Abstract
Aims
Determining the aetiology of left ventricular hypertrophy (LVH) can be challenging due to the similarity in clinical presentation and cardiac morphological features of diverse causes of disease. In particular, distinguishing individuals with hypertrophic cardiomyopathy (HCM) from the much larger set of individuals with manifest or occult hypertension (HTN) is of major importance for family screening and the prevention of sudden death. We hypothesized that an artificial intelligence method based joint interpretation of 12-lead electrocardiograms and echocardiogram videos could augment physician interpretation.
Methods and results
We chose not to train on proximate data labels such as physician over-reads of ECGs or echocardiograms but instead took advantage of electronic health record derived clinical blood pressure measurements and diagnostic consensus (often including molecular testing) among physicians in an HCM centre of excellence. Using more than 18 000 combined instances of electrocardiograms and echocardiograms from 2728 patients, we developed LVH-fusion. On held-out test data, LVH-fusion achieved an F1-score of 0.71 in predicting HCM, and 0.96 in predicting HTN. In head-to-head comparison with human readers LVH-fusion had higher sensitivity and specificity rates than its human counterparts. Finally, we use explainability techniques to investigate local and global features that positively and negatively impact LVH-fusion prediction estimates providing confirmation from unsupervised analysis the diagnostic power of lateral T-wave inversion on the ECG and proximal septal hypertrophy on the echocardiogram for HCM.
Conclusion
These results show that deep learning can provide effective physician augmentation in the face of a common diagnostic dilemma with far reaching implications for the prevention of sudden cardiac death.
Keywords: Artificial Intelligence, Multimodal data, Electrocardiogram, Echocardiogram, Hypertrophic cardiomyopathy, Hypertension
Graphical Abstract
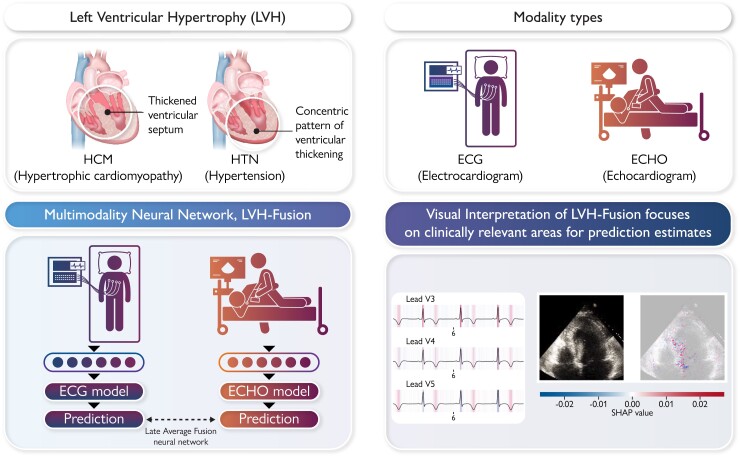
Graphical Abstract.
We present LVH-fusion, a multi-modal deep learning method to jointly model electrical (ECG) and ultrasound-based (ECHO) time series data of the heart. We demonstrate its potential with application to the diagnosis of left ventricular hypertrophy. Furthermore, we explored explainability techniques to investigate local and global features that positively/negatively impact predictions to provide actionable insight to model estimates.
Introduction
Hypertrophic cardiomyopathy (HCM) is the most common cardiac genetic disease with an estimated prevalence in the general population of 1:500 to 1:200.1 Hypertrophic cardiomyopathy is an autosomal dominant mendelian disease that can be associated with significant morbidity in the form of heart failure and sudden death.2 Thus, identifying patients with HCM has significance well beyond the individual, with many proband diagnoses leading to screening of several generations of a family. Diagnosis of HCM can be difficult due to the high prevalence of manifest hypertension in the general population, present in up to 45% of US adults3 (this before counting the occult disease). Thus, a common diagnostic dilemma for clinicians when faced with left ventricular hypertrophy (LVH) on the ECG or echocardiogram is how to rule out HCM. In a small study, the rates of misclassification of HCM were as high as 30% with hypertension being the most common misdiagnosis.4 Although the American Heart Association provides guidelines for the diagnosis of hypertension and HCM separately, distinguishing between them is a task that most physicians may feel ill equipped to perform. This provides an opportunity for physician augmentation through artificial intelligence (AI).
New advances in AI have led to rapid expansion of medical deep learning applications with an emphasis on medical specialties that hold a high degree of visual pattern recognition tasks such as radiology, pathology, ophthalmology, dermatology, and most notably cardiology.5 Imaging and electrical phenotypes of HCM6,7 are the first line clinical tools.
Interpretation of the ECG relies on direct visual assessment making it ideal for deep learning approaches. Previous work has demonstrated that demographic and medical data can be learned including detection of low ejection fraction, something typically requiring echocardiography to confirm.8–11 Our prior work using video computation of echocardiograms has demonstrated efficient detection of left ventricular hypertrophy and the identification of a broad range of cardiovascular disease.12,13
Combining data sources as human diagnosticians do, has the potential to provide an AI algorithm with greater diagnostic power.14 We focus here on the two most frequent diagnostic modalities in cardiology. To date, no published work has explored the benefits of a multimodal deep learning model using electrocardiogram and echocardiogram data, although there has been some exploration of combining separately trained diagnostic models in a single pipeline.15,16 We hypothesize that multimodal deep learning may provide added benefit in distinguishing patterns that are not easily discernible from individual modalities. We present LVH-fusion, the first model to jointly model electrical and ultrasound-based time series data of the heart. We demonstrate its potential with application to the diagnosis of left ventricular hypertrophy.
