Abstract
Artificial intelligence (AI) solutions that automatically extract information from digital histology images have shown great promise for improving pathological diagnosis. Prior to routine use, it is important to evaluate their predictive performance and obtain regulatory approval. This assessment requires appropriate test datasets. However, compiling such datasets is challenging and specific recommendations are missing. A committee of various stakeholders, including commercial AI developers, pathologists, and researchers, discussed key aspects and conducted extensive literature reviews on test datasets in pathology. Here, we summarize the results and derive general recommendations on compiling test datasets. We address several questions: Which and how many images are needed? How to deal with low-prevalence subsets? How can potential bias be detected? How should datasets be reported? What are the regulatory requirements in different countries? The recommendations are intended to help AI developers demonstrate the utility of their products and to help pathologists and regulatory agencies verify reported performance measures. Further research is needed to formulate criteria for sufficiently representative test datasets so that AI solutions can operate with less user intervention and better support diagnostic workflows in the future.
Introduction
The application of artificial intelligence techniques to digital tissue images has shown great promise for improving pathological diagnosis1–3. They can not only automate time-consuming diagnostic tasks and make analyses more sensitive and reproducible, but also extract new digital biomarkers from tissue morphology for precision medicine4.
Pathology involves a large number of diagnostic tasks, each being a potential application for AI. Many of these involve the characterization of tissue morphology. Such tissue classification approaches have been developed for identifying tumors in a variety of tissues, including lung5,6, colon7, breast8,9, and prostate9 but also in non-tumor pathology, e.g., kidney transplants10. Further applications include predicting outcomes11,12 or gene mutations5,13,14 directly from tissue images. Similar approaches are also employed to detect and classify cell nuclei, e.g., to quantify the positivity of immunohistochemistry markers like Ki67, ER/PR, Her2, and PD-L115,16.
Testing AI solutions is an important step to ensure that they work reliably and robustly on routine laboratory cases. AI algorithms run the risk of exploiting feature associations that are specific to their training data17. Such “overfitted” models tend to perform poorly on previously unseen data. To obtain a realistic estimate of the prediction performance on real-word data, it is common practice to apply AI solutions to a test dataset. The results are then compared with reference results in terms of task-specific performance metrics, e.g., sensitivity, specificity, or area under the receiver operating characteristic curve (ROC-AUC).
Test datasets may only be used once to evaluate the performance of a finalized AI solution17. They may not be considered during development. This can be considered a consequence of Goodhart’s law stating that measures cease to be meaningful when used as targets18: If AI solutions are optimized for test datasets, they cannot provide realistic performance estimates for real-world data. Test datasets are also referred to as “hold-out datasets” or “(external) validation datasets.” The term “validation,” however, is not used consistently in the machine learning community and can also refer to model selection during development17.
Besides overfitting, AI methods are prone to “shortcut learning”19. Many datasets used in the development of AI methods contain confounding variables (e.g., slide origin, scanner type, patient age) that are spuriously correlated with the target variable (e.g., tumor type)20. AI methods often exploit features that are discriminative for such confounding variables and not for the target variable21. Despite working well for smaller datasets containing similar correlations, such methods fail in more challenging real-world scenarios in ways humans never would22. To minimize the likelihood of spurious correlations between confounding variables and the target variable, test datasets must be large and diversified20. At the same time, test datasets must be small enough to be acquired with realistic effort and cost. Finding a good balance between these requirements is a major challenge for AI developers.
Comparatively little attention has been paid to compiling test datasets for AI solutions in pathology. Datasets for training, on the other hand, were considered frequently9,23–28. Training data are collected with a different goal than test datasets: While training datasets should produce the best possible AI models, test datasets should provide the most realistic performance assessment for routine use, which presents unique challenges.
Some publications address individual problems in compiling test datasets in pathology, e.g., how to avoid bias in the performance evaluation caused by site-specific image features in test datasets29. Other publications provide general recommendations for evaluating AI methods for medical applications without considering the specific challenges of pathology30–34.
Appropriate test datasets are critical to demonstrate the utility of AI solutions as well as to obtain regulatory approval. However, the lack of guidance on how to compile test datasets is a major barrier to the adoption of AI solutions in laboratory practice.
This article gives recommendations for test datasets in pathology. It summarizes the results of extensive literature reviews and discussions by a committee of various stakeholders, including commercial AI developers, pathologists, and researchers. This committee was established as part of the EMPAIA project (Ecosystem for Pathology Diagnostics with AI Assistance), aiming to facilitate the adoption of AI in pathology35.
Results
The next sections discuss and provide recommendations on various aspects that must be considered when creating test datasets. For meaningful performance estimates, test datasets must be both diverse enough to cover the variability of data in routine diagnostics and large enough to allow statistically meaningful analyses. Relevant subsets must be covered, and test datasets should be unbiased. Moreover, test datasets must be sufficiently independent of datasets used in the development of AI solutions. Comprehensive information about test datasets must be reported and regulatory requirements must be met when evaluating the clinical applicability of AI solutions.
Target population of images
Compiling a test dataset requires a detailed description of the intended use of the AI solution to be tested. The intended use must clearly indicate for which diagnostic task(s) the solutions may be used, and whether the use is limited to images with certain characteristics. All images an AI solution may encounter in its intended use constitute its “target population of images.” A test dataset must be an adequate sample of this target population (see Fig. 1) to provide a reasonable estimate of the prediction performance of the AI solution. For all applications in pathology, the target population is distributed across multiple dimensions of variability, see Table 1.
Fig. 1. Schematic overview of sampling regimes for performance assessment in the entire target population of images or in specific subsets.
Overall performance assessment requires a representative sample along all dimensions of variability, relevant subsets are typically limited along one dimension (e.g., age range or scanner type).
Table 1.
| Origin | Variabilities |
|---|---|
| Patient |
• Patient ethnicity • Patient demographics • Disease stage/severity • Rare cases of disease • Comorbidities • Biological differences (genetic, transcriptional, epigenetic, proteomic, and metabolomic) |
| Specimen sampling |
• Tissue heterogeneity • Size of tissue section • Coverage of diseased/healthy/boundary regions • Tissue damage, e.g., torn, cauterized • Surgical ink present |
| Slide processing |
• Inter-material and device differences • Preparation differences (fixation, dehydration; freezing; mechanical handling) • Cutting artifacts (torn, folded, deformed, thick or inhomogeneously thick tissue) • Foreign matter/floaters in specimen • Over-/under-staining, inhomogeneous staining • Foreign objects on slide/cover slip (dirt, stain residue, pen markings, fingerprint) • Cracks, air bubbles, scratches • Slide age |
| Imaging/image processing |
• Inter- and intra-scanner differences • Out-of-focus images, heterogeneous focus • Amount of background in analyzed image region • Magnification/image resolution • Heterogeneous illumination • Grid noise, stitching artifacts • Lossy image compression |
| Ground truth annotation |
• Inter- and intra-observer differences • Ambiguous cases |
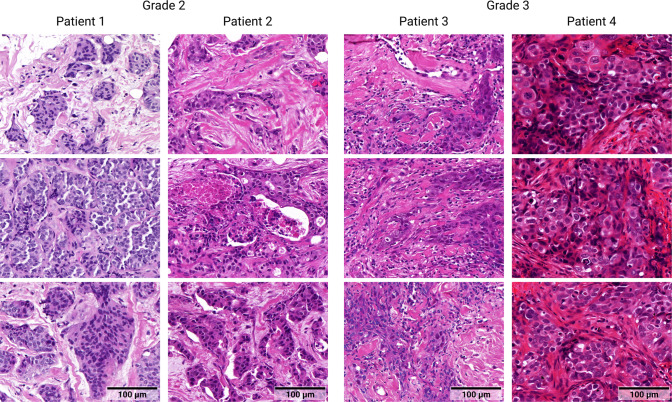
Biological variability: The visual appearance of tissue varies between normal and diseased states. This is what AI solutions are designed to detect and characterize. But even tissue of the same category can look very different (see Fig. 2). The appearance is influenced by many factors (e.g., genetic, transcriptional, epigenetic, proteomic, and metabolomic) that differ between patients as well as between demographic and ethnic groups36. These factors often vary spatially (e.g., different parts of organs are differently affected) and temporally (e.g., the pathological alterations differ based on disease stage) within a single patient37.
