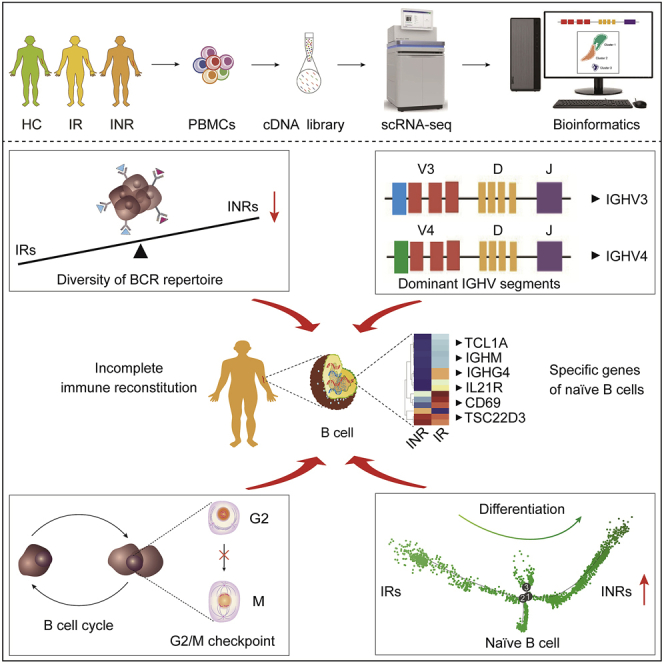
Summary
Incomplete immune reconstitution happens in some HIV-infected patients who have achieved persistent viral suppression under antiretroviral therapy (ART). We performed single-cell RNA sequencing for peripheral blood mononuclear cells to analyze B cell receptor (BCR) repertoire and B cell subtypes in health controls (non-HIV-infected, HCs), HIV-infected immunological responders (IRs), and immunological nonresponders (INRs). We found that the dominant usage of IGHV gene segments of naïve B cells and memory B cells were IGHV3 and IGHV4, and the diversity of BCR repertoire was decreased in INRs. Differentiation trajectory analysis showed that the low differentiation of naïve B cells was related to satisfactory immune status. The cell cycle of B cells with immune-specific genes of IgD+ B cells was degraded in INRs, which was mediated by the anaphase-promoting complex/cyclosome pathway in the phase of G2/M checkpoints. These findings provide significant insights to understand the function of B cell-mediated immune response in immune reconstitution after HIV infection.
Subject areas: Immunology, Immune response, Transcriptomics
Graphical abstract
Highlights
-
•
Different immune responses show distinctive BCR repertoire
-
•
IGHV3 and IGHV4 are dominant genes expressed in naive and memory B cells of IRs
-
•
Naive B cells mediate immune reconstitution in HIV-infected individuals on ART
Immunology ; Immune response; Transcriptomics
Introduction
The introduction of antiretroviral treatment (ART) prevents HIV replication, thereby reducing the viral load to an undetectable level and gradually increasing CD4+ T cells to the normal level (>500 cells/μL) simultaneously. However, around 20%–40% HIV-infected patients failed to restore their CD4+ T cell counts (<350 cells/μL) after receiving ART;1 these patients are classified as immunological nonresponders (INRs). Compared with HIV-infected individuals whose CD4+ T cell counts have reached more than 350 cells/μL (immunological responders, IRs), INRs have severely compromised immunity, leaving them susceptible to AIDS and opportunistic infections.2,3 It was reported that deficient immune reconstitution may relate to reduced production of progenitor cells in bone marrow, reduced thymic output, and abnormal immune activation.4,5 However, these studies are not sufficient to reveal the mechanisms of failed immune reconstitution after HIV infection.
B cell-mediated immune response is sustained by HIV-specific memory B cells and plasma cells,6 suggesting an important role of B cells in immune reconstitution after HIV infection. HIV infection alters the memory B cell compartment and reduces naïve B cells by increasing cell apoptosis and exhaustion, which leads to a decrease in the quality of response to HIV.6 The B cell receptors (BCRs) can react to antigens, thus leading to B cell clone expansion and secretion of high-affinity antibodies.7 The variety of BCR is determined by the usage and diversity of V, D, and J genes in complementary determining region 3 (CDR3). BCR rearrangement corresponds to diversity of immunoglobulin (Ig) segments during B cell development.7 There is a linkage between the B cell clone and stereotypical immune response by analyzing BCR repertoires.8,9 Therefore, we intend to investigate the role of B cells in failed immune reconstitution after HIV infection.
Single-cell RNA sequencing (scRNA-seq) provides an effective way to explore cellular heterogeneity, differentiation state, and gene expression models of cell populations.10 In the present study, BCR repertoire and corresponding characteristics of B cell subtypes were detected. We showed that a high ratio of naïve B cells with low differentiation was related to satisfactory immune reconstitution. Furthermore, cell cycle of B cells was impaired and mediated by the anaphase-promoting complex/cyclosome (APC/C) pathway in G2/M checkpoints.
Results
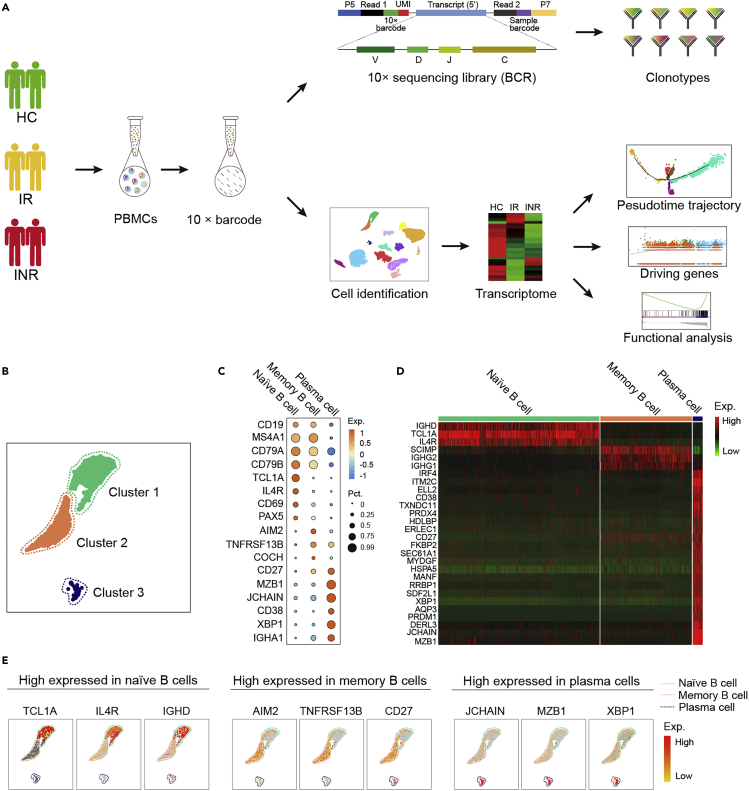
Three B cell subsets were identified by scRNA-Seq
We isolated peripheral blood mononuclear cells (PBMCs) of 13 participants to construct cDNA libraries and performed sc-RNA sequencing and sc-BCR sequencing using a 10x Genomics single-cell-based platform (Figure 1A). Three cluster (cluster 1, 2, and 3) cells were identified as B cells after data filtering (Figures 1B and S1). The B cell subsets corresponding to cluster 1 and cluster 2 were further annotated according to highly expressed CD19, CD79A, CD79B, and MS4A1 (Figure 1C). Furthermore, cluster 1 distinguishingly expressed high levels of TCL1A, IGHD, and IL4R (Figures 1D and 1E) and was defined as naïve B cells. Cluster 2 distinguishingly expressed IGHG1, IGHG2, AIM2, and TNFRSF13B and was defined as memory B cells (Figures 1D and 1E). Cluster 3 distinguishingly expressed IGHA1, CD38, JCHAIN, and MZB1 and was defined as plasma cells (Figures 1C–1E). By analyzing the expression and distribution of selected marker genes, we identified and confirmed phenotypes of naïve B cells, memory B cells, and plasma cells.
