Abstract
Torreya grandis is a characteristic rare economic tree species in subtropical mountainous areas. The kernels of T. grandis have rich content of organic acids, and malate is the predominant organic acid in T. grandis kernels. However, the contents, biosynthesis/metabolism pathway and transcriptional regulation of malate in developing T. grandis kernels remain completely unknown. Here, the organic acid composition in developing T. grandis kernels was first analyzed. The results showed that the content of malate was increased during the maturation T. grandis kernels. A malate synthase (TgMLS) gene might be involved in the accumulation of malate based on transcriptome data, gene expression and enzyme activity analysis. Transient expression of TgMLS in tobacco resulted in the high malate synthase activity and malate content. Furthermore, a basic helix-loop-helix transcription factor (bHLH), TgbHLH87 was identified to positively regulate the TgMLS expression via directly binding the TgMLS promoter. Our finding contributes to mechanism underlying malate accumulation in T. grandis kernels.
Keywords: Torreya grandis, Kernel, Malate, TgMLS, TgbHLH87
Graphical abstract
Highlights
-
•
The organic acids composition was first explored in T. grandis nuts.
-
•
The malate content was increased during the maturation T. grandis nuts.
-
•
A malate synthase (TgMLS) gene was involved in malate accumulation.
-
•
TgbHLH87 positively regulates the TgMLS expression.
1. Introduction
Torreya grandis belongs to the family of Taxaceae. It is a rare economic tree species characteristic for the subtropical mountainous areas in China (Yuan et al., 2017). The kernels of T. grandis have rich contents of lipids, tocopherols, polyphenols, amino acids and flavonoids, resulting in high nutritional and medicinal effects as well as economic values (He et al., 2016; Ni and Shi, 2014). Our previous studies have investigated the synthesis/metabolism of fatty acids, tocopherols, polyphenols, amino acids and flavonoids (Ding et al., 2020; Lou et al., 2019, 2022; Song et al., 2021, 2022; Suo et al., 2019; Wu et al., 2018; Zhang et al., 2022a). However, to our knowledge, there are only few studies concerning the content and biosynthesis of organic acids in T. grandis kernels.
Organic acids not only play vital roles in influencing the organoleptic characteristics of fruit, but also have protective roles against various diseases due to their antioxidant activity and antimicrobial activity (Silva et al., 2004; Vaughan and Geissler, 2009; Cruz-Romero et al., 2013). There are many kinds of organic acids, such as malate, citrate, citramalate, citraconic acid, etc. Among them, malate is the predominant organic acids in most plants. For example, in apple, 85% of organic acids is malate (Zhang et al., 2010). Malate has multiple vital roles in the whole life cycle of plants. Malate is a key intermediate in the tricarboxylic acid (TCA) cycle and is imported into mitochondria as a respiratory substrate (Sweetman et al., 2009). Malate can act as a pH regulator (Hu et al., 2016) and an essential storage carbon molecule (Martinoia and Rentsch, 1994). It is also play an important role in regulating the stomatal function (Lee et al., 2008). Notably, malate acts together with soluble sugars to adjust the edible quality of fruits. Cristofori et al. (2008) showed that malic acid is the predominant organic acids (about 80%) in hazelnut (Corylus avellana L.) nuts. However, the question about which is the predominant organic acids in T. grandis nuts is need to be answered.
The abundance of malate in plants is mainly determined by the synthesis, transportation and degradation of the compound. Many key enzymes are believed to be directly involved in this process, such as malate synthase (MLS), NAD-dependent malate dehydrogenase (NAD-MDH), NADP-dependent malic enzyme (NADP-ME), Al-activated malate transporter (ALMT), etc (Brito et al., 2020; Comai et al., 1992; Pua et al., 2003). MLS and NAD-MDH are the key enzymes involved in malate synthesis, while NADP-ME functions in malate degradation. ALMT mediates the transport of malate from the cytosol to the vacuole (Wang et al., 2018; J. Yan et al., 2021). Control of the above malate synthesis/metabolism-related genes at the transcription level is apparent in a variety of plant species. In apple, MdMYB73 regulates malate transportation by directly modulating ALMT9 expression (Hu et al., 2017), while MdWRKY126 or MdbHLH3 modulates the malate synthesis via directly activating the MdMDH5 or MdNAD-MDH expressions, separately (Yu et al., 2021; Zhang et al., 2022b). In maize, a helix-loop-helix transcription factor ZmHLH128 could regulates the ZmNADP-ME via directly binding to its promoter (Borba et al., 2018). In Orchidaceae, a homeobox transcription factor PaHB5 positively regulates the expression of PaMLS (Wu et al., 2020). However, the malate biosynthesis/metabolism pathway and transcriptional regulation on malate accumulation in T. grandis kernels remain completely unknown.
In this study, the organic acid composition in the developmental T. grandis kernels was analyzed. Furthermore, the molecular mechanism of the biosynthesis of malate, which is the predominant organic acid in developing T. grandis kernels, was unraveled. This study can help us to use the molecular biotechnology to develop the new health products rich in malate based on T. grandis.
