Summary
Mutations and defects in nuclear lamins can cause major pathologies, including inflammation and inflammatory diseases. Yet, the underlying molecular mechanisms are not known. We now report that the pro-inflammatory activation of macrophages, as induced by LPS or pathogenic E. coli, reduces Lamin-A/C levels thereby augmenting pro-inflammatory gene expression and cytokine secretion. We show that the activation of bone-marrow-derived macrophages (BMDMs) causes the phosphorylation and degradation of Lamin-A/C, as mediated by CDK1 and Caspase-6, respectively, necessary for upregulating IFN-β expression. Enhanced IFN-β expression subsequently increases pro-inflammatory gene expression via the IFN-β-STAT axis. Pro-inflammatory gene expression was also amplified in the complete absence of Lamin-A/C. Alternatively, pharmacological inhibition of either Lamin-A/C phosphorylation or degradation significantly downregulated pro-inflammatory gene expression, as did the targeting of IFN-β-STAT pathway members, i.e. phospho-STAT1 and phospho-STAT3. As Lamin-A/C is a previously unappreciated regulator of the pro-inflammatory macrophage response, our findings suggest novel opportunities to treat inflammatory diseases.
Subject areas: Immunology, Immune response, Cell biology, Functional aspects of cell biology
Graphical abstract

Highlights
-
•
Proinflammatory activation results in Lamin-A/C phosphorylation and degradation
-
•
Inhibiting Lamin-A/C phosphorylation & degradation reduce pro-inflammatory response
-
•
Lamin-A/C degradation regulates pro-inflammatory response via IFN-β-STAT axis
-
•
New anti-inflammatory targets, as bacterial infection causes Lamin-A/C degradation
Immunology; Immune response; Cell biology; Functional aspects of cell biology
Introduction
While the cell nucleus serves as the site of signal processing and transcription, the nuclear envelope, comprising the nuclear lamina, along with other cellular components1 regulates the signal transmission from the cytoplasm to the nucleus and the structural organization of the chromatin.2 The nuclear lamina is composed of a dense meshwork assembled from nuclear A-type lamins (Lamin-A and C), B-type lamins (Lamin-B1 and-B2) and of associated proteins located on the inner side of the nuclear envelope.3,4 Along with providing mechanical stability to the nucleus, the nuclear lamina also plays a central role in the 3-dimensional organization of chromatin, and thereby regulates gene expression profiles by modulating the activity of chromatin remodeling enzymes and transcription factors.5,6 Owing to its central role in maintaining nuclear architecture and function, it has been shown that perturbations to the nuclear lamina, such as mutations, contribute to the broad spectrum of disease manifestations in humans, collectively termed laminopathies.7 Thus, understanding the assembly and disassembly of the nuclear lamina and associated proteins, and underlying molecular mechanisms has become an important biomedical challenge.
The involvement of nuclear lamina and associated changes of the nuclear architecture in the immune response was implicated recently.8 For instance, T-cell activation results in a significant upregulation of Lamin-A, which is required for the activation of the T-cell receptor.9,10 Mutations in the Lamin-A gene have been linked to abnormal inflammatory processes in patients with progeria and laminopathy.11 Although it has been suggested that nuclear lamins may be important mediators in the coordination of immune responses,12,13,14 as the serum levels of inflammatory cytokines like IL-6 and TNF-α are increased in Lamin-A mutant mice,14 a regulatory relationship between nuclear lamins and the inflammatory state of the immune cell remains unexplored. Finding novel molecular mechanisms that control inflammatory processes to reduce chronic inflammation, and to improve on the resolution of inflammation are destined to increase the variety of therapeutic strategies and molecular targets.
When asking whether nuclear lamins might play a crucial role during the inflammatory activation of macrophages, we discovered that the pro-inflammatory activation of macrophages results in a significant reduction of Lamin-A/C mRNA and protein levels. Towards gaining mechanistic insights, we found that macrophage pro-inflammatory activation uses part of the molecular machinery involved in mitosis15,16,17 to phosphorylate Lamin-A/C by CDK1 and to degrade Lamin-A/C by Caspase-6, leading to an overall reduction of Lamin-A/C levels. During mitosis, the entire nuclear lamina gets dissolved due to the phosphorylation of Lamins, as phosphorylated Lamins cannot polymerize to form a nuclear lamina meshwork.18 However, unlike during mitosis, Lamin-A/C phosphorylation during the pro-inflammatory macrophage activation does not lead to a complete breakdown of the nuclear envelope. This decrease in Lamin-A/C levels during macrophage activation, in part by its degradation and paralleled by its downregulation, is opposite to the Lamin-A/C increase observed during T-cell activation.9,10 This suggests potential and unexplored opposing roles of Lamin-A/C levels during macrophage and T-cell inflammatory activation, which needs further explorations. Functionally, we find here that Lamin-A/C reduction also augments the pro-inflammatory gene expression and cytokine secretion.
Revealing whether nuclear lamins might play regulatory roles in pro-inflammatory macrophage activation, and perhaps also in the resolution of inflammation, is not only essential to advance our basic science knowledge but might also provide novel opportunities to develop therapeutic strategies to treat macrophage-induced inflammatory diseases. By using selective pharmacological inhibitors which are under clinical trials against kinases and enzymes responsible for Lamin-A/C phosphorylation and degradation, respectively, we asked whether Lamin-A/C reduction is necessary to stage an inflammatory response.
Despite our expanding knowledge of macrophage pro-inflammatory activation and involved molecular players,19,20,21,22 clear limitations remain as macrophage inflammatory onset and progression are tightly co-regulated by many diverse transcriptional and epigenetic processes. We found that Lamin-A/C reduction impinges on enhanced pro-inflammatory gene and cytokines expression via the augmentation of the IFN-β-STAT pathway. Lastly, as a proof-of-concept, we explored whether pro-inflammatory macrophage activation by bacterial infection also follows the same route of Lamin-A/C reduction, or not. Validating the clinical relevance of our findings, we show that the pharmaceutical inhibition of Lamin-A/C degradation can tame the inflammatory response of macrophages when exposed to heat-killed or active E. coli.
Our findings show a completely unexpected behavior, whereby macrophages, mitotic or non-mitotic, use enzymes of the mitotic and apoptotic machinery, but to promote inflammation, suggesting a fundamentally new mechanistic understanding by which the nuclear lamina regulates macrophage pro-inflammatory response. This may enable the future development of drugs for treating diseases associated with excessive secretion of pro-inflammatory cytokines and other macrophage-derived metabolites, such as chronic inflammatory diseases, by targeting Lamin-A/C phosphorylation, degradation, and associated molecular mechanisms.
Results
Macrophage pro-inflammatory activation results in lamin-A/C downregulation
To examine whether macrophage pro-inflammatory activation alters Lamin-A/C expression in vivo, we first re-examined a previously published RNA-Sequencing data set of microglia from lipopolysaccharide (LPS) injected wild-type (WT) mice, where microglia were isolated 24 h post-LPS-injection (Figure 1A).23 As expected, the pro-inflammatory genes were highly upregulated, including those encoding for cytokines (e.g., Il6, Il10, I12b, and Tnf-α), chemokines (e.g., Ccl4, Ccl3 and Ccl5) and enzymes necessary for pro-inflammatory marker synthesis (Nos2 and Ptgs2). Yet, the same data set also revealed that the mRNA expression of Lamin-A/C in these pro-inflammatory activated microglia was significantly downregulated (Figure 1A). When re-examining other previously published RNA-Sequencing datasets of in vitro LPS-activated tissue resident macrophages, we found that the same correlation holds true for macrophages from different organs in mice and humans- peritoneal,24 microglia,25 bone marrow-derived macrophages (BMDMs) from two different mouse strains,26 as well as for human monocytes27 and human alveolar macrophages28 (Figure 1A). The same holds true for LPS-activated BMDMs across species, including rat, buffalo, cow, goat, and horse29 (Figures 1A and S1) and for two different macrophage cell lines J774A.1 (mouse)30 and THP-1 (human)31 (Figure 1A). In all these described data sets, macrophages were isolated from the respective tissues, cultured according to the respective protocols, and treated with LPS for 2 to 6 h.24,25,26,27,28,29,30,31 Remarkably, the downregulation of Lamin-A/C mRNA levels occurred universally in all species, cell lines, and primary tissue-resident macrophages, without any sexual dimorphism, in vivo and in vitro (Figures 1A and S1). This suggests a previously unknown inverse-correlation between pro-inflammatory genes and Lamin-A/C mRNA expression levels. It should be noted that all of this was done on the mRNA level, and that effect on the Lamin-A/C protein level needs to be determined.
Figure 1.
Pro-inflammatory macrophage activation results in a Lamin-A/C downregulation in mice and humans, independent of the organ or sex of the donors
(A) Color coded map shows the mRNA expression levels of various pro-inflammatory genes and Lamin-A/C in microglia of LPS-injected mice,23 in LPS-treated mouse peritoneal macrophages,24 microglia,25 bone marrow-derived macrophages isolated from rodents of different species (mouse and rat29) and of different mouse strains26 (BALB/C and C57BL/6), as well as in LPS-treated human monocyte27 and alveolar macrophage.28 The same is seen in LPS-treated macrophage cell line J774.A1 (mouse)30 and THP-1 (human).31 Expression data were obtained from public repositories as cited.
(B) Scheme of the experimental setup, BMDMs were treated with LPS for different durations of time.
(C) Violin plots show the normalized levels of Lamin-A/C mRNA in Untreated (UT), and LPS-treated BMDMs as determined by qPCR. Levels were further normalized to the UT BMDM conditions to find the fold change.
(D) Scheme of experimental setup to illustrate the reversibility of the process. BMDMs were treated with LPS for 24 h for pro-inflammatory activation followed by culturing in media without LPS for 24 h (Wash out) to shed light on inflammation and resolution in macrophages.
(E) Time course quantification shows a reversal in the gene expression levels of pro-inflammatory gene IL-6 in UT, 24 h LPS-treated BMDMs, and after 24 h LPS Wash out. Data are presented as Mean ± S.E and normalized to the UT BMDM condition to find the fold change.
(F) Box plots show reversal of secreted IL-6 cytokine levels in UT, 24 h LPS-treated BMDMs, and after 24 h LPS Wash out. Levels were normalized to the UT BMDM condition to find the fold change.
(G) Time course quantification shows a reversal in the expression levels of Lamin-A/C in UT, 24 h LPS-treated BMDMs and after 24 h LPS Wash out. Data are presented as Mean ± S.E and normalized to the UT BMDM condition to find the fold change.
(H) Representative orthogonal views of nucleus in UT and LPS-treated (24 h) BMDM nuclei stained with Lamin-A/C (green) antibody. Scale Bar = 5 μm.
(I) Line-intensity profiles (corresponding to white lines in H) of Lamin-A/C in UT, and 24 h LPS-treated BMDMs.
(J) Box plots show normalized protein levels of Lamin-A/C in UT, and LPS-treated (24 h) BMDMs determined using antibodies binding to different epitopes of the Lamin-A/C protein, as shown in the cartoon. Levels were quantified by immunofluorescence and normalized to the UT BMDM condition. All the plots p-values were obtained with the two-sided Student’s t-test. In all the box plots, the boxes show 25th and 75th percentiles, the middle horizontal lines show the median, small open squares show the mean, and whiskers indicate S.D. All the experiments were independently repeated three or more times.
Downregulation of lamin-A/C mRNA expression levels correlates inversely with the expression of pro-inflammatory genes
To confirm the observed decline in the Lamin-A/C mRNA expression levels, we stimulated BMDMs with LPS and performed quantitative PCR (qPCR). As early as 3 h after LPS treatment, levels of Lamin-A/C mRNA declined by 40% and after 24 h, a steady decline to as much as 80% of gene expression was observed (Figures 1B and 1C). We further confirmed this by using LPS-treated alveolar macrophages, which also show a significant reduction in Lamin-A/C mRNA levels which inversely correlated with an upregulated pro-inflammatory gene expression (Figure S2A). Only one exception to this observed inverse correlative relationship between Lamin-A/C downregulation and the pro-inflammatory gene expression of macrophages could be identified here: for the immortalized RAW264.7 macrophage cell line, we were able to confirm that LPS activation results in an increase in Lamin-A/C mRNA level (Figure S2B) shown by qPCR, as previously shown in published studies.32,33 This is contrary to our observations made in different primary tissue-resident macrophages and in other immortalized macrophage cell lines (Figures 1A, 1C, and S1). However, this contradictory observation might not be surprising as multiple papers highlight significant cell signalling differences between primary and RAW264.7 macrophages.34,35 Altogether, our data (Figure 1C) taken together with the available literature on primary macrophages (Figures 1A and S1), thus suggest that the previously proposed role of Lamin-A/C during pro-inflammation observed in RAW264.7 macrophages33 cannot entirely be translated to primary BMDMs and consequently, as Figure 1A illustrates, to organ-specific tissue-resident macrophages in vivo and in vitro.
Despite the fundamental physiological role of the pro-inflammatory macrophage response to stage a defence against infection, the proper resolution of inflammation is essential for functional tissue repair after pathogen or injury challenge.36 Thus, one of the central causes of chronic inflammatory diseases has been attributed to macrophages remaining in a continuous pro-inflammatory phenotype state.37 To test whether Lamin-A/C levels revert to their homeostatic levels during inflammation resolution, we mimicked the resolution phase by first treating BMDMs with LPS for 24 h followed by washing out LPS and culturing BMDMs in media without LPS for another 24 h (Figure 1D). As expected, there was a sharp decline in the transcription of pro-inflammatory genes such as IL-6 (Figure 1E) and IL-6 cytokine secretion (Figure 1F), whereas the mRNA expression levels of Lamin-A/C returned to near homeostatic levels (Figure 1G) post-24 h of LPS washout. These observations further confirm the inverse correlative relationship between Lamin-A/C and pro-inflammatory gene expression.
