Abstract
G protein-coupled receptors (GPCRs) form the largest class of membrane receptors in the mammalian genome with nearly 800 human genes encoding for unique subtypes. Accordingly, GPCR signaling is implicated in nearly all physiological processes. However, GPCRs have been difficult to study due in part to the complexity of their function which can lead to a plethora of converging or diverging downstream effects over different time and length scales. Classic techniques such as pharmacological control, genetic knockout and biochemical assays often lack the precision required to probe the functions of specific GPCR subtypes. Here we describe the rapidly-growing set of optogenetic tools, ranging from methods for optical control of the receptor itself to optical sensing and manipulation of downstream effectors. These tools permit the quantitative measurements of GPCRs and their downstream signaling with high specificity and spatiotemporal precision.
I. Introduction
G protein-coupled receptors (GPCRs) serve as critical membrane receptors that mediate many fundamental aspects of biological function, including sensory transduction, neuromodulation, respiration, regulation of heart rate and control of blood pressure (Pierce et al., 2002). GPCRs respond to a wide range of extracellular stimuli including neurotransmitters, peptides, hormones, ions, or lipids to initiate complex intracellular signaling pathways with precise spatial and temporal dynamics. The GPCR superfamily forms the largest class of receptors with over 800 human genes encoding distinct subtypes, and, accordingly, serves as the largest class of drug targets for the treatment of diseases (Hauser et al., 2017). Despite a long-standing appreciation of the physiological importance of GPCR signaling and recent breakthroughs at the structural level (Erlandson et al., 2018; Venkatakrishnan et al., 2013), a relatively limited understanding of the functional mechanisms of GPCRs at the cell biological and systems levels currently exists, in part due to the challenge of directly manipulating and sensing receptors in physiological settings with higher spatiotemporal precision than classical pharmacological or genetic approaches. This challenge has motivated new, high resolution optical techniques, which are the topic of this review.
Following agonist binding, GPCRs undergo a series of conformational changes that allow them to activate heterotrimeric G proteins which are comprised of Gα, Gβ, and Gγ proteins (Fig. 1A). The active conformation of the receptor is recognized by the C-terminal helix of Gα (Rasmussen et al., 2011b) (Flock et al., 2017; Liu et al., 2019), which subsequently exchanges GDP for GTP to promote dissociation of the heterotrimer into a GTP-bound Gα subunit and an active Gβγ complex. The liberated G proteins have a wide-ranging pallet of target effectors, including membrane-embedded enzymes and ion channels, depending on the G protein subtype that is engaged. In addition, G protein-mediated signals are thought to be amplified over time due to the ability of one stimulated GPCR to activate multiple heterotrimeric G proteins, as has been most clearly demonstrated for rhodopsin which can activate thousands of G proteins per second (Ernst et al., 2007). Further non-linearity of signaling can come from the generation of second messengers which can spread with unique spatiotemporal parameters to relay the signal to different cellular locations. G protein signaling is terminated following hydrolysis of the Gα-bound GTP, which can be accelerated by regulator of G protein signaling (RGS) proteins, which leads to the reassembly of the heterotrimeric G protein.
Figure 1.
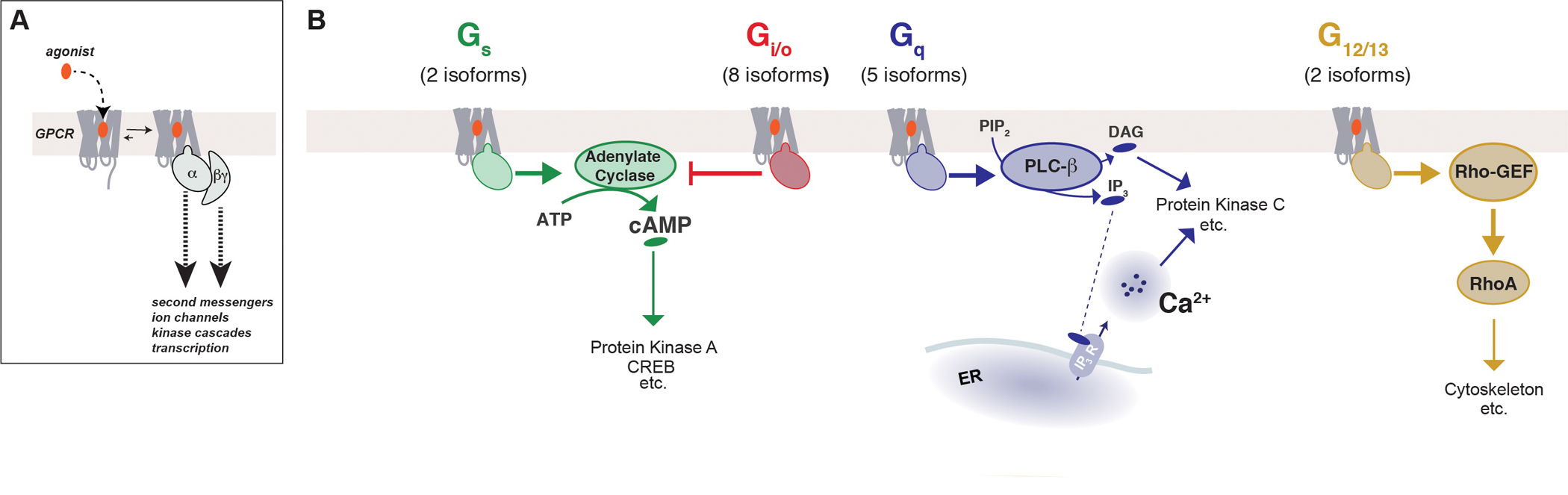
Basic mechanisms of GPCR signaling
(A) GPCRs bind ligands which initiate a conformational change in the receptor that allows the receptor to couple to and signal through heterotrimeric G proteins, which activates various second messengers and effectors.
(B) Canonical signaling pathways mediated by the four families of Gα protein subtypes.
A given GPCR typically has a preferred Gα target, but in many cases can target multiple G protein families (Flock et al., 2017). The four families of Gα proteins include the Gs, Gi/o, Gq, and G12/13 subtypes which all contain multiple members and produce distinct, canonical responses (Fig. 1B). Generally, activated Gαs proteins activate adenylyl cyclase to generate cAMP from ATP which can, in turn, lead to activation of protein kinase A (PKA). PKA phosphorylates a wide range of proteins, including other GPCRs. On the other hand, activated Gαi/o proteins inhibit adenylyl cyclase which leads to reduced levels of cAMP and active PKA. Activated Gαq proteins activate phospholipase C-beta (PLC-ß), which produces inositol trisphosphate (IP3) and diacylglycerol (DAG) from phosphatidylinositol 4,5-bisphosphate (PIP2) and leads to the release of calcium from intracellular stores via IP3 receptors of the endoplasmic reticulum. Elevations in intracellular calcium can lead to complex oscillatory responses and target myriad other signaling proteins including kinases, such as protein kinase C (PKC) and calcium-calmodulin dependent kinase II (CaMKII), and phosphatases, such as calcineurin (Grundmann and Kostenis, 2017). Gα12/13, the least frequently targeted family, activates Rho GTPases to regulate the actin cytoskeleton which can control cell mobility. Meanwhile, the Gβγ complex has direct targets of its own including, most prominently, the opening and closing of ion channels such as the G protein-coupled inwardly-rectifying potassium (GIRK) channels and voltage-gated calcium channels (VGCCs) (Khan et al., 2013). While no clear preference of different beta or gamma subtypes for interaction with Gα subtypes has been observed, many Gβγ-driven processes have been shown to be specific for Gi/o-coupled receptors. The mechanism for GPCR subtype-specificity of Gβγ-driven signaling remains debated, but a recent study proposed that Gi/o-coupled signaling has an intrinsically higher rate of G protein activation which produces a greater local concentration of free Gβγ than other receptor subtypes (Touhara and MacKinnon, 2018).
The aforementioned upstream signaling pathways and effector molecules only scratch the surface of the complex cellular consequences of GPCR activation. Other direct targets of G proteins likely exist and many downstream cellular responses have been observed without clear information on the intermediate signaling events that link the response to the initial GPCR activation. For example, GPCR activation often leads to activation of the MAPK/ERK pathway which produces long-lasting effects on the cell by modulating gene transcription, cell proliferation and differentiation. Importantly, in addition to their coupling to G proteins themselves, GPCRs can couple and signal through β-arrestins (DeWire et al., 2007). Canonically, β-arrestins are recruited to the C-terminal tail of activated GPCRs following phosphorylation by G protein-coupled receptor kinases (GRKs). β-arrestin interaction with the GPCR sterically blocks interactions with G proteins while also serving as an endocytic adaptor to promote the internalization of the receptor to desensitize signaling (Rajagopal and Shenoy, 2018). Alternatively, following binding to the ligand-bound receptor, β-arrestin can be allosterically activated and serve as a scaffold to initiate various signaling pathways (Weis and Kobilka, 2018). In particular, β-arrestins have been shown to initiate the MAPK/ERK pathway, independent of G proteins, either indirectly via activation of Src or through the formation of a complex with RAF-1, MEK and ERK (Luttrell et al., 1999; Luttrell et al., 2001; Strungs and Luttrell, 2014). However, ongoing debate exists over the relative contributions of G proteins and β-arrestins to ERK activation by GPCRs (O’Hayre et al., 2017) (Luttrell et al., 2018) (Grundmann et al., 2018). Interestingly, the identity of the GPCR agonist can bias the receptor to preferentially couple and signal through either G proteins or β-arrestins, in a phenomenon termed “biased agonism” that is promising clinically but poorly understood mechanistically (Smith et al., 2018). Finally, there is a growing body of evidence that internalized GPCRs can initiate signals from endosomes and other organelles, permitting functional diversity and an additional layer of complexity to signal transduction (Eichel and von Zastrow, 2018).
Ultimately, a single GPCR can initiate a mosaic of independent, converging or diverging signaling pathways that consists of a multitude of effectors and second messengers spread throughout space to achieve an assortment of short- (i.e. hundreds of milliseconds) and long-lasting (i.e. hours to days) effects on the cell and, ultimately, organism. To complicate matters, an individual receptor can serve different functions depending on its subcellular localization or the cell type in which it’s expressed. Furthermore, multiple GPCR subtypes are typically expressed in any given cell, some of which can share the same endogenous ligand. Thus, signaling pathways mediated by one GPCR can crosstalk with that of a different GPCR, heightening the difficulty of studying one receptor or pathway in isolation. Finally, GPCR signaling often occurs within multi-cellular circuits where different cell-types have distinct responses that can influence other cells within the network. This immense complexity presents a challenge to studying GPCRs: how can we achieve the precision required to thoroughly study one receptor or one component of a signaling pathway in order to dissect their direct contribution to a biological process? In this review we describe techniques that utilize optical probes to control and observe GPCR activity and signaling with high spatiotemporal precision and genetic targeting. First, we discuss existing methods for the optical manipulation (section II) and detection (section III) of GPCRs themselves. We highlight important examples of the use of these tools to decipher mechanistic aspects about GPCR function. We then discuss methods for the optical manipulation (section IV) and detection (section V) of different intracellular molecules that contribute to GPCR signaling. This survey provides a framework for the further design and application of optogenetic tools for GPCRs.
II. Optical control of GPCRs
One of the most successful realms of optogenetics has been the development of techniques for the optical control of GPCRs. Ideally, optical control allows one to target a specific GPCR subtype with spatial and temporal resolution in a genetically-defined cell type. Such precise targeting should allow one to determine the relative contribution of a receptor subpopulation to a physiological process, including those that take place at the organismal level such as behavior, and better define the receptor’s response to specific temporal patterns of activity in defined subcellular regions. A number of complementary techniques have been developed to allow for control of a range of GPCRs and recent studies have begun to exploit these tools to gain meaningful biological insight. In this section, we summarize the main categories of approaches, assess their relative advantages and caveats, and describe illustrative examples of their application.
Typically, GPCRs have been manipulated using a combination of application of native or synthetic pharmacology. A vast library of agonists, antagonists, and modulators exists for different receptor subtypes which represent the basis for many clinical approaches (Wacker et al., 2017). However, when applying these drugs to biological samples, especially in tissue or in vivo, many limitations emerge including lack of receptor subtype specificity, lack of cell-type specificity, and the inability to rapidly apply and remove the compound to defined locations within the tissue or cell. Taking advantage of the plethora of well-characterized ligands, photopharmacology has blossomed into a flexible and powerful field of chemical biology. The general principle of photopharmacology is to incorporate light-sensitive groups into biologically-active ligands in order to control their activity with light (Figure 2A). Early studies used “caged” ligands, including most frequently caged glutamate, which are released from a chemical enclosure (i.e. 4-methoxy-7-nitroindolinyl, MNI) upon illumination (Callaway and Katz, 1993; Reiner et al., 2015). Various caging strategies have been developed, some of which have been applied to GPCR ligands. Banghart et al developed caged agonists and antagonists(Banghart et al., 2018; Banghart and Sabatini, 2012; Banghart et al., 2013) for opioid receptors which have been used to probe the spatial extent of enkephalin signaling in the locus coeruleus (Banghart and Sabatini, 2012) and to define specific aspects of receptor kinetics (Banghart et al., 2013; Williams, 2014).
Figure 2.
