Abstract
Conformational changes in viral membrane proteins drive membrane fusion, a critical step in virus entry and infection. Here we describe a simple and rapid virus blotting immunoassay to define conformational changes with a panel of monoclonal antibodies to distinct sites across a viral glycoprotein. This dot blot technique has been utilized to define low pH-triggered changes in the prefusion form of the herpesviral fusogen gB. At pH of <6.2 there are specific changes in herpes simplex virus 1 gB domains I and V. This corresponds broadly to host cell endosomal pH. Many of the identified changes are at least partially reversible. This method can be adapted to document changes in viral proteins that are not fusion proteins, including those induced by alternate triggers such as receptor-binding or protease cleavage.
Keywords: Immunoassay, Dot blot, Antibodies, Nitrocellulose membrane, Virus entry, Glycoproteins, Herpesvirus, Herpes simplex virus, Conformational change, Membrane fusion, Low pH
1. Introduction
Virus entry is a key process in the viral replication cycle by which the incoming viral particle gains initial access to the host cell. All enveloped viruses must fuse with the target cell membrane to initiate successful entry [1]. The energetically unfavorable merging of two stable lipid bilayer membranes is thought to be driven by the conformational transition from prefusion to postfusion forms of the viral fusion glycoprotein. This major structural rearrangement, or conformational change, results in exposure of hydrophobic fusion peptide sequences. The exposed hydrophobic regions then interact with lipids of the target membrane, which is an essential destabilizing step in the fusion reaction [2]. Conformational changes in viral fusion proteins can be triggered by host cell cues, the most common of which is the low pH environment of an endosomal compartment. Enfuvirtide, a HIV antiviral, blocks a critical conformational change in the viral fusion protein gp41 [3].
There are biochemical, biophysical, and structural approaches to detecting and evaluating protein conformational change. The binding of a specific monoclonal antibody, for example, can depend on the primary, secondary, or quaternary structure of the antigen. Thus antigenicity as defined by antibody reactivity can be employed to distinguish different conformations of a single protein. Antibody–antigen interactions can be assessed by many immunological techniques, including flow cytometry, enzyme-linked immunosorbent assay (ELISA), Western blotting, and dot blotting.
The Herpesviridae family of large, enveloped DNA viruses causes significant morbidity and mortality. Many different virus-encoded membrane proteins, glycosylated and nonglycosylated, decorate the surface of herpesvirions. They comprise the multicomponent machinery that orchestrates receptor-binding, membrane fusion, and entry. Glycoprotein B (gB) is the core fusion protein, but it typically requires two or more additional viral membrane proteins to execute fusion. Glycoprotein B is thought to undergo major structural rearrangements during membrane fusion. The other required glycoproteins play critical triggering and regulatory roles and may also assume multiple conformations, particularly when comparing prefusion and postfusion states. In addition to gB, the gH/gL heterodimer and gD are necessary for herpes simplex virus (HSV) entry and membrane fusion [4-8].
Here we describe a rapid virus blotting (dot blot) immunoassay to define conformational changes with a panel of monoclonal antibodies to distinct sites across a viral glycoprotein. Scientists have taken experimental advantage of the ability of proteins to bind to nitrocellulose membranes for many years. In the simplest example of a dot blot, a protein is added to the membrane substrate and passively binds. The dot blot method (also known as slot blot) mirrors Western blotting (or immunoblotting), except that proteins are not first separated by electrophoresis. Thus dot blot may allow proteins to be analyzed in a more native state.
The prefusion form of HSV-1 gB undergoes low pH-triggered conformational changes that are thought to be fusion-associated [9-12]. These changes are at least partially reversible. In addition to the dot blot approach, gB conformation changes have been detected by polyacrylamide gel electrophoresis, tryptophan fluorescence spectroscopy, and immunofluorescence microscopy [13-17]. The dot blot approach described here can be adapted to document changes in viral proteins that are not fusion proteins, including those induced by alternate triggers such as receptor-binding or protease cleavage.
2. Materials
Dot blot apparatus (such as Minifold I Dot-Blot System, Schleicher and Schuell).
Nitrocellulose membrane (such as Protran Nitrocellulose Blotting Membrane, 0.45 μM pore size).
Filter paper (such as Whatman 3MM Chromatography Paper).
Vacuum source (central or pump) (see Note 1).
Source of virus antigen (such as extracellular HSV-1 strain KOS virions, ~105–106 PFU per dot).
Mildly acidic pH medium: Dulbecco’s modified Eagle’s medium (DMEM) (bicarbonate-free) solid, 0.2% bovine serum albumin (BSA), 5 mM succinate, 5 mM 2-(N-morpholino)ethanesulfonic acid (MES), 5 mM HEPES. Make up to 1 L with sterile, cell-culture grade water. Filter and store at 4 °C.
