Abstract
The recent clinical success of multiple mRNA-based SARS-CoV-2 vaccines has proven the potential of RNA formulated in lipid nanoparticles (LNPs) in humans, and products based on base-modified RNA, sequence-optimized RNA, and self-replicating RNAs formulated in LNPs are all in various stages of clinical development. However, much remains to be learned about critical parameters governing the manufacturing and use of LNP-RNA formulations. One important issue that has received limited attention in the literature to date is the identification of optimal storage conditions for LNP-RNA that preserve long-term activity of the formulations. Here, we analyzed the physical structure, in vivo expression characteristics, and functional activity of alphavirus-derived self-replicating RNA (repRNA)-loaded LNPs encoding HIV vaccine antigens following storage in varying temperatures, buffers, and in the presence or absence of cryoprotectants. We found that for lipid nanoparticles with compositions similar to clinically-used LNPs, storage in RNAse-free PBS containing 10% (w/v) sucrose at −20 °C was able to maintain vaccine stability and in vivo potency at a level equivalent to freshly prepared vaccines following 30 days of storage. LNPs loaded with repRNA could also be lyophilized with retention of bioactivity.
Keywords: Lipid nanoparticle, Vaccine storage, RNA delivery, Freeze-storage, Lyophilization
Graphical abstract
1. Introduction
One of the first experiments on the delivery of RNA molecules dates back to 1978, when mouse lymphocytes were transfected in vitro with mRNA encoding rabbit globin using liposomes [1]. Since then, advances in ionizable lipids and RNA loading techniques led to the approval of the first siRNA-based lipid nanoparticle (LNP) therapeutic, Onpattro (Patisiran) by the US Food and Drug Administration (FDA) in 2018 [[2], [3], [4], [5]]. By early 2020 when the COVID-19 pandemic occurred, several mRNA-loaded LNP formulations had reached clinical trials [6]. In December 2020, the FDA issued Emergency Use Authorization (EUA) for COVID-19 vaccines from Pfizer-BioNTech (BNT162b2) and Moderna (mRNA-1273), both of which utilize LNPs loaded with mRNA encoding the viral spike protein as the immunogen [7,8]. Currently, LNP formulations of diverse RNA products including oligonucleotides (e.g., siRNA, miRNA, anti-sense oligonucleotides, etc.) [[9], [10], [11], [12], [13]], base-modified RNA [[14], [15], [16]], sequence-optimized RNA [[17], [18], [19], [20]], and self-replicating RNAs [[21], [22], [23], [24]] derived from alphaviruses, flaviviruses, measles viruses, and rhabdoviruses are in various stages of clinical development for applications ranging from vaccines to cancer therapy. Most recently, a COVID-19 vaccine based on LNP-formulated self-replicating RNA (ARCT-154) achieved promising phase 3 efficacy data in Vietnam [25,26].
While both of the approved mRNA vaccines for COVID-19 are similar in structure, BNT162b2 uses phosphate-buffered saline (PBS) as the solvent (with 20% w/v sucrose), whereas mRNA-1273 uses tris-HCl-buffered saline (TBS; with 8% w/v sucrose) [[27], [28], [29], [30], [31], [32]]. Both formulations have reported reliable maintenance of vaccine stability and efficacy in these sucrose buffers, but the range of storage temperature and lifetime is broad; mRNA-1273 is stored at −20 °C for up to 6 months, whereas BNT162b2 is concentrated and stored at −70 °C for up to 6 months, with more recent data indicating storage at −25 °C to −15 °C for two weeks is also stable [27,33,34]. There is limited public knowledge regarding the impact of key storage parameters (e.g., cryoprotectant, dispersant, temperature, etc.) on LNP-RNA vaccines, as noted by many authors in light of the recent pandemic [28,33,[35], [36], [37], [38]]. In the limited number of published studies, several common parameters are found to affect storage outcomes. Sugar-based cryoprotectants, such as sucrose, trehalose, and mannitol, improve the stability of LNPs during freeze-thawing and lyophilization [35,39,40]. Constituting the LNPs in different aqueous solvents (e.g., water, saline) or in a mix with organic solvents (e.g., ethanol) displayed minor effects in improving the stability of LNP-RNA [35,39,40]. However, the most dominant variable seems to be the storage temperature, ranging from flash freezing in liquid nitrogen to refrigeration at 4 °C, where different formulations seem to favor different temperatures. We previously reported a formulation comprised of TT3 (a lipid-like ionizable molecule), DOPE, cholesterol, and DMG-PEG2k loaded with mRNA encoding luciferase that was successfully flash frozen in liquid nitrogen, and stored for up to 3 months with 5% w/v sucrose or trehalose [39]. On the other hand, a formulation composed of an unidentified ionizable lipid, DSPC, cholesterol, and DMG-PEG2k loaded with mRNA encoding the receptor binding domain (RBD) of SARS-CoV-2 mouse-adapted strain was maintained at room temperature for at least 7 days, and potentially longer at 4 °C [41]. Another work presented imidazole-modified lipids that seem to form highly stable RNA-loaded structures by addition of ether bonds and amine head groups that allow for pi-stacking [42]; impressively, this LNP was able to retain RNA integrity and function for out to 25 weeks at 4 °C, 18 weeks at 25 °C, and 3 weeks at 37 °C in PBS without the need for cryoprotectants.
The exact reason for the wide range of shelf lives reported for LNP-RNA vaccines remains unknown. In addition, many studies of LNP-RNA stability have focused on functional measures using mRNA encoding for a reporter gene (e.g., luciferase, GFP) rather than direct assessments of vaccine immunogenicity, and analyses linking structural integrity of LNPs to vaccine activity are lacking. To help fill this knowledge gap, here we investigated the effects of cryoprotectants, buffer type (phosphate- and tris-buffered saline), and storage temperature on both the structural and functional maintenance of self-replicating RNA-loaded LNP (LNP-RNA) vaccines.
2. Material and methods
2.1. Materials
N1,N3,N5-tris(3-(didodecylamino)propyl)benzene-1,3,5-tricarboxamide (TT3) was synthesized as previously described [43]; (6Z,9Z,28Z,31Z)-Heptatriaconta-6,9,28,31-tetraen-19-yl 4-(dimethylamino) butanoate (DLin-MC3-DMA) was purchased from MedChemExpress (CAT#HY-112251); 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE; CAT#850725), Cholesterol (CAT#700100), 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG2k; CAT#88015) were purchased from Avanti Polar Lipids. HIV Env trimer N332-GT2 was prepared as previously described [44]. Citrate buffer (pH 3; CAT#J61391-AK) was purchased from Alfa Aesar. For dialysis, 20 K MWCO Slide-A-Lyzer™ MINI Dialysis Device (ThermoFisher Scientific), RNAse-free PBS (AM9625; ThermoFisher Scientific), Tris Buffer (ThermoFisher Scientific), and D-(+)-Sucrose, Ultrapure DNAse-, RNAse-free (97061–432; VWR) were used. mRNA encoding for GFP was purchased from APExBIO (CAT# R1007). For bioluminescence studies, luciferin was purchased from GoldBio (CAT# LUCK-1G).
2.2. RNA synthesis
Venezuelan equine encephalitis virus (VEE) replicon plasmid DNA was prepared based on mutant constructs previously described [[45], [46], [47]]. Firefly luciferase or HIV immunogen N332-GT2 (a stabilized SOSIP trimer of the HIV envelope glycoprotein spike [44]) were cloned after the subgenomic promoter as previously described [45,46]. Replicon RNAs were in vitro transcribed (IVT) from templates of linearized VEE DNA constructs using the HiScribe T7 Quick High Yield RNA Synthesis Kit (New England Biolabs) following the manufacturer's instructions. The resulting replicon RNAs were capped and methylated using the ScriptCap Cap 1 Capping System (Cellscript) according to the manufacturer's instructions. RNA was purified in water with 300,000 PES columns (Sartorius), and purity was assessed by gel electrophoresis.
2.3. Lipid nanoparticle synthesis
Lipid nanoparticles (LNPs) were synthesized using a microfluidic organic-aqueous precipitation method. The organic phase was prepared by solubilizing the lipids TT3, Dlin-MC3-DMA, DOPE, Cholesterol, and DMG-PEG2k in ethanol at a molar ratio of 10:25:20:40:5. The aqueous phase of RNA was prepared by diluting the RNA (stored in RNAse-free water) with 10 mM citrate buffer at pH 3.0 (CAT#J61391-AK; Alfa Aesar). Lipids were stored in ethanol at −20 °C, and RNA constructs were stored in RNAse-free water at −80 °C and were thawed on ice before use. The two phases were prepared at an ethanol:aqueous volume ratio of 1:2, and RNA and lipids combined at an N:P ratio of 2:1. Each phase was loaded into a syringe (BD), and locked onto the NxGen microfluidic cartridge for mixing using a NanoAssemblr Ignite instrument (Precision Nanosystems). The Ignite was set to operate with the following settings: volume ratio- 2:1; flow rate- 12 mL/min; waste volume- 0 mL. The resulting LNPs were dialyzed against predetermined buffers (0–30% w/v sucrose in pH 7.4 PBS or TBS) using 20 K MWCO Slide-A-Lyzer™ MINI Dialysis casettes (ThermoFisher Scientific) at 25 °C for 90 min, with an exchange of the buffer reservoir after 45 min. The dialyzed LNPs were then stored under predetermined test conditions.
2.4. LNP-RNA lyophilization
LNP-RNAs were synthesized and diluted (50, 20, 10, or 3.3 ng/μL) in different buffers (PBS, 10%, or 30% w/v sucrose) and frozen (−20 °C, −80 °C or − 200 °C) for 24 h according to the test conditions. The frozen samples were then lyophilized for 24 h in a LabConco Freezone 4.5 Liter Benchtop Freeze Dry System. Eppendorf tubes containing the frozen samples were opened, covered with perforated parafilm, and placed in a pre-chilled 50 mL conical tube during lyophilization. Lyophilization was conducted at the default fixed setting of −60 °C. Lyophilized samples were stored at 4 °C unless hydrated immediately, and were reconstituted in deionized water before use.
