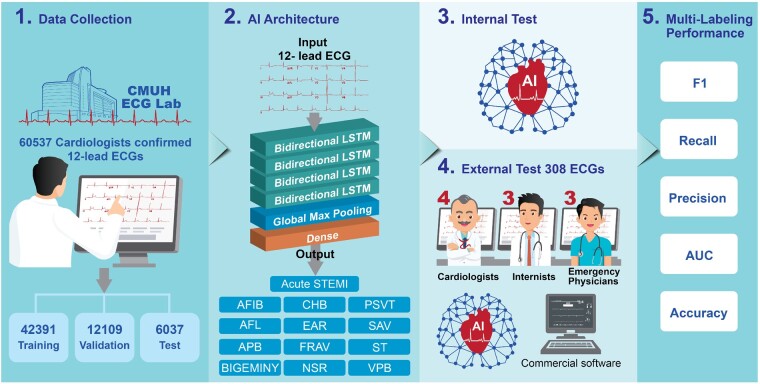
Abstract
Aims
To develop an artificial intelligence-based approach with multi-labelling capability to identify both ST-elevation myocardial infarction (STEMI) and 12 heart rhythms based on 12-lead electrocardiograms (ECGs).
Methods and results
We trained, validated, and tested a long short-term memory (LSTM) model for the multi-label diagnosis of 13 ECG patterns (STEMI + 12 rhythm classes) using 60 537 clinical ECGs from 35 981 patients recorded between 15 January 2009 and 31 December 2018. In addition to the internal test above, we conducted a real-world external test, comparing the LSTM model with board-certified physicians of different specialties using a separate dataset of 308 ECGs covering all 13 ECG diagnoses. In the internal test, the area under the curves (AUCs) of the LSTM model in classifying the 13 ECG patterns ranged between 0.939 and 0.999. For the external test, the LSTM model for multi-labelling of the 13 ECG patterns evaluated by AUC was 0.987 ± 0.021, which was superior to those of cardiologists (0.898 ± 0.113, P < 0.001), emergency physicians (0.820 ± 0.134, P < 0.001), internists (0.765 ± 0.155, P < 0.001), and a commercial algorithm (0.845 ± 0.121, P < 0.001). Of note, the LSTM model achieved an accuracy of 0.987, AUC of 0.997, and precision, recall, and F1 score of 0.952, 0.870, and 0.909, respectively, in detecting STEMI.
Conclusions
We demonstrated the usefulness of an LSTM model in the multi-labelling detection of both rhythm classes and STEMI in competitive testing against board-certified physicians. This AI tool exceeding the cardiologist-level performance in detecting STEMI and rhythm classes on 12-lead ECG may be useful in prioritizing chest pain triage and expediting clinical decision-making in healthcare.
Keywords: Artificial intelligence (AI), ST-elevation myocardial infarction (STEMI)
Graphical Abstract
Introduction
Cardiovascular disease is the leading cause of death due to non-communicable disease globally,1 with ischaemic heart disease accounting for the largest number of deaths among cardiovascular diseases. Acute ST-segment elevation myocardial infarction (STEMI), one of the most serious ischaemic heart diseases, is a medical emergency, which requires early diagnosis to initiate timely coronary reperfusion therapy to reduce morbidity and mortality. The resting 12-lead electrocardiogram (ECG) is a simple and non-invasive tool that is routinely used to screen for many cardiovascular diseases including rhythm disorders and acute STEMI. However, accurate interpretations of 12-lead ECGs by primary care physicians, including emergency physicians and internists, are often limited due to a lack of experience and knowledge as compared to those of cardiologists. Accordingly, a delay in providing appropriate patient triage and timely interventional therapies could occur worldwide in daily practice.
Computerized diagnosis has been used to assist the interpretation of ECGs to improve clinical efficiency. However, the sensitivities and specificities of different computerized algorithms for the detection of STEMI range 0.62–0.93 and 0.89–0.99, respectively, which vary significantly and remain to be improved.2,3 This aforementioned yet unmet need has motivated researchers to develop rapid and reliable ECG diagnostic algorithms for STEMI to initiate early life-saving intervention. Recently, artificial intelligence (AI) using machine learning or deep learning technologies have revolutionized traditional diagnostic procedures in medical practice, particularly in the automatic interpretation of medical images, such as mammographs,4 chest X-rays,5,6 ultrasound,7 and magnetic resonance imaging.8 For cardiovascular images, deep learning has been developed to interpret the results of ECG, echocardiography, coronary computed tomography, and single-photon emission computed tomography for the evaluation of myocardial perfusion.9
The usefulness of machine learning technology in detecting different cardiac arrhythmias has been shown to surpass the performance of conventional computerized ECG diagnosis reaching the cardiologist’s performance level.10 The most common AI-based approaches for identifying different heart rhythms were based on annotating single-lead ECG tracings in previous studies,10,11 which provided limited information as compared with 12-lead ECG signals. Furthermore, it remains challenging for an AI model to interpret all different types of rhythm disorders on 12-lead ECG signals, and for multi-label diagnosis. Additionally, STEMI from ECG signals is more difficult than the identification of arrhythmias because a higher detection sensitivity and essential 12-lead ECG signals for machine learning are both required.12 Zhao et al.12 developed a machine learning-based diagnostic algorithm to identify STEMI using 12-lead ECG signals with a sensitivity of 97% and specificity of 99% which outperformed a commercial auto-diagnostic model. However, this AI-based algorithm was solely designed to identify whether the ECG signal was STEMI or not STEMI.
In a real-world scenario, a 12-lead ECG usually may contain multiple diagnostic features including both electrical and structural information. Compared to the single-labelling model that can only classify one specific ECG pattern, in multi-labelling classification, each ECG sample can be associated with multiple labels. The objective of the current study was to develop a machine learning-based algorithm with multi-labelling capability that could identify both STEMI and 12 different heart rhythm disorders using 60 537 clinical 12-lead ECG signals for training, validation, and internal testing. We also conducted real-world clinical testing for external validation, which compared the performance of the AI model with that of board-certified physicians of different specialties, including cardiologists, emergency physicians, and internists. We believe that the results of the current study may further advance the utility of AI-based approaches in assisting the diagnosis of clinically relevant cardiovascular diseases from 12-lead ECGs.
