Abstract
Background
We have demonstrated that a neural network is able to predict a person’s age from the electrocardiogram (ECG) [artificial intelligence (AI) ECG age]. However, some discrepancies were observed between ECG-derived and chronological ages. We assessed whether the difference between AI ECG and chronological age (Age-Gap) represents biological ageing and predicts long-term outcomes.
Methods and results
We previously developed a convolutional neural network to predict chronological age from ECGs. In this study, we used the network to analyse standard digital 12-lead ECGs in a cohort of 25 144 subjects ≥30 years who had primary care outpatient visits from 1997 to 2003. Subjects with coronary artery disease, stroke, and atrial fibrillation were excluded. We tested whether Age-Gap was correlated with total and cardiovascular mortality. Of 25 144 subjects tested (54% females, 95% Caucasian) followed for 12.4 ± 5.3 years, the mean chronological age was 53.7 ± 11.6 years and ECG-derived age was 54.6 ± 11 years (R2 = 0.79, P < 0.0001). The mean Age-Gap was small at 0.88 ± 7.4 years. Compared to those whose ECG-derived age was within 1 standard deviation (SD) of their chronological age, patients with Age-Gap ≥1 SD had higher all-cause and cardiovascular disease (CVD) mortality. Conversely, subjects whose Age-Gap was ≤1 SD had lower all-cause and CVD mortality. Results were unchanged after adjusting for CVD risk factors and other survival influencing factors.
Conclusion
The difference between AI ECG and chronological age is an independent predictor of all-cause and cardiovascular mortality. Discrepancies between these possibly reflect disease independent biological ageing.
Keywords: ECG age, Biological ageing, Artificial intelligence, Mortality
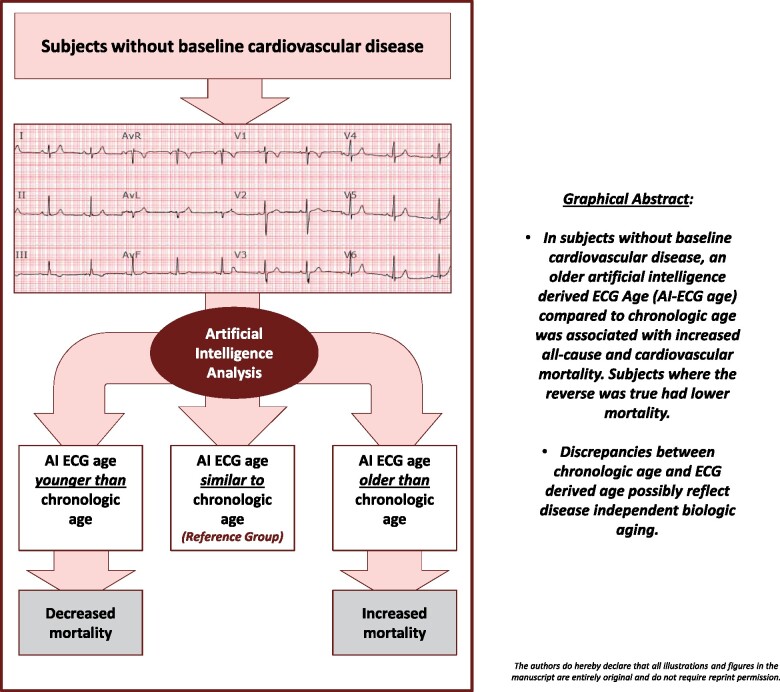
Graphical Abstract
In subjects without baseline cardiovascular disease, an older artificial intelligence (AI)-derived electrocardiogram (ECG) Age (AI-ECG age) compared to chronological age was associated with increased all-cause and cardiovascular mortality. Subjects, where the reverse was true, had lower mortality. Discrepancies between chronological age and ECG-derived age possibly reflect disease independent biological ageing.
Lay Summary
Biologic ageing is the main risk factor for many cardiovascular conditions and a determinant of long-term outcomes. We have shown that a person’s age can be predicted with an electrocardiogram (ECG) (a medical test that records the electrical activity of the heart) using an artificial intelligence (AI). However, in some subjects, differences were observed between the ages derived from the ECG and the chronological age.
Our objective was to evaluate whether these differences represent biological ageing and if they would predict long-term death.
Our hypothesis was that those deemed older by the ECG (positive difference between ECG and chronological age) would be associated with higher mortality, and that people deemed younger (negative difference) would live longer, regardless of other cardiovascular risk factors.
From the results, we obtained, we can interpret that the differences between chronological age and that of the ECG obtained with AI possibly reflect biological ageing and predicts cardiovascular mortality regardless of the person’s disease and other causes.
The ECG contains a wealth of valuable information and has been used to diagnose a variety of heart diseases. ECG AI analysis can provide information beyond the state of a specific disease and can be used to identify biological ageing.
Introduction
Ageing has been defined as a persistent decline in an organism’s age-specific fitness components due to internal physiologic deterioration.1 It is characterized by the accumulation of molecular and cellular damage, which drive progressive functional decline and increases vulnerability for disease and mortality. Considering chronological age as an exposure domain in ageing, several factors including genetics, diet, environmental interactions, mental health, and a host of other known and unknown variables affect ageing providing a cumulative effect measured by time. As such, persons of the same chronological age will not necessarily experience ageing to the same extent. Chronological age simply represents how long an organism has been alive. Biological age, also referred to as physiologic age, on the other hand, refers to the gradual decline in an organism’s functional status—the clearest measure of which is mortality. The concept of biological age has therefore been used as a more holistic description of ageing to factor in the aforementioned ageing contributors. However, we typically want to measure ageing before its irreversible endpoint. Several ageing biomarkers have been put forward, but there remains no consensus as to which one most accurately measures ageing. Indeed, there is a push to measure ageing based on multiple biochemical, molecular, and genetic biomarkers.2
The heart age has been used as a means of assessing the biological age of the cardiovascular system.3 It represents the age implied by a person’s calculated cardiovascular risk assuming all other modifiable risk factors are within ideal ranges.4 For example, a 50-year-old person who smokes, has dyslipidaemia and uncontrolled hypertension could have the same cardiovascular risk profile as a 69-year-old that has none of these conditions. His heart age would therefore be 19 years older than his chronological age. Several heart age prediction tools exist and typically require a mix of physical [e.g. chronological age, body mass index (BMI), blood pressure, etc.] and biochemical (e.g. serum cholesterol levels, diabetes diagnosis, etc.) inputs in order to estimate the heart age. Others have incorporated non-invasive parameters such as coronary artery calcification, and carotid intima-media thickness into their cardiovascular risk assessments.3,5,6 A major shortcoming of all these tools is their inability to account for ageing independent of known cardiovascular factors which are assumed to be reliable proxies for heart age. Ageing however remains the more compelling predictor of both heart age and cardiovascular events. As such, a better estimate of heart age would place more emphasis on capturing ageing.
