Abstract
Background:
Both trichloroethylene (TCE) and tetrachloroethylene (PCE) are high-priority chemicals subject to numerous human health risk evaluations by a range of agencies. Metabolism of TCE and PCE determines their ultimate toxicity; important uncertainties exist in quantitative characterization of metabolism to genotoxic moieties through glutathione (GSH) conjugation and species differences therein.
Objectives:
This study aimed to address these uncertainties using novel in vitro liver models, interspecies comparison, and a sensitive assay for quantification of GSH conjugates of TCE and PCE, S-(1,2-dichlorovinyl)glutathione (DCVG) and S-(1,2,2-trichlorovinyl) glutathione (TCVG), respectively.
Methods:
Liver in vitro models used herein were suspension, 2-D culture, and micropatterned coculture (MPCC) with primary human, rat, and mouse hepatocytes, as well as human induced pluripotent stem cell (iPSC)-derived hepatocytes (iHep).
Results:
We found that, although efficiency of metabolism varied among models, consistent with known differences in their metabolic capacity, formation rates of DCVG and TCVG generally followed the patterns , and primary . Data derived from MPCC were most consistent with estimates from physiologically based pharmacokinetic models calibrated to in vivo data.
Discussion:
For TCE, the new data provided additional empirical support for inclusion of GSH conjugation-mediated kidney effects as critical for the derivation of noncancer toxicity values. For PCE, the data reduced previous uncertainties regarding the extent of TCVG formation in humans; this information was used to update several candidate kidney-specific noncancer toxicity values. Overall, MPCC-derived data provided physiologically relevant estimates of GSH-mediated metabolism of TCE and PCE to reduce uncertainties in interspecies extrapolation that constrained previous risk evaluations, thereby increasing the precision of risk characterizations of these high-priority toxicants. https://doi.org/10.1289/EHP12006
Introduction
Trichloroethylene (TCE) and tetrachloroethylene (PCE) are chlorinated solvents with a broad range of applications in industrial and other settings. TCE and PCE are used in the production of other solvents, dry cleaning of fabrics, and metal degreasing.1–3 Due to their volatility, human exposures to TCE and PCE are assumed to be mainly through inhalation (either through direct contact or vapor intrusion); both chemicals are also commonly found in ground and drinking water.4 TCE is the most commonly detected compound among contaminants that are found at National Priority List (NPL) sites in the United States, and concurrent contamination of TCE with PCE is widespread.2,5
TCE has been classified as a known human carcinogen with kidney and liver as primary targets,4,6–8 whereas PCE has been classified as a probable human carcinogen with strongest evidence for bladder cancer.4,9,10 These chemicals remain high priorities for risk assessment and risk management, especially for noncancer risks associated with chronic exposures in residences or office buildings where vapor intrusion from groundwater contamination is suspected. The United States Environmental Protection Agency (U.S. EPA) listed TCE and PCE among 10 “high-priority” existing chemical substances under Section 6 of the Toxic Substances Control Act as amended by the Frank R. Lautenberg Chemical Safety for the 21st Century Act.11 In addition, the U.S. Army Public Health Center is conducting reanalysis of the TCE data to update an occupational exposure limit (OEL) because it is a ubiquitous contaminant on military installations where both residential and office buildings may have vapor intrusion from contaminated groundwater.12
Both cancer and noncancer effects associated with TCE and PCE are known to be mediated by their metabolites.2,4,10,13,14 These compounds are metabolized through two pathways: oxidation and glutathione (GSH) conjugation.4,15–18 The major oxidative metabolites for TCE are trichloroacetic acid (TCA) and trichloroethanol (TCOH); for PCE, only TCA has been consistently detected from oxidation in both human and experimental animals.1,9,19–21 Major metabolites through GSH conjugation for TCE include S-(1,2-dichlorovinyl)glutathione (DCVG), S-(1,2-dichlorovinyl)-L-cysteine (DCVC), and N-acetyl-S-(1,2-dichlorovinyl)-L-cysteine (NAcDCVC). For PCE, GSH metabolites include S-(1,2,2-trichlorovinyl)glutathione (TCVG), S-(1,2,2-trichlorovinyl)-L-cysteine (TCVC), and N-acetyl-S-(1,2,2-trichlorovinyl)-L-cysteine (NAcTCVC).22–26 Even though the metabolic flux through oxidation predominates for both TCE and PCE, their GSH conjugates are thought to be of critical importance as the metabolites derived from cysteine conjugates, formed in kidneys, are known to be highly reactive and are genotoxic.2,17
The extent of metabolism of TCE and PCE to their GSH conjugates and subsequent kidney effects has been regarded as a key uncertainty in previous risk assessments, especially because of the inconsistency among previous studies of GSH metabolism that used hepatocytes and subcellular fractions from human and rodent liver.15,17,22,24,26–30 Recent advancements in liver-based in vitro models,31 including engineered human liver models,32–34 have improved the capabilities of in vitro systems to mimic physiologically relevant metabolic function of the liver. These models also improved our ability to collect and interpret pharmacokinetic data.35–38 One liver model that showed great potential for in vitro metabolism studies is a micropatterned coculture (MPCC) system.39 The MPCC platform has been shown to maintain the viability and metabolic activity of hepatocytes from rodents and humans for several weeks.39–42 These improvements further highlight the potential for using MPCC to investigate xenobiotic metabolism, especially in cases where uncertainties are to be addressed through direct comparisons of species-specific metabolism.
Thus, because there is a critical need to characterize GSH conjugation metabolism of TCE and PCE, we used several liver in vitro models, including suspension, 2-D culture, and MPCC. We used primary human hepatocytes (PHH), human induced pluripotent stem cell (iPSC)-derived hepatocytes (iHep), primary rat hepatocytes (PRH), and primary mouse hepatocytes (PMH) and determined formation of GSH conjugation metabolites DCVG and TCVG. We compared our results with those published previously to resolve existing uncertainties and used new data to reevaluate assumptions made by the U.S. EPA in deriving noncancer candidate reference toxicity values in their most recent assessments.7,9 The results of this study provide critical new information pertaining to the utility of in vitro liver models and show that MPCC model provides physiologically relevant estimates of TCE and PCE metabolism. These data reduce interspecies extrapolation uncertainties in chemical risk evaluations and confirm the validity of kidney toxicity data for both TCE and PCE.
Materials and Methods
Chemicals
TCE, PCE, GSH, and distilled water with 0.1% formic acid were obtained from Sigma-Aldrich. Methanol and acetonitrile were from Fisher Scientific. DCVC (purity ), S-(1,2-dichlorovinyl)- (DCVC*, purity , isotopic purity ), DCVG (), and (DCVG* purity , isotopic purity ) were obtained from TLC Pharmaceutical Standards. N-acetyl-S-(1,2-dichlorovinyl)-L-cysteine (NAcDCVC, purity 99.8%), , (, purity: 97.6%, isotopic purity: 99.0%), and N-acetyl-S-(1,2,2-trichlorovinyl)-L-cysteine (NAcTCVC, purity: 99.7%) were purchased from Toronto Research Chemicals. TCVC (purity: 98.4%), (, purity: 97.5%), S-(1,2,2-trichlorovinyl)glutathione (TCVG, purity: 98.9%), (TCVG*, purity: 90.4%), and (NAcTCVC* purity: 99.0%), were synthesized by Dr. Avram Gold at the University of North Carolina at Chapel Hill as detailed previously.43
Cells
Cryopreserved primary human hepatocytes (PHHs) from two different sources were used in this study. Single donor PHH (35-y-old Asian male) were obtained from Lonza (Lot No. HUM4122) and 10-donor PHH pooled sample was obtained from Gibco (Lot No. HPP1825348). Induced pluripotent stem cell (iPSC)-derived hepatocytes (iHep) were obtained from FujiFilm Cellular Dynamics International (Lot No. 103664). Primary rat (Sprague Dawley; Charles River Laboratories) hepatocytes (PRH) and primary mouse (C57Bl/6J or CD-1; Charles River Laboratories) hepatocytes (PMH) were isolated following the protocol reported by Seglen44 for rat hepatocytes and the protocol of Lee et al.45 for mouse hepatocytes; these experiments were performed in accordance with the institutional guidelines (Protocol 21-046, University of Illinois at Chicago Institutional Animal Care and Use Committee).
iHep Differentiation
Culture conditions for experiments with iHep were previously described.46 Briefly, iHep were seeded with plating media (DMEM/F12; Thermo Fisher) supplemented with 2% B27 supplement (Thermo Fisher), dexamethasone, gentamicin, and oncostatin M (R&D Systems) at a density of cells/well on a 6-well plate, which was precoated with collagen (VWR International, LLC). Six hours after incubation at 37°C, , media were replaced to new plating media to remove unattached cells. Black-walled, clear-bottom, tissue cultured 96-well plates (Corning Inc.) were used for 2-D sandwich culture. This plate was precoated overnight at 37°C, with collagen type acetic acid and rinsed 3 times with PBS before cell seeding. Cells were differentiated for 5 d with daily plating media changes.
