Graphical abstract
Keywords: Biomedical engineering, Convolution neural networks (CNN), Ensemble deep learning, Chronic Obstructive Pulmonary Diseases (COPD), COVID-19, Diagnosis & Classification, Transfer learning, Medical Imaging
Abstract
Background and Objective
In the current COVID-19 outbreak, efficient testing of COVID-19 individuals has proven vital to limiting and arresting the disease's accelerated spread globally. It has been observed that the severity and mortality ratio of COVID-19 affected patients is at greater risk because of chronic pulmonary diseases. This study looks at radiographic examinations exploiting chest X-ray images (CXI), which have become one of the utmost feasible assessment approaches for pulmonary disorders, including COVID-19. Deep Learning(DL) remains an excellent image classification method and framework; research has been conducted to predict pulmonary diseases with COVID-19 instances by developing DL classifiers with nine class CXI. However, a few claim to have strong prediction results; because of noisy and small data, their recommended DL strategies may suffer from significant deviation and generality failures.
Methods
Therefore, a unique CNN model(PulDi-COVID) for detecting nine diseases (atelectasis, bacterial-pneumonia, cardiomegaly, covid19, effusion, infiltration, no-finding, pneumothorax, viral-Pneumonia) using CXI has been proposed using the SSE algorithm. Several transfer-learning models: VGG16, ResNet50, VGG19, DenseNet201, MobileNetV2, NASNetMobile, ResNet152V2, DenseNet169 are trained on CXI of chronic lung diseases and COVID-19 instances. Given that the proposed thirteen SSE ensemble models solved DL's constraints by making predictions with different classifiers rather than a single, we present PulDi-COVID, an ensemble DL model that combines DL with ensemble learning. The PulDi-COVID framework is created by incorporating various snapshots of DL models, which have spearheaded chronic lung diseases with COVID-19 cases identification process with a deep neural network produced CXI by applying a suggested SSE method. That is familiar with the idea of various DL perceptions on different classes.
Results
PulDi-COVID findings were compared to thirteen existing studies for nine-class classification using COVID-19. Test results reveal that PulDi-COVID offers impressive outcomes for chronic diseases with COVID-19 identification with a 99.70% accuracy, 98.68% precision, 98.67% recall, 98.67% F1 score, lowest 12 CXIs zero-one loss, 99.24% AUC-ROC score, and lowest 1.33% error rate. Overall test results are superior to the existing Convolutional Neural Network(CNN). To the best of our knowledge, the observed results for nine-class classification are significantly superior to the state-of-the-art approaches employed for COVID-19 detection. Furthermore, the CXI that we used to assess our algorithm is one of the larger datasets for COVID detection with pulmonary diseases.
Conclusion
The empirical findings of our suggested approach PulDi-COVID show that it outperforms previously developed methods. The suggested SSE method with PulDi-COVID can effectively fulfill the COVID-19 speedy detection needs with different lung diseases for physicians to minimize patient severity and mortality.
1. Introduction
An unexpected death and debilitating infection worldwide, are caused by SARS-CoV-2, which patients have detected with COVID-19. One-to-one COVID-19 transmission occurs quickly among two persons in intimate exposure via plumes or minute droplets produced by talk, coughing, and sneezing. After becoming infectious, patients commonly experience these signs: flu, coughing, shortness of breath, smell, and taste [1]. As of 07 January 2022, there are still more than 298,915,721 infected cases, 5,469,303 people have died, and 9,118,223,397 vaccine doses have been administered worldwide [5].
In general, the RT-PCR test is utilized to diagnose COVID-19. It can detect infectious RNA in a nasopharyngeal swab [15]. It necessitates specialized materials and kits that are not readily available. It requires at least 8–12 h, which is inconvenient because COVID-19 + Ve patients should be detected and followed as soon as feasible. Some investigations discovered that RT-PCR findings from many tests performed on the same individuals at different stages throughout the illness were varied, resulting in a significant FNR [16]. As a result, it may misclassify COVID-19 patients as uninfected. The Author proposed combining the RT-PCR test with additional clinical procedures such as radiography imaging [50].
In parallel to RT-PCR, medical imaging assessment is a powerful clinical method for identifying COVID-19 quickly. Clinicians study and evaluate CXI and CT scans to assess whether or not a suspectable subject has been infected with SARS-CoV-2 [27], [26]. Subsequent investigations have shown atypical abnormalities in radiographic imaging of COVID-19 patients, and it was frequently employed during the early stages of the global pandemic [11], [14], [24].
Although CXI is extremely efficient, it contains a professional radiologist to physically judge for COVID19 case detection, which is not a time-saving method that loads medical professional skills. However, the proportion of radiologists is far smaller than that of individuals undergoing monitoring. An AI-aided diagnosing tool is required to help the radiologist in detecting COVID19 occurrences in a much quicker, more immediate, and accurate manner. Otherwise, infected persons may not be recognized and isolated as fast as practicable and may not undergo appropriate treatments [25], [26].
Agreeing to American Lung Association [34], Lancet report [35], and Geng et al. [24], the severity and mortality rates are increased due to chronic pulmonary diseases in COVID-19-confirmed patients[23]. So, we consider this as a challenge and opportunity for further research. Moreover, the studies [36], [39], [41], [42], [43] reported higher performance, whereas their used COVID-19 samples size are small. So, to urge this need and minimize the mortality rates because of pulmonary diseases with COVID-19 in this work, we expanded dl-based automatic detection of pulmonary disease with COVID-19 (PulDi-COVID) using CXI for nine class classifications. Moreover, we also investigate how to use eight CNN for identifying pulmonary disease with COVID-19 cases with thirteen probable selective snapshots stacked ensemble (SSE) models. Our outcomes highlight the need to utilize proper computing tools to analyze COVID-19 concerns and assist associated decision-making. The following precise research questions (RQ) motivated our study:
RQ1) How important chronic pulmonary diseases can affect the COVID-19 diagnosis? Are mortality rates increasing due to the existence of chronic pulmonary disorders along with COVID-19 diagnosis?
RQ2) How can we obtain the highest performance with nine-class classification using CNN?
RQ3) What is the comparative performance of various DL algorithms for pulmonary disease classification with COVID-19 cases detection, and which classification algorithm performs better?
RQ4) Is there any common automated tool for detecting maximum diseases with the assistance of clinicians and radiologists?
We focused on searching chronic pulmonary diseases with COVID-19–related datasets to address the above questions to achieve multi-class classification from online public repositories. Specifically, we used pulmonary diseases with COVID-19-related datasets from NIH, Kaggle, GitHub, and dl-based networks to model various issues related to COVID-19 from the radiographic domain. This paper's primary contributions are as follows:
-
1)
We used the eight most popular transfer learning CNN models based on state-of-the-art DL architectures.
-
2)
Applied nine-class classification using chronic lung diseases with COVID-19 cases detection.
-
3)
Proposed an SSE strategy with the awareness of varied class-level accuracies for different DL models. SSE models achieve superior performance by minimizing the variance of prediction errors to the competing base learners.
-
4)
Evaluated individual DL models and proposed PulDi-COVID experimentally, showing the promising results of PulDi-COVID. The main aim of SSE is to reduce the error rate and enhance accuracy.
-
5)
To evaluate the performance of this framework, three publicly available datasets of CXI have been used, which are both more widely available tests to perform, and more sensitive to COVID-19. The obtained results outperform the existing methods by a significant margin.
-
6)
This research will be helpful for clinicians and radiologists to minimize the workload, severity, and deaths of COVID-19 patients because the mortality rate may increase as chronic lung diseases present in COVID-19 affected individuals.
Our experiment results highlight the necessity of detecting chronic pulmonary disorders with COVID-19 as well as appropriate simulation tools for understanding COVID-19 concerns and guiding associated decision-making. The following is the general layout of the article. First, we'll give a quick overview of online healthcare forums. Section 2 discusses COVID-19-related concerns as well as some comparative studies. Section 3 describes the preprocessing data methods used in our research, and the DL approaches for detecting chronic pulmonary illness with COVID-19. Section 4 then describes the results. Lastly, sections 5 and 6 explore future studies and draw conclusions based on DL techniques for evaluating the COVID-19 radiology society. The set of acronyms used is shown in Table 1 .
Table 1.
List of abbreviations.
| Sr. No. | Abbreviation | Full Form |
|---|---|---|
| 1. | AI | Artificial Intelligence. |
| 2. | AUC | Area under the ROC Curve. |
| 3. | BM | Base-Model. |
| 4. | CAD | Computer-Aided Diagnostic. |
| 5. | CNN | Convolution Neural Network. |
| 7. | CXI | Chest-X-ray Images. |
| 8. | CL | Convolution Layer. |
| 9. | CT | Computed Tomography. |
| 10. | DL | Deep Learning. |
| 11. | COVID-19 | Coronavirus-Disease-2019. |
| 12. | FC | Fully Connected Layer. |
| 13. | FN | False Negative. |
| 14. | FNR | False Negative Rate. |
| 15. | FP | False Positive. |
| 16. | FPR | False-Positive-Rate. |
| 17. | HPC | High-Performance-Computing. |
| 18. | ML | Machine Learning. |
| 19. | MM | Meta-Model. |
| 20. | MSE | Mean Squared Error. |
| 21. | PulDi-COVID | Pulmonary Diseases with COVID-19. |
| 22. | SSE | Selective-snapshots Stacked Ensemble. |
| 23. | TL | Transfer Learning. |
| 24. | ResNet | residual neural network. |
| 25. | ReLU | Rectified Linear Unit. |
| 26. | RT-PCR | Reverse Transcription–Polymerase Chain Reaction. |
| 26. | RNN | Recurrent Neural Networks. |
| 27. | ROC | Receiver Operating Characteristic. |
| 28. | ROC | Receiver Operating Characteristic. |
| 29. | VGG | Visual Geometry Group. |
| 30. | TN | True Negative. |
| 31. | TP | True Positive. |
| 32. | TPR | True-Positive-Rate. |
| 33. | VGG | Visual Geometry Group. |
2. Background and related work
For a clearer insight into the PulDi-COVID model, we describe the ensemble DL and CNN for pulmonary illnesses with COVID-19 instance identification.
2.1. Ensemble Deep learning
Being a robust ML approach, DL is extensively used in various disciplines, including computer vision, voice translation, healthcare image assessment, pharmaceutical research, and so on [7], [8]. This includes a multi-layer neural network capable of extracting top-level characteristics from primary input, including images, and producing predictive outcomes depending on those characteristics. A DL modeling algorithm is divided into two steps: training and inference. Training is a random and repeated calculation that generates a model depending on training inputs. Different parameters must be initialized during the training process, particularly learning rate, epoch number, and batch size, with alternative setups resulting in models with varying accuracies. Inference involves the procedure of making predictions using the learned DL model. Other prominent DL approaches are accessible, including CNN and RNN. In contrast, CNN is widely used, and it is helpful for image identification, classification, and healthcare image analysis [6].
However, because of outliers in the dataset and unpredictability in the DL method, it is prone to high variation and generalization error [10]. Despite specific strategies, like data augmentation and regularisation [9], the difficulties with DL models are still not effectively tackled.
Ensemble learning is a viable strategy for overcoming these issues for multiple ML models [11]. This offers a hybrid learning framework capable of producing highly accurate and resilient predicting outcomes than a single model by intelligently merging several ML models. There are other ensemble algorithms available, such as averaging[11], random forest[12], boosting[13], and stacking[14]. To render the ensemble victorious, we should ensure that the simulations included in the combination are varied. We investigate ensemble DL for pulmonary illnesses using COVID-19 case identification in the suggested project by integrating different DL models using an ensemble technique.