Figure 1.
Three B cell subsets were identified by scRNA-seq
(A) A schematic workflow of study design. PBMCs were collected and separated into three groups: healthy control (non-HIV-infected, HC), immunological responders (IRs), and immunological nonresponders (INRs).
(B) The UMAP plots of the B cell subsets distribution. A single dot represented a single cell.
(C) The bubble diagram of selected marker genes expression to distinguish the B cell subtypes from other PBMCs: naïve B cells (cluster 1), memory B cells (cluster 2), and plasma cells (cluster 3). The red bubble represented the higher expression of the marker genes; the large bubble represented a higher proportion of the B cell subset expressed marker genes.
(D) The heatmap of the selected marker genes expression to distinguish naïve B cells, memory B cells, and plasma cells.
(E) The UMAP plots denote the distribution of subtype-specific marker genes in naïve B cells, memory B cells, and plasma cells. The single dot represents a single cell, and the red dots represent cells with a higher expression level of marker genes.
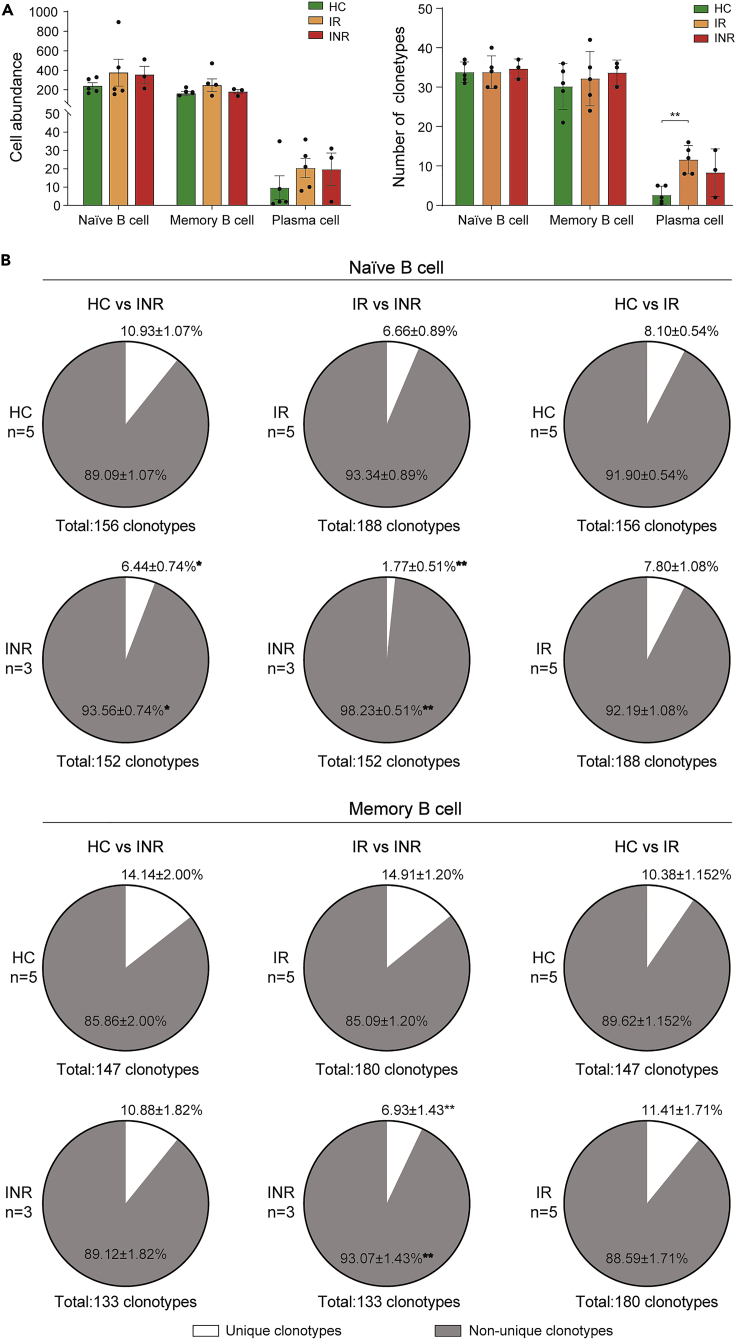
Proportions of unique BCR clonotypes of naïve B cells and memory B cells decreased in INRs
To reveal the BCR repertoire dynamic during immune reconstitution after HIV infection, we assessed the BCR repertoire among the HC, IR, and INR groups. Our results indicated that the abundance of naïve B cells and memory B cells with IGH was higher in the IR and INR groups than that in the HC group. The number of IGHV clonotypes of plasma cells was significantly higher in the IRs than that in the HC group (p < 0.01) (Figure 2A). Furthermore, we counted up the proportion of unique (detected in one group) and nonunique (detected in two groups) BCR clonotypes of naïve B cells and memory B cells in the comparison groups of HC vs. INR, HC vs. IR, and IR vs. INR. The results showed that the proportion of unique BCR clonotypes of naïve B cells and memory B cells was decreased in the INR group (p < 0.05) (Figure 2B), suggesting a decreased diversity of BCR repertoire in INRs. We were unable to analyze the BCR clonotypes of plasma cells due to their low percentage.
Figure 2.
Proportions of unique BCR clonotypes of naïve B cells and memory B cells decreased in INRs
(A) The abundance of B cell subsets with IGH and the number of clonotypes of IGHV.
(B) The proportion of B cells with unique BCR clonotypes in the comparisons of the HC and INR groups, the HC and IR groups, and the IR and INR groups, respectively. Data were presented as mean ± SEM. Unpairwise comparisons of values were performed using a Student’s t test. ∗p < 0.05, ∗∗p < 0.01.