Methods
Data acquisition and study population
The HCM cohort was derived from patients with diagnosed HCM and followed at the Stanford Center for Inherited Cardiovascular Disease. Patients were diagnosed with HCM with consideration of the 2020 ACC/AHA guideline for HCM17 and inclusive of multimodality imaging, family history, and genetic screening. The definition of hypertension was based on the 2017 ACC/AHA guidelines,18 SBP >130 mmHg and DBP >89 mmHg based on an average of ≥ 2 readings, on ≥ 2 occasions at least 2 weeks apart. The hypertension cohort was derived from patients selected based on medical record evidence of at least five separate, consecutive outpatient systolic blood pressure readings >150 mmHg, at least 2 weeks apart. Exclusion criteria included any ECG clinical annotations of ventricular-pacing, left bundle branch block, and patients who had concurrent presence of both HCM and hypertension. In addition, we excluded any data from both electrocardiograms and echocardiograms data sets if the date acquired was after a documented myectomy procedure.
We retrieved 15 761 electrocardiograms (ECGs) and 3234 transthoracic echocardiograms from 2728 unique individuals at Stanford Health Care. Standard 12-lead ECGs were divided into training, validation, and test partitions based on a unique patient identification number to ensure that no patient overlap existed across data partitions. Echocardiogram videos from Stanford Medicine were curated for apical four-chamber view videos.
Data processing and selection
Electrocardiogram signals were filtered to remove any baseline wander and powerline interference. Normalization of 12-lead ECGs was performed by lead over a random subset of the study sample population, using mean and standard deviation. Echocardiogram videos were processed identical fashion as Oyuang et al.13 Single apical-4-chamber 2D greyscale videos were identified by unique patient identifiers. Preprocessing of echocardiogram videos to standard resolution and removal of identifiable information outside of the ultrasound sector such as text, ECG and respirometer data was removed according to previously described methods.13 Given multiple electrocardiograms and echocardiograms per individual present within our dataset, we examined the effects of different data selection methods on model training and performance metrics. We selected three different data selection methods to understand the impacts of incorporating different timepoints into model training and evaluation; (i) first clinical presentation for all data partitions, (ii) all clinical presentations in the training partition with only first clinical presentation selected for the validation and test partitions, and (iii) all clinical presentations for all partitions. Extended details of each selection method can be found in Supplementary material online, Table S1.
Overview of model training framework
Training for the single-modal and multimodal neural network models were executed independently.
Models were trained using a two-stage grid search approach to find the optimal hyperparameters. In the initial hyperparameter search, evaluation metrics from the validation set can be found in the Supplementary material online, Tables S2 and S3. The hyperparameters that yielded the best-performing models were selected for additional training and hyperparameter search considering various loss functions, loss weighting for minority class and minority class oversampling. Final models were selected from the lowest validation loss.
Single-modal model training
For electrocardiogram single-modal model training, the following hyperparameters included: model architecture: {VGG11, VGG13, VGG16, VGG19, densenet169, densenet121, densenet201, densenet161 resnet18, resnet34, resnet50, resnet101, resnet152, resnext50_32 × 4d, resnext101_32 × 8d, wide_resnet50_2 wide_resnet101_2}; batch size: {32, 64, 75}; Optimizer: {SGD, Adam}, and Hz: {500, 250}. The first hyperparameter search involved training all combinations of hyperparameters above for 100 epochs and saving results from the epoch with the lowest loss. Furthermore, we explored a second hyperparameter search which explored class weighted loss functions, oversampling minority class samples and setting final bias term to the expected class ratios from top performing models from the initial hyperparameters search. We examined expanding training to 150 epochs and considering both loss and auPRC results for selection of the final model. The selected hyperparameters that resulted in best performance on the validation set were the following: ResNet 34 model, oversampling minority class, Adam optimizer, batch size of 64, and sampling rate of 500.
For echocardiogram unimodal model training, the following hyperparameters included: Model architecture: {r2plus1d_18, mc3_18, r3d_18}, Number of frames: {96, 64, 32, 16, 8, 4, 1}; Period: {2, 4}; Pre-trained weights: {True, False}. For pre-trained models, weights trained on the Kinetics-400 data set were used.19 The first hyperparameter search involved training all combinations of hyperparameters above for 100 epochs and saving results from the epoch with the lowest loss. Furthermore, we explored a second hyperparameter search which explored class weighted loss functions, oversampling minority class samples and setting final bias term to the expected class ratios from top performing models from the initial hyperparameters search. We examined expanding training to 300 epochs and considering both loss and auPRC results for selection of the final model. The selected hyperparameters that resulted in best performance on the validation set were the following: r2plus1d_18 model, pre-trained weights, weighted minority class, Adam optimizer, batch size of 20, and frames 16 with sampling period of 4.
Multimodal model training
For multimodal training models, the electrocardiogram and echocardiogram data were paired according to unique patient identifiers. Data selection for the earliest clinical encounter was selected for all training, validation, and test set partitions; this resulted in a total of 1414 training, 176 validation, and 168 internal test samples. The detailed demographic characteristics of the data set can be found in Table 1. We hypothesized that using the learned weights from the trained single-modal models would benefit training so we explored both pre-trained late fusion and random late fusion models. All multimodal models were trained to 300 epochs, and we considered both loss and auPRC results for selection of the final multimodal model. We implemented LVH-Fusion using PyTorch on the Stanford University Research cluster, Sherlock. The selected hyperparameters that resulted in best performance on the validation set were the following: r2plus1d_18 model + ResNet 34, pre-trained weights, weighted minority class, Adam optimizer, batch size of 10, and frames 16 with sampling period of 4.
