Abstract
New vaccine strategies are needed for prevention of leptospirosis, a widespread human and veterinary disease caused by invasive spirochetes belonging to the genus Leptospira. We have examined the immunoprotective capacity of the leptospiral porin OmpL1 and the leptospiral outer membrane lipoprotein LipL41 in the Golden Syrian hamster model of leptospirosis. Specialized expression plasmids were developed to facilitate expression of leptospiral proteins in Escherichia coli as the membrane-associated proteins OmpL1-M and LipL41-M. Although OmpL1-M expression is highly toxic in E. coli, this was accomplished by using plasmid pMMB66-OmpL1, which has undetectable background expression without induction. LipL41-M expression and processing were enhanced by altering its lipoprotein signal peptidase cleavage site to mimic that of the murein lipoprotein. Active immunization of hamsters with E. coli membrane fractions containing a combination of OmpL1-M and LipL41-M was found to provide significant protection against homologous challenge with Leptospira kirschneri serovar grippotyphosa. At 28 days after intraperitoneal inoculation, survival in animals vaccinated with both proteins was 71% (95% confidence interval [CI], 53 to 89%), compared with only 25% (95% CI, 8 to 42%) in the control group (P < 0.001). On the basis of serological, histological, and microbiological assays, no evidence of infection was found in the vaccinated survivors. The protective effects of immunization with OmpL1-M and LipL41-M were synergistic, since significant levels of protection were not observed in animals immunized with either OmpL1-M or LipL41-M alone. In contrast to immunization with the membrane-associated forms of leptospiral proteins, hamsters immunized with His6-OmpL1 and His6-LipL41 fusion proteins, either alone or in combination, were not protected. These data indicate that the manner in which OmpL1 and LipL41 associates with membranes is an important determinant of immunoprotection.
Leptospirosis is considered to be the most widespread zoonotic disease in the world (12). Reservoir hosts with chronic renal tubular infection transmit pathogenic Leptospira species to new hosts through urinary shedding. Leptospiral infection in humans may result in a fulminant, life-threatening illness characterized by liver and kidney failure (21). Leptospirosis appears to be emerging in both developed and underdeveloped regions of the world. A recent study found a 16% seropositivity rate among inner city black males (15). Urban residents are at particular risk for leptospirosis if homelessness results in percutaneous exposure to urine of rats shedding pathogenic leptospires (57). In tropical areas of the world, acute leptospirosis accounts for roughly 10% of hospitalizations for acute febrile illness (19), and leptospirosis epidemics occur predictably after periods of heavy rain and flooding (13). Leptospirosis also remains prevalent in domestic cattle, pigs, and dogs despite widespread vaccination (6, 55).
Commercial leptospiral vaccines rely on “bacterins,” or inactivated whole cells, an approach that has been used since the development of leptospiral culture media in the early part of the 20th century (37, 46). Efforts to reduce the incidence of local reactions to whole-cell vaccines have been directed toward elimination of proteins from culture media (5, 17) and utilization of subcellular fractionation. Studies showing that isolated outer membrane retained most, if not all, of the protective antigens found in whole-cell vaccines helped to focus interest on the leptospiral surface (2, 4). Subsequently, it was shown that a major component of the immunity resulting from whole-cell and outer membrane vaccines involves a humoral immune response to the serovar-specific carbohydrate antigens of leptospiral lipopolysaccharide (LPS) (1, 42). Though the immunity generated by whole-cell vaccines is readily demonstrable in the hamster model of leptospirosis, a number of deficiencies are evident when this strategy is applied to domestic animals: (i) cross-protection against many of the 250 different serovars of pathogenic Leptospira species is lacking, (ii) annual or biannual revaccination is necessary to maintain immunity, (iii) local reactions are common after vaccination, and (iv) protection in cattle is difficult to demonstrate even under optimal conditions (8–10).
These problems have led to an examination of leptospiral outer membrane proteins (OMPs) because of their potential usefulness as subunit vaccines. The leptospiral outer membrane contains both transmembrane proteins, such as the porin OmpL1 (29, 50), and lipoproteins, such as LipL41 (51) and LipL36 (31). OMPs that are exposed on the leptospiral surface are potentially relevant in pathogenesis because of their location at the interface between leptospires and the mammalian host. Results of surface immunoprecipitation studies suggest that OmpL1 and LipL41 are surface exposed (32, 51). Exposure of OmpL1 on the leptospiral surface has also been demonstrated by immunoelectron microscopy (29). Topological considerations also suggest that OmpL1, a transmembrane porin, should have surface-exposed epitopes (29). Unlike leptospiral LPS, OmpL1 and LipL41 are antigenically conserved among pathogenic Leptospira species (28, 51). Their promise as vaccine candidates was also enhanced by the finding that OmpL1 and LipL41 are expressed during infection of the mammalian host (3).
Because of the difficulties involved in purifying native proteins free of other leptospiral membrane components, the clearest demonstration of protective efficacy should come from studies involving recombinant OMPs. There is a growing body of evidence that recombinant transmembrane (41, 44, 48, 59, 60) and lipoprotein (11, 22, 23, 27, 34, 45, 47) OMPs can provide degrees of protection against a variety of bacterial infections. In this study, we developed systems for expression of recombinant OmpL1 and LipL41 as membrane proteins and examined their immunoprotective potential alone and in combination. Our findings demonstrate that when expressed as membrane proteins, OmpL1 and LipL41 together provide a significant level of protection against homologous challenge in the hamster model of leptospirosis. These results provide the first evidence for the feasibility of using recombinant OMPs as vaccines for prevention of leptospirosis.
MATERIALS AND METHODS
Bacterial strains, media, and plasmids.
Leptospira kirschneri serovar grippotyphosa strain RM52 (hereafter referred to as L. kirschneri RM52) (32, 56) was obtained from the National Leptospirosis Reference Center (National Animal Disease Center, Agricultural Research Service, U.S. Department of Agriculture, Ames, Iowa). Leptospires were cultivated in Johnson-Harris bovine serum albumin Tween 80 medium (Bovuminar PLM-5 Microbiological Media; Intergen) (38). Escherichia coli DH5α [supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] was used as the host strain for transformations of recombinant DNA. E. coli XL1-Blue [recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi-1 lac (F′ proAB lacIqZΔM15 Tn10 [Tetr])c] (Stratagene) was used as the host strain for plasmid pMMB66-OmpL1. The DE3 lysogen of E. coli JM109 [recA1 supE44 endA1 hsdR17 gyrA96 relA1 thiΔ(lac-proAB) (F′ traD36 proAB+ lacIq lacZΔM15)] (Promega) was used as the host strain for derivatives of the expression plasmid pET-15b (Novagen). E. coli BLR(DE3)pLysS [F− ompT hsdSB (rB− mB−) gal dcm Δ(srl-recA)306::Tn10(TcR) (DE3) pLysS(CmR)] (Novagen) was used as the host strain for the expression plasmid pRSET (Invitrogen). E. coli cells were routinely grown in Luria-Bertani (LB) broth or on LB agar, unless otherwise mentioned (49).
Gel electrophoresis and immunoblotting.
Samples for sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) were solubilized in final sample buffer, composed of 62.5 mM Tris hydrochloride (pH 6.8), 10% glycerol, 5% 2-mercaptoethanol, and 2% SDS. Proteins were separated on a 12% gel with a discontinuous buffer system (39) and stained with Coomassie brilliant blue or were transferred to nitrocellulose (Schleicher & Schuell) for immunoblotting. For antigenic detection on immunoblots, the nitrocellulose was blocked with 5% nonfat dry milk in 0.1 M phosphate-buffered saline (PBS; pH 7.4)–0.1% Tween 20 (PBS-T), incubated for 1 h with antiserum diluted 1:5,000 (unless otherwise noted) in PBS-T, and probed with donkey anti-rabbit antibody conjugated to horseradish peroxidase (Amersham). Antigen-antibody binding was detected by using the Amersham enhanced chemiluminescence (ECL) system. Blots were incubated in ECL reagents for 1 min and then exposed to Hyperfilm (Amersham). Densitometry of gels was performed with an AMBIS Imager and QuantProbe software (Scanalytics Inc., Billerica, Mass.).
Preparation of recombinant OmpL1-M and LipL41-M.
