Abstract
Aims
We sought to investigate whether artificial intelligence (AI) and specifically deep neural networks (NNs) for electrocardiogram (ECG) signal analysis can be explained using human-selected features. We also sought to quantify such explainability and test if the AI model learns features that are similar to a human expert.
Methods and results
We used a set of 100 000 ECGs that were annotated by human explainable features. We applied both linear and non-linear models to predict published ECG AI models output for the detection of patients’ age and sex. We further used canonical correlation analysis to quantify the amount of shared information between the NN features and human-selected features. We reconstructed single human-selected ECG features from the unexplained NN features using a simple linear model. We noticed a strong correlation between the simple models and the AI output ( of 0.49–0.57 for the linear models and of 0.69–0.70 for the non-linear models). We found that the correlation of the human explainable features with either 13 of the strongest age AI features or 15 of the strongest sex AI features was above 0.85 (for comparison, the first 14 principal components explain 90% of the human feature variance). We linearly reconstructed single human-selected ECG features from the AI features with up to 0.86.
Conclusion
This work shows that NNs for ECG signals extract features in a similar manner to human experts and that they also generate additional novel features that help achieve superior performance.
Keywords: Electrocardiography, Cardiac age, Artificial intelligence, Explainable AI, Deep learning, Neural networks
Graphical Abstract
Introduction
Deep learning and specifically convolutional neural networks (NNs)1,2 enable computers to develop data-derived rules to solve complex classification problems without human knowledge regarding the structure of the input. Examples include detecting asymptomatic left ventricular dysfunction from an electrocardiogram (ECG),3 and determining age, sex, and cardiovascular risk from fundus photography.4 The same network architecture that is used to distinguish between images of dogs and cats can be used to classify Chest X-rays for pneumonia.5,6 Similarly, the same network structure used to identify the presence of life-threatening diseases from an ECG can be used to determine whether a person is male or female from a given ECG. The only difference is that during model training, the ground truth labels represent the specific characteristic that the network is to learn. In a convolutional NN, instead of using human-selected features for signal processing, network features are created by projecting the input on a set of weights, and optimizing the weights in a non-linear manner using labels during the training phase, with the objective of lowering the overall estimation or classification error. Through an iterative process,7 the network learns relevant rules and applies them to extract pertinent features for the specific test it is trained to solve. Because deep learning replaces human-engineered, hardcoded rules with computer-generated dynamically created rules based on data, biases in feature selection are possibly removed and human limitations have been overcome. However, deep learning is currently unexplainable. Moreover, it obfuscates the signal features used by its model and may allow the model to learn false association rules5 that may later be used for adversarial attacks.6,8
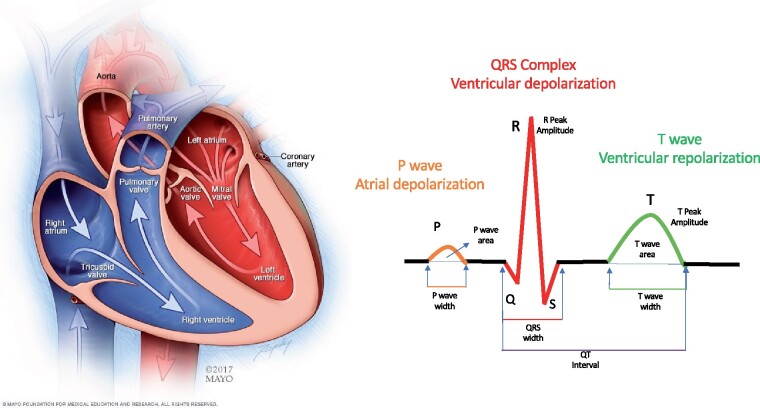
In this work, we aim to understand the features selected by NNs for the analysis of the ECG. The ECG is the recording of the heart’s electrical activity at a distance, i.e. from the body’s surface. It was first recorded by Augustus Waller in 18879 and was later fully developed by Willem Einthoven in 1895.10 It is routinely used to detect cardiovascular diseases and abnormal heart rhythms. The rules used for these diagnoses are based on temporal changes in the signal. The ECG signal results from the activation of myocytes during different phases of the cardiac cycle. Since its discovery, the ECG has been used to record a number of physiologic and pathologic conditions, and with research and physician experience, the presence of specific features on the ECG tracing have been used to designate the presence or absence of specific biological conditions and disease states.11–14 We refer to the ECG features (such as ST-segment elevation and T-wave amplitude) as the ‘vocabulary’ for signal components fed into the model (i.e. the information the model uses to create its output), where the level of explanation depends on the volume and variety of features in the vocabulary. Some features are demonstrated in Figure 1. It is recognized that multiple medical conditions may affect any individual feature, and any individual condition usually impacts multiple features. For diagnosis, clinicians are trained to recognize the most salient features associated with a given condition, while other changes, due to their small magnitude or variability are ignored. Human-crafted models weigh-selected features to classify the absence or presence of a disease state, such as acute myocardial infarction, associated with the features of ST-segment elevation. It is not known whether a NN trained to detect the same condition from the same set of ECGs would use similar signal features (Figure 2A).