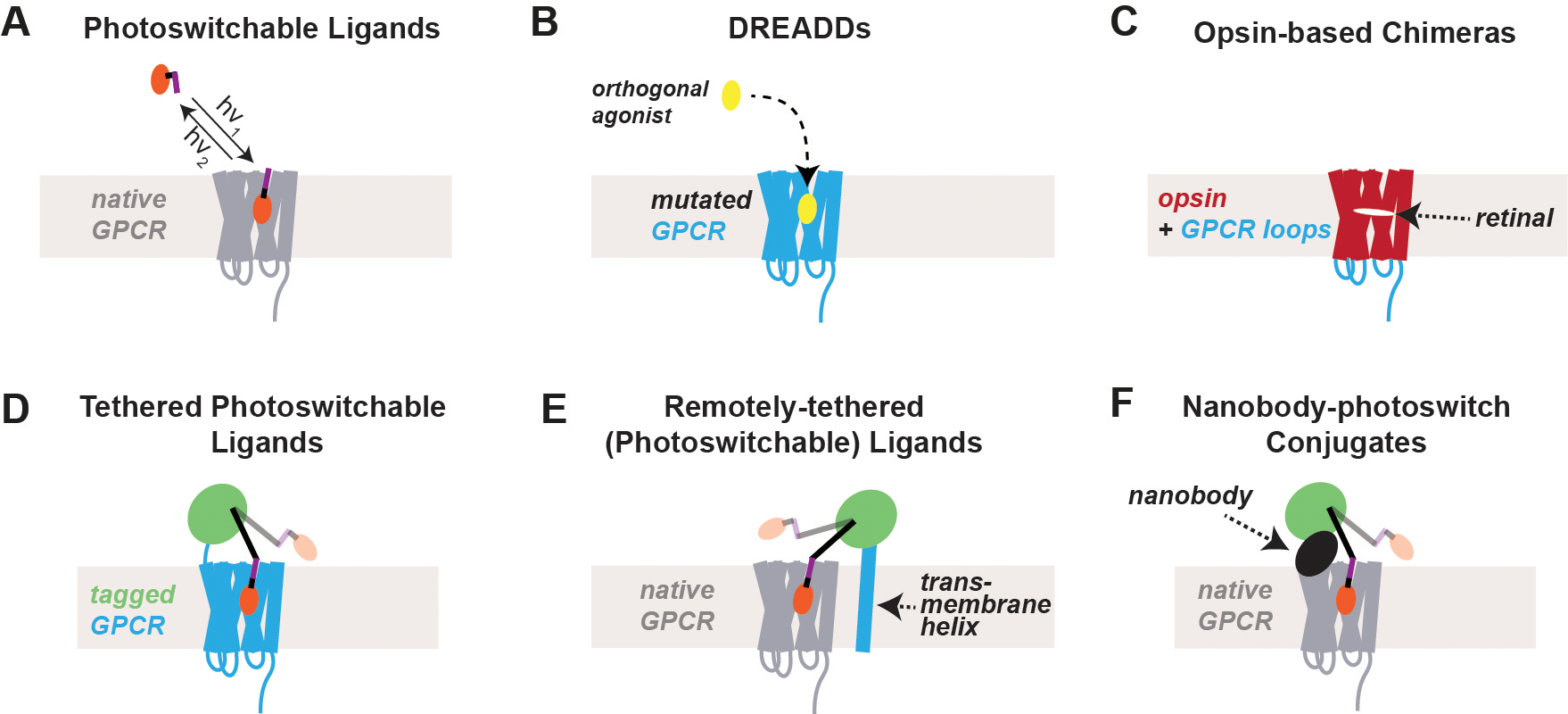
Chemical and optical methods for the targeted control of GPCRs
(A) Photoswitchable ligands are typically composed of a well-characterized ligand (orange) and a light-sensitive moiety (purple) that either isomerizes or releases the ligand in the presence of light. This system allows the control of native GPCRs by controlling ligand availability with high time resolution. The temporal resolution of reversal is limited by ligand unbinding and spatial targeting is limited by diffusion.
(B) DREADDs are mutated GPCRs that are engineered to respond to a synthetic agonist but to no longer respond to a native ligand. By targeting DREADD expression, one can achieve genetically-targeted chemical (i.e. “chemogenetic”) control of GPCR signaling.
(C) Opsins are naturally-occurring light-activatable GPCRs that have been taken advantage of for the optical control of GPCR activation. To study and manipulate signaling pathways associated with specific GPCRs, chimeras may be designed that incorporate the intracellular loops and/or the C-terminal tail of the GPCR of interest.
(D) Photoactivation of specific GPCRs can be achieved by tethering a photoswitchable ligand to a self-labeling tag, such as SNAP (green). This system requires heterologous expression of a tagged GPCR which allows the incorporation of receptor mutants or variants but can lead to overexpression. Tethered photoswitchable ligands allow the highest temporal resolution of both ligand binding and un-binding.
(E) Photoactivation of native GPCRs with genetic targeting can be accomplished by the expression of a tagged transmembrane domain that is tethered to a photoswitchable ligand.
(F) Native GPCRs may also be optically-controlled via photoswitchable ligands tethered via self-labeling tags (green) to nanobodies that recognizes an extracellular site on the receptor. Nanobody-tethered photoswitchable ligands can either be delivered directly or can be genetically-encoded for heterologous expression and cell-type targeting.
While applicable to many compounds and a clear enhancement over normal drugs, uncaging is irreversible which limits the types of experiments that can be designed. To overcome this, over the last 15 years the photoswitchable ligands have been developed for a wide range of signaling proteins, including GPCRs. Photoswitchable ligands are typically built around the azobenzene chemical photoswitch, which toggles between cis and trans configurations in response to light and can, thus, modulate the functional properties of attached compounds (Beharry and Woolley, 2011). Dirk Trauner and others have developed azobenzene-based photoswitchable agonists for class A and B GPCRs, including mu-opioid receptors (Schonberger and Trauner, 2014), muscarinic acetylcholine receptors (Agnetta et al., 2017), cannabinoid receptors (Westphal et al., 2017) and the glucagon-like peptide receptor (Broichhagen et al., 2015b). While agonists have been the main focus of photopharmacological development, the flexibility of the approach allows other types of ligands to be developed. For example, photoswitchable allosteric modulators of metabotropic glutamate receptors (mGluRs) have been developed (Pittolo et al., 2014) and applied in vivo to probe the basis of mGluR4-mediated pain processing (Zussy et al., 2018). While the photopharmacological toolset is constantly expanding and in use for applications, soluble photopharmacological ligands have shortcomings due to the lack of genetic targeting, the typical lack of true subtype-specificity and limitations in both the speed and spatial precision of receptor targeting due to diffusion.
One of the most successful strategies to improve the cell-type precision of GPCR activation has been with the so-called “chemogenetic” approach. Most prominently, Bryan Roth and colleagues have developed “Designer Receptors Exclusively Activated by Designer Drugs” (DREADDs) which are engineered muscarinic receptors that no longer retain sensitivity to their native agonist, acetylcholine, but are able to be activated by clozapine N-oxide (CNO), a synthetic agonist. DREADDs have been used extensively for in vivo studies in rodents to probe the cell-type and circuit basis of behavior but their application for detailed analysis of specific GPCR-driven signaling processes has been limited (Urban and Roth, 2015). However, engineering of DREADDs is ongoing and recent years have seen the report of new DREADDs that allow multiplexing (Vardy et al., 2015), G protein or arrestin-biased DREADDs (Hu et al., 2016; Nakajima and Wess, 2012), and subcellularly-targeted DREADDs (Stachniak et al., 2014). Overall, the genetically-targeted GPCR activation enabled by chemogenetics makes this an extremely valuable approach, but optical approaches are needed for applications which require high spatiotemporal precision.
One robust approach to the optical control of GPCR activation is to employ opsins, the visual photoreceptors which use a native retinal chromophore as a light-sensing ligand (Palczewski, 2006). A pioneering optogenetic demonstration co-expressed a G protein-coupled opsin, a Gα subunit and an arrestin from the drosophila visual system to allow for optical control of neural activity in cultured hippocampal neurons (Zemelman et al., 2002). While channelrhodopsins are now typically used for direct optical excitation of neurons (Fenno et al., 2011), various G protein-coupled opsins have been employed for optical control of G protein pathways via heterologous expression. This includes rhodopsin (Li et al., 2005) or cone opsins (Masseck et al., 2014) for Gi/o activation, jellyfish opsin (Bailes et al., 2012) for Gs activation and melanopsin (Qiu et al., 2005; Spoida et al., 2016) for Gq activation. Notably, Karunarathne et al (Karunarathne et al., 2013) used spatially-targeted activation of a Gi/o-coupled opsin to locally manipulate PIP3 levels and induce neurite extension in cultured neurons and, recently, Makowka et al (Makowka et al., 2019) used jellyfish opsin to compare the effects of targeted Gs activation in the left versus right atrium of the heart. While wild-type opsins are powerful for optical control of defined G protein-mediated signaling cascades, various groups have introduced the intracellular loops and/or C-terminal tails of different GPCRs to attempt to mimic the signaling of different GPCRs while maintaining light activation (Figure 2C) (Kim et al., 2005; Tichy et al., 2019). This approach has been used for a variety of circuit neuroscience applications (Airan et al., 2009; Gunaydin et al., 2014) (Siuda et al., 2015), but has not yet been employed for more precise cell biological studies.
Despite the utility of opsin-based optical control of GPCR signaling, the temporal precision of such approaches remains limited due the intrinsically slow off kinetics of opsins and the ability to probe receptor-specific function via the chimeric technique remains unclear. As a means of achieving optical control of full-length GPCRs with both spatiotemporal and genetic targeting, photoswitchable tethered ligands (PTLs) have been developed for both ion channels and GPCRs. This strategy has been most thoroughly developed for metabotropic glutamate receptors (mGluRs) and is based on the covalent attachment of a photoswitchable agonist to the receptor. Initial versions of this were based on introduction of a cysteine mutation into the ligand binding domain a full-length mGluR for attachment of a maleimide-conjugated azobenzene-glutamate compound termed “MAG” (Carroll et al., 2015; Levitz et al., 2013). The photophysical properties of the azobenzene allow rapid switching of the ligand on the millisecond time scale between inactive trans and active cis states and since the mutated receptor needs to be heterologously expressed, genetic targeting is enabled. Importantly, since the PTL is covalently-attached to the receptor of interest, this system facilitates complete subtype specificity even if the ligand moiety itself is non-specific, as is the case with MAG. Levitz et al (Levitz et al., 2016) took advantage of this high degree of control to probe the relative contribution to downstream activation of one or two agonists binding within homo and heterodimeric mGluRs. Recently, PTLs were also developed for D1 and D2 dopamine receptors (Donthamsetti et al., 2017), showing the generalizability of this approach to class A GPCRs. While maleimide-based PTLs have shown utility for biophysical and cellular applications, the cysteine-maleimide chemistry is undesirable in complex preparations due to potential off-target effects and the need to identify an optimal cysteine on the receptor is a major challenge for PTL engineering. To overcome this, “photoswitchable, orthogonal, remotely-tethered ligands” (PORTLs) were developed which genetically fuse the photoswitchable ligand to a self-labelling tag (i.e. SNAP) at the N-terminus of the receptor (Figure 2D). This method improves the orthogonality and efficiency of the system for mGluRs (Broichhagen et al., 2015a), enables multiplexing using SNAP and CLIP tags (Levitz et al., 2017) and works efficiently in vivo (Berry et al., 2017). Furthermore, branched PORTLs have recently been reported which enhance photoswitching efficiency and allow the incorporation of a fluorophore for simultaneous detection of the targeted receptor (Acosta-Ruiz et al, bioRxiv, 2019).
The optogenetic approaches to the manipulation of GPCRs described in this section have opened up new avenues of research and many applications remain to be pursued from molecular biophysics to cell biology and systems neuroscience. However, the challenge of many of the aforementioned techniques is that they require heterologous expression of a receptor. This provides the ability to genetically target specific cell types and the ability to introduce biologically-relevant mutations or variants to the GPCR of interest, but it also raises the possibility of overexpression which can alter the fundamental biology of the system and compromise the physiological relevance of the study. One possibility is to express a genetically-encoded protein that can be labeled with a tethered ligand to then target nearby native receptors (Figure 2E). Such an approach has been demonstrated for GPCRs with a tethered antagonist for muscarinic receptors (Shields et al., 2017) or with a glutamate PORTL for mGluR2 (Donthamsetti et al., 2019). Alternatively, receptor-targeting single-chain antibodies (i.e. nanobodies) may provide a means of delivery of a PORTL to a native GPCR (Figure 2F). A recent proof-of-principle study showed that SNAP-tagged anti-GFP nanobodies can enable optical control of a GFP-tagged mGluR (Farrants et al., 2018). Both of these techniques have great promise for optical control of native receptors but will require further engineering and characterization to be widely applicable. Ultimately, a key aspect of future optogenetic studies of GPCRs will be the ability to apply these tools in conjunction with optical control (section III) and measurement (section IV) of other aspects of GPCR signaling to answer biological questions.
III. Optical control of GPCR effectors and related signaling pathways
Activation of a single GPCR can lead to modulation of a diverse set of downstream effectors such as small GTPases, ion channels, kinases and other enzymes, regulators of translation and transcription factors. This divergent signaling makes it difficult to determine the relative contribution of specific GPCR effectors to subsequent physiological processes. Bypassing the receptor to control specific signaling nodes or regulatory proteins can permit the probing of the spatiotemporal properties and role of that specific component while limiting confounding effects from other signaling proteins. Although pharmacology can be used to examine the role of a particular signaling protein where available, this approach can be non-specific and is often irreversible, especially when targeting enzymes. Recently, optogenetic tools have been developed in an effort to manipulate specific intracellular signaling proteins with high precision. While synthetic chemical photoswitches have been the typical approach to the optical control of GPCRs themselves (section II), naturally-occurring light-sensitive proteins have been the core component in the design of optical control of intracellular signaling. The different configurations and mechanisms of such photoactivatable proteins have been reviewed extensively elsewhere (Repina et al., 2017) (Goglia and Toettcher, 2019), but will be described where relevant below.