2.2. CNN-Related work
The radiography imaging community makes significant contributions to medical data processing and studies, which eventually aids in advancing health technologies. Various AI-aided diagnosis techniques relying on DL have already been suggested to minimize the load of diagnosis from CT and CXI for radiologists [26], [28], [29], [30], [31], [32]. CXI has become a prominent and widely used source of information for COVID-19 early diagnosis because of various advantages of CXI, particularly mobility, affordability, ease of access, and quick screening compared to CT.
It has been found that the majority of authors implemented binary [38], [39], [42], [45] and three [36], [37], [41], [43], [44], [48] and multi [40], [46] class classification with COVID instances. Wang et al. [41] combined ResNet-101 and ResNet-152 models to classify COVID-19 from pneumonia and normal CXI. Developed fusion system attained 96.1 % accuracy. However, used COVID-19 samples are small (i.e., 128). Narin et al. [42] compared CNN variants trained on CXI (ResNet50, Inception V3, and Inception-ResNetV2) for COVID-19 identification, finding that ResNet-50 leads the other two approaches with 98 % accuracy. Chowdhury et al. [12] conducted a comparison of various DL networks(AlexNet, ResNet18, DenseNet201, SqueezeNet) to binary classification (COVID-19, normal) for CXI, indicating that SqueezeNet outpaces with 98.3 % accuracy. Rahimzadeh et al. [37] made a chain of Xception + ResNet50V2 to classify 80 CXI of COVID-19, 6054 CXI of pneumonia, and 8851 CXI of normal instances and were able to achieve a 91.4 % accuracy. Alqudah et al.[38] developed AOCT-NET for binary classification (Covid19, normal) with a 95.2 % accuracy. Hemdan et al.[39] proposed COVIDX-Net(DenseNet201, Inceptionv3, VGG19, MobileNetv2, Xception, InceptionResNetv2 and ResNetv2) using 25 cases/class for COVID-19, no infections, attained F1-score of 0.89 % for normal and 91 % for COVID-19. Mishra et al. [45] developed binary classification of DL model (CovAI-Net) using CNN variants (Inception, DenseNet, Xception). CoviAI-Net attained 100 % precision and specificity for ‘COVID + Ve’ class.
Asnaoui et al. [40] experimented comparative study of DL variants (IncpetionResNetV2, VGG16/19, DenseNet201, InceptionResNetV2, InceptionV3, Resnet50, MobileNetV2) using CXI of 2780 for bacterial, 1493 for viral, 231 of Covid19, and 1583 normal instances. IncpetionResNetV2 outperformed with 92.18 % accuracy, 96.06 % specificity, 92.38 % precision, and 92.07 % F1-score. Another study by Ucar and Korkmaz [43] implemented SqueezeNet-Bayes for Covid19(76), normal(1583), and pneumonia(4290) classification with an accuracy of 98.30 %. DarkCovidNet is CNN based DL model created by Ozturk et al. [44] for Covid19(1 2 5), normal(5 0 0), and pneumonia(5 0 0) classification with an accuracy of 87.02 %. Tang et al. [47] developed an ensemble dl-based(EDL-COVID) model for the classification of COVID(5 7 3) instances from pneumonia(6053) and normal(8851) from CXI. EDL-COVID attained 95 % accuracy and 96 % sensitivity. Zhou et al. [48] developed an ensemble dl-based model (EDL_COVID) to classify Covid19, lung tumors, and normal cases from CT scans. EDL_COVID utilized 2500 samples per class for experimentation. They achieved 99.05 % accuracy, 99.6 % specificity, and 98.59 % F1-score.
3. Materials and methods
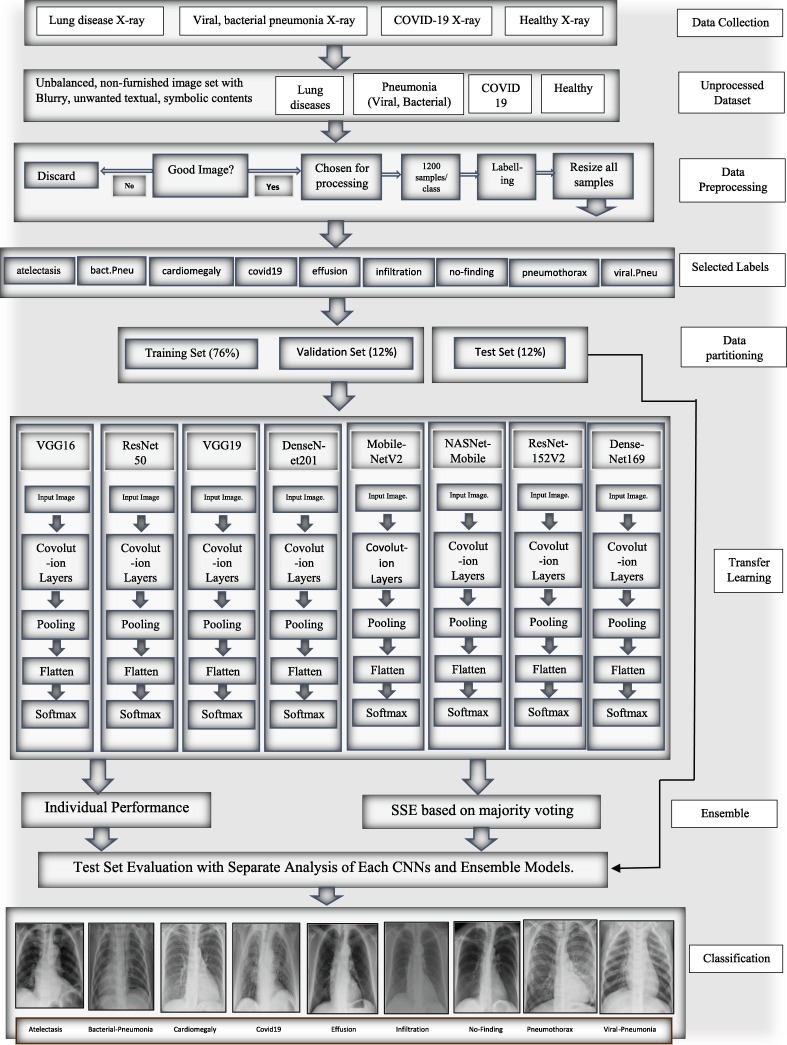
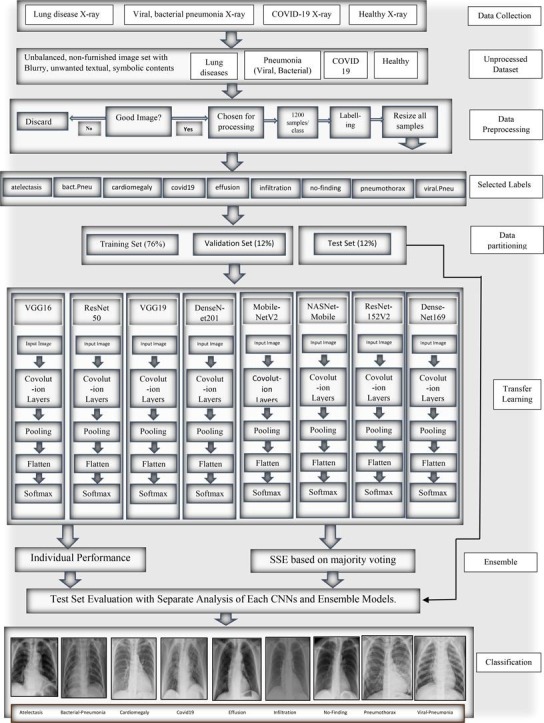
This section discusses the datasets, preprocessing, and the proposed PulDi-COVID, an SSE algorithm depending on eight cutting-edge DL architectures. The entire flowchart for PulDi-COVID, as shown in Fig. 1 , comprises two stages: snapshot DL models and ensembling. The training step of DL models is responsible for creating several snapshots, which are subsequently integrated for a meta-learner prediction in the SSE algorithm of the ensemble step (Section 3.3). The PulDi-COVID application source code is described in Appendix A.
Fig. 1.
Flow chart of proposed PulDi-Covid framework.
3.1. Dataset
Until related pulmonary disorders are involved, measuring the efficiency of any classification method in identifying COVID-19 infection is critical. As a result, the entire dataset includes a subset of CXI associated with various lung illnesses with COVID-19 and healthy cases. The online public repository of NIH ChestX-ray8 [2] dataset initially contains fourteen class labels of 112,120 CXI from 30,805 unique patients with chronic lung diseases. Viral and bacterial pneumonia samples are collected from the Kaggle dataset [26]. At the same time, COVID-19 instances are collected from the Kaggle COVID-19 detection challenge [27]. Furthermore, the collected dataset was undergone data preprocessing, which is discussed in the next section.
3.2. Pre-processing
For experimentation purposes, we choose six disorders from fourteen ChestX-ray8 [2] labels: atelectasis, cardiomegaly, effusion, infiltration, no-finding/healthy, and pneumothorax. The composed Kaggle dataset contains viral and bacterial pneumonia samples [3]. Simultaneously, COVID-19 cases from the COVID-19 detection challenge [4] are collected. The assembled dataset of CXI from three repositories contains noisy samples, which further need to preprocess. The noisy and blurry CXI has been discarded manually from the collected datasets. To avoid data imbalance, each class is retained with an equal number of samples. The assembled dataset has been partitioned to the 76 %:12 %:12 % ratios for train, val, and test sets. For nine-class detection, we used 10,800 observations from three repositories integrations. For each labeled class in training, 1000 CXIs are evaluated, 100 CXIs for each validation class, and 100 CXIs for each test set class. The pixel data of the given samples were normalized within 0 and 1 for normalization. The CXI used in the sets of data under examination were gray, and the rescale is done by converting 1./255 to adjacent pixels. The train set is augmented online with 'imagedatagenerator,' which increases the collection and adds robustness to the neural model, minimizing the likelihood of overfitting concerns. Shear-range to 0.2, zoom-range to 0.2 are augmentation approaches utilized for cardiomegaly and bacterial pneumonia. Equal sample sizes are allocated for each class after augmentation to avoid data imbalance. This contributes to the creation of the network and improves test imaging performance. Table 2 shows the label data distribution for each class. The major issue was with the database itself. We have programmatically selected those samples whose image id contains a single label instead of multi-labels. Our preferred database faced class imbalance issues, especially for certain categories such as ‘cardiomegaly’. There were no other large diverse databases suitable for real-time CAD implementable framework development.
Table 2.
Multisource dataset used after preprocessing.
| Source | ChestX-ray8[2] |
Kaggle Pneumonia[3] | Covid19[4] | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Labels | Criteria | Atelectasis | Cardiomegaly | Effusion | Infiltration | Healthy | Pneumothorax | Viral | Bacterial | Covid-19 |
| Sample Size | Actual | 4215 | 1093 | 3955 | 9547 | 60,361 | 2194 | 2780 | 1493 | 6054 |
| Eliminated | 774 | 351 | 557 | 1346 | 4731 | 832 | 586 | 434 | 1893 | |
| Remaining | 3441 | 742 | 3398 | 8201 | 55,630 | 1362 | 2194 | 1059 | 4161 | |
| Balanced class | Real | 1100 | 742 | 1100 | 1100 | 1100 | 1100 | 1100 | 1059 | 1100 |
| Augmented | 0 | 358 | 0 | 0 | 0 | 0 | 0 | 41 | 0 | |
The creators of certain openly available data sets increase the dataset by augmenting it. Several photos in these databases may be duplicated. If we split those photographs in train, val, and test sets, the pictures in the tests and val-sets likely replicate. Assume a photograph is additionally augmented into 5 photos, three of them remain inside the train pool whereas the remaining percentage is divided among val and test sets. Performance may be misleading. Although it already previously recognizes the significant pictures in the training phase, the classifier can readily identify items. During this case, however, the simulation might collapse if evaluated using real-world photos. To deal with such a problem & verify that there is zero information leaks throughout network train and assessment, we employed a sourced dataset [2], [26], [27] in which verified there are no redundant CXR images. Furthermore, the dataset has been meticulously separated into train, val, and final testing/model assessment data. To prevent database leaks, we initially divided the dataset and augmented just the train set. To check the duplicate we used images translation to numeric data and compare each numeric value. Another technique using MSE, where sum of the squared difference between the two images should be lower to check similar images.
