation of reticulocytes. Blood 1998, 91, 2573–2580. 10.1182/blood.V91.7.2573. [DOI] [PubMed] [Google Scholar]
- Cheng L.; Zhao W.; Hill A. F. Exosomes and their role in the intercellular trafficking of normal and disease associated prion proteins. Mol. Aspects Med. 2018, 60, 62–68. 10.1016/j.mam.2017.11.011. [DOI] [PubMed] [Google Scholar]
- Qi J.; Zhou Y.; Jiao Z.; Wang X.; Zhao Y.; Li Y.; Chen H.; Yang L.; Zhu H.; Li Y. Exosomes Derived from Human Bone Marrow Mesenchymal Stem Cells Promote Tumor Growth Through Hedgehog Signaling Pathway. Cell Physiol Biochem 2017, 42, 2242–2254. 10.1159/000479998. [DOI] [PubMed] [Google Scholar]
- Demory Beckler M.; Higginbotham J. N.; Franklin J. L.; Ham A. J.; Halvey P. J.; Imasuen I. E.; Whitwell C.; Li M.; Liebler D. C.; Coffey R. J. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol. Cell Proteomics 2013, 12, 343–355. 10.1074/mcp.M112.022806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y.; Wu N.; Gan X.; Yan W.; Morrell J. C.; Gould S. J. Higher-Order Oligomerization Targets Plasma Membrane Proteins and HIV Gag to Exosomes. PLoS Biol. 2007, 5, e158 10.1371/journal.pbio.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley J.; Cordy B.; Lucia D.; Fradkin L. G.; Budnik V.; Thomson T. Retrovirus-like Gag Protein Arc1 Binds RNA and Traffics across Synaptic Boutons. Cell 2018, 172, 262–274. 10.1016/j.cell.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuzyn E. D.; Day C. E.; Kearns R. B.; Kyrke-Smith M.; Taibi A. V.; McCormick J.; Yoder N.; Belnap D. M.; Erlendsson S.; Morado D. R.; Briggs J. A. G.; Feschotte C.; Shepherd J. D. The Neuronal Gene Arc Encodes a Repurposed Retrotransposon Gag Protein that Mediates Intercellular RNA Transfer. Cell 2018, 172, 275–288. 10.1016/j.cell.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J. C.; Zelarayán L. C. The Mingle-Mangle of Wnt Signaling and Extracellular Vesicles: Functional Implications for Heart Research. Front Cardiovasc Med. 2018, 5, 10. 10.3389/fcvm.2018.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.; Sun Y.; Zhang Q. Emerging Role of Extracellular Vesicles in Bone Remodeling. J. Dent. Res. 2018, 97, 859–868. 10.1177/0022034518764411. [DOI] [PubMed] [Google Scholar]
- Borges F. T.; Melo S. A.; Özdemir B. C.; Kato N.; Revuelta I.; Miller C. A.; Gattone V. H.; LeBleu V. S. 2nd; Kalluri R. TGF-β1-containing exosomes from injured epithelial cells activate fibroblasts to initiate tissue regenerative responses and fibrosis. J. Am. Soc. Nephrol. 2013, 24, 385–392. 10.1681/ASN.2012101031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampey G. C.; Saifuddin M.; Schwab A.; Barclay R.; Punya S.; Chung M. C.; Hakami R. M.; Zadeh M. A.; Lepene B.; Klase Z. A.; El-Hage N.; Young M.; Iordanskiy S.; Kashanchi F. Exosomes from HIV-1-infected Cells Stimulate Production of Pro-inflammatory Cytokines through Trans-activating Response (TAR) RNA. J. Biol. Chem. 2016, 291, 1251–1266. 10.1074/jbc.M115.662171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough I. J.; Vincent J. P. Exosomes in developmental signalling. Development 2016, 143, 2482–2493. 10.1242/dev.126516. [DOI] [PubMed] [Google Scholar]
- Zheng J.; Hernandez J. M.; Doussot A.; Bojmar L.; Zambirinis C. P.; Costa-Silva B.; van Beek E.; Mark M. T.; Molina H.; Askan G.; Basturk O.; Gonen M.; Kingham T. P.; Allen P. J.; D’Angelica M. I.; DeMatteo R. P.; Lyden D.; Jarnagin W. R. Extracellular matrix proteins and carcinoembryonic antigen-related cell adhesion molecules characterize pancreatic duct fluid exosomes in patients with pancreatic cancer. HPB (Oxford) 2018, 20, 597–604. 10.1016/j.hpb.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santasusagna S.; Moreno I.; Navarro A.; Castellano J. J.; Martinez F.; Hernández R.; Muñoz C.; Monzo M. Proteomic Analysis of Liquid Biopsy from Tumor-Draining Vein Indicates that High Expression of Exosomal ECM1 Is Associated with Relapse in Stage I-III Colon Cancer. Transl. Oncol. 2018, 11, 715–721. 10.1016/j.tranon.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima K.; Aoki N.; Kato T.; Kitajima K.; Matsuda T. Secretion of a peripheral membrane protein, MFG-E8, as a complex with membrane vesicles. Eur. J. Biochem. 2002, 269, 1209–1218. 10.1046/j.1432-1033.2002.02758.x. [DOI] [PubMed] [Google Scholar]
- Shen B.; Fang Y.; Wu N.; Gould S. J. Biogenesis of the posterior pole is mediated by the exosome/microvesicle protein-sorting pathway. J. Biol. Chem. 2011, 286, 44162–44176. 10.1074/jbc.M111.274803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisitkun T.; Shen R.-F.; Knepper M. A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 13368–13373. 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friand V.; David G.; Zimmermann P. Syntenin and syndecan in the biogenesis of exosomes. Biol. Cell 2015, 107, 331–341. 10.1111/boc.201500010. [DOI] [PubMed] [Google Scholar]
- Baietti M. F.; Zhang Z.; Mortier E.; Melchior A.; Degeest G.; Geeraerts A.; Ivarsson Y.; Depoortere F.; Coomans C.; Vermeiren E.; Zimmermann P.; David G. Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- Juan T.; Fürthauer M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin Cell Dev Biol. 2018, 74, 66–77. 10.1016/j.semcdb.2017.08.022. [DOI] [PubMed] [Google Scholar]
- Taha E. A.; Ono K.; Eguchi T. Roles of Extracellular HSPs as Biomarkers in Immune Surveillance and Immune Evasion. Int. J. Mol. Sci. 2019, 20, 4588. 10.3390/ijms20184588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopf F. H.; Biebl M. M.; Buchner J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017, 18, 345–360. 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- Anderson H. C. Vesicles associated with calcification in the matrix of epiphyseal cartilage. J. Cell Biol. 1969, 41, 59–72. 10.1083/jcb.41.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhshian Nik A.; Hutcheson J. D.; Aikawa E. Extracellular Vesicles As Mediators of Cardiovascular Calcification. Front Cardiovasc Med. 2017, 4, 78. 10.3389/fcvm.2017.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C.; Boussac M.; Véron P.; Ricciardi-Castagnoli P.; Raposo G.; Garin J.; Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001, 166, 7309–7318. 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- Kamerkar S.; LeBleu V. S.; Sugimoto H.; Yang S.; Ruivo C. F.; Melo S. A.; Lee J. J.; Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi K. B.; Fijalkowski N.; Cano M.; Handa J. T. Decreased membrane complement regulators in the retinal pigmented epithelium contributes to age-related macular degeneration. Journal of Pathology 2013, 229, 729–742. 10.1002/path.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak J.; Miekus K.; Kucia M.; Zhang J.; Reca R.; Dvorak P.; Ratajczak M. Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006, 20, 847–856. 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- Pegtel D. M.; Cosmopoulos K.; Thorley-Lawson D. A.; van Eijndhoven M. A.; Hopmans E. S.; Lindenberg J. L.; de Gruijl T. D.; Würdinger T.; Middeldorp J. M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 6328–6333. 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasso J. M.; Ambrose B. J. B.; Tenchov R.; Datta R. S.; Basel M. T.; DeLong R. K.; Zhou Q. A. The Progress and Promise of RNA Medicine—An Arsenal of Targeted Treatments. J. Med. Chem. 2022, 65, 6975–7015. 10.1021/acs.jmedchem.2c00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z.; Batagov A. O.; Schinelli S.; Wang J.; Wang Y.; El Fatimy R.; Rabinovsky R.; Balaj L.; Chen C. C.; Hochberg F.; Carter B.; Breakefield X. O.; Krichevsky A. M. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 2017, 8, 1145. 10.1038/s41467-017-01196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang P.; Shah M.; Beljanski V. Exosomal RNAs in diagnosis and therapies. Non-coding RNA Research 2022, 7, 7–15. 10.1016/j.ncrna.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.; Yuan T.; Tschannen M.; Sun Z.; Jacob H.; Du M.; Liang M.; Dittmar R. L.; Liu Y.; Liang M.; Kohli M.; Thibodeau S. N.; Boardman L.; Wang L. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 2013, 14, 319. 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlert C.; Melo S. A.; Protopopov A.; Tang J.; Seth S.; Koch M.; Zhang J.; Weitz J.; Chin L.; Futreal A.; Kalluri R. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 2014, 289, 3869–3875. 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur B. K.; Zhang H.; Becker A.; Matei I.; Huang Y.; Costa-Silva B.; Zheng Y.; Hoshino A.; Brazier H.; Xiang J.; Williams C.; Rodriguez-Barrueco R.; Silva J. M.; Zhang W.; Hearn S.; Elemento O.; Paknejad N.; Manova-Todorova K.; Welte K.; Bromberg J.; et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone P.; Savini C.; Kurelac I.; Chang Q.; Amato L. B.; Strillacci A.; Stepanova A.; Iommarini L.; Mastroleo C.; Daly L.; Galkin A.; Thakur B. K.; Soplop N.; Uryu K.; Hoshino A.; Norton L.; Bonafé M.; Cricca M.; Gasparre G.; Lyden D.; et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E9066–E9075. 10.1073/pnas.1704862114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanam J.; Chetty V. K.; Barthel L.; Reinhardt D.; Hoyer P.-F.; Thakur B. K. DNA in extracellular vesicles: from evolution to its current application in health and disease. Cell Biosci. 2022, 12, 37. 10.1186/s13578-022-00771-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi A.; Villar-Prados A.; Oliphint P. A.; Zhang J.; Song X.; De Hoff P.; Morey R.; Liu J.; Roszik J.; Clise-Dwyer K.; Burks J. K.; O’Halloran T. J.; Laurent L. C.; Sood A. K. Mechanisms of nuclear content loading to exosomes. Sci. Adv. 2019, 5, eaax8849 10.1126/sciadv.aax8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista B. S.; Eng W. S.; Pilobello K. T.; Hendricks-Muñoz K. D.; Mahal L. K. Identification of a conserved glycan signature for microvesicles. J. Proteome Res. 2011, 10, 4624–4633. 10.1021/pr200434y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.; Royo F.; Aizpurua-Olaizola O.; Pazos R.; Boons G. J.; Reichardt N. C.; Falcon-Perez J. M. Glycosylation of extracellular vesicles: current knowledge, tools and clinical perspectives. J. Extracell Vesicles 2018, 7, 1442985. 10.1080/20013078.2018.1442985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Rosa G.; Ruggeri C.; Aloisi A. From Exosome Glycobiology to Exosome Glycotechnology, the Role of Natural Occurring Polysaccharides. Polysaccharides 2021, 2, 311–338. 10.3390/polysaccharides2020021. [DOI] [Google Scholar]
- Costa J. Glycoconjugates from extracellular vesicles: Structures, functions and emerging potential as cancer biomarkers. Biochim Biophys Acta Rev. Cancer 2017, 1868, 157–166. 10.1016/j.bbcan.2017.03.007. [DOI] [PubMed] [Google Scholar]
- Tumor Markers. https://www.cancer.gov/about-cancer/diagnosis-staging/diagnosis/tumor-markers-fact-sheet (accessed July 27, 2022).
- Exosome Diagnostics and Therapeutics Market Size, Share, Growth, and Forecast to 2029 | DataMIntelligence. https://www.medgadget.com/2022/04/exosome-diagnostics-and-therapeutics-market-size-share-growth-and-forecast-to-2029-datamintelligence.html (accessed August 16, 2022).
- Brennan K.; Martin K.; FitzGerald S. P.; O’Sullivan J.; Wu Y.; Blanco A.; Richardson C.; Mc Gee M. M. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 2020, 10, 1039. 10.1038/s41598-020-57497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L. L.; Zhu J.; Liu J. X.; Jiang F.; Ni W. K.; Qu L. S.; Ni R. Z.; Lu C. H.; Xiao M. B. A Comparison of Traditional and Novel Methods for the Separation of Exosomes from Human Samples. Biomed Res. Int. 2018, 2018, 3634563. 10.1155/2018/3634563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momen-Heravi F.; Balaj L.; Alian S.; Trachtenberg A. J.; Hochberg F. H.; Skog J.; Kuo W. P. Impact of biofluid viscosity on size and sedimentation efficiency of the isolated microvesicles. Front. Physiol. 2012, 3, 162. 10.3389/fphys.2012.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. Z.; Ma Z. J.; Kang X. W. Current status and outlook of advances in exosome isolation. Anal Bioanal Chem. 2022, 414, 7123–7141. 10.1007/s00216-022-04253-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Wu H. J.; Fine D.; Schmulen J.; Hu Y.; Godin B.; Zhang J. X.; Liu X. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab Chip 2013, 13, 2879–2882. 10.1039/c3lc41343h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bioo Scientific Corporation Releases The ExoMir(TM) Kit To Facilitate RNA Analysis in Cell-Free Fluids. https://www.biospace.com/article/releases/bioo-scientific-corporation-releases-the-exomir-tm-kit-to-facilitate-rna-analysis-in-cell-free-fluids-/ (accessed August 16, 2022).
