Abstract
A mutation in the msbB gene of Escherichia coli results in the synthesis of E. coli lipopolysaccharide (LPS) that lacks the myristic acid moiety of lipid A. Although such mutant E. coli cells and their purified LPS have a greatly reduced ability to stimulate human immune cells, a minor reduction in the mouse inflammatory response is observed. When the msbB mutation is transferred into a clinical isolate of E. coli, there is a significant loss in virulence, as assessed by lethality in BALB/c mice. When a cloned msbB gene is provided to functionally complement the msbB mutant, virulence returns, providing direct evidence that the msbB gene product is an important virulence factor in a murine model of E. coli pathogenicity. In the genetic background of the clinical E. coli isolate, the msbB mutation also results in filamentation of the cells at 37°C but not at 30°C, a reduction in the level of the K1 capsule, an increase in the level of complement C3 deposition, and an increase in both opsonic and nonopsonic phagocytosis of the msbB mutant, phenotypes that can help to explain the loss in virulence. The demonstration that the inhibition of msbB gene function reduces the virulence of E. coli in a mouse infection model warrants further investigation of the msbB gene product as a novel target for antibiotic therapy.
The lipopolysaccharide (LPS) of gram-negative bacteria is a complex amphipathic molecule composed of lipids and sugars. Both of these constituents of LPS have been shown to play critical roles during the interaction between the gram-negative pathogen and the innate host defense (1, 22, 36). LPS is best known for its strong immunostimmulatory ability, yet the role of this activity in virulence is poorly understood. It was not until the lipid A component of LPS was chemically synthesized and shown to have as potent a stimulatory capability as highly purified LPS that the lipid A moiety became generally accepted as the primary stimulatory component of LPS (9). Since that time, numerous studies have attempted to define the essential structural components of lipid A that are necessary for immunomodulation (25, 28, 34). The fact that the host inflammatory response is so profoundly affected by LPS, which is often found among the normal flora of the human body, has also led to the suggestion that the host uses this interaction as a surveillance mechanism to detect any breach of a host barrier (37). Recently, it has been suggested that some gram-negative bacteria have adapted the structure of their lipid A in such a way that it may provide a means to evade host surveillance and thus allow colonization to occur (7, 19, 27). Such an event could be critical to the establishment of chronic inflammatory disease.
Although there is a clearly defined activity for LPS as an immunomodulator, little is known about the role of the lipid A component of LPS as a virulence determinant in the processes of infection and invasion. This lack of knowledge is due to the fact that most mutations in lipid A biosynthesis result in a conditionally lethal phenotype (24), obviating their use in virulence studies. We recently reported that a mutation in the msbB gene of Escherichia coli results in a strain that no longer adds the myristic acid moiety to the lipid A of the LPS (33). In the E. coli K-12 background used, the msbB strain had no obvious defect in growth phenotype; this mutation therefore represented the first mutation in lipid A biosynthesis that permitted studies on the role of LPS when viable bacteria are presented to host defense cells. The msbB strain and the nonmyristoylated LPS (nmLPS) isolated from the msbB strain both showed a dramatically reduced ability to stimulate either myeloid or nonmyeloid immune cells (33). Khan et al. (12) recently showed that a Salmonella typhimurium msbB strain could achieve wild-type growth kinetics in vivo yet was avirulent, demonstrating a role for LPS in the virulence of this organism.
The role of the msbB gene product in the virulence of other bacteria remains unknown, however. General conclusions about the role of the msbB gene product cannot be drawn from mouse studies with S. typhimurium, since this bacterium is a mouse pathogen that has evolved complex regulatory mechanisms to adapt to the localized environments of the host. Therefore, although a functional msbB gene is necessary for S. typhimurium virulence, it remains to be determined if it has a role in the virulence of the more common nosocomial bacteria. In this study, the msbB mutation was transferred into a clinical isolate of E. coli to evaluate its role in virulence and its potential as a target for novel antimicrobial therapies. In a mouse model of infection commonly used to test the efficacy of novel antibacterial agents, the msbB mutation resulted in an increased 50% lethal dose (LD50). The reduction in virulence could not be accounted for solely by a reduction in LPS toxicity. Rather, the msbB strain displayed a pleiotropic phenotype; the mutation altered cell surface interactions with complement and rendered the strain more susceptible to opsonization. These results indicate that normal msbB gene function may be necessary for bacterial resistance to host defense factors and that targeting msbB could be a successful strategy for novel antimicrobial therapies.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phages.
E. coli JM83 F− ara Δ(lac-proAB) rpsL (Strr) [φ80 dlacΔ(lacZ)M15] was the K-12 strain used for preliminary site-directed mutagenesis in this study (39). E. coli JM109 was used for plasmid propagation, as a host for pBIP3 derivatives, and for the generation of M13 transducing stocks (39). E. coli BMS67C12 was derived earlier from JM83 and contains a transposon Tn5 insertion in its msbB gene (33). E. coli H16 was originally isolated from the blood of a meningitis patient, has the serotype O18:K1:H7, and is from the collection of clinical isolates maintained by Richard Darveau at the University of Washington, Seattle. Vector plasmids pUC18 (39) and pBR322 (2) were used in this study. Plasmid pBIP3 and the helper phage f1R189 were described earlier (32). Bacteriophage P1vir was obtained from the American Type Culture Collection, Manassas, Va.
Bacterial media and growth conditions.
E. coli was routinely grown in LB medium (30) or T-Soy broth (TSB) (BBL, Cockeysville, Md.) at 30 or 37°C. Salt-free LB agar with 5% sucrose was used for the selection of double recombinants as described previously (32). T-Soy agar (TSA) is TSB supplemented with 1.5% agar. When needed as selective agents, antibiotics were added at the following final concentrations: ampicillin, 100 μg/ml; kanamycin sulfate, 75 μg/ml; streptomycin sulfate, 20 μg/ml; spectinomycin sulfate, 20 μg/ml; and tetracycline HCl, 25 μg/ml. Precise enumeration of viable cell counts (CFU per milliliter) during growth studies, serum susceptibility testing, and animal studies and for stimulation experiments was done as described earlier (13).
Recombinant DNA methods.