Fig. 2. Examples of variability between biopsy images, illustrating a combination of inter- and intraindividual biological variability (tissue structure) and inter-individual technical variability (staining).
The images show H&E-stained breast tissue of female patients with invasive carcinomas of no special type, scanned at 40× objective magnification.
Technical variability: Processing and digitization of tissue sections consists of several steps (e.g., tissue fixation, processing, cutting, staining and digitization) all of which can contribute to image variability38. Differences in section thickness and staining solutions can lead to variable staining appearances39. Artifacts frequently occur during tissue processing, including elastic deformations, inclusion of foreign objects, and cover glass scratches40. Differences in illumination, resolution, and encoding algorithms of slide scanner models also affect the appearance of tissue images38.
Observer variability: Images in test datasets are commonly associated with a reference label like a disease category or score determined by a human observer. It is well known that the assessment of tissue images is subject to intra- and inter-observer variability41–47. This variability results from subjective biases (e.g., caused by training, specialization, and experience) but also from inherent ambiguities in the images48,49.
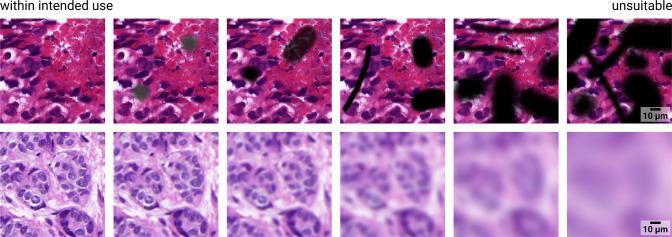
Routine laboratory work occasionally produces images that are unsuitable for the intended use of an AI solution, e.g., because they are ambiguous or of insufficient quality. Most AI solutions require prior quality assurance steps to ensure that solutions are only applied to suitable images50,51. The boundary between suitable and unsuitable images is usually fuzzy and there are difficult images that cannot be clearly assigned to either category (see Fig. 3).
Fig. 3. Examples of different severity levels of imaging artifacts.
The leftmost images are clearly within the intended use of algorithms for analyzing breast cancer histologies, whereas the rightmost images are clearly unsuitable. However, it is not obvious where to draw the line between those two regimes. The top row shows simulated foreign objects, the bottom row shows simulated focal blur, the original tissue images show H&E-stained breast tissue of female patients with invasive carcinomas of no special type, scanned at 40× objective magnification (same as in Fig. 1).
Defining the target population is challenging and presumes a clear definition of the intended use by the AI developer. The target population of images must be defined before test data are collected. It must be clearly stated which subsets of images fall under the intended use. Such subsets may consist of specific disease variants, demographic characteristics, ethnicities, staining characteristics, artifacts, or scanner types. These subsets typically overlap, e.g., the subset of images of one scanner type contains images from different patient age groups. A particular challenge is to define where the target population ends. Examples of images within and outside the intended use can help human observers sort out unsuitable images as objectively as possible.
Data collection
Test datasets must be representative of the entire target population of images, i.e., sufficiently diverse and unbiased. To minimize spurious correlations between confounding variables and the target variable and to uncover shortcut learning in AI methods, all dimensions of biological and technical variability must be adequately covered for the classes considered20,28, also reflecting the variability of negative cases without visible pathology28,52.
All images encountered in the normal laboratory workflow must be considered. One way to achieve this is to collect all cases that occurred over a given time period52 long enough for a sufficient number of cases to be collected (e.g., one year9). These cases should be collected from multiple international laboratories, since they differ in their spectra of patients and diseases, technical equipment and operating procedures. Data should be collected at the point in the workflow where the AI solution would be applied, taking into account possible prior quality assurance steps in the workflow.
All data in a test dataset must be collected according to a consistent acquisition protocol (see “Reporting”). The best way to ensure this is to prospectively collect test data according to this protocol. Retrospective datasets were typically compiled for a different purpose and are thus likely to be subject to selection bias, that is difficult to adjust for53. If retrospective data are used in a test dataset, a comprehensive description of the acquisition protocol must be available so that potential issues can be identified54.
Annotation
Test datasets for AI solutions contain not only images, but also annotations representing the expected analysis result, e.g., slide-level labels or delineations of tissue regions. In most cases, such reference annotations must be prepared by human observers with sufficient experience in the diagnostic use case. Since humans are prone to intra- and inter-observer variability, annotations in test datasets should be created by multiple observers from different hospitals or laboratories. For unequivocal results, it can be helpful to organize consensus conferences and to use standardized electronic reporting formats41. Any remaining disagreement should be documented with justification (e.g., suboptimal sample quality) and considered when evaluating AI solutions. Semi-automatic annotation methods can help reduce the effort required for manual annotation55,56. However, they can introduce biases themselves and should therefore be monitored by human observers.
Curation
Unsuitable data that does not fit the intended use of an AI solution should not be included in a test dataset. Such data usually must be detected by human observers, e.g., in a dedicated data curation step or during the generation of reference annotations. To avoid selection bias, it is important not to exclude artifacts or atypical images that are part of the intended use of the product9,52,57.
There are automated tools to support the detection of unsuitable data58. Some approaches detect unsuitable images based on basic image features such as brightness, predominant colors, and sharpness59,60 or by detecting typical artifacts like tissue folds and air bubbles61,62. Other methods analyze domain shifts63–65 or use dedicated neural networks trained for outlier detection66. There are also approaches for detecting outliers depending on the tested AI solution63,67–70. Although these approaches can help exclude unsuitable images from test datasets, they do not yet appear to be mature enough to be used entirely without human supervision.
Synthetic data
There are a variety of techniques for extending datasets with synthetic data. Some techniques alter existing images in a generic (e.g., rotation, mirroring) or histology-specific way (e.g., stain transformations26 or emulation of image artifacts40,71–76). Other techniques create fully synthetic images from scratch77–81. These techniques are useful for data augmentation1,2,82, i.e., enriching development data in order to avoid overfitting and increase robustness. However, they cannot replace original real-world data for test datasets. Because all of these techniques are based on simplified models of real-world variability, they are likely to introduce biases into a test dataset and make meaningful performance measurement impossible.
Sample size
Any test dataset is a sample from the target population of images, thus any performance metric computed on a test dataset is subject to sampling error. In order to draw reliable conclusions from evaluation results, the sampling error must be sufficiently small. Larger samples generally result in lower sampling error, but are also more expensive to produce. Therefore, the minimum sample size required to achieve a maximum allowable sampling error should be determined prior to data collection.
Many different methods have been proposed for sample size determination. Most of them refer to statistical significance tests which are used to test a prespecified hypothesis about a population parameter (e.g., sensitivity, specificity, ROC-AUC) on the basis of an observed data sample83–85. Such sample size determination methods are commonly used in clinical trial planning and available in many statistical software packages70.
When evaluating AI solutions in pathology, the goal is more often to estimate a performance metric with a sufficient degree of precision than to test a previously defined hypothesis. Confidence intervals (CIs) are a natural way to express the precision of an estimated metric and should be reported instead of or in addition to test results86. A CI is an interval around the sample statistic that is likely to cover the true population value at some confidence level, usually 95%87. The sample statistic can either be the performance metric itself or a difference between the performance metrics of two methods, e.g., when comparing performance to an established solution.
When using CIs, the sample size calculation can be based on the targeted width of the CI which is inversely proportional to the precision of the performance estimation86. Several approaches have been proposed for that matter88–92. To determine a minimum sample size, assumptions regarding the sample statistic, its variability, and usually also its distributional form must be made. The open-source software “presize” implements several of these methods and provides a simple web-based user interface to perform CI-based sample size calculations for common performance metrics93.