A limited body of work has pursued optical control of the direct effectors and regulators of GPCRs but such tools could be extremely valuable for parsing the relative roles, dynamics and regulation of G protein or β-arrestin-mediated signaling. Notably, an optogenetic β-arrestin2 was recently reported which utilizes the cryptochrome CRY2 system (Takenouchi et al., 2018). CRY2 is a photoreceptor from A. thaliana that contains a flavin adenine dinucleotide (FAD) chromophore which is reduced following illumination with blue (405–488 nm) light which enables it to both homo-oligomerize and to bind its native interaction partner, CIB (Kennedy et al., 2010; Liu et al., 2008). CIB and CRY were fused to the C-terminal tail of the β2-adrenergic receptor (β2AR) and the N-terminus of β-arrestin2, respectively (Takenouchi et al., 2018). Upon illumination with blue light, β-arrestin2 was translocated to the surface which was shown to be sufficient for the endocytosis of β2AR, even in the absence of agonist. Importantly, the CRY-β-arrestin2 and β2AR-CIB interaction is reversible; in the dark, β-arrestin2 dissociated from the receptor and redistributed throughout the cytosol over the span of ten minutes. The CRY2 system has also been used to enable photoactivation of RGS proteins (O’Neill and Gautam, 2014) (Hannanta-Anan and Chow, 2018). RGS proteins regulate GPCR signaling by accelerating the intrinsic GTPase activity of the Gα subunit to promote its re-association with the Gβγ complex to tune the frequency, duration, and strength of signaling (De Vries et al., 2000). In one case (Hannanta-Anan and Chow, 2018), optical control was achieved by splitting RGS2 into its catalytic box and N-terminal amphipathic helix, and fusing each part to CRY2 and a truncated version of CIB (CIBN), respectively (Figure 3B). Blue light illumination initiates the dimerization of CRY2 and CIBN, translocating the RGS catalytic box to the surface, terminating Gαq-induced calcium signaling. This tool was used to establish the role of RGS2 in maintaining a negative feedback loop on calcium oscillations mediated by activation of the M3 muscarinic acetylcholine receptor (M3R), a Gq-coupled receptor. The CRY2-CIB system has also been used to recruit the C-terminal domain of GRKs to the membrane to bind and sequester Gβγ (O’Neill and Gautam, 2014). A similar approach has also been reported to optically recruit Gα proteins to the membrane to initiate signaling (Yu et al., 2016).
Figure 3.
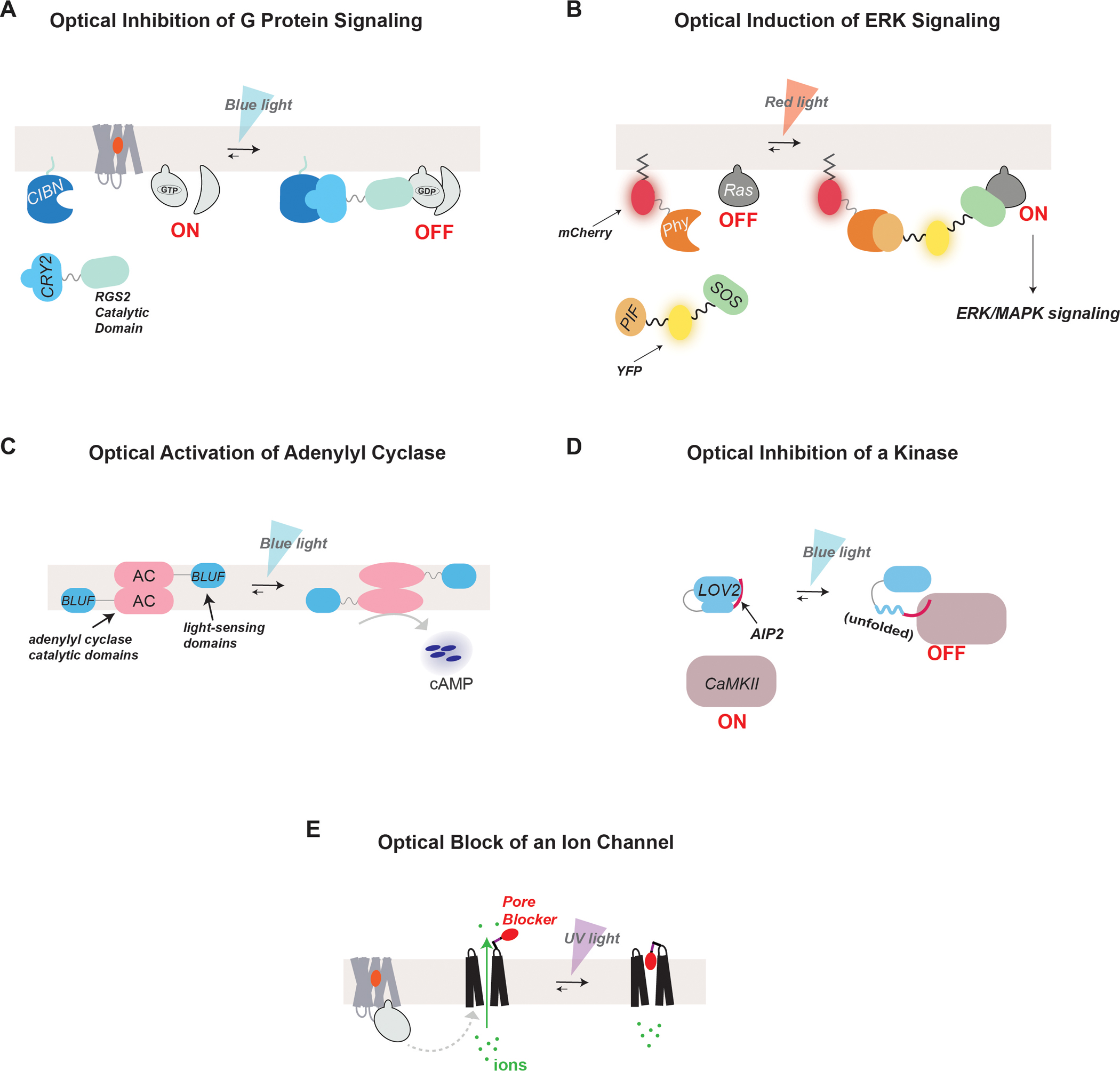
Optical methods for the control of GPCR effectors
(A) The CRY-CIB system can be used for blue light-induced association of two proteins. For example, optical inhibition of G protein-mediated signaling has been achieved by utilizing the CRY-CIB system to induce the translocation of RGS2 to the plasma membrane with blue light. This recruitment brings RGS2 in close proximity to membrane-bound Gα-GTP which allows it to effectively inhibit G protein signaling.
(B) The Phy-PIF system can be used for red light-induced association of two proteins. For example, photocontrol of ERK signaling can be achieved through the induction of the surface translocation of SOS using the Phy-PIF system. Phy was fused to a CAAX motif for membrane incorporation, while SOS was fused to PIF. Red light induces dimerization of Phy and PIF, translocating SOS to the membrane to promote interaction with and activation of Ras GTPase and initiate ERK signaling. Fluorescent proteins were incorporated into the constructs to observe their expression and dynamic localization.
(C) Optical control of cAMP production can be achieved by heterologously expressing bPAC, a photoactivatable adenylyl cyclase from Beggiatoa, which has increased catalytic activity in the presence of blue light.
(D) The LOV domain has been utilized to gain photocontrol of the activity of various signaling proteins. Following blue light illumination, the Jalpha helix of the LOV domain unfolds allowing it to release functional peptides or proteins for light-gated activity. In the illustrated case, LOV2 was fused to a CaMKII inhibitory peptide, AIP2, which becomes released in the presence of blue light.
(E) Photoswitchable tethered ligands can be employed for the optical control of ion channels. For example, a photoswitchable pore blocker may be attached to an ion channel using cysteine chemistry. The pore blocker is designed such that it fits into the opening of the channel only in the presence of light to effectively block ion conductance.
Further downstream from the receptors, optical control has been used to manipulate the levels of cAMP, a key second messenger controlled by both Gi/o and Gq-coupled receptors (Figure 1). Naturally-occurring photoactivated adenylyl cyclases (PACs) have been identified (Patel and Gold, 2015), including euPAC from Euglena gracilis (Iseki et al., 2002), bPAC from Beggiatoa (Stierl et al., 2011), and OaPAC from Oscillatoria acuminate (Ohki et al., 2016). PACs contain “blue light sensor using FAD” (BLUF) domains conjugated to one or two adenylyl-cyclase domains, whereby illumination with blue light (~455 nm) induces a conformational change which activates the enzymatic activity of the adenylyl cyclase domains (Figure 3C). Activation induces rapid and reversible generation of cAMP, and can be employed in heterologous cells or organisms to study the effects of cAMP signaling with spatiotemporal precision. Optical control of adenylyl cyclase activity has been used to manipulate cAMP concentration in a variety of contexts (Bellmann et al., 2010; Nagahama et al., 2007; Schroder-Lang et al., 2007) (Tsvetanova and von Zastrow, 2014; Weissenberger et al., 2011; Zhou et al., 2016). In particular, Zhou et al (Zhou et al., 2016) utilized PAC in cultured neurons to determine that transient elevations of cAMP levels increased axonal length and that this increase was dependent on the duration of increased cAMP production. Moreover, a critical caveat of utilizing PACs to control cAMP levels is that PACs exhibit some dark activity (Schroder-Lang et al., 2007), which can alter the basal cAMP levels in the cell or organism in which the tool is employed.
A more extensive toolset of photoactivatable proteins has been developed for the study of various aspects of kinase signaling (Leopold et al., 2018). For example, a number of strategies have been employed for optogenetic perturbation of the ERK/MAPK pathway, which is typically initiated following either GPCR or, more canonically, receptor tyrosine kinase (RTK) activation (Wetzker and Bohmer, 2003) (Kolch, 2005). Toettcher et al (Toettcher et al., 2013) used the phytochrome B (Phy)-PIF light-induced dimerization system (Levskaya et al., 2009) to recruit the small GTPase SOS to the membrane following 650 nm illumination to initiate ERK activation following interaction with membrane-anchored Ras GTPases (Figure 3D). Similarly, Zhang et al (Zhang et al., 2014) used the CRY2-CIB pair to optically initiate ERK signaling by recruiting RAF-1, a vital kinase in the initiation of the ERK/MAPK cascade, which led to differentiation and neurite outgrowth in PC12 cells. Photoswitchable MEK1 and MEK2 have also been developed based on light-induced uncaging via the fluorescent protein Dronpa (Zhou et al., 2017) and a photo-inhibitable SRC kinase was developed using light-induced allosteric control (Dagliyan et al., 2016). Together these tools allow different points of the MAPK/ERK signaling cascade to be precisely perturbed depending on the relevant application.
Phosphoinositide 3-kinase (PI3K) and Akt (Protein Kinase B) signaling, additional downstream targets of GPCR signaling and regulators of cell growth, survival, and proliferation, have also been targeted for optogenetic control. Classically, PI3K phosphorylates PIP2 to generate PIP3, recruiting Akt to the cell surface, which initiates signaling to mTOR (Manning and Toker, 2017). Optogenetic control of PI3K signaling was first achieved using the Phy-PIF system to recruit PI3K to the surface and induce PIP2 phosphorylation at specific regions of the membrane with targeted 650 nm light (Toettcher et al., 2011). Alternatively, manipulation of this pathway can also be achieved by using the CRY2/CIB system to optically recruit phosphatases to the plasma membrane to manipulate PIP3 levels (Idevall-Hagren et al., 2012) or to optically recruit Akt itself (Katsura et al., 2015; Ong et al., 2016; Xu et al., 2016). Receptor tyrosine kinases, which activate many of the same effectors as GPCRs and also crosstalk with GPCRs (Wang et al., 2018), have also been targeted for optogenetic control using light-induced dimerization systems (Chang et al., 2014; Grusch et al., 2014; Kainrath et al., 2017; Reichhart et al., 2016).
As an alternative to optical activation of heterologous constructs, optogenetic kinase inhibitors have also been developed to enable silencing of native kinases (Leopold et al., 2018). Most of these have been based on the LOV domain, which is a photoreceptor domain that binds a flavin chromophore and following absorption of blue light undergoes a conformational change including unfolding of the Jα helix(Harper et al., 2003). This unfolding event can be coupled sterically or allosterically to other proteins or peptides to produce the desired output(Dagliyan and Hahn, 2019). For example, an optogenetic inhibitor was recently designed for CaMKII (Murakoshi et al., 2017), a kinase that is activated by calcium and has been shown to regulate the function of several GPCRs via phosphorylation (Liu et al., 2009) (Chen et al., 2013b; Guetg et al., 2010) (Jin et al., 2013) (Mockett et al., 2011). The photoactivatable inhibitor of CAMKII was termed paAIP2 and is based on the fusion of a LOV domain to autocamtide inhibitory peptide 2 (AIP2), a potent CaMKII inhibitor (Ishida et al., 1998). Upon illumination with blue light, AIP2 is released and becomes available to inhibit CAMKII activity. This tool was used to inhibit long-term potentiation, a form of synaptic plasticity, in dendritic spines, and define the critical timing of CAMKII activation required for the induction of synaptic plasticity (Murakoshi et al., 2017). Similarly, Yi et al (Yi et al., 2014) fused a LOV domain to an inhibitory peptide to enable optical inhibition of PKA activity and Melero-Fernandez de Mera et al (Melero-Fernandez de Mera et al., 2017) fused a LOV domain to an inhibitory domain to enable the optical inhibition of JNK, a downstream target of the MAPKs. Alternatively, unnatural amino acid incorporation combined with conjugation of a tethered, photoswitchable inhibitor has been used to optically control MEK1 (Tsai et al., 2015). While optogenetic control of kinases have not been applied explicitly in the context of GPCR signaling, these tools should enable the probing of the functional roles of specific pathways following the activation of a particular GPCR.
A number of other optogenetic tools utilize the same families of light-sensitive proteins to permit manipulation of cellular processes and may have utility in the context of GPCR signaling including the optical control of Rho GTPases (Yazawa et al., 2009), transcription (Motta-Mena et al., 2014; Wang et al., 2012), protein clustering and phase transition (Bugaj et al., 2013; Shin et al., 2017; Taslimi et al., 2014), organelle localization (Niopek et al., 2014; van Bergeijk et al., 2015), protein degradation (Bonger et al., 2014; Renicke et al., 2013), protein secretion (Chen et al., 2013a) and direct recruitment of proteins to membranes (Glantz et al., 2018). Further engineering efforts and new applications will be needed to advance this field and allow for these techniques to be used for studying GPCR signaling on the relatively fast time scales needed.