Total DNA was isolated by use of an Easy-DNA Kit from Invitrogen (San Diego, Calif.) according to the manufacturer's instructions, except that following treatment of the DNA with RNase, the DNA was extracted with phenol and chloroform to remove any residual nucleases, ethanol precipitated, and resuspended in TE buffer (10 mM Tris, 1 mM EDTA [pH 8.0]). Restriction endonucleases and DNA modification enzymes were obtained from commercial sources and used according to the manufacturer's instructions. Blotting of DNA to nitrocellulose was done by a modification of the Southern blotting technique (30). Plasmid pBMS66 (Fig. 1) contains an intact and functional msbB gene cloned into the high-copy-number vector pUC18 (12). Supercoiled pBMS66 DNA to be labeled for hybridization was first treated with an ATP-dependent DNase (Plasmid-Safe; Epicentre Technologies, Madison, Wis.) to eliminate any E. coli chromosomal DNA contamination. The 1,630-bp msbB DNA fragment from pBMS66 was then isolated following digestion with EcoRI and BglII and electrophoresis in a 0.9% agarose gel (31). Incorporation of 32P-dCTP into the linear DNA fragment was done by use of a Random Priming Kit from Boehringer Mannheim Biochemicals, (Indianapolis, Ind.). The msbB gene was transferred from pBMS66 into the lower-copy-number vector pBR322 to create pBMS76 by use of the native EcoRI site at the 5′ end of msbB and the BamHI site 3′ of the msbB gene, which was created in the original PCR cloning of msbB (Fig. 1). Transformation of E. coli H16 was accomplished by standard CaCl2 shock methodology (30).
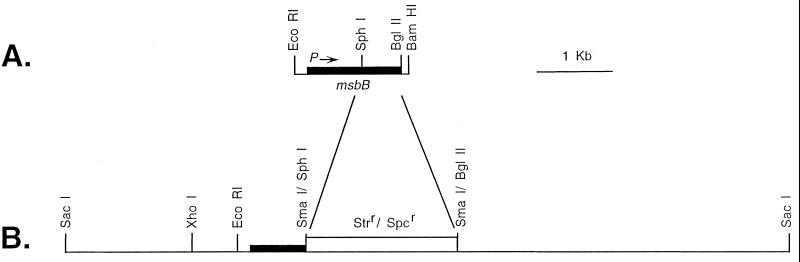
FIG. 1.
Genetic organization of the cloned msbB gene and a mutated form of the msbB gene as recombined into a clinical isolate of E. coli. (A) Structure of the 1,630-bp region of E. coli K-12 DNA which contains the functional msbB gene that was cloned into the high-copy-number vector pUC18 to form pBMS66 or into the lower-copy-number vector pBR322 to form pBMS76. (B) Structure of the mutated msbB gene in plasmid pBMS71 that was used for directed mutagenesis of E. coli JM83. This mutated form of the msbB gene was subsequently transferred by P1 transduction to E. coli H16 to form strain M600.
Mutagenesis of E. coli JM83 by use of the M13-pBIP3 system.
Site-directed mutagenesis of E. coli JM83 was done essentially as described by Slater and Maurer (32), except that the pBIP3 vector was first modified to allow more rapid identification of single recombinants. The Ampr gene was isolated from pBR322 by PCR with the primers 5′-ATCGATCCGCATATGTGCAAGCAGCAGATTAC and 5′-ATCGATCCTCATATGCCTCGTGATACGCCTAT. The Ampr gene was then cloned into the NdeI site located near the M13 origin in pBIP3 to form vector pBMS70. Upon infection with the f1R189 helper phage and M13 replication, the Ampr gene is lost. Single recombinants can then be rapidly differentiated from isolates that contain replicating plasmids by screening for sensitivity to ampicillin.
For mutagenesis of the msbB gene, plasmid pBMS71 was constructed (Fig. 1). To construct pBMS71, the 3′-terminal portion of the msbB gene in pBMS69, a subclone of pBMS67 (33), was removed between the SphI and BglII sites, and blunt ends were created with the Klenow fragment. The Strr Spcr SmaI fragment from pHP45Ω (23) was cloned into this construct to give pBMS69.S. The SstI fragment containing the mutagenized msbB gene was transferred from pBMS69.S into the single SstI site of pBMS70 to give pBMS71. Plasmid pBMS71 was used for site-directed mutagenesis of E. coli JM83 with Strr and Spcr in combination as a selectable marker. Single recombinants identified as described above were then grown without antibiotic selection and plated on salt-free LB agar with 5% sucrose to select for double recombinants as described previously (32). Total DNA was purified from each sucrose-resistant Kans Strr Spcr isolate, digested with BglII and PstI, and analyzed by Southern hybridization to confirm the recombination event. Potential mutants were also phenotypically tested for the reduced ability to stimulate the expression of E-selectin by human umbilical vein endothelial cells (HUVEC) (33). LPS was also isolated from each mutant and analyzed as described earlier (33). All mutagenesis experiments were done at 30°C.
Mutagenesis of E. coli H16 by use of P1vir.
Strains derived from JM83 and with the appropriate chromosomal insertional replacements were used to generate P1vir transducing lysates (17). E. coli H16 was grown overnight in 3 ml of LB broth at 37°C. The cells were harvested by centrifugation and washed vigorously by vortexing in 5 ml of phosphate-buffered saline (PBS). Following centrifugation and removal of the PBS, the cells were washed vigorously a second time in 5 ml of MC buffer (0.1 M MgSO4, 0.005 M CaCl2) (17), centrifuged, and finally resuspended in 3 ml of MC buffer for transduction. A 0.25-ml quantity of H16 cells was mixed with each serial dilution of P1vir (msbB::Strr Spcr) lysate, and the phage were allowed to adsorb for 20 min at 30°C. An equal volume of 0.1 M sodium citrate (pH 7.0) was added and mixed, and 1 ml of LB broth with 0.05 M sodium citrate was also added. The infected cells were placed in a 30°C incubator shaker for 3 h to allow recombination and expression of markers to occur. Dilutions of the cells were inoculated onto LB agar plates that contained streptomycin sulfate and spectinomycin sulfate and were incubated at 30°C for 2 days. Potential transductants were streaked for isolation and replica picked to test for Kans or temperature sensitivity. Total DNA was purified from each Kans Strr Spcr isolate, digested with BglII and PstI, and analyzed by Southern hybridization to confirm that recombination had occurred at msbB. Potential mutants were also phenotypically tested for the reduced ability to stimulate the expression of E-selectin by HUVEC (33). LPS was also isolated from each mutant and analyzed as described earlier (33).
