Abstract
Phosphatidylinositol 3-kinases (PI3Ks) are lipid kinases involved in cellular growth and division. Somatic mutations in one of the PI3K catalytic subunit genes, PIK3CA, are frequently found in numerous malignancies, including colorectal cancer (CRC). Several PIK3CA inhibitors are approved for the treatment of breast cancer and lymphoma. Activating mutations in PIK3CA tend to occur in exons 9 and 20, with mutations in other exons 1, 4, and 7 being less common. Most test systems for PIK3CA mutation screening are designed to detect mutations in exons 9 and 20, leaving exons 1–7 overlooked. We have developed a multiplex AS-PCR to screen for PIK3CA mutations in exons 1, 4, 7, 9, and 20. Validation was performed on 515 CRC samples of patients from Siberia and the Far East of Russia. The assay sensitivity was 0.05–0.5% of mutant DNA, and the overall PIK3CA mutation frequency was 13.01%, with 9.32% of mutations in exon 9, 1.94% in exon 20, and 1.74% in exons 1–7. The assay designed is suitable for the analysis of activating PIK3CA mutations in formalin-fixed paraffin-embedded tissue samples. The present work is the first study characterizing the PIK3CA mutation frequency in CRC patients from the eastern part of Russia.
Keywords: PIK3CA, Somatic mutations, CRC, Multiplex amplification, Allele-specific PCR, ARMS
PIK3CA; Somatic mutations; CRC; Multiplex amplification; Allele-specific PCR; ARMS.
1. Introduction
Colorectal cancer (CRC) is the third most common and second most deadly malignancy worldwide, with over 1.9 million new cases and 935,000 deaths in 2020 [1]. CRC is defined as a group of malignancies originating from epithelial cells of various parts of the large intestine. Despite a 9-fold difference in the CRC incidence rate between world regions, a steady increase in CRC cases has been registered in Eastern Europe, South Eastern and South Central Asia, and South America. In addition, the number of early-onset CRC cases (age at diagnosis <50 years) is increasing in many countries, including the developed ones.
Molecular markers of CRC that can predict response to a target therapy or treatment regimen are known as predictive biomarkers [2]. Among the predictive biomarkers recommended by ASCP (American Society for Clinical Pathology) guidelines, activating mutations in KRAS, NRAS, and BRAF genes in the EGFR signaling pathway and the deficiency status of DNA mismatch repair are routinely screened in clinical practice. However, other components of the EGFR signaling pathway, such as PIK3CA and PTEN, could also affect the response of CRC to anti-EGFR antibody therapy and first-line chemotherapy [3, 4, 5, 6]. However, the data on the effect of PIK3CA mutations on target therapy are still inconsistent [7, 8].
Phosphatidylinositol 3-kinases (PI3Ks) are heterodimeric lipid kinases that phosphorylate phosphatidylinositol and participate in signaling pathways essential for cell proliferation, adhesion, survival, and motility [9]. PIK3s are known to be involved in the molecular pathogenesis of CRC [10]. PIK3 heterodimers consist of a regulatory and a catalytic subunit encoded by several genes: PIK3R1, PIK3R2, and PIK3R3—isoforms of p85 regulatory subunit, and PIK3CA, PIK3CB, and PIK3CD—p110α, p110β, p110γ catalytic subunits, respectively [11]. Somatic mutations activating PI3Ks are most frequent in the PIK3CA gene [12], occurring in around 36% of breast cancer [13] and 13–25% of CRC cases [14, 15]. The most common mutation hotspots are exon 9, protein helical domain (codons 542, 545), and exon 20, protein kinase domain (codon 1047). Mutations in the helical domain disrupt interaction with the regulatory subunit [16], while mutations in the kinase domain facilitate interactions with membranes, bypassing the need for association with RAS protein [17]. Mutated PIK3CA has become a target for several specific drugs that inhibit the activated PIK3CA protein, such as FDA-approved alpelisib [18] for breast cancer and idelalisib [19] for lymphoma treatment.
To date, several methods have been developed for somatic mutation testing, such as allele-specific PCR (AS-PCR) [20, 21], allele-specific LAMP (AS-LAMP) [22], next-generation sequencing (NGS) [23, 24], digital PCR [25], PCR-restriction fragment length polymorphism analysis (PCR-RFLP) [26], primer extension assay (SNaPshot) [27], and variations of the mentioned methods. The most popular approach in routine clinical practice is real-time AS-PCR. This relatively fast, simple, and low-cost technique allows mutations to be screened in standard diagnostic laboratories. Numerous test systems have been developed and approved for testing activating PIK3CA mutations. Despite being relatively straightforward, the method has caused several concerns. Formalin-fixed paraffin-embedded (FFPE) tissues are the main source of DNA for the screening. While preserving DNA for a prolonged time, the process of FFPE preparation leads to DNA degradation and multiple chemical modifications. DNA purified from FFPE tissues is fragmented into short stretches with numerous modifications of sugar-phosphate backbone and nucleobases [28]. Chemical modifications result in pauses of DNA polymerase during amplification, while the short length of template fragments limits the length of amplicons. All these factors reduce the PCR efficiency of FFPE-derived DNA.
The overall amount of template DNA from FFPE is limited, limiting the number of available molecular tests. Given the above, multiplex PCR is one possible solution to overcome the issues related to the analysis of FFPE DNA. Multiplex PCR simultaneously amplifies several DNA targets in a single tube, allowing multiple mutations to be detected simultaneously. Thus, not only does this method provide valuable DNA samples, but it also can save personnel and machine time. The most common approach to amplify more than one target in a single reaction is to use hydrolyzing probes (e.g., TaqMan probes). However, several other techniques have been introduced: high-resolution melting (HRM) [29], fluorescent melting curve analysis (FMCA) [30], molecular beacons [31], and others.
Multiplex PCR has posed several issues, including primer interference, reduced amplification efficiency, and decreased mutation detection sensitivity [32]. Thus, primers can form energetically favorable heterodimers, which may deplete the overall primer pool and concur with intended amplicons for DNA polymerases, reducing the amplification rate. The intended amplicons could also have drastically different amplification efficiencies, provided that one of the amplicons accumulates faster and inhibits the amplification of other targets. Thus, it is necessary to accurately optimize assay parameters, with primer concentration being the main variable to solve the problems arising from using multiple primer sets in the same reaction. Altering the primer concentration affects the amplification efficacy directly and ensures uniform amplification rates of different targets, preventing possible inhibition and decreasing overall analysis sensitivity.
Numerous multiplex PCR test systems have been designed to screen for the activating somatic mutation in the PIK3CA gene. Most of them are intended to detect the frequent mutations in exons 9 and 20, leaving aside less abundant mutations in exons 1, 4, and 7. In breast cancer, mutations in exons 1–7 were reported to account for up to 11% of mutated samples or 4.25% of all tested specimens [13]. The frequency of the additional PIK3CA mutation in CRC remains almost unstudied [24], affecting the overall understanding of PIK3CA mutations in CRC pathogenesis and treatment. It should be mentioned that the mutational spectrum of the PIK3CA gene in several groups of patients with CRC is virtually unknown. Thus, there are no data on PIK3CA mutations in patients with CRC from Siberia and the Far East, while the other regions of the Russian Federation have been studied scarcely [33, 34, 35].
