Vaccination represents the key strategy to control the COVID-19 pandemic through induction of neutralising antibody responses and T cell-associated immunity that substantially decrease the risk of developing severe disease.1, 2 However, individuals who are immunocompromised (eg, because of comorbidities, high age, or immunosuppressive treatment) might not mount a full adaptive immune response and thus remain susceptible. For individuals at high risk, individual monoclonal antibodies (mAbs) or cocktails of mAbs are administered as prophylaxis or therapy.3, 4 All mAbs currently approved by the US Food and Drug Administration (FDA) or European Medicines Agency (EMA) target the spike (S) protein (appendix pp 1–2).5 During the course of the COVID-19 pandemic, several SARS-CoV-2 lineages evolved mutations that confer partial or full resistance against some mAbs.6, 7, 8, 9 Consequently, only few mAbs remain suitable for treatment of individuals at high risk, and only bebtelovimab shows high efficacy against multiple omicron sublineages.8 However, novel omicron sublineages have been detected, harbouring additional S protein mutations within the epitopes of bebtelovimab and other mAbs (figure A ; appendix p 11). Novel sublineages include BA.4.6 (with increasing incidence in several countries worldwide), BA.2.75.2 (with increasing incidence in India), BJ.1 (mainly observed in India and Bangladesh; notably BJ.1 is one parental lineage of the currently increasing XBB recombinant), and BQ.1.1 (with increasing incidence in the USA and Europe).
Figure.
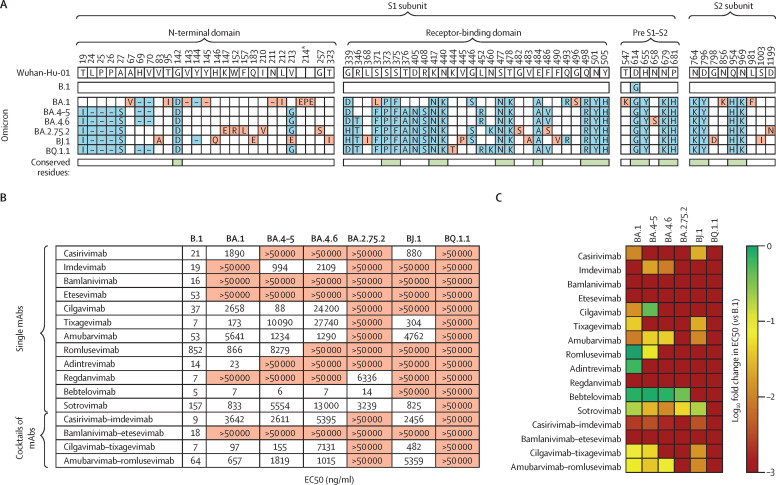
Extensive resistance of omicron sublineage B.Q.1.1 to neutralisation by mAbs
(A) Location of mutations (blue and red) in the spike proteins of SARS-CoV-2 lineages B.1, BA.1, and BA.4–5 (which are identical at the amino acid level), BA.4.6, BA.2.75.2, BJ.1, and BQ.1.1 (numbered according to the spike protein of SARS-CoV-2 Wuhan-Hu-01). Mutations that are unique to only one of the omicron sublineages are highlighted in red and conserved mutations among omicron sublineages are indicated beneath the sequences in green. (B) Pseudovirus particles carrying the indicated S proteins were preincubated with different concentrations of single mAbs or cocktails of mAbs, before being inoculated onto Vero cells. Pseudovirus entry was analysed at 16–18 h post-inoculation, by measuring firefly luciferase activity in cell lysates, and was normalised against samples without any antibodies (0% inhibition). The EC50 was calculated by use of a non-linear regression model. Data represent the mean of three biological replicates (performed with four technical replicates). For additional information see the appendix (p 12). (C) Heatmap indicating the fold change in EC50 compared with B.1 pseudovirus particles. EC50=the concentration required for 50% of maximum inhibition. mAbs=monoclonal antibodies. Pre S1–S2=the domain between the receptor-binding domain and the S1–S2 cleavage site. S=spike. *The BA.1 spike protein contains a unique insertion at position 214 (EPE).