- Nordin J. Z.; Lee Y.; Vader P.; Mäger I.; Johansson H. J.; Heusermann W.; Wiklander O. P.; Hällbrink M.; Seow Y.; Bultema J. J.; Gilthorpe J.; Davies T.; Fairchild P. J.; Gabrielsson S.; Meisner-Kober N. C.; Lehtiö J.; Smith C. I.; Wood M. J.; El Andaloussi S. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine 2015, 11, 879–883. 10.1016/j.nano.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Oksvold M. P.; Neurauter A.; Pedersen K. W. Magnetic bead-based isolation of exosomes. Methods Mol. Biol. 2015, 1218, 465–481. 10.1007/978-1-4939-1538-5_27. [DOI] [PubMed] [Google Scholar]
- Pedersen K. W.; Kierulf B.; Neurauter A. Specific and Generic Isolation of Extracellular Vesicles with Magnetic Beads. Methods Mol. Biol. 2017, 1660, 65–87. 10.1007/978-1-4939-7253-1_7. [DOI] [PubMed] [Google Scholar]
- Greening D. W.; Xu R.; Ji H.; Tauro B. J.; Simpson R. J. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. 2015, 1295, 179–209. 10.1007/978-1-4939-2550-6_15. [DOI] [PubMed] [Google Scholar]
- Mizutani K.; Terazawa R.; Kameyama K.; Kato T.; Horie K.; Tsuchiya T.; Seike K.; Ehara H.; Fujita Y.; Kawakami K.; Ito M.; Deguchi T. Isolation of prostate cancer-related exosomes. Anticancer Res. 2014, 34, 3419–3423. [PubMed] [Google Scholar]
- Woo J.; Sharma S.; Gimzewski J. The Role of Isolation Methods on a Nanoscale Surface Structure and its Effect on the Size of Exosomes. J. Circ Biomark 2016, 5, 11. 10.5772/64148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exosome Isolation Kit Pan, human. https://www.miltenyibiotec.com/US-en/products/exosome-isolation-kit-pan-human.html#gref (accessed August 16, 2022).
- Lohmann L. J.; Strube J. Accelerating Biologics Manufacturing by Modeling: Process Integration of Precipitation in mAb Downstream Processing. Processes 2020, 8, 58. 10.3390/pr8010058. [DOI] [Google Scholar]
- Wang X.; Kwak K. J.; Yang Z.; Zhang A.; Zhang X.; Sullivan R.; Lin D.; Lee R. L.; Castro C.; Ghoshal K.; Schmidt C.; Lee L. J. Extracellular mRNA detected by molecular beacons in tethered lipoplex nanoparticles for diagnosis of human hepatocellular carcinoma. PLoS One 2018, 13, e0198552 10.1371/journal.pone.0198552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliescu F. S.; Vrtačnik D.; Neuzil P.; Iliescu C. Microfluidic Technology for Clinical Applications of Exosomes. Micromachines (Basel) 2019, 10, 392. 10.3390/mi10060392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.; Wijerathne H.; Godwin A. K.; Soper S. A. Isolation and analysis methods of extracellular vesicles (EVs). Extracell Vesicles Circ Nucl. Acids 2021, 2, 80–103. 10.20517/evcna.2021.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijerathne H.; Witek M. A.; Jackson J. M.; Brown V.; Hupert M. L.; Herrera K.; Kramer C.; Davidow A. E.; Li Y.; Baird A. E.; Murphy M. C.; Soper S. A. Affinity enrichment of extracellular vesicles from plasma reveals mRNA changes associated with acute ischemic stroke. Commun. Biol. 2020, 3, 613. 10.1038/s42003-020-01336-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang B.; Zhu Y.; Ni J.; Thompson J.; Malouf D.; Bucci J.; Graham P.; Li Y. Extracellular vesicles: the next generation of biomarkers for liquid biopsy-based prostate cancer diagnosis. Theranostics 2020, 10, 2309–2326. 10.7150/thno.39486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.; Ouyang Y.; Wang Z.; Zhang R.; Huang P. H.; Chen C.; Li H.; Li P.; Quinn D.; Dao M.; Suresh S.; Sadovsky Y.; Huang T. J. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 10584–10589. 10.1073/pnas.1709210114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibsen S. D.; Wright J.; Lewis J. M.; Kim S.; Ko S. Y.; Ong J.; Manouchehri S.; Vyas A.; Akers J.; Chen C. C.; Carter B. S.; Esener S. C.; Heller M. J. Rapid Isolation and Detection of Exosomes and Associated Biomarkers from Plasma. ACS Nano 2017, 11, 6641–6651. 10.1021/acsnano.7b00549. [DOI] [PubMed] [Google Scholar]
- Sidhom K.; Obi P. O.; Saleem A. A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option?. Int. J. Mol. Sci. 2020, 21, 6466. 10.3390/ijms21186466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. D.; Zacharias W.; Gercel-Taylor C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol. Biol. 2011, 728, 235–246. 10.1007/978-1-61779-068-3_15. [DOI] [PubMed] [Google Scholar]
- Taylor D. D.; Lyons K. S.; Gerçel-Taylor C. Shed membrane fragment-associated markers for endometrial and ovarian cancers. Gynecol. Oncol. 2002, 84, 443–448. 10.1006/gyno.2001.6551. [DOI] [PubMed] [Google Scholar]
- Lozano-Ramos I.; Bancu I.; Oliveira-Tercero A.; Armengol M. P.; Menezes-Neto A.; Del Portillo H. A.; Lauzurica-Valdemoros R.; Borràs F. E. Size-exclusion chromatography-based enrichment of extracellular vesicles from urine samples. J. Extracell Vesicles 2015, 4, 27369. 10.3402/jev.v4.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira D.; Moreira J. N.; Rodrigues L. R. New advances in exosome-based targeted drug delivery systems. Critical Reviews in Oncology/Hematology 2022, 172, 103628. 10.1016/j.critrevonc.2022.103628. [DOI] [PubMed] [Google Scholar]
- Yang D.; Zhang W.; Zhang H.; Zhang F.; Chen L.; Ma L.; Larcher L. M.; Chen S.; Liu N.; Zhao Q.; Tran P. H. L.; Chen C.; Veedu R. N.; Wang T. Progress, opportunity, and perspective on exosome isolation - efforts for efficient exosome-based theranostics. Theranostics 2020, 10, 3684–3707. 10.7150/thno.41580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala-Mar S.; Donoso-Quezada J.; Gallo-Villanueva R. C.; Perez-Gonzalez V. H.; González-Valdez J. Recent advances and challenges in the recovery and purification of cellular exosomes. Electrophoresis 2019, 40, 3036–3049. 10.1002/elps.201800526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraunhofer W.; Winter G. The use of asymmetrical flow field-flow fractionation in pharmaceutics and biopharmaceutics. Eur. J. Pharm. Biopharm. 2004, 58, 369–383. 10.1016/j.ejpb.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Zhang K.; Yue Y.; Wu S.; Liu W.; Shi J.; Zhang Z. Rapid Capture and Nondestructive Release of Extracellular Vesicles Using Aptamer-Based Magnetic Isolation. ACS Sensors 2019, 4, 1245–1251. 10.1021/acssensors.9b00060. [DOI] [PubMed] [Google Scholar]
- Batrakova E. V.; Kim M. S. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J. Controlled Release 2015, 219, 396–405. 10.1016/j.jconrel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargo Loading into Exosomes. https://www.creative-biolabs.com/exosome/cargo-loading-into-exosomes.htm?gclid=Cj0KCQjw3eeXBhD7ARIsAHjssr8jAvBATTlmPXiuVhOO0cHVvbQ925eoWMXRUiPSWYtJQtQCze12NYQaAlwoEALw_wcB%20;%2010.3389/fbioe.2020.586130 (accessed August 18, 2022).
- Xi X. M.; Xia S. J.; Lu R. Drug loading techniques for exosome-based drug delivery systems. Pharmazie 2021, 76, 61–67. 10.1691/ph.2021.0128. [DOI] [PubMed] [Google Scholar]
- Ran N.; Gao X.; Dong X.; Li J.; Lin C.; Geng M.; Yin H. Effects of exosome-mediated delivery of myostatin propeptide on functional recovery of mdx mice. Biomaterials 2020, 236, 119826. 10.1016/j.biomaterials.2020.119826. [DOI] [PubMed] [Google Scholar]
- Vakhshiteh F.; Rahmani S.; Ostad S. N.; Madjd Z.; Dinarvand R.; Atyabi F. Exosomes derived from miR-34a-overexpressing mesenchymal stem cells inhibit in vitro tumor growth: A new approach for drug delivery. Life Sci. 2021, 266, 118871. 10.1016/j.lfs.2020.118871. [DOI] [PubMed] [Google Scholar]
- Pascucci L.; Coccè V.; Bonomi A.; Ami D.; Ceccarelli P.; Ciusani E.; Viganò L.; Locatelli A.; Sisto F.; Doglia S. M.; Parati E.; Bernardo M. E.; Muraca M.; Alessandri G.; Bondiolotti G.; Pessina A. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: a new approach for drug delivery. J. Controlled Release 2014, 192, 262–270. 10.1016/j.jconrel.2014.07.042. [DOI] [PubMed] [Google Scholar]
- Fitts C. A.; Ji N.; Li Y.; Tan C. Exploiting Exosomes in Cancer Liquid Biopsies and Drug Delivery. Adv. Healthcare Mater. 2019, 8, 1801268. 10.1002/adhm.201801268. [DOI] [PubMed] [Google Scholar]
- Pignatello R.Biological membranes and their role in physio-pathological conditions. In Drug-Biomembrane Interaction Studies; Pignatello R., Ed.; Woodhead Publishing: Cambridge, U.K., 2013; pp 1–46. [Google Scholar]
- Agrawal A. K.; Aqil F.; Jeyabalan J.; Spencer W. A.; Beck J.; Gachuki B. W.; Alhakeem S. S.; Oben K.; Munagala R.; Bondada S.; Gupta R. C. Milk-derived exosomes for oral delivery of paclitaxel. Nanomedicine 2017, 13, 1627–1636. 10.1016/j.nano.2017.03.001. [DOI] [PubMed] [Google Scholar]
- Giassafaki L.-P. N.; Siqueira S.; Panteris E.; Psatha K.; Chatzopoulou F.; Aivaliotis M.; Tzimagiorgis G.; Müllertz A.; Fatouros D. G.; Vizirianakis I. S. Towards analyzing the potential of exosomes to deliver microRNA therapeutics. J. Cell. Physiol. 2021, 236, 1529–1544. 10.1002/jcp.29991. [DOI] [PubMed] [Google Scholar]
- Wang J.; Chen D.; Ho E. A. Challenges in the development and establishment of exosome-based drug delivery systems. J. Controlled Release 2021, 329, 894–906. 10.1016/j.jconrel.2020.10.020. [DOI] [PubMed] [Google Scholar]
- Luan X.; Sansanaphongpricha K.; Myers I.; Chen H.; Yuan H.; Sun D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 2017, 38, 754–763. 10.1038/aps.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane T. N.; Raiker R. S.; Jay S. M. Exogenous DNA Loading into Extracellular Vesicles via Electroporation is Size-Dependent and Enables Limited Gene Delivery. Mol. Pharmaceutics 2015, 12, 3650–3657. 10.1021/acs.molpharmaceut.5b00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M.; Sawada K.; Miyamoto M.; Shimizu A.; Yamamoto M.; Kinose Y.; Nakamura K.; Kawano M.; Kodama M.; Hashimoto K.; Kimura T. Exploring the potential of engineered exosomes as delivery systems for tumor-suppressor microRNA replacement therapy in ovarian cancer. Biochemical and biophysical research communications 2020, 527, 153–161. 10.1016/j.bbrc.2020.04.076. [DOI] [PubMed] [Google Scholar]
- Lv Q.; Deng J.; Chen Y.; Wang Y.; Liu B.; Liu J. Engineered Human Adipose Stem-Cell-Derived Exosomes Loaded with miR-21-5p to Promote Diabetic Cutaneous Wound Healing. Mol. Pharmaceutics 2020, 17, 1723–1733. 10.1021/acs.molpharmaceut.0c00177. [DOI] [PubMed] [Google Scholar]
- Hajipour H.; Farzadi L.; Roshangar L.; Latifi Z.; Kahroba H.; Shahnazi V.; Hamdi K.; Ghasemzadeh A.; Fattahi A.; Nouri M. A human chorionic gonadotropin (hCG) delivery platform using engineered uterine exosomes to improve endometrial receptivity. Life Sci. 2021, 275, 119351. 10.1016/j.lfs.2021.119351. [DOI] [PubMed] [Google Scholar]
- Donoso-Quezada J.; Ayala-Mar S.; González-Valdez J. State-of-the-art exosome loading and functionalization techniques for enhanced therapeutics: a review. Crit. Rev. Biotechnol. 2020, 40, 804–820. 10.1080/07388551.2020.1785385. [DOI] [PubMed] [Google Scholar]
- Kooijmans S. A. A.; Stremersch S.; Braeckmans K.; de Smedt S. C.; Hendrix A.; Wood M. J. A.; Schiffelers R. M.; Raemdonck K.; Vader P. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J. Controlled Release 2013, 172, 229–238. 10.1016/j.jconrel.2013.08.014. [DOI] [PubMed] [Google Scholar]
- Sato Y. T.; Umezaki K.; Sawada S.; Mukai S.-a.; Sasaki Y.; Harada N.; Shiku H.; Akiyoshi K. Engineering hybrid exosomes by membrane fusion with liposomes. Sci. Rep. 2016, 6, 21933. 10.1038/srep21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P. H. L.; Wang T.; Yin W.; Tran T. T. D.; Nguyen T. N. G.; Lee B.-J.; Duan W. Aspirin-loaded nanoexosomes as cancer therapeutics. Int. J. Pharm. 2019, 572, 118786. 10.1016/j.ijpharm.2019.118786. [DOI] [PubMed] [Google Scholar]