Standard recombinant DNA procedures were performed as described elsewhere (49). Restriction endonuclease digests were performed as recommended by the suppliers (New England Biolabs and Promega). The membrane-associated forms of OmpL1 and LipL41 are designated OmpL1-M and LipL41-M, respectively. OmpL1-M was expressed in E. coli XL1-Blue (Stratagene), using plasmid pMMB66-OmpL1. Expression plasmid pMMB66-OmpL1 was constructed by using the intact ompL1 gene, including the region encoding its native signal peptide, as described previously (50). In brief, the ompL1 gene was obtained from the original pBluescript SK (−) isolate, pIBL2, containing a 2.5-kb EcoRI fragment with the ompL1 gene of L. kirschneri RM52 (29). PCR was used to amplify the ompL1 gene with a HindIII restriction endonuclease site at its 5′ end. A 378-bp HindIII-NcoI fragment of the amplified ompL1 gene, containing the 5′ end of the gene, was ligated into pIBL2, yielding plasmid pIBL2-HN. A 1.7-kb HindIII-EcoRI fragment of pIBL2-HN was ligated into pMMB66HE placing expression of the ompL1 gene under control of the tac promoter. Plasmid pMMB66-OmpL1 was sequenced in the region of the ompL1 gene and the region upstream of the ompL1 gene. DNA sequencing was performed at the University of California-Los Angeles Core DNA Sequencing Facility by the dideoxy chain termination method with fluorescein-labeled dideoxy nucleotides (Applied Biosystems).
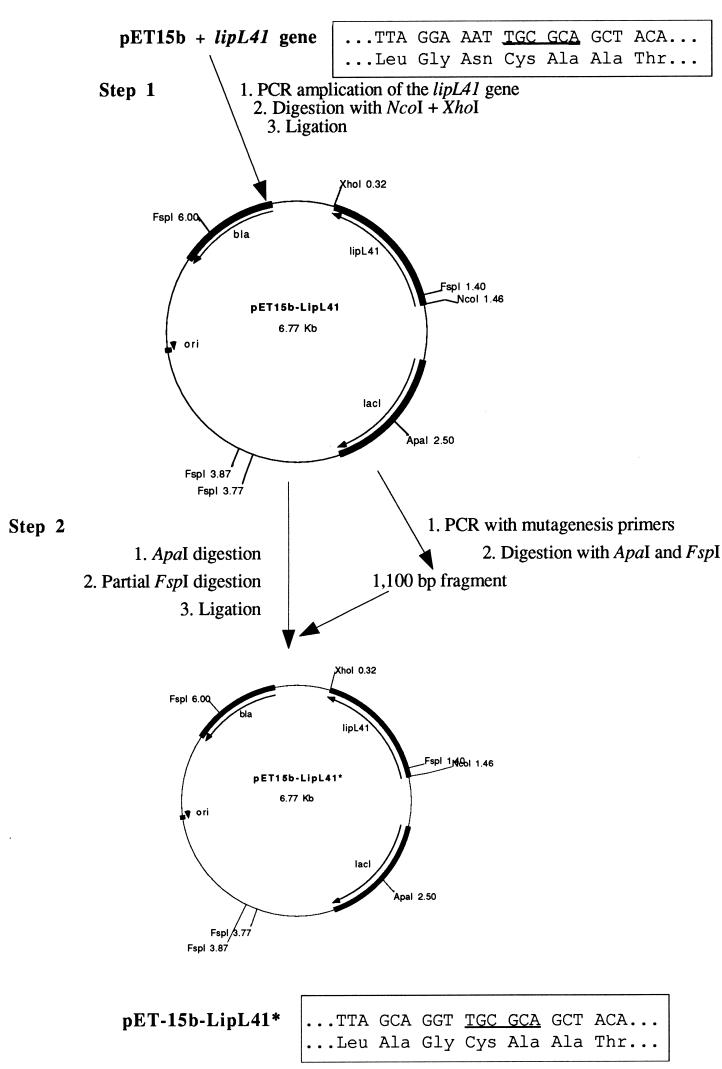
LipL41-M was expressed in E. coli JM109(DE3) (Promega) by using plasmid pET-15b-LipL41*, in which the lipoprotein signal peptidase cleavage site of LipL41 was changed from its native sequence (LGNC) to a sequence preferred by E. coli lipoprotein signal peptidase (LAGC). As shown in Fig. 1, construction of pET-15b-LipL41* was accomplished in two steps. The first step was to construct the pET-15b-LipL41 expression plasmid by PCR amplifying the intact lipL41 gene and cloning it in pET-15b (Novagen). The forward oligonucleotide primer contained the nucleotide sequence coding for the six amino acids of LipL41 (beginning with the N-terminal methionine), including an NcoI restriction endonuclease site (underlined): 5′-TG TTA CCC ATG GGG AGA AAA TTA TCT TCT CT-3′. The reverse oligonucleotide primer contained the nucleotide sequence coding for the five C-terminal amino acids of LipL41 and the TAA stop codon, including an XhoI restriction endonuclease site (underlined): 5′-AAA GGA CTC GAG TTA CTT TGC GTT GCT TTC-3′. L. kirschneri RM52 genomic DNA was used as the template. The 1,077-bp NcoI-XhoI fragment of the amplified lipL41 gene was ligated to pET-15b digested with NcoI and XhoI. The resulting construct was designated pET-15b-LipL41.
FIG. 1.
PCR mutagenesis of the lipoprotein signal peptidase cleavage site of LipL41. In step 1, the lipL41 gene of L. kirschneri was PCR amplified and inserted into pET15b. In step 2, a portion of pET15b-LipL41 plasmid was amplified by using primers that alter the sequence encoding the two amino acids preceding cysteine. The mutagenized FspI-ApaI fragment was then inserted into pET15b-LipL41. The location of the FspI site is underlined in the boxed sequences, which also show the amino acid sequences commencing with the leucine of the lipoprotein signal peptidase cleavage site before (LGNC) and after (LAGC) PCR mutagenesis.
The second step in the construction of pET-15b-LipL41* was to alter plasmid pET-15b-LipL41 at the lipoprotein signal peptidase cleavage site by PCR mutagenesis. The forward oligonucleotide primer contained the nucleotide sequence coding for a region of pET-15b upstream of a unique ApaI restriction endonuclease site: 5′-TGA ATT TGA TTG CGA GTG AGA TAT-3′. The reverse oligonucleotide primer contained the nucleotide sequence coding for an altered LipL41 lipoprotein signal peptidase cleavage site and the adjacent FspI restriction endonuclease site (underlined): 5′-TGT AGC TGC GCA ACC TGC TAA GAA CAT AAG GAG AAC TAA-3′. Plasmid pET-15b-LipL41 was used as the template. The 1,200-bp amplification product was digested with ApaI and FspI and ligated to pET-15b-LipL41 digested to completion with ApaI and then partially digested with FspI. Partial digestion with FspI was necessary since pET-15b-LipL41 contains four FspI sites. After digestion of pET-15b-LipL41 with ApaI at 25°C for 1 h followed by FspI at 37°C for 1 min, 10 mM EDTA was added to stop the reaction. The 5,500-bp fragment of pET-15b-LipL41, digested at only the correct FspI site, was isolated by electrophoresis in a 0.9% agarose gel. The resulting construct, pET-15b-LipL41*, was transformed into E. coli JM109(DE3).
Fresh E. coli transformants were used for expression of OmpL1-M and LipL41-M. One hundred milliliters of SOC medium (49) plus ampicillin (100 μg/ml) was inoculated with 2 ml of overnight culture. Cultures were incubated in a shaking incubator at 37°C. When the optical density at 600 nm reached 0.5, expression was achieved by isopropylthio-β-d-galactoside (IPTG; Sigma) induction. Cultures were incubated in a shaking incubator at 37°C for an additional 3 to 4 h. Cells were pelleted by centrifugation and resuspended in 2.5 ml of ice-cold TEN buffer (50 mM Tris hydrochloride [pH 8.0], 1 mM EDTA, 100 mM NaCl). The cell suspension was passed two times through a French press (12,000 lb/in2). Unbroken cells were removed by centrifugation at 5,000 × g for 10 min. The membrane fraction was isolated by centrifugation at top speed in a microcentrifuge for 30 min. The membrane pellet was washed two times in 2.5 ml of TEN buffer. The recovery of recombinant OmpL1-M and LipL41-M was then evaluated by densitometry of SDS-polyacrylamide gels and determination of protein concentration by using bicinchoninic acid (54).
Preparation of His6-OmpL1 and His6-LipL41.