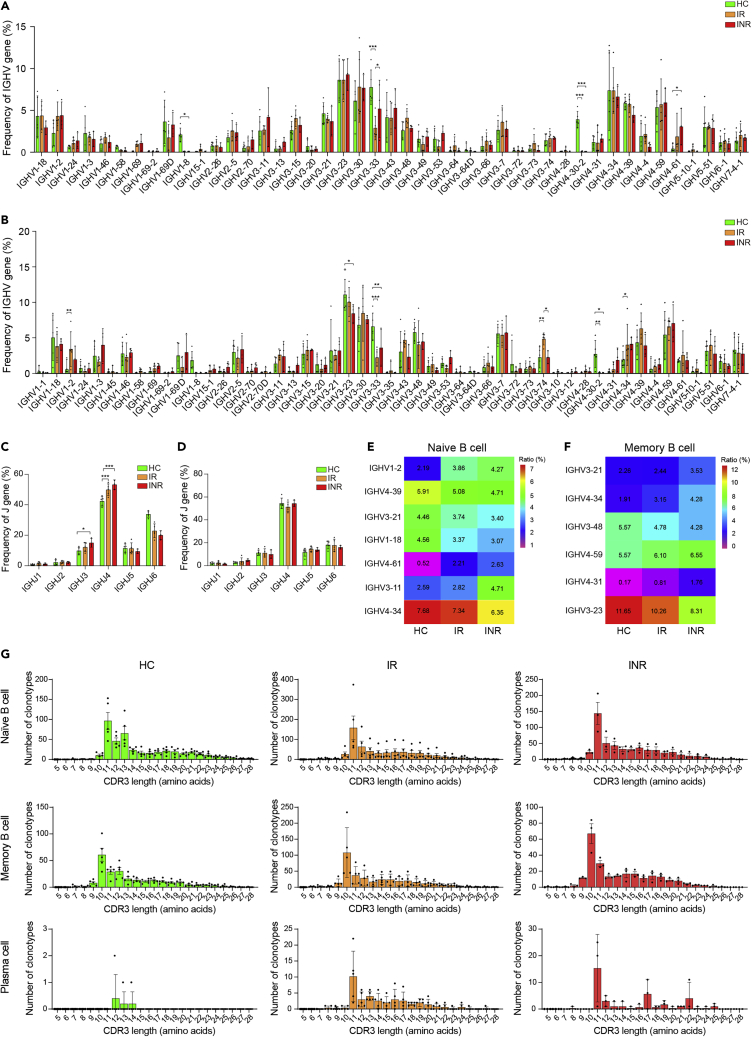
A high proportion of IGHV3 and IGHV4 gene usage in naïve B cells and memory B cells
We counted the frequency of BCR IGHV and IGHJ gene usage in naïve B cells and memory B cells among the HC, IR, and INR groups (Figure 3). The results showed that the dominant usage of the IGHV gene segment in naïve B cells (Figure 3A) and memory B cells (Figure 3B) were IGHV3 and IGHV4, and the dominant usage of the IGHJ gene segment was IGHJ4 (Figures 3C and 3D). The frequency of IGHJ4 in naïve B cells was higher in the IR and INR groups than that in the HC group (Figure 3C). Furthermore, the high frequency of IGHV3-11 and low frequency of IGHV4-34 and IGHV4-39 in naïve B cells were observed in the INR group compared with the IR and HC groups Figure 3E). The high frequency of IGHV3-21, IGHV4-31, IGHV4-34, and IGHV4-59 and low frequency of IGHV3-23 and IGHV3-48 in memory B cells were observed in the INR group compared with the IR and HC groups (Figure 3F).
Figure 3.
A high proportion of IGHV3 and IGHV4 gene usage in naïve B cells and memory B cells
(A) The distribution of BCR IGHV gene segment usage in naïve B cells.
(B) The distribution of BCR IGHV segment usage in memory B cells.
(C) The distribution of BCR IGHJ gene segment usage in naïve B cells.
(D) The distribution of BCR IGHJ gene segment usage in memory B cells. Data are presented as mean ± SEM (Tables S2–S5).
(E) The different proportions of IGHV gene segments usage in naïve B cells among the HC, IR, and INR groups.
(F) The different proportions of IGHV gene segments usage in memory B cells among the HC, IR, and INR groups.
(G) The distribution of CDR3 amino acids length in naïve B cells, memory B cells, and plasma cells, respectively. Data were presented as mean ± SEM Unpairwise comparisons of values were performed using a Student’s t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
We further analyzed the CDR3 amino acids length distribution in three B cell subsets among the HC, IR, and INR groups. The results showed that the length of CDR3 was mainly distributed at 11–13 amino acids for most BCR clonotypes in naïve B cells, the dominant length of CDR3 was 10 amino acids for most BCR clonotypes in memory B cells, and the dominant length of CDR3 was 11 amino acids for most BCR clonotypes in plasma cells in the IR and INR groups (Figure 3G). There were no statistical differences in CDR3 length among the IR, INR, and HC groups.