To check whether Lamin-A/C reduction upon LPS treatment holds true at the Lamin-A/C protein levels as well, BMDMs were treated with LPS (for 24 h) and stained for Lamin-A/C (Figure 1H). Quantitative fluorescence imaging demonstrated a significant decline in the Lamin-A/C protein levels, as detected by antibody staining (Figures 1I and S3), i.e. by more than 50% upon 24 h LPS treatment (Figure 1J). To exclude the possibility that the decline of the Lamin-A/C levels is owed to the inaccessibility of epitopes due to structural changes in the lamina meshwork,38 and to validate that the observed reduction of antibody binding can be attributed to an authentic Lamin-A/C protein decline, we used multiple antibodies to stain for Lamin-A/C by targeting either its N or C termini (Figure 1J) and observed a similar and significant decline in Lamin-A/C levels.
Since Lamin-A and -C co-assemble into the dense fibrillar network of the nuclear lamina, together with the other isoforms Lamin-B1 and -B2 and other lamin-associated proteins, we asked whether the observed downregulation is specific for Lamin-A/C, or also observable for the evolutionary older Lamin-B1 and Lamin-B2 isoforms. Quantifications of the fluorescence imaging of Lamin-B1 (Figures S4A and S4B) and Lamin-B2 (Figures S4D and S4E) demonstrated that there were no significant differences in the protein levels between untreated and 24 h LPS-treated BMDMs, as detected by antibody staining (Figures S4C and S4F). We thus focused here on examining the role of Lamin-A/C degradation during the pro-inflammatory macrophage response.
Lamin-A/C protein downregulation in macrophages is accompanied by its active degradation
To distinguish between the potential effects on Lamin-A versus Lamin-C, a time course study using immunoblotting was performed that indeed showed a significant reduction in the levels of both isoforms within 6 h of LPS treatment, which was further reduced to almost ∼50% within a 24 h LPS treatment period (Figures 2A and 2B). To further strengthen the hypothesis that a reduction in Lamin-A and C levels is a general hallmark of pro-inflammatory activation, we treated BMDMs with TNF-α (another pro-inflammatory phenotype-inducing chemical agent) (Figure S5A) and observed a similar decrease of Lamin-A and C protein levels.
Figure 2.
Pro-inflammatory macrophage activation induces Lamin-A/C phosphorylation and degradation
(A) Immunoblot shows total levels of Lamin-A and Lamin-C in Untreated (UT), and LPS-treated BMDMs. α-tubulin served as a loading control.
(B) Time course quantifications of Lamin-A and Lamin-C normalized with α-tubulin over three independent immunoblotting experiments. Calculated values were further normalized to the UT BMDM condition to find the fold change. Data are presented as Mean ± S.E.
(C) Longer exposure of immunoblots against Lamin-A/C in UT, and LPS-treated BMDMs to detect lower fragmented Lamin-A/C bands (50kDa and 40kDa). α-tubulin served as a loading control.
(D) Box plots show degraded Lamin-A/C fragments (50kDa and 40kDa) normalized to α-tubulin between UT and LPS-treated BMDMs. Calculated values were further normalized to the UT BMDM condition to find the fold change.
(E) Immunoblots against Lamin-A/C in UT, LPS-treated, LPS Wash-out BMDMs to detect fragmented Lamin-A/C bands (∼50kDa and ∼40kDa). α-tubulin served as a loading control.
(F) Immunoblot shows total levels of p-(Ser22)-Lamin-A and p-(Ser22)-Lamin-C along with a phosphorylated fragmented band of Lamin-A/C (∼28kDa) in UT, and LPS-treated BMDMs. α-tubulin served as a loading control.
(G) Time course quantification of total p-(Ser22)-Lamin-A+C normalized to the total Lamin-A/C over three independent immunoblotting experiments. Calculated values were further normalized to the UT BMDM condition to find the fold change.
(H) (Top) Schematic shows the direct or indirect CDK1-mediated Lamin-A/C phosphorylation, which is known from other cells to be blocked by PP2A during the cell cycle. Also shown are the drugs used in this study to selectively inhibit the activity of the respective enzymes. (Bottom) Schematic shows Caspase-6 mediated Lamin-A/C degradation as inhibited by Z-VEID-FMK.
(I) Box plots show the lowering of pro-inflammatory gene expression (qPCR) (Left) and secreted cytokine levels (ELISA) (Right) in LPS-treated BMDMs, also treated with RO-3306 or Z-VEID-FMK to inhibit Lamin-A/C phosphorylation and degradation, respectively. Levels were normalized to the 6 h LPS-treated BMDM condition to find the fold change.
(J) Box plots show an increase in the pro-inflammatory gene expression (qPCR) (Left) and secreted cytokine levels (ELISA) (Right) in LPS-treated BMDMs, also treated with LB-100, a PP2A inhibitor, as compared to BMDMs treated with only LPS for 24 h. Levels were normalized to the 24 h LPS-treated BMDM condition to find the fold change. For all the plots p-values were obtained with the two-sided Student’s t-test. In all the box plots, the boxes show 25th and 75th percentiles, the middle horizontal lines show the median, small open squares show the mean, and whiskers indicate S.D. All the experiments were independently repeated three or more times.
Previous studies have quantified the turnover rates of lamins in quiescent fibroblasts, uncovering that around 10% of Lamin-A/C is replaced in the lamina meshwork with newly synthesized Lamin-A/C proteins within 24 h.39 Since these Lamin-A/C turnover rates are not sufficient to explain the decrease of Lamin-A/C protein levels of up to 50% in LPS-treated BMDMs, an active process to more rapidly reduce or downregulate Lamin-A/C protein levels must take place. Interestingly, higher exposed immunoblots showed a significant fragmentation of Lamin-A/C (∼50 kDa and ∼40 kDa) in 24 h LPS-treated macrophages (Figures 2C and 2D), suggesting their degradation. This also suggests a potential combinatorial effect of both in reducing the total Lamin-A/C levels during the LPS treatment (Figures 1H–1J and 2A and 2B), i.e., lower transcription (Figure 1C) and enhanced degradation of Lamin-A/C (Figures 2C and 2D). Interestingly, along with the reversal in the expression of pro-inflammatory genes and Lamin-A/C mRNA levels during inflammatory resolution (Figures 1D–1G), 24 h LPS washout reduces the levels of degraded Lamin-A/C fragments (Figure 2E). These observations suggest a restoration of the Lamin-A/C levels upon LPS withdrawal, strengthening our hypothesis of a potential role of Lamin-A/C during pro-inflammatory activation of macrophages.
Lamin-A/C degradation is preceded by lamin-A/C phosphorylation
The compelling observation that Lamin-A/C protein levels are reduced, and that Lamin-A/C gets degraded during LPS treatment motivated us to explore in more depth the events that lead to Lamin-A/C degradation.
As Lamin-A/C phosphorylation by CDK1 (at residues Serine 22 and 392) triggers Lamin-A/C depolymerization at the onset of nuclear envelope breakdown during mitosis in other cell types,40 and precedes the enzymatic cleavage of Lamin-A/C,41 mostly due to Caspase-6 enzyme activity,42 we conducted time course immunoblotting experiments with BMDMs of phospho-Serine 22 Lamin-A/C (pSer22-Lamin-A/C). Quantification of immunoblots shows a significant increase in the pSer22-Lamin-A/C/total Lamin-A/C ratio already within 1 h of LPS treatment (Figure S5B), peaking at 6 h of LPS-treatment, and then declined as the total pSer22-Lamin-A/C levels diminished over the subsequent 12 and 24 h timepoints (Figures 2F and 2G). Confocal microscopy imaging further revealed significant nucleoplasmic accumulation of pSer22-Lamin-A/C upon 6 h LPS treatment (Figures S5C and S5D), possibly as fragments, followed by a decline at 24 h of LPS treatment (Figure S5C). We also examined the levels of active CDK1 (phospho-Threonine 161 CDK1) in BMDMs during the first 6 h of LPS treatment by immunoblotting (Figure S5E). The phosphorylated CDK1 fraction increased significantly within 1 h post-LPS treatment, peaked at 2 h and then returned to homeostatic levels by the end of 3–6 h (Figures S5E and S5F), confirming significant activation of CDK1 post-LPS treatment suggesting that this increase in CDK1 activity induces Lamin-A/C phosphorylation.
Pharmacological inhibition of CDK1 and Caspase-6 reduces the expression of pro-inflammatory genes and cytokine secretion
To verify whether Lamin-A/C phosphorylation and degradation by the enzymes CDK1 and Caspase-6, respectively, is required to initiate pro-inflammatory gene expression, these two enzymes were targeted by RO-3306 (a selective ATP-competitive inhibitor of CDK1)43 and Z-VEID-FMK (a selective irreversible inhibitor of Caspase-6),42 respectively (Figure 2H). RO-3306 is one of the many cyclin-dependent kinase inhibitors, which have been identified as potential anti-inflammatory agents44 and are in human phase-II clinical trials to treat adjunctive therapy for rheumatoid arthritis.45 Z-VEID-FMK is one of the multiple caspase inhibitors which have been tested in various preclinical studies for the treatment of cell death related pathologies.46 Immunoblotting of LPS and either RO-3306 or Z-VEID-FMK treated BMDMs indeed confirmed the efficacy of RO-3306 and Z-VEID-FMK in inhibiting either Lamin-A/C phosphorylation (Figure S6A) or degradation (Figure S6B), respectively. Strikingly, inhibiting either Lamin-A/C phosphorylation or degradation, with RO-3306 or Z-VEID-FMK, respectively, significantly decreased IL-6 and TNF-α mRNA expression, as well as IL-6 and TNF-α cytokine secretion upon pro-inflammatory activation (Figure 2I). Interestingly, CDK1 inhibition had a stronger effect than Caspase-6 inhibition. This suggests for the first time, to the best of our knowledge, that CDK1 and Caspase-6 inhibition prevents Lamin-A/C degradation, and with this pro-inflammatory gene expression. Suggesting that the staging of an efficient pro-inflammatory response of BMDMs thus requires Lamin-A/C degradation.
CDK1 and Caspase-6 inhibition has little effect on pro-inflammatory gene expression in lamin-A/C knockout bone-marrow-derived macrophages
Since pharmacological inhibitors can have unanticipated side and off-target effects, we used a mouse model with a deletion in the Lamin-A gene47 to assess the transcriptional response of pro-inflammatory genes in the absence and presence of LPS, as well as in the presence of the inhibitors RO-3306 or Z-VEID-FMK.
In Lamin-A/C knockout BMDMs (here after referred as KO-BMDMs), the absence of Lamin-A/C post-isolation from bone marrow was confirmed here by immunoblotting (Figure S7A). We also validated that KO-BMDMs differentiated and polarized in the same manner as WT-BMDMs, which are known to polarize upon LPS treatment towards M1 phenotype, which is characterized by a significant increase in cell and nuclear spreading areas along with higher cell circularity.48 The insignificant cellular or nuclear morphological differences between WT- and KO-BMDMs before and after LPS treatment (Figures S7B–S7G) confirmed that the KO-BMDMs differentiated normally into functional macrophages. However, KO-BMDMs showed marginally higher cell circularity (Figure S7F). It is also important to note that Lamin-B1 and Lamin-B2 levels remain unaltered in all the KO-BMDMs.47
Asking how pro-inflammatory processes proceed even in the absence of Lamin-A/C by the use of these KO-BMDMs circumvents potential off target effects of inhibitors like RO-3306 and Z-VEID-FMK, as RO-3306 might inhibit other CDKs in vitro besides CDK1, and Z-VEID-FMK might inhibit other caspases potentially involved in the pro-inflammatory response. To test our hypothesis further, KO-BMDMs were treated with LPS in the presence or absence of RO-3306. In contrast to WT-BMDMs (Figure 2I), an insignificant downregulation was observed in pro-inflammatory gene expression (IL-6) in LPS-treated KO-BMDMs (Figure S6C). Z-VEID-FMK treatment resulted in only a mild upregulation of pro-inflammatory gene expression (Figure S6C) in LPS-treated KO-BMDMs.
Conversely, to increase Lamin-A/C phosphorylation which can be accomplished by keeping CDK1 constitutively active, we restrained protein phosphatase 2A (PP2A) activity (Figure 2H)49 by using LB-100, a PP2A selective competitive inhibitor,50 also known to have anti-cancer activity.51 PP2A is a CDK1 counteracting phosphatase, which restrains CDK1 activity during e.g. interphase49 by dephosphorylation. Indeed, IL-6 and TNF-α mRNA expression levels were significantly increased, as well as the IL-6 and TNF-α cytokine secretion levels when treating with LPS and LB-100 (Figure 2J). However, IL-6 mRNA expression was solely increased by 2-fold in LPS and LB-100 treated KO-BMDMs compared to solely LPS-treated KO-BMDMs (Figure S6D), while in WT-BMDMs IL-6 mRNA expression increases by 20-fold in LPS and LB-100 treated WT-BMDMs compared to solely LPS-treated WT-BMDMs (Figure 2J). TNF-α was insignificantly increased upon LPS and LB-100 treatment in KO-BMDMs (Figures S6D and S2J), whereas in WT-BMDMs TNF-α mRNA expression increased by 2-fold in LPS and LB-100 treated WT-BMDMs compared to solely LPS-treated WT-BMDMs (Figure 2J). Taken together, these data suggest that CDK1 and Caspase-6 inhibition, and with this Lamin-A/C phosphorylation and degradation, exerts a previously undescribed role in increasing the pro-inflammatory macrophage response. Thereby, also ruling out the dominant negative effect of a truncated Lamin-A/C, which has been found in the Lamin-A/C knockout mice used in this study.52 The potential regulatory mechanism of Lamin-A/C degradation regulated pro-inflammatory macrophage response was probed next.