- Le Saux S.; Aarrass H.; Lai-Kee-Him J.; Bron P.; Armengaud J.; Miotello G.; Bertrand-Michel J.; Dubois E.; George S.; Faklaris O.; Devoisselle J. M.; Legrand P.; Chopineau J.; Morille M. Post-production modifications of murine mesenchymal stem cell (mMSC) derived extracellular vesicles (EVs) and impact on their cellular interaction. Biomaterials 2020, 231, 119675. 10.1016/j.biomaterials.2019.119675. [DOI] [PubMed] [Google Scholar]
- Shin K.; Fogg V. C.; Margolis B. Tight junctions and cell polarity. Annu. Rev. Cell Dev Biol. 2006, 22, 207–235. 10.1146/annurev.cellbio.22.010305.104219. [DOI] [PubMed] [Google Scholar]
- Yang H.; Fu H.; Wang B.; Zhang X.; Mao J.; Li X.; Wang M.; Sun Z.; Qian H.; Xu W. Exosomal miR-423-5p targets SUFU to promote cancer growth and metastasis and serves as a novel marker for gastric cancer. Mol. Carcinog. 2018, 57, 1223–1236. 10.1002/mc.22838. [DOI] [PubMed] [Google Scholar]
- Wu J.; Li S.; Zhang P. Tumor-derived exosomes: immune properties and clinical application in lung cancer. Cancer Drug Resistance 2022, 5, 102–113. 10.20517/cdr.2021.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciravolo V.; Huber V.; Ghedini G. C.; Venturelli E.; Bianchi F.; Campiglio M.; Morelli D.; Villa A.; Della Mina P.; Menard S.; Filipazzi P.; Rivoltini L.; Tagliabue E.; Pupa S. M. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J. Cell. Physiol. 2012, 227, 658–667. 10.1002/jcp.22773. [DOI] [PubMed] [Google Scholar]
- Qiao L.; Hu S.; Huang K.; Su T.; Li Z.; Vandergriff A.; Cores J.; Dinh P. U.; Allen T.; Shen D.; Liang H.; Li Y.; Cheng K. Tumor cell-derived exosomes home to their cells of origin and can be used as Trojan horses to deliver cancer drugs. Theranostics 2020, 10, 3474–3487. 10.7150/thno.39434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramini B.; Masciale V.; Haider K. H. Defining lung cancer stem cells exosomal payload of miRNAs in clinical perspective. World J. Stem Cells 2020, 12, 406–421. 10.4252/wjsc.v12.i6.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.; Yan C.; Mu L.; Huang K.; Li X.; Tao D.; Wu Y.; Qin J. Fibroblast-Derived Exosomes Contribute to Chemoresistance through Priming Cancer Stem Cells in Colorectal Cancer. PLoS One 2015, 10, e0125625 10.1371/journal.pone.0125625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R.; Wang S.; Zhao R. C. Exosomes from human adipose-derived mesenchymal stem cells promote migration through Wnt signaling pathway in a breast cancer cell model. Mol. Cell. Biochem. 2013, 383, 13–20. 10.1007/s11010-013-1746-z. [DOI] [PubMed] [Google Scholar]
- Regmi S.; Pathak S.; Kim J. O.; Yong C. S.; Jeong J. H. Mesenchymal stem cell therapy for the treatment of inflammatory diseases: Challenges, opportunities, and future perspectives. Eur. J. Cell Biol. 2019, 98, 151041. 10.1016/j.ejcb.2019.04.002. [DOI] [PubMed] [Google Scholar]
- Adelipour M.; Babaei F.; Mirzababaei M.; Allameh A. Correlation of micro vessel density and c-Myc expression in breast tumor of mice following mesenchymal stem cell therapy. Tissue Cell 2017, 49, 315–322. 10.1016/j.tice.2017.01.007. [DOI] [PubMed] [Google Scholar]
- Kim G. B.; Shon O. J.; Seo M. S.; Choi Y.; Park W. T.; Lee G. W. Mesenchymal Stem Cell-Derived Exosomes and Their Therapeutic Potential for Osteoarthritis. Biology (Basel) 2021, 10, 285. 10.3390/biology10040285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue R.; Lu S.; Luo Y.; Zeng J.; Liang H.; Qin D.; Wang X.; Wang T.; Pu J.; Hu H. Mesenchymal stem cell-derived exosomal microRNA-182-5p alleviates myocardial ischemia/reperfusion injury by targeting GSDMD in mice. Cell Death Discov 2022, 8, 202. 10.1038/s41420-022-00909-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Direct Biologics clinical trials. https://clinicaltrials.gov/ct2/results?cond=&term=exoflo&cntry=&state=&city=&dist=&Search=Search (accessed August 18, 2022).
- A Global Expanded Access Protocol on Bone Marrow Mesenchymal Stem Cell Derived Extracellular Vesicle Infusion Treatment for Patients With COVID-19 Associated ARDS. https://clinicaltrials.gov/ct2/show/NCT04657458 (accessed August 20, 2022).
- Sengupta V.; Sengupta S.; Lazo A.; Woods P.; Nolan A.; Bremer N. Exosomes Derived from Bone Marrow Mesenchymal Stem Cells as Treatment for Severe COVID-19. Stem Cells Dev. 2020, 29, 747–754. 10.1089/scd.2020.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Study of ExoFlo for the Treatment of Medically Refractory Ulcerative Colitis. https://clinicaltrials.gov/ct2/show/NCT05176366 (accessed August 20, 2022).
- A Phase I Study of ExoFlo, an ex Vivo Culture-expanded Adult Allogeneic Bone Marrow Mesenchymal Stem Cell Derived Extracellular Vesicle Isolate Product, for the Treatment of Medically Refractory Crohn’s Disease. https://clinicaltrials.gov/ct2/show/NCT05130983 (accessed August 20, 2022).
- Intermediate Size Expanded Access for the Use of ExoFlo in the Treatment of Abdominal Solid Organ Transplant Patients Who Are at Risk of Worsening Allograft Function With Conventional Immunosuppressive Therapy Alone. https://clinicaltrials.gov/ct2/show/NCT05215288 (accessed August 20, 2022).
- Bone Marrow Mesenchymal Stem Cell Derived Extracellular Vesicles Infusion Treatment for Mild-to-Moderate COVID-19: A Phase II Clinical Trial. https://clinicaltrials.gov/ct2/show/NCT05125562 (accessed August 20, 2022).
- Sridharan R.; Cameron A. R.; Kelly D. J.; Kearney C. J.; O’Brien F. J. Biomaterial based modulation of macrophage polarization: a review and suggested design principles. Mater. Today 2015, 18, 313–325. 10.1016/j.mattod.2015.01.019. [DOI] [Google Scholar]
- Poh A. R.; Ernst M. Targeting Macrophages in Cancer: From Bench to Bedside. Front. Oncol. 2018, 8, 49. 10.3389/fonc.2018.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.; Zhang Q.; Zhang X.; Han Q.; Li H.; Mao Y.; Wang X.; Guo H.; Irwin D. M.; Niu G.; Tan H. Exosomes from Macrophages Exposed to Apoptotic Breast Cancer Cells Promote Breast Cancer Proliferation and Metastasis. J. Cancer 2019, 10, 2892–2906. 10.7150/jca.31241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R.; Gao L.; Li Y.; Li G.; Qin P.; Wei Z.; Li D.; Qian C.; Li J.; Yang G. M2 macrophage-derived exosomes carry miR-1271-5p to alleviate cardiac injury in acute myocardial infarction through down-regulating SOX6. Mol. Immunol. 2021, 136, 26–35. 10.1016/j.molimm.2021.05.006. [DOI] [PubMed] [Google Scholar]
- Kosaka N.; Iguchi H.; Yoshioka Y.; Takeshita F.; Matsuki Y.; Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010, 285, 17442–17452. 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelbrunn M.; Gutiérrez-Vázquez C.; Villarroya-Beltri C.; González S.; Sánchez-Cabo F.; González M.; Bernad A.; Sánchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z.; Yang F.; Yu L.; Yu Z.; Jiang L.; Wang Q.; Yang Y.; Wang L.; Cao X.; Wang J. Activated T cell exosomes promote tumor invasion via Fas signaling pathway. J. Immunol. 2012, 188, 5954–5961. 10.4049/jimmunol.1103466. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Xie Y.; Li W.; Chibbar R.; Xiong S.; Xiang J. CD4(+) T cell-released exosomes inhibit CD8(+) cytotoxic T-lymphocyte responses and antitumor immunity. Cell. Mol. Immunol. 2011, 8, 23–30. 10.1038/cmi.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.; Huang C.; Song B.; Xiao Y.; Fang M.; Feng J.; Wang P. CD4+CD25+ regulatory T cells-derived exosomes prolonged kidney allograft survival in a rat model. Cell. Immunol. 2013, 285, 62–68. 10.1016/j.cellimm.2013.06.010. [DOI] [PubMed] [Google Scholar]
- COVID-19 Specific T Cell Derived Exosomes (CSTC-Exo). https://clinicaltrials.gov/ct2/show/NCT04389385 (accessed August 17, 2022).
- Webb R. L.; Kaiser E. E.; Scoville S. L.; Thompson T. A.; Fatima S.; Pandya C.; Sriram K.; Swetenburg R. L.; Vaibhav K.; Arbab A. S.; Baban B.; Dhandapani K. M.; Hess D. C.; Hoda M. N.; Stice S. L. Human Neural Stem Cell Extracellular Vesicles Improve Tissue and Functional Recovery in the Murine Thromboembolic Stroke Model. Translational Stroke Research 2018, 9, 530–539. 10.1007/s12975-017-0599-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.; Zhuang X.; Xiang X.; Liu Y.; Zhang S.; Liu C.; Barnes S.; Grizzle W.; Miller D.; Zhang H. G. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol. Ther. 2010, 18, 1606–1614. 10.1038/mt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. S.; Haney M. J.; Zhao Y.; Mahajan V.; Deygen I.; Klyachko N. L.; Inskoe E.; Piroyan A.; Sokolsky M.; Okolie O.; Hingtgen S. D.; Kabanov A. V.; Batrakova E. V. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine 2016, 12, 655–664. 10.1016/j.nano.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney M. J.; Klyachko N. L.; Zhao Y.; Gupta R.; Plotnikova E. G.; He Z.; Patel T.; Piroyan A.; Sokolsky M.; Kabanov A. V.; Batrakova E. V. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J. Controlled Release 2015, 207, 18–30. 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E.; Nam G. H.; Hong Y.; Kim Y. K.; Kim D. H.; Yang Y.; Kim I. S. Comparison of exosomes and ferritin protein nanocages for the delivery of membrane protein therapeutics. J. Controlled Release 2018, 279, 326–335. 10.1016/j.jconrel.2018.04.037. [DOI] [PubMed] [Google Scholar]
- Hong Y.; Nam G.-H.; Koh E.; Jeon S.; Kim G. B.; Jeong C.; Kim D.-H.; Yang Y.; Kim I.-S. Exosome as a Vehicle for Delivery of Membrane Protein Therapeutics, PH20, for Enhanced Tumor Penetration and Antitumor Efficacy. Adv. Funct. Mater. 2018, 28, 1703074. 10.1002/adfm.201703074. [DOI] [Google Scholar]
- Momen-Heravi F.; Bala S.; Bukong T.; Szabo G. Exosome-mediated delivery of functionally active miRNA-155 inhibitor to macrophages. Nanomedicine 2014, 10, 1517–1527. 10.1016/j.nano.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L.; Gu C.; Gan Y.; Shao L.; Chen H.; Zhu H. Exosome-mediated siRNA delivery to suppress postoperative breast cancer metastasis. J. Controlled Release 2020, 318, 1–15. 10.1016/j.jconrel.2019.12.005. [DOI] [PubMed] [Google Scholar]
- Jin Y.; Lee J. S.; Min S.; Park H.-J.; Kang T. J.; Cho S.-W. Bioengineered Extracellular Membranous Nanovesicles for Efficient Small-Interfering RNA Delivery: Versatile Platforms for Stem Cell Engineering and In Vivo Delivery. Adv. Funct. Mater. 2016, 26, 5804–5817. 10.1002/adfm.201601430. [DOI] [Google Scholar]
- Ran F. A.; Hsu P. D.; Wright J.; Agarwala V.; Scott D. A.; Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. M.; Yang Y.; Oh S. J.; Hong Y.; Seo M.; Jang M. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J. Controlled Release 2017, 266, 8–16. 10.1016/j.jconrel.2017.09.013. [DOI] [PubMed] [Google Scholar]
- Srivastava A.; Amreddy N.; Babu A.; Panneerselvam J.; Mehta M.; Muralidharan R.; Chen A.; Zhao Y. D.; Razaq M.; Riedinger N.; Kim H.; Liu S.; Wu S.; Abdel-Mageed A. B.; Munshi A.; Ramesh R. Nanosomes carrying doxorubicin exhibit potent anticancer activity against human lung cancer cells. Sci. Rep. 2016, 6, 38541. 10.1038/srep38541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. S.; Haney M. J.; Zhao Y.; Yuan D.; Deygen I.; Klyachko N. L.; Kabanov A. V.; Batrakova E. V. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: in vitro and in vivo evaluations. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 195–204. 10.1016/j.nano.2017.09.011. [DOI] [PubMed] [Google Scholar]
- Osterman C. J. D.; Lynch J. C.; Leaf P.; Gonda A.; Ferguson Bennit H. R.; Griffiths D.; Wall N. R. Curcumin Modulates Pancreatic Adenocarcinoma Cell-Derived Exosomal Function. PLoS One 2015, 10, e0132845 10.1371/journal.pone.0132845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.; Martin P.; Fogarty B.; Brown A.; Schurman K.; Phipps R.; Yin V. P.; Lockman P.; Bai S. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm. Res. 2015, 32, 2003–2014. 10.1007/s11095-014-1593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G.; Han Y.; An Y.; Ding Y.; He C.; Wang X.; Tang Q. NRP-1 targeted and cargo-loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo. Biomaterials 2018, 178, 302–316. 10.1016/j.biomaterials.2018.06.029. [DOI] [PubMed] [Google Scholar]