Suspension Cell Culture
In vitro hepatocyte metabolism was evaluated using suspension cultures of PHH (pooled sample; Lot No. HPP1825348), iHep, PRH, and PMH (see Figure S1 for study design). In brief, cells were suspended in William’s E medium (Gibco) and adjusted to the cell concentration of . Five hundred microliters of TCE or PCE ( in 1% acetone and William’s E medium) was spiked in of the cell working stock with or without GSH to a final chemical concentration of in 0.5% acetone (with or without GSH) and cell number of . One hundred microliters of the reaction mixture were removed subsequently at 0, 60, 120, and 240 min to individual Eppendorf tubes for further sample extraction and chemical analysis detailed below.
2-D Monolayer Cultures of Hepatocytes
To create hepatocyte monolayers, 24- (for PMH) or 96- (for PHH and PRH) well plates were coated with rat tail collagen I (Corning). PMH were seeded at 300,000 cells/well in 24-well plates. PHH (both pooled sample and single donor sample) and PRH were seeded at 50,000 cells/well in 96-well plates (see Figure S1 for study design). All monolayer cultures were maintained in hepatocyte medium containing DMEM (Corning) supplemented with HEPES [4-(2-hydroxyethyl)-1-piperazineethane-sulfonic acid] buffer (Corning), 1% penicillin/streptomycin, 10% bovine serum, and glucagon (Sigma-Aldrich). PHH medium (both pooled sample and single donor sample) also contained dexamethasone (Sigma-Aldrich) and 1% Premix Universal Culture Supplement, which contains insulin, human transferrin, and selenous acid (Corning). PRH medium also contained epidermal growth factor, hydrocortisone, and insulin. PMH medium also contained dexamethasone and insulin. The media was replaced in all cultures every 48 h and collected at each time point for further analysis. On day 7 of culture, monolayer cultures of PHH (both pooled and sample and single donor sample) and PRH were treated with stock solution of TCE or PCE, with and without GSH in 100% acetone for the final concentration of TCE or PCE, and 0 or GSH, in 0.5% acetone. Media samples were collected for analytical chemistry 1 h after this treatment and fresh media without chemicals was added. Cultures were incubated for an additional 47 h when media was collected for measurements of albumin and urea (see below). Cultures of PMH were treated with chemicals on day 8 of culture and samples were collected exactly as detailed above.
2-D Sandwich Culture of iHep
Differentiated iHep were collected with StemPro Accutase (Thermo Fisher) from the 6-well plate. The cell clusters were centrifuged (, 3 min) and resuspended with plating media. iHep were plated in 96-well plates at a density of and incubated to attach for 6 h at 37°C, (see Figure S1 for study design). Media were replaced to of plating media with Matrigel (Corning). After incubation overnight at 37°C, 5% CO2, media were exchanged to the maintenance medium (DMEM/F12, 2% B27, dexamethasone, gentamicin). Media was collected and exchanged every 48 h. On day 4 of culture, cells were treated with stock solution of TCE or PCE, with and without GSH in 100% acetone for the final concentration of TCE or PCE, and 0 or GSH, in 0.5% acetone. Media samples were collected for analytical chemistry 1 h after this treatment and fresh media without chemicals was added. Cultures were incubated for an additional 47 h when media was collected for measurements of albumin and urea (see below).
MPCC Experiments
Previous studies have reported the development of the MPCC platform containing liver parenchymal cells arranged onto collagen-coated islands of experimentally optimized dimensions and subsequently surrounded by 3T3-J2 murine embryonic fibroblasts that are known to sustain robust functions in hepatocytes from numerous species.39,40 In addition to adapting the MPCC technique for primary human, rat and mouse hepatocytes, we used optimized conditions that were previously reported for iHep in MPCC.41 Briefly, rat tail collagen I was adsorbed onto tissue culture plastic 24- (for PMH) or 96- (for PHH and PRH) well plates followed by patterning lithographically via oxygen plasma treatment to create diameter circular domains spaced apart from center-to-center. PHHs, iHep, PRH, or PMH were seeded into the corresponding micropatterned plates in serum-free culture medium (see Figure S1 for study design). The hepatocytes preferentially attached to the circular collagen domains, leaving approximately 4,500 iHep, 5,000 PHH, and 3,200 PRH per well in 96-well plates, or 25,000 PMH per well in 24-well plates. After the circular domains were fully seeded, the cultures were rinsed to remove the unattached hepatocytes, and medium containing fetal bovine serum (FBS; Gibco) was added to the cultures overnight. Separately, 3T3-J2 murine embryonic fibroblasts (gift of Dr. Howard Green, Harvard University)47 were cultured in medium containing 10% bovine calf serum, 1% penicillin/streptomycin, and 89% DMEM. After the fibroblasts reached 85%–90% confluence, they were passaged with 0.25% trypsin/EDTA (Corning) that was neutralized with medium containing 10% serum and pelleted via centrifugation, and cells were then used for further experimentation. All 3T3-J2 murine embryonic fibroblasts were used between passage 7 and 10. After 5 d of culture for iHep or 18–24 h for PHH, PRH, and PMH, the 3T3-J2 murine embryonic fibroblasts were seeded in hepatocyte medium at a density of cells/24-well or 15,000 cells/96-well to create MPCC. iHep hepatocyte medium was composed of DMEM supplemented with B-27 supplement, HEPES buffer, , penicillin/streptomycin, 1% bovine calf serum, dexamethasone, glucagon, oncostatin M (R&D Systems), and FPH2 small molecule (Sigma-Aldrich). Media was replaced every 48 h and collected for further analyses as detailed below. MPCC cultures of PHH (both pooled and sample and single donor sample) and PRH were treated on day 7 with stock solution of TCE or PCE, with and without GSH in 100% acetone for the final concentration of TCE or PCE, and 0 or GSH, in 0.5% acetone. For PMH, treatments were performed on day 8. For iHep, treatments were performed on day 12 of culture. Media samples were collected for analytical chemistry 1 h after this treatment, and fresh media without chemicals was added. Cultures were incubated for additional 47 h when media was collected for measurements of albumin and urea (see below).
Morphological and Functional Assessments
The morphology of the monolayers and MPCC was assessed via an IX83 automated microscope (Olympus America) with a high-sensitivity 4.2MP sCMOS camera (ORCA-Flash4.0 ) using phase contrast objectives (). Urea production in culture supernatants was quantified via a colorimetric end point assay kit (Cat. No. 0580-250; Stanbio Labs). Culture supernatants were also assayed for albumin production by using a competitive enzyme-linked immunosorbent assay (human ELISA kit E80-129, rat ELISA kit E111-125, mouse ELISA kit E99-134; Bethyl Laboratories) with horseradish peroxidase detection and 3,3′,5,5′-tetramethylbenzidine (TMB; Rockland Immunochemicals) as the substrate. The absorbance measurement for both albumin and urea assays were detected on the Synergy H1 multimode plate reader (Biotech).
Analytical Methods for TCE and PCE Metabolites
TCE and PCE produce various metabolites through both oxidation and glutathione conjugation pathways.4,23,48 To analyze a subset of all the potential metabolites produced from TCE or PCE exposure, we used liquid chromatography–tandem mass spectrometry (LC-MS/MS). These metabolites and information on their analytical detection can be found in Table 1. Each media sample () was mixed with of chilled acetonitrile containing mixture of DCVG, DCVC, and NAcDCVC internal standards or mixture of TCVG, TCVC, and NAcTCVC internal standards. Next, samples were centrifuged at for 10 min. The supernatant was dried under vacuum using a SpeedVac (Speed SPD1010; Thermo Scientific) and reconstituted with of aqueous mobile phase prior to LC-MS/MS analyses.
Table 1.
TCE and PCE Metabolites evaluated in this study.
| Metabolite | Analytical assay | Ionization mode |
Mass transition or quantitation ion (m/z) |
Collision energy (eV) |
|---|---|---|---|---|
| DCVG | LC-MS/MS | 13 | ||
| † | 25 | |||
| 37 | ||||
| DCVGa | LC-MS/MS | † | 25 | |
| 25 | ||||
| DCVC | LC-MS/MS | † | 8 | |
| 25 | ||||
| 45 | ||||
| DCVCa | LC-MS/MS | † | 25 | |
| 10 | ||||
| NAcDCVC | LC-MS/MS | 9 | ||
| † | 17 | |||
| 5 | ||||
| NAcDCVCa | LC-MS/MS | 9 | ||
| † | 17 | |||
| TCVG | LC-MS/MS | 13 | ||
| 25 | ||||
| † | 13 | |||
| TCVGa | LC-MS/MS | † | 17 | |
| 13 | ||||
| TCVC | LC-MS/MS | † | 9 | |
| 21 | ||||
| 9 | ||||
| TCVCa | LC-MS/MS | † | 9 | |
| 21 | ||||
| NAcTCVC | LC-MS/MS | † | 9 | |
| 17 | ||||
| 9 | ||||
| NAcTCVCa | LC-MS/MS | † | 9 | |
| 17 |
Note: DCVC, S-(1,2-dichlorovinyl)-L-cysteine; DCVG, S-(1,2-dichlorovinyl)glutathione; LC-MS/MS, liquid chromatography–tandem mass spectrometry; :NAcDCVC, N-acetyl-S-(1,2-dichlorovinyl)-L-cysteine; NAcTCVC, N-acetyl-S-(1,2,2-trichlorovinyl)-L-cysteine; PCE, tetrachloroethylene; TCE, trichloroethylene; TCVC, S-(1,2,2-trichlorovinyl)-L-cysteine.
