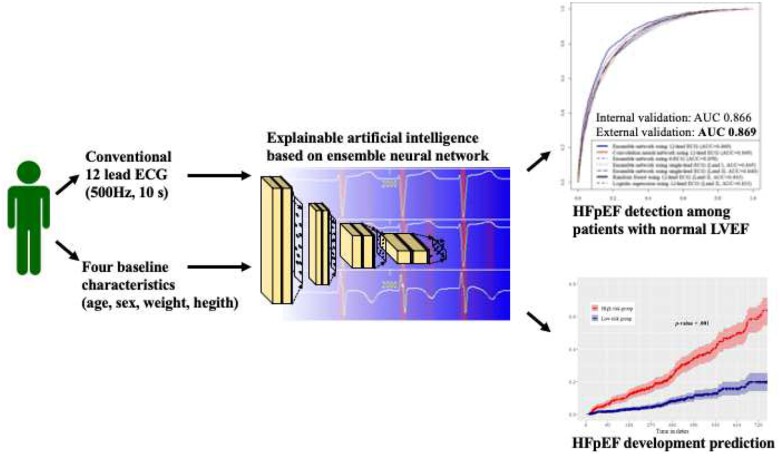
Abstract
Aims
Although heart failure with preserved ejection fraction (HFpEF) is a rapidly emerging global health problem, an adequate tool to screen it reliably and economically does not exist. We developed an interpretable deep learning model (DLM) using electrocardiography (ECG) and validated its performance.
Methods and results
This retrospective cohort study included two hospitals. 34 103 patients who underwent echocardiography and ECG within 1 week and indicated normal left ventricular systolic function were included in this study. A DLM based on an ensemble neural network was developed using 32 671 ECGs of 20 169 patients. The internal validation included 1979 ECGs of 1979 patients. Furthermore, we conducted an external validation with 11 955 ECGs of 11 955 patients from another hospital. The endpoint was to detect HFpEF. During the internal and external validation, the area under the receiver operating characteristic curves of a DLM using 12-lead ECG for detecting HFpEF were 0.866 (95% confidence interval 0.850–0.883) and 0.869 (0.860–0.877), respectively. In the 1412 individuals without HFpEF at initial echocardiography, patients whose DLM was defined as having a higher risk had a significantly higher chance of developing HFpEF than those in the low-risk group (33.6% vs. 8.4%, P < 0.001). Sensitivity map showed that the DLM focused on the QRS complex and T-wave.
Conclusion
The DLM demonstrated high performance for HFpEF detection using not only a 12-lead ECG but also 6- single-lead ECG. These results suggest that HFpEF can be screened using conventional ECG devices and diverse life-type ECG machines employing the DLM, thereby preventing disease progression.
Keywords: Heart failure, Electrocardiography, Deep learning, Artificial intelligence
Graphical Abstract
Introduction
The prevalence of heart failure (HF) is estimated to be 1.1–5.5% in the general population.1 It is one of the most prominent causes of morbidity and health care expenditure worldwide and continues to increase in prevalence at an alarming rate. Almost half of all patients with HF have a normal ejection fraction (EF). The prevalence of this syndrome, termed heart failure with preserved ejection fraction (HFpEF), continues to increase in the developed world, likely because of the increasing prevalence of typical risk factors, including older age, female gender, hypertension, metabolic syndrome, renal dysfunction, and obesity.2–4 Epidemiological data revealed that the prevalence of HFpEF relative to heart failure with reduced ejection fraction (HFrEF) is increasing at a rate of 1% per year, indicating that HFpEF is becoming the most prevalent type of HF.
Because HFpEF is a complex syndrome that can result from structural and functional cardiac disorders, rather than a single disease entity, its correct diagnosis can be challenging even for HF specialists. This is caused by multiple pathophysiologic processes, but diagnostic criteria remain general, including dyspnoea and fluid overload, normal left ventricular (LV) ejection fraction, elevated natriuretic peptides, and evidence of HF or diastolic dysfunction.5,6 The echocardiographic assessment of LV diastolic function is important in the routine evaluation of patients with HF. More importantly, LV diastolic dysfunction can develop without any clinical symptoms and is not uncommon even in the general population.7 Diagnosis is often delayed, as the condition can be asymptomatic. The routine use of echocardiography for screening diastolic dysfunction is expensive and time consuming. As such, echocardiography is often performed only in patients with suspected HFpEF for the early detection of asymptomatic patients. Hence, cost-effective strategies that quantify aspects of the diastolic function or hemodynamic changes associated with the LV diastolic dysfunction are urgently required to establish a diagnosis of HFpEF.
The majority of patients with HFpEF undergo the electrical remodelling of the myocardium, manifested as ECG abnormalities. However, it is not easy to detect subtle ECG changes; therefore, the current state of ECG is not useful for detecting HFpEF. To develop a reliable HFpEF detection method based on ECG, we used a deep learning model (DLM) based on an ensemble neural network. Recently, deep learning has demonstrated high accuracy and applicability in computer vision, speech recognition, and signal processing.8 Furthermore, deep learning has been applied in medical domains, and studies regarding the DLM have been performed where left systolic dysfunction, valvular heart disease, hyperkalaemia, and anaemia have been diagnosed, and the occurrence of atrial fibrillation predicted using ECG.9–13 In recent studies, several deep learning algorithms were developed for detecting HF using electrocardiography. The endpoint in most of the previous studies was HFrEF.14–16 In this study, we developed and validated a DLM for detecting HFpEF using a diverse type of ECG. Furthermore, we used a sensitivity map to visualize the important part of ECG for detecting HFpEF using the DLM for making interpretation and comparing with the previously available medical information.