2.5. Particle characterization
Dynamic light scattering (DLS; Malvern Panalytical) was performed to determine the hydrodynamic size, polydispersity index, and zeta-potential of the LNPs. For hydrodynamic size measurements, 10 μL of particles in different buffers were diluted in 800 μL of deionized water and placed into the 1.5 mL cuvette (Fisher Scientific) for measurement. The same sample was transferred to the folded capillary zeta cell (Malvern Panalytical) to measure the zeta-potential. Electron microscopy was conducted to qualitatively assess the LNP size and polydispersity. In sample preparation for cryogenic electron microscopy (cryo-EM), 3 μL of the particles sample in buffer containing solution was dropped on a lacey copper grid coated with a continuous carbon film and blotted to remove excess sample without damaging the carbon layer by Gatan Cryo Plunge III. Grid was then mounted on a Gatan 626 single tilt cryo-holder equipped in the TEM column. The specimen and holder tip were cooled down by liquid‑nitrogen, and the temperature was maintained during transfer into the microscope and subsequent imaging. For negative stained-transmission electron microscopy (TEM), 10 μL of the particle sample in buffer containing solution was dropped on a 200 mesh copper grid coated with a continuous carbon film and excess solution was removed after 60 s of waiting by blotting with a wipe. Then 10 μL of negative staining solution, phosphotungstic acid as a 1% aqueous solution was dropped on the TEM grid and immediately removed by blotting. Another 10 μL of the stain is applied to the grid with blotted removal after 30 s, and the grid is dried at room temperature. The dried grid was mounted on a JEOL single tilt holder equipped in the TEM column. The specimen was cooled down by liquid‑nitrogen. Imaging on a JEOL 2100 FEG microscope was conducted using a minimum dose method that is essential to avoid sample damage under the electron beam. The microscope was operated at 200 kV and with a magnification in the ranges of 10,000–60,000 for assessing particle size and distribution. All images were recorded on a Gatan 2kx2k UltraScan CCD camera.
2.6. Mice
Female Balb/C (JAX Stock No. 000651) mice 6–8 weeks of age were maintained in the animal facility at the Massachusetts Institute of Technology (MIT). All animal studies and procedures were carried out following federal, state, and local guidelines under an IACUC-approved animal protocol.
2.7. In vivo mouse imaging
To qualitatively assess the efficacy of thawed LNPs compared with freshly prepared samples using repRNA encoding for luciferase (repLuc) as a reporter, we simulated vaccination in cohorts of Balb/C mice by intramuscularly (i.m.) injecting 1 μg RNA doses of the LNPs loaded with repLuc in each of the left and right gastrocnemius muscle. At days 1, 3, 7, 10, and 15 post-i.m. injection, the mice were intraperitoneally (i.p.) administered 200 μL of luciferin (50 mg/mL in PBS), and imaged using the In Vivo Imaging System (Xenogen IVIS 200; PerkinElmer) 10 min post-i.p. injection.
2.8. Serum antibody titer quantification
To quantitatively assess the efficacy of thawed LNPs compared with freshly prepared samples using repRNA encoding for the HIV env trimer, we vaccinated healthy Balb/C mice by injecting 1 μg RNA doses of the LNP-RNA i.m. in each of the left and right gastrocnemius muscle. At weeks 2 and 4 post-i.m. injection, the mice underwent retro-orbital bleeding; blood was collected in Z-gel PP tubes for blood serum collection (CAT#41.1500.005; Sarstedt). Serum was collected by centrifuging blood at 10,000 ×g for 4 min, and stored at −80 °C prior to use. To conduct ELISAs, NUNC MaxiSorp plates were coated overnight with 1 μg/mL purified HIV antigen in PBS, then blocked for 2 h with 10% BSA in PBS. Mouse sera were initially diluted 50× in blocking buffer, followed by 3× serial dilutions. Diluted sera were transferred to blocked plates and incubated for 2 h. HRP-conjugated immunoglobulins (e.g., IgG, IgG1, IgG2a, IgG2b, IgG3, IgM; Bio-Rad) were used as detection antibodies at 1:5000 for endpoint titer assessments, with gp120-specific monoclonal antibody VRC01 used as a positive control. 3,3′,5,5’-Tetramethylbenzidine (TMB) signal was read using a microplate reader by subtracting the absorbance at 450 nm by that at 550 nm.
2.9. Germinal center (GC) assay
Balb/C mice were immunized as described above, and at 2 weeks post-vaccination, popliteal lymph nodes were collected and mechanically dissociated to obtain single cell suspensions. Zombie Aqua (BioLegend) in PBS was used to stain for cell viability, and antibody staining was performed in fluorescence-activated cell sorting (FACS) buffer (PBS, 1% bovine serum albumin (BSA), 0.02% NaN3, and 2 mM EDTA). Fc-mediated binding was blocked using purified anti-CD16/32 (2.5 μg/mL; 93; BioLegend) at 4 °C for 15 min, on top of which we added primary antibodies for cell surface staining at 4 °C for 30 min. GC B cells were stained using anti-GL7 PerCPCy5.5 (GL7; BioLegend), anti-CD38 AF488 (90; BioLegend), anti-B220 PECy7 (RA3-6B2; BioLegend), and anti-CD4 BV711(GK1.5; BioLegend). Follicular helper T cells were assessed using anti-CD4-BV711 (GK1.5; BioLegend), anti-B220-PECy7 (RA3-6B2; BioLegend), anti-CXCR5-PE (phycoerythrin) (L138D7; BioLegend), and anti-PD1-BV421 (29F.1A12; BioLegend). The HIV env trimer used as the antigen was conjugated to either BV605 (streptavidin-conjugated; BioLegend) or APC-Cy7 (streptavidin-conjugated; BioLegend), and both probes were used to detect antigen-specific B cells. Stained cells were fixed with 4% paraformaldehyde (ThermoFisher Scientific) for 10 min at 25 °C, washed and resuspended in FACS buffer for flow cytometric analysis on a BD FACSymphony™ A3 Cell Analyzer (BD Biosciences).
2.10. Enzyme-linked immunosorbent spot (ELISpot) assay
Spleens were harvested 2 weeks after mice had been intramuscularly vaccinated with LNPs loaded with repRNA encoding for the HIV immunogen in the left and right gastrocnemius muscles of mice. Splenocytes were isolated by mechanical dissociation of the spleen and erythrocytes were removed using the Gibco Ammonium-Chloride-Potassium lysing buffer (Thermo Fisher Scientific). ELISpot was conducted using the mouse IFN-γ ELISPOT Kit (BD Biosciences). Cells were seeded on IFN-γ-coated wells at 106 cells/well in triplicate for three pools of the HIV env trimer peptides, which were added to the cells at 2 μg/mL. Cells were stimulated by wrapping the plate in foil and incubating overnight at 37 °C. Plates were developed and detected according to manufacturer's instructions. Plates were scanned using the CTL-ImmunoSpot Plate Reader, and data were analyzed using CTL ImmunoSpot Software.
2.11. Bead-based ELISA cytokine quantification
Mouse muscles were analyzed using the Legendplex mouse antivirus response panel (Biolegend) following the manufacturer's suggested protocol and analyzed using the LEGENDplex Data Analysis Software Suite. At 24 h post-vaccination, gastrocnemius muscles were collected from mice. Tissues were homogenized in GentleMACS M Tubes (Miltenyi Biotec) filled with 1.5 mL of lysis buffer (20 mM Tris (pH 7.8), 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 1% Triton X-100, 10% (w/v) glycerol, 1 mM EDTA, 1 mM dithiothreitol, HALT Protease Cocktail (ThermoFisher)) using the GentleMACS Octo Dissociator (Miltenyi Biotec). The tissues were homogenized and then further incubated in the lysis buffer overnight at room temperature on a rotor shaker before being analyzed or flash frozen and stored at −80 °C until later analysis.
3. Results
3.1. Effect of sucrose on maintenance of structural and functional integrity of LNP-RNA
Lipid nanoparticles (LNPs) were synthesized using a microfluidic system mixing an organic phase of lipids in ethanol with aqueous phase repRNA in water to induce self-assembly of LNPs encapsulating the replicon (LNP-RNA). LNPs were prepared using a 2:1 ratio of ionizable lipid amine groups to RNA phosphates (equivalent to a 2.9:1 ionizable lipid:RNA mass ratio). The resulting particles were dialyzed into pH 7.4 phosphate-buffered saline (PBS) or tris-buffered saline (TBS) containing 0%, 5%, 10%, or 30% w/v sucrose to test the effects of buffer and cryoprotectant. The particles were then stored at 4 °C, −20 °C, −80 °C, or − 200 °C (flash freezing in liquid nitrogen). After 7 days, the LNP-RNA were thawed at 25 °C, and their hydrodynamic size and polydispersity were measured using dynamic light scattering (DLS). Freshly prepared LNPs in their respective buffer and sucrose concentrations served as controls. As shown in Table 1 , regardless of buffer and sucrose concentration, storage of LNPs at −80 °C resulted in particle aggregation and high polydispersity, while storage at other temperatures (4 °C, −20 °C, and − 200 °C) maintained LNP size distributions comparable to fresh particles. Notably, the absence of sugar in the buffer for both PBS- and TBS-stored LNPs resulted in a 20–50 nm increase in the particle size distribution, hinting at the cryopreservative effects of sucrose. Interestingly, structural integrity was well maintained at all tested temperatures when mRNA was loaded into the same LNPs instead of repRNA (Supplementary Fig. S1).
Table 1.
Dynamic light scattering (DLS) assessment of lipid nanoparticles. (n = 3).
| PBS | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0% sucrose | 5% sucrose | 10% sucrose | 30% sucrose | |||||||||||||||||
| Temp | Fresh | 4 °C | −20 °C | −80 °C | −200 °C | Fresh | 4 °C | −20 °C | −80 °C | −200 °C | Fresh | 4 °C | −20 °C | −80 °C | −200 °C | Fresh | 4 °C | −20 °C | −80 °C | −200 °C |
| PDI ± st. dev |
0.233 ± 0.030 |
0.401 ± 0.016 |
0.273 ± 0.012 |
0.912 ± 0.153 |
0.282 ± 0.041 |
0.294 ± 0.060 |
0.357 ± 0.050 |
0.293 ± 0.072 |
0.818 ± 0.261 |
0.212 ± 0.018 |
0.292 ± 0.061 |
0.280 ± 0.027 |
0.273 ± 0.056 |
0.980 ± 0.035 |
0.171 ± 0.062 |
0.283 ± 0.049 |
0.336 ± 0.027 |
0.286 ± 0.063 |
0.688 ± 0.209 |
0.280 ± 0.092 |
|
Z-avg diam. ± st. dev (nm) |
114 ± 3 |
107 ± 40 |
125 ± 1 |
4037 ± 2267 |
141 ± 5 |
92 ± 18 |
95 ± 7 |
96 ± 6 |
1448 ± 781 |
124 ± 51 |
90 ± 7 |
88 ± 10 |
88 ± 13 |
3927 ± 4238 |
93 ± 21 |
96 ± 8 |
97 ± 24 |
86 ± 14 |
741 ± 498 |
83 ± 7 |
| TBS | ||||||||||||||||||||
| PDI ± st. dev |
0.293 ± 0.016 |
0.225 ± 0.008 |
0.246 ± 0.01 |
0.784 ± 0.201 |
0.255 ± 0.016 |
0.251 ± 0.021 |
0.257 ± 0.060 |
0.239 ± 0.071 |
0.787 ± 0.188 |
0.228 ± 0.047 |
0.231 ± 0.012 |
0.247 ± 0.036 |
0.225 ± 0.054 |
0.866 ± 0.155 |
0.235 ± 0.043 |
0.230 ± 0.036 |
0.281 ± 0.103 |
0.239 ± 0.088 |
0.907 ± 0.161 |
0.255 ± 0.042 |
| Z-avg diam. ± st. dev (nm) | 113 ± 11 |
159 ± 7 |
162 ± 5 |
1959 ± 786 |
155 ± 8 |
123 ± 17 |
113 ± 7 |
120 ± 17 |
1140 ± 590 |
120 ± 14 |
118 ± 12 |
114 ± 10 |
120 ± 17 |
1558 ± 1302 |
121 ± 24 |
119 ± 13 |
120 ± 15 |
122 ± 19 |
4856 ± 3625 |
118 ± 13 |
A similar overall trend was seen in the polydispersity index of the LNPs (Table 1 and Supplementary Fig. S2), where the average PDI of LNPs increased significantly to ∼0.9 when LNPs were thawed from storage at −80 °C, versus a baseline of ∼0.3 when freshly prepared. The zeta-potential of the LNPs also increased significantly from +3 mV to ∼24 mV after storage at −80 °C, suggesting reorganization of the particle surface composition (Supplementary Fig. S3). On the other hand, retention of encapsulated RNA by the LNPs showed a different trend (Supplementary Fig. S4); while LNP-RNA that were freshly prepared, stored at −20 °C, −80 °C, and − 200 °C maintained an average encapsulation efficiency of approximately 90%, particles that were stored at 4 °C displayed a significant drop to below 70% (p < 0.001), with an increase in RNA detectable outside of the particles (p < 0.001). Overall, temperature seemed to play a more dominant role in determining structural retention of replicon-loaded LNPs over sucrose concentrations in the 0–30% w/v range.