In the present work, a multiplex qPCR approach has been developed to test the most common mutations of PIK3CA in CRC samples by optimizing the assay conditions (AS-primers types and concentrations, elongations temperatures). The multiplex qPCR was intended to evaluate PIK3CA mutation frequencies in patients with CRC from Siberia and the Far East of Russia, previously unstudied on the matter.
2. Materials and methods
2.1. Clinical samples and DNA extraction
DNA was extracted from FFPE tumor tissue sections using QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. FFPE sections were obtained from 515 colorectal cancer patients operated on in 14 regional cancer centers across the Siberian and the Far Eastern Federal Districts of Russia. The study was conducted according to the guidelines of the Declaration of Helsinki and was approved by the Local Medical Ethical Committee of ICBFM SB RAS (N1 meeting, January 11, 2022). Written informed consent was obtained from all subjects involved in the study. Detailed information about the study group is provided in Table 1.
Table 1.
Descriptive characteristics of patients with colorectal cancer (N = 515) and the prevalence of KRAS mutations.
| Parameter | Value | Percent |
|---|---|---|
| Women, number of patients | 294 | 57.1 |
| Men, number of patients | 221 | 42.9 |
| Age, years, median (lower quartile, upper quartile) | 59 (53, 65) | |
| T∗, number of patients | T1: 8 | 1.55 |
| T2: 52 | 10.10 | |
| T3: 159 | 30.9 | |
| T4: 289 | 56.1 | |
| Tx: 7 | 1.4 | |
| N∗, number of patients | N0: 181 | 35.2 |
| N1: 173 | 33.6 | |
| N2: 100 | 19.4 | |
| N3: 4 | 0.8 | |
| Nx: 57 | 11.1 | |
| M∗, number of patients | M0: 294 | 57.1 |
| M1: 198 | 38.4 | |
| Mx: 24 | 4.7 | |
| KRAS mutations, number of patients | 167 | 32.43 |
| G12A, number of patients | 16 | 3.11 |
| G12C, number of patients | 13 | 2.52 |
| G12D, number of patients | 61 | 11.84 |
| G12R, number of patients | 1 | 0.19 |
| G12V, number of patients | 58 | 11.26 |
| G13D, number of patients | 18 | 3.50 |
| WT, number of patients | 348 | 67.57 |
- staging according to the TNM classification of malignant tumors (7th edition). T, size, and extent of the primary tumor; N, degree of spread to regional lymph nodes; M, presence of distant metastasis, number of patients.
2.2. Detection of common KRAS gene mutations
Common KRAS mutations were analyzed using the therascreen KRAS RGQ PCR Kit, Version 1 (Qiagen, Hilden, Germany), according to the manufacturer's instructions. The amplification procedure was conducted using CFX96 Thermal Cycler (Bio-Rad, Hercules, CA, USA).
2.3. Detection of PIK3CA mutations
PIK3CA mutation analysis was performed using an in-house allele-specific qPCR assay with hydrolyzing probes (TaqMan). Reactions were performed in 20 μL volume containing 65 mM Tris-HCl, pH 8.9, 24 mM (NH4)2SO4, 0.05% Tween-20, 3 mM MgSO4, 0.2 mM dNTPs, 300 nM primers, 100 nM hydrolyzing probe (Table 2) and 1 U of Taq-polymerase. Amplification was carried out in CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA), according to the following program: 95 °C for 3 min followed by 45 cycles of 95 °C for 10 s, and 60 °C for 40 s with fluorescent signals collected on FAM, HEX, and ROX channels, unless otherwise indicated. The data obtained were analyzed with CFX Manager software (Bio-Rad, Hercules, CA, USA).
Table 2.
List of oligonucleotide primers and probes.
| Primer | 5′-Sequence-3′ |
|---|---|
| Lac-U | CGTCGTTTGGTATGGCTTCATTC |
| Lac-R | AGGACCGAAGGAGCTAACCG |
| Lac-P | HEX-CGGTTCCCAACGATCAAGGCGAG-BHQ2 |
| ALB-F | GACTTGCCAAGACATATGAAACC |
| ALB-R | TCCAACAATAAACCTACCACTTTG |
| ALB-P | ROX-TGCTGTGCCGCTGCAGATCC-BHQ2 |
| R88Q-U | TTCTTACATTTTCGTAAGTGTTACTCAAG |
| R88Q-R | GAAAAAGCCGAAGGTCACAAAATT |
| R88Q-P | ROX-AGCAGAAAGGGAAGAATTTTTTGATGAAAC-BHQ2 |
| N345K–U | CCTTTGGGTTATAAATAGTCCACTCAGAA |
| N345K–P | FAM-AAAAATTCTTTGTGCAACCTACGTGA-BHQ |
| N345K-R | GACTTTACCTTATCAATGTCTCGAATATTTCCT |
| C420R–U | AAGTGTTTTGAAATGTGTTTTATAATTTAG |
| C420R-R | TATTTCCCCATGCCAATGGTCG |
| C420R–P | HEX-CTAGTGAATATTTTTCTTTGTTTTTTAAGGAAC-BHQ |
| E542-F | GGAAAATGACAAAGAACAGCTCA |
| E542K–P | FAM-CAATTTCTACACGAGATCCTCTCTC-BHQ1 |
| E542K-1 | CTTTCTCCTGCTCAGTGATTTT |
| E542K-2 | CTTTCTCCTGCTCAGTGATTCT |
| E542K-3 | CTTTCTCCTGCTCAGTGAATCT |
| E545K-1 | CATAGAAAATCTTTCTCCTGCTT |
| E545K-2 | CCATAGAAAATCTTTCTCCTGCCT |
| E545K-3 | CCATAGAAAATCTTTCTCCTCCCT |
| Q546K-R | TGACTCCATAGAAAATCTTTCTCCCT |
| Ex20-F1 | CTGAGCAAGAGGCTTTGGAGTA |
| Ex20-P1 | HEX-TCATGAAACAAATGAATGATGCA-BHQ2 |
| H1047R-R | TGTTGTCCAGCCACCATGTC |
| M1043I-R | CTTTGGAGTATTTCATGAAACAAACT |
| PIC-1047L | TTGTTGTCCAGCCACCATGGA |
| H1047Y-R | TCATGAAACAAATGAATGATGCCT |
| Ex20-F2 | CATGCTGTTTAATTGTGTGGAAG |
| Ex20-P2 | HEX-TGGCTGGACAACAAAAATGG-BHQ2 |
2.4. Positive controls
Plasmids carrying PIK3CA and ALB gene fragments served as positive controls and were used to assess method sensitivity. For the PIK3CA gene, plasmids carried either wild-type or mutated fragments of exons 1, 4, 7, 9, 20. All control plasmids used in this study were made in Shanghai RealGene Bio-tech, Inc. (Shanghai, China), purified, sequenced (SB RAS Genomics Core Facility, ICBFM SB RAS, Novosibirsk, Russia), and quantified using NanoDrop Lite A4 spectrophotometer (Thermo Fisher Scientific, Waltham, MA USA).