Methods
Study design and population
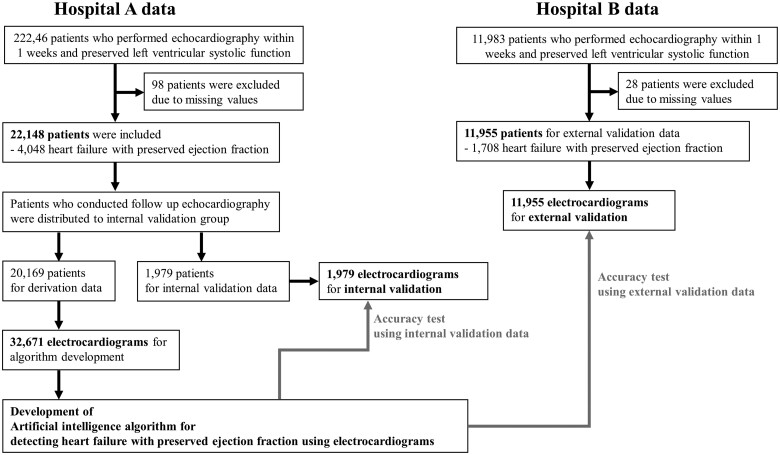
This multicentre retrospective cohort study performed in this investigation involved data from two hospitals—A and B—to develop and validate an ensemble neural network-based DLM for detecting HFpEF. The eligible study population included adult patients (aged ≥ 15 years) who underwent both ECG and echocardiography within 1 week for clinical evaluation or health examination and had been confirmed to have a normal or near-normal LV ejection fraction, defined as an ejection fraction of 50% or more. We excluded subjects whose demographic, ECG, or echocardiographic information was not available. As shown in Figure 1, patients who were treated at hospital A (October 2016–May 2020) were split into DLM development and internal validation datasets. Patients who underwent follow-up echocardiography after an initial evaluation were distributed to an internal validation dataset. Patients who had no follow-up echocardiography were distributed to a development dataset that was used to develop the DLM. Subsequently, we evaluated the accuracy of the DLM using the internal validation dataset. Furthermore, we used data from hospital B (March 2017–May 2020) as an external validation dataset to verify the applicability of the DLM across centres. Because the purpose of the validation data was to assess the accuracy of the DLM, we only used the most recent ECG signal before their first echocardiography in the study period for the internal and external validation datasets.
Figure 1.
Study flowchart.
The Institutional Review Board of Sejong General Hospital (2019-0057) and Mediplex Sejong Hospital (2019-008) approved this study protocol and waived the requirement for informed consent due to impracticality and minimal harm.
Endpoint and predictive variables
The primary endpoint was the presence of HFpEF, which was defined as left ventricular diastolic dysfunction (LVDD) in the presence of normal or near-normal LV ejection fraction and had symptoms and signs of HFpEF. Normal or near-normal LV ejection fraction defined as an ejection fraction of 50% or more. LVDD was defined in accordance with the most recent guidelines with the following cut-off values suggesting abnormal diastolic function: (i) septal e′ < 7 cm/s and/or lateral e′ <10 cm/s; (ii) averaged E/e′ > 14; (iii) tricuspid regurgitation velocity > 2.8 m/s; and (iv) left atrial volume index > 34 mL/m2. Patients who satisfied ≥ more than one-half of these criteria were defined as having an abnormal diastolic dysfunction.17 We defined the symptoms and signs of HFpEF as chest discomfort, palpitation, exercise intolerance, fatigue, oedema, dyspnoea, syncope, and general weakness, and confirmed the information from the echocardiography report, in which the cardiologist has mentioned the reason for conducting echocardiography. The echocardiographic findings were obtained from comprehensive two-dimensional (2D) Doppler echocardiography. Acquisitions and measurements were performed by licensed sonographers and cardiologists who were blinded to any other study data. We used demographic information and ECG as predictive variables. We used demographic information and ECG as predictive variables. We used four variables (age, sex, weight, and height) as demographic information. These four variables are simple and can be objectively collected consistently during the screening evaluation. We did not use past medical history, because the information relied on the patient’s memory, and there was a possibility of error and undetected disease. Only definite epidemiologic information was used for the evaluation.
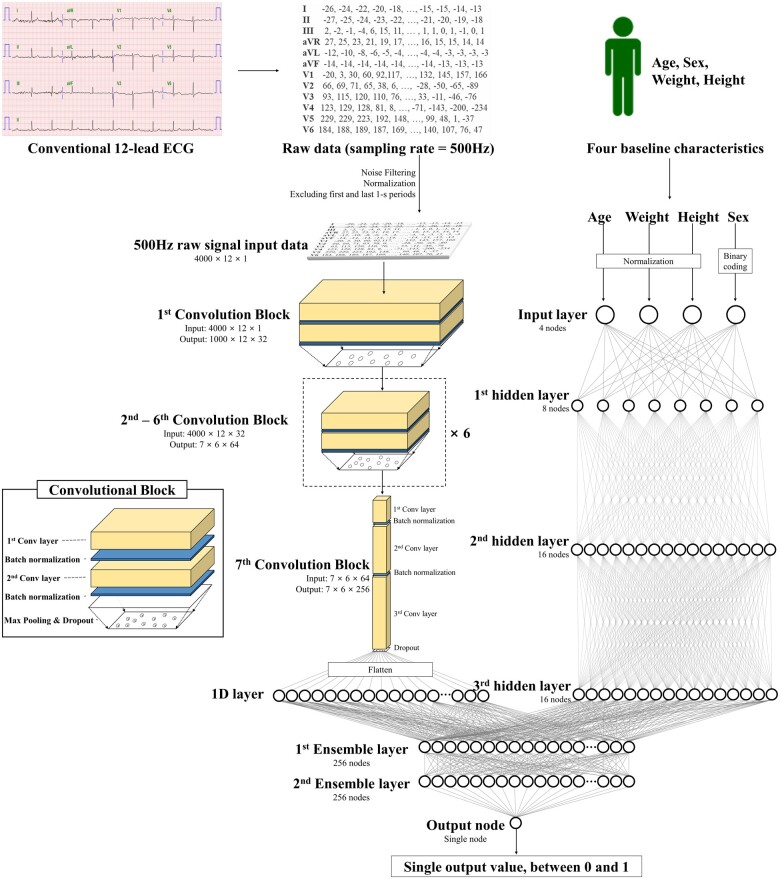
We used the raw data from each 12-lead ECG, amounting to 5000 data points for each lead, recorded over 10 s (500 Hz), and 60 000 data points from each ECG. We used 8 s of ECG data by excluding the first and last 1-s periods because more artefacts were contained within this range. We created a dataset using the entire 12-lead ECG data. Furthermore, we used partial datasets from the 12-lead ECG data, such as the limb six-lead (I, II, III, aVL, aVR, and aVF) and single lead (I or II). We selected those leads as they can be easily recorded by wearable and lifestyle devices in contact with the patient’s limbs.18 Consequently, when we developed and validated an algorithm using 12-lead ECG, we used a dataset comprising 12 × 4000 2D data points. Similarly, for the six-lead and single-lead ECG signals, we used datasets comprising 6 × 4000 and 1 × 4000 data points, respectively.