Finally, the GPCR effectors most commonly targeted for optogenetic control are ion channels, which have typically been manipulated via similar photopharmacological techniques to those targeting GPCRs (Hull et al., 2018; Paoletti et al., 2019). A wide range of mammalian ion channels of relevance to GPCR signaling, including voltage-gated potassium channels (Banghart et al., 2004), ligand-gated cation channels (Volgraf et al., 2006), leak potassium channels (Sandoz et al., 2012), and G protein-gated GIRK channels (Barber et al., 2016) have been successfully targeted with photoswitchable ligands. A common approach has been to either use a photoswitchable pore blocker (Fig. 3E) or photoswitchable agonist conjugated to an engineered extracellular cysteine which affords the system with both optical and genetic control. The ability to directly target potential ion channel effectors with light should allow a better understanding of the coupling properties of GPCRs and ion channels. For example, Sandoz et al (Sandoz et al., 2012) used a photoswitchable quaternary ammonium pore blocker to find that TREK1 channels are non-canonical downstream targets of native GABAB receptors in hippocampal neurons. While the PORTL and NB-based approaches described in section II have not yet been applied to ion channels, they hold great promise for further enhancing this efficiency and applicability of optical control of ion channels. The ongoing development of optogenetic techniques should continue to illuminate the coordinated signaling of GPCRs and ion channels.
IV. Optical sensing of GPCR signaling
While optogenetic control of GPCRs (section II) and GPCR effectors (section III) are powerful methods, the ability to observe the spatiotemporal dynamics of individual pathway components provides a complementary approach to aid in the understanding of the precise function of GPCR subtypes. In this section we will survey existing methods for detection of GPCR activation and signaling using fluorescence-based reporters. These sensors, which have been developed for nearly every node of GPCR signaling, allow for the direct observation and measurement of the intensity, duration, frequency, compartmentalization, and spread of signaling in a given stimulated pathway.
Förster resonance energy transfer (FRET) has been utilized extensively as a tool to measure the initial events of GPCR-mediated activation and signaling (Lohse et al., 2012). FRET describes the distance-dependent energy transfer between a donor fluorophore and an acceptor fluorophore that can be easily measured in live cells on standard microscopes (Miyawaki, 2011). Beginning in the mid-2000s a number of sensors have been produced to sense the ligand-induced conformational changes of GPCRs (Vilardaga et al., 2003; Vilardaga et al., 2005). Such sensors have typically been based around the introduction of fluorescent proteins into the intracellular loops of the seven helix transmembrane domains of GPCRs (Figure 4A). More recently, organic fluorophores have also been introduced into the extracellular ligand binding domains of class C GPCRs using self-labelling tags, such as SNAP and CLIP (Xue et al., 2015), to sense the inter-subunit rearrangements that mediate activation of these dimeric receptors (Doumazane et al., 2013; Doumazane et al., 2011; Vafabakhsh et al., 2015) (Gutzeit et al., 2019; Lecat-Guillet et al., 2017). One limitation of this approach is that it is difficult to find fluorophore attachment positions that give clear signals and fluorescent proteins often disrupt function of the GPCR. Unnatural amino acid incorporation has emerged as an alternative approach to fluorophore attachment without the bulk of fluorescent proteins but have so far been most valuable for biophysical studies and have been limited in their use in physiological settings (Tian et al., 2017).
Figure 4.
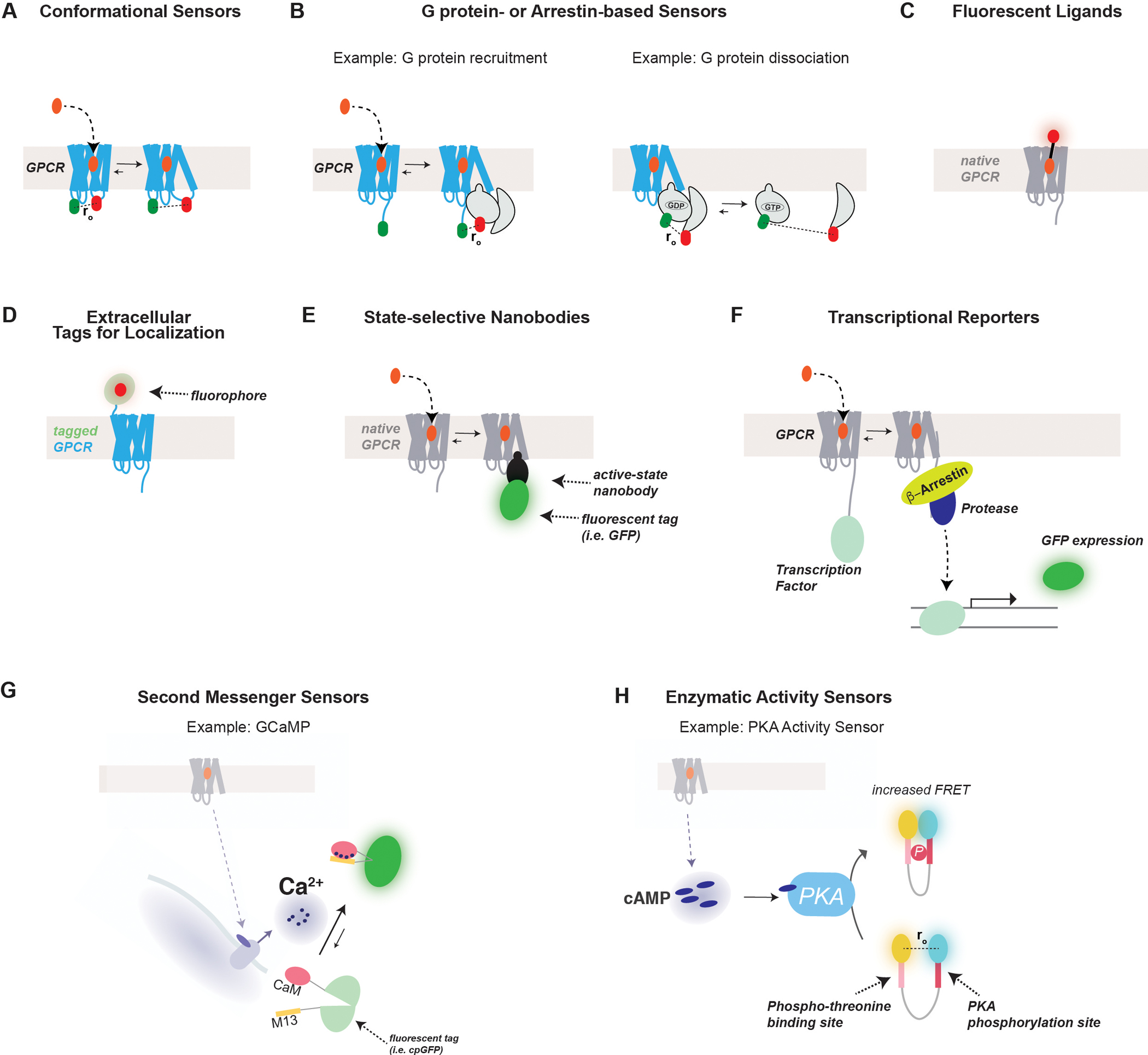
Optical sensing of GPCRs and their downstream effectors.
(A) FRET-based sensors may be used to detect the conformational changes associated with GPCR activation following ligand binding. Typically, donor and acceptor fluorophores (i.e. fluorescent proteins) are introduced into the intracellular loops of the receptor which move relative to each other during activation. Often these fluorophores disrupt coupling to G proteins making them more typically used for understanding receptor pharmacology or structure/function relationships rather than signaling properties.
(B) FRET-based sensors have also been developed to sense G protein recruitment to the receptor (left) or dissociation of G protein heterotrimers (right). For G protein recruitment, a fluorophore is typically fused to the C-terminal tail of the receptor while the Gα subunit is tagged with a fluorophore in an internal site to avoid disrupting function and receptor specificity. To sense G protein dissociation, the Gα subunit is tagged as described above, while another fluorophore is typically placed on the N terminus of either Gβ or Gγ. Similar sensors have also been developed to sense the recruitment or conformational changes of β-arrestins.
(C) Ligands conjugated to fluorophores have been developed to aid in their visualization both in vitro and in vivo.
(D) GPCRs can be tagged with a fluorophore, either a fluorescent protein or dye conjugation to a tag (i.e. SNAP, CLIP, or Halo) to permit the observation of their surface and subcellular localizations in living cells.
(E) Fluorophore-tagged nanobodies have been developed which can be genetically encoded and used to detect the conformational state of a receptor, G protein or β-arrestin. Accumulation of fluorescence in a given location (i.e. plasma membrane or endosomes) indicates a population of receptor or effectors in a state recognized by the NB.
(F) GPCR activation can also be sensed using a transcriptional reporter, such as the Tango assay. In this assay a transcription factor and a protease site are incorporated into the C-terminal tail of the receptor, while β-arrestin is tagged with a protease. Receptor activation leads to β-arrestin-protease recruitment, leading to the cleavage and release of the transcription factor which will permit expression of a reporter gene (i.e. GFP).
(G) A classic sensor for the detection of the second messenger, Ca2+, is GCaMP. Ca2+ binding to CaM permits its association with the M13 peptide which induce the closure and increased fluorescence of cpGFP. Many permutations of GCaMP and other related calcium sensors exist with variable kinetics, sensitivity, subcellular targeting and spectral properties to allow the ideal construct to be used for the relevant application.
(H) FRET-based enzymatic activity sensors are widely used to detect kinase activity. In the case of the PKA sensor, AKAR, the sensor contains N- and C-terminal donor and acceptor fluorophores, a PKA phosphorylation site, and a domain that binds to the phosphorylated residue. Following phosphorylation by active PKA, association of the two sites leads to increased FRET between the fluorophores.
FRET-based approaches have also been developed to monitor the coupling of an activated GPCR with G proteins (Gales et al., 2005; Hein et al., 2005; Hein et al., 2006; Nobles et al., 2005), and well as the dissociation of Gα subunit and Gβγ complex following receptor activation (Gales et al., 2005) (Bunemann et al., 2003) (Figure 4B). Similar FRET-based or bioluminescence resonance energy transfer (BRET)-based approaches have also been used to measure β-arrestin recruitment to GPCRs (Vilardaga et al., 2003) and β-arrestin conformational changes following receptor activation (Nuber et al., 2016) (Lee et al., 2016). These FRET-based tools have been used in living cells to define the millisecond time scale of receptor activation, probe structural models of receptor and effector activation, and as powerful assays for drug screening (Lohse et al., 2012).
While deciphering the details of receptor activation and effector coupling in living cells remains a major goal, a parallel issue in the field of GPCR biology is to detect receptor localization dynamics within the cell. One long-standing approach is to use fluorescently-labeled ligands which bind to receptors and allow their visualization (Figure 4C). This approach is compatible with native receptors and has been used in tissue to detect receptor multimers (Albizu et al., 2010). However, the development of fluorescent agonists or antagonists for a given GPCR is challenging and the efficacy of the ligand makes it difficult to determine the effects of receptor activation on localization. An alternative has been to label the GPCR with a fluorophore directly without the need for inclusion of a pharmacophore that recognizes a binding site (Figure 4D). A number of labeling techniques have been developed which allow one to attach fluorophores to heterologous receptors, including the SNAP, CLIP, and Halo-tags. A major advantage of this approach versus simply tagging a receptor with a fluorescent protein is that extracellular tags can be added to the N-terminus of the receptor and membrane-impermeable fluorophores may be used to limit visualization to surface receptors. For example, Thomsen et al (Thomsen et al., 2016) labeled an N-terminally SNAP-tagged β2AR with an impermeable fluorophore and used this technique to track activation-induced internalization which led to co-localization of receptors, G proteins and ß-arrestins in endosomes. In a particularly striking study, a GPCR and a Gα were labeled with SNAP- and CLIP-, respectively, and single molecule imaging was used to decipher the timing of receptor/G protein coupling in live cells (Sungkaworn et al., 2017).
The ideal optogenetic sensors for GPCRs would be able to effectively sense receptor or effector conformation and localization of native, untagged receptors. One promising avenue for this is to use nanobodies (NBs) which recognize specific states of the GPCR, G protein or arrestin. NBs were crucial to the GPCR structural revolution, but have also been tagged with fluorophores and expressed in living cells to sense GPCR signaling (Heukers et al., 2019; Manglik et al., 2017). State-selective nanobodies that stabilize active or inactive states of GPCRs (Che et al., 2018; Rasmussen et al., 2011a; Scholler et al., 2017), G proteins (Gulati et al., 2018; Westfield et al., 2011) or arrestins (Cahill et al., 2017) exist and their accumulation in specific cellular sites can, in principle, provide information on both protein localization and conformation (Figure 4E). In a pioneering study, Irannejad et al used (Irannejad et al., 2013) an NB that recognizes the active state of the ß2AR (Nb80) and a NB that recognizes the active state of Gαs (Nb37) to show that internalized receptors can couple to G proteins from endosomes. Improved engineering methods continue to increase the rate of development of new NBs which should facilitate the widespread use of these approaches (McMahon et al., 2018). However, one caveat is that state-selective NBs themselves can serve as pharmacological agonists, antagonists or modulators and can, thus, perturb the system in a concentration-dependent manner.
As an alternative to sensing the GPCR activation process directly, transcription-coupled fluorescent sensors have also been pursued. The transcriptional activation following ß-arrestin translocation (“Tango”) system has been developed and modified to allow ß-arrestin recruitment to a specific GPCR to be coupled to protease-induced release of a transcription factor to drive the transcription of luciferase or a fluorescent protein (Figure 4F) (Barnea et al., 2008; Kroeze et al., 2015). While this approach lacks temporal precision, the amplification and time-integration it allows for detection of receptor activation over extended time periods. In an impressive study, this system was adapted for use in vivo in mice where coincident light-activation and ligand binding are necessary for protease activity and reporter gene expression. This “iTango” system was used to report on dopamine receptor activation in different cell types over a defined time period to identify reward- and locomotion-related dopamine-sensitive neurons in the striatum (Lee et al., 2017).