In contrast to recombinant OmpL1-M and LipL41-M, His6 fusion proteins were expressed without signal peptides. The His6-OmpL1 and His6-LipL41 fusion proteins were prepared by a modification of the methods described previously (29). Briefly, plasmid pRSET (Invitrogen) containing the ompL1 or lipL41 gene (without the signal peptide) was transformed into E. coli BLR(DE3)pLysS (Novagen). Expression of the His6 fusion proteins was achieved by IPTG induction. The His6 fusion proteins were solubilized in 6 M guanidine and purified by affinity chromatography using Ni2+-nitrilotriacetic acid-agarose (Qiagen). The His6-OmpL1 fusion protein was dialyzed in PBS containing 10% glycerol, 1.0% Triton X-100, and 0.025% sodium azide. The His6-LipL41 fusion protein was dialyzed in PBS containing 10% glycerol, 0.3% Triton X-100, and 0.025% sodium azide. Purity was determined by SDS-PAGE. Protein concentration was determined by using bicinchoninic acid (54).
Immunization and challenge experiments.
Virulent L. kirschneri RM52 was passaged less than five times prior to intraperitoneal (i.p.) inoculation of Golden Syrian hamsters (Harlan Sprague-Dawley). Leptospires were enumerated by dark-field microscopy as described by Miller (43). An initial series of challenge studies was performed on unimmunized 3- and 9-week-old hamsters to standardize the animal model. Nine-week-old hamsters (in groups of eight) and 3-week-old hamsters (in groups of four) were challenged i.p. with 10, 100, or 1,000 L. kirschneri cells.
Three protection experiments were conducted with immunized hamsters. In the initial protection experiment, hamsters in groups of nine were immunized three times i.p. at 4-week intervals with E. coli membrane fractions containing 50 μg of either OmpL1-M, LipL41-M, or a combination of E. coli membrane fractions containing 50 μg of OmpL1-M and 50 μg of LipL41-M. Each arm of the experiment included a control group immunized with membrane fractions from the same strain of E. coli transformed with the same expression plasmid (without the leptospiral gene) prepared in the same manner as the test material. Two weeks after administration of the final immunization, hamsters were challenged i.p. with 100 virulent L. kirschneri RM52, cells. The format of the second protection experiment was the same as that for the first experiment, with the following exceptions: (i) there were six animals per group in the second experiment instead of 9, (ii) the level of recombinant leptospiral protein achieved in the second experiment was 3 to 5% of the total protein instead of 5 to 10%, and (iii) an additional control group of unimmunized hamsters of the same age was included in the experiment. In the third protection experiment, both the membrane-associated and the His6 fusion protein forms of OmpL1 and LipL41 were tested alone and in combination. Hamsters in groups of nine were immunized with 50 μg of recombinant leptospiral protein at 4-week intervals. To assess the effects of adjuvant and an alternative immunization route, the third protection experiment involved immunization given subcutaneously with aluminum hydroxide (alum) adjuvant prepared as previously described (35). There was one control group for the animals immunized with membrane-associated proteins and another control group for the animals immunized with the His6 fusion proteins. The formats of the three protection experiments are summarized in Table 1.
TABLE 1.
Summary of hamster immunization and challenge experimentsa
| Exptl design parameter | Expt 1 | Expt 2 | Expt 3 |
|---|---|---|---|
| No. of groups | 6 | 6 | 8 |
| No. of animals/group | 9 | 6 | 9 |
| Route of immunization | i.p. | i.p. | s.q. |
| Immunization dose (μg of recombinant) | 50 | 50 | 50 |
| Adjuvant | None | None | Aluminum hydroxide |
| Purity of recombinant (%) | 5–10 | 3–5 | 3–5 |
| Immunization schedule (day) | 0, 28, 55 | 0, 28, 56 | 0, 28, 56 |
| Challenge day | 69 | 70 | 70 |
| Challenge dose (no. of L. kirschneri cells i.p.) | 100 | 100 | 500 |
| Groups (host:antigen) | XL:OmpL1-M | XL:OmpL1-M | XL:OmpL1-M |
| XL (control) | XL (control) | JM:LipL41-M | |
| JM:LipL41-M | JM:LipL41-M | XL:OmpL1-M, JM:LipL41-M | |
| JM (control) | JM (control) | XL + JM (control) | |
| XL:OmpL1-M, JM:LipL41-M | XL:OmpL1-M, JM:LipL41-M | His6-OmpL1 | |
| XL + JM (control) | XL + JM (control) | His6-LipL41 | |
| His6-OmpL1, His6-LipL41 | |||
| Saline control |
Abbreviations: XL, E. coli XL1-Blue with pMMB66 or pMMB66-OmpL1; JM, E. coli JM109(DE3) with pET-15b or pET-15b-LipL41*; s.q., subcutaneous.
Enzyme-linked immunosorbent assay (ELISA).
Two weeks after each immunization, representative hamsters from each group were bled from the retro-orbital venous plexus to assess the response to immunization. Immulon microtiter plates (Dynatech) were coated at 37°C overnight with 125 ng of purified His6 fusion proteins in 0.05 M sodium carbonate buffer (pH 9.6). The plates were washed three times with PBS containing 0.05% Tween 20, 200 μl of blocking buffer (PBS containing 0.05% Tween 20 and 1% nonfat dried milk) was added, and the plates were incubated at 37°C for 1 h. After removal of the blocking buffer, 100 μl of antiserum diluted 1:100 with blocking buffer was added, and the plates were incubated at 37°C for 2 h. After washing three times, 100 μl of 1:5,000 diluted sheep anti-hamster immunoglobulin G conjugated to horseradish peroxidase (Amersham), was added and the plates were incubated at 37°C for 2 h. After washing three more times, 100 μl of 3,3′,5,5′-tetramethylbenzidine substrate was added, and the plates were incubated at 37°C for 20 min on an orbital shaker. Then 100 μl of a 1 M solution of sulfuric acid was added to stop the reaction. Absorbance was read at a wavelength of 450 nm with a microplate reader (model 550; Bio-Rad). Each serum sample was tested four separate times. Absorbance readings were normalized by subtracting the value obtained from antigen-negative control wells. Statistical analysis was performed by using Student's t test for two independent means.
Analyses of response to challenge with L. kirschneri.
Hamsters were examined daily to monitor the response to challenge. Moribund hamsters were sacrificed, and kidneys were removed for culture and histopathology. At 28 days after challenge, blood and kidney tissue was obtained under sterile conditions for serological studies, culture, and histopathology. Serological analysis was performed by using the microscopic agglutination titer (MAT) as described elsewhere (20), using L. kirschneri RM52 as the antigen. One kidney was disrupted in sterile PBS and inoculated into leptospiral medium containing 100 μg of 5-fluorouracil per ml to inhibit growth of bacterial contaminants. The other kidney was fixed in formalin and paraffin embedded. Tissue sections were stained with hematoxylin and eosin and examined for evidence of interstitial nephritis. Statistical analyses were performed by using the two-tailed Student t test for samples with unequal variance.
RESULTS
Expression of OmpL1-M in E. coli.
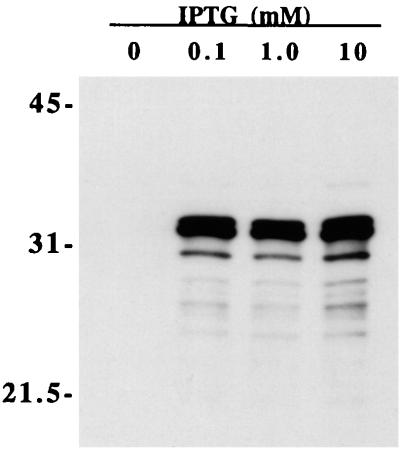
We have previously described expression of recombinant OmpL1 by using plasmid pMMB66-OmpL1, which results in localization of recombinant OmpL1 to the E. coli outer membrane (50). Plasmid pMMB66 encodes the lacIq gene so that E. coli strains transformed with this plasmid overproduce the lac repressor, which binds to the lac operator region of the tac promoter (26). Nevertheless, there are usually relatively high levels of background expression of cloned genes when this plasmid is used (33). Viability of cells transformed with the pMMB66-OmpL1 construct was not expected, because expression of OmpL1 is extremely toxic in E. coli, resulting in a 50-fold decrease in host cell viability within 30 min after IPTG induction (50). In part, this is explained by the lack of OmpL1 expression without induction (Fig. 2). To evaluate the reason for the lack of background expression, we obtained the DNA sequence of the ompL1 gene and the upstream regulatory region of plasmid pMMB66-OmpL1. Comparison to the sequence of pMMB66EH (GenBank accession no. X15234) revealed a single nucleotide difference, resulting in conversion of the sequence of the ribosome-binding site from -AGGA- to -AAGA-. Although we have not formally tested its effect, it seems plausible that this point mutation could account for the observed phenotype through reduced complementarity to E. coli 16S rRNA. Perhaps as a result of its toxicity, the level of OmpL1-M expression varied from 5 to 10%, as determined by densitometry of Coomassie blue-stained acrylamide gels (Fig. 3). For the OmpL1-M material used in the first protection experiment, we were able to achieve levels of expression of 10% of total membrane protein. However, subsequent yields were typically 5% or less. Expression of OmpL1-M was studied in assays using a variety of different host strains, and E. coli XL1-Blue was found to produce the most consistent results.