Internal standard, see “Materials and Methods” section.
Selected quantifier for analysis by LC-MS/MS analysis.
Determination of TCE glutathione conjugation metabolites via LC-MS/MS was performed as previously described by Luo et al.48 In brief, analysis was performed using a 1290 Infinity II LC system and a 6470 triple-quadrupole mass spectrometer (both from Agilent Technologies). Samples () were automatically injected and chromatographed on a ZORBAX SSHD Eclipse Plus C18 column (, ; Agilent Technologies) with a guard column (, ; Agilent Technologies). Column temperature was maintained at 25°C. Initial chromatographic condition was maintained at 90% solvent A (water with 0.1% acetic acid, v/v) and 10% solvent B (methanol with 0.1% acetic acid, v/v) for 1 min, then increased to 90% solvent B by 3 min, then to 98% solvent B by 4 min, and then returned to initial condition until 7 min for sufficient equilibration prior to next run. Flow rate was set at . Metabolite levels in media were quantified by using the peak area ratios of standards to isotopically labeled internal standards in an 8-point calibration curve (0, 1.25, 2.5, 5, 12.5, 25, 50, 125 pmol). In these experiments, the detection limit for TCE GSH metabolites was , the limit of quantitation (LOQ) was , and the recovery of metabolites ranged from 67% to 83%.
Determination of PCE glutathione conjugation metabolites via LC-MS/MS was performed as previously described by Luo et al.23 In brief, analysis was performed using 1290 Infinity II LC system and 6470 triple-quadrupole mass spectrometer (Agilent Technologies). Samples () were automatically injected and chromatographed on a ZORBAX SSHD Eclipse Plus C18 column (, ; Agilent Technologies) with a guard column (, ; Agilent Technologies). Column temperature was controlled at 25°C. For each sample analysis, initial chromatographic conditions were 80% solvent A (water with 0.1% formic acid) and 20% solvent B (methanol with 0.1% formic acid). Conditions were maintained from 0–1 min, then increased to 90% of solvent B by 3 mins, then to 98% of solvent B by 4 mins, then to 20% of solvent B by 4.2 mins. Next, 20% of solvent B was maintained until 7 mins to allow sufficient equilibration time prior to the next injection. Flow rate was maintained at . Metabolite levels in media were quantified by using the peak area ratios of standards to isotopically labeled internal standards in an 8-point calibration curve (0, 1.25, 2.5, 5, 12.5, 25, 50, 125 pmol). In these experiments, the detection limit for PCE GSH metabolites was , the LOQ was , and the recovery of metabolites ranged from 56% to 88%.
Comparisons with Previous Studies
Data on GSH conjugation metabolism of TCE and PCE has been previously measured both in vitro and in vivo, as well as incorporated into physiologically based pharmacokinetic (PBPK) models. For comparison with current study results, both in vitro and in vivo GSH conjugation rates were converted to in vitro intrinsic clearance rates in nmol/h/million hepatocytes at the tested concentration of . For TCE, previous in vitro studies included experiments in subcellular fractions (cytosol or microsomes)28,29 and in both subcellular fractions and hepatocyte suspensions.15,17,22,30 The PBPK model for TCE18 estimated GSH conjugation rates in mice, rats, and humans based on in vivo data on TCE conjugation as well as overall mass balance.15,49–51 For PCE, previous relevant in vitro liver metabolism studies are experiments in subcellular fractions26,27 and in both subcellular fractions and hepatocyte suspensions.17,24 The PBPK model for PCE52 estimated in vivo GSH conjugation rates in mice, rats, and humans based on in vivo data on GSH conjugation metabolites in rats, as well as overall mass balance.
Miscellaneous
The number of replicates in each condition is listed in the legend. Statistical analyses were conducted using GraphPad Prism (version 9.4.1; GraphPad Software). Figures were prepared using GraphPad Prism, Adobe Photoshop (Adobe) and/or PowerPoint (Microsoft).
Results
This study used several liver in vitro models to evaluate TCE and PCE metabolism through the GSH conjugation pathway: suspension, 2-D culture, and MPCC (Figure 1 and Figure S1). Various sources of liver parenchymal cells were used including PHHs, iHep, PRH, and PMH. Cells were treated with TCE or PCE () with and without GSH () following the methods used in previous studies.17 Formation of GSH conjugation pathway metabolites was evaluated. These results were taken together in combination with previously reported metabolite in vitro data to facilitate liver model comparison and determine species differences.
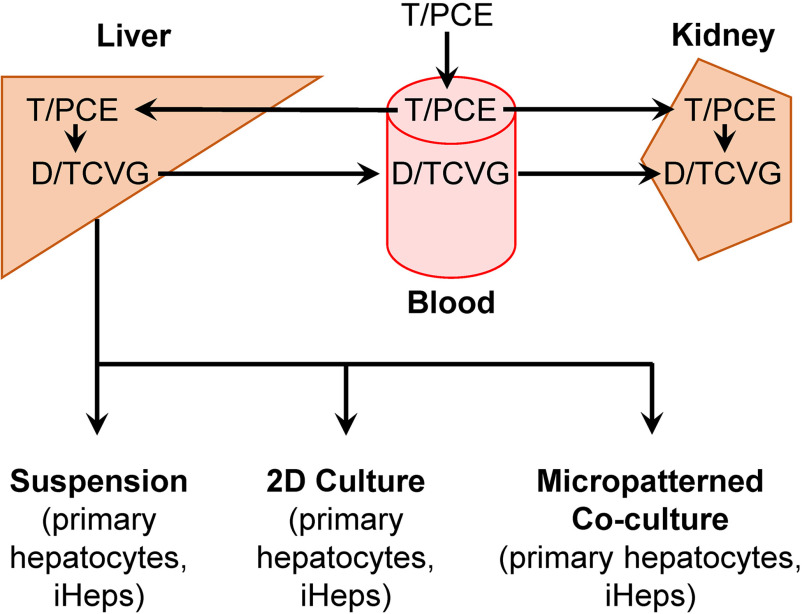
Figure 1.
Schematic of TCE and PCE GSH conjugation metabolism with liver in vitro models. The diagram summarizes tissue localization of metabolic reactions and transport of TCE and PCE GSH conjugation metabolites, DCVG, and TCVG, respectively. A majority of D/TCVG formation occurs in the liver; however, formation of D/TCVG does occur, although to a lesser extent, in the kidneys. Each of the in vitro models used in this study are shown beneath the liver. Note: DCVG, S-(1,2-dichlorovinyl)-glutathione; GSH, glutathione; PCE, tetrachloroethylene; TCE, trichloroethylene; TCVG, S-(1,2,2-trichlorovinyl)glutathione.
Hepatic Morphological and Functional Assessments in MPCC
Morphology and basic function of cells in MPCC was evaluated prior to chemical treatment (Figure 2). We observed compact, concentric islands of healthy hepatocytes surrounded by 3T3-J2 fibroblasts across multiple species. In addition to morphology, albumin production and urea secretion were evaluated. Chemical treatments were performed on day 7–8 to allow for the model to reach its optimal function as demonstrated in previous studies.39,40 Following chemical treatment, supernatant media was assessed for albumin and urea at the final timepoint of day 9–10. Albumin production showed variability between single donor of PHHs, iHep, and primary rat hepatocytes. Albumin was detected at the highest levels in PRH MPCC on days 7 and 9; the amounts were approximately between 500 to hep/d for the three testing conditions. In PMH MPCC, albumin production increased over the culture period from approximately hep/d on day 2, to hep/d on day 10. In PHH MPCC, albumin production peaked on day 7 with a range of 60 to hep/d before decreasing to a range of 60 to hep/d on day 9. In iHep MPCC, albumin amounts steadily increased from day 3 to day 9. The highest levels were detected on day 9 at approximately hep/d for all three conditions. Estimated human liver output for albumin is hep/d.34
Figure 2.
Representative baseline morphology and hepatic functional data collected before and after chemical treatment (final concentration of for TCE and PCE, and for GSH, in 0.5% acetone) from different sources of hepatocytes in MPCCs with 3T3-J2 fibroblasts. (A) Primary human hepatocytes (Lot No. HUM4122), (B) induced hepatocyte-like cells (iHep), (C) primary rat hepatocytes, (D) primary mouse hepatocytes patterned onto collagen islands in culture media. Following seeding of 3T3-J2 fibroblasts, hepatic morphology was maintained in MPCCs for 7 d prior to chemical exposure. Red dashed ovals indicate hepatocyte areas. Phase contrast objectives () were used to obtain the images. Normalized hepatic functional data for albumin secretion and urea synthesis are shown to the right of each corresponding MPCC hepatocyte model. The asterisk symbol indicates the day of chemical exposure in each model. No significant differences were observed among treatments at each time point, with the exception of the data for urea in primary rat hepatocytes where vehicle-treatment-designated wells had overall greater values both before and after TCE or PCE treatments (one-way ANOVA comparing vehicle with and ). See Excel Tables S1 (for albumin) and S2 (for urea) for the raw data plotted in this Figure. Note: ANOVA, analysis of variance; GSH, glutathione; MPCC, micropatterned coculture; PCE, tetrachloroethylene; TCE, trichloroethylene.