The electrocardiogram (ECG) is a graphic representation of cardiac functioning that is known to reflect several features including normal ageing, disease states, and subject-specific characteristics. For example, age-related ECG changes, independent of cardiovascular disease (CVD), such as changes in QRS and T-wave amplitudes, cardiac activation patterns, and leftward axis shifts have been described.7 Similarly, specific ECG patterns in congenital and acquired CVDs (e.g. myocardial infarction, hypertension, etc.) have also been described.8–10 Finally, utilization of the ECG has been proposed as a biometric sensor similar to a retinal scan or a fingerprint, given the subject-specific features it contains.11,12 Given these factors, several non-artificial intelligence (AI) methods have been described to estimate the ‘heart age’ (in years) using the ECG as an input. These have typically relied on specific ECG features and statistical models alone or in conjunction with other physical characteristics e.g. BMI.13,14 A higher predicted ECG age compared to chronological age was noted in subjects with CVD and risk factors. However, the correlation between this ECG-chronological age discrepancy to all-cause or cardiovascular mortality has not been previously reported. It is also unknown whether the discrepancy reflects biological ageing, could be explained by the presence of cardiovascular risk factors alone or if it extends beyond the presence of traditional cardiovascular risk factors.
AI using machine learning has been shown to outperform traditional cardiovascular risk estimation models when provided with similar inputs.15,16 Reasons for this improved performance include machine learning’s non-confinement to linearity assumptions and its ability to utilize a larger range of continuous variables without the need to dichotomize them. Our previous work demonstrated that AI could use standard 12-lead ECGs to predict chronological age and sex, left ventricular dysfunction, and atrial fibrillation.17–19 Discrepancies between AI-enabled ECG predicted and actual chronological age were associated with individual health status.17 Whether AI analysis of standard 12-lead digital ECGs is associated with future long-term events and reflects biological ageing is currently unknown. In this work, we also explore whether AI analysis of ECGs can provide information beyond specific disease states to identify biological ageing.
Methods
Study population
Subjects were selected from a population-based, historical cohort, in Olmsted County, Minnesota which was developed using resources from the Rochester Epidemiology Project (REP).20,21 The REP is a federally funded linkage system that indexes medical records, medications, procedures, and other health-related information from the primary providers of medical care in Olmsted County: Olmsted Medical Center, the Mayo Clinic, and a few other individual private providers.20,21
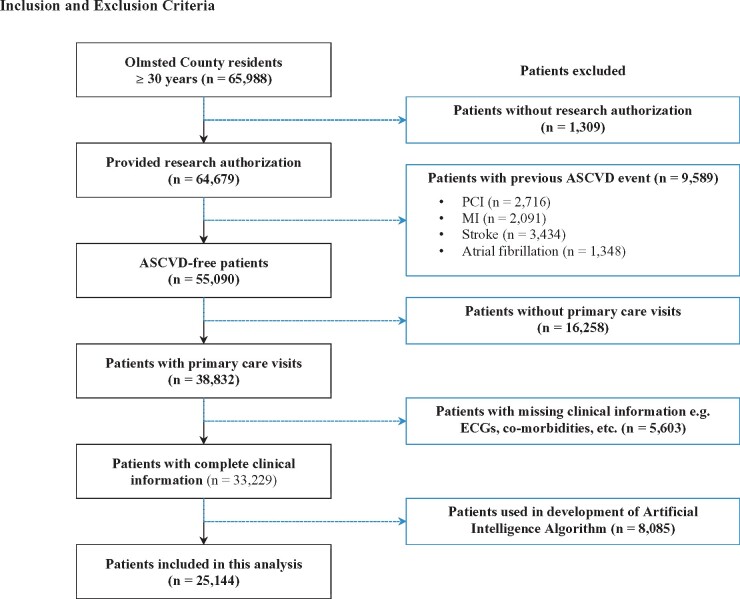
Using the population census and the REP database, all county residents aged ≥30 years with primary care outpatient clinic visits between 1997 and 2003 and standard, digital, resting 10-s, 12-lead supine ECGs were identified. Subjects were restricted to ≥30 years as individuals younger than this are expected to have less mortality and CVD. We excluded subjects with a baseline history of atherosclerotic CVD (defined as coronary artery disease, angina, myocardial infarction, and stroke at the time of or within 30 days of the ECG), atrial fibrillation, those who did not provide research authorization and those whose ECGs were used to develop the AI age estimation model (Figure 1). Subjects with a history of heart failure were not excluded. This approach was adopted in order to have our population approximate a primary prevention cohort and to better understand how comorbid conditions might influence the association between Age-Gap and mortality. We did however also perform a subgroup analyses excluding all subjects with heart failure diagnoses. When subjects had multiple ECGs, only the earliest ECG was selected.
Figure 1.
Inclusion and exclusion criteria.
Baseline data and outcomes
Baseline data including subject demographics, comorbidities, clinical diagnoses (using the International Classification of Diseases-9th), laboratory values, and vital signs at the time of the index primary care outpatient clinic visit were abstracted from the REP database using previously validated methodology.22 Primary outcomes were all-cause and cardiovascular mortality. These were obtained through 31 December 2016 directly from the REP, which records vital status from federal and state vital statistics offices as well as the National Death Index.23,24 Cause of death was defined using International Classification of Diseases, Ninth Revision (ICD-9), codes from 1997 to 1998 and International Classification of Diseases, 10th Revision (ICD-10), codes from 1999 to 2016. All-cause mortality was defined as any death. Cardiovascular mortality was defined as ICD-9 codes 390 to 398, 402, and 404 to 429 and ICD-10 codes I00 to I09, I11, I13, and I20 to I51. These represent diagnoses covering chronic rheumatic heart disease, hypertensive disease, ischaemic heart disease, pulmonary circulation diseases, and other forms of heart disease. Follow-up was considered to be complete. The study protocol was approved by the institutional review boards of both the Mayo Clinic and Olmsted Medical Center.