Figure 1.
Cardiac anatomy and the electrocardiogram signal. The heart has four chambers. The upper chambers (the atria) are activated by the signal reflected in the electrocardiogram as the P-wave. The lower chambers (the ventricles) are rapidly activated resulting in the QRS complex; the relaxation of the ventricles (repolarization) is represented by the smoother T-wave. A number of human-selected features, such as the peak amplitude of the various waves, the areas and widths of the different waves, deviation from baseline, and other morphological characteristics have a known biological mechanism and associations with specific pathologies.
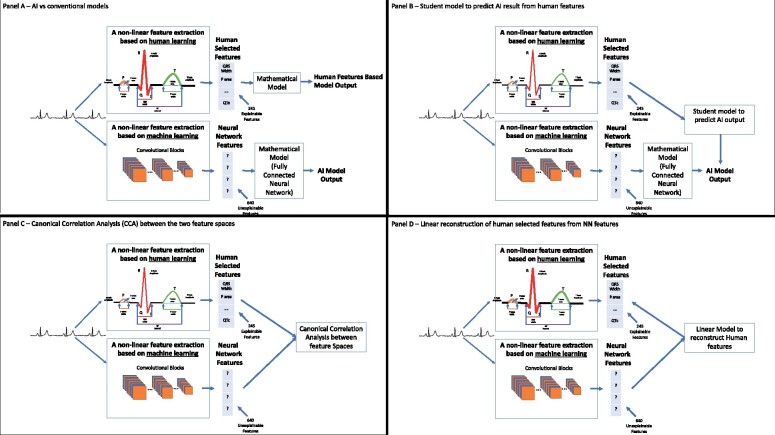
Figure 2.
Basic design of classifiers using human-selected or neural network-selected features, and an approach to quantify the relationship between these features. (A) Top: Human-selected features are directly used for classification, for example, features measuring the ST-elevation across leads can be used to classify electrocardiogram signals with or without cardiac injury; the term ‘mathematical model’ in this figure refers to a classifier. Bottom: A neural network uses convolutional layers to extract signal features, and then feeds those inscrutable features into the model. (B) Use of human-selected features in a student model to predict the neural networks output. The extent to which the student model predicts the neural network output is indicative of the extent to which human-selected features may be used by the neural network. (C) Use of canonical correlation analysis to assess the overall correlation between human-selected features and the features selected by the convolutional layers (feature extraction layers) of the neural network. (D) Use of a linear model to reconstruct single human-selected features from neural network features.
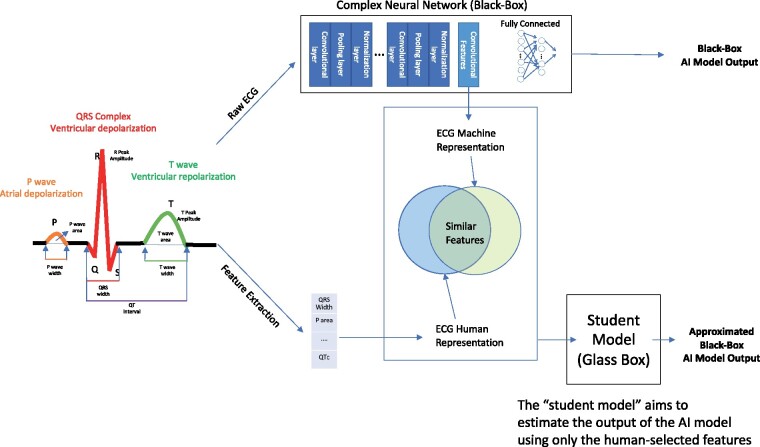
We hypothesized that convolutional NNs extract similar, linearly correlated signal features, to those identified by humans, including features that are hardly correlated with the expected output of the model. We further hypothesized that the human-recognizable features can be used to explain to some extent the output of the NNs and that the ability to explain the model will improve when using these human-recognizable features in a non-linear way. To test these hypotheses, we developed methods to extract NN features and applied quantitative methods to measure the correlations between these features and the human extracted features. We also further explained the output of the NNs with student models; such models aim to estimate the output by using only the human-selected features (see Graphical Abstract). All of these methods help determine the explainability of the NN in terms of human-selected features, and whether the network may find novel features that are not identified by humans.
Methods
The role of vocabulary and human explanation
We defined a reasonable explanation as the translation of the rules used by a model for output determination to a language that a human expert can understand and replicate. These rules are specific to the problem one tries to solve. In order to define an explainable model for understanding NNs for ECG processing, we identified the domain-specific vocabulary of human-selected features and basic methods for explainability and correlation. Figure 2 provides a conceptual diagram of the proposed scheme, whose details are explained below.
