Abstract
Machine learning (ML) is a sub-field of artificial intelligence that uses computer algorithms to extract patterns from raw data, acquire knowledge without human input, and apply this knowledge for various tasks. Traditional statistical methods that classify or regress data have limited capacity to handle large datasets that have a low signal-to-noise ratio. In contrast to traditional models, ML relies on fewer assumptions, can handle larger and more complex datasets, and does not require predictors or interactions to be pre-specified, allowing for novel relationships to be detected. In this review, we discuss the rationale for the use and applications of ML in heart failure, including disease classification, early diagnosis, early detection of decompensation, risk stratification, optimal titration of medical therapy, effective patient selection for devices, and clinical trial recruitment. We discuss how ML can be used to expedite implementation and close healthcare gaps in learning healthcare systems. We review the limitations of ML, including opaque logic and unreliable model performance in the setting of data errors or data shift. Whilst ML has great potential to improve clinical care and research in HF, the applications must be externally validated in prospective studies for broad uptake to occur.
Keywords: Machine learning, Heart failure, Artificial intelligence
Graphical Abstract
Graphical abstract.
Introduction
One of the most common causes of hospitalization among older patients, heart failure (HF) poses challenges in diagnosis, management, organization of health services, and risk prediction.1,2 HF is one of the most expensive healthcare conditions to manage in high-income countries. Patients receive numerous diagnostic tests, invasive procedures, and therapies over the course of their illness, generating large amounts of data that can be aggregated into registries or other institutional databases to evaluate healthcare utilization, quality and cost of care, and disease progression.3 The size, complexity, and dynamic nature of these ‘big data’ can be challenging for traditional analytical methods to make sense of.4
Machine learning (ML) encompasses computational techniques that can extract patterns from data, acquire knowledge, and apply this knowledge to tasks such as risk prediction.5 ML methods can handle temporal, large-volume, and multi-modality data [e.g., sound, language, tabular electronic health record (EHR), imaging, and metabolomic data].6 In HF, delivering the right care to the right patient is challenged by diagnostic uncertainty, variation in treatment and safety response due to suboptimal generalizability of clinical trial results, complexity in risk stratification, and limited integration of information at the point of care. ML can play an important role in bridging these gaps in HF and has important advantages over traditional human-derived models.
What is artificial intelligence and machine learning?
Artificial intelligence (AI), the imitation of human cognition by technology, can be used to guide clinical care and decision-making without human involvement in the process. One sub-field of AI is ML, which provides computers with the capacity to evaluate data beyond programmatic algorithms, identifying patterns within data, mapping learned patterns to unseen data, and improving the performance of computational tasks beyond human capaibilities (Box 1).7
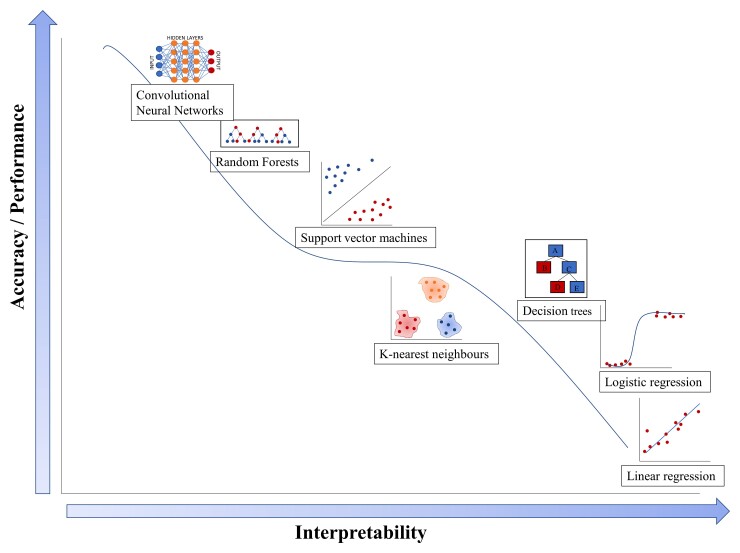
ML and traditional statistical approaches have several unique as well as overlapping capabilities (Table 1). ML methods are well-equipped to handle high-dimensional datasets with a very large number of variables that make traditional statistical approaches such as regression challenging. ML can also handle correlated or collinear data points and assess complex interactions between predictors. ML does not typically isolate the ‘effect’ of a single variable and does not require that predictor variables be selected a priori10 (Table 1). ML can also generate dynamic models, where the training data are continuously updated to account for temporal changes in data. For example, ‘baseline’ characteristics such as haemodynamics, laboratory values, and comorbidities may evolve during a study. The evolution of these characteristics may be important in predicting outcomes, but traditional statistical approaches are often not equipped to handle them. ML algorithms allow for higher performing, accurate computation of non-linear relationships, but the higher accuracy comes at the cost of interpretability13 (Figure 1).
Table 1.
Differences between traditional statistical models and machine learning algorithms
| Method | Traditional regression models | Machine learning |
|---|---|---|
| Assumptions | ||
| ȃIndependence | Predictors are assumed to be independent of each other. | Predictors do not need to be independent. |
| ȃMulticollinearity | No multicollinearity—predictors should not correlate with each other. | Multicollinearity allowed. |
| Predictors | ||
| ȃSelection of predictors | Prespecified. | Does not have to be prespecified. |
| Data structure | ||
| ȃReasoning | Inductive—derives a rule for the relationship between the input and the outcome.11 | Transductive—can predict outcomes using inputs from the training set without deriving a general rule.11 |
| ȃDimensionality | Performs well with low signal-to-noise ratio, but poorly with high-dimensional data. | Performs well with high-dimensional data with high signal-to-noise ratio.12 |
| ȃSample size | Smaller sample size, fewer events required per predictor. | Larger sample size, more events required per predictor. |
| Performance | ||
| ȃInteractions | Can test for a limited number of prespecified interactions.12 | Can handle large number of non-prespecified interactions.12 |
| ȃEffect size | The effect of individual predictors is of interest. | The effect of individual predictors is not of interest, prediction is prioritized. |
| ȃPerformance | Lower accuracy. | Higher accuracy, particularly for non-linear, non-smooth relationships. |
| ȃInterpretability | Models are easier to interpret and explain. | Models are more challenging to interpret, can be a ‘black box’. |
| ȃDichotomization | Calibration poor with dichotomized predictor and outcome variables. | Better calibration with dichotomous predictor and outcome variables. |
Figure 1.
Comparison of the accuracy and interpretability of statistical vs. machine learning models. Traditional regression models demonstrate poor accuracy but are easier to interpret. Machine learning models with non-linear relationships offer superior accuracy but are harder to interpret. Adapted from Stewart 2020.14
ML can be supervised or unsupervised (see Supplementary material online, Figure S1). Supervised ML uses human-labelled data to learn the underlying patterns in a process called ‘model training’. Data labelling of outcomes—for example, ‘hospitalized’ or ‘not hospitalized’—requires human input. The algorithms then learn the relationships between variables and outcomes. As new input data is fed into the model, weights are adjusted until the model has been fitted appropriately. Using this method, a model can be trained to predict events (e.g. hospitalization) in new datasets.
