Abstract
Background & objectives:
Infections caused by vancomycin-resistant Enterococci are difficult to treat given the limited therapeutic alternatives. Different gene clusters are known to confer vancomycin resistance. vanA and vanB genes are transferable and are clinically relevant. This cross-sectional study aimed to identify the vancomycin-resistant genotypes in the strains causing urinary tract infection and also to test the in vitro efficacy of linezolid and pristinamycin against the vancomycin-resistant isolates.
Methods:
Antimicrobial resistance profile of 118 enterococcal isolates was evaluated. Minimum inhibitory concentration of vancomycin, teicoplanin and high-level gentamicin (HLG) was determined by micro broth dilution. The vancomycin-resistant isolates were tested against linezolid and pristinamycin by micro-broth dilution and E strip method. The presence of vancomycin-resistant genes was detected by multiplex polymerase chain reaction and was sequenced and analyzed.
Results:
Most commonly isolated species were Enterococcus faecalis (76.9%) and Enterococcus faecium (16.9%). It was found that 43 per cent of the isolates were resistant to HLG and 16.9 per cent to vancomycin. Higher resistance was seen against fluoroquinolones, erythromycin, tetracycline and β-lactam drugs. However, 5.08 per cent strains were resistant to tigecycline. All vancomycin-resistant strains were sensitive to pristinamycin and one was resistant to linezolid. vanA and vanB gene were found in 15 and five isolates, respectively. The gene sequences were submitted to NCBI gene bank and accession numbers were obtained.
Interpretation &conclusions:
The present study showed prevalence of vanA and vanB genes carrying Enterococcus in a tertiary care centre in north India. The emergence of resistance against drugs such as tigecycline and linezolid is a topic of concern as it will be a therapeutic challenge for physicians.
Keywords: Enterococcus, linezolid –resistance –tigecyline, vanA, vanB, vancomycin
Enterococci are commensal of the intestine but can be responsible for serious infections at other body sites, most commonly the urinary tract1. They show intrinsic as well as acquired resistance to a wide range of antibiotics2. A glycopeptide antibiotic like vancomycin is considered as the ‘last resort’ for treatment and is used only in case of gram positive infections where there is resistance against penicillin and cephalosporins3. However, over the last two decades, there has been a steep increase in cases of vancomycin resistance in Enterococci4. There is so far no composite data available about the trend in India, but as per sporadic reports, the prevalence of vancomycin-resistant Enterococci (VRE) in India was about one per cent in 2003, which got inflated up to 8.7 per cent in 20135,6,7. Similarly, increase in prevalence has been reported from Uttar Pradesh8 and Maharashtra9,10 and Sikkim11,12.
Vancomycin binds to amide bond of the D-alanyl-D-alanine sequence of the muramyl pentapeptide of the peptidoglycan and stops its elongation, ultimately inhibiting the microbial cell wall synthesis. VRE synthesizes altered binding sites like D-ala-D-lactate or D-ala-D-serine to which the vancomycin molecule fails to bind2. Nine different gene clusters (vanA, vanB, vanC, vanD, vanE, vanG, vanL, vanM and vanN) have been identified so far that are responsible for vancomycin resistance. These genotypes differ from each other in terms of transferability, resistance towards other glycopeptides and minimum inhibitory concentration (MIC) levels13,14. However, of these, vanA and vanB are most clinically relevant15. Both vanA and vanB genes are plasmid borne and confer acquired inducible high-level resistance to vancomycin. Unlike vanA, vanB strains are sensitive to teicoplanin. vanC is seen only in Enterococcus gallinarum, E. casseliflavus and E. flavescens. It is chromosomally encoded and bestows non-transmissible, low-level resistance towards vancomycin16. vanD is also chromosomally located and when present, strains show moderate-level resistance towards vancomycin and teicoplanin. vanE, vanG, vanL and vanN strains display low-level resistance to vancomycin but are sensitive to teicoplanin. Resistance profile of vanM is similar to that of vanA17. Studies done in India have shown the prevalence of vanA, vanB and vanC among the Enterococci isolated from clinical samples11,18,19.
Other than urinary tract infections (UTI), Enterococcus is also associated with bloodstream intra-abdominal, intra-pelvic and wound infections1. Glycopeptide resistance makes treatment of such infections challenging as it limits the therapeutic options. In this context, linezolid and pristinamycin are two safe and effective available alternatives20. An effective treatment for VRE warrants accurate and quick detection of type of resistance that may help in determining a worthy alternative. Early detection of resistance profile may help in preventing spread of the VRE in a hospital setup as well as in the community. The present study, hence, aimed to determine the antimicrobial resistance profile and detect the prevalence of six (vanA, vanB, vanC, vanD, vanE and vanG) main vancomycin-resistant genotypes in the Enterococci isolated from clinical samples. In vitro activity of linezolid and pristinamycin against the VRE isolates was also evaluated.