Downstream of receptor activation, genetically-encoded fluorescent sensors have been developed for the detection of the canonical second messengers that are generated following GPCR activation including, most prominently, Ca2+ ions. Typically, Gq-coupled GPCR activation leads to the release of calcium from the endoplasmic reticulum (ER) (Figure 1B). Additionally, Gi/o-coupled GPCRs can inhibit VGCCs, to reduce the influx of calcium, which is a particularly common form of presynaptic inhibition in the nervous system. While early calcium sensors were based on FRET (Miyawaki et al., 1997), the most commonly used calcium sensors are those in the family of GCaMP (Nakai et al., 2001), which has been optimized for maximal signal to noise and engineered to produce a wide range of spectral and kinetic variants (Greenwald et al., 2018) for simultaneous sensing of additional effectors. The general design is based on a circularly-permuted GFP (cpGFP) fused to calmodulin and the M13 domain from myosin light chain kinase. Ca2+ binding brings calmodulin and M13 in close proximity, enclosing the GFP chromophore that is otherwise exposed to the cytosol and therefore quenched. Thus, an increase in GFP fluorescence intensity is associated with an increase in Ca2+ binding, enabling one color imaging of relative calcium concentration with time resolution on the order of milliseconds (Figure 4G). Extensive protein engineering work has also been applied to the development of FRET-based sensors for cAMP (Paramonov et al., 2015), a second messenger regulated by Gi/o- and Gs-coupled receptors (Figure 1B). Most recently, a 1-color sensor based on a similar design principle to GCaMP has been developed, termed cAMPr (Hackley et al., 2018). cAMPr is also based on a cpGFP, where one end is fused to the catalytic subunit of PKA (PKA-C) and the other end is tethered to a regulatory subunit of PKA (PKA-R). cAMP binding induces a conformational change which separates PKA-C and PKA-R from one another, permitting GFP to fluoresce efficiently.
Beyond the fluorescent sensors for Ca2+ and cAMP, genetically-encoded fluorescent sensors have also been reported for voltage (Platisa and Pieribone, 2018), phospholipids, pH, and other second messengers(Greenwald et al., 2018) and further design optimization promises to improve the sensitivity and ease with which these tools can be applied in complex systems. In an especially exciting series of breakthroughs, the cpGFP-based sensor design has been applied to develop sensitive fluorescence reporters for a variety of GPCR-targeting neurotransmitters and neuromodulators. Patriarchi et al (Patriarchi et al., 2018) reported a suite of tools based on the insertion of cpGFP into intracellular loop 3 of class A GPCRs to develop sensors for dopamine, norepinephrine, melatonin, opioids, and serotonin. The dopamine sensor, “dLight1” was validated in vivo where it’s been used for studies of behavior-associated dopamine dynamics in the striatum (Mohebi et al., 2019; Patriarchi et al., 2018). Given that these constructs are unable to couple to downstream effectors their main utility will likely be as sensors for the extracellular ligand rather than GPCR-specific signaling properties. Furthermore, similarly-designed sensors have also been reported for dopamine (Sun et al., 2018) and norepinephrine (Feng et al., 2019) and bacterial binding domains have been fused to cpGFP for optimized sensors of glutamate (Marvin et al., 2013; Marvin et al., 2018) or GABA (Marvin et al., 2019). While extremely valuable and increasingly used throughout neuroscience, there is a major caveat to the use of fluorescent biosensors in the study of the dynamics of extracellular signals or intracellular second messengers that may limit these tools for precise, quantitative study of GPCR signaling. For example, in the case of calcium sensors, it has been reported that at high expression levels they can act as calcium buffers, making it difficult to precisely interpret the magnitude and length of signaling and requiring appropriate controls (McMahon and Jackson, 2018).
Downstream of second messengers, recent years have seen the development of improved FRET-based sensors for protein effectors including for the GPCR-relevant kinases PKA, PKC, ERK and CaMKII (Greenwald et al., 2018). Such sensors typically either monitor the activation-associated conformational changes of the kinase itself or employ a separate construct containing a specific peptide substrate to measure enzymatic activity of the kinase. For example, a large body of work has developed “Camui” sensors of CaMKII activation based on the introduction of fluorescent proteins to the N- and C-termini to report on the Ca2+/Calmodulin-associated conformational changes of the CaMKII holoenzyme (Takao et al., 2005) (Kwok et al., 2008; Lam et al., 2012). Camui has been used to assess the activation kinetics and spatial spread of CaMKII at single dendritic spines (Chang et al., 2017; Lee et al., 2009). However, Camui highlights one of the shortcomings of conformational biosensors in that the precise conformational changes being sensed are often unclear given the complex tertiary and quaternary structural rearrangements that are known to take place upon CaMKII activation (Chao et al., 2011; Stratton et al., 2014). In addition, approaches such as this also require over-expression of an active enzyme, which alters the endogenous protein concentration and can lead to non-physiological effects.
Using the enzymatic activity-based strategy, a series of FRET-based sensors, termed A kinase activity reporters (“AKAR”), have been developed to monitor PKA activity (Allen and Zhang, 2006; Barbagallo et al., 2016; Chen et al., 2014; Demeautis et al., 2017; Mo et al., 2017; Tang and Yasuda, 2017). An example is AKAR3, which consists of a truncated CFP as the donor fluorophore, a phosphopeptide binding domain (FHA), a consensus region of PKA substrates, and a Venus as the acceptor fluorophore. Activated PKA phosphorylates the consensus region, which then can bind to FHA, bringing CFP and Venus in close proximity results in higher FRET (Figure 4H). Using this approach combined with 2-photon lifetime fluorescence lifetime measurements Chen et al (Chen et al., 2014 )observed increased PKA activity in hippocampal CA1 neurons following activation of β2AR. In a more recent study, the same authors used AKAR to find that, unexpectedly, Gq-coupled receptors, including M1 muscarinic receptors, mGluR5 and a Gq-coupled DREADD, can lead to PKA activation in hippocampal neurons (Chen et al., 2017).
The peptide-phosphorylation based strategy has also been applied to develop FRET sensors for the activity of ERK (“EKAR”) (Harvey et al., 2008; Tang and Yasuda, 2017 ), PKC (“CKAR”) (Violin and Newton, 2003), Src (Ouyang et al., 2008; Wang et al., 2005), AKT (Gao and Zhang, 2008) and, most recently, CaMKII (“FRESCA”) (Ardestani et al., 2019). A recent re-design of kinase activity sensors using a cpGFP-based design to enable 1-color sensing of PKA, PKC, ERK or AKT activity with enhanced dynamics range across spectral variants (Mehta et al., 2018) should facilitate multiplexing-based studies and further applications of these sensors in complex systems. Importantly, similarly to overexpression of active enzymes, overexpression of enzyme substrates can alter the cellular signaling landscape and distort the system thus requiring rigorous controls for the application of these sensors.
Given the frequent inability of peptide substrates to distinguish between related kinase subtypes and the difficulty of indirect activity sensors to provide high temporal or spatial resolution information on the enzyme itself, sensors based on the kinase of interest itself remain an important part of the toolkit. For example, Colgan et al developed sensors to distinguish between the activities of three PKC subtypes (PKCα, PKCβ, PKCγ) in dendritic spines (Colgan et al., 2018). Two separate FRET-based sensors were designed for each isozyme; one to monitor enzyme translocation to the surface (ITRACK) and one to monitor enzyme activity based on binding to a substrate (IDOCKS). Remarkably, these tools permitted the tracking of the activities of specific PKC isozymes during plasticity of dendritic spines of CA1 pyramidal neurons in organotypic hippocampal slices where the authors found that PKCα is the only subtype required for structural plasticity.
Crucially, the aforementioned genetically-encoded biosensors can be targeted to measure signaling at specific organelles or discrete sub-regions of the cell, a perspective of increasing importance for understanding GPCR function (Halls and Canals, 2018). For instance, GCaMP variants have been tagged with an endoplasmic reticulum (ER) signal peptide to measure the behavior of axonal ER calcium during evoked synaptic activity (de Juan-Sanz et al., 2017) and have been targeted to the nucleus to reveal GPCR-induced calcium responses (Vincent et al., 2016). Similarly, targeting of ERK activity sensors to the cytosol or the nucleus revealed bias between different mu-opioid receptor agonists in their potential to produce transient versus sustained ERK activation in the cytosol and nucleus (Halls et al., 2016). Ultimately, studies of this nature will provide a crucial means of dissecting the subcellular dynamics of GPCR signaling.
The toolkit described in this review should continue to expand and enable the mechanistic study of GPCR signaling in a variety of contexts ranging from single cells to behaving animals. The continued multiplexing of optical control and optical sensing should facilitate these applications but will require continual engineering to optimize the underlying tools and uncover new design principles. As other techniques that are suited to probe GPCR function with high precision continue to advance, such as cryo-electron microscopy (Cheng, 2018), gene-editing (Gao et al., 2019; Nishiyama et al., 2017), tissue and in vivo imaging (Alon et al., 2019; Yang and Yuste, 2017), proteomics (Han et al., 2018; Lobingier et al., 2017; Paek et al., 2017) and super-resolution imaging (von Diezmann et al., 2017), the number of creative applications which require the high precision of optogenetic tools should only increase and allow for new aspects of GPCR biology to be revealed.
Acknowledgements:
The authors thank David Simon and Margaret Stratton for helpful feedback. NA is supported by an NSF Graduate Research Fellowship and JL is supported by an R35 grant (1 R35 GM124731) from NIGMS and the Rohr Family Research Scholar Award.
References:
- Agnetta L, Kauk M, Canizal MCA, Messerer R, Holzgrabe U, Hoffmann C, and Decker M (2017). A Photoswitchable Dualsteric Ligand Controlling Receptor Efficacy. Angew Chem Int Ed Engl 56, 7282–7287. [DOI] [PubMed] [Google Scholar]
- Airan RD, Thompson KR, Fenno LE, Bernstein H, and Deisseroth K (2009). Temporally precise in vivo control of intracellular signalling. Nature 458, 1025–1029. [DOI] [PubMed] [Google Scholar]
- Albizu L, Cottet M, Kralikova M, Stoev S, Seyer R, Brabet I, Roux T, Bazin H, Bourrier E, Lamarque L, et al. (2010). Time-resolved FRET between GPCR ligands reveals oligomers in native tissues. Nat Chem Biol 6, 587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MD, and Zhang J (2006). Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochem Biophys Res Commun 348, 716–721. [DOI] [PubMed] [Google Scholar]
- Alon S, Huynh GH, and Boyden ES (2019). Expansion microscopy: enabling single cell analysis in intact biological systems. FEBS J 286, 1482–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardestani G, West MC, Maresca TJ, Fissore RA, and Stratton MM (2019). FRET-based sensor for CaMKII activity (FRESCA): A useful tool for assessing CaMKII activity in response to Ca(2+) oscillations in live cells. J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailes HJ, Zhuang LY, and Lucas RJ (2012). Reproducible and sustained regulation of Galphas signalling using a metazoan opsin as an optogenetic tool. PLoS One 7, e30774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banghart M, Borges K, Isacoff E, Trauner D, and Kramer RH (2004). Light-activated ion channels for remote control of neuronal firing. Nat Neurosci 7, 1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banghart MR, He XJ, and Sabatini BL (2018). A Caged Enkephalin Optimized for Simultaneously Probing Mu and Delta Opioid Receptors. ACS Chem Neurosci 9, 684–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banghart MR, and Sabatini BL (2012). Photoactivatable neuropeptides for spatiotemporally precise delivery of opioids in neural tissue. Neuron 73, 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banghart MR, Williams JT, Shah RC, Lavis LD, and Sabatini BL (2013). Caged naloxone reveals opioid signaling deactivation kinetics. Mol Pharmacol 84, 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbagallo F, Xu B, Reddy GR, West T, Wang Q, Fu Q, Li M, Shi Q, Ginsburg KS, Ferrier W, et al. (2016). Genetically Encoded Biosensors Reveal PKA Hyperphosphorylation on the Myofilaments in Rabbit Heart Failure. Circ Res 119, 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber DM, Schonberger M, Burgstaller J, Levitz J, Weaver CD, Isacoff EY, Baier H, and Trauner D (2016). Optical control of neuronal activity using a light-operated GIRK channel opener (LOGO). Chem Sci 7, 2347–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnea G, Strapps W, Herrada G, Berman Y, Ong J, Kloss B, Axel R, and Lee KJ (2008). The genetic design of signaling cascades to record receptor activation. Proc Natl Acad Sci U S A 105, 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beharry AA, and Woolley GA (2011). Azobenzene photoswitches for biomolecules. Chem Soc Rev 40, 4422–4437. [DOI] [PubMed] [Google Scholar]