Subsets
AI solutions that are very accurate on average often perform much worse on certain subsets of their target population of images94, a phenomenon known as “hidden stratification.” Such differences in performance can exceed 20%22. Hidden stratification occurs particularly in low-prevalence subsets, but may also occur in subsets with poor label quality or subtle distinguishing characteristics22. There are substantial differences in cancer incidence, e.g., by gender, socioeconomic status, and geographic region95. Hence, hidden stratification may result in disproportionate harm to patients in less common demographic groups and jeopardize the clinical applicability of AI solutions22. Common performance measures computed on the entire test dataset can be dominated by larger subsets and do not indicate whether there are subsets for which an AI solution underperforms96.
To detect hidden stratification, AI solutions must be evaluated independently on relevant subsets of the target population of images (e.g., certain medical characteristics, patient demographics, ethnicities, scanning equipment)22,94, see Fig. 1. This means in particular that the metadata for identifying the subsets must be available30. Performance evaluation on subsets is an important requirement to obtain clinical approval by the FDA (see “Regulatory requirements”). Accordingly, such subsets should be specifically delineated within test datasets. Each subset needs to be sufficiently large to allow statistically meaningful results (see “Sample size”). It is important to provide information on why and how subsets were collected so that any issues AI solutions may have with specific subsets can be specifically tracked (see “Reporting”). Identifying subsets at risk of hidden stratification is a major challenge and requires extensive knowledge of the use case and the distribution of possible input images22. As an aid, potentially relevant subsets can also be detected automatically using unsupervised clustering approaches such as k-means22. If a detected cluster underperforms compared to the entire dataset, this may indicate the presence of hidden stratification that needs further examination.
Bias detection
Biases can make test datasets unsuitable for evaluating the performance of AI algorithms. Therefore, it is important to identify potential biases and to mitigate them early during data acquisition28. Bias, in this context, refers to sampling bias, i.e., the test dataset is not a randomly drawn sample from the target population of images. Subsets to be evaluated independently may be biased by construction with respect to particular features (e.g., patient age). Here, it is important to ensure that the subsets do not contain unexpected biases with respect to other features. For example, the prevalence of slide scanners should be independent of patient age, whereas the prevalence of diagnoses may vary by age group. Bias detection generally involves comparing the feature distributions of the test dataset and the target population of images. Similar methods can also be used to test the diversity or representativeness of a test dataset.
For features represented as metadata (e.g., patient age, slide scanner, or diagnosis), the distributions of the test dataset and target population can be compared using summary statistics (e.g., mean and standard deviation, percentiles, or histograms) or dedicated representativeness metrics97,98. Detection of bias in an entire test dataset requires a good estimate of the feature distribution of the target population of images. Bias in subsets can be detected by comparing the subset distribution to the entire dataset. Several toolkits for measuring bias based on metadata have been proposed99,100 and evaluated101.
Detecting bias in the image data itself is more challenging. Numerous features can be extracted from image data and it is difficult to determine the distribution of these features in the target population of images. Similar to automatic detection of unsuitable data, there are automatic methods to reveal bias in image data. Domain shifts63 can be detected either by comparing the distributions of basic image features (e.g., contrast) or by more complex image representations learned through specific neural network models63,66,102. Another approach is to train trivial machine learning models with modified images from which obvious predictive information has been removed (e.g., tumor regions): If such models perform better than chance, this indicates bias in the dataset103,104.
Independence
In the development of AI solutions, it is common practice to split a given dataset into two sets, one for development (e.g., a training and a validation set for model selection) and one for testing17. AI methods are prone to exploit spurious correlations in datasets as shortcut opportunities19. In this case, the methods perform well on data with similar correlations, but not on the target population. If both development and test datasets are drawn from the same original dataset, they are likely to share spurious correlations, and the performance on the test dataset may overestimate the performance on the target population. Therefore, datasets used for development and testing need to be sufficiently independent. As explained below, it is not sufficient for test datasets to merely contain different images than development datasets17,19.
To account for memory constraints, histologic whole-slide images (WSIs) are usually divided into small sub-images called “tiles.” AI methods are then applied to each tile individually, and the result for the entire WSI is obtained by aggregating the results of the individual tiles. If tiles are randomly assigned, tiles from the same WSI can end up in both the development and the test datasets, possibly inflating performance results. A substantial number of published research studies are affected by this problem105. Therefore, to avoid any risk of bias, none of the tiles in a test dataset may originate from the same WSI as the tiles in the development set105.
Datasets can contain site-specific feature distributions29. If these site-specific features are correlated with the outcome of interest, AI methods might use these features for classification rather than the relevant biological features (e.g., tissue morphology) and be unable to generalize to new datasets. A comprehensive evaluation based on multi-site datasets from The Cancer Genome Atlas (TCGA) showed that including data from one site in development and test datasets often leads to overoptimistic estimates of model accuracy29. This study also found that commonly used color normalization and augmentation methods did not prevent models from learning site-specific features, although stain differences between laboratories appeared to be a primary source of site-specific features. Therefore, the images in development and test datasets must originate not only from different subjects, but should also from different clinical sites31,106,107.
As described in the Introduction section, a given AI solution should only be evaluated once against a given test dataset17. Datasets published in the context of challenges or studies (many of which are based on TCGA4 and have regional biases108) should generally not be used as test datasets: it cannot be ruled out that they were taken into account in some form during development, e.g., inadvertently or as part of pretraining. Ideally, test datasets should not be published at all and the evaluation should be conducted by an independent body with no conflicts of interest30.
Reporting
Adequate reporting of test datasets is essential to determine whether a particular dataset is appropriate for a particular AI solution. Detailed metadata on the coverage of various dimensions of variability is required for detecting bias and identifying relevant subsets. Data provenance must be tracked to ensure that test data are sufficiently disjoint from development data28,29. Requirements for the test data109 and acquisition protocols110 should also be reported so that further data can be collected later. Accurate reporting of test datasets is important in order to submit evaluation results traceable to the test data for regulatory approval111.
Various guidelines for reporting clinical research and trials, including diagnostic models, have been published112. Some of these have been adapted specifically for machine learning approaches113,114 or such adaptation is under development115–118. However, only very few guidelines elaborate on data reporting119, and there is not yet consensus on structured reporting of test datasets, particularly for computational pathology.
Data acquisition protocols should comprehensively describe how and where the test dataset was acquired, handled, processed, and stored109,110. This documentation should include precise details of the hardware and software versions used and also cover the creation of reference annotations. Moreover, quality criteria for rejecting data and procedures for handling missing data119 should be reported, i.e., aspects of what is not in the dataset. To facilitate data management and analysis, individual images should be referenced via universally unique identifiers120 and image metadata should be represented using standard data models121,122. Protocols should be defined prior to data acquisition when prospectively collecting test data. Completeness and clarity of the protocols should be verified during data acquisition.
Reported information should characterize the acquired dataset in a useful way. For example, summary statistics allow an initial assessment whether a given dataset is an adequate sample of the target population (see section “Bias detection”). Relevant subsets and biases identified in the dataset should be reported as well. Generally, one should collect and report as much information as feasible with the available resources, since retrospectively obtaining missing metadata is hard or impossible. If there will be multiple versions of a dataset, e.g., due to iterative data acquisition or review of reference annotations, versioning is needed. Suitable hashing can guarantee integrity of the entire dataset as well as its individual samples, and identify datasets without disclosing contents.
Regulatory requirements
AI solutions in pathology are in vitro diagnostic medical devices (IVDMDs) because they evaluate tissue images for diagnostic purposes outside the human body. Therefore, regulatory approval is required for sale and use in a clinical setting123. The U.S. Food and Drug Administration (FDA) and European Union (EU) impose similar requirements to obtain regulatory approval. This includes compliance with certain quality management and documentation standards, a risk analysis, and a comprehensive performance evaluation. The performance evaluation must demonstrate that the method provides accurate and reliable results compared to a gold standard (analytical performance) and that the method provides real benefit in a clinical context (clinical performance). Good test datasets are an essential prerequisite for a meaningful evaluation of analytical performance.
EU + UK
In the EU and UK, IVDMDs are regulated by the In vitro Diagnostic Device Regulation (IVDR, formally “Regulation 2017/746”)124. After a transition period, compliance with the IVDR will be mandatory for novel routine pathology diagnostics as of May 26, 2022. The IVDR does not impose specific requirements on test datasets used in the analytical performance evaluation. However, the EU has put forward a proposal for an EU-wide regulation on harmonized rules for the assessment of AI125.