Laboratory vortex mixer.
Laboratory rocker.
Primary antibody (monoclonal or polyclonal) to viral envelope protein. Mouse monoclonal antibodies to HSV gB, H126, and H1817 (Virusys) are examples used here.
Horseradish peroxidase (HRP)-conjugated secondary antibody (e.g., goat anti-mouse HRP).
Phosphate buffered saline (PBS).
PBS-T wash buffer: 0.2% Tween 20 in PBS.
Blocking buffer: 5% nonfat dry milk prepared in PBS-T. Prepare just prior to use.
Distilled water.
0.05 N HCl.
0.05 N NaOH.
Water bath, 37 °C.
Chemiluminescence substrate for HRP.
Film cassette.
X-ray film.
Plastic transparency sheets, tweezer, cutting board, and low-lint, delicate task wipes.
Automatic film developer in darkroom.
3. Methods
3.1. Assembly of Dot Blot Apparatus
Measure and cut nitrocellulose membrane to the desired size. Handle membrane with forceps and gloved hands.
Place nitrocellulose membrane in a clean dish, and add PBS to activate.
Rock membrane for 20 min at room temperature.
Wet filter paper with PBS.
Attach filtration plate to vacuum plenum (Fig. 1). Back the nitrocellulose membrane with filter paper and place on top of the filtration plate. Place sample well plate with silicone O-rings facing down on top of the membrane. Clamp the sandwich in place with the adjustable latches (see Note 2).
Attach vacuum hose to the vacuum plenum on the assembled dot blot apparatus (see Note 3).
Turn vacuum on for 2 min to test. Turn off vacuum. Remove the sample plate, and verify that there are uniform O-ring indentations on the membrane. This is an indication that proper filtration is achieved.
Turn vacuum off until samples are ready to be applied.
Fig. 1.
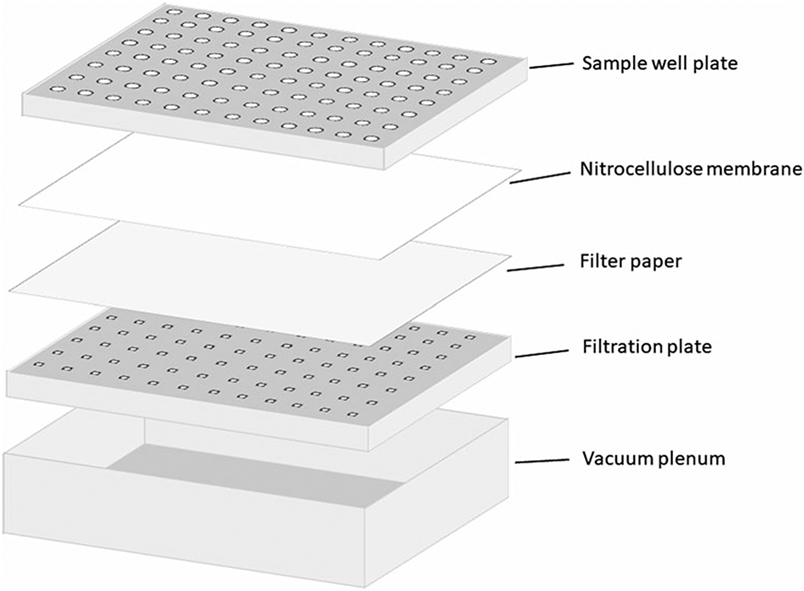
Schematic of assembly for dot blot immunoassay of conformational change of viral entry glycoproteins
3.2. Antigen Preparation
Adjust mildly acidic pH medium to the target pH by adding a predetermined volume of 0.05 N HCl (see Note 4).
Add HSV-1 to adjusted pH medium to a final concentration of ~105 PFU per 200 μL sample to be dotted. Vortex briefly.
Incubate pH-treated virus in 37 °C water bath for 10 min.
To measure for reversibility of conformational changes, add predetermined amount of 0.05 N NaOH (see Note 5), then incubate virus for another 10 min in a 37 °C water bath. Otherwise, virus is added to nitrocellulose membrane directly.
3.3. Antigen Blotting
Turn the vacuum on. Vortex samples briefly and apply sample (200–500 μL) to appropriate well.
Add 200 μL of sample per dot using a micropipette to allow rapid and even dispersal of sample to the membrane.
Let sample sit with vacuum on while preparing to add blocking buffer.
3.4. Membrane Blocking
Turn off vacuum. Disassemble dot blot apparatus. Release latches, and remove the sample well plate. With forceps, transfer nitrocellulose membrane to parafilm. Clean the apparatus properly (see Note 6).
On a clean cutting board, use scalpel and ruler to trim membrane as necessary. Mark with pencil to orient the position of samples.