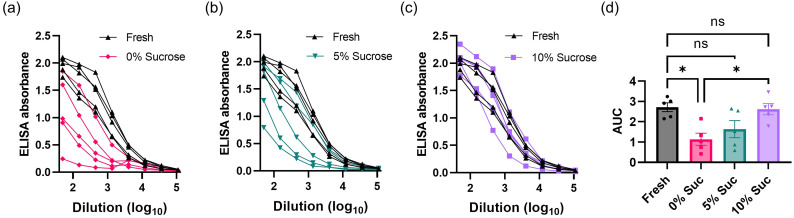
As storage in 0% w/v sucrose seemed to maintain the overall physical integrity of the particles despite a slight shift in the hydrodynamic size, we aimed to determine whether the presence of sugar was necessary for preservation of vaccine potency in vivo. To this end, we loaded LNPs with repRNA encoding for a stabilized HIV Env SOSIP trimer immunogen (termed N332-GT2) designed for priming B cells targeting the N332 supersite of the HIV Env spike [44]. The vaccine particles were stored in PBS containing 0%, 5%, or 10% w/v sucrose for a week at −20 °C, then thawed and injected i.m. into mice, and serum antibody titers were evaluated 4 weeks later by ELISA (Fig. 1 ). Overall, there was a trend for increased antibody production with increasing concentrations of sucrose: While LNPs stored in PBS with 0% w/v sucrose (Fig. 1a) were able to induce antibody production against the immunogen, the level was significantly lower than that of freshly prepared vaccines (p = 0.0118) and the vaccine stored with 10% w/v sucrose (p = 0.0185, Fig. 1d). Storage in 5% w/v sucrose elicited antibody titers that were not statistically significantly different from that of the fresh vaccines (p = 0.1069) but showed a trend toward a weaker response in some animals (Fig. 1b). However, animals vaccinated with LNP-RNAs stored in 10% w/v sucrose generated serum antibody dilution curves that overlapped well with those of mice vaccinated with freshly prepared LNP-RNAs, with no statistically significant difference (p = 0.9959, Fig. 1c, d).
Fig. 1.
Effects of buffer and sucrose concentration on storage of lipid nanoparticle vaccines. (a-d) Groups of balb/C mice (n = 5 animals/group) were immunized i.m. with 1 μg RNA in each leg using LNPs that had been stored under the indicated conditions at −20 °C for 7 days prior to vaccination. Shown are raw ELISA absorbances vs. serum dilution curves for each animal for LNP-RNA in PBS containing: (a) 0% w/v sucrose; (b) 5% w/v sucrose; and (c) 10% w/v sucrose. The area under the curve (AUC) of the ELISA absorbance vs. dilution curves are shown in (d) as means ± s.e.m. Statistics represent one-way ANOVA and Tukey's HSD Test (ns = not significant; *, p < 0.05).
As we observed the lowest PDI measurements in both PBS and TBS, and the most reliable antibody responses storing in 10% w/v sucrose, we decided to move forward using this concentration of sucrose for subsequent studies.
3.2. Effect of buffer on the in vivo performance of LNP-RNAs
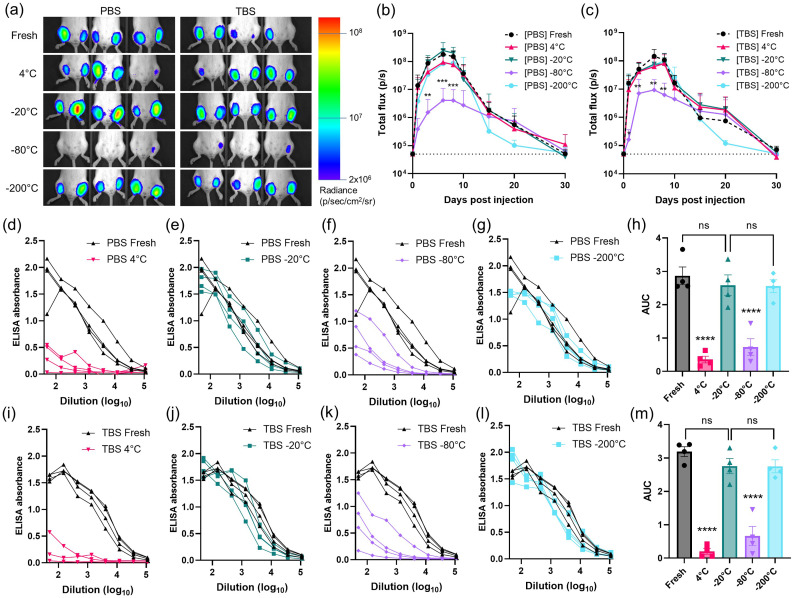
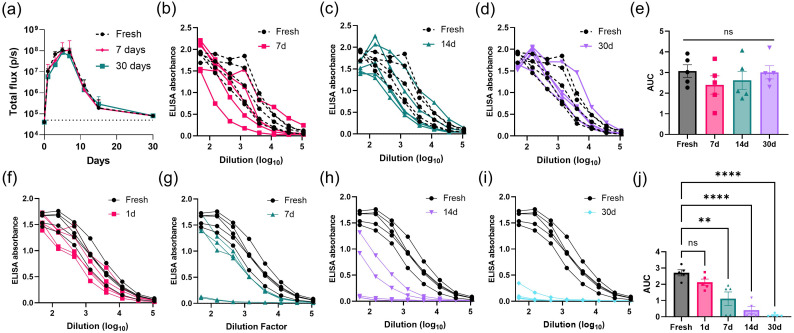
Next, we carried out measurements of in vivo RNA expression for LNPs stored under different conditions. LNPs were prepared carrying repRNA encoding for firefly luciferase, and the LNP-RNA were dialyzed in 10% w/v sucrose in PBS or TBS. The LNPs were then stored at 4 °C, −20 °C, −80 °C, or − 200 °C (liquid nitrogen) for 7 days prior to thawing for injection. On the day of injection, fresh samples were synthesized and dialyzed against 10% w/v sucrose in PBS or TBS. LNPs were administered i.m. into the left and right gastrocnemius muscles of mice, followed by longitudinal bioluminescence imaging of luciferase expression using an IVIS Spectrum whole-animal imaging system (Fig. 2a-c). As expected from our prior studies of this replicon system [48], luciferase expression rapidly climbed for ∼7–10 days, then slowly decayed toward baseline after ∼30 days. However, in alignment with the particle distribution data shown in Fig. 1, LNPs stored at −80 °C in either PBS or TBS exhibited 10 to 100-fold lower peak luciferase signals, indicating significantly diminished RNA delivery (p peak = 0.0003). LNPs stored at other temperatures in PBS and TBS generally produced approximately 2 to 4-fold lower signal compared to the freshly prepared particles at the peak timepoint of day 7 post-injection, despite having demonstrated particle distributions similar to the fresh samples. In addition, samples stored at −200 °C showed an early decay in expression between days 15 and 20. The preparation that performed most closely to the fresh sample was LNPs prepared in 10% w/v sucrose in PBS and stored at −20 °C (p peak = 0.8777), suggesting this condition may provide optimal retention of LNP structure and function.
Fig. 2.
In vivo transfection and antibody titer response following administration of LNPs loaded with RNA stored under different conditions. (a-c) Groups of balb/c mice (n = 3 animals/group averaged across a total of 6 legs/group) were injected i.m. in both the left and right gastrocnemius muscles with 1 μg replicon RNA in LNPs stored in 10% sucrose under the indicated conditions. (a) shows photographs of mice at day 6 post-injection with luciferin channel overlay and a log-scale gradient in radiance (p/s/cm2/sr). Luciferase reporter signals over time of vaccines in (b) PBS; or (c) TBS are plotted, where dotted lines indicate background signal of untreated mice; shown are means ± standard deviation (n = 12). Statistics represent two-way ANOVA and Tukey's HSD Test (*, p < 0.05; **, p < 0.01; ***, p < 0.001). (d-m) Groups of balb/C mice (n = 4 animals/group) were immunized i.m. in each leg with 1 μg RNA loaded in LNPs that had been stored under the indicated conditions for 7 days prior to vaccination, and serum antibody responses were quantified by ELISA assay conducted on mouse sera collected at 4 weeks post-vaccination. Shown are raw ELISA signal vs. serum dilution curves for each animal for PBS- (d-g) and TBS- (i-l) stored samples. Area-under-the-curve (AUC) values for each absorbance vs. dilution data set are shown for PBS- (h) and TBS- (m) stored samples with means ± s.e.m. Statistics represent one-way ANOVA and Tukey's HSD Test (ns = not significant; ****, p < 0.0001).
