Abstract
The attenuated expression of virulence genes found in a group A streptococcal strain that is naturally pathogenic for mice was postulated to result from a defect in the strain's multigene regulator, Mga. The sequence of the mga gene reveals three amino acid changes in the gene product that might affect protein function. The defect in the mga gene was complemented by providing either the closely similar mga4 allele or a more divergent mga1 allele in trans. Complementation increased the amount of emm50 transcript and the quantity of surface-extractable M protein, restoring virulence function.
Streptococcus pyogenes (the group A streptococcus [GAS]) is an important human pathogen that commonly causes nasopharyngitis (strep throat), impetigo, and tonsillitis, as well as severe infections, such as necrotizing fasciitis, pyomyositis, bacterial sepsis, and streptococcal toxic shock syndrome. In addition, some GAS infections result in the nonsuppurative sequelae of rheumatic fever and acute glomerulonephritis. Although GAS is almost exclusively a human pathogen, a serotype M50 strain was isolated from infections in mice (7). This strain, B514, has recently been used in mouse models to investigate virulence factors of the GAS that contribute to disease (8, 9).
Mga is a multiple-gene regulator of GAS which activates several virulence genes, including those encoding the M protein (emm) and its relatives, the complement C5a peptidase (scpA), and the secreted inhibitor of complement (sic) (3, 10, 16, 19, 22, 23). Mga is also required for transcription of its own gene (mga) (15). The Mga sequence shows some similarity to those of response regulators of two-component sensor-transducer systems that are required for the expression of many genes in bacteria. Like response regulators, Mga is environmentally regulated and it has been shown to bind DNA upstream of several of the genes which it activates (11, 12). However, Mga is about twice as large as most proteins of this type and has three potential helix-turn-helix (HTH) motifs in its N terminus and two potential response regulator motifs at its C-terminal end (16, 18).
The mga gene exists in two major types that diverge from each other by approximately 22%. One type, characterized by mga1, is generally associated with mga regulons containing emm genes of subfamily 1, which includes emm1, emm6, emm5, and emm24, among others. The second type, characterized by mga4, is generally associated with mga regulons containing emm genes of subfamily 2, which includes emm2, emm4, emm49, and emm5, among others. These two forms of mga regulons appear to reflect a differing evolutionary history for the virulence loci and, in most instances, are concordant with the preferred tissue site for colonization in a host (2, 5, 6). Despite the considerable divergence between the major mga types, it has been shown that the two types have overlapping functions by the functional complementation of a deleted mga1 allele with a cloned mga4 allele (1).
In the M50 mouse-virulent GAS strain, there is very little transcription of either mrp (M-related protein) or emm and, consequently, there is a low level of M protein in this strain (24). Because the strain is M deficient, it is not surprising that a deletion of the emm gene had no consequences for virulence in the mouse models for long-term throat colonization or for pneumonia (9). We suggested that a defect in the mga gene in this strain might be the cause of this low transcription level. To test this idea, we have sequenced the mga gene from the M50 strain and determined that there are amino acid differences from other Mga proteins that could make it dysfunctional. We then used wild-type mga alleles from two different GAS strains to complement the mga allele resident in the M50 strain.
Sequencing and analysis of the mga gene from B514.
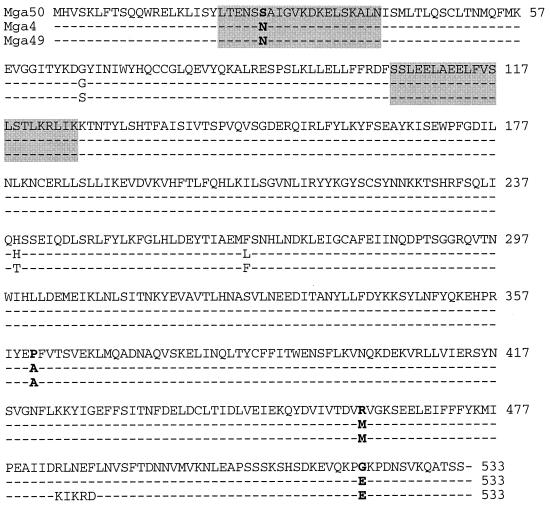
The complete nucleotide sequence of the structural gene for mga from strain B514, called mga50, was determined from PCR-generated DNA fragments made with B514 chromosomal DNA as the template. The open reading frame encodes 533 amino acids. As expected, mga50 differs substantially (22% divergence) from the mga1 gene of the JRS4 M6 serotype strain (mga regulon type 1) (16). It is closely related to mga genes that are derived from the second form of mga (2% divergence from mga4 and mga49). To decipher potential deficiencies in this particular mga50 allele, it was compared to the homologous mga4 and mga49 alleles from strains AP4 and CS101, respectively (Fig. 1) (1, 18).