Transfer nitrocellulose to a clean dish. Add blocking buffer to cover the surface of the membrane (see Note 7).
Place dish on a rocker, and rock for at least 20 min at room temperature.
3.5. Addition of Primary and Secondary Antibody
Prepare primary antibody dilution in blocking buffer. Dilution (ascites or serum) is to be determined empirically, and can range from 1:500 to 1:30,000. If using purified IgG, optimal concentration must also be determined.
Replace blocking buffer with primary antibody solution.
Place dish on a rocker and rock overnight at room temperature.
Discard primary antibody. Perform 3–4 initial washes, by quickly adding and replacing PBS-T wash buffer.
Wash the membrane by adding wash buffer to dish and rocking for 10 min at room temperature. Remove buffer, and repeat three times.
Prepare secondary (HRP-conjugated) antibody dilution in blocking buffer. Dilutions typically range from 1:10,000 to 1:30,000. Add sufficient volume to cover surface of nitrocellulose membrane.
Rock for 20–30 min at room temperature.
Discard secondary antibody. Perform 3–4 initial washes, by quickly adding and replacing PBS-T wash buffer.
Wash the membrane, by adding PBS-T wash buffer to dish and rocking for 10 min at room temperature. Remove buffer, and repeat three times.
3.6. Developing Blot
Immediately prior to use, prepare chemiluminescence HRP substrate according to manufacturer’s instructions. Prepare ~20 μL substrate per dot.
Discard wash buffer. Using forceps, gently blot membrane with wipe to remove excess substrate.
Transfer membrane to a plastic transparency sheet.
Pipette substrate evenly across the surface of membrane.
Quickly place another transparency sheet on top, and use gloved fingers to press across the membrane gently and thoroughly to allow even distribution of substrate. Avoid air bubbles.
With forceps, transfer membrane to between two new transparency sheets. Place in film cassette.
In darkroom, place film on top and seal cassette. Expose for desired period of time. Develop film in automatic processor.
Completed dot blot experiments have indicated that HSV-1 gB undergoes conformational changes in response to mildly acidic pH (Fig. 2), and that these changes are partially reversible (Fig. 3).
Fig. 2.
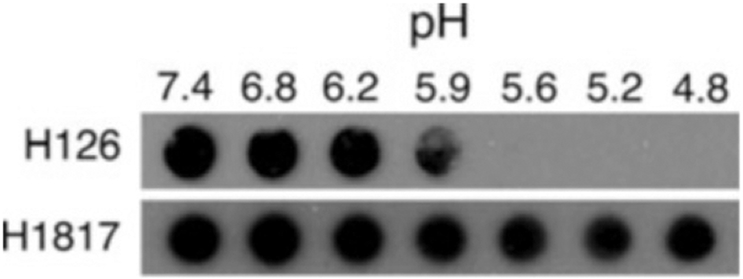
Dot blots of HSV-1 strain KOS probed with monoclonal antibodies to gB. Virions were treated for 10 min at 37 °C with medium buffered to the indicated pHs and were blotted immediately to membrane. Blots were probed at neutral pH with the indicated gB-specific antibodies, followed by horseradish peroxidase-conjugated goat secondary antibody. Decreased reactivity of acid-treated HSV with H126 indicates conformational change (see Note 8) (Reproduced from ref. 9 with permission from the American Society for Microbiology)
Fig. 3.
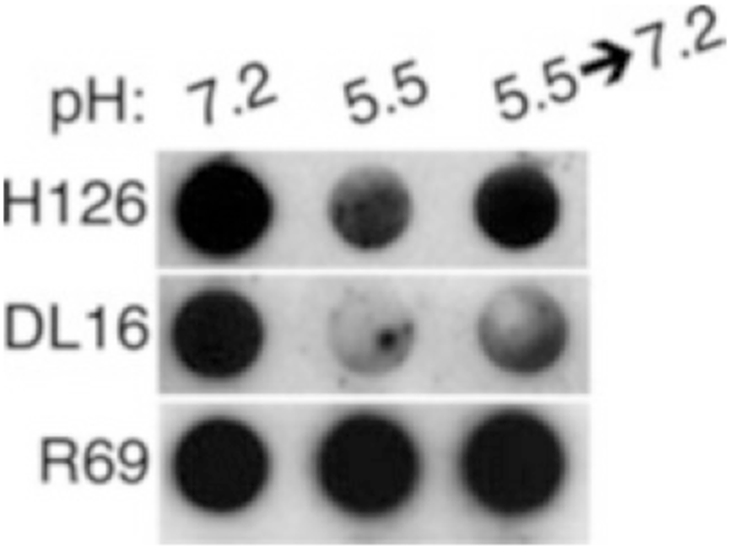
Dot blot illustrating partial reversibility of conformational change in HSV-1 gB. HSV-1 KOS virions were treated with medium buffered to pH 7.2 or 5.5. For the indicated samples, pH was neutralized back to 7.2 for 10 min at 37 °C. Membranes were probed at neutral pH with the indicated antibodies, followed by horseradish peroxidase-conjugated secondary antibody (Reproduced from ref. 9 with permission from the American Society for Microbiology)
4. Notes
A reliable, strong vacuum source is recommended for quality dots that are well-defined and reproducible.