3.3. Proposed methodology
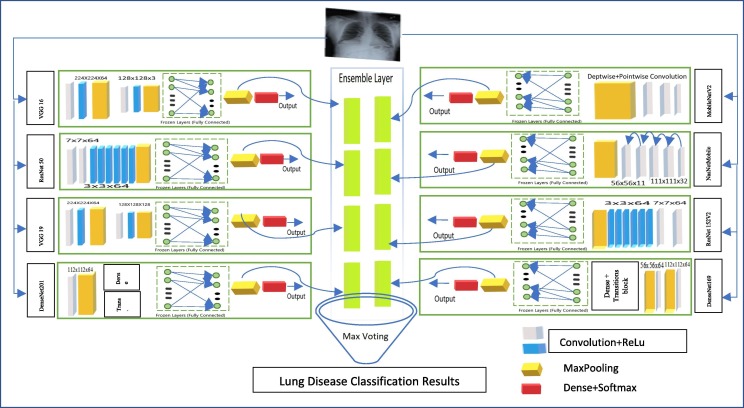
Our framework includes a 2-phase transfer learning (TL): a training approach and an ensemble technique. We employed eight TL-CNNs for feature extraction in the first phase: VGG16[39], ResNet50[26], VGG19[39], DenseNet201[12], MobileNetV2, NASNetMobile, ResNet152V2[41], and DenseNet169 [51]. Custom build layers replace the classifier and fully connected layers. TL is the process of adopting the weights of a pretrained network and applying recently learned characteristics to decide on a unique class name. In TL, a model that has been pretrained on the 'imagenet' is employed, and this model has learned to detect top-level image features in the early layers [53]. A dense layer was appended to the CNN structure for TL. The model then determines which feature groupings will aid in identifying features in new data gathering. In this kind of case, the usage of pretrained networks based on the notion of TL might be beneficial. In TL, information obtained by a network trained on a massive dataset is utilized to tackle a similar problem using a lesser dataset. It assists in eliminating the requirement for an enormous dataset and a considerably more extended period, which are needed by DL algorithms that are learned from scratch [22], [23], [24]. Fig. 1 depicts the recommended architecture's block diagram.
VGG16 or VGG19 consisting of 16 and 19 convolutional layers. It’s a standard CNN [52] architecture with multiple channels. The numbers “16″ and ”19″ represent the networks weight levels(i.e. CL). VGG19 thus has “3″ extra convolutional layers over VGG16. The image size of 224x224x3 set to first input layer. ReLU is employed in all of the VGG channel's hidden tiers. First CL set to 224x224 with kernel size 64. Followed by 128x128 with kernel size of 3. The remaining layers from VGG16/19 are set to trainable = false. A dense layer attained with a size of 25088-neurons. ResNet50 and ResNet152V2 designed to solve the problem of the disappearing gradient. Residual block networks accepts the input size with 224x224 and 299x299 pixels. The skip connection between 7x7x64 and 3x3x64 allow the network to fit undermapping. If any layer hurt the performance of architecture then it will be skipped by regularization. The filter size set to 64 and kernel set to 3 for the second layer. The remaining layers froze and global pooling and dense and softmax(9) for last layer. Form Fig. 2 MobileNet uses depthwise(DW) + pairwise(PW) separable-CL. In comparison to the structure with ordinary convolutions to the equal depths inside the networks, it greatly decreases the range of parameters. As a consequence, DNNs are compact. Filter parameters for DW and PW are 32 and PW 64 with a stride of 3. After that, we use a 1x1 filter to cover the depth and point dimension. The last flatten + dense contains 62,720 neurons for 9-class classification. DenseNet performs channelwise concatenations instead of passing individual layer results to next layer. It uses concatenated denseBlock + transitionBlock for each layer is receiving a “collective knowledge” from all earlier layers. The default input size of 224x224x3 set to first CL. First and second CLs are of size (112x112x64) and (56x56x64) followed by maxpooling. For both DenseNet169/201 initial 2 blocks are trained while remaining set to false for training. The last layer of flatten + dense deals with 94080-neurons for 9-disease classifications. The default input accepted by NasNetMobile with a size of 224x224x3. In CL(111x111x32) return a feature map of the same dimension. Whereas, reduction block(56x56x32) returns a feature map where the feature map height and width are reduced by a factor of two. The last flatten + dense contains 51,744 neurons for 9 class-classification.
Fig. 2.
Proposed PulDi-Covid SSE ensemble.
The following are the drawbacks of utilizing a single model and the benefits of adopting multi-modal fusion in disease categorization. However a mono-shape permutation operation can decrease the number of dimensions and increase the model's operating acceleration, but this couldn't assure adequate mining of features, particularly in complicated environments, where crucial data is easily lost as well as the feature impact of the final disease detection is reduced. The aforesaid challenges can be efficiently solved using multi-scale information retrieved by different classifiers of convolutional units. However, the previous transmission of every convolutional network could simply verify that the features are transmitted to the outer filter and can't be fused with the model's shallower information. As a consequence, the illness-detection CXIs have inadequate explanations. Our multi-branch merger achieves pattern extraction using several parallel convolutional filters and emphasizes meaningful data using a feature fusing that achieves feature interactions without expanding the model's complexities. In comparison to serial fusing, parallel fusing across convolutions can take advantage of differing convolution kernel sizes, ensuring the complete retrieval of both higher and lower frequencies features and making the integration layered feature include additional important data. Fusion block contains 8 layers(individual feature-vectors), construction of stacked layers, and final fusion. To create a fusion layer in phase 2, we have collected all the 8 models features(8*[9-lables*64]) individually to make stacking ensemble; finally, all feature-vectors are embedded in stacked constructed layer(Fig. 2). This requires additional improvement in the extraction of desirable feature’s as well as the correlation of various aspects from BMs to MMs.
We used the SSE technique with thirteen probable ensembles(i.e., base models-BM are preferred based on minimum error rate) for experimentation in the second phase. The possibility of generating ensembles from eight BMs is not only chosen thirteen but also 2n-1 possible ensembles. So, we try ensemble best BMs without skipping. After producing several DL networks & their weights from the main step, now we proceed to the proposed assembly step for developing PulDi-COVID by stacking various models, as shown in Fig. 1. Several ensembling ways exist for model ensemble, as detailed in section 2.1. Stacking is a prominent ensembling approach for snapshot ensemble learning [20]. It finds the maximum class probabilities from all models for each class for an input image. Let M denote the total classifiers (eight DL models). Let pm, x(si) is the class likelihood of xth class output by mth classifier concerning the source image si. Then the greatest predicated class predicted is for × ∈ [1, X] of the source image si, with X indicating the total classes. The conventional ensembling technique (stacking, bagging, boosting, and averaging) implies that almost all classifiers have equal weights. Furthermore, three introductory remarks are made.
-
1)
Overall testing accuracy rates for individual classes in a DL model are often varied.
-
2)
Several DL models have varying levels of accuracy for every classification.
-
3)
It shows that we cannot treat every model equivalently throughout the model assembly process.
We developed an SSE technique for TL ensemble related to the above three findings, as shown in Algorithm-1. Let is the test accuracy of ith model for jth class for test CXI. The measured weights of ith model for jth class are represented as . For each source image si, we begin by obtaining the results of each classification probabilities from mth model for ∀m ∈ M. The classification probabilities may then be assessed as of PulDi-COVID by a maximum of the stacked class likelihood of all M for ∀x ∈ X. Furthermore, we obtain the classification results by reporting the class number with the highest-class probabilities for every input image. Overview of the proposed PulDi-COVID model is presented in algorithm 1.
Algorithm 1
Proposed dl-based SSE Algorithm for PulDi-COVID.
| a. | Parameter: [train, test, valid] = split(Dataset), a = argmax, m1 = model, t = train, v = valid, tt = test, b = batch, w = weights, em = ensemble-model, si = ith sample input of CXI dataset, X = class. |
| b. | Input: Dataset D = sum |
| c. |
Output: ensemble meta-classifier, c(si): predicted class index for ith image with PulDi-COVID. Model Training, Validation, testing: |
| 1. | Action 1: Base model classification |
| 2. | for c = [VGG16, ResNet50, VGG19, DenseNet201, MobileNetV2, NASNetMobile, ResNet152V2, |
| 3. | DenseNet169] |
| 4. | for j = 1:150 // epoch range from 1 to 150 |
| 5. | [t(j), v(j)] = partition(t, v) |
| 6. | for k=(t/b, v/b) // sample/batch |
| 7. | model(m1, c, j) = train(c(m1), a, t(j), v(j)) // training |
| 8. | valid(m1, c, j) = valid((c(m1), a, j), v(j)) // validation |
| 9. | end |
| 10. | end |
| 11. | tt(c(m1), a) = predict(max(test(c(m1), a, tt))) // prediction on test |
| 12. | c(si) = tt(c(m1), a) // predicted class label with index |
| 13. | end |
| 14. | acquireWeights = model(w) // |
| 15. | Action 2: Create SSE ensemble-classifier(meta-model) and load feature vectors. |
| 16. | for i = 1:8 |
| 17. | em = model(a). append(c(i))// input as base model structure to create meta-ensemble-learner |
| 18. | em(w) = loadWeights(acquireWeights(c[1…8])) |
| 19. | Action 3: Test ensemble classifier |
| 20. | tt(em, a) = predict(max(test(em, a, tt))) // estimate the class probability |
| 21. | = tt(em, a) |
| 22. | c(si) = // predicted specific class label with index |
| 23. | end; |
3.4. Stacking ensemble
A set of features considered a group instead of individually is an ensemble. An ensemble technique generates various models and then merges them to address a problem. Ensemble approaches to aid in improving the model's strength. Some of the ensemble techniques are averaging [11], max voting, GBM, XGBM, adaboost[33], stacking[14], blending[17], bagging[18], and boosting[13]. A stacked ensemble is an ML algorithm that practices a MM/classifier to merge several base-classifiers(BMs). Stacking learns the optimum way to aggregate predictions from several high-performing models. The BMs are trained on the whole dataset, followed by the MM, which is trained upon features returned (as output) by the BM. In stacking, the BMs are often distinct. The MM aids in the extraction of features from BM to attain the highest level of accuracy. The advantage of stacking is that it may combine the expertise of several high-performing models on a classification/regression job to create recommendations that outperform any one model in the ensemble [17]. It employs a meta-learning strategy to find the optimum way to aggregate predictions from two or more underlying ML techniques. In contrast to boosting, multiple models are utilized in stacking to learn how to merge the predicted results from the participating models. A stacking model's framework consists of two or more BM, also known as 0th step models, and a MM that integrates the predictions of the BM, known as a 1st step model.
Step-0 Models (Base-Models): Models fit the training data and collected predictions.
Step-1 Model (meta-Model): Model that learns how to best combine the predictions of the base models.
