Abstract
Aims
To improve short-and long-term predictions of mortality and atrial fibrillation (AF) among patients with congenital heart disease (CHD) from a nationwide population using neural networks (NN).
Methods and results
The Swedish National Patient Register and the Cause of Death Register were used to identify all patients with CHD born from 1970 to 2017. A total of 71 941 CHD patients were identified and followed-up from birth until the event or end of study in 2017. Based on data from a nationwide population, a NN model was obtained to predict mortality and AF. Logistic regression (LR) based on the same data was used as a baseline comparison. Of 71 941 CHD patients, a total of 5768 died (8.02%) and 995 (1.38%) developed AF over time with a mean follow-up time of 16.47 years (standard deviation 12.73 years). The performance of NN models in predicting the mortality and AF was higher than the performance of LR regardless of the complexity of the disease, with an average area under the receiver operating characteristic of >0.80 and >0.70, respectively. The largest differences were observed in mortality and complexity of CHD over time.
Conclusion
We found that NN can be used to predict mortality and AF on a nationwide scale using data that are easily obtainable by clinicians. In addition, NN showed a high performance overall and, in most cases, with better performance for prediction as compared with more traditional regression methods.
Keywords: Congenital heart disease, Mortality, Atrial fibrillation, Deep learning, Artificial intelligence
Introduction
Congenital heart disease (CHD) is the most common congenital malformation currently affecting almost 1% of all live-births worldwide.1,2 For most patients, CHD is a lifelong condition with varying severity depending on the congenital malformation per se, or the accompanying interventions, comorbidities, risk factors, or life style.3 Currently, 90% of patients with mild, 75% with moderate, and 40% with the complex disease will reach the age of 60 years and increasingly experience the risk of acquired cardiovascular disease.3–5 Previous knowledge on the impact of common cardiovascular comorbidities including hypertension, diabetes, atrial fibrillation (AF), and heart failure on mortality have mostly been performed on a general population without CHD. Recent reports have indicated a substantially higher risk of mortality among patients with CHD as a consequence of such comorbidities, as compared with controls.6–9
In more traditional epidemiological cohort studies using medical data, the most common practice to analysis and compare risk is the use of e.g. logistic regression (LR) or survival analysis methods such as Kaplan–Meier (survival probability) and Cox proportional hazard regression models for comparing risk between groups. An important aspect of these models is the medical and practical decisions to be considered for the models to be valid. For some cases, this could be a challenge for researchers to consider all possible aspects, especially for more complex diseases. The complexity and long-term perspectives of CHD may potentially benefit from improved analysis methods such as neural networks (NN) and deep learning but so far there is limited data from patients with CHD.10 In an analysis of a large contemporary single-centre cohort of 10 019 CHD patients, the authors reported good performance of a disease severity score derived from deep learning models in the prediction of mortality during an 8-year follow-up period.11 Additionally, in a recent study about auscultation of heart sound among CHD patients using an AI-AA platform showed high accuracy in detection of abnormal heart sound with the good concordance with auscultation from physicians.12 Improvements in computational power over the past two decades have led to an increase in the use of deep learning-driven algorithms in the field of healthcare science.13–16 Neural networks are deep learning algorithms that are one of the most successful tools for machine learning. They consist of a series of connected layers that, when appropriately trained, output increasingly meaningful representations of the input data leading to the sought-after result.17 Previous models have been created using data from multi-imaging techniques or wide-spectrum biomarkers. However, this will be less useful for the regular clinician coming across many patients with CHD where such data may not be routinely available. Prediction of morbidity such as AF and mortality may influence both the use and timing of preventive medical treatment such as anticoagulation as well as the planning of lifetime management. However, few studies have reported on the long-term predictability of mortality and AF among patients with CHD using commonly attainable comorbidities as risk factors.
The aim of this study was to investigate the possibility of improving the prediction of short- and long-term mortality and AF from birth using easily attainable variables through the use of a NN that would analyse Swedish registries containing data of a complete nationwide population of CHD patients born between 1970 and 2017 as well as to compare performances against traditional regression methods.