2. Materials and methods
2.1. Plant materials
Torreya grandis cv. “Merrillii” grafting tree (Panmugang, Linan City, Zhejiang Province, China) was used in this study. The kernels were collected from seeds at two typical stages during the filling stage (mid-July, named “Early stage”) and mature stage (mid-September, named “Later stage”), then immediately frozen in liquid nitrogen and stored at -80 °C for further analysis.
2.2. Analysis of organic acids profiling
Metabolites analysis was performed by the Metware Biotechnology Co. Ltd. (Wuhan, China). In briefly, the grounded vacuum freeze-drying samples were dissolved in 1.0 mL methanol at 4 °C for 12 h. After centrifugation, the filtrated extracts were performed to LC-MS analysis. The detailed analysis was same as our previous study (Lou et al., 2022). Fold change (FC) and variable importance in projection (VIP) value of orthogonal partial least squares discriminant analysis (OPLS-DA) model were used to screen the differential organic acids. Screening criteria (≤ FC ≤ 0.5, and P-value < 0.05) were used to screen the significant enrichments.
For analysis of malate contents in nuts and tobacco leaves, the harvested samples were ground with liquid nitrogen, and homogenized with 1 mL of 80% (v/v) ethanol, and vortexed thoroughly. After centrifuging, the supernatant was used to measure malate content using an enzymatic method as describe by Delhaize et al. (1993) and Wang et al. (2010).
2.3. Isolation of total RNA and real-time PCR analysis
Total RNA was isolated from T. grandis kernels using the RNAprep Pure Plant Kit (Polysaccharides & Polyphenolics-rich) (TIANGEN, China). Real-time PCR was performed using EvaGreen 2 × qPCR MasterMix-No Dye (abm, China) according to the manufacturer's instructions. Expression levels for target genes were measured using the 2–ΔΔCT method with Tgactin as described previously (Livak and Schmittgen, 2001; L. Yan et al., 2021).
2.4. Luciferase assays
The full length of screened transcription factors was cloned into pGreenII 62-SK vector. The promoter of TgMLS was separately cloned into the pGreenII 0800-LUC vector. Then, the above vectors were introduced into Agrobacterium tumefaciens strain GV3101. The strain was transformed into the four-week-old Nicotiana benthamiana leaves as described by Lou et al. (2022). After infiltration for 3 days, the LUC/REN was calculated according to the manufacturer's instructions of Dual-Lumi™ II Luciferase Reporter Gene Assay Kit,Dual-Lumi™ II Luciferase Assay Kit (Beyotime, China).
2.5. Transient expression of TgMLS in tobacco leaves
The Agrobacterium tumefaciens strain GV3101 carrying TgMLS fused with green fluoresecent protein (TgMLS-GFP) or GFP empty (GFP) was transformed into the four-week-old Nicotiana benthamiana leaves as described by Lou et al. (2022). After culturing for 3 days, the leaves transiently expressed TgMLS-GFP or GFP were collected to analyze the malate synthase activity and the malate content.
2.6. Yeast one hybrid (Y1H)
The fragments of TgMLS promoter were cloned into the vector pLacZi (Table S2), and the full-length coding sequence of TgbHLH87 was inserted into the pB42AD vector using the specific primers listed in Table S3. The constructs were co-transformed into the yeast strain EGY48 and cultured on a synthetic dropout/-Trp-Ura/Gal/Raf/X-Gal (80 μg/mL) plate.
2.7. Analysis of malate synthase activity
The harvested tobacco leaves ground with liquid nitrogen were homogenized with PBS buffer, and vortexed thoroughly. After centrifuging for twice, the supernatant was used to measure the malate synthase activity using the Plant MLS EMSA kit (Meimian, China).
2.8. Pearson correlation analysis
The correlation of TRINITY_DN123721_c0_g1 with other transporters were calculated with Person correlation coefficient. In addition, the top 10 transcription factors with highest correlation were selected, and their relative expressions and inner correlation were visualized by calling ‘pheatmap’ and ‘corrplot’ function in R language (v4.0.5), respectively.
3. Results
3.1. Organic acid composition analysis in T. grandis seeds
The seed development of T. grandis generally includes four stages: slow-growing stage (May/year), expansion stage (June/year), filling stage (July to August/year) and mature stage (September/year). Our previous studies have shown that material transformation and accumulation of T. grandis seed occurred in the periods from the filling stage (named “Early stage”) to mature stage (named “Later stage”). To explore the organic acids accumulation in maturation of T. grandis seeds, the Early stage and Later stage of T. grandis seeds were used to analyze the organic acids in T. grandis seeds. A total of 10 organic acids, including citrate, malate, citraconic acid, were increased, while a total of 27 organic acids, including kynurenic acid, methylglutaric acid, were decreased, during the period of the Early stage to Later stage (Fig. 1 and Table S1).
Fig. 1.
Identification of differentially accumulated organic acid in T. grandis seeds. A heatmap showing different organic acid from two kernel stages. Three biological replicates were performed for each sample.