- Salarpour S.; Forootanfar H.; Pournamdari M.; Ahmadi-Zeidabadi M.; Esmaeeli M.; Pardakhty A. Paclitaxel incorporated exosomes derived from glioblastoma cells: comparative study of two loading techniques. Daru 2019, 27, 533–539. 10.1007/s40199-019-00280-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooshabadi V. T.; Khanmohammadi M.; Shafei S.; Banafshe H. R.; Malekshahi Z. V.; Ebrahimi-Barough S.; Ai J. Impact of atorvastatin loaded exosome as an anti-glioblastoma carrier to induce apoptosis of U87 cancer cells in 3D culture model. Biochem Biophys Rep 2020, 23, 100792. 10.1016/j.bbrep.2020.100792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y.; Li S.; Song J.; Ji T.; Zhu M.; Anderson G. J.; Wei J.; Nie G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 2014, 35, 2383–2390. 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- Lai N.; Wu D.; Liang T.; Pan P.; Yuan G.; Li X.; Li H.; Shen H.; Wang Z.; Chen G. Systemic exosomal miR-193b-3p delivery attenuates neuroinflammation in early brain injury after subarachnoid hemorrhage in mice. J. Neuroinflammation 2020, 17, 74. 10.1186/s12974-020-01745-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Chopp M.; Meng Y.; Katakowski M.; Xin H.; Mahmood A.; Xiong Y. Effect of exosomes derived from multipluripotent mesenchymal stromal cells on functional recovery and neurovascular plasticity in rats after traumatic brain injury. J. Neurosurg. 2015, 122, 856–867. 10.3171/2014.11.JNS14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Sui H.; Zheng Y.; Jiang Y.; Shi Y.; Liang J.; Zhao L. Curcumin-primed exosomes potently ameliorate cognitive function in AD mice by inhibiting hyperphosphorylation of the Tau protein through the AKT/GSK-3β pathway. Nanoscale 2019, 11, 7481–7496. 10.1039/C9NR01255A. [DOI] [PubMed] [Google Scholar]
- Qi Y.; Guo L.; Jiang Y.; Shi Y.; Sui H.; Zhao L. Brain delivery of quercetin-loaded exosomes improved cognitive function in AD mice by inhibiting phosphorylated tau-mediated neurofibrillary tangles. Drug Delivery 2020, 27, 745–755. 10.1080/10717544.2020.1762262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.; Li W.; Mao L.; Peng W.; Long D.; Li D.; Zhou R.; Dang X. Genetically engineered exosomes display RVG peptide and selectively enrich a neprilysin variant: a potential formulation for the treatment of Alzheimer’s disease. J. Drug Target. 2021, 29, 1128–1138. 10.1080/1061186X.2021.1929257. [DOI] [PubMed] [Google Scholar]
- Qu M.; Lin Q.; Huang L.; Fu Y.; Wang L.; He S.; Fu Y.; Yang S.; Zhang Z.; Zhang L.; Sun X. Dopamine-loaded blood exosomes targeted to brain for better treatment of Parkinson’s disease. J. Controlled Release 2018, 287, 156–166. 10.1016/j.jconrel.2018.08.035. [DOI] [PubMed] [Google Scholar]
- Kojima R.; Bojar D.; Rizzi G.; Hamri G. C.; El-Baba M. D.; Saxena P.; Ausländer S.; Tan K. R.; Fussenegger M. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat. Commun. 2018, 9, 1305. 10.1038/s41467-018-03733-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izco M.; Blesa J.; Schleef M.; Schmeer M.; Porcari R.; Al-Shawi R.; Ellmerich S.; de Toro M.; Gardiner C.; Seow Y.; Reinares-Sebastian A.; Forcen R.; Simons J. P.; Bellotti V.; Cooper J. M.; Alvarez-Erviti L. Systemic Exosomal Delivery of shRNA Minicircles Prevents Parkinsonian Pathology. Mol. Ther. 2019, 27, 2111–2122. 10.1016/j.ymthe.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. T.; Im W.; Ban J. J.; Lee M.; Jung K. H.; Lee S. K.; Chu K.; Kim M. Exosome-Based Delivery of miR-124 in a Huntington’s Disease Model. J. Mov Disord 2017, 10, 45–52. 10.14802/jmd.16054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiot M. C.; Hall L. M.; Coles A. H.; Haraszti R. A.; Godinho B. M.; Chase K.; Sapp E.; Ly S.; Alterman J. F.; Hassler M. R.; Echeverria D.; Raj L.; Morrissey D. V.; DiFiglia M.; Aronin N.; Khvorova A. Exosome-mediated Delivery of Hydrophobically Modified siRNA for Huntingtin mRNA Silencing. Mol. Ther. 2016, 24, 1836–1847. 10.1038/mt.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcayaga-Miranda F.; Cuenca J.; Khoury M. Antimicrobial Activity of Mesenchymal Stem Cells: Current Status and New Perspectives of Antimicrobial Peptide-Based Therapies. Front. Immunol. 2017, 8, 339. 10.3389/fimmu.2017.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrani M. I.; Bellio M. A.; Sagel A.; Saylor M.; Kapp W.; VanOsdol K.; Haskell G.; Stewart D.; Abdullah Z.; Santos I.; Milberg J.; Arango A.; Mitrani A.; Shapiro G. C. Case Report: Administration of Amniotic Fluid-Derived Nanoparticles in Three Severely Ill COVID-19 Patients. Frontiers in Medicine 2021, 8, 583842. 10.3389/fmed.2021.583842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera-Heredia M. I.; Morales L. C.; Vidal O. M.; Barceló E.; Silvera-Redondo C.; Vélez J. I.; Garavito-Galofre P. Exosomes: Potential Disease Biomarkers and New Therapeutic Targets. Biomedicines 2021, 9, 1061. 10.3390/biomedicines9081061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T.; Takahashi Y.; Takakura Y. Possibility of Exosome-Based Therapeutics and Challenges in Production of Exosomes Eligible for Therapeutic Application. Biol. Pharm. Bull. 2018, 41, 835–842. 10.1248/bpb.b18-00133. [DOI] [PubMed] [Google Scholar]
- Bashyal S.; Thapa C.; Lee S. Recent progresses in exosome-based systems for targeted drug delivery to the brain. J. Controlled Release 2022, 348, 723–744. 10.1016/j.jconrel.2022.06.011. [DOI] [PubMed] [Google Scholar]
- Dougherty J. A.; Mergaye M.; Kumar N.; Chen C.-A.; Angelos M. G.; Khan M. Potential Role of Exosomes in Mending a Broken Heart: Nanoshuttles Propelling Future Clinical Therapeutics Forward. Stem Cells Int. 2017, 2017, 5785436. 10.1155/2017/5785436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoodzadeh Hosseini H.; Ali Imani Fooladi A.; Reza Nourani M.; Ghanezadeh F. The Role of Exosomes in Infectious Diseases. Inflammation & Allergy - Drug Targets (Discontinued) 2013, 12, 29–37. 10.2174/1871528111312010005. [DOI] [PubMed] [Google Scholar]
- Schorey J. S.; Harding C. V. Extracellular vesicles and infectious diseases: new complexity to an old story. J. Clin. Invest. 2016, 126, 1181–1189. 10.1172/JCI81132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolini A.; Ferrari P.; Biava P. M. Exosomes and Cell Communication: From Tumour-Derived Exosomes and Their Role in Tumour Progression to the Use of Exosomal Cargo for Cancer Treatment. Cancers (Basel) 2021, 13, 822. 10.3390/cancers13040822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y.; Ding X.; Zhou L.; Zhang L.; Yang X. Mesenchymal stem cells-derived exosomal microRNA-139-5p restrains tumorigenesis in bladder cancer by targeting PRC1. Oncogene 2021, 40, 246–261. 10.1038/s41388-020-01486-7. [DOI] [PubMed] [Google Scholar]
- Liu D.; Chen C.; Cui M.; Zhang H. miR-140-3p inhibits colorectal cancer progression and its liver metastasis by targeting BCL9 and BCL2. Cancer Medicine 2021, 10, 3358–3372. 10.1002/cam4.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue S.; Ye X.; Zhou T.; Gan D.; Qian H.; Fang W.; Yao M.; Zhang D.; Shi H.; Chen T. PGRN(−/−) TAMs-derived exosomes inhibit breast cancer cell invasion and migration and its mechanism exploration. Life Sci. 2021, 264, 118687. 10.1016/j.lfs.2020.118687. [DOI] [PubMed] [Google Scholar]
- Tang Y.; Zong S.; Zeng H.; Ruan X.; Yao L.; Han S.; Hou F. MicroRNAs and angiogenesis: a new era for the management of colorectal cancer. Cancer Cell Int. 2021, 21, 221. 10.1186/s12935-021-01920-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M.; Shen Y.; Wang P.; Xie Z.; Xu S.; Zhu Z.; Wang Y.; Lyu Y.; Wang D.; Xu L.; Bi J.; Yang H. Exosomes Isolated From Human Umbilical Cord Mesenchymal Stem Cells Alleviate Neuroinflammation and Reduce Amyloid-Beta Deposition by Modulating Microglial Activation in Alzheimer’s Disease. Neurochem. Res. 2018, 43, 2165–2177. 10.1007/s11064-018-2641-5. [DOI] [PubMed] [Google Scholar]
- Nakano M.; Fujimiya M. Potential effects of mesenchymal stem cell derived extracellular vesicles and exosomal miRNAs in neurological disorders. Neural Regen Res. 2021, 16, 2359–2366. 10.4103/1673-5374.313026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Wang Z.; Xing H.; Wang Y.; Guo Y. Exosomes derived from miR-188-3p-modified adipose-derived mesenchymal stem cells protect Parkinson’s disease. Molecular Therapy - Nucleic Acids 2021, 23, 1334–1344. 10.1016/j.omtn.2021.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Couto G.; Gallet R.; Cambier L.; Jaghatspanyan E.; Makkar N.; Dawkins J. F.; Berman B. P.; Marban E. Exosomal MicroRNA Transfer Into Macrophages Mediates Cellular Postconditioning. Circulation 2017, 136, 200–214. 10.1161/CIRCULATIONAHA.116.024590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J.-S.; Schumacher S. M.; Gao E.; Chuprun J. K.; Ibetti J.; Roy R.; Khan M.; Kishore R.; Koch W. J. Characterization of βARKct engineered cellular extracellular vesicles and model specific cardioprotection. American Journal of Physiology-Heart and Circulatory Physiology 2021, 320, H1276–H1289. 10.1152/ajpheart.00571.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrazzo P.; Pizzuti V.; Zia S.; Sargenti A.; Gazzola D.; Roda B.; Bonsi L.; Alviano F. Microfluidic Tools for Enhanced Characterization of Therapeutic Stem Cells and Prediction of Their Potential Antimicrobial Secretome. Antibiotics 2021, 10, 750. 10.3390/antibiotics10070750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P.; Zhou J.; Zhang J.; Dong Y.; Liu Y. Exosome: The Regulator of the Immune System in Sepsis. Front. Pharmacol. 2021, 12, 671164. 10.3389/fphar.2021.671164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprecher E.A Phase II Randomized, Double-blind, Placebo-controlled Study to Evaluate the Safety and Efficacy of Exosomes Overexpressing CD24 to Prevent Clinical Deterioration in Patients With Moderate or Severe COVID-19 Infection. https://www.clinicaltrials.gov/ct2/show/NCT04969172?term=exosomes&cond=covid-19&draw=2&rank=1 (accessed August 17, 2022).
- Papageorgiou A.; Papadopetraki A.; A P.; Ikonomidis I.; Koynova R.; Tenchov B. The effect of remote ischemic conditioning on blood plasma heat capacity profiles. A differential scanning calorimetry study. Archives of Hellenic Medicine 2021, 38, 43–48. [Google Scholar]
- Wang D.; Hu B.; Hu C.; Zhu F.; Liu X.; Zhang J.; Wang B.; Xiang H.; Cheng Z.; Xiong Y.; Zhao Y.; Li Y.; Wang X.; Peng Z. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazini L.; Rochette L.; Malka G. Exosomes contribution in COVID-19 patients’ treatment. J. Transl. Med. 2021, 19, 234. 10.1186/s12967-021-02884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athens Medical Society. Safety and Efficacy of Exosomes Overexpressing CD24 in Two Doses for Patients with Moderate or Severe COVID-19. https://www.clinicaltrials.gov/ct2/show/NCT04902183?term=exosomes&cond=covid-19&draw=2 (accessed August 17, 2022).
- Tel-Aviv Sourasky Medical Center. Evaluation of the Safety of CD24-Exosomes in Patients With COVID-19 Infection. https://www.clinicaltrials.gov/ct2/show/NCT04747574?term=exosomes&cond=covid-19&draw=2 (accessed August 17, 2022).
- Tyumina O.Safety and Efficiency of Method of Exosome Inhalation in COVID-19 Associated Pneumonia (COVID-19EXO2). https://clinicaltrials.gov/ct2/show/NCT04602442 (accessed August 17, 2022).
- Evaluation of Safety and Efficiency of Method of Exosome Inhalation in SARS-CoV-2 Associated Pneumonia. (COVID-19EXO). https://clinicaltrials.gov/ct2/show/NCT04491240 (accessed August 17, 2022).