Table 1.
Demographics of hypertrophic cardiomyopathy subjects
| Sex | Race/ethnicity | % of total | % of sex |
|---|---|---|---|
| Female | American Indian or Native American or Alaskan Native | 0.264% | 0.654% |
| Asian/Asian-American | 5.898% | 14.597% | |
| Black/African-American | 1.144% | 2.832% | |
| Latina/Latino/Hispanic | 3.609% | 8.932% | |
| Middle Eastern | 0.264% | 0.654% | |
| Native Hawaiian or other pacific islander | 0.880% | 2.179% | |
| Other | 4.313% | 10.675% | |
| South Asian-Indian/Pakistani/Bangladeshi | 0.880% | 2.179% | |
| White/European-American | 23.151% | 57.298% | |
| Male | American Indian or Native American or Alaskan Native | 0.264% | 0.443% |
| Asian/Asian-American | 5.370% | 9.010% | |
| Black/African-American | 3.081% | 5.170% | |
| Latina/Latino/Hispanic | 4.225% | 7.090% | |
| Middle Eastern | 0.704% | 1.182% | |
| Native Hawaiian or other pacific islander | 1.232% | 2.068% | |
| Other | 9.067% | 15.214% | |
| South Asian-Indian/Pakistani/Bangladeshi | 1.144% | 1.920% | |
| White/European-American | 34.507% | 57.903% |
Comparison to feature-based models
Standard reported features from TraceMaster electrocardiogram machines were extracted for each ECG considered in this study. We used these features for input into a XGboost model to determine if a feature-based method would exceed the performance metrics of the unimodal neural network models, Supplementary material online, Table S4. The list of ECG features used were modelled from Kwon et al. 2020.10
Comparison with normal samples
In order to explore how our neural networks perform on non-left ventricular hypertrophy individuals, we sampled electrocardiograms with clinical annotations of sinus rhythm and echocardiograms with a normal ejection fraction >45. We took the best-performing single-modal model and retrained them to include an additional non-LVH class; details of sample size and performance metrics can be found in Supplementary material online, Table S5 and Supplementary material online, Table S6, respectively.
Ablation experiments
To further understand how the neural networks make their predictions, we explored various ablation studies.
We retrained the single-modal echo model with data ablated in the following ways:
a single randomly selected frame of each echo, repeated for the length of the original video to compare with the best-performing unimodal model.
The end-diastolic frame from each echo, repeated for the length of the original video to fairly compare with the best-performing unimodal model. The end-diastolic frame was identified by a trained sonographer from EchoNet-dynamic.13
Using the estimated left ventricular segmentation from EchoNet-dynamic,13 we set all pixels to zero except a segmented box around the left ventricle.
For electrocardiogram, we retrained the single-modal models for the following experiments:
Using eight of the 12 leads, to compare with the best-performing unimodal model.
Masking out each lead independently to compare with the best-performing single-modal model and understand impacts each lead holds on performance.
Echocardiogram models were trained to 300 epochs and electrocardiogram models were trained for 150 epochs.
SHAP interpretation experiments
SHAP GradientExplainer20 uses an extension of integrated gradient values and SHAP values, which aims to attribute an importance value to each input feature by integrating the gradients of all interpolations between a foreground sample (test samples) and a provided background samples (training data). The importance scores sum up to approximately the difference between the expected value of all background samples and the individual prediction estimate of interest. We applied this method to both ECG and echocardiogram models; 1500 samples were used to build the background distribution for the ECG model and 80 samples were used to build the background distribution for the echocardiogram model. In both cases, the full test set was used as foreground samples.
Results
We developed a multimodal deep learning framework, LVH-fusion, that takes as input time-based electrical and echocardiographic data of the heart. We applied this framework in a common clinical challenge: the determination of the aetiology of left ventricular hypertrophy. Motivated by prior work on deep learning applied to electrocardiogram signals and echocardiogram videos,9,13,21 LVH-fusion jointly models both electrocardiogram and echocardiogram data. It is trained not with proximate human-derived ECG and echocardiogram labels but rather via a gold standard diagnosis independently derived from the Electronic Health Records (HTN) or through the consensus diagnosis of HCM within a centre of excellence.
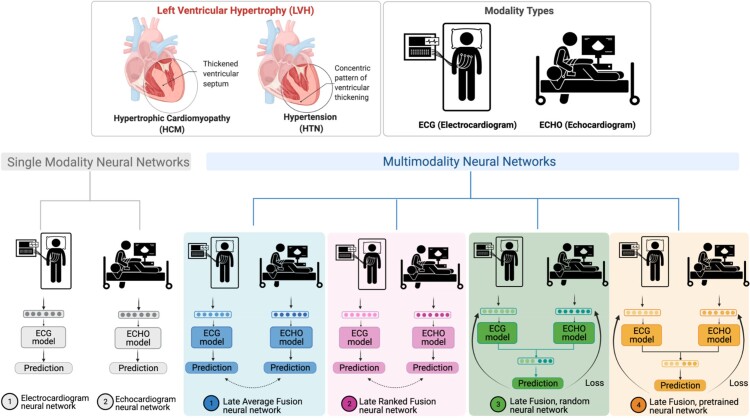
In this study, both single-modal and multimodal neural network models were examined (Figure 1). Four different multimodal fusion architectures were explored, combining ECG and echocardiogram information in different ways. For both late-average fusion and late-ranked fusion models, decision level fusion was used to combine the outputs of electrocardiogram and echocardiogram classifiers.22 In the late-average fusion model, soft voting is performed by computing the average probability for each class from the individual ECG and echocardiogram classifiers and predicts the class with maximal average probability. In the late-ranked fusion model, the probabilities for each class from the individual ECG and echocardiogram classifiers are ranked and a prediction is determined from the highest ranked probability. For the late fusion models, both pre-trained and random, the learned feature representations from each modality were concatenated together before the final classification layer. In this situation the fusion model considers both inputs and during training and the loss is calculated jointly. We explored the benefits of randomly initialized weights and pre-trained weights in the late fusion model. Finally, the single-modal models provide a benchmark against which to compare multimodal models that jointly consider the paired electrocardiogram and echocardiogram data, demonstrating the benefit of a combined approach.