Methods
Data collection and labelling
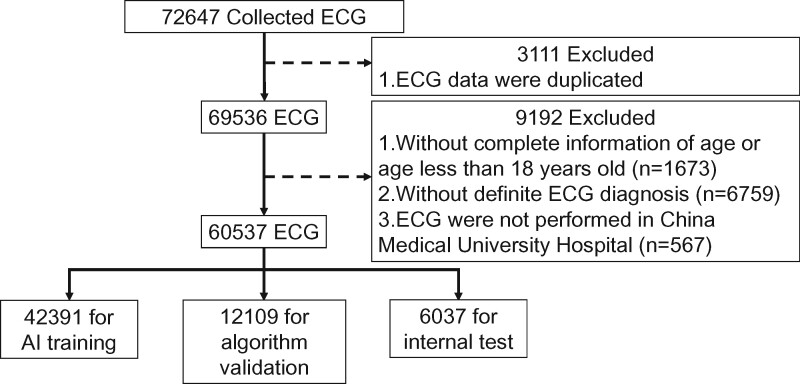
In this study, we retrieved 12-lead ECG data reflecting normal sinus rhythm (NSR) and 12 types of cardiovascular diseases, which included acute STEMI, rhythm disorders and conduction defects. We retrieved the 12-lead ECGs which have been diagnosed by 37 experienced board-certified cardiologists and stored in the digital ECG database of China Medical University, Taichung, recorded between 15 January 2009 and 31 December 2018 for machine learning. The 12-lead ECG was recorded according to a standardized protocol and lead position at a sampling rate of 500 Hz using a computerized ECG machine (GE Healthcare MAC 2000/3500/5500, USA).13 The ECG machine recorded 10-s resting 12-lead ECG signals, which were extracted in Extensible Markup Language (XML) format and converted into arrays of numerical values. Rhythm disorders included atrial fibrillation (AFIB), atrial flutter (AFL), atrial premature beat, ventricular bigeminy (BIGEMINY), ectopic atrial rhythm (EAR), paroxysmal supraventricular tachycardia (PSVT), sinus tachycardia (ST), and ventricular premature beat, and conduction defects included complete heart block (CHB), first degree AV block (FRAV), and second degree AV block (SAV). Because 2 to 1 AV block could either be Mobitz type 1 or Mobitz type 2 AV block,14 we therefore decided to use a broader definition of SAV in this study. The digital ECG was transmitted and stored at the ECG core laboratory of CMUH. In total, 72 647 12-lead ECGs were retrospectively retrieved in an extensible markup language (XML) format. The study protocol was reviewed and approved by the Research Ethics Committee of China Medical University Hospital (CMUH109-REC2-076).
We preprocessed the ECG data before model training. All ECGs of the 12 types of cardiovascular disease and NSR were confirmed by board-certified cardiologists before the study. ECGs were excluded for the following reasons: duplicate data (n = 3111), incomplete information regarding patient age or age less than 18 years old (n = 1673), absence of a definite diagnosis (n = 6759), and ECG examination not performed at CMUH (n = 567) (Figure 1). A total of 60 537 ECGs from 35 981 patients were included in this study, which were separated into training, validation, and testing sets at a ratio of 7:2:1 for developing the proposed AI model. Thus, all the ECG signals including those for the internal test were confirmed and labelled by experienced cardiologists before starting the research. The mean age of patients was 65.06 ± 17.9 years, and 44% of them were female.
Figure 1.
Data collection and labelling. In total, 72 647 12-lead electrocardiograms were retrospectively retrieved. Electrocardiograms with duplicate data (n = 3111), incomplete information of age or age less than 18 years old (n = 1673), absence of definite diagnosis (n = 6759), and those not performed at China Medical University Hospital (n = 567) were excluded. The remaining 60 537 electrocardiogram signals from 35 981 patients were included in this study.
Table 1 presents the number of 12-lead ECGs used for training, validation, and internal testing for each of the 12 cardiac rhythms and acute STEMI. A total of 1889 STEMI ECGs diagnosed by board-certified cardiologists before the study were divided into 1344 ECGs for algorithm training, 338 ECGs for validation, and 207 ECGs for internal testing. Among the 13 ECG diagnoses, AFIB had the largest ECG sample size of 16 366 signals to generate 11 430, 3348, and 1588 signals for training, validation, and testing, respectively. EAR had the smallest sample size with 346 ECG samples, which were divided into 247, 69, and 30 samples for the training, validation, and testing sets, respectively.
Table 1.
Data set for training, validation, and testing of the LSTM model
| No. | Type of ECG diagnoses | Abbreviation | ECG no. total | ECG no. training | ECG no. validation | ECG no. testing |
|---|---|---|---|---|---|---|
| 1 | Acute ST-elevation myocardial infarction | Acute STEMI | 1889 | 1344 | 338 | 207 |
| 2 | Atrial fibrillation | AFIB | 16 366 | 11 430 | 3348 | 1588 |
| 3 | Atrial flutter | AFL | 5953 | 4164 | 1189 | 618 |
| 4 | Atrial premature beat | APB | 5685 | 4004 | 1107 | 574 |
| 5 | Ventricular bigeminy | BIGEMINY | 1812 | 1275 | 366 | 171 |
| 6 | Complete heart block | CHB | 398 | 277 | 79 | 42 |
| 7 | Ectopic atrial rhythm | EAR | 346 | 247 | 69 | 30 |
| 8 | First-degree AV block | FRAV | 2719 | 1903 | 534 | 282 |
| 9 | Normal sinus rhythm | NSR | 9258 | 6511 | 1815 | 932 |
| 10 | Paroxysmal supraventricular tachycardia | PSVT | 3681 | 2560 | 740 | 381 |
| 11 | Second-degree AV block | SAV | 803 | 569 | 166 | 68 |
| 12 | Sinus tachycardia | ST | 11 718 | 8167 | 2416 | 1135 |
| 13 | Ventricular premature beat | VPB | 4892 | 3430 | 954 | 508 |
| Total | 65 520 | 45 863 | 13 121 | 6536 | ||
AV, atrioventricular; ECG, electrocardiogram; LSTM, long short-term memory.
The distribution of the multi-labelling tasks is shown in Table 2. Multi-labelling means that one ECG might contain more than one diagnosis labels. The label counts ranged from 0 to 4. Zero was for the ECG signals that did not belong to any of the 12 heart rhythm classes or acute STEMI analysed in the current study, and 4 was for those which contained four of the 13 ECG labels. Of all the 60 537 signals used in this study, 91.032% of them (55 108 samples) contained one of the 13 ECG labels, and there were 4262 (7.040%), 176 (0.291%), and 4 (0.007%) samples with two, three, and four types of the 13 ECG diagnoses, respectively.
Table 2.
Distribution of multi-labelling analysis
| Label counts | Total |
Training set |
Validation set |
Testing set |
||||
|---|---|---|---|---|---|---|---|---|
| Data no. | % | Data no. | % | Data no. | % | Data no. | % | |
| 0 | 987 | 1.630 | 900 | 2.123 | 54 | 0.446 | 33 | 0.547 |
| 1 | 55 108 | 91.032 | 38 567 | 90.979 | 11 042 | 91.188 | 5499 | 91.088 |
| 2 | 4262 | 7.040 | 2821 | 6.655 | 961 | 7.936 | 480 | 7.951 |
| 3 | 176 | 0.291 | 102 | 0.241 | 51 | 0.421 | 23 | 0.381 |
| 4 | 4 | 0.007 | 1 | 0.002 | 1 | 0.008 | 2 | 0.033 |
| Total | 60 537 | 100 | 42 391 | 100 | 12 109 | 100 | 6037 | 100 |
Proposed deep learning model and internal test
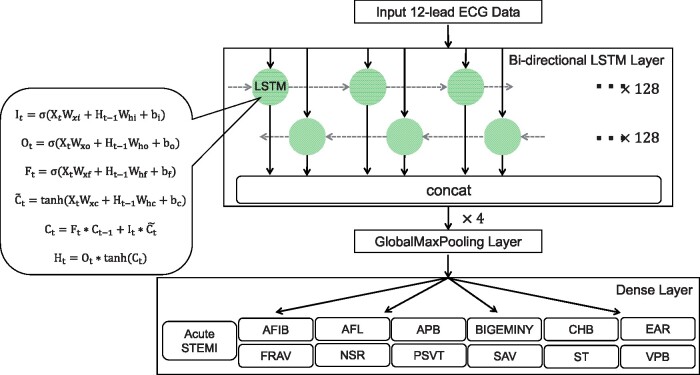
Recurrent neural network (RNN) is a special neural network structure, which can pass the output of a certain layer back to the layer itself as input, so it is suitable for processing data of a sequential nature, such as ECG. The long short-term memory (LSTM) model is an improved version of the conventional RNN with four major components: memory cell, input gate, output gate, and forget gate. Using certain mathematical functions, these cells and gates allow information to be propagated from one layer to the next in the neural network in ways that the information can be discarded in whole or in parts, or updated anew to retain and propagate only the information gained during the learning process. Previous studies have shown the feasibility and performance of the LSTM architecture in dealing with ECG signals15,16 and more technical details of LSTM can be found in the references therein. The bidirectional LSTM model used in our previous study for the single-label detection of the same 12 rhythm classes17 was extended in the present work. The activation function was modified to be sigmoid and 13 different labels were used in the output dense layer to enable multi-label detection. A diagram of the model architecture used in the current study is shown in Figure 2.