Lamin-A/C reduction in wild-type bone-marrow-derived macrophages upregulates pro-inflammatory genomic programs mostly belonging to the interferon-beta pathway, as enhanced in lamin-A/C knockout bone-marrow-derived macrophages
Along with IL-6 and TNF-α, macrophage pro-inflammatory activation leads to the upregulation of different pro-inflammatory gene clusters.22 To check if either Lamin-A/C reduction upon LPS treatment, or its elimination has an influence on these other clusters, we performed RNA-Sequencing experiments using WT- and KO-BMDMs (Figure 3A).
Figure 3.
Lamin-A/C knockout augments pro-inflammatory genomic programs
(A) Scheme shows the experimental RNA-Sequencing and ELISA setup between wild-type (WT) and Lamin-A/C knockout (KO) BMDMs.
(B) Heatmap shows the top 25 upregulated and downregulated genes between WT- and KO- BMDMs (Fold change>2 and p-value<0.05).
(C) Heatmap shows the top 25 upregulated and downregulated genes between LPS-treated (6 h) WT- and KO-BMDMs (Fold change>2 and p-value<0.05).
(D) Heat map shows the levels of selected pro-inflammatory genes upregulated in LPS-treated KO-BMDMs as compared to LPS-treated WT-BMDMs.
(E) Box plots show the differences in secreted cytokine levels of TNF-α and IL-6 between WT vs. WT + LPS, and WT + LPS vs. KO + LPS-treated BMDMs. Levels were normalized to the untreated WT-BMDM condition to find the fold change.
(F) Venn-diagram shows the number of overlapped genes (Fold change>2 and p < 0.05) between LPS induced genes in WT-BMDMs and genes which get further induced (241) or repressed (160) in LPS-treated KO-BMDMs.
(G) Tables show Gene Ontology analysis of different gene clusters.
(H) Venn-diagram shows the number of overlapped genes (Fold change>2 and p < 0.05) between LPS repressed genes in WT-BMDMs and genes which further get repressed (140) or induced (153) in LPS-treated KO-BMDMs.
(I) Venn-diagram shows the overlapped genes (Fold change>2 and p < 0.05) between LPS-induced and Lamin-A/C dependent (Cluster-I) and Interferon-β (IFN-β) dependent genes.
(J) Scheme shows the experimental setup of RNA-Sequencing and data analysis to used to identify how Lamin-A/C reduction influenced pro-inflammatory transcription factors. For all the plots, p values were obtained with the two-sided Student’s t-test. In all the box plots, the boxes show 25th and 75th percentiles, the middle horizontal lines show the median, small open squares show the mean, and whiskers indicate S.D. Triplicate samples were used for RNA-Sequencing. All the other experiments were independently repeated two or more times.
Unsupervised cluster analysis segregated WT- and KO-BMDMs and revealed prominent differences between their transcriptomes at baseline (Figure S8A) and in response to 6 h LPS stimulation (Figure S8B). Figure 3B shows top 25 up- and down-regulated genes between WT- and KO-BMDMs, suggesting that KO-BMDMs already have higher expression of pro-inflammatory genes like Ccl8 as compared to WT-BMDMs (Figure 3B). This clearly suggests that pro-inflammatory genomic programs are potentiated in KO-BMDMs. Gene ontology (GO) analysis of WT- vs. KO-BMDMs also suggested that upregulated functional clusters in KO-BMDMs largely belong to the category of inflammation, whereas downregulated functional clusters largely belong to the category of cell cycle and DNA damage and repair (Figure S9A).
While looking at differences in LPS-treated WT- versus KO-BMDMs, checking the status of the Ccl8 gene suggested that Lamin-A/C reduction also augments the LPS response in KO-BMDMs (Figure 3C). Further analysis of all the genes upregulated in LPS-treated KO-BMDMs vs. LPS-treated WT-BMDMs revealed several critical and well-characterized pro-inflammatory gene clusters (Figure 3D). These include chemokine related (e.g. Cxcl1, Cxcl10), Interferon related (e.g. Ifit1, 2, and 3), Interleukins (e.g. Il-6, Il-1a), NF-κB related (e.g. NFκb2, NFκbiz) and TNF related (e.g. TNF-α) genes (Figure 3D). These pro-inflammatory genes were not only upregulated at the mRNA levels, but also upregulated at their cytokine secretion levels in LPS-treated KO-BMDMs (Figures 3D and 3E).
To fully characterize these up- and down- regulated genes, and the associated signaling pathways and biological functions, we further analyzed the RNA-Sequencing data. RNA-Sequencing data sets revealed that 1518 genes were upregulated upon the LPS treatment of WT-BMDMs for 6 h, the expression of several of these genes was also dependent on Lamin-A/C reduction (Figure 3F). 241 genes (Cluster-I) were further upregulated in LPS-treated KO-BMDMs, whereas 160 genes (Cluster-II) were downregulated in LPS-treated KO-BMDMs (Figure 3F). GO analysis of Cluster-I genes revealed clusters involved in the pro-inflammatory response, innate immune response, response to LPS, response to IFN-β, Nitric Oxide synthesis, and IL-6 productions, and several other critical inflammation-related clusters (Figure 3G). In contrast, genes in Cluster-II only belonged to the immune response and cellular proliferation (Figure 3G). GO analysis revealed that genes exclusively upregulated in LPS-treated KO-BMDMs (Cluster-III) did not belong to any significant processes, whereas genes exclusively downregulated in LPS-treated KO-BMDMs (Cluster-IV) belonged to apoptotic pathways (Figure 3G). Contrary to this, 1820 genes were downregulated upon LPS treatment of WT-BMDMs for 6 h (Figure 3H). Several of these genes were further either downregulated (Cluster-V) or upregulated (Cluster-VI) in LPS-treated KO-BMDMs (Figure 3H). GO analysis of Cluster-V revealed processes mainly involved in the cell cycle, whereas Cluster-VI did not belong to any major biological processes. GO analysis further revealed that genes in cluster Cluster-VII (exclusively downregulated in KO- LPS-treated BMDMs) and Cluster-VIII (exclusively upregulated in LPS-treated KO-BMDMs) did not belong to any major biological processes (Figure 3G). To understand the association of pathways and genes, that were upregulated in LPS-treated KO-BMDMs vs. LPS-treated WT-BMDMs, KEGG pathway analysis was performed. Gene sets mainly associated with TNF-α and NF-κB signaling, two central pathways involved in macrophage pro-inflammatory activation (Figure S9B), were upregulated when comparing LPS-treated KO-BMDMs vs. LPS-treated WT-BMDMs, whereas the downregulated gene sets were mainly associated with the MAP-Kinase signaling pathway (Figure S9B).
Cluster-I is particularly interesting (Figures 3F and G), as it is the dominant gene cluster involved in regulating the pro-inflammatory response of BMDMs, and at the same time is seen to be upregulated by Lamin-A/C reduction or knockout (Figure 3I). Most importantly and relevant to the pro-inflammatory macrophages response, a total of 168 genes (70%) in Cluster-I are regulated by the Interferon-beta (IFN-β) pathway (Figure 3I) and are therefore under the control of similar transcriptional regulation and factors (Figure 3J). These results suggest that a reduction or complete deletion of Lamin-A/C significantly upregulates IFN-β related pro-inflammatory gene expression. Lamin-A/C reduction might drive the pro-inflammatory gene expression via the IFN-β pathway, which we tested next.
Augmentation of the IFN-β-STAT axis in lipopolysaccharide activated lamin-A/C knockout bone-marrow-derived macrophages
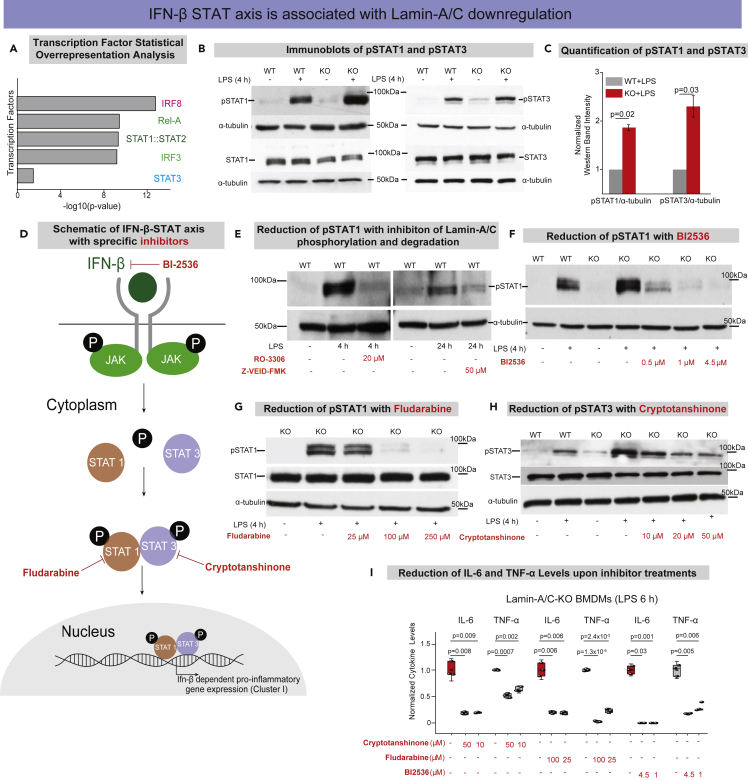
To understand how Lamin-A/C reduction might regulate changes in pro-inflammatory gene expression, we subsequently focused on identifying associated transcription factors. Therefore, the promoters of 168 Lamin-A/C- and IFN-β- dependent genes were subjected to a statistical overrepresentation analysis to detect enriched transcription factor binding sites (Figure 3J). The analysis showed that the most overrepresented matrices corresponded to IRFs, p65, and STATs sites (Figures 4A, S10A, and S10B). This result gave a crucial hint that IRFs, NF-κB, and STATs are potential transcription factors that might be influenced by Lamin-A/C degradation or deletion, which was probed in subsequent steps.
Figure 4.
Degradation or knockout of Lamin-A/C augments the IFN-β-STAT axis during pro-inflammatory activation
(A) Bar graphs show transcription factors with corresponding -log10(p-value) found by statistical overrepresentation analysis to detect enriched transcription factor binding sites in the promoter of the 168 genes (LPS induced, and Lamin-A/C influenced Interferon-Beta dependent Genes).
(B) Immunoblots show p-(Tyr701)-STAT1 (upper) and total-STAT1 (lower) (Left), and p-(Tyr705)-STAT3 (upper), and total-STAT3 (lower) (Right) levels in wild-type (WT), WT + LPS, Lamin-A/C knockout (KO), and KO + LPS-treated BMDMs.
(C) Bar graphs show the quantification of pSTAT1 and pSTAT3 levels normalized with α-tubulin. Calculated values were further normalized to the WT + LPS-treated BMDM condition over three independent experiments. Data are presented as Mean ± S.E.
(D) Scheme shows the IFN-β-STAT axis and how the selective inhibitors used in this study inhibit the phosphorylation of different STATs and thus inhibit pro-inflammatory gene expression.
(E) Immunoblots show pSTAT1 levels in WT, WT + LPS-treated BMDMs with RO-3306 and Z-VEID-FMK.
(F) Immunoblot shows pSTAT1 levels in WT, WT + LPS, KO, KO + LPS and KO + LPS-treated BMDMs with different concentrations of BI-2536.
(G) Immunoblot shows pSTAT1 and total STAT1 levels in KO, KO + LPS and KO + LPS-treated BMDMs with different concentrations of Fludarabine.
(H) Immunoblot shows pSTAT3 and total STAT3 levels in WT, WT + LPS, KO, KO + LPS, and KO + LPS-treated BMDMs with different concentrations of Cryptotanshinone.
(I) Box plots show the lowering of secreted cytokine levels (IL-6 and TNF-α) in LPS-treated KO-BMDMs, treated with multiple concentration of Cryptotanshinone, Fludarabine, and BI-2536, as compared to only LPS-treated KO-BMDMs. Levels were normalized to the 6 h LPS-treated BMDM condition to find the fold change. In all the Immunoblots, α-tubulin served as loading control. In all the box plots, boxes show 25th and 75th percentiles, the middle horizontal lines show the median, small open squares show the mean, and whiskers indicate S.D. For all the plots p-values were obtained with the two-sided Student’s t-test. All experiments were independently repeated three or more times.
As a majority of the pro-inflammatory genes, whose upregulation correlates with Lamin-A/C reduction or knockout, is expected to be under the regulation of the STAT family of transcription factors (Figures 4A, S10A&B), we first probed the phosphorylation of STAT1 and STAT3.53 We could indeed confirm that the differential activity of the STAT family of transcription factors underlies the observed upregulation of pro-inflammatory genes. As expected, levels of pSTAT1(Tyr701) and pSTAT3(Tyr705) are significantly increased upon LPS treatment in WT-BMDMs, and these levels were further augmented in LPS-treated KO-BMDMs (Figures 4B and 4C). Hence, STAT1/3 phosphorylation is modulated by Lamin-A/C reduction, but not the overall STAT1 or STAT3 levels (Figure 4B). To probe whether Lamin-A/C reduction not only affects the phosphorylation status of STAT transcription factors, but also their recruitment to the promoter region of inflammatory genes, ChIP-qPCR experiments were conducted. Higher enrichment of pSTAT1 at the promoter regions of IL-6 and TNF-α genes in LPS-treated KO-BMDMs (Figure S11A) confirmed higher transcriptional activity of pSTAT1 in LPS-treated KO-BMDMs compared to WT-BMDMs, using IgG as control (Figure S11B). To confirm that the STAT phosphorylation is indeed dependent on Lamin-A/C reduction, we show that the inhibition of Lamin-A/C phosphorylation and degradation with the clinical pharmaceutical drugs RO-3306 and Z-VEID-FMK, respectively, could indeed reduce the LPS induced increase in STAT1 phosphorylation during the macrophage inflammatory response (Figure 4E). This gives first hints that the pro-inflammatory response of macrophages can indeed be tuned down by targeting Lamin-A/C degradation.