Figure 1.
LVH-fusion study design.
Data acquisition and selection
With the approval of Stanford Institutional Review Board, we retrieved electrocardiograms and echocardiograms from patients between 2006 and 2018 at Stanford Medicine. The data were split into training, validation, and test sets with no patient overlap between sets. Owing to the fact that multiple electrocardiograms and echocardiograms are present within the healthcare system record, we explored various data selection scenarios to understand what selection methods are best suited for this specific task. The quantitative comparison of all data selection used can be found in Supplementary material online, Table S1. The final model was trained using a patient’s first ECG and first echocardiogram in the system.
Model performance
Four multimodal fusion models were explored: late-average, late-ranked, pre-trained late fusion, and random late fusion (Figure 1). The performance metrics of each model is detailed in Table 2.
Table 2.
Performance of LVH-fusion compared with alternative deep learning architectures
| Models | auROC | auPRC | F1-score | Sensitivity | Specificity | Precision | NPV | FPV | FNR | FDR |
|---|---|---|---|---|---|---|---|---|---|---|
| Multimodal: late-averaged fusion (LVH-fusion) | 0.92 (0.862–0.965) | 0.80 (0.665–0.907) | 0.73 (0.585–0.842) | 0.73 (0.562–0.882) | 0.96 (0.929–0.985) | 0.73 (0.562–0.882) | 0.96 (0.929–0.985) | 0.04 (0.014–0.071) | 0.27 (0.120–0.438) | 0.27 (0.118–0.440) |
| Multimodal: late-ranked fusion | 0.92 (0.870–0.961) | 0.76 (0.628–0.878) | 0.49 (0.361–0.598) | 0.82 (0.667–0.950) | 0.76 (0.701–0.818) | 0.35 (0.239–0.458) | 0.96 (0.933–0.991) | 0.24 (0.181–0.301) | 0.18 (0.050–0.323) | 0.65 (0.542–0.760) |
| Multimodal: late fusion random | 0.89 (0.832–0.942) | 0.64 (0.481–0.803) | 0.56 (0.409–0.679) | 0.68 (0.500–0.846) | 0.88 (0.838–0.925) | 0.47 (0.323–0.615) | 0.95 (0.915–0.978) | 0.12 (0.075–0.162) | 0.32 (0.158–0.481) | 0.53 (0.385–0.677) |
| Multimodal: late fusion pre-trained | 0.89 (0.829–0.944) | 0.62 (0.461–0.779) | 0.45 (0.333–0.558) | 0.86 (0.737–0.966) | 0.71 (0.642–0.768) | 0.31 (0.213–0.407) | 0.97 (0.943–0.992) | 0.29 (0.233–0.357) | 0.14 (0.032–0.267) | 0.69 (0.594–0.789) |
| Single-modal: ECG | 0.87 (0.785–0.937) | 0.62 (0.461–0.776) | 0.51 (0.367–0.633) | 0.68 (0.500–0.842) | 0.85 (0.793–0.894) | 0.41 (0.273–0.543) | 0.94 (0.910–0.976) | 0.15 (0.107–0.207) | 0.32 (0.154–0.483) | 0.59 (0.462–0.730) |
| Single-modal: Echocardiogram | 0.88 (0.803–0.941) | 0.70 (0.549–0.833) | 0.63 (0.486–0.746) | 0.73 (0.562–0.882) | 0.91 (0.868–0.946) | 0.55 (0.400–0.706) | 0.96 (0.924–0.984) | 0.09 (0.053–0.132) | 0.27 (0.118–0.435) | 0.45 (0.292–0.607) |
The late-average model achieved the highest F1-score and specificity rates 0.73 and 0.96, respectively, on the held-out test set. We conducted experiments to study the performance of single-modal models trained on only ECG and echocardiogram to demonstrate the benefit of multimodal models. The multimodal models outperform single-modal model F1-scores, which increase from 0.51 to 0.73. Furthermore, the false-discovery rates are significantly reduced from 0.59 to 0.27. To provide context for these results, we also trained the single-modal models to predict left ventricular aetiology using standard quantitative features from the electrocardiogram. This baseline model achieved sensitivity rates of 0.50 for predicting HCM which is considerably lower than LVH-fusion (see Supplementary material online, Table S4). These results show that the proposed electrocardiogram signals model discover novel characteristics not accounted for with the quantitative features. Finally, to examine the discriminatory power of our methodology, we performed a sensitivity analysis for predicting LVH aetiology including the additional classification task of ‘normal.’ In this context, LVH-fusion maintains high discriminatory power in predicting LVH from normal ECG and echocardiogram videos, suggesting that false positive rates of hypertension or HCM would be low if the model was extended to this use case (see Supplementary material online, Table S5 and S6).