Figure 2.
Algorithm processing and developing. We used a bidirectional, four-layer LSTM model, with each layer containing 128 neurons. It, input gate; Ot, output gate; Ft, forget gate; σ, logistic sigmoid function; Xt, input sequence; Ht-1. Wxi, Wxo, Wxf, Wxc, and Whi, the previous block output; Who, Whf, Whc, weight parameters; bi, bo, bf, bc, bias parameters; , candidate memory cell similar to the three gates but using a tanh activation function; Ct-1, the previous LSTM block memory; Ht, the final block output. AFIB, atrial fibrillation; AFL, atrial flutter; APB, atrial premature beat; BIGEMINY, ventricular bigeminy; CHB, complete heart block; EAR, ectopic atrial rhythm; FRAV, first degree AV block; LSTM, long short-term memory; NSR, normal sinus rhythm; PSVT, paroxysmal supraventricular tachycardia; SAV, second degree AV block; ST, sinus tachycardia; STEMI, ST-segment elevation myocardial infarction; VPB, ventricular premature beat.
The model’s main hyperparameter settings included binary cross-entropy as the loss function, Adam optimizer with 0.001 learning rate, 128 batch size, and 75 epochs. Accuracy, F1, precision, and recall were used as metrics to monitor training and validation results. The internal test set was part of the data collected (n = 6536) and was used to assess the AI model’s performance after completion of model’s training and validation.
Comparative test (external test)
To establish the comparative test dataset, we randomly retrieved 12-lead ECGs of the pre-specified 13 diagnosis labels in which the diagnoses had been confirmed by cardiologists. The selected ECGs were randomly presented to three experienced cardiologists for a consensus labelling to serve as ground truth. To determine the minimal sample size required to detect differences of 13 ECG patterns by the LSTM model, we performed a power analysis.18 Assuming the averaged accuracy of each group in annotating the 13 ECG classes (with ≥10 positive labels in each class) ranging between 0.55 and 0.9 according to our previously published data,17 a minimal testing annotation number of 317 was required to reach a statistical power of >0.80. Therefore, collection of ECGs for external test was completed when the amount of ECGs with multi-labelling testing annotations exceeded the minimum required number of 317 with at least 10 positive labels for each class. Following this strategy, 308 ECGs were collected from both CMUH (n = 154) and Asia University Hospital (n = 154). All the ECGs, and the diagnosis labels, for the external test were not included in the original testing set. The three experienced committee members simultaneously annotated all the 308 computerized ECGs one by one and the consensus of each ECG diagnosis was used as the ground truth for external testing. After completing the ground truth annotations, the external test was carried out 4 days later with four other cardiologists, three internists, and three emergency physicians being tested simultaneously at the designated room to interpret the 308 ECGs using a web-based online system as described previously.17 A closed-domain online platform was set up for doctors to label the 308 12-lead ECGs’ data (http://private.ip/ecgtest/). The platform page was prepared in advance by staff before the physicians arrived at the designated room. All participating physicians received a full explanation of the rules and demonstration of the web-based testing if necessary before starting the test. The 308 test ECG records were shown in random order to each of the physicians after they entering their personal identification number and clicking the Get Data button. The physicians could actively click any index ECG number to retrieve the first 12-lead ECG recording. The next 12-lead ECG, randomly picked by algorithm, jumped out automatically after clicking the ‘Submit’ button when finishing annotation of the first ECG.
Their performance against the ground truth was compared to that of the LSTM model. We also compared AI performance with diagnosis based on a commercial algorithm (Data management software MUSE™, GE Healthcare, USA). The study protocol for the comparative test was reviewed and approved by the Research Ethics Committee of CMUH (CMUH109-REC3-020).
Statistical analysis
As in our previous study,17 accuracy, the area under the curve (AUC), precision, recall (sensitivity), and F1 score were used to evaluate model performance in both internal and external tests. The accuracy, precision, and recall of each ECG annotation were respectively calculated according to the following formula:
The accuracy of each rhythm class and STEMI presented was the averaged accuracy value from all ECG signals in that particular class annotated by the proposed AI model as well as the comparators.
F 1 metric (F1 score) was calculated from the precision and recall of the test, which was a harmonic mean of recall and precision to see the performance of the model. The formula of F1 is shown as follows:
Furthermore, a confusion matrix was used to analyse the differences between the model prediction and ground truth for each of the 13 types of ECG patterns. To assess the multi-labelling performance in comparative testing, each of the 13 ECG diagnoses was classified as correct or incorrect against the ground truth, resulting in a total of 359 annotations of the 13 ECG patterns from 308 12-lead ECG recordings. The Mann–Whitney U test was performed to investigate differences in AUC between the different groups of physicians and the LSTM model in the comparative test. Statistical analyses were performed using IBM SPSS Statistics version 22 (IBM, Armonk, NY, USA) or R (version 3.6.2) and Python (version 3.8) as appropriate.
Results
Table 3 represents the performance of the LSTM model for the classification of the 12 heart rhythms and acute STEMI, where the accuracies of the LSTM model ranged between 0.942 and 0.998. The accuracy, AUC, precision, recall, and F1 score of the LSTM model in detecting acute STEMI were 0.983, 0.957, 0.818, 0.652, and 0.726, respectively. The accuracies of the LSTM model in detecting BIGEMINY, CHB, EAR, SAV were all higher than 0.990 with the highest accuracy of 0.998 for BIGEMINY. The proposed model achieved an AUC of >0.955 for all rhythm classes, which ranged from 0.955 for AFL to 0.999 for CHB. The precision and recall of the LSTM model in detecting the 12 heart rhythm classes ranged from 0.706 to 0.944 and from 0.400 to 0.977, respectively. The F1 scores, which represent the harmonic mean of the precision and recall of the LSTM model in detecting the 12 heart rhythm classes, ranged from 0.511 for EAR to 0.960 for BIGEMINY.
Table 3.