The EU proposal125 considers AI-based IVDMDs as “high-risk AI systems” (preamble (30)). For test datasets used in the evaluation of such systems, the proposal imposes certain quality criteria: test datasets must be “relevant, representative, free of errors and complete” and “have the appropriate statistical properties” (Article 10.3). Likewise, it requires test datasets to be subject to “appropriate data governance and management practices” (preamble (44)) with regard to design choices, suitability assessment, data collection, and identification of shortcomings.
USA
In the US, IVDMDs are regulated in the Code of Federal Regulations (CFR) Part 809126. Just like the IVDR, the CFR does not impose specific requirements on test datasets used in the analytical performance evaluation. However, the CFR states that products should be accompanied by labeling stating specific performance characteristics (e.g., accuracy, precision, specificity, and sensitivity) related to normal and abnormal populations of biological specimens.
In 2021, the FDA approved the first AI software for pathology127. In this context, the FDA has established a definition and requirements for approval of generic AI software for pathology, formally referred to as “software algorithm devices to assist users in digital pathology”128.
Test datasets used in analytical performance studies are expected to contain an “appropriate” number of images. To be “representative of the entire spectrum of challenging cases” (3.ii.A. and B. of source128) that can occur when the product is used as intended, test datasets should cover multiple operators, slide scanners, and clinical sites and contain “clinical specimens with defined, clinically relevant, and challenging characteristics.”(3.ii.B. of source128) In particular, test datasets should be stratified into relevant subsets (e.g., by medical characteristics, patient demographics, scanning equipment) to allow separate determination of performance for each subset. Case cohorts considered in clinical performance studies (e.g., evaluating unassisted and software-assisted evaluation of pathology slides with intended users) are expected to adhere to similar specifications.
Product labeling according to CFR 809 was also defined in more detail. In addition to the general characteristics of the dataset (e.g., origin of images, annotation procedures, subsets, …), limitations of the dataset (e.g., poor image quality or insufficient sampling of certain subsets) that may cause the software to fail or operate unexpectedly should be specified.
In summary, there are much more specific requirements for test datasets in the US than in the EU. However, none of the regulations clearly specify how the respective requirements can be achieved or verified.
Discussion
Our recommendations on compiling test datasets are summarized in Fig. 4. They are intended to help AI developers demonstrate the robustness and practicality of their solutions to regulatory agencies and end users. Likewise, the advice can be used to check whether test datasets used in the evaluation of AI solutions were appropriate and reported performance measures are meaningful. Much of the advice can also be transferred to image analysis solutions without AI and to similar domains where solutions are applied to medical images, such as radiology or ophthalmology.
Fig. 4. Overview of recommendations on compiling test datasets.
Prior to data acquisition, the acquisition process must be thoroughly planned. In particular, the intended use of the AI solution must be precisely understood in order to derive the requirements for test datasets.
A key finding of the work is that it remains challenging to compile test datasets and that there are still many unanswered questions. The current regulatory requirements remain vague and do not specify in detail important aspects such as the required diversity or size of a test dataset. In principle, the methods described in the “Bias detection” and “Sample size” sections can be used to assess whether a sample is sufficiently diverse or large, respectively. These methods depend on a precise and comprehensive definition of the target population of images. However, since this population usually cannot be formally specified but only roughly described, it can be difficult to apply these methods in a meaningful way in practice.
For regulatory approval, a plausible justification is needed why the test dataset used was good enough. Besides following the advice in this paper, it can also be helpful to refer to published studies in which AI solutions have been comprehensively evaluated. Additional guidance can be found in the summary documents of approved AI solutions published by the FDA, which include information on their evaluation106. It turns out that many of the AI devices approved by the FDA were evaluated only at a small number of sites106 with limited geographic diversity129. Test sets used in current studies typically involved thousands of slides, hundreds of patients, less than five sites, and less than five scanner types50,52,130,131.
Today, AI solutions in pathology may not be used for primary diagnosis, but only in conjunction with a standard evaluation by the pathologist128. Therefore, compared to a fully automated usage scenario, requirements for robustness are considerably lower. This also applies to the expected confidence in the performance measurement and the scope of the test dataset used. In a supervised usage scenario, the accuracy of an AI solution determines how often the user needs to intervene to correct results, and thus its practical usefulness. End users are interested in the most meaningful evaluation of the accuracy of AI solutions to assess their practical utility. Therefore, a comprehensive evaluation of the real-world performance of a product, taking into account the advice given in this paper, can be an important marketing tool.
Limitations and outlook
Some aspects of compiling test datasets were not considered in this article. One aspect is how to collaborate with data donors, i.e., how to incentivize or compensate them for donating data. Other aspects include the choice of software tools and data formats for the compilation and storage of datasets or how the use of test datasets should be regulated. These aspects must be clarified individually for each use case and the AI solution to be tested. Furthermore, we do not elaborate on legal aspects of collecting test data, e.g., obtaining consent from patients, privacy regulations, licensing, and liability. For more details on these topics, we refer to other works132. The present paper focuses exclusively on compiling test datasets. For advice on other issues related to validating AI solutions in pathology, such as how to select an appropriate performance metric, how to make algorithmic results interpretable, or how to conduct a clinical performance evaluation with end users, we also refer to other works30,31,33,34,133,134.
For AI solutions to operate with less user intervention and to better support diagnostic workflows, real-world performance must be assessed more accurately than is currently possible. The key to accurate performance measures is the representativeness of the test dataset. Therefore, future work should focus on better characterizing the target population of images and how to collect more representative samples. Empirical studies should be conducted on how different levels of coverage of the variability dimensions (e.g., laboratories, scanner types) affect the quality of performance evaluation for common use cases in computational pathology.
In addition, clear criteria should be developed to delineate the target population from unsuitable data. Currently, the assessment of the suitability of data is typically done by humans, which might introduce subjective bias. Automated methods can help to make the assessment of suitability more objective (see “Curation”) and should therefore be further explored. However, such automated methods must be validated on dedicated test datasets themselves.
Another open challenge is how to deal with changes in the target population of images. Since the intended use for a particular product is fixed, in theory the requirements for the test datasets should also be fixed. However, the target distribution of images is influenced by several factors that change over time. These include technological advances in specimen and image acquisition, distribution of scanner systems used, and shifting patient populations133,135. As part of post-market surveillance, AI solutions must be continuously monitored during their entire lifecycle111. Clear processes are required for identifying changes in the target population of images and adapting performance estimates accordingly.
Conclusions
Appropriate test datasets are essential for meaningful evaluation of the performance of AI solutions. The recommendations provided in this article are intended to help demonstrate the utility of AI solutions in pathology and to assess the validity of performance studies. The key remaining challenge is the vast variability of images in computational pathology. Further research is needed on how to formalize criteria for sufficiently representative test datasets so that AI solutions can operate with less user intervention and better support diagnostic workflows in the future.
Author contributions
A.H. and C.G. organized the committee work. A.H., C.G., and L.O.S. conceived the manuscript. A.H., C.G., L.O.S., F.Z., T.E., K.S., M.W., and R.D.B. wrote the manuscript. All authors participated in the committee work and contributed to the literature review. The final version of the paper was reviewed and approved by all authors.
Funding
A.H., C.G., L.O.S., F.Z., T.E., K.S., A.K., C.O.R., T.S., R.C., P.B., P.H., and N.Z. were supported by the German Federal Ministry for Economic Affairs and Climate Action via the EMPAIA project (grant numbers 01MK20002A, 01MK20002B, 01MK20002C, 01MK20002E). M.Ka., M.P., and H.M. received funding from the Austrian Science Fund (FWF), Project P-32554 (Explainable Artificial Intelligence), the Austrian Research Promotion Agency (FFG) under grant agreement No. 879881 (EMPAIA), and the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 857122 (CY-Biobank). P.S. was funded by Helmholtz Association’s Initiative and Networking Fund through Helmholtz AI. R.B. was supported by the START Program of the Faculty of Medicine of the RWTH Aachen University (Grant-Nr. 148/21). P.B. was also supported by the German Research Foundation (DFG, Project IDs 322900939, 454024652, 432698239, and 445703531), the European Research Council (ERC, CoG AIM.imaging.CKD No. 101001791), the German Federal Ministries of Health (Deep Liver, No. ZMVI1-2520DAT111), Education and Research (STOP- SGS-01GM1901A). The funders had no role in the committee work, discussions, literature research, decision to publish, or preparation of the manuscript. Open Access funding enabled and organized by Projekt DEAL.