Understanding model performance
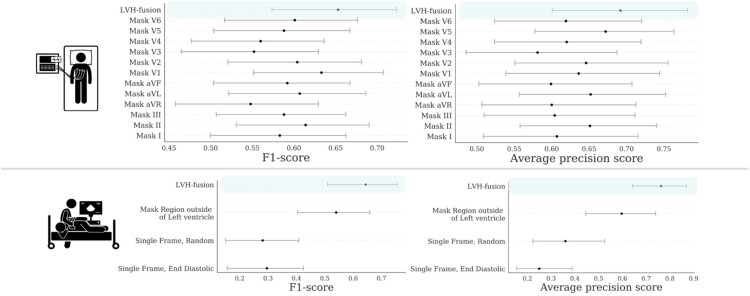
In order to improve our understanding of how LVH-fusion classifies left ventricular aetiology, we implemented a series of ablation studies similar to Hughes et al.23 to determine what information models rely on to make predictions. For electrocardiogram single-modal models we examined the impact of varying the number of leads from the standard 12 leads to 8 leads, and masking each lead to understand the impact each lead holds for prediction estimates. We find that although no single-lead harbours a statistically significant impact on the overall model performance, masking out lead V3 and aVR had the highest negative impact on prediction estimates, Figure 2. Next, since the standard 12-lead ECG contains eight algebraically independent leads, we considered the impact of masking multiple leads combinations. We observe an overall reduction in classification metrics when masking multiple leads at a time with no significant difference between masking the four dependent leads (III, aVL, aVF, aVR) and a random subselection of four leads, Supplementary material online, Figure S1. These results suggest LVH-fusion benefits from the complete 12-lead input and classification metrics are negatively impacted with any non-specific reduction in leads.
Figure 2.
Ablation studies impact on model performance. Bootstrap 95% CI for performance metrics, F1-score and average precision score, for each model trained on ablated input data. for each prediction metric is shown. (TOP row) Results from ablating ECG input. (BOTTOM row) Results from ablating echocardiogram input. For each ablation setting, a separate model was trained on that type of ablated data to quantify the information content in the data.
For the echocardiogram single-modal model, we examined segmentation, restricting the prediction algorithm to (i) only the region around the left ventricle, (ii) random single frames, and (iii) single end-diastolic frames. Restricting the echocardiogram model to the area around the left ventricle caused a decrease in accuracy, showing the model relies on information outside of that region to make classifications. This is interesting given the focus of clinicians on the left ventricle when considering LVH, even despite the fact that hypertension could impact the left atrium by causing restriction and HCM affects all four chambers. Restricting the model’s input to a single frame further decreases accuracy, demonstrating that motion information is important in distinguishing between HCM and hypertension. Figure 2 details the performance of each ablation experiment.
Model interpretations
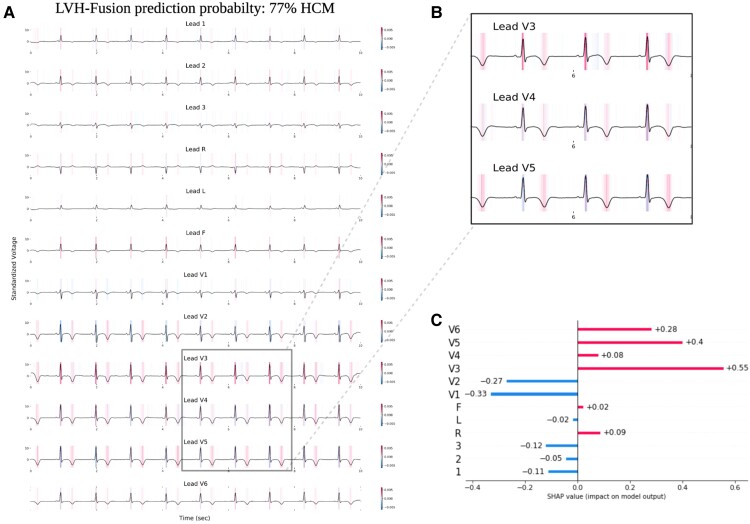
To improve our understanding of how LVH-fusion classifies left ventricular aetiology, we implemented SHAP GradientExplainer, a game theory approach to explain the output of a machine learning algorithm.20 Relating this method to the ECG model, this approach takes the prediction of a model and estimates the gradient with respect to each individual timestep for every lead from the input signal. For echocardiogram videos, an analogous methodology applies: the gradient of the model's prediction was calculated with respect to every pixel from the input video. In each case, the calculated value is then compared to a provided background distribution, the training data. The value of the calculated gradients for each timestep/pixel is then assigned an importance score such that highly impactful scores (denoted in red) hold positive impacts on prediction estimates. Values with low importance scores negatively influence prediction estimates (denoted in blue), Figure 3 and Figure 4.
Figure 3.
LVH-fusion ECG interpretations. SHAP explanations of one true positive, HCM sample (A). Red areas indicate timesteps that hold a positive impact on prediction, whereas blue timesteps indicate a negative impact on prediction, no colour is neutral. (B) Selected regions of ECG leads denote timesteps of high estimated importance, focusing on inverted T-waves and lead V3 R peaks. (C) Local explanations of the cumulative SHAP values on prediction output across leads. Lead V3 overall contains the highest values of SHAP values for this sample presented.
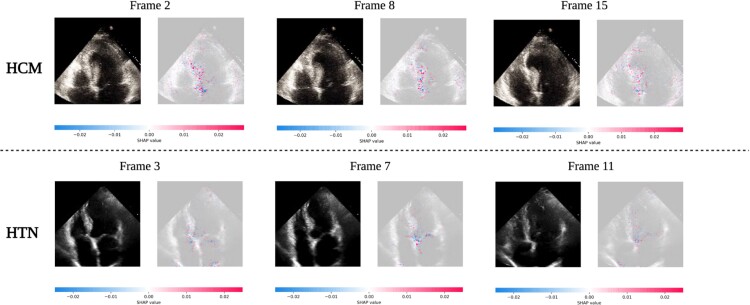
Figure 4.