In contrast to supervised learning, unsupervised ML algorithms are exploratory and discover patterns without human-labelled data.10 By recognizing clinically relevant patterns or phenotypes that may not be evident to the clinician, ML can unearth disease mechanisms and improve the accuracy of diagnosis, management, and risk prediction in cardiovascular medicine.10
Deep learning (DL) is a subset of ML that uses multiple layers of artificial neural networks to identify patterns or make predictions of patterns (see Supplementary material online, Table S1).9 There is a hierarchy in the arrangement of layers, from learning simple representations of data to more complex relationships as the data is passed through deeper layers. DL is particularly useful with big data, as it does not require variable selection or rely as much on feature engineering to learn from big data sources such as EHRs.15 For example, DL models can predict incident HF by examining temporal relations amongst a large number of evolving variables (i.e., comorbidities, physiologic measures, laboratory indices, medication prescriptions, invasive procedures).16
Machine learning applications in HF
Prediction of incident HF
ML algorithms can identify risk factors for incident HF. In a prospective cohort of over 500 000 individuals in the United Kingdom, a supervised ML model confirmed leg bioimpedance as a major risk factor in addition to known risk factors for HF; lower leg bioimpedance values were associated with HF incidence during the 9.8-year follow-up.17 The resulting ML model, comprising leg bioimpedance, age, sex, and self-reported myocardial infarction provided a highly accurate prediction of incident HF without the variables being prespecified. This demonstrates that ML can identify novel HF risk factors for HF that may not otherwise be considered. A further application of ML algorithms may be the prediction of disease in populations that are not represented in registries or trials in which traditional clinical prediction models were derived or validated; utilizing a large set of unspecified variables to predict disease instead of limiting variables to those generated from homogenous research populations may mitigate historical structural biases and research inequities.18,19
In a prospective study of patients enrolled in the Action to Control Cardiovascular Risk in Diabetes trial, a risk score for 5-year HF incidence was created using ML techniques.20 The supervised ML model demonstrated better discrimination than a traditional Cox-proportional hazards model in a predicting incident HF within the cohort in the 4.9 years of study follow-up.20 Models that predict HF tend to treat race or ethnicity as a covariate, rather than developing race-specific models, which may be more appropriate due to variations in risk factors across racial or ethnic groups.21 In a retrospective pooled analysis of cohort studies, ML was used to develop a race-specific model to predict HF incidence,21 and it outperformed a traditional, non-race specific model.21
Diagnosis of HF
ML algorithms could assist physicians in early diagnosis of HF in at-risk patients. An electrocardiogram (ECG) is a non-invasive, widely available tool that can be used for early diagnosis of HF.22 A DL algorithm for ECG-based HF identification (DEHF) was developed and validated for this purpose,22 using data including demographic information and ECG features from EHRs.22 The DEHF algorithm was superior in detecting HF with reduced ejection fraction (HFrEF) (C-statistic 0.843, 95% CI 0.840–0.845) compared to logistic regression (C-statistic 0.800, 95% CI 0.797–0.803) and random forest ML algorithms (C-statistic 0.807, 95% CI 0.804–0.810).22
AI when combined with expert knowledge may be superior than AI alone. An AI-Clinical Decision Support System that was developed using expert knowledge combined with a ML approach for the diagnosis of HF with reduced, mildly reduced, and preserved ejection fraction improved diagnostic accuracy over an expert-driven or ML approach alone.23 The variables in the ML model included left ventricular ejection fraction, left atrial volume index, left ventricular mass index, ECG features, clinical features and physical exam features.
Earlier recognition and improved diagnostic accuracy of HF may allow for more timely investigations for the underlying aetiology and earlier initiation of guideline-indicated therapy to delay disease progression. The use of ML as a diagnostic aide in HF is a nascent field, however. In the absence of external validation and prospective testing of interventions based on ML outputs, the clinical impact remains to be realized.
Classification of HF phenotypes
ML may improve the current classification of HF. Compared to HFrEF, the underlying phenotypic heterogeneity is more complex in HF with preserved ejection fraction (HFpEF).24–26 A prospective study of 397 ambulatory patients with HFpEF performed phenotype mapping using ML algorithms with data from EHRs.25 This technique resulted in a novel classification method for HFpEF, which clusters study participants into phenotypes according to clinical characteristics, ECG and echocardiographic parameters, invasive haemodynamics, and outcomes (Table 2).25 These findings are important, as the improved classification of HFpEF may facilitate recruitment of patients most likely to benefit from a given intervention in randomized trials.25
Table 2.
Heart failure phenotypes identified in machine learning models
| Study and population | Group 1 | Group 2 | Group 3 | Group 4 | Group 5 | Group 6 | |
|---|---|---|---|---|---|---|---|
| HF phenogroups and clinical outcomes | |||||||
| ȃShah et al. 201425 ȃHFpEF |
Characteristics | Younger, least electrical and myocardial remodelling, lower BNP. | Higher prevalence of obesity, diabetes, OSA with worst LV relaxation, highest PCWP, highest PVR. | Oldest, with worst electrical (wide QRS) and myocardial remodelling (thickest LV, worst RV function), more CKD, highest BNP. | — | — | — |
| Outcomes | Lower risk of CV death, HF hospitalization, or all-cause death. | Intermediate risk of CV death, HF hospitalization, or all-cause death. | Highest risk of CV death, HF hospitalization, or all-cause death. | — | — | — | |
| ȃGevaert et al. 202127 ȃHFrEF, HFmEF, HfpEF |
Characteristics | Coronary artery disease as the predominant comorbidity. | Valvular heart disease as the predominant comorbidity. | Fewest comorbidities (hypertension and diabetes). | Atrial fibrillation as the predominant comorbidity. | COPD as the predominant comorbidity. | OSA as the predominant comorbidity. |
| Outcomes | Fourth highest risk of composite all-cause death or hospitalization at 12 months. | Second highest risk of composite all-cause death or hospitalization at 12 months. | Lowest risk of composite all-cause death or hospitalization at 12 months. | Fifth highest risk of composite all-cause death or hospitalization at 12 months. | Highest risk of composite all-cause death or hospitalization at 12 months | Third highest risk of composite all-cause death or hospitalization at 12 month. | |
| ȃBose et al. 201828 ȃHFrEF, HFmEF, HFpEF patients enrolled in a home telehealth programme |
Characteristics | Younger patients, with the lowest proportion of CAD, greatest number of comorbidities, highest number of prescribed medications. | Oldest patients, primary women, lowest proportion of CKD, diabetes, fewest comorbidities. | Older patients, highest proportion with CAD, CKD, lowest number of prescribed medications. | — | — | — |