We next assessed the immunogenicity of LNP-replicon vaccines that were loaded with repRNA encoding the N332-GT2 HIV immunogen, and stored in 10% sucrose in PBS or TBS for 7 days at 4 °C, −20 °C, −80 °C, or − 200 °C (liquid nitrogen). Mice were immunized i.m. with recovered LNPs, and serum antibody responses were evaluated 4 weeks later by ELISA (Fig. 2d-m). Interestingly, vaccines stored either at 4 °C or − 80 °C showed a dramatic decrease in antibody responses compared to the other storage conditions or fresh LNPs (Fig. 2d, f, i and k, p < 0.0001). By contrast, PBS- and TBS-prepared vaccines stored at −20 °C (Fig. 2e and j, p -20 °C /PBS = 0.9113; p -20 °C /TBS = 0.5545) and − 200 °C (Fig. 2g and l, p -200 °C /PBS = 0.8791; p -200 °C /TBS = 0.5431) induced antibody titers that were not statistically different from freshly prepared vaccines. While very similar in outcome, we narrowed our focus to analysis of LNPs prepared in PBS for downstream analyses, rather than TBS based on the luminescence reporter and antibody response statistics (Fig. 2h and m).
3.3. Qualitative assessment of LNP stability
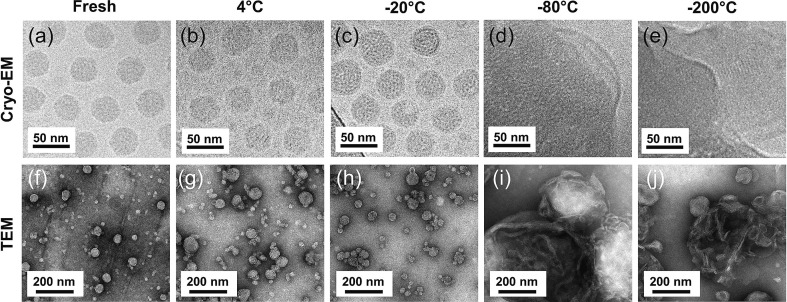
While dynamic light scattering (DLS) offers quantitative information about particle size distribution and hydrodynamic size, the information tends to be skewed toward larger particles or aggregates in polydisperse samples [[49], [50], [51]]. To gain further insights into particle structure, we next carried out transmission electron microscopy (TEM) and cryogenic electron microscopy (cryo-EM) imaging of samples recovered from different storage temperatures. Both the TEM and cryo-EM images show relatively monodispersed LNPs of approximately 45 nm in diameter in the freshly prepared samples and those stored at 4 °C or − 20 °C for a week (Fig. 3a-c and f-h). However, samples thawed after storing at −80 °C or − 200 °C (flash frozen in liquid nitrogen) showed significant aggregation and/or fusion of LNPs into large structures (Fig. 3d, e and i, j).
Fig. 3.
Cryogenic electron microscope (Cryo-EM; top row) and transmission electron microscope (TEM; bottom row) images of LNPs stored for 7 days. LNPs freshly prepared or stored at indicated temperatures in PBS containing 10% w/v sucrose for 7 days followed by thawing at 25 °C were imaged: (a,f) fresh synthesis; (b,g) 4 °C; (c,h) -20 °C; (d.i) -80 °C; and (e,j) -200 °C. TEM images were obtained with negative stain using 1% phosphotungstic acid (PTA).
3.4. Immunogenicity of LNP-replicon vaccines stored at different temperatures
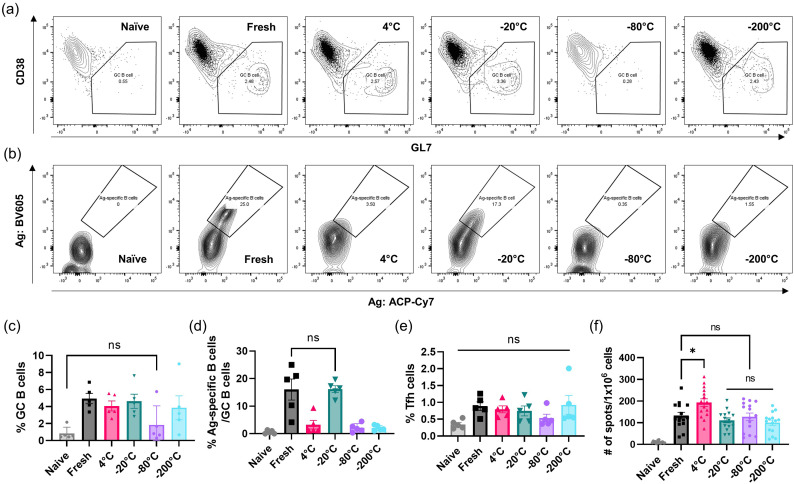
To gain further insight regarding storage effects on LNP-RNA vaccine efficacy, we evaluated additional readouts of the immune response following immunization with freshly prepared particles vs. LNPs stored at different temperatures. To this end, LNPs were loaded with repRNA encoding the N332-GT2 HIV immunogen, stored in 10% w/v sucrose in PBS at either 4 °C, −20 °C, −80 °C, −200 °C for 7 days, and then thawed and administered to balb/C mice alongside freshly prepared samples. At week 2 post-injection, mice were sacrificed and popliteal lymph nodes (draining nodes from the gastrocnemius injection site) were harvested for evaluating germinal center (GC) responses (Fig. 4a-e) and splenocytes were collected for assessing T cell responses using enzyme-linked immune absorbent spot (ELISpot) (Fig. 4f). Flow cytometric analyses of cells recovered from the draining popliteal lymph nodes revealed that while vaccines stored at 4 °C, −20 °C, and − 200 °C induced prominent GC B cell responses, −80 °C stored vaccines did not elicit a response statistically different from naïve control animals (p = 0.9619, Fig. 4a and c). Interestingly, despite inducing GC B cell differentiation, vaccines stored at 4 °C or − 200 °C failed to prime a meaningful population of antigen-specific GC B cells that could bind to recombinant HIV Env trimer probes, and only vaccines stored at −20 °C elicited a strong antigen-specific GC B cell response matching that elicited by freshly-prepared vaccines (Fig. 4b and d). Follicular helper T cell (Tfh) responses were modest and not statistically different among any of the groups (Fig. 4e). In contrast to the GC data, IFN-γ-producing antigen-specific T cell responses in the spleen were induced by all of the vaccines, with modest differences between groups (Fig. 4f and Supplementary Fig. S5). Based on these results, we selected the −20 °C freezing as the optimum storage condition for our LNP-RNA vaccine formulation.
Fig. 4.
Evaluation of storage temperatures on vaccine immunogenicity. Mice were immunized for GC assay of popliteal lymph nodes and ELISpot of splenocytes at 2-weeks post-vaccination. Gating for GC B cells are shown in (a) and antigen-specific B cells are shown in (b). Shown are frequency of total GC B cells (c), antigen-specific B cells (d), follicular helper T cells (e), and number of spots per 106 cells (f). Bar graphs are geometric means ± s.e.m. Statistics represent one-way ANOVA and Tukey's HSD Test (ns = not significant; *, p < 0.05).
We also assessed endpoint titers of diverse mouse antibody isotypes (IgG1, IgG2a, IgG2b, IgG3, and IgM) to detect any differences in antibody class switching elicited by this optimal storage condition. While the low overall antibody titer induced by the 4 °C stored vaccines rendered near background levels of other Ig isotypes, the −20 °C stored vaccines produced IgG1, IgG2a, IgG2b, IgG3, and IgM levels that were not statistically different from those of the freshly prepared vaccine (Supplementary Fig. S6).
Lastly, we investigated whether LNPs thawed from different storage conditions elicit different inflammatory responses at the injection site, by profiling cytokine production in the muscle at 24 h post-vaccination (Supplementary Fig. S7). Overall, we found similar increases in IFN-γ, KC, MCP-1, RANTES, IP-10, IL-10, IFN-β, IFN-α, and IL-6 levels in the muscle following immunization with vaccines that were freshly prepared, stored at −20 °C, and stored at −200 °C. In contrast, vaccines that were stored at 4 °C or − 80 °C show more muted responses.
3.5. Shelf-life of frozen LNPs at −20 °C and thawed LNPs at 4 °C
We next evaluated the stability of LNPs over a more extended period of storage time. LNPs were loaded with repRNA encoding firefly luciferase, and stored for 7 or 30 days in 10% w/v sucrose in PBS at −20 °C. Then, we administered the thawed particles or a freshly synthesized batch i.m. in groups of mice and evaluated bioluminescence signals in the muscles over 30 days by IVIS imaging. As shown in Fig. 5a and Supplementary Fig. S8, there was no statistical significance between freshly prepared batch and stored batches (p > 0.2). Next, we prepared LNPs loaded with replicons encoding the N332-GT2 immunogen, and stored them for 7, 14, or 30 days before thawing for intramuscular injection in mice. Mice were then retro-orbitally bled six-weeks post-vaccination for ELISA quantification of antibody titer in the sera. Vaccines stored out to 30 days elicited equivalent antibody responses against the HIV immunogen as the freshly prepared vaccines (Fig. 5b-e).
Fig. 5.
Shelf-life of LNP-RNA vaccines stored long-term in frozen or thawed states. (a) Groups of balb/c mice (n = 3 animals/group averaged across a total of 6 legs/group) were injected i.m. in both the left and right gastrocnemius muscles with 1 μg replicon RNA in LNPs stored in 10% w/v sucrose at −20 °C for indicated durations. Shown are luciferase reporter signals over time; dotted lines indicate background signal of untreated mice, shown are means ± standard deviation. (b-e) balb/c mice were i.m. vaccinated in both the left and right gastrocnemius muscles with 1 μg replicon RNA in LNPs that were recovered from indicated duration of storage at −20 °C: (b) 7 days; (c) 14 days; (d) 30 days. (f-j) balb/C mice were vaccinated i.m. (1 μg LNP-RNA per animal, 5 animals/group) with vaccines that had been thawed from −20 °C and refrigerated at 4 °C for: (f) 1 day; (g) 7 days; (h) 14 days; (i) 30 days. Shown are serum IgG dilution curves of individual mouse at 4-weeks post-vaccination. Error bar represents standard error mean in n = 5. Statistics represent one-way ANOVA and Tukey's HSD Test (ns = not significant; **, p < 0.01; ****, p < 0.0001).
Once we validated that LNP-RNA vaccines in 10% w/v sucrose in PBS can be stored for at least 30 days at −20 °C without losing potency, we then investigated the shelf-life of the LNPs when stored under refrigeration (4 °C) post-thawing. This experimental setup simulates a clinical setting, in which frozen vaccines are thawed for patient dosing, and remaining doses are stored at 4 °C for dosing at another time. To this end, LNPs loaded with repRNA encoding the N332-GT2 immunogen were prepared in 10% w/v sucrose in PBS were stored at −20 °C for 7 days, then thawed at room temperature briefly before being placed in the refrigerator at 4 °C for 7, 14, or 30 days. The thawed LNPs at 4 °C and a freshly prepared set of LNP vaccines were intramuscularly administered to mice. Mouse sera were collected at 4-weeks post-vaccination for ELISA quantification of antibody titer. As shown in Fig. 5f-j, there is a strong negative correlation between vaccine-induced antibody titer and increasing storage time at 4 °C post-thawing from −20 °C. A single day of storage at 4 °C post-thawing generates strong antibody titers that are statistically not different from that of the fresh sample, although already trending downward (Fig. 5f; p 1d = 0.4615). LNPs kept for 7 days post-thaw failed to seroconvert 2 out of 5 animals (Fig. 5g; p 7d = 0.0014), and this decay in antibody responses continued with further 4 °C storage for 14 or 30 days (Fig. 5h-i).