We compared neutralisation of omicron sublineages BA.1, BA.4–5 (in which the amino acid sequence of the S protein is identical), BA.4.6, BA.2.75.2, BJ.1 and BQ.1.1 by single mAbs or mAb cocktails that are currently in clinical use, mAbs for which clinical use has been restricted or discontinued, and mAbs currently being evaluated in clinical trials. We used pseudovirus particles (pp) that represent a suitable model to investigate SARS-CoV-2 cell entry and its neutralisation.10 As we expected, pseudovirus particles bearing the BA.1 S protein (BA.1pp) were efficiently neutralised by bebtelovimab, adintrevimab, and cilgavimab–tixagevimab (50% effective concentration [EC50] <100 ng/ml), moderately neutralised by tixagevimab, romlusevimab, sotrovimab, and amubarvimab–romlusevimab (EC50 100–1000 ng/ml), and poorly neutralised by casirivimab, cilgavimab, amubarvimab, and casirivimab–imdevimab (EC50 1000–10 000 ng/ml).7 Furthermore, BA4–5pp were efficiently neutralised by bebtelovimab and cilgavimab, moderately neutralised by imdevimab and cilgavimab–tixagevimab, and poorly neutralised by amubarvimab, romlusevimab, sotrovimab, casirivimab–imdevimab, and amubarvimab–romlusevimab, in line with expectations.8 For BA.4.6pp, bebtelovimab caused efficient neutralisation, whereas poor neutralisation was noted for imdevimab, amubarvimab, casirivimab–imdevimab, cilgavimab–tixagevimab, and amubarvimab–romlusevimab. With BA.2.75.2pp, bebtelovimab caused efficient neutralisation, whereas regdanvimab and sotrovimab caused poor neutralisation. For BJ.1pp, none of the tested mAbs or mAb cocktails caused high neutralisation, whereas casirivimab, tixagevimab, sotrovimab, and cilgavimab–tixagevimab showed moderate neutralisation, and amubarvimab, casirivimab–imdevimab, and amubarvimab–romlusevimab caused poor neutralisation. Finally, none of the tested mAbs or mAb cocktails caused appreciable neutralisation of BQ.1.1pp (figure B–C; appendix p 12).
Our data reveal that emerging omicron sublineages are resistant to most (ie, BA.4.6, BA.2.75.2, and BJ.1) or all (BQ.1.1) clinically used mAbs. As a consequence, in patients at high risk, treatment with mAbs alone might not provide a therapeutic benefit in regions of the globe in which BQ.1.1 is spreading, suggesting that additional treatment options (eg, paxlovid or molnupiravir) should be considered. Furthermore, novel, broadly active mAbs are urgently needed for prophylactic or therapeutic treatment, or both, in patients at high risk.
This online publication has been corrected. The corrected version first appeared at thelancet.com/infection on November 29, 2022
AK, IN, and MH do contract research (testing of vaccinee sera for neutralising activity against SARS-CoV-2) for Valneva, unrelated to this Correspondence. SP does contract research (testing of vaccinee sera for neutralising activity against SARS-CoV-2) for Valneva and served as advisor for BioNTech, unrelated to Correspondence, and acknowledges funding by the Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung [BMBF]; 01KI2006D, 01KI20328A, and 01KX2021), the EU project UNDINE (grant agreement number 101057100), the Ministry for Science and Culture of Lower Saxony (14–76103–184, MWK HZI COVID-19), and the German Research Foundation (DFG; PO 716/11–1 and PO 716/14–1). H-MJ received funding from BMBF (01KI2043 and NaFoUniMedCovid19-COVIM: 01KX2021), the Bavarian State Ministry for Science and the Arts and DFG through the research training groups RTG1660 and TRR130, the Bayerische Forschungsstiftung (Project CORAd), and the Kastner Foundation. All other authors declare no competing interests.
Supplementary Material
References
- 1.Thompson MG, Yoon SK, Naleway AL, et al. Association of mRNA Vaccination With Clinical and Virologic Features of COVID-19 Among US Essential and Frontline Workers. JAMA. 2022;328:1523–1533. doi: 10.1001/jama.2022.18550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olson SM, Newhams MM, Halasa NB, et al. Effectiveness of BNT162b2 vaccine against critical COVID-19 in adolescents. N Engl J Med. 2022;386:713–723. doi: 10.1056/NEJMoa2117995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milam AN, Doan DT, Childress DT, Durham SH. Evaluation of monoclonal antibodies in preventing hospitalizations, emergency department visits, and mortality in high-risk COVID-19 patients. J Pharm Technol. 2022;38:169–173. doi: 10.1177/87551225221080027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levin MJ, Ustianowski A, De Wit S, et al. Intramuscular AZD7442 (tixagevimab–cilgavimab) for prevention of COVID-19. N Engl J Med. 2022;386:2188–2200. doi: 10.1056/NEJMoa2116620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Focosi D, McConnell S, Casadevall A, Cappello E, Valdiserra G, Tuccori M. Monoclonal antibody therapies against SARS-CoV-2. Lancet Infect Dis. 2022;22:e311–e326. doi: 10.1016/S1473-3099(22)00311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheward DJ, Kim C, Fischbach J, et al. Omicron sublineage BA.2.75.2 exhibits extensive escape from neutralising antibodies. Lancet Infect Dis. 2022;22:1538–1540. doi: 10.1016/S1473-3099(22)00663-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arora P, Zhang L, Krüger N, et al. SARS-CoV-2 omicron sublineages show comparable cell entry but differential neutralization by therapeutic antibodies. Cell Host Microbe. 2022;30:1103. doi: 10.1016/j.chom.2022.04.017. 11.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arora P, Kempf A, Nehlmeier I, et al. Augmented neutralisation resistance of emerging omicron subvariants BA.2.12.1, BA.4, and BA.5. Lancet Infect Dis. 2022;22:1117–1118. doi: 10.1016/S1473-3099(22)00422-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao Y, Jian F, Wang J, et al. Imprinted SARS-CoV-2 humoral immunity induces convergent omicron RBD evolution. bioRxiv. 2022 doi: 10.1101/2022.09.15.507787. published online Oct 30. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt F, Weisblum Y, Muecksch F, et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med. 2020;217:e20201181. doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.