Diagnostic performance of the LSTM model for 12 individual heart rhythms and acute STEMI
| ECG diagnoses | Accuracy | AUC | Precision | Recall | F 1 |
|---|---|---|---|---|---|
| Acute STEMI | 0.983 | 0.957 | 0.818 | 0.652 | 0.726 |
| AFIB | 0.942 | 0.985 | 0.887 | 0.892 | 0.890 |
| AFL | 0.946 | 0.955 | 0.779 | 0.662 | 0.716 |
| APB | 0.966 | 0.979 | 0.854 | 0.777 | 0.814 |
| BIGEMINY | 0.998 | 0.987 | 0.944 | 0.977 | 0.960 |
| CHB | 0.997 | 0.999 | 0.795 | 0.738 | 0.765 |
| EAR | 0.996 | 0.939 | 0.706 | 0.400 | 0.511 |
| FRAV | 0.979 | 0.978 | 0.826 | 0.691 | 0.753 |
| NSR | 0.973 | 0.959 | 0.889 | 0.939 | 0.913 |
| PSVT | 0.984 | 0.992 | 0.883 | 0.856 | 0.869 |
| SAV | 0.994 | 0.977 | 0.837 | 0.529 | 0.649 |
| ST | 0.968 | 0.990 | 0.916 | 0.915 | 0.916 |
| VPB | 0.978 | 0.981 | 0.878 | 0.852 | 0.865 |
AFIB, atrial fibrillation; AFL, atrial flutter; APB, atrial premature beat; AUC, area under curve; BIGEMINY, Ventricular bigeminy; CHB, complete heart block; EAR, ectopic atrial rhythm; FRAV, first-degree AV block; LSTM, long short-term memory; NSR, normal sinus rhythm; PSVT, paroxysmal supraventricular tachycardia; SAV, second-degree AV block; ST, sinus tachycardia; STEMI, ST-elevation myocardial infarction; VPB, ventricular premature beat.
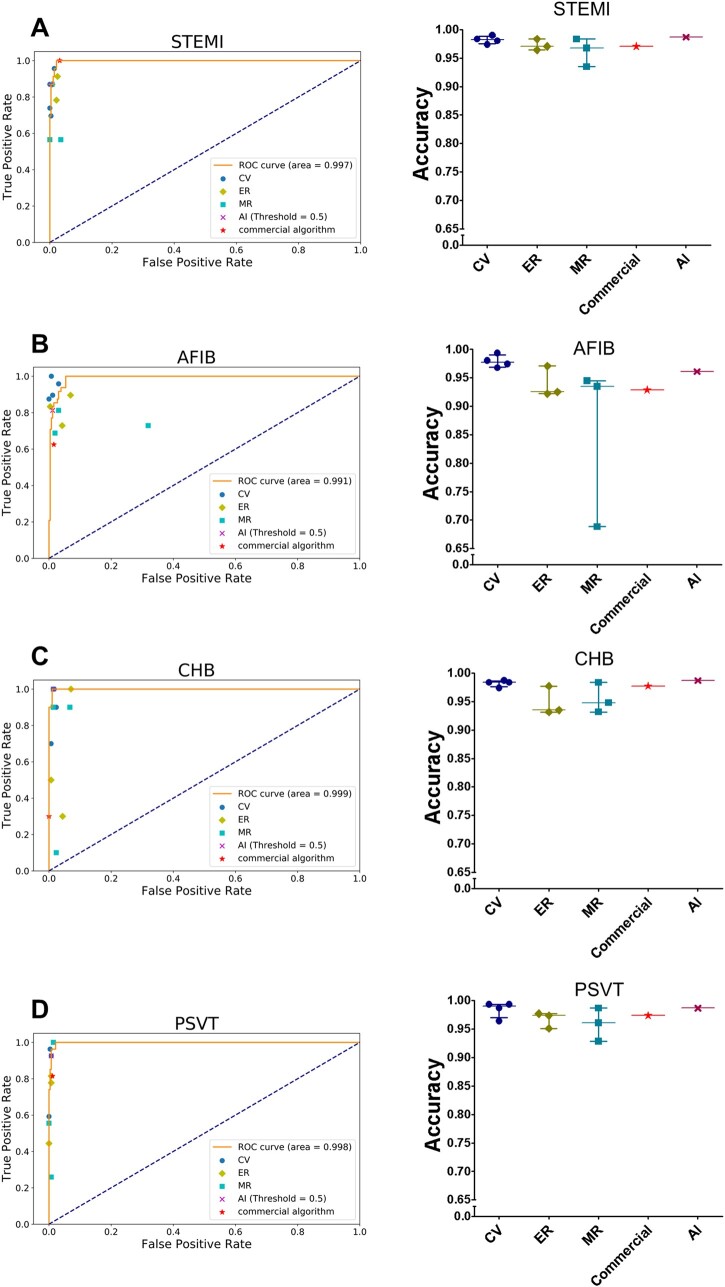
In external testing, we compared the accuracy, AUC, precision, recall, and F1 score of the LSTM model with those of a commercial ECG algorithm and different broad-certified doctors (four cardiologists, three emergency physicians, and three internists) for classifying the 12 heart rhythms and acute STEMI using another 308 randomly collected ECGs. The LSTM model essentially outperformed board-certified physicians and the commercial algorithm (Supplementary material online, Table S1). Taking four types of important cardiovascular disease as an example: for detecting AFIB, the AUC of the LSTM model was 0.991, which was higher than the mean AUC of 0.960, 0.810, 0.890, and 0.805 respectively for cardiologists, internists, emergency physicians, and the commercial algorithm; for detecting CHB, the AUC of the LSTM model was 0.999, which was superior to the mean AUC of 0.942, 0.799, 0.780, and 0.650 respectively for cardiologists, internists, emergency physicians, and the commercial algorithm; and for detecting PSVT, the AUC of the LSTM model was 0.998, which was greater than the mean AUC of 0.929, 0.799, 0.837, and 0.902 respectively for cardiologists, internists, emergency physicians, and the commercial algorithm. Of note, the AUC of the LSTM model for detecting acute STEMI was 0.997, which was also higher than the mean AUC of 0.905, 0.826, 0.919, and 0.984, respectively for cardiologists, internists, emergency physicians, and the commercial algorithm. Two representative ECGs in the external testing are shown in Figure 3. Taking all 13 ECG diagnoses together, the overall mean AUC of the LSTM model (0.987 ± 0.021) was superior to that of cardiologists (0.898 ± 0.113, P < 0.001), emergency physicians (0.820 ± 0.134, P < 0.001), internists (0.765 ± 0.155, P < 0.001), and the commercial algorithm (0.845 ± 0.121, P < 0.001) (Table 4). Figure 4 and Supplementary material online, Figure S1 depict the individual AUCs and accuracies of the 13 ECG patterns for the LSTM model in comparison to those for the board-certified physicians of varying specialties and the commercial algorithm in detecting acute STEMI and 12 heart rhythms.