Competing interests
F.Z. is a shareholder of asgen GmbH. P.S. is a member of the supervisory board of asgen GmbH. All other authors declare that they have no conflict of interest.
Footnotes
The original online version of this article was revised: The original version of this article was unfortunately missing the competing interests section. The "Competing interests" section should read: F.Z. is a shareholder of asgen GmbH. P.S. is a member of the supervisory board of asgen GmbH. All other authors declare that they have no conflict of interest.
These authors contributed equally: André Homeyer, Christian Geißler, Lars Ole Schwen, Falk Zakrzewski, Theodore Evans, Klaus Strohmenger, Max Westphal, Roman David Bülow.
Change history
9/23/2022
A Correction to this paper has been published: 10.1038/s41379-022-01163-y
References
- 1.Serag A, Ion-Margineanu A, Qureshi H, McMillan R, Martin M-JS, Diamond J, et al. Translational AI and deep learning in diagnostic pathology. Front Med 6, (2019). [DOI] [PMC free article] [PubMed]
- 2.Abels E, Pantanowitz L, Aeffner F, Zarella MD, Laak J, Bui MM, et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: A white paper from the digital pathology association. J Pathol 249, 286–294 (2019). [DOI] [PMC free article] [PubMed]
- 3.Moxley-Wyles B, Colling R, Verrill C. Artificial intelligence in pathology: An overview. Diagn Histopathol 26, 513–520 (2020).
- 4.Echle A, Rindtorff NT, Brinker TJ, Luedde T, Pearson AT, Kather JN. Deep learning in cancer pathology: A new generation of clinical biomarkers. Br J Cancer 124, 686–696 (2021). [DOI] [PMC free article] [PubMed]
- 5.Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 24, 1559–1567 (2018). [DOI] [PMC free article] [PubMed]
- 6.Wang X, Chen H, Gan C, Lin H, Dou Q, Tsougenis E, et al. Weakly supervised deep learning for whole slide lung cancer image analysis. IEEE Trans Cybern 50, 3950–3962 (2020). [DOI] [PubMed]
- 7.Iizuka O, Kanavati F, Kato K, Rambeau M, Arihiro K, Tsuneki M. Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Sci Rep 10, (2020). [DOI] [PMC free article] [PubMed]
- 8.Cruz-Roa A, Gilmore H, Basavanhally A, Feldman M, Ganesan S, Shih NNC, et al. Accurate and reproducible invasive breast cancer detection in whole-slide images: A deep learning approach for quantifying tumor extent. Sci Rep 7, (2017). [DOI] [PMC free article] [PubMed]
- 9.Campanella G, Hanna MG, Geneslaw L, Miraflor A, Krauss Silva VW, Busam KJ, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med 25, 1301–1309 (2019). [DOI] [PMC free article] [PubMed]
- 10.Kers J, Bülow RD, Klinkhammer BM, Breimer GE, Fontana F, Abiola AA, et al. Deep learning-based classification of kidney transplant pathology: A retrospective, multicentre, proof-of-concept study. Lancet Digit Health 4, e18–e26 (2022). [DOI] [PubMed]
- 11.Skrede O-J, De Raedt S, Kleppe A, Hveem TS, Liestøl K, Maddison J, et al. Deep learning for prediction of colorectal cancer outcome: A discovery and validation study. Lancet 395, 350–360 (2020). [DOI] [PubMed]
- 12.Saillard C, Schmauch B, Laifa O, Moarii M, Toldo S, Zaslavskiy M, et al. Predicting survival after hepatocellular carcinoma resection using deep learning on histological slides. Hepatology 72, 2000–2013 (2020). [DOI] [PubMed]
- 13.Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 25, 1054–1056 (2019). [DOI] [PMC free article] [PubMed]
- 14.Couture HD, Williams LA, Geradts J, Nyante SJ, Butler EN, Marron JS, et al. Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. npj Breast Cancer 4, (2018). [DOI] [PMC free article] [PubMed]
- 15.Höfener H, Homeyer A, Weiss N, Molin J, Lundström CF, Hahn HK. Deep learning nuclei detection: A simple approach can deliver state-of-the-art results. Comput Med Imaging Graph 70, 43–52 (2018). [DOI] [PubMed]
- 16.Balkenhol MCA, Ciompi F, Świderska-Chadaj Ż, Loo R van de, Intezar M, Otte-Höller I, et al. Optimized tumour infiltrating lymphocyte assessment for triple negative breast cancer prognostics. Breast 56, 78–87 (2021). [DOI] [PMC free article] [PubMed]
- 17.Lever J, Krzywinski M, Altman N. Model selection and overfitting. Nat Methods 13, 703–704 (2016).
- 18.Strathern M. “Improving ratings”: Audit in the British university system. Eur Rev 5, 305–321 (1997).
- 19.Geirhos R, Jacobsen J-H, Michaelis C, Zemel R, Brendel W, Bethge M, et al. Shortcut learning in deep neural networks. Nat Mach Intell 2, 665–673 (2020).
- 20.Schmitt M, Maron RC, Hekler A, Stenzinger A, Hauschild A, Weichenthal M, et al. Hidden variables in deep learning digital pathology and their potential to cause batch effects: Prediction model study. J Med Internet Res 23, e23436 (2021). [DOI] [PMC free article] [PubMed]
- 21.Wallis D, Buvat I. Clever Hans effect found in a widely used brain tumour MRI dataset. Med Image Anal 77, 102368 (2022). [DOI] [PubMed]
- 22.Oakden-Rayner L, Dunnmon J, Carneiro G, Re C. Hidden stratification causes clinically meaningful failures in machine learning for medical imaging. In Proceedings of the ACM conference on health, inference, and learning 151–159 (2020). [DOI] [PMC free article] [PubMed]
- 23.Nagpal K, Foote D, Liu Y, Chen P-HC, Wulczyn E, Tan F, et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. npj Digit Med 2, (2019). [DOI] [PMC free article] [PubMed]
- 24.Tang H, Sun N, Shen S. Improving generalization of deep learning models for diagnostic pathology by increasing variability in training data: Experiments on osteosarcoma subtypes. J Pathol Inform 12, 30 (2021). [DOI] [PMC free article] [PubMed]
- 25.Vali-Betts E, Krause KJ, Dubrovsky A, Olson K, Graff JP, Mitra A, et al. Effects of image quantity and image source variation on machine learning histology differential diagnosis models. J Pathol Inform 12, 5 (2021). [DOI] [PMC free article] [PubMed]
- 26.Tellez D, Litjens G, Bándi P, Bulten W, Bokhorst J-M, Ciompi F, et al. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med Image Anal 58, 101544 (2019). [DOI] [PubMed]
- 27.Anghel A, Stanisavljevic M, Andani S, Papandreou N, Rüschoff JH, Wild P, et al. A high-performance system for robust stain normalization of whole-slide images in histopathology. Front Med 6, (2019). [DOI] [PMC free article] [PubMed]
- 28.Marée R. The need for careful data collection for pattern recognition in digital pathology. J Pathol Inform 8, 19 (2017). [DOI] [PMC free article] [PubMed]
- 29.Howard FM, Dolezal J, Kochanny S, Schulte J, Chen H, Heij L, et al. The impact of site-specific digital histology signatures on deep learning model accuracy and bias. Nat Commun 12, (2021). [DOI] [PMC free article] [PubMed]
- 30.Oala L, Fehr J, Gilli L, Balachandran P, Leite AW, Calderon-Ramirez S, et al. ML4H auditing: From paper to practice. In Proceedings of the machine learning for health NeurIPS workshop vol. 136 280–317 (2020).