LVH-fusion echocardiogram interpretations. SHAP explanations for two true positive samples, HCM (top row) and HTN (bottom row). Each class has three frames selected with SHAP values overlaid. Red areas indicate pixels that hold a positive impact on prediction, whereas blue pixels indicate a negative impact on prediction, no colour is neutral. We observe red areas of importance converging on the asymmetric septal wall in the HCM example.
We emphasize samples of ECG and echocardiograms from the test partition to deduce regions the model found most impactful to prediction estimates, Figure 3 and 4. In Figure 3, the ECG interpretation results highlight an overall focus on V3 and T-wave inversion in leads V1-V6. Both the observed early R wave progression and T-wave inversion are indications of HCM. Summarized local interpretations for each lead provides explanations of the overall impact each lead has on prediction estimates. Additional examples of ECG interpretation tracings can be found in the Supplementary material online, Figure S2. Comparably, the interpretation results of the echocardiogram videos, Figure 4, clearly depicts asymmetric proximal septal thickness, a hallmark distinction of HCM across all frames of the video. Next, to examine local summary interpretations, we segmented the left ventricle on each frame for duration of a video's length. This allowed us to quantitatively compare the positive and negative impacts the estimated LV size had on overall prediction estimates, Supplementary material online, Figure S3.
To further examine if the regions of importance identified in distinct samples are globally similar across all predictions, a summation or averaging across all local instances was performed. This approach provides a highly compressed, global insight into the model's behaviour. We considered per lead contributions to predictions in ECGs and left ventricular segmentation in echocardiogram videos. Global summary results for ECG corroborates our results from the ablation studies, lead V3 and aVR holds valuable information for model’s prediction estimates, Supplementary material online, Figure S4.
Comparison against physician interpretation
We had two expert readers review ECG tracings and echocardiogram videos and asked them to make a diagnosis of HTN or HCM. We selected 45 samples (40 HTN and 5 HCM) from the test set to compare LVH-fusion. The LVH-fusion model outperformed these expert cardiologists (one of whom has 20 years of experience in diagnosing HCM). LVH-fusion correctly classified three of the five ECG and echocardiogram HCM samples. Variability between cardiologists varied greatly, with one cardiologist matching LVH-fusion sensitivity estimates but with a reduction in specificity, while cardiologist two failed to correctly classify any of the HCM ECG samples provided.
Discussion
In this study, we report the first multimodal (ECG and echocardiogram based) deep learning model in clinical cardiology and use it to predict the aetiology of left ventricular hypertrophy. Combining complementary knowledge from multiple modalities can improve diagnostic performance in clinical practice. The trained model demonstrates high discriminatory ability in distinguishing HCM from hypertension with an AUC of 0.91, AUPRC of 0.78. Furthermore, ablation studies provided independent support from unsupervised analysis for clinicians’ focus on ECG lateral repolarization and echocardiographic proximal septal hypertrophy for the diagnosis of HCM. Combining complementary information from multiple modalities is intuitively appealing for improving the performance of learning-based approaches. Our results can be directly applied in general medical and cardiology clinics where exposure to rare conditions such as HCM limits confidence in human diagnostic prediction alone.
Deep learning models specifically focused on single modalities in cardiology have shown impressive results for arrhythmia detection, age, and other clinical actionable insights.8,10,21 Previously Ko et al., focused on using convolutional neural networks (CNN) for ECG interpretation with respect to HCM.24 They showed high discriminatory power in classifying HCM against a background population of left ventricular hypertrophy by ECG alone. However, approximately 28-30% of HCM cases had concurrent hypertension, inhibiting a direct comparison of possible distinction between HCM and hypertension. Zhang et al16 focuses exclusively on echocardiograms in a fully automated approach to disease detection. Our method differs in three important ways, first we consider both ECG and echocardiogram jointly to make a classification prediction in differentiating between HCM and hypertension. Secondly, LVH-fusion model architecture differs significantly from the aforementioned study. We explored model architectures with variable integration of temporal convolutions instead of an image-based 2D CNN which operates on individual frames of the video. Empirical studies have shown the benefits of different spatiotemporal convolutions for video-based classification over 2D CNNs which are unable to model temporal information and motion patterns, which one would deem to be critical aspects for correct video analysis.25 Additionally, two different video views were necessary for detection of HCM, our method holds high discriminatory power using only one video view. To date, deep learning research addressing non-pulmonary hypertension detection using both electrocardiogram and echocardiogram was unknown.
One previous approach successfully used both ECG and echocardiogram data individually with a stepwise approach to diagnosis of cardiac amyloidosis,15 whereas here we focus on fusion method applications of multimodal deep learning of electrocardiograms and echocardiograms together.