Urea secretion exhibited similar trends as albumin production. The greatest amounts of urea were observed in PRH MPCC. Peak levels occurred on day 9 for all three conditions; however, wells designated for vehicle produced about hep/d, whereas wells designated for TCE and PCE with GSH produced approximately hep/d; however, the above differences in urea secretion across the wells were likely not due to the compounds, because trends were observed both before and after treatment with vehicle or compounds. In comparison, PMH MPCC produced approximately hep/d at day 10 of culture. In PHH MPCC, urea secretion was highest on day 3 and then decreased on day 5, followed by increasing amounts on day 7 and 9. The amount of urea secreted on day 3 ranged between 30– hep/d. Urea secretion in iHep MPCC was lower compared to PHH and PRH MPCCs. Estimated human liver output for urea is hep/d.34
Metabolite Formation Rates across in Vitro Models and Sources of Hepatocytes
Multiple in vitro liver models and sources of hepatocytes were used in this study to characterize TCE and PCE GSH conjugation metabolism. Following chemical addition for 1 h, media samples were collected and analyzed with quantitative LC-MS/MS methods to screen for GSH conjugation metabolites. For TCE, three metabolites were screened for: DCVG, DCVC, and NAcDCVC. Similarly, for PCE we screened for TCVG, TCVC, and NAcTCVC. In this study, we detected only DCVG and TCVG, which is consistent with previous data that demonstrated a majority of DCVG and TCVG formation occurs in liver, whereas the cysteine metabolites are primarily formed and found in the kidney.14
The concentrations of DCVG and TCVG varied by in vitro model and hepatocyte source (Figure 3; left graphs). DCVG and TCVG concentrations ranged from 0.07 to and 0.10 to , respectively. Both metabolites were detected at the highest concentrations using pooled PHHs in suspension cultures; however, the lowest concentration of DCVG was reported with PMH in 2-D culture, whereas the lowest TCVG concentration occurred with PMH in MPCC.
Figure 3.
Comparison of DCVG and TCVG concentrations and rates of formation across different in vitro liver models and sources of hepatocytes used in this study. (A) Plotted are detected concentrations (left, ) and normalized rates of formation (right, ) for TCE GSH conjugation metabolite, DCVG. (B) Same data for PCE GSH conjugation metabolite, TCVG. The vertical red dotted lines represent the analytical method limit of quantification for each metabolite. The horizontal dotted lines separate data from different models. The following number of replicates were analyzed: for suspension cultures of PHH (pooled) and iHep; for suspension cultures of PRH and PMH; for 2-D cultures of PHH (pooled and HUM4122A), iHep, and PRH; for PMH 2-D cultures. for MPCC cultures of PHH (HUM4122A) and PMH; for MPCC cultures of PHH (pooled) and iHep; and for MPCC cultures of PRH. See Excel Tables S3 (for TCE) and S4 (for PCE) for the raw data plotted in this Figure. Note: DCVG, S-(1,2-dichlorovinyl)glutathione; GSH, glutathione; MPCC, micropatterned coculture; PCE, tetrachloroethylene; PHH, primary human hepatocytes; PMH, primary mouse hepatocytes; PRH, primary rat hepatocytes; SD, standard deviation; TCE, trichloroethylene; TCVG, S-(1,2,2-trichlorovinyl)glutathione.
Next, to enable comparison of DCVG and TCVG formation across models and to other studies, we normalized our data by accounting for cell density and incubation volume to derive metabolite formation rates (Figure 3; right graphs). DCVG and TCVG formation rates ranged from 0.07 to hep and 0.11 to hep, respectively. In TCE suspension cultures, human hepatocytes created DCVG at rates 18- to 45-fold higher than PRH and 46- to 120-fold higher than PMH. Human hepatocytes in 2-D culture produced DCVG at rates 4- to 7-fold higher than PRH and 12- to 21-fold higher than PMH. PHH in MPCC yielded DCVG formation rates that were 2- to 3-fold greater than PRH and 6- to 11-fold greater than PMH. In PCE suspension cultures, human hepatocytes created TCVG at rates 77- to 186-fold higher than PRH and 128- to 277-fold higher than PMH. Human hepatocytes produced TCVG at rates equal to 7-fold higher than PRH and 3- to 25-fold higher than PMH. In MPCC cultures, iHep generated TCVG at a rate of 1.5-fold less than PRH; however, pooled PHHs produced TCVG at approximately a 2-fold higher rate than PRH. PHH and iHep in MPCC generated TCVG at rates 3- to 7-fold greater than PMH. Overall, we observed the greatest species differences in DCVG and TCVG formation in suspension cultures and least in MPCCs.
Comparison of Metabolite Formation Rates with Historical Data
Figure 4 compares the results of the current study with previously reported rates of GSH conjugation either measured in vitro or estimated based on PBPK modeling calibrated to in vivo data (Excel Tables S5 and S6). With respect to in vitro comparisons, in humans, for both TCE and PCE, there was a clear trend across in vitro models of suspension Lash et al.15,17,22,30 Green et al.28 and Dekant et al.29 data (Figure 4A). For rats and mice, the pattern differed with respect to suspension and Green et al. and Dekant et al. data, with Lash et al.15,17,22,30 for both TCE and PCE, Green et al.28 and Dekant et al.29 similar to 2-D for TCE, and Green et al. and Dekant et al. intermediate between MPCC and 2-D for PCE (Figure 4B–C).
Figure 4.
GSH conjugation formation rates for TCE and PCE across various studies. (A) Box and whiskers plots are shown comparing TCE and PCE GSH conjugation metabolite formation rates using human data, (B) rat data, and (C) mouse data. In each box, the black vertical lines inside the box denote median values; boxes extend from the 25th to the 75th percentile of each group’s distribution of values; vertical extending lines indicate the minimum and maximum values. Individual values are shown as black dots. Studies that did not test the specified conditions are identified as NA. All TCE data points plotted can be found in Excel Table S3. All PCE data points plotted can be found in Excel Table S4. Note: GSH, glutathione; NA, not available; PCE, tetrachloroethylene; TCE, trichloroethylene.
When comparing our study results with the TCE and PCE PBPK modeling results calibrated in vivo GSH data (humans and rats for TCE, rats for PCE), the MPCC results were the most consistent, followed by the data from suspension cultures in our study. An interesting finding was that, for mice, for which the PBPK model estimated GSH conjugation rates indirectly based on mass balance, the MPCC results are close to the median PBPK-based estimates. For PCE in humans, the indirect, mass balance-based estimates of GSH conjugation from the PBPK were highly uncertain, spanning around 3,000-fold.52 All our results were more consistent with PBPK model-derived higher-end estimates, contrary to the data reported by Green et al.28 and Dekant et al.29
Discussion
It is not unusual that species concordance overall, and characterization of species-specific metabolism in particular,53 are the areas where major uncertainties exist that prevent regulatory risk assessments from being as precise in their estimates of safe exposure levels as would be preferred. Although the so-called “uncertainty factors” have been used traditionally to express the degree to which default assumptions are to be used when the reliable data are lacking,54 calls have been made to use all available information, including new approach methods (NAMs) data,55 to reduce uncertainties and increase both confidence and precision in risk characterization. Indeed, development of NAMs that can help fill important information gaps in chemical safety assessments is one of the primary goals of the U.S. EPA work plan to reduce use of vertebrate animals in chemical testing.56 One type of a NAM that may assist with addressing the uncertainties in species-specific metabolism of drugs and chemicals is microphysiological systems for the liver.34 A number of such models have been developed by researchers in academia and the private sector,57 and their use in chemical risk assessment has been proposed in next-generation risk assessments.58 Although the opportunities are many, the challenges are also formidable, including the cost, complexity, and low throughput of most of the available models.38,59,60
With these considerations in mind and the desire to address the specific key gaps in GSH metabolism of TCE and PCE across species, we chose to use the MPCC model,39 together with more traditional suspension and 2-D cultures of hepatocytes,31 to enable these experiments with sufficient replication. Although conventional 2-D platforms (i.e., hepatocyte monocultures on collagen-adsorbed polystyrene or glass) decline in functionality over time, controlling homotypic interactions between hepatocytes with circular domains of empirically optimized dimensions and heterotypic interactions with supportive 3T3-J2 murine embryonic fibroblasts has been shown to induce high levels of stable functions in hepatocytes from various species, including human, rat, and mouse, as well as human iHep.33 To this end, 3T3-J2 fibroblasts have been found to express various liver developmental signals, including T-cadherin and decorin, allowing hepatocytes to maintain in vivo-like morphology, polarity, and functionality for several weeks in vitro. Furthermore, MPCCs also appear to be more consistent with metabolism estimates based on PBPK models calibrated to in vivo GSH conjugation data, providing addition empirical support for their quantitative validity.