3.2. The expression and activity of MLS during maturation of T. grandis seeds
Our previous study have shown that malate was the predominant organic acids in T. grandis kernels (Lou et al., 2022). Thus, we focused on the malate synthesis in this study. To explore the molecular mechanism of malate accumulation in T. grandis kernels, the genes involved in malate synthesis were analyzed based on the previous RNA-seq data unraveling the expression profiles during the maturation of T. grandis seeds. Only one unigene for TgMLS (TRINITY_DN123304_c2_g4) in Later stage was expressed at a much higher level compared to those in Early stage during the maturation of T. grandis seed, which was further confirmed via the quantitative real-time PCR (qRT-PCR) assay (Fig. 2A and B). Next, we detected the malate synthesis activity and malate content during the maturation of T. grandis seed. As shown in Fig. 2C, the activity of malate synthase and malate content in Later stage was much higher compared to those in Early stage during the maturation of T. grandis seed. Taken together, these results suggest that TgMLS might contribute to the malate accumulation during the maturation of T. grandis seed.
Fig. 2.
The expression and activity of MLS during maturation of T. grandis seeds. (A) The FPKM value of TgMLS during maturation of T. grandis seeds. (B) The relative expression level of TgMLS in two kernel stages. (C) The malate synthase activity and (D) malate content in two kernel stage. Error bars in (A–D) indicate SD (n = 3). The asterisk in (A–D) shows a significant difference between different maturation stage using the unpaired Student's t-test (*P < 0.05).
3.3. Sequence alignment and phylogenetic analysis of TgMLS
According to the annotated results from the previous transcriptome sequencing (RNA-seq) (Zhang et al., 2022a), the full-length of TgMLS was obtained. To further investigate the amino acid sequence homology between TgMLS and the other MLS, a phylogenetic analysis was generated by the neighbor-joining method (Saitou and Nei, 1987). The result showed that TgMLS displayed a close relationship with CaMLS from Capsicum annuum, ApMLS from Abrus precatorius and PvMLS from Pistacia vera (Fig. 3A). Sequence alignment indicated that TgMLS exhibited high identities (44.6%–83.7%) with its close MLS proteins. TgMLS had a pfam01274 superfamily conserved domain (16–540 region), which also extensively existed in other MLS proteins (Fig. 3B).
Fig. 3.
Sequence alignment and phylogenetic analysis of TgMLS. (A) Phylogenetic analysis of TgMLS and closely related MLS protein in other species using MEGA6 program. The accession numbers of these MLS proteins are as follows: SmMLS (Selaginella moellendorffii, XP_002967078.1), ApMLS (Abrus precatorius, XP_027336875.1), McMLS (Momordica charantia, XP_022137538.1), PvMLS (Pistacia vera, XP_031276974.1), CaMLS (Capsicum annuum, KAF3683365.1), TpMLS (Trifolium pratense, PNY13237.1), MtMLS (Medicago truncatula, XP_013444720.1) and AtMLS (Arabidopsis thaliana, NP_001190219.1). Numbers in (A) indicate the bootstrap values after 1000 replicates. (B) Alignment of the TgMLS with closely related MLS protein in other species using DNAMAN V6. The putative malate synthase domain (approximate 524 amino acids) is marked by a red arrow. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. Analysis of MLS activity and malate accumulation after transient overexpression of TgMLS in tobacco leaves
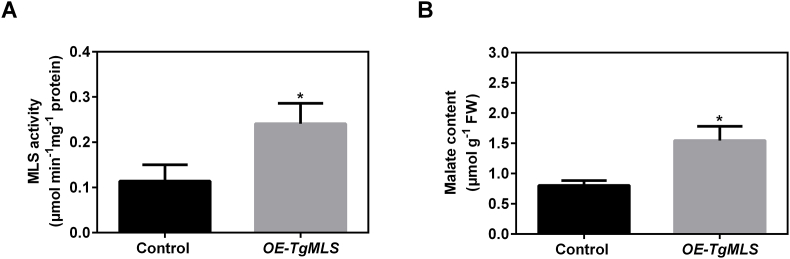
To prove the role of TgMLS in malate synthesis, TgMLS-GFP or GFP driven by CaMV35S promoter was transiently expressed in tobacco leaves. Then, the malate synthesis activity and malate content in the tobacco leaves transiently expressed TgMLS-GFP or GFP were analyzed. As shown in Fig. 4A, the activity of malate synthase in the leaves transiently expressed TgMLS-GFP was much stronger than that in the leaves transiently expressed with GFP. In agreement with the MLS activity, the malate content was significantly higher in the leaves transiently expressed TgMLS-GFP. These results indicate that TgMLS is involved in malate synthesis via enhancing the MLS activity in tobacco leaves.
Fig. 4.
Transient overexpression of TgMLS in tobacco leaves resulted in increased MLS activity and malate accumulation. (A) The malate synthase activity in transiently overexpressed tobacco leaves. (B) The malate contents in transiently overexpressed tobacco leaves. Error bars in (A) and (B) indicate SD (n = 3). The asterisk in (A) and (B) shows a significant difference compared to the control using the unpaired Student's t-test (*P < 0.05).