ECG: background and structure
The ECG is the recording of the heart’s electrical activity from the body’s surface. Each individual myocyte has a resting negative electrical potential relative to the outside of the cell membrane due to the distribution of ions across it.11 Highly regulated voltage changes, controlled by membrane ion-channels, permit individual myocytes to depolarize, allowing electrical signals to propagate across the myocardial syncytium, which through electrical–mechanical coupling result in coordinated mechanical contraction. Each myocyte then repolarizes (recovers its resting negative potential) in preparation for the impulse to follow. The ECG is the summation in space and time of all of the individual myocyte voltage changes and depicts the progression of electrical activation through the cardiac chambers (Figure 1). Since the progression of cardiac wave fronts occur in three-dimensional space, the recording acquired from any given skin electrode will reflect the projection of the electrical vector at that particular point in space, so that a given signal will have a different appearance when recorded from different sites. Conversely, recording from multiple surface locations permits characterization of the cardiac site or origin of a given impulse. In the conventional ECG, 12 leads are recorded. The electrical activity in each heartbeat is divided into 5 main temporal waves (features), the P, Q, R, S and T waves (Figure 1). The P-wave represents atrial depolarization, the Q, R and S waves (typically referred to as the QRS complex) represent ventricular depolarization, and the T-wave reflects ventricular repolarization.
Human-selected, explainable ECG features
When the ECG is acquired during normal rhythm, the morphology of each complex tends to have substantial homology among beats, so that an averaged beat is often used for morphologic feature extraction.15–17 The human-engineered process of feature extraction from ECG is non-trivial and non-linear. It entails selection of specific signal components (e.g. the ST-segment) which is useful if associated with specific conditions. For the present study, we used the human-defined features extracted and stored by the MUSE system. The system begins with the detection of each QRS complex in a segment and selection of a window of time around it, aligning the windows using a fiducial point in the QRS and averaging the complexes to a single representative beat. The features (Figure 1) are extracted by finding the onset and offset of each component and identifying human-selected characteristics such as areas, maximum amplitudes, slopes, durations, and so on for each constitutive element, creating a descriptive vocabulary for signal characteristics. The Muse system that we use includes a matrix of human-selected features that are automatically extracted from each lead in a 12-lead ECG.
Experimental setting
We used two previously described deep convolutional NN,18 which were trained to classify ECGs for two different tasks: classification of sex and estimation of age. Using these networks, we conducted experiments with ECG signals from the Mayo Clinic digital data vault collected between January 1994 and February 2017 with institutional review board approval. ECGs were randomly selected from all-comers including cardiac and non-cardiac patients; 57.4% were male and the mean age was 58.7 ± 15.7 years. The cohort used for these experiments was selected in a similar way to the cohorts used to train and validate the original models we sought to explain; however, the current cohort is independent of the latter ones.18 Among the 100 000 ECG signals, were used to train the student models (denoted as the student model training set) and were used to evaluate the student models (denoted as the student model testing set).
In the training of the previous age and sex models,18 each ECG signal was zero padded from 5000 × 12 (10 seconds sampled at 500 Hz) to 5120 × 12 (i.e. for each of the 12 leads, the padded signal length was 5120), and no additional inputs were used. For the sex classification problem, labels of patient sex were provided as binary variables (0/1 for female/male) and the predicted output for the testing data obtained values in [0,1] indicating the probability of being a male. For the age estimation problem, labels of patient ages between 18 and 100 were provided and the predicted output for the testing data obtained values in [18,100].
The architecture of the age convolutional NN and the sex convolutional NN was the same except for the final output layer’s activation [linear for age regression and SoftMax (binary classification) for sex]. In both networks, the first component is composed of convolutional blocks,19 which reduce the dimension of each 512012 signal to 640. This was the feature extraction component of the network (Figure 2). We thus defined the NN-selected features as the 640 outputs of the last convolutional layer. The next network component was the mathematical model; in this case, fully connected layers that received the 640 features selected by the convolutional layers and manipulated them to obtain the desired output (sex classification or age estimation, Figure 2A, bottom). Additionally, a total of 245 human-selected features derived from the median beat of each of the ECGs was extracted using the Muse database (Figure 2A, top). Some of the features were based on the morphology of a single lead and were extracted for each lead separately, but others, such as intervals (QT, RR, QRS) were calculated based on all 12 leads.20
We used the following notation, where for brevity, we did not distinguish between sex classification and age estimation, as their models are identical except for the final output layer’s activation:
were the student model training and testing matrices of NN features;
were the student model training and testing matrices of human-selected features; and
were the student model training and testing output of the NN with the trained parameters.
We used the NN outputs to train and test the student model and not the given labels since we sought to explain the NN output rather than create human features-based models.