To illustrate the potential for MPCC to quantitatively impact human health risk assessment, we consider the implications of this work on reducing uncertainties in interspecies differences in metabolism characterized in the U.S. EPA IRIS Toxicological Reviews for TCE7 and PCE.9 With respect to TCE, the U.S. EPA7 used five studies in mice and rats to derive candidate RfDs61–65 and three studies to derive candidate RfCs61,62,64 (Figure 5A–B). The U.S. EPA noted7 that “There remains substantial uncertainty in the extrapolation of GSH conjugation from rodents to humans due to limitations in the available data.” The GSH conjugation data for TCE reported in the available studies are highly discordant, with the studies by Lash et al. reporting values several orders of magnitude higher than those reported by Green and Dekant et al. The U.S. EPA Science Advisory Board suggested that the discordance between these in vitro studies and PBPK model estimates constituted uncertainties in the rate of GSH conjugation in humans and thus recommended against relying on the PBPK modeling-based estimates of GSH conjugation for toxicity value derivation based on kidney end points in the U.S. EPA TCE assessment.7 As a result, the candidate RfDs and RfCs for kidney toxicity,64,65 which used the GSH conjugation predictions from the PBPK model for interspecies, intraspecies, and route-to-route extrapolation, were not considered of sufficient confidence to be used as a primary basis for the overall RfD and RfC. Instead, the final was based on developmental immune,63 immune,61 and fetal cardiac effects,62 and the final was based on immune61 and fetal cardiac effects.62 However, the MPCC data from our study provide independent corroboration for both the TCE PBPK model18 used by the U.S. EPA, as well as the in vitro measurements by Lash et al., increasing the confidence in the candidate RfDs of 64 and 65 and candidate RfC of 64 for kidney effects (Figures 5A–B). Because these are very close to the candidate RfD and RfC based on other critical end points, they further strengthen the basis for the U.S. EPA’s overall toxicity values for TCE noncancer effects by oral or inhalation exposures.
Figure 5.
Comparison of cRfD (panels A and C) and cRfC (panels B and D) for TCE (A,B) and PCE (C,D) with corresponding uncertainty factors. Plotted are points of departure (open diamonds) and candidate toxicity values (open circles) as well as uncertainty factors for study-specific effects as indicated by the references. The vertical red dotted line represents the final RfD or RfC values for TCE7 and PCE.9 , animal to human; , database; , human variability; , LOAEL to NOAEL. Red dashed-line rectangles identify original (as identified in U.S. EPA9) and alternative (based on the refined estimates of GSH conjugation using the data from this study) cRfD and cRfC for PCE. All data points for TCE and PCE cRfD and cRfC, including uncertainty factor values, can be found in Excel Tables S7–S9. Note: cRfC, candidate reference concentration; cRfD, candidate reference dose; LOAEL, lowest observed adverse effect level; NOAEL, no observed adverse effect level; PCE, tetrachloroethylene; TCE, trichloroethylene.
With respect to PCE, the U.S. EPA9 used three studies in humans66–68 and a study in rats and mice69 to derive candidate RfDs and RfCs (Figures 5C–D). Based on conflicting previous in vitro data, uncertainties of many orders of magnitude were noted for PCE GSH conjugation in humans, and thus again the U.S. EPA did not rely on the PBPK modeling-based estimates of GSH conjugation for toxicity value derivation based on kidney end points in the PCE assessment.9 In particular, the PBPK model52 predictions for GSH conjugation exhibited uncertainties in humans for this pathway, with few human in vitro data and no human in vivo data on this pathway. Because of these uncertainties, interspecies extrapolation of kidney effects68,69 was based on AUC of PCE in blood instead of a more mechanistically supported dose metric related to GSH conjugation. In this case, by analogy to the success of MPCC data for TCE being consistent with in vivo GSH conjugation data, the MPCC data can be used to substantially reduce the uncertainties from the PBPK modeling, which spanned such a wide range due to the lack of sufficient in vivo calibration data on GSH conjugation metabolites. In particular, restricting the range of PBPK model-based GSH conjugation predictions to those consistent with MPCC data (see Excel Tables S7–S9 for details), we can use the results both for interspecies extrapolation from rats to humans and route-to-route extrapolation from inhalation to oral exposure. This approach shifts the PODs based on kidney effects to lower values as follows. For the RfD, the resulting kidney-specific RfD values are based on either human or rat data, which is in the range of the neurotoxicity study-specific RfDs of 66,67 that formed the basis of the overall RfD of (Figure 5C). Similarly, for the RfC, the resulting kidney-specific RfD value is based on rat data, which is in the range of the neurotoxicity study-specific RfCs of 66,67 that formed the basis of the overall RfC of (Figure 5D). Moreover, use of the MPCC to restrict the PBPK model predictions also leads to greater overall concordance between the human- and rat-based kidney-specific reference values.
We note a few limitations in our study. First, our testing concentration of for TCE and PCE is well above estimated environmental exposure levels.70,71 The estimated GSH conjugation values for TCE and PCE range from 0.2 to .7,52 Therefore, testing at is expected to saturate hepatocytes and limit metabolic capacity at . We also acknowledge that our MPCC configuration does not contain other liver nonparenchymal cells (i.e., liver sinusoidal endothelial cells, hepatic stellate cells, Kupffer cells/macrophages) that are known to regulate hepatocyte functions72,73; however, in vitro cultured/passaged liver nonparenchymal cells, including liver sinusoidal endothelial cells,74 hepatic stellate cells,75 and Kupffer cells76 are not able to induce functions in primary hepatocytes or iHep in MPCC to the same level and duration as cultured 3T3-J2 fibroblasts.33 Additionally, hepatocytes are cultured on rat tail collagen I rather than other mixtures of extracellular matrix proteins found in the liver, which can be challenging to source in substantial quantities and are significantly more expensive than rat collagen I.32 Nonetheless, the MPCC model has exhibited tremendous success in drug screening and recapitulating in vivo phenomena.77–80
In summary, TCE and PCE remain high-priority substances for U.S. EPA risk evaluation and engender significant public health concern, but key uncertainties have remained with respect to GSH conjugation-mediated toxicity. Traditional in vitro systems such as hepatocyte suspensions or subcellular fractions have led to disparate results that have reduced confidence in PBPK modeling of this pathway. Here we hypothesized that MPCC, which takes advantage of advancements in engineered human liver platforms to better mimic physiologically relevant conditions, can provide in vitro data to fill these data gaps and reduce their uncertainties. Our MPCC metabolism data support higher levels of TCE and PCE GSH conjugation flux in humans in comparison with rats or mice, corroborating previous TCE PBPK modeling and reducing uncertainty in previous PCE PBPK modeling. These data thereby facilitate the inclusion of kidney-specific effects for toxicity value derivation, resulting in greater confidence in the U.S. EPA’s noncancer toxicity values for these compounds. Overall, these data suggest that MPCCs can provide physiologically relevant estimates of metabolism to reduce interspecies extrapolation uncertainties, particularly for substances whose toxicity involves bioactivation.
Supplementary Material
Acknowledgments
This work was supported, in part, by grants from the National Institute of Environmental Health Sciences (P42 ES027704) and the U.S. EPA (RD84003201 and RD84045001). The views expressed in this manuscript do not reflect those of the funding agencies. The use of specific commercial products in this work does not constitute endorsement by the funding agencies.