Development of deep learning model
The DLM was developed using many hidden layers of neurons to learn complex hierarchical nonlinear representations from the data.8 As a block comprising six stages, it has two convolutional layers, two batch normalization layers, one max pooling layer, and one dropout layer (Figure 2).19,20 The last layer of the seventh block was fully connected to a one-dimensional (1D) layer composed of 256 nodes. The input layer of epidemiology (age, sex, weight, and height) was concatenated with the 1D layer. There were two fully connected 1D layers after the flattened layer, and the second layer was connected to the output node, which was composed of one node. The values of the output node represent the possibility of detecting HFpEF, and the output node uses a sigmoid function as an activation function, as the output of the sigmoid function is between 0 and 1. The final number of layers of convolutional part, multilayer perceptron part, and ensemble part are 44, 4, and 3, respectively. We confirmed the final architecture of the DLM using a grid search. We used TensorFlow’s open-source software library (Google LLC, Mountain View, CA, USA) as the backend and conducted our experiment with Python (version 3.6; Python Software Foundation, Beaverton, OR, USA).21
Figure 2.
Architecture of deep learning-based model for detecting HFpEF. Conv, convolutional neural network; ECG, electrocardiography; HFpEF, heart failure with preserved ejection fraction.
Furthermore, we developed an additional machine learning model to compare it with the ensemble network-based DLM. Hence, we used a logistic regression model (LR), random forest (RF) model, and convolutional neural network only model (CNN) developed with R (R developed Core Team, Vienna, Austria) and Python.8,22,23 These machine learning methods performed better than traditional methods in several medical domains in previous studies.
Statistical analysis
Continuous variables were presented as means and standard deviations and were compared using the unpaired Student’s t-test or Mann–Whitney U test (Table 1). Categorical variables were expressed as frequencies and percentages and then compared using the χ2 test. At each input of the validation data, each DLM calculated the possibility of HFpEF in the range from 0 (non-HFpEF, normal) to 1 (HFpEF). To confirm the performance of the developed DLM, we compared the possibility calculated by the DLM with the presence of HFpEF in the validation dataset. We used the area under the receiver operating characteristic curve (AUC) to measure the performance of the DLM. Furthermore, we used the sensitivity, specificity, positive predictive value, negative predictive value, accuracy, and F-measure as comparative metrics. We selected the cut-off point using the Youden J statistics of development datasets and confirmed the results. A two-sided P-value of <0.001 was considered significant for all tests. We evaluated the 95% confidence interval using bootstrapping (10 000 times resampling with replacement). All statistical analyses were performed using R.24
Table 1.
Baseline characteristics
| Characteristics |
Hospital A (derivation and internal validation data)
N = 22 148 |
Hospital B (external validation data)
N = 11 955 |
|||||
|---|---|---|---|---|---|---|---|
| Non-HFpEF | HFpEF | P a | Non-HFpEF | HFpEF | P a | P b | |
| Study subjects, N (%) | 18 100 (81.7%) | 4048 (18.3%) | 10 247 (85.7%) | 1708 (14.3%) | <0.001 | ||
| Baseline characteristics | |||||||
| Age | 56.70 (14.99) | 70.44 (11.29) | <0.001 | 55.13 (14.06) | 70.59 (11.75) | <0.001 | <0.001 |
| Male, N (%) | 56.70 (14.99) | 70.44 (11.29) | <0.001 | 5199 (50.7) | 581 (34.0) | 0.001 | 0.027 |
| Weight | 65.10 (12.02) | 62.85 (12.27) | <0.001 | 66.07 (12.53) | 63.44 (12.95) | <0.001 | <0.001 |
| Height | 163.15 (9.29) | 157.90 (9.60) | <0.001 | 163.84 (9.24) | 157.65 (9.70) | <0.001 | <0.001 |
| BSA | 1.70 (0.19) | 1.63 (0.19) | <0.001 | 1.72 (0.19) | 1.64 (0.20) | <0.001 | <0.001 |
| LVSD | 28.63 (4.58) | 29.19 (5.69) | <0.001 | 29.64 (3.82) | 29.82 (4.81) | 0.080 | <0.001 |
| LVDD | 46.64 (4.66) | 47.60 (5.95) | <0.001 | 47.90 (3.88) | 48.61 (4.93) | <0.001 | <0.001 |
| LAD | 37.35 (5.83) | 45.50 (7.30) | <0.001 | 35.80 (4.96) | 41.40 (6.83) | <0.001 | <0.001 |
| LAVI | 24.72 (7.15) | 33.60 (9.70) | <0.001 | 20.06 (5.63) | 27.04 (7.75) | <0.001 | |
| E | 62.41 (17.18) | 71.01 (23.12) | <0.001 | 64.87 (16.85) | 72.98 (21.49) | <0.001 | 0.003 |
| A | 67.90 (17.99) | 84.99 (24.33) | <0.001 | 68.52 (17.96) | 89.36 (22.67) | <0.001 | 0.010 |
| DT | 196.43 (46.93) | 219.07 (60.44) | <0.001 | 213.72 (43.76) | 232.60 (54.24) | <0.001 | <0.001 |
| E′ | 7.38 (2.57) | 4.88 (1.55) | <0.001 | 7.30 (2.39) | 4.49 (1.40) | <0.001 | 0.470 |
| A′ | 8.99 (1.84) | 7.91 (2.09) | <0.001 | 8.72 (1.84) | 7.96 (1.99) | <0.001 | <0.001 |
| E over E′ | 8.98 (2.70) | 14.27 (4.24) | <0.001 | 9.35 (2.49) | 15.70 (3.53) | <0.001 | 0.984 |
| TR velocity | 2.20 (0.25) | 2.44 (0.37) | <0.001 | 2.19 (0.22) | 2.43 (0.36) | <0.001 | <0.001 |