3.5. Identification of candidate transcriptional modulators involved in regulating TgMLS expression
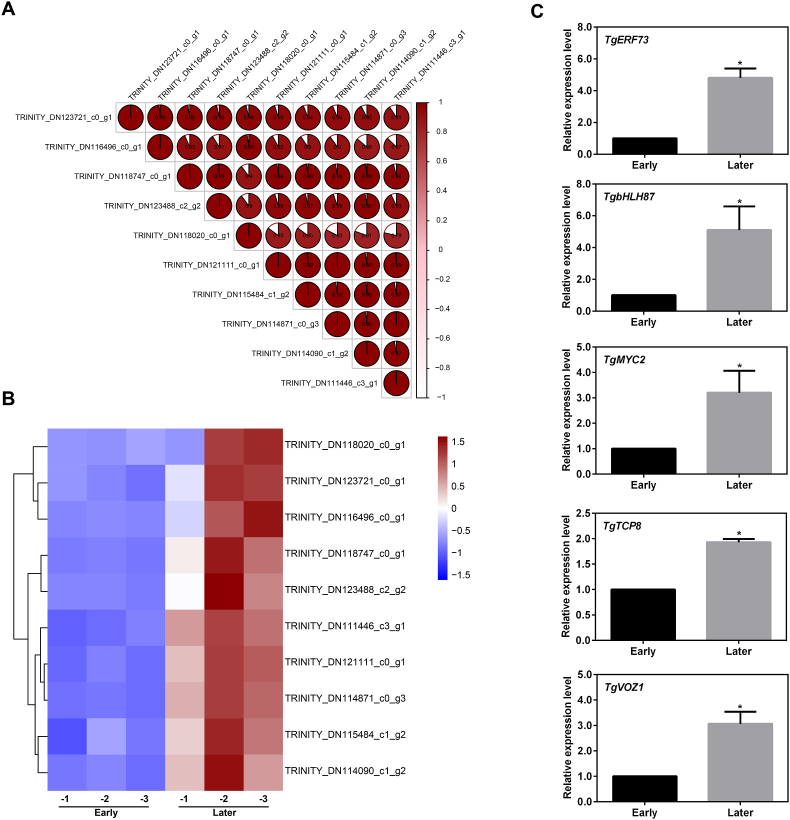
The above results have shown that the expression of TgMLS was related to the malate accumulation, implying that the transcriptional regulation on TgMLS might exist during the maturation of T. grandis seed. Our previous study has analyzed the expression profiles during the maturation of T. grandis seed (Zhang et al., 2022a). Thus, in the current study, to unravel the key transcription factors involving TgMLS expression, co-expression analysis was first performed based on the previous RNA-seq data. As shown in Fig. 5A, a total of 10 transcription factors significantly correlated to TgMLS were identified with the Pearson correlation coefficient ≥0.90. The top 5 transcription factors, named TgbHLH87 (TRINITY_DN96370_c1_g2), TgERF73 (TRINITY_DN116496_c0_g1), TgMYC2 (TRINITY_DN114152_c0_g1), TgTCP8 (TRINITY_DN107834_c0_g1) and TgVOZ1 (TRINITY_DN116340_c0_g3), were selected and further confirmed by qRT-PCR (Fig. S1). As expected, the qRT-PCR and previous RNA-Seq data were highly correlated (Fig. 5B and C), implying that these transcription factors might regulate the TgMLS expression in T. grandis seed.
Fig. 5.
Identification of candidate transcriptional modulators involved in regulating TgMLS expression. (A) Pearson's correlation analysis between the expression of TgMLS level and top transcription factors (ten) from transcriptome data. (B) A heatmap of the above transcription factors during maturation of T. grandis seeds. (C) The expression of the top 5 transcription factors during maturation of T. grandis seeds via qRT-PCR assay. Error bars in (C) indicate SD (n = 3). The asterisk in (C) shows a significant difference between different maturation stage using the unpaired Student's t-test (*P < 0.05).
3.6. The regulation of TgbHLH87 on TgMLS expression
To further explore the regulation of the above screened transcription factors on TgMLS expression, the dual-luciferase analysis was first performed. The promoter of TgMLS (2000 bp) was cloned and fused to firefly luciferase protein (Fluc) at the N-terminus, which also had a renilla luciferase (Rluc). The effector plasmids contain screened transcription factor or empty vector. The Fluc/Rluc ratio represents the ability of transcription factor to transcriptionally activate the TgMLS promoter. As shown in Fig. 6, only TgbHLH87 significantly enhanced the activity of LUC driven by TgMLS promoter, but the other transcription factors did not affect the activities of LUC driven by TgMLS promoter. The results imply that TgbHLH87 might positively regulate the TgMLS expression.
Fig. 6.