Methods
Study population
The present study uses data from the Swedish National Patient Register and the Cause of Death Register. The National Patient Register was started in 1964 and has obtained a full nationwide coverage of all in-hospital admissions and contributory diagnoses since 1987. All hospitals in Sweden are required to report to the register with the exact date of hospitalization. As such, if a patient was not registered (stated as ‘N/A’ and coded as ‘0’ in the database) it was regarded as the patient did not have a certain event. Because of this, there are no missing data for the diagnoses used in the current study. Thus, starting 2001, all diagnoses from outpatient clinics were also recorded in the National Patient Register. The Cause of Death Register records the mortality data of Swedish citizens nationwide starting from 1961. Diagnoses are coded according to the International Classification of Disease (ICD) system, ICD-8 (1968–86), ICD-9 (1987–96), and ICD-10 (1996 onwards). The National Patient Register and the Cause of Death Register were linked through the unique Swedish 10-digit personal identifier. All CHD patients were divided according to their birth year into five different groups: (i) 1970–79, (ii) 1980–89, (iii) 1990–99, (iv) 2000–09, and (v) 2010–17.
Definitions of diagnosis
In the current cohort, all patients with at least one outpatient visit or a discharged diagnosis of CHD from the hospital and were born between 1970 and 2017 were identified in the National Patient Register and Cause of Death register and followed-up until death or the end of the study duration in December 2017. Currently, CHD patients were followed-up from birth rather than diagnosis date. A majority of all diagnoses of CHD cases were identified at birth. Because of the very long follow-up time, CHD patients were divided into birth decades in order to capture potential treatment effect over time. Diagnoses were identified through ICD 8, 9, and 10 codes. Supplementary material online, Table S1 lists the full ICD-codes used for the identification of CHD in the National Patient Register and the Cause of Death Register. In addition, the CHD population were grouped into six groups according to a hierarchical classification system based on lesion severity according to the Botto/Liu classification. Lesion Groups 1 and 2 represent the most complex conditions, while Lesion Groups 3, 4, and 5 represent those with coarctations of the aorta, ventricular septal defects, and atrial septal defects, respectively. Lesion Group 6 represents those with CHD not included in the other lesion groups.18,19 Corresponding ICD-codes for each lesion groups are shown in Supplementary material online, Table S2. Comorbidities including hypertension, diabetes mellitus, heart failure, myocardial infarction, and AF were defined by ICD-codes as described in Supplementary material online, Table S3. Congenital cardiac intervention was defined as when a CHD patient underwent at least one cardiovascular surgical procedure or a cardiac interventional catheterization according to the Swedish Classification of Operations (6th edition, Swedish version) or following the classification of surgical procedures (1.9th edition, Swedish version).
Statistical analysis
Baseline characteristics of the study population are shown as categorical and continuous data. Categorical variables are shown as numbers with percentages, while continuous variables are presented as means with standard deviations and medians with interquartile ranges. A NN was used to predict the 1-, 3-, 5-, 10-, 20-, and 30-year mortality and development of AF, using clinically relevant input variables obtained from the national registries such as age, decade of birth, sex, lesion groups and year of onset of AF (excluded for the prediction of AF), heart failure, hypertension, diabetes, myocardial infarction, and congenital cardiac intervention. For performance, the averaged area under the receiver operating characteristics (AUROC) with 95% confidence interval (CI) was estimated. Logistic regression (LR) was used for baseline comparison. An overview of the study design can be seen in Figure 1, while a more detailed description of the data processing, algorithm training, and evaluations for NN methods used are available in the Supplementary material online, Methods section. For model validation and prediction, the TRIPOD checklist was used as reference and confirmation.
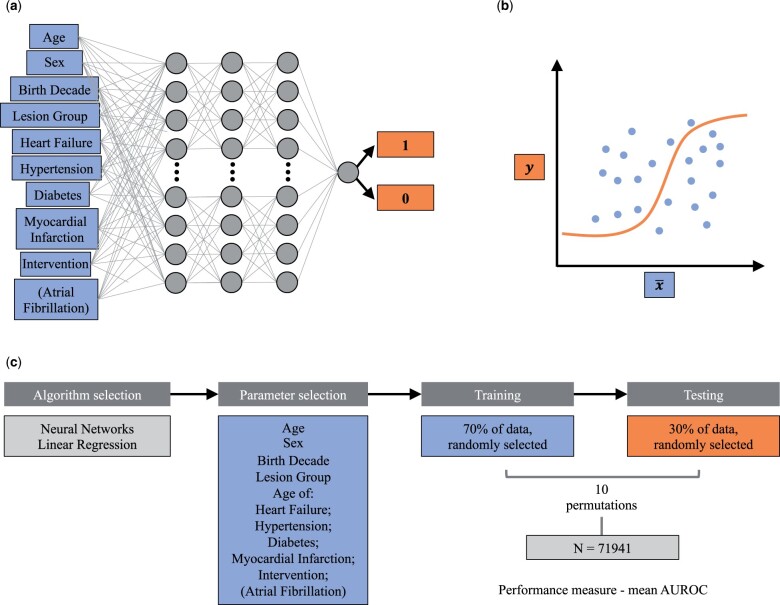
Figure 1.