FIG. 1.
Comparison of amino acid sequences from different GAS mga gene sequences. Mga50 was aligned via the Clustal W algorithm with Mga4 (1) and Mga49 (VirR49) (18). Positions of identical residues are marked with a dash. Polymorphic residues are noted, with bold type indicating those residues at which Mga50 varies from both Mga4 and Mga49. Two potential HTH motifs at the amino terminus are marked as boxes. Two possible domains for sensor proteins to recognize are located at the carboxy terminus, and they are at residues 172 to 300 and residue 404 to the end of the protein, as described by Perez-Casal et al. (16). Residues 484 to 499 differ due to a frameshift in mga49 relative to the sequences of the other two alleles.
In the 608 bp upstream of the open reading frame, mga50 differs from mga4 and mga49 at nine sites (1, 18). Six of these are transitional changes, two are transversions, and one is a single-base-pair deletion. No differences were at bases that would predict deficiencies in the ability of mga50 to be transcribed or regulated properly. None of the polymorphic sites correspond to the −35, −10, or promoter regions as defined by primer extension start sites (11, 19) or to Mga binding regions as defined by footprinting analysis (11).
Within the coding region, in addition to the single-amino-acid substitutions described below, there is one frameshift near the carboxy terminus of the protein-encoded by mga49 relative to those encoded by mga4 and mga50. If this sequence is correct, it would suggest that the extreme C terminus of the protein is not required for Mga function. A similar conclusion was drawn from the ability of mga4 and mga1 to complement each other although they differ completely in sequence in this area (1).
Among the three mga genes, there are seven polymorphic sites in the coding region (Fig. 1). At three of these, the mga50 allele is synonymous with one of the other alleles. At residue 26, which lies in a predicted HTH motif, there is a serine in the mga50 product and an asparagine in both the mga4 and mga49 products. Although the change in amino acid is not dramatic, this change has the potential to affect the DNA binding function of the Mga50 protein and to lead to the decreased expression of Mga-activated genes that was observed in strain B514/Sm (24). A second nonsynonymous site, at residue 361, may have significant potential to affect protein function. This residue is a proline in Mga50 and an alanine in the other Mga proteins. Because proline can disrupt a protein's secondary structure, this change could influence overall protein conformation. Two additional nonsynonymous changes at residues 461 and 521 fall roughly within one of Mga's two predicted response regulator sites, but neither changes a residue that is considered to be key to the operation of a response regulator site. Because this general region differs greatly between the divergent mga alleles which can cross-complement each other, its functional significance is not yet understood. At this time, we do not know which, if any, of these nonsynonymous changes contribute to the defective function of Mga50.
Analysis of transcripts following complementation.
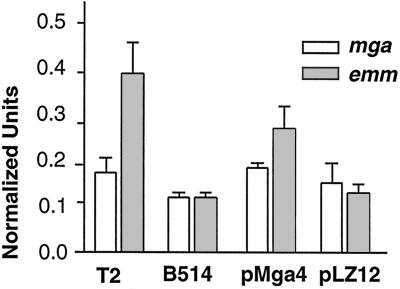
To establish whether a functional defect in mga50 is indeed responsible for the attenuated expression of the mga-regulated genes, a complementation test was performed. The mga gene (mga4) from a serotype 4 strain, AP4, was cloned into the shuttle vector pLZ12-Spec (1) and electroporated into the M50 strain B514. Quantitative RNA slot blotting was performed to measure the amounts of the transcripts of emm50 and mga50. Methods were previously described (13, 24). RNA was isolated from different strains during exponential growth. RNA in 2- and 0.4-μg amounts was loaded onto filters in duplicate. Blots were reacted with emm- or mga-specific DNA and then stripped for hybridization with the recA probe. Hybridized counts were detected by a phosphorimager. The amounts of RNA in the blots were normalized with a recA transcript and compared to those in a standard strain, T2/44/RB4, which was previously found to make normal levels of M protein (24).