The MM is trained on the predictions made by BM on out-of-sample data. Data that was not utilized for training the BM is given to them; predictions are formed; these predictions, along with the expected outcomes, constitute the input and output sets of the training dataset required to fit the MM. In the case of regression, the results of the BM given as input to the MM may be actual values, and in the case of classification, they may be probability values and class names.
4. Experiments & results
PulDi-COVID uses CNN to retrieve features from CXI and COVID-19 to classify chronic pulmonary illnesses using an ensemble of eight distinct DL classifiers(based on the identical input size, i.e., 224x224). The TL models were trained for a total of 150 epochs. The stacking approach was used to modify the hyperparameters of the DL classifiers. Individual models and the proposed SSE classifier's performance for each class label were evaluated using the confusion matrix, ROC curve, precision, recall, F1-score, accuracy, error-rate, AUC-ROC-score, and zero-one loss and test time per input image.
4.1. Simulation Requirements.
The experiment was run-on HPC. The platform cast for the simulation experiment was an Intel-XeonE5-2630v3@2.40 GHz, 64.0 GB RAM, and a 48 TB hard disk with 16 child nodes and one central node. CentOS6, Python3.8, Tensorflow, Keras, Jupyter-notebook, matplotlib, numpy, and pandas are employed for conducting tests. It provides an optimized runtime for DL research and high-end computation access to a dependable GPU.
4.2. Experimental setup
Experiments have been carried out in two-phase TL training and an SSE scheme. In the first stage, the individual DL models were tested using CXI datasets from the CXR-lung-disease [2], viral and bacterial pneumonia dataset [3], and COVID-19 dataset [4]. The input CXI were first set to 224 × 224 utilizing VGG16, ResNet50, VGG19, DenseNet201, MobileNetV2, NASNetMobile, ResNet152V2, and DenseNet169. From Table 3 , training for each network with target epochs is set to 150. Based on early-stoppage and callback to escape overfitting and constant performance [52] (if there is no improvement at the training and validation phase). The parameters applied to early stopping and callback are provided in Table 3. Initial ‘l_r’ assigned as ‘0.0001′. The epoch/iteration size will be decided automatically. Training operation of every DCNNs will be stopped robotically based on Table 3 criteria. The 'adam' optimizer was applied for training, and the ‘l_r’ was well-ordered internally. Whereas 64 batch size for train and val set and 32 for the test set. Lastly, 9-class expectation outcomes for each dl-BMs from the 'softmax' layer (Table 4 ). With the help of early stopping. Eventually, individual DL models experimental performance accumulated.
Table 3.
Early-stopping & callback hyperparamters.
| Sr. No. | Parameters | Value |
|---|---|---|
| 1 | Input Size | 224x224 |
| 2 | Target epoch | 150 |
| 3 | Patience. | 10 |
| 4 | Mindelta | 0.0001 |
| 5 | Verbose | 1 |
| 6 | Learning rate | 0.0001 |
| 7 | Mode | “auto” |
| 8 | Monitor | “val_loss” |
| 9 | Baseline | “none” |
| 10 | min_lr | 0.000001 |
| 11 | Restoreweights | “true” |
| 12 | Batch_size | 64 |
| 13 | Optimizer | ‘adam’ |
| 14 | Dynamic_l_r | ‘auto’ |
Table 4.
Obtained architectural details and results of individual BM-CNNs.
| Model | Trainable parameters | Training Time(Hrs) | Macro avg. Precision | Macro avg. Recall | Macro avg. F1-Score | Macro avg. Accuracy | Zero-one Loss (Out of 900 test samples) | Error Rate (%) |
|---|---|---|---|---|---|---|---|---|
| VGG16 | 225,801 | 18.54 | 77.10 | 77.11 | 76.71 | 94.91 | 206 | 22.88 |
| ResNet50 | 903,177 | 26.10 | 44.72 | 44.44 | 41.31 | 87.65 | 500 | 55.55 |
| VGG19 | 225,801 | 19.27 | 66.98 | 66.78 | 66.10 | 92.61 | 299 | 33.22 |
| DenseNet201 | 846,729 | 31.42 | 88.11 | 86.44 | 85.92 | 96.98 | 122 | 13.55 |
| MobileNetV2 | 564,489 | 25.27 | 92.15 | 92.00 | 91.97 | 98.22 | 72 | 8 |
| NASNetMobile | 465,705 | 23.36 | 65.92 | 57.33 | 52.89 | 90.51 | 384 | 42.66 |
| ResNet152V2 | 903,177 | 28.53 | 93.46 | 93.22 | 93.15 | 98.49 | 61 | 6.77 |
| DenseNet169 | 733,833 | 24.49 | 89.66 | 89.00 | 88.92 | 97.55 | 99 | 11 |
Note: Bold values are optimal results.
In the second phase, we have composed the proposed stacking ensemble model (i.e., PulDi-COVID) to evaluate nine-class classification. The possible thirteen SSE models are prepared using received weights from the base learners. Ensemble models benefit from aggregating relevant data from multiple classification techniques to create a highly reliable model. Variance and bias are also minimized, resulting in a more minor anticipated error. Furthermore, a feature vector block that was wrongly learned by classification can still be successfully categorized by leveraging the pattern acquired by other classifiers, which the ensemble model exploits. Because of these properties, ensemble models are an excellent choice for tackling difficult classification and regression tasks [21]. All received base-learners weights files are united based on two to eight individual TL models to create a selective stacked ensemble model. Then we estimated the class probability of PulDi-COVID by the maximum of the stacked class probabilities of all models. Next, we identified the class by providing the class id with the highest-class possibility for every input image. We have executed these networks using TensorFlow and Keras.
4.3. Performance metrics
The parameters employed in this article to evaluate the performance of PulDi-COVID were accuracy(Acc.), precision(Pre.), recall(Rec.), specificity(Spe.), F1score, zero-one loss(z), and error rate(e). Criteria for assessment were obtained from the confusion matrix concerning the CNNs' classification task as follows: (a) Positive instances that were properly recognized (TP), (b) Negative cases that were wrongly categorized (FN), (c) Negative cases that were correctly identified (TN), and (d) Positive cases that were misclassified (FP).
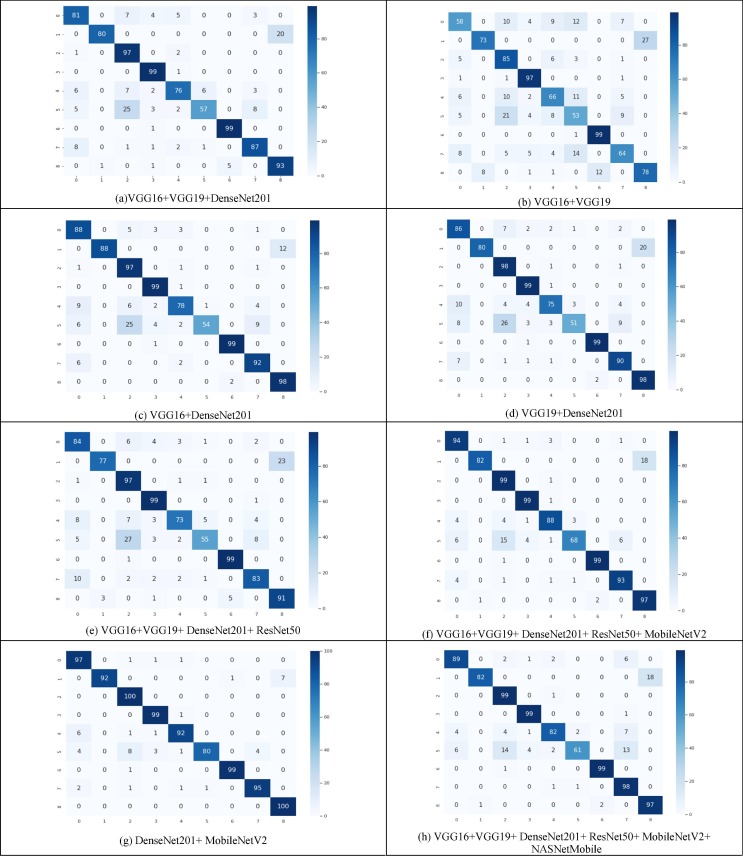
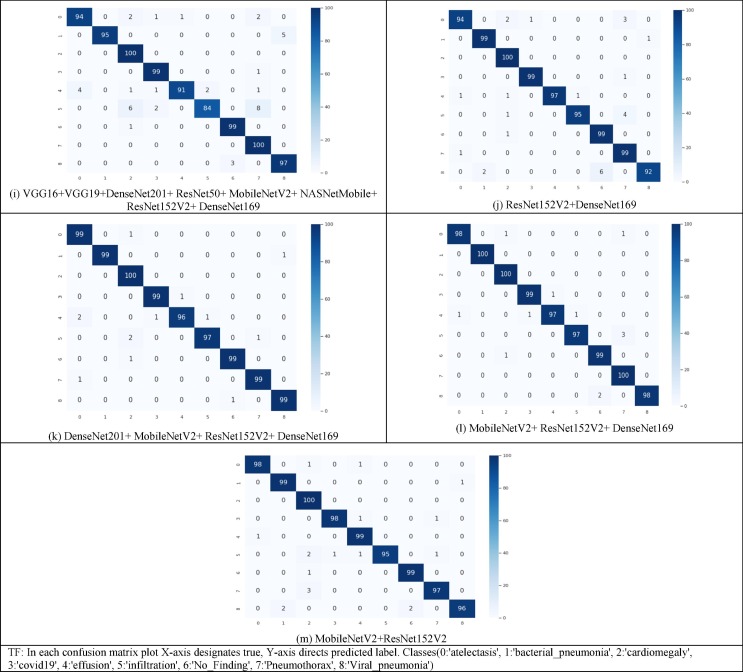
Fig. 3 depicts the confusion matrix of all thirteen SSE ensembled models. The formulae used to calculate the values of these metrics are listed below:
-
1.
Acc. = (TP + TN)/(TP + FP + TN + FN)
-
2.
Pre. = TP/(TP + FP)
-
3.
Rec. = TP/(TP + FN)
-
4.
Spe. = TN/(FP + TN)
-
5.
F1-score = 2 × (Pre. × Rec.)/(Pre. + Rec.)
-
6.
e =// sum (not-equal (pred, true)) / sum(all-classes(total-test-samples))
-
7.
z(i, j)=
Fig. 3.
Confusion matrix attained for all SSE models at the test time.
4.4. Results
Table 4, Table 5, Table 6 summarize the comprehensive classification outcomes achieved across all models regarding various metrics. The assessment methods mentioned in Table 4, Table 5, Table 6 were the more commonly used to measure classification efficiency. The performance for all individual DL networks has been shown in Table 4; we processed the PulDi-COVID performance for the separate classes in Table 5. The process is repeated for thirteen probable ensemble models. It can be observed that the ResNet152 attained the maximum performance followed by MobileNetV2 from individual CNNs (Table 4). ResNet152V2 reached the highest accuracy of 98.49 %, the precision of 93.46 %, recall of 93.22 %, F1-score of 93.15 %, zero-one loss of 61, and the lowest error rate of 6.77 % among 8 CNNs. From Table 5, chronic pulmonary diseases with nine-class classification the highest attained accuracy of 99.77 % for cardiomegaly, and pneumothorax class, 99.66 % for atelectasis and effusion class, 99.55 % for infiltration class by proposed ensemble model; for atelectasis, cardiomegaly, effusion, infiltration, and pneumothorax highest attained precision of 98.98 %, 98.03 %, 100 %, 100 % and 99 % respectively; and recall of 99 %, 100 %, 99 %, 97 %, and 100 % respectively; specificity of 99.87 %, 99.75 %, 100 %, 100 %, and 99.87 % respectively; F1-score of 98.49 %, 99.01 %, 98.47 %, 97.98 %, and 99 % respectively; and AUC of 99.31 %, 99.87 %, 99.31 %, 98.43 %, and 99.75 % respectively. However, for the comparison of pneumonia and COVID-19, the bacterial pneumonia class attained the uppermost 100 % performance for all metrics (this class may lead to overfit). However, the accuracy of 99.77 %, precision of 99 %, recall of 99 %, specificity of 99.87 %, F1-score of 99 %, and AUC of 99.43 % were attained by Covid19 and no-finding classes. For viral pneumonia class, achieved an accuracy of 99.77 %, precision of 100 %, recall of 100 %, specificity of 100 %, F1-score of 99 %, and AUC of 99.56 %.