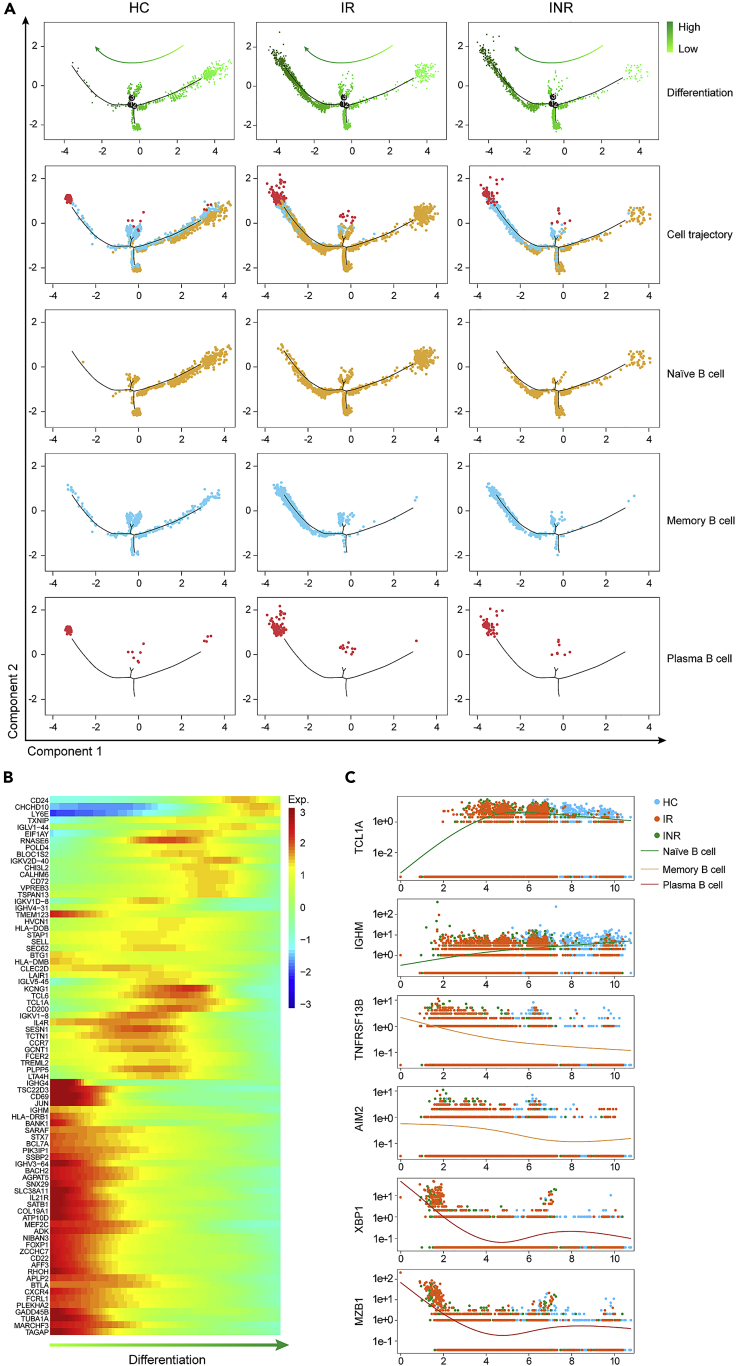
Lower differentiation of naïve B cells in IRs
We further investigated the differentiation trajectories of naïve B cells, memory B cells, and plasma cells. The results showed that naïve B cells, memory B cells, and plasma cells formed into a relative process with different cell differentiation statuses in pseudo-time (Figure 4A). The diagrams showed a higher density of cell populations in high differentiation statuses in the IR and INR groups than that in the HC group, and the cells with low differentiation statuses were decreased in the IR and INR groups compared with the HC group (Figure 4A). By exploring the distribution of naïve B cells, memory B cells, and plasma cells in pseudo-time trajectories, we showed a higher density of memory B cells and plasma cells were distributed on the high differentiation statuses in the IR and INR groups than that in the HC group, but there was no significant difference in memory B cells and plasma cells distribution between the INR and IR groups (Figure 4A). For naïve B cells, the decreased density was observed on the low differentiation statuses in the INR group compared to the IR group, while most naïve B cells were distributed on low differentiation statuses in the HC group, which suggested that lower differentiation of naïve B cells was related to satisfactory immune status.
Figure 4.
Decreased naïve B cells with low differentiation in INRs
(A) The pseudo-time differentiation status of three B subtypes among the HC, IR, and INR groups. Different colors represent the different directions of cell differentiation. The darker of green color is, the higher differentiation of B cells is. A single dot represents a single cell.
(B) The expression tendency of differentiation-specific genes during B cell differentiation.
(C) The expression tendency of subtype-specific marker genes for naïve B cells, memory B cells, and plasma cells among the HC, IR, and INR groups. Each dot represented a single cell that expressed a corresponding gene. The x axis was pseudo-time and the y axis was the level of gene expression. The green lines indicate subtype-specific marker genes for naïve B cells, the orange lines indicate subtype-specific marker genes for memory B cells, and the red lines indicate subtype-specific marker genes for plasma cells.
We next analyzed the signature genes during cell differentiation statuses in pseudo-time. The results showed that B cells highly expressed IGHG4, TSC22D3, CD69, IL21R, FOXP1, CD22, and BACH2 at the beginning of B cell differentiation. The above genes levels were gradually decreased, and the expression of KCNG1, CD200, IGKV1-8, SESN1, GCNT1, and PLPP5 were gradually increased during B cell differentiation, while these genes were gradually downregulated at the late stage of B cell differentiation (Figure 4B).
We further analyzed signature genes of naïve B cells, memory B cells, and plasma cells (Figure 4C). The diagram showed that the expression tendency of TCL1A and IGHM were increased in naïve B cells and there was a significant distinction of cell distribution in pseudo-time between the HC and IR groups, while cell distribution was mixed between the INR and IR groups. In addition, there was a high ratio of memory B cells with expression of AIM2 and TNFRSF13B and a high ratio of plasma cells with expression of XBP1 and MZB1 in the IR group compared with the the HC group, which suggested different characteristics of naïve B cells, memory B cells, and plasma cells among the HC, IR, and INR groups. The distribution of naïve B cells with TCL1A and IGHM, memory B cells with AIM2 and TNFRSF13B expression, and plasma cells with XBP1 and MZB1 expression were highly overlapped in the IR and INR groups rather than in the HC group, suggesting the similarities of naïve B cells, memory B cells, and plasma cells between the IR and INR groups (Figure 4C).
Taken together, naïve B cells had a lower degree of cell differentiation and expressed a higher level of IGHM in the HC group than that in the IR and INR groups. Naïve B cells had a higher degree of cell differentiation in IRs and INRs and had a differentiation relationship with memory B cells and plasma cells, suggesting a positive correlation between satisfactory immune status and the high abundance of naïve B cells with low differentiation.