Overview of artificial intelligence model
A convolutional neural network (CNN) model using Keras with a Tensorflow (Google, Mountain View, CA, USA) backend was previously developed and validated. A total of 774 783 unique subjects with ECGs were used to develop the neural network: 399 750 in the training, 99 977 in the internal validation, and 275 056 in the holdout testing sets. The network contained stacked blocks of convolutional, max pooling, and batch normalization.25 Each block was followed by a non-linear activation function. After the first group of blocks extracted temporal features, another spatial block was used to fuse data from all leads, and then the extracted features were used in a fully connected network. A detailed description of the network has previously been published.17 The network utilized 10 s samples of resting, digital, standard 12-lead ECGs of patients with data in the Mayo Clinic digital vault. Its output was the AI-enabled ECG predicted age as continuous number. Our current study used the previous network with no additional retraining using ECGs from the current study population.
Statistical analysis
We summarized subject characteristics with frequencies and percentages or means ± standard deviations (SDs) as appropriate. Age-Gap was defined as the algebraic difference between AI-enabled ECG predicted age and chronological age and was calculated by subtracting the chronological age from the AI ECG age. Therefore, a positive Age-Gap would reflect a person identified as being older by AI compared to his/her actual chronological age, and a negative Age-Gap will reflect a person that is recognized as younger by the computer compared to his/her chronological age. Chronological age and the AI-enabled ECG predicted age (in years) were assessed and correlated with the Pearson correlation coefficient (R2). The functional form Age-Gap was operationalized as both a continuous and categorical (≥2 SD below, 1–2 SD below, within 1 SD, 1–2 SD above, ≥2 SD above) variable as appropriate for different analyses. The continuous form was used to evaluate its relationship to all-cause and cardiovascular mortality. To graphically illustrate the association between Age-Gap and mortality, polynomial smoothing splines were utilized. Kaplan–Meier curves with the log-rank test and Cox proportional hazards models, were adjusted for factors associated with Age-Gap (e.g. chronological age, sex, BMI, ongoing smoking, heart failure history and dyslipidaemia history) and additional factors know to be associated with mortality (e.g. diabetes history, hypertension history, systolic blood pressure, and total cholesterol). Findings were summarized using hazard ratios (HRs) and 95% confidence intervals (CIs). Multiplicative interactions between Age-Gap and select subject characteristics were calculated. The assumption of proportionality for the Cox proportional hazards models was assessed graphically (and fulfilled).
Logistic regression analyses were used to investigate the effects of covariates on Age-Gap. The primary analysis compared those with an Age-Gap ≥1 SD to those within 1 SD. Each sociodemographic and clinical characteristic was examined for association with and without chronological age adjustment. Findings were summarized using odds ratios (ORs) and 95% CIs. A multinomial logistic regression analysis was also applied where the ≥1 SD group was broken into 1–2 SD and 2+ SD. A contrast was constructed within the multinomial regression framework to test for heterogeneity of association across SD groups and this test is reported as Phet. Two-sided P-values <0.05 were considered statistically significant. All analyses were completed using SAS version 9.4 (SAS Institute Inc., Cary, NC, USA) and R (www.r-porject.org).
Results
Of 25 144 subjects tested (54% females, 95% Caucasian) and followed for 12.5 ± 5.3 years, the mean chronological and AI ECG ages were 53.7 ± 11.6 years and 54.6 ± 11 years, respectively (R2 = 0.79, P < 0.0001). The neural network performed well in predicting the age of most subjects in this cohort with a mean Age-Gap of 0.88 ± 7.4 years. Other baseline characteristics of our study population are shown in Table 1.
Table 1.
Baseline characteristics of the study population
| Variable | Study population (N = 25 144) |
|---|---|
| Demographics | |
| Race/ethnicity, n (%) | |
| White non-Hispanic | 22 983 (91.4%) |
| White Hispanic | 805 (3.2%) |
| Black or African American | 699 (2.8%) |
| Other | 657 (2.6%) |
| Female sex, n (%) | 13 548 (54%) |
| Chronological age, years | 53.71 ± 11.59 |
| AI ECG age, years | 54.59 ± 10.99 |
| Age-Gap, years | 0.88 ± 7.41 |
| Clinical history | |
| Body mass index | 28.71 ± 6.30 |
| Heart failure history, n (%) | 2371 (9%) |
| COPD history, n (%) | 1334 (5%) |
| Chronic kidney disease history, n (%) | 1209 (5%) |
| Current smoker, n (%) | 3969 (16%) |
| Diabetes history n (%) | 2551 (10%) |
| Hypertension history, n (%) | 4215 (17%) |
| Dyslipidaemia history, n (%) | 5438 (22%) |
| Alcoholism history, n (%) | 1267 (5%) |
Data are n (%) or mean (SD).
AI ECG Age, artificial intelligence enabled ECG age; Age-Gap, AI ECG age minus chronological age.