| EF | 59.00 (6.26) | 57.14 (6.43) | <0.001 | 62.35 (6.30) | 62.22 (7.24) | 0.467 | <0.001 |
| Heart rate | 71.78 (14.10) | 71.68 (16.04) | 0.706 | 70.38 (13.29) | 72.18 (16.18) | <0.001 | <0.001 |
| PR interval | 156.55 (48.26) | 143.52 (77.02) | <0.001 | 160.29 (38.24) | 154.08 (65.09) | <0.001 | <0.001 |
| QT interval | 398.26 (36.74) | 416.96 (46.14) | <0.001 | 399.30 (35.40) | 412.06 (46.57) | <0.001 | 0.205 |
| QTc | 430.70 (28.91) | 449.02 (35.64) | <0.001 | 428.10 (27.47) | 445.22 (34.84) | <0.001 | <0.001 |
| QRSd | 94.78 (14.68) | 98.79 (20.16) | <0.001 | 94.62 (13.35) | 97.65 (17.89) | <0.001 | 0.008 |
| P axis | 66.31 (82.62) | 105.36 (133.91) | <0.001 | 54.96 (61.12) | 76.11 (110.36) | <0.001 | <0.001 |
| R axis | 39.64 (41.18) | 32.51 (44.63) | <0.001 | 39.90 (38.24) | 29.34 (38.93) | 0.001 | 0.911 |
| T axis | 39.91 (38.97) | 58.50 (67.44) | <0.001 | 37.65 (32.40) | 55.79 (59.90) | <0.001 | <0.001 |
A, late diastolic mitral inflow velocity; A′, late diastolic mitral annular tissue velocity; BSA, body surface area; DT, deceleration time; E, early diastolic mitral inflow velocity; E′, early diastolic mitral annular tissue velocity; EF, ejection fraction; LAD, left atrial dimension; LAVI, left atrial volume index; LVDD, left ventricular diastolic dimension; LVSD, left ventricular systolic dimension; QRSd QRS duration; TR, tricuspid regurgitation.
The alternative hypothesis for this P-value was that there is a difference between the heart failure with preserved ejection fraction and non-heart failure with preserved ejection fraction data group for each variable.
The alternative hypothesis for this P-value was that there is a difference between the hospital A (development and internal validation data group) and B (external validation group) for each variable.
To understand the model and perform a comparison with existing medical knowledge, it is important to identify the region that significantly affects the DLM decision. We employed a sensitivity map using a saliency method.25 The map was computed using the first-order gradients of the classifier probabilities with respect to the input signals. If the probability of a classifier is sensitive to a specific region of the signal, the region would be considered significant in the model. We used a gradient class activation map as a sensitivity map and a guided gradient backpropagation method. We confirmed the variable importance of each developed model using the Akaike information criterion and the mean decreased Gini.
Confirming deep learning model performance to predict heart failure with preserved ejection fraction development as subgroup analysis
We hypothesized that the ECGs would display subtle abnormal patterns in the pre-HFpEF phase, and that the developed DLM would classify some of these cases as abnormal, yielding a false positive test (a study subject classified as having HFpEF but considered as non-HFpEF) as the initial result. We conducted a subgroup analysis of patients who underwent follow-up echocardiography in the internal and external validation datasets. The difference in date between the initial and follow-up echocardiography data was over 14 days. Among those patients, we confirmed the development of HFpEF in patients who were initially considered non-HFpEF based on the initial echocardiography. The DLM was categorized into high- and low-risk groups based on the risk score using cut-off values, which were determined using the Youden’s J statistic with the development dataset.26 We used the Kaplan–Meier method to analyse HFpEF development for 24 months using an AI algorithm.
Confirming deep learning model performance to predict left ventricular diastolic dysfunction among asymptomatic patients as subgroup analysis
As the purpose of the DLM was to use it in all situations (admitted patients, outpatient department, and general check-up), we decided to study the population without considering any specific situation. Furthermore, one of the purposes of this developed DLM was to screen for LVDD among patients who had no symptoms, we confirmed the performance of the DLM in the subgroup analysis. Patients who underwent echocardiography without symptoms as a general health check-up in validation datasets were selected as the study population for subgroup analysis. The endpoint of the subgroup analysis was LVDD in the presence of normal or near-normal LV ejection fraction, defined as an ejection fraction of 50% or more.
Results
Among the 34 229 patients who were eligible for this study, 126 were excluded owing to missing values (Figure 1). The study included 34 103 patients, of whom 5756 had HFpEF. A DLM was developed using a development dataset comprising 32 671 12-lead ECG from 20 169 patients. The performance of the algorithm was then confirmed using 1979 ECG data points from the 1979 patients in the internal validation dataset from hospital A, and 11 955 ECGs from the 11 955 patients in the external validation dataset from hospital B (Table 1).