Material & Methods
This cross-sectional study included aseptically collected mid-stream urine samples from symptomatic patients, which were sent to the Microbiology laboratory, Hakeem Abdul Hamid Centenary Hospital, a tertiary care centre in New Delhi, from January 2016 to December 2018, for bacteriological analysis. Permission to conduct the study was obtained from the Institutional Ethics Committee of Jamia Hamdard, New Delhi. The samples were subjected to routine microscopy and culture by plating onto five per cent sheep blood agar and MacConkey agar (Hi-Media, Mumbai) and incubated for 16-48 h in presence of 5-10 per cent CO2 at 37°C21. The enterococcal colonies on blood agar were identified as per colony characteristics and Gram staining and confirmed on the basis of catalase test, hydrolysis of bile esculin, growth in the presence of 6.5 per cent NaCl brain-heart infusion broth and resistance to bacitracin21. Species identification was done using GP ID card, VITEK-2 system (Biomerieux, France).
Antibiotic sensitivity test (AST) was performed by VITEK-2 system using AST-P628 card and Kirby-Bauer disc diffusion method as per CLSI guidelines22 using amoxycillin (10 μg), penicillin (10 units/disc), norfloxacin (10 µg), erythromycin (15 μg), high-level gentamicin (HLG) (120 µg), vancomycin (30 μg), teicoplanin (30 μg), linezolid (30 µg), ciprofloxacin (5 µg), levofloxacin (5 μg), ampicillin-clavulanic acid (20/10 μg), nitrofurantoin (300 μg), tigecycline (15 μg), tetracycline (30 μg) and chloramphenicol (30 μg)22. A reference strain of E. faecalis ATCC 29212 was used as a control.
MIC for high level gentamycin (HLG), vancomycin and teicoplanin was determined using micro broth dilution method following the protocol and MIC breakpoints as per CLSI guidelines22. The round bottom microdilution plate was inoculated with 50 µl of inoculum in each well, followed by 50 µl of various dilutions of the antibiotic. One well containing 100 µl of inoculum (without antibiotic) and another with 100 µl of sterile broth were used as positive and negative control, respectively. The tray was sealed with plastic tape and incubated at 35±2°C for 16-20 h in an incubator. The lowest concentration of the antibiotic that completely inhibited growth in the well as detected by the naked eye was considered as the MIC22. The in vitro activity of linezolid and pristinamycin against the VRE isolates was checked by micro broth dilution and E test ( Ezy MIC strip, Hi media), respectively.
Bacterial DNA was extracted from isolates as previously described23. Presence of the six main genes encoding the vancomycin-resistance determinants were investigated by PCR using previously published specific primers18,24,25. ATCC 51299 was used as positive control.
PCR reactions were carried out in 25 µl volume using 10 µl commercially available ready to use master mix, 0.5 µl of each forward and reverse primers, 6 µl of molecular grade water and 3 µl of DNA template (10 µg/ml). DNA amplification was done in a PCR thermocycler (2720 thermal cycler, Applied Biosystems) with the following thermal cycling profile: an initial denaturation step at 94°C for two minutes, followed by 25 cycles of amplification (94°C for 60 sec, 55°C for 60 sec and 72°C for 60 sec) and an extension at 72°C for 5 min. PCR products were resolved on a 1.5 per cent agarose gel stained with ethidium bromide, using a Sub-Cell GT electrophoresis system (Bio-Rad, USA). A 100 bp DNA ladder (Vivantis Technologies Sdn. Bhd., Malaysia) was run in every gel and the size of each VRE genotype was determined26.
The PCR products were sequenced using the ABI Big Dye Terminator version 3.1” Cycle sequencing kit (Applied Biosystems, Life Technologies Co., USA) in a Micro Amp Optical 96-Well Reaction plate with ABI 3500 Genetic Analyzer (Applied Biosystems). The obtained sequences were subjected to BLAST searches (ncbi.nlm.nih.gov/BLAST/). Multiple sequence alignment was performed using Clustal Omega software (http://www.clustal.org/omega/). Data generated was analyzed using the SPSS software (IBM SPSS Statistics for Windows, Version 25.0., IBM Corp., Armonk, NY, US). Statistical significance was determined among variables by using Chi-square test. Significance was defined as P<0.05.