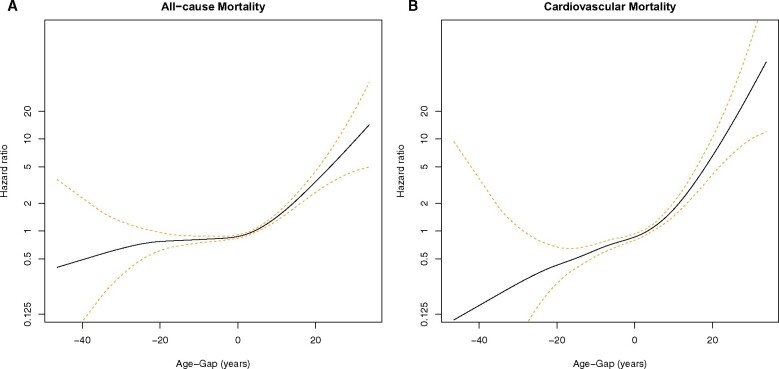
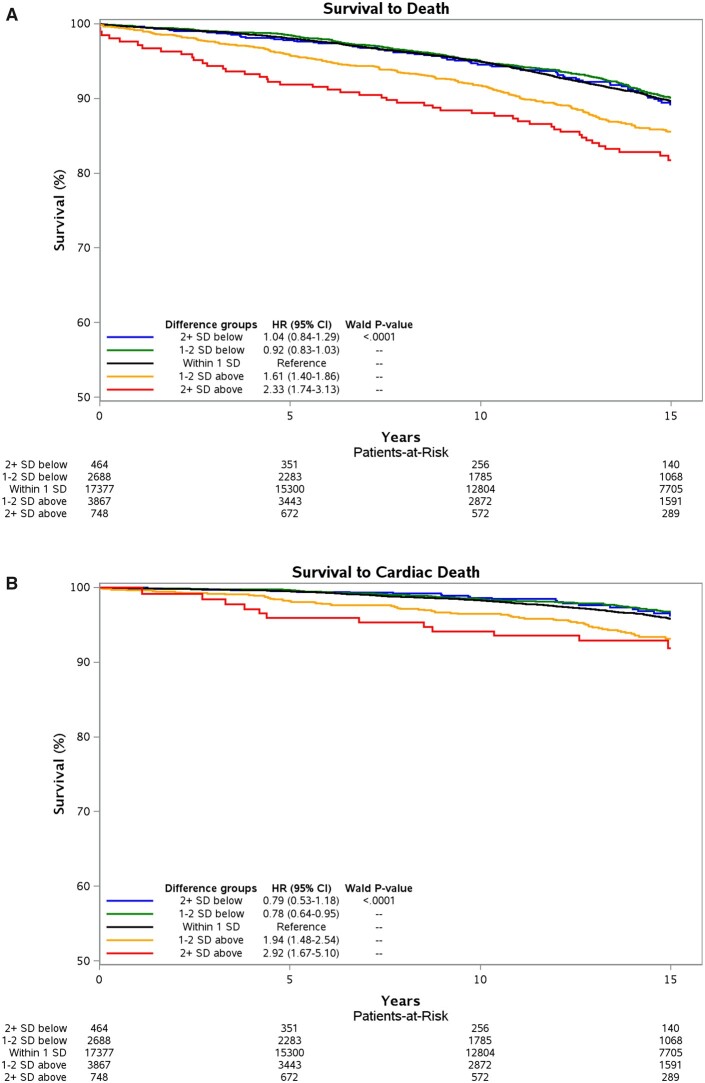
Subjects with positive Age-Gap had ECG-derived age older than their chronological age while those with negative Age-Gap had ECG-derived age younger than their chronological age. Primary outcomes were all-cause and cardiovascular mortality. There was a clear relationship between Age-Gap and both primary outcomes as shown in Figures 2A and B (as well as in Supplementary material online, Table S1 and Figures S1a–c and S2a–c). Compared to those whose AI-enabled ECG-derived age was within 1 SD (±7.4 years) of their chronological age, subjects with Age-Gap greater than +1 SD (considered older) had increased risk for all-cause mortality, while those with Age-Gap less than −1 SD (considered younger) had decreased risk (Figure 3A). After multivariate adjustment, those with Age-Gap between +1 and +2 SD and those with Age-Gap >+2 SD had increased risk of all-cause mortality (HR 1.61, 95% CI 1.40–1.86; HR 2.33, 95% CI 1.74–3.13, respectively). Those with Age-Gap between −1 and −2 SD and Age-Gap <−2 SD had trends towards decreased risk for all-cause mortality, P-value for trend <0.0001 (Figure 3A, Supplementary material online, Table S1).
Figure 2.
(A and B) Spline curves showing the relationship between Age-Gap and mortality [all-cause (A) and cardiovascular (B)].
Figure 3.
(A) All-cause mortality across different Age-Gap categories. Curves adjusted for chronological age, sex, current smoking, diabetes history, hypertension history, systolic blood pressure, dyslipidaemia history, total cholesterol, heart failure history, and body mass index. A positive Age-Gap means subjects’ electrocardiogram-derived ages were older than their chronological ages. In this figure, this includes 2+ SD above and 1–2 SD above. A negative Age-Gap means subjects’ electrocardiogram-derived ages were younger than their chronological ages. In this figure, this includes 2+ SD below and 1–2 SD below. (B) Cardiovascular mortality across different Age-Gap categories. Curves adjusted for chronological age, sex, current smoking, diabetes history, hypertension history, systolic blood pressure, dyslipidaemia history, total cholesterol, heart failure history, and body mass index. A positive Age-Gap means subjects’ electrocardiogram-derived ages were older than their chronological ages. In this figure, this includes 2+ SD above and 1–2 SD above. A negative Age-Gap means subjects’ electrocardiogram-derived ages were younger than their chronological ages. In this figure, this includes 2+ SD below and 1–2 SD below.
There was a more robust association between Age-Gap and cardiovascular mortality across both sides of the SD spectrum (Figure 3B and Table 2). After multivariate adjustment, those with Age-Gap > +2 SD had the highest risk (HR 2.92, 95% CI 1.67–5.10) followed by those with Age-Gap between +1 and +2 SD (HR 1.94, 95% CI 1.48–2.54). Those with Age-Gap <−2 SD had the lowest risk for cardiovascular mortality (HR 0.79, 95% CI 0.53–1.18) followed by those with Age-Gap between −1 and −2 SD (HR 0.78, 95% CI 0.64–0.95), P-value for trend <0.0001 (Figure 3B). The absolute risk difference between these last two groups was small at ∼1%. The decreased cardiovascular mortality for those with Age-Gap <−2 SD was not statistically significant likely due to insufficient sample size.
Table 2.