- Fu S.; Wang Y.; Xia X.; Zheng J. C. Exosome engineering: Current progress in cargo loading and targeted delivery. NanoImpact 2020, 20, 100261. 10.1016/j.impact.2020.100261. [DOI] [Google Scholar]
- Soltani F.; Parhiz H.; Mokhtarzadeh A.; Ramezani M. Synthetic and Biological Vesicular Nano-Carriers Designed for Gene Delivery. Curr. Pharm. Des. 2015, 21, 6214–6235. 10.2174/1381612821666151027153410. [DOI] [PubMed] [Google Scholar]
- Das C. K.; Jena B. C.; Banerjee I.; Das S.; Parekh A.; Bhutia S. K.; Mandal M. Exosome as a Novel Shuttle for Delivery of Therapeutics across Biological Barriers. Mol. Pharmaceutics 2019, 16, 24–40. 10.1021/acs.molpharmaceut.8b00901. [DOI] [PubMed] [Google Scholar]
- Karpman D.; Stahl A.-l.; Arvidsson I. Extracellular vesicles in renal disease. Nature Reviews Nephrology 2017, 13, 545–562. 10.1038/nrneph.2017.98. [DOI] [PubMed] [Google Scholar]
- Fiandaca M. S.; Kapogiannis D.; Mapstone M.; Boxer A.; Eitan E.; Schwartz J. B.; Abner E. L.; Petersen R. C.; Federoff H. J.; Miller B. L.; Goetzl E. J. Identification of preclinical Alzheimer’s disease by a profile of pathogenic proteins in neurally derived blood exosomes: A case-control study. Alzheimers Dement 2015, 11, 600–607. 10.1016/j.jalz.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saman S.; Kim W.; Raya M.; Visnick Y.; Miro S.; Saman S.; Jackson B.; McKee A. C.; Alvarez V. E.; Lee N. C. Y.; Hall G. F. Exosome-associated Tau Is Secreted in Tauopathy Models and Is Selectively Phosphorylated in Cerebrospinal Fluid in Early Alzheimer Disease. J. Biol. Chem. 2012, 287, 3842–3849. 10.1074/jbc.M111.277061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Chen Y.; Pei F.; Zeng C.; Yao Y.; Liao W.; Zhao Z. Extracellular Vesicles in Liquid Biopsies: Potential for Disease Diagnosis. Biomed Res. Int. 2021, 2021, 6611244. 10.1155/2021/6611244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D.; Li Y.; Wang M.; Gu J.; Xu W.; Cai H.; Fang X.; Zhang X. Exosomes as a new frontier of cancer liquid biopsy. Mol. Cancer 2022, 21, 56. 10.1186/s12943-022-01509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukouris S.; Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin. Appl. 2015, 9, 358–367. 10.1002/prca.201400114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larssen P.; Wik L.; Czarnewski P.; Eldh M.; Löf L.; Ronquist K. G.; Dubois L.; Freyhult E.; Gallant C. J.; Oelrich J.; Larsson A.; Ronquist G.; Villablanca E. J.; Landegren U.; Gabrielsson S.; Kamali-Moghaddam M. Tracing Cellular Origin of Human Exosomes Using Multiplex Proximity Extension Assays. Mol. Cell Proteomics 2017, 16, 502–511. 10.1074/mcp.M116.064725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Bi J.; Huang J.; Tang Y.; Du S.; Li P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomedicine 2020, 15, 6917–6934. 10.2147/IJN.S264498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryani A.; Denecke B. Exosomes as a Nanodelivery System: a Key to the Future of Neuromedicine?. Mol. Neurobiol. 2016, 53, 818–834. 10.1007/s12035-014-9054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.; Xu K.; Zheng X.; Chen T.; Wang J.; Song Y.; Shao Y.; Zheng S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduction and Targeted Therapy 2020, 5, 144. 10.1038/s41392-020-00258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B.; Li Y.; Zhou Y.; Ng T. K.; Zhao C.; Gan Q.; Gu X.; Xiang J. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J. Cell. Physiol. 2019, 234, 1416–1425. 10.1002/jcp.26936. [DOI] [PubMed] [Google Scholar]
- Wong C. H.; Chen Y. C. Clinical significance of exosomes as potential biomarkers in cancer. World J. Clin Cases 2019, 7, 171–190. 10.12998/wjcc.v7.i2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R.; LeBleu V. S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, aau6977. 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A.; Zhang T.; Zheng M.; Liu Y.; Chen Z. Exosomal proteins as potential markers of tumor diagnosis. J. Hematol. Oncol. 2017, 10, 175. 10.1186/s13045-017-0542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.; Jiang W.; Lv M.; Fan S.; Lu Y.; Wu Q.; Pi J. Potentiality of Exosomal Proteins as Novel Cancer Biomarkers for Liquid Biopsy. Front. Immunol. 2022, 13, 792046. 10.3389/fimmu.2022.792046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T.; Wong D. T. W. Saliva-Exosomics in Cancer: Molecular Characterization of Cancer-Derived Exosomes in Saliva. Enzymes 2017, 42, 125–151. 10.1016/bs.enz.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logozzi M.; De Milito A.; Lugini L.; Borghi M.; Calabrò L.; Spada M.; Perdicchio M.; Marino M. L.; Federici C.; Iessi E.; Brambilla D.; Venturi G.; Lozupone F.; Santinami M.; Huber V.; Maio M.; Rivoltini L.; Fais S. High Levels of Exosomes Expressing CD63 and Caveolin-1 in Plasma of Melanoma Patients. PLoS One 2009, 4, e5219 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka Y.; Konishi Y.; Kosaka N.; Katsuda T.; Kato T.; Ochiya T. Comparative marker analysis of extracellular vesicles in different human cancer types. Journal of Extracellular Vesicles 2013, 2, 20424. 10.3402/jev.v2i0.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker M.-W.; Reichert D.; Susser S.; Sarrazin C.; Martinez Y.; Herrmann E.; Zeuzem S.; Piiper A.; Kronenberger B. Soluble Serum CD81 Is Elevated in Patients with Chronic Hepatitis C and Correlates with Alanine Aminotransferase Serum Activity. PLoS One 2012, 7, e30796 10.1371/journal.pone.0030796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao M.; Huang Y.; Lang Z.; Zhao H.; Saito Y.; Nagano T.; Kawagoe I.; Divisi D.; Hu X.; Jiang G. Proteomic analysis of plasma exosomes in patients with non-small cell lung cancer. Translational Lung Cancer Research 2022, 11, 1434–1452. 10.21037/tlcr-22-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graner M. W.; Alzate O.; Dechkovskaia A. M.; Keene J. D.; Sampson J. H.; Mitchell D. A.; Bigner D. D. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009, 23, 1541–1557. 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran L.; Honsho M.; Zahn T. R.; Keller P.; Geiger K. D.; Verkade P.; Simons K. Alzheimer’s disease β-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 11172–11177. 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exosome Sciences. https://www.exosomesciences.com/ (accessed August 23, 2022).
- Stern R. A.; Tripodis Y.; Baugh C. M.; Fritts N. G.; Martin B. M.; Chaisson C.; Cantu R. C.; Joyce J. A.; Shah S.; Ikezu T.; Zhang J.; Gercel-Taylor C.; Taylor D. D. Preliminary Study of Plasma Exosomal Tau as a Potential Biomarker for Chronic Traumatic Encephalopathy. J. Alzheimers Dis. 2016, 51, 1099–1109. 10.3233/JAD-151028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aethlon Medical’s Exosome Sciences To Participate In Clinical Research Study To Diagnose Chronic Traumatic Encephalopathy (CTE). https://www.prnewswire.com/news-releases/aethlon-medicals-exosome-sciences-to-participate-in-clinical-research-study-to-diagnose-chronic-traumatic-encephalopathy-cte-300202532.html (accessed August 23, 2022).
- The DIAGNOSE-CTE Research Project (DIAGNOSE-CTE). https://clinicaltrials.gov/ct2/show/NCT02798185 (accessed August 23, 2022).
- Alvarez-Erviti L.; Seow Y.; Schapira A. H.; Gardiner C.; Sargent I. L.; Wood M. J.; Cooper J. M. Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol. Dis. 2011, 42, 360–367. 10.1016/j.nbd.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.; Cheruvanky A.; Hu X.; Matsumoto T.; Hiramatsu N.; Cho M. E.; Berger A.; Leelahavanichkul A.; Doi K.; Chawla L. S. Urinary exosomal transcription factors, a new class of biomarkers for renal disease. Kidney Int. 2008, 74, 613–621. 10.1038/ki.2008.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley D. M.; Sheman N. E.; Nelson K.; Theodorescu D. Isolation and identification of potential urinary microparticle biomarkers of bladder cancer. J. Proteome Res. 2008, 7, 2088–2096. 10.1021/pr700775x. [DOI] [PubMed] [Google Scholar]
- Nilsson J.; Skog J.; Nordstrand A.; Baranov V.; Mincheva-Nilsson L.; Breakefield X.; Widmark A. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br. J. Cancer 2009, 100, 1603–1607. 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-L.; Lai Y.-F.; Tang P.; Chien K.-Y.; Yu J.-S.; Tsai C.-H.; Chen H.-W.; Wu C.-C.; Chung T.; Hsu C.-W. Comparative and targeted proteomic analyses of urinary microparticles from bladder cancer and hernia patients. J. Proteome Res. 2012, 11, 5611–5629. 10.1021/pr3008732. [DOI] [PubMed] [Google Scholar]
- Khan S.; Jutzy J. M.; Valenzuela M. M. A.; Turay D.; Aspe J. R.; Ashok A.; Mirshahidi S.; Mercola D.; Lilly M. B.; Wall N. R. Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS One 2012, 7, e46737 10.1371/journal.pone.0046737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noto G.; Paolini L.; Zendrini A.; Radeghieri A.; Caimi L.; Ricotta D. C-src enriched serum microvesicles are generated in malignant plasma cell dyscrasia. PLoS One 2013, 8, e70811 10.1371/journal.pone.0070811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandfeld-Paulsen B.; Aggerholm-Pedersen N.; Bæk R.; Jakobsen K. R.; Meldgaard P.; Folkersen B. H.; Rasmussen T. R.; Varming K.; Jørgensen M. M.; Sorensen B. S. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol. Oncol. 2016, 10, 1595–1602. 10.1016/j.molonc.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. H.; Xue L.; Hsu C. C.; Paez J. S.; Pan L.; Andaluz H.; Wendt M. K.; Iliuk A. B.; Zhu J. K.; Tao W. A. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 3175–3180. 10.1073/pnas.1618088114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S.; Tian H.; Li X.; Jin D.; Li X.; Kong J.; Yang C.; Yang X.; Lu Y.; Luo Y.; Lin B.; Niu W.; Liu T. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS One 2017, 12, e0175050 10.1371/journal.pone.0175050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Chen Y.; Guo X.; Zhou L.; Jia Z.; Peng Z.; Tang Y.; Liu W.; Zhu B.; Wang L.; Ren C. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J. Cell Mol. Med. 2017, 21, 838–847. 10.1111/jcmm.12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S.; Takeuchi A.; Yamaguchi S.; Mitani Y.; Watanabe T.; Matsuda K.; Hotta T.; Shively J. E.; Yamaue H. Clinical implications of carcinoembryonic antigen distribution in serum exosomal fraction-Measurement by ELISA. PLoS One 2017, 12, e0183337 10.1371/journal.pone.0183337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbelaiz A.; Azkargorta M.; Krawczyk M.; Santos-Laso A.; Lapitz A.; Perugorria M. J.; Erice O.; Gonzalez E.; Jimenez-Agüero R.; Lacasta A.; Ibarra C.; Sanchez-Campos A.; Jimeno J. P.; Lammert F.; Milkiewicz P.; Marzioni M.; Macias R. I. R.; Marin J. J. G.; Patel T.; Gores G. J.; et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 2017, 66, 1125–1143. 10.1002/hep.29291. [DOI] [PubMed] [Google Scholar]
- Logozzi M.; Angelini D. F.; Iessi E.; Mizzoni D.; Di Raimo R.; Federici C.; Lugini L.; Borsellino G.; Gentilucci A.; Pierella F.; Marzio V.; Sciarra A.; Battistini L.; Fais S. Increased PSA expression on prostate cancer exosomes in in vitro condition and in cancer patients. Cancer Lett. 2017, 403, 318–329. 10.1016/j.canlet.2017.06.036. [DOI] [PubMed] [Google Scholar]
- Kawakami K.; Fujita Y.; Matsuda Y.; Arai T.; Horie K.; Kameyama K.; Kato T.; Masunaga K.; Kasuya Y.; Tanaka M.; Mizutani K.; Deguchi T.; Ito M. Gamma-glutamyltransferase activity in exosomes as a potential marker for prostate cancer. BMC Cancer 2017, 17, 316. 10.1186/s12885-017-3301-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.; Yang Y.; Zeng Y.; He M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip 2016, 16, 489–496. 10.1039/C5LC01117E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K.; Ishikawa N.; Tatsuguchi A.; Saichi N.; Fujii R.; Nakagawa H. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Sci. Rep. 2014, 4, 6232. 10.1038/srep06232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandfeld-Paulsen B.; Jakobsen K. R.; Bæk R.; Folkersen B. H.; Rasmussen T. R.; Meldgaard P.; Varming K.; Jørgensen M. M.; Sorensen B. S. Exosomal proteins as diagnostic biomarkers in lung cancer. J. Thorac. Oncol. 2016, 11, 1701–1710. 10.1016/j.jtho.2016.05.034. [DOI] [PubMed] [Google Scholar]
- Wang X.; Zhong W.; Bu J.; Li Y.; Li R.; Nie R.; Xiao C.; Ma K.; Huang X.; Li Y. Exosomal protein CD82 as a diagnostic biomarker for precision medicine for breast cancer. Mol. Carcinog. 2019, 58, 674–685. 10.1002/mc.22960. [DOI] [PubMed] [Google Scholar]
- Yoshioka Y.; Kosaka N.; Konishi Y.; Ohta H.; Okamoto H.; Sonoda H.; Nonaka R.; Yamamoto H.; Ishii H.; Mori M. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat. Commun. 2014, 5, 3591. 10.1038/ncomms4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan B.; Yue S.; Galli U.; Rana S.; Gross W.; Müller M.; Giese N. A.; Kalthoff H.; Becker T.; Büchler M. W. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int. J. Cancer 2015, 136, 2616–2627. 10.1002/ijc.29324. [DOI] [PubMed] [Google Scholar]
- Zhou H.; Pisitkun T.; Aponte A.; Yuen P. S.; Hoffert J. D.; Yasuda H.; Hu X.; Chawla L.; Shen R.-F.; Knepper M. A. Exosomal Fetuin-A identified by proteomics: a novel urinary biomarker for detecting acute kidney injury. Kidney Int. 2006, 70, 1847–1857. 10.1038/sj.ki.5001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde-Vancells J.; Rodriguez-Suarez E.; Gonzalez E.; Berisa A.; Gil D.; Embade N.; Valle M.; Luka Z.; Elortza F.; Wagner C. Candidate biomarkers in exosome-like vesicles purified from rat and mouse urine samples. Prot. Clin. Appl. 2010, 4, 416–425. 10.1002/prca.200900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales P. A.; Pisitkun T.; Hoffert J. D.; Tchapyjnikov D.; Star R. A.; Kleta R.; Wang N. S.; Knepper M. A. Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. 2009, 20, 363–379. 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan M. J.; Noerholm M.; Bentink S.; Belzer S.; Skog J.; O’Neill V.; Cochran J. S.; Brown G. A. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis. 2015, 18, 370–375. 10.1038/pcan.2015.40. [DOI] [PubMed] [Google Scholar]
- Keller S.; König A.-K.; Marmé F.; Runz S.; Wolterink S.; Koensgen D.; Mustea A.; Sehouli J.; Altevogt P. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett. 2009, 278, 73–81. 10.1016/j.canlet.2008.12.028. [DOI] [PubMed] [Google Scholar]
- Li J.; Sherman-Baust C. A.; Tsai-Turton M.; Bristow R. E.; Roden R. B.; Morin P. J. Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer 2009, 9, 1–11. 10.1186/1471-2407-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winck F. V.; Prado Ribeiro A. C.; Ramos Domingues R.; Ling L. Y.; Riaño-Pachón D. M.; Rivera C.; Brandão T. B.; Gouvea A. F.; Santos-Silva A. R.; Coletta R. D.; Paes Leme A. F. Insights into immune responses in oral cancer through proteomic analysis of saliva and salivary extracellular vesicles. Sci. Rep. 2015, 5, 16305. 10.1038/srep16305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.; Xia Z.; Shang Z.; Sun K.; Niu X.; Qian L.; Fan L. Y.; Cao C. X.; Xiao H. Facile preparation of salivary extracellular vesicles for cancer proteomics. Sci. Rep. 2016, 6, 24669. 10.1038/srep24669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keryer-Bibens C.; Pioche-Durieu C.; Villemant C.; Souquère S.; Nishi N.; Hirashima M.; Middeldorp J.; Busson P. Exosomes released by EBV-infected nasopharyngeal carcinoma cells convey the viral latent membrane protein 1 and the immunomodulatory protein galectin 9. BMC Cancer 2006, 6, 283. 10.1186/1471-2407-6-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L.; Zhang T.; Ma H.; Pan Y.; Wang S.; Liu X.; Dai X.; Zheng Y.; Lee L. P.; Liu F. Discovering the Secret of Diseases by Incorporated Tear Exosomes Analysis via Rapid-Isolation System: iTEARS. ACS Nano 2022, 16, 11720. 10.1021/acsnano.2c02531. [DOI] [PubMed] [Google Scholar]
- Lässer C. Exosomal RNA as biomarkers and the therapeutic potential of exosome vectors. Expert Opin. Biol. Ther. 2012, 12, S189–197. 10.1517/14712598.2012.680018. [DOI] [PubMed] [Google Scholar]
- Taylor D. D.; Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Mitchell P. S.; Parkin R. K.; Kroh E. M.; Fritz B. R.; Wyman S. K.; Pogosova-Agadjanyan E. L.; Peterson A.; Noteboom J.; O’Briant K. C.; Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 10513–10518. 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter M. P.; Ismail N.; Zhang X.; Aguda B. D.; Lee E. J.; Yu L.; Xiao T.; Schafer J.; Lee M.-L. T.; Schmittgen T. D. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One 2008, 3, e3694 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowits G.; Gerçel-Taylor C.; Day J. M.; Taylor D. D.; Kloecker G. H. Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer 2009, 10, 42–46. 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- Health, H. M. Prostasomes as Diagnostic Tool for Prostate Cancer Detection. https://clinicaltrials.gov/ct2/show/NCT03694483 (accessed August 17, 2022).