Results
A total of 118 Enterococcus strains were isolated from all the urine samples processed within the duration of the study. Of these 28 per cent of the presenting patients were male. The most commonly isolated species was E. faecalis (76.9%). About 16.9 per cent of the isolates were identified as E. faecium. Among other species, E. avium was detected in two samples, while E. durans and E. gallinarum were detected in one specimen each. Table I shows antibiotic resistance profile of the isolates. Most of the isolates were resistant to penicillin (82.2%). High resistance was also seen towards fluoroquinolones. Least resistance was seen towards tigecycline (5.08%), linezolid (0.8%) and none were resistant to daptomycin. 43 per cent isolates were HLG resistant (HLGR). Five isolates which appeared resistant to HLG by disc detection method were found to be sensitive when tested by micro broth dilution method as well as by VITEK2 system. Resistance towards HLG was significantly higher in E. faecium than in E. faecalis (P≤ 0.01) and also in case of ampicillin-clavulanic acid (P=0.003). Glycopeptide resistance was limited to E. faecalis and E. faecium and all the other isolated species were sensitive. A total of 15 isolates were resistant to both vancomycin and teicoplanin whereas five species were resistant only to vancomycin, indicating vanA and vanB phenotype, respectively.
Table I.
Antibiotic resistance profile of Enterococcal isolates
| Antibiotic | All species (n=118) % | E. faecalis (n=94) % | E. faecium (n=20) % | P |
|---|---|---|---|---|
| Amoxicillin | 79.6 | 76.5 | 100 | 0.06 |
| Ampicillin-clavulanic acid | 31.3 | 26.5 | 60 | 0.003 |
| Chloramphenicol | 32.2 | 31.9 | 40 | 0.48 |
| Ciprofloxacin | 77.1 | 77.6 | 85 | 0.46 |
| Daptomycin | 0 | 0 | 0 | - |
| Erythromycin | 73.7 | 72.3 | 80 | 0.47 |
| High level Gentamicin | 43.2 | 39.3 | 70 | 0.01 |
| Levofloxacin | 72.8 | 72.3 | 85 | 0.23 |
| Linezolid | 0.8 | 0 | 5 | - |
| Nitrofurantoin | 27.9 | 25.5 | 45 | 0.08 |
| Norfloxacin | 77.9 | 77.6 | 90 | 0.21 |
| Penicillin | 82.2 | 79.7 | 100 | 0.1 |
| Teicoplanin | 12.7 | 11.7 | 20 | 0.31 |
| Tetracycline | 73.7 | 75.3 | 65 | 0.33 |
| Tigecycline | 5.08 | 4.25 | 10 | 0.29 |
| Vancomycin | 16.9 | 14.8 | 30 | 0.1 |
For the VRE isolates, the MIC of vancomycin was in the range 32 µg/ml to ≥512 µg/ml (Table II). For one isolate, vancomycin MIC was ≥512 µg/ml. For isolates showing vanB phenotype, vancomycin MIC was up to ≥64 µg/ml and teicoplanin MIC was ≤2 µg/ml. All VRE isolates were sensitive to pristinamycin (MIC ≤1 µg/ml). The linezolid MIC for the one resistant VRE isolate was ≥8 µg/ml.
Table II.
Minimum inhibitory concentration of glycopeptide antibiotics (by micro-broth dilution method)
| Phenotype detected | Enterococcal spp. | Total isolates | MIC (µg/ml%) | Number of isolates | |
|---|---|---|---|---|---|
|
| |||||
| Vancomycin | Teicoplanin | ||||
| Van A | E. faecium | 4 | ≥512 | ≥64 | 1 |
| ≥256 | ≥32 | 1 | |||
| ≥128 | ≥128 | 2 | |||
| E. faecalis | 11 | ≥256 | ≥128 | 2 | |
| ≥128 | ≥128 | 1 | |||
| ≥128 | ≥64 | 1 | |||
| ≥128 | ≥32 | 1 | |||
| ≥64 | ≥64 | 2 | |||
| ≥64 | ≥32 | 4 | |||
| Van B | E. faecium | 2 | ≥64 | ≤2 | 2 |
| E. faecalis | 3 | ≥64 | ≤2 | 2 | |
| ≥32 | ≤2 | 1 | |||
MIC: minimum inhibitory concentration
Figure 1 depicts antibiotic-resistant profile of VREs compared to that of vancomycin-sensitive Enterococci (VSE). All the VREs were resistant to penicillin group and 95 per cent were resistant to fluoroquinolones. Around 80 per cent VRE were also HLGR but only 35 per cent of the VSE strains expressed resistance towards HLG. The VRE (17%) showed significantly higher resistance than the VSE (83%) against all antibiotics tested (P< 0.05).
Fig. 1.