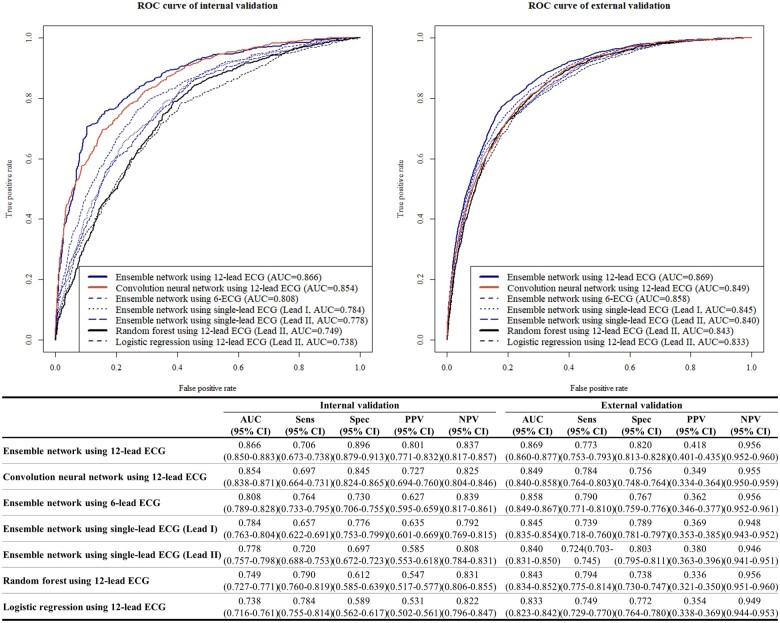
During the internal and external validations, the AUC of the DLM was 0.866 [95% confidence interval (CI) 0.850–0.883] and 0.869 (95% CI 0.860–0.877), respectively (Figure 3). These values imply that the DLM performed better than the other machine learning algorithms. During the internal and external validations, the AUC of the DLM using six-lead ECG was 0.808 (95% CI 0.789–0.828) and 0.858 (95% CI 0.849–0.858), respectively. The AUC of the single-lead AI algorithm during the internal and external validation using lead I was 0.784 (95% CI 0.763–0.804) and 0.845 (95% CI 0.835–0.854), respectively; all results are shown in Figure 3.
Figure 3.
Performance of model for detecting heart failure with preserved ejection fraction. AUC, area under the receiver operating characteristic curve; CI, confidence interval; ECG, electrocardiography; NPV, negative predictive value; PPV, positive predictive value; ROC, receiver operating characteristic curve; Sens, sensitivity; Spec, specificity.
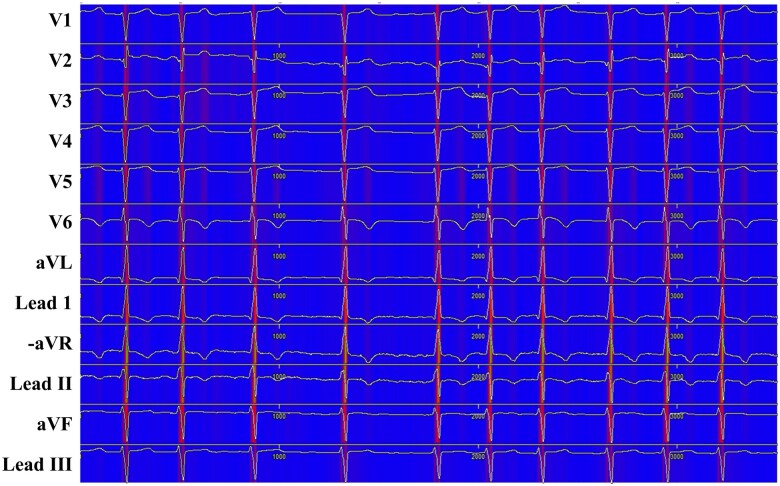
We used a sensitivity map to visualize the ECG region used in the DLM to detect HFpEF (Figure 4). The map shows that the DLM focused on the QRS complex, particularly the R-wave, in most patients. The DLM focused on not only the QRS complex but also the T-wave in patients. As shown in Table 2, the variable importance differed for each prognostic model. Whereas the LR and RF used the T-wave axis as an important predictive variable, the DLM used the QRS duration as an important predictive variable.
Figure 4.
Sensitivity map of deep learning model for detecting HFpEF. HFpEF, heart failure with preserved ejection fraction.
Table 2.
Importance of variables in development data for each prediction model
| Variable importance rank | Logistic regression (deviance difference) | Random forest (mean decreased Gini) | Deep learning (difference in AUC) |
|---|---|---|---|
| 1 | Age | Age | Age |
| (2450) | (2135) | (0.069) | |
| 2 | T-wave axis | T-wave axis | R-wave axis |
| (521) | (1712) | (0.059) | |
| 3 | Height | QT interval | T-wave axis |
| (249) | (1270) | (0.022) | |
| 4 | Weight | Height | Weight |
| (147) | (1148) | (0.021) | |
| 5 | Presence of atrial fibrillation or flutter | R-wave axis | QRS duration |
| (144) | (1120) | (0.019) | |
| 6 | P-wave axis | P-wave axis | Height |
| (56) | (1043) | (0.017) | |
| 7 | QT interval | Weight | QT interval |
| (44) | (1009) | (0.016) | |
| 8 | QRS duration | QRS duration | P-wave axis |
| (32) | (997) | (0.015) | |
| 9 | PR interval | PR interval | PR interval |
| (24) | (988) | (0.014) | |
| 10 | R-wave axis | Heart rate | Heart rate |
| (15) | (893) | (0.012) | |
| 11 | Heart rate | Sex | Sex |
| (10) | (144) | (0.011) | |
| 12 | Sex | Presence of atrial fibrillation and flutter (132) | Presence of atrial fibrillation and flutter (0.005) |
| (1) |
AUC, area under the receiver operating characteristic curve.