- ExoDx Prostate Test. https://www.exosomedx.com/physicians/exodx-prostate-test (accessed August 17, 2022).
- FDA Grants Breakthrough Device Designation to Bio-Techne’s ExoDx Prostate (IntelliScore) (EPI) Test. https://www.exosomedx.com/news-events/fda-grants-breakthrough-device-designation-bio-technes-exodx-prostate-intelliscore-epi (accessed August 23, 2022).
- Tanaka Y.; Kamohara H.; Kinoshita K.; Kurashige J.; Ishimoto T.; Iwatsuki M.; Watanabe M.; Baba H. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer 2013, 119, 1159–1167. 10.1002/cncr.27895. [DOI] [PubMed] [Google Scholar]
- Takeshita N.; Hoshino I.; Mori M.; Akutsu Y.; Hanari N.; Yoneyama Y.; Ikeda N.; Isozaki Y.; Maruyama T.; Akanuma N. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br. J. Cancer 2013, 108, 644–652. 10.1038/bjc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-F.; Mandel E. M.; Thomson J. M.; Wu Q.; Callis T. E.; Hammond S. M.; Conlon F. L.; Wang D.-Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara Y.; Ono K.; Horie T.; Nishi H.; Nagao K.; Kinoshita M.; Watanabe S.; Baba O.; Kojima Y.; Shizuta S. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ. Cardiovasc. Genet. 2011, 4, 446–454. 10.1161/CIRCGENETICS.110.958975. [DOI] [PubMed] [Google Scholar]
- Lv L.-L.; Cao Y.-H.; Ni H.-F.; Xu M.; Liu D.; Liu H.; Chen P.-S.; Liu B.-C. MicroRNA-29c in urinary exosome/microvesicle as a biomarker of renal fibrosis. American journal of physiology-renal physiology 2013, 305, F1220–F1227. 10.1152/ajprenal.00148.2013. [DOI] [PubMed] [Google Scholar]
- Lv L.-L.; Cao Y.-H.; Pan M.-M.; Liu H.; Tang R.-N.; Ma K.-L.; Chen P.-S.; Liu B.-C. CD2AP mRNA in urinary exosome as biomarker of kidney disease. Clin. Chim. Acta 2014, 428, 26–31. 10.1016/j.cca.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Hong B. S.; Cho J.-H.; Kim H.; Choi E.-J.; Rho S.; Kim J.; Kim J. H.; Choi D.-S.; Kim Y.-K.; Hwang D. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics 2009, 10, 556. 10.1186/1471-2164-10-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J.; Chen R.; Zhao L.; Xu Y.; Cao Z.; Xu H.; Chen X.; Shi X.; Zhu Y.; Lyu J.; Jiang J.; Wang Y.; Zhou T.; He J.; Wei X.; Wu J. B.; Yang B.; Wang F. Circulating exosomal mRNA profiling identifies novel signatures for the detection of prostate cancer. Mol. Cancer 2021, 20, 58. 10.1186/s12943-021-01349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Xu H. Serum exosomal miR-378 upregulation is associated with poor prognosis in non-small-cell lung cancer patients. J. Clin. Lab. Anal. 2020, 34, e23237 10.1002/jcla.23237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janpipatkul K.; Trachu N.; Watcharenwong P.; Panvongsa W.; Worakitchanon W.; Metheetrairut C.; Oranratnachai S.; Reungwetwattana T.; Chairoungdua A. Exosomal microRNAs as potential biomarkers for osimertinib resistance of non-small cell lung cancer patients. Cancer Biomark. 2021, 31, 281–294. 10.3233/CBM-203075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q.; Yu L.; Lin X.; Zheng Q.; Zhang S.; Chen D.; Pan X.; Huang Y. Combination of Serum miRNAs with Serum Exosomal miRNAs in Early Diagnosis for Non-Small-Cell Lung Cancer. Cancer Manag. Res. 2020, 12, 485–495. 10.2147/CMAR.S232383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S.; Zheng S.; Lu Z.; Wang X.; Wang Y.; Zhang G.; Xu H.; Huang J.; Lei Y.; Liu C.; Sun N.; He J. Exosomal miR-375-3p breaks vascular barrier and promotes small cell lung cancer metastasis by targeting claudin-1. Transl Lung Cancer Res. 2021, 10, 3155–3172. 10.21037/tlcr-21-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.; Zhang R.; Zhu Y.; Hao R. Exosome-derived microRNA-433 inhibits tumorigenesis through incremental infiltration of CD4 and CD8 cells in non-small cell lung cancer. Oncol. Lett. 2021, 22, 607. 10.3892/ol.2021.12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.; Wang J.; Ma L. J.; Yang H. B.; Jing J. F.; Jia M. M.; Zhang X. J.; Guo F.; Gao J. N. Identification of serum exosomal miR-148a as a novel prognostic biomarker for breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7303–7309. 10.26355/eurrev_202007_21889. [DOI] [PubMed] [Google Scholar]
- Hirschfeld M.; Rücker G.; Weiß D.; Berner K.; Ritter A.; Jäger M.; Erbes T. Urinary Exosomal MicroRNAs as Potential Non-invasive Biomarkers in Breast Cancer Detection. Mol. Diagn. Ther. 2020, 24, 215–232. 10.1007/s40291-020-00453-y. [DOI] [PubMed] [Google Scholar]
- Han M.; Hu J.; Lu P.; Cao H.; Yu C.; Li X.; Qian X.; Yang X.; Yang Y.; Han N.; Dou D.; Zhang F.; Ye M.; Yang C.; Gu Y.; Dong H. Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell Death Dis. 2020, 11, 43. 10.1038/s41419-020-2250-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jiang M.; Zhang W.; Zhang R.; Liu P.; Ye Y.; Yu W.; Guo X.; Yu J. Cancer exosome-derived miR-9 and miR-181a promote the development of early-stage MDSCs via interfering with SOCS3 and PIAS3 respectively in breast cancer. Oncogene 2020, 39, 4681–4694. 10.1038/s41388-020-1322-4. [DOI] [PubMed] [Google Scholar]
- Guo T.; Wang Y.; Jia J.; Mao X.; Stankiewicz E.; Scandura G.; Burke E.; Xu L.; Marzec J.; Davies C. R.; Lu J. J.; Rajan P.; Grey A.; Tipples K.; Hines J.; Kudahetti S.; Oliver T.; Powles T.; Alifrangis C.; Kohli M.; et al. The Identification of Plasma Exosomal miR-423-3p as a Potential Predictive Biomarker for Prostate Cancer Castration-Resistance Development by Plasma Exosomal miRNA Sequencing. Front Cell Dev Biol. 2021, 8, 602493. 10.3389/fcell.2020.602493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S.; Park Y. H.; Jung S.-H.; Jang S.-H.; Kim M. Y.; Lee J. Y.; Chung Y. J. Urinary exosome microRNA signatures as a noninvasive prognostic biomarker for prostate cancer. npj Genomic Medicine 2021, 6, 45. 10.1038/s41525-021-00212-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Dong Y.; Wang K. J.; Deng Z.; Zhang W.; Shen H. F. Plasma exosomal miR-125a-5p and miR-141-5p as non-invasive biomarkers for prostate cancer. Neoplasma 2021, 67, 1314–1318. 10.4149/neo_2020_191130N1234. [DOI] [PubMed] [Google Scholar]
- Li Z.; Li L. X.; Diao Y. J.; Wang J.; Ye Y.; Hao X. K. Identification of Urinary Exosomal miRNAs for the Non-Invasive Diagnosis of Prostate Cancer. Cancer Manag. Res. 2021, 13, 25–35. 10.2147/CMAR.S272140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che Y.; Shi X.; Shi Y.; Jiang X.; Ai Q.; Shi Y.; Gong F.; Jiang W. Exosomes Derived from miR-143-Overexpressing MSCs Inhibit Cell Migration and Invasion in Human Prostate Cancer by Downregulating TFF3. Mol. Ther Nucleic Acids 2019, 18, 232–244. 10.1016/j.omtn.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yu L.; Sui B.; Fan W.; Lei L.; Zhou L.; Yang L.; Diao Y.; Zhang Y.; Li Z.; Liu J.; Hao X. Exosomes derived from osteogenic tumor activate osteoclast differentiation and concurrently inhibit osteogenesis by transferring COL1A1-targeting miRNA-92a-1-5p. J. Extracell Vesicles 2021, 10, e12056 10.1002/jev2.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L.; Ping F.; Fan Z.; Zhang C.; Deng M.; Cheng B.; Xia J. Salivary exosomal miR-24-3p serves as a potential detective biomarker for oral squamous cell carcinoma screening. Biomed. Pharmacother. 2020, 121, 109553. 10.1016/j.biopha.2019.109553. [DOI] [PubMed] [Google Scholar]
- He T.; Guo X.; Li X.; Liao C.; Wang X.; He K. Plasma-Derived Exosomal microRNA-130a Serves as a Noninvasive Biomarker for Diagnosis and Prognosis of Oral Squamous Cell Carcinoma. J. Oncol. 2021, 2021, 5547911. 10.1155/2021/5547911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni B.; Gondaliya P.; Kirave P.; Rawal R.; Jain A.; Garg R.; Kalia K. Exosome-mediated delivery of miR-30a sensitize cisplatin-resistant variant of oral squamous carcinoma cells via modulating Beclin1 and Bcl2. Oncotarget 2020, 11, 1832–1845. 10.18632/oncotarget.27557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W.; Wang Y.; Chen Y.; Guo Y.; Li Q.; Wei X. Exosomal miR-130b-3p Promotes Progression and Tubular Formation Through Targeting PTEN in Oral Squamous Cell Carcinoma. Frontiers in Cell and Developmental Biology 2021, 9, 616306. 10.3389/fcell.2021.616306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.; Zhuang Y.; Zhang J.; Chen M.; Wu S. Four circulating exosomal miRNAs as novel potential biomarkers for the early diagnosis of human colorectal cancer. Tissue Cell 2021, 70, 101499. 10.1016/j.tice.2021.101499. [DOI] [PubMed] [Google Scholar]
- Liu H.; Liu Y.; Sun P.; Leng K.; Xu Y.; Mei L.; Han P.; Zhang B.; Yao K.; Li C.; Bai J.; Cui B. Colorectal cancer-derived exosomal miR-106b-3p promotes metastasis by down-regulating DLC-1 expression. Clin. Sci. (London) 2020, 134, 419–434. 10.1042/CS20191087. [DOI] [PubMed] [Google Scholar]
- Chen X.; Jia M.; Ji J.; Zhao Z.; Zhao Y. Exosome-Derived Non-Coding RNAs in the Tumor Microenvironment of Colorectal Cancer: Possible Functions, Mechanisms and Clinical Applications. Front. Oncol. 2022, 12, 887532. 10.3389/fonc.2022.887532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. J.; Lim J. W.; Moritz R. L.; Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev. Proteomics 2009, 6, 267–283. 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- Silva J.; Garcia V.; Rodriguez M.; Compte M.; Cisneros E.; Veguillas P.; Garcia J. M.; Dominguez G.; Campos-Martin Y.; Cuevas J.; Peña C.; Herrera M.; Diaz R.; Mohammed N.; Bonilla F. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer 2012, 51, 409–418. 10.1002/gcc.21926. [DOI] [PubMed] [Google Scholar]
- Friel A. M.; Corcoran C.; Crown J.; O’Driscoll L. Relevance of circulating tumor cells, extracellular nucleic acids, and exosomes in breast cancer. Breast Cancer Res. Treat. 2010, 123, 613–625. 10.1007/s10549-010-0980-2. [DOI] [PubMed] [Google Scholar]
- Rashed M. H.; Bayraktar E.; Helal G. K.; Abd-Ellah M.; Amero P.; Chavez-Reyes A.; Rodriguez-Aguayo C. Exosomes: From Garbage Bins to Promising Therapeutic Targets. Int. J. Mol. Sci. 2017, 18, 538. 10.3390/ijms18030538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G.; Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai K.; Tanaka N.; Nakano T.; Kakazu E.; Kondo Y.; Inoue J.; Shiina M.; Fukushima K.; Hoshino T.; Sano K.; Ueno Y.; Shimosegawa T.; Sugamura K. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem. Biophys. Res. Commun. 2010, 399, 384–390. 10.1016/j.bbrc.2010.07.083. [DOI] [PubMed] [Google Scholar]