Table 5.
PulDi-COVID MM-classification performance assessment based on each class label.
| Ensemble Model | Metrics | Atelecta-sis | Bacterial pneumonia | Cardio-megaly | Covid19 | Effusion | Infiltrat-ion | No-Finding | Pneumo-thorax | Viral Pneumonia |
|---|---|---|---|---|---|---|---|---|---|---|
| VGG16 + VGG19 + DenseNet201 | Accuracy (%) | 95.66 | 97.66 | 95.22 | 98.55 | 96 | 94.44 | 99.33 | 97 | 97 |
| Precision (%) | 80.19 | 98.76 | 70.8 | 89.18 | 86.36 | 89.06 | 95.19 | 86.13 | 82.3 | |
| Recall (%) | 81 | 80 | 97 | 99 | 76 | 57 | 99 | 87 | 93 | |
| Specificity (%) | 97.5 | 99.87 | 95 | 98.5 | 98.5 | 99.12 | 99.37 | 98.25 | 97.5 | |
| F1-score (%) | 80.59 | 88.39 | 81.85 | 93.83 | 80.85 | 69.51 | 97.05 | 86.56 | 87.32 | |
| AUC (%) | 89.25 | 89.93 | 96 | 98.75 | 87.25 | 78.06 | 99.18 | 92.62 | 95.25 | |
| VGG16 + VGG19 | Accuracy (%) | 92.55 | 96.11 | 93.11 | 97.88 | 93.11 | 90.22 | 98.55 | 93.44 | 94.55 |
| Precision (%) | 69.87 | 90.12 | 64.39 | 85.84 | 70.21 | 56.38 | 89.18 | 73.56 | 74.28 | |
| Recall (%) | 58 | 73 | 85 | 97 | 66 | 53 | 99 | 64 | 78 | |
| Specificity (%) | 96.87 | 99 | 94.12 | 98 | 96.5 | 94.87 | 98.5 | 97.12 | 96.62 | |
| F1-score (%) | 63.38 | 80.66 | 73.27 | 91.08 | 68.04 | 54.63 | 93.83 | 68.44 | 76.09 | |
| AUC (%) | 77.43 | 86 | 89.56 | 97.5 | 81.25 | 73.93 | 98.75 | 80.56 | 87.31 | |
| VGG16 + DenseNet201 | Accuracy (%) | 96.22 | 98.66 | 95.66 | 98.77 | 96.55 | 94.77 | 99.66 | 97.44 | 98.44 |
| Precision (%) | 80 | 100 | 72.93 | 90.82 | 89.65 | 98.18 | 98.01 | 85.98 | 89.09 | |
| Recall (%) | 88 | 88 | 97 | 99 | 78 | 54 | 99 | 92 | 98 | |
| Specificity (%) | 97.25 | 100 | 95.5 | 98.75 | 98.87 | 99.87 | 99.75 | 98.12 | 98.5 | |
| F1-score (%) | 83.81 | 93.61 | 83.26 | 94.73 | 83.42 | 69.67 | 98.5 | 88.88 | 93.33 | |
| AUC (%) | 92.62 | 94 | 96.25 | 98.87 | 88.43 | 76.93 | 99.37 | 95.06 | 98.25 | |
| VGG19 + DenseNet201 | Accuracy (%) | 95.66 | 97.77 | 95.55 | 98.66 | 96.33 | 94.11 | 99.66 | 97.11 | 97.55 |
| Precision (%) | 77.47 | 100 | 72.05 | 90 | 90.36 | 92.72 | 98.01 | 84.9 | 83.05 | |
| Recall (%) | 86 | 80 | 98 | 99 | 75 | 51 | 99 | 90 | 98 | |
| Specificity (%) | 96.87 | 100 | 95.25 | 98.62 | 99 | 99.5 | 99.75 | 98 | 97.5 | |
| F1-score (%) | 81.51 | 88.88 | 83.05 | 94.28 | 81.96 | 65.8 | 98.5 | 87.37 | 89.9 | |
| AUC (%) | 91.43 | 90 | 96.62 | 98.81 | 87 | 75.25 | 99.37 | 94 | 97.75 | |
| VGG16 + VGG19 + DenseNet201 + ResNet50 | Accuracy (%) | 95.55 | 97.11 | 94.88 | 98.44 | 96.11 | 94.11 | 99.33 | 96.44 | 96.44 |
| Precision (%) | 77.77 | 96.25 | 69.28 | 88.39 | 90.12 | 87.3 | 95.19 | 84.69 | 79.82 | |
| Recall (%) | 84 | 77 | 97 | 99 | 73 | 55 | 99 | 83 | 91 | |
| Specificity (%) | 97 | 99.62 | 94.62 | 98.37 | 99 | 99 | 99.37 | 98.12 | 97.12 | |
| F1-score (%) | 80.76 | 85.55 | 80.83 | 93.39 | 80.66 | 67.48 | 97.05 | 83.83 | 85.04 | |
| AUC (%) | 90.5 | 88.31 | 95.81 | 98.68 | 86 | 77 | 99.18 | 90.56 | 94.06 | |
| VGG16 + VGG19 + DenseNet201 + ResNet50 + MobileNetV2 | Accuracy (%) | 97.77 | 97.88 | 97.44 | 99.22 | 97.88 | 96 | 99.66 | 98.44 | 97.66 |
| Precision (%) | 87.03 | 98.79 | 81.81 | 94.28 | 92.63 | 94.44 | 98.01 | 93 | 84.34 | |
| Recall (%) | 94 | 82 | 99 | 99 | 88 | 68 | 99 | 93 | 97 | |
| Specificity (%) | 98.25 | 99.87 | 97.25 | 99.25 | 99.125 | 99.5 | 99.75 | 99.125 | 97.75 | |
| F1-score (%) | 90.38 | 89.61 | 89.59 | 96.58 | 90.25 | 79.07 | 98.5 | 93 | 90.23 | |
| AUC (%) | 96.12 | 90.93 | 98.12 | 99.12 | 93.56 | 83.75 | 99.37 | 96.06 | 97.37 | |
| DenseNet201 + MobileNetV2 | Accuracy (%) | 98.33 | 99.11 | 98.66 | 99.33 | 98.66 | 97.66 | 99.77 | 99 | 99.22 |
| Precision (%) | 88.99 | 100 | 89.28 | 95.19 | 95.83 | 98.76 | 99 | 95.95 | 93.45 | |
| Recall (%) | 97 | 92 | 100 | 99 | 92 | 80 | 99 | 95 | 100 | |
| Specificity (%) | 98.5 | 100 | 98.5 | 99.37 | 99.5 | 99.87 | 99.87 | 99.5 | 99.12 | |
| F1-score (%) | 92.82 | 95.83 | 94.34 | 97.05 | 93.87 | 88.39 | 99 | 95.47 | 96.61 | |
| AUC (%) | 97.75 | 96 | 99.25 | 99.18 | 95.75 | 89.93 | 99.43 | 97.25 | 99.56 | |
| VGG16 + VGG19 + DenseNet201 + ResNet50 + MobileNetV2 + NASNetMobile | Accuracy (%) | 97.66 | 97.88 | 97.55 | 99.22 | 97.33 | 95.33 | 99.66 | 96.77 | 97.66 |
| Precision (%) | 89.89 | 98.79 | 82.5 | 94.28 | 93.18 | 95.31 | 98.01 | 78.4 | 84.34 | |
| Recall (%) | 89 | 82 | 99 | 99 | 82 | 61 | 99 | 98 | 97 | |
| Specificity (%) | 98.75 | 99.87 | 97.37 | 99.25 | 99.25 | 99.62 | 99.75 | 96.62 | 97.75 | |
| F1-score (%) | 89.44 | 89.61 | 90 | 96.58 | 87.23 | 74.39 | 98.5 | 87.11 | 90.23 | |
| AUC (%) | 93.87 | 90.93 | 98.18 | 99.12 | 90.62 | 80.31 | 99.37 | 97.31 | 97.37 | |
| VGG16 + VGG19 + DenseNet201 + ResNet50 + MobileNetV2 + NASNetMobile + ResNet152V2 + DenseNet169 | Accuracy (%) | 98.88 | 99.44 | 98.88 | 99.44 | 98.88 | 98 | 99.55 | 98.66 | 99.11 |
| Precision (%) | 95.91 | 100 | 90.9 | 96.11 | 98.91 | 97.67 | 97.05 | 89.28 | 95.09 | |
| Recall (%) | 94 | 95 | 100 | 99 | 91 | 84 | 99 | 100 | 97 | |
| Specificity (%) | 99.5 | 100 | 98.75 | 99.5 | 99.87 | 99.75 | 99.62 | 98.5 | 99.37 | |
| F1-score (%) | 94.94 | 97.43 | 95.23 | 97.53 | 94.79 | 90.32 | 98.02 | 94.34 | 96.04 | |
| AUC (%) | 96.75 | 97.5 | 99.37 | 99.25 | 95.43 | 91.87 | 99.31 | 99.25 | 98.18 | |
| ResNet152V2 + DenseNet169 | Accuracy (%) | 99.11 | 99.66 | 99.44 | 99.77 | 99.66 | 99.33 | 99.22 | 99 | 99 |
| Precision (%) | 97.91 | 98.01 | 95.23 | 99 | 100 | 98.95 | 94.28 | 92.52 | 98.92 | |
| Recall (%) | 94 | 99 | 100 | 99 | 97 | 95 | 99 | 99 | 92 | |
| Specificity (%) | 99.75 | 99.75 | 99.37 | 99.87 | 100 | 99.87 | 99.25 | 99 | 99.87 | |
| F1-score (%) | 95.91 | 98.5 | 97.56 | 99 | 98.47 | 96.93 | 96.58 | 95.65 | 95.33 | |
| AUC (%) | 96.87 | 99.37 | 99.68 | 99.43 | 98.5 | 97.43 | 99.12 | 99 | 95.93 | |
| DenseNet201 + MobileNetV2 + ResNet152V2 + DenseNet169 | Accuracy (%) | 99.55 | 99.88 | 99.55 | 99.77 | 99.44 | 99.55 | 99.77 | 99.77 | 99.77 |
| Precision (%) | 97.05 | 100 | 96.15 | 99 | 98.96 | 98.97 | 99 | 99 | 99 | |
| Recall (%) | 99 | 99 | 100 | 99 | 96 | 97 | 99 | 99 | 99 | |
| Specificity (%) | 99.62 | 100 | 99.5 | 99.87 | 99.87 | 99.87 | 99.87 | 99.87 | 99.87 | |
| F1-score (%) | 98.02 | 99.49 | 98.03 | 99 | 97.46 | 97.98 | 99 | 99 | 99 | |
| AUC (%) | 99.31 | 99.5 | 99.75 | 99.43 | 97.93 | 98.43 | 99.43 | 99.43 | 99.43 | |
| MobileNetV2 + ResNet152V2 + DenseNet169 | Accuracy (%) | 99.66 | 100 | 99.77 | 99.77 | 99.55 | 99.55 | 99.66 | 99.55 | 99.77 |
| Precision (%) | 98.98 | 100 | 98.03 | 99 | 98.97 | 98.97 | 98.01 | 96.15 | 100 | |
| Recall (%) | 98 | 100 | 100 | 99 | 97 | 97 | 99 | 100 | 98 | |
| Specificity (%) | 99.87 | 100 | 99.75 | 99.87 | 99.87 | 99.87 | 99.75 | 99.5 | 100 | |
| F1-score (%) | 98.49 | 100 | 99.01 | 99 | 97.98 | 97.98 | 98.5 | 98.03 | 98.99 | |
| AUC (%) | 98.93 | 100 | 99.87 | 99.43 | 98.43 | 98.43 | 99.37 | 99.75 | 99 | |
| MobileNetV2 + ResNet152V2 | Accuracy (%) | 99.66 | 99.66 | 99.22 | 99.66 | 99.55 | 99.44 | 99.66 | 99.44 | 99.44 |
| Precision (%) | 98.98 | 98.01 | 93.45 | 98.98 | 97.05 | 100 | 98.01 | 97.97 | 98.96 | |
| Recall (%) | 98 | 99 | 100 | 98 | 99 | 95 | 99 | 97 | 96 | |
| Specificity (%) | 99.87 | 99.75 | 99.12 | 99.87 | 99.62 | 100 | 99.75 | 99.75 | 99.87 | |
| F1-score (%) | 98.49 | 98.5 | 96.61 | 98.49 | 98.02 | 97.43 | 98.5 | 97.48 | 97.46 | |
| AUC (%) | 98.93 | 99.37 | 99.56 | 98.93 | 99.31 | 97.5 | 99.37 | 98.37 | 97.93 |
Table 6.