Hazard ratios for cardiovascular mortality
| Chronological age and sex adjusted |
Multivariate model |
|||
|---|---|---|---|---|
| HR | P-value | HR | P-value | |
| Chronological age, per 1 year increase | 1.13 (1.12–1.14) | <0.0001 | 1.11 (1.10–1.11) | <0.0001 |
| Female sex | 0.56 (0.49–0.64) | <0.0001 | 0.57 (0.48–0.70) | <0.0001 |
| Current smoker | 1.88 (1.55–2.28) | <0.0001 | 1.73 (1.43–2.09) | <0.0001 |
| Diabetes history | 3.03 (2.60–3.54) | <0.0001 | 1.97 (1.69–2.32) | <0.0001 |
| Hypertension history | 5.18 (4.50–5.98) | <0.0001 | 4.83 (4.16–5.60) | <0.0001 |
| Systolic blood pressure, per mmHg increase | 1.05 (1.03–1.07) | <0.0001 | 1.04 (1.02–1.06) | <0.0001 |
| Dyslipidaemia history | 1.60 (1.38–1.85) | <0.0001 | 1.37 (1.16–1.62) | 0.0002 |
| Total cholesterol, per 1 mg/dL increase | 0.99 (0.98–1.00) | 0.5192 | 1.00 (0.99–1.01) | 0.7210 |
| Heart failure history | 3.32 (2.86–3.85) | <0.0001 | 3.02 (2.59–3.53) | <0.0001 |
| BMI, per 5 kg/m2 increase | 1.13 (1.07–1.20) | <0.0001 | 1.00 (0.99–1.07) | 0.7957 |
| ECG age, per 1 year increase | 1.05 (1.04–1.07) | <0.0001 | 1.04 (1.03–1.06) | <0.0001 |
| Δ ECG-chronological age, years | 1.05 (1.04–1.06) | <0.0001 | 1.04 (1.03–1.06) | <0.0001 |
| Grouped Δ ECG-chronological age | ||||
| 2+ SD below | 0.61 (0.41–0.92) | 0.0188 | 0.79 (0.53–1.18) | 0.2462 |
| 1–2 SD below | 0.66 (0.54–0.80) | <0.0001 | 0.78 (0.64–0.95) | 0.0119 |
| Within 1 SD (Reference group) | ||||
| 1–2 SD above | 2.23 (1.71–2.90) | <0.0001 | 1.94 (1.48–2.54) | <0.0001 |
| 2+ SD above | 4.84 (2.78–8.43) | <0.0001 | 2.92 (1.67–5.10) | 0.0002 |
Data are hazard ratios (95% CI).
Multivariate model adjusts for chronological age, sex, current smoking, diabetes history, hypertension history, systolic blood pressure, dyslipidaemia history, total cholesterol, heart failure history, and body mass index.
BMI, body mass index; COPD, chronic obstructive pulmonary disease.
There was an increased prevalence of some comorbidities with increasing Age-Gap. After adjusting for chronological age, several comorbidities were associated with Age-Gap ≥1 SD (Table 3): current smoking (OR 1.19, 1.09–1.29), BMI per 5 kg/m2 increase (OR 1.03, 1.02–1.03), dyslipidaemia history (OR 1.27, 1.17–1.37), and heart failure history (OR 1.70, 1.49–1.94). Multivariable analysis showed that chronological age, sex, BMI, heart failure history, current smoking, and dyslipidaemia history were independently associated with Age-Gap ≥1 SD. When analysed between SD subgroups (1–2 and 2+ SD), there were increased odds of having heart failure with increasing Age-Gap: Age-Gap between +1 and +2 SD (OR 1.52, 1.33–1.75) and Age-Gap> +2 SD (OR 3.05, 2.33–3.99), Phet < 0.001. While not statistically significant, other comorbidities demonstrated a trend to increased odds of being present across increasing Age-Gap subgroups (Table 4).
Table 3.
Chronological age-adjusted logistic regression analysis evaluating the odds for having comorbidities and Age-Gap ≥1 SD compared to Age-Gap within 1 SD
| Variable | OR | P-value |
|---|---|---|
| Chronological age adjusted | ||
| Female sex | 0.86 (0.81–0.92) | <0.0001 |
| Current smoker | 1.19 (1.09–1.29) | <0.0001 |
| Diabetes history | 1.03 (0.92–1.16) | 0.58 |
| Hypertension history | 1.12 (1.02–1.22) | 0.02 |
| Dyslipidaemia history | 1.27 (1.17–1.37) | <0.0001 |
| BMI, per 5 kg/m2 increase | 1.03 (1.02–1.03) | <0.0001 |
| COPD history | 1.02 (0.88–1.18) | 0.78 |
| Chronic kidney disease history | 1.06 (0.91–1.23) | 0.48 |
| Heart failure history | 1.70 (1.49–1.94) | <0.0001 |
| Multivariate adjusted | ||
| Chronological age, per 5 year increase | 0.71 (0.70–0.73) | <0.0001 |
| Female sex | 0.94 (0.87–1.00) | 0.06 |
| BMI, per 5 kg/m2 increase | 1.13 (1.10–1.16) | <0.0001 |
| Heart failure history | 1.55 (1.36–1.77) | <0.0001 |
| Current smoker | 1.17 (1.07–1.28) | 0.0003 |
| Dyslipidaemia history | 1.12 (1.03–1.22) | 0.007 |
Data are odds ratios (95% CI).
BMI, body mass index; COPD, chronic obstructive pulmonary disease.
Table 4.
Multinomial logistic regression (within 1 SD, 1–2 SD above, 2+ SD above); chronological age adjusted
| Age-Gap | Variable | OR | P-value | P het |
|---|---|---|---|---|
|
1–2 SD 2+ SD |
Female sex |
0.86 (0.80–0.93) 0.86 (0.74–0.99) |
<0.0001 0.04 |
0.90 |
|
1–2 SD 2+ SD |
Current smoker |
1.16 (1.05–1.27) 1.35 (1.13–1.62) |
0.002 0.0008 |
0.10 |
|
1–2 SD 2+ SD |
Diabetes history |
1.00 (0.88–1.13) 1.20 (0.94–1.52) |
0.99 0.15 |
0.18 |
|
1–2 SD 2+ SD |
Hypertension history |
1.09 (0.99–1.20) 1.23 (1.02–1.50) |
0.07 0.03 |
0.25 |
|
1–2 SD 2+ SD |
Dyslipidaemia history |
1.24 (1.14–1.35) 1.44 (1.22–1.69) |
<0.0001 <0.0001 |
0.09 |
|
1–2 SD 2+ SD |
BMI, per 5 kg/m2 increase |
1.03 (1.02–1.03) 1.03 (1.02–1.04) |
<0.0001 <0.0001 |
0.81 |
|
1–2 SD 2+ SD |
COPD history |
1.02 (0.87–1.19) 1.04 (0.77–1.42) |
0.84 0.78 |
0.87 |
|
1–2 SD 2+ SD |
Chronic kidney disease history |
1.01 (0.85–1.19) 1.32 (0.97–1.80) |
0.92 0.07 |
0.10 |
|
1–2 SD 2+ SD |
Heart failure history |
1.52 (1.33–1.75) 3.05 (2.33–3.99) |
<0.0001 <0.0001 |
<0.0001 |
Data are odds ratios (95% CI).
BMI, body mass index; COPD, chronic obstructive pulmonary disease.