The regulation of transcription factors on TgMLS promoter. (A) The schematic image of the double-reporter and effector plasmids. (B–F) Dual-luciferase analysis. Error bars in (B–F) indicate SD (n = 3). Scale bar corresponds to 1 cm. The asterisk in shows a significant difference between empty and transcription factor using the unpaired Student's t-test (*P < 0.05). (G) TgbHLH87 directly binds the TgMLS promoter in yeast. Yeast EGY48 strain co-transformed the fragments of TgMLS promoter and pB42AD-TgbHLH87 or empty pB42AD vector were grown on a synthetic dropout/-Trp-Ura/Gal/Raf/X-Gal (80 μg/mL) plate for three days. The number at the top represents the dilutions times of an optical density at 600 nm. (H) A working model of TgMLS involvement in malate biosynthesis.
Since TgbHLH87 positively regulated TgMLS expression (Fig. 6), we wondered whether TgbHLH87 directly regulated the expression of TgMLS. HLH proteins can regulate the expression of their target genes by recognizing E-box (5′-CANNTG-3′) or G-box (5′-CACGTG-3′) motifs. Then, we searched and identified seven E-box or G-box motifs in the TgMLS promoter (Fig. S2). To determine whether TgbHLH87 protein could bind to the promoter of TgMLS, yeast one-hybrid assay was used. The promoter 1 (-1 to -400 bp) and promoter 2 (-1500 to -1900 bp) fragments of TgMLS were constructed into the pLacZi vector, while TgbHLH87 was inserted into the pB42AD. As shown in Fig. 6G, the yeast EGY48 transformed with TgbHLH87 and promoter 1 showed blue colour, indicating that TgbHLH87 directly bound the promoter of TgMLS in yeast. Taken together, these results indicate that TgbHLH87 positively regulates the TgMLS expression via directly binding TgMLS promoter.
4. Discussion
As a valuable economic tree species characteristic for subtropical mountainous areas in China (Yuan et al., 2017), T. grandis seeds not only have high nutrition values, but also provide health promoting and medicinal effects for humans. At present, researches focus on exploring the composition, synthesis/metabolism, molecular mechanisms of fatty acid, tocopherols, polyphenols, amino acids and flavonoids in T. grandis seeds (Ding et al., 2020; Lou et al., 2019, 2022; Song et al., 2021, 2022; Suo et al., 2019; Wu et al., 2018; Zhang et al., 2022a). In addition, organic acids also are believed to be an important bioactive component (Silva et al., 2004; Vaughan and Geissler, 2009; Cruz-Romero et al., 2013). Though our previous study has shown that malate was the predominant organic acids in T. grandis seeds (Lou et al., 2022), the change of organic acids during the maturation of T. grandis seeds is still unknown. Here, we found a total of 10 organic acids, including citrate, malate, citraconic acid, were increased, while a total of 27 organic acids, including kynurenic acid, methylglutaric acid, were decreased, during the period of the Early stage to Later stage (Fig. 1 and Table S1). In most ripe fruits, such as apple, peach and tomato, the most abundant organic acids are malate and citrate (Etienne, Génard, Lobit, Mbeguié-A-Mbéguié, & Bugaud, 2013). However, the accumulation patterns of malate and citrate are very different during the fruit maturation of these plant species (Etienne et al., 2002). For example, the citrate content obviously increased during the fruit development in peach, while the malate content was unchanged (Zheng et al., 2021). In apple, the concentration of malate was increased after blooming, while the accumulation of citrate was decreased (Zhang et al., 2022b; Zhang et al., 2010). Interestingly, our study found that the content of both malate and citrate increased during the maturation of T. grandis seeds. To our knowledge, this is the first time to illustrate the changes in the composition of organic acids during the maturation of T. grandis seeds, which provides important guidance for developing products with ideal flavor and texture from T. grandis seeds.
The accumulation of malate is determined by several processes, synthesis regulated by MLS or NAD-MDH enzymes, degradation mediated by NADP-ME enzyme, transportation regulated by ALMT (Brito et al., 2020; Comai et al., 1992; Pua et al., 2003). In peach, PpNAD-MDH1, PpALMT9, were the key genes directly involved in the biosynthesis of malate (Zheng et al., 2021). In apple, MdALMT9, MdMDH5 and MdNAD-MDH play vital roles in directly regulating the synthesis of malate (Hu et al., 2017; Yu et al., 2021; Zhang et al., 2022b). However, the expressions of homologous genes of these above genes in T. grandis were not affected during the maturation of T. grandis seeds. Here, our study first proposes a new insight that TgMLS might be the most vital gene in malate synthesis in T. grandis based on the following reasons: first, the expression of TgMLS was significantly increased during the maturation of T. grandis seeds based on the findings of the RNA-seq and qRT-PCR assay (Fig. 2A and B). Secondly, the activity of malate synthase was significantly raised in this process (Fig. 2C). Thirdly, transiently overexpression of TgMLS in tobacco leaves significantly enhanced the activity of malate synthase and the malate content in the leaves (Fig. 4). This phenomenon is very different from the mechanism of malate synthesis in other fruits. The gluconeogenic role of the glyoxylate cycle is important for seed development in oils-rich plants, such as T. grandis nuts, and MLS is responsible for malate synthesis by catalysing the Claisen condensation of glyoxylate with acetyl-CoA in the glyoxysome (Eastmond and Graham, 2001). This might help to explain why TgMLS play a vital role in malate accumulation during the maturation of T. grandis nuts.