Limitations of this study
We introduce a combined method to include both ECG and echocardiogram videos, in line with common clinical practice to initiate a new approach in AI applied to cardiology. It is important to note the limitations of this study despite the benefits it may provide. Ascertainment bias may exist within our study due to selecting HCM patients from our centre of excellence. In addition, we lack the availability to test our model at an external validation site. To mitigate this limitation, we have open source the code and released the trained models to facilitate reproducibility and further research on multimodal research. Medical decision making is complex, often relying on a combination of physician's judgment, experience, diagnostic and screening test results, and longitudinal follow-up. In the case of a patient presenting with anything other than severe, grossly asymmetric LVH, suspicion for HCM would be higher for patients who do not obviously have hypertension. However, occult hypertension is common and challenging to rule out and with mild ‘grey zone’ hypertrophy, it is not uncommon to make this assumption. Similarly, for patients who present with LVH and manifest hypertension, the question is always ‘is hypertension alone enough to explain this degree of LVH?’ Given the implications of missing a diagnosis of HCM—a mendelian disease associated with heart failure and sudden death—most generalists do not feel confident ignoring the possibility of HCM. In these cases, aggressively treating hypertension and re-reviewing the patient can help but challenges in follow-up, adherence, and effectiveness of therapy make the window of equipoise long. This process extends the critical and necessary process of evaluation for at-risk relatives enabling early diagnosis and identification of patients with the overall goal of contributing to improvement in clinical care.
These are the clinical scenarios into which LVH-fusion will have the most benefit. Yet, this is merely the first application of the approach. A similar approach to the identification of other causes of LVH such as Fabry disease or cardiac amyloidosis can be applied using similar ‘gold standard’ diagnostic labels to those we use here. The future of deep learning in medicine is a move beyond reproducing human-derived label features to capitalizing on unsupervised machine learned features vs. a gold standard diagnostic or prognostic label. This will allow machine augmentation of the human led diagnostic journey.
Conclusion
In summary, we develop a deep learning model incorporating ECG and echocardiogram time series data and apply it to help identify HCM patients from within the much larger group of patients presenting with LVH due to hypertension or unknown causes. We present various well known fusion methods of combining data streams from multiple modalities and compare these comprehensively to single-modal models. Further studies should explore the real world application of physician augmentation approaches such as LVH-fusion in medical practice.
Supplementary Material
Acknowledgements
We thank our academic partners who helped with data acquisition, James Tooley and A.J. Rogers. Some of the computing for this project was performed on the Sherlock cluster. We would like to thank Stanford University and the Stanford Research Computing Centre for providing computational resources and support that contributed to these research results.
Contributor Information
Jessica Torres Soto, Department of Biomedical Data Science, Stanford University, USA.
J Weston Hughes, Department of Computer Science, Stanford University, USA.
Pablo Amador Sanchez, Department of Medicine, Division of Cardiology, Stanford University, Stanford, California, USA.
Marco Perez, Department of Medicine, Division of Cardiology, Stanford University, Stanford, California, USA.
David Ouyang, Department of Cardiology, Smidt Heart Institute, Cedars-Sinai Medical Center, USA; Division of Artificial Intelligence in Medicine, Department of Medicine, Cedars-Sinai Medical Center, USA.
Euan A Ashley, Department of Medicine, Division of Cardiology, Stanford University, Stanford, California, USA.
Supplementary material
Supplementary material is available at European Heart Journal – Digital Health.
Funding
Institutional Funds, Stanford University, Stanford, CA.
Data Availability
All the code for LVH-fusion will be available at https://github.com/AshleyLab/lvh-fusion/upon publication. The data that support the findings of this study are available on request from the corresponding author upon approval of data sharing committees of the respective institutions.
References
- 1. Semsarian C, Ingles J, Maron MS, Maron BJ. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol 2015;65:1249–1254. [DOI] [PubMed] [Google Scholar]
- 2. Ho CY, Day SM, Ashley EA, Michels M, Pereira AC, Jacoby D, Cirino AL, Fox JC, Lakdawala NK, Ware JS, Caleshu CA, Helms AS, Colan SD, Girolami F, Cecchi F, Seidman CE, Sajeev G, Signorovitch J, Green EM, Olivotto I. Genotype and Lifetime Burden of Disease in Hypertrophic Cardiomyopathy: Insights from the Sarcomeric Human Cardiomyopathy Registry (SHaRe). Circulation 2018;138:1387–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Whelton PK, Carey RM, Aronow WS, Casey DE, Collins KJ, Dennison Himmelfarb C, DePalma SM, Gidding S, Jamerson KA, Jones DW, MacLaughlin EJ. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension 2018;71:e13–e115. [DOI] [PubMed] [Google Scholar]