- Hoshino D.; Kirkbride K. C.; Costello K.; Clark E. S.; Sinha S.; Grega-Larson N.; Tyska M. J.; Weaver A. M. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 2013, 5, 1159–1168. 10.1016/j.celrep.2013.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco F.; Perrotta C.; Novellino L.; Francolini M.; Riganti L.; Menna E.; Saglietti L.; Schuchman E. H.; Furlan R.; Clementi E.; Matteoli M.; Verderio C. Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J. 2009, 28, 1043–1054. 10.1038/emboj.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K.; Hsu C.; Chiantia S.; Rajendran L.; Wenzel D.; Wieland F.; Schwille P.; Brügger B.; Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- Li B.; Antonyak M. A.; Zhang J.; Cerione R. A. RhoA triggers a specific signaling pathway that generates transforming microvesicles in cancer cells. Oncogene 2012, 31, 4740–4749. 10.1038/onc.2011.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savina A.; Fader C. M.; Damiani M. T.; Colombo M. I. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic 2005, 6, 131–143. 10.1111/j.1600-0854.2004.00257.x. [DOI] [PubMed] [Google Scholar]
- Middleton R. C.; Rogers R. G.; De Couto G.; Tseliou E.; Luther K.; Holewinski R.; Soetkamp D.; Van Eyk J. E.; Antes T. J.; Marbán E. Newt cells secrete extracellular vesicles with therapeutic bioactivity in mammalian cardiomyocytes. J. Extracell Vesicles 2018, 7, 1456888. 10.1080/20013078.2018.1456888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N.; Iguchi H.; Hagiwara K.; Yoshioka Y.; Takeshita F.; Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A.; Kim H.; McGee L.; Johnson A. E.; Talwar S.; Marugan J.; Southall N.; Hu X.; Lal M.; Mondal D.; Ferrer M.; Abdel-Mageed A. B. High-throughput screening identified selective inhibitors of exosome biogenesis and secretion: A drug repurposing strategy for advanced cancer. Sci. Rep. 2018, 8, 8161. 10.1038/s41598-018-26411-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrie A.; Krumeich S.; Reyal F.; Recchi C.; Moita L. F.; Seabra M. C.; Ostrowski M.; Théry C. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012, 72, 4920–4930. 10.1158/0008-5472.CAN-12-0925. [DOI] [PubMed] [Google Scholar]
- Hendrix A.; De Wever O. Rab27 GTPases distribute extracellular nanomaps for invasive growth and metastasis: implications for prognosis and treatment. Int. J. Mol. Sci. 2013, 14, 9883–9892. 10.3390/ijms14059883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso R.; Mazzeo C.; Rodriguez M. C.; Marsh M.; Fraile-Ramos A.; Calvo V.; Avila-Flores A.; Merida I.; Izquierdo M. Diacylglycerol kinase α regulates the formation and polarisation of mature multivesicular bodies involved in the secretion of Fas ligand-containing exosomes in T lymphocytes. Cell Death Differ. 2011, 18, 1161–1173. 10.1038/cdd.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader C. M.; Sánchez D. G.; Mestre M. B.; Colombo M. I. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim. Biophys. Acta 2009, 1793, 1901–1916. 10.1016/j.bbamcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Federici C.; Petrucci F.; Caimi S.; Cesolini A.; Logozzi M.; Borghi M.; D’Ilio S.; Lugini L.; Violante N.; Azzarito T.; Majorani C.; Brambilla D.; Fais S. Exosome release and low pH belong to a framework of resistance of human melanoma cells to cisplatin. PLoS One 2014, 9, e88193 10.1371/journal.pone.0088193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L.; Kalimuthu S.; Gangadaran P.; Oh J. M.; Lee H. W.; Baek S. H.; Jeong S. Y.; Lee S. W.; Lee J.; Ahn B. C. Exosomes Derived From Natural Killer Cells Exert Therapeutic Effect in Melanoma. Theranostics 2017, 7, 2732–2745. 10.7150/thno.18752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzner D.; Schnaars M.; van Rossum D.; Krishnamoorthy G.; Dibaj P.; Bakhti M.; Regen T.; Hanisch U. K.; Simons M. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J. Cell Sci. 2011, 124, 447–458. 10.1242/jcs.074088. [DOI] [PubMed] [Google Scholar]
- Feng D.; Zhao W. L.; Ye Y. Y.; Bai X. C.; Liu R. Q.; Chang L. F.; Zhou Q.; Sui S. F. Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010, 11, 675–687. 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- Tian T.; Zhu Y. L.; Zhou Y. Y.; Liang G. F.; Wang Y. Y.; Hu F. H.; Xiao Z. D. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J. Biol. Chem. 2014, 289, 22258–22267. 10.1074/jbc.M114.588046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escrevente C.; Keller S.; Altevogt P.; Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 2011, 11, 108. 10.1186/1471-2407-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima L. G.; Chammas R.; Monteiro R. Q.; Moreira M. E.; Barcinski M. A. Tumor-derived microvesicles modulate the establishment of metastatic melanoma in a phosphatidylserine-dependent manner. Cancer Lett. 2009, 283, 168–175. 10.1016/j.canlet.2009.03.041. [DOI] [PubMed] [Google Scholar]
- Christianson H. C.; Svensson K. J.; van Kuppevelt T. H.; Li J. P.; Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 17380–17385. 10.1073/pnas.1304266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R.; Rai A.; Chen M.; Suwakulsiri W.; Greening D. W.; Simpson R. J. Extracellular vesicles in cancer — implications for future improvements in cancer care. Nature Reviews Clinical Oncology 2018, 15, 617–638. 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- Barnie P. A.; Afrifa J.; Gyamerah E. O.; Amoani B.. Extracellular Vesicles as Biomarkers and Therapeutic Targets in Cancers. In Extracellular Vesicles; Paul M. K., Ed.; IntechOpen: London, 2022. [Google Scholar]
- Naito Y.; Yoshioka Y.; Yamamoto Y.; Ochiya T. How cancer cells dictate their microenvironment: present roles of extracellular vesicles. Cell. Mol. Life Sci. 2017, 74, 697–713. 10.1007/s00018-016-2346-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marleau A. M.; Chen C.-S.; Joyce J. A.; Tullis R. H. Exosome removal as a therapeutic adjuvant in cancer. J. Transl. Med. 2012, 10, 134. 10.1186/1479-5876-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A. T.; Somoza V. Extracellular Vesicles as Vehicles for the Delivery of Food Bioactives. J. Agric. Food Chem. 2019, 67, 2113–2119. 10.1021/acs.jafc.8b06369. [DOI] [PubMed] [Google Scholar]
- Li L.; Ngo H. T. T.; Hwang E.; Wei X.; Liu Y.; Liu J.; Yi T. H. Conditioned Medium from Human Adipose-Derived Mesenchymal Stem Cell Culture Prevents UVB-Induced Skin Aging in Human Keratinocytes and Dermal Fibroblasts. Int. J. Mol. Sci. 2020, 21, 49. 10.3390/ijms21010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X.; Song S.; Chen N.; Liao J.; Zeng L. Stem cell-derived exosomes: A supernova in cosmetic dermatology. J. Cosmet. Dermatol. 2021, 20, 3812–3817. 10.1111/jocd.14438. [DOI] [PubMed] [Google Scholar]
- Oh M.; Lee J.; Kim Y. J.; Rhee W. J.; Park J. H. Exosomes Derived from Human Induced Pluripotent Stem Cells Ameliorate the Aging of Skin Fibroblasts. Int. J. Mol. Sci. 2018, 19, 1715. 10.3390/ijms19061715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.; Bai X.; Zhao B.; Li Y.; Zhang Y.; Li Z.; Wang X.; Luo L.; Han F.; Zhang J.; Han S.; Cai W.; Su L.; Tao K.; Shi J.; Hu D. Cell-free therapy based on adipose tissue stem cell-derived exosomes promotes wound healing via the PI3K/Akt signaling pathway. Exp. Cell Res. 2018, 370, 333–342. 10.1016/j.yexcr.2018.06.035. [DOI] [PubMed] [Google Scholar]
- Munir J.; Lee M.; Ryu S. Exosomes in Food: Health Benefits and Clinical Relevance in Diseases. Adv. Nutr. 2020, 11, 687–696. 10.1093/advances/nmz123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y.; Gao J.; He Y.; Jiang L. Plant extracellular vesicles. Protoplasma 2020, 257, 3–12. 10.1007/s00709-019-01435-6. [DOI] [PubMed] [Google Scholar]
- Plant Exosomes ± Curcumin to Abrogate Symptoms of Inflammatory Bowel Disease. https://clinicaltrials.gov/ct2/show/NCT04879810 (accessed August 19, 2022).
- Plant Exosomes and Patients Diagnosed With Polycystic Ovary Syndrome (PCOS) 17. https://clinicaltrials.gov/ct2/show/NCT03493984 (accessed August 20, 2022).
- Edible Plant Exosome Ability to Prevent Oral Mucositis Associated With Chemoradiation Treatment of Head and Neck Cancer. https://clinicaltrials.gov/ct2/show/NCT01668849 (accessed August 19, 2022).
- Ciloa - A Breakthrough Exosome Technology. https://www.ciloa.fr/technology/ (accessed August 18, 2022).
- Exosome Research Services. https://clarabio.tech/pages/exosome-research (accessed August 18, 2022).
- Creative Biolabs - Services. https://www.creative-biolabs.com/exosome/services.htm.
- Exosome service company. https://everzom.com/ (accessed August 18, 2022).
- MSC derived EV. http://exosomeplus.com/product/list.php?ca_id=1010 (accessed August 18, 2022).
- Exosomics services. https://www.exosomics.eu/services/ (accessed August 18, 2022).
- Exosomics products. https://www.exosomics.eu/products/.
- FUJIFILM Wako Chemicals. https://labchem-wako.fujifilm.com/us/category/lifescience/exosome_research/index.html (accessed August 18, 2022).
- HansaBioMed Life Sciences. https://www.hansabiomed.eu/index.php/about-us (accessed August 18, 2022).
- Lonza. https://www.lonza.com/company-overview (accessed August 18, 2022).
- NanoFCM. https://www.nanofcm.com/category/products/ (accessed August 18, 2022).
- Nanoview Biosciences. https://www.nanoviewbio.com/#our-products (accessed August 18, 2022).
- ReNeuron. https://www.reneuron.com/ctx-derived-exosomes/ (accessed August 18, 2022).
- RoosterBio. https://www.roosterbio.com/ev-exosome-analytical-services/ (accessed August 18, 2022).
- RoosterBio Exosomes/Extracellular Vesicles. https://www.roosterbio.com/product-category/extracellular-vesicles-exosomes/ (accessed August 18, 2022).
- System Biosciences - EXOSOME RESEARCH PRODUCTS. https://www.systembio.com/products/exosome-research (accessed August 23, 2022).
- ThermoFisher Scientific - Exosome Products for Testing and Research. https://www.thermofisher.com/us/en/home/life-science/cell-analysis/exosomes.html?SID=fr-exosome-main (accessed August 18, 2022).
- Anjarium Biosciences. https://www.anjarium.com/ (accessed August 18, 2022).
- Aruna Bio. https://www.arunabio.com/aruna-bio-our-platform (accessed August 18, 2022).
- Capricor https://capricor.com/exosomes/ (accessed August 18, 2022).
- Carmine Therapeutics https://www.carminetherapeutics.com/ (accessed August 18, 2022).