Transcriptional regulation is an important mechanism in regulating malate accumulation in plants. For example, MdMYB73, MdWRKY126 or MdbHLH3 regulates malate synthesis by directly activating ALMT9, MdMDH5 or MdNAD-MDH expression in apple (Hu et al., 2017; Yu et al., 2021; Zhang et al., 2022b). PaHB5 could positively regulate the expression of PaMLS in orchidaceae (Wu et al., 2020). AtWRKY46 directly inhibits the AtALMT1 to modulate the malate secretion (Ding et al., 2013). The TgMLS expression was obviously increased during the maturation of T. grandis seeds (Fig. 2A and B). Therefore, it is very reasonable to expect that TgMLS was affected largely by transcriptional regulation. Pearson correlation analysis revealed that the expression levels of some transcription factors were closely correlated to the TgMLS expression, indicating that these transcription factors might regulate the TgMLS expression to mediate the malate in developmental T. grandis kernel. Dual-luciferase analysis and yeast one-hybrid assay suggested that TgbHLH87 directly regulated the TgMLS expression via binding the TgMLS promoter (Fig. 6). Based on these results, our work unraveled the molecular mechanism of TgMLS during the accumulation of malate in T. grandis kernel.
5. Conclusions
In this study, the organic acids composition in developmental T. grandis seeds was analyzed. The results showed that the content of malate, a predominant organic acids in T. grandis kernels, was increased during the seed maturation. According to transcriptome data, gene expression and enzyme activity analysis, we proposed that a malate synthase (TgMLS) gene might be involved in malate accumulation. Furthermore, transient expression of TgMLS in tobacco resulted in the high malate synthase activity and malate content. Notably, TgbHLH87 was identified to directly regulate the TgMLS expression. The illustration of biosynthesis/metabolism pathway and regulation of malate during the maturation of T. grandis kernels not only provides new insights into the mechanism underlying malate accumulation in T. grandis kernels, but also provide useful guidance to further develop new products based on T. grandis.
CRediT authorship contribution statement
Jingwei Yan: Methodology, Validation, Investigation, Resources, Writing – original draft, Funding acquisition. Weijie Chen: Methodology, Validation, Investigation. Hao Zeng: Methodology, Validation. Hao Cheng: Resources. Jinwei Suo: Resources. Chenliang Yu: Methodology. Baoru Yang: Writing – review & editing. Heqiang Lou: Writing – review & editing, Supervision. Lili Song: Resources, Writing – review & editing, Supervision. Jiasheng Wu: Resources, Writing – review & editing, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by the Key Research and Development Program of Zhejiang Province (2021C02001 and 2022C02061); the National Natural Science Foundation of China (32001349); the Scientific Research Startup Fund Project of Zhejiang A&F University (2022LFR033).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crfs.2022.11.017.
Contributor Information
Heqiang Lou, Email: 20170030@zafu.edu.cn.
Lili Song, Email: lilisong@zafu.edu.cn.
Jiasheng Wu, Email: wujs@zafu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Borba A.R., Serra T.S., Górska A., Gouveia P., Cordeiro A.M., Reyna-Llorens I.…Oliveira M.M. Synergistic binding of bHLH transcription factors to the promoter of the maize NADP-ME gene used in C4 photosynthesis is based on an ancient code found in the ancestral C3 state. Mol. Biol. Evol. 2018;35(7):1690–1705. doi: 10.1093/molbev/msy060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito V.C., de Almeida C.P., Barbosa R.R., Carosio M.G.A., Ferreira A.G., Fernandez L.G.…Ribeiro P.R. Overexpression of Ricinus communis L. malate synthase enhances seed tolerance to abiotic stress during germination. Ind. Crop. Prod. 2020;145 doi: 10.1016/j.indcrop.2020.112110. [DOI] [Google Scholar]
- Comai L., Matsudaira K.L., Heupel R.C., Dietrich R.A., Harada J.J. Expression of a Brassica napus malate synthase gene in transgenic tomato plants during the transition from late embryogeny to germination. Plant Physiol. 1992;98(1):53–61. doi: 10.1104/pp.98.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofori V., Ferramondo S., Bertazza G., Bignami C. Paper Presented at the VII International Congress on Hazelnut 845. 2008. Nut quality and sensory evaluation of hazelnut cultivars. [Google Scholar]