Defining a student model and an explainability score
We used a secondary student model designed to predict the output of the NN using the human-selected features to explain the NN. For simplicity, we first considered a linear regression model. That is, we defined a vector and a real number and fit a standard least-squares linear regression model where is an vector of ones. The corresponding R2 statistic, which incorporated the testing data, was interpreted as the linear explainability score. It has values between 0 and 1, where 1 designates perfect linear explanation and 0 an irrelevant vocabulary for linear explanation. It was computed as follows
where for a vector and denote the mean and Euclidean norms, respectively.
We also used a non-linear model to explain the output using the human-selected features. This model used a fully connected network with two layers of 128 and 64 neurons and ReLU activation functions, followed by linear regression. The model was trained using a small set of hyperparameters and internally validated on a subset of the training data. Using matrices of parameters and , a vector of size and a scalar , the non-linear model was expressed as . We use the following statistic as the non-linear explainability score:
The difference between the non-linear and linear explainability scores quantified the improved performance of a non-linear versus a linear model (Figure 2B).
Canonical correlation between the feature spaces
We used canonical correlation analysis (CCA)21 to assess the overall correlation between the spaces of the human-selected and NN features (Figure 2C). CCA searches for linear transformations of the two sets of variables that maximize the cross-correlation between the transformed sets. In our case, we aimed to quantify the correlation between the rows of the matrices and that represent NN and human-selected features, respectively, and we pursued CCA as follows. We first subtracted from each row of each matrix the mean of all rows of that matrix, so the variables were centred. For , we sought matrices and of coefficients of linear transformations, with respective sizes and, such that and maximize the Frobenius norm of their cross-correlation matrix. The singular values of this maximal cross-correlation matrix are the canonical correlation coefficients. We computed them as follows. Let and be the matrices of left singular column vectors (arranged by descending order of singular values) of and , respectively. Then the canonical correlation coefficients are the singular values of the matrix . These numbers are between zero and 1, where higher numbers indicate higher correlation. Due to redundancies, one expects that many of these coefficients should be close to zero. However, existence of coefficients sufficiently large, where indicate a sufficiently close -dimensional subspaces of human-selected and NN features. In order to reliably assess the amount of shared information between the two feature spaces, we compared the number of pairs with a high correlation coefficient discovered by CCA to the reduced number of features obtained by principal component analysis22 that explained most of the variance.
Extraction of selected human features from neural network features
We tried to represent single human-selected features as linear combinations of NN features (Figure 2D). We identified the ith training and testing student model human-selected features with the ith rows of the matrices and , which we denote by and , respectively. We linearly regressed against the rows of . That is, we found a vector and a real number and fit a standard least-squares linear regression model where is an vector of ones. The corresponding R2 statistic, which incorporated the testing data, was interpreted as the linear explainability score. It has values between 0 and 1, where 1 designates perfect linear explanation and 0 an irrelevant vocabulary for linear explanation. It is computed as follows
For human-selected features that were extracted from each of the leads (e.g. T amplitude), we also tested the ability to reconstruct the averaged feature value across leads.
To verify that the network ability to reproduce the human features is not derived from a simple correlation between the human-selected features and the patient age and sex we calculated the corresponding statistics as well as the area under the curve (AUC) for detecting the patient’s sex using that single feature alone.
Final comment on methods
We did not report p-values, since they rely on strong model assumptions. Such models are not clear in our setting and we noted various obstacles in determining them. We thus preferred to use methods that do not rely on model assumptions, such as CCA and statistics. For the same reason, we avoided multiple testing.
Results
Using human features in a student model to explain neural network output
We predicted the output of the two NNs (age and sex) using human features via linear and non-linear student models. We quantified the variance information explained by these models via their statistic. For example, of value 1 means that we can explain 100% of the NN outputs using human features. For age estimation, the linear student model explained 57.1% of the variance (). A non-linear student NN with two layers explained 70.2% of the variance (). The difference between the two (13.1%) is evidence of the non-linear use of these features by the deep NN. In fact, the NN uses a similar non-linear model after its convolutional blocks.
For sex classification, the linear student model explained 49.4% of the variance (). The non-linear student model explained 68.5% of the variance (), where the difference between the non-linear and linear explainability (19.3%) was even greater. Indeed, a linear model is often less useful for a binary classification than continuous regression.
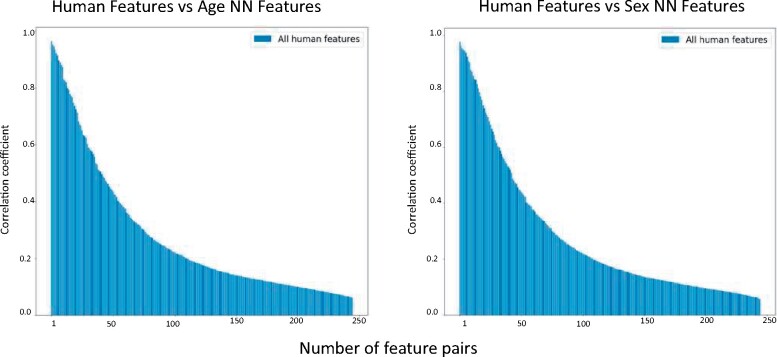
Using canonical correlation analysis to assess the overall correlation between the feature spaces
The canonical correlation coefficients for both sex classification and age estimation is shown in Figure 3. In the age model, 13 of the 245 feature pairs had canonical correlation coefficients of 0.85 or higher and 8 of those had a coefficient of 0.9 or higher. For the sex model, 15 of 245 of the feature pairs had canonical correlation coefficients of 0.85 or higher and 10 of those had coefficients of 0.9 or higher.