| Outcomes | Longest home health length of stay, most hospitalizations. | Lower than average home health length of stay, similar hospitalizations to overall cohort. | Shortest home health length of stay, fewest hospitalizations. | — | — | — | |
| HF phenogroups and response to device therapy | |||||||
| ȃCikes et al. 2019.29 ȃHFrEF patients with CRT-D |
Characteristics | Non-ischaemic cardiomyopathy, LBBB, longest QRS, youngest, high proportion female, most remodelled LA, LV, RV, lowest LVEF. | Ischaemic cardiomyopathy, low proportion of LBBB, largest proportion male, hypertension, diabetes, least remodelled LA, LV, high LVEF. | Non-ischaemic cardiomyopathy, LBBB, highest proportion female, less remodelled LA, LV, high LVEF | Ischaemic cardiomyopathy, low proportion LBBB, high proportion male, HTN, diabetes, remodelled LV, RV, low LVEF, extensive apical scar. | — | — |
| Outcomes | CRT-D responders. | CRT-D non-responders. | CRT-D responders. | CRT-D non-responders. | — | — | |
| HF phenogroups and response to pharmacotherapy | |||||||
| ȃAhmad et al. 201830 ȃHFrEF, HFmEF, HFpEF |
Characteristics | Oldest, largest proportion female, largest proportion HFpEF, highest BP, highest proportion non-ischaemic, highest BNP, lowest use of BB and ACEi. | Younger age, mainly male, highest BMI, greatest prevalence of diabetes, hypertension, dyslipidaemia, high proportion of HFrEF, ischaemic cardiomyopathy, lowest BNP. | Older age, lower prevalence of HFpEF, lowest BMI, most comorbidities including OSA, CKD, atrial fibrillation, COPD, high proportion of ischaemic cardiomyopathy, highest NT-proBNP, low use of BB, ACEi. | Youngest, mainly male, high BMI, highest proportion of HFrEF, highest proportion of ischaemic cardiomyopathy, fewest comorbidities, low BNP. | — | — |
| Outcomes | Derived the least benefit from BB. Moderate 1-year survival. | Derived the greatest benefit from ARB. Highest 1-year survival. | Derived the greatest benefit from BB, benefited from nitrate therapy. Lowest 1-year survival. | Derived the greatest benefit from BB, worst outcomes among patients on diuretics, worst outcomes on nitrates. High 1-year survival. | — | — | |
ACEi, ACE inhibitor; BB, beta-blocker; BNP, brain-derived natriuretic peptide; CAD, coronary artery disease; CKD, chronic kidney disease; COPD, chronic obstructive pulmonary disease; CRT-D, cardiac resynchronization therapy-defibrillator; CV, cardiovascular; HFmEF, heart failure with mid-range ejection fraction; HFpEF, heart failure with preserved ejection fraction; HFrEF, heart failure with reduced ejection fraction; LA, left atrium; LV, left ventricle; LVEF, left ventricular ejection fraction; NT-proBNP, N-terminal prohormone of brain-derived natriuretic hormone; OSA, obstructive sleep apnoea; PCWP, pulmonary capillary wedge pressure; PVR, pulmonary vascular resistance; RV, right ventricle. Cells marked with a "—" indicate that there was a limited number of phenogroups (<6).
Another unsupervised ML analysis of 1693 patients hospitalized with HF across the left vebtricular ejection fraction (LVEF) spectrum revealed 6 discrete phenogroups based on common comorbidities: coronary artery disease, valvular heart disease, atrial fibrillation, chronic obstructive pulmonary disease (COPD), obstructive sleep apnoea (OSA), or few comorbidities (Table 2).27 Phenogroups were LVEF-independent, with each group encompassing a wide range of LVEF. The groups stratified risk of composite all-cause death or hospitalization as well as a composite cardiovascular (CV) death or HF hospitalization at 6 and 12 months post-discharge more effectively than LVEF.27
Similarly, unsupervised ML can be used to establish clinical phenogroups with predictive values based on transcriptomic or metabolomic profiles.31 Such phenogroups or subgroups may have differential response to therapies,32 but this needs to be proven in prospective studies.
Prediction of outcomes following HF diagnosis
Existing HF prediction models are underused among cardiologists due to their complexity, lack of integration with work flow, and limited knowledge on how risk prediction can be used to improve outcomes.33 Risk prediction models aim to identify patients who are at risk of adverse events1 or who may benefit from closer follow-up and post-discharge services. From a systems level, risk stratification is important in light of the readmission penalties imposed by the Medicare Hospital Readmissions Reduction Programme.34 However, accurate risk prediction remains an unmet need which may be met through ML. The DL algorithm for predicting mortality of patients with Acute HF (DAHF), is a risk stratification model for predicting in-hospital and long-term mortality. Evidence from a large retrospective cohort study in Korea demonstrated the ability of DAHF to outperform mortality risk prediction models for HF such as Get with the Guidelines-Heart Failure Score (GWTG) and Meta-Analysis Global Group for Heart Failure (MAGGIC).35 The DAHF predicted in-hospital through to 36-month mortality with greater discrimination than the GWTG risk score for in-hospital mortality (C-statistic 0.880; 95% CI 0.876–0.884 vs. 0.728; 95% CI 0.720–0.737) and the MAGGIC risk score for 36-month mortality (C-statistic 0.813; 95% CI 0.810–0.816 vs. 0.729; 95% CI 0.726–0.733).35 This may be because DL algorithms do not limit the number of input predictive factors, preventing the unintended loss of data that comes from restricting analyses to known associations.35
Data from cardiac monitoring, either external or implantable, can be used in real time to develop algorithms for risk prediction. The Multisensor Non-invasive Remote Monitoring for Prediction of Heart Failure Exacerbation (LINK-HF) study examined the `accuracy of a remote monitoring system using a ML algorithm to predict hospitalization (unplanned non-trauma hospitalization).36 The platform was able to predict precursors to hospitalization with a median alert time of 6.5 days in advance of the readmission.36 Implantable haemodynamic monitoring systems of pulmonary artery pressures (PAP) have demonstrated conflicting effects on clinical endpoints in the CardioMEMS Heart Sensor Allows Monitoring of Pressures to Improve Outcomes in New York Heart Association (NYHA) Functional Class III Heart Failure Patients (CHAMPION) and the Haemodynamic-guided Management of Heart Failure (GUIDE-HF) trials.37,38 Unlike the CHAMPION trial in which implantable haemodynamic-guided HF therapy decreased the rate of HF hospitalizations, PAP monitoring did not reduce the primary composite endpoint or component endpoints of all-cause mortality or HF hospitalization in the GUIDE-HF trial.38 In a sensitivity analysis of GUIDE-HF, a significant treatment effect was observed with PAP-guided HF therapy prior to, but not during, the COVID-19 pandemic.38 Event rates decreased during the COVID-19 pandemic and there was no longer a difference in guideline-directed medical therapy (GDMT) changes in the treatment relative to control groups during the pandemic; this, along with changes in patient behaviour (e.g. improved medication adherence, better nutrition, and hospital avoidance) may have attenuated the estimated treatment effect of PAP-guided care during the pandemic in GUIDE-HF.38 While the breadth of evidence does not favour the use of PAP monitoring overall, ML algorithms with haemodynamic and EHR data from PAP-monitored patients may help determine which patients benefit the most from this intervention.