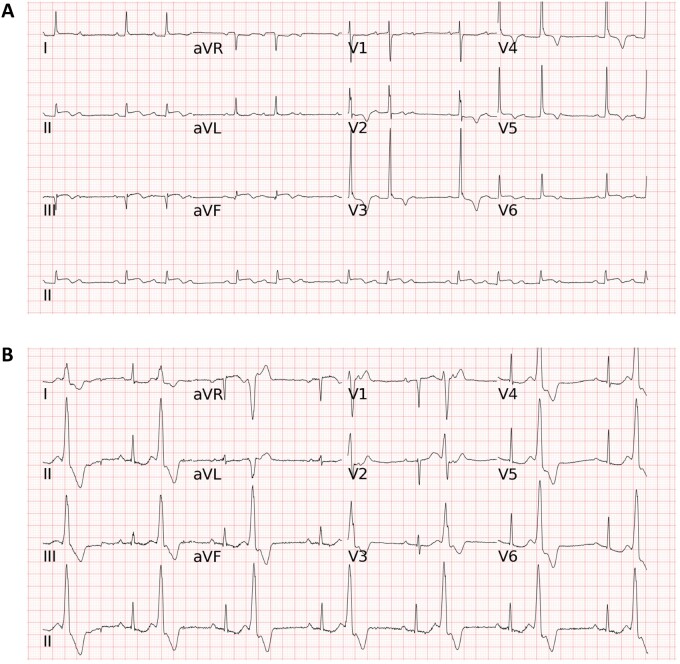
Figure 3.
Two representative electrocardiograms in the external testing. (A) The long short-term memory model, all of the four cardiologists, one of the three emergency physicians, and the commercial algorithm correctly classified the electrocardiogram as second degree AV block and acute STEMI, whereas two emergency physicians and all of the three internists annotated either second degree AV block or ST-elevation myocardial infarction but not both for this electrocardiogram. (B) The long short-term memory model correctly classified the electrocardiogram as BIGEMINY and first degree AV block, while most doctors (8 of the 10 physicians) and the commercial algorithm only annotated BIGEMINY but not first degree AV block. Abbreviations for the electrocardiogram diagnoses as in Figure 2.
Table 4.
Performance by area under curve (AUC) for the proposed model, board-certified doctors, and the commercial algorithm for 308 testing ECGs
| Groups | AUC | P-valuesa |
|---|---|---|
| LSTM model | 0.987 ± 0.021 | reference |
| Board-certified doctors | ||
| Internists | 0.765 ± 0.155 | <0.001 |
| Emergency physicians | 0.820 ± 0.134 | <0.001 |
| Cardiologists | 0.898 ± 0.113 | <0.001 |
| Commercial algorithm | 0.845 ± 0.121 | <0.001 |
Mann–Whitney U test.
LSTM, long short-term memory.
Figure 4.
Performance of the long short-term memory model and different groups of board-certified doctors in detecting acute ST-elevation myocardial infarction and different heart rhythms. These are the accuracies and receiver operating characteristic curves in detecting (A) ST-elevation myocardial infarction (B) atrial fibrillation (C) complete heart block (D) paroxysmal supraventricular tachycardia of our artificial intelligence model and the results of a commercial algorithm and different groups of doctors in the comparative external tests. The orange line was the receiver operating characteristic curve of the long short-term memory model. The different colour points represent different groups of board-certified doctors. AI, artificial intelligence; CV, cardiologists; ER, emergency physicians; LSTM, long short-term memory; MR, internists; abbreviations for the electrocardiogram diagnoses are as in Figure 2. Only the four important classes, discussed in the main text are shown here, the rest was presented in Supplementary material online, Figure S1.
To assess the multi-labelling performance in comparative testing, each of the 13 ECG diagnoses may appear alone (single labelling, n = 260) or in combination with other classes (multi-labelling, n = 99) on an ECG recording, resulting in a total of 359 class annotations from 308 ECG recordings for the external test. Table 5 shows the accuracies of single labelling and multi-labelling for STEMI and 12 heart rhythm classes (including AV blocks) of the LSTM model in the external testing. The diagnoses on each of the 308 ECGs by the proposed AI model and the comparators were classified as correct or incorrect against the ground truth for comparison. The accuracies of single labelling and multi-labelling for STEMI and 12 heart rhythms were in general similar or only a few hundredths off except for FRAV. Of the 308 ECGs for external testing, 8 ECGs were STEMI only and 15 ECGs were STEMI co-existing with rhythm disorders. Accuracies of single labelling for STEMI and multi-labelling for STEMI co-existing with rhythm classes were 0.9962 and 0.9375, respectively. The accuracies, sensitivities, specificities, and F1 scores of single labelling and multi-labelling for STEMI are shown in the Supplementary material online, Table S2. The representative STEMI ECG images of false negative cases missed by humans and the computer in the external test are presented in Figure 5.
Table 5.
Accuracies of single labelling and multi-labelling for STEMI and 12 heart rhythms of the LSTM model
| ECG diagnoses | Accuracy of single labelling (n = 260) | Accuracy of multi-labelling (n = 99) |
|---|---|---|
| Acute STEMI | 0.9962 (n = 8) | 0.9375 (n = 15) |
| AFIB | 0.9577 (n = 46) | 0.9792 (n = 2) |
| AFL | 0.9577 (n = 18) | 1.0000 (n = 0)a |
| APB | 0.9846 (n = 17) | 0.9167 (n = 16) |
| BIGEMINY | 0.9962 (n = 21) | 1.0000 (n = 4) |
| CHB | 0.9885 (n = 8) | 0.9792 (n = 2) |
| EAR | 0.9462 (n = 17) | 0.9375 (n = 5) |
| FRAV | 0.9808 (n = 15) | 0.8125 (n = 20) |
| NSR | 0.9462 (n = 32) | 1.0000 (n = 0)a |
| PSVT | 0.9846 (n = 27) | 1.0000 (n = 0)a |
| SAV | 0.9692 (n = 31) | 0.9167 (n = 5) |
| ST | 0.9769 (n = 14) | 0.9792 (n = 14) |
| VPB | 0.9923 (n = 6) | 0.9375 (n = 16) |
Abbreviations for the ECG diagnoses are as in Table 1.
The diagnostic class did not appear in the multi-labelling ECGs; thus, the accuracy means the true negative rate.
Figure 5.
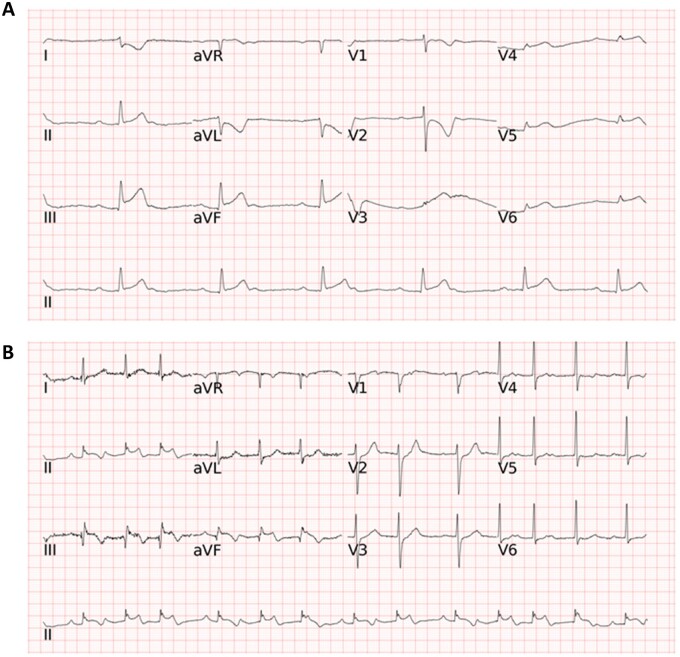
The representative ST-elevation myocardial infarction electrocardiogram images of false negative cases missed by humans and the computer in the external test. (A) The artificial intelligence model correctly annotated ST-elevation myocardial infarction, whereas one of the four cardiologists labelled ‘Not STEMI’ resulting in a false negative annotation. (B) All the four cardiologists correctly diagnosed ST-elevation myocardial infarction, while the artificial intelligence model annotated ‘Not STEMI’ and it was counted as a false negative.