Figure 3.
Canonical correlation between human-selected and neural network-selected features. The canonical correlation analysis describes the correlation between the human-selected features and the age estimation neural network-selected features (left) and between the human-selected and neural network-selected features of the sex classification network (right). Each bar represents the canonical correlation coefficient between one pair of features from both spaces (neural network feature space and human-selected feature space).
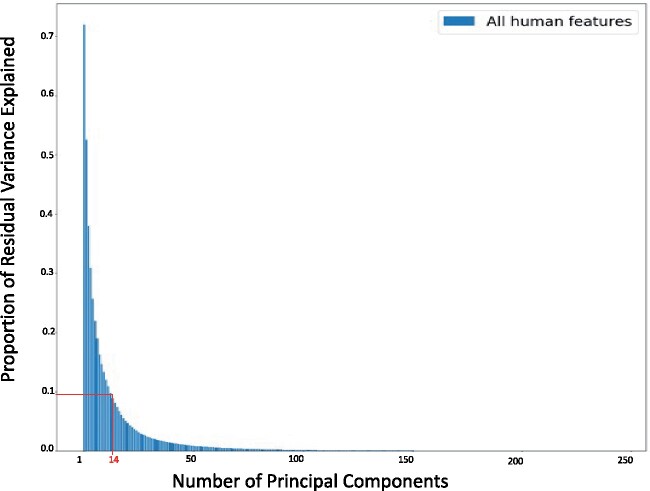
While 13 and 15 out of 245 may seem like a small number of pairs, it is important to note that human-selected features are linearly correlated to one another due to biological reasons. Indeed, Figure 4 depicts the proportion of residual variance explained as a function of principal components. It emphasizes that the first 14 principal components explain 90% of the human feature variance (see red lines in this figure).
Figure 4.
Proportion of residual variance of human features as a function of the number of principal components. Since the human features have inherent biological correlations, we used principal component analysis to quantify the number of unique features. As seen in the figure, 14 features explain 90% of the information in the human-selected feature space.
Human features extraction from the neural network features
To further understand the relationship between the two kinds of features, we created linear models to reconstruct single human-selected features from NN features . Table 1 reports statistics as a measure of variance explainability for human features in the two networks (sex or age). If the feature is computed for each lead separately, and not derived from all 12 leads, then the table reports the maximal value of the statistics from all leads and the statistics of the average feature value across leads. Supplementary material online further presents the statistics of all features including all leads, and the statistics between each human-selected feature and patient age and sex, as well as the AUC for detecting the patient’s sex using that single feature alone. The feature with the highest correlation with output and highest AUC was ‘Maximum R Amplitude’; its statistic for age estimation is 0.13 and its AUC for detection of sex is 0.68.
Table 1.
statistic as a measure of variance explainability for single human features in the two networks (sex or age)
| Feature name | Highest R2 among leads | Lead with the highest R2 | R 2 of average feature among leads |
|---|---|---|---|
| Based on the age network | |||
| Average RR interval | 0.835 | 0.835 | |
| Max R amplitude | 0.824 | avL | 0.800 |
| Max S amplitude | 0.805 | III | 0.746 |
| R-wave peak amplitude | 0.803 | I | 0.789 |
| QRS complex area | 0.777 | III | 0.778 |
| T-wave peak amplitude | 0.774 | I | 0.742 |
| T-wave area | 0.766 | I | 0.729 |
| T-wave full area | 0.763 | I | 0.725 |
| R-wave area | 0.756 | I | 0.727 |
| P-wave full area | 0.729 | avR | 0.662 |
| P-wave area | 0.725 | avR | 0.663 |
| QRS complex duration | 0.670 | 0.670 | |
| P-wave peak amplitude | 0.658 | avR | 0.496 |
| S-wave peak amplitude | 0.654 | avL | 0.616 |
| QT interval | 0.620 | 0.620 | |
| S-wave area | 0.596 | V5 | 0.634 |