Abnormal function of B cells involved in failed immune reconstitution
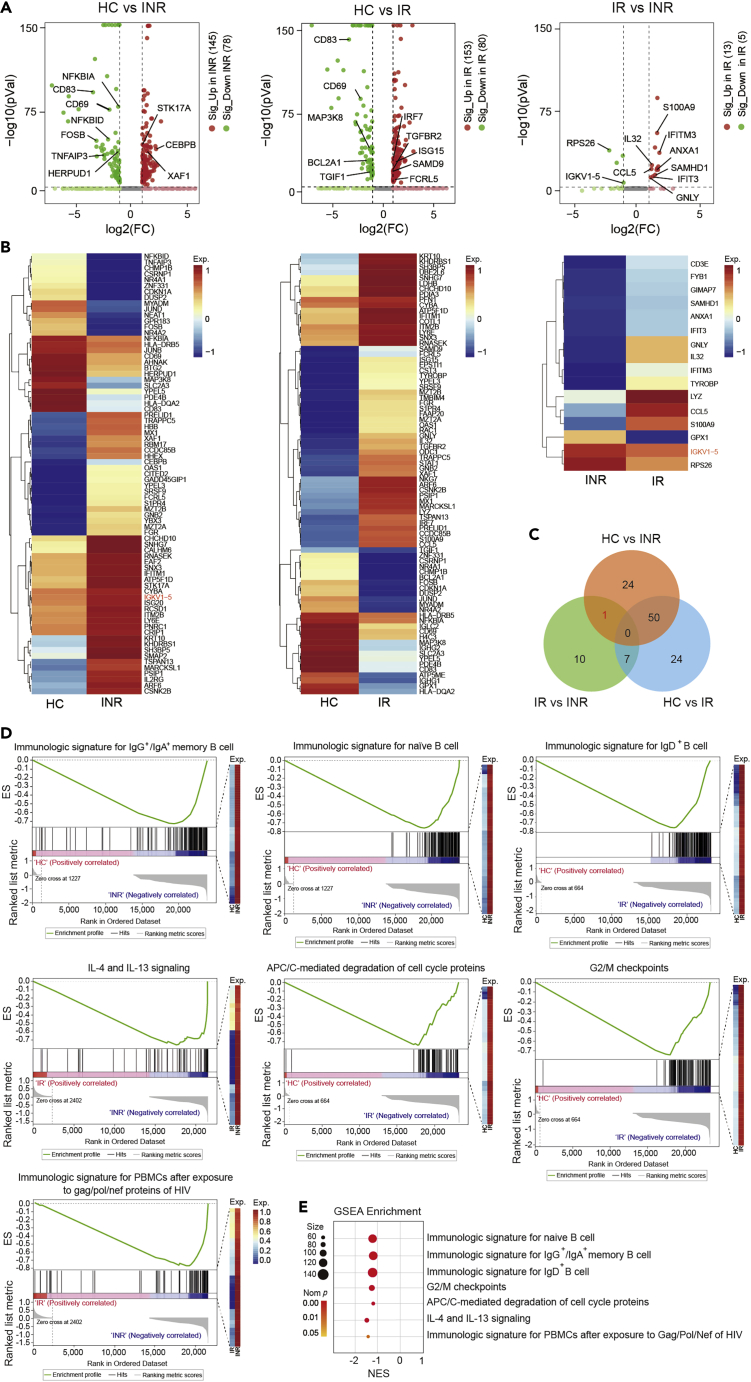
To analyze the function of B cells in immune reconstitution after HIV infection, we expanded the sample size to perform single-cell transcriptome sequencing. We detected the differential expressed genes (DEGs) of B cells among the 8 HC individuals, 10 IR individuals, and 8 INR individuals. A total of 75 significant DEGs were identified in B cells between the HC and INR groups, 81 significant DEGs between the HC and IR groups, and 18 significant DEGs between the IR and INR groups (Figures 5A and 5B). The top 10 DEGs were annotated in volcano plots (Figure 5A). A total of 50 significant DEGs were identified both in the comparison group of HCs vs. INRs and the comparison group of HCs vs. IRs, which may be related to HIV infection. A DEG of IGKV1-5 was identified in the comparison group of HCs vs. INRs and the comparison group of IRs vs. INRs, but not in the comparison group of HCs vs. IRs (Figures 5B and 5C), which may be related to the failed immune reconstitution after HIV infection.
Figure 5.
Abnormal function of B cells involved in failed immune reconstitution
(A) The volcano plot of DEGs expressed in B cells in comparisons between the HC and INR groups, the HC and IR groups, and the INR and IR groups, respectively. DEGs were defined as log2 FC ≥ 1 and p < 0.05.
(B) Heatmap showed the DEGs in B cells among the HC, IR, and INR groups.
(C) Venn diagram showed the amount of overlapping and nonoverlapping DEGs between the HC and INR groups, the HC and IR groups, or the INR and IR groups.
(D) GSEA-scoring plots of gene expression profile in B cells from the HC, IR, and INR groups. Seven plots of representative gene set relevant to immunologic function that were significantly enriched. The gene list corresponding to significantly enriched pathways was shown in Figure S2.
(E) The bubble diagram showed the p value and enrichment score of significantly enriched pathways.
We further analyzed our data with gene set enrichment analysis (GSEA) (Table S1). Compared to the HC group, the increased gene sets in the INR groups were associated with immunological signatures for IgG+/IgA+ memory B cells and naïve B cells. Compared to the HC group, the increased genes in the IR group were associated with immunological signature for IgD+ B cells, APC/C-mediated degradation of cell cycle proteins, and G2/M checkpoints. Compared to the IR group, the increased genes in the INR group were associated with IL-4/IL-13 signaling and immunologic signature for PBMCs after exposure to Gag/Pol/Nef proteins of HIV (Figures 5D and 5E). The relative expression of immunologic signature genes for the above B cell subtypes was shown in Figure S2. Furthermore, the immunological signature genes for IgD+ B cells were mainly enriched in the process of cell cycle and involved in the early phase of the HIV life cycle, while these functions were not enriched in immunological signature genes for naïve B cells (Figure S3). Together, these results suggested an abnormal function of memory B cells and naïve B cells, including IL-4 and IL-13 pathway and APC/C-mediated G2/M cell cycle, involved in failed immune reconstitution after HIV-1 infection.
We further analyzed DE-Gs in naïve B cells, memory B cells, and plasma cells that related to HIV infection under the criteria of significantly differential expressed in the comparison of HC vs. INR and comparison of HC vs. IR, but not in the comparison of IR vs. INR. The results showed that 147 DEGs in naïve B cells, 113 DEGs in memory B cells, and 22 DEGs in plasma cells were identified. Functional analysis demonstrated that DEGs that related to HIV infection in naïve B cells enriched the pathway of cytokine signaling in the immune system and DEGs that related to HIV infection in memory B cells enriched the pathway of interferon signaling (Figure S4).
Discussion
Failed restoration of CD4+ T cell level leads to increased morbidity and mortality of patients with HIV infection.11 The abnormal B cell function is a critical factor for the progression of HIV infection, including excessive activation of immature/transitional and short-life plasma B cells, declining the resident memory B cells response, and increased expression of Fas to accelerate apoptosis progression of B cells.12,13,14,15 Moreover, higher expression of IL-10 of Breg or Treg cells could reduce or inhibit viral-specific effector T cells response in HIV-infected individuals.16 The results of the present study indicate a potential way in which B cells affect immune status after HIV infection.
Our study demonstrated that the BCR repertoire diversity decreased in INRs, and there was a similar feature of CDR3 amino acids length between memory B cells and naïve B cells among HCs, IRs, and INRs. Furthermore, the dominant IGHV gene usage of memory B cells and naïve B cells were IGHV3 and IGHV4, and the dominant IGHJ gene usage was IGHJ4. However, the frequency of IGHV3 and IGHV4 gene usage was changed obviously after virus infection.17 Previous studies provided evidence that COVID-19 infection resulted in changed IGHV and IGHJ gene usage.8,18 Our data indicated a low level of IGHV4-34 and IGHV4-39 gene usage of naïve B cells and a low level of IGHV3-23 and IGHV3-48 gene usage of memory B cells in the INR group. HIV infection could reduce the response of memory B cells, which may correspond to the reduced frequency of usage of paired IGHJ4 to most IGHV genes in INRs compared to HCs.6 Overall, we assumed that B cells experienced obvious specific V(D)J rearrangements in INR individuals, which played an impaired role in HIV infection.