2.5. Droplet digital PCR
The ddPCR was performed using the QX100 system (Bio-Rad, Hercules, CA, USA) according to the manufacturer's recommendations. The reaction mixture in a volume of 20 μL contained 1× ddPCR master mix (Bio-Rad, USA), 0.9 μM Lac-U/R primers, 0.25 μM Lac-P probe (Table 2), and approximately 104–103 copies of the tested plasmid standard. The entire reaction mixtures with 70 μL of droplet generation oil (Bio-Rad, Hercules, CA, USA) were loaded into a disposable plastic cartridge (Bio-Rad, Hercules, CA, USA) and placed in the droplet generator. After processing, the droplets obtained from each sample were transferred to a 96-well PCR plate (Eppendorf, Hamburg, Germany). The amplification was carried out using T100TM Thermal Cycler (Bio-Rad, Hercules, CA, USA) according to the program: DNA polymerase activation at 95 °C for 10 min followed by 45 cycles of PCR amplification (94 °C for 30 s and 57 °C for 60 s), and 98 °C for 10 min, 2 °C/s ramp rate at all steps. After the PCR, the droplets were counted with the QX100 Droplet Reader. The data obtained were analyzed with QuantaSoft software (Bio-Rad, Hercules, CA, USA).
3. Results
3.1. Mutation selection and control sample preparation
In the present work, we have designed an approach to detect several common activating mutations in the PIK3CA gene based on multiplex allele-specific quantitative PCR (Figure 1). Together with the most frequent activating mutations E542K, E545K, H1047L, and H1047R, several additional mutations were selected for screening based on their frequency in previously published studies and confirmed functional significance [36]. Using the criteria mentioned above, mutations R88Q [37], N345K [37], C420R [38], Q546K [3], M1043I [39], H1047Y [38] (Table 3) were tested in the present study, as they were previously identified in samples from patients with CRC. A mutation E545A [40], characterized as activating, was discarded from the present study, as it was detected in the PIK3CA pseudogene [41].
Figure 1.
Schematic overview of the experimental workflow.
Table 3.
Tested somatic mutation in the PIK3CA gene.
| Exon | Nucleotide | Amino Acid | COSMIC MutationID |
|---|---|---|---|
| 1 | c.263G>A | p.R88Q | COSV55874568 |
| 4 | c.1035T>A | p.N345K | COSV55873276 |
| 7 | c.1258T>C | p.C420R | COSV55874020 |
| 9 | c.1624G>A | p.E542K | COSV55873227 |
| c.1633G>A | p.E545K | COSV55873239 | |
| c.1636C>A | p.Q546K | COSV55873527 | |
| 20 | c.3129G>T | p.M1043I | COSV55878974 |
| c.3139C>T | p.H1047Y | COSV55876499 | |
| c.3140A>T | p.H1047L | COSV55873401 | |
| c.3140A>G | p.H1047R | COSV55873195 |
A set of plasmids was constructed based on the pBlueScript II SK (+) vector and 200 bp fragments of the PIK3CA gene as control samples for evaluating the multiplex qPCR efficiency. The control plasmids harbored fragments with one of the PIK3CA mutations selected for screening and partial wild-type sequences of the respective PIK3CA exons. All standards were quantified by droplet digital PCR using the QX200 platform (Bio-Rad, USA), primers, and TaqMan probe to detect the β-lactamase gene.
3.2. Optimization of multiplex qPCR assay
3.2.1. Testing allele-specific primers designed by different strategies
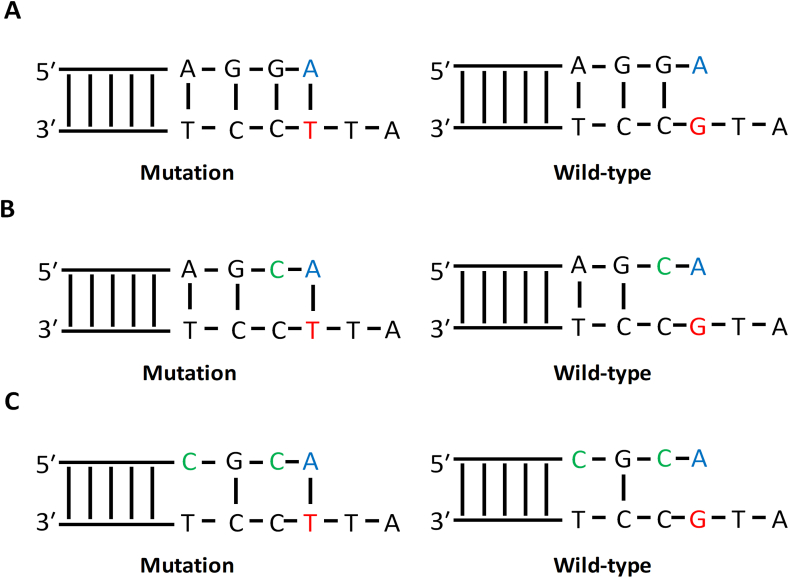
Several approaches were used to design allele-specific primers for multiplex qPCR. Specifically, we compared classical allele-specific primers with one mismatch, AS-primers designed by a method described by Li et al. [42] with one additional mismatch at -2 position from primer's 3′-end, and AS-primers with two additional mismatches at -2 and -4 position from 3′-end [Figure 2(A, B, C)].
Figure 2.
Different designs of allele-specific primers. (A) Classical design with one mismatch at 3′-end of allele-specific primer. (B) Additional mismatch at -2 position from 3′-end of allele-specific primer. (C) Additional mismatches at -2 and -4 position from the 3′-end of allele-specific primer. The mutated nucleotide at the template is marked by red, the allele-specific nucleotide of the primer—by blue, additional mismatches in the primer—by green.
Allele-specific primers designed with different approaches were tested on the model of E542K and E545K mutations: classical AS-primers E542K-1, E545K-1; AS-primers with one additional mismatch E542K-2, E545K-2; AS-primers with two additional mismatches (Table 2). Forward primer E542K-R and TaqMan probe E542K–P were used as common primers for all AS-primers.
From this point onward, a set of primers targeting a fragment of the ALB gene was used to estimate the overall DNA amount in an analyzed specimen. The ALB gene, with no hereditary chromosomal aberrations reported, was chosen for this task, making quantification robust. Control samples prepared using plasmids with fragments of the PIK3CA and ALB genes were used to assess the PCR efficacy and discrimination ability for different AS-primers. The concentrations of plasmids were 105 copies/μL with 5% of mutated DNA or with wild-type DNA only. A cycle of quantification values (Cq) was used to compare AS-primers: each sample was run in triplicate, and mean Cq values were computed for further analyses. For each control sample, the difference (ΔCq) between Cq with ALB primers (CqALB) and Cq for the AS-primers (CqAS) was calculated using Eq. (1):
| ΔCq = CqAS − CqALB | (1) |
The results of the assessment are presented in Table 4. For both mutations, AS-primers with two additional mismatches demonstrated the lowest efficacy of amplification and the highest Cq values. At the same time, the classical AS-primers had the highest PCR efficacy and the lowest Cq values. However, the classical AS-primers demonstrated the most inferior discrimination ability between wild-type and mutated DNA due to the lowest difference between ΔCq values for wild-type DNA and mutated DNA. Thus, primers with one additional mismatch were chosen for further work since they demonstrated a reasonable compromise between amplification efficacy and discrimination ability. For all other PIK3CA gene mutations studied here, the approach of Li et al. was applied with one additional mismatch in AS-primers.