Discussion
The strengths of the current study include (i) use of a large amount of clinically collected 12-lead ECG data (60 537 ECGs) for machine learning to generate a novel AI model capable of multi-label diagnosis; (ii) demonstration of the effectiveness of the AI model in detecting 12 cardiac rhythms and acute STEMI, surpassing the performances of a commercial algorithm and board-certified physicians including cardiologists, emergency physicians, and internists in a real-world external validation test; and (iii) the ultrafast and accurate auto-diagnosis capability of the AI model will be useful in prioritizing chest pain triage and expediting the clinical decision-making process for primary percutaneous coronary intervention (PCI).
AI-based approach for detection of acute STEMI
Acute STEMI is a medical emergency resulting in high morbidity and mortality that requires timely and accurate diagnosis to initiate early reperfusion therapy. Thus the current guidelines suggest that the time from first medical contact-to-device should be ≤90 min for STEMI patients to undergo primary PCI, and the door-to-balloon time should be within 60 min in primary PCI-capable institutes.19–21 A 12-lead ECG is a key diagnostic tool for identifying acute STEMI. Therefore, a number of AI-based automatic diagnostic tools have been developed to facilitate timely and precise ECG diagnosis of STEMI. Although the performance of most of these computerized algorithms has an accuracy, sensitivity, specificity, and F1 score >0.9, the usefulness of the AI models has not been confirmed in a clinically relevant scenario. Recently, Zhao et al. employed a Res-Net AI algorithm to detect STEMI or not STEMI and found that the AI model performance was better than the average performance of 15 medical doctors including cardiologists, medical residents, and medical interns using a dataset of 667 STEMI ECGs and 7571 control ECGs for algorithm development; and 50 STEMI and 50 non-STEMI ECGs for clinical testing. In the current study, we developed a multi-labelling LSTM model by using a dataset of 65 520 ECGs including 1889 STEMIs, along with 63 631 ECGs with 12 cardiac rhythms, to develop an algorithm capable of annotating not only STEMI but also rhythm classes. Of note, coexistence of STEMI with one or more rhythm classes appears to have a comparable accuracy rate with single labelling STEMI diagnosis in our study (0.9375 vs. 0.9962). Further large-scaled studies are needed to confirm this finding. The performance of the LSTM model was tested against three groups of board-certified physicians including four cardiologists, three internists, and three emergency physicians using 308 ECGs, where the physicians evaluated the ECGs in a web-based testing environment at the same place and time. We believe that the multi-labelling design of our AI model and its superiority in performance over primary care physicians shows that it performs at a clinically useful level in the auto-diagnosis of STEMI from 12-lead ECG.
Recent guidelines recommend that a 12-lead ECG should be transmitted to physicians who take care of patients suspected of having STEMI in a timely manner.22 However, primary physicians, including emergency doctors and internists, may not have similar level of experience as cardiologists in interpreting STEMI ECG, leading to an unintentional delay in STEMI diagnosis. The current study showed that the overall accuracy rate in interpreting STEMI ECG was 0.962 for internists, 0.973 for emergency physicians, 0.971 for the commercial algorithm, 0.982 for cardiologists, and 0.987 for the LSTM model. The results provide evidence reinforcing the importance of the active participation of cardiologists in a team to expedite the diagnosis of acute STEMI. Since the data indicate that the performance of the LSTM model was superior to that of cardiologists, the AI-based approach can be a useful alternative in providing timely and accurate diagnosis of STEMI in clinical practice. Furthermore, the performance of our LSTM model in identifying STEMI ECG was far superior to that of the commercial algorithm. All these findings support the notion that the AI-based approach, which reaches a cardiologist-level in diagnosing STEMI based on ECG on an all-day basis, is a useful tool in accelerating the triage of patients presenting with chest pain, and may play a role in preventing avoidable delay in STEMI patients undergoing reperfusion therapy.
AI model capable of multi-labelling to detect rhythm classes and STEMI
Accumulating evidence has demonstrated the feasibility and efficacy of using deep learning technologies to classify and detect common cardiac arrhythmias based on single-lead or 12-lead ECG signals.10,12 The MIT-BIH arrhythmia database23 is the most common single-lead ECG dataset used for machine learning approaches to classify cardiac rhythms based on ECG signal fragments from a small number of patients. Hannun et al.10 developed a useful deep neural network (DNN) model to classify 12 rhythm classes, using 91 232 single-lead ECGs from a large sample of patients with a high overall accuracy rate surpassing the average performance of cardiologists. Recently, we employed an LSTM model using 65 932 12-lead ECG signals from 38 899 patients to detect 12 cardiac rhythms with superior overall performance when compared with the performance of cardiologists, internists, and emergency physicians in a clinical testing competition.17 However, all of these AI models were only able to classify one specific cardiac rhythm and were not designed to detect other critical or potentially life-threatening cardiac diseases, such as acute myocardial infarction or ventricular fibrillation.
Recently, Mostayed et al.24 developed a deep learning approach to detect the ECG changes of ST elevation or depression in addition to classifying seven rhythm disorders using the China Physiological Signal Challenges (CPSC) dataset. The model can only detect one class of ECG and was not tested in a clinically relevant scenario. Recently, Chen et al.25 employed a convolutional neural network (CNN) model to classify both rhythm disorders and ST changes using the CPSC dataset. Although this model appears to be able to detect both single and multiple ECG classes, the design of the model was not primarily for the multiple labelling of ECG classes and was not specific for detecting STEMI-related ST elevation. Similarly, their model was not tested against different levels of physicians in a head-to-head competition in a real-world situation.
In the current study, we developed a specialized LSTM architecture in a deep learning neural network that enabled the AI algorithm to perform multi-labelling diagnosis to detect 12 rhythm classes and acute STEMI. The external validation of the AI model against physicians of different specialties, including cardiologist, internists, and emergency physicians who must interpret 12-lead ECGs during their daily practice, was carried out as a clinically relevant competition. The overall performance of the AI model on 13 ECG diagnoses evaluated by AUC was superior to that of board-certified internists, emergency physicians, and cardiologists. With the ultrafast annotation time of the AI (4 s for 308 12-lead ECGs) compared to a much longer interpretation time (69–146 min for board-certified physicians), we believe that this approach may expedite the diagnostic process for computerized 12-lead ECG interpretation, leading to more effective patient triage, thus accelerating decision-making for subsequent interventions, particularly for patients with acute STEMI.