In our study, high levels of TCL1A and IGHD in naïve B cells, high levels of AIM2 and TNFRSF13B in memory B cells, and high levels of XBP1 and MZB1 in plasma cells were observed, which were consistent with previous studies.19,20,21 Furthermore, we identified naïve B cells as the starting point of differentiation to memory B cells by cellular trajectory analysis, which was coincidental with the similar distribution of BCR V/J gene usage between naïve B cells and memory B cells in the present study. In addition, we showed a reduced naïve B cell set with low differentiation in INRs compared with IRs. It has been shown that the excessive activation and apoptosis of immature B cells observed in advancing HIV-infected patients could explain the less differentiated degree of naïve B cells,13,22 and the correlation between declined immune response and a low percentage of naïve T cells was observed in HIV-infected individuals on ART.23,24,25 The high differentiation in memory B cells and plasma cells was associated with persistent or chronic HIV infection, even in HIV patients with restored CD4+ T cells after ART.6 The present study displayed a decreased percentage of memory B cells and low differentiation of naïve B cells in INRs, indicating a vital role of naïve B cells in immune reconstitution after HIV infection.
We further analyzed the changed function of B cells using GSEA. We revealed a gene set associated with immunological signatures for IgG+/IgA+ memory B cells was highly expressed in INRs rather than in HCs. A recent study demonstrated that IgA and IgG antibodies of B cell lineages were related to the development of vulnerability to the HIV-1 site.26 The level of memory B cells was correlated with CD4+ T cell counts in patients, which could influence HIV-infected disease progression.27 Abnormal memory B cells could contribute to the ineffectiveness of the antibody response in HIV-infected individuals.28 The less differentiation of naïve B cells in INRs than in IRs suggests naïve B cells could be influenced by immune reconstruction. In addition, abnormal expression of the gene sets enriched in the function of APC/C-mediated degradation of cell cycle proteins and G2/M checkpoints were observed in IRs rather than in HCs. HIV-1 Vif degrades essential phosphorylation regulators to induce G2/M cell-cycle arrest,29 and HIV-1 Vpr promotes cell-cycle arrest through the depletion of CCDC137.30 Combining the results of the present study, it was indicated that the abnormal cell cycle of B cells affected immune status after HIV-1 infection. Furthermore, a gene set associated with the signaling pathway of IL-4/IL-13 was highly expressed in INRs rather than in IRs. Our results indicated that low differentiation of naïve B cells may be correlated with abnormal B cell (IgG+/IgA+ memory B cells and IgD+ B cells) response, and the altered cell cycle of these abnormal B cells may promote failed immune restoration after HIV infection.
In conclusion, our BCR repertoire and B cell subsets transcriptome analysis provides comparative information on the role of B cell-mediated immune response in immune reconstitution after HIV infection. We demonstrated that the BCR clonotypes were decreased in INRs, low differentiation of naïve B cells with special immunological signature genes for IgD+ B cells was related to the immune status of HIV-1-infected patients, and the altered cell cycle of these B cells may promote failed immune restoration after HIV infection. Future studies are necessary to investigate the development and function of naïve B cells after HIV infection.
Limitations of the study
We would like to unravel the potential function of B cells in HIV infection in the present study. Although, we performed function annotation for B cells among different groups, and an altered cell cycle of B cells was identified in the INR group, more specialized experiments need to be conducted to verify the altered functions of B cells in INRs. Also, it will be important to determine the relationship between heterogeneity of B cell differentiation and immune reconstitution.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Chromium Single Cell 5′ Library Kit | 10× GENOMICS | PN-1000002 |
| Chromium Single Cell 5′ Gel Bead Kit | 10× GENOMICS | PN-1000003 |
| Chromium Single Cell 3’/5′ Library Construction Kit | 10× GENOMICS | PN-1000020 |
| Chromium Single Cell A Chip Kits | 10× GENOMICS | PN-1000009 |
| Chromium i7 Multiplex Kit | 10× GENOMICS | PN-120262 |
| Dynabeads MyOneTM SILANE | 10× GENOMICS | PN-2000048 |
| Ficoll-Paque™ PREMIUM density gradient media | GE Health | 17-5446-02 |
| Buffer EB | Qiagen | Cat#19086 |
| SPRIselect Reagent Kit | Beckman Coulter | Cat#B23318 |
| High Sensitivity DNA assay Kit | Agilent | SD-UF0000034 Rev. D |
| Deposited data | ||
| scRNA-seq for HIV-infected patients | This paper | BioProject: PRJNA783363 |
| scRNA-seq for heathy controls | Ref. [20] | Genome Sequence Archive: HRA001149 |
| Software and algorithms | ||
| R | R Development Core Team | cran.r-project.org |
| SingleR | R package | http://www.bioconductor.org/ |
| Seurat 3.1.1 | Ref.31 | http://satijalab.org/seurat |
| Cell Ranger | 10x Genomics | https://www.10xgenomics.com/support |
| Monocle | R package | http://cole-trapnell-lab.github.io/monocle-release/ |
| GSEA | GSEA and MSigDB Team | https://www.gseamsigdb.org/ |
| GraphPad Prism | GraphPad Software LLC | https://www.graphpad-prism.cn/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yi-Qun Kuang (yq610433@hotmail.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human subjects
HIV-infected IR and INR were enrolled under the following criteria: (1) HIV-1 positive; (2) aged 30–50 years old; (3) male; (4) Han ethnicity; (5) sustainable viral load suppression (<50 copies/mL plasma) over 2 years after ART; (6) no HIV-1 viral load blipped during ART; (7) no immune reconstitution inflammatory syndrome (IRIS) occurred during ART; and (8) no hepatitis C virus (HCV)/HBV/tuberculosis (TB) co-infection. Ultimately, 10 HIV-infected IR (mean age of 39.40 ± 9.94, CD4+ T cells >500 cells/μL) and 8 HIV-infected INR (mean age of 43.67 ± 8.74, CD4+ T cells <350 cells/μl) were enrolled at the Third People’s Hospital of Kunming City, the Zhengzhou Sixth People’s Hospital, and the Guangzhou Eighth People’s Hospital. In addition, 8 male Han ethnicity healthy controls (HC) with a mean age of 38.00 ± 7.12 were used as the control group, and the transcriptome and BCR sequencing data of the 5 HC subjects were obtained from selected samples under a 10× Genomics single-cell platform by Illumina sequencing in previously published work.20
Ethics statement
This study was approved by the Ethics Committee of the First Affiliated Hospital of Kunming Medical University (No. 2018-L-42).
Method details
Preparation for PBMC suspension
Approximately 5 mL of anti-coagulated venous blood of 26 participants (8 HCs, 10 IRs and 8 INRs) was obtained and processed within 1 hour of collection. PBMCs were prepared by using Ficoll PaqueTM PREMIUM density gradient media (GE Health, 17-5446-02) according to the manufacturer’s instructions.