The HeartLogic Index™, a proprietary algorithm derived from Boston Scientific Cardiac resynchronization-defibrillator (CRT-D) device data, is an example of applied ML.39 The algorithm—using heart sounds, thoracic impedance, respiratory rate, tidal volume, heart rate and patient activity—delivers an alert when a certain threshold is reached. This index was demonstrated to predict HF events (hospitalization or need for IV diuretics)39 and can potentially also be used to detect subclinical decompensation.40 At a cut-point of 16, the HeartLogic Index provides clinicians with an alert signaling an increased risk of HF hospitalization (modest performance with sensitivity 70% and positive predictive value 11.3%). As in the case of non-ML models, there is a paucity of randomized clinical trial evidence on the effect of care pathways guided by these alerts. Thus, the impact of the HeartLogic Index on clinical outcomes has yet to be established.
Optimization of medical and device therapy
Medical therapy
A majority of patients do not receive target doses of evidence-based medical therapies in HF, possibly due to under-prescribing by clinicians, barriers to access, or intolerance to medications.41 ML algorithms have been used to improve HF medical management by assessing for heterogeneity in response to HF therapies. For example, ML methods were applied to EHRs of 44 886 patients in the Swedish HF Registry assess for heterogeneity in response to HF pharmacotherapy across propensity-matched clusters.30 Four clusters—based on demographic, NYHA class, LVEF, comorbidities and lab indices—were identified with marked differences in 1-year survival and response to therapies (Table 2).30 Thus, ML may be used to better classify HF patients into high- and low-risk subgroups and identify those most likely to derive benefit with the least side effects in GDMT.
ML can potentially be used to optimize GDMT prescription in HF and identify patients at risk for adverse drug reactions. By extracting data from EHRs, ML algorithms could be used to provide recommendations to clinicians regarding optimal sequencing and dosing of evidence-based therapies.42 This approach could help reach a wider range of patients who may not otherwise have access to multidisciplinary HF clinics that are often concentrated in urban centres.
Device therapy—patient selection and care
ML may also be used to optimize patient selection for device therapy in HF. Depending on the definition used, up to 40% of HF patients are non-responders to cardiac resynchronization therapy (CRT)43–45 A post-hoc analysis of the Multicentre Automatic Defibrillator Implantation Trial with CRT trial assessed whether an ML algorithm could accurately identify patients who respond to CRT.29 Input for the unsupervised ML algorithm included 50 variables selected at baseline such as demographic and laboratory data, ECG and echocardiography measurements, and data on medication use and recruitment centre.29 The ML algorithm identified four phenogroups of HF patients, including patients most likely to respond to CRT-D vs. implantable-cardioverter defibrillators (ICDs) (Table 2).29 Another ML model using device data from the Comparison of Medical Therapy, Pacing, and Defibrillation in Heart Failure trial46 improved the ability to discriminate outcomes after CRT relative to the referral criteria of bundle branch block (BBB) morphology and QRS duration46 The ML model produced more precise results, indicating an eight-fold difference in survival between those with the highest and lowest predicted probability for death.46 These examples highlight that ML models may improve patient selection for CRT by highlighting patients who are most likely to derive benefit.46
ICDs reduce the risk of sudden cardiac death (SCD) in patients with left ventricular dysfunction, and 3–5% of HF patients with primary prophylactic ICDs receive shocks with ensuing psychological distress.47 In a post-hoc analysis of the SCD-Heart Failure Trial, an ML model used heart rate variability data to predict ventricular tachycardia (VT) with good discrimination, creating a 10-second and 5-minute warning system.48 Such a warning system could potentially avert injury by giving patients the opportunity to pull over to park if driving or to find a place to sit or recline if walking. With more advance notice, it could potentially also allow for medical optimization to avert VT or change anti-tachycardia pacing settings to minimize ICD shocks.
The possible applications of ML in selecting patients for medical and device therapies and optimizing care require prospective testing in a randomized controlled trial (RCT) prior to clinical uptake.
ML and clinical trial design
Clinical trials that test cardiovascular interventions are limited by sub-optimal patient selection, under-enrolment of certain demographic groups, non-adherence49 to the intervention, and study withdrawals, but ML can address some of these. A significant barrier to trial success is identifying a study population that has high enough event rates to potentially demonstrate treatment effect. Risk enrichment strategies are commonly used to include higher risk patients,50 but this is limited by the human selection of inclusion criteria. ML can be used to identify patient phenotypes most likely to respond to a given treatment based on prior clinical trials as well as to identify predictors of clinical endpoints of interest. Once the optimal profile has been described for inclusion, the next challenge is in identifying suitable patients. This is difficult for both patients and clinicians who may not be aware of available clinical trials, or if the inclusion and exclusion criteria, which may be numerous, have been met. DL, a form of ML, can be used to connect eligible patients to ongoing trials through the use of natural language processing (NLP). NLP is a powerful tool for automated text interpretation that could be used to analyze clinical trial databases and EMRs to identify eligible patients based on phenotypes and to connect them to the right trials.51
The average proportion of patients who drop out of clinical trials is up to 30%.52 Identifying which patients are at risk of non-adherence or dropping out of a trial is another potential application of ML, especially in an era of wearable and mobile devices. In one study, a smartphone-based platform was used to record patients consuming their medication while DL for image recognition flagged non-adherencen.53 The ML algorithm was then able to predict future non-adherence, which could be used to screen participants in a trial run-in phase. Similar approaches can be used to predict and mitigate trial drop-outs and losses to follow-up.