Limitations
This study has several limitations. First, neither the pre-labelled STEMI ECGs for machine learning nor the STEMI ECGs used for external testing were verified based on coronary angiographic findings or levels of cardiac enzyme elevation. There could have been some false positive computer reported cases of STEMI. However, the annotation of 12-lead ECG as acute STEMI was according to international gold standard criteria26 judged by board-certified cardiologists who would make the decision whether to perform primary PCI based on the diagnostic criteria for acute STEMI, i.e. (i) typical/atypical symptoms of chest pain; (ii) 12-lead ECG findings; and (iii) cardiac enzyme elevation, but not coronary angiography. It should be noted that given a 12-lead ECG fulfilling the diagnosis of acute STEMI, coronary angiography may disclose no culprit coronary lesions, which can be due to a variety of causes, such as coronary artery spasm, acute pericarditis, Takotsubo cardiomyopathy, or spontaneous reperfusion.27,28 We believe that it is clinically relevant to select typical STEMI ECGs as labelled by board-certified cardiologists for machine learning to develop a cardiologist-level AI model, such as the current algorithm, to accelerate the triage of patients with acute chest pain. Second, our LSTM model was trained to detect whether there was STEMI present or not, which included different STEMI patterns such as anterior, lateral, inferior, and other STEMIs. Of the 1889 ECGs of STEMI in the training and validation sets, 470 ECGs were anterior STEMI (24.9%), 86 ECGs were lateral STEMI (4.6%), 704 ECGs were inferior STEMI (37.3%), 587 ECGs were combination of anterior, lateral or inferior STEMI (31.1%), and 42 ECG were other STEMIs (2.2%). It is a limitation that the LSTM model was not trained to identify different STEMI patterns. However, in clinical practice, the most important procedure after identifying STEMI is to arrange emergency PCI irrespective of the different types of STEMI. We will train the LSTM model to identify different types of STEMI in the future. Third, we did not specifically incorporate 12-lead ECGs with baseline drifting, motion artefacts, or electromagnetic interference as input data for training. Therefore, the current AI model may not work in patients with the aforementioned ECG noise. Fourth, the current LSTM model was not designed to annotate more complex ECG patterns, such as those in patients with wide QRS complex rhythms, bundle branch block, ventricular tachycardia/fibrillation, multifocal atrial tachycardia, junctional rhythm, and wandering atrial pacemaker, primarily because we did not have a sufficient number of these ECGs for machine learning thus far. Indeed, further efforts should be devoted to improving our model in differentiating complex wide QRS tachycardias in the future. Fifth, class unbalancing might have bias in training the AI model. Because the prevalence of certain cardiac arrhythmias such as EAR and CHB was relatively uncommon in our patient cohort, we were not able to collect the same numbers of ECGs for all classes from our digital ECG database. To reduce the bias caused by class unbalancing, we have assigned different class weights to the 13 types of ECG diagnoses in the training program to ensure the training efficacy of classes (e.g. EAR and CHB) with limited ECG signals. With this approach, we found that, the accuracies of the LSTM model on EAR and CHB with a relatively lower ECG number for training were actually comparable to those of some ECG classes with a much higher ECG number, such as AFIB, in the internal and the external tests. Lastly, it is possible that there are more limitations not mentioned above to the current study. For example, it is difficult to moderate electrode position in every recording, the ideal dimensionality remains to be defined, there are still other important diagnoses not included in the algorithm, i.e. old myocardial infarction etc. And of course the AI model is still a black box lacking explainability. Indeed, further improvement in the AI modelling is still needed to tackle the aforementioned limitations.
Conclusions
We demonstrated the usefulness of an LSTM AI model in multiple labelling to detect both rhythm classes and acute STEMI using a large quantity of 12-lead ECG signals by conducting competitive testing against board-certified physicians. This AI-based approach exceeding an average performance of cardiologists in the auto-diagnosis of STEMI ECGs on an all-day basis may be a useful tool in expediting the triage process in patients presenting with acute chest pain, and may play a role in preventing delay for reperfusion therapy in STEMI patients.
Supplementary material
Supplementary material is available at European Heart Journal is available at online.
Funding
This study was supported in part by the Taiwan Ministry of Science and Technology [MOST 109-2314-B-039-045, MOST 108-2314-B-039-055, and MOST 107-2314-B-039-061]; and China Medical University Hospital [DMR-CELL-1802, DMR108-180, DMR-108-013, DMR-109-012, and DMR-110-012]. None of these funding sources had a further role in study design; collection, analysis, or interpretation of data; writing the report; or decision to submit the paper for publication.
Data availability
The data underlying this article are available in the article and in its online supplementary material.
Conflict of interest: none declared.
Supplementary Material
References
- 1.GBD 2016 Causes of Death Collaborators. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017;390:1151–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Garvey JL, Zegre-Hemsey J, Gregg R, Studnek JR.. Electrocardiographic diagnosis of ST segment elevation myocardial infarction: an evaluation of three automated interpretation algorithms. J Electrocardiol 2016;49:728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bosson N, Sanko S, Stickney RE, Niemann J, French WJ, Jollis JG, Kontos MC, Taylor TG, Macfarlane PW, Tadeo R, Koenig W, Eckstein M.. Causes of prehospital misinterpretations of ST elevation myocardial infarction. Prehosp Emerg Care 2017;21:283–290. [DOI] [PubMed] [Google Scholar]
- 4. Becker AS, Marcon M, Ghafoor S, Wurnig MC, Frauenfelder T, Boss A.. Deep learning in mammography: diagnostic accuracy of a multipurpose image analysis software in the detection of breast cancer. Invest Radiol 2017;52:434–440. [DOI] [PubMed] [Google Scholar]
- 5. Becker AS, Bluthgen C, Phi van VD, Sekaggya-Wiltshire C, Castelnuovo B, Kambugu A, Fehr J, Frauenfelder T.. Detection of tuberculosis patterns in digital photographs of chest X-ray images using deep learning: feasibility study. Int J Tuberc Lung Dis 2018;22:328–335. [DOI] [PubMed] [Google Scholar]