Macro average(overall) results obtained on ensembling models.
| Ensemble Model | Macro avg. Precision (%) | Macro avg. Recall (%) | Macro avg. F1-score (%) | Macro avg. Accuracy (%) | Zero-one Loss (Out of 900 test samples) | AUC-ROC-Score (%) | Error Rate (%) | Test Time |
|---|---|---|---|---|---|---|---|---|
| VGG16 + VGG19 + DenseNet201 | 86.45 | 85.44 | 85.11 | 96.76 | 131 | 91.81 | 14.55 | 4.130 s |
| VGG16 + VGG19 | 74.87 | 74.78 | 74.39 | 94.39 | 227 | 85.81 | 25.22 | 0.569 s |
| VGG16 + DenseNet201 | 89.41 | 88.11 | 87.69 | 97.35 | 107 | 93.31 | 11.88 | 3.518 s |
| VGG19 + DenseNet201 | 87.62 | 86.22 | 85.70 | 96.93 | 124 | 92.24 | 13.77 | 3.458 s |
| VGG16 + VGG19 + DenseNet201 + ResNet50 | 85.43 | 84.22 | 83.85 | 96.49 | 142 | 91.12 | 15.77 | 4.560 s |
| VGG16 + VGG19 + DenseNet201 + ResNet50 + MobileNetV2 | 91.60 | 91.00 | 90.81 | 98 | 81 | 94.93 | 9 | 11.203 s |
| DenseNet201 + MobileNetV2 | 95.17 | 94.89 | 94.83 | 98.86 | 46 | 97.12 | 5.11 | 3.918 s |
| VGG16 + VGG19 + DenseNet201 + ResNet50 + MobileNetV2 + NASNetMobile | 90.53 | 89.56 | 89.24 | 97.67 | 94 | 94.12 | 10.44 | 9.923 s |
| VGG16 + VGG19 + DenseNet201 + ResNet50 + MobileNetV2 + NASNetMobile + ResNet152V2 + DenseNet169 | 95.66 | 95.44 | 95.41 | 98.98 | 41 | 97.43 | 4.55 | 18.254 s |
| ResNet152V2 + DenseNet169 | 97.21 | 97.11 | 97.11 | 99.35 | 26 | 98.37 | 2.88 | 5.400 s |
| DenseNet201 + MobileNetV2 + ResNet152V2 + DenseNet169 | 98.57 | 98.56 | 98.56 | 99.67 | 13 | 99.18 | 1.44 | 11.087 s |
| MobileNetV2 + ResNet152V2 + DenseNet169 | 98.68 | 98.67 | 98.67 | 99.70 | 12 | 99.24 | 1.33 | 8.590 s |
| MobileNetV2 + ResNet152V2 | 97.94 | 97.89 | 97.89 | 99.53 | 19 | 98.81 | 2.11 | 3.515 s |
| TF: Bold values are observed as optimal results. | ||||||||
Table 6 shows the overall(average) outcomes found for thirteen ensembling possibilities, comprising zero-one loss (all nine-class classification), AUCscore, e-rate, and testing time per CXI, etc. Among all thirteen ensembled, MobileNetV2 + ResNet152V2 + DenseNet169 achieved the overall highest outcomes. Proposed PulDi-COVID (MobileNetV2 + ResNet152V2 + DenseNet169) attained highest overall accuracy of 99.70 %, precision of 98.68 %, recall of 98.67 %, F1-score of 98.67 %, minimum zero-one loss to 12, and lowest error rate of 1.33 % among thirteen SSE. Even after ensembling the worst performance targeted by VGG16 + VGG19 with 227 samples of misclassification for nine class classifications. However, 94.39 % accuracy was attained by the VGG16 + VGG19 model but attained the lowest precision of 74.87 %, recall of 74.78 %, F1-score of 74.39 %, and the error rate of 25.22 %(which is the highest and it's not recommended). For testing individual images, 0.569 s were taken by VGG16 + VGG19 and 8.590 s taken by MobileNetV2 + ResNet152V2 + DenseNet169 models.
Ensembling of (VGG16 + VGG19 + DenseNet201), (VGG16 + VGG19), (VGG16 + DenseNet201), (VGG19 + DenseNet201), (VGG16 + VGG19 + DenseNet201 + ResNet50), (VGG16 + VGG19 + DenseNet201 + ResNet50 + MobileNetV2) are used to forecast the significant misclassified instances, as shown in Fig. 3. The misconception was most likely caused by the comparable imaging findings of the disease cases.
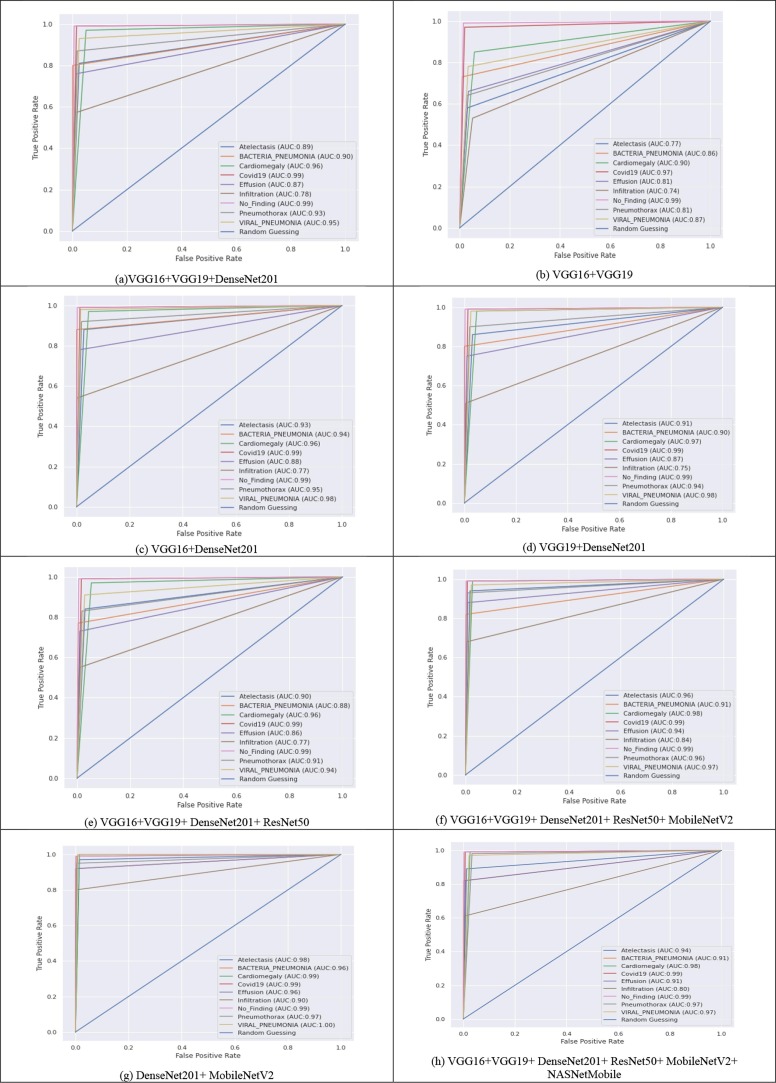
The ROC curves of all thirteen models are shown in Fig. 4 . ROC is a 2-D chart that compares the TPR as opposed to the FPRThe ROC curve illustrates the sensitivity and specificity. TPR on the y-axis and FPR on the x-axis are used to plot the ROCs. Higher AUC scores are important in medical diagnoses. As a result, its simulations in medical analytics aid data analysts in their diagnostic investigation predicting analysis. From Fig. 4, the highest ROCAUCscore of 99.24 % for all lung diseases attained by MobileNetV2 + ResNet152V2 + DenseNet169, followed by 99.18 % of DenseNet201 + MobileNetV2 + ResNet152V2 + DenseNet169.
Fig. 4.
ROC curves obtained for all ensembling models at the testing phase.
4.5. Comparing PulDi-COVID with cutting-edge systems
Table 7 compares the results of PulDi-COVID with several other current studies for the automated identification of COVID-19. The methods suggested in [37], [39], [40], [44], [46] obtained accuracy of 91.40 %, 90 %, 92.18 %, 87.02 %, and 80.60 %, respectively. However, their utilized CXI is small[37], [39], [42], [43], [46] and applied for three-class classification only. The systems shown in [36], [41], [43] have enhanced the accuracy to 99.4 %, 96.10 %, and 98.30 %, respectively. The methods suggested in [48] show a COVID-19 detection accuracy of 99.05 %. PulDi-COVID beat all of these classifiers in terms of accuracy. PulDi-COVID obtained 99.70 % accuracy, indicating that it could be a useful tool for early diagnosis and nine-class classification for lung disease detection using CXIs. However, due to the multiheaded ensembles, the MobileNetV2 + ResNet152V2 + DenseNet169 model takes 8.59 s to test individual CXI.
Table 7.