Testing the toxicity of nmLPS and the lethality of E. coli strains in mice.
Toxicity studies with purified LPS (obtained as described in reference 17) or live bacteria were conducted with BALB/c mice inoculated intraperitoneally with 0.2 ml. LPS was made as a 2-mg/ml stock in PBS with the aid of sonication in a bath sonicator. LPS was then diluted further in PBS for use as an inoculum. For testing the potency of purified LPS, animals were injected with and without the coadministration of galactosamine, which has been shown to significantly reduce the LPS LD50. Galactosamine (800 mg/kg of body weight) administration was performed as described previously (3).
Bacterial studies were performed with seed cultures of E. coli strains that were inoculated from freezer stocks and grown overnight in TSB with antibiotic selection. Inoculum cultures were started with a 1/100 dilution of the seed cultures in 50 ml of TSB with antibiotics. Inoculum cultures were grown to mid-log phase (A660, 0.6 to 0.8), chilled on ice for 10 min, and harvested by centrifugation. The cells were washed once in cold PBS and resuspended in immunogen diluent (0.1% TSB, 0.1% HEPES buffer, 0.85% NaCl [pH 7.2]). Cell densities were adjusted based on A660 values by use of conversion factors empirically derived for each strain during earlier growth studies at each temperature (30 and 37°C). Cell densities were confirmed for each strain by plating dilutions on TSA plates, and culture purity was checked by testing the highest-density dilution of each strain.
Detection of murine C3 deposition and the presence of the K1 capsule.
Detection of complement C3 deposition on the surface of viable bacterial cells was done essentially as described earlier (20). Bacterial cells were grown as described above and washed once with PBS, and the density was set at an A550 of 0.4 in PBS. For C3 studies, the cells were mixed with an equal volume of fresh pooled normal murine serum (BALB/c) or pooled normal human serum and incubated for 30 min at 37°C. The bacterial cells were washed three times with cold PBS and resuspended in PBS containing either a fluorescein-conjugated goat anti-murine complement C3 antibody or a fluorescein-conjugated goat anti-human complement C3 antibody (each from Cappel/Organon Teknika Corp., Durham, N.C.). Both antibodies were diluted 1/200 and incubated with the cells at room temperature for 30 min with gentle agitation. Following three washes with cold PBS, the cells were suspended in PBS and analyzed by flow cytometry. A sample of the stained cells was dried on a slide and mounted for microscopic examination.
Cells used for detection of the K1 capsule were grown as described above but, upon harvest, were pelleted and resuspended gently in PBS, the cell density was set at an A550 of 0.4 in PBS, and the E. coli K1 capsule was detected by incubation with a human monoclonal immunoglobulin M antibody with specificity to E. coli K1 capsular antigen (26) for 30 min at 37°C. Following two gentle washes with cold PBS, the binding of this antibody was detected with a fluorescein-conjugated goat anti-human immunoglobulin M antibody (Cappel/Organon Teknika) diluted to a final concentration of 10 μg/ml and incubated for 30 min at room temperature with gentle agitation. Cells were gently washed twice with cold PBS and suspended in PBS for analysis by flow cytometry. A sample of the stained cells was dried on a slide and mounted for microscopic examination.
Opsonization of E. coli strains and uptake by polymorphonuclear phagocytes.
Blood was obtained from normal healthy human volunteers by venipuncture with heparin-containing syringes. Neutrophils were isolated by density gradient centrifugation with Polymorphprep (Nycomed Pharma AS, Oslo, Norway) as described by the manufacturer.
Bacterial cells were grown as described above; following a PBS wash, the number of cells were set at either 4 × 107 CFU/ml or equal densities, as determined spectrophotometrically (A660) with GVBB buffer (35). Opsonization and phagocytosis of each bacterial strain were then determined by a previously described chemiluminescence assay (35). Normal human serum was treated at 56°C for 30 min to inactivate residual complement activity. To replace complement activity, C5-depleted serum (Quidel Laboratories, San Diego, Calif.) was used (35).
RESULTS
nmLPS is a potent agonist in BALB/c mice.
E. coli nmLPS has a greatly reduced ability to stimulate either human myeloid or human nonmyeloid immune cells (33). However, significantly different endotoxin activities between human and murine systems have been demonstrated. Specifically, underacylated E. coli lipid IVA, which is similar in structure to nmLPS (nmLPS contains one additional secondary fatty acid), is an antagonist in human in vitro systems yet is a potent agonist in murine systems (8). Therefore, the toxicity of nmLPS in two different in vivo murine systems was examined. As shown in Table 1, either with or without prior administration of galactosamine, little or no significant difference in the toxicity of nmLPS or wild-type LPS was observed. These data are consistent with previous reports that underacylated E. coli LPS is a potent agonist in mice. The values found for wild-type LPS and nmLPS compare well with previously reported toxicity levels for purified LPS in both galactosamine-sensitized (3) and normal (29) mice. These data demonstrate that the loss of one secondary fatty acid does not significantly reduce the toxicity of E. coli LPS in mice.
TABLE 1.
Toxicity of purified LPSa
| Galactosamine | Source of LPS | No. of survivors
at the indicated LPS dose (μg/animal):
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 400 | 200 | 100 | 50 | 125 | 25 | 5 | 1 | ||
| Absent | JM83 | 0 | 0 | 2 | 5 | ||||
| BMS67C12 | 0 | 0 | 5 | 5 | |||||
| Present | JM83 | 0 | 0 | 2 | 5 | ||||
| BMS67C12 | 0 | 0 | 0 | 5 | |||||
Purified LPSs from the parent E. coli K-12 strain JM83 and the msbB mutant strain BMS67C12 were each inoculated into five BALB/c mice at the doses indicated. Purified LPS was suspended at a concentration of 2 mg/ml in pyrogen-free PBS with the aid of a bath sonicator. Dilutions of each LPS were made in PBS and inoculated by intraperitoneal injection in a volume of 0.2 ml. Galactosamine was administered intraperitoneally at 800 mg/kg simultaneously with a dose of LPS as described previously (3). Animals were observed for 7 days, with all deaths being observed within the first 5 days postinoculation.