- Usman W. M.; Pham T. C.; Kwok Y. Y.; Vu L. T.; Ma V.; Peng B.; Chan Y. S.; Wei L.; Chin S. M.; Azad A.; He A. B.-L.; Leung A. Y. H.; Yang M.; Shyh-Chang N.; Cho W. C.; Shi J.; Le M. T. N. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 2018, 9, 2359. 10.1038/s41467-018-04791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EV Therapeutics - gallery. https://www.evtherapeutics.com/gallery (accessed August 18, 2022).
- EV Therapeutics - platform. https://www.evtherapeutics.com/about-us (accessed August 18, 2022).
- Evora BioSciences. https://www.evorabio.com/ (accessed August 18, 2022).
- Evox Pipeline. https://www.evoxtherapeutics.com/Pipeline (accessed August 18, 2022).
- Exocel Bio - Exosome Products. https://www.exocelbio.com/products (accessed August 18, 2022).
- Exocel Bio. https://www.exocelbio.com/ (accessed August 18, 2022).
- Cho B. S.; Kim J. O.; Ha D. H.; Yi Y. W. Exosomes derived from human adipose tissue-derived mesenchymal stem cells alleviate atopic dermatitis. Stem Cell. Res. Ther. 2018, 9, 187. 10.1186/s13287-018-0939-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ExoCoBio. http://www.exocobio.com/default/eng/02/02.php (accessed August 18, 2022).
- Exogenus Therapeutics. https://www.exogenus-t.com/pipeline-2/ (accessed August 18, 2022).
- Henriques-Antunes H.; Cardoso R. M. S.; Zonari A.; Correia J.; Leal E. C.; Jiménez-Balsa A.; Lino M. M.; Barradas A.; Kostic I.; Gomes C.; Karp J. M.; Carvalho E.; Ferreira L. The Kinetics of Small Extracellular Vesicle Delivery Impacts Skin Tissue Regeneration. ACS Nano 2019, 13, 8694–8707. 10.1021/acsnano.9b00376. [DOI] [PubMed] [Google Scholar]
- Florica Therapeutics. https://floricatherapeutics.com/ (accessed August 18, 2022).
- Ilias Biologics - Platform Technology. https://www.iliasbio.com/our/platform.php (accessed August 18, 2022).
- Ilias Biologics - Pipline - ILB-202. https://www.iliasbio.com/pipeline/srIkB.php (accessed August 18, 2022).
- Innocan Pharma. https://innocanpharma.com/cell-therapy/ (accessed August 18, 2022).
- Kimera Labs. https://kimeralabs.com/ (accessed August 18, 2022).
- Kimera Labs products. https://kimeralabs.com/products/ (accessed August 18, 2022).
- MDimune - BioDrone Platform. http://www.mdimune.com/en/m62.php (accessed August 18, 2022).
- MDimune Pipeline. http://www.mdimune.com/en/m71.php (accessed August 18, 2022).
- OmniSpirant Therapeutics. https://www.omnispirant.com/ (accessed August 18, 2022).
- Regen Suppliers. https://www.regensuppliers.com/rebellaxo.html (accessed August 18, 2022).
- Xollent Biotech. https://www.xollentbio.com/pipeline (accessed August 18, 2022).
- Craif. https://craif.com/en/science/ (accessed August 18, 2022).
- Mercy Bioanalytics. https://mercybio.com/the-mercy-halo-test/ (accessed August 18, 2022).
- Bortolin L. T.; Salem D. P.; Banerjee S.. Preliminary results for a novel single extracellular vesicle assay for early stage ovarian cancer: The power of co-localized detection of surface biomarkers. In AACR Anual Meeting, New Orleans, LA, 2022. [Google Scholar]
- Salem D. P.; Bortolin L. T.; Banerjee S.. Preliminary results for a novel single extracellular vesicle assay for early lung cancer: The power of co-localized detection of surface biomarkers. In AACR Annual Meeting, New Orleans, LA, 2022. [Google Scholar]
- Matsumoto S.; Sakata Y.; Suna S.; Nakatani D.; Usami M.; Hara M.; Kitamura T.; Hamasaki T.; Nanto S.; Kawahara Y.; Komuro I. Circulating p53-Responsive MicroRNAs Are Predictive Indicators of Heart Failure After Acute Myocardial Infarction. Circ. Res. 2013, 113, 322–326. 10.1161/CIRCRESAHA.113.301209. [DOI] [PubMed] [Google Scholar]
- Morishita M.; Takahashi Y.; Nishikawa M.; Takakura Y. Pharmacokinetics of Exosomes-An Important Factor for Elucidating the Biological Roles of Exosomes and for the Development of Exosome-Based Therapeutics. J. Pharm. Sci. 2017, 106, 2265–2269. 10.1016/j.xphs.2017.02.030. [DOI] [PubMed] [Google Scholar]
- Dudzik D.; Macioszek S.; Struck-Lewicka W.; Kordalewska M.; Buszewska-Forajta M.; Waszczuk-Jankowska M.; Wawrzyniak R.; Artymowicz M.; Raczak-Gutknecht J.; Siluk D.; Markuszewski M. J. Perspectives and challenges in extracellular vesicles untargeted metabolomics analysis. TrAC, Trends Anal. Chem. 2021, 143, 116382. 10.1016/j.trac.2021.116382. [DOI] [Google Scholar]
- Rajagopal C.; Harikumar K. B. The Origin and Functions of Exosomes in Cancer. Front. Oncol. 2018, 8, 66. 10.3389/fonc.2018.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- iExosomes in Treating Participants With Metastatic Pancreas Cancer With KrasG12D Mutation. https://clinicaltrials.gov/ct2/show/NCT03608631 (accessed August 19, 2022).
- The Use of Exosomes In Craniofacial Neuralgia. https://clinicaltrials.gov/ct2/show/NCT04202783 (accessed August 19, 2022).
- Organicell. https://organicell.com/ (accessed August 18, 2022).
- Direct Biologics - ExoFlo. https://directbiologics.com/exoflo/ (accessed August 18, 2022).
- PEP on a Skin Graft Donor Site Wound. https://clinicaltrials.gov/ct2/show/NCT04664738 (accessed August 19, 2022).
- The Safety and the Efficacy Evaluation of Allogenic Adipose MSC-Exos in Patients With Alzheimer’s Disease. https://clinicaltrials.gov/ct2/show/NCT04388982 (accessed August 19, 2022).
- Rion - Clinical pipeline. https://riontx.com/pipeline/ (accessed August 19, 2022).
- Shi A.; Li J.; Qiu X.; Sabbah M.; Boroumand S.; Huang T. C.-T.; Zhao C.; Terzic A.; Behfar A.; Moran S. L. TGF-β loaded exosome enhances ischemic wound healing in vitro and in vivo. Theranostics 2021, 11, 6616–6631. 10.7150/thno.57701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisby C. K.; Shadrin I. Y.; Rolland T. J.; Stalboerger P. G.; Zhou B.; Trabuco E. C.; Behfar A.; Occhino J. A. Exosome-Induced Vaginal Tissue Regeneration in a Porcine Mesh Exposure Model. Female Pelvic Med. Reconstr. Surg. 2021, 27, 609–615. 10.1097/SPV.0000000000001005. [DOI] [PubMed] [Google Scholar]
- Qi J.; Liu Q.; Reisdorf R. L.; Boroumand S.; Behfar A.; Moran S. L.; Amadio P. C.; Gingery A.; Zhao C. Characterization of a purified exosome product and its effects on canine flexor tenocyte biology. Journal of Orthopaedic Research 2020, 38, 1845–1855. 10.1002/jor.24587. [DOI] [PubMed] [Google Scholar]
- Shi G.; Wang Y.; Wang Z.; Thoreson A. R.; Jacobson D. S.; Amadio P. C.; Behfar A.; Moran S. L.; Zhao C. A novel engineered purified exosome product patch for tendon healing: An explant in an ex vivo model. Journal of Orthopaedic Research 2021, 39, 1825–1837. 10.1002/jor.24859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Study Investigating the Ability of Plant Exosomes to Deliver Curcumin to Normal and Colon Cancer Tissue. https://clinicaltrials.gov/ct2/show/NCT01294072 (accessed August 19, 2022).
- Aegle Therapeutics. https://www.aegletherapeutics.com/technology.html (accessed August 19, 2022).
- MSC EVs in Dystrophic Epidermolysis Bullosa. https://clinicaltrials.gov/ct2/show/NCT04173650 (accessed August 19, 2022).
- Evaluation of Personalized Nutritional Intervention on Wound Healing of Cutaneous Ulcers in Diabetics. https://clinicaltrials.gov/ct2/show/NCT05243368 (accessed August 19, 2022).
- A Study of exoASO-STAT6 (CDK-004) in Patients With Advanced Hepatocellular Carcinoma (HCC) and Patients With Liver Metastases From Primary Gastric Cancer and Colorectal Cancer (CRC). https://clinicaltrials.gov/ct2/show/NCT05375604 (accessed August 19, 2022).
- Codiak - pipeline. https://www.codiakbio.com/pipeline-programs/pipeline (accessed August 19, 2022).
- Kamerkar S.; Leng C.; Burenkova O.; Jang S. C.; McCoy C.; Zhang K.; Dooley K.; Kasera S.; Zi T.; Sisó S.; Dahlberg W.; Sia C. L.; Patel S.; Schmidt K.; Economides K.; Soos T.; Burzyn D.; Sathyanarayanan S. Exosome-mediated genetic reprogramming of tumor-associated macrophages by exoASO-STAT6 leads to potent monotherapy antitumor activity. Sci. Adv. 2022, 8, eabj7002 10.1126/sciadv.abj7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitti Labs clinical trials. https://clinicaltrials.gov/ct2/results?cond=&term=vitti+labs&cntry=&state=&city=&dist=&Search=Search (accessed August 19, 2022).
- Vitti Labs. https://www.vittilabs.com/research (accessed August 19, 2022).
- LRRK2 and Other Novel Exosome Proteins in Parkinson’s Disease. https://clinicaltrials.gov/ct2/show/NCT01860118 (accessed August 19, 2022).
- Exosome Diagnostics. https://www.exosomedx.com/physicians/exodx-prostate-test (accessed August 19, 2022).
- Detection of Either the EML4-ALK Gene Rearrangements or the T790M EGFR Mutation in the Plasma of Advanced NSCLC Patients. https://clinicaltrials.gov/ct2/show/NCT03236675 (accessed August 19, 2022).
- Urine Biomarker for Kidney Transplant Rejection. https://clinicaltrials.gov/ct2/show/NCT05072951 (accessed August 19, 2022).
- Exosome Diagnostics Obviates Tissue Biopsies. https://www.exosomedx.com/sites/default/files/2020-02/2018_04_17_aacr_final.pdf (accessed August 19, 2022).
- El Fekih R.; Hurley J.; Tadigotla V.; Alghamdi A.; Srivastava A.; Coticchia C.; Choi J.; Allos H.; Yatim K.; Alhaddad J.; Eskandari S.; Chu P.; Mihali A. B.; Lape I. T.; Lima Filho M. P.; Aoyama B. T.; Chandraker A.; Safa K.; Markmann J. F.; Riella L. V.; et al. Discovery and Validation of a Urinary Exosome mRNA Signature for the Diagnosis of Human Kidney Transplant Rejection. J. Am. Soc. Nephrol. 2021, 32, 994–1004. 10.1681/ASN.2020060850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor Y. L.Identification of Plasma-Derived, EV-Based Biomarkers for Glioblastoma. https://www.exosomedx.com/sites/default/files/2020-07/2020-07-20_ISEV2020_GBM_Khodor.pdf (accessed August 19, 2022).
- Chakrabortty S. K.Identification of a saliva exosomal RNA signature for Sjogren’s syndrome. https://www.exosomedx.com/sites/default/files/2020-07/2020-07-20_ISEV2020_Sjogrens_SKC.pdf (accessed August 19, 2022).
- miR Scientific. https://www.mirscientific.com/science-technology (accessed August 19, 2022).
- miR Scientific announces FDA Breakthrough Device Designation for its Prostate Cancer Liquid Biopsy Test. https://www.prnewswire.com/news-releases/mir-scientific-announces-fda-breakthrough-device-designation-for-its-prostate-cancer-liquid-biopsy-test-301151403.html (accessed August 20, 2022).
- Evaluation of Renal Sodium Excretion After Salt Loading in Heart Failure With Preserved Ejection Fraction (ERES-HFpEF). https://clinicaltrials.gov/ct2/show/NCT03837470 (accessed August 19, 2022).
- Rheo-Erythrocrine Dysfunction as a Biomarker for RIC Treatment in Acute Ischemic Stroke (ENOS). https://clinicaltrials.gov/ct2/show/NCT04266639 (accessed August 19, 2022).
- Non-coding RNAs Analysis of Eosinophil Subtypes in Asthma. https://clinicaltrials.gov/ct2/show/NCT04542902 (accessed August 19, 2022).
- Explore Potential Plasma and BALF Immunometabolic and Lipidomic Biomarkers for Identifying ARDS Endotypes. https://clinicaltrials.gov/ct2/show/NCT05451342 (accessed August 19, 2022).
- Aetholon. https://www.aethlonmedical.com/the-hemopurifier/the-hemopurifier-in-infectious-disease (accessed August 19, 2022).
- Hemopurifier Plus Pembrolizumab in Head and Neck Cancer. https://clinicaltrials.gov/ct2/show/NCT04453046 (accessed August 19, 2022).
- Gooldy M.; Roux C. M.; LaRosa S. P.; Spaulding N.; Fisher C. J. Jr. Removal of clinically relevant SARS-CoV-2 variants by an affinity resin containing Galanthus nivalis agglutinin. PLoS One 2022, 17, e0272377 10.1371/journal.pone.0272377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aethlon Medical Announces Publication of Peer-Reviewed Journal Article Describing Hemopurifier Resin Binding of Seven COVID-19 Variants. https://www.prnewswire.com/news-releases/aethlon-medical-announces-publication-of-peer-reviewed-journal-article-describing-hemopurifier-resin-binding-of-seven-covid-19-variants-301595507.html (accessed August 19, 2022).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.