Study methodology. (A,B) Schematic representation of the machine learning algorithms employed in this study: (A) neural network and (B) logistic regression. (C) Schematic overview of the methodology employed in the study.
Neural network
Neural networks are deep learning algorithms that consist of a series of connected layers. After appropriate training, these networks put out increasingly meaningful representations of the input data eventually leading to the sought-after result. Each layer is composed of computational units that simulate the function of biological neurons, whose connection weights were adjusted in the training phase to learn how to calculate the desired output from the input data,17,20 as schematically shown in Figure 1.
We used a feed-forward NN based on three dense 32-neuron layers and an output layer providing a single value between 0 and 1. The model takes the input parameters (the variables defined in data pre-processing) and a ground truth value of 0 (dies/no AF) or 1 (lives/AF) in the training phase. The batch size in each training set was 100, and the number of epochs was optimized to prevent overfitting by halting the training when the validation accuracy of the hold-out validation set from the training dataset had not increased for 10 epochs. This was repeated five times to find the optimal number of epochs before re-training of the model on all of the training data with the optimal number of epochs. We implemented this model using the Python-based Keras library with a TensorFlow backend.21
Logistic regression
Logistic regression is a predictive model that models a binary-dependable variable, producing an appropriate baseline for other machine learning models. The model was fitted using the same variables used for the NN algorithm. The model was implemented using the Python-based Scikit-learn package for machine learning.
Ethics
This study was approved by the Regional Ethical Review Board of Gothenburg University (Gbg 912-16, T619-18) and complies with all tenets of the Declaration of Helsinki. All personal identifiers were replaced with a unique code for anonymization in the final data set.
Results
Characteristics of the study population
Table 1 shows the baseline characteristics of patients with CHD born between 1970 and 2017 obtained from the registries. A total of 71 941 CHD patients were identified from the registries. During the study period, a total of 5768 (8%) patients with CHD died, with a median follow-up of 13.5 years (interquartile range of 5.81–25.50 years). Furthermore, a total of 995 (1.38%) developed AF, and 18 109 (25.17%) underwent at least one congenital cardiac intervention during follow-up (Table 1). Baseline characteristics of the study population were divided by lesion groups and birth decades are shown in Supplementary material online, Tables S4 and S5. For the most complex lesion groups (Lesion Groups 1 and 2) mortality and AF were considerably higher compared to the less complex groups (Lesion Groups 3–6). In addition, they also had considerably more interventions e.g. surgical procedures compared to other groups.
Table 1.
Baseline characteristics of patients with congenital heart disease born in 1970–2017 (n = 71 941)
| Patients with congenital heart disease | |
|---|---|
| Sex | |
| Male | 36 102 (50.18%) |
| Female | 35 839 (49.82%) |
| Born in Sweden | 67 814 (94.26%) |
| Birth decades | |
| 1970–79 | 7545 (10.49%) |
| 1980–89 | 9814 (13.64%) |
| 1990–99 | 13 997 (19.46%) |
| 2000–09 | 21 459 (29.83%) |
| 2010–17 | 19 126 (26.59%) |
| Lesion groups | |
| Lesion Group 1a | 5421 (7.54%) |
| Lesion Group 2b | 3855 (5.36%) |
| Lesion Group 3c | 3358 (4.67%) |
| Lesion Group 4d | 22 950 (31.90%) |
| Lesion Group 5e | 14 635 (20.34%) |
| Lesion Group 6f | 21 722 (30.19%) |
| All-cause mortality | 5768 (8.02%) |
| Atrial fibrillation | 995 (1.38%) |
| Myocardial infarction | 205 (0.28%) |
| Heart failure | 2714 (3.77%) |
| Hypertension | 1399 (1.94%) |
| Diabetes | 713 (0.99%) |
| Congenital cardiac intervention | 18 109 (25.17%) |
| Mean follow-up time, years (SD) | 16.47 ± 12.73 |
| Median follow-up time, years (IQR) | 13.50 (5.81–25.50) |
IQR, interquartile range; SD, standard deviation.