3.6. Lyophilization of LNP-RNA vaccines
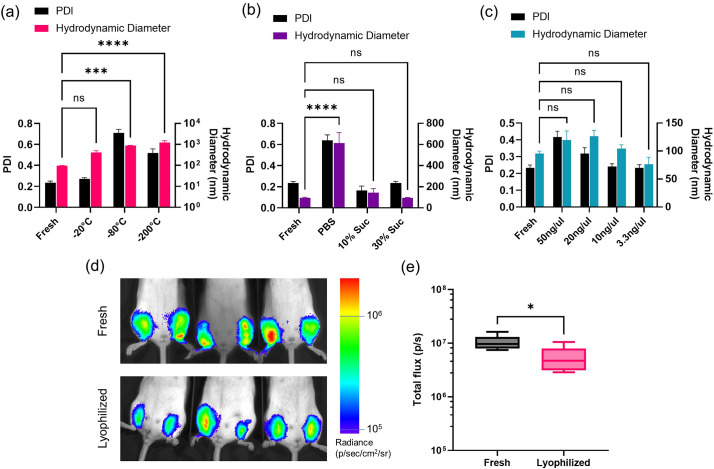
While we have established a reliable freezing protocol for long-term storage of LNP-formulated replicon vaccines, lyophilization to keep vaccines in a dry powder form is another clinically-relevant storage strategy. Dry solids simplify aseptic handling procedures, decrease sample weight and volume for easier shipping, improve stability, and allow easy dose adjustments by dissolution at desired concentrations [[52], [53], [54]]. However, freeze drying has proven to be challenging for LNP-RNA formulations [29,35,39]. To determine key factors that affect stability of lyophilized LNP-RNA particles, we selected three parameters to test in lyophilization: freezing temperature, cryoprotectant concentration, and sample concentration. To this end, the formulations were frozen, lyophilized, and re-hydrated for DLS assessment of particle distribution and hydrodynamic diameter. We tested initial freezing at −20 °C along with −80 °C and flash freezing at −200 °C using liquid nitrogen, which are the more commonly used freezing temperatures for lyophilization. Particles that were lyophilized and resuspended after being frozen at −20 °C were able to retain a polydispersity index (PDI) of 0.272, whereas those of the −80 °C and − 200 °C groups showed aggregation with PDI reaching 0.710 (−80 °C) and 0.517 (−200 °C) (Fig. 6a). The size of the particles also increased from an average hydrodynamic diameter (by intensity distribution) of 421 nm (−20 °C) to 877 nm (−80 °C) and 1219 nm (−200 °C). While none of the freezing temperatures were able to retain the correct Z-average size of approximately 90 nm by intensity distribution on DLS (equivalent to approximately 45 nm by number distribution and cryo-EM), we observed a trend toward smaller particles with increasing temperature. The −20 °C stored vaccines displayed an average hydrodynamic diameter of 421 nm, but was statistically not different from the fresh batch (p = 0.1858). We next tested the effects of cryoprotectant concentration on lyophilization of LNP-RNA (Fig. 6b). The vaccine particles were prepared in PBS, 10% w/v sucrose, or 30% w/v sucrose, and frozen at −20 °C prior to undergoing lyophilization. The dry particles were then rehydrated in deionized water for DLS assessment. Here, we found a striking difference between samples that were lyophilized in PBS versus samples that were cryoprotected with sucrose. Compared with freshly prepared LNPs that had an average PDI of 0.233, the PBS samples presented an average PDI of 0.638, the 10% w/v sucrose samples 0.165, and the 30% w/v sucrose samples 0.235, indicating a more monodisperse population of particles in the presence of sucrose. Moreover, the average hydrodynamic size of the particles in PBS significantly increased to an average of 613 nm compared to the fresh samples, which average at 96 nm (p < 0.0001). Lyophilization in 10% w/v sucrose led to a slight increase in the average particle size to 144 nm, but this difference was not statistically significant (p = 0.7957). Lastly, the 30% w/v sucrose samples averaged at 94.0 nm, which was statistically the closest to the fresh samples (p > 0.9999). As a final parameter, we assessed the effect of LNP-RNA concentration on lyophilization. Here, we froze and lyophilized the same mass of LNP-RNA in increasing volumes of 10% w/v sucrose in PBS at −20 °C, and resuspended the lyophilized particles in equal volumes for DLS assessment. The 10% w/v sucrose concentration was selected over the 30% w/v sucrose to accommodate for the final concentration of sucrose appropriate for in vivo administration after resuspension; in fact, the presence of excessive amounts of sucrose in low resuspension volumes resulted in viscous consistency in the hydrated vaccine that was ill-suited for downstream analyses and in vivo administration. Fig. 6c shows that while there were no statistically significant differences in the formulations (p 50 μg /uL = 0.2223; p 20 μg /uL = 0.0688; p 10ug/uL = 0.9319; p 3.3 μg /uL = 0.4302), there was a trend toward lower PDI and hydrodynamic size with decreasing LNP concentration.
Fig. 6.
Evaluation of lyophilized LNP-RNA formulations. (a-c) LNP-RNA was synthesized (n = 3 samples/condition), lyophilized under indicated conditions, then rehydrated and analyzed by DLS. (a) LNPs were frozen at the indicated temperatures in PBS without sucrose. (b) LNPs were frozen at −20 °C in indicated buffers prior to lyophilization. (c) LNPs at the indicated concentrations (in terms of RNA amount) were frozen at −20 °C in PBS with 10% sucrose prior to lyophilization. Error bars represent standard deviation, and statistics indicate Two-way ANOVA and Tukey's HSD Test (ns = not significant; ***, p < 0.001; ****p < 0.0001). Black columns show PDI on the left y-axis, while the colored bars show the average hydrodynamic diameter from intensity distribution on the right y-axis. (d-e) IVIS bioluminescence imaging in mice that were administered i.m. freshly prepared or lyophilized LNPs loaded with RNA encoding luciferase. Lyophilized LNPs were placed in 10% w/v sucrose and frozen at a concentration of 3.3 ng/uL in −20 °C prior to undergoing lyophilization. Shown are representative photograph/false-color overlays (d) and total bioluminescence signal 1 day post-LNP administration (e). Statistics represent unpaired two-tailed t-test with *, p < 0.05.
Finally, we intramuscularly administered freshly prepared and lyophilized LNPs loaded with repRNA encoding for firefly luciferase to evaluate the in vivo transfection efficiency using the IVIS bioluminescence imaging system (Fig. 6d-e). For this study, we selected the lyophilization protocol that was found to best maintain the particle structure – LNPs were placed in 10% w/v sucrose and frozen at a concentration of 3.3 ng/μL in −20 °C prior to undergoing lyophilization. At day 1 post administration, both freshly prepared and lyophilized LNP-replicons showed strong luciferase expression, but the signal of lyophilized particles was ∼50% that of the freshly prepared samples (p = 0.0191). In summary, we investigated three parameters that contribute to maintenance of size and distribution of lyophilized particles. By optimizing and combining these factors, we were able to improve the retention of LNP-RNA structure and dispersity post-lyophilization and resuspension.
4. Discussion
The present study was conducted with the intention of shedding light on physical changes occurring with self-replicating RNA-loaded lipid nanoparticles (LNP-RNA) in commonly employed storage conditions (by cryoprotectant concentration, buffer type, and maintenance temperature), and how these differences relate to in vivo transfection and vaccine potency. The particular LNP formulation that we employed in this study includes two ionizable lipids (TT3 and DLin-MC3-DMA), which we found gave effective transfection in the muscle and humoral immune responses in combination, and 5 mol% of DMG-PEG to supplement the stability of the near-neutrally charged LNPs. Using this formulation, we found that both phosphate and tris buffers were suitable for storing LNPs, and that sucrose was an important cryoprotectant for the maintenance of the structural integrity, physical stability, or activity of the repRNA payload. The dominating factor was the storage temperature.
The optimal sucrose concentration was found to be 10% w/v (Fig. 1), similar to other published work on cryoprotection of biological materials [[55], [56], [57], [58], [59]]. It is believed that for a set material concentration, there is a minimum number of sugar molecules required to sufficiently disrupt interactions between the polar water molecules (by formation of water-sucrose hydrogen bonds [60]) to slow down the freezing rate, and form larger ice crystal with minimal ice-water interface wherein the materials tend to localize [61,62]. In fact, rather than being homogenously dispersed throughout the sample, sucrose is reported to form a thin sheet in a nonfrozen state along the surface of lipid bilayers, and keeps materials from making direct contact with ice [63,64]. Empirically, 10% w/v sucrose enabled these processes to effectively promote retention of the particles' structure without aggregation.
Dynamic light scattering (DLS) is a technique that is commonly used to assess particle stability and hydrodynamic size distribution. However, relying solely on scattering data provided by DLS can be misleading in terms of accuracy, particularly with polydisperse sub-100 nm particles where neither the intensity nor number distributions offer accurate information [[49], [50], [51]]. Thus, we also carried out transmission electron microscopy (TEM) and cryogenic electron microscopy (cryo-EM) to fully capture the physical state of the LNPs upon thawing from storage. Interestingly, storage at −20 °C was able to maintain LNP structural integrity, while irreversible aggregation was observed following thawing of particles from −80 °C. But this seems to be the case only for repRNA; when the same LNPs were loaded with mRNA and stored, the particles retained their structural integrity at all tested temperatures (Supplementary Fig. S1). Other studies have also reported LNP-mRNA formulations that are stable at these lower temperatures (e.g., BNT162b2's − 70 °C storage of COVID-19 vaccine [7,28,29]). We hypothesize these differences between the behavior of LNPs carrying repRNA vs. mRNA may reflect potential differences in the packing structure of the RNA molecules with the lipids in the nanoparticle at a molecular level and/or differences in particle stability associated with differences in the N:P ratios used for mRNA vs. repRNA, leading to different sensitivities to the freezing/thawing process. Alphavirus-based self-replicating RNA is ∼10-fold larger than typical mRNA used in vaccines, and may organize with the LNP core in a distinct manner from shorter mRNAs. Further, we employed a 2:1 N:P ratio for effectively packaging repRNA in LNPs with ∼90% RNA encapsulation efficiency and effective in vivo delivery, whereas literature reports the same N:P ratio can reduce the encapsulation efficiency of mRNA molecules down to only 40% [65]). Additionally, −80 °C offers an intermediate between −20 °C slow freezing and − 200 °C flash freezing. While cooling from room temperature to −20 °C is expected to induce slow but short cooling that yields large ice crystals dispersed in unfrozen liquid and sheets of sucrose (liquid + ice phase), −200 °C flash freezing is expected to instantaneously form solid ice with little to no ice nucleation (solid phase) [66,67]. In contrast, freezing down to −80 °C from room temperature is expected to have an accelerated cooling rate compared to −20 °C, as water is able to freeze without nucleation below −40 °C [67]. Thus, the resulting state will have a mix of smaller ice crystals intermixed with glass phase in which the particles and sucrose molecules are concentrated. Smaller ice crystals have greater interfacial area compared to large crystals, which is not favorable for cryopreservation [62]. Moreover, concentration of particles in the glass phase may further facilitate aggregation of unstable particles [68]. Thus, we hypothesize that repRNA-loaded LNPs are more vulnerable to aggregation during freezing to or thawing from −80 °C due to a mix of the particles' unique structural properties when loaded with repRNA over mRNA and the type of ice phase that is formed at this temperature and cooling rate.