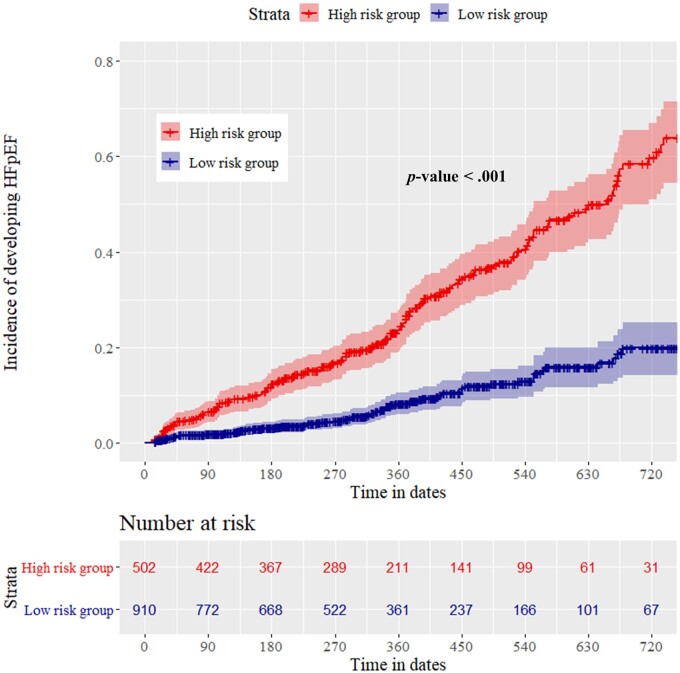
Our study comprised 2231 patients (1979 and 252 patients in the internal and external validation datasets, respectively) with follow-up echocardiographic results. Among them, 1412 patients were normal (non-HFpEF) at initial echocardiography. We conducted a subgroup analysis of HFpEF development after initial echocardiography in these 1412 patients, of whom 246 developed HFpEF within 24 months. The high-risk group of the DLM showed a significantly higher hazard (Figure 5) and higher development rate of HFpEF than the low-risk group (33.6% vs. 8.4%, P < 0.001).
Figure 5.
Cumulative hazard of developing HFpEF in patients with an initially normal. HFpEF, denoted heart failure with preserved ejection fraction.
In the subgroup analysis of 2566 patients who underwent echocardiography without any symptom as a general check-up in the validation dataset. Among these patients, 128 had LVDD without symptom or sign. The AUC of the DLM for detecting LVDD among the study population was 0.837 (0.805–0.870) in validation, respectively. We described the detailed performance of DLB for detecting LVDD among the asymptomatic population in Supplementary material online.
Discussion
We developed and validated a DLM based on an ensemble network for HFpEF detection using 12-, 6-, and single-lead ECG, and it demonstrated reasonable performance. Subsequently, we visualized our DLM to determine the regions and characteristics of the ECG that were used for HFpEF prediction and confirmed the important variable for the decision in another machine learning model, such as LR, RF, and CNN. We conducted a subgroup analysis for patients with non-HFpEF (normal) at initial echocardiography; it was demonstrated that the DLM can predict the development of HFpEF. To our knowledge, this study is the first to develop a DLM for detecting and predicting HFpEF and show interpretable patterns of decision making using the DLM.
Developing a reliable screening tool for detecting and predicting HFpEF is crucial as LV diastolic dysfunction can develop without any clinical symptoms or exhibit minimal symptoms. Echocardiographic assessment of left ventricular diastolic function is an integral part of the routine evaluation of patients presenting with symptoms of dyspnoea. Guidelines for diastolic function assessment were comprehensive, including several 2D and Doppler parameters to grade diastolic dysfunction and to estimate LV filling pressures.17 However, the inclusion of many parameters in the guidelines was perceived to render diastolic function assessment too complex and echocardiography is expensive and time consuming. If HFpEF is detectable using the conventional 12-lead ECG or a diverse life-type ECG device, patients can then be referred for echocardiography and early diagnosis. ECG is a non-invasive, inexpensive, and requires a short time to perform. Hence, we developed a DLM as a reliable HFpEF screening tool. Deep learning includes feature learning, which is a set of methods that allow feature extraction and a model to be created from various data types, such as images, 2D data, and waveform signals.8 In this study, we used raw ECG data (2D numerical data). In previous studies, Attia et al. and our group developed a DLM for screening LV systolic dysfunction, arrhythmia, valvular heart disease, LV hypertrophy, hyperkalaemia, and anaemia.9–13 However, deep learning is not an optimal method owing to the unreliability of its outcomes due to the low interpretability of the decision process.8
A major limitation of deep learning technology in medicine is that the decision-making process remains unpredictable. Therefore, we adopted a sensitivity map and variable importance in the DLM. The sensitivity map showed that the DLM-focused QRS complex and variable importance result showed that the R-wave axis was high variable importance in the DLM. In a previous study, the Cornell product [amplitude of R-wave in aVL + depth of S-wave in V3) * QRS] was used; it is an easily applicable ECG marker of HFpEF and predicts poor prognosis by reflecting the severity of diastolic dysfunction.27 Another study showed that scoring system derived from this study, including the presence or absence of left atrial hypertrophy, QRS duration > 100 ms, right bundle branch block, ST-T segment changes and prolongation of the QT interval can be used to predict the type of HF.28 And the QRS axis was associated with HFpEF and prognosis, and HFpEF and QRS prolongation were harmful in HFpEF for mortality.29 And QRS prolongation and axis change are associated with the pathophysiology of HFpEF (cellular and ventricular hypertrophy, fibrosis of the conduction system and myocardium, ischaemia, and diastolic dysfunction). 29 The DLM focused not only on the QRS complex but also on the T-wave. A previous study pertaining to the detection of diastolic dysfunction using ECG based on machine learning showed that the T-wave is an important predictor of HFpEF.30 In our study, T-waves were used in LR and RF as important variables for detecting HFpEF, whereas the QRS duration indicated a highly variable importance in the DLM.