- 31.Maleki F, Muthukrishnan N, Ovens K, Reinhold C, Forghani R. Machine learning algorithm validation. Neuroimaging Clin N Am 30, 433–445 (2020). [DOI] [PubMed]
- 32.Cabitza F, Campagner A, Soares F, García de Guadiana-Romualdo L, Challa F, Sulejmani A, et al. The importance of being external. Methodological insights for the external validation of machine learning models in medicine. Comput Methods Programs Biomed 208, 106288 (2021). [DOI] [PubMed]
- 33.Park SH, Choi J, Byeon J-S. Key principles of clinical validation, device approval, and insurance coverage decisions of artificial intelligence. Korean J Radiol 22, 442 (2021). [DOI] [PMC free article] [PubMed]
- 34.Hond AAH de, Leeuwenberg AM, Hooft L, Kant IMJ, Nijman SWJ, Os HJA van, et al. Guidelines and quality criteria for artificial intelligence-based prediction models in healthcare: A scoping review. npj Digit Med 5, (2022). [DOI] [PMC free article] [PubMed]
- 35.Hufnagl P. EMPAIA – Ökosystem zur Nutzung von KI in der Pathologie. Pathologe 42, 135–141 (2021). [DOI] [PubMed]
- 36.Ramón y Cajal S, Sesé M, Capdevila C, Aasen T, De Mattos-Arruda L, Diaz-Cano SJ, et al. Clinical implications of intratumor heterogeneity: Challenges and opportunities. J Mol Med 98, 161–177 (2020). [DOI] [PMC free article] [PubMed]
- 37.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 15, 81–94 (2017). [DOI] [PubMed]
- 38.Chen Y, Zee J, Smith A, Jayapandian C, Hodgin J, Howell D, et al. Assessment of a computerized quantitative quality control tool for whole slide images of kidney biopsies. J Pathol 253, 268–278 (2021). [DOI] [PMC free article] [PubMed]
- 39.Focke CM, Bürger H, Diest PJ van, Finsterbusch K, Gläser D, Korsching E, et al. Interlaboratory variability of Ki67 staining in breast cancer. Eur J Cancer 84, 219–227 (2017). [DOI] [PubMed]
- 40.Schömig-Markiefka B, Pryalukhin A, Hulla W, Bychkov A, Fukuoka J, Madabhushi A, et al. Quality control stress test for deep learning-based diagnostic model in digital pathology. Mod Pathol 34, 2098–2108 (2021). [DOI] [PMC free article] [PubMed]
- 41.Allison KH, Reisch LM, Carney PA, Weaver DL, Schnitt SJ, O’Malley FP, et al. Understanding diagnostic variability in breast pathology: Lessons learned from an expert consensus review panel. Histopathology 65, 240–251 (2014). [DOI] [PMC free article] [PubMed]
- 42.El-Badry AM, Breitenstein S, Jochum W, Washington K, Paradis V, Rubbia-Brandt L, et al. Assessment of hepatic steatosis by expert pathologists. Ann Surg 250, 691–697 (2009). [DOI] [PubMed]
- 43.Martinez AE, Lin L, Dunphy CH. Grading of follicular lymphoma: Comparison of routine histology with immunohistochemistry. Arch Path Lab Med 131, 1084–1088 (2007). [DOI] [PubMed]
- 44.Kujan O, Khattab A, Oliver RJ, Roberts SA, Thakker N, Sloan P. Why oral histopathology suffers inter-observer variability on grading oral epithelial dysplasia: An attempt to understand the sources of variation. Oral Oncol 43, 224–231 (2007). [DOI] [PubMed]
- 45.Boiesen P, Bendahl P-O, Anagnostaki L, Domanski H, Holm E, Idvall I, et al. Histologic grading in breast cancer: Reproducibility between seven pathologic departments. Acta Oncol 39, 41–45 (2000). [DOI] [PubMed]
- 46.Oni L, Beresford MW, Witte D, Chatzitolios A, Sebire N, Abulaban K, et al. Inter-observer variability of the histological classification of lupus glomerulonephritis in children. Lupus 26, 1205–1211 (2017). [DOI] [PubMed]
- 47.Furness PN, Taub N, Assmann KJM, Banfi G, Cosyns J-P, Dorman AM, et al. International variation in histologic grading is large, and persistent feedback does not improve reproducibility. Am J Surg Pathol 27, 805–810 (2003). [DOI] [PubMed]
- 48.Tizhoosh HR, Diamandis P, Campbell CJV, Safarpoor A, Kalra S, Maleki D, et al. Searching images for consensus. Am J Pathol 191, 1702–1708 (2021). [DOI] [PubMed]
- 49.Homeyer A, Nasr P, Engel C, Kechagias S, Lundberg P, Ekstedt M, et al. Automated quantification of steatosis: Agreement with stereological point counting. Diagn Pathol 12, (2017). [DOI] [PMC free article] [PubMed]
- 50.Perincheri S, Levi AW, Celli R, Gershkovich P, Rimm D, Morrow JS, et al. An independent assessment of an artificial intelligence system for prostate cancer detection shows strong diagnostic accuracy. Mod Pathol 34, 1588–1595 (2021). [DOI] [PMC free article] [PubMed]
- 51.Silva LM da, Pereira EM, Salles PGO, Godrich R, Ceballos R, Kunz JD, et al. Independent real-world application of a clinical-grade automated prostate cancer detection system. J Pathol 254, 147–158 (2021). [DOI] [PMC free article] [PubMed]
- 52.Ianni JD, Soans RE, Sankarapandian S, Chamarthi RV, Ayyagari D, Olsen TG, et al. Tailored for real-world: A whole slide image classification system validated on uncurated multi-site data emulating the prospective pathology workload. Sci Rep 10, (2020). [DOI] [PMC free article] [PubMed]
- 53.Talari K, Goyal M. Retrospective studies – utility and caveats. J R Coll Physicians Edinb 50, 398–402 (2020). [DOI] [PubMed]
- 54.Gianfrancesco MA, Tamang S, Yazdany J, Schmajuk G. Potential biases in machine learning algorithms using electronic health record data. JAMA Intern Med 178, 1544 (2018). [DOI] [PMC free article] [PubMed]
- 55.Gamper J, Koohbanani NA, Benes K, Graham S, Jahanifar M, Khurram SA, et al. PanNuke dataset extension, insights and baselines. (Preprint arXiv:2003.10778 [q-bio.QM]). (2020).
- 56.Graham S, Jahanifar M, Azam A, Nimir M, Tsang Y-W, Dodd K, et al. Lizard: A large-scale dataset for colonic nuclear instance segmentation and classification. (Preprint arXiv:2108.11195 [cs.LG]). (2021).
- 57.Freeman K, Geppert J, Stinton C, Todkill D, Johnson S, Clarke A, et al. Use of artificial intelligence for image analysis in breast cancer screening programmes: Systematic review of test accuracy. BMJ 374, n1872 (2021). [DOI] [PMC free article] [PubMed]
- 58.Janowczyk A, Zuo R, Gilmore H, Feldman M, Madabhushi A. HistoQC: An open-source quality control tool for digital pathology slides. JCO Clin Cancer Inform 3, 1–7 (2019). [DOI] [PMC free article] [PubMed]
- 59.Ameisen D, Deroulers C, Perrier V, Bouhidel F, Battistella M, Legrès L, et al. Towards better digital pathology workflows: Programming libraries for high-speed sharpness assessment of whole slide images. Diagn Pathol 9, (2014). [DOI] [PMC free article] [PubMed]
- 60.Senaras C, Niazi MKK, Lozanski G, Gurcan MN. DeepFocus: Detection of out-of-focus regions in whole slide digital images using deep learning. PLOS ONE 13, e0205387 (2018). [DOI] [PMC free article] [PubMed]
- 61.Avanaki ARN, Espig KS, Xthona A, Lanciault C, Kimpe TRL. Automatic image quality assessment for digital pathology. In Breast imaging 431–438 (Springer International Publishing, 2016).