| P-wave duration | 0.573 | avR | 0.589 |
| R-wave duration | 0.553 | avL | 0.469 |
| QTc interval (corrected by Bazzet) | 0.548 | 0.548 | |
| S-wave duration | 0.545 | V6 | 0.471 |
| Q-wave area | 0.443 | avR | 0.433 |
| PR interval | 0.429 | 0.429 | |
| Q peak amplitude | 0.412 | I | 0.363 |
| Q-wave duration | 0.411 | avR | 0.409 |
| T-wave duration | 0.248 | avR | 0.294 |
| Based on the sex network | |||
| Max R amplitude | 0.862 | V4 | 0.840 |
| Max S amplitude | 0.855 | avR | 0.759 |
| R-wave peak amplitude | 0.852 | V4 | 0.833 |
| R-wave area | 0.840 | V4 | 0.784 |
| Average RR interval | 0.818 | 0.818 | |
| T-wave peak amplitude | 0.794 | I | 0.809 |
| QRS complex area | 0.783 | V4 | 0.800 |
| T-wave full area | 0.780 | I | 0.775 |
| T-wave area | 0.779 | I | 0.777 |
| QRS complex duration | 0.753 | 0.753 | |
| P-wave full area | 0.679 | avR | 0.646 |
| P-wave area | 0.676 | avR | 0.643 |
| S-wave peak amplitude | 0.651 | V4 | 0.628 |
| P-wave peak amplitude | 0.606 | I | 0.509 |
| QT interval | 0.604 | 0.604 | |
| S-wave area | 0.590 | V4 | 0.633 |
| QTc interval (corrected by Bazzet) | 0.581 | 0.581 | |
| R-wave duration | 0.559 | V3 | 0.465 |
| P-wave duration | 0.523 | V4 | 0.541 |
| S-wave duration | 0.516 | V6 | 0.459 |
| Q-wave area | 0.414 | avR | 0.441 |
| Q-wave duration | 0.379 | avR | 0.398 |
| PR interval | 0.368 | 0.368 | |
| Q peak amplitude | 0.364 | I | 0.359 |
| T-wave duration | 0.255 | avR | 0.313 |
We report only the maximalvalues among leads andvalues of the averaged features across leads, since the human features across leads are correlated (values across leads are reported in the Supplementary material online). Features that were derived from all 12 leads together are present as is (clearly, for these features the third column does not assign anything and the second and fourth columans assign the same value). The features are sorted according to a descending maximal value.
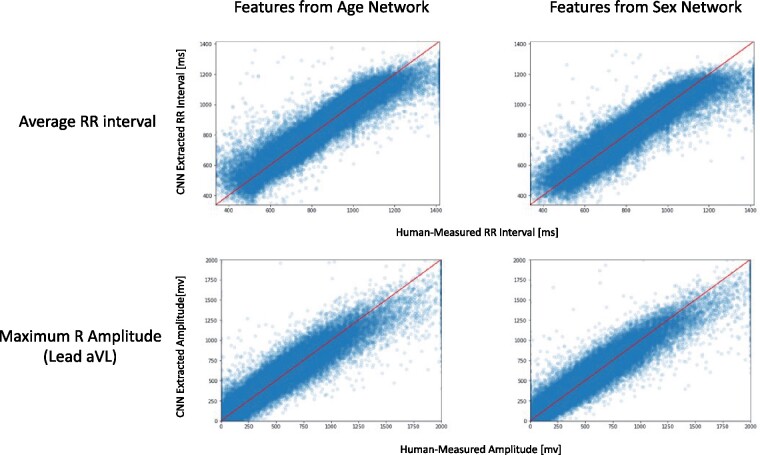
Figure 5 demonstrates the strong correlation between each feature value (depicted on the x axis) and its reconstruction from the NN using the linear regression model (depicted on its y axis) for two features (average RR interval and maximal R amplitude) in both networks. Interestingly, for the age network, the feature with the highest statistic was the patient heart rate (average RR interval) even though there is practically no correlation between the patients’ age and their heart rate ( < 0.001). In addition, even though the age and sex networks were trained separately, each with a different objective, and had different NN feature spaces, when extracting the human-selected features from the two different NN feature spaces, in both cases the same set of features had high values.
Figure 5.
Two examples of human-selected features that were reconstructed in a linear manner from the neural network feature space: age estimation neural network features (left) and sex classification neural network features (right). Even though the networks were trained separately, both networks possess a similar ability to reconstruct specific human identifiable features, which are non-linear in nature (average RR interval in the upper panels and maximum R-wave amplitude in the lower panels).