Subgroup analysis showed that our results did not significantly change when we excluded subjects with a history of heart failure (Supplementary material online, Tables S2 and S3). It also showed that the algorithm performed well across self-identified racial/ethnic groups with similar HRs for each racial/ethnic group when assessing Age-Gap and all-cause mortality (Supplementary material online, Table S4). A test for the interaction between racial/ethnic subgroups and Age-Gap was non-significant (P Wald = 0.47). A similar situation was seen when exploring potential gender bias (Supplementary material online, Table S5). HRs for all-cause mortality were not significantly different by sex and the interaction between sex and Age-Gap was also not significant (P Wald = 0.60).
Discussion
In this study, we provide a proof of concept that AI ECG age obtained using a standard 12-lead ECG is associated with all-cause and cardiovascular mortality, beyond what would be expected from chronological age alone. This association persists even after adjusting for comorbidities known to predict survival, indicating that AI analysis of standard 12-lead ECGs could be employed as a marker of either advanced or slower biological ageing. The purpose of our work is not to predict chronological age from the 12-lead ECG but to demonstrate that AI-enabled ECG predicted age identifies signals related to the biological ageing of the cardiovascular system by demonstrating that the gap between chronological age and what the ECG predicts reflects different ageing rates. To the best of our knowledge, no other studies have assessed the correlation between Age-Gap and long-term outcomes or proposed Age-Gap as a biomarker of the ageing process
Our AI model to predict chronological age performed well (r = 0.837) for most subjects as previously described.17 However, age prediction was imperfect, as several subjects had discrepancies between their chronological and ECG-derived ages. We refer to this discrepancy as Age-Gap and it is conceptually similar to excess heart age (EHA).26,27 In this study, we show that rather than representing random error, Age-Gap derived from the ECG is correlated with both all-cause and cardiovascular mortality in a population-based historical cohort of subjects with no known baseline cardiovascular comorbidities. All-cause and cardiovascular mortality risks are demonstrated to increase in a step-wise manner with Age-Gap. We also show a correlation between Age-Gap and known comorbidities.
Ball et al.13 and Starc et al.14 used Bayesian and linear regression models to determine heart age using a several predefined variables based on P-wave, QRS complex, and T-wave characteristics extracted from standard and signal averaged ECGs in small cohorts. We have employed AI interpretation of standard 12-lead ECGs to predict heart age in a much larger subject cohort. The nature of our AI analysis allows a wider range of ECG variables to be used in heart age estimation. Hirsch et al.27 showed that mean EHA was associated with cardiovascular mortality and race. The heart age they used in calculating EHA was based on seven sex-specific, non-laboratory based Framingham risk prediction inputs. The correlation between EHA and cardiovascular mortality was not surprising as cardiovascular risk factors were used to derive the heart age estimates. A major limitation of this study was that they did not explore the correlation between EHA and total mortality. Our study estimated heart age using a standard 12-lead ECG as the singular input. We showed a correlation not only with cardiovascular mortality but also with all-cause mortality indicating that Age-Gap captures facets beyond traditional cardiovascular risk factors. Finally, Raghunath et al.28 used the ECG to predict short term (1 year) mortality, but without any attempt to use the ECG as a measure of biological age or to predict long-term outcomes.
It is important to note that while several methods (e.g. multiple linear regression, principal component analysis, the Hochschild’s method, and the Klemera and Doubal’s method) for assessing biological age exist, they require categorical, binary, or continuous variables as inputs.29–32 Our approach represents a paradigm shift in two ways. First, the raw ECG signals, which we utilize as our sole input, do not fit into any of these input categories. Second, our analysis of this non-traditional input is done using deep neural networks that can consider linear and non-linear dynamics that other methods typically haven’t been able to fully account for. Allowing AI to explore the ECG on its own potentially allows it to sidestep investigator biases and to identify and use ECG features which may not be readily identified by cardiologists. While this approach provides an optimal representation of the data, it is not readily explainable using tools available today.
Exploring potential bias
A recurring concern about deployment of AI in healthcare is about potential racial and sex bias. The original AI algorithm that was tested in this study’s cohort was developed using a predominantly Caucasian population.17 Subgroup analysis based on this study’s cohort showed that despite this, the algorithm performed well across racial groups with the interaction between Age-Gap and racial subgroups being non-significant. Both sexes were about evenly represented in the cohort used to develop the AI algorithm.17 As expected, subgroup analysis showed that the model performed well across both sexes with the interaction between sex and Age-Gap also not being significant.
Excess and reduced risk
We demonstrated increased risk for all-cause and cardiovascular mortality with increasing Age-Gap. This association with excess risk was even more robust when exploring cardiovascular mortality (although this endpoint was largely based on ICD diagnoses). This is expected as Age-Gap is based on the ECG which is a direct output from a critical component of the cardiovascular system. There was also lower all-cause and cardiovascular mortality with a negative Age-Gap. For all-cause mortality, this effect was attenuated after one Age-Gap SD (7.4 years) and is possibly due to the small sample size of the >−2 SD subgroup.
Potential applications
A fundamental goal of medicine is to prevent morbidity and mortality. Ageing is the most consistent risk factor for morbidity and mortality but measuring it has proved challenging. A potential use of the Age-Gap is to identify individuals who are ageing beyond what would be expected from their chronological age. Along with medical histories and other clinical data, it could potentially be used to identify subjects with excess risk that may represent preclinical or undiagnosed disease for which interventions can be deployed and perhaps create opportunities for prevention. A future state integrating Age-Gap into patients’ ECG reports could help facilitate this. A potential criticism of this approach is that all patients should be advised to adopt a healthy lifestyle, undertake appropriate screening, and take anti-ageing measures and that the information provided by the Age-Gap is therefore redundant. However, one does not need to look too far to conclude that such an approach has not been universally successful. We believe Age-Gap is an addition to the healthcare toolkit that can be used to drive increased patient motivation to make sustained positive lifestyle changes. It could also serve as a reminder for healthcare providers to screen for pre-symptomatic disease. We recognize that our work is largely a proof of concept, and as such, it will be pretentious to extensively talk about immediate applicability. We do however hope it may also be useful in assessing frailty, and, by serial Age-Gap assessment, the effectiveness of anti-ageing drugs and/or of adoption of a healthy lifestyle. Frailty is associated with biological age although we acknowledge that they are not the same. We do however hope that further research into Age-Gap can help elucidate whether this hypothesized association with ageing holds true.