We found that maintenance of LNP structure (as determined by light scattering and morphological analysis by TEM) is not necessarily a faithful indicator of in vivo functionality post storage. For example, storage of LNP-RNA at 4 °C was able to retain the physical state of the particles based on DLS and electron microscopy results compared with freshly prepared samples (Table 1 and Figs. 3b and g), but failed to function as reliably when it came to activity studies using either the reporter repRNA or the antigen-encoding repRNA (Fig. 2, Fig. 4). This outcome indicates that while 4 °C storage is sufficient to keep the LNP delivery vehicles intact, the encapsulated repRNA molecules are negatively affected [29]. This idea is further supported by results from a test of post-thaw shelf life in Fig. 5f-j, where vaccines that were stored effectively and thawed for 4 °C storage showed decreased vaccine efficacy beyond one day at 4 °C. A study carried out to calculate the theoretical cleavage rate of the RNA molecules predicted that an mRNA molecule of 4000 nucleotides would have a half-life of 941 days when stored at 5 °C under RNAse-free conditions, but that longer repRNAs would be more susceptible to hydrolytic cleavage [69]. While such reactions are not a concern at freezing temperatures, prevention of RNA degradation is a must in refrigeration despite the protective coating the LNPs may offer RNA molecules; thus, it may be possible that the decreased and variable efficacies seen in our LNP-RNA samples kept at 4 °C may have been due to such degradative reactions [31]. Indeed, we found a significant amount of RNA leakage only from the LNPs that were stored at 4 °C for a week (Supplementary Fig. S4). Another observation we noted is that while 4 °C storage is able to retain a relatively high transfection (Fig. 2b and c), its downstream efficacy in generating humoral responses is significantly reduced (Fig. 2i). It may be possible that the RNA sequence has an influence on transfection, especially in the long repRNAs where differences in nucleotide sequence may affect its packing within LNPs, and in turn its stability during freezing and thawing. Moreover, Supplementary Fig. S7 shows that vaccines stored at 4 °C generate decreased cytokine production in the muscle compared to vaccines that are freshly prepared or stored at −20 °C or − 200 °C; thus, we believe that LNPs thawed from 4 °C may have reduced adjuvant activity despite maintaining sufficient ability to transfect cells. We hypothesize RNA leakage out of the particles following 4 °C storage may lead to changes to the internal packing structure of the LNPs to a state that is less inflammatory, and/or decreased repRNA delivered into cells at the injection site may have led to a diminished innate immune response.
In contrast to 4 °C storage, LNPs flash frozen to −200 °C performed moderately well in terms of in vivo gene expression (Fig. 2), although this storage regimen showed severe aggregation by electron microscopy (Fig. 3e and j). This aggregation may potentially be the result of sample preparation steps in electron microscopy – such as drying on the grid for TEM or undergoing another round of flash freezing for cryo-EM. This fragility or sensitivity of flash frozen LNP-RNA to post-thaw handling may suggest unknown material properties that we do not observe for freshly prepared, 4 °C stored, or − 20 °C stored samples. Coupled with the fact that liquid nitrogen maintenance for cold-chain transport is impractical, flash freezing does not appear to be a suitable storage option.
Overall, LNP-RNA vaccine in 10% w/v sucrose in PBS was maintained well both physically (Fig. 1, Fig. 3c and h) and functionally (Fig. 2, Fig. 4, Fig. 5) in −20 °C storage for at least 30 days. This temperature offers a feasible solution for cold-chain transport of vaccines, as demonstrated by Moderna for their COVID-19 vaccine (mRNA-1273) [27,33,34]. For our LNP-RNA system, −20 °C may be a sufficiently high temperature to offer an ideal ice nucleation temperature and a slow cooling rate to help retain particle structure, while also being a sufficiently low temperature to inhibit RNA degradation by hydrolytic cleavage events.
A more ideal solution would be the lyophilization of LNP-RNA vaccines to a dry powder form that is well maintained at room temperature. However, this endeavor has proven to be challenging, with little to no successful lyophilization of LNP-repRNA formulations yet reported in the literature. Our results in Fig. 6 show that while optimization of several parameters is able to improve the structural maintenance of the formulations, in vivo transfection fully equivalent to fresh LNPs remains difficult to achieve, which echoes findings from another prior study of LNPs carrying mRNA [39]. It may be possible that freeze-drying the vaccine leaves RNA molecules more vulnerable to hydrolytic degradation during resuspension or to interactions with serum proteins in vivo. On the other hand, Ball et al. flash froze LNPs loaded with siRNA for 30 min in liquid nitrogen prior to lyophilization, and found that reconstitution of the lyophilized LNPs in deionized water substantially reduced their in vitro gene silencing capability to 35%, whereas the addition of 22% ethanol during reconstitution successfully maintained gene silencing at a similar level to that of freshly prepared LNPs (80% vs 90%) [35]. Though recovery was successful, administration of formulations in ethanol or an added process of removing the ethanol prior to administration would be difficult to translate to clinical settings [29]. Nonetheless, successful lyophilization of LNP-RNA formulations may require greater control over freezing and drying temperatures than is generally available in the basic benchtop freeze-dryers that are used in academic labs. In industry, Moderna has reported that their cytomegalovirus vaccine (mRNA-1647) currently in phase 2 clinical trial can be successfully lyophilized and stored for over 18 months at 5 °C [29,70,71]. In addition, Arcturus Therapeutics in collaboration with Duke-NUS Medical School in Singapore have also reported successful lyophilization of their repRNA-encapsulating LNP vaccine (ARCT-021) [29,72]. Unfortunately, details of the processes are not publicly available. More recently, an effective lyophilization method for an mRNA vaccine was reported to stably store them at 4 °C in dry powder form for out to 24-weeks using an optimized freezing step with two separate sublimation and desorption dry cycles [73].
5. Conclusions
In light of the COVID-19 pandemic, long-term storage and cold-chain transport of vaccines have become a critical step in the successful translation and use of RNA-loaded lipid nanoparticles (LNP-RNA). At the same time, we came to realize that there is very scarce information on the relationship between LNP-RNA's physical properties, storage conditions, and vaccine efficacy. Here, we investigated what physical changes the vaccines undergo in different storage conditions (by concentration of cryoprotectant, type of buffer, and storage temperature), and how those changes relate to the vaccine efficacy in vivo using both RNA encoding for reporter proteins (e.g. luciferase) and for an actual HIV immunogen. Ultimately, we found that LNP-RNA vaccines are stably stored in 10% w/v sucrose in PBS at −20 °C for at least 30 days. Further, we found that replicon-carrying LNPs could also be lyophilized and retain substantial in vivo bioactivity.
Funding
This work was supported by the NIH (awards AI161297, AI144462, and AI048240 to DJI, and CA265706 to DJI and YD), the Marble Center for Nanomedicine, and the Ragon Institute of MGH, MIT, and Harvard. DJI is an investigator of the Howard Hughes Medical Institute.
Author contributions
BK and DJI conceptualized the study and wrote the manuscript. BK also designed experiments, obtained experimental data, and analyzed results. RRH obtained experimental data, and contributed to data analyses and manuscript writing. TR, DY, NL, WA, MBM, MC, BL, YZ, and YZ contributed to obtaining experimental data and editing of the manuscript.
CRediT authorship contribution statement
Byungji Kim: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Visualization, Supervision. Ryan R. Hosn: Validation, Investigation, Writing – original draft, Writing – review & editing. Tanaka Remba: Investigation, Writing – review & editing. Dongsoo Yun: Investigation, Writing – review & editing. Na Li: Investigation, Writing – review & editing. Wuhbet Abraham: Investigation, Writing – review & editing. Mariane B. Melo: Investigation, Writing – review & editing. Manuel Cortes: Investigation, Writing – review & editing. Bridget Li: Investigation, Writing – review & editing. Yuebao Zhang: Resources, Writing – review & editing. Yizhou Dong: Resources, Writing – review & editing. Darrell J. Irvine: Conceptualization, Resources, Supervision, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgements
We thank the Koch Institute's Robert A. Swanson (1969) Biotechnology Center for technical support. We also thank Dr. William R. Schief from The Scripps Research Institute (TSRI) for providing the sequence for the HIV immunogen N332-GT2 used in the study. Biorender was used for generation of graphical abstract.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jconrel.2022.11.022.
Appendix A. Supplementary data
Supplementary figures
Data availability
Data will be made available on request.