The message level for mga50 in strain B514 was approximately 70% of the message level for mga2 in strain T2/44/RB4 (Fig. 2). However, this difference did not appear to be significant, based on the nearness of the standard deviation values. Neither the strain transformed with vector-only plasmid pLZ12-Spec nor the strain transformed with the complementing plasmid pMga4 differed in transcript levels from those of the wild-type representative strain, T2/44/RB4 (Fig. 2).
FIG. 2.
Transcript levels in B514 compared to those in T2/44/RB4 and complemented strains. Transcripts specific for mga, emm, and recA were detected in strains T2/44/RB4 (T2), B514, B514/pMga4 (pMga4), and B514/pLZ12-Spec (pLZ12). Normalized units are defined as the band intensity measured for mga or emm probe at a particular RNA concentration divided by the band intensity of a recA internal control probe at the same RNA concentration (13).
The message level for emm50 in strain B514 was approximately 25% of the message level for emm2 in strain T2/44/RB4 (Fig. 2). Addition of the vector-only plasmid pLZ12-Spec to strain B514 did not increase the emm50 transcript level significantly. Complementation of B514 with mga4 in pMga4 increased the emm transcript a little more than twofold, to a level approximately 70% of that in T2/44/RB4. Thus, we conclude that a functional defect in the mga50 gene or Mga50 protein can be complemented by the presence of the mga4 gene and Mga4 protein in trans. Moreover, as had been found for Mga4 and the gene emm6, the heterologous Mga4 protein functions to activate the emm50 gene. The less-than-twofold increase in the mga transcript level in B514 before complementation argues that the defect is probably in protein function.
Restoration of wild-type levels of M family proteins in strain B514 by complementation with mga-containing plasmids.
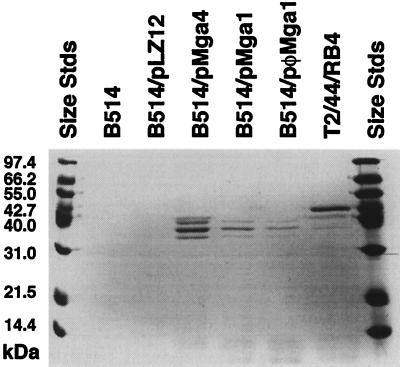
It had been suggested that the decreased transcription of the emm gene results in a lower level of M protein on the surface of B514 (24). This was tested by comparing the amount of M protein in CNBr extracts of strain B514 with strains complemented with different mga alleles expressed under different promoters (Fig. 3). Complementation by the mga4 allele under its own promoter has been described (1). The divergent mga1 gene from a serotype M6 (mga1 from strain D471) GAS strain was cloned into the shuttle vector pLZ12-Spec and expressed either under its own promoter or under one derived from the Lactococcus lactis phage SKIIG (1, 17). As previously described (24), M50 was barely detectable in B514 and in B514/pLZ12-Spec (Fig. 3, lanes 2 and 3). However, both the mga4 and mga1 genes complement the mga50 allele for the production of M50. For mga1, complementation occurs both from the native mga1 promoter (Fig. 3, lanes 4 and 5) and from a constitutive phage promoter (13) (lane 6). CNBr extracts were made by the method of Raeder et al. (21) from in vitro-grown cultures with an absorbance of 0.6 at 600 nm. The number of cells per extract was normalized, and the bands in each extract were visualized on sodium dodecyl sulfate gels by Coomassie blue staining. The detection of measurable amounts of M proteins in complemented strains shows that the heterologous Mga proteins are functional and the amount of M50 protein on the cell surface increases when the amount of emm transcript increases.
FIG. 3.
CNBr extracts of surface proteins from B514, T2/44/RB4, and plasmid-transformed B514 cells. Stds, standards. B514/pLZ12, B514/pLZ12-Spec.
Antiphagocytic function of M50 protein.
The antiphagocytic function of the M protein of GAS is considered to be a critical virulence factor because it enables the bacteria to resist killing by polymorphonuclear leukocytes. The hyaluronic capsule of GAS also performs an antiphagocytic function in some strains (4, 14). Complementation of Mga increased the levels of M and M-related proteins in B514 from the low basal level to substantial levels approximately equivalent to those in fresh human clinical isolates. To demonstrate that complementation also restores resistance to opsonophagocytosis, we used a direct bactericidal assay in whole human blood. For this, we used a derivative of B514, B514.039, in which the hasA gene required to form a capsule had been insertionally inactivated (9). This allowed the test of function to be completed in the absence of a capsule, a second potential mediator of antiphagocytic resistance.