- 62.Smit G, Ciompi F, Cigéhn M, Bodén A, Laak J van der, Mercan C. Quality control of whole-slide images through multi-class semantic segmentation of artifacts. (2021).
- 63.Stacke K, Eilertsen G, Unger J, Lundstrom C. Measuring domain shift for deep learning in histopathology. IEEE J Biomed Health Inform 25, 325–336 (2021). [DOI] [PubMed]
- 64.Bozorgtabar B, Vray G, Mahapatra D, Thiran J-P. SOoD: Self-supervised out-of-distribution detection under domain shift for multi-class colorectal cancer tissue types. In 2021 IEEE/CVF international conference on computer vision workshops (ICCVW) 3317–3326 (IEEE, 2021).
- 65.Linmans J, Laak J van der, Litjens G. Efficient out-of-distribution detection in digital pathology using multi-head convolutional neural networks. In Proceedings of the third conference on medical imaging with deep learning MIDL 2020 vol. 121 465–478 (PMLR, 2020).
- 66.Guha Roy A, Ren J, Azizi S, Loh A, Natarajan V, Mustafa B, et al. Does your dermatology classifier know what it doesn’t know? Detecting the long-tail of unseen conditions. Med Image Anal 75, 102274 (2022). [DOI] [PubMed]
- 67.Çallı E, Murphy K, Sogancioglu E, Ginneken B van. FRODO: Free rejection of out-of-distribution samples: Application to chest X-ray analysis. (Preprint arXiv:1907.01253 [cs.LG]). (2019).
- 68.Cao T, Huang C-W, Hui DY-T, Cohen JP. A benchmark of medical out of distribution detection. (Preprint arXiv:2007.04250 [stat.ML]). (2020).
- 69.Berger C, Paschali M, Glocker B, Kamnitsas K. Confidence-based out-of-distribution detection: A comparative study and analysis. (Preprint arXiv:2107.02568 [cs.CV]). (2021).
- 70.Zhang O, Delbrouck J-B, Rubin DL. Out of distribution detection for medical images. In Uncertainty for safe utilization of machine learning in medical imaging, and perinatal imaging, placental and preterm image analysis 102–111 (Springer International Publishing, 2021).
- 71.Wang NC, Kaplan J, Lee J, Hodgin J, Udager A, Rao A. Stress testing pathology models with generated artifacts. J Pathol Inform 12, 54 (2021). [DOI] [PMC free article] [PubMed]
- 72.Sinha A, Ayush K, Song J, Uzkent B, Jin H, Ermon S. Negative data augmentation. (Preprint arXiv:2102.05113 [cs.AI]). (2021).
- 73.Lehmussola A, Ruusuvuori P, Selinummi J, Huttunen H, Yli-Harja O. Computational framework for simulating fluorescence microscope images with cell populations. IEEE Trans Med Imaging 26, 1010–1016 (2007). [DOI] [PubMed]
- 74.Ulman V, Svoboda D, Nykter M, Kozubek M, Ruusuvuori P. Virtual cell imaging: A review on simulation methods employed in image cytometry. Cytometry A 89, 1057–1072 (2016). [DOI] [PubMed]
- 75.Gadermayr M, Gupta L, Appel V, Boor P, Klinkhammer BM, Merhof D. Generative adversarial networks for facilitating stain-independent supervised and unsupervised segmentation: A study on kidney histology. IEEE Trans Med Imaging 38, 2293–2302 (2019). [DOI] [PubMed]
- 76.Moghadam AZ, Azarnoush H, Seyyedsalehi SA, Havaei M. Stain transfer using generative adversarial networks and disentangled features. Comput Biol Med 142, 105219 (2022). [DOI] [PubMed]
- 77.Niazi MKK, Abas FS, Senaras C, Pennell M, Sahiner B, Chen W, et al. Nuclear IHC enumeration: A digital phantom to evaluate the performance of automated algorithms in digital pathology. PLOS ONE 13, e0196547 (2018). [DOI] [PMC free article] [PubMed]
- 78.Levine AB, Peng J, Farnell D, Nursey M, Wang Y, Naso JR, et al. Synthesis of diagnostic quality cancer pathology images by generative adversarial networks. J Pathol 252, 178–188 (2020). [DOI] [PubMed]
- 79.Quiros AC, Murray-Smith R, Yuan K. PathologyGAN: Learning deep representations of cancer tissue. (Preprint arXiv:1907.02644 [stat.ML]). (2019).
- 80.Jose L, Liu S, Russo C, Nadort A, Ieva AD. Generative adversarial networks in digital pathology and histopathological image processing: A review. J Pathol Inform 12, 43 (2021). [DOI] [PMC free article] [PubMed]
- 81.Deshpande S, Minhas F, Graham S, Rajpoot N. SAFRON: Stitching across the frontier network for generating colorectal cancer histology images. Med Image Anal 77, 102337 (2022). [DOI] [PubMed]
- 82.Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J Pathol Inform 7, 29 (2016). [DOI] [PMC free article] [PubMed]
- 83.Adcock CJ. Sample size determination: A review. J R Stat Soc Ser D 46, 261–283 (1997).
- 84.Pepe MS. The statistical evaluation of medical tests for classification and prediction. (Oxford University Press, 2004).
- 85.Flahault A, Cadilhac M, Thomas G. Sample size calculation should be performed for design accuracy in diagnostic test studies. J Clin Epidemiol 58, 859–862 (2005). [DOI] [PubMed]
- 86.Bland JM. The tyranny of power: Is there a better way to calculate sample size? BMJ 339, b3985 (2009). [DOI] [PubMed]
- 87.Hazra A. Using the confidence interval confidently. J Thorac Dis 9, 4124–4129 (2017). [DOI] [PMC free article] [PubMed]
- 88.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143, 29–36 (1982). [DOI] [PubMed]
- 89.Simel DL, Samsa GP, Matchar DB. Likelihood ratios with confidence: Sample size estimation for diagnostic test studies. J Clin Epidemiol 44, 763–770 (1991). [DOI] [PubMed]
- 90.Kelley K, Maxwell SE, Rausch JR. Obtaining power or obtaining precision. Eval Health Prof 26, 258–287 (2003). [DOI] [PubMed]
- 91.Riley RD, Debray TPA, Collins GS, Archer L, Ensor J, Smeden M, et al. Minimum sample size for external validation of a clinical prediction model with a binary outcome. Stat Med 40, 4230–4251 (2021). [DOI] [PubMed]
- 92.Pavlou M, Qu C, Omar RZ, Seaman SR, Steyerberg EW, White IR, et al. Estimation of required sample size for external validation of risk models for binary outcomes. Stat Methods Med Res 30, 2187–2206 (2021). [DOI] [PMC free article] [PubMed]
- 93.Haynes A, Lenz A, Stalder O, Limacher A. Presize: An R-package for precision-based sample size calculation in clinical research. J Open Source Softw 6, 3118 (2021).
- 94.Echle A, Laleh NG, Quirke P, Grabsch HI, Muti HS, Saldanha OL, et al. Artificial intelligence for detection of microsatellite instability in colorectal cancer—a multicentric analysis of a pre-screening tool for clinical application. ESMO Open 7, 100400 (2022). [DOI] [PMC free article] [PubMed]
- 95.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin 71, 209–249 (2021). [DOI] [PubMed]
- 96.Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLOS ONE 10, e0118432 (2015). [DOI] [PMC free article] [PubMed]
- 97.Qi M, Cahan O, Foreman MA, Gruen DM, Das AK, Bennett KP. Quantifying representativeness in randomized clinical trials using machine learning fairness metrics. JAMIA Open 4, (2021). [DOI] [PMC free article] [PubMed]
- 98.Cabitza F, Campagner A, Sconfienza LM. As if sand were stone. New concepts and metrics to probe the ground on which to build trustable AI. BMC Med Inform Decis Mak 20, (2020). [DOI] [PMC free article] [PubMed]
- 99.Saleiro P, Kuester B, Hinkson L, London J, Stevens A, Anisfeld A, et al. Aequitas: A bias and fairness audit toolkit. (Preprint arXiv:1811.05577 [cs.LG]). (2018).