To provide a potential mechanism of how the reduction of a nuclear envelope protein, i.e., Lamin-A/C might drive the phosphorylation of cytoplasmic transcription factors, such as members of the STAT family, we checked the levels of IFN-β and Janus Kinase (JAK). Upon LPS activation, it is well known that macrophages upregulate the expression levels of Type I interferons, such as interferon alpha and beta (IFN-α and -β)54 (Figures 4D and S12A). Secreted IFNs bind their receptors, subsequently in response to IFNs binding to their receptors, JAKs get phosphorylated and in turn phosphorylate different STATs, typically STAT1, 2 & 3, which then stimulate pro-inflammatory gene expression54 (Figures 4D and S12A). Thus, we checked whether the reduction in Lamin-A/C levels might influence the expression levels of IFN-β, or not. We confirmed a higher expression of IFN-β mRNA in LPS-treated KO-BMDMs compared to WT-BMDMs (Figures S12B and S12C). At the same time, we showed that the expression of IFN-β mRNA correlates with Lamin-A/C phosphorylation, as WT-BMDMs treated with LPS in the presence of, the CDK1 inhibitor, RO-3306 showed significantly lower levels of IFN-β mRNA compared to only LPS-treated WT-BMDMs (Figure S12D). This higher mRNA expression of IFN-β in KO-BMDMs is expected to lead to higher secretion of IFN-β into the medium, which in turn should activate the IFN-β-STAT-axis.
Therefore, we hypothesized that Lamin-A/C phosphorylation and degradation might enhance IFN-β expression, and with this the pro-inflammatory gene expression. To further confirm this, we pretreated LPS stimulated WT-BMDMs with RO-3306 to block Lamin-A/C degradation, and subsequently supplemented the cell medium with IFN-β (Figure S12E), to compensate for the Lamin-A/C degradation-induced reduction of IFN-β expression. We found that IL-6 mRNA expression levels were indeed decreased upon RO-3306 treatment, as expected (Figures 2I and S12E). Upon subsequent IFN-β supplementation, however, the IL-6 mRNA expression levels downstream of IFN-β-STAT-axis were significantly increased (Figure S12E). To further strengthen this hypothesis, we confirmed higher phosphorylation of JAK1 (Tyr1034/1035) in LPS-treated KO BMDMs as compared to LPS-treated WT-BMDMs (Figure S13A). Altogether, this provides a plausible mechanism of how Lamin-A/C degradation might regulate STAT1/3 activity, i.e. via increased IFN-β expression, thereby increasing JAK activity and subsequently the downstream phosphorylation of STAT1/3.
To exclude that the amplified transcriptional responses in the KO-BMDMs do not reflect an increased hypersensitivity to LPS, but are primarily mediated by the increased phosphorylation of STAT1/3, we also checked the levels of other crucial transcription factors (pSTAT5 (Tyr694), pIRF3 (Ser396), and p65), as activated during the LPS treatment of macrophages, and found them to remained unchanged (Figures S13B–S13D).
Pharmaceutical inhibition of the IFN-β-STAT axis reverses the enhanced lipopolysaccharide response in lamin-A/C KO-bone-marrow-derived macrophages
To further test whether the enhanced LPS response in KO-BMDMs could be reversed by the pharmaceutical inhibition of STAT pathway members, KO-BMDMs were treated with inhibitors to suppress the expression and activity of IFN-β and phosphorylation of the STAT family of transcription factors (Figures 4D and 4F–4I). We thereby took advantage of BI-2536, a selective drug to reduce IFN-β levels by inhibiting its transcription55 and which is in clinical trials against various cancers.56,57 Indeed, BI-2536 treatment was able to reduce the levels of pSTAT1 in LPS-treated KO-BMDMs in a concentration-dependent manner, finally to similar levels as only LPS-treated WT-BMDMs (Figure 4F). BI-2536 inhibition also resulted in reduced mRNA levels of pro-inflammatory genes IL-6 and TNF-α (Figure S14), along with secreted IL-6 and TNF-α cytokines in LPS-treated KO-BMDMs (Figure 4I). To confirm this also for STAT1, we used Fludarabine, a clinically approved specific inhibitor of pSTAT1,58 also used clinically to treat chronic lymphocytic leukemia (CLL)59 and non-Hodgkins lymphoma (NHL).60 Fludarabine treatment was able to counteract the hyper-upregulation of pSTAT1 levels in LPS-treated KO-BMDMs (Figure 4G). When selectively blocking STAT3 phosphorylation using the drug Cryptotanshinone,61 which is known for its anticancer activities,61 the normal activation levels of pSTAT3 in LPS-treated KO-BMDMs were restored (Figure 4H). Upon different drug treatments, levels of total STAT1 and STAT3, however. remained largely similar (Figures 4G and 4H). Lastly, using these drugs in LPS-treated KO-BMDMs, we could significantly reduce the levels of pro-inflammatory cytokines (IL-6 and TNF-α) in LPS-treated KO-BMDMs (Figure 4I). Altogether, these observations suggest that indeed KO-BMDMs have a hyper-activated IFN-β pathway and that the IFN-β-STAT axis can be targeted by inhibiting Lamin-A/C phosphorylation and degradation.
Inhibiting lamin-A/C phosphorylation downregulates the inflammatory response of E. coli infected macrophages
Gram-negative bacteria, such as E. coli, activate macrophages due to the presence of LPS on their outer cell membrane. To validate that the mechanism described in this study is independent of whether macrophages get exposed to LPS in soluble form or exposed to gram-negative bacteria, we asked whether bacterial infection of macrophages also prompts the activation of inflammatory programs via the above-described Lamin-A/C phosphorylation and degradation route. Therefore, BMDMs were exposed to heat-inactivated killed E. coli or active living E. coli and the expression status of Lamin-A/C was monitored 6 h post-infection (Figure 5A). Again, exposure to E. coli resulted in a significant reduction of Lamin-A/C mRNA levels within the first 6 h of exposure (Figure 5B). This observation was accompanied by a significant Lamin-A/C degradation post-24 h heat-inactivated killed or active living E. coli infection (Figure 5C). This confirms that LPS induced degradation of Lamin-A/C is inherent to macrophage activation by soluble LPS or cell wall bound LPS of gram-negative bacteria.
Figure 5.
Inhibiting Lamin-A/C phosphorylation downregulates the inflammatory response of E. coli infected macrophages
(A) Scheme shows the experimental setup: wild-type BMDMs were infected with either heat inactivated (HI) or active (Act) E. coli.
(B) Box plot shows mRNA levels of Lamin-A/C in uninfected (UI) or E. coli infected BMDMs (HI or Act) as determined by qPCR. Levels were normalized to UI BMDMs condition to find the fold change.
(C) Immunoblot shows Lamin-A/C degradation in E. coli infected BMDMs (HI or Act) as compared to UI BMDMs. α-tubulin served as a loading control.
(D) Scheme shows the experimental setup: wild-type BMDMs were infected with E. coli (HI or Act) and treated with RO-3306.
(E) Box plots show normalized mRNA levels of IL-6 and TNF-α in UI and E. coli infected BMDMs (HI or Act) also treated with RO-3306. Values were further normalized to the UI BMDM condition to find the fold change.
(F) Box plots show normalized secreted IL-6 and TNF-α cytokine levels in UI and E. coli infected BMDMs (HI or Act) also treated with RO-3306. Values were normalized to the UI BMDM condition to find the fold change.
(G) Immunoblots show pSTAT1 and pSTAT3 in BMDMs infected with E. coli (HI or Act) and treated with RO-3306 as compared to only E. coli infected BMDMs (HI or Act). α-tubulin served as loading control.
(H) Schematic overview shows how pro-inflammatory activation by LPS or infection with E. coli of primary bone-marrow derived macrophages results in the phosphorylation and degradation of Lamin-A/C and its regulatory role in the pro-inflammatory gene expression and cytokine secretion via the IFN-β-STAT axis. In all the box plots, boxes show 25th and 75th percentiles, the middle horizontal lines show the median, small open squares show the mean, and whiskers indicate S.D. p values were obtained with the two-sided Student’s t-test. All experiments were independently repeated three or more times.
Towards therapeutic applications, we finally asked whether the pro-inflammatory response of macrophages upon E. coli infection could also be scaled down by inhibiting Lamin-A/C phosphorylation with the pharmaceutical inhibitor RO-3306. Infecting BMDMs with heat-inactivated killed E. coli, or with active living E. coli, while inhibiting Lamin-A/C phosphorylation by RO-3306 pretreatment (Figures 2H and 5D), not only decreased the expression of pro-inflammatory genes (IL-6, and TNF-α) (Figure 5E), but also the production and secretion of inflammatory cytokines (IL-6, and TNF-α) (Figure 5F). Also, E. coli induced phosphorylation of STAT1 and STAT3, as required for pro-inflammatory gene expression and cytokine synthesis can be completely blocked by inhibiting CDK1 and Caspase-6 and with this Lamin-A/C phosphorylation and degradation (Figure 5G). Taken together, we discovered here a new molecular route by which the cytokine production and secretion induced by gram-negative bacteria infections of macrophages might be reduced.
Discussion and outlook
As chronic inflammatory diseases are thought to be stimulated by the excessive secretion of pro-inflammatory cytokines and of other macrophage-derived metabolites, the discovery of additional potent regulators that control this excessive secretion is thus destined to initiate the research and development of novel therapeutic strategies and molecular targets. It had not been reported before that the degradation of nuclear lamins are potential regulators of pro-inflammatory macrophage response, including the subsequent secretion of inflammatory stimulants. Our data now suggest that Lamin-A/C phosphorylation and degradation are a prerequisite for the pro-inflammatory response of macrophages (Figures 2 and 5). Inflammation could indeed be reduced by pharmaceutical inhibition of either the Lamin-A/C degradation by either targeting CDK1 or Caspase-6 (Figures 2 and 5), or by inhibiting members of the IFN-β-STAT axis, particularly pSTAT1 or pSTAT3 (Figure 4). Several human diseases and pathological conditions, including muscular dystrophy and human inflammageing, have been shown to be associated with mutations or low levels of Lamin-A/C along with higher levels of inflammatory metabolites circulating in the blood. Here, we discovered novel Lamin-A/C regulated events that modulate the pro-inflammatory response of macrophages, as discussed sequentially below (Figure 5H).
First, we here discovered that proinflammatory macrophage activation correlates with reduced expression of Lamin-A/C at the mRNA level across species (Figure 1). However, Lamin-A/C is not only reduced at the expression level, but Lamin-A/C gets actively degraded upon LPS activation by phosphorylation and subsequent degradation, mediated by CDK1 and Caspase-6, respectively (Figure 5H). These events ultimately lead to a subsequent upregulation of pro-inflammatory gene expression and cytokine secretion (Figure 2). Even though targeting CDKs has previously been shown to inhibit LPS induced activation of macrophages,62 the molecular events leading to the inhibitory effect were not known. Our pharmaceutical inhibition studies provide insights into the regulatory mechanisms, suggesting that CDK1 induced Lamin-A/C phosphorylation subsequently triggers Lamin-A/C protein degradation. As Lamin-A/C phosphorylation has recently been implicated in accelerated age-associated pathologies like progeria,63 with potential connections to inflammatory gene expression, the actual role of Lamin-A/C has remained an unsolved mystery. Our data also shed light into another unsolved question, namely why Caspase-6 deficient mice show reduced TNF-α levels in plasma upon LPS-injection.64 This question was raised as Caspase-6 is classified as an effector or executioner caspase, based on its pro-apoptotic activity in cleaving other caspases and structural proteins like Lamin-A/C.65,66,67 With our inhibitory experiments we now uncover the missing link suggesting that the degradation of Lamin-A/C by Caspase-6 controls pro-inflammatory gene expression and cytokine secretion. Yet, it is still unclear how Lamin-A/C downregulation and degradation are mechanistically induced upon pro-inflammatory activation. Foremost, the downregulation of Lamin-A/C mRNA expression might be due to alterations in the retinoic acid signaling pathways, with retinoic acid (RA) functioning as a ligand for nuclear RA receptors (RARs). RA-RARs are crucial for the transcription of key developmental genes and it has been shown that RA treatment represses Lamin-A/C transcription in adult granulocyte differentiation68 and in mesenchymal stem cells.69
Second, while the IFN-β-activated STAT family of transcription factors is known to drive the expression of pro-inflammatory genes, we discovered here that the Lamin-A/C phosphorylation and degradation upon pro-inflammatory activation of macrophages augments the phosphorylation of STAT1 and STAT3, but not of STAT5 (Figure 4). We found that Lamin-A/C degradation leads to enhanced IFN-β expression. It was shown recently that Emerin, another nuclear envelope protein bound to Lamin-A/C, binds STAT3 and by this has an impact on STAT3 retention to the nuclear membrane. Since Lamin-A/C knockout is known to inhibit Emerin localization to the nuclear envelope,47 we would like to suggest that Lamin-A/C degradation might further regulate the spatio-temporal distribution of STAT3 in the cell nucleus and thus the activity of STAT3. However, further studies are required to clarify whether and how other nuclear envelope proteins might regulate the IFN-β-STAT axis during the pro-inflammatory response. With the identified key players here, we hope that our findings stimulate many further explorations of the underpinning mechanisms and of potential therapeutic innovations. Yet, not only transcription factors might be retained in the nuclear lamina meshwork but also other factors such as histone methyltransferases or histone acetyltransferases. As histone modifications also co-regulate the transcriptional machinery and since a link has been established between WDR5 (a member of the COMPASS-complex family of histone methyltransferases) and Lamin-A/C70 and HDAC2 and Lamin-A/C,71 nuclear lamina proteins might regulate the pro-inflammatory gene expression via alterations of the epigenetic landscape. However, the mechanism of how Lamin-A/C is downregulated and degraded upon pro-inflammatory activation and how this leads to the full pro-inflammatory response needs further investigation.