Table 4.
Assessment of different AS-primers design.
| Mutation | AS-primer | CqALB-WT | CqAS-WT | ΔCqWT | CqALB-5% | CqAS-5% | ΔCq5% |
|---|---|---|---|---|---|---|---|
| E542K | E542K-1 | 22.34 | 35.73 | 13.39 | 22.48 | 31.88 | 9.40 |
| E542K-2 | 22.34 | 42.88 | 20.54 | 22.48 | 32.40 | 9.92 | |
| E542K-3 | 22.34 | N/A∗ | -† | 22.48 | 41.13 | 18.65 | |
| E545K | E545K-1 | 22.34 | 35.18 | 12.84 | 22.42 | 26.54 | 4.12 |
| E545K-2 | 22.34 | 43.54 | 21.2 | 22.42 | 27.06 | -† | |
| E545K-3 | 22.34 | N/A | -† | 22.42 | 39.80 | 17.38 |
N/A—no amplification, no Cq value has been obtained.
no ΔCq could be calculated.
3.2.2. Selecting optimal concentration for primers in multiplex qPCR
After selecting optimal AS-primers, AS-primers and primers for the control ALB gene fragment were combined in several multiplexes to save machine time, labor contribution of personnel, and DNA amount of the specimens analyzed (Table 5).
Table 5.
Multiplexes composition for PIK3CA mutations analysis.
| Multiplex | Forward primer | Probe | Reverse primer |
|---|---|---|---|
| ALB/R88Q/N345K/C420R | ALB-F | ALB-P | ALB-R |
| R88Q-F | R88Q-P | R88Q-R | |
| N345K–F | N345K–P | N345-R | |
| C420–F | C420–P | C420-R | |
| ALB/E542K/H1047L | ALB-F | ALB-P | ALB-R |
| E542K–F | E542K-R | E542K-2 | |
| Ex20-F1 | Ex20-P1 | H1047L-R | |
| ALB/E545K/H1047R | ALB-F | ALB-P | ALB-R |
| E542K–F | E542K-R | E545K-2 | |
| Ex20-F1 | Ex20-P1 | H1047R-R | |
| ALB//M1043I | ALB-F | ALB-P | ALB-R |
| Ex20-F2 | Ex20-P2 | M1043I-R | |
| ALB/Q546K/H1049Y | ALB-F | ALB-P | ALB-R |
| E542K–F | E542K-R | Q546K-R | |
| Ex20-F2 | Ex20-P2 | H1047Y-R |
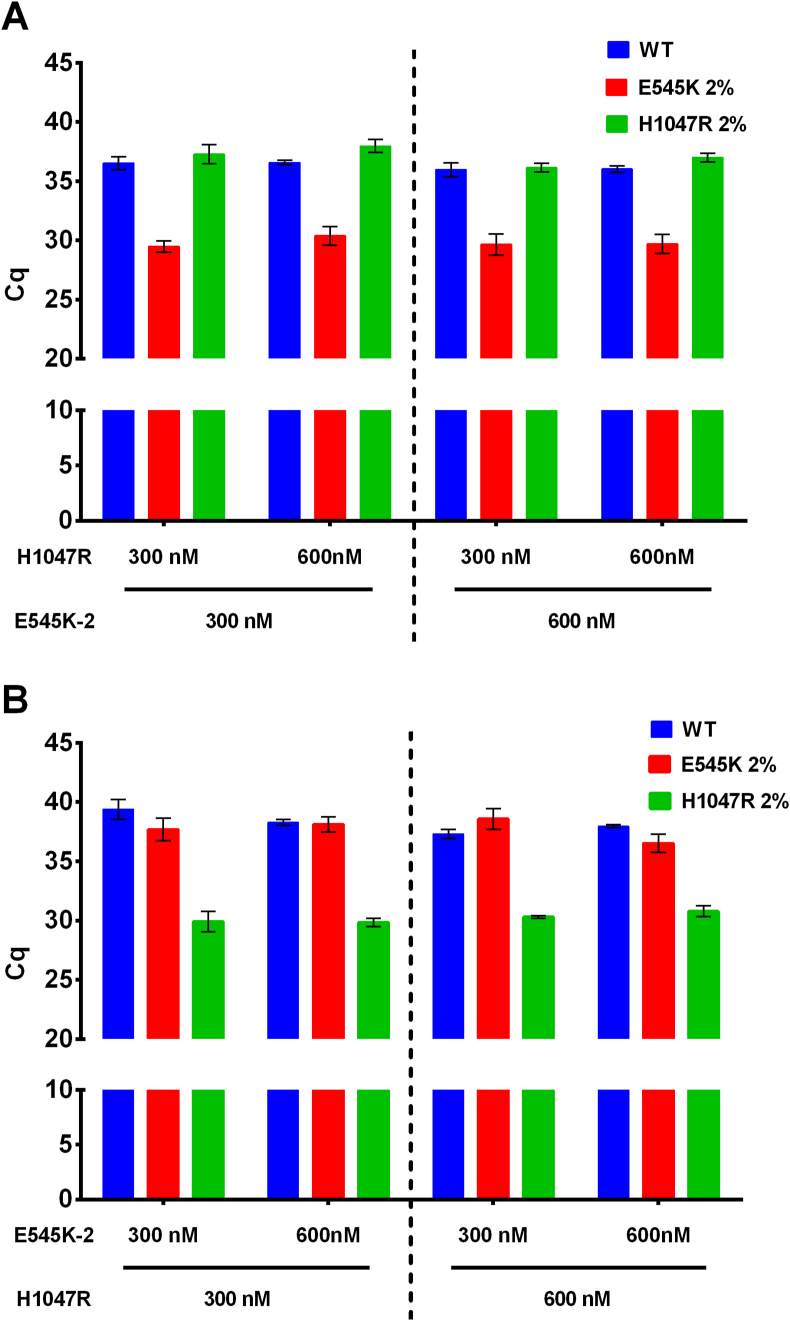
To increase the efficacy of multiplex PCR, we tested different concentrations of AS-primers (300 and 600 nM) with a fixed concentration of the control ALB primers (300 nM) on the model of ALB/E545K/H1047R multiplex. The concentrations of control samples were 104 copies/μL with 2% of mutated DNA or with wild-type DNA only. Cq values were used to compare AS-primers, each specimen was run in triplicate, and mean Cq values were computed for further analyses. The results obtained are presented in Figure 3(A, B).
Figure 3.
Comparison of different AS-primers concentrations. (A) Cq values for AS-primer E545K-2. (B) Cq values for AS-primer H1047R. Y-axis represents Cq values, X-axis—AS-primers concentration, the shade indicates different control samples as it is defined in the legend.
A direct comparison of different AS-primers concentrations revealed 300 nM and 600 nM AS-primers E545K-2 and H1047R-R to have a similar amplification efficacy and not interfere with each other. No difference in Cq or amplitude of end-point fluorescence was noted. AS-primers were added to a final concentration of 300 nM in other multiplexes to reduce the primer's consumption.