Lesion Group 1 was defined as conotruncal defects [common arterial trunk, aortopulmonary septal defect, transposition of the great arteries (unrepaired lesions and surgically repaired), tetralogy of Fallot, double-outlet left ventricle, double-outlet right ventricle and congenitally corrected transposition/discordant atrioventricular and ventriculoatrial connection].
Lesion Group 2 was defined as severe non-conotruncal defects (common ventricle, and hypoplastic left heart syndrome endocardial cushion defect/atrioventricular septal defect). In addition this group contains univentricular heart defects.
Lesion Group 3 was defined as coarctation of the aorta.
Lesion Group 4 was defined as ventricular septal defect.
Lesion Group 5 was defined as atrial septal defect.
Lesion Group 6 was defined as all other heart and circulatory system anomalies that were not included in the other lesion groups.
Overall performance of mortality and atrial fibrillation among congenital heart disease patients
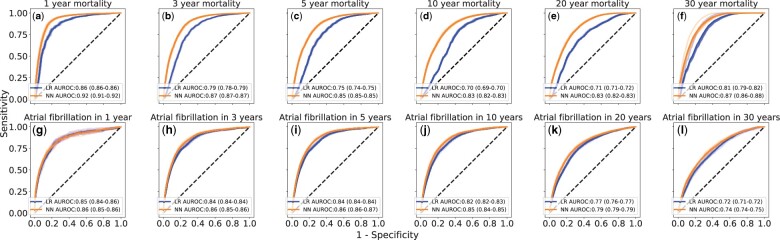
Figure 2 shows the resulting performance in the prediction of mortality and AF. Overall, the average performance for mortality among CHD patients was higher for NN than LR from the first to the last year with an AUROC of 0.92 (95% CI 0.91–0.92) to 0.87 (95% CI 0.86–0.88) compared with 0.86 (95% CI 0.86–0.86) to 0.81 (95% CI 0.79–0.82), respectively. For AF, a similar trend in performance was observed with an AUROC of 0.86 (95% CI 0.85–0.86) to 0.74 (95% CI 0.74–0.75) for NN and 0.85 (95% CI 0.84–0.86) to 0.72 (95% CI 0.71–0.72) for LR.
Figure 2.
Performance of the machine learning algorithms on the internal test data set. Average receiver operating characteristics curves and area under the receiver operating characteristics for neural network (NN) (orange lines) and logistic regression (LR) (blue lines) for the predicted the short- and long-term prognosis on (A–F) mortality and (G–L) atrial fibrillation. The receiver operating characteristics curves for 10 resamples are shown as the shaded region around the average receiver operating characteristics curves, and the corresponding 95% CI for the area under the receiver operating characteristics is shown in parentheses.
For the short-term performance of the 1- and 5-year mortality, the NN was observed to outperform LR. The AUROC after 1 year was 0.92 (95% CI 0.91–0.92) for NN as compared with the 0.86 (95% CI 0.86–0.86) for LR. Corresponding results for the 5-year mortality was 0.85 (95% CI 0.85–0.85) and 0.75 (95% CI 0.74–0.75), respectively. For the long-term performance, a similar trend was observed with a decreasing AUROC, except for the 30-year mortality. The 10- and 30-year mortality showed an AUROC of 0.83 (95% CI 0.82–0.83) to 0.87 (95% CI 0.86–0.88) for NN as compared with 0.70 (95% CI 0.69–0.70) to 0.81 (95% CI 0.79–0.82) for LR, respectively.
Regarding the short-term performance of AF, the results were similar after 1 and 5 years with an AUROC of 0.86 (95% CI 0.85–0.86 and 0.86–0.87, respectively) for NN. The results were also similar at 0.85 and 0.84 for LR over time. Regarding the long-term performance, the result after 10 and 30 years differed slightly, with an AUROC of 0.84 (95% CI 0.84–0.85) down to 0.74 (95% 0.74–0.75 CI) and 0.82 (95% CI 0.82–0.83) to 0.72 (95% 0.71–0.72 CI), respectively.