The purpose of the developed DLM is to screen for HFpEF. The developed DLM achieved an AUC of 0.866–0.869. The DLM performed better than other typically used screening methods in clinical settings, e.g., mammography for breast cancer screening (AUC = 0.78; positive predictive value: 3–12%) and faecal occult blood testing for detecting colorectal neoplasia (AUC = 0.71; overall sensitivity: 29%).31,32 Although the performance of the developed DLM was unsatisfactory, the possibility of applying deep learning to ECG for screening HFpEF was demonstrated in this study.
Several limitations were present in our study. First, as this study was only conducted in two hospitals in Korea, the algorithm used must be further validated in patients with HFrEF in other countries. Second, as this was a retrospective study, a prospective study must be conducted in clinical settings, embedding the algorithm in the hospital electronic health record or ECG machine. These two tasks will be attempted in our future studies. Third, as we had confirmed the DLB (deep learning based model) using single-lead ECG in part of the data of 12-lead ECG, we should conduct further study to confirm the performance of the DLM in wearable devices. This is the next study subject of our study group. Fourth, the decision process of the algorithm must be further investigated based on deep learning. For example, additional experiments must be performed to understand the deep learning process and determine the characteristics of the QRS complex that affect DLM’s decision. This will be attempted in our next study.
Conclusion
A DLM based on an ensemble neural network demonstrated accurate performance in detecting HFpEF using ECG and successfully predicted the development of HFpEF.
Supplementary material
Supplementary material is available at European Heart Journal – Digital Health online.
Supplementary Material
Acknowledgements
This research was result of a study on the ‘High Performance Computing Support’ and ‘AI voucher’ Project, supported by the ‘Ministry of Science and ICT and National IT Industry Promotion Agency of South Korea.
Funding
The National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2020R1F1A1073791).
Conflict of interest; K.H.K., H.J.E., K.H.J., S.Y.L., and B.H.O. declare that they have no competing interests. J.K. and J.P. are co-founder and stakeholder in Medical AI Co., a medical artificial intelligence company. J.K. and Y.C. are researchers of Body friend Co. There are no products in development or marketed products to declare. This does not alter our adherence to European Heart Journal-Digital Health.
Data availability
No new data were generated or analysed in support of this research.
References
- 1. Owan TE, Hodge DO, Herges RM, Jacobsen SJ, Roger VL, Redfield MM. Trends in prevalence and outcome of heart failure with preserved ejection fraction. N Engl J Med 2006;355:251–259. [DOI] [PubMed] [Google Scholar]
- 2. Reddy YNV, Borlaug BA. Heart failure with preserved ejection fraction. Curr Probl Cardiol 2016;41:145–188. [DOI] [PubMed] [Google Scholar]
- 3. Unger ED, Dubin RF, Deo R, Daruwalla V, Friedman JL, Medina C, Beussink L, Freed BH, Shah SJ. Association of chronic kidney disease with abnormal cardiac mechanics and adverse outcomes in patients with heart failure and preserved ejection fraction. Eur J Heart Fail 2016;18:103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meta-analysis Global Group in Chronic Heart Failure (MAGGIC). The survival of patients with heart failure with preserved or reduced left ventricular ejection fraction: an individual patient data meta-analysis. Eur Heart J 2012;33:1750–1757. [DOI] [PubMed] [Google Scholar]
- 5. Ponikowski P, Voors AA,, Anker SD, Bueno H, Cleland JGF, Coats AJS, Falk V, González-Juanatey JR, Harjola V-P, Jankowska EA, Jessup M, Linde C, Nihoyannopoulos P, Parissis JT, Pieske B, Riley JP, Rosano GMC, Ruilope LM, Ruschitzka F, Rutten FH, van der Meer P; ESC Scientific Document Group. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC)Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur Heart J 2016;37:2129–2200. [DOI] [PubMed] [Google Scholar]
- 6. Webb J, Fovargue L, Tøndel K, Porter B, Sieniewicz B, Gould J, Rinaldi CA, Ismail T, Chiribiri A, Carr-White G. The emerging role of cardiac magnetic resonance imaging in the evaluation of patients with HFpEF. Curr Heart Fail Rep 2018;15:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dargie HJ. Effect of carvedilol on outcome after myocardial infarction in patients with left-ventricular dysfunction: the CAPRICORN randomised trial. Lancet (Lond, Engl ) 2001;357:1385–1390. [DOI] [PubMed] [Google Scholar]
- 8. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–444. [DOI] [PubMed] [Google Scholar]
- 9. Attia ZI, Kapa S, Lopez-Jimenez F, McKie PM, Ladewig DJ, Satam G, Pellikka PA, Enriquez-Sarano M, Noseworthy PA, Munger TM, Asirvatham SJ, Scott CG, Carter RE, Friedman PA. Screening for cardiac contractile dysfunction using an artificial intelligence–enabled electrocardiogram. Nat Med 2019;25:70–74. [DOI] [PubMed] [Google Scholar]
- 10. Attia ZI, Noseworthy PA, Lopez-Jimenez F, Asirvatham SJ, Deshmukh AJ, Gersh BJ, Carter RE, Yao X, Rabinstein AA, Erickson BJ, Kapa S, Friedman PA. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet 2019;394:861–867. [DOI] [PubMed] [Google Scholar]