References
- 1.Dimitriadis G.J. Translation of rabbit globin mRNA introduced by liposomes into mouse lymphocytes. Nature. 1978;274:923–924. doi: 10.1038/274923a0. [DOI] [PubMed] [Google Scholar]
- 2.Fyfe I. Treatment success in hereditary transthyretin amyloidosis. Nat. Rev. Neurol. 2018;14:509. doi: 10.1038/s41582-018-0048-1. [DOI] [PubMed] [Google Scholar]
- 3.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W., Stebbing D., Crosley E.J., Yaworski E., Hafez I.M., Dorkin J.R., Qin J., Lam K., Rajeev K.G., Wong K.F., Jeffs L.B., Nechev L., Eisenhardt M.L., Jayaraman M., Kazem M., Maier M.A., Srinivasulu M., Weinstein M.J., Chen Q., Alvarez R., Barros S.A., De S., Klimuk S.K., Borland T., Kosovrasti V., Cantley W.L., Tam Y.K., Manoharan M., Ciufolini M.A., Tracy M.A., de Fougerolles A., MacLachlan I., Cullis P.R., Madden T.D., Hope M.J. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 4.Akinc A., Zumbuehl A., Goldberg M., Leshchiner E.S., Busini V., Hossain N., Bacallado S.A., Nguyen D.N., Fuller J., Alvarez R., Borodovsky A., Borland T., Constien R., de Fougerolles A., Dorkin J.R., Jayaprakash K.N., Jayaraman M., John M., Koteliansky V., Manoharan M., Nechev L., Qin J., Racie T., Raitcheva D., Rajeev K.G., Sah D.W., Soutschek J., Toudjarska I., Vornlocher H.P., Zimmermann T.S., Langer R., Anderson D.G. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akinc A., Maier M.A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., Ansell S., Du X., Hope M.J., Madden T.D., Mui B.L., Semple S.C., Tam Y.K., Ciufolini M., Witzigmann D., Kulkarni J.A., van der Meel R., Cullis P.R. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 6.Crommelin D.J.A., Anchordoquy T.J., Volkin D.B., Jiskoot W., Mastrobattista E. Addressing the cold reality of mRNA vaccine stability. J. Pharm. Sci. 2021;110:997–1001. doi: 10.1016/j.xphs.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliver S.E., Gargano J.W., Marin M., Wallace M., Curran K.G., Chamberland M., McClung N., Campos-Outcalt D., Morgan R.L., Mbaeyi S., Romero J.R., Talbot H.K., Lee G.M., Bell B.P., Dooling K. The advisory committee on immunization practices’ interim recommendation for use of Pfizer-BioNTech COVID-19 vaccine - United States, December 2020. Morb. Mortal. Wkly Rep. 2020;69:1922–1924. doi: 10.15585/mmwr.mm6950e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliver S.E., Gargano J.W., Marin M., Wallace M., Curran K.G., Chamberland M., McClung N., Campos-Outcalt D., Morgan R.L., Mbaeyi S., Romero J.R., Talbot H.K., Lee G.M., Bell B.P., Dooling K. The advisory committee on immunization practices’ interim recommendation for use of Moderna COVID-19 vaccine - United States, December 2020. MMWR Morb. Mortal. Wkly Rep. 2021;69:1653–1656. doi: 10.15585/mmwr.mm695152e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim B., Park J., Sailor M.J. Rekindling RNAi therapy: materials design requirements for in vivo siRNA delivery. Adv. Mater. 2019;31:1903637. doi: 10.1002/adma.201903637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu B., Zhong L., Weng Y., Peng L., Huang Y., Zhao Y., Liang X.-J. Therapeutic siRNA: state of the art. Signal Transduct. Target. Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 12.Roberts T.C., Langer R., Wood M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020;19:673–694. doi: 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinaldi C., Wood M.J.A. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 2017;14:9–21. doi: 10.1038/nrneurol.2017.148. [DOI] [PubMed] [Google Scholar]
- 14.Minnaert A.-K., Vanluchene H., Verbeke R., Lentacker I., Smedt S.C.D., Raemdonck K., Sanders N.N., Remaut K. Strategies for controlling the innate immune activity of conventional and self-amplifying mRNA therapeutics: getting the message across. Adv. Drug Deliv. Rev. 2021;176:113900. doi: 10.1016/j.addr.2021.113900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chien K.R., Zangi L., Lui K.O. Synthetic chemically modified mRNA (modRNA): toward a new technology platform for cardiovascular biology and medicine. Cold Spring Harb. Perspect. Med. 2014;5 doi: 10.1101/cshperspect.a014035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S.C., Sekhon S.S., Shin W.-R., Ahn G., Cho B.-K., Ahn J.-Y., Kim Y.-H. Modifications of mRNA vaccine structural elements for improving mRNA stability and translation efficiency. Mol. Cell. Toxicol. 2021;18:1–8. doi: 10.1007/s13273-021-00171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahin U., Oehm P., Derhovanessian E., Jabulowsky R.A., Vormehr M., Gold M., Maurus D., Schwarck-Kokarakis D., Kuhn A.N., Omokoko T., Kranz L.M., Diken M., Kreiter S., Haas H., Attig S., Rae R., Cuk K., Kemmer-Brück A., Breitkreuz A., Tolliver C., Caspar J., Quinkhardt J., Hebich L., Stein M., Hohberger A., Vogler I., Liebig I., Renken S., Sikorski J., Leierer M., Müller V., Mitzel-Rink H., Miederer M., Huber C., Grabbe S., Utikal J., Pinter A., Kaufmann R., Hassel J.C., Loquai C., Türeci Ö. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature. 2020;585:107–112. doi: 10.1038/s41586-020-2537-9. [DOI] [PubMed] [Google Scholar]
- 18.Gebre M.S., Rauch S., Roth N., Yu J., Chandrashekar A., Mercado N.B., He X., Liu J., McMahan K., Martinot A., Martinez D.R., Giffin V., Hope D., Patel S., Sellers D., Sanborn O., Barrett J., Liu X., Cole A.C., Pessaint L., Valentin D., Flinchbaugh Z., Yalley-Ogunro J., Muench J., Brown R., Cook A., Teow E., Andersen H., Lewis M.G., Boon A.C.M., Baric R.S., Mueller S.O., Petsch B., Barouch D.H. Optimization of non-coding regions for a non-modified mRNA COVID-19 vaccine. Nature. 2021;601:410–414. doi: 10.1038/s41586-021-04231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leppek K., Byeon G.W., Kladwang W., Wayment-Steele H.K., Kerr C.H., Xu A.F., Kim D.S., Topkar V.V., Choe C., Rothschild D., Tiu G.C., Wellington-Oguri R., Fujii K., Sharma E., Watkins A.M., Nicol J.J., Romano J., Tunguz B., Participants E., Barna M., Das R. Biorxiv Prepr. Serv. Biology; 2021. Combinatorial Optimization of mRNA Structure, Stability, and Translation for RNA-Based Therapeutics. 2021.03.29.437587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster H., Sharp P.S., Athanasopoulos T., Trollet C., Graham I.R., Foster K., Wells D.J., Dickson G. Codon and mRNA sequence optimization of microdystrophin transgenes improves expression and physiological outcome in dystrophic mdx mice following AAV2/8 gene transfer. Mol. Ther.: J. Am. Soc. Gene Ther. 2008;16:1825–1832. doi: 10.1038/mt.2008.186. [DOI] [PubMed] [Google Scholar]
- 21.L. K Replicon RNA viral vectors as vaccines. Nato Adv. Sci. Inst. Se. 2016;4:39. doi: 10.3390/vaccines4040039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundstrom K. Self-replicating RNA viruses for RNA therapeutics. Molecules. 2018;23:3310. doi: 10.3390/molecules23123310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blakney A.K., Ip S., Geall A.J. An update on self-amplifying mRNA vaccine development. Nato Adv. Sci. Inst. Se. 2021;9:97. doi: 10.3390/vaccines9020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pollock K.M., Cheeseman H.M., Szubert A.J., Libri V., Boffito M., Owen D., Bern H., McFarlane L.R., O’Hara J., Lemm N.-M., McKay P., Rampling T., Yim Y.T.N., Milinkovic A., Kingsley C., Cole T., Fagerbrink S., Aban M., Tanaka M., Mehdipour S., Robbins A., Budd W., Faust S., Hassanin H., Cosgrove C.A., Winston A., Fidler S., Dunn D., McCormack S., Shattock R.J., Adams K., Amini F., Atako N.B., Bakri A., Barclay W., Brodnicki E., Brown J.C., Byrne R., Chilvers R., Coelho S., Day S., Desai M., Dorman E., Elliott T., Flight K.E., Fletcher J., Galang J., Gohil J., Gupta A., Harlow C., Hu K., Kalyan M., Lagrue D., Liscano E., Njenga C., Polra K., Powlette D.A., Randell P., Rauchenberger M., Redknap I., Ricamara M., Rogers P., Sallah H., Samnuan K., Schumacher M., Shah Z., Shaw R., Shaw T., Sivapatham S., Slater S., Sorley K., Storch R., Tan E., Tan T., Thielemans L., Whitely S., Valentine C., Varghese J., Vikraman A., Wilkins M. Safety and immunogenicity of a self-amplifying RNA vaccine against COVID-19: COVAC1, a phase I, dose-ranging trial. Eclinicalmedicine. 2022;44 doi: 10.1016/j.eclinm.2021.101262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.A Trial Evaluating the Safety and Immunogenicity of 3 COVID-19 SARS-CoV-2 RNA Vaccines in Healthy Adults. 2021. https://clinicaltrials.gov/ct2/show/NCT05037097
- 26.The ARCT-154 Self-Amplifying RNA Vaccine Efficacy Study (ARCT-154-01) 2022. https://clinicaltrials.gov/ct2/show/NCT05012943
- 27.Hatziantoniou S., Maltezou H.C., Tsakris A., Poland G.A., Anastassopoulou C. Anaphylactic reactions to mRNA COVID-19 vaccines: a call for further study. Vaccine. 2021;39:2605–2607. doi: 10.1016/j.vaccine.2021.03.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uddin M.N., Roni M.A. Challenges of storage and stability of mRNA-based COVID-19 vaccines. Nato Adv. Sci. Inst. Se. 2021;9:1033. doi: 10.3390/vaccines9091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G., Jiskoot W., Crommelin D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int. J. Pharm. 2021;601 doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., Mihai C., Lynn A., McFadyen I., Moore M., Senn J., Stanton M.G., Almarsson Ö., Ciaramella G., Brito L.A. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brader M.L., Williams S.J., Banks J.M., Hui W.H., Zhou Z.H., Jin L. Encapsulation state of messenger RNA inside lipid nanoparticles. Biophys. J. 2021;120:2766–2770. doi: 10.1016/j.bpj.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.COVID-19 Vaccine Moderna. European Medicines Agency; 2021. [Google Scholar]
- 33.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6:1–17. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.EMA Approves New Storage Option for Pfizer-BioNTech Vaccine, Easing Distribution and Storage of Doses across European Union. 2021. [Google Scholar]