Comparison of model performance of mortality and atrial fibrillation by lesion groups, birth decades, and congenital cardiac surgery
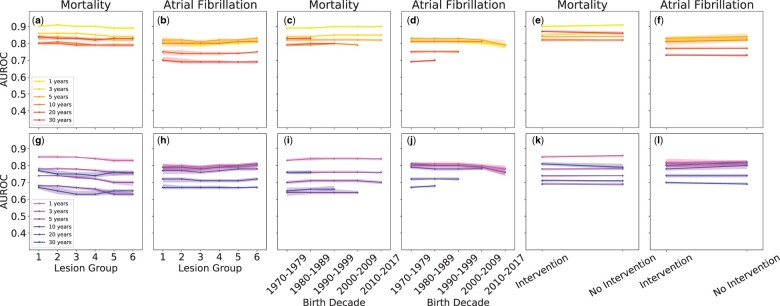
Figure 3 and Supplementary material online, Tables S6–S8 show a detailed comparison of prediction performance for mortality and AF by lesions groups, birth decades, and congenital cardiac intervention. The performance in predicting mortality was higher for NN compared with LR. The highest performances within lesion groups were observed for 1-year predictions, with an overall AUROC of ∼0.89–0.91 and ∼0.83–0.85 for NN and LR, respectively. Over time, performance was observed to decrease but remained higher for NN, with an AUROC of ∼0.80–0.83 compared with ∼0.74–0.77 for LR for 30-year predictions. From the first to the last birth decades, an AUROC of ∼0.89–0.90 for NN and ∼0.83–0.84 for LR was observed for 1-year predictions. For 30-year predictions, the AUROC within birth decades was ∼0.83 and ∼0.76, respectively. Among CHD patients who underwent congenital cardiac intervention, an AUROC of 0.90 (95% CI 0.89–0.91) and 0.87 (95% CI 0.0.87–0.88) was found for 1- and 30-year predictions for NN, respectively. Corresponding results for LR were 0.85 (95% CI 0.85–0.86) and 0.81 (95% CI 0.80–0.82), respectively.
Figure 3.
Performance dependence on the specific input variables. Average area under the receiver operating characteristics for mortality and atrial fibrillation for neural network (yellow to red lines for increasing number of years) and logistic regression (magenta to dark blue lines for increasing number of years) for the input variables: (A, B, G, H) lesion groups (C, D, I, J) birth decades, and (E, F, K, L) whether the patient had undergone congenital surgery or not. The 95% CI is shown as the shaded regions around the lines. Note that birth decades are only included in appropriate long-term predictions relative to the end of the study in 2017.
The prediction performance of AF was similar between models but was slightly higher overall for NN. Among the lesion groups, an AUROC of ∼0.80 was observed for 1-year predictions for both models. For 30-year predictions, performance decreased to ∼0.69 for NN and ∼0.67 for LR. For birth decades, an overall AUROC of ∼0.81 was found for both models for 1-year predictions. This result decreased to ∼0.70 and ∼0.68 for 30-year predictions for NN and LR, respectively. Among congenital cardiac surgery an AUROC of 0.83 (95% CI 0.82–0.84) and 0.82 (95% CI 0.81–0.84) was observed for NN and LR, respectively, for 1-year predictions. The performance for both models was observed to decrease for 30-year predictions with AUROC of 0.73, 95% CI 0.73–0.74 for NN and 0.70 (95% CI 0.69-0.70) for LR.
Discussion
In the present study, we have developed and evaluated risk prediction score models to predict mortality and AF specific to CHD patients using easily attainable data through administrative medical registers over a long period of follow-up. A NN was used to predict mortality and development of AF over a short-and long-term perspective using a nationwide population that contained all patients with CHD born between 1970 and 2017. When compared with a simpler LR model, NN showed a higher predictive performance over time, most notably in mortality.
Prediction of mortality on the individual— or at least a smaller group— level is important to focus preventive action and treatments, including anticoagulation for patients with AF or repeat surgery for patients with moderately malfunctioning valves. The issue of acting proactively and not simply reactively has been the focus for the treatment of CHD in recent years. As important as it is not to intervene too late, it may be just as important to refrain from intervening too early in young individuals, who have many life years ahead of them and may face multiple interventions. In contrast, for the elderly patient with acquired heart disease, an intervention that is predicted to last 10–15 years is often enough to last their lifetime.