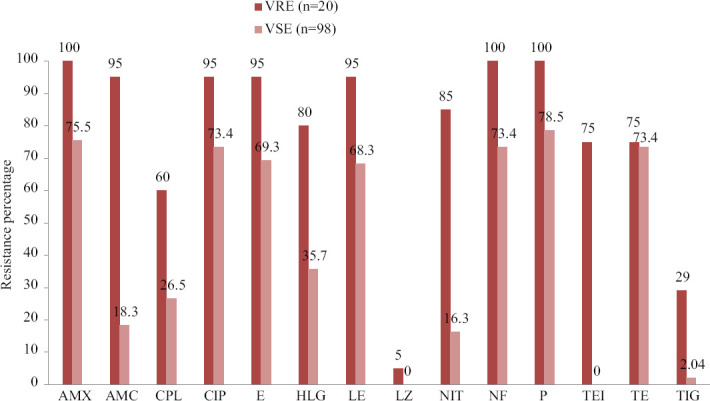
Antibiotic resistance pattern of VRE and non-VRE strains. AMX, amoxicillin; AMC, ampicillin- clavulanic acid; CPL, chloramphenicol; CIP, ciprofloxacin; E, erythromycin; HLG, high-level gentamicin; LE, levofloxacin; LZ, linezolid; NIT, nitrofurantoin; NF, Norfloxacin, P, penicillin; TEI, Teicoplanin; TE, tetracycline; TIG, tigecycline
Vancomycin-resistant gene was detected in 20 isolates by PCR. Fifteen of these strains carried vanA gene and five carried vanB gene. None of the other genotypes were detected and none of the strains carried multiple resistance genes. Table III depicts the distribution of PCR confirmed VRE genotypes, their association with gender, age and MIC of HLG. About 35 per cent of the VRE strains were isolated from male patients (not significant). The infected patients were from all different age groups, ranging from nine months to 79 years.
Table III.
Distribution of vancomycin-resistant enterococci confirmed by polymerase chain reaction
| Isolate | Species | Patient details | HLG MIC | Antibiotic sensitivity | Genotype confirmed by PCR | ||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Sex | Age | Teicoplanin | Vancomycin | ||||
| 1 | E. faecalis | Male | 29 | >500 | S | R | Van B |
| 2 | E. faecalis | Female | 26 | <500 | R | R | Van A |
| 3 | E. faecalis | Male | 9 | >500 | R | R | Van A |
| 4 | E. faecium | Male | 10 | >1000 | R | R | Van A |
| 5 | E. faecium | Female | 28 | >2000 | R | R | Van A |
| 6 | E. faecalis | Female | 79 | >500 | R | R | Van A |
| 7 | E. faecalis | Female | 40 | >500 | S | R | Van B |
| 8 | E. faecalis | Male | 24 | <500 | R | R | Van A |
| 9 | E. faecalis | Female | 6 | >1000 | R | R | Van A |
| 10 | E. faecium | Female | 35 | >500 | R | R | Van A |
| 11 | E. faecalis | Female | 19 | >500 | R | R | Van A |
| 12 | E. faecalis | Male | 65 | >500 | R | R | Van A |
| 13 | E. faecium | Male | 38 | >1000 | S | R | Van B |
| 14 | E. faecalis | Male | 41 | >500 | S | R | Van B |
| 15 | E. faecium | Female | 40 | >500 | S | R | Van B |
| 16 | E. faecalis | Female | 24 | <500 | R | R | Van A |
| 17 | E. faecalis | Female | 37 | <500 | R | R | Van A |
| 18 | E. faecalis | Female | 12 | >500 | R | R | Van A |
| 19 | E. faecalis | Female | 20 | >500 | R | R | Van A |
| 20 | E. faecium | Female | 37 | >2000 | R | R | Van A |
HLG: high-level gentamicin; VRE: vancomycin-resistant Enterococci; S: Sensitive; R: Resistant
All the PCR products were subjected to Sanger sequencing. The size of the 15 vanA gene sequences ranged from 655 to 685 bp. The sequences were compared with E. faecium transposon Tn1546 (complete cds, gene bank accession ID: M97297.1). On BLASTn analysis seven isolates showed 100 per cent identity to the subject sequence, whereas eight isolates showed 99 per cent identity. In six sequences, there was a single mismatch. At the position corresponding to bp 7658 of Tn1546 complete cds, the base thymine was replaced by cytosine in the test sequence. On comparing the resultant amino acid chain of both the subject and test sequence, it was found that in the test sequence, alanine appeared instead of valine (as present in the subject sequence at the corresponding position), due to the alternation of base pair. In gene sequence of another isolate, at the position corresponding to bp 7777 of Tn1546 complete cds, the base guanine was replaced by thymine resulting in a stop codon, truncating the protein at that position. The nucleotide sequence of one isolate showed several stop codons and two gaps when compared with the subject sequence.