Discussion
In this work, we sought to determine whether the features selected by NNs designed for ECG analysis are human understandable features. We also tried to assess whether the difference between the classification capabilities of NNs and humans stem from the use of different signal features, the non-linear nature of NNs, or both. We summarize our findings as follows: (i) NNs for ECG signals predominantly use features that are correlated with human understandable features; (ii) human-selected features, however, explain only part of the NN model output. For sex classification, we found a 70.2% variance explanation with a non-linear model and for age estimation, it was 68.5%. Thus, identification of novel features (signal components not part of the current vocabulary used to describe ECG signals) by the network seems to contribute to the superior performance of NNs; (iii) the non-linear nature of NNs also contributes to their superior performance. Indeed, the linear student models for both age estimation and sex classification were able to explain less than the non-linear student models. In summary, NNs predominantly use human-recognizable features, but then add additional non-human labelled features and non-linearity, accounting for their superior performance compared to traditional methods. Additionally, as the NN features were extracted without any specific feature engineering, errors in human feature creation may be eliminated and extraction time significantly shortened, as it does not involve manual review of each tracing. On the other hand, there is a voluminous body of literature describing methods to optimally extract-selected ECG features, such as ‘QRS Width’, ‘T Wave Area’, and ‘QT Interval’.15–17
The demonstrated ability to derive known ECG features with biological meaning from NN features in a linear way may mean that these features are not unique to human intelligence. Indeed, two different NNs (age and sex classifiers) seem to utilize the same human-selected features without any a priori knowledge of what an ECG signal should look like, including the detection of features that are uncorrelated with the model labels. For example, the age estimation model demonstrated strong ability to estimate the ECG heart rate from the NN features (= 0.835) with almost no correlation between the patient age and their heart rate ( = 0.0009). This supports the hypothesis that some of the NN features are natural in ECGs and are not specific to the outcome the network is trained to detect. Not all human-identified features were used by the NNs. This might be considered a limitation, but we believe it is another sign that each network underwent a meaningful learning process resulting in the selection of features that have a direct association with the classification task it was assigned.
Furthermore, we were not able to perfectly explain the output of the model using the vocabulary of human-selected features, that is, the score was less than 1. There are three potential explanations for this finding. The first is that the NN found features that reflect components of the signals not defined by most humans, including features that are often described as ‘gestalt’.23 These almost invisible features that appear to expert physicians might be hard to explain using any natural language and hard coded rules. The second is that the vocabulary used by humans to describe signal features is somehow ambiguous and the definitions of some feature elements lack sufficient accuracy to provide robust classification. The last is that the network found false associations, for example, a feature that was present in the training set but was not generalizable or relevant for common instances. Such features represent a bias in the training set and might be exploited to permit a simple adversarial attack. To improve explainability in such cases one may apply adversarial training and possibly noise injection.24,25
While our work is focused on ECG analysis, and ECG-based features, we present a general framework to extract and compare NN features and human-selected features. In particular, we suggest student models and simple quantitative methods of correlating and explaining human-selected features using NN features. We thus expect our methods to apply to other fields, where human-engineered features exist.
We developed our framework using ECGs for several reasons. First, the use of NNs to classify ECGs is well established due to the availability of large, well-annotated digital data sets. Second, these networks have achieved human expert level capabilities with regards to reading ECG rhythms and have superseded humans in detecting a number of otherwise occult pathologies such as left ventricular dysfunction, hypertrophic cardiomyopathy, and subject age and sex based on the ECG alone. These are tasks humans are incapable of, and understanding how these networks accomplish them might yield new medical knowledge. And lastly, ECG analysis has been performed for many years resulting in a very rich, biologically meaningful vocabulary of features that is carefully recorded. As the mechanism behind the features in the vocabulary is known, translating the NN rules to these human features provides a direct link to the biology that drives the NN decision.
Understanding human-selected features that artificial intelligence (AI) models are looking at is important for the adoption of the technology in clinical medicine. Given the high stakes, the potential for novel or unexpected recommendations, the risk of implicit bias and false associations, and the possibility of legal liability, clinicians may be hesitant to respond to medical diagnoses or therapies proposed by NNs without a general understanding of the specific features or characteristics they process. The ability to explain predictive AI models may enhance the ability to improve their performance and to predict appropriate use cases for their adoption. Furthermore, as much as AI models may identify novel signal components in creating their classifications, new insights may be derived regarding the signal and its association with health and disease, leading to fundamentally novel insights into disease pathogenesis.
Supplementary material
Supplementary material is available at European Heart Journal – Digital Health online.
Funding
The corresponding author was partially supported by the National Science Foundation (DMS-1830418).
Conflict of interest: none declared.
Data availability
All requests for raw and analyzed data and related materials will be reviewed by the Mayo Clinic legal department and Mayo Clinic Ventures to verify whether the request is subject to any intellectual property or confidentiality obligations. Requests for patient-related data not included in the paper will not be considered. Any data and materials that can be shared will be released via a Material Transfer Agreement.
Supplementary Material
References
- 1. LeCun Y, Boser B, Denker JS, Henderson D, Howard RE, Hubbard W, and Jackel LD. Backpropagation applied to handwritten zip code recognition. Neural Comput 1989; 1:541–551. [Google Scholar]
- 2. LeCun Y, Huang FJ, Bottou L.. Learning methods for generic object recognition with invariance to pose and lighting. In: Proceedings of the 2004 IEEE Computer Society Conference on Computer Vision and Pattern Recognition. Washington, DC, USA: IEEE Computer Society; 2004, pp. 97–104. [Google Scholar]
- 3.Attia ZI, et al. Screening for cardiac contractile dysfunction using an artificial intelligence–enabled electrocardiogram. Nat Med 2019;25:70–74. [DOI] [PubMed] [Google Scholar]
- 4. Poplin R, Poplin R, Varadarajan AV, Blumer K, Liu Y, McConnell MV, Corrado GS, Peng L and Webster DRet al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng 2018; 2:158. [DOI] [PubMed] [Google Scholar]
- 5. Zech JR, Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ and Oermann EKet al. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med 2018;15:e1002683–e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Narodytska N, Kasiviswanathan S. Simple black-box adversarial attacks on deep neural networks. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW). 2017, pp. 1310–1318.