Finally, data obtained from our study advocate the efficacy of targeting nuclear lamina proteins and associated molecular mechanism to protect macrophages from virulent E. coli infection-induced inflammation. Moreover, the ability of these targets to tame the pro-inflammatory macrophage response could be attributed to their ability to downregulate the TLR4 and STAT pathways (Figure 5). Therefore, targeting nuclear lamina could be an ideal candidate immunomodulator and anti-inflammatory agent against diseases caused by E. coli infections. However, further studies using animal infection models will be needed to warrant the ex vivo and in vivo efficacy of these targets. Especially, the development of animal infection models using other pathogens than E. coli would further broaden our discovery, as already in tuberculosis-infected mice Lamin-A/C mRNA levels are significantly downregulated in alveolar macrophages.72
Towards the clinical significance of our findings, we confirmed that several different tissue-resident macrophages, and not only BMDMs, exhibit reduced Lamin-A/C mRNA levels upon LPS activation or in chronically inflamed tissues (Figure 1), suggesting a major yet generalized role of the nuclear lamina during macrophage activation. These findings indicate that the nuclear lamina and associated proteins might be able to serve as novel therapeutic targets to reduce inflammation in multiple tissues across several acute or chronic inflammatory diseases. The generalization of these findings, showing that inflammation finally results in a reduced Lamin-A/C expression also in other primary macrophages of rodents, humans, and other species (Figure 1), is essential for the exploitation of this knowledge for clinical applications. Therefore, targeting the nuclear lamina could be a novel strategy to immunomodulate the inflammatory response caused either by bacterial infections, as shown here with E. coli infection, or by chronic inflammatory processes. We demonstrate here that the pro-inflammatory response of macrophages can be reduced by targeting the enzymes involved in Lamin-A/C degradation, CDK1, and Caspase-6 (Figures 2 and 5). The ability of these targets to tame the pro-inflammatory macrophage response are suggested here to be attributed to their ability to downregulate the expression of IFN-β, and thereby potentially decreasing the autocrine and paracrine signaling leading to the activation of the IFN-β-STAT axis. Exploiting the ability of these targets to downregulate the STAT pathways either directly, or by downregulating IFN-β expression (Figure 4), provides new opportunities to develop immunomodulator and anti-inflammatory agents to cope with acute bacterial infections or chronic inflammation. This study provides mechanistic insights into how macrophage inflammatory response during several pathological conditions could potentially be controlled by therapeutically targeting molecular intermediates involved in Lamin-A/C phosphorylation (CDKs), proteases necessary for Lamin-A/C degradation (Caspase-6) and in the IFN-β-STAT expression axis. Using different pharmacological inhibitors to inhibit Lamin-A/C phosphorylation, degradation, and phosphorylation of STAT1/3 within the IFN-β-STAT-axis, we have identified possible molecular targets in proof-of-concept experiments (Figure 4). As it is still unclear how Lamin-A/C downregulation and degradation is mechanistically induced upon pro-inflammatory activation of the TLR4 receptor, additional targets might be revealed in the future once this mechanism has been fully elucidated. In support of the clinical significance in the context of chronic inflammation, it is highly relevant that tissue-resident macrophages in a variety of inflamed human tissues show a Lamin-A/C mRNA downregulation, concomitant with an upregulation of pro-inflammatory genes (Figure 1). As some of these targets also influence the cell cycle, a major consortium has recently started a phase II trial to target CDKs as an adjunctive therapy in rheumatoid arthritis.45 Our data now suggest that these targets could directly regulate the proinflammatory response of macrophages. Lastly, our work also opens novel avenues to explore the potential role of other nuclear lamina-related proteins in taming pro-inflammatory reactions and the far less well-understood processes of resolving inflammatory episodes.
Limitations of the study
The present study demonstrates that Lamin-A/C phosphorylation and degradation are necessary to drive pro-inflammatory gene expression in macrophages. Even with all the controls presented in this study, further experiments could express mutant Lamin A/Cs in macrophages that cannot be phosphorylated on the CDK1 site and/or that cannot be degraded by Caspase 6. Even though such experiments go beyond the scope of this study, they could further test the hypothesis that certain mutations might prevent the pro-inflammatory macrophage response, even if challenged by LPS. Since our study suggests a significant degradation of the nuclear lamina upon LPS treatment, future research could explore how Lamin-A/C degradation might reduce the mechanical stability of the nuclear lamina. Softening of the nucleus might help activated macrophages to squeeze through tiny spatial confinements, however, such squeezing has been shown for other cell types to cause DNA damage. It also remains unknown how the creation of Lamin A/C fragments might further tune nuclear functions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Lamin-A/C antibody | Abcam | Cat#ab8980; RRID: AB_306909 |
| Mouse monoclonal anti-Lamin-A/C antibody | Abcam | Cat#ab8984; RRID: AB_306913 |
| Mouse monoclonal anti-Lamin-A/C antibody | Cell Signaling | Cat#4777; RRID: AB_10545756 |
| Rabbit polyclonal ani-phospho-Ser22-Lamin-A/C antibody | Cell Signaling | Cat#2026; RRID: AB_2136155 |
| Mouse monoclonal anti-NFκB/p65 antibody | Santa Cruz | Cat#sc-8008; RRID: AB_628017 |
| Rabbit monoclonal anti-phospho-STAT1 (Tyr701) antibody | Cell Signaling | Cat#7649; RRID: AB_10950970 |
| Rabbit monoclonal anti-phospho-STAT3 (Tyr705) antibody | Cell Signaling | Cat#9145; RRID: AB_2491009 |
| Rabbit monoclonal anti-phospho-STAT5 (Tyr694) antibody | Cell Signaling | Cat#9359; RRID: AB_823649 |
| Rabbit polyclonal anti-STAT1 antibody | Cell Signaling | Cat#9172; RRID: AB_2198300 |
| Rabbit monoclonal anti -STAT3 antibody | Cell Signaling | Cat#4904; RRID: AB_331269 |
| Rabbit monoclonal anti-STAT5 antibody | Cell Signaling | Cat#94205; RRID: AB_2737403 |
| Rabbit polyclonal anti-phospho-JAK1 (Tyr1034/1035) antibody | Cell Signaling | Cat#3331; RRID: AB_2265057 |
| Rabbit monoclonal anti-phospho-IRF3 (Ser396) antibody | Cell Signaling | Cat#4947; RRID: AB_823547 |
| Rabbit polyclonal anti-phospho-CDK1 (Thr161) antibody | Abcam | Cat#ab194874 |
| Rabbit polyclonal anti-alpha-tubulin antibody | Abcam | Cat#ab15246; RRID: AB_301787 |
| Rabbit polyclonal anti-IgG antibody | Merck | Cat#12–370 |
| Bacterial and virus strains | ||
| Escherichia coli strain K-12 | ATCC | Cat#12435 |
| Escherichia coli | ATCC | Cat#35218 |
| Chemicals, peptides, and recombinant proteins | ||
| BI-2536 | Axon Medchem | Cat#1129; CAS: 755038-02-9 |
| Cryptotanshinone | Cayman Chemical | Cat#16987; CAS: 35825-57-1 |
| Fludarabine | R&D Systems | Cat#3495; CAS: 21679-14-1 |
| RO-3306 | AdipoGen | Cat#AG-CR1-3515; CAS: 872573-93-8 |
| Caspase-6 Inhibitor Z-VEID-FMK | R&D Systems | Cat#FMK006 |
| IFN-β | Pbl assay science | Cat#12400–1 |
| Critical commercial assays | ||
| Murine IL-6 Mini TMB ELISA Development Kit | PeproTech | Cat#900-TM50 |
| Murine TNF-α Mini TMB ELISA Development Kit | PeproTech | Cat#900-TM54 |
| Deposited data | ||
| Raw and analyzed RNA-Seq data | This paper | GEO: GSE144094 |
| RNA-Seq microglia of LPS-injected mice | Bennett et al. 2016 | BioProject: PRJNA30727 |
| RNA-Seq LPS-treated peritoneal macrophages | Fleming et al. 2015 | SRA: SRR1918864, SRR1918994, SRR1918999-SRR1919012 and SRR1919014-SRR1919018 |
| RNA-Seq LPS-treated microglia (in vitro) | Pulido-Salgado et al. 2018 | GEO: GSE90046 |
| RNA-Seq LPS-treated murine bone-marrow derived macrophages (in vitro) | Raza et al. 2014 | GEO: GSE44292 |
| RNA-Seq LPS-treated human monocytes (in vitro) | Novakovic et al. 2017 | GEO: GSE85243 |
| RNA-Seq LPS-treated human alveolar macrophages (in vitro) | Pinilla-Vera et al. 2016 |
https://doi.org/10.1371/journal.pone.0159329.s003 https://doi.org/10.1371/journal.pone.0159329.s004 |
| RNA-Seq LPS treated buffalo bone marrow derived macrophages (in vitro) | Bush et al. 2020 | BioProject: PRJEB25226 |
| RNA-Seq LPS treated cow bone marrow derived macrophages (in vitro) | Bush et al. 2020 | BioProject: PRJEB22535 |
| RNA-Seq LPS treated goat bone marrow derived macrophages (in vitro) | Bush et al. 2020 | BioProject: PRJEB23196 |
| RNA-Seq LPS treated horse bone marrow derived macrophages (in vitro) | Bush et al. 2020 | BioProject: PRJEB24920 |
| RNA-Seq LPS treated rat bone marrow derived macrophages (in vitro) | Bush et al. 2020 | BioProject: PRJEB22553 |
| RNA-Seq LPS treated J774A1 (in vitro) | Kitamura et al. 2008 | GEO: GSE4288 |
| RNA-Seq LPS treated THP.1 (in vitro) | Leyva-Illades et al. 2010 | GEO: GSE19315 |
| Experimental models: Cell lines | ||
| RAW264.7 macrophages | ATCC | Cat#TIB-71; RRID: CVCL_0493 |
| Experimental models: Organisms/strains | ||
| Lamin-A KO (Lamin-A−/−) C57BL/6 mice | Sullivan et al. 1999 | N/A |
| C57BL/6 mice | The Laboratory of Annette Oxenius | N/A |
| Oligonucleotides | ||
| qPCR Primers Tnf-α, IL6, 18srRNA, Lamin-A/C and IFN-β | This paper | Table S1 |
| ChIP-qPCR Primers Tnf-α and IL6 | This paper | Table S2 |
| Software and algorithms | ||
| ImageJ | Davarinejad | http://www.yorku.ca/yisheng/Internal/Protocols/ImageJ.pdf |
| Matlab | This paper | N/A |
Resource availability
Lead contact
Information and requests for resources should be directed to and will be fulfilled by the lead contact Nikhil Jain (nikhilvision@gmail.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Bone marrow derived macrophages (BMDMs) and alveolar macrophages were obtained from Lamin-A knockout (Lamin-A−/−), which have have been described previously,47 and/or 5–7 weeks old C57BL/6 mice. Experiments were done with both and female mice. Lamin-A mutant mice were provided with gel diet supplement (Nutri-Gel Diet, BioServe) to improve hydration and metabolism following the onset of phenotypic decline. Maintenance and euthanasia of animals were performed in accordance with relevant guidelines and ethical regulations approved by the Cornell University Institutional Animal Care and Use Committee, protocol nos. 2011–0099 and 2012–0115.
Further RAW264.7 macrophage cells were used as noted in the key resources table.
Method details
Animals
Lamin-A knockout mice (Lamin-A−/−), which have been described previously.47
Bone marrow isolation, cell culture and primary macrophage differentiation
Femurs were isolated from post-mortem healthy mice (5–7 weeks old C57BL/6) and bone marrow was flushed with PBS. Bone marrow was further passed through a 7 μm cell strainer to obtain a single-cell suspension. Bone marrow was centrifuged and suspended in BMDM medium. Four million cells were seeded in 60 mm nontreated plastic dishes for 7 days in 5% CO2 at 37°C. Equal volumes of medium were again added on day 4. BMDMs were used on day 7 for further experiments. BMDM culture medium contained Dulbecco’s modified Eagle’s medium (DMEM) (Cat No. 31966–021, Gibco), 10% (v/v) fetal bovine serum (FBS), glutamine (Cat No. 35050–038, Gibco), non-essential amino acids (Cat No. 11140–035, Gibco), sodium pyruvate (Cat No. 11360–039, Gibco), β-mercaptoethanol (Cat No. 31350–010, Gibco), penicillin–streptomycin (Cat No. 15140–122, Gibco) and L929 medium. L929 medium was prepared by culturing 2 million L929 cells in 200 mL medium in a 300 cm2 flask for 8 days without changing the medium. The medium used for L929 cell culture contained RPMI, 10% FBS, glutamine, nonessential amino acids, sodium pyruvate, HEPES, and β-mercaptoethanol. L929 fibroblast cells were cultured and maintained in 5% CO2 at 37°C.