The vanB gene sequences were compared with E. faecalis vancomycin resistance genes, complete cds (gene bank accession ID: U35369.1). The size of the five gene sequences ranged from 471 to 590 bp. Four out of five sequences showed 100 per cent identity with the subject sequence. In sequence of one isolate, there was an extra base (alanine) between the position corresponding to bp 5771 and 5772 of the subject sequence. As the mismatch was close to the end of the chromatogram, it could not be concluded if it was a mutation or sequencing artefact. There was, however, no observable difference in glycopeptide MIC or resistance towards other antibiotics among these isolates with mutations. The phylogenetic tree of isolates with vanA and vanB genes, prepared using the clustal omega software (http://www.clustal.org/omega/), is depicted in Figure 2.
Fig. 2.
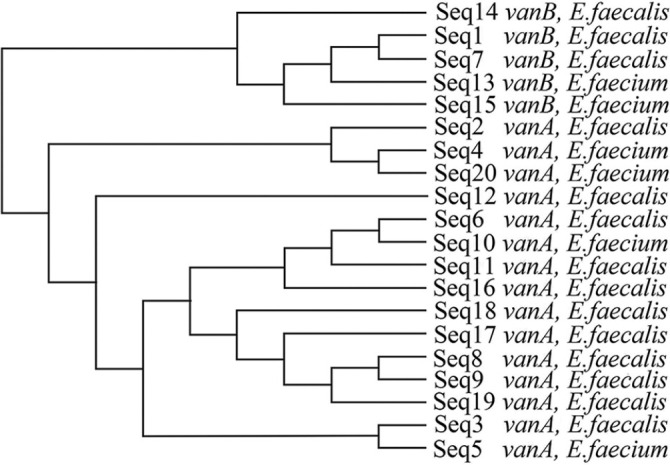
Phylogenetic tree prepared using clustal omega software.
Discussion
Enterococci are important pathogens causing a wide range of infections. E. faecalis is the most prevalent species worldwide, whereas E. faecium is known to be more resistant to antimicrobials13. In concurrence with other reports, the isolation rate of E. faecalis strains was much higher than E. faecium in the present study. Phukan et al19 in 2016 reported 81 per cent isolated species as E. faecalis. The difference in isolation rate between the two species was, however, much less in the 2019 study from Pune9. On the contrary, in a similar study published in 2017 from Delhi, 64.8 per cent isolates were E. faecium and only 32.8 per cent isolates were E. faecalis6.
The resistance towards antibiotics may be chromosomal in some species, and/or plasmid mediated and transferable in others2. Resistance may be low or high depending on the species and the gene responsible. Hence, the knowledge of the causative species, antibiotic susceptibility and the gene involved become important factors to determine the presence, spread and nature of drug resistance. In accordance to reports published from other parts of the country, high level of resistance was seen towards β-lactam drugs in the present study. The article published from Katihar, Bihar by Biswas et al11 reported 86 per cent resistance towards penicillin in contrast to considerably low resistance (58%) that was reported from Guwahati19. Most isolates (97.4%) were found to be resistant to penicillin by Maradia et al27. The present study reports high level of resistance towards fluoroquinolones which is in tandem with previous reports8. Purohit et al6 reported 96 per cent resistance towards ciprofloxacin. Akin to present report, high degree of resistance towards tetracycline and erythromycin was noticed by Biswas et al11. Complete resistance towards erythromycin was reported by Raj et al28 in 2019 from Kolkata.
Resistance towards HLG was found in 43 per cent strains in the present study. In 2014 study from Chennai, the HLG resistance reported was 30 per cent29. Many studies have reported even lower rate of resistance toward HLG27,30. However, the studies conducted in 2017 at Delhi6 and at Kolkata in 201928 reported 74.2 and 68.75 per cent HLGR, respectively. HLGR among E. faecium were significantly higher than E. faecalis (P=0.032). Shridhar and Dhanashree31 also reported similar trend but their finding was not statistically significant.
While comparing the two pathogenic species, resistance rate among E. faecium towards ampicillin-clavulanic acid, nitrofurantoin and HLG was found to be significantly higher than that of E. faecalis (P<0.05). This could be because E. faecium has a higher capacity to modify its metabolism under selective pressure and the presence of many chromosomally encoded enzymes making them more resistant to antibiotics than other members of the genus6,30.