3.2.3. Selecting the optimal elongation temperature for multiplex qPCR
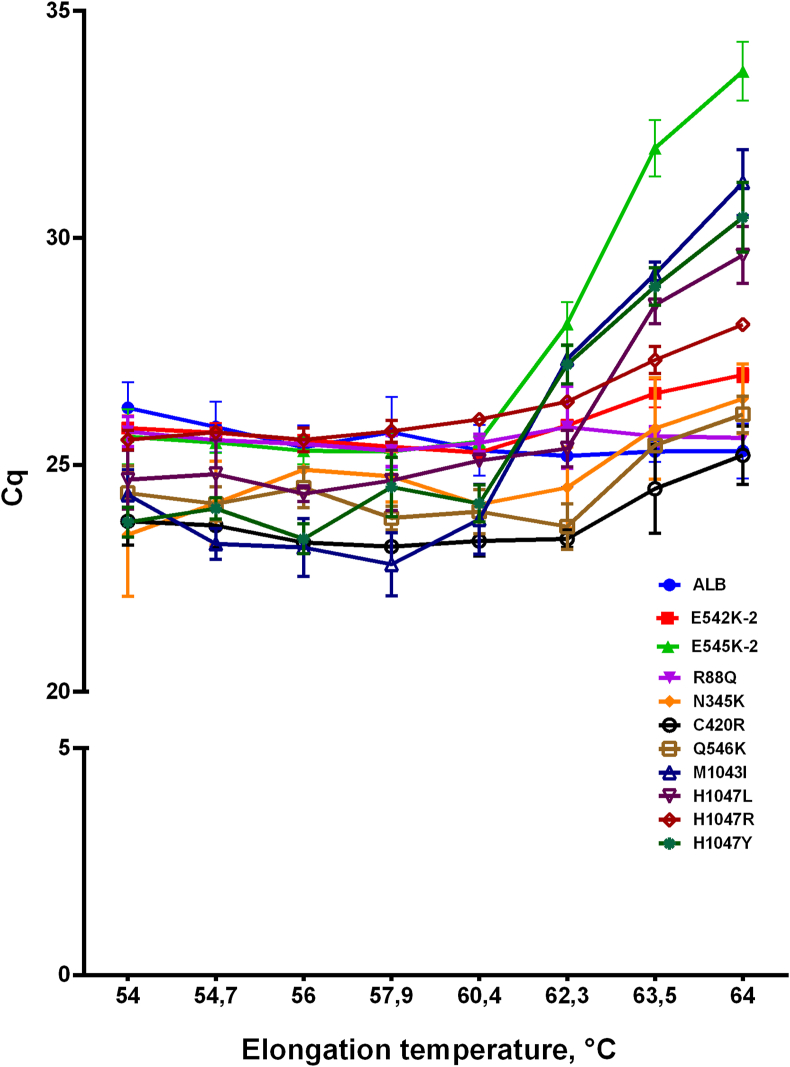
To ensure the highest amplification efficacy, we varied the elongation temperature of PCR with all multiplexes at the range from 54 to 64 °C. The concentrations of control samples were 104 copies/μL with 100% of mutated DNA or with wild-type DNA only. Cq values were used to compare AS-primers, with each specimen run in triplicate, and mean Cq values were computed for further analyses. The results obtained are presented in Figure 4.
Figure 4.
Elongation temperature for primers in multiplex PCR. Y-axis represents Cq values, X-axis—elongation temperature in PCR, primers are indicated by colors and symbols as it is defined in the legend.
For all sets of primers and probes analyzed, the optimal temperature was 60 °C because the lowest Cq and the highest end-point fluorescence were observed at this temperature. Therefore, all further experiments were conducted at this temperature.
3.3. Sensitivity assessment of multiplex qPCR assay
The sensitivity of the multiplex qPCR assay developed was defined as the lowest detectable percent of mutations on the background of wild-type DNA. The sensitivity of multiplex qPCR was evaluated using control samples with a variable percentage of each mutation separately, ranging from 0.05 to 2% of mutated DNA. The overall DNA concentrations of control samples were 104 copies/μL. Cq values were used to compare AS-primers, each specimen was run in triplicate, and mean Cq values were computed for further analyses. For each control sample, the difference (ΔCq) between Cq with ALB primers (CqALB) and Cq for the AS-primers (CqAS) was calculated using Eq. (1).
The results of the assessment are presented in Table 6. The multiplex qPCR assay designed detected at least 0.05% of N345K, E542K, E545K, H1047L, and H1047R mutations as the differences between ΔCq for wild-type sample and the sample with 0.05% or the respective mutation were more than 1. For other mutations (R88Q, N345K, C420R, Q546R, M1043I, H1047Y), 0.5% of mutated DNA on the background of wild-type DNA could be found.
Table 6.
Sensitivity of multiplex qPCR assay for different mutations.
| Mutation | Mutation % | CqALB | CqAS | ΔCq | Mutation | Mutation % | CqALB | CqAS | ΔCq |
|---|---|---|---|---|---|---|---|---|---|
| R88Q | 0.05 | 26.54 | 31.24 | 4.70 | N345K | 0.05 | 26.65 | 34.01 | 7.36 |
| 0.5 | 26.97 | 30.78 | 3.81 | 0.5 | 26.85 | 33.18 | 6.33 | ||
| 1 | 26.93 | 29.84 | 2.91 | 1 | 26.36 | 32.68 | 6.32 | ||
| 2 | 27.05 | 29.06 | 2.01 | 2 | 26.17 | 31.45 | 5.28 | ||
| WT | 26.60 | 31.88 | 5.28 | WT | 26.11 | 35.59 | 9.48 | ||
| C420R | 0.05 | 26.37 | 32.04 | 5.67 | E542K | 0.05 | 26.19 | 31.38 | 5.19 |
| 0.5 | 26.69 | 31.22 | 4.53 | 0.5 | 26.60 | 30.52 | 3.92 | ||
| 1 | 26.59 | 30.56 | 3.97 | 1 | 26.28 | 29.65 | 3.37 | ||
| 2 | 26.83 | 29.82 | 2.99 | 2 | 26.61 | 29.42 | 2.81 | ||
| WT | 26.64 | 32.86 | 6.22 | WT | 26.27 | 32.22 | 5.95 | ||
| E545K | 0.05 | 26.28 | 29.66 | 5.19 | Q546K | 0.05 | 26.37 | 31.97 | 5.60 |
| 0.5 | 26.36 | 30.96 | 3.92 | 0.5 | 26.20 | 31.19 | 3.45 | ||
| 1 | 26.48 | 29.18 | 3.37 | 1 | 26.40 | 30.99 | 4.59 | ||
| 2 | 26.60 | 28.60 | 2.81 | 2 | 26.79 | 29.65 | 4.40 | ||
| WT | 26.37 | 31.62 | 6.45 | WT | 26.59 | 32.16 | 5.57 | ||
| M1043I | 0.05 | 26.39 | 33.80 | 7.41 | H1047L | 0.05 | 26.70 | 33.10 | 6.40 |
| 0.5 | 26.11 | 31.09 | 4.98 | 0.5 | 26.44 | 31.73 | 5.29 | ||
| 1 | 26.26 | 30.01 | 3.75 | 1 | 26.49 | 29.58 | 3.09 | ||
| 2 | 26.20 | 27.70 | 1.50 | 2 | 26.87 | 27.61 | 0.74 | ||
| WT | 25.86 | 33.74 | 7.88 | WT | 26.72 | 33.24 | 6.52 | ||
| H1047R | 0.05 | 26.40 | 30.22 | 3.82 | H1047Y | 0.05 | 26.06 | 31.63 | 5.57 |
| 0.5 | 26.38 | 28.72 | 2.34 | 0.5 | 26.29 | 30.57 | 4.28 | ||
| 1 | 26.32 | 28.17 | 1.85 | 1 | 26.26 | 29.77 | 3.51 | ||
| 2 | 26.50 | 26.97 | 0.47 | 2 | 26.18 | 27.81 | 1.63 | ||
| WT | 26.49 | 32.55 | 6.06 | WT | 25.25 | 31.11 | 5.86 |
3.4. Testing clinical samples
The multiplex qPCR assay for PIK3CA mutation screening was validated on clinical samples of FFPE-derived DNA from 515 patients with colorectal cancer operated on in 14 regional cancer centers across Siberia and the Far East of Russia. All patients participating in the study signed informed consent. DNA was extracted from FFPE tumor tissue sections using QIAamp DNA FFPE Tissue Kit. Common KRAS mutations were analyzed using the therascreen KRAS RGQ PCR Kit, Version 1. The details of the study group and the results of KRAS testing are presented in Table 1.