Alveolar macrophage isolation
Alveolar macrophage isolation: Mice were euthanized by overdose (400 mg/kg body weight) of sodium pentobarbital by intraperitoneal injection. Following anesthesia, a tracheostomy tube was placed, and mouse lungs were washed four times with 10 mL of sterile cold PBS (pH 7.4) containing 2 mM EDTA. The recovered Bronchoalveolar lavage was centrifuged at 1500 rpm for 7 min and the cells were harvested. The cell pellet was then resuspended in sterile medium and cultured for 2 h before activating with LPS.
RAW264.7 macrophage cell culture
RAW264.7 macrophages (ATCC Cat #TIB-71) were cultured in DMEM (Cat No. 31966–021, Gibco), containing 10% (v/v) FBS and 1% penicillin–streptomycin (Cat No. 15140–122, Gibco) at 37 °C, 5% CO2.
Macrophage activation
All studies were performed with primary mouse BMDMs unless stated differently. Unlike the frequently used immortalized cell lines, BMDMs more closely mimic macrophage behaviour in vivo. Pro-inflammatory activation was achieved by stimulating macrophages with LPS (50 ng/mL) (L3129 Sigma). TNF-α treatment was done by treating BMDMs with 100 ng/mL TNF-α (410-MT-010, R&D Systems).
Drug treatments
Phosphorylation of STAT1 was inhibited by supplementing the BMDMs medium with BI-2536 (1129 Axon Medchem). BMDMs were treated with different concentration of BI-2536 for 1 h before activating with LPS. BI-2536 was kept in the culture-media during LPS treatment. Phosphorylation of STAT3 was inhibited by treating BMDMs with Cryptotanshinone. BMDMs were treated with different concentration of Cryptotanshinone for 2 h before activating with LPS. Cryptotanshinone (16987 Cayman Chemical) was kept in the culture-media during LPS treatment. For Fludarabine (3495 R&D Systems) treatment, BMDMs were treated with different concentration of drug for 24 h before activating with LPS and Fludarabine was kept in the culture-media during LPS treatment. Phosphorylation of Lamin-A/C was inhibited with RO-3306 (AG-CR1-3515-M005 AdipoGen). BMDMs were treated with different concentration of RO-3306 for 2 h before activating with LPS. RO-3306 was kept in the culture-media during LPS treatment (50 ng/mL) (L3129 Sigma). Degradation of Lamin-A/C was inhibited by Caspase-6 Inhibitor Z-VEID-FMK (FMK006 R&D Systems). BMDMs were treated with different concentration of Caspase-6 Inhibitor Z-VEID-FMK for 2 h before activating with LPS. Caspase-6 Inhibitor Z-VEID-FMK was kept in the culture-media during LPS treatment. LB-100 (S7537 Lubio Science) was used to inhibit PP2A activity. BMDMs were treated with 2.5μM LB-100 for 2h before activating with LPS. LB-100 was kept in the culture-media during LPS-treatment (50 ng/mL) (L3129 Sigma). For IFN-β treatment, IFN-β (12400–1, pbl assay science) was supplemented to the media after 30 min of LPS-treatment.
Immunostaining
BMDMs were rinsed twice with 1X PBS, followed by fixation using 4% paraformaldehyde (18814–20, Polysciences Inc.) in 1X PBS for 20 min. BMDMs were washed and permeabilized with 0.3% Triton X-100 (Sigma) in 1X PBS for 10 min. After washing twice, BMDMs were treated with 2% BSA (blocking solution) for 60 min before incubating with primary antibody (diluted in 2% BSA) at 4°C overnight. Primary antibodies against Lamin-A/C (1:100, ab8980 Abcam; 1:100, ab8984 Abcam; 1:100 4777S Cell Signalling), phospho-Ser22-Lamin-A/C (pSer22) (1:100, 2026 Cell Signalling) and p65 (NFκB p65 1:300, sc-8008 Santa Cruz) were used for staining. BMDMs were then washed with blocking solution and incubated with the corresponding secondary antibodies, diluted in blocking solution, along with the nuclear stain Hoechst-33342 (1:1000, 62249 Life Technologies) for 45 min. Filamentous actin (F-actin) was labelled using phalloidin Alexa-Fluor 568 (1:200, Life Technologies). All antibodies used in this study have previously been extensively used and validated for immunofluorescence studies.
Confocal imaging
Images of single BMDM were taken using a Leica SP8 confocal microscope. To estimate spreading areas, 2D images were taken using a 40X (0.7 NA) air objective. To estimate protein expression levels, BMDMs were stained with the desired antibodies and 3D confocal images were captured rather than projected images. Fluorescence images were captured using a 63X (1.25 NA) oil objective with a z-step of 0.5 μm and a confocal pinhole of 1 A.U. Imaging conditions were kept the same during all experiments. See quantification and statistical analysis for downstream analyses.
Cytokine secretion
Cell-culture medium was collected post drug treatment or LPS stimulation for desired time (mentioned in the respective figures). To remove cell debris, medium was centrifuged, and the supernatant was stored at −80°C till further processing. ELISA kits were purchased from Peprotech TNF-α (900-54), and IL-6 (900-TM50) and experiment was performed according to the manufacturer’s protocol. See quantification and statistical analysis for downstream analyses.
Immunoblotting
Briefly, BMDMs were washed once with ice-cold PBS and lysed for 20 min with lysis buffer (20mM TRIS,150mM NaCl, 1mM EGTA, 1mM EDTA, 1% Triton-X-100) supplemented with phosphatase inhibitor (1:100 P5726, Sigma Aldrich) and protease inhibitor (1:10 11836170001, Sigma-Aldrich) on ice. BMDMs were scratched and lysate was collected, homogenized and centrifuged at 4°C. Protein concentrations were determined with a BCA protein assay kit (23225, Thermo Fischer Scientific) and cell lysates were incubated with 5X Reducing Sample Buffer (0.3M Tris-HCL pH6.8, 100mM DTT, 5% SDS, 50% Glycerol, Pyronin G) for 5 min at 95°C. Protein lysate was separated using 10% Bis-Tris polyacrylamide gel. Proteins were transferred by wet tank transfer (110V 1h) on to a nitrocellulose membrane. Membranes were blocked for 1 h using 5% BSA in TBS-T (0.01% Triton-X) and incubated overnight with primary antibodies (1:500 p-(Tyr701)-STAT1, 7649 Cell Signaling; 1:500 p-(Tyr705)-STAT3, 9145 Cell Signaling; 1:500 p-(Tyr694)-STAT5, 9359S Cell Signaling; 1:500 STAT1, 9172 Cell Signaling; 1:500 STAT3, 4904 Cell Signaling; 1:500 STAT5, 94205 Cell Signaling; 1:100 p-(Tyr1034/1035)-JAK1, 3331S Cell Signaling; 1:250 p-(Ser396)-IRF3, 4947 Cell Signaling; 1:100 Lamin-A/C, 4777S Cell Signalling; 1:100 p-(Ser22)-Lamin-A/C, 2026 Cell Signalling; 1:1000 alpha-tubulin, 15246 Abcam; 1:100 p-(Thr161)-Cdk1, 194874 Abcam) in 5% BSA TBST (0.01% Triton-X). Membranes were washed, incubated for 1 h with 1:10000 (donkey anti-rabbit secondary HRP Jackson or respectively donkey anti-mouse secondary HRP Jackson) in 5% BSA TBS-T (0.01% Triton-X) and washed again before exposing to Immunoblotting Substrate (Pierce ECL Immunoblotting Substrate 32209 Thermo Fisher Scientific). See quantification and statistical analysis for downstream analyses.
RNA isolation and real-time qPCR
RNA was isolated with a Qiagen RNA Isolation Kit according to the manufacturer’s protocol. cDNA was prepared using iScript Advanced cDNA Synthesis Kit (172–5038 BioRad). Real-time qPCR experiments were performed using SsoAdvanced™ Universal SYBR® Green Supermix (1725271 BioRad) in a CFX96 Real-Time PCR Detection System (Bio-Rad). Primers are listed in the key resources table and in Table S1. See quantification and statistical analysis for downstream analyses.
RNA sequencing
RNA was isolated with the Qiagen RNA Isolation Kit according to the manufacturer’s protocol. For library preparation, the quantity and quality of the isolated RNA were determined with a Qubit (1.0) Fluorometer (Life Technologies) and a Tapestation 4200 (Agilent). The TruSeq Stranded HT mRNA Sample Prep Kit (Illumina) was used in subsequent steps following the manufacturer’s protocol. Briefly, total RNA samples (1 μg) were ribosome-depleted and then reverse-transcribed into double-stranded cDNA with actinomycin added during the first-strand synthesis. The cDNA samples were fragmented, end-repaired and polyadenylated before ligation of TruSeq adapters. The adapters contain the index for dual multiplexing. Fragments containing TruSeq adapters on both ends were selectively enriched with PCR. The quality and quantity of the enriched libraries were validated using a Qubit (1.0) fluorometer and the Tapestation 4200 (Agilent). The product is a smear with an average fragment size of ∼360 bp. The libraries were normalized to 10 nM in Tris-Cl 10 mM, pH 8.5 with 0.1% Tween 20. For cluster generation and sequencing, a Hiseq 4000 SR Cluster Kit (Illumina) was used using 8 pM of pooled normalized libraries on the cBOT V2. Sequencing was performed on the Illumina HiSeq with single-end 125 bp using the HiSeq 3000/4000 SBS Kit (Illumina). Reads quality was checked with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and sequencing adapters were trimmed using Trimmomatic63. Reads at least 20 bases long, and an overall average Phred quality score greater than 10 were aligned to the reference genome and transcriptome (FASTA and GTF files, respectively, downloaded from Ensemble, genome build GRCm38) with STAR v2.5.164, with default settings for single end reads. The distribution of reads across genomic features was quantified using the R package Genomic Ranges from Bioconductor Version 3.065. Differentially expressed genes were identified using the R package edgeR from Bioconductor Version 3.066. See quantification and statistical analysis for downstream analyses.
ChIP-qPCR
10 million BMDMs per condition were cultured and treated accordingly. After 6 h of LPS treatment 16% Formaldehyde (18814–20, Polysciences Inc.) was added to the medium in the dish to a concentration of 1% and incubated (gently mixing) for 10 min 1 mL of ice-cold 1.25 M glycine was added and incubated (gently mixing) for 5 min. The medium was discarded, and the dishes transferred onto ice. BMDMs on the dishes were washed with 5 mL of ice-cold PBS supplemented with protease inhibitor and phosphatase inhibitor (1:10 11836170001, Sigma-Aldrich, 1:100 P5726, Sigma Aldrich – PBS/c) and scratched in 2 mL PBS/c, which was collected. The dishes were washed with 1mL PBS/c and pooled with the 2 mL. Scratched BMDMs were centrifuged for 8 min at 300 g and 4°C. The supernatant was aspirated, and the dry pellet snap frozen on dry ice. The pellet was thawed on ice and resuspended in 50 μL SLB/c buffer (10 mM Tris-HCL pH 8, 10 mM EDTA, 1% SDS supplemented with protease inhibitor (11836170001, Sigma-Aldrich)). Chromatin was sheared with a Covaris S220 AFA System (Covaris) with the following settings (Peak Power 140, Duty Factor 10.0, Cycles Burst 2000, Repetitions 15, 30 s ON/30 s OFF) at 4°C. Samples were centrifuged for 5 min at 20000 rpm and 4°C and the supernatant transferred into low-binding tubes. 9 volumes (450 μL) of CDB/c (16.7 mM Tris-HCl pH8, 167 mM NaCl, 1.2 mM EDTA, 1.1% Triton X-100, 0.01% SDS supplemented with protease inhibitor and phosphatase inhibitor (1:10 11836170001, Sigma-Aldrich, 1:100 P5726, Sigma Aldrich) was added to the sample and incubated for at least 30 min with 25 μL Dynabeads A (10001D, Thermo Fischer Scientific) resuspended in CDB/c. After incubation the supernatant was transferred and 1% of the volume was stored separately at −20°C (input). The remaining sample was incubated o/n on a rotary shaker at 4°C with 2 μg of the appropriate antibody (IgG, 12–370 Merck; pSTAT1, 7649 Cell Signaling). The next day 25 μL Dynabeads resuspended in CDB/c were added and incubated for at least 1 h at 4°C on a rotary shaker. Beads were washed sequentially with low-salt wash buffer (20 mM Tris-Hcl pH 8, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS), high-salt wash buffer (20 mM Tris-HCl pH 8, 500 mM NaCl, 2 mM EDTA, 1% Triton-X-100, 0.1% SDS), LiCl wash buffer (10 mM Tris-HCl pH 8, 250 mM LiCl, 1 mM EDTA, 1% sodium deoxycholate, 1% Igepal-CA630) and TE (10 mM Tris-HCl pH8, 1 mM EDTA). Immuno-precipitants were eluted from beads with Elution Buffer (100 mM NaHCO3, 1% SDS) incubating 15 min on a rotary shaker. 20 μL of 5 M NaCl was added to the elutions and to the with EB diluted 1% input sample and incubated at 65°C and 95 rpm for 2 h. 10 μL o f 0.5 M EDTA, 20 μL 1 M Tris-HCl pH 6.5 and 1 μL of 20 mg/mL proteinase K was added to each sample and incubated for 1 h at 45°C. DNA was extracted with the phenol chloroform extraction protocol. After DNA extraction the DNA-concentrations of the samples were measured. DNA samples were diluted, and the immunoprecipitation reaction assessed by qPCR. Primers are listed in the key resources table and in Table S2. See quantification and statistical analysis for downstream analyses.