A recent study from a single tertiary centre that used deep learning algorithms estimated that the prognosis and the potential of guiding therapy in adult patients with CHD were high.11 That study included patients above the age of 18 years old and analysed over 44 000 medical records with an accuracy over 90%. Other studies also using deep learning algorithms and with more clinical details have shown similar results with high predictability.22 Our study, which used NN, demonstrated an overall good prediction of mortality and AF from birth up to the age of 47 years old. This study included both children and young adults with CHD, which is especially important, as the highest relative mortality in patients with CHD has been reported to occur during childhood and, more specifically, during the first 5 years of age.23 In another study, using machine learning methods and NN to predict operative mortality among patients who underwent cardiac surgery using medical records showed no distinct advantages in model performance over more traditional methods.24 However, in our study involving a CHD population, we observed a notably favourable performance of NN as compared with LR for mortality after congenital cardiac intervention.
A challenge with CHD patients is the complexity of the disease, where each individual heart defect requires different and individualized treatment over time. As such, predictive modelling using more traditional regression models need to take into account several factors, such as potential interactions with risk factors, the complexity of CHD, and improved survival over time. Acquiring these data along with the necessary detailed clinical information, which is commonly needed for modelling along with a long follow-up time, is difficult. Therefore, an advantage of NN is the ability to accept a wide range of data sources as input variables, making it possible to account and adapt for different scenarios without the need of complex and detailed information when modelling CHD patients. Currently, the largest difference in model performance of NN and LR was observed regarding the complexity of CHD patients, especially for mortality within lesion groups and among patients that had previously undergone congenital cardiac intervention. Similar findings have been previously observed when comparing more advanced deep learning models and more traditional regression methods for the prediction of mortality in CHD patients.25 However, most of these studies have used complex and detailed data with a shorter follow-up time. Our models show good performance regarding predictability on mortality and AF, both in the relatively short- and long-term periods. Most importantly, we found that the models were better than the conventional regression methods, while using simple and easily attainable risk factors and comorbidities.
In summary, we have demonstrated that the use of NN shows good predictability of mortality and AF as compared with more traditional methods when using only a few variables easily attainable through medical records. The largest differences in the performance of the models over time were found in the complexity of CHD patients. A combination of administrative and clinical data will therefore be promising for future use in NN in this complex, heterogeneous, and vulnerable patient group.
Strengths and limitations
Our study has several strengths and limitations. A major strength is the use of data from a nationwide registry, which includes all patients with CHD in Sweden born between 1970 and 2017. In addition, since healthcare system is mainly funded by the Swedish government, it is available to all citizens, which helps in minimizing the selection bias as opposed to a single-centre study or other private healthcare systems. Thus, our results can be deemed representative of the CHD patients in Sweden. A limitation of this study is the sole use of administrative data with no clinical or detailed medical records like blood pressure levels, cholesterol levels, or potentially detailed medical treatments e.g. the use of OAC or information on surgical techniques and peri-operative clinical care, as they were unavailable at the time of the study. The prediction models are therefore not able to take into account any improvements in the different techniques for congenital cardiac surgery or intervention over time as well as other confounding biases. However, to counter for this, we performed separate calculations by birth decades to capture the major effects of intervention and medical improvements over time. Nevertheless, the performance of our models for NN was good despite the ever-changing landscape of congenital heart interventions. Additionally, a limitation in all registry-based studies is the risk of misclassification or incorrectly coded diagnoses. However, the validation of diagnoses in the National Patient Register and the Cause of Death Register, especially for cardiovascular disease, has previously been shown to be high, with a positive predictive value of 85%–95%. 26
Conclusions
The complexity of CHD combined with increasing survival is a challenge when making accurate risk models. In the current study, we used only simple and easily attainable variables found in medical records commonly available for most clinicians. Neural networks showed a high performance overall and, in most cases, with better performance for prediction as compared with more traditional regression methods.
Supplementary material
Supplementary material is available at European Heart Journal – Digital Health
Supplementary Material
Acknowledgements
We thank Edanz Group (https://en-author-services.edanz.com/ac) for editing a draft of this manuscript.