Adjust the tightness of the latches so proper tension is achieved. Overtightening or undertightening will affect how the vacuum is distributed to the sample well plate.
Before blotting samples, apply a volume of PBS to a control well to verify that vacuum filtration is working properly.
Prior to experiment, determine the volume of 0.05 N HCl necessary to adjust the medium to the pH to be tested.
Prior to experiment, determine the volume of 0.05 N NaOH necessary to return the sample to the initial pH, typically pH 7.2–7.6.
Rinse wells thoroughly with 70% ethanol using a squirt bottle. Wash thoroughly with soapy water. Do not soak, as warping of plates and O-rings may occur. Rinse thoroughly with distilled water. Air-dry apparatus.
Use as small a container as possible to reduce the volume of antibody solution needed. This will increase the liquid to membrane surface ratio.
Immunoassays such as the dot blot benefit greatly from monoclonal antibody development efforts and extensive epitope mapping studies (see ref. 18).
Acknowledgments
This work was supported by Public Health Service grants AI119159 and GM008336 from the National Institutes of Health.
References
- 1.Nicola AV, Aguilar HC, Mercer J, Ryckman B, Wiethoff CM (2013) Virus entry by endocytosis. Adv Virol 2013:469538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White JM, Whittaker GR (2016) Fusion of enveloped viruses in endosomes. Traffic 17(6):593–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warnke D, Barreto J, Temesgen Z (2007) Antiretroviral drugs. J Clin Pharmacol 47(12):1570–1579 [DOI] [PubMed] [Google Scholar]
- 4.Cai WH, Gu B, Person S (1988) Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol 62(8):2596–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forrester A et al. (1992) Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol 66(1):341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson DC, Ligas MW (1988) Herpes simplex viruses lacking glycoprotein D are unable to inhibit virus penetration: quantitative evidence for virus-specific cell surface receptors. J Virol 62(12):4605–4612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicola AV, Straus SE (2004) Cellular and viral requirements for rapid endocytic entry of herpes simplex virus. J Virol 78(14):7508–7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weed DJ, Nicola AV (2017) Herpes simplex virus membrane fusion. Adv Anat Embryol Cell Biol 223:29–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dollery SJ, Delboy MG, Nicola AV (2010) Low pH-induced conformational change in herpes simplex virus glycoprotein B. J Virol 84(8):3759–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roller DG, Dollery SJ, Doyle JL, Nicola AV (2008) Structure-function analysis of herpes simplex virus glycoprotein B with fusion-from-without activity. Virology 382(2):207–216 [DOI] [PubMed] [Google Scholar]
- 11.Weed DJ, Pritchard SM, Gonzalez F, Aguilar HC, Nicola AV (2017) Mildly acidic pH triggers an irreversible conformational change in the fusion domain of herpes simplex virus 1 glycoprotein B and inactivation of viral entry. J Virol 91(5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicola AV (2016) Herpesvirus entry into host cells mediated by endosomal low pH. Traffic 17(9):965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siekavizza-Robles CR, Dollery SJ, Nicola AV (2010) Reversible conformational change in herpes simplex virus glycoprotein B with fusion-from-without activity is triggered by mildly acidic pH. Virol J 7:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dollery SJ, Wright CC, Johnson DC, Nicola AV (2011) Low-pH-dependent changes in the conformation and oligomeric state of the prefusion form of herpes simplex virus glycoprotein B are separable from fusion activity. J Virol 85(19):9964–9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weed DJ, Dollery SJ, Komala Sari T, Nicola AV (2018) Acidic pH mediates changes in antigenic and oligomeric conformation of herpes simplex virus gB and is a determinant of cell-specific entry. J Virol 92(17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wudiri GA, Schneider SM, Nicola AV (2017) Herpes simplex virus 1 envelope cholesterol facilitates membrane fusion. Front Microbiol 8:2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dollery SJ, Lane KD, Delboy MG, Roller DG, Nicola AV (2010) Role of the UL45 protein in herpes simplex virus entry via low pH-dependent endocytosis and its relationship to the conformation and function of glycoprotein B. Virus Res 149(1):115–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bender FC et al. (2007) Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J Virol 81(8):3827–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]