- 6. Baltruschat IM, Nickisch H, Grass M, Knopp T, Saalbach A.. Comparison of deep learning approaches for multi-label chest X-ray classification. Sci Rep 2019;9:6381–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pehrson LM, Lauridsen C, Nielsen MB.. Machine learning and deep learning applied in ultrasound. Ultraschall Med 2018;39:379–381. [DOI] [PubMed] [Google Scholar]
- 8. Akkus Z, Galimzianova A, Hoogi A, Rubin DL, Erickson BJ.. Deep learning for brain MRI segmentation: state of the art and future directions. J Digit Imaging 2017;30:449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Al'Aref SJ, Anchouche K, Singh G, Slomka PJ,, Kolli KK,, Kumar A, Pandey M, Maliakal G, van Rosendael AR, Beecy AN, Berman DS, Leipsic J, Nieman K, Andreini D, Pontone G, Schoepf UJ, Shaw LJ, Chang HJ, Narula J, Bax JJ, Guan Y, Min JK.. Clinical applications of machine learning in cardiovascular disease and its relevance to cardiac imaging. Eur Heart J 2019;40:1975–1986. [DOI] [PubMed] [Google Scholar]
- 10. Hannun AY, Rajpurkar P, Haghpanahi M, Tison GH, Bourn C, Turakhia MP, Ng AY.. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med 2019;25:65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mathews SM, Kambhamettu C, Barner KE.. A novel application of deep learning for single-lead ECG classification. Comput Biol Med 2018;99:53–62. [DOI] [PubMed] [Google Scholar]
- 12. Zhao Y, Xiong J, Hou Y, Zhu M, Lu Y, Xu Y, Teliewubai J, Liu W, Xu X, Li X, Liu Z, Peng W, Zhao X, Zhang Y, Xu Y.. Early detection of ST-segment elevated myocardial infarction by artificial intelligence with 12-lead electrocardiogram. Int J Cardiol 2020;317:223–230. [DOI] [PubMed] [Google Scholar]
- 13. Chang KC,, Huang CL, Liang HY, Chang SS, Wang YC,, Liang WM, Lane HY, Chen CH, Stephen Huang SK.. Gender-specific differences in susceptibility to low-dose methadone-associated QTc prolongation in patients with heroin dependence. J Cardiovasc Electrophysiol 2012;23527–533. [DOI] [PubMed] [Google Scholar]
- 14. Barold SS, Hayes DL.. Second-degree atrioventricular block: a reappraisal. Mayo Clin Proc 2001;76:44–57. [DOI] [PubMed] [Google Scholar]
- 15. Gao J, Zhang H, Lu P, Wang Z.. An effective LSTM recurrent network to detect arrhythmia on imbalanced ECG dataset. J Healthc Eng 2019;2019:6320651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Saadatnejad S, Oveisi M, Hashemi M.. LSTM-based ECG classification for continuous monitoring on personal wearable devices. IEEE J Biomed Health Inform 2020;24:515–523. [DOI] [PubMed] [Google Scholar]
- 17. Chang KC, Hsieh PH, Wu MY, Wang YC, Chen JY, Tsai FJ, Shih ESC, Hwang MJ, Huang TC.. Usefulness of machine learning-based detection and classification of cardiac arrhythmias with 12-lead electrocardiograms. Can J Cardiol 2021;37:94–104. [DOI] [PubMed] [Google Scholar]
- 18. Faul F, Erdfelder E, Lang AG, Buchner A. G. Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods 2007;39:175–191. [DOI] [PubMed] [Google Scholar]
- 19. Levine GN, Bates ER, Blankenship JC,, Bailey SR, Bittl JA, Cercek B, Chambers CE, Ellis SG, Guyton RA, Hollenberg SM,, Khot UN, Lange RA, Mauri L, Mehran R, Moussa ID, Mukherjee D, Ting HH, O'Gara PT, Kushner FG, Ascheim DD,, Brindis RG, Casey DE Jr, Chung MK, de Lemos JA, Diercks DB, Fang JC, Franklin BA, Granger CB, Krumholz HM, Linderbaum JA, Morrow DA, Newby LK, Ornato JP, Ou N, Radford MJ, Tamis-Holland JE, Tommaso CL, Tracy CM, Woo YJ, Zhao DX.. 2015 ACC/AHA/SCAI focused update on primary percutaneous coronary intervention for patients with ST-elevation myocardial infarction: an update of the 2011 ACCF/AHA/SCAI guideline for percutaneous coronary intervention and the 2013 ACCF/AHA guideline for the management of ST-elevation myocardial infarction. J Am Coll Cardiol 2016;67:1235–1250. [DOI] [PubMed] [Google Scholar]
- 20. Ibanez B, James S, Agewall S, Antunes MJ, Bucciarelli-Ducci C, Bueno H, Caforio ALP, Crea F, Goudevenos JA, Halvorsen S, Hindricks G, Kastrati A, Lenzen MJ, Prescott E, Roffi M, Valgimigli M, Varenhorst C, Vranckx P, Widimsky PESC Scientific Document Group.. 2017 ESC Guidelines for the management of acute myocardial infarction in patients presenting with ST-segment elevation: The Task Force for the management of acute myocardial infarction in patients presenting with ST-segment elevation of the European Society of Cardiology (ESC). Eur Heart J 2018;39:119–177. [DOI] [PubMed] [Google Scholar]
- 21. Li YH, Lee CH, Huang WC, Wang YC, Su CH, Sung PH, Chien SC, Hwang JJ.. 2020 focused update of the 2012 guidelines of the Taiwan Society of Cardiology for the management of ST-segment elevation myocardial infarction. Acta Cardiol Sin 2020;36:285–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arnett DK, Blumenthal RS, Albert MA,, Buroker AB, Goldberger ZD, Hahn EJ, Himmelfarb CD,, Khera A, Lloyd-Jones D, McEvoy JW, Michos ED, Miedema MD, Munoz D, Smith SC Jr, Virani SS, Williams KA Sr, Yeboah J, Ziaeian B.. 2019 ACC/AHA guideline on the primary prevention of cardiovascular disease: executive summary: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol 2019;74:1376–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moody GB, Mark RG.. The impact of the MIT-BIH arrhythmia database. IEEE Eng Med Biol Mag 2001;20:45–50. [DOI] [PubMed] [Google Scholar]
- 24. Mostayed A, Luo J, Shu X, Wee W. Classification of 12-lead ECG signals with bi-directional LSTM network. 2018;arXiv:1811.02090. https://arxiv.org/abs/1811.02090
- 25. Chen TM, Huang CH, Shih ESC,, Hu YF, Hwang MJ.. Detection and classification of cardiac arrhythmias by a challenge-best deep learning neural network model. iScience 2020;23:100886–100894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thygesen K,, Alpert JS, Jaffe AS, Simoons ML,, Chaitman BR, White HDJoint ESC/ACCF/AHA/WHF Task Force for the Universal Definition of Myocardial InfarctionKatus HA, Lindahl B, Morrow DA, Clemmensen PM, Johanson P, Hod H, Underwood R, Bax JJ, Bonow RO, Pinto F, Gibbons RJ, Fox KA,, Atar D, Newby LK, Galvani M, Hamm CW, Uretsky BF, Steg PG, Wijns W, Bassand JP,, Menasche P, Ravkilde J, Ohman EM, Antman EM, Wallentin LC, Armstrong PW, Simoons ML, Januzzi JL, Nieminen MS, Gheorghiade M, Filippatos G, Luepker RV, Fortmann SP, Rosamond WD, Levy D, Wood D, Smith SC, Hu D, Lopez-Sendon JL, Robertson RM, Weaver D, Tendera M, Bove AA, Parkhomenko AN, Vasilieva EJ, Mendis S.. Third universal definition of myocardial infarction. Circulation 2012;126:2020–2035.22923432 [Google Scholar]
- 27. Villablanca PA, Briceno DF, Jagannath AD, Cohen M, Pyo R.. Coronary artery spasm: is ST-elevation key for diagnosis? Acute Card Care 2016;18:11–12. [DOI] [PubMed] [Google Scholar]
- 28. Zhong-qun Z, Chong-quan W, Sclarovsky S, Nikus KC, Chao-rong H, Shan M.. ST-segment deviation pattern of takotsubo cardiomyopathy similar to acute pericarditis: diffuse ST-segment elevation. J Electrocardiol 2013;46:84–89. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.
Conflict of interest: none declared.