Strengths and limitations
Our study has several strengths. Our AI model to predict age from standard 12 ECGs was developed using a very large sample of subjects. This model was then applied to another large and widely used cohort of subjects with validated outcomes and extended follow-up. This notwithstanding, there are several limitations to our study. First, we hypothesize that Age-Gap reflects biological ageing but acknowledge that no clear gold standard exists to assess biological ageing independent of disease states.33 Second, several comorbidities and cardiovascular outcomes were based on ICD codes which may not always be accurate. We also did not have a mortality adjudication committee as part of this work. While cause of death using ICD codes has been used extensively in epidemiologic studies, we recognize it is imperfect and can lead to misclassification. However, the information from the REP is routinely reviewed for accuracy, has been widely used and has been shown to be reliable.23,24 Furthermore, our main primary outcome was all-cause mortality which is less prone to bias. Third, while subjects were free of known CVD at baseline, their point of entry into the database was via a primary care visit. Therefore, and it is not unreasonable to assume ECGs were prompted by clinical concerns in some subjects. Including diagnoses up to 30 days after the incident ECG and then excluding subjects with known baseline cardiovascular conditions, attempts to mitigate this concern. Fourth, subjects with a baseline history of heart failure were not excluded and it can be argued that the AI algorithm is picking this up. However, only a small percentage (∼9%) of patients had this history. Furthermore, the association between Age-Gap and our primary outcome measures remained significant after removing all patients with a heart failure history. Fifth, while we tried to adjust for known variables that could affect our outcome, we acknowledge that this is not perfect. We were only able to adjust for validated variables captured in our dataset and other confounders may exist that may continue to affect our output despite our diligent efforts. Sixth, while every effort was made to determine the cause of mortality, we cannot exclude cause of death apportioning errors made in vital statistics databases used to ascertain mortality. Finally, our population is predominantly Caucasian and not racially diverse. It does however reflect the general population of Minnesota and the Midwest region of the USA.34 Our subgroup analysis also shows that the algorithm performs well across racial/ethnic groups.
Future research
Further research is needed to assess whether Age-Gap will be associated with long-term outcomes in community, non-patient and more ethnically diverse populations. Other areas for further research include comparing AI ECG age to other estimators of heart age (e.g. CDC heart age), comparing risk profile outputs from neural network ECG analysis with other risk prediction algorithms (e.g. Charlson comorbidity index, multi-morbidity index, etc.), determining which ECG parameters drive excess risk and whether neural network predicted age correlates with known markers of accelerated ageing.
Lead author biography

Dr Francisco Lopez-Jimenez is a Professor of Medicine at Mayo College of Medicine, the chair of the Division of Preventive Cardiology and the Co-Director of Artificial Intelligence in Cardiology at Mayo Clinic. Dr Lopez-Jimenez did his cardiology fellowship at Mount Sinai Medical Center in Miami, Florida and at Brigham and Women’s Hospital, Harvard Medical School. He trained in preventive cardiology at the VA Boston Healthcare System and Harvard Medical School and holds a Master’s of Science from Harvard School of Public Health and an MBA degree from Augsburg University. He was the Program Director for the first CME course focused on artificial intelligence in Cardiology organized by Mayo Clinic in July of 2019. The artificial intelligence programme in cardiology at Mayo Clinic has been very productive publishing several articles in the last 12 months primarily using ECG signals but has extended to outcome-based machine learning and image processing. He has published more than 300 scientific publications, including invited editorials in journals such as Lancet, and his scientific work has been cited more than 58,000 times. He was a guest speaker at the Global Submit on Circulatory Health Focused on Digital Health and AI in 2019, a meeting organized by the World Heart Federation to determine the next steps national and international organizations need to take to develop, implement, scale, and regulate artificial intelligence related to cardiovascular diseases.
Supplementary material
Supplementary material is available at European Heart Journal - Digital Health online.
Funding
This publication was made possible in part by funding from the department of Cardiovascular Medicine, Mayo Clinic, Rochester MN, and Resources of the Rochester Epidemiology Project (REP), which is supported by the National Institute of Aging under Award Number R01AG034676. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding sources.
Conflict of interest: P.A.F., Z.I.A., F.L.-J., and S.K. have filed intellectual property related to the AI algorithm used here to detect biological age from the ECG.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author. All requests for raw and analysed data and related materials, excluding programming code, will be reviewed by the Mayo Clinic legal department and Mayo Clinic Ventures to verify whether the request is subject to any intellectual property or confidentiality obligations. Requests for patient-related data not included in the paper will not be considered. Any data and materials that can be shared will be released via a Material Transfer Agreement.
Supplementary Material
References
- 1. Rose M. Evolutionary Biology of Aging. New York: Oxford University Press; 1991. [Google Scholar]
- 2. Sebastiani P, Thyagarajan B, Sun F, Schupf N, Newman AB, Montano M, Perls TT.. Biomarker signatures of aging. Aging Cell 2017;16:329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Groenewegen KA, den Ruijter HM, Pasterkamp G, Polak JF, Bots ML, Peters SA.. Vascular age to determine cardiovascular disease risk: a systematic review of its concepts, definitions, and clinical applications. Eur J Prev Cardiol 2016;23:264–274. [DOI] [PubMed] [Google Scholar]
- 4. D'Agostino RB Sr, Vasan RS, Pencina MJ, Wolf PA, Cobain M, Massaro JM, Kannel WB.. General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation 2008;117:743–753. [DOI] [PubMed] [Google Scholar]