- Bellmann D, Richardt A, Freyberger R, Nuwal N, Schwarzel M, Fiala A, and Stortkuhl KF (2010). Optogenetically Induced Olfactory Stimulation in Drosophila Larvae Reveals the Neuronal Basis of Odor-Aversion behavior. Front Behav Neurosci 4, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MH, Holt A, Levitz J, Broichhagen J, Gaub BM, Visel M, Stanley C, Aghi K, Kim YJ, Cao K, et al. (2017). Restoration of patterned vision with an engineered photoactivatable G protein-coupled receptor. Nat Commun 8, 1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonger KM, Rakhit R, Payumo AY, Chen JK, and Wandless TJ (2014). General method for regulating protein stability with light. ACS Chem Biol 9, 111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broichhagen J, Damijonaitis A, Levitz J, Sokol KR, Leippe P, Konrad D, Isacoff EY, and Trauner D (2015a). Orthogonal Optical Control of a G Protein-Coupled Receptor with a SNAP-Tethered Photochromic Ligand. ACS Cent Sci 1, 383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broichhagen J, Podewin T, Meyer-Berg H, von Ohlen Y, Johnston NR, Jones BJ, Bloom SR, Rutter GA, Hoffmann-Roder A, Hodson DJ, et al. (2015b). Optical Control of Insulin Secretion Using an Incretin Switch. Angew Chem Int Ed Engl 54, 15565–15569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, and Schaffer DV (2013). Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods 10, 249–252. [DOI] [PubMed] [Google Scholar]
- Bunemann M, Frank M, and Lohse MJ (2003). Gi protein activation in intact cells involves subunit rearrangement rather than dissociation. Proc Natl Acad Sci U S A 100, 16077–16082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill TJ 3rd, Thomsen AR, Tarrasch JT, Plouffe B, Nguyen AH, Yang F, Huang LY, Kahsai AW, Bassoni DL, Gavino BJ, et al. (2017). Distinct conformations of GPCR-beta-arrestin complexes mediate desensitization, signaling, and endocytosis. Proc Natl Acad Sci U S A 114, 2562–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway EM, and Katz LC (1993). Photostimulation using caged glutamate reveals functional circuitry in living brain slices. Proc Natl Acad Sci U S A 90, 7661–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll EC, Berlin S, Levitz J, Kienzler MA, Yuan Z, Madsen D, Larsen DS, and Isacoff EY (2015). Two-photon brightness of azobenzene photoswitches designed for glutamate receptor optogenetics. Proc Natl Acad Sci U S A 112, E776–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JY, Parra-Bueno P, Laviv T, Szatmari EM, Lee SR, and Yasuda R (2017). CaMKII Autophosphorylation Is Necessary for Optimal Integration of Ca(2+) Signals during LTP Induction, but Not Maintenance. Neuron 94, 800–808 e804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KY, Woo D, Jung H, Lee S, Kim S, Won J, Kyung T, Park H, Kim N, Yang HW, et al. (2014). Light-inducible receptor tyrosine kinases that regulate neurotrophin signalling. Nat Commun 5, 4057. [DOI] [PubMed] [Google Scholar]
- Chao LH, Stratton MM, Lee IH, Rosenberg OS, Levitz J, Mandell DJ, Kortemme T, Groves JT, Schulman H, and Kuriyan J (2011). A mechanism for tunable autoinhibition in the structure of a human Ca2+/calmodulin- dependent kinase II holoenzyme. Cell 146, 732–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che T, Majumdar S, Zaidi SA, Ondachi P, McCorvy JD, Wang S, Mosier PD, Uprety R, Vardy E, Krumm BE, et al. (2018). Structure of the Nanobody-Stabilized Active State of the Kappa Opioid Receptor. Cell 172, 55–67 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Gibson ES, and Kennedy MJ (2013a). A light-triggered protein secretion system. J Cell Biol 201, 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Granger AJ, Tran T, Saulnier JL, Kirkwood A, and Sabatini BL (2017). Endogenous Galphaq-Coupled Neuromodulator Receptors Activate Protein Kinase A. Neuron 96, 1070–1083 e1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Saulnier JL, Yellen G, and Sabatini BL (2014). A PKA activity sensor for quantitative analysis of endogenous GPCR signaling via 2-photon FRET-FLIM imaging. Front Pharmacol 5, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Oldfield S, Butcher AJ, Tobin AB, Saxena K, Gurevich VV, Benovic JL, Henderson G, and Kelly E (2013b). Identification of phosphorylation sites in the COOH-terminal tail of the mu-opioid receptor. J Neurochem 124, 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y (2018). Single-particle cryo-EM-How did it get here and where will it go. Science 361, 876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan LA, Hu M, Misler JA, Parra-Bueno P, Moran CM, Leitges M, and Yasuda R (2018). PKCalpha integrates spatiotemporally distinct Ca(2+) and autocrine BDNF signaling to facilitate synaptic plasticity. Nat Neurosci 21, 1027–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagliyan O, and Hahn KM (2019). Controlling protein conformation with light. Curr Opin Struct Biol 57, 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagliyan O, Tarnawski M, Chu PH, Shirvanyants D, Schlichting I, Dokholyan NV, and Hahn KM (2016). Engineering extrinsic disorder to control protein activity in living cells. Science 354, 1441–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Juan-Sanz J, Holt GT, Schreiter ER, de Juan F, Kim DS, and Ryan TA (2017). Axonal Endoplasmic Reticulum Ca(2+) Content Controls Release Probability in CNS Nerve Terminals. Neuron 93, 867–881 e866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries L, Zheng B, Fischer T, Elenko E, and Farquhar MG (2000). The regulator of G protein signaling family. Annu Rev Pharmacol Toxicol 40, 235–271. [DOI] [PubMed] [Google Scholar]
- Demeautis C, Sipieter F, Roul J, Chapuis C, Padilla-Parra S, Riquet FB, and Tramier M (2017). Multiplexing PKA and ERK1&2 kinases FRET biosensors in living cells using single excitation wavelength dual colour FLIM. Sci Rep 7, 41026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWire SM, Ahn S, Lefkowitz RJ, and Shenoy SK (2007). Beta-arrestins and cell signaling. Annu Rev Physiol 69, 483–510. [DOI] [PubMed] [Google Scholar]
- Donthamsetti PC, Broichhagen J, Vyklicky V, Stanley C, Fu Z, Visel M, Levitz JL, Javitch JA, Trauner D, and Isacoff EY (2019). Genetically Targeted Optical Control of an Endogenous G Protein-Coupled Receptor. J Am Chem Soc 141, 11522–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donthamsetti PC, Winter N, Schonberger M, Levitz J, Stanley C, Javitch JA, Isacoff EY, and Trauner D (2017). Optical Control of Dopamine Receptors Using a Photoswitchable Tethered Inverse Agonist. J Am Chem Soc 139, 18522–18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumazane E, Scholler P, Fabre L, Zwier JM, Trinquet E, Pin JP, and Rondard P (2013). Illuminating the activation mechanisms and allosteric properties of metabotropic glutamate receptors. Proc Natl Acad Sci U S A 110, E1416–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumazane E, Scholler P, Zwier JM, Trinquet E, Rondard P, and Pin JP (2011). A new approach to analyze cell surface protein complexes reveals specific heterodimeric metabotropic glutamate receptors. FASEB J 25, 66–77. [DOI] [PubMed] [Google Scholar]
- Eichel K, and von Zastrow M (2018). Subcellular Organization of GPCR Signaling. Trends Pharmacol Sci 39, 200–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandson SC, McMahon C, and Kruse AC (2018). Structural Basis for G Protein-Coupled Receptor Signaling. Annu Rev Biophys. [DOI] [PubMed] [Google Scholar]
- Ernst OP, Gramse V, Kolbe M, Hofmann KP, and Heck M (2007). Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proc Natl Acad Sci U S A 104, 10859–10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrants H, Gutzeit VA, Acosta-Ruiz A, Trauner D, Johnsson K, Levitz J, and Broichhagen J (2018). SNAP-Tagged Nanobodies Enable Reversible Optical Control of a G Protein-Coupled Receptor via a Remotely Tethered Photoswitchable Ligand. ACS Chem Biol 13, 2682–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhang C, Lischinsky JE, Jing M, Zhou J, Wang H, Zhang Y, Dong A, Wu Z, Wu H, et al. (2019). A Genetically Encoded Fluorescent Sensor for Rapid and Specific In Vivo Detection of Norepinephrine. Neuron 102, 745–761 e748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno L, Yizhar O, and Deisseroth K (2011). The development and application of optogenetics. Annu Rev Neurosci 34, 389–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flock T, Hauser AS, Lund N, Gloriam DE, Balaji S, and Babu MM (2017). Selectivity determinants of GPCR-G-protein binding. Nature 545, 317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gales C, Rebois RV, Hogue M, Trieu P, Breit A, Hebert TE, and Bouvier M (2005). Real-time monitoring of receptor and G-protein interactions in living cells. Nat Methods 2, 177–184. [DOI] [PubMed] [Google Scholar]
- Gao X, and Zhang J (2008). Spatiotemporal analysis of differential Akt regulation in plasma membrane microdomains. Mol Biol Cell 19, 4366–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Hisey E, Bradshaw TWA, Erata E, Brown WE, Courtland JL, Uezu A, Xiang Y, Diao Y, and Soderling SH (2019). Plug-and-Play Protein Modification Using Homology-Independent Universal Genome Engineering. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glantz ST, Berlew EE, Jaber Z, Schuster BS, Gardner KH, and Chow BY (2018). Directly light-regulated binding of RGS-LOV photoreceptors to anionic membrane phospholipids. Proc Natl Acad Sci U S A 115, E7720–E7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goglia AG, and Toettcher JE (2019). A bright future: optogenetics to dissect the spatiotemporal control of cell behavior. Curr Opin Chem Biol 48, 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald EC, Mehta S, and Zhang J (2018). Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks. Chem Rev 118, 11707–11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann M, and Kostenis E (2017). Temporal Bias: Time-Encoded Dynamic GPCR Signaling. Trends Pharmacol Sci 38, 1110–1124. [DOI] [PubMed] [Google Scholar]
- Grundmann M, Merten N, Malfacini D, Inoue A, Preis P, Simon K, Ruttiger N, Ziegler N, Benkel T, Schmitt NK, et al. (2018). Lack of beta-arrestin signaling in the absence of active G proteins. Nat Commun 9, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusch M, Schelch K, Riedler R, Reichhart E, Differ C, Berger W, Ingles-Prieto A, and Janovjak H (2014). Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. EMBO J 33, 1713–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetg N, Abdel Aziz S, Holbro N, Turecek R, Rose T, Seddik R, Gassmann M, Moes S, Jenoe P, Oertner TG, et al. (2010). NMDA receptor-dependent GABAB receptor internalization via CaMKII phosphorylation of serine 867 in GABAB1. Proc Natl Acad Sci U S A 107, 13924–13929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati S, Jin H, Masuho I, Orban T, Cai Y, Pardon E, Martemyanov KA, Kiser PD, Stewart PL, Ford CP, et al. (2018). Targeting G protein-coupled receptor signaling at the G protein level with a selective nanobody inhibitor. Nat Commun 9, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, Lammel S, Mirzabekov JJ, Airan RD, Zalocusky KA, et al. (2014). Natural neural projection dynamics underlying social behavior. Cell 157, 1535–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzeit VA, Thibado J, Stor DS, Zhou Z, Blanchard SC, Andersen OS, and Levitz J (2019). Conformational dynamics between transmembrane domains and allosteric modulation of a metabotropic glutamate receptor. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackley CR, Mazzoni EO, and Blau J (2018). cAMPr: A single-wavelength fluorescent sensor for cyclic AMP. Sci Signal 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halls ML, and Canals M (2018). Genetically Encoded FRET Biosensors to Illuminate Compartmentalised GPCR Signalling. Trends Pharmacol Sci 39, 148–157. [DOI] [PubMed] [Google Scholar]
- Halls ML, Yeatman HR, Nowell CJ, Thompson GL, Gondin AB, Civciristov S, Bunnett NW, Lambert NA, Poole DP, and Canals M (2016). Plasma membrane localization of the mu-opioid receptor controls spatiotemporal signaling. Sci Signal 9, ra16. [DOI] [PubMed] [Google Scholar]
- Han S, Li J, and Ting AY (2018). Proximity labeling: spatially resolved proteomic mapping for neurobiology. Curr Opin Neurobiol 50, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannanta-Anan P, and Chow BY (2018). Optogenetic Inhibition of Galphaq Protein Signaling Reduces Calcium Oscillation Stochasticity. ACS Synth Biol 7, 1488–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SM, Neil LC, and Gardner KH (2003). Structural basis of a phototropin light switch. Science 301, 1541–1544. [DOI] [PubMed] [Google Scholar]
- Harvey CD, Ehrhardt AG, Cellurale C, Zhong H, Yasuda R, Davis RJ, and Svoboda K (2008). A genetically encoded fluorescent sensor of ERK activity. Proc Natl Acad Sci U S A 105, 19264–19269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser AS, Attwood MM, Rask-Andersen M, Schioth HB, and Gloriam DE (2017). Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov 16, 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein P, Frank M, Hoffmann C, Lohse MJ, and Bunemann M (2005). Dynamics of receptor/G protein coupling in living cells. EMBO J 24, 4106–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein P, Rochais F, Hoffmann C, Dorsch S, Nikolaev VO, Engelhardt S, Berlot CH, Lohse MJ, and Bunemann M (2006). Gs activation is time-limiting in initiating receptor-mediated signaling. J Biol Chem 281, 33345–33351. [DOI] [PubMed] [Google Scholar]
- Heukers R, De Groof TWM, and Smit MJ (2019). Nanobodies detecting and modulating GPCRs outside in and inside out. Curr Opin Cell Biol 57, 115–122. [DOI] [PubMed] [Google Scholar]
- Hu J, Stern M, Gimenez LE, Wanka L, Zhu L, Rossi M, Meister J, Inoue A, Beck-Sickinger AG, Gurevich VV, et al. (2016). A G Protein-biased Designer G Protein-coupled Receptor Useful for Studying the Physiological Relevance of Gq/11-dependent Signaling Pathways. J Biol Chem 291, 7809–7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull K, Morstein J, and Trauner D (2018). In Vivo Photopharmacology. Chem Rev 118, 10710–10747. [DOI] [PubMed] [Google Scholar]
- Idevall-Hagren O, Dickson EJ, Hille B, Toomre DK, and De Camilli P (2012). Optogenetic control of phosphoinositide metabolism. Proc Natl Acad Sci U S A 109, E2316–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irannejad R, Tomshine JC, Tomshine JR, Chevalier M, Mahoney JP, Steyaert J, Rasmussen SG, Sunahara RK, El-Samad H, Huang B, et al. (2013). Conformational biosensors reveal GPCR signalling from endosomes. Nature 495, 534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseki M, Matsunaga S, Murakami A, Ohno K, Shiga K, Yoshida K, Sugai M, Takahashi T, Hori T, and Watanabe M (2002). A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature 415, 1047–1051. [DOI] [PubMed] [Google Scholar]
- Ishida A, Shigeri Y, Tatsu Y, Uegaki K, Kameshita I, Okuno S, Kitani T, Yumoto N, and Fujisawa H (1998). Critical amino acid residues of AIP, a highly specific inhibitory peptide of calmodulin-dependent protein kinase II. FEBS Lett 427, 115–118. [DOI] [PubMed] [Google Scholar]