FIG. 2.
Immunoblot showing OmpL1 expression at different IPTG concentrations. A culture of E. coli XL1-Blue containing plasmid pMMB66-OmpL1 was grown to log phase, separated into four equal volumes, and treated with various concentrations of IPTG. Cells were incubated for an additional 3 h in a shaker incubator at 37°C, pelleted in a microcentrifuge, resuspended in final sample buffer, and analyzed by SDS-PAGE and immunoblotting. A companion SDS-polyacrylamide gel loaded with the same four samples was stained with Coomassie brilliant blue to verify that all four lanes contained comparable amounts of material (data not shown). The locations of molecular size standards are shown (in kilodaltons) on the left. Even though use of the tac promoter typically results in relatively high background expression levels, expression using plasmid pMMB66-OmpL1 was not detectable without IPTG induction.
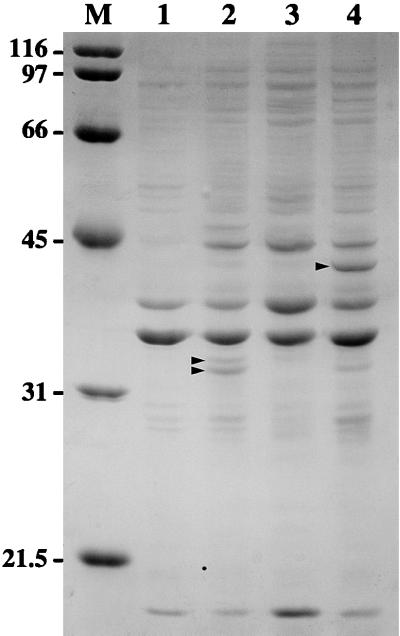
FIG. 3.
Coomassie brilliant blue-stained SDS-polyacrylamide gel showing material representative of E. coli membrane fractions used to immunize hamsters. Lanes: 1 and 2, E. coli XL1-Blue with plasmids pMMB66 and pMMB66-OmpL1, respectively; 3 and 4, E. coli JM109(DE3) with plasmids pET15b and pET15b-LipL41*, respectively. Locations of the OmpL1 doublet in lane 2 and of LipL41 in lane 4 are shown (arrowheads). Positions of molecular size markers (M) are given in kilodaltons.
Expression of LipL41-M in E. coli.
Expression of LipL41-M in E. coli by using plasmid pET-15b-LipL41, which contains the native lipL41 gene including the atypical lipoprotein signal peptidase cleavage site (LGNC), results in inefficient processing by E. coli lipoprotein signal peptidase (51). For this reason, we used PCR mutagenesis to produce a plasmid, pET-15b-LipL41*, in which the lipoprotein signal peptidase cleavage site was altered to that of the E. coli murein lipoprotein (LAGC). Both the processing of LipL41 by E. coli lipoprotein signal peptidase and the total amount of protein expressed were significantly improved with use of pET-15b-LipL41* (Fig. 4). Yields of LipL41-M were determined by densitometry of Coomassie blue-stained acrylamide gels to be 3 to 5% of the total protein in the E. coli membrane fraction (Fig. 3).
FIG. 4.
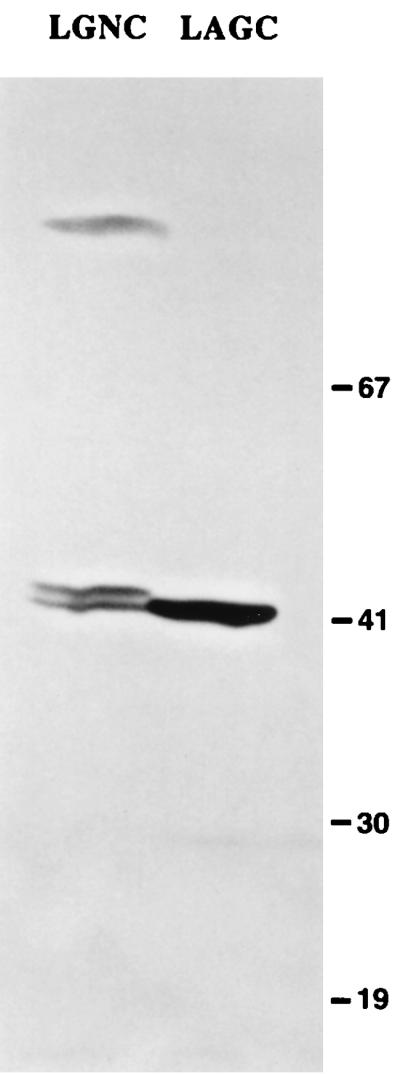
Immunoblot showing recombinant LipL41 expression before and after site-specific mutagenesis of the lipL41 gene. Expression studies were performed with plasmids pET15b-LipL41 (containing the native lipoprotein signal peptidase cleavage site, LGNC) and pET15b-LipL41* (containing the modified lipoprotein signal peptidase cleavage site, LAGC). E. coli JM109(DE3) was transformed with pET15b-LipL41 (left) or pET15b-LipL41* (right). In the left-hand lane, incomplete processing of LipL41 by E. coli lipoprotein signal peptidase results in formation of a doublet. The doublet's lower band represents processed LipL41, while the doublet's upper band represents unprocessed LipL41. As shown in the right-hand lane, expression and processing of LipL41 were much more efficient after modification of the lipoprotein signal peptidase cleavage site. The locations of molecular mass markers are shown on the right in kilodaltons.
Antibody response of hamsters to immunization with recombinant leptospiral proteins.
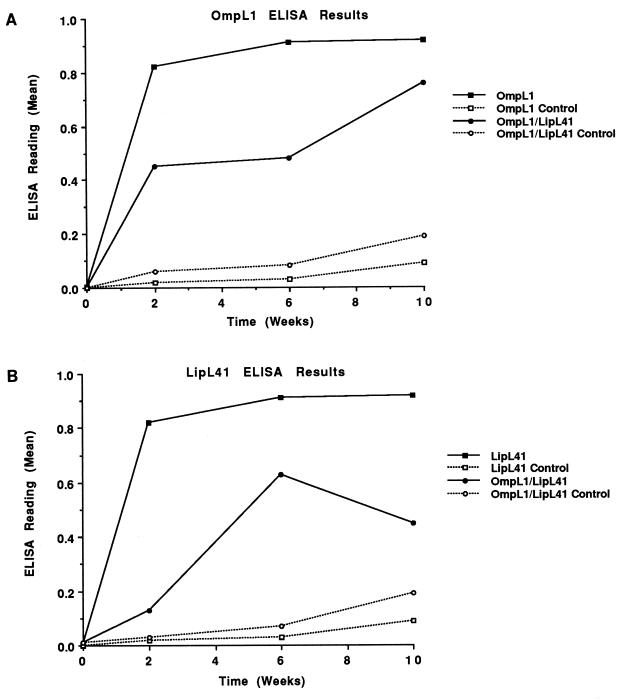
Hamsters were immunized three times at 4-week intervals with E. coli membrane fractions with and without OmpL1-M, LipL41-M, or a combination of both proteins. A humoral immune response to these proteins was documented by ELISA measurement of antibodies in blood samples obtained from representative animals 2 weeks after each immunization (Fig. 5). Antibody levels after the first and second immunizations were higher in animals immunized with a single protein than in those immunized with both OmpL1-M and LipL41-M. A significant ELISA booster response was not obtained after the second and third immunizations.
FIG. 5.