BCR sequencing and data processing
The construction and sequencing of the BCR library were performed on platforms of Chromium™ Single Cell V(D)J Solution (Gene Denovo), which used GemCode to package barcoded single-cell 5′ gel beads (10× genomics, PN-1000020, PN-1000009, PN-1000003), a master mix with cells, and partitioning oil on a microfluidic chip to generate single-cell Gel Bead-In-EMlusion (GEMs). Then GEMs were recycled and purified by Dynabeads MyOneTM SILANE (10× genomics, PN-2000048), the single-cell 5′ gel bead was dissolved and any co-partitioned cell was lysed by Buffer EB (Qiagen, Cat#19086) to release mRNAs and performed reverse transcription to produce barcoded, full-length cDNA from poly-adenylated mRNAs. After incubation, the GEMs were broken and pooled post-GEM-RT were recovered, barcoded cDNAs were extracted and purified by SPRIselect Reagent Kit (Beckman Coulter, Cat#B23318), then full-length V(D)J segments can then be enriched and amplified by PCR to generate sufficient mass for library construction.
The amplification products were enzymatic fragmentation and the sizes were selected by using SPRIselect Reagent Kit (Beckman Coulter, Cat#B23318) to generate variable length fragments that collectively span the V(D)J segments of the enriched BCR transcripts. By adding primer sequences and sample index, the enriched transcripts were used to construct the library via end repair, A-tailing, adaptor ligation, and PCR amplification. The single-cell V(D)J reagent kit protocol produces V(D)J enriched and 5′ gene expression Illumina-ready sequencing libraries, a library comprising standard Illumina paired-end constructs which begin and end with P5 and P7, the libraries were quality controlled by Agilent 2100 Bioanalyzer using High Sensitivity DNA assay Kit (Agilent, Cat# SD-UF0000034 Rev. D). The final libraries contained the P5 and P7 priming sites were used in Illumina bridge amplification. After sequencing, we used Cell Ranger (https://support.10xgenomics.com/single-cell-vdj/software/overview/welcome) to assemble, quantify, and annotate paired V(D)J transcript sequences.
Identification of clonotyping and diversity
Cell barcodes were grouped into clonotypes if they share the same set of productive CDR3 nucleotide sequences. A three-layer donut chart was used to visualize the repertoire clonality. Rarefaction analysis was used to compare the repertoire diversity, rarefaction curves were interpolated from 0 to the current sample size and then extrapolated up to the size of the largest of the samples. Repertoire information of all samples was exhibited using a set of basic features: CDR3 abundance, CDR3 length, Variable (V) and Joining (J) segment usage, the length of V/J gene in the CDR3 region, and V-J gene paired frequency in CDR3 junctions.
Single-cell RNA sequencing and data processing
The PBMC suspensions of 26 participants were loaded on a 10× Genomics GemCode Single-cell instrument that generates single-cell Gel Bead-In-EMlusion (GEMs). Single-cell capturing and downstream library constructions were performed using Chromium i7 Multiplex Kit (10× GENOMICS, PN-120262) and Chromium Single Cell 5′ Library Kit (10× GENOMICS, PN-1000002). After dissolution of the Gel Bead in a GEM, primers containing Illumina® R1 sequence, a 16-nt 10 × barcode, a 10-nt Unique Molecular Identifier (UMI), and a poly-dT primer sequence were released and mixed with cell lysate and Master Mix. Barcoded full-length cDNAs were then reverse-transcribed from poly-adenylated mRNAs. Residual biochemical reagents and primers were removed by using Dynabeads MyOneTM SILANE (10× GENOMICS, PN-2000048) in a post-GEM reaction mixture. Then, the barcoded cDNAs were amplified by PCR to construct the library. Read primers R1 and R2, sample index P5 and P7 were then added during library construction via End Repair, A-tailing, Adaptor Ligation, and PCR. The final libraries containing the P5 and P7 primers were used in Illumina bridge amplification.
ScRNA-seq was performed on the Illumina platform by Gene Denovo Biotechnology Co. 10× Genomics Cell Ranger was used to convert raw data files to FASTQ files. Reads with low-quality barcodes and UMIs were filtered out and then clean reads mapped to reference genome, and reads mapped to transcriptome uniquely and intersecting an exon at least 50% were considered for UMI counting. Quantification was performed based on corrected UMI sequences and valid barcodes, the cells by gene matrices were produced via UMI counting and cell barcode calling. The cells with gene matrices for each sample were individually imported to Seurat31 version 3.1.1. Cells with a high number of UMIs (≥8000), a high proportion of mitochondrial genes (≥10%), and abnormal gene numbers <500 or >4000 were filtered out. Then, we used “LogNormalize” to normalize gene expression and used Seurat to minimize the effects of batch effect and behavioral conditions on clustering. We performed canonical correspondence analysis, and used Mutual Nearest Neighbors (MNN) to construct correspondence relationships among cells, the correspondence relationship was used as anchors to complete data integration and batch normalization. Furthermore, we measured the correlation between each cell subset, then used Uni-form Mani-fold Approximation and Projection for Dimension Reduction (UMAP) to visualize cell clusters. Cell type annotation was performed by the SingleR R packages.
Trajectory analysis of cell differentiation
Cell trajectory was analyzed using the matrix of cells and gene expressions by Monocle. Monocle reduced the space down to one with two dimensions and ordered the cells (sigma = 0.001, lambda = NULL, param. gamma = 10, tol = 0.001).32 The trajectory has a tree-like structure, including tips and branches. The branches occur because cells execute alternative gene expression programs. Monocle develops BEAM to test for branch-dependent gene expression by formulating the problem as a contrast between two negative binomial GLMs.33 In addition, monocle can find genes that are differentially expressed between groups of cells and assess the statistical significance of those changes, we identified key genes related to the development and differentiation process with FDR <10−5.
Differential gene expression and function annotation
Differentially expressed genes (DEGs) were analyzed between HCs and IRs, HCs and INRs, IRs and INRs. DE-Gs were selected under the condition of log2 fold change (FC) ≥ 1 and adjusted p < 0.05. For functional annotation, gene set enrichment analysis (GSEA) was performed with 1000 permutations for each test. Nom p < 0.05 and |NES| > 1 was statistically significant for GSEA.
Quantification and statistical analysis
The statistical analysis was performed using GraphPad Prism 8.0 (GraphPad Software LLC) with an unpaired Student’s t test in the comparison groups. p < 0.05 was considered statistically significant, values denoted with ∗ reflected significance levels as follows: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Results were shown as the mean ± standard error of the mean (SEM).
Acknowledgments
We thank Dr. Chiyu Zhang at the Shanghai Public Health Clinical Center, Fudan University, Shanghai and Dr. Xianwen Ren at the Changping Laboratory, Beijing, for insightful discussions. This work was partly supported by the National Natural Science Foundation of China (32060177, 82260397 and 81660094), the Fund of Yunling Scholar (YLXL20170002), the General Joint Project from the Department of Science and Technology of Yunnan Province and Kunming Medical University (2017FE467(-038) and 2017FE467(-130)), the Project for Innovation Team of the Department of Science and Technology of Yunnan Province (2018HC005), the Department of Education of Yunnan Province (2019Y0352), the Health Commission of Yunnan Province (2018NS0085), the Fund of Yunnan Provincial Clinical Research Center for General Surgical Diseases and Yunnan Provincial Clinical Research Center for Skin Immune Diseases (2019ZF012) from the Department of Science and Technology of Yunnan Province, Yunnan Fundamental Research Projects (202201AT070292 and 202201AU070202), and the Open Research Fund of Beijing Key Laboratory for HIV/AIDS Research (BJYAHKF2021001). We would like to thank the participants at the Third People’s Hospital of Kunming City from whom the study samples were obtained and the staff involved in all the sample collection and processing.