References
- 1.Reitz RH, Gargas ML, Mendrala AL, Schumann AM. 1996. In vivo and in vitro studies of perchloroethylene metabolism for physiologically based pharmacokinetic modeling in rats, mice, and humans. Toxicol Appl Pharmacol 136(2):289–306, PMID: , 10.1006/taap.1996.0036. [DOI] [PubMed] [Google Scholar]
- 2.Cichocki JA, Guyton KZ, Guha N, Chiu WA, Rusyn I, Lash LH. 2016. Target organ metabolism, toxicity, and mechanisms of trichloroethylene and perchloroethylene: key similarities, differences, and data gaps. J Pharmacol Exp Ther 359(1):110–123, PMID: , 10.1124/jpet.116.232629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guha N, Loomis D, Grosse Y, Lauby-Secretan B, El Ghissassi F, Bouvard V, et al. 2012. Carcinogenicity of trichloroethylene, tetrachloroethylene, some other chlorinated solvents, and their metabolites. Lancet Oncol 13(12):1192–1193, PMID: , 10.1016/S1470-2045(12)70485-0. [DOI] [PubMed] [Google Scholar]
- 4.IARC (International Agency for Research on Cancer) Working Group on the Evaluation of Carcinogenic Risks to Humans. 2014. Trichloroethylene, tetrachloroethylene, and some other chlorinated agents. IARC Monogr Eval Carcinog Risks Hum 106:1–512, PMID: . [PMC free article] [PubMed] [Google Scholar]
- 5.Fay RM, Mumtaz MM. 1996. Development of a priority list of chemical mixtures occurring at 1188 hazardous waste sites, using the HazDat database. Food Chem Toxicol 34(11–12):1163–1165, PMID: , 10.1016/s0278-6915(97)00090-2. [DOI] [PubMed] [Google Scholar]
- 6.Rusyn I, Chiu WA, Lash LH, Kromhout H, Hansen J, Guyton KZ. 2014. Trichloroethylene: mechanistic, epidemiologic and other supporting evidence of carcinogenic hazard. Pharmacol Ther 141(1):55–68, PMID: , 10.1016/j.pharmthera.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.U.S. EPA (U.S. Environmental Protection Agency). 2011. Toxicological Review of Trichloroethylene (CAS No. 79-01-6): In Support of Summary Information on the Integrated Risk Information System (IRIS). Washington, DC: U.S. EPA. [Google Scholar]
- 8.NTP (National Toxicology Program). 2021. Report on Carcinogens, Fifteenth Edition. Research Triangle Park: U.S. Department of Health and Human Services. [Google Scholar]
- 9.U.S. EPA. 2012. Toxicological Review of Tetrachloroethylene (CAS No. 127-18-4): In Support of Summary Information on the Integrated Risk Information System (IRIS). Washington, DC: U.S. Environmental Protection Agency. [Google Scholar]
- 10.NTP. 1986. NTP toxicology and carcinogenesis studies of tetrachloroethylene (perchloroethylene) (CAS No. 127-18-4) in F344/N rats and B6C3F1 mice (inhalation studies). Natl Toxicol Program Tech Rep Ser 311:1–197, PMID: . [PubMed] [Google Scholar]
- 11.U.S. EPA. 2016. EPA Names First Chemicals for Review under New TSCA Legislation. https://19january2017snapshot.epa.gov/newsreleases/epa-names-first-chemicals-review-under-new-tsca-legislation_.html [accessed 24 November 2022].
- 12.National Academies of Sciences Engineering and Medicine. 2019. Review of DOD’s Approach to Deriving an Occupational Exposure Level for Trichloroethylene. Washington, DC: The National Academies Press. [PubMed] [Google Scholar]
- 13.Richard AM, Huang R, Waidyanatha S, Shinn P, Collins BJ, Thillainadarajah I, et al. 2021. The Tox21 10K compound library: collaborative chemistry advancing toxicology. Chem Res Toxicol 34(2):189–216, PMID: , 10.1021/acs.chemrestox.0c00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lash LH, Chiu WA, Guyton KZ, Rusyn I. 2014. Trichloroethylene biotransformation and its role in mutagenicity, carcinogenicity and target organ toxicity. Mutat Res Rev Mutat Res 762:22–36, PMID: , 10.1016/j.mrrev.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lash LH, Lipscomb JC, Putt DA, Parker JC. 1999. Glutathione conjugation of trichloroethylene in human liver and kidney: kinetics and individual variation. Drug Metab Dispos 27(3):351–359, PMID: . [PubMed] [Google Scholar]
- 16.Lash LH, Qian W, Putt DA, Hueni SE, Elfarra AA, Sicuri AR, et al. 2002. Renal toxicity of perchloroethylene and S-(1,2,2-trichlorovinyl)glutathione in rats and mice: sex- and species-dependent differences. Toxicol Appl Pharmacol 179(3):163–171, PMID: , 10.1006/taap.2001.9358. [DOI] [PubMed] [Google Scholar]
- 17.Lash LH, Putt DA, Huang P, Hueni SE, Parker JC. 2007. Modulation of hepatic and renal metabolism and toxicity of trichloroethylene and perchloroethylene by alterations in status of cytochrome P450 and glutathione. Toxicology 235(1–2):11–26, PMID: , 10.1016/j.tox.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiu WA, Okino MS, Evans MV. 2009. Characterizing uncertainty and population variability in the toxicokinetics of trichloroethylene and metabolites in mice, rats, and humans using an updated database, physiologically based pharmacokinetic (PBPK) model, and Bayesian approach. Toxicol Appl Pharmacol 241(1):36–60, PMID: , 10.1016/j.taap.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 19.Völkel W, Friedewald M, Lederer E, Pähler A, Parker J, Dekant W. 1998. Biotransformation of perchloroethene: dose-dependent excretion of trichloroacetic acid, dichloroacetic acid, and N-acetyl-S-(trichlorovinyl)-L-cysteine in rats and humans after inhalation. Toxicol Appl Pharmacol 153(1):20–27, PMID: , 10.1006/taap.1998.8548. [DOI] [PubMed] [Google Scholar]
- 20.Lash LH, Parker JC. 2001. Hepatic and renal toxicities associated with perchloroethylene. Pharmacol Rev 53(2):177–208, PMID: . [PubMed] [Google Scholar]
- 21.Chiu WA, Jinot J, Scott CS, Makris SL, Cooper GS, Dzubow RC, et al. 2013. Human health effects of trichloroethylene: key findings and scientific issues. Environ Health Perspect 121(3):303–311, PMID: , 10.1289/ehp.1205879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lash LH, Xu Y, Elfarra AA, Duescher RJ, Parker JC. 1995. Glutathione-dependent metabolism of trichloroethylene in isolated liver and kidney cells of rats and its role in mitochondrial and cellular toxicity. Drug Metab Dispos 23(8):846–853, PMID: . [PubMed] [Google Scholar]
- 23.Luo YS, Cichocki JA, McDonald TJ, Rusyn I. 2017. Simultaneous detection of the tetrachloroethylene metabolites S-(1,2,2-trichlorovinyl) glutathione, S-(1,2,2-trichlorovinyl)-L-cysteine, and N-acetyl-S-(1,2,2-trichlorovinyl)-L-cysteine in multiple mouse tissues via ultra-high performance liquid chromatography electrospray ionization tandem mass spectrometry. J Toxicol Environ Health A 80(9):513–524, PMID: , 10.1080/15287394.2017.1330585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lash LH, Qian W, Putt DA, Desai K, Elfarra AA, Sicuri AR, et al. 1998. Glutathione conjugation of perchloroethylene in rats and mice in vitro: sex-, species-, and tissue-dependent differences. Toxicol Appl Pharmacol 150(1):49–57, PMID: , 10.1006/taap.1998.8402. [DOI] [PubMed] [Google Scholar]
- 25.Cristofori P, Sauer AV, Trevisan A. 2015. Three common pathways of nephrotoxicity induced by halogenated alkenes. Cell Biol Toxicol 31(1):1–13, PMID: , 10.1007/s10565-015-9293-x. [DOI] [PubMed] [Google Scholar]
- 26.Dekant W, Birner G, Werner M, Parker J. 1998. Glutathione conjugation of perchloroethene in subcellular fractions from rodent and human liver and kidney. Chem Biol Interact 116(1–2):31–43, PMID: , 10.1016/s0009-2797(98)00077-5. [DOI] [PubMed] [Google Scholar]
- 27.Green T, Odum J, Nash JA, Foster JR. 1990. Perchloroethylene-induced rat-kidney tumors—an investigation of the mechanisms involved and their relevance to humans. Toxicol Appl Pharmacol 103(1):77–89, PMID: , 10.1016/0041-008x(90)90264-u. [DOI] [PubMed] [Google Scholar]
- 28.Green T, Dow J, Ellis MK, Foster JR, Odum J. 1997. The role of glutathione conjugation in the development of kidney tumours in rats exposed to trichloroethylene. Chem Biol Interact 105(2):99–117, PMID: , 10.1016/s0009-2797(97)00040-9. [DOI] [PubMed] [Google Scholar]
- 29.Dekant W, Koob M, Henschler D. 1990. Metabolism of trichloroethene–in vivo and in vitro evidence for activation by glutathione conjugation. Chem Biol Interact 73(1):89–101, PMID: , 10.1016/0009-2797(90)90110-9. [DOI] [PubMed] [Google Scholar]
- 30.Lash LH, Qian W, Putt DA, Jacobs K, Elfarra AA, Krause RJ, et al. 1998. Glutathione conjugation of trichloroethylene in rats and mice: sex-, species-, and tissue-dependent differences. Drug Metab Dispos 26(1):12–19, PMID: . [PubMed] [Google Scholar]