Funding
The study was financed by grants from the Swedish state under the agreement between the Swedish government and the country councils, the ALF-agreement (grant number: 917361 and 236611) and the Swedish Heart-Lung Foundation (grant number: 20180644). The funders had no role in the design of the study, collection and analysis of data or interpretation of the results.
Data availability
This study is based on data from the Swedish Inpatient register held by the National Board of Health and Welfare (http://www.socialstyrelsen.se). The dataset contains sensitive personal information and is subjected to secrecy in accordance with the Swedish Public Access to Information and Secrecy Act (OSL, 2009:400). The data used for this study is located at the institution of the authors, and is available given that an ethical approval can be obtained from the Swedish Ethical Review Authority at the University of Gothenburg. A formal request can be made to corresponding author Dr. Giang KW at Gothenburg University, Institution of Medicine, Department of Molecular and Clinical medicine (contact: wai.giang.kok@gu.se).
Conflict of interest: none declared.
References
- 1. Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol 2002;39:1890–900. [DOI] [PubMed] [Google Scholar]
- 2. Khoshnood B, Lelong N, Houyel L, Thieulin A, Jouannic J, Magnier S, Delezoide A, Magny J, Rambaud C, Bonnet D, Goffinet F; EPICARD Study Group. Prevalence, timing of diagnosis and mortality of newborns with congenital heart defects: a population-based study. Heart 2012;98:1667–1673. [DOI] [PubMed] [Google Scholar]
- 3. Mandalenakis Z, Rosengren A, Skoglund K, Lappas G, Eriksson P, Dellborg M. Survivorship in children and young adults with congenital heart disease in Sweden. JAMA Intern Med 2017;177:224–230. [DOI] [PubMed] [Google Scholar]
- 4. Moons P, Bovijn L, Budts W, Belmans A, Gewillig M. Temporal trends in survival to adulthood among patients born with congenital heart disease from 1970 to 1992 in Belgium. Circulation 2010;122:2264–2272. [DOI] [PubMed] [Google Scholar]
- 5. van der Bom T, Mulder BJ, Meijboom FJ, van Dijk AP, Pieper PG, Vliegen HW, Konings TC, Zwinderman AH, Bouma BJ. Contemporary survival of adults with congenital heart disease. Heart 2015;101:1989–1995. [DOI] [PubMed] [Google Scholar]
- 6. Björk A, Mandalenakis Z, Giang KW, Rosengren A, Eriksson P, Dellborg M. Incidence of Type 1 diabetes mellitus and effect on mortality in young patients with congenital heart defect - a nationwide cohort study. Int J Cardiol 2020;310:58–63. [DOI] [PubMed] [Google Scholar]
- 7. Dellborg M, Bjork A, Pirouzi Fard MN, Ambring A, Eriksson P, Svensson AM, Gudbjornsdottir S. High mortality and morbidity among adults with congenital heart disease and type 2 diabetes. Scand Cardiovasc J 2015;49:344–350. [DOI] [PubMed] [Google Scholar]
- 8. Gilljam T, Mandalenakis Z, Dellborg M, Lappas G, Eriksson P, Skoglund K, Rosengren A. Development of heart failure in young patients with congenital heart disease: a nation-wide cohort study. Open Heart 2019;6:e000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mandalenakis Z, Rosengren A, Lappas G, Eriksson P, Gilljam T, Hansson PO, Skoglund K, Fedchenko M, Dellborg M. Atrial fibrillation burden in young patients with congenital heart disease. Circulation 2018;137:928–937. [DOI] [PubMed] [Google Scholar]
- 10. Diller GP, Babu-Narayan S, Li W, Radojevic J, Kempny A, Uebing A, Dimopoulos K, Baumgartner H, Gatzoulis MA, Orwat S. Utility of machine learning algorithms in assessing patients with a systemic right ventricle. Eur Heart J Cardiovasc Imaging 2019;20:925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diller GP, Kempny A, Babu-Narayan SV, Henrichs M, Brida M, Uebing A, Lammers AE, Baumgartner H, Li W, Wort SJ, Dimopoulos K, Gatzoulis MA. Machine learning algorithms estimating prognosis and guiding therapy in adult congenital heart disease: data from a single tertiary centre including 10 019 patients. Eur Heart J 2019;40:1069–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lv J, Dong B, Lei H, Shi G, Wang H, Zhu F, Wen C, Zhang Q, Fu L, Gu X, Yuan J, Guan Y, Xia Y, Zhao L, Chen H. Artificial intelligence-assisted auscultation in detecting congenital heart disease, Eur Heart J Dig Health 2021;2:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krittanawong C, Zhang H, Wang Z, Aydar M, Kitai T. Artificial intelligence in precision cardiovascular medicine. J Am Coll Cardiol 2017;69:2657–2664. [DOI] [PubMed] [Google Scholar]