Vancomycin resistance is attributed to altered cell wall receptors. Many genes are responsible for the change and confer varied degree of resistance14. In the present study, nearly 17 per cent of the isolates were vancomycin resistant. This is similar to several studies conducted across the country in the past decade where the isolation percentage of VRE was found to be consistently on the lower side6,9,19,28,31-33. In the present study, incidence of vancomycin resistance among E. faecium was much higher (30%, E. faecalis -14.8%) but not statistically significant (P=0.1). This is in accordance with findings of many other studies done in India and abroad29,34,35. In general, the incidence of VRE infection was found to be higher in critically ill patients with prolonged hospital stay or elderly patients9. In the present study, however, VRE was isolated from patients of all age group.
The resistance percentage towards all other antibiotics in the present study was found to be significantly higher in the case of vancomycin-resistant strains than the vancomycin-sensitive ones (P<0.05). Similar result was found by Tripathi et al8 and Biswas et al11 but the findings were not statistically significant. In majority of cases, this may be due sharing of a common transferable plasmid by the resistance genes. Conversely, the VRE isolates tested by Phukan et al19 did not express higher resistance towards other antibiotics, except for erythromycin. Likewise, Bera et al36 found the VRE strains to be less resistant to other antimicrobials.
There are only limited therapeutic options available against VRE and chief among these are daptomycin, tigecycline, linezolid and pristinamycin37. All the VRE strains tested were sensitive to daptomycin and pristinamycin. One isolate was found to be resistant to linezolid (MIC ≥8 µg/ml). Linezolid resistance in Enterococci has been reported infrequently from India and when reported, the incidence was found to be low11,19. In two studies conducted in 2015 in Chandigarh38 and Jaipur39, all the VRE strains were sensitive to linezolid. In 2017, Purohit et al6 reported two Linezolid-resistant VRE from Delhi, and nearly five per cent of all Enterococci and 20 per cent VRE tested were resistant to tigecycline. The increasing trend of resistance towards linezolid and tigecycline needs to be evaluated further.
The MIC of vancomycin for the VRE isolates was as high as ≥512 in one isolate. Highest teicoplanin MIC was ≥128 µg/ml as seen in five isolates. The vancomycin and teicoplanin MIC was found comparable to previous reports9,11,36.
In the present study, out of the 20 VRE strains, 15 were also resistant to teicoplanin indicating vanA phenotype. Among these, 11 were E. faecalis and four were E. faecium, and five strains were sensitive to teicoplanin with MIC ≤2 μg/ml, suggesting vanB phenotype. The phenotypic criterion for detection of the resistant type was found comparable to the genotypic method in the present study. Through PCR, the same 15 strains were shown to carry vanA gene and the other five strains had vanB gene present and no other genotypes were detected. This is in concordance with previous reports all the isolates where were found to carry vanA gene only8,23. There are, however, other reports as well where one of the study isolates of E. gallinarum carried vanC1 and another carried both vanC1 and vanA gene7. In another study, vanC gene was found in seven E. gallinarum strains40. Overall, the incidence of vanA gene was significantly higher than vanB gene in the present study, similar to other reports11 suggesting that vanA appears to be the prevalent strain in the Indian context, followed by vanB. As vanC is chromosomal and restricted to only three species, it has been reported infrequently. Other vancomycin-resistant genotypes are not frequently reported from India till date.
Overall, the knowledge about resistance genotypes is important to understand the epidemiology of the infection and to formulate treatment and prevention strategies. The study emphasizes the prevalence of vanA and vanB gene carrying Enterococci in the region. The capacity of the two primary vancomycin-resistant genes to cross over (even to other species) makes their prevalence in Enterococci a significant cause of concern. As the treatment options for VRE infection are limited, the emergence of resistance against drugs such as tigecycline and linezolid should be taken seriously. Studies like the present one should be conducted in frequent intervals to detect the spread of VRE and effectiveness of the available treatment options.
Footnotes
Financial support & sponsorship: None.
Conflicts of Interest: None.