Construction of msbB mutations in E. coli K-12 and transfer into clinical isolate E. coli H16.
E. coli K-12 laboratory strains are generally not virulent in mice, making it difficult to evaluate the potential of any inactivated gene as a target for novel antimicrobial therapies (for example, the LD50 for E. coli K-12 strain JM83 was greater than 108 bacteria/mouse; data not shown). Therefore, to more accurately access the effect of the msbB gene on mouse survival, a human clinical isolate which displays an approximate 3-log reduction in the LD50 for mice was used (Table 2).
TABLE 2.
Pathogenicity of E. coli strains in BALB/c micea
| E. coli strain | No. of survivors/total no. of mice
infected at the following bacterial challenge (CFU/animal):
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 1 × 104 | 5 × 104 | 1 × 105 | 5 × 105 | 1 × 106 | 5 × 106 | 1 × 107 | 5 × 107 | |
| H16 | 5/5 | 9/10 | 5/10 | 0/10 | 0/5 | 0/5 | ||
| H16(pBMS76) | 5/5 | 7/10 | 7/10 | 1/10 | 0/5 | 0/5 | ||
| M600 (msbB) | 5/5 | 5/5 | 10/10 | 2/10 | 0/10 | 0/5 | ||
| M600(pBMS76) | 5/5 | 9/10 | 5/10 | 0/10 | 0/5 | 0/5 | ||
Data are compiled from several experiments with groups of 5 or 10 mice 8 to 10 weeks old. Seed cultures of E. coli strains were inoculated from freezer stocks and grown overnight in TSB with antibiotic selection. Inoculum cultures were started with a 1/100 dilution of the seed cultures in 50 ml of TSB with antibiotics. Inoculum cultures were grown to mid-log phase (A660, 0.6 to 0.8), chilled on ice for 10 min, and harvested by centrifugation. The cells were washed once in cold PBS and resuspended in immunogen diluent. Cell densities were adjusted based on A660 values by use of conversion factors empirically derived from each strain during earlier growth studies. Cell densities were confirmed for each strain by plating dilutions targeted at 100 CFU/plate on TSA, and culture purity was checked by testing the highest-density dilution of each strain. Plate counts were within 10% of targeted CFU per milliliter for each of the strains, except for M600. Accurate density adjustment for strain M600 was hindered by the filamentation of the cells grown at 37°C. Actual CFU per animal for each of two experiments at inocula of 1 × 106, 5 × 106, and 1 × 107 were 1.76 × 106 and 1.24 × 106, 8.8 × 106 and 6.2 × 106, 1.76 × 107 and 1.24 × 107, respectively. Mice were inoculated with a 0.2-ml volume administered by intraperitoneal injection. Animal lethality was monitored for 5 days, although the majority of deaths occurred within the first 48 h.
A stable and selectable msbB mutation was first generated in an E. coli K-12 background by use of plasmid pBMS71 (Fig. 1) and the site-directed mutagenesis method of Slater and Maurer (32). In early experiments with this system, we found that in spite of taking the suggested precautions, a significant number of potential single recombinants carried replicating plasmid DNA and had not undergone integration. To ease the identification of single recombinants, an Ampr marker was inserted into a region of the vector plasmid that is lost when integration has occurred (pBMS70).
Once the msbB mutation was constructed in the K-12 strain, a P1vir lysate was generated and used to transduce the mutation into the clinical isolate E. coli H16. E. coli H16 has a K1 capsule that normally prevents infection by bacteriophage P1. To make the strain amenable to P1 transduction, vigorous washing was done to remove the capsule and expose the appropriate bacteriophage receptors (see Materials and Methods). Several potential P1 transductants were recovered by selection of the Strr Spcr marker. Total DNA was isolated from six of these isolates and analyzed by Southern blot hybridization to confirm the genetic exchange of the mutated msbB gene for the normal msbB gene (as diagrammed in Fig. 1). All six of the isolates chosen were confirmed to have undergone the correct recombination events. Each of the six isolates was tested for the ability to stimulate E-selectin expression by HUVEC. Every strain had a stimulation profile similar to that found for the E. coli K-12 msbB mutant strain BMS67C12 (33). LPS purified from each of the msbB mutant strains also lacked myristic acid in its fatty acid profile.
The high-copy-number plasmid pBMS66 carries an intact copy of the msbB gene and can functionally complement an msbB mutation (33). It has been previously reported that the msbB gene can suppress the temperature-sensitive phenotype of an htrB mutant, but only when provided on a high-copy-number vector (11). To remove potential artifacts due to the very high copy number of the pUC18 vector in pBMS66, an intact copy of the msbB gene was transferred into the lower-copy-number vector pBR322 to create pBMS76 (Fig. 1). When either pBMS66 or pBMS76 was transformed into the H16 msbB mutant strain M600, the myristic acid moiety returned to the lipid A of the LPS, along with the ability to stimulate E-selectin expression by HUVEC.
A mutation in msbB significantly reduces the virulence of E. coli H16 in a murine system.
E. coli H16 and the derivative strains created above were used to assess the pathogenic contribution made by the product of the msbB gene of E. coli. As shown in Table 2, H16 is a relatively potent pathogen when injected intraperitoneally into BALB/c mice, with an LD50 of approximately 2.5 × 105 CFU/animal. When a mutation in the msbB gene is present (strain M600), there is a significant loss in pathogenicity (Table 2). With the transfer of an intact copy of the functional msbB gene into the msbB mutant, M600(pBMS76), the pathogenicity of E. coli returns to a level similar to that of the parental strain H16 (Table 2). In the control, in which H16 carries the cloned msbB gene on a low-copy-number vector, H16(pBMS76), no significant change in pathogenicity is observed (Table 2). Additional LD50 experiments with the bacterial strains grown at 30°C also demonstrated a similar loss of virulence for strain M600 (data not shown). Since it was found that strain M600 forms filaments when grown at 37°C (see below), each CFU of strain M600 represents a significantly larger bacterial load based on cell density; therefore, the differences observed in the LD50 results presented in Table 2 are likely to be somewhat conservative.
The msbB mutant is resistant to serum but is still opsonized by complement component C3.