- 35.Ball R.L., Bajaj P., Whitehead K.A. Achieving long-term stability of lipid nanoparticles: examining the effect of pH, temperature, and lyophilization. Int. J. Nanomedicine. 2017;12:305–315. doi: 10.2147/ijn.s123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muralidhara B.K., Baid R., Bishop S.M., Huang M., Wang W., Nema S. Critical considerations for developing nucleic acid macromolecule based drug products. Drug Discov. Today. 2016;21:430–444. doi: 10.1016/j.drudis.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 37.Muralidhara B.K., Wong M. Critical considerations in the formulation development of parenteral biologic drugs. Drug Discov. Today. 2020;25:574–581. doi: 10.1016/j.drudis.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 38.Gerhardt A., Voigt E., Archer M., Reed S., Larson E., Hoeven N.V., Kramer R., Fox C., Casper C. BioRxiv; 2021. A Thermostable, Flexible RNA Vaccine Delivery Platform for Pandemic Response. [Google Scholar]
- 39.Zhao P., Hou X., Yan J., Du S., Xue Y., Li W., Xiang G., Dong Y. Long-term storage of lipid-like nanoparticles for mRNA delivery. Bioact. Mater. 2020;5:358–363. doi: 10.1016/j.bioactmat.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirane D., Tanaka H., Nakai Y., Yoshioka H., Akita H. Development of an alcohol dilution-lyophilization method for preparing lipid nanoparticles containing encapsulated siRNA. Biol. Pharm. Bull. 2018;41:1291–1294. doi: 10.1248/bpb.b18-00208. [DOI] [PubMed] [Google Scholar]
- 41.Zhang N.N., Li X.F., Deng Y.Q., Zhao H., Huang Y.J., Yang G., Huang W.J., Gao P., Zhou C., Zhang R.R., Guo Y., Sun S.H., Fan H., Zu S.L., Chen Q., He Q., Cao T.S., Huang X.Y., Qiu H.Y., Nie J.H., Jiang Y., Yan H.Y., Ye Q., Zhong X., Xue X.L., Zha Z.Y., Zhou D., Yang X., Wang Y.C., Ying B., Qin C.F. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182:1271–1283. doi: 10.1016/j.cell.2020.07.024. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ripoll M., Bernard M.-C., Vaure C., Bazin E., Commandeur S., Perkov V., Lemdani K., Nicolaï M.-C., Bonifassi P., Kichler A., Frisch B., Haensler J. An imidazole modified lipid confers enhanced mRNA-LNP stability and strong immunization properties in mice and non-human primates. Biomaterials. 2022;286 doi: 10.1016/j.biomaterials.2022.121570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.L. B., L. X., D. B., W. J., M. W. D., S. Y., G. M. K., T. X., D. L. A., K. A. B., D. Y An orthogonal array optimization of lipid-like nanoparticles for mRNA delivery in vivo. Nano Lett. 2015;15:8099–8107. doi: 10.1021/acs.nanolett.5b03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steichen J.M., Lin Y.-C., Havenar-Daughton C., Pecetta S., Ozorowski G., Willis J.R., Toy L., Sok D., Liguori A., Kratochvil S., Torres J.L., Kalyuzhniy O., Melzi E., Kulp D.W., Raemisch S., Hu X., Bernard S.M., Georgeson E., Phelps N., Adachi Y., Kubitz M., Landais E., Umotoy J., Robinson A., Briney B., Wilson I.A., Burton D.R., Ward A.B., Crotty S., Batista F.D., Schief W.R. A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses. Science. 2019;366 doi: 10.1126/science.aax4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y., Su Z., Zhao W., Zhang X., Momin N., Zhang C., Wittrup K.D., Dong Y., Irvine D.J., Weiss R. Multifunctional oncolytic nanoparticles deliver self-replicating IL-12 RNA to eliminate established tumors and prime systemic immunity. Nat. Can. 2020;1:882–893. doi: 10.1038/s43018-020-0095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y., Teague B., Zhang Y., Su Z., Porter E., Dobosh B., Wagner T., Irvine D.J., Weiss R. In vitro evolution of enhanced RNA replicons for immunotherapy. Sci. Rep. UK. 2019;9:6932. doi: 10.1038/s41598-019-43422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wroblewska L., Kitada T., Endo K., Siciliano V., Stillo B., Saito H., Weiss R. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 2015;33:839–841. doi: 10.1038/nbt.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melo M., Porter E., Zhang Y., Silva M., Li N., Dobosh B., Liguori A., Skog P., Landais E., Menis S., Sok D., Nemazee D., Schief W.R., Weiss R., Irvine D.J. Immunogenicity of RNA replicons encoding HIV Env immunogens designed for self-assembly into nanoparticles. Mol. Ther. 2019;27:2080–2090. doi: 10.1016/j.ymthe.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malm A.V., Corbett J.C.W. Improved dynamic light scattering using an adaptive and statistically driven time resolved treatment of correlation data. Sci. Rep. UK. 2019;9:13519. doi: 10.1038/s41598-019-50077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wishard A., Gibb B.C. Dynamic light scattering - an all-purpose guide for the supramolecular chemist. Supramol. Chem. 2019;31:608–615. doi: 10.1080/10610278.2019.1629438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farkas N., Kramar J.A. Dynamic light scattering distributions by any means. J. Nanopart. Res. 2021;23:120. doi: 10.1007/s11051-021-05220-6. [DOI] [Google Scholar]
- 52.Lale S.V., Goyal M., Bansal A.K. Development of lyophilization cycle and effect of excipients on the stability of catalase during lyophilization. Int. J. Pharm. Investig. 2011;1:214–221. doi: 10.4103/2230-973x.93007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Molnar A., Lakat T., Hosszu A., Szebeni B., Balogh A., Orfi L., Szabo A.J., Fekete A., Hodrea J. Lyophilization and homogenization of biological samples improves reproducibility and reduces standard deviation in molecular biology techniques. Amino Acids. 2021;53:917–928. doi: 10.1007/s00726-021-02994-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patel S.M., Pikal M.J. Emerging freeze-drying process development and scale-up issues. AAPS PharmSciTech. 2011;12:372–378. doi: 10.1208/s12249-011-9599-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rumsey S., Galeano N., Arad Y., Deckelbaum R. Cryopreservation with sucrose maintains normal physical and biological properties of human plasma low density lipoproteins. J. Lipid Res. 1992;33:1551–1561. doi: 10.1016/s0022-2275(20)41409-9. [DOI] [PubMed] [Google Scholar]
- 56.Wang L., Ma Y., Gu Y., Liu Y., Zhao J., Yan B., Wang Y. Cryoprotectant choice and analyses of freeze-drying drug suspension of nanoparticles with functional stabilisers. J. Microencapsul. 2018;35:1–26. doi: 10.1080/02652048.2018.1462416. [DOI] [PubMed] [Google Scholar]
- 57.Voci S., Gagliardi A., Salvatici M.C., Fresta M., Cosco D. Influence of the dispersion medium and cryoprotectants on the physico-chemical features of gliadin- and zein-based nanoparticles. Pharm. 2022;14:332. doi: 10.3390/pharmaceutics14020332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alkilany A.M., Abulateefeh S.R., Mills K.K., Yaseen A.I.B., Hamaly M.A., Alkhatib H.S., Aiedeh K.M., Stone J.W. Colloidal stability of citrate and mercaptoacetic acid capped gold nanoparticles upon lyophilization: effect of capping ligand attachment and type of cryoprotectants. Langmuir. 2014;30:13799–13808. doi: 10.1021/la504000v. [DOI] [PubMed] [Google Scholar]
- 59.Doktorovova S., Shegokar R., Fernandes L., Martins-Lopes P., Silva A.M., Müller R.H., Souto E.B. Trehalose is not a universal solution for solid lipid nanoparticles freeze-drying. Pharm. Dev. Technol. 2013;19:922–929. doi: 10.3109/10837450.2013.840846. [DOI] [PubMed] [Google Scholar]
- 60.Roozen M.J.G.W., Hemminga M.A. Molecular motion in sucrose-water mixtures in the liquid and glassy state as studied by spin probe ESR. J. Phys. Chem. 1990;94:7326–7329. doi: 10.1021/j100381a068. [DOI] [Google Scholar]
- 61.Passot S., Trelea I.C., Marin M., Galan M., Morris G.J., Fonseca F. Effect of controlled ice nucleation on primary drying stage and protein recovery in vials cooled in a modified freeze-dryer. J. Biomech. Eng. 2009;131:74511. doi: 10.1115/1.3143034. [DOI] [PubMed] [Google Scholar]
- 62.Arsiccio A., Pisano R. The ice-water interface and protein stability: a review. J. Pharm. Sci. 2020;109:2116–2130. doi: 10.1016/j.xphs.2020.03.022. [DOI] [PubMed] [Google Scholar]
- 63.Yu G., Li R., Hubel A. Interfacial interactions of sucrose during cryopreservation detected by Raman spectroscopy. Langmuir. 2019;35:7388–7395. doi: 10.1021/acs.langmuir.8b01616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mathlouthi M., Genotelle J. Role of water in sucrose crystallization1 presented at the second international meeting of the Portuguese carbohydrate chemistry group.1. Carbohydr. Polym. 1998;37:335–342. doi: 10.1016/s0144-8617(98)00079-4. [DOI] [Google Scholar]
- 65.Carrasco M.J., Alishetty S., Alameh M.-G., Said H., Wright L., Paige M., Soliman O., Weissman D., Cleveland T.E., Grishaev A., Buschmann M.D. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun. Biol. 2021;4:956. doi: 10.1038/s42003-021-02441-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reiser P., Birch G.G., Mathlouthi M. 1995. Sucrose, Properties and Applications; pp. 186–222. [DOI] [Google Scholar]
- 67.Roos Y., Karel M. Amorphous state and delayed ice formation in sucrose solutions. Int. J. Food Sci. Technol. 1991;26:553–566. doi: 10.1111/j.1365-2621.1991.tb02001.x. [DOI] [Google Scholar]
- 68.Chung N.-O., Lee M.K., Lee J. Mechanism of freeze-drying drug nanosuspensions. Int. J. Pharm. 2012;437:42–50. doi: 10.1016/j.ijpharm.2012.07.068. [DOI] [PubMed] [Google Scholar]
- 69.Wayment-Steele H.K., Kim D.S., Choe C.A., Nicol J.J., Wellington-Oguri R., Watkins A.M., Sperberg R.A.P., Huang P.S., Participants E., Das R. Biorxiv; 2021. Theoretical Basis for Stabilizing Messenger RNA through Secondary Structure Design. 2020.08.22.262931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.A Study to Evaluate the Efficacy, Safety, and Immunogenicity of mRNA-1647 Cytomegalovirus (CMV) Vaccine in Healthy Participants 16 to 40 Years of Age. 2021. https://clinicaltrials.gov/ct2/show/nct05085366
- 71.Kon E., Elia U., Peer D. Principles for designing an optimal mRNA lipid nanoparticle vaccine. Curr. Opin. Biotechnol. 2021;73:329–336. doi: 10.1016/j.copbio.2021.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Machado B.A.S., Hodel K.V.S., Fonseca L., Mascarenhas L.A.B., Andrade L., Rocha V.P.C., Soares M.B.P., Berglund P., Duthie M.S., Reed S.G., Badaro R. The importance of RNA-based vaccines in the fight against COVID-19: an overview. Nato Adv. Sci. Inst. Se. 2021;9:1345. doi: 10.3390/vaccines9111345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Muramatsu H., Lam K., Bajusz C., Laczkó D., Karikó K., Schreiner P., Martin A., Lutwyche P., Heyes J., Pardi N. Lyophilization provides long-term stability for a lipid nanoparticle-formulated, nucleoside-modified mRNA vaccine. Mol. Ther. 2022;30:1941–1951. doi: 10.1016/j.ymthe.2022.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Data Availability Statement
Data will be made available on request.