Bacterial infection
For active E. coli infection, the bacterial strain MG1655 was used.48 For heat inactivated E. coli infection, the bacterial strain ATCC35218 was used.73 E. coli strains were grown in LB medium and diluted in antibiotic-free BMDM culture medium before experiments after overnight growth to an optical density of 0.02. For heat inactivated E. coli infection, heat inactivated E. coli were added to the BMDMs in a concentration of 2.5x106 bacteria/mL.
Quantification and statistical analysis
Confocal imaging analysis
All the quantifications per cell were carried out using MATLAB2019b (MathWorks) or ImageJ (NIH). Imaging conditions were kept the same during all experiments. Briefly as previously described,48 total levels of selected proteins in a cell were defined as the sum of the nuclear and cytoplasmic fractions of the proteins.
Cytokine secretion analysis
All data fitting and quantifications were done using OriginPro8.1 and MATLAB2019b (MathWorks), respectively.
Quantification of immunoblots
The films were scanned and converted into 8-bit (grayscale) images. For each protein of interest single regions of interest were defined and the mean gray value was measured for the protein and for the background. To get relative amounts as a ratio of each protein band relative to the lane’s loading control, single regions were as well measured for the single bands of the loading control and the background of the loading control and protein of interest. The mean gray values were inverted (255-mean gray value) and the inverted background was subtracted from the inverted protein value to get the net protein value. The net protein value was divided by the net loading control value to get relative amounts of the protein of interest.
Real-time qPCR analysis
The qPCR analysis is explained here with exemplary (triplicate average) Ct values produced during real-time q-PCR analysis:
| Raw Ct values 18s rRNA | Raw Ct values gene of interest | |
|---|---|---|
| WT | 6.59 | 26.42 |
| WT + LPS | 7.22 | 17.84 |
Step 1 normalization of Ct-values
Raw Ct values (18s rRNA) were subtracted of the Raw Ct values (gene of interest) of the sample.
| Normalized Raw Ct values | |
|---|---|
| WT | 26.42–6.59 = 19.83 |
| WT + LPS | 17.84–7.22 = 10.62 |
Step 2 normalization to WT sample
| Normalized Ct values | |
|---|---|
| WT | 19.83–19.83 = 0 |
| WT + LPS | 10.62–19.83 = −9.21 |
Step 3 Calculation of Fold-Change
| Fold Change | |
|---|---|
| WT | 2ˆ0 = 1 |
| WT + LPS | 2ˆ-9.21 = 592.2 |
In this case the calculated fold change would be 592.2 when comparing the gene expression between the WT and WT + LPS treated BMDMs samples. This calculated fold change was then plotted in Origin Pro.
Functional analysis of genes
Gene Ontology and KEGG pathway analysis of differentially regulated genes was performed using DAVID software. Prediction of Transcription factors and their binding analysis was performed using PSCAN and Interferome.
Reanalysis of previously published RNA-Sequencing data
The data downloaded from the public repositories was formatted in different manners. To give an insight of our analysis we will explain how our analysis was done based on one example. In the following, we will explain how we extracted the data for murine microglia treated with LPS for 6 h (GSE90046).
Step 1 downloading and filtering the data
We downloaded the respective data from the GEO repository (GSE90046) and subsequently filtered the data for the genes of interest (Lmna and pro-inflammatory genes). The Sequencing data was given, in this case, in counts per gene (in triplicates).
Step 2 normalizing data to total counts
To normalize with the total counts, we calculated to number of total counts of the RNA-Sequencing per sample (by adding up all counts across the sample) and the divided the respective counts of our gene of interest by the total number of counts.
Step 3 calculating the log2fold-change
Subsequently we took the mean of the triplicates (of the gene of interest) and calculated the log2fold change comparing untreated and 6 h LPS treated murine microglia. This was done by making the ratio of the means (Mean fragments of Control samples/Mean fragments of Control 6 h LPS samples) and then calculating the log2 of the resulting ratio.
Step 3 plotting the data
To plot the log2fold changes in one big panel as done in Figure 1A, we gathered all the log2fold-changes of the different species and cell types and plotted these in a heatmap by using Matlab2020.
Chip-qPCR analysis
The ChIP fold enrichments were calculated with the % Input–Method. This method is described in the following:
The signals obtained from ChIP are divided by signals obtained from an Input sample. This Input sample represents the amount of chromatin used in the ChIP. In these experiments a 1% of starting chromatin was used as Input. The calculation was done as following, example is illustrated below:
Step 1 Adjusting the input
| Adjusted input to 100% | ||
|---|---|---|
| Raw Ct | Ct Input – 6.644 | |
| Input 1% (WT + LPS sample) | 32.7 | 26.1 |
Step 2 Percent Input calculation
With the Adjusted Input (26.1) calculated the Percent Input of the sample was calculated.
| Percent Input | ||
|---|---|---|
| Triplicate average Ct | 100∗2ˆ(26.1 – Ct(IP) | |
| pSTAT1 (WT + LPS sample) | 31.3 | 2.7 |
These calculations were done for the different samples with their respective Input samples.
Step 3 Fold Change calculation
To calculate the fold change enrichment the single samples were divided by the calculated Percent Input of the WT + LPS sample (2.7).
| Percent input | Fold Change enrichment (Percent Input/2.7) | |
|---|---|---|
| pSTAT1 (WT + LPS sample) | 2.7 | 1 |
| pSTAT1 (KO + LPS sample) | 7.2 | 2.6 |
This calculated fold change enrichment was subsequently plotted in OriginPro.
Statistical analysis
The cell number assayed for quantification is given in individual figures. Comparisons were performed by means of a two-tailed Student t-test. If not described differently p values were obtained with the two-sided Student's t-test or and the boxes, presented in the plots, show 25th and 75th percentiles, the middle horizontal lines show the median, small open squares show the mean, and whiskers indicate S.D or data was presented as Mean ± S.E. All the statistic and data plotting were done in OriginPro software.
Acknowledgments
The authors thank the Mice Facility at ETH Zurich and Prof. Annette Oxenius (ETH Zurich) for donating post-mortem animals for bone marrow isolation and Chantel Spencer for technical support in experiments like ELISA. The authors thank Prof. Manfred Kopf (ETH Zurich) for providing alveolar macrophages. We also want to thank Cornell University Institutional Animal Care and Use Committee and Hannah Fong for isolating and sending bone marrow from KO mice. We further thank the ScopeM facility at ETH Zurich for access to confocal microscopy and the Genetic Diversity Center at ETH. This work was supported by ETH Zurich, SNF NCCR ‘Molecular Systems Engineering, SNF CR32I3_156931 (VV co-PI), an ETH Research Grant ETH-24 18-1 grant to V.V., the National Institutes of Health (NIH) awards R01HL082792, R01GM137605, and U54CA210184 to J.L., University of Birmingham (UoB), and SNF SPARK award CRSK-3_195952 to N.J.
Author contributions
NJ conceived of the project. JM and NJ designed and performed the experiments. JM and NJ analyzed the data. AE isolated and provided the bone marrow isolated from Lamin-A/C-KO mice, and VV, JL and MM contributed to the work through discussions. JM, VV, and NJ wrote the article. All authors edited and approved the article.
Declaration of interests
The authors declare that there is no conflict of interest regarding the publication of this article.
Published: December 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105528.
Contributor Information
Viola Vogel, Email: viola.vogel@hest.ethz.ch.
Nikhil Jain, Email: nikhilvision@gmail.com.
Supplemental information
Data and code availability
Generated RNA-Seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper analyses existing, publicly available data. The accession numbers for the datasets are listed in the key resources table. All other data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyse the data reported in this paper is available from the lead contact.
References
- 1.Osmanagic-Myers S., Dechat T., Foisner R. Lamins at the crossroads of mechanosignaling. Genes Dev. 2015;29:225–237. doi: 10.1101/gad.255968.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lammerding J. Mechanics of the nucleus. Compr. Physiol. 2011;1:783–807. doi: 10.1002/cphy.c100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Leeuw R., Gruenbaum Y., Medalia O. Nuclear lamins: thin filaments with major functions. Trends Cell Biol. 2018;28:34–45. doi: 10.1016/j.tcb.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Ho C.Y., Lammerding J. Lamins at a glance. J. Cell Sci. 2012;125:2087–2093. doi: 10.1242/JCS.087288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrés V., González J.M. Role of A-type lamins in signaling, transcription, and chromatin organization. J. Cell Biol. 2009;187:945–957. doi: 10.1083/JCB.200904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Steensel B., Belmont A.S. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell. 2017;169:780–791. doi: 10.1016/J.CELL.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobrzynska A., Gonzalo S., Shanahan C., Askjaer P., Doisy E.A. The nuclear lamina in health and disease. Nucleus. 2016;7:233–248. doi: 10.1080/19491034.2016.1183848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta S., Marcel N., Sarin A., Shivashankar G.V. Role of actin dependent nuclear deformation in regulating early gene expression. PLoS One. 2012;7 doi: 10.1371/JOURNAL.PONE.0053031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rocha-Perugini V., González-Granado J.M. Nuclear envelope lamin-A as a coordinator of T cell activation. Nucleus. 2014;5 doi: 10.4161/NUCL.36361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.González-Granado J.M., Silvestre-Roig C., Rocha-Perugini V., Trigueros-Motos L., Cibrián D., Morlino G., Blanco-Berrocal M., Osorio F.G., Freije J.M.P., López-Otín C., et al. Nuclear envelope lamin-A couples actin dynamics with immunological synapse architecture and T cell activation. Sci. Signal. 2014;7:ra37. doi: 10.1126/scisignal.2004872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran J.R., Chen H., Zheng X., Zheng Y. Lamin in inflammation and aging. Curr. Opin. Cell Biol. 2016;40:124–130. doi: 10.1016/j.ceb.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kristiani L., Kim M., Kim Y. Role of the nuclear lamina in age-associated nuclear reorganization and inflammation. Cells. 2020;9:718. doi: 10.3390/CELLS9030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H., Zheng X., Zheng Y. Lamin-B in systemic inflammation, tissue homeostasis, and aging. Nucleus. 2015;6 doi: 10.1080/19491034.2015.1040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osorio F.G., Bárcena C., Soria-Valles C., Ramsay A.J., de Carlos F., Cobo J., Fueyo A., Freije J.M.P., López-Otín C. Nuclear lamina defects cause ATM-dependent NF-κB activation and link accelerated aging to a systemic inflammatory response. Genes Dev. 2012;26:2311–2324. doi: 10.1101/GAD.197954.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Güttinger S., Laurell E., Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 2009;10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- 16.Kochin V., Shimi T., Torvaldson E., Adam S.A., Goldman A., Pack C.G., Melo-Cardenas J., Imanishi S.Y., Goldman R.D., Eriksson J.E. Interphase phosphorylation of lamin A. J. Cell Sci. 2014;127:2683–2696. doi: 10.1242/jcs.141820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ottaviano Y., Gerace L. Phosphorylation of the nuclear lamins during interphase and mitosis. J. Biol. Chem. 1985;260:624–632. doi: 10.1016/s0021-9258(18)89778-2. [DOI] [PubMed] [Google Scholar]
- 18.Heald R., McKeon F. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell. 1990;61:579–589. doi: 10.1016/0092-8674(90)90470-Y. [DOI] [PubMed] [Google Scholar]
- 19.Ivashkiv L.B. Epigenetic regulation of macrophage polarization and function. Trends Immunol. 2013;34:216–223. doi: 10.1016/J.IT.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain N., Moeller J., Vogel V. Mechanobiology of macrophages: how physical factors coregulate macrophage plasticity and phagocytosis. Annu. Rev. Biomed. Eng. 2019;21:267–297. doi: 10.1146/annurev-bioeng-062117-121224. [DOI] [PubMed] [Google Scholar]
- 21.Jain N., Shahal T., Gabrieli T., Gilat N., Torchinsky D., Michaeli Y., Vogel V., Ebenstein Y. Global modulation in DNA epigenetics during pro-inflammatory macrophage activation. Epigenetics. 2019;14:1183–1193. doi: 10.1080/15592294.2019.1638700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence T., Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol. 2011;11:750–761. doi: 10.1038/NRI3088. [DOI] [PubMed] [Google Scholar]