Performance comparison of PulDi-COVID classifier based on an already developed system.
| Reference | Model | Class with sample size | Performance | Test Time/ Image(S) |
|---|---|---|---|---|
| Zabirul [36] | CNN-LSTM | Covid19:613, Pneumonia:1252, Normal:1252. | Accuracy:99.4 %, AUC:99.9 %, Specificity:99.2 %, Sensitivity:99.3 %, F1-score:98.9 %. | 113 s |
| Rahimzadeh et al. [37] | Xception + ResNet50V2 | Covid19:80, Pneumonia:6054, Normal:8851. | Accuracy:91.4 %. | – |
| Alqudah et al. [38] | AOCT-NET | Covid19, Normal. | Accuracy:95.2 %. | 6.3 s |
| Hemdan et al. [39] | COVIDX-Net | Covid19:25, Normal:25. | Accuracy: 90.0 %. | 4.0 s |
| Asnaoui et al. [40] | IncpetionRes NetV2 | Bacterial: 2780, Virus:1493, Covid19:231, Normal:1583. | Accuracy: 92.18 %, Sensitivity:92.11 %, Specificity: 96.06 %, Precision:92.38 %, F1-score: 92.07 %. | 262 s |
| Wang et al. [41] | ResNet-101 + ResNet-152 | Covid19:140, Pneumonia:8620, Normal:7966. | Accuracy: 96.1 %. | – |
| Narin et al. [42] | ResNet-50 | Covid19:50, Normal:50. | Accuracy:98.00 %. | – |
| Ucar and Korkmaz [43] | SqueezeNet- Bayes | Covid19:76, Normal:1583, Pneumonia: 4290. | Accuracy: 98.30 %. | – |
| Ozturk et al. [44] | DarkCovidNet | Covid19:125, Normal:500, Pneumonia:500. | Accuracy:87.02 %. | – |
| Mishra et al. [45] | CovAI-Net | Covid + Ve:369, Covid -Ve:309. | Precision: 100 %, Sensitivity:96.74 %, Specificity:100 %. | – |
| Loey et al. [46] | GoogleNet | Covid19:69, Normal:79, Bacterial:79, virus:79. | Accuracy:80.6 %. | – |
| Tang et al. [47] | EDL-COVID | Covid19:573, Pneumonia:6053, Normal:8851. | Accuracy:95 %, Sensitivity:96 %, PPV:94.1 %. | 450 s/100 image Apx. |
| Zhou et al. [48] | EDL_COVID | Covid19:2500, Lung tumors:2500, Normal:2500. | Accuracy:99.05 %, Specificity:99.6 %, F1-score:98.59 %. | 2251 s |
| Ilhan et al. [54] | Deep Feature Fusion | Covid19:125, Pneumonia:500, Normal:500. | Accuracy:90.84 %, Precision:100 %, Recall:97.6 %. | – |
| Proposed | PulDi-COVID (SSE) | Atelectasis, Bacterial Pneumonia, Covid19, Cardiomegaly, Effusion, Infiltration, No-Finding, Pneumothorax, Viral Pneumonia. (1200 cxi/class = 10800 images). | Accuracy:99.70 %, Specificity:99.91 %, Precision:98.68 %, Recall:98.67 %, F1-score:98.67 %, AUC-ROC-score:99.24 %, Error rate:1.33 %, Zero-one loss:12. | 8.59 s |
5. Discussion
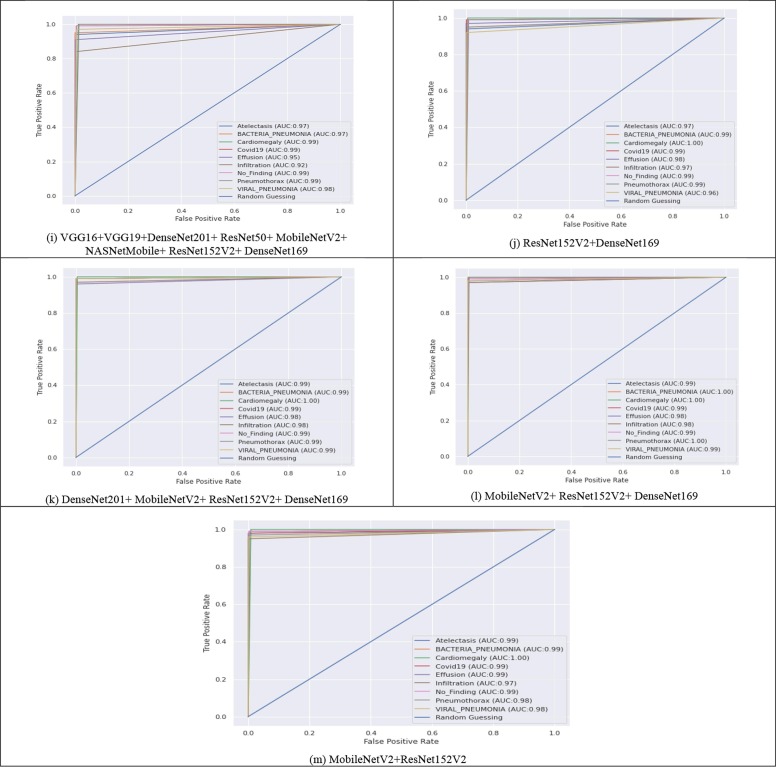
Despite the availability of datasets on online public platforms, the research of CXI for the accurate assessment of COVID-19 infection has attracted a lot of interest. Following that, various efforts were made to create an exact diagnostic model employing DL approaches. The notion of TL has been widely applied in CNNs. However, most of the older approaches were assessed using minimal data. The studies [37], [39], [42], [43], [46] attained remarkable performance but used a very tiny sample size for the COVID-19 class. Furthermore, in certain circumstances, the data is skewed. Grad-CAM graphical representations are also used to confirm the accuracy of the outcomes. Fig. 6 depicts the infographics that are equated to the predicted results/ class. The CAD's recommendations were examined by analyzing the binding of thorax illnesses as depicted in obtained heatmaps (Fig. 5 ) depict several examples of GradCAM mapping of image data from the set of data. A feature vector histogram is overlaid on the actual picture to show how the embedding design recognizes it and sheds more light on specific areas of the pixel. The framework pays more emphasis to the location outlined in orange-red (ROI), while the section featured in light-blue receives less recognition. This aesthetic depiction of focus engages end-users in identifying or confirming ROI where symptoms exist and can be confined.
Fig. 6.
Grad-CAM Visualisation of thoracic abnormalities with heatmaps (highlighting essential regions for the model prediction and its source CXRs.).
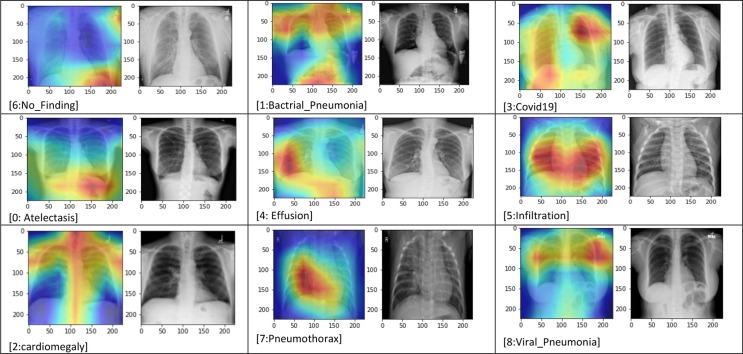
Fig. 5.
Training-Validation accuracy and loss plots of each DCNNs.
In the first phase, we systematically assessed the eight most popular DL models' effectiveness: VGG16, ResNet50, VGG19, DenseNet201, MobileNetV2, NASNetMobile, ResNet152V2, DenseNet169 for the prediction of chronic pulmonary diseases with COVID-19 infections from CXI. Extensive tests were carried out on a rather big dataset, considering a variety of criteria to establish the best functioning model for automatic disease diagnosis. The CXI of the various lung disorders, COVID-19, pneumonia, and normals, were obtained from three different sources [2], [3], [4]. To address the issue of data imbalance, an equal size of samples was chosen for all classes. Experimented findings and extensive comparative analysis of all approaches revealed that the PulDi-COVID model outperformed eight models and state-of-the-art methods.
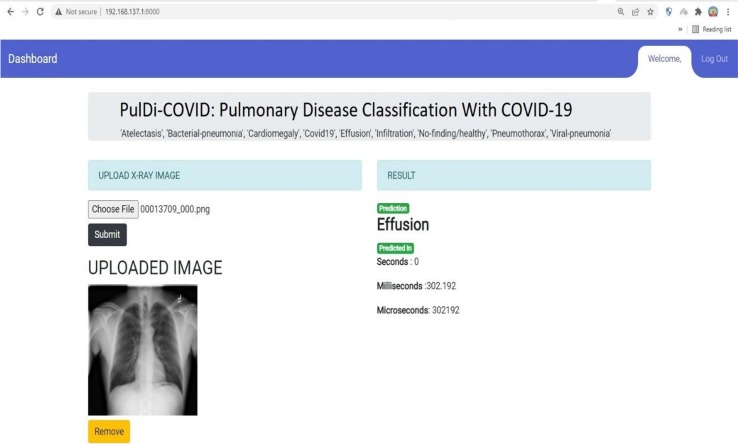
This research aims to find the possible biomarkers from chronic lung disease with COVID-19 to minimize the mortality rates and provide assistance to healthcare staff. Also, reduce the error rate to enhance the accuracy. From the obtained experimented results of individual transfer learning, it has been observed that it attained the lowest error rate by ResNet152V2 of 6.77 %(61 misclassified CXI) and proposed PulDi-COVID(MobileNetV2 + ResNet152V2 + DenseNet169) by 1.33 %(12 misclassified CXI) for nine class classification. This difference error rate of 5.44 %(49 misclassified CXI) shows that the proposed PulDi-COVID model is robust and efficient for chronic pulmonary diseases with COVID-19 cases detection in this pandemic era using a developed GUI application (Fig. 7 ). Limitation of ensemble model deals with model overfitting as seen for bacterial pneumonia class (shows 100 % performance for all metrics. Further research will look at radiography pictures to discover COVID-19 variations such as Beta, Delta, Omicron, and IHU[49].
Fig. 7.
Deployed GUI Web-application for Pulmonary(Lung) disease detection and classification with COVID-19.
6. Conclusion
COVID-19 has substantially detrimental influences on our daily lives, extending from public healthcare services to the entire economic system. This study introduced a PulDi-COVID model that uses CXI to diagnose chronic pulmonary disease with COVID-19. PulDi-COVID, the suggested model, effectively delivers accurate diagnoses for 9 class classifications (atelectasis, bacterial-pneumonia, cardiomegaly, covid19, effusion, infiltration, no-finding, pneumothorax, viral-pneumonia). The suggested framework has a classification accuracy of 99.70 %, a precision of 98.68 %, recall of 98.67 %, F1-score of 98.67 %, a minimum zero-one loss of 12, and the lowest error rate of 1.33 %, which is the maximum attained accuracy on the datasets utilized in the experimentations to the best of knowledge. Another addition to the study is the compilation of the largest dataset for the assessment of classification methods. In terms of accuracy and other metrics, the effectiveness of PulDi-COVID is proven to be superior to 14 current approaches. The result of our suggested technique demonstrates its improvement over previous methods. Our SSE model's empirical explanation is offered. The model's outcomes were described, and physicians may adopt it in the future. Our long-term objective is to combine COVID-19 cases with 14 illness classifications from the NIH [19] dataset (Chest X-ray Dataset of 14 Common Thorax Diseases). We also utilized large datasets to train our proposed PulDi-COVID approach and evaluate its efficiency with a broader range of current techniques. We hope that the purpose of this proposed dl-based model can benefit healthcare workers in detecting pulmonary diseases with COVID-19 to minimize severity and deaths. Also, this research with the deployment of a web application will be helpful to radiology assistance in spotting COVID19 and pulmonary illnesses. Future studies will identify different COVID19 variants using multimodal radiography imaging.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A.
Data availability.
No datasets were generated during the current study. The datasets analyzed during this work are made publicly available in this published article.
Compliance with Ethical Standards.
-
•
No funding received for this work.
-
•
All methods in papers including individual subjects were carried out in line with the institutions and/or national scientific board's ethical principles, as well as the 1964 Helsinki statement and its subsequent revisions or equivalent ethical criteria.
-
•
We also affirm that any component of the study reported in this publication that involves human subjects was carried out with the appropriate permission of all applicable entities, and that such authorization is acknowledged in the article.
Data availability
The authors do not have permission to share data.