- 7. Lecun Y, Bottou L, Bengio Y, Haffner P.. Gradient-based learning applied to document recognition. Proc IEEE 1998; 86:2278–2324. [Google Scholar]
- 8.Han X, Hu Y, Foschini L, Chinitz L, Jankelson L and Ranganath R Deep learning models for electrocardiograms are susceptible to adversarial attack. Nat Med 2020;26:360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barold SS. Willem Einthoven and the birth of clinical electrocardiography a hundred years ago. Card Electrophysiol Rev 2003;7:99–104. [DOI] [PubMed] [Google Scholar]
- 10. Fisch C. Evolution of the clinical electrocardiogram. Card Electrophysiol Rev 2003;7: 99–104.12766530 [Google Scholar]
- 11. Becker DE. Fundamentals of electrocardiography interpretation. Anesth Prog 2006;53:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robb GP, Marks HH.. Postexercise electrocardiogram in arteriosclerotic heart disease: its value in diagnosis and prognosis. JAMA 1967;200:918–926. [PubMed] [Google Scholar]
- 13. Wellens HJ, Bär FW, Lie K.. The value of the electrocardiogram in the differential diagnosis of a tachycardia with a widened QRS complex. Am J Med 1978, 64: 27–33. [DOI] [PubMed] [Google Scholar]
- 14. Blackburn H., Keys A., Simonson E., Rautaharju P., Punsar S.. The electrocardiogram in population studies: a classification system. Circulation 1960;21:1160–1175. [DOI] [PubMed] [Google Scholar]
- 15. Attia ZI, DeSimone CV, Dillon JJ, Sapir Y, Somers VK, Dugan JL, Bruce CJ, Ackerman MJ, Asirvatham SJ, Striemer BL, Bukartyk J, Scott CG, Bennet KE, Ladewig DJ, Gilles EJ, Sadot D, Geva AB and Friedman PA. et al. Novel bloodless potassium determination using a signal-processed single-lead ECG. J Am Heart Assoc 2016;5:e002746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jesus S, Rix H.. High resolution ECG analysis by an improved signal averaging method and comparison with a beat-to-beat approach. J Biomed Eng 1988;10:25–32. [DOI] [PubMed] [Google Scholar]
- 17. Karpagachelvi S, Arthanari M, Sivakumar M. ECG Feature Extraction Techniques - A Survey Approach. Int J Comput Sci Inf Secur 2010;8: 76–80. [Google Scholar]
- 18. Attia ZI, et al. Age and sex estimation using artificial intelligence from standard 12-lead ECGs. Circ Arrhythm Electrophysiol 2019;12:e007284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lawrence S, Giles CL, Tsoi AC, Back AD.. Face recognition: a convolutional neural-network approach. IEEE Trans Neural Netw 1997;8:98–113. [DOI] [PubMed] [Google Scholar]
- 20. Garson A. How to measure the QT interval—what is normal? Am J Cardiol 1993;72:B14–B16. [DOI] [PubMed] [Google Scholar]
- 21. Stewart D, Love W.. A general canonical correlation index. Psychol Bull 1968;70:160. [DOI] [PubMed] [Google Scholar]
- 22. Wold S, Esbensen K, Geladi P.. Principal component analysis. Chemometr Intell Lab Syst 1987;2:37–52. [Google Scholar]
- 23. deSouza IS, Sinert R.. Is experienced physician gestalt with an electrocardiogram sufficient to accurately exclude acute myocardial infarction in a patient with suspected acute coronary syndrome? Acad Emerg Med 2020;27:83–84. [DOI] [PubMed] [Google Scholar]
- 24. Wang B, Yuan B, Shi Z, Osher S.. EnResNet: ResNet ensemble via the Feynman-Kac formalism to improve deep neural network robustness. Adv Neural Inf Process Syst 2019;32. [Google Scholar]
- 25. Rakin AS, He Z, Fan D. Parametric noise injection: trainable randomness to improve deep neural network robustness against adversarial attack. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). 2019, pp. 588–597.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All requests for raw and analyzed data and related materials will be reviewed by the Mayo Clinic legal department and Mayo Clinic Ventures to verify whether the request is subject to any intellectual property or confidentiality obligations. Requests for patient-related data not included in the paper will not be considered. Any data and materials that can be shared will be released via a Material Transfer Agreement.