Antibody response of hamsters immunized with leptospiral OMPs. Hamsters were immunized with E. coli membrane fractions with (solid lines) and without (broken lines) OmpL1-M, LipL41-M, or a combination of both. Antibody levels were determined by ELISA against purified His6-OmpL1 (A) and His6-LipL41 (B) fusion proteins. Antibody levels were lower in hamsters immunized with both antigens than in those immunized with single antigens.
Determination of LD50 for i.p. challenge of hamsters with L. kirschneri.
Since lethality of some leptospiral strains can be variable in older hamsters and we planned to use prolonged immunization schedules, it was important to standardize our animal model and determine whether the results would be affected by the age of the hamster at the time of challenge. We found that both weanling and young adult hamsters were exquisitely susceptible to lethal infection after i.p. inoculation with low-passage isolates of L. kirschneri RM52. Both 3- and 9-week-old hamsters were challenged with 10, 100, or 1,000 organisms by the i.p. route. The survival at 28 days after challenge was 20% or less for all doses tested, indicating that the 50 lethal dose (LD50) for i.p. challenge of hamsters with L. kirschneri RM52 is less than 10 organisms (data not shown). The mean time to death after challenge was approximately 9 days for 3-week-old hamsters and approximately 10 days for 9-week-old hamsters. The mean time to death also appeared to be affected slightly by inoculum size. Hamster age and inoculum did not affect the LD50 at 28 days after challenge.
Immunoprotection of hamsters with OmpL1 and LipL41.
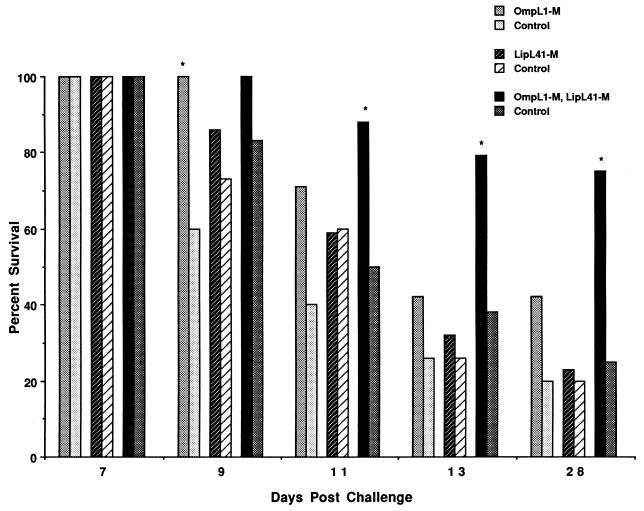
Hamsters immunized with a combination of OmpL1-M- and LipL41-M-containing E. coli membrane fractions had better survival than controls in all three experiments (Table 2 and Fig. 6). At 28 days after i.p. inoculation, survival in animals vaccinated with both proteins was 71% (95% confidence interval [CI], 53% to 89%), compared with only 25% (95% CI, 8 to 42%) in the control group. This difference was statistically significant in the first experiment (P < 0.004), and the statistical power of this effect increased when the results of all three experiments were combined (P < 0.001). The survival benefit of immunization was demonstrated despite a strong i.p. challenge with ≥100 organisms (≥10 times the LD50). As shown in Table 2, survival at 28 days postchallenge in animals immunized with a combination of OmpL1-M and LipL41-M varied from 50 to 100% in the three experiments. By comparison, survival in the control groups varied from 16 to 33%. The consistent level of lethal infection in the control groups demonstrated the reproducibility of the conditions used in the three experiments, including the virulence of the challenge isolate, while controlling for any possible effects of immunization with E. coli membrane fractions. The consistency of the results in the control groups supports the approach of analyzing the combined results of the three protection experiments. As shown in Fig. 6, the survival benefit of immunization with both proteins was recognizable by day 11 postchallenge and was maintained thereafter.
TABLE 2.
Protective effect of immunization with a combination of OmpL1-M and LipL41-M
| Group | No. (%) of surviving
animals/groupa
|
|||
|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | Total | |
| OmpL1-M | 9/9 (100) | 0/6 (0) | 1/9 (11) | 10/24 (42) |
| Control | 3/9 (33) | 0/6 (0) | 3/15 (20) | |
| Significance | P < 0.004 | NS | NSb | NS |
| LipL41-M | 2/7 (29) | 1/6 (17) | 2/9 (22) | 5/22 (23) |
| Control | 3/9 (33) | 0/6 (0) | 3/15 (20) | |
| Significance | NS | NS | NSb | NS |
| OmpL1-M + LipL41-M | 9/9 (100) | 3/6 (50) | 5/9 (55) | 17/24 (71) |
| Control | 3/9 (33) | 1/6 (17) | 2/9 (22) | 6/24 (25) |
| Significance | P < 0.004 | NS | NS | P < 0.001 |
Number of surviving animals at 28 days after challenge/number of animals challenged. Statistical analysis was performed by using Student's t test for two independent means (significance, P < 0.05). NS, not significant.
Significance determined by comparison to control group for OmpL1-M + LipL41-M arm of experiment 3.
FIG. 6.
Survival of hamsters challenged with L. kirschneri after immunization with leptospiral OMPs. Hamsters were immunized with E. coli membrane fractions with and without OmpL1-M, LipL41-M, or a combination of both. Bars denote percent survival at various time points after i.p. challenge with virulent L. kirschneri. Asterisks denote a significant difference in survival versus control (P < 0.05). Data represent the sum of three separate experiments.
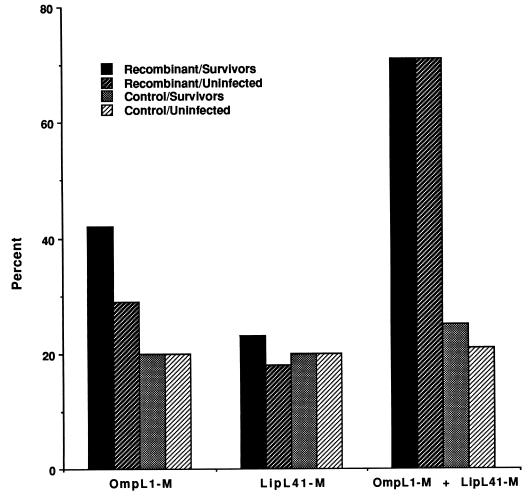
Serological, microbiological, and histological assays were used to examine 28-day survivors for evidence of sublethal infection. The MAT test, a sensitive serological measure of sublethal infection, was positive in 14% of survivors. Sublethal infection usually occurs in the kidneys, and so leptospiral culture and histological examination of survivor kidneys were also performed. Positive cultures and abnormal histopathology were found only in animals with positive MAT tests. No evidence of sublethal infection was detected in any of the surviving recipients of dual immunization (Fig. 7).
FIG. 7.
No evidence of infection in hamsters immunized with a combination of OmpL1-M and LipL41-M. Hamsters were immunized with E. coli membrane fractions with and without OmpL1-M, LipL41-M, or a combination of both. Bars denote percentages of animals surviving with or without evidence of infection 28 days after i.p. challenge with virulent L. kirschneri. Data represent the sum of three separate experiments. The largest number of infected survivors was seen in hamsters immunized with OmpL1-M alone. In no group were the differences between the total number of survivors and the number of uninfected survivors significant (P < 0.05).
In contrast to the results with dual immunization, immunization with OmpL1-M alone demonstrated protection only in the first experiment. Each of the nine hamsters immunized with OmpL1-M alone survived challenge in experiment 1, and the difference in survival compared to controls (33% survival) was statistically significant (Table 2). However, in the second and third experiments, survival of hamsters immunized with OmpL1-M alone was less than or equal to that of the control group. Combined analysis of the results from all three experiments did not demonstrate significant protection with OmpL1-M alone at 28 days after challenge. No evidence of protection in hamsters immunized with LipL41-M alone was observed in any of the three experiments.
No synergy and little, if any, protection was observed in hamsters immunized with either His6-OmpL1 (44% survival), His6-LipL41 (22% survival), or a combination of both fusion proteins (33% survival) compared to saline-immunized controls (11% survival). These challenge results might have been different had the immunization with either membrane or His6 fusion proteins been performed with another adjuvant, a greater amount of antigen, or a different immunization time course.