- Cruz-Romero M.C., Murphy T., Morris M., Cummins E., Kerry J.P. Antimicrobial activity of chitosan, organic acids and nano-sized solubilisates for potential use in smart antimicrobially-active packaging for potential food applications. Food Control. 2013;34(2):393–397. doi: 10.1016/j.foodcont.2013.04.042. [DOI] [Google Scholar]
- Delhaize E., Ryan P.R., Randall P.J. Aluminum tolerance in wheat (Triticum aestivum L.)(II. Aluminum-stimulated excretion of malic acid from root apices) Plant Physiol. 1993;103(3):695–702. doi: 10.1104/pp.103.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M., Lou H., Chen W., Zhou Y., Zhang Z., Xiao M.…Zhang F. Comparative transcriptome analysis of the genes involved in lipid biosynthesis pathway and regulation of oil body formation in Torreya grandis kernels. Ind. Crop. Prod. 2020;145 doi: 10.1016/j.indcrop.2019.112051. [DOI] [Google Scholar]
- Ding Z., Yan J., Xu X., Li G., Zheng S. WRKY 46 functions as a transcriptional repressor of ALMT 1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. 2013;76(5):825–835. doi: 10.1111/tpj.12337. [DOI] [PubMed] [Google Scholar]
- Eastmond P.J., Graham I.A. Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 2001;6(2):72–78. doi: 10.1016/S1360-1385(00)01835-5. [DOI] [PubMed] [Google Scholar]
- Etienne A., Génard M., Lobit P., Mbeguié-A-Mbéguié D., Bugaud C. What controls fleshy fruit acidity? A review of malate and citrate accumulation in fruit cells. J. Exp. Bot. 2013;64(6):1451–1469. doi: 10.1093/jxb/ert035. [DOI] [PubMed] [Google Scholar]
- Etienne C., Moing A., Dirlewanger E., Raymond P., Monet R., Rothan C. Isolation and characterization of six peach cDNAs encoding key proteins in organic acid metabolism and solute accumulation: involvement in regulating peach fruit acidity. Physiol. Plantarum. 2002;114(2):259–270. doi: 10.1034/j.1399-3054.2002.1140212.x. [DOI] [PubMed] [Google Scholar]
- He Z., Zhu H., Li W., Zeng M., Wu S., Chen S.…Chen J. Chemical components of cold pressed kernel oils from different Torreya grandis cultivars. Food Chem. 2016;209:196–202. doi: 10.1016/j.foodchem.2016.04.053. [DOI] [PubMed] [Google Scholar]
- Hu D., Li Y., Zhang Q., Li M., Sun C., Yu J., Hao Y. The R2R3-MYB transcription factor MdMYB73 is involved in malate accumulation and vacuolar acidification in apple. Plant J. 2017;91(3):443–454. doi: 10.1111/tpj.13579. [DOI] [PubMed] [Google Scholar]
- Hu D., Sun C.H., Ma Q.J., You C.X., Cheng L., Hao Y.J. MdMYB1 regulates anthocyanin and malate accumulation by directly facilitating their transport into vacuoles in apples. Plant Physiol. 2016;170(3):1315–1330. doi: 10.1104/pp.15.01333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Choi Y., Kim Y.Y., Jeon B., Maeshima M., Yoo J.Y., Martinoia E., Lee Y. The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nat. Cell Biol. 2008;10:1217–1223. doi: 10.1038/ncb1782. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lou H., Ding M., Wu J., Zhang F., Chen W., Yang Y.…Song L. Full-length transcriptome analysis of the genes involved in tocopherol biosynthesis in Torreya grandis. J. Agric. Food Chem. 2019;67(7):1877–1888. doi: 10.1021/acs.jafc.8b06138. [DOI] [PubMed] [Google Scholar]
- Lou H., Yang Y., Zheng S., Ma Z., Chen W., Yu C.…Wu J. Identification of key genes contributing to amino acid biosynthesis in Torreya grandis using transcriptome and metabolome analysis. Food Chem. 2022 doi: 10.1016/j.foodchem.2022.132078. [DOI] [PubMed] [Google Scholar]
- Martinoia E., Rentsch D. Malate compartmentation-responses to a complex metabolism. Annu. Rev. Plant Biol. 1994;45(1):447–467. doi: 10.1146/annurev.pp.45.060194.002311. [DOI] [Google Scholar]
- Ni L., Shi W. Composition and free radical scavenging activity of kernel oil from Torreya grandis, Carya Cathayensis, and Myrica Rubra. Iran. J. Pharm. Res. (IJPR): IJPR. 2014;13(1):221. [PMC free article] [PubMed] [Google Scholar]
- Pua E., Chandramouli S., Han P., Liu P. Malate synthase gene expression during fruit ripening of Cavendish banana (Musa acuminata cv. Williams) J. Exp. Bot. 2003;54(381):309–316. doi: 10.1093/jxb/erg030. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Silva B.M., Andrade P.B., Valentão P., Ferreres F., Seabra R.M., Ferreira M.A. Quince (Cydonia oblonga Miller) fruit (pulp, peel, and seed) and jam: antioxidant activity. J. Agric. Food Chem. 2004;52(15):4705–4712. doi: 10.1021/jf040057v. [DOI] [PubMed] [Google Scholar]