All FFPE-derived DNA samples from patients with CRC were screened for the presence of the PIK3CA gene mutations using the multiplex qPCR assay developed. The results of the testing are presented in Table 7. According to our results, the most frequent PIK3CA mutations in the CRC samples studied were mutations E542K and E545K in the exon 9, found in 7.77% of all patients or 59.7% of all found samples with PIK3CA mutations. All mutations in exon 9 were found in 9.32% of the samples studied, accounting for 84.21% of mutated samples. Mutations in the exon 20 were much less abundant, accounting for 1.94% of CRC samples or 14.92% of all mutated samples. Mutations in exons 1, 4, and 7 were found in 0.58% of all samples or 4.48% of mutated samples for each exon. No correlation was noticed between the PIK3CA status and age, biological sex, and mutations in the KRAS gene exon 2. However, a moderate positive correlation was observed between the KRAS positive status and T status (correlation coefficient of 0.426).
Table 7.
Prevalence of the PIK3CA mutations in CRC samples.
| Samples | PIK3CA |
KRAS WT |
KRAS mutated |
|||
|---|---|---|---|---|---|---|
| Number | Percent | Number | Percent | Number | Percent | |
| All | 515 | 100 | 347 | 67.38 | 168 | 32.62 |
| WT | 448 | 86.99 | 309 | 60.00 | 139 | 26.99 |
| Mut | 67 | 13.01 | 38 | 7.38 | 29 | 5.63 |
| R88Q | 3 | 0.58 | 2 | 0.39 | 1 | 0.19 |
| N345K | 3 | 0.58 | 0 | 0.00 | 3 | 0.58 |
| C420R | 3 | 0.58 | 1 | 0.19 | 2 | 0.39 |
| Exon 9 | 48 | 9.32 | 32 | 6.21 | 16 | 3.11 |
| E542K | 26 | 5.05 | 18 | 3.50 | 8 | 1.55 |
| E545K | 14 | 2.72 | 8 | 1.55 | 6 | 1.17 |
| E545G | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| Q546K | 5 | 0.97 | 3 | 0.58 | 2 | 0.39 |
| Q546L | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| E542K/E545K | 2 | 0.39 | 2 | 0.39 | 0 | 0.00 |
| E542K/H1047R | 1 | 0.19 | 1 | 0.19 | 0 | 0.00 |
| Exon 20 | 10 | 1.94 | 3 | 0.58 | 7 | 1.36 |
| M1043I | 1 | 0.19 | 1 | 0.19 | 0 | 0.00 |
| H1047Y | 1 | 0.19 | 0 | 0.00 | 1 | 0.19 |
| H1047L | 3 | 0.58 | 0 | 0.00 | 3 | 0.58 |
| H1047R | 5 | 0.97 | 2 | 0.39 | 3 | 0.58 |
4. Discussion
After PIK3CA inhibitors, alpelisib [18], and idelalisib [19], were introduced into clinical practice, PIK3CA testing has become indispensable in molecular diagnostics of several malignancies, such as breast cancer and lymphoma. Numerous test systems have been designed to detect activating mutations in PIK3CA based on allele-specific PCR, allele-specific LAMP, high-resolution melting, digital PCR, NGS, and various modifications of these methods. Most assays designed are intended to screen for the most frequent mutations in exons 9 and 20 in PIK3CA. However, activating mutations in other exons of PIK3CA could also form a significant fraction of mutated PIK3CA samples, as shown for breast cancer.
Various AS-PCR-based approaches have been developed to detect the most common PIK3CA mutations. Thus, Board et al. combined multiplex AS-PCR and Scorpions primers for an assay suitable for identifying more than 0.1% of mutant DNA [43]. Alvarez-Garcia et al. designed an AS-PCR-based assay with internal control and phosphate blocking primers, detecting 5–10% of E542K, E545K, and H1047R mutations [21]. Patel et al. developed a multiplex assay determining 2 ng of mutant genomic DNA [44]. Zeng et al. used a PNA-PCR-based assay detecting 0.1–0.2% of mutations in exons 9 and 20 in cfDNA from CRC patients [45]. Among FDA-approved commercial kits, the therascreen PIK3CA RGQ PCR Kit detects in exons 1–7 only C420R mutation, while cobas PIK3CA Mutation Test also detects R88Q and N345K mutations. Therefore, there is still a need for a simple and robust assay to detect activating mutations in PIK3CA, as mutations in exons 1–7 are overlooked. The assay described in the present work is intended to fill this gap. It allows screening for the most frequent mutations in exons 1–7 along with classical mutations in exons 9 and 20 with a sensitivity of 0.05–0.5%, which is suitable for testing FFPE DNA samples.
It should be noted that the assay developed has several limitations. Thus, allele-specific PCR is a targeted approach intended to search only for known mutations while leaving other alterations unnoticed. Thus, AS-PCR-based tests would miss a measurable fraction of activating mutations. The need for ample DNA for the analysis is another limitation that arises when the number of assay tubes increases with the number of mutations to be analyzed. Multiplexing allows the number of AS-PCR reactions to be reduced. However, there is a tendency for the number of molecular biomarkers studied to increase, leading to a growing demand for DNA quantity. Another is the AS-PCR sensitivity in the range of one percent, which is insufficient to screen for somatic mutations in circulating tumor DNA. Alternative approaches could be applied, such as NGS to screen for novel mutations or digital PCR to detect mutations with higher sensitivity. In other words, a suitable method should be applied for each task according to the specific features of a technique and the capabilities of the diagnostic laboratory.
The PIK3CA mutation frequency observed in patients with CRC from Siberia and Far East of Russia is consistent with the previous studies. Thus, 13% of CRC samples studied were PIK3CA-mutated. Most mutations, 9.32 %, were found in exon 9, while only 1.94% were found in exon 20, with a ratio of mutations in exon 9 and 20 equal to 5:1 [6, 10, 14, 15, 24]. Mutations E542K, E545K, and H1047R were found to be the most frequent. Surprisingly, mutations R88Q, N345K, and C420R in exons 1–4 comprised 1.74% of all samples studied, almost the same as in exon 20, which is similar to a fraction of mutations in exons 1–7 in breast cancer [13]. These findings indicate the presence of a sizeable PIK3CA mutation fraction in CRC patients that was unaccounted for previously. It should be noted that despite different genetic backgrounds and mixed origins of patients from previously published data, the present study has found the overall PIK3CA mutation frequency and the ratio between mutations in exons to be similar across different populations.