Additionally, ML may be useful in clinical trial design, particularly in adaptive trials where trial procedures such as treatment allocation can be modified after trial initiation, allowing for trials to be conducted with fewer patients and resources.54 Another common application of adaptive design is in dose-finding, such that the dose can be modified over the trial duration as information becomes available on the toxicity of each dose.55 The trade-off between the changes required to find the optimal dose and treating as many patients with the optimal dose55 in a single trial can be managed by ML methods such as the multi-armed bandit model.55
Limitations of ML and areas for improvement
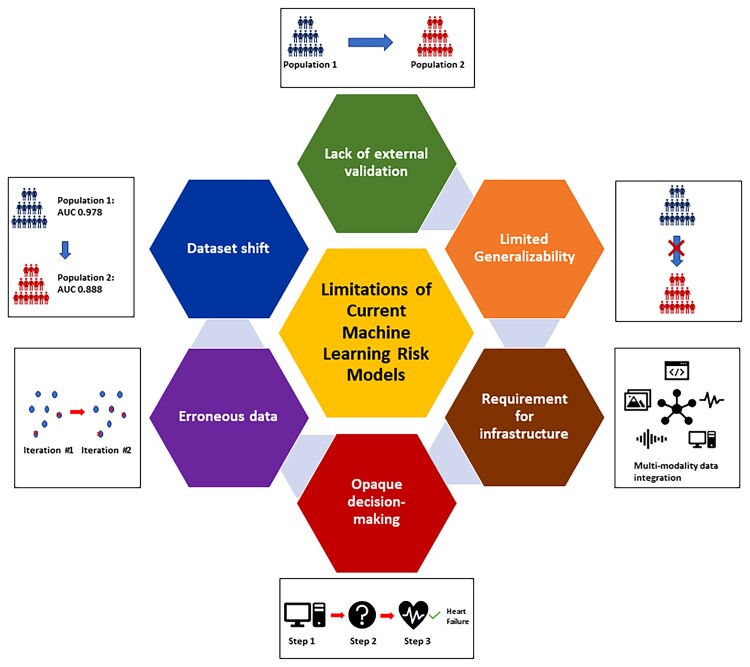
Despite the great potential of AI applications in medicine, there are several barriers to their uptake in HF management (Figure 2). Recent guidelines such as Consolidated Standards of Reporting Trials - Artificial Intelligence (CONSORT-AI), Standard Protocol Items: Recommendations for Interventional Trials - Artificial Intelligence (SPIRIT-AI), and Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis - Machine Learning (TRIPOD-ML) offer recommendations for AI model developers with a view to enhancing adaptability, scalability, and interpretability.56 Robustness and generalization error plague all insights or predictions of computational models but, when deployed in safety-critical settings, the lack of confidence intervals on a model’s output can be particularly problematic; research into generating accurate confidence intervals for prediction is ongoing.49
Figure 2.
Limitations of machine learning models. Machine learning models face several limitations including lack of external validation, limited generalizability, opaque decision-making, logistical challenges in implementation due to reliance on digital infrastructure, error propagation between iterations, and dataset shift. The effectiveness of machine learning models in improving outcomes at the point of care needs to be tested prospectively in randomized controlled trials.
The opaque logic and lack of explainability behind why a ML model outputs a particular prediction is a persistent limitation in the uptake of ML algorithms at the point of care; if clinicians do not understand the process that resulted in a recommendation, then they are more likely to ignore it. Owing to the large number of variables in the datasets in which ML is employed, algorithms may incorporate variables and interaction terms that are statistically significant but not clinically relevant. A recent example of these limitations arose during the COVID-19 pandemic; most ML models to predict pneumonia or COVID-19 in chest x-rays (CXR) relied on meaningless features within the CXR, suggesting that these models would fail when used in contexts different than that of training (e.g. in a different hospital or location).50
The performance of an ML algorithm is only as reliable as the data used to derive it, and errorneous or missing training data can degrade model performance over time.57 This propagation of errors is particularly problematic when ML algorithms are dynamic and rely on continuous data inputs such as EHRs. Data used to develop ML algorithms must be cleaned and validated for reliable predictions, with detection of out-of-range values and identification of skew.57 This process is computationally demanding and challenging to automate, but essential for ML to provide clinicians high-quality, accurate predictions or classifications. Missing data can also adversely affect model performance; however, ML algorithms and techniques can be used to impute missing data, and preserve algorithm performance.58
Clinical AI applications must adequately address dataset shift, a phenomenon of degrading performance when the training data for a model differs from the data used to provide clinical advice.59 This can occur through covariate shift, where the distribution of covariates differs between training and test datasets; prior probability shift, where the distribution of outcomes differs between datasets; and concept drift, where the relationship between the covariates and the outcome differs over time. While the degradation of generalizability due to dataset shift can be mitigated through appropriate sample selection, feature selection, and re-weighting, it remains an important limitation to the performance of ML models in validation cohorts.
One of the largest barriers to uptake of ML algorithms is the lack of assessment of their clinical impact in prospective studies such that the benefits of ML approaches remain theoretical. Although there are validated models for the early diagnosis of HF,22 current studies focus on the performance of each model and there are no studies of the economic or health systems benefits of population-level screening for HF using ML algorithms. Similarly, the effect of predictive models in guiding pharmacotherapy and device interventions has not been evaluated in randomized controlled trials.32,60 As a result, it is not known whether ML-driven risk stratification and patient selection for interventions leads to improved clinical and patient-reported outcomes, although this is a limitation of both traditional statistical and ML approaches.
Logistic difficulties are important barriers to the use of ML in HF, as most healthcare data are not readily available in a form suitable for ML applications.61 Programmes are needed to organize and aggregate the data from EHRs to allow for application of ML algorithms at the point of care.61
Conclusion
The applications of ML in HF are expanding. ML algorithms can be applied in HF diagnosis, classification, and prognosis (Box 2). The potential of ML to select patients for medical and device therapies needs to be harnessed through prospective testing and validation in clinical studies. As ML tools become more widely available, validated, and implemented in clinical practice, these novel algorithms will positively influence HF care and outcomes.
Lead author biography
 Dr. Harriette Van Spall is a cardiologist and scientist at McMaster University with appointments in the Department of Medicine and the Department of Health Research Methods, Evidence, and Impact. Dr. Van Spall earned her Doctor of Medicine degree and postgraduate fellowships at the University of Toronto. She then earned a Master of Public Health degree at Harvard University. Funded by the Canadian Institutes of Health Research, she leads clinical trials and big data research related to Heart Failure.
Dr. Harriette Van Spall is a cardiologist and scientist at McMaster University with appointments in the Department of Medicine and the Department of Health Research Methods, Evidence, and Impact. Dr. Van Spall earned her Doctor of Medicine degree and postgraduate fellowships at the University of Toronto. She then earned a Master of Public Health degree at Harvard University. Funded by the Canadian Institutes of Health Research, she leads clinical trials and big data research related to Heart Failure.
Supplementary Material
Contributor Information
Tauben Averbuch, Department of Medicine, McMaster University, Hamilton, Ontario, Canada.
Kristen Sullivan, Department of Medicine, McMaster University, Hamilton, Ontario, Canada.
Andrew Sauer, Department of Cardiology, University of Kansas Health System, Kansas City, KS, USA.
Mamas A Mamas, Keele Cardiovascular research group, Keele University, Stoke on Trent, Staffordshire.
Adriaan A Voors, University of Groningen, Groningen, The Netherlands.
Chris P Gale, Department of Cardiology, University of Leeds, Leeds, West Yorkshire.
Marco Metra, Azienda Socio Sanitaria Territoriale Spedali Civili and University of Brescia, Brescia, Italy.