- 31.Soldatow VY, Lecluyse EL, Griffith LG, Rusyn I. 2013. In vitro models for liver toxicity testing. Toxicol Res (Camb) 2(1):23–39, PMID: , 10.1039/C2TX20051A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khetani SR, Berger DR, Ballinger KR, Davidson MD, Lin C, Ware BR. 2015. Microengineered liver tissues for drug testing. J Lab Autom 20(3):216–250, PMID: , 10.1177/2211068214566939. [DOI] [PubMed] [Google Scholar]
- 33.Monckton CP, Brown GE, Khetani SR. 2021. Latest impact of engineered human liver platforms on drug development. APL Bioeng 5(3):031506, PMID: , 10.1063/5.0051765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baudy AR, Otieno MA, Hewitt P, Gan J, Roth A, Keller D, et al. 2020. Liver microphysiological systems development guidelines for safety risk assessment in the pharmaceutical industry. Lab Chip 20(2):215–225, PMID: , 10.1039/c9lc00768g. [DOI] [PubMed] [Google Scholar]
- 35.Sarkar U, Ravindra KC, Large E, Young CL, Rivera-Burgos D, Yu J, et al. 2017. Integrated assessment of diclofenac biotransformation, pharmacokinetics, and Omics-Based toxicity in a three-dimensional human liver-immunocompetent coculture system. Drug Metab Dispos 45(7):855–866, PMID: , 10.1124/dmd.116.074005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsamandouras N, Kostrzewski T, Stokes CL, Griffith LG, Hughes DJ, Cirit M. 2017. Quantitative assessment of population variability in hepatic drug metabolism using a perfused three-dimensional human liver microphysiological system. J Pharmacol Exp Ther 360(1):95–105, PMID: , 10.1124/jpet.116.237495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakolish C, Luo YS, Valdiviezo A, Vernetti LA, Rusyn I, Chiu WA. 2021. Prediction of hepatic drug clearance with a human microfluidic four-cell liver acinus microphysiology system. Toxicology 463:152954, PMID: , 10.1016/j.tox.2021.152954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valdiviezo A, Kato Y, Baker ES, Chiu WA, Rusyn I. 2022. Evaluation of metabolism of a defined pesticide mixture through multiple in vitro liver models. Toxics 10(10):566, PMID: , 10.3390/toxics10100566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khetani SR, Bhatia SN. 2008. Microscale culture of human liver cells for drug development. Nat Biotechnol 26(1):120–126, PMID: , 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 40.Ukairo O, Kanchagar C, Moore A, Shi J, Gaffney J, Aoyama S, et al. 2013. Long-term stability of primary rat hepatocytes in micropatterned cocultures. J Biochem Mol Toxicol 27(3):204–212, PMID: , 10.1002/jbt.21469. [DOI] [PubMed] [Google Scholar]
- 41.Berger DR, Ware BR, Davidson MD, Allsup SR, Khetani SR. 2015. Enhancing the functional maturity of induced pluripotent stem cell-derived human hepatocytes by controlled presentation of cell-cell interactions in vitro. Hepatology 61(4):1370–1381, PMID: , 10.1002/hep.27621. [DOI] [PubMed] [Google Scholar]
- 42.Ware BR, Brown GE, Soldatow VY, LeCluyse EL, Khetani SR. 2019. Long-Term engineered cultures of primary mouse hepatocytes for strain and species comparison studies during drug development. Gene Expr 19(3):199–214, PMID: , 10.3727/105221619X15638857793317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim S, Collins LB, Boysen G, Swenberg JA, Gold A, Ball LM, et al. 2009. Liquid chromatography electrospray ionization tandem mass spectrometry analysis method for simultaneous detection of trichloroacetic acid, dichloroacetic acid, S-(1,2-dichlorovinyl)glutathione and S-(1,2-dichlorovinyl)-L-cysteine. Toxicology 262(3):230–238, PMID: , 10.1016/j.tox.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seglen PO. 1976. Preparation of isolated rat liver cells. Methods Cell Biol 13:29–83, PMID: . [DOI] [PubMed] [Google Scholar]
- 45.Lee SM, Pusec CM, Norris GH, De Jesus A, Diaz-Ruiz A, Muratalla J, et al. 2021. Hepatocyte-specific loss of PPARγ protects mice from NASH and increases the therapeutic effects of rosiglitazone in the liver. Cell Mol Gastroenterol Hepatol 11(5):1291–1311, PMID: , 10.1016/j.jcmgh.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grimm FA, Iwata Y, Sirenko O, Bittner M, Rusyn I. 2015. High-Content assay multiplexing for toxicity screening in induced pluripotent stem cell-derived cardiomyocytes and hepatocytes. Assay Drug Dev Technol 13(9):529–546, PMID: , 10.1089/adt.2015.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rheinwald JG, Green H. 1975. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell 6(3):331–343, PMID: , 10.1016/S0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- 48.Luo YS, Furuya S, Chiu W, Rusyn I. 2018. Characterization of inter-tissue and inter-strain variability of TCE glutathione conjugation metabolites DCVG, DCVC, and NAcDCVC in the mouse. J Toxicol Environ Health A 81(1–3):37–52, PMID: , 10.1080/15287394.2017.1408512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Birner G, Vamvakas S, Dekant W, Henschler D. 1993. Nephrotoxic and genotoxic N-acetyl-S-dichlorovinyl-L-cysteine is a urinary metabolite after occupational 1,1,2-trichloroethene exposure in humans: implications for the risk of trichloroethene exposure. Environ Health Perspect 99:281–284, PMID: , 10.1289/ehp.9399281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bernauer U, Birner G, Dekant W, Henschler D. 1996. Biotransformation of trichloroethene: dose-dependent excretion of 2,2,2-trichloro-metabolites and mercapturic acids in rats and humans after inhalation. Arch Toxicol 70(6):338–346, PMID: , 10.1007/s002040050283. [DOI] [PubMed] [Google Scholar]
- 51.Lash LH, Putt DA, Brashear WT, Abbas R, Parker JC, Fisher JW. 1999. Identification of S-(1,2-dichlorovinyl)glutathione in the blood of human volunteers exposed to trichloroethylene. J Toxicol Environ Health A 56(1):1–21, PMID: , 10.1080/009841099158204. [DOI] [PubMed] [Google Scholar]
- 52.Chiu WA, Ginsberg GL. 2011. Development and evaluation of a harmonized physiologically based pharmacokinetic (PBPK) model for perchloroethylene toxicokinetics in mice, rats, and humans. Toxicol Appl Pharmacol 253(3):203–234, PMID: , 10.1016/j.taap.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 53.Green T. 1990. Species differences in carcinogenicity: the role of metabolism in human risk evaluation. Teratog Carcinog Mutagen 10(2):103–113, PMID: , 10.1002/tcm.1770100206. [DOI] [PubMed] [Google Scholar]
- 54.Meek ME, Renwick A, Ohanian E, Dourson M, Lake B, Naumann BD, et al. 2002. Guidelines for application of chemical-specific adjustment factors in dose/concentration-response assessment. Toxicology 181–182:115–120, PMID: , 10.1016/s0300-483x(02)00265-2. [DOI] [PubMed] [Google Scholar]
- 55.Rusyn I, Chiu WA. 2022. Decision-making with new approach methodologies: time to replace default uncertainty factors with data. Toxicol Sci 189(1):148–149, PMID: , 10.1093/toxsci/kfac033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.U.S. EPA. 2021. New Approach Methods Work Plan: Reducing Use of Vertebrate Animals in Chemical Testing. Washington, DC: U.S. EPA. [Google Scholar]
- 57.Messelmani T, Morisseau L, Sakai Y, Legallais C, Le Goff A, Leclerc E, et al. 2022. Liver organ-on-chip models for toxicity studies and risk assessment. Lab Chip 22(13):2423–2450, PMID: , 10.1039/d2lc00307d. [DOI] [PubMed] [Google Scholar]
- 58.Nitsche KS, Müller I, Malcomber S, Carmichael PL, Bouwmeester H. 2022. Implementing organ-on-chip in a next-generation risk assessment of chemicals: a review. Arch Toxicol 96(3):711–741, PMID: , 10.1007/s00204-022-03234-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rusyn I, Sakolish C, Kato Y, Stephan C, Vergara L, Hewitt P, et al. 2022. Microphysiological systems evaluation: experience of TEX-VAL tissue chip testing consortium. Toxicol Sci 188(2):143–152, PMID: , 10.1093/toxsci/kfac061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kato Y, Lim AY, Sakolish C, Valdiviezo A, Moyer HL, Hewitt P, et al. 2022. Analysis of reproducibility and robustness of OrganoPlate® 2-lane 96, a liver microphysiological system for studies of pharmacokinetics and toxicological assessment of drugs. Toxicol In Vitro 85:105464, PMID: , 10.1016/j.tiv.2022.105464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keil DE, Peden-Adams MM, Wallace S, Ruiz P, Gilkeson GS. 2009. Assessment of trichloroethylene (TCE) exposure in murine strains genetically-prone and non-prone to develop autoimmune disease. J Environ Sci Health A Tox Hazard Subst Environ Eng 44(5):443–453, PMID: , 10.1080/10934520902719738. [DOI] [PubMed] [Google Scholar]