DISCUSSION
The focus of our research is to identify proteins that are relevant to leptospiral pathogenesis and host immunity. The most direct method available for assessing the relevance of a protein in leptospiral pathogenesis is to determine whether immunization with that protein is able to alter the disease in an animal model. This approach carries with it the caveat that it is not possible to anticipate which method of immunization or which formulation of the immunogen is most likely to produce a relevant immune response. In evaluating the immunoprotective capacity of leptospiral OMPs, a potentially important aspect of immunogenicity is their association with a bacterial membrane. We have therefore focused our efforts on developing expression systems that allow leptospiral OMPs to be produced as recombinant proteins in ways that preserve membrane association.
OmpL1 is a transmembrane OMP which functions as a porin in the leptospiral outer membrane. Based on crystallography studies of E. coli OMPs and analysis of its amino acid sequence, OmpL1 is predicted to contain transmembrane segments that traverse the outer membrane in beta conformation. Alternating amino acids face the hydrophobic interior of the lipid bilayer and the hydrophilic interior of the porin channel. The conformation of a recombinant OMP is most likely to resemble that of the native protein when it is allowed to associate with a lipid bilayer. Amphipathic detergents solubilize membrane proteins by reproducing this arrangement but may simultaneously denature other elements of protein conformation. Another concern involves the potential need for interactions with cytoplasmic or periplasmic chaperonins in order for proper folding to occur. We felt that the best way to address these uncertainties is to immunize hamsters with recombinant OmpL1 localized to the E. coli outer membrane. For this reason, plasmid pMMB66-OmpL1 was constructed by using the intact ompL1 gene, including the region encoding its signal peptide, in order to allow export beyond the cytoplasmic membrane. We have previously documented that OmpL1 expressed by plasmid pMMB66-OmpL1 is associated with the E. coli outer membrane. While inclusion of a single peptide allows membrane association to occur, it also creates difficulties with toxicity and plasmid stability. Nevertheless, we believe that obtaining high levels of OmpL1-M were crucial to the success of these experiments. The spontaneous mutation within the ribosome-binding site of plasmid pMMB66-OmpL1 was fortuitous in that toxic background expression was eliminated while still allowing relatively high-level expression after IPTG induction (Fig. 2). This modification of a tac promoter-containing expression plasmid may be superior to other approaches for expression of proteins that are toxic to E. coli. As evidence for this, our attempts at expressing OmpL1 with its signal peptide in E. coli DE3 host strains by using plasmids whose expression is regulated by the T7 promoter have been unsuccessful.
An important aspect of membrane association by LipL41 is modification of its amino-terminal cysteine by fatty acids. The evidence that LipL41 is a lipoprotein include three established criteria: (i) the deduced amino acid sequence of LipL41 includes an N-terminal signal peptide with an LXYC lipoprotein signal peptidase cleavage site, (ii) LipL41 is labeled by palmitate intrinsic labeling of L. kirschneri, and (iii) processing of LipL41 is inhibited by globomycin (36, 51). Because lipid modification of LipL41 is essential for membrane association, we felt that it was important to ensure that lipidation occurred when LipL41 was expressed as a recombinant protein in E. coli. This would be consistent with studies involving the Borrelia burgdorferi lipoprotein, OspA, whose immunogenicity and immunoprotective capacity are enhanced by lipidation (18). However, previous experiments had shown that LipL41 is relatively inefficiently processed by E. coli lipoprotein signal peptidase (51). Like most bacterial lipoproteins, LipL41 has a leucine at the −3 position relative to cysteine. However, we wondered whether the amino acids between the leucine and the cysteine were responsible for the poor processing of LipL41 in E. coli. The arginine at the −1 position was of particular concern, since this position is usually occupied by amino acids such as alanine or glycine with small neutral side chains. We used a PCR mutagenesis strategy to alter the pET-15b-LipL41 plasmid sequence so as to change the LipL41 lipoprotein signal peptidase cleavage site from LGNC to LAGC. Alanine and glycine are the most common amino acids at the −2 and −1 positions, respectively, relative to cysteine in the lipoprotein signal peptidase cleavage site of E. coli lipoproteins including the abundant murein lipoprotein (36, 58). These changes resulted in much more efficient expression and processing of LipL41 in E. coli (Fig. 4). This is the first demonstration that we are aware of for the role of amino acids in the −1 and −2 positions relative to cysteine in the efficiency of processing by E. coli lipoprotein signal peptidase. It is worth noting that optimized expression of LipL41 by using plasmid pET-15b-LipL41* produces a recombinant form of LipL41 with the same amino acid sequence as native LipL41.
Immunization of hamsters with a combination of OmpL1-M and LipL41-M resulted in significant protection against i.p. challenge with virulent L. kirschneri. Survival in animals vaccinated with both proteins was 71% (95% CI, 53 to 89%), compared with only 25% (95% CI, 8 to 42%) in the control group. Despite some variation from experiment to experiment, survival rates in groups receiving dual immunization were consistently higher than those in the control groups. In the first experiment nine of nine hamsters survived to 28 days postchallenge, compared with only three of nine hamsters in the control group (P < 0.004). As shown in Table 2, the lowest survival rate in dually immunized hamsters (50%) was observed in experiment 2, which also had the lowest survival rate in controls (17%), perhaps suggesting a difference in challenge strength or hamster susceptibility in that experiment. One potential explanation for the differences in protection observed in the three experiments was that the percentage of OmpL1-M, relative to total E. coli membrane protein, used for immunization of animals was higher in experiment 1 (10%) than in experiments 2 and 3 (3 to 5%). Although the difference in survival between the animals immunized with both proteins and the controls was statistically significant only in the first experiment, combining the results of all three experiments improved the significance of the result from P < 0.004 to P < 0.001, indicating that a protective effect was being observed throughout all three experiments.
In contrast to immunization with a combination of OmpL1-M and LipL41-M, immunization with OmpL1-M alone did not afford a significant level of protection at 28 days after challenge. However, analysis of the time course of hamster survival indicated that protection following immunization with OmpL1-M alone was statistically significant at 9 days after challenge, when the lethal effects of the challenge were just beginning to become evident in the controls (Fig. 6). This result suggests that OmpL1-M immunization resulted in a delay in the time course of infection without significant clearance of organisms. This hypothesis is consistent with the finding that 3 of the 10 animals immunized with OmpL1-M alone that survived to 28 days postchallenge had sublethal infection (Fig. 7).
Given the relative ease with which the His6 fusion proteins were expressed and purified, it was disappointing that a protective benefit was not observed. A variety of different explanations are available to explain why OmpL1-M and LipL41-M were immunoprotective, in contrast to the His6 fusion proteins. First, as in other spirochetal diseases, humoral immune mechanisms may be more important than cell-mediated immunity for protection against leptospirosis. Recognition of conformational epitopes may be more important in antibody binding to native OmpL1 and/or LipL41 on the leptospiral surface. OmpL1 and LipL41 conformational epitopes may require membrane association, or at least association with phospholipids, LPS, or other membrane components. LPS may also be enhancing immunoprotection in our experiments by functioning as an adjuvant. Two other examples of the potential relevance of conformational epitopes are OmpH of Pasteurella multocida and PorA of Neisseria meningitidis. In the former case, purified native OmpH was protective whereas recombinant OmpH expressed as a His6 fusion protein without its signal peptide was not protective (40). In the latter case, immunization with a His6-PorA fusion protein was unable to elicit bactericidal antibodies unless it was incorporated into liposomes (16). A mechanism for reducing the immunoprotective capacity of the His6 fusion proteins may be that the His6 fusion partner is inhibitory. Several attempts were made to remove the fusion partner with enterokinase, but this proved to be technically difficult, perhaps due to inactivation of enterokinase by urea or Triton X-100, which were needed to maintain solubility of His6-OmpL1 and His6-LipL41. Another mechanism may be the denaturation of conformational epitopes by detergent and/or the denaturing conditions used to purify the His6 fusion proteins. This possibility is supported by the finding that Triton X-100-solubilized His6-OmpL1 produces an abnormally large pore size in the black lipid bilayer assay compared with OmpL1-M (30). An alternative approach would be to use inclusion bodies, which have the advantage of avoiding detergent and can also be produced without the His6 fusion partner.
We do not understand the mechanism(s) for the synergistic immunoprotection elicited by OmpL1-M and LipL41-M. Protection of immunocompromised mice against lethal infection with Pseudomonas aeruginosa is enhanced by immunization with a hybrid OprF-OprI fusion protein (59). As is the case for OmpL1 and LipL41, OprF and OprI differ with respect to their mode of membrane integration. Another potentially pertinent similarity is the fact that OprF, like OmpL1, is a transmembrane protein while OprI, like LipL41, is a lipoprotein.