Neal Ravindra, Department of Computer Science, Yale University, New Haven, CT, USA.
Harriette G C Van Spall, Department of Medicine, McMaster University, Hamilton, Ontario, Canada; Population Health Research Institute, Hamilton, Ontario, Canada; Department of Health Research Methods, Evidence, and Impact, McMaster University, Hamilton, Ontario, Canada.
Supplementary material
Supplementary material is available at European Heart Journal – Digital Health.
Authors’ contributions
H.G.C.V. conceived the study idea. T.A., K.S., N.R., and H.G.C.V. drafted the manuscript. All authors edited the manuscript. H.G.C.V. assumes responsibility for the scientific integrity of this work. All involved authors approved the final article.
Box 1. Glossary of machine learning terms.
Artificial neural networks: A non-linear machine learning algorithm that is modelled after human neurons, such that it is able to learn from data and provide responses in the form of predictions or classifications.8
ML: Uses computer algorithms to extract patterns from raw data, acquire knowledge without human input, and use this for various tasks.9
Supervised ML: Uses labelled datasets with human input to predict outcomes or classify observations.9,10
Unsupervised ML: Uses datasets to identify hidden patterns.9
Deep neural network: A subset of ML that uses many layers of artificial neural networks to make predictions from data sets.9
ML, machine learning.
Box 2. Take home points.
ML offers important advantages over typical methods: it is able to handle larger data sets, datasets that evolve over time, and collinearity.
ML can be used to identify patterns in data, such as classification of phenogroups (unsupervised ML), or to predict outcomes (supervised ML).
ML may improve early diagnosis of HF, classification of HF, device selection, medication titration, risk prognostication, and clinical trial enrolment and retention
ML is limited by the lack of transparency in how models make decisions, lack of prospective validation of models, and degrading performance over time.
ML is a rapidly growing field that may improve HF care; however, further work on validation and interpretability is needed before it is incorporated in routine care.
HF, heart failure; ML, machine learning.
Funding
H.G.C.V. receives grant support from the Canadian Institutes of Health Research and Heart and Stroke Foundation of Canada. No other sources of funding were required for this review.
Conflict of interest: None declared.
Data availability
No new data were generated or analyzed in support of this research.
References
- 1. Averbuch T, Lee SF, Mamas MA, Oz UE, Perez R, Connolly SJ, Ko DT-, Van Spall HGC. Derivation and validation of a two-variable index to predict 30-day outcomes following heart failure hospitalization. ESC Heart Fail. Published online May 1, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Van Spall HG, Averbuch T, Lee SF, Oz UE, Mamas MA, Januzzi JL, Ko DT. The LENT index predicts 30 day outcomes following hospitalization for heart failure. ESC Heart Fail 2021;8:518–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cook JA, Collins GS. The rise of big clinical databases. Br J Surg 2015;102:e93–e101. [DOI] [PubMed] [Google Scholar]
- 4. Docherty AB, Lone NI. Exploiting big data for critical care research. Curr Opin Crit Care 2015;21:467–472. [DOI] [PubMed] [Google Scholar]
- 5. Choi RY, Coyner AS, Kalpathy-Cramer J, Chiang MF, Campbell JP. Introduction to machine learning, neural networks, and deep learning. Transl Vis Sci Technol 2020;9:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Demchenko Y, Grosso P, de Laat C, Membrey P. Addressing big data issues in scientific data infrastructure. In: 2013 International Conference on Collaboration Technologies and Systems (CTS). IEEE 2013:48–55. [Google Scholar]
- 7. Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med 2019;380:1347–1358. [DOI] [PubMed] [Google Scholar]
- 8. Artificial Neural Networks for Machine Learning - Every aspect you need to know about. DataFlair. Published July 15, 2017. Accessed December 28, 2020. https://data-flair.training/blogs/artificial-neural-networks-for-machine-learning/
- 9. Noorbakhsh-Sabet N, Zand R, Zhang Y, Abedi V. Artificial intelligence transforms the future of health care. Am J Med 2019;132:795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson KW, Torres Soto J, Glicksberg BS, Shameer K, Miotto R, Ali M, Ashley E, Dudley JT. Artificial intelligence in cardiology. J Am Coll Cardiol 2018;71:2668–2679. [DOI] [PubMed] [Google Scholar]
- 11. Kukar M, Grošelj C. Transductive machine learning for reliable medical diagnostics. J Med Syst 2005;29:13–32. [DOI] [PubMed] [Google Scholar]
- 12. Levy JJ, O’Malley AJ. Don’t dismiss logistic regression: the case for sensible extraction of interactions in the era of machine learning. BMC Med Res Methodol 2020;20:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weller DL, Love TMT, Wiedmann M. Interpretability versus accuracy: a comparison of machine learning models built using different algorithms, performance measures, and features to predict E. coli levels in agricultural water. Front Artif Intell 2021;4:628441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stewart M. Guide to Interpretable Machine Learning. Towards data science. Published March 19, 2020. Accessed September 1, 2021. https://towardsdatascience.com/guide-to-interpretable-machine-learning-d40e8a64b6cf
- 15. Goodfellow I, Bengio Y, Courville A. Deep learning: The MIT Press; 2016. [Google Scholar]
- 16. Rao S, Li Y, Ramakrishnan R, Canoy D, Cleland J, Lukasiewicz T, Salimi-Khorshidi G, Rahimi K. An explainable transformer-based deep learning model for the prediction of incident heart failure. arXiv:210111359 [cs]. Published online January 27, 2021. Accessed September 2, 2021. http://arxiv.org/abs/2101.11359 [DOI] [PubMed] [Google Scholar]
- 17. Lindholm D, Fukaya E, Leeper NJ, Ingelsson E. Bioimpedance and new-onset heart failure: a longitudinal study of >500 000 individuals from the general population. J Am Heart Assoc 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu JW, Le N, Wei S, Zühlke L, Lopes R, Zannad F, Van Spall HGC. Global representation of heart failure clinical trial leaders and collaborators: a systematic bibliometric review 2000-2020. SSRN J. Published online 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moledina SM, Kontopantelis E, Wijeysundera HC, Banerjee S, Van Spall HGC, Gale CP, Shah BN, Mohamed MO, Weston C, Shoaib A, Mamas MA. Ethnicity-dependent performance of the global registry of acute coronary events risk score for prediction of non-ST-segment elevation myocardial infarction in-hospital mortality: nationwide cohort study. Eur Heart J. Published online February 2022;24:ehac052. [DOI] [PubMed] [Google Scholar]