- 23.Bennett M.L., Bennett F.C., Liddelow S.A., Ajami B., Zamanian J.L., Fernhoff N.B., Mulinyawe S.B., Bohlen C.J., Adil A., Tucker A., et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA. 2016;113:E1738–E1746. doi: 10.1073/PNAS.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleming B.D., Chandrasekaran P., Dillon L.A.L., Dalby E., Suresh R., Sarkar A., El-Sayed N.M., Mosser D.M. The generation of macrophages with anti-inflammatory activity in the absence of STAT6 signaling. J. Leukoc. Biol. 2015;98:395–407. doi: 10.1189/JLB.2A1114-560R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pulido-Salgado M., Vidal-Taboada J.M., Barriga G.G.-D., Solà C., Saura J. RNA-Seq transcriptomic profiling of primary murine microglia treated with LPS or LPS + IFNγ. Sci. Rep. 2018;8:16096. doi: 10.1038/S41598-018-34412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raza S., Barnett M.W., Barnett-Itzhaki Z., Amit I., Hume D.A., Freeman T.C. Analysis of the transcriptional networks underpinning the activation of murine macrophages by inflammatory mediators. J. Leukoc. Biol. 2014;96:167–183. doi: 10.1189/JLB.6HI0313-169R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novakovic B., Habibi E., Wang S.-Y., Arts R.J.W., Davar R., Megchelenbrink W., Kim B., Kuznetsova T., Kox M., Zwaag J., et al. β-Glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell. 2016;167:1354–1368.e14. doi: 10.1016/J.CELL.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinilla-Vera M., Xiong Z., Zhao Y., Zhao J., Donahoe M.P., Barge S., Horne W.T., Kolls J.K., McVerry B.J., Birukova A., et al. Full spectrum of LPS activation in alveolar macrophages of healthy volunteers by whole transcriptomic profiling. PLoS One. 2016;11:01593299–e159422. doi: 10.1371/journal.pone.0159329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bush S.J., McCulloch M.E.B., Lisowski Z.M., Muriuki C., Clark E.L., Young R., Pridans C., Prendergast J.G.D., Summers K.M., Hume D.A. Species-specificity of transcriptional regulation and the response to lipopolysaccharide in mammalian macrophages. Front. Cell Dev. Biol. 2020;8:661. doi: 10.3389/FCELL.2020.00661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitamura H., Ito M., Yuasa T., Kikuguchi C., Hijikata A., Takayama M., Kimura Y., Yokoyama R., Kaji T., Ohara O. Genome-wide identification and characterization of transcripts translationally regulated by bacterial lipopolysaccharide in macrophage-like J774.1 cells. Physiol. Genomics. 2008;33:121–132. doi: 10.1152/PHYSIOLGENOMICS.00095.2007. [DOI] [PubMed] [Google Scholar]
- 31.Leyva-Illades D., Cherla R.P., Galindo C.L., Chopra A.K., Tesh V.L. Global transcriptional response of macrophage-like THP-1 cells to Shiga toxin type 1. Infect. Immun. 2010;78:2454–2465. doi: 10.1128/IAI.01341-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das A., Yang C.-S., Arifuzzaman S., Kim S., Kim S.Y., Jung K.H., Lee Y.S., Chai Y.G. High-resolution mapping and dynamics of the transcriptome, transcription factors, and transcription Co-factor networks in classically and alternatively activated macrophages. Front. Immunol. 2018;9:22. doi: 10.3389/fimmu.2018.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y., Bayona P.W., Kim M., Chang J., Hong S., Park Y., Budiman A., Kim Y.J., Choi C.Y., Kim W.S., et al. Macrophage lamin A/C regulates inflammation and the development of obesity-induced insulin resistance. Front. Immunol. 2018;9:696. doi: 10.3389/fimmu.2018.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng A.Y., Tu C., Shen S., Xu D., Oursler M.J., Qu J., Yang S. Comparative characterization of osteoclasts derived from murine bone marrow macrophages and RAW 264.7 cells using quantitative proteomics. JBMR plus. 2018;2:328–340. doi: 10.1002/JBM4.10058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norris P.C., Reichart D., Dumlao D.S., Glass C.K., Dennis E.A. Specificity of eicosanoid production depends on the TLR-4-stimulated macrophage phenotype. J. Leukoc. Biol. 2011;90:563–574. doi: 10.1189/JLB.0311153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fullerton J.N., Gilroy D.W. Resolution of inflammation: a new therapeutic Frontier. Nat. Rev. Drug Discov. 2016;15:551–567. doi: 10.1038/nrd.2016.39. [DOI] [PubMed] [Google Scholar]
- 37.Parisi L., Gini E., Baci D., Tremolati M., Fanuli M., Bassani B., Farronato G., Bruno A., Mortara L. Macrophage polarization in chronic inflammatory diseases: killers or builders? J. Immunol. Res. 2018;2018:8917804. doi: 10.1155/2018/8917804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ihalainen T.O., Aires L., Herzog F.A., Schwartlander R., Moeller J., Vogel V. Differential basal-to-apical accessibility of lamin A/C epitopes in the nuclear lamina regulated by changes in cytoskeletal tension. Nat. Mater. 2015;14:1252–1261. doi: 10.1038/NMAT4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buchwalter A., Hetzer M.W. Nucleolar expansion and elevated protein translation in premature aging. Nat. Commun. 2017;8:328–413. doi: 10.1038/s41467-017-00322-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peter M., Nakagawa J., Dorée M., Labbé J.C., Nigg E.A. In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell. 1990;61:591–602. doi: 10.1016/0092-8674(90)90471-P. [DOI] [PubMed] [Google Scholar]
- 41.Buxboim A., Swift J., Irianto J., Spinler K.R., Dingal P.C.D.P., Athirasala A., Kao Y.-R.C., Cho S., Harada T., Shin J.W., Discher D.E. Matrix elasticity regulates lamin-A, C phosphorylation and turnover with feedback to actomyosin. Curr. Biol. 2014;24:1909–1917. doi: 10.1016/j.cub.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruchaud S., Korfali N., Villa P., Kottke T.J., Dingwall C., Kaufmann S.H., Earnshaw W.C. Caspase-6 gene disruption reveals a requirement for lamin A cleavage in apoptotic chromatin condensation. EMBO J. 2002;21:1967–1977. doi: 10.1093/EMBOJ/21.8.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vassilev L.T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D.C., Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. USA. 2006;103:10660–10665. doi: 10.1073/PNAS.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leitch A.E., Haslett C., Rossi A.G. Cyclin-dependent kinase inhibitor drugs as potential novel anti-inflammatory and pro-resolution agents. Br. J. Pharmacol. 2009;158:1004–1016. doi: 10.1111/J.1476-5381.2009.00402.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siebert S., Pratt A.G., Stocken D.D., Morton M., Cranston A., Cole M., Frame S., Buckley C.D., Ng W.F., Filer A., et al. Targeting the rheumatoid arthritis synovial fibroblast via cyclin dependent kinase inhibition: an early phase trial. Medicine (Baltim.) 2020;99 doi: 10.1097/MD.0000000000020458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dhani S., Zhao Y., Zhivotovsky B. A long way to go: caspase inhibitors in clinical use. Cell Death Dis. 2021;12:949–1013. doi: 10.1038/s41419-021-04240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C.L., Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999;147:913–920. doi: 10.1083/JCB.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jain N., Vogel V. Spatial confinement downsizes the inflammatory response of macrophages. Nat. Mater. 2018;17:1134–1144. doi: 10.1038/s41563-018-0190-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Forester C.M., Maddox J., Louis J.V., Goris J., Virshup D.M. Control of mitotic exit by PP2A regulation of Cdc25C and Cdk1. Proc. Natl. Acad. Sci. USA. 2007;104:19867–19872. doi: 10.1073/PNAS.0709879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong C.S., Ho W., Zhang C., Yang C., Elder J.B., Zhuang Z. LB100, a small molecule inhibitor of PP2A with potent chemo- and radio-sensitizing potential. Cancer Biol. Ther. 2015;16:821–833. doi: 10.1080/15384047.2015.1040961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D’Arcy B.M., Swingle M.R., Papke C.M., Abney K.A., Bouska E.S., Prakash A., Honkanen R.E. The antitumor drug LB-100 is a catalytic inhibitor of protein phosphatase 2A (PPP2CA) and 5 (PPP5C) coordinating with the active-site catalytic metals in PPP5C. Mol. Cancer Ther. 2019;18:556–566. doi: 10.1158/1535-7163.MCT-17-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jahn D., Schramm S., Schnölzer M., Heilmann C.J., de Koster C.G., Schütz W., Benavente R., Alsheimer M. A truncated lamin A in the Lmna −/− mouse line. Nucleus. 2012;3:463–474. doi: 10.4161/nucl.21676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Majoros A., Platanitis E., Kernbauer-Hölzl E., Rosebrock F., Müller M., Decker T. Canonical and non-canonical aspects of JAK-STAT signaling: lessons from interferons for cytokine responses. Front. Immunol. 2017;8:29. doi: 10.3389/FIMMU.2017.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/NRI3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Malik N., Vollmer S., Nanda S.K., Lopez-Pelaez M., Prescott A., Gray N., Cohen P. Suppression of interferon β gene transcription by inhibitors of bromodomain and extra-terminal (BET) family members. Biochem. J. 2015;468:363–372. doi: 10.1042/BJ20141523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sebastian M., Reck M., Waller C.F., Kortsik C., Frickhofen N., Schuler M., Fritsch H., Gaschler-Markefski B., Hanft G., Munzert G., von Pawel J. The efficacy and safety of BI 2536, a novel plk-1 inhibitor, in patients with stage IIIB/IV non-small cell lung cancer who had relapsed after, or failed, chemotherapy: results from an open-label, randomized phase II clinical trial. J. Thorac. Oncol. 2010;5:1060–1067. doi: 10.1097/JTO.0B013E3181D95DD4. [DOI] [PubMed] [Google Scholar]
- 57.Ciceri P., Müller S., O’Mahony A., Fedorov O., Filippakopoulos P., Hunt J.P., Lasater E.A., Pallares G., Picaud S., Wells C., et al. Dual kinase-bromodomain inhibitors for rationally designed polypharmacology. Nat. Chem. Biol. 2014;10:305–312. doi: 10.1038/nchembio.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frank D.A., Mahajan S., Ritz J. Fludarabine-induced immunosuppression is associated with inhibition of STAT1 signaling. Nat. Med. 1999;5:444–447. doi: 10.1038/7445. [DOI] [PubMed] [Google Scholar]
- 59.Casak S.J., Lemery S.J., Shen Y.L., Rothmann M.D., Khandelwal A., Zhao H., Davis G., Jarral V., Keegan P., Pazdur R. U.S. Food and drug administration approval: rituximab in combination with fludarabine and cyclophosphamide for the treatment of patients with chronic lymphocytic leukemia. Oncol. 2011;16:97–104. doi: 10.1634/THEONCOLOGIST.2010-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anderson V.R., Perry C.M. Fludarabine: a review of its use in non-Hodgkin’s lymphoma. Drugs. 2007;67:1633–1655. doi: 10.2165/00003495-200767110-00008. [DOI] [PubMed] [Google Scholar]
- 61.Shin D.-S., Kim H.N., Shin K.D., Yoon Y.J., Kim S.J., Han D.C., Kwon B.M. Cryptotanshinone inhibits constitutive signal transducer and activator of transcription 3 function through blocking the dimerization in DU145 prostate cancer cells. Cancer Res. 2009;69:193–202. doi: 10.1158/0008-5472.CAN-08-2575. [DOI] [PubMed] [Google Scholar]
- 62.Du J., Wei N., Guan T., Xu H., An J., Pritchard K.A., Shi Y. Inhibition of CDKS by roscovitine suppressed LPS-induced •NO production through inhibiting NFκB activation and BH4 biosynthesis in macrophages. Am. J. Physiol. Cell Physiol. 2009;297:C742–C749. doi: 10.1152/ajpcell.00138.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ikegami K., Secchia S., Almakki O., Lieb J.D., Moskowitz I.P. Phosphorylated lamin A/C in the nuclear interior binds active enhancers associated with abnormal transcription in progeria. Dev. Cell. 2020;52:699–713.e11. doi: 10.1016/j.devcel.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ladha S., Qiu X., Casal L., Caron N.S., Ehrnhoefer D.E., Hayden M.R. Constitutive ablation of caspase-6 reduces the inflammatory response and behavioural changes caused by peripheral pro-inflammatory stimuli. Cell Death Discov. 2018;4:40. doi: 10.1038/s41420-018-0043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cowling V., Downward J. Caspase-6 is the direct activator of caspase-8 in the cytochrome c-induced apoptosis pathway: absolute requirement for removal of caspase-6 prodomain. Cell Death Differ. 2002;9:1046–1056. doi: 10.1038/SJ.CDD.4401065. [DOI] [PubMed] [Google Scholar]
- 66.Hirata H., Takahashi A., Kobayashi S., Yonehara S., Sawai H., Okazaki T., Yamamoto K., Sasada M. Caspases are activated in a branched protease cascade and control distinct downstream processes in fas-induced apoptosis. J. Exp. Med. 1998;187:587–600. doi: 10.1084/JEM.187.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McIlwain D.R., Berger T., Mak T.W. Caspase functions in cell death and disease. Cold Spring Harb. Perspect. Biol. 2013;5:0086566–a8728. doi: 10.1101/cshperspect.a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Olins A.L., Herrmann H., Lichter P., Kratzmeier M., Doenecke D., Olins D.E. Nuclear envelope and chromatin compositional differences comparing undifferentiated and retinoic acid- and phorbol ester-treated HL-60 cells. Exp. Cell Res. 2001;268:115–127. doi: 10.1006/excr.2001.5269. [DOI] [PubMed] [Google Scholar]
- 69.Swift J., Ivanovska I.L., Buxboim A., Harada T., Dingal P.C.D.P., Pinter J., Pajerowski J.D., Spinler K.R., Shin J.W., Tewari M., et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shin J.-W. Squeezing cells through the epigenetic machinery. Proc. Natl. Acad. Sci. USA. 2018;115:8472–8474. doi: 10.1073/PNAS.1811184115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mattioli E., Andrenacci D., Garofalo C., Prencipe S., Scotlandi K., Remondini D., Gentilini D., Di Blasio A.M., Valente S., Scarano E., et al. Altered modulation of lamin A/C-HDAC2 interaction and p21 expression during oxidative stress response in HGPS. Aging Cell. 2018;17 doi: 10.1111/acel.12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rothchild A.C., Olson G.S., Nemeth J., Amon L.M., Mai D., Gold E.S., Diercks A.H., Aderem A. Alveolar macrophages generate a noncanonical NRF2-driven transcriptional response to Mycobacterium tuberculosis in vivo. Sci. Immunol. 2019;4:eaaw6693. doi: 10.1126/sciimmunol.aaw6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quintin J., Saeed S., Martens J.H.A., Giamarellos-Bourboulis E.J., Ifrim D.C., Logie C., Jacobs L., Jansen T., Kullberg B.J., Wijmenga C., et al. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe. 2012;12:223–232. doi: 10.1016/J.CHOM.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Generated RNA-Seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper analyses existing, publicly available data. The accession numbers for the datasets are listed in the key resources table. All other data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyse the data reported in this paper is available from the lead contact.