It is our hope that these studies will lead to improved vaccines for prevention of leptospirosis. Leptospiral outer membrane proteins have a number of advantages over current leptospiral vaccines. Unlike the carbohydrate leptospiral serovar determinants, OmpL1 and LipL41 are highly conserved among pathogenic Leptospira species and are expressed both in cultivated organisms and during mammalian infection. Protection against heterologous challenge is likely to be successful given the high degree of LipL41 and OmpL1 sequence conservation among pathogenic Leptospira species. The LipL41 amino acid sequences of L. interrogans serovar pomona and L. kirschneri serovar grippotyphosa (GenBank accession no. U31426 and L46794, respectively) are 99% identical. Preliminary studies suggest that the amino acid OmpL1 sequence is also highly conserved among pathogenic Leptospira species (28).
The goal of this study was to provide evidence that when used in combination, OmpL1 and LipL41 are protective immunogens. Follow-up studies are needed to determine whether the combination of OmpL1 and LipL41 is cross-protective against heterologous challenge. Since less than 100% of the dually immunized animals were protected, there is a need for studies directed toward defining how the immunity resulting from OMP immunization can be improved. Determination of the relative contributions of cellular and humoral immunity to the protection that we observed would provide insight into how to improve immunity. In addition, more reliable and efficient expression systems are needed for production of OmpL1-M and LipL41-M. Studies are needed to determine whether OmpL1-M and LipL41-M can be further purified while retaining their immunoprotective properties. Definition of the mechanism of immunity and the location of immunoprotective epitopes could also make it possible to use systems for recombinant protein expression which are more efficient than membrane protein expression. The results of these studies should have relevance to vaccine development in spirochetal and other bacterial diseases in which OMPs are being explored as potential vaccinogens (7, 14, 24, 25, 52, 53).
ACKNOWLEDGMENTS
This work was supported by funding from VA Medical Research Funds (D.A.H.), a UCLA School of Medicine Frontiers of Science Award (to D.A.H.), Public Health Service grant AI-34431 (to D.A.H.), and the UCLA-Merial Collaborative Research Project.
We thank C. Bolin for invaluable assistance in developing the L. kirschneri challenge model of leptospirosis. We thank S. K. Haake, J. N. Miller, and D. R. Blanco for helpful suggestions and critical review of the manuscript.
REFERENCES
- 1.Adler B, Faine S. Serological and protective-antibody responses of rabbits to leptospiral antigens. J Med Microbiol. 1978;11:401–409. doi: 10.1099/00222615-11-4-401. [DOI] [PubMed] [Google Scholar]
- 2.Auran N E, Johnson R C, Ritzi D M. Isolation of the outer sheath of Leptospiraand its immunogenic properties in hamsters. Infect Immun. 1972;5:968–975. doi: 10.1128/iai.5.6.968-975.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett J K, Barnett D, Bolin C A, Summers T A, Wagar E A, Cheville N F, Hartskeerl R A, Haake D A. Expression and distribution of leptospiral outer membrane components during renal infection of hamsters. Infect Immun. 1999;67:853–861. doi: 10.1128/iai.67.2.853-861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bey R F, Auran N E, Johnson R C. Immunogenicity of whole cell and outer envelope leptospiral vaccines in hamsters. Infect Immun. 1974;10:1051–1056. doi: 10.1128/iai.10.5.1051-1056.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bey R F, Johnson R C. Protein-free and low-protein media for the cultivation of Leptospira. Infect Immun. 1978;19:562–569. doi: 10.1128/iai.19.2.562-569.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birnbaum N, Barr S C, Center S A, Schermerhorn T, Randolph J F, Simpson K W. Naturally acquired leptospirosis in 36 dogs: serological and clinicopathological features. J Small Anim Pract. 1998;39:231–236. doi: 10.1111/j.1748-5827.1998.tb03640.x. [DOI] [PubMed] [Google Scholar]
- 7.Blanco D R, Champion C I, Exner M M, Shang E S, Skare J T, Hancock R E, Miller J N, Lovett M A. Recombinant Treponema pallidum rare outer membrane protein 1 (Tromp1) expressed in Escherichia colihas porin activity and surface antigenic exposure. J Bacteriol. 1996;178:6685–6692. doi: 10.1128/jb.178.23.6685-6692.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolin C A, Cassells J A, Zuerner R L, Trueba G. Effect of vaccination with a monovalent Leptospira interrogansserovar hardjo type hardjo-bovis vaccine on type hardjo-bovis infection of cattle. Am J Vet Res. 1991;52:1639–1643. [PubMed] [Google Scholar]
- 9.Bolin C A, Thiermann A B, Handsaker A L, Foley J W. Effect of vaccination with a pentavalent leptospiral vaccine on Leptospira interrogansserovar hardjo type hardjo-bovis infection of pregnant cattle. Am J Vet Res. 1989;50:161–165. [PubMed] [Google Scholar]
- 10.Bolin C A, Zuerner R L, Trueba G. Effect of vaccination with a pentavalent leptospiral vaccine containing Leptospira interrogansserovar hardjo type hardjo-bovis on type hardjo-bovis infection of cattle. Am J Vet Res. 1989;50:2004–2008. [PubMed] [Google Scholar]
- 11.Cameron C E, Castro C, Lukehart S A, Van Voorhis W C. Function and protective capacity of Treponema pallidum subsp. pallidumglycerophosphodiester phosphodiesterase. Infect Immun. 1998;66:5763–5770. doi: 10.1128/iai.66.12.5763-5770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention 13 April 1999, posting date. Leptospirosis fact sheet. [Online.] http://www.cdc.gov/ncidod/dbmd/leptofact.htm. [19 May 1999, last date accessed.]
- 13.Centers for Disease Control and Prevention. Outbreak of acute febrile illness and pulmonary hemorrhage—Nicaragua, 1995. Morbid Mortal Weekly Rep. 1995;44:841–843. [Google Scholar]
- 14.Champion C I, Blanco D R, Exner M M, Erdjument-Bromage H, Hancock R E, Tempst P, Miller J N, Lovett M A. Sequence analysis and recombinant expression of a 28-kilodalton Treponema pallidumsubsp. pallidum rare outer membrane protein (Tromp2) J Bacteriol. 1997;179:1230–1238. doi: 10.1128/jb.179.4.1230-1238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Childs J E, Schwartz B S, Ksiazek T G, Graham R R, LeDuc J W, Glass G E. Risk factors associated with antibodies to leptospires in inner-city residents of Baltimore: a protective role for cats. Am J Public Health. 1992;82:597–599. doi: 10.2105/ajph.82.4.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christodoulides M, Brokks J L, Rattue E, Heckels J E. Immunization with recombinant class 1 outer membrane protein from Neisseria meningitidis: influence of liposomes and adjuvants on antibody avidity, recognition of native protein and the induction of a bactericidal immune response against meningococci. Microbiology. 1998;144:3027–3037. doi: 10.1099/00221287-144-11-3027. [DOI] [PubMed] [Google Scholar]
- 17.Christopher W L, Adler B, Faine S. Immunogenicity of leptospiral vaccines grown in protein-free medium. J Med Microbiol. 1982;15:493–501. doi: 10.1099/00222615-15-4-493. [DOI] [PubMed] [Google Scholar]
- 18.Erdile L F, Brandt M A, Warakomski D J, Westrack G J, Sadziene A, Barbour A G, Mays J P. Role of attached lipid in immunogenicity of Borrelia burgdorferiOspA. Infect Immun. 1993;61:81–90. doi: 10.1128/iai.61.1.81-90.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everard C O, Fraser-Chanpong G M, Everard J D. The incidence of severe leptospirosis in Trinidad. Trop Geogr Med. 1987;39:126–132. [PubMed] [Google Scholar]
- 20.Faine S. Guidelines for the control of leptospirosis. Offset publication no. 67. Geneva, Switzerland: World Health Organization; 1982. [PubMed] [Google Scholar]
- 21.Farr R W. Leptospirosis. Clin Infect Dis. 1995;21:1–8. doi: 10.1093/clinids/21.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Fikrig E, Barthold S W, Kantor F S, Flavell R A. Protection of mice against the Lyme disease agent by immunizing with recombinant OspA. Science. 1990;250:553–556. doi: 10.1126/science.2237407. [DOI] [PubMed] [Google Scholar]
- 23.Finke M, Duchene M, Eckhardt A, Domdey H, von Specht B U. Protection against experimental Pseudomonas aeruginosa infection by recombinant P. aeruginosa lipoprotein I expressed in Escherichia coli. Infect Immun. 1990;58:2241–2244. doi: 10.1128/iai.58.7.2241-2244.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Venter J C, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 25.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Venter J C, et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 26.Furste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 27.Gerlach G F, Anderson C, Klashinsky S, Rossi-Campos A, Potter A A, Willson P J. Molecular characterization of a protective outer membrane lipoprotein (OmlA) from Actinobacillus pleuropneumoniaeserotype 1. Infect Immun. 1993;61:565–572. doi: 10.1128/iai.61.2.565-572.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haake, D. A. Unpublished data.