- 20. Segar MW, Vaduganathan M, Patel KV, Butler J, Fonarow GC, Basit M, Kannan V, Grodin JL, Everett B, Willett D, Berry J, Pandey A. Machine learning to predict the risk of incident heart failure hospitalization among patients with diabetes: the WATCH-DM risk score. Diabetes Care 2019;42:2298–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Segar MW, Jaeger BC, Patel KV, Nambi Vijay, Ndumele CE, Correa A, Butler J, Chandra A, Ayers C, Rao S, Lewis AA, Raffield LM, Rodriguez CJ, Michos ED, Ballantyne CM, Hall ME, Mentz RJ, de Lemos JA, Pandey A. Development and validation of machine learning–based race-specific models to predict 10-year risk of heart failure: a multicohort analysis. Circulation 2021;143:2370–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kwon JM, Kim KH, Jeon KH, Kim HM, Kim MJ, Lim S-M, Song PS, Park J, Choi RK, Oh B-H. Development and validation of deep-learning algorithm for electrocardiography-based heart failure identification. Korean Circ J 2019;49:629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Choi DJ, Park JJ, Ali T, Lee S. Artificial intelligence for the diagnosis of heart failure. npj Digit Med 2020;3:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gevaert AB, Kataria R, Zannad F, Sauer AJ, Damman K, Sharma K, Shah SJ, Van Spall HGC. Heart failure with preserved ejection fraction: recent concepts in diagnosis, mechanisms and management. Heart. Published online January 12, 2022:heartjnl-2021-319605. [DOI] [PubMed] [Google Scholar]
- 25. Shah SJ, Katz DH, Selvaraj S, Burke MA, Yancy CW, Gheorghiade M, Bonow RO, Huang C-C, Deo RC. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation 2015;131:269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Segar MW, Patel KV, Ayers C, Basit M, Tang WHW, Willett D, Berry J, Grodin JL, Pandey A. Phenomapping of patients with heart failure with preserved ejection fraction using machine learning-based unsupervised cluster analysis. Eur J Heart Fail 2020;22:148–158. [DOI] [PubMed] [Google Scholar]
- 27. Gevaert AB, Tibebu S, Mamas MA, Ravindra NG, Lee SF, Ahmad T, Ko DT, Januzzi JL, Van Spall HGC. Clinical phenogroups are more effective than left ventricular ejection fraction categories in stratifying heart failure outcomes. ESC Heart Fail 2021;8:2741–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bose E, Radhakrishnan K. Using unsupervised machine learning to identify subgroups among home health patients with heart failure using telehealth. Comput Inform Nurs 2018;36:242–248. [DOI] [PubMed] [Google Scholar]
- 29. Cikes M, Sanchez-Martinez S, Claggett B, Duchateau N, Piella G, Butakoff C, Pouleur AC, Knappe D, Biering-Sørensen T, Kutyifa V, Moss A, Stein K, Solomon SD, Bijnens B. Machine learning-based phenogrouping in heart failure to identify responders to cardiac resynchronization therapy. Eur J Heart Fail 2019;21:74–85. [DOI] [PubMed] [Google Scholar]
- 30. Ahmad T, Lund LH, Rao P, Ghosh R, Warier P, Vaccaro B, Dahlström U, O’Connor CM, Felker GM, Desai NR. Machine learning methods improve prognostication, identify clinically distinct phenotypes, and detect heterogeneity in response to therapy in a large cohort of heart failure patients. J Am Heart Assoc 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Woolley RJ, Ceelen D, Ouwerkerk W, Tromp J, Figarska SM, Anker SD, Dickstein K, Filippatos G, Zannad F, Metra M, Ng L, Samani N, Veldhuisen DJ, Lang C, Lam CS, Voors AA. Machine learning based on biomarker profiles identifies distinct subgroups of heart failure with preserved ejection fraction. Eur J Heart Fail 2021;23:983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tromp J, Ouwerkerk W, Demissei BG, Anker SD, Cleland JG, Dickstein K, Filippatos G, van der Harst P, Hillege HL, Lang CC, Metra M, Ng LL, Ponikowski P, Samani NJ, van Veldhuisen DJ, Zannad F, Zwinderman AH, Voors AA, van der Meer P. Novel endotypes in heart failure: effects on guideline-directed medical therapy. Eur Heart J 2018;39:4269–4276. [DOI] [PubMed] [Google Scholar]
- 33. Rahimi K, Bennett D, Conrad N, Williams TM, Basu J, Dwight J, Woodward M, Patel A, McMurray J, MacMahon S. Risk prediction in patients with heart failure: a systematic review and analysis. JACC Heart Fail 2014;2:440–446. [DOI] [PubMed] [Google Scholar]
- 34. Wadhera RK, Joynt Maddox KE, Wasfy JH, Haneuse S, Shen C, Yeh RW. Association of the hospital readmissions reduction program with mortality among medicare beneficiaries hospitalized for heart failure, acute myocardial infarction, and pneumonia. JAMA 2018;320:2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kwon JM, Kim KH, Jeon KH, Lee SE, Lee H-Y, Cho H-J, Choi JO, Jeon E-S, Kim M-S, Kim J-J, Hwang K-K, Chae SC, Baek SH, Kang S-M, Choi D-J, Yoo B-S, Kim KH, Park H-Y, Cho M-C, Oh B-H, Abete P. Artificial intelligence algorithm for predicting mortality of patients with acute heart failure. PLoS One 2019;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stehlik J, Schmalfuss C, Bozkurt B, Nativi-Nicolau J, Wohlfahrt P, Wegerich S, Rose K, Ray R, Schofield R, Deswal A, Sekaric J, Anand S, Richards D, Hanson H, Pipke M, Pham M. Continuous wearable monitoring analytics predict heart failure hospitalization. Circ Heart Fail 2020;13:e006513. [DOI] [PubMed] [Google Scholar]
- 37. Abraham WT, Stevenson LW, Bourge RC, Lindenfeld JA, Bauman JG, Adamson PB. Sustained efficacy of pulmonary artery pressure to guide adjustment of chronic heart failure therapy: complete follow-up results from the CHAMPION randomised trial. Lancet 2016;387:453–461. [DOI] [PubMed] [Google Scholar]
- 38. Lindenfeld J, Zile MR, Desai AS, Bhatt K, Ducharme A, Horstmanshof D, Krim SR, Maisel A, Mehra MR, Paul S, Sears SF, Sauer AJ, Smart F, Zughaib M, Castaneda P, Kelly J, Johnson N, Sood P, Ginn G, Henderson J, Adamson PB, Costanzo MR. Haemodynamic-guided management of heart failure (GUIDE-HF): a randomised controlled trial. Lancet 2021;398:991–1001. [DOI] [PubMed] [Google Scholar]