- Jin DZ, Guo ML, Xue B, Fibuch EE, Choe ES, Mao LM, and Wang JQ (2013). Phosphorylation and feedback regulation of metabotropic glutamate receptor 1 by calcium/calmodulin-dependent protein kinase II. J Neurosci 33, 3402–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainrath S, Stadler M, Reichhart E, Distel M, and Janovjak H (2017). Green-Light-Induced Inactivation of Receptor Signaling Using Cobalamin-Binding Domains. Angew Chem Int Ed Engl 56, 4608–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunarathne WK, Giri L, Kalyanaraman V, and Gautam N (2013). Optically triggering spatiotemporally confined GPCR activity in a cell and programming neurite initiation and extension. Proc Natl Acad Sci U S A 110, E1565–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura Y, Kubota H, Kunida K, Kanno A, Kuroda S, and Ozawa T (2015). An optogenetic system for interrogating the temporal dynamics of Akt. Sci Rep 5, 14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, and Tucker CL (2010). Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods 7, 973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SM, Sleno R, Gora S, Zylbergold P, Laverdure JP, Labbe JC, Miller GJ, and Hebert TE (2013). The expanding roles of Gbetagamma subunits in G protein-coupled receptor signaling and drug action. Pharmacol Rev 65, 545–577. [DOI] [PubMed] [Google Scholar]
- Kim JM, Hwa J, Garriga P, Reeves PJ, RajBhandary UL, and Khorana HG (2005). Light-driven activation of beta 2-adrenergic receptor signaling by a chimeric rhodopsin containing the beta 2-adrenergic receptor cytoplasmic loops. Biochemistry 44, 2284–2292. [DOI] [PubMed] [Google Scholar]
- Kolch W (2005). Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol 6, 827–837. [DOI] [PubMed] [Google Scholar]
- Kroeze WK, Sassano MF, Huang XP, Lansu K, McCorvy JD, Giguere PM, Sciaky N, and Roth BL (2015). PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat Struct Mol Biol 22, 362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok S, Lee C, Sanchez SA, Hazlett TL, Gratton E, and Hayashi Y (2008). Genetically encoded probe for fluorescence lifetime imaging of CaMKII activity. Biochem Biophys Res Commun 369, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AJ, St-Pierre F, Gong Y, Marshall JD, Cranfill PJ, Baird MA, McKeown MR, Wiedenmann J, Davidson MW, Schnitzer MJ, et al. (2012). Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods 9, 1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecat-Guillet N, Monnier C, Rovira X, Kniazeff J, Lamarque L, Zwier JM, Trinquet E, Pin JP, and Rondard P (2017). FRET-Based Sensors Unravel Activation and Allosteric Modulation of the GABAB Receptor. Cell Chem Biol 24, 360–370. [DOI] [PubMed] [Google Scholar]
- Lee D, Creed M, Jung K, Stefanelli T, Wendler DJ, Oh WC, Mignocchi NL, Luscher C, and Kwon HB (2017). Temporally precise labeling and control of neuromodulatory circuits in the mammalian brain. Nat Methods 14, 495–503. [DOI] [PubMed] [Google Scholar]
- Lee MH, Appleton KM, Strungs EG, Kwon JY, Morinelli TA, Peterson YK, Laporte SA, and Luttrell LM (2016). The conformational signature of beta-arrestin2 predicts its trafficking and signalling functions. Nature 531, 665–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Escobedo-Lozoya Y, Szatmari EM, and Yasuda R (2009). Activation of CaMKII in single dendritic spines during long-term potentiation. Nature 458, 299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold AV, Chernov KG, and Verkhusha VV (2018). Optogenetically controlled protein kinases for regulation of cellular signaling. Chem Soc Rev 47, 2454–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz J, Broichhagen J, Leippe P, Konrad D, Trauner D, and Isacoff EY (2017). Dual optical control and mechanistic insights into photoswitchable group II and III metabotropic glutamate receptors. Proc Natl Acad Sci U S A 114, E3546–E3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz J, Habrian C, Bharill S, Fu Z, Vafabakhsh R, and Isacoff EY (2016). Mechanism of Assembly and Cooperativity of Homomeric and Heteromeric Metabotropic Glutamate Receptors. Neuron 92, 143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz J, Pantoja C, Gaub B, Janovjak H, Reiner A, Hoagland A, Schoppik D, Kane B, Stawski P, Schier AF, et al. (2013). Optical control of metabotropic glutamate receptors. Nat Neurosci 16, 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A, Weiner OD, Lim WA, and Voigt CA (2009). Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gutierrez DV, Hanson MG, Han J, Mark MD, Chiel H, Hegemann P, Landmesser LT, and Herlitze S (2005). Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci U S A 102, 17816–17821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, and Lin C (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539. [DOI] [PubMed] [Google Scholar]
- Liu X, Xu X, Hilger D, Aschauer P, Tiemann JKS, Du Y, Liu H, Hirata K, Sun X, Guixa-Gonzalez R, et al. (2019). Structural Insights into the Process of GPCR-G Protein Complex Formation. Cell 177, 1243–1251 e1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XY, Mao LM, Zhang GC, Papasian CJ, Fibuch EE, Lan HX, Zhou HF, Xu M, and Wang JQ (2009). Activity-dependent modulation of limbic dopamine D3 receptors by CaMKII. Neuron 61, 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobingier BT, Huttenhain R, Eichel K, Miller KB, Ting AY, von Zastrow M, and Krogan NJ (2017). An Approach to Spatiotemporally Resolve Protein Interaction Networks in Living Cells. Cell 169, 350–360 e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MJ, Nuber S, and Hoffmann C (2012). Fluorescence/bioluminescence resonance energy transfer techniques to study G-protein-coupled receptor activation and signaling. Pharmacol Rev 64, 299–336. [DOI] [PubMed] [Google Scholar]
- Luttrell LM, Ferguson SS, Daaka Y, Miller WE, Maudsley S, Della Rocca GJ, Lin F, Kawakatsu H, Owada K, Luttrell DK, et al. (1999). Beta-arrestin-dependent formation of beta2 adrenergic receptor-Src protein kinase complexes. Science 283, 655–661. [DOI] [PubMed] [Google Scholar]
- Luttrell LM, Roudabush FL, Choy EW, Miller WE, Field ME, Pierce KL, and Lefkowitz RJ (2001). Activation and targeting of extracellular signal-regulated kinases by beta-arrestin scaffolds. Proc Natl Acad Sci U S A 98, 2449–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrell LM, Wang J, Plouffe B, Smith JS, Yamani L, Kaur S, Jean-Charles PY, Gauthier C, Lee MH, Pani B, et al. (2018). Manifold roles of beta-arrestins in GPCR signaling elucidated with siRNA and CRISPR/Cas9. Sci Signal 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowka P, Bruegmann T, Dusend V, Malan D, Beiert T, Hesse M, Fleischmann BK, and Sasse P (2019). Optogenetic stimulation of Gs-signaling in the heart with high spatio-temporal precision. Nat Commun 10, 1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglik A, Kobilka BK, and Steyaert J (2017). Nanobodies to Study G Protein-Coupled Receptor Structure and Function. Annu Rev Pharmacol Toxicol 57, 19–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, and Toker A (2017). AKT/PKB Signaling: Navigating the Network. Cell 169, 381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Borghuis BG, Tian L, Cichon J, Harnett MT, Akerboom J, Gordus A, Renninger SL, Chen TW, Bargmann CI, et al. (2013). An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat Methods 10, 162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Scholl B, Wilson DE, Podgorski K, Kazemipour A, Muller JA, Schoch S, Quiroz FJU, Rebola N, Bao H, et al. (2018). Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR. Nat Methods 15, 936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Shimoda Y, Magloire V, Leite M, Kawashima T, Jensen TP, Kolb I, Knott EL, Novak O, Podgorski K, et al. (2019). A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nat Methods. [DOI] [PubMed] [Google Scholar]
- Masseck OA, Spoida K, Dalkara D, Maejima T, Rubelowski JM, Wallhorn L, Deneris ES, and Herlitze S (2014). Vertebrate cone opsins enable sustained and highly sensitive rapid control of Gi/o signaling in anxiety circuitry. Neuron 81, 1263–1273. [DOI] [PubMed] [Google Scholar]
- McMahon C, Baier AS, Pascolutti R, Wegrecki M, Zheng S, Ong JX, Erlandson SC, Hilger D, Rasmussen SGF, Ring AM, et al. (2018). Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat Struct Mol Biol 25, 289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SM, and Jackson MB (2018). An Inconvenient Truth: Calcium Sensors Are Calcium Buffers. Trends Neurosci 41, 880–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Zhang Y, Roth RH, Zhang JF, Mo A, Tenner B, Huganir RL, and Zhang J (2018). Single-fluorophore biosensors for sensitive and multiplexed detection of signalling activities. Nat Cell Biol 20, 1215–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melero-Fernandez de Mera RM, Li LL, Popinigis A, Cisek K, Tuittila M, Yadav L, Serva A, and Courtney MJ (2017). A simple optogenetic MAPK inhibitor design reveals resonance between transcription-regulating circuitry and temporally-encoded inputs. Nat Commun 8, 15017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A (2011). Development of probes for cellular functions using fluorescent proteins and fluorescence resonance energy transfer. Annu Rev Biochem 80, 357–373. [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, and Tsien RY (1997). Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388, 882–887. [DOI] [PubMed] [Google Scholar]
- Mo GC, Ross B, Hertel F, Manna P, Yang X, Greenwald E, Booth C, Plummer AM, Tenner B, Chen Z, et al. (2017). Genetically encoded biosensors for visualizing live-cell biochemical activity at super-resolution. Nat Methods 14, 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockett BG, Guevremont D, Wutte M, Hulme SR, Williams JM, and Abraham WC (2011). Calcium/calmodulin-dependent protein kinase II mediates group I metabotropic glutamate receptor-dependent protein synthesis and long-term depression in rat hippocampus. J Neurosci 31, 7380–7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohebi A, Pettibone JR, Hamid AA, Wong JT, Vinson LT, Patriarchi T, Tian L, Kennedy RT, and Berke JD (2019). Dissociable dopamine dynamics for learning and motivation. Nature 570, 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, Lynch KW, and Gardner KH (2014). An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol 10, 196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi H, Shin ME, Parra-Bueno P, Szatmari EM, Shibata ACE, and Yasuda R (2017). Kinetics of Endogenous CaMKII Required for Synaptic Plasticity Revealed by Optogenetic Kinase Inhibitor. Neuron 94, 37–47 e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahama T, Suzuki T, Yoshikawa S, and Iseki M (2007). Functional transplant of photoactivated adenylyl cyclase (PAC) into Aplysia sensory neurons. Neurosci Res 59, 81–88. [DOI] [PubMed] [Google Scholar]
- Nakai J, Ohkura M, and Imoto K (2001). A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat Biotechnol 19, 137–141. [DOI] [PubMed] [Google Scholar]
- Nakajima K, and Wess J (2012). Design and functional characterization of a novel, arrestin-biased designer G protein-coupled receptor. Mol Pharmacol 82, 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niopek D, Benzinger D, Roensch J, Draebing T, Wehler P, Eils R, and Di Ventura B (2014). Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells. Nat Commun 5, 4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama J, Mikuni T, and Yasuda R (2017). Virus-Mediated Genome Editing via Homology-Directed Repair in Mitotic and Postmitotic Cells in Mammalian Brain. Neuron 96, 755–768 e755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobles M, Benians A, and Tinker A (2005). Heterotrimeric G proteins precouple with G protein-coupled receptors in living cells. Proc Natl Acad Sci U S A 102, 18706–18711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuber S, Zabel U, Lorenz K, Nuber A, Milligan G, Tobin AB, Lohse MJ, and Hoffmann C (2016). beta-Arrestin biosensors reveal a rapid, receptor-dependent activation/deactivation cycle. Nature 531, 661–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hayre M, Eichel K, Avino S, Zhao X, Steffen DJ, Feng X, Kawakami K, Aoki J, Messer K, Sunahara R, et al. (2017). Genetic evidence that beta-arrestins are dispensable for the initiation of beta2-adrenergic receptor signaling to ERK. Sci Signal 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill PR, and Gautam N (2014). Subcellular optogenetic inhibition of G proteins generates signaling gradients and cell migration. Mol Biol Cell 25, 2305–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki M, Sugiyama K, Kawai F, Tanaka H, Nihei Y, Unzai S, Takebe M, Matsunaga S, Adachi S, Shibayama N, et al. (2016). Structural insight into photoactivation of an adenylate cyclase from a photosynthetic cyanobacterium. Proc Natl Acad Sci U S A 113, 6659–6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong Q, Guo S, Duan L, Zhang K, Collier EA, and Cui B (2016). The Timing of Raf/ERK and AKT Activation in Protecting PC12 Cells against Oxidative Stress. PLoS One 11, e0153487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang M, Sun J, Chien S, and Wang Y (2008). Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. Proc Natl Acad Sci U S A 105, 14353–14358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek J, Kalocsay M, Staus DP, Wingler L, Pascolutti R, Paulo JA, Gygi SP, and Kruse AC (2017). Multidimensional Tracking of GPCR Signaling via Peroxidase-Catalyzed Proximity Labeling. Cell 169, 338–349 e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K (2006). G protein-coupled receptor rhodopsin. Annu Rev Biochem 75, 743–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P, Ellis-Davies GCR, and Mourot A (2019). Optical control of neuronal ion channels and receptors. Nat Rev Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramonov VM, Mamaeva V, Sahlgren C, and Rivero-Muller A (2015). Genetically-encoded tools for cAMP probing and modulation in living systems. Front Pharmacol 6, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N, and Gold MG (2015). The genetically encoded tool set for investigating cAMP: more than the sum of its parts. Front Pharmacol 6, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarchi T, Cho JR, Merten K, Howe MW, Marley A, Xiong WH, Folk RW, Broussard GJ, Liang R, Jang MJ, et al. (2018). Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, and Lefkowitz RJ (2002). Seven-transmembrane receptors. Nat Rev Mol Cell Biol 3, 639–650. [DOI] [PubMed] [Google Scholar]