In order to understand the relationship between reduced virulence and inactivation of the msbB gene, the relevant bacterial strains were tested for susceptibility to killing by normal human serum. H16, M600, H16(pBMS76), and M600(pBMS76) were all resistant to incubation in the presence of 80% normal human serum at either 30 or 37°C for 3 h. Resistance was defined as an increase in the bacterial titer in comparison to the initial inoculum. In contrast, the K-12 strains JM83 and BMS67C12 were both sensitive to as little as 5% pooled normal human serum.
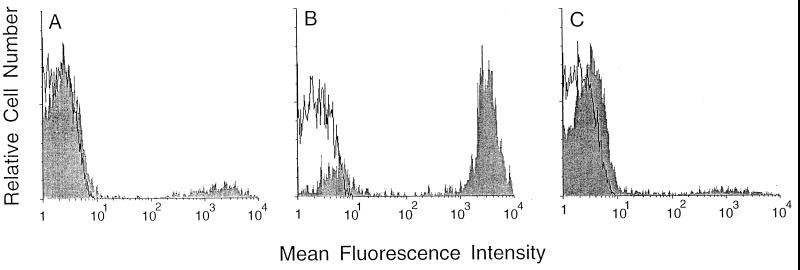
Since strain H16 and its derivatives were found to be serum resistant, we suspected that the loss of virulence might be associated with an increase in the clearance of the bacteria via opsonization and phagocytosis. Immunostaining of the bacterial cells with an antibody directed against complement component C3 following treatment of the cells with normal mouse serum demonstrated a significant increase in C3 deposition on the msbB mutant (Fig. 2; see Fig. 5). This increase in C3 deposition is dependent on the msbB gene product, since the complemented strain, M600(pBMS76), stained in a manner similar to that of wild-type strain H16 (Fig. 2). Bacteria were also stained in additional experiments with normal human serum and an antibody to human complement component C3, with similar results. E. coli K-12 strain JM83, which was exquisitely sensitive to serum, stained positively in all of the above experiments. In experiments with heat-inactivated serum, the cells were not stained by any of the antibodies to C3. The lack of C3 deposition for strain H16 or M600(pBMS76) is consistent with earlier reports that the K1 capsule plays a role in the prevention of the deposition of complement components on the surface of these cells (5).
FIG. 2.
Detection of murine complement component C3 on the surface of bacterial cells. The open curves indicate the epifluorescence of cells that were treated only with fluorescein-conjugated anti-C3 antibody. The shaded curves indicate the epifluorescence of cells treated with fluorescein-conjugated anti-C3 antibody following treatment with normal mouse serum. (A) E. coli H16. (B) E. coli M600. (C) E. coli M600(pBMS76).
FIG. 5.
Detection of the K1 capsule on E. coli strains grown to log phase in LB broth at 37°C. (A) Bright-field microscopy of strain M600 stained with anti-K1 antibody. (B) Fluorescence microscopy of the same field as that shown in panel A. (C and D) Fluorescence microscopy of strains H16 (C) and M600(pBMS76) (D) also stained with anti-K1 antibody. (E) For comparison, fluorescence staining of M600 with anti-C3 antibody. Bar, 10 μm.
The msbB mutant has an increased susceptibility to opsonic and nonopsonic phagocytosis.
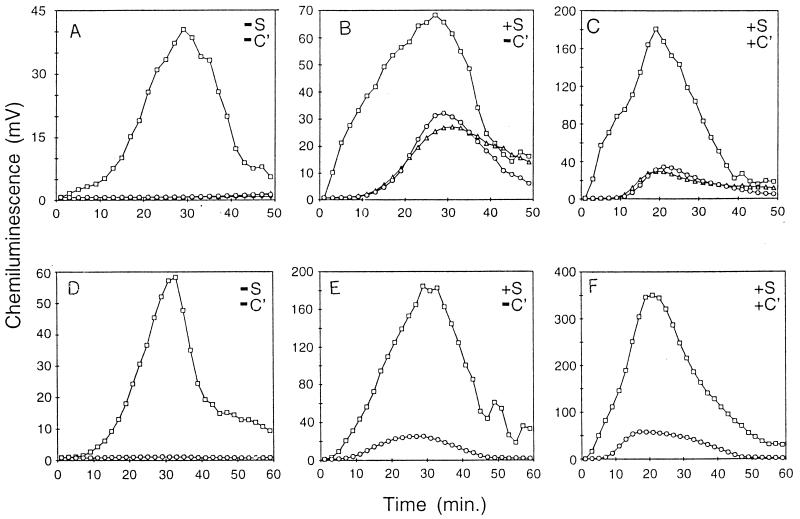
An increase in the opsonization of the msbB mutant was functionally confirmed by a method in which the phagocytosis of bacteria by human neutrophils is monitored with a chemoluminescent dye (35). As shown in Fig. 3, under all of the conditions examined, the msbB mutant strain M600 was more readily phagocytized than either the parental strain H16 or the complemented mutant strain M600(pBMS76). When no serum or complement was present, detectable phagocytosis of mutant strain M600 was significant compared to that of H16 or complemented mutant strain M600(pBMS76) (Fig. 3A). We interpret this observation as an example of nonopsonic phagocytosis (16). The addition of heat-inactivated serum increased the level of phagocytosis of all three strains (Fig. 3B). The addition of a functional complement system to strains H16 and M600(pBMS76) provided no significant increase in phagocytosis (Fig. 3C). This result is as would be expected for strains that have an intact K1 capsule preventing C3 deposition. However, with the addition of complement, there was at least a twofold increase in the phagocytosis of strain M600 (Fig. 3C). This result indicates that the addition of complement increased C3 deposition (as found above), in turn providing an increase in the level of phagocytosis of strain M600.
FIG. 3.
Phagocytosis of E. coli strains by polymorphonuclear leukocytes (PMNs). (A to C) E. coli strains grown at 37°C and added to PMNs based on cell titers. (D to F) E. coli strains grown at 30°C and added to PMNs at equal titers and densities. Data show PMNs with E. coli H16 (open circles), PMNs with E. coli M600 (open squares), and PMNs with E. coli M600(pBMS76) (open triangles). (A and D) Buffer only. (B and E) With heat-treated serum (S). (C and F) With heat-treated serum and complement (C′).