- 11. Galloway CD, Valys A V., Shreibati JB, Treiman DL, Petterson FL, Gundotra VP,, Albert DE, Attia ZI, Carter RE, Asirvatham SJ, Ackerman MJ, Noseworthy PA, Dillon JJ, Friedman PA. Development and validation of a deep-learning model to screen for hyperkalemia from the electrocardiogram. JAMA Cardiol 2019;4:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kwon J, Lee SY, Jeon K, Lee Y, Kim K, Park J, Oh B, Lee M. Deep learning-based algorithm for detecting aortic stenosis using electrocardiography. J Am Heart Assoc 2020;9:e014717. doi: 10.1161/JAHA.119.014717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kwon J, Cho Y, Jeon K-H, Cho S, Kim K-H, Baek SD, Jeung S, Park J, Oh B-H. A deep learning algorithm to detect anaemia with ECGs: a retrospective, multicentre study. Lancet Digit Heal 2020;2:e358–e367. [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y, Xia M. Application of deep neural network for congestive heart failure detection using ECG signals. J Phys Conf Ser 2020;1642:012021. [Google Scholar]
- 15. Samuel OW, Yang B, Geng Y, Asogbon MG, Pirbhulal S, Mzurikwao D, Idowu OP, Ogundele TJ, Li X, Chen S, Naik GR, Fang P, Han F, Li G. A new technique for the prediction of heart failure risk driven by hierarchical neighborhood component-based learning and adaptive multi-layer networks. Futur Gener Comput Syst 2020;110:781–794. [Google Scholar]
- 16. Sbrollini A, De Jongh MC, Ter Haar CC, Treskes RW, Man S, Burattini L, Swenne CA. Serial electrocardiography to detect newly emerging or aggravating cardiac pathology: a deep-learning approach. Biomed Eng Online 2019;18:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nagueh SF, Smiseth OA, Appleton CP, Byrd BF, Dokainish H, Edvardsen T, Flachskampf FA, Gillebert TC, Klein AL, Lancellotti P, Marino P, Oh JK, Popescu BA, Waggoner AD. Recommendations for the evaluation of left ventricular diastolic function by echocardiography: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J Am Soc Echocardiogr 2016;29:277–314. [DOI] [PubMed] [Google Scholar]
- 18. Walsh JA, Topol EJ, Steinhubl SR. Novel wireless devices for cardiac monitoring. Circulation 2014;130:573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 2014;15:1929–1958. [Google Scholar]
- 20. Jayalakshmi T, Santhakumaran A. Statistical normalization and backpropagation for classification. Int J Comput Theory Eng 2011;3:89–93. [Google Scholar]
- 21. Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Kudlur M, Levenberg J, Monga R, Moore S, Murray DG, Steiner B, Tucker P, Vasudevan V, Warden P, Wicke M, Yu Y, Zheng X, Brain G. TensorFlow: a system for large-scale machine learning. 12th USENIX Symp Oper Syst Des Implement (OSDI ’16) 2016; pp. 265–284. [Google Scholar]
- 22. Calcagno V, De Mazancourt C. glmulti: an R package for easy automated model selection with (generalized) linear models. J Stat Softw 2010;34:1–29. [Google Scholar]
- 23. Khalilia M, Chakraborty S, Popescu M. Predicting disease risks from highly imbalanced data using random forest. BMC Med Inform Decis Making 2011;11:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carpenter J, Bithell J. Bootstrap confidence intervals: when, which, what? A practical guide for medical statisticians. Stat Med 2000;19:1141–1164. [DOI] [PubMed] [Google Scholar]
- 25. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. Proceedings of the IEEE International Conference on Computer Vision; 2017. pp. 1;618–626. [Google Scholar]
- 26. Schisterman EF, Perkins NJ, Liu A, Bondell H. Optimal cut-point and its corresponding Youden index to discriminate individuals using pooled blood samples. Epidemiology 2005;16:73–81. [DOI] [PubMed] [Google Scholar]
- 27. Tan ES, Chan SP, Xu CF, Yap J, Richards AM, Ling LH, Sim D, Jaufeerally F, Yeo D, Loh SY, Ong HY, Leong KTG, Ng TP, Nyunt SZ, Feng L, Okin P, Lam CS, Lim TW. Cornell product is an ECG marker of heart failure with preserved ejection fraction. Heart Asia 2019;11:e011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hendry PB, Krisdinarti L, Erika M. Scoring system based on electrocardiogram features to predict the type of heart failure in patients with chronic heart failure. Cardiol Res 2016;7:110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lund LH, Jurga J, Edner M, Benson L, Dahlström U, Linde C, Alehagen U. Prevalence, correlates, and prognostic significance of QRS prolongation in heart failure with reduced and preserved ejection fraction. Eur Heart J 2013;34:529–539. [DOI] [PubMed] [Google Scholar]
- 30. Kagiyama N, Piccirilli M, Yanamala N, Shrestha S, Farjo PD, Casaclang-Verzosa G, Tarhuni WM, Nezarat N, Budoff MJ, Narula J, Sengupta PP. Machine learning assessment of left ventricular diastolic function based on electrocardiographic features. J Am Coll Cardiol 2020;76:930–941. [DOI] [PubMed] [Google Scholar]
- 31. Pisano ED, Gatsonis C, Hendrick E, Yaffe M, Baum JK, Acharyya S, Conant EF, Fajardo LL, Bassett L, D’Orsi C, Jong R, Rebner M; Digital Mammographic Imaging Screening Trial (DMIST) Investigators Group. Diagnostic performance of digital versus film mammography for breast-cancer screening. N Engl J Med 2005;353:1773–1783. [DOI] [PubMed] [Google Scholar]
- 32. Haug U, Kuntz KM, Knudsen AB, Hundt S, Brenner H. Sensitivity of immunochemical faecal occult blood testing for detecting left- vs right-sided colorectal neoplasia. Br J Cancer 2011;104:1779–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were generated or analysed in support of this research.