- Pittolo S, Gomez-Santacana X, Eckelt K, Rovira X, Dalton J, Goudet C, Pin JP, Llobet A, Giraldo J, Llebaria A, et al. (2014). An allosteric modulator to control endogenous G protein-coupled receptors with light. Nat Chem Biol 10, 813–815. [DOI] [PubMed] [Google Scholar]
- Platisa J, and Pieribone VA (2018). Genetically encoded fluorescent voltage indicators: are we there yet? Curr Opin Neurobiol 50, 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Kumbalasiri T, Carlson SM, Wong KY, Krishna V, Provencio I, and Berson DM (2005). Induction of photosensitivity by heterologous expression of melanopsin. Nature 433, 745–749. [DOI] [PubMed] [Google Scholar]
- Rajagopal S, and Shenoy SK (2018). GPCR desensitization: Acute and prolonged phases. Cell Signal 41, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, et al. (2011a). Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature 469, 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. (2011b). Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature 477, 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichhart E, Ingles-Prieto A, Tichy AM, McKenzie C, and Janovjak H (2016). A Phytochrome Sensory Domain Permits Receptor Activation by Red Light. Angew Chem Int Ed Engl 55, 6339–6342. [DOI] [PubMed] [Google Scholar]
- Reiner A, Levitz J, and Isacoff EY (2015). Controlling ionotropic and metabotropic glutamate receptors with light: principles and potential. Curr Opin Pharmacol 20, 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renicke C, Schuster D, Usherenko S, Essen LO, and Taxis C (2013). A LOV2 domain-based optogenetic tool to control protein degradation and cellular function. Chem Biol 20, 619–626. [DOI] [PubMed] [Google Scholar]
- Repina NA, Rosenbloom A, Mukherjee A, Schaffer DV, and Kane RS (2017). At Light Speed: Advances in Optogenetic Systems for Regulating Cell Signaling and Behavior. Annu Rev Chem Biomol Eng 8, 13–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoz G, Levitz J, Kramer RH, and Isacoff EY (2012). Optical control of endogenous proteins with a photoswitchable conditional subunit reveals a role for TREK1 in GABA(B) signaling. Neuron 74, 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholler P, Nevoltris D, de Bundel D, Bossi S, Moreno-Delgado D, Rovira X, Moller TC, El Moustaine D, Mathieu M, Blanc E, et al. (2017). Allosteric nanobodies uncover a role of hippocampal mGlu2 receptor homodimers in contextual fear consolidation. Nat Commun 8, 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonberger M, and Trauner D (2014). A photochromic agonist for mu-opioid receptors. Angew Chem Int Ed Engl 53, 3264–3267. [DOI] [PubMed] [Google Scholar]
- Schroder-Lang S, Schwarzel M, Seifert R, Strunker T, Kateriya S, Looser J, Watanabe M, Kaupp UB, Hegemann P, and Nagel G (2007). Fast manipulation of cellular cAMP level by light in vivo. Nat Methods 4, 39–42. [DOI] [PubMed] [Google Scholar]
- Shields BC, Kahuno E, Kim C, Apostolides PF, Brown J, Lindo S, Mensh BD, Dudman JT, Lavis LD, and Tadross MR (2017). Deconstructing behavioral neuropharmacology with cellular specificity. Science 356. [DOI] [PubMed] [Google Scholar]
- Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, and Brangwynne CP (2017). Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell 168, 159–171 e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siuda ER, Copits BA, Schmidt MJ, Baird MA, Al-Hasani R, Planer WJ, Funderburk SC, McCall JG, Gereau R.W.t., and Bruchas MR (2015). Spatiotemporal control of opioid signaling and behavior. Neuron 86, 923–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Lefkowitz RJ, and Rajagopal S (2018). Biased signalling: from simple switches to allosteric microprocessors. Nat Rev Drug Discov 17, 243–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoida K, Eickelbeck D, Karapinar R, Eckhardt T, Mark MD, Jancke D, Ehinger BV, Konig P, Dalkara D, Herlitze S, et al. (2016). Melanopsin Variants as Intrinsic Optogenetic On and Off Switches for Transient versus Sustained Activation of G Protein Pathways. Curr Biol 26, 1206–1212. [DOI] [PubMed] [Google Scholar]
- Stachniak TJ, Ghosh A, and Sternson SM (2014). Chemogenetic synaptic silencing of neural circuits localizes a hypothalamus-->midbrain pathway for feeding behavior. Neuron 82, 797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierl M, Stumpf P, Udwari D, Gueta R, Hagedorn R, Losi A, Gartner W, Petereit L, Efetova M, Schwarzel M, et al. (2011). Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J Biol Chem 286, 1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton M, Lee IH, Bhattacharyya M, Christensen SM, Chao LH, Schulman H, Groves JT, and Kuriyan J (2014). Activation-triggered subunit exchange between CaMKII holoenzymes facilitates the spread of kinase activity. Elife 3, e01610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strungs EG, and Luttrell LM (2014). Arrestin-dependent activation of ERK and Src family kinases. Handb Exp Pharmacol 219, 225–257. [DOI] [PubMed] [Google Scholar]
- Sun F, Zeng J, Jing M, Zhou J, Feng J, Owen SF, Luo Y, Li F, Wang H, Yamaguchi T, et al. (2018). A Genetically Encoded Fluorescent Sensor Enables Rapid and Specific Detection of Dopamine in Flies, Fish, and Mice. Cell 174, 481–496 e419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungkaworn T, Jobin ML, Burnecki K, Weron A, Lohse MJ, and Calebiro D (2017). Single-molecule imaging reveals receptor-G protein interactions at cell surface hot spots. Nature 550, 543–547. [DOI] [PubMed] [Google Scholar]
- Takao K, Okamoto K, Nakagawa T, Neve RL, Nagai T, Miyawaki A, Hashikawa T, Kobayashi S, and Hayashi Y (2005). Visualization of synaptic Ca2+ /calmodulin-dependent protein kinase II activity in living neurons. J Neurosci 25, 3107–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenouchi O, Yoshimura H, and Ozawa T (2018). Unique Roles of beta-Arrestin in GPCR Trafficking Revealed by Photoinducible Dimerizers. Sci Rep 8, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, and Yasuda R (2017). Imaging ERK and PKA Activation in Single Dendritic Spines during Structural Plasticity. Neuron 93, 1315–1324 e1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, Kennedy MJ, and Tucker CL (2014). An optimized optogenetic clustering tool for probing protein interaction and function. Nat Commun 5, 4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ARB, Plouffe B, Cahill TJ 3rd, Shukla AK, Tarrasch JT, Dosey AM, Kahsai AW, Strachan RT, Pani B, Mahoney JP, et al. (2016). GPCR-G Protein-beta-Arrestin Super-Complex Mediates Sustained G Protein Signaling. Cell 166, 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Furstenberg A, and Huber T (2017). Labeling and Single-Molecule Methods To Monitor G Protein-Coupled Receptor Dynamics. Chem Rev 117, 186–245. [DOI] [PubMed] [Google Scholar]
- Tichy AM, Gerrard EJ, Sexton PM, and Janovjak H (2019). Light-activated chimeric GPCRs: limitations and opportunities. Curr Opin Struct Biol 57, 196–203. [DOI] [PubMed] [Google Scholar]
- Toettcher JE, Gong D, Lim WA, and Weiner OD (2011). Light-based feedback for controlling intracellular signaling dynamics. Nat Methods 8, 837–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher JE, Weiner OD, and Lim WA (2013). Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touhara KK, and MacKinnon R (2018). Molecular basis of signaling specificity between GIRK channels and GPCRs. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YH, Essig S, James JR, Lang K, and Chin JW (2015). Selective, rapid and optically switchable regulation of protein function in live mammalian cells. Nat Chem 7, 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetanova NG, and von Zastrow M (2014). Spatial encoding of cyclic AMP signaling specificity by GPCR endocytosis. Nat Chem Biol 10, 1061–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban DJ, and Roth BL (2015). DREADDs (designer receptors exclusively activated by designer drugs): chemogenetic tools with therapeutic utility. Annu Rev Pharmacol Toxicol 55, 399–417. [DOI] [PubMed] [Google Scholar]
- Vafabakhsh R, Levitz J, and Isacoff EY (2015). Conformational dynamics of a class C G-protein-coupled receptor. Nature 524, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bergeijk P, Adrian M, Hoogenraad CC, and Kapitein LC (2015). Optogenetic control of organelle transport and positioning. Nature 518, 111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy E, Robinson JE, Li C, Olsen RHJ, DiBerto JF, Giguere PM, Sassano FM, Huang XP, Zhu H, Urban DJ, et al. (2015). A New DREADD Facilitates the Multiplexed Chemogenetic Interrogation of Behavior. Neuron 86, 936–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, and Babu MM (2013). Molecular signatures of G-protein-coupled receptors. Nature 494, 185–194. [DOI] [PubMed] [Google Scholar]
- Vilardaga JP, Bunemann M, Krasel C, Castro M, and Lohse MJ (2003). Measurement of the millisecond activation switch of G protein-coupled receptors in living cells. Nat Biotechnol 21, 807–812. [DOI] [PubMed] [Google Scholar]
- Vilardaga JP, Steinmeyer R, Harms GS, and Lohse MJ (2005). Molecular basis of inverse agonism in a G protein-coupled receptor. Nat Chem Biol 1, 25–28. [DOI] [PubMed] [Google Scholar]
- Vincent K, Cornea VM, Jong YI, Laferriere A, Kumar N, Mickeviciute A, Fung JST, Bandegi P, Ribeiro-da-Silva A, O’Malley KL, et al. (2016). Intracellular mGluR5 plays a critical role in neuropathic pain. Nat Commun 7, 10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violin JD, and Newton AC (2003). Pathway illuminated: visualizing protein kinase C signaling. IUBMB Life 55, 653–660. [DOI] [PubMed] [Google Scholar]
- Volgraf M, Gorostiza P, Numano R, Kramer RH, Isacoff EY, and Trauner D (2006). Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat Chem Biol 2, 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Diezmann A, Shechtman Y, and Moerner WE (2017). Three-Dimensional Localization of Single Molecules for Super-Resolution Imaging and Single-Particle Tracking. Chem Rev 117, 7244–7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker D, Stevens RC, and Roth BL (2017). How Ligands Illuminate GPCR Molecular Pharmacology. Cell 170, 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Qiao Y, and Li Z (2018). New Insights into Modes of GPCR Activation. Trends Pharmacol Sci 39, 367–386. [DOI] [PubMed] [Google Scholar]
- Wang X, Chen X, and Yang Y (2012). Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods 9, 266–269. [DOI] [PubMed] [Google Scholar]
- Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, and Chien S (2005). Visualizing the mechanical activation of Src. Nature 434, 1040–1045. [DOI] [PubMed] [Google Scholar]
- Weis WI, and Kobilka BK (2018). The Molecular Basis of G Protein-Coupled Receptor Activation. Annu Rev Biochem 87, 897–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenberger S, Schultheis C, Liewald JF, Erbguth K, Nagel G, and Gottschalk A (2011). PACalpha--an optogenetic tool for in vivo manipulation of cellular cAMP levels, neurotransmitter release, and behavior in Caenorhabditis elegans. J Neurochem 116, 616–625. [DOI] [PubMed] [Google Scholar]
- Westfield GH, Rasmussen SG, Su M, Dutta S, DeVree BT, Chung KY, Calinski D, Velez-Ruiz G, Oleskie AN, Pardon E, et al. (2011). Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proc Natl Acad Sci U S A 108, 16086–16091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal MV, Schafroth MA, Sarott RC, Imhof MA, Bold CP, Leippe P, Dhopeshwarkar A, Grandner JM, Katritch V, Mackie K, et al. (2017). Synthesis of Photoswitchable Delta(9)-Tetrahydrocannabinol Derivatives Enables Optical Control of Cannabinoid Receptor 1 Signaling. J Am Chem Soc 139, 18206–18212. [DOI] [PubMed] [Google Scholar]
- Wetzker R, and Bohmer FD (2003). Transactivation joins multiple tracks to the ERK/MAPK cascade. Nat Rev Mol Cell Biol 4, 651–657. [DOI] [PubMed] [Google Scholar]
- Williams JT (2014). Desensitization of functional micro-opioid receptors increases agonist off-rate. Mol Pharmacol 86, 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Nan D, Fan J, Bogan JS, and Toomre D (2016). Optogenetic activation reveals distinct roles of PIP3 and Akt in adipocyte insulin action. J Cell Sci 129, 2085–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Karpenko IA, Hiblot J, and Johnsson K (2015). Imaging and manipulating proteins in live cells through covalent labeling. Nat Chem Biol 11, 917–923. [DOI] [PubMed] [Google Scholar]
- Yang W, and Yuste R (2017). In vivo imaging of neural activity. Nat Methods 14, 349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa M, Sadaghiani AM, Hsueh B, and Dolmetsch RE (2009). Induction of protein-protein interactions in live cells using light. Nat Biotechnol 27, 941–945. [DOI] [PubMed] [Google Scholar]
- Yi JJ, Wang H, Vilela M, Danuser G, and Hahn KM (2014). Manipulation of endogenous kinase activity in living cells using photoswitchable inhibitory peptides. ACS Synth Biol 3, 788–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Onodera H, Aono Y, Kawano F, Ueda Y, Furuya A, Suzuki H, and Sato M (2016). Optical manipulation of the alpha subunits of heterotrimeric G proteins using photoswitchable dimerization systems. Sci Rep 6, 35777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemelman BV, Lee GA, Ng M, and Miesenbock G (2002). Selective photostimulation of genetically chARGed neurons. Neuron 33, 15–22. [DOI] [PubMed] [Google Scholar]
- Zhang K, Duan L, Ong Q, Lin Z, Varman PM, Sung K, and Cui B (2014). Light-mediated kinetic control reveals the temporal effect of the Raf/MEK/ERK pathway in PC12 cell neurite outgrowth. PLoS One 9, e92917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XX, Fan LZ, Li P, Shen K, and Lin MZ (2017). Optical control of cell signaling by single-chain photoswitchable kinases. Science 355, 836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Tanaka KF, Matsunaga S, Iseki M, Watanabe M, Matsuki N, Ikegaya Y, and Koyama R (2016). Photoactivated adenylyl cyclase (PAC) reveals novel mechanisms underlying cAMP-dependent axonal morphogenesis. Sci Rep 5, 19679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zussy C, Gomez-Santacana X, Rovira X, De Bundel D, Ferrazzo S, Bosch D, Asede D, Malhaire F, Acher F, Giraldo J, et al. (2018). Dynamic modulation of inflammatory pain-related affective and sensory symptoms by optical control of amygdala metabotropic glutamate receptor 4. Mol Psychiatry 23, 509–520. [DOI] [PubMed] [Google Scholar]