As shown in Table 1, the parent strain (B514.039) and the parent strain transformed with the vector alone (B514.039/pLZ12-Spec) were killed in human blood (80% reduction over a 3-h period) while the complemented strain, B514.039/pMga4, survived and grew 10-fold over the same period. We conclude that one or more of the M or M-like proteins in B514 are capable of conferring resistance to phagocytosis. Moreover, this result demonstrates that the presence of adequate amounts of surface M protein can function to provide resistance to phagocytosis in the absence of a capsule. This protection could be mediated in part by the M-related proteins Mrp50 and Enn50 because the amounts of all three are increased when the defective Mga protein is complemented (data not shown); Mrp2, a homolog of Mrp50, was recently shown to participate in antiphagocytic protection in another streptococcal strain (20).
TABLE 1.
Phagocytosis of capsule-negative B514 strains
| Strain | Phenotypeab | Concn
(CFU/ml)
|
Growthc | ||
|---|---|---|---|---|---|
| Input | In plasma | In blood | |||
| B514.039 | Mgaatt | 5.7 × 102 | 2.5 × 103 | 1.2 × 102 | 0.2 |
| B514.039/pLZ12-Spec | Mgaatt | 5.0 × 102 | 1.4 × 103 | 1.6 × 102 | 0.2 |
| B514.039/pMga4 | Mgawt | 1.7 × 102 | 5.0 × 102 | 1.5 × 103 | 10 |
Mgaatt, attenuated Mga activity; Mgawt, wild-type Mga activity.
Determined after 3-h rotation.
Calculated as follows: (CFU/ml of blood)/(input CFU/ml).
Taken together, our findings support the hypothesis that a defect in the Mga50 protein was the source of previously noted M protein-related deficiencies in the virulence of strain B514. Four polymorphic sites were identified as possible contributors to the dysfunction of Mga50. Of the four, the change in residue 26, which lies within one of the HTH motifs, is thought to be the most significant of these changes.
Nucleotide sequence accession number.
The sequence of mga50 was deposited in GenBank under accession no. AF071802.
Acknowledgments
This work was supported at the University of Alabama at Birmingham by an American Heart Association award to S.K.H. and by NIH grants AI40645 and AI21548. The work at Emory University was supported by NIH grant AI20723, and K.S.M. was supported in part by NIH fellowship AI09460. The DNA Sequencing Core Facility at UAB is supported in part by a grant from the Howard Hughes Medical Institute to the School of Medicine and by a grant from the UAB Health Services Foundation.
REFERENCES
- 1.Andersson G, McIver K S, Heden L-O, Scott J R. Complementation of divergent mga genes in group A Streptococcus. Gene. 1996;175:77–81. doi: 10.1016/0378-1119(96)00124-2. [DOI] [PubMed] [Google Scholar]
- 2.Bessen D E, Sotir C M, Readdy T L, Hollingshead S K. Genetic correlates of throat and skin isolates of group A streptococci. J Infect Dis. 1996;173:896–900. doi: 10.1093/infdis/173.4.896. [DOI] [PubMed] [Google Scholar]
- 3.Caparon M G, Scott J R. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc Natl Acad Sci USA. 1987;84:8677–8681. doi: 10.1073/pnas.84.23.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dale J B, Washburn R G, Marques M B, Wessels M R. Hyaluronate capsule and surface M protein in resistance to opsonization of group A streptococci. Infect Immun. 1996;64:1495–1501. doi: 10.1128/iai.64.5.1495-1501.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollingshead S K, Arnold J, Readdy T L, Bessen D E. Molecular evolution of a multi-gene family in group A streptococci. Mol Biol Evol. 1994;11:208–219. doi: 10.1093/oxfordjournals.molbev.a040103. [DOI] [PubMed] [Google Scholar]
- 6.Hollingshead S K, Readdy T L, Yung D L, Bessen D E. Structural heterogeneity of the emmgene cluster in group A streptococci. Mol Microbiol. 1993;8:707–717. doi: 10.1111/j.1365-2958.1993.tb01614.x. [DOI] [PubMed] [Google Scholar]
- 7.Hook E W, Wagner R R, Lancefield R C. An epizootic in Swiss mice caused by a group A streptococcus, newly designated type 50. Am J Hyg. 1960;72:111–119. doi: 10.1093/oxfordjournals.aje.a120127. [DOI] [PubMed] [Google Scholar]