References
- 1.Goel V, Kumar D, Kumar R, Mathur P, Singh S. Community acquired enterococcal urinary tract infections and antibiotic resistance profile in North India. J Lab Physicians. 2016;8:50–4. doi: 10.4103/0974-2727.176237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller WR, Munita JM, Arias CA. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther. 2014;12:1221–36. doi: 10.1586/14787210.2014.956092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Truman AW, Kwun MJ, Cheng J, Yang SH, Suh JW, Hong HJ. Antibiotic resistance mechanisms inform discovery:Identification and characterization of a novel amycolatopsis strain producing ristocetin. Antimicrob Agents Chemother. 2014;58:5687–95. doi: 10.1128/AAC.03349-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benamu E, Deresinski S. Vancomycin-resistant enterococcus infection in the hematopoietic stem cell transplant recipient:An overview of epidemiology, management, and prevention. F1000Res. 2018;7:3. doi: 10.12688/f1000research.11831.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mamtora D, Saseedharan S, Bhalekar P, Katakdhond S. Microbiological profile and antibiotic susceptibility pattern of Gram-positive isolates at a tertiary care hospital. J Lab Physicians. 2019;11:144–8. doi: 10.4103/JLP.JLP_173_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Purohit G, Gaind R, Dawar R, Verma PK, Aggarwal KC, Sardana R, et al. Characterization of vancomycin resistant Enterococci in hospitalized patients and role of gut colonization. J Clin Diagn Res. 2017;11:C01–5. doi: 10.7860/JCDR/2017/25988.10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Praharaj I, Sujatha S, Parija SC. Phenotypic &genotypic characterization of vancomycin resistant Enterococcus isolates from clinical specimens. Indian J Med Res. 2013;138:549–56. [PMC free article] [PubMed] [Google Scholar]
- 8.Tripathi A, Shukla SK, Singh A, Prasad KN. Prevalence, outcome and risk factor associated with vancomycin-resistant Enterococcus faecalis and Enterococcus faecium at a tertiary care hospital in Northern India. Indian J Med Microbiol. 2016;34:38–45. doi: 10.4103/0255-0857.174099. [DOI] [PubMed] [Google Scholar]
- 9.Shete VC, Grover N, Kumar M, Bhatt P. Phenotypic detection and molecular characterization of vancomycin-resistant Enterococci. J Nat Sci Biol Med. 2019;10:34–7. [Google Scholar]
- 10.Shinde RS, Koppikar GV, Oommen S. Characterization and antimicrobial susceptibility pattern of clinical isolates of Enterococci at a tertiary care hospital in Mumbai, India. Ann Trop Med Public Health. 2012;5:85–8. [Google Scholar]
- 11.Biswas PP, Dey S, Adhikari L, Sen A. Detection of vancomycin resistance in Enterococcus species isolated from clinical samples and feces of colonized patients by phenotypic and genotypic methods. Indian J Pathol Microbiol. 2016;59:188–93. doi: 10.4103/0377-4929.182015. [DOI] [PubMed] [Google Scholar]
- 12.Yangzom T, Kumar Singh TS. Study of vancomycin and high-level aminoglycoside-resistant Enterococcus species and evaluation of a rapid spot test for enterococci from Central Referral Hospital, Sikkim, India. J Lab Physicians. 2019;11:192–9. doi: 10.4103/JLP.JLP_5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci:From commensals to leading causes of drug resistant infection. Boston: Massachusetts Eye and Ear Infirmary; 2014. [PubMed] [Google Scholar]
- 14.Sujatha S, Praharaj I. Glycopeptide resistance in gram-positive cocci:A review. Interdiscip Perspect Infect Dis. 2012;2012:781679. doi: 10.1155/2012/781679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mak A, Miller MA, Chong G, Monczak Y. Comparison of PCR and culture for screening of vancomycin-resistant enterococci:Highly disparate results for vanA and vanB. J Clin Microbiol. 2009;47:4136–7. doi: 10.1128/JCM.01547-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faron ML, Ledeboer NA, Buchan BW. Resistance mechanisms, epidemiology, and approaches to screening for vancomycin-resistant Enterococcus in the health care setting. J Clin Microbiol. 2016;54:2436–47. doi: 10.1128/JCM.00211-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett JE, Dolin R, Blaser MJ, editors. Mandell, Douglas, and Bennett's principles and practice of infectious diseases. 7th ed. Philadelphia: Elsevier/Saunders; 2015. [Google Scholar]
- 18.Mathur P, Kapil A, Chandra R, Sharma P, Das B. Antimicrobial resistance in Enterococcus faecalis at a tertiary care centre of northern India. Indian J Med Res. 2003;118:25–8. [PubMed] [Google Scholar]