We found that strain M600 forms filaments when grown at 37°C (see below). In the above experiments (Fig. 3A to C), the cultures were all set at equivalent cell titers. Additional experiments done with cultures grown at 37°C and adjusted to equivalent culture densities showed similar results (data not shown). In Fig. 3D to F, the cultures were grown at 30°C, at which the same density of each culture has a similar number of bacteria, as defined by CFU. As shown in Fig. 3D to F, the phagocytic trend is similar to that for cells grown at 37°C.
The msbB mutant forms filaments when grown at 37°C but not at 30°C.
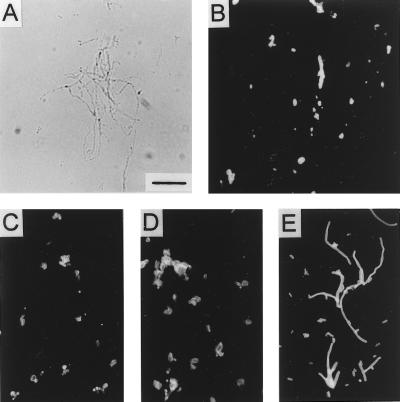
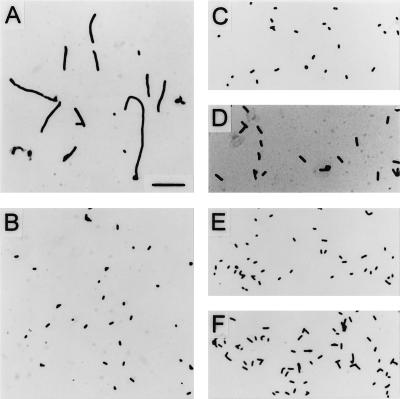
When experimentally determining appropriate culture density conversion factors for animal experiments, we observed that strain M600 required a much higher culture density to give a culture titer (CFU) similar to that of parental strain H16. Microscopic observation of the cultures revealed that the msbB mutant formed filaments when grown at 37°C but not when grown at 30°C or when functionally complemented by the cloned msbB gene [M600(pBMS76)] (Fig. 4). This result is in contrast to that obtained with the msbB mutation in a K-12 background (Fig. 4) (11).
FIG. 4.
Bright-field microscopy of E. coli strains grown to log phase in LB broth cultures at 30 or 37°C. (A) M600, 37°C. (B) M600, 30°C. (C) H16, 37°C. (D) M600(pBMS76), 37°C. (E) JM83, 37°C. (F) BMS67C12, 37°C. Bar, 10 μm.
K1 capsule accumulation on the surface of the msbB mutant is impaired.
Since previous reports described an important role for the K1 capsule in the prevention of C3 deposition and opsonization and since we observed an increase in both C3 deposition and phagocytosis for strain M600, we examined the K1 capsule of M600 to look for any major changes as a result of the msbB mutation. An antibody to the K1 capsule was used to stain bacterial cells grown at 30 and 37°C. As shown in Fig. 5, both H16 and M600(pBMS76) cells stained positively for the K1 capsule, whereas only discrete portions of the M600 filaments were stained. In contrast, M600 cells were completely stained with the antibody to murine C3 (Fig. 5E). Both H16 and M600(pBMS76) cells stained the same at 30°C as at 37°C (data not shown). However, no staining was seen with M600 cells grown at 30°C, indicating a complete lack of a detectable K1 capsule (data not shown). No staining was observed with any of the K-12 strains used as controls.
DISCUSSION
Determining the role of the lipid A moiety of LPS as a virulence determinant in the process of colonization and infection has proven difficult. All previously known mutations in the lipid A biosynthetic pathway have been conditionally lethal, a phenotype which precludes definitive in vivo studies with viable bacteria (24). Recently, however, a mutation in the msbB gene in E. coli K-12, a gene involved in one of the last steps in lipid A biosynthesis, was described and was not conditionally lethal for growth. This finding has provided the opportunity to examine the role of an incomplete lipid A structure in virulence studies. For example, examination of an S. typhimurium msbB mutant has provided the first evidence that lethality after infection may be due to the toxic effects of LPS (12). In this report, the effects of an msbB mutation in a virulent E. coli strain were examined to determine the role of lipid A in a generalized bacterial infection model and to help determine the potential of the gene product as a new therapeutic target.
When the msbB mutation was placed into a clinical isolate of E. coli (H16), a significant loss (slightly greater than 1 log unit) in virulence was observed. Complementation with a plasmid containing an intact msbB gene restored the virulence phenotype. However, evidence suggests that the effect of the LPS structural mutation on virulence was not direct. LPS toxicity studies performed by two different methods revealed at best only a threefold reduction in the toxicity of the purified nmLPS compared to its wild-type parent. Therefore, msbB-encoded LPS which is structurally similar to lipid IVA is also an agonist in mice and an antagonist in human systems (8). Rather, more likely, a cell surface change associated with the msbB mutation in the E. coli K-1 strain explains the reduced virulence of this strain. Associated with the msbB mutation in the clinical isolate were filament formation and a reduced level of the K1 capsule. We suspect that these changes rendered the cells much more susceptible to the initial innate host defense phagocytic clearance mechanisms, such as nonopsonic clearance and C3 opsonization.
Therefore, although not directly examined, it is likely that the initial mouse clearance of the msbB strain was enhanced, reducing virulence by decreasing the number of bacteria able to seed vital organs. In contrast, as mentioned above, it has been proposed that the reduced virulence of an msbB strain of S. typhimurium was due to the lower toxicity of the altered LPS produced by this strain. These observations are not in conflict, however. S. typhimurium, as a natural pathogen of mice (as few as 100 bacteria are sufficient to kill 100% of the mice infected), has specialized virulence mechanisms. The inability to synthesize a fully aceylated LPS could have hampered its pathogenesis at one or several different steps during the development of disease (for example, in the experiments with the msbB mutant, it took 8 days for the wild-type parental strain to kill the mouse host). In contrast, in our study with E. coli, most deaths occurred in the first 2 days, reflecting the fact that with this strain and an animal model, virulence is essentially a measure of the ability of the mouse to initially clear the infecting bacterial dose.