5. Conclusions
This paper describes the multiplex AS-PCR-based assay for screening PIK3CA mutations in exons 1, 4, 7, 9, and 20, with a sensitivity of 0.05–0.5% of mutant DNA against wild-type DNA. The assay designed was validated on 515 CRC samples of patients from Siberia and the Far East of Russia, with an overall PIK3CA-mutated fraction of 13%. To conclude, this work is the first study characterizing PIK3CA mutation frequency in this region.
Declarations
Author contribution statement
Igor P. Oscorbin: Analyzed and interpreted the data; Wrote the paper.
Oguljan P. Beginyazova; Inna V. Khlistun; Darya V. Shamovskaya; Natalia A. Oskina: Performed the experiments.
Maxim L. Filipenko: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by Russian State Budget project for ICBFM SB RAS [121031300045-2].
Data availability statement
Data will be made available on request.
Declaration of interest’s statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Sepulveda A.R., Hamilton S.R., Allegra C.J., Grody W., Cushman-Vokoun A.M., Funkhouser W.K., Kopetz S.E., Lieu C., Lindor N.M., Minsky B.D., et al. Molecular biomarkers for the evaluation of colorectal cancer: guideline from the American society for clinical Pathology, college of American pathologists, association for molecular Pathology, and American society of clinical oncology. J. Mol. Diagn. 2017;19:187–225. doi: 10.1016/j.jmoldx.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu J.-M., Wang Y., Wang Y.-L., Wang Y., Liu T., Ni M., Li M.-S., Lin L., Ge F.-J., Gong C., et al. PIK3CA mutations contribute to acquired cetuximab resistance in patients with metastatic colorectal cancer. Clin. Cancer Res. 2017;23:4602–4616. doi: 10.1158/1078-0432.CCR-16-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao C., Yang Z.Y., Hu X.F., Chen Q., Tang J.L. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2012;23:1518–1525. doi: 10.1093/annonc/mdr464. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y., Shi Y., Lin J., Ye Y.-B., Wang X.-J., Chen G., Guo Z.-Q. Combined analysis of EGFR and PTEN status in patients with KRAS wild-type metastatic colorectal cancer. Medicine (Baltim.) 2015;94:e1698. doi: 10.1097/MD.0000000000001698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q., Shi Y., Zhou K., Wang L., Yan Z., Liu Y., Xu L., Zhao S., Chu H., Shi T., et al. PIK3CA mutations confer resistance to first-line chemotherapy in colorectal cancer. Cell Death Dis. 2018;9:739. doi: 10.1038/s41419-018-0776-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prenen H., De Schutter J., Jacobs B., De Roock W., Biesmans B., Claes B., Lambrechts D., Van Cutsem E., Tejpar S. PIK3CA mutations are not a major determinant of resistance to the epidermal growth factor receptor inhibitor cetuximab in metastatic colorectal cancer. Clin. Cancer Res. 2009;15:3184–3188. doi: 10.1158/1078-0432.CCR-08-2961. [DOI] [PubMed] [Google Scholar]
- 8.Lièvre A., Ouine B., Canet J., Cartier A., Amar Y., Cacheux W., Mariani O., Guimbaud R., Selves J., Lecomte T., et al. Protein biomarkers predictive for response to anti-EGFR treatment in RAS wild-type metastatic colorectal carcinoma. Br. J. Cancer. 2017;117:1819–1827. doi: 10.1038/bjc.2017.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fruman D.A., Chiu H., Hopkins B.D., Bagrodia S., Cantley L.C., Abraham R.T. The PI3K pathway in human disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cathomas G. PIK3CA in colorectal cancer. Front. Oncol. 2014;4 doi: 10.3389/fonc.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X., Xu Y., Zhou Q., Chen M., Zhang Y., Liang H., Zhao J., Zhong W., Wang M. PI3K in cancer: its structure, activation modes and role in shaping tumor microenvironment. Future Oncol. 2018;14:665–674. doi: 10.2217/fon-2017-0588. [DOI] [PubMed] [Google Scholar]
- 12.Samuels Y., Wang Z., Bardelli A., Silliman N., Ptak J., Szabo S., Yan H., Gazdar A., Powell S.M., Riggins G.J., et al. “High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 13.Martínez-Sáez O., Chic N., Pascual T., Adamo B., Vidal M., González-Farré B., Sanfeliu E., Schettini F., Conte B., Brasó-Maristany F., et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. 2020;22:45. doi: 10.1186/s13058-020-01284-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin J., Shi Y., Zhang S., Yang S. PIK3CA mutation and clinicopathological features of colorectal cancer: a systematic review and Meta-Analysis. Acta Oncol. (Madr). 2020;59:66–74. doi: 10.1080/0284186X.2019.1664764. [DOI] [PubMed] [Google Scholar]
- 15.Voutsadakis I.A. The landscape of PIK3CA mutations in colorectal cancer. Clin. Colorectal Cancer. 2021 doi: 10.1016/j.clcc.2021.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Burke J.E., Perisic O., Masson G.R., Vadas O., Williams R.L. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110α (PIK3CA) Proc. Natl. Acad. Sci. U. S. A. 2012;109:15259–15264. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burke J.E., Williams R.L. Synergy in activating class I PI3Ks. Trends Biochem. Sci. 2015;40:88–100. doi: 10.1016/j.tibs.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 18.André F., Ciruelos E., Rubovszky G., Campone M., Loibl S., Rugo H.S., Iwata H., Conte P., Mayer I.A., Kaufman B., et al. Alpelisib for PIK3CA -mutated, hormone receptor–positive advanced breast cancer. N. Engl. J. Med. 2019;380:1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 19.Cheah C.Y., Fowler N.H. Idelalisib in the management of lymphoma. Blood. 2016;128:331–336. doi: 10.1182/blood-2016-02-702761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt H., Kulasinghe A., Allcock R., Tan L., Mokany E., Kenny L., Punyadeera C. A pilot study to non-invasively track PIK3CA mutation in head and neck cancer. Diagnostics. 2018;8:79. doi: 10.3390/diagnostics8040079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alvarez-Garcia V., Bartos C., Keraite I., Trivedi U., Brennan P.M., Kersaudy-Kerhoas M., Gharbi K., Oikonomidou O., Leslie N.R. A simple and robust real-time qPCR method for the detection of PIK3CA mutations. Sci. Rep. 2018;8:4290. doi: 10.1038/s41598-018-22473-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalofonou M., Malpartida-Cardenas K., Alexandrou G., Rodriguez-Manzano J., Yu L.-S., Miscourides N., Allsopp R., Gleason K.L.T., Goddard K., Fernandez-Garcia D., et al. A novel hotspot specific isothermal amplification method for detection of the common PIK3CA p.H1047R breast cancer mutation. Sci. Rep. 2020;10:4553. doi: 10.1038/s41598-020-60852-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jauhri M., Bhatnagar A., Gupta S., Bp M., Minhas S., Shokeen Y., Aggarwal S. Prevalence and coexistence of KRAS, BRAF, PIK3CA, NRAS, TP53, and APC mutations in Indian colorectal cancer patients: next-generation sequencing–based cohort study. Tumor Biol. 2017;39 doi: 10.1177/1010428317692265. [DOI] [PubMed] [Google Scholar]