- 29.Haake D A, Champion C I, Martinich C, Shang E S, Blanco D R, Miller J N, Lovett M A. Molecular cloning and sequence analysis of the gene encoding OmpL1, a transmembrane outer membrane protein of pathogenic Leptospiraspp. J Bacteriol. 1993;175:4225–4234. doi: 10.1128/jb.175.13.4225-4234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haake, D. A., M. M. Exner, and R. E. W. Hancock. Unpublished data.
- 31.Haake D A, Martinich C, Summers T A, Shang E S, Pruetz J D, McCoy A M, Mazel M K, Bolin C A. Characterization of leptospiral outer membrane lipoprotein LipL36: downregulation associated with late log-phase growth and mammalian infection. Infect Immun. 1998;66:1579–1587. doi: 10.1128/iai.66.4.1579-1587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haake D A, Walker E M, Blanco D R, Bolin C A, Miller M N, Lovett M A. Changes in the surface of Leptospira interrogansserovar grippotyphosa during in vitro cultivation. Infect Immun. 1991;59:1131–1140. doi: 10.1128/iai.59.3.1131-1140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haake S K, Wang X. Cloning and expression of FomA, the major outer-membrane protein gene from Fusobacterium nucleatum T18. Arch Oral Biol. 1997;42:19–24. doi: 10.1016/s0003-9969(96)00105-7. [DOI] [PubMed] [Google Scholar]
- 34.Hanson M S, Cassatt D R, Guo B P, Patel N K, McCarthy M P, Dorward D W, Hook M. Active and passive immunity against Borrelia burgdorferidecorin binding protein A (DbpA) protects against infection. Infect Immun. 1998;66:2143–2153. doi: 10.1128/iai.66.5.2143-2153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 36.Hayashi S, Wu H C. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 37.Ido Y, Hoki R, Ito H, Wani H. The prophylaxis of Weil's disease (spirochaetosis icterohaemorrhagica) J Exp Med. 1916;24:471–483. doi: 10.1084/jem.24.5.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson R C, Harris V G. Differentiation of pathogenic and saprophytic leptospires. I. Growth at low temperatures. J Bacteriol. 1967;94:27–31. doi: 10.1128/jb.94.1.27-31.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 40.Luo Y, Glisson J R, Jackwood M W, Hancock R E, Bains M, Cheng I H, Wang C. Cloning and characterization of the major outer membrane protein gene (ompH) of Pasteurella multocidaX-73. J Bacteriol. 1997;179:7856–7864. doi: 10.1128/jb.179.24.7856-7864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin D, Cadieux N, Hamel J, Brodeur B R. Highly conserved Neisseria meningitidis surface protein confers protection against experimental infection. J Exp Med. 1997;185:1173–1183. doi: 10.1084/jem.185.7.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Midwinter A, Faine S, Adler B. Vaccination of mice with lipopolysaccharide (LPS) and LPS-derived immuno-conjugates from Leptospira interrogans. J Med Microbiol. 1990;33:199–204. doi: 10.1099/00222615-33-3-199. [DOI] [PubMed] [Google Scholar]
- 43.Miller J N. Spirochetes in body fluids and tissues: manual of investigative methods. Springfield, Ill: Charles C. Thomas; 1971. [Google Scholar]
- 44.Murphy T F, Kyd J M, John A, Kirkham C, Cripps A W. Enhancement of pulmonary clearance of Moraxella (Branhamella) catarrhalis following immunization with outer membrane protein CD in a mouse model. J Infect Dis. 1998;178:1667–1675. doi: 10.1086/314501. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen T P, Lam T T, Barthold S W, Telford III S R, Flavell R A, Fikrig E. Partial destruction of Borrelia burgdorferiwithin ticks that engorged on OspE- or OspF-immunized mice. Infect Immun. 1994;62:2079–2084. doi: 10.1128/iai.62.5.2079-2084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noguchi H. A comparative study of experimental prophylactic inoculation against Leptospira icterohaemorrhagiae. J Exp Med. 1918;28:561. doi: 10.1084/jem.28.5.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Preac-Mursic V, Wilske B, Patsouris E, Jauris S, Will G, Soutschek E, Rainhardt S, Lehnert G, Klockmann U, Mehraein P. Active immunization with pC protein of Borrelia burgdorferi protects gerbils against B. burgdorferi infection. Infection. 1992;20:342–349. doi: 10.1007/BF01710681. [DOI] [PubMed] [Google Scholar]
- 48.Ruffolo C G, Adler B. Cloning, sequencing, expression, and protective capacity of the oma87 gene encoding the Pasteurella multocida87-kilodalton outer membrane antigen. Infect Immun. 1996;64:3161–3167. doi: 10.1128/iai.64.8.3161-3167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 50.Shang E S, Exner M M, Summers T A, Martinich C, Champion C I, Hancock R E W, Haake D A. The rare outer membrane protein, OmpL1, of pathogenic Leptospiraspecies is a heat-modifiable porin. Infect Immun. 1995;63:3174–3181. doi: 10.1128/iai.63.8.3174-3181.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shang E S, Summers T A, Haake D A. Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospiraspecies. Infect Immun. 1996;64:2322–2330. doi: 10.1128/iai.64.6.2322-2330.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skare J T, Champion C I, Mirzabekov T A, Shang E S, Blanco D R, Erdjument-Bromage H, Tempst P, Kagan B L, Miller J N, Lovett M A. Porin activity of the native and recombinant outer membrane protein Oms28 of Borrelia burgdorferi. J Bacteriol. 1996;178:4909–4918. doi: 10.1128/jb.178.16.4909-4918.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skare J T, Mirzabekov T A, Shang E S, Blanco D R, Erdjument-Bromage H, Bunikis J, Bergstrom S, Tempst P, Kagan B L, Miller J N, Lovett M A. The Oms66 (p66) protein is a Borrelia burgdorferiporin. Infect Immun. 1997;65:3654–3661. doi: 10.1128/iai.65.9.3654-3661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. . (Erratum, 163:279, 1987.) [DOI] [PubMed] [Google Scholar]
- 55.Thiermann A B. Leptospirosis: current developments and trends. J Am Vet Med Assoc. 1984;184:722–725. [PubMed] [Google Scholar]
- 56.Thiermann A B, McClellan R D, Hill H T. Improved techniques for the isolation of leptospires from swine abortion cases. Ann Proc Am Assoc Vet Lab Diagn. 1984;27:233–244. [Google Scholar]
- 57.Vinetz J M, Glass G E, Flexner C E, Mueller P, Kaslow D C. Sporadic urban leptospirosis. Ann Intern Med. 1996;125:794–798. doi: 10.7326/0003-4819-125-10-199611150-00002. [DOI] [PubMed] [Google Scholar]
- 58.von Heijne G. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 1989;2:531–534. doi: 10.1093/protein/2.7.531. [DOI] [PubMed] [Google Scholar]
- 59.von Specht B U, Knapp B, Muth G, Broker M, Hungerer K D, Diehl K D, Massarrat K, Seemann A, Domdey H. Protection of immunocompromised mice against lethal infection with Pseudomonas aeruginosa by active or passive immunization with recombinant P. aeruginosaouter membrane protein F and outer membrane protein I fusion proteins. Infect Immun. 1995;63:1855–1862. doi: 10.1128/iai.63.5.1855-1862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Y, Thomas W R, Chong P, Loosmore S M, Klein M H. A 20-kilodalton N-terminal fragment of the D15 protein contains a protective epitope(s) against Haemophilus influenzaetype a and type b. Infect Immun. 1998;66:3349–3354. doi: 10.1128/iai.66.7.3349-3354.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]