- 19.Phukan C, Lahkar M, Ranotkar S, Saikia KK. Emergence of vanA gene among vancomycin-resistant enterococci in a tertiary care hospital of North-East India. Indian J Med Res. 2016;143:357–61. doi: 10.4103/0971-5916.182627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Driscoll T, Crank CW. Vancomycin-resistant enterococcal infections:Epidemiology, clinical manifestations, and optimal management. Infect Drug Resist. 2015;8:217–30. doi: 10.2147/IDR.S54125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koneman EW, Allen SD, Janda WM, Schreckenberger PC. The gram positive cocci:Staphylococci and related organisms. In: Koneman EW, Allen SD, Janda WM, Schreckenberger PC, Propcop GW, Woods GL, et al., editors. Color atlas and textbook of diagnostic microbiology. 6th ed. Philadelphia: Lippincott-Raven; 2006. pp. 624–62. [Google Scholar]
- 22.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, CLSI supplement M100S. 26th ed. Wayne, Pennsylvania: CLSI; 2016. [Google Scholar]
- 23.Tripathi A, Shukla SK, Singh A, Prasad KN. A new approach of real time polymerase chain reaction in detection of vancomycin-resistant enterococci and its comparison with other methods. Indian J Med Microbiol. 2013;31:47–52. doi: 10.4103/0255-0857.108721. [DOI] [PubMed] [Google Scholar]
- 24.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–7. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Depardieu F, Perichon B, Courvalin P. Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J Clin Microbiol. 2004;42:5857–60. doi: 10.1128/JCM.42.12.5857-5860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pérez-Hernández X, Méndez-Alvarez S, Delgado T, Moreno A, Reyes-Darias A, Sierra López A, et al. Low prevalence of vancomycin-resistant enterococci in clinical samples from hospitalized patients of the Canary Islands, Spain. Int Microbiol. 2002;5:117–20. doi: 10.1007/s10123-002-0075-y. [DOI] [PubMed] [Google Scholar]
- 27.Maradia MR, Mehta K, Prajapati K, Vadsmiya M, Shah P, Vegad M. Prevalence of multi drug resistant Enterococcus species isolated from urine samples in a tertiary care hospital, Western India. Int J Med Sci Public Health. 2017;6:715–9. [Google Scholar]
- 28.Raj HJ, Das (Sarkar) M, Mondal S. Prevalence of enterococcal infection and the antimicrobial susceptibility profile of the organism with special reference to vancomycin:A study in a rural medical college hospital in Eastern India. IOSR J Dent Med Sci. 2019;18:9–15. [Google Scholar]
- 29.Gangurde N, Mane M, Phatale S. Prevalence of multidrug resistant enterococci in a tertiary care hospital in India:A growing threat. Open J Med Microbiol. 2014;4:11–5. [Google Scholar]
- 30.Kapoor L, Randhawa VS, Deb M. Antimicrobial resistance of enterococcal blood isolates at a pediatric care hospital in India. Jpn J Infect Dis. 2005;58:101–3. [PubMed] [Google Scholar]
- 31.Shridhar S, Dhanashree B. Antibiotic susceptibility pattern and biofilm formation in clinical isolates of Enterococcus spp. Interdiscip Perspect Infect Dis. 2019;2019:7854968. doi: 10.1155/2019/7854968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ligitha SG, Sasikumari O. Vancomycin resistant enterococcal infections in infants and neonates. J Acad Clin Microbiol. 2018;20:105–7. [Google Scholar]
- 33.Yadav G, Thakuria B, Madan M, Agwan V, Pandey A. Linezolid and vancomycin resistant enterococci:A therapeutic problem. J Clin Diagn Res. 2017;11:C07–11. doi: 10.7860/JCDR/2017/27260.10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pol S, Devhare D, Bharadwaj R. Vancomycin resistant enterococci:An emerging problem in a tertiary care hospital, Pune, India. Int J Med Microbiol Trop Dis. 2017;3:27–30. [Google Scholar]
- 35.Wang QY, Li RH, Shang XH. Urinary tract infection caused by Enterococcus isolates:Aetiology and antimicrobial resistance patterns. J Chemother. 2015;27:117–9. doi: 10.1179/1973947814Y.0000000192. [DOI] [PubMed] [Google Scholar]
- 36.Bera SS, Mehta S, Dwivedi MB, Singh VA, Paul R, Prabhas R. Emergence of vanA and vanB gene among vancomycin-resistant enterococci in faecal samples from ICU patients. Int J Adv Res. 2017;5:117–24. [Google Scholar]
- 37.Barber KE, King ST, Stover KR, Pogue JM. Therapeutic options for vancomycin-resistant enterococcal bacteremia. Expert Rev Anti Infect Ther. 2015;13:363–77. doi: 10.1586/14787210.2015.1001839. [DOI] [PubMed] [Google Scholar]
- 38.Gupta V, Singla N, Behl P, Sahoo T, Chander J. Antimicrobial susceptibility pattern of vancomycin resistant enterococci to newer antimicrobial agents. Indian J Med Res. 2015;141:483–6. doi: 10.4103/0971-5916.159309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta S. Emergence of linezolid resistance in clinical isolates of vancomycin-resistant enterococci. Int J Adv Med Health Res. 2016;3:107–8. [Google Scholar]
- 40.Banerjee T, Anupurba S, Filgona J, Singh DK. Vancomycin-resistance enterococcal colonization in hospitalized patients in relation to antibiotic usage in a tertiary care hospital of North India. J Lab Physicians. 2015;7:108–11. doi: 10.4103/0974-2727.163123. [DOI] [PMC free article] [PubMed] [Google Scholar]