- 5. Khalil Y, Mukete B, Durkin MJ, Coccia J, Matsumura ME.. A comparison of assessment of coronary calcium vs carotid intima media thickness for determination of vascular age and adjustment of the Framingham Risk Score. Prev Cardiol 2010;13:117–121. [DOI] [PubMed] [Google Scholar]
- 6. Shaw LJ, Raggi P, Berman DS, Callister TQ.. Coronary artery calcium as a measure of biologic age. Atherosclerosis 2006;188:112–119. [DOI] [PubMed] [Google Scholar]
- 7. Simonson E. The effect of age on the electrocardiogram. Am J Cardiol 1972;29:64–73. [DOI] [PubMed] [Google Scholar]
- 8. Khairy P, Marelli AJ.. Clinical use of electrocardiography in adults with congenital heart disease. Circulation 2007;116:2734–2746. [DOI] [PubMed] [Google Scholar]
- 9. Lanza GA. The electrocardiogram as a prognostic tool for predicting major cardiac events. Prog Cardiovasc Dis 2007;50:87–111. [DOI] [PubMed] [Google Scholar]
- 10. Thygesen K, Alpert JS, Jaffe AS, Chaitman BR, Bax JJ, Morrow DA, White HD.. Fourth universal definition of myocardial infarction (2018). J Am Coll Cardiol 2018;72(18):2231–2264. [DOI] [PubMed] [Google Scholar]
- 11. Lourenço A, Silva H, Fred A.. Unveiling the biometric potential of finger-based ECG signals. Comput Intell Neurosci 2011;2011:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pinto J, Cardoso J, Lourenço A, Carreiras C.. Towards a continuous biometric system based on ECG signals acquired on the steering wheel. Sensors 2017;17:2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ball R, Feiveson A, Schlegel T, Starc V, Dabney A.. Predicting “heart age” using electrocardiography. J Pers Med 2014;4:65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Starc V, Leban MA, Šinigoj P, Vrhovec M, Potočnik N, Fernlund E, Liuba P, Schlegel TT.. Can functional cardiac age be predicted from the ECG in a normal healthy population? In: 2012 Computing in Cardiology. IEEE; 2012, pp. 101–104. [Google Scholar]
- 15. Mortazavi BJ, Bucholz EM, Desai NR, Huang C, Curtis JP, Masoudi FA, Shaw RE, Negahban SN, Krumholz HM.. Comparison of machine learning methods with national cardiovascular data registry models for prediction of risk of bleeding after percutaneous coronary intervention. JAMA Netw Open 2019;2:e196835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rymer JA, Rao SV.. Enhancement of risk prediction with machine learning: rise of the machines. JAMA Netw Open 2019;2:e196823. [DOI] [PubMed] [Google Scholar]
- 17. Attia ZI, Friedman PA, Noseworthy PA, Lopez-Jimenez F, Ladewig DJ, Satam G, Pellikka PA, Munger TM, Asirvatham SJ, Scott CG, Carter RE, Kapa S.. Age and sex estimation using artificial intelligence from standard 12-lead ECGs. Circ Arrhythm Electrophysiol 2019;12:e007284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Attia ZI, Kapa S, Lopez-Jimenez F, McKie PM, Ladewig DJ, Satam G, Pellikka PA, Enriquez-Sarano M, Noseworthy PA, Munger TM, Asirvatham SJ, Scott CG, Carter RE, Friedman PA.. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram. Nat Med 2019;25:70–74. [DOI] [PubMed] [Google Scholar]
- 19. Attia ZI, Noseworthy PA, Lopez-Jimenez F, Asirvatham SJ, Deshmukh AJ, Gersh BJ, Carter RE, Yao X, Rabinstein AA, Erickson BJ, Kapa S, Friedman PA.. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet 2019;394:861–867. [DOI] [PubMed] [Google Scholar]
- 20. Melton LJ 3rd. History of the Rochester Epidemiology Project. Mayo Clin Proc 1996;71:266–274. [DOI] [PubMed] [Google Scholar]
- 21. Rocca WA, Yawn BP, St Sauver JL, Grossardt BR, Melton LJ 3rd. History of the Rochester Epidemiology Project: half a century of medical records linkage in a US population. Mayo Clin Proc 2012;87:1202–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Melton LJ 3rd, Rocca WA, Roger VL.. Development of population research at Mayo Clinic. Mayo Clin Proc 2014;89:e17–e20. [DOI] [PubMed] [Google Scholar]
- 23. St Sauver JL, Grossardt BR, Yawn BP, Melton LJ 3rd, Pankratz JJ, Brue SM, Rocca WA.. Data resource profile: the Rochester Epidemiology Project (REP) medical records-linkage system. Int J Epidemiol 2012;41:1614–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. St Sauver JL, Grossardt BR, Yawn BP, Melton LJ 3rd, Rocca WA.. Use of a medical records linkage system to enumerate a dynamic population over time: the Rochester epidemiology project. Am J Epidemiol 2011;173:1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. In: Francis B, David B, (eds). IProceedings of the 32nd International Conference on Machine Learning. Image Proceedings of Machine Learning Research: PMLR; 2015, 448–456.
- 26. Yang Q, Zhong Y, Ritchey M, Cobain M, Gillespie C, Merritt R, Hong Y, George MG, Bowman BA.. Vital signs: predicted heart age and racial disparities in heart age among US adults at the state level. MMWR Morb Mortal Wkly Rep 2015;64:950–958. [DOI] [PubMed] [Google Scholar]
- 27. Hirsch JR, Waits G, Li Y, Soliman EZ.. Racial differences in heart age and impact on mortality. J Natl Med Assoc 2018;110:169–175. [DOI] [PubMed] [Google Scholar]
- 28. Raghunath S, Cerna AEU, Jing L, vanMaanen DP, Stough J, Hartzel DN, Leader JB, Kirchner HL, Good CW, Patel AA.. Deep neural networks can predict mortality from 12-lead electrocardiogram voltage data. Nat Med 2020;26:886–891. [DOI] [PubMed] [Google Scholar]
- 29. Klemera P, Doubal S.. A new approach to the concept and computation of biological age. Mech Ageing Dev 2006;127:240–248. [DOI] [PubMed] [Google Scholar]
- 30. Krøll J, Saxtrup O.. On the use of regression analysis for the estimation of human biological age. Biogerontology 2000;1:363–368. [DOI] [PubMed] [Google Scholar]
- 31. Levine ME. Modeling the rate of senescence: can estimated biological age predict mortality more accurately than chronological age? J Gerontol A Biol Sci Med Sci 2013;68:667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakamura E, Miyao K, Ozeki T.. Assessment of biological age by principal component analysis. Mech Ageing Dev 1988;46:1–18. [DOI] [PubMed] [Google Scholar]
- 33. Wagner K-H, Cameron-Smith D, Wessner B, Franzke B.. Biomarkers of aging: from function to molecular biology. Nutrients 2016;8:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sauver JLS, Grossardt BR, Leibson CL, Yawn BP, Melton LJ III, Rocca WA.. Generalizability of epidemiological findings and public health decisions: an illustration from the Rochester Epidemiology Project. Mayo Clin Proc 2012;87):151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author. All requests for raw and analysed data and related materials, excluding programming code, will be reviewed by the Mayo Clinic legal department and Mayo Clinic Ventures to verify whether the request is subject to any intellectual property or confidentiality obligations. Requests for patient-related data not included in the paper will not be considered. Any data and materials that can be shared will be released via a Material Transfer Agreement.