- 24.Li W., Qiu T., Dong L., Zhang F., Guo L., Ying J. Prevalence and characteristics of PIK3CA mutation in mismatch repair-deficient colorectal cancer. J. Cancer. 2020;11:3827–3833. doi: 10.7150/jca.37437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borkowska E.M., Barańska M., Kowalczyk M., Pietruszewska W. Detection of PIK3CA gene mutation in head and neck squamous cell carcinoma using droplet digital PCR and RT-qPCR. Biomolecules. 2021;11:818. doi: 10.3390/biom11060818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W.-M., Hu T.-T., Zhou L.-L., Feng Y.-M., Wang Y.-Y., Fang J. Highly sensitive detection of the PIK3CA H1047R mutation in colorectal cancer using a novel PCR-RFLP method. BMC Cancer. 2016;16:454. doi: 10.1186/s12885-016-2493-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurst C.D., Zuiverloon T.C.M., Hafner C., Zwarthoff E.C., Knowles M.A. A SNaPshot assay for the rapid and simple detection of four common hotspot codon mutations in the PIK3CA gene. BMC Res. Notes. 2009;2:66. doi: 10.1186/1756-0500-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenze D., Müller H.-H., Hummel M. Considerations for the use of formalin-fixed and paraffin-embedded tissue specimens for clonality analysis. J. Hematop. 2012;5:27–34. [Google Scholar]
- 29.Helsmoortel C., Kooy R.F., Vandeweyer G. Multiplexed high resolution melting assay for versatile sample tracking in a diagnostic and research setting. J. Mol. Diagnostics. 2016;18:32–38. doi: 10.1016/j.jmoldx.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Botezatu I.V., Nechaeva I.O., Stroganova А.М., Senderovich A.I., Kondratova V.N., Shelepov V.P., Lichtenstein A.V. Asymmetric real-time PCR and multiplex melting curve analysis with TaqMan probes for detecting PIK3CA mutations. Data Brief. 2015;5:913–917. doi: 10.1016/j.dib.2015.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marras S.A., Kramer F.R., Tyagi S. Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal. 1999;14:151–156. doi: 10.1016/s1050-3862(98)00018-7. [DOI] [PubMed] [Google Scholar]
- 32.Sint D., Raso L., Traugott M. Advances in multiplex PCR: balancing primer efficiencies and improving detection success. Methods Ecol. Evol. 2012;3:898–905. doi: 10.1111/j.2041-210X.2012.00215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emelyanova, M.A.; Amossenko, F.A.; Semyanikhina, A. V; Aliev, V.A.; Barsukov, Y.A.; Lyubchenko, L.N.; Nasedkina, T. V [Biochip detection of KRAS, BRAF, and PIK3CA somatic mutations in colorectal cancer patients]. Mol. Biol. (Mosk). 49, 617–627. [DOI] [PubMed]
- 34.Yanus G.A., Belyaeva A.V., Ivantsov A.O., Kuligina E.S., Suspitsin E.N., Mitiushkina N.V., Aleksakhina S.N., Iyevleva A.G., Zaitseva O.A., Yatsuk O.S., et al. Pattern of clinically relevant mutations in consecutive series of Russian colorectal cancer patients. Med. Oncol. 2013;30:686. doi: 10.1007/s12032-013-0686-5. [DOI] [PubMed] [Google Scholar]
- 35.Fedyanin M.Y., Strogonova A.M., Senderovich A.I., Dranko S.L., Kozlov N.A., Tryakin A.A., Sehina O.V., Elsnukaeva H.H.M., Bulanov A.A., Pokataev I.A., et al. Concordance of KRAS, NRAS, BRAF, PIK3CA mutation status between the primary tumor and metastases in patients with colorectal cancer. Malig. tumours. 2017:6–13. [Google Scholar]
- 36.Gymnopoulos M., Elsliger M.-A., Vogt P.K. Rare cancer-specific mutations in PIK3CA show gain of function. Proc. Natl. Acad. Sci. USA. 2007;104:5569–5574. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong J., Cho M., Sy M., Salgia R., Fakih M. Molecular profiling of metastatic colorectal tumors using next-generation sequencing: a single-institution experience. Oncotarget. 2017;8:42198–42213. doi: 10.18632/oncotarget.15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reggiani Bonetti L., Barresi V., Bettelli S., Caprera C., Manfredini S., Maiorana A. Analysis of KRAS, NRAS, PIK3CA, and BRAF mutational profile in poorly differentiated clusters of KRAS-mutated colon cancer. Hum. Pathol. 2017;62:91–98. doi: 10.1016/j.humpath.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 39.Kato S., Iida S., Higuchi T., Ishikawa T., Takagi Y., Yasuno M., Enomoto M., Uetake H., Sugihara K. PIK3CA mutation is predictive of poor survival in patients with colorectal cancer. Int. J. Cancer. 2007;121:1771–1778. doi: 10.1002/ijc.22890. [DOI] [PubMed] [Google Scholar]
- 40.Zhu Y.-F., Yu B.-H., Li D.-L., Ke H.-L., Guo X.-Z., Xiao X.-Y. PI3K expression and PIK3CA mutations are related to colorectal cancer metastases. World J. Gastroenterol. 2012;18:3745–3751. doi: 10.3748/wjg.v18.i28.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Müller C.I., Miller C.W., Hofmann W.-K., Gross M.E., Walsh C.S., Kawamata N., Luong Q.T., Koeffler H.P. Rare mutations of the PIK3CA gene in malignancies of the hematopoietic system as well as endometrium, ovary, prostate and osteosarcomas, and discovery of a PIK3CA pseudogene. Leuk. Res. 2007;31:27–32. doi: 10.1016/j.leukres.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 42.Li B., Kadura I., Fu D.-J., Watson D.E. Genotyping with TaqMAMA. Genomics. 2004;83:311–320. doi: 10.1016/j.ygeno.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Board R.E., Thelwell N.J., Ravetto P.F., Little S., Ranson M., Dive C., Hughes A., Whitcombe D. Multiplexed assays for detection of mutations in PIK3CA. Clin. Chem. 2008;54:757–760. doi: 10.1373/clinchem.2007.098376. [DOI] [PubMed] [Google Scholar]
- 44.Patel R., Tsan A., Tam R., Desai R., Spoerke J., Schoenbrunner N., Myers T.W., Bauer K., Smith E., Raja R. Mutation scanning using MUT-MAP, a high-throughput, microfluidic chip-based, multi-analyte panel. PLoS One. 2012;7 doi: 10.1371/journal.pone.0051153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng Q., Xie L., Zhou N., Liu M., Song X. Detection of PIK3CA mutations in plasma DNA of colorectal cancer patients by an ultra-sensitive PNA-mediated PCR. Mol. Diagn. Ther. 2017;21:443–451. doi: 10.1007/s40291-017-0269-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.