- 8.Husmann L K, Dillehay D L, Jennings V M, Scott J R. Streptococcus pyogenes infection in mice. Microb Pathog. 1996;20:213–224. doi: 10.1006/mpat.1996.0020. [DOI] [PubMed] [Google Scholar]
- 9.Husmann L K, Yung D L, Hollingshead S K, Scott J R. Role of putative virulence factors of Streptococcus pyogenesin mouse models of long-term throat colonization and pneumonia. Infect Immun. 1997;65:1422–1430. doi: 10.1128/iai.65.4.1422-1430.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kihlberg B M, Cooney J, Caparon M G, Olsen A, Bjorck L. Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb Pathog. 1995;19:299–315. doi: 10.1016/s0882-4010(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 11.McIver K S, Heath A S, Green B D, Scott J R. Specific binding of the activator Mga to promoter sequences of the emm and scpAgenes in the group A streptococcus. J Bacteriol. 1995;177:6619–6624. doi: 10.1128/jb.177.22.6619-6624.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McIver K S, Heath A S, Scott J R. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emmtranscription. Infect Immun. 1995;63:4540–4542. doi: 10.1128/iai.63.11.4540-4542.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McIver K S, Scott J. Role of mgain growth phase regulation of virulence genes of group A streptococcus. J Bacteriol. 1997;179:5178–5187. doi: 10.1128/jb.179.16.5178-5187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moses A E, Wessels M R, Zalcman K, Alberti S A, Natanson-Yaron S, Menes T, Hanski E. Relative contributions of hyaluronic acid capsule and M protein to virulence in a mucoid strain of the group A streptococcus. Infect Immun. 1997;65:64–71. doi: 10.1128/iai.65.1.64-71.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okada N, Geist R T, Caparon M G. Positive transcriptional control of mryregulates virulence in the group A streptococcus. Mol Microbiol. 1993;7:893–903. doi: 10.1111/j.1365-2958.1993.tb01180.x. [DOI] [PubMed] [Google Scholar]
- 16.Perez-Casal J, Caparon M G, Scott J R. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogeneswith similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Platteeuw C, Simons G, de Vos W M. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1994;60:587–593. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Podbielski A, Flosdorff A, Weber-Heynemann J. The group A streptococcal virR49 gene controls expression of four structural virregulon genes. Infect Immun. 1995;63:9–20. doi: 10.1128/iai.63.1.9-20.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Podbielski A, Peterson J A, Cleary P. Surface protein-CAT reporter fusions demonstrate differential gene expression in the virregulon of Streptococcus pyogenes. Mol Microbiol. 1992;6:2253–2265. doi: 10.1111/j.1365-2958.1992.tb01401.x. [DOI] [PubMed] [Google Scholar]
- 20.Podbielski A, Schnitzler N, Beyhs P, Boyle M D P. M-related protein (Mrp) contributes to group A streptococcal resistance to phagocytosis by human granulocytes. Mol Microbiol. 1996;19:429–441. doi: 10.1046/j.1365-2958.1996.377910.x. [DOI] [PubMed] [Google Scholar]
- 21.Raeder R, Otten R, Chamberlin L, Boyle M. Functional and serological analysis of type II immunoglobulin G-binding proteins expressed by pathogenic group A streptococci. J Clin Microbiol. 1992;30:3074–3081. doi: 10.1128/jcm.30.12.3074-3081.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robbins J C, Spanier J G, Jones S J, Simpson W J, Cleary P P. Streptococcus pyogenestype 12 M protein gene regulation by upstream sequences. J Bacteriol. 1987;169:5633–5640. doi: 10.1128/jb.169.12.5633-5640.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott J R, Cleary P P, Caparon M G, Kehoe M, Heden L, Musser J M, Hollingshead S K, Podbielski A. New name for the positive regulator of the M protein of group A Streptococcus. Mol Microbiol. 1995;17:799. doi: 10.1111/j.1365-2958.1995.mmi_17040799.x. [DOI] [PubMed] [Google Scholar]
- 24.Yung D L, Hollingshead S K. DNA sequencing and gene expression of the emmgene cluster in an M50 mouse virulent group A streptococci. Infect Immun. 1996;64:2193–2200. doi: 10.1128/iai.64.6.2193-2200.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]