The pleiotropic phenotype of the msbB mutation in the clinical E. coli background was not predicted from the phenotype observed for the E. coli K-12 genetic background. Our findings that an msbB mutation in the H16 genetic background not only changed the myristoylization of lipid A but also resulted in filamentation of the cells at 37°C and a reduction in the presence of the K1 capsule are in contrast to the limited phenotype thus far described for an msbB mutation in an E. coli K-12 genetic background. It has been reported, however, that mutations in genes such as envA or firA have also been found to have pleotropic phenotypes beyond their direct roles in lipid A biosynthesis, so our findings may not be unusual (25, 38). Interactions between the K1 capsule and LPS have also been suggested, but such interactions have not yet been clearly defined (15). From our data it might be suggested that the anchoring of the K1 capsule may be dependent upon the structure of the lipid A moiety of LPS or that the product of the msbB gene may have an influence upon the synthesis of the K1 capsule. Clearly, further study is required to define the apparent relationship between these two outer membrane components.
It is interesting to note that although the msbB strain demonstrated a significant loss of the K1 capsule, it was resistant to the bactericidal action of serum. Earlier reports have implicated the K1 capsule as an important virulence determinant for pathogenic E. coli strains, and it has been concluded that the K1 capsule aids in the resistance of such strains to the bactericidal activity of normal serum as well as resistance to neutrophil killing (5, 6, 14). As mentioned above, our data support the proposed role of the K1 capsule in the prevention of serum opsonization. From our results, it would appear that the msbB strain has an insufficient level of the K1 capsule to prevent opsonization, yet enough K1 capsule may be associated with the outer membrane such that serum killing by the C5b-C9 complex is still prevented. Failure to alter resistance to the bactericidal action of serum is a characteristic previously reported for a mutation which reduces fatty acid aceylation in Haemophilus influenzae (18).
It appears that the msbB gene product would be a likely new therapeutic target, due to the multiple and severe compromises observed both in bacterial morphology and cell wall components required for resistance to host defense mechanisms. In addition, the significant reduction of nmLPS toxicity in human in vitro systems renders the inhibition of this gene product even more desirable. The clinical utility of any therapeutic agent directed against the msbB gene product, when and if developed, remains to be determined, however. It is not possible at this time to predict how many different strains against which these putative inhibitors would need to have activity to make them clinically useful.
ACKNOWLEDGMENTS
We thank Laura Houston and Sheila Lukehart for carrying out the opsonization studies, Brian Bainbridge for purification of LPS and gas chromatographic analysis, Bob Reife for advice on microscopy, Grace Maloney for guidance with the animal experiments, Walt Shuford for supplying the anti-K1 antibody, and Jennifer Price for assistance in preparing the manuscript.
REFERENCES
- 1.Baker P J, Hraba T, Taylor C E, Stashak P W, Fauntleroy M B, Zahringer U, Takayama K, Sievert T R, Hronowski X, Cotter R J, Perez-Perez G. Molecular structures that influence the immunomodulatory properties of the lipid A and inner core region oligosaccharides of bacterial lipopolysaccharides. Infect Immun. 1994;62:2257–2269. doi: 10.1128/iai.62.6.2257-2269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 3.Bucklin S E, Morrison D C. Bacteremia versus endotoxemia in experimental mouse leukopenia—role of antibiotic chemotherapy. J Infect Dis. 1996;174:1249–1254. doi: 10.1093/infdis/174.6.1249. [DOI] [PubMed] [Google Scholar]
- 4.Clementz T, Bednarski J, Raetz C R H. Escherichia coligenes encoding KDO dependent acyltransferases that incorporate laurate and myristate into lipid A. FASEB J. 1995;9:A1311. . (Abstract.) [Google Scholar]
- 5.Cross A, Asher L, Seguin M, Yuan L, Kelly N, Hammack C, Sadoff J, Gemski P. The importance of a lipopolysaccharide-initiated, cytokine-mediated host defense mechanism in mice against extraintestinally invasive Escherichia coli. J Clin Investig. 1995;96:676–686. doi: 10.1172/JCI118110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross A S, Gemski P, Sadoff J C, Ørskov F, Ørskov I. The importance of the K1 capsule in invasive infections caused by Escherichia coli. J Infect Dis. 1984;149:184–193. doi: 10.1093/infdis/149.2.184. [DOI] [PubMed] [Google Scholar]
- 7.Darveau R P, Cunningham M D, Bailey T, Seachord C, Ratcliffe K, Bainbridge B, Dietsch M, Page R C, Aruffo A. Ability of bacteria associated with chronic inflammatory disease to stimulate E-selectin expression and promote neutrophil adhesion. Infect Immun. 1995;63:1311–1317. doi: 10.1128/iai.63.4.1311-1317.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delude R L, Savedra R, Jr, Zhao H, Thieringer R, Yamamoto S, Fenton M J, Golenbock D T. CD14 enhances cellular responses to endotoxin without imparting ligand-specific recognition. Proc Natl Acad Sci USA. 1995;92:9288–9292. doi: 10.1073/pnas.92.20.9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galanos C, Luderitz O, Rietschel E T, Westphal O, Brade H, Brade L, Freudenberg M, Schade U, Imoto M, Yoshimura H, Kusumoto S, Shiba T. Synthetic and natural Escherichia colifree lipid A express identical endotoxic activities. Eur J Biochem. 1985;148:1–5. doi: 10.1111/j.1432-1033.1985.tb08798.x. [DOI] [PubMed] [Google Scholar]
- 10.Hammond S M. Inhibitors of lipopolysaccharide biosynthesis impair the virulence potential of Escherichia coli. FEMS Microbiol Lett. 1992;100:293–298. doi: 10.1111/j.1574-6968.1992.tb14055.x. [DOI] [PubMed] [Google Scholar]