- 14. Johnson KW, Torres Soto J, Glicksberg BS, Shameer K, Miotto R, Ali M, Ashley E, Dudley JT. Artificial intelligence in cardiology. J Am Coll Cardiol 2018;71:2668–2679. [DOI] [PubMed] [Google Scholar]
- 15. Chen JH, Asch SM. Machine learning and prediction in medicine - beyond the peak of inflated expectations. N Engl J Med 2017;376:2507–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lown M, Brown M, Brown C, Yue AM, Shah BN, Corbett SJ, Lewith G, Stuart B, Moore M, Little P. Machine learning detection of atrial fibrillation using wearable technology. PLoS One 2020;15:e0227401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521(7553):436–444. [DOI] [PubMed] [Google Scholar]
- 18. Botto LD, Lin AE, Riehle-Colarusso T, Malik S, Correa A. Seeking causes: classifying and evaluating congenital heart defects in etiologic studies. Birth Defects Res A Clin Mol Teratol 2007;79:714–727. [DOI] [PubMed] [Google Scholar]
- 19. Liu S, Joseph KS, Luo W, León JA, Lisonkova S, Van den Hof M, Evans J, Lim K, Little J, Sauve R, Kramer MS, Canadian Perinatal Surveillance System (Public Health Agency of Canada). Effect of folic acid food fortification in Canada on congenital heart disease subtypes. Circulation 2016;134:647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mehlig B. Artificial neural networks (arXiv:1901.05639 [cs.LG]). https://arxiv.org/abs/1901.05639 (15 December 2020).
- 21. Keras CF. Keras. https://keras.io (18 July 2019).
- 22. Luo Y, Li Z, Guo H, Cao H, Song C, Guo X, Zhang Y. Predicting congenital heart defects: a comparison of three data mining methods. PLoS One 2017;12:e0177811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mandalenakis Z, Giang KW, Eriksson P, Liden H, Synnergren M, Wåhlander H, Fedchenko M, Rosengren A, Dellborg M. Survival in children with congenital heart disease: have we reached a peak at 97%? J Am Heart Assoc 2020;9:e017704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benedetto U, Sinha S, Lyon M, Dimagli A, Gaunt TR, Angelini G, Sterne J. Can machine learning improve mortality prediction following cardiac surgery? Eur J Cardiothorac Surg 2020;58:1130–1136. [DOI] [PubMed] [Google Scholar]
- 25. Benedetto U, Dimagli A, Sinha S, Cocomello L, Gibbison B, Caputo M, Gaunt T, Lyon M, Holmes C, Angelini GD. Machine learning improves mortality risk prediction after cardiac surgery: systematic review and meta-analysis. J Thorac Cardiovasc Surg 2020. doi: 10.1016/j.jtcvs.2020.07.105. [DOI] [PubMed] [Google Scholar]
- 26. Ludvigsson JF, Andersson E, Ekbom A, Feychting M, Kim JL, Reuterwall C, Heurgren M, Olausson PO. External review and validation of the Swedish national inpatient register. BMC Public Health 2011;11:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study is based on data from the Swedish Inpatient register held by the National Board of Health and Welfare (http://www.socialstyrelsen.se). The dataset contains sensitive personal information and is subjected to secrecy in accordance with the Swedish Public Access to Information and Secrecy Act (OSL, 2009:400). The data used for this study is located at the institution of the authors, and is available given that an ethical approval can be obtained from the Swedish Ethical Review Authority at the University of Gothenburg. A formal request can be made to corresponding author Dr. Giang KW at Gothenburg University, Institution of Medicine, Department of Molecular and Clinical medicine (contact: wai.giang.kok@gu.se).
Conflict of interest: none declared.