- 4. Magnusson P, Palm A, Branden E, Mörner S. Misclassification of hypertrophic cardiomyopathy: validation of diagnostic codes. Clin Epidemiol 2017;9:403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Esteva A, Chou K, Yeung S, Naik N, Madani A, Mottaghi A, Liu Y, Topol E, Dean J, Socher R. Deep learning-enabled medical computer vision. NPJ Digit Med 2021;4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pennacchini E, Musumeci MB, Fierro S, Francia P, Autore C. Distinguishing hypertension from hypertrophic cardiomyopathy as a cause of left ventricular hypertrophy. J Clin Hypertens 2015;17:239–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doi YL, Deanfield JE, McKenna WJ, Dargie HJ, Oakley CM, Goodwin JF. Echocardiographic differentiation of hypertensive heart disease and hypertrophic cardiomyopathy. Br Heart J 1980;44:395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Attia ZI, Friedman PA, Noseworthy PA, Lopez-Jimenez F, Ladewig DJ, Satam G, Pellikka PA, Munger TM, Asirvatham SJ, Scott CG, Carter RE, Kapa S. Age and sex estimation using artificial intelligence from standard 12-Lead ECGs. Circ Arrhythm Electrophysiol 2019;12:e007284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hannun AY, Rajpurkar P, Haghpanahi M, Tison GH, Bourn C, Turakhia MP, Ng AY. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med 2019;25:65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kwon J-M, Cho Y, Jeon K-H, Cho S, Kim K-H, Baek SD, Jeung S, Park J, Oh B-H. A deep learning algorithm to detect anaemia with ECGs: a retrospective, multicentre study. Lancet Digit Health 2020;2:e358–e367. [DOI] [PubMed] [Google Scholar]
- 11. Yao X, Rushlow DR, Inselman JW, McCoy RG, Thacher TD, Behnken EM, Bernard ME, Rosas SL, Akfaly A, Misra A, Molling PE, Krien JS, Foss RM, Barry BA, Siontis KC, Kapa S, Pellikka PA, Lopez-Jimenez F, Attia ZI, Shah ND, Friedman PA, Noseworthy PA. Artificial intelligence–enabled electrocardiograms for identification of patients with low ejection fraction: a pragmatic, randomized clinical trial. Nature Medicine 2021. [DOI] [PubMed] [Google Scholar]
- 12. Madani A, Ong JR, Tibrewal A, Mofrad MRK. Deep echocardiography: data-efficient supervised and semi-supervised deep learning towards automated diagnosis of cardiac disease. NPJ Digit Med 2018;1:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ouyang D, He B, Ghorbani A, Yuan N, Ebinger J, Langlotz CP, Heidenreich PA, Harrington RA, Liang DH, Ashley EA, Zou JY. Video-based AI for beat-to-beat assessment of cardiac function. Nature 2020;580:252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang S-C, Pareek A, Zamanian R, Banerjee I, Lungren MP. Multimodal fusion with deep neural networks for leveraging CT imaging and electronic health record: a case-study in pulmonary embolism detection. Sci Rep 2020;10:22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goto S, Mahara K, Beussink-Nelson L, Ikura H, Katsumata Y, Endo J, Gaggin HK, Shah SJ, Itabashi Y, MacRae CA, Deo RC. Artificial intelligence-enabled fully automated detection of cardiac amyloidosis using electrocardiograms and echocardiograms. Nat Commun 2021;12:2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J, Gajjala S, Agrawal P, Tison GH, Hallock LA, Beussink-Nelson L, Lassen MH, Fan E, Aras MA, Jordan C, Fleischmann KE, Melisko M, Qasim A, Shah SJ, Bajcsy R, Deo RC. Fully Automated Echocardiogram Interpretation in Clinical Practice. Circulation 2018;138:1623–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Null N, Ommen SR, Mital S, Burke MA, Day SM, Deswal A, Elliott P, Evanovich LL, Hung J, Joglar JA, Kantor P, Kimmelstiel C, Kittleson M, Link MS, Maron MS, Martinez MW, Miyake CY, Schaff HV. 2020 AHA/ACC guideline for the diagnosis and treatment of patients with hypertrophic cardiomyopathy. Circulation 2020;142:e558–e631. [DOI] [PubMed] [Google Scholar]
- 18. Whelton PK, Carey RM, Aronow WS, Casey DE, Collins KJ, Dennison Himmelfarb C, DePalma SM, Gidding S, Jamerson KA, Jones DW, MacLaughlin EJ, Muntner P, Ovbiagele B, Smith SC, Spencer CC, Stafford RS, Taler SJ, Thomas RJ, Williams KA, Williamson JD, Wright JT. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: Executive Summary: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension 2018;71:1269–1324. [DOI] [PubMed] [Google Scholar]
- 19. Kay W, Carreira J, Simonyan K, Zhang B, Hillier C, Vijayanarasimhan S, Viola F, Green T, Back T, Natsev P, Suleyman M. The kinetics human action video dataset. arXiv [cs.CV] 2017. [Google Scholar]
- 20. Sundararajan M, Taly A, Yan Q. Axiomatic attribution for deep networks. arXiv [cs.LG] 2017. [Google Scholar]
- 21. Attia ZI, Noseworthy PA, Lopez-Jimenez F, Asirvatham SJ, Deshmukh AJ, Gersh BJ, Carter RE, Yao X, Rabinstein AA, Erickson BJ, Kapa S, Friedman PA. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet 2019;394:861–867. [DOI] [PubMed] [Google Scholar]
- 22. Sagi O, Rokach L. Ensemble learning: a survey. Wiley Interdiscip Rev Data Min Knowl Discov 2018;8:e1249. [Google Scholar]
- 23. Hughes JW, Yuan N, He B, Ouyang J, Ebinger J, Botting P, Lee J, Theurer J, Tooley JE, Neiman K, Lungren MP, Liang D, Schnittger I, Harrington B, Chen JH, Ashley EA, Cheng S, Ouyang D, Zou JY. Deep learning prediction of biomarkers from echocardiogram videos. bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ko W-Y, Siontis KC, Attia ZI, Carter RE, Kapa S, Ommen SR, Demuth SJ, Ackerman MJ, Gersh BJ, Arruda-Olson AM, Geske JB, Asirvatham SJ, Lopez-Jimenez F, Nishimura RA, Friedman PA, Noseworthy PA. Detection of hypertrophic cardiomyopathy using a convolutional neural network-enabled electrocardiogram. J Am Coll Cardiol 2020;75:722–733. [DOI] [PubMed] [Google Scholar]
- 25. Tran D, Wang H, Torresani L, Ray J, LeCun Y, Paluri M. A closer look at spatiotemporal convolutions for action recognition. Proc IEEE Conf Comput Vision Pattern Recognition 2018:6450–6459. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the code for LVH-fusion will be available at https://github.com/AshleyLab/lvh-fusion/upon publication. The data that support the findings of this study are available on request from the corresponding author upon approval of data sharing committees of the respective institutions.