- 100.Bellamy RKE, Dey K, Hind M, Hoffman SC, Houde S, Kannan K, et al. AI Fairness 360: An extensible toolkit for detecting, understanding, and mitigating unwanted algorithmic bias. (Preprint arXiv:1810.01943 [cs.AI]). (2018).
- 101.Lee MSA, Singh J. The landscape and gaps in open source fairness toolkits. In Proceedings of the 2021 CHI conference on human factors in computing systems 1–13 (2021).
- 102.Roohi A, Faust K, Djuric U, Diamandis P. Unsupervised machine learning in pathology. Surg Pathol Clin 13, 349–358 (2020). [DOI] [PubMed]
- 103.Model I, Shamir L. Comparison of data set bias in object recognition benchmarks. IEEE Access 3, 1953–1962 (2015).
- 104.Shamir L. Evaluation of face datasets as tools for assessing the performance of face recognition methods. Int J Comput Vision 79, 225–230 (2008). [DOI] [PMC free article] [PubMed]
- 105.Bussola N, Marcolini A, Maggio V, Jurman G, Furlanello C. AI slipping on tiles: Data leakage in digital pathology. In Pattern recognition. ICPR international workshops and challenges 167–182 (Springer International Publishing, 2021).
- 106.Wu E, Wu K, Daneshjou R, Ouyang D, Ho DE, Zou J. How medical AI devices are evaluated: Limitations and recommendations from an analysis of FDA approvals. Nat Med 27, 582–584 (2021). [DOI] [PubMed]
- 107.König IR, Malley JD, Weimar C, Diener H-C, and AZ. Practical experiences on the necessity of external validation. Stat Med 26, 5499–5511 (2007). [DOI] [PubMed]
- 108.Celi LA, Cellini J, Charpignon M-L, Dee EC, Dernoncourt F, Eber R, et al. Sources of bias in artificial intelligence that perpetuate healthcare disparities—a global review. PLOS Digit Health 1, e0000022 (2022). [DOI] [PMC free article] [PubMed]
- 109.ITU-T Focus Group on AI for Health. DEL05.4: Training and test data specification. (2020).
- 110.ITU-T Focus Group on AI for Health. DEL05.1: Data requirements. (2020).
- 111.Medical Device Coordination Group. Report MDCG 2022-2: Guidance on general principles of clinical evidence for in vitro diagnostic medical devices (IVDs). (2022).
- 112.Moons KGM, Altman DG, Reitsma JB, Ioannidis JPA, Macaskill P, Steyerberg EW, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): Explanation and elaboration. Ann Intern Med 162, W1–W73 (2015). [DOI] [PubMed]
- 113.Liu X, Rivera SC, Moher D, Calvert MJ, Denniston AK. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: The CONSORT-AI extension. BMJ 370, m3164 (2020). [DOI] [PMC free article] [PubMed]
- 114.Norgeot B, Quer G, Beaulieu-Jones BK, Torkamani A, Dias R, Gianfrancesco M, et al. Minimum information about clinical artificial intelligence modeling: The MI-CLAIM checklist. Nat Med 26, 1320–1324 (2020). [DOI] [PMC free article] [PubMed]
- 115.Wiegand T, Krishnamurthy R, Kuglitsch M, Lee N, Pujari S, Salathé M, et al. WHO and ITU establish benchmarking process for artificial intelligence in health. Lancet 394, 9–11 (2019). [DOI] [PubMed]
- 116.Wenzel M, Wiegand T. Toward global validation standards for health AI. IEEE Commun Stand Mag 4, 64–69 (2020).
- 117.Sounderajah V, Ashrafian H, Golub RM, Shetty S, De Fauw J, Hooft L, et al. Developing a reporting guideline for artificial intelligence-centred diagnostic test accuracy studies: The STARD-AI protocol. BMJ Open 11, e047709 (2021). [DOI] [PMC free article] [PubMed]
- 118.Collins GS, Dhiman P, Navarro CLA, Ma J, Hooft L, Reitsma JB, et al. Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ Open 11, e048008 (2021). [DOI] [PMC free article] [PubMed]
- 119.Stevens LM, Mortazavi BJ, Deo RC, Curtis L, Kao DP. Recommendations for reporting machine learning analyses in clinical research. Circ Cardiovasc Qual Outcomes 13, (2020). [DOI] [PMC free article] [PubMed]
- 120.Leach P, Mealling M, Salz R. A Universally Unique IDentifier (UUID) URN namespace (2005).
- 121.Herrmann MD, Clunie DA, Fedorov A, Doyle SW, Pieper S, Klepeis V, et al. Implementing the DICOM standard for digital pathology. J Pathol Inform 9, 37 (2018). [DOI] [PMC free article] [PubMed]
- 122.Goldberg IG, Allan C, Burel J-M, Creager D, Falconi A, Hochheiser H, et al. The open microscopy environment (OME) data model and XML file: Open tools for informatics and quantitative analysis in biological imaging. Genome Biol 6, R47 (2005). [DOI] [PMC free article] [PubMed]
- 123.Homeyer A, Lotz J, Schwen L, Weiss N, Romberg D, Höfener H, et al. Artificial intelligence in pathology: From prototype to product. J Pathol Inform 12, 13 (2021). [DOI] [PMC free article] [PubMed]
- 124.European Commission. Regulation (EU) 2017/746 of the European Parilament and of the Council of 5 April 2017 on in vitro diagnostic medical devices and repealing Directive 98/79/EC and Commission Decision 2010/227/EU. (2017).
- 125.European Commission. Proposal for a regulation of the European parliament and of the council laying down harmonised rules on artificial intelligence (artificial intelligence act) and amending certain union legislative acts. (2021).
- 126.Code of Federal Regulations, Title 21, Chapter I, Subchapter H, Part 809 – in vitro diagnostic products for human use. (2021).
- 127.U.S. Food & Drug Administration. FDA authorizes software that can help identify prostate cancer. (2021).
- 128.U.S. Food & Drug Administration. DEN200080.Letter.DENG.pdf. (2021).
- 129.Kaushal A, Altman R, Langlotz C. Geographic distribution of US cohorts used to train deep learning algorithms. JAMA 324, 1212 (2020). [DOI] [PMC free article] [PubMed]
- 130.Bulten W, Kartasalo K, Chen P-HC, Ström P, Pinckaers H, Nagpal K, et al. Artificial intelligence for diagnosis and Gleason grading of prostate cancer: The PANDA challenge. Nat Med 28, 154–163 (2022). [DOI] [PMC free article] [PubMed]
- 131.Dudgeon SN, Wen S, Hanna MG, Gupta R, Amgad M, Sheth M, et al. A pathologist-annotated dataset for validating artificial intelligence: A project description and pilot study. J Pathol Inform 12, 45 (2021). [DOI] [PMC free article] [PubMed]
- 132.Rodrigues R. Legal and human rights issues of AI: Gaps, challenges and vulnerabilities. J Responsib Technol 4, 100005 (2020).
- 133.Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G, King D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med 17, (2019). [DOI] [PMC free article] [PubMed]
- 134.Evans T, Retzlaff CO, Geißler C, Kargl M, Plass M, Müller H, et al. The explainability paradox: Challenges for xAI in digital pathology. Future Gener Comput Syst 133, 281–296 (2022).
- 135.Finlayson SG, Subbaswamy A, Singh K, Bowers J, Kupke A, Zittrain J, et al. The clinician and dataset shift in artificial intelligence. N Engl J Med 385, 283–286 (2021). [DOI] [PMC free article] [PubMed]
- 136.Taqi S, Sami S, Sami L, Zaki S. A review of artifacts in histopathology. J Oral Maxillofac Surg Med Pathol 22, 279 (2018). [DOI] [PMC free article] [PubMed]
- 137.Chatterjee S. Artefacts in histopathology. J Oral Maxillofac Surg Med Pathol 18, 111 (2014). [DOI] [PMC free article] [PubMed]
- 138.Pursnani D, Arora S, Katyayani P, C A, Yelikar BR. Inking in surgical pathology: Does the method matter? A procedural analysis of a spectrum of colours. Turk Patoloji Derg (2016). [DOI] [PubMed]