- Song L., Meng X., Yang L., Ma Z., Zhou M., Yu C.…Lou H. Identification of key genes and enzymes contributing to nutrition conversion of Torreya grandis nuts during post-ripening process. Food Chem. 2022;384 doi: 10.1016/j.foodchem.2022.132454. [DOI] [PubMed] [Google Scholar]
- Song L., Wen S., Ye Q., Lou H., Gao Y., Bajpai V.K., Xiao J. Advances on delta 5-unsaturated-polymethylene-interrupted fatty acids: Resources, biosynthesis, and benefits. Crit. Rev. Food Sci. Nutr. 2021:1–23. doi: 10.1080/10408398.2021.1953960. [DOI] [PubMed] [Google Scholar]
- Suo J., Tong K., Wu J., Ding M., Chen W., Yang Y.…Song L. Comparative transcriptome analysis reveals key genes in the regulation of squalene and β-sitosterol biosynthesis in Torreya grandis. Ind. Crop. Prod. 2019;131:182–193. doi: 10.1016/j.indcrop.2019.01.035. [DOI] [Google Scholar]
- Sweetman C., Deluc L.G., Cramer G.R., Ford C.M., Soole K.L. Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry. 2009;70:1329–1344. doi: 10.1016/j.phytochem.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Vaughan J., Geissler C. OUP; Oxford: 2009. The New Oxford Book of Food Plants. [Google Scholar]
- Wang L., Ma M., Zhang Y., Wu Z., Guo L., Luo W.…Zhang S. Characterization of the genes involved in malic acid metabolism from pear fruit and their expression profile after postharvest 1-MCP/ethrel treatment. J. Agric. Food Chem. 2018;66(33):8772–8782. doi: 10.1021/acs.jafc.8b02598. [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhao Y., Yi Q., Li K., Yu Y., Chen L. Overexpression of malate dehydrogenase in transgenic tobacco leaves: enhanced malate synthesis and augmented Al-resistance. Acta Physiol. Plant. 2010;32(6):1209–1220. doi: 10.1007/s11738-010-0522-x. [DOI] [Google Scholar]
- Wu J., Huang J., Hong Y., Zhang H., Ding M., Lou H.…Song L. De novo transcriptome sequencing of Torreya grandis reveals gene regulation in sciadonic acid biosynthesis pathway. Ind. Crop. Prod. 2018;120:47–60. doi: 10.1016/j.indcrop.2018.04.041. [DOI] [Google Scholar]
- Wu W., Hsiao Y., Lu H., Liang C., Fu C., Huang T.…Tsai W. Expression regulation of MALATE SYNTHASE involved in glyoxylate cycle during protocorm development in Phalaenopsis aphrodite (Orchidaceae) Sci. Rep. 2020;10(1):1–16. doi: 10.1038/s41598-020-66932-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Liu Y., Yang L., He H., Huang Y., Fang L.…Zhang A. Cell wall beta-1,4-galactan regulated by the BPC1/BPC2-GALS1 module aggravates salt sensitivity in Arabidopsis thaliana. Mol. Plant. 2021;14(3):411–425. doi: 10.1016/j.molp.2020.11.023. https://doi:10.1016/j.molp.2020.11.023 [DOI] [PubMed] [Google Scholar]
- Yan L., Zheng H., Liu W., Liu C., Jin T., Liu S., Zheng L. UV-C treatment enhances organic acids and GABA accumulation in tomato fruits during storage. Food Chem. 2021;338 doi: 10.1016/j.foodchem.2020.128126. [DOI] [PubMed] [Google Scholar]
- Yu J., Gu K., Sun C., Zhang Q., Wang J., Ma F.…Hao Y. The apple bHLH transcription factor MdbHLH3 functions in determining the fruit carbohydrates and malate. Plant Biotechnol. J. 2021;19(2):285–299. doi: 10.1111/pbi.13461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Zhao L., Qiu L., Xu D., Tong Y., Guo W.…Zheng B. Transcriptome and hormonal analysis of grafting process by investigating the homeostasis of a series of metabolic pathways in. Torreya grandis cv. Merrillii. Industrial Crops and Products. 2017;108:814–823. doi: 10.1016/j.indcrop.2017.07.026. [DOI] [Google Scholar]
- Zhang F., Ma Z., Qiao Y., Wang Z., Chen W., Zheng S.…Wu J. Transcriptome sequencing and metabolomics analyses provide insights into the flavonoid biosynthesis in Torreya grandis kernels. Food Chem. 2022;374 doi: 10.1016/j.foodchem.2021.131558. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Li P., Cheng L. Developmental changes of carbohydrates, organic acids, amino acids, and phenolic compounds in ‘Honeycrisp’apple flesh. Food Chem. 2010;123(4):1013–1018. doi: 10.1016/j.foodchem.2010.05.053. [DOI] [Google Scholar]
- Zhang L., Ma B., Wang C., Chen X., Ruan Y., Yuan Y.…Li M. MdWRKY126 modulates malate accumulation in apple fruit by regulating cytosolic malate dehydrogenase (MdMDH5) Plant Physiol. 2022;188(4):2059–2072. doi: 10.1093/plphys/kiac023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Zhao L., Jiang X., Cherono S., Liu J., Ogutu C.…Han Y. Assessment of organic acid accumulation and its related genes in peach. Food Chem. 2021;334 doi: 10.1016/j.foodchem.2020.127567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.