- 62.Johnson PD, Goldberg SJ, Mays MZ, Dawson BV. 2003. Threshold of trichloroethylene contamination in maternal drinking waters affecting fetal heart development in the rat. Environ Health Perspect 111(3):289–292, PMID: , 10.1289/ehp.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peden-Adams MM, Eudaly JG, Heesemann LM, Smythe J, Miller J, Gilkeson GS, et al. 2006. Developmental immunotoxicity of trichloroethylene (TCE): studies in B6C3F1 mice. J Environ Sci Health A Tox Hazard Subst Environ Eng 41(3):249–271, PMID: , 10.1080/10934520500455289. [DOI] [PubMed] [Google Scholar]
- 64.NTP. 1988. Toxicology and carcinogenesis studies of trichloroethylene (CAS No. 79-01-6) in four strains of rats (ACI, August, Marshall, Osborne-Mendel) (gavage studies). Natl Toxicol Program Tech Rep Ser 273:1–299, PMID: . [PubMed] [Google Scholar]
- 65.Woolhiser M, Krieger S, Thomas J, Hotchkiss J. 2006. Trichloroethylene (TCE): Immunotoxicity potential in CD rats following a 4-week vapor inhalation exposure. Midland, MI: Dow Chemical Company. 2006(031020). [Google Scholar]
- 66.Echeverria D, White RF, Sampaio C. 1995. A behavioral evaluation of PCE exposure in patients and dry cleaners: a possible relationship between clinical and preclinical effects. J Occup Environ Med 37(6):667–680, PMID: , 10.1097/00043764-199506000-00008. [DOI] [PubMed] [Google Scholar]
- 67.Cavalleri A, Gobba F, Paltrinieri M, Fantuzzi G, Righi E, Aggazzotti G. 1994. Perchloroethylene exposure can induce colour vision loss. Neurosci Lett 179(1–2):162–166, PMID: , 10.1016/0304-3940(94)90959-8. [DOI] [PubMed] [Google Scholar]
- 68.Mutti A, Alinovi R, Bergamaschi E, Biagini C, Cavazzini S, Franchini I, et al. 1992. Nephropathies and exposure to perchloroethylene in dry-cleaners. Lancet 340(8813):189–193, PMID: , 10.1016/0140-6736(92)90463-d. [DOI] [PubMed] [Google Scholar]
- 69.JISA (Japan Industrial Safety Association). 1993. Carcinogenicity Study of Tetrachloroethlene by Inhalation in Rats and Mice. Hadano, Japan: JISA. [Google Scholar]
- 70.ATSDR (Agency for Toxic Substances and Disease Registry). 1997. Toxicological Profile for Trichloroethylene (TCE). Atlanta, GA: Agency for Toxic Substances and Disease Registry, U.S. Department of Health and Human Services. [Google Scholar]
- 71.ATSDR. 2019. Toxicological Profile for Tetrachloroethylene. Atlanta, GA: Agency for Toxic Substances and Disease Registry, Division of Toxicology and Human Health Sciences. [PubMed] [Google Scholar]
- 72.Kmiec Z. 2001. Cooperation of liver cells in health and disease. Adv Anat Embryol Cell Biol 161:1–151. [DOI] [PubMed] [Google Scholar]
- 73.Milosevic N, Schawalder H, Maier P. 1999. Kupffer cell-mediated differential down-regulation of cytochrome P450 metabolism in rat hepatocytes. Eur J Pharmacol 368(1):75–87, PMID: , 10.1016/s0014-2999(98)00988-1. [DOI] [PubMed] [Google Scholar]
- 74.Ware BR, Durham MJ, Monckton CP, Khetani SR. 2018. A cell culture platform to maintain long-term phenotype of primary human hepatocytes and endothelial cells. Cell Mol Gastroenterol Hepatol 5(3):187–207, PMID: , 10.1016/j.jcmgh.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davidson MD, Kukla DA, Khetani SR. 2017. Microengineered cultures containing human hepatic stellate cells and hepatocytes for drug development. Integr Biol (Camb) 9(8):662–677, PMID: , 10.1039/c7ib00027h. [DOI] [PubMed] [Google Scholar]
- 76.Nguyen TV, Ukairo O, Khetani SR, McVay M, Kanchagar C, Seghezzi W, et al. 2015. Establishment of a hepatocyte-Kupffer cell coculture model for assessment of proinflammatory cytokine effects on metabolizing enzymes and drug transporters. Drug Metab Dispos 43(5):774–785, PMID: , 10.1124/dmd.114.061317. [DOI] [PubMed] [Google Scholar]
- 77.March S, Ramanan V, Trehan K, Ng S, Galstian A, Gural N, et al. 2015. Micropatterned coculture of primary human hepatocytes and supportive cells for the study of hepatotropic pathogens. Nat Protoc 10(12):2027–2053, PMID: , 10.1038/nprot.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ware BR, Berger DR, Khetani SR. 2015. Prediction of Drug-Induced liver injury in micropatterned co-cultures containing iPSC-derived human hepatocytes. Toxicol Sci 145(2):252–262, PMID: , 10.1093/toxsci/kfv048. [DOI] [PubMed] [Google Scholar]
- 79.Khetani SR, Kanchagar C, Ukairo O, Krzyzewski S, Moore A, Shi J, et al. 2013. Use of micropatterned cocultures to detect compounds that cause drug-induced liver injury in humans. Toxicol Sci 132(1):107–117, PMID: , 10.1093/toxsci/kfs326. [DOI] [PubMed] [Google Scholar]
- 80.Wang WW, Khetani SR, Krzyzewski S, Duignan DB, Obach RS. 2010. Assessment of a micropatterned hepatocyte coculture system to generate major human excretory and circulating drug metabolites. Drug Metab Dispos 38(10):1900–1905, PMID: , 10.1124/dmd.110.034876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





![Figure 4A are set of two box and whiskers plots titled human, plotting This study [Micropatterned Co-Cultures, primary human hepatocyte, and induced hepatocyte-like cells; 2-D; primary human hepatocyte pooled and single; suspension; primary human hepatocyte and induced hepatocyte-like cells], Chiu and others 2009 [P B P K], Lash and others 1995, 1998, 1999, 2007 [suspension and subcellular fractions], and Green and others 1997, Dekant and others 1990 [subcellular fractions] and This study [Micropatterned Co-Cultures, primary human hepatocyte, and induced hepatocyte-like cells; 2-D; primary human hepatocyte pooled and single; suspension; primary human hepatocyte and induced hepatocyte-like cells], Chiu and Ginsberg 2011 [P B P K], Lash and others 1998, 2007, [suspension and subcellular fractions], Dekant and others, 1998 [subcellular fractions], and Green and others 1990 (y-axis) across trichloroethylene glutathione conjugation (nanomole per hour per 1 molar hepatocyte, ranging from 0.01 to 0.1 in increments of 0.09, 0.1 to 1 in increments of 0.9, 1 to 10 in increments of 9, 10 to 100 in increments of 90, 100 to 1000 in increments of 900 and tetrachloroethylene glutathione conjugation (nanomole per hour per 1 molar hepatocyte, ranging from 0.01 to 0.1 in increments of 0.09, 0.1 to 1 in increments of 0.9, 1 to 10 in increments of 9, 10 to 100 in increments of 90 (x-axis). Figure 4B are set of two box and whiskers plots titled rat, plotting This study [Micropatterned Co-Cultures, 2 D, suspension], Chiu and others 2009 [P B P K], Lash and others 1995, 1998, 1999, 2007 [suspension and subcellular fractions], and Green and others 1997, Dekant and others 1990 [subcellular fractions] and This study [Micropatterned Co-Cultures, 2 D, suspension], Chiu and Ginsberg 2011 [P B P K], Lash and others 1998, 2007, [suspension and subcellular fractions], Dekant and others, 1998 [subcellular fractions], and Green and others 1990 (y-axis) across trichloroethylene glutathione conjugation (nanomole per hour per 1 molar hepatocyte, ranging from 0.01 to 0.1 in increments of 0.09, 0.1 to 1 in increments of 0.9, 1 to 10 in increments of 9, 10 to 100 in increments of 90, 100 to 1000 in increments of 900 and tetrachloroethylene glutathione conjugation (nanomole per hour per 1 molar hepatocyte, ranging from 0.01 to 0.1 in increments of 0.09, 0.1 to 1 in increments of 0.9, 1 to 10 in increments of 9, 10 to 100 in increments of 90 (x-axis). Figures 4C are set of two box and whiskers plots titled rat, plotting This study [Micropatterned Co-Cultures, 2 D, suspension], Chiu and others 2009 [P B P K], Lash and others 1995, 1998, 1999, 2007 [suspension and subcellular fractions], and Green and others 1997, Dekant and others 1990 [subcellular fractions] and This study [Micropatterned Co-Cultures, 2 D, suspension], Chiu and Ginsberg 2011 [P B P K], Lash and others 1998, 2007, [suspension and subcellular fractions], Dekant and others, 1998 [subcellular fractions], and Green and others 1990 (y-axis) across trichloroethylene glutathione conjugation (nanomole per hour per 1 molar hepatocyte, ranging from 0.01 to 0.1 in increments of 0.09, 0.1 to 1 in increments of 0.9, 1 to 10 in increments of 9, 10 to 100 in increments of 90, 100 to 1000 in increments of 900 and tetrachloroethylene glutathione conjugation (nanomole per hour per 1 molar hepatocyte, ranging from 0.01 to 0.1 in increments of 0.09, 0.1 to 1 in increments of 0.9, 1 to 10 in increments of 9, 10 to 100 in increments of 90 (x-axis).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/0845/9707501/0d55c9fcb98f/ehp12006_f4.jpg)