- 39. Boehmer JP, Hariharan R, Devecchi FG, Smith AL, Molon G, Capucci A, An Q, Averina V, Stolen CM, Thakur PH, Thompson JA, Wariar R, Zhang Y, Singh JP. A multisensor algorithm predicts heart failure events in patients with implanted devices: results from the MultiSENSE study. JACC Heart Fail 2017;5:216–225. [DOI] [PubMed] [Google Scholar]
- 40. Bachtiger P, Plymen CM, Pabari PA, Howard JP, Whinnett ZI, Opoku F, Janering S, Faisal AA, Francis DP, Peters NS. Artificial intelligence, data sensors and interconnectivity: future opportunities for heart failure. Card Fail Rev 2020;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peri-Okonny PA, Mi X, Khariton Y, Patel KK, Thomas L, Fonarow GC, Sharma PP, Duffy CI, Albert NM, Butler J, Hernandez AF, McCague K, Williams FB, DeVore AD, Patterson JH, Spertus JA. Target doses of heart failure medical therapy and blood pressure: insights from the CHAMP-HF registry. JACC Heart Fail 2019;7:350–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sullivan K, Mamas MA, Van Spall HGC. Machine learning could facilitate optimal titration of guideline-directed medical therapy in heart failure. J Am Coll Cardiol 2019;74:1424–1425. [DOI] [PubMed] [Google Scholar]
- 43. Daubert C, Behar N, Martins RP, Mabo P, Leclercq C. Avoiding non-responders to cardiac resynchronization therapy: a practical guide. Eur Heart J 2017;38:1463–1472. [DOI] [PubMed] [Google Scholar]
- 44. Hoogslag GE, Höke U, Thijssen J, Auger D, Marsan NA, Wolterbeek R, Holman ER, Schalij MJ, Bax JJ, Verwey HF, Delgado V. Clinical, echocardiographic, and neurohormonal response to cardiac resynchronization therapy: are they interchangeable? Pacing Clin Electrophysiol 2013;36:1391–1401. [DOI] [PubMed] [Google Scholar]
- 45. Delnoy PP, Ritter P, Naegele H, Orazi S, Szwed H, Zupan I, Goscinska-Bis K, Anselme F, Martino M, Padeletti L. Association between frequent cardiac resynchronization therapy optimization and long-term clinical response: a post hoc analysis of the clinical evaluation on advanced resynchronization (CLEAR) pilot study. Europace 2013;15:1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kalscheur MM, Kipp RT, Tattersall MC, Mei C, Buhr KA, DeMets DL, Field ME, Eckhardt LL, Page CD. Machine learning algorithm predicts cardiac resynchronization therapy outcomes: lessons from the COMPANION trial. Circ Arrhythm Electrophysiol 2018;11:e005499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stecker EC, Vickers C, Waltz J, Socoteanu C, John BT, Mariani R, McAnulty JH, Gunson K, Jui J, Chugh SS. Population-based analysis of sudden cardiac death with and without left ventricular systolic dysfunction: two-year findings from the oregon sudden unexpected death study. J Am Coll Cardiol 2006;47:1161–1166. [DOI] [PubMed] [Google Scholar]
- 48. Au-Yeung WTM, Reinhall PG, Bardy GH, Brunton SL. Development and validation of warning system of ventricular tachyarrhythmia in patients with heart failure with heart rate variability data. PLoS One 2018;13:e0207215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Murali KM, Mullan J, Chen JHC, Roodenrys S, Lonergan M. Medication adherence in randomized controlled trials evaluating cardiovascular or mortality outcomes in dialysis patients: A systematic review. BMC Nephrol 2017;18:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Van Spall HGC, Averbuch T, Damman K, Voors AA. Risk and risk reduction in trials of heart failure with reduced ejection fraction: absolute or relative? Eur J Heart Fail. Published online June 16, 2021:ejhf.2248. [DOI] [PubMed] [Google Scholar]
- 51. Helgeson J, Rammage M, Urman A, Roebuck MC, Coverdill S, Pomerleau K, Dankwa-Mullan I, Liu L-I, Sweetman RW, Chau Q, Williamson MP, Vinegra M, Haddad TC, Goetz MP. Clinical performance pilot using cognitive computing for clinical trial matching at Mayo Clinic. JCO 2018;36:e18598–e18598. [Google Scholar]
- 52. Harrer S, Shah P, Antony B, Hu J. Artificial intelligence for clinical trial design. Trends Pharmacol Sci 2019;40:577–591. [DOI] [PubMed] [Google Scholar]
- 53. Koesmahargyo V, Abbas A, Zhang L, Guan L, Feng S, Yadav V, Galatzer-Levy IR. Accuracy of machine learning-based prediction of medication adherence in clinical research. Psychiatry Clin Psychol 2020. [DOI] [PubMed] [Google Scholar]
- 54. Mahajan R, Gupta K. Adaptive design clinical trials: methodology, challenges and prospect. Indian J Pharmacol 2010;42:201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Aziz M, Kaufmann E, Riviere MK. On multi-armed bandit designs for dose-finding clinical trials. arXiv: 190307082 [cs, stat]. Published online April 7, 2020. Accessed March 9, 2022. http://arxiv.org/abs/1903.07082
- 56. Cruz Rivera S, Liu X, Chan A-W, Denniston AK, Calvert MJ, Darzi A, Holmes C, Yau C, Moher D, Ashrafian H, Deeks JJ, Ferrante di Ruffano L, Faes L, Keane PA, Vollmer SJ, Lee AY, Jonas A, Esteva A, Beam AL, Panico MB, Lee CS, Haug C, Kelly CJ, Yau C, Mulrow C, Espinoza C, Fletcher J, Moher D, Paltoo D, Manna E, Price G, Collins GS, Harvey H, Matcham J, Monteiro J, ElZarrad MK, Ferrante di Ruffano L, Oakden-Rayner L, McCradden M, Keane PA, Savage R, Golub R, Sarkar R, Rowley S, The SPIRIT-AI and CONSORT-AI Working Group, SPIRIT-AI and CONSORT-AI Steering Group, SPIRIT-AI and CONSORT-AI Consensus Group . Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. Nat Med 2020;26:1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Breck E, Polyzotis N, Roy S, Whang S, Zinkevich M. DATA VALIDATION FOR MACHINE LEARNING. In: Proceedings of the 2nd SysML Conference; 2019.
- 58. Emmanuel T, Maupong T, Mpoeleng D, Semong T, Mphago B, Tabona O. A survey on missing data in machine learning. J Big Data 2021;8:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Finlayson SG, Subbaswamy A, Singh K, Bowers J, Kupke A, Zittrain J, Kohane IS, Saria S. The clinician and dataset shift in artificial intelligence. N Engl J Med 2021;385:283–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cikes M, Sanchez-Martinez S, Claggett B, Duchateau N, Piella G, Butakoff C, Pouleur AC, Knappe D, Biering-Sørensen T, Kutyifa V, Moss A, Stein K, Solomon SD, Bijnens B. Machine learning-based phenogrouping in heart failure to identify responders to cardiac resynchronization therapy: machine learning-based approach to patient selection for CRT. Eur J Heart Fail 2019;21:74–85. [DOI] [PubMed] [Google Scholar]
- 61. Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G, King D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med 2019;17:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were generated or analyzed in support of this research.