- 11.Karow M, Georgopoulos C. Isolation and characterization of the Escherichia coli msbB gene, a multicopy suppressor of null mutations in the high-temperature-requirement gene htrB. J Bacteriol. 1992;174:702–710. doi: 10.1128/jb.174.3.702-710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan S A, Everest P, Servos S, Foxwell N, Zähringer U, Brade H, Rietschel E T, Dougan G, Charles I G, Maskell D J. A lethal role for lipid A in Salmonellainfections. Mol Microbiol. 1998;29:571–579. doi: 10.1046/j.1365-2958.1998.00952.x. [DOI] [PubMed] [Google Scholar]
- 13.Koch A L. Growth measurement: colony counts. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C: American Society for Microbiology; 1981. pp. 185–188. [Google Scholar]
- 14.Leying H, Suerbaum S, Kroll H-P, Stahl D, Opferkuch W. The capsular polysaccharide is a major determinant of serum resistance in K-1-positive blood culture isolates of Escherichia coli. Infect Immun. 1990;58:222–227. doi: 10.1128/iai.58.1.222-227.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacLachlan P R, Keenleyside W J, Dodgson C, Whitfield C. Formation of the K30 (group I) capsule in Escherichia coliO9:K30 does not require attachment to lipopolysaccharide lipid A-core. J Bacteriol. 1993;175:7515–7522. doi: 10.1128/jb.175.23.7515-7522.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahenthiralingam E, Speert D P. Nonopsonic phagocytosis of Pseudomonas aeruginosaby macrophages and polymorphonuclear leukocytes requires the presence of the bacterial flagellum. Infect Immun. 1995;63:4519–4523. doi: 10.1128/iai.63.11.4519-4523.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 268–274. [Google Scholar]
- 18.Nichols W A, Raetz C R H, Clementz T, Smith A L, Hanson J A, Ketterer M R, et al. htrB of Haemophilus influenzae: determination of biochemical activity and effects on virulence and lipooligosaccharide toxicity. J Endotoxin Res. 1997;4:163–172. [Google Scholar]
- 19.Ogawa T, Uchida H, Amino K. Immunobiological activities of chemically defined lipid A from lipopolysaccharides of Porphyromonas gingivalis. Microbiology. 1994;140:1209–1216. doi: 10.1099/13500872-140-5-1209. [DOI] [PubMed] [Google Scholar]
- 20.Ohno A, Isii Y, Tateda K, Matumoto T, Miyazaki S, Yokota S, Yamaguchi K. Role of LPS length in clearance rate of bacteria from the bloodstream in mice. Microbiology. 1995;141:2749–2756. doi: 10.1099/13500872-141-10-2749. [DOI] [PubMed] [Google Scholar]
- 21.Onishi H R, Pelak B A, Gerckens L S, Silver L L, Kahan F M, Chen M-H, Patchett A A, Galloway S M, Hyland S A, Anderson M S, Raetz C R H. Antibacterial agents that inhibit lipid A biosynthesis. Science. 1996;274:980–982. doi: 10.1126/science.274.5289.980. [DOI] [PubMed] [Google Scholar]
- 22.Pier G B, Grout M, Zaidi T S, Olsen J C, Johnson L G, Yankaskas J R, Goldberg J B. Role of mutant CFTR in hypersusceptibility of cystic fibrosis patients to lung infections. Science. 1996;271:64–67. doi: 10.1126/science.271.5245.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 24.Raetz C R H. Bacterial endotoxins: extraordinary lipids that activate eucaryotic signal transduction. J Bacteriol. 1993;175:5745–5753. doi: 10.1128/jb.175.18.5745-5753.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raetz C R H. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1035–1063. [Google Scholar]
- 26.Raff H V, Devereux D, Shuford W, Abbott-Brown D, Maloney G. Human monoclonal antibody with protective activity for Escherichia coli K1 and Neiseria meningitidisgroup B infections. J Infect Dis. 1988;157:118–126. doi: 10.1093/infdis/157.1.118. [DOI] [PubMed] [Google Scholar]
- 27.Reife R A, Shapiro R A, Bamber B A, Berry K K, Mick G E, Darveau R P. Porphyromonas gingivalislipopolysaccharide is poorly recognized by molecular components of innate host defense in a mouse model of early inflammation. Infect Immun. 1995;63:4686–4694. doi: 10.1128/iai.63.12.4686-4694.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rietschel E T, Kirkae T, Schade F U, Mamat U, Schmidt G, Loppnow H, Ulmer A J, Zähringer U, Seydel U, Padova F D, Schreier M, Brade H. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- 29.Rudbach J A, Keegan D S, Sowell C G. Calculating therapeutic indices and therapeutic advantages for endotoxins and monophosphoral lipid A: an evaluation of data from the scientific literature. J Endotoxin Res. 1995;2:301–310. [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Schwartz H, Whitton J L. A rapid, inexpensive method for eluting DNA from agarose or acrylamide gel slices without using toxic or chaotropic materials. BioTechniques. 1992;13:205–206. [PubMed] [Google Scholar]
- 32.Slater S, Maurer R. Simple phagemid-based system for generating allele replacements in Escherichia coli. J Bacteriol. 1993;175:4260–4262. doi: 10.1128/jb.175.13.4260-4262.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Somerville J E, Jr, Cassiano L, Bainbridge B, Cunningham M D, Darveau R P. A novel Escherichia colilipid A mutant that produces an antiinflammatory lipopolysaccharide. J Clin Investig. 1996;97:359–365. doi: 10.1172/JCI118423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takada H, Kotani S. Structure-function relationships of lipid A. In: Morrison D C, Ryan J L, editors. Bacterial endotoxic lipopolysaccharides. I. Molecular biochemistry and cellular biology. Boca Raton, Fla: CRC Press, Inc.; 1992. pp. 107–130. [Google Scholar]
- 35.Underwood K, Sjöstrom K, Darveau R, Lamont R, Schenkein H, Gunsolley J, Page R, Engel D. Serum antibody opsonic activity against Actinobacillus actinomycetemcomitans in human periodontal diseases. J Infect Dis. 1993;168:1436–1443. doi: 10.1093/infdis/168.6.1436. [DOI] [PubMed] [Google Scholar]
- 36.van Putten J P M. Phase variation of lipopolysaccharide directs interconversion of invasive and immuno-resistant phenotypes of Neisseria gonorrhoeae. EMBO J. 1993;12:4043–4051. doi: 10.1002/j.1460-2075.1993.tb06088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogel S N, Hogan M M. Role of cytokines in endotoxin-mediated host responses. In: Oppenheim J J, Shevach E M, editors. Immunophysiology: the role of cells and cytokines in immunity and inflammation. Oxford, England: Oxford University Press; 1990. pp. 238–252. [Google Scholar]
- 38.Vuorio R, Vaara M. Comparison of the phenotypes of the lpxA and lpxD mutants of Escherichia coli. FEMS Microbiol Lett. 1995;134:227–232. doi: 10.1111/j.1574-6968.1995.tb07942.x. [DOI] [PubMed] [Google Scholar]
- 39.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]