References
- 1.Coronavirus Disease 2019. [Online]. Available: https://en.wikipedia.org/wiki/Coronavirus_disease_2019 .
- 2.Summers, Ronald (Lung Disease X-Ray Dataset). CXR8 - National Institutes of Health - Clinical Center. Accessed On 12 September 2021. Online Available https://nihcc.app.box.com/v/ChestXray-NIHCC/folder/37178474737.
- 3.Paul Mooney (Chest X-Ray Pneumonia Dataset). Accessed On 12 September 2021. [Online]. Available: https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia.
- 4.SIIM-FISABIO-RSNA COVID-19 Detection Challenge. Accessed On 12 September 2021. [Online]. Available: https://www.kaggle.com/c/siim-covid19-detection.
- 5.Who coronavirus disease (covid-19) dashboard, (https://covid19.who.int/). Accessed On 09 January 2022.
- 6.Albawi S., Mohammed T.A., Al-Zawi S. Proc. Int. Conf. Eng. Technol; 2017. Understanding of a convolutional neural network; pp. 1–6. [Google Scholar]
- 7.Cao Z., Liao T., Song W., Chen Z., Li C. Detecting the shuttlecock for a badminton robot: A YOLO based approach. Expert Systems with Applications. 2021;164:113833. [Google Scholar]
- 8.Ma C., Liu Z., Cao Z., Song W., Zhang J., Zeng W. Cost-sensitive deep forest for price prediction. Pattern Recognition. 2020;107:107499. [Google Scholar]
- 9.S. Chen, E. Dobriban, and J. H. Lee, “Invariance reduces variance: Understanding data augmentation in deep learning and beyond,” 2019, arXiv:1907.10905.
- 10.D. Jakubovitz, R. Giryes, and M. R. Rodrigues, “Generalization error in deep learning,” in Compressed Sensing and Its Applications, Springer, 2019, pp. 153–193.
- 11.Polikar R. In: Ensemble Machine Learning. Zhang C., Ma Y., editors. Springer US; Boston, MA: 2012. “Ensemble learning,” in Ensemble Machine Learning; pp. 1–34. [Google Scholar]
- 12.Chowdhury M.E.H., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Emadi N.A., Reaz M.B.I., Islam M.T. Can AI Help in Screening Viral and COVID-19 Pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- 13.Svetnik V., Wang T., Tong C., Liaw A., Sheridan R.P., Song Q. “Boosting: An ensemble learning tool for compound classification and QSAR modeling. J. Chem. Inf. Model. 2005;45(3):786–799. doi: 10.1021/ci0500379. [DOI] [PubMed] [Google Scholar]
- 14.F. Divina, A. Gilson, F. Goméz-Vela, M. García Torres, and J. F. Torres, “Stacking ensemble learning for short-term electricity consumption forecasting,” Energies, vol. 11, no. 4, 2018, Art. No. 949.
- 15.H. Wong et al., “Frequency and distribution of chest radiographic findings in COVID-19 positive patients”, Radiology, 2020. [DOI] [PMC free article] [PubMed]
- 16.Li Y., Yao L., Li J., Chen L., Song Y., Cai Z., Yang C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020;92(7):903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ensemble techniques in python. Accessed on: 15 December 2021. [Online]. Available: https://www.geeksforgeeks.org/ensemble-methods-in-python/.
- 18.Stacked Ensemble technique. Accessed on: 15 December 2021. [Online]. Available: https://machinelearningmastery.com/stacking-ensemble-machine-learning-with-python/.
- 19.NIH Chest X-ray Dataset of 14 Common Thorax Disease Categories. Accessed On 2 December 2021. [Online]. Available: https://nihcc.app.box.com/v/ChestXray-NIHCC.
- 20.G. Huang, Y. Li, G. Pleiss, Z. Liu, J. E. Hopcroft, and K. Q. Weinberger, “Snapshot ensembles: Train 1, get M for free,” 2017, arXiv:1704.00109.
- 21.Geron A. second ed. O'Reilly Media, Inc; 2019. In: Hands-on machine learning with scikit-learn, keras, and TensorFlow. [Google Scholar]
- 22.Ledezma C.A., Zhou X., Rodriguez B., Tan P.J., Diaz-Zuccarini V. A modeling and machine learning approach to ECG feature engineering for the detection of ischemia using pseudo-ECG. PLoS ONE. 2019;14(8):PMC6690680. doi: 10.1371/journal.pone.0220294. https://doi:10.1371/journal.pone.0220294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semenzato L., Botton J., Drouin J., Cuenot F., Dray-Spira R., Weill A., Zureik M. Chronic diseases, health conditions and risk of COVID-19-related hospitalization and in-hospital mortality during the first wave of the epidemic in France: a cohort study of 66 million people. The Lancet Regional Health - Europe. 2021;8:100158. doi: 10.1016/j.lanepe.2021.100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geng J., Yu X., Bao H., Feng Z., Yuan X., Zhang J., Chen X., Chen Y., Li C., Yu H. Chronic Diseases as a Predictor for Severity and Mortality of COVID-19: A Systematic Review with Cumulative Meta-Analysis. Front. Med. 2021;8 doi: 10.3389/fmed.2021.588013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.H. S. Maghdid, A. T. Asaad, K. Z. Ghafoor, A. S. Sadiq, and M. K. Khan, “Diagnosing COVID-19 pneumonia from x-ray and CT images using deep learning and transfer learning algorithms,” 2020, arXiv:2004.00038.
- 26.Wang L., Lin Z.Q., Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdel-Basset M., Chang V., Hawash H., Chakrabortty R.K., Ryan M. FSS-2019-nCov: A deep learning architecture for semi-supervised few-shot segmentation of COVID-19 infection. Knowledge-Based Systems. 2021;212:106647. doi: 10.1016/j.knosys.2020.106647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall L.O., Paul R., Goldgof D.B., Goldgof G.M.. Finding covid-19 from chest x-rays using deep learning on a small dataset. arXiv preprint arXiv:2004020602020.
- 29.Singh G.A.P., Gupta P. Performance analysis of various machine learning-based approaches for detection and classification of lung cancer in humans. Neural Computing Applications. 2019;31(10):6863–6877. [Google Scholar]
- 30.Klang E. Deep learning and medical imaging. J Thorac Dis. 2018;10(3):1325–1328. doi: 10.21037/jtd.2018.02.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shan F., Gao Y., Wang J., Shi W., Shi N., Han M., et al. Lung infection quantification of covid-19 in ct images with deep learning. arXiv preprint arXiv:2003046552020.
- 32.Liu J., Pan Y., Li M., Chen Z., Tang L., Lu C., et al. Applications of deep learning to mri images: asurvey. Big Data Min Anal. 2018;1(1):1–18. [Google Scholar]
- 33.Rajaraman S., Siegelman J., Alderson P.O., Folio L.S., Folio L.R., Antani S.K. Iteratively Pruned Deep Learning Ensembles for COVID-19 Detection in Chest X-Rays. IEEE Access. 2020;8:115041–115050. doi: 10.1109/ACCESS.2020.3003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.American Lung Association. Accessed On 12 September 2021. [Online]. Available: https://www.lung.org/lung-health-diseases/lung-disease-lookup/covid-19/chronic-lung-diseases-and-covid.
- 35.Aveyard P., Gao M., Lindson N., Hartmann-Boyce J., Watkinson P., Young D., Coupland C.A.C., Tan P.S., Clift A.K., Harrison D., Gould D.W., Pavord I.D., Hippisley-Cox J. Association between pre-existing respiratory disease and its treatment, and severe COVID-19: a population cohort study. The Lancet Respiratory Medicine. 2021;9(8):909–923. doi: 10.1016/S2213-2600(21)00095-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zabirul Islam, Milon Islam,, Amanullah Asraf. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. [DOI] [PMC free article] [PubMed]
- 37.Rahimzadeh M, Attar A. A new modified deep convolutional neural network for detecting COVID-19 from X-ray images. http://arxiv.org/abs/2004.08052; 2020. [DOI] [PMC free article] [PubMed]
- 38.Alqudah AM, Qazan S, Alquran HH, Alquran H, Qasmieh IA, Alqudah A. Covid-2019 detection using X-ray images and artificial intelligence hybrid systems. https://doi.org/10.13140/RG.2.2.16077.59362/1 ; 2020.
- 39.Hemdan EE-D, Shouman MA, Karar ME. COVIDX-net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images. http://arxiv.org/abs/2003.11055 ; 2020.
- 40.El Asnaoui K., Chawki Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J Biomol Struct Dyn. 2021;39(10):3615–3626. doi: 10.1080/07391102.2020.1767212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ningwei Wang, Hongzhe Liu*, Cheng Xu. Deep Learning for The Detection of COVID-19 Using Transfer Learning and Model Integration. IEEE Conference (ICEIEC); https://doi.org/10.1109/ICEIEC49280.2020.9152329 ; 2020.
- 42.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. Pattern Anal Applic. 2021;24(3):1207–1220. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.F. Ucar, D. Korkmaz, COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diag nosis of the coronavirus disease 2019 (COVID-19) from X-Ray images, Med.Hypotheses (2020) 109761. [DOI] [PMC free article] [PubMed]
- 44.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohit Mishra, Varun Parashar, Rushikesh Shimpi. Development and evaluation of an AI System for early detection of Covid-19 pneumonia using X-ray. https://doi.org/10.1109/BigMM50055.2020.00051 .
- 46.Loey M., Smarandache F., Khalifa N.E.M. Within the lack of chest COVID-19 X-ray dataset: a novel detection model based on GAN and deep transfer learning. Symmetry. 2020;12:651. [Google Scholar]
- 47.Tang S., Wang C., Nie J., Kumar N., Zhang Y., Xiong Z., Barnawi A. EDL-COVID: Ensemble Deep Learning For COVID-19 Cases Detection From Chest X-ray Iimages. IEEE Transactions On Industrial. Info. Sept. 2021;17(9) doi: 10.1109/TII.2021.3057683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.T. Zhou, H.L. Lu, Z. Yang et al. The ensemble deep learning model for novel COVID-19 on CT images. https://doi.org/10.1016/j.asoc.2020.1068851568-4946/ . [DOI] [PMC free article] [PubMed]
- 49.Covid-19: Omicron less severe than Delta variant, UK studies find. Accessed On 12 September 2021. [Online]. Available: https://www.business-standard.com/article/current-affairs/covid-19-omicron-less-severe-than-delta-variant-uk-studies-find-121122300641_1.html.
- 50.Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Rev. Mol. Diagn. 2020;10(5):453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhosale YH, Patnaik KS. Application of Deep Learning Techniques in Diagnosis of Covid-19 (Coronavirus): A Systematic Review. Neural Process Lett. 2022 Sep 16:1-53. DOI: 10.1007/s11063-022-11023-0. Epub ahead of print. PMID: 36158520; PMCID: PMC9483290. [DOI] [PMC free article] [PubMed]
- 52.Bhosale Y.H., Patnaik K.S, IoT Deployable Lightweight Deep Learning Application For COVID-19 Detection With Lung Diseases Using RaspberryPi; IEEE-ICIBT-2022, pp. 1-6.
- 53.Y. H. Bhosale, et al., Deep Convolutional Neural Network Based Covid-19 Classification From Radiology X-Ray Images For IoT Enabled Devices; IEEE-ICACCS-2022, pp. 1398-1402.
- 54.Ilhan H.O., Serbes G., Aydin N. Decision and feature level fusion of deep features extracted from public COVID-19 data-sets. Applied Intelligence. 2022;52(8):8551–8571. doi: 10.1007/s10489-021-02945-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors do not have permission to share data.