Author contributions
Conceptualization, Y.Q.K., K.H.W., and J.H.W.; Methodology, J.J. and Y.Z.; Resources, D.L., J.Q.Y., J.H.M., J.H.W., and X.L.Z.; Investigation, J.J., Y.Z., D.L., and J.H.M.; Formal Analysis, J.J., Y.Z., D.L., J.H.W., and Y.Q.K.; Writing – Original Draft, J.J. and Y.Z.; Writing – Review & Editing, Y.Q.K.; Supervision, Y.Q.K.
Declaration of interests
The authors declare no competing interests.
Published: December 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105559.
Supplemental information
Data and code availability
-
•
Single-cell RNA-seq data have been deposited at Sequence Read Archive (SRA) database in National Center for Biotechnology Information (NCBI) and are publicly available as of the date of publication. This paper does not report original code. The accession number for the scRNA-seq data reported in this paper is PRJNA783363 (BioProject).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
Codes of preprocessing, normalization, clustering and plotting of single-cell datasets are available on Zenodo: https://doi.org/10.5281/zenodo.7262476.
References
- 1.Nakanjako D., Kiragga A.N., Musick B.S., Yiannoutsos C.T., Wools-Kaloustian K., Diero L., Oyaro P., Lugina E., Ssali J.C., Kambugu A., Easterbrook P. Frequency and impact of suboptimal immune recovery on first-line antiretroviral therapy within the International Epidemiologic Databases to Evaluate AIDS in East Africa. Aids. 2016;30:1913–1922. doi: 10.1097/QAD.0000000000001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang X., Su B., Zhang X., Liu Y., Wu H., Zhang T. Incomplete immune reconstitution in HIV/AIDS patients on antiretroviral therapy: challenges of immunological non-responders. J. Leukoc. Biol. 2020;107:597–612. doi: 10.1002/JLB.4MR1019-189R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shete A., Dhayarkar S., Sangale S., Medhe U., Panchal N., Rahane G., Yelgate R., Dhamanage A., Gangakhedkar R. Incomplete functional T-cell reconstitution in immunological non-responders at one year after initiation of antiretroviral therapy possibly predisposes them to infectious diseases. Int. J. Infect. Dis. 2019;81:114–122. doi: 10.1016/j.ijid.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Utay N.S., Hunt P.W. Role of immune activation in progression to AIDS. Curr. Opin. HIV AIDS. 2016;11:131–137. doi: 10.1097/COH.0000000000000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massanella M., Negredo E., Clotet B., Blanco J. Immunodiscordant responses to HAART--mechanisms and consequences. Expet Rev. Clin. Immunol. 2013;9:1135–1149. doi: 10.1586/1744666X.2013.842897. [DOI] [PubMed] [Google Scholar]
- 6.Moir S., Fauci A.S. B-cell responses to HIV infection. Immunol. Rev. 2017;275:33–48. doi: 10.1111/imr.12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cyster J.G., Allen C.D.C. B cell responses: cell interaction dynamics and decisions. Cell. 2019;177:524–540. doi: 10.1016/j.cell.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montague Z., Lv H., Otwinowski J., DeWitt W.S., Isacchini G., Yip G.K., Ng W.W., Tsang O.T., Yuan M., Liu H., et al. Dynamics of B cell repertoires and emergence of cross-reactive responses in patients with different severities of COVID-19. Cell Rep. 2021;35:109173. doi: 10.1016/j.celrep.2021.109173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galson J.D., Schaetzle S., Bashford-Rogers R.J.M., Raybould M.I.J., Kovaltsuk A., Kilpatrick G.J., Minter R., Finch D.K., Dias J., James L.K., et al. Deep sequencing of B cell receptor repertoires from COVID-19 patients reveals strong convergent immune signatures. Front. Immunol. 2020;11:605170. doi: 10.3389/fimmu.2020.605170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luecken M.D., Theis F.J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019;15:e8746. doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh S., Toor J.S., Sharma A., Arora S.K. Signature genes associated with immunological non-responsiveness to anti-retroviral therapy in HIV-1 subtype-c infection. PLoS One. 2020;15:e0234270. doi: 10.1371/journal.pone.0234270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vinuesa C.G. HIV and T follicular helper cells: a dangerous relationship. J. Clin. Investig. 2012;122:3059–3062. doi: 10.1172/JCI65175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malaspina A., Moir S., Ho J., Wang W., Howell M.L., O'Shea M.A., Roby G.A., Rehm C.A., Mican J.M., Chun T.W., Fauci A.S. Appearance of immature/transitional B cells in HIV-infected individuals with advanced disease: correlation with increased IL-7. Procd. Natl. Acad. Sci. USA. 2006;103:2262–2267. doi: 10.1073/pnas.0511094103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Titanji K., Chiodi F., Bellocco R., Schepis D., Osorio L., Tassandin C., Tambussi G., Grutzmeier S., Lopalco L., De Milito A. Primary HIV-1 infection sets the stage for important B lymphocyte dysfunctions. Aids. 2005;19:1947–1955. doi: 10.1097/01.aids.0000191231.54170.89. [DOI] [PubMed] [Google Scholar]
- 15.Rethi B., Sammicheli S., Amu S., Pensieroso S., Hejdeman B., Schepis D., Thang P.H., Chiodi F. Concerted effect of lymphopenia, viraemia and T-cell activation on Fas expression of peripheral B cells in HIV-1-infected patients. Aids. 2013;27:155–162. doi: 10.1097/QAD.0b013e32835b8c5e. [DOI] [PubMed] [Google Scholar]
- 16.Amu S., Ruffin N., Rethi B., Chiodi F. Impairment of B-cell functions during HIV-1 infection. Aids. 2013;27:2323–2334. doi: 10.1097/QAD.0b013e328361a427. [DOI] [PubMed] [Google Scholar]
- 17.Xiang H., Zhao Y., Li X., Liu P., Wang L., Wang M., Tian L., Sun H.X., Zhang W., Xu Z., et al. Landscapes and dynamic diversifications of B-cell receptor repertoires in COVID-19 patients. Hum. Immunol. 2022;83:119–129. doi: 10.1016/j.humimm.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J.Y., Wang X.M., Xing X., Xu Z., Zhang C., Song J.W., Fan X., Xia P., Fu J.L., Wang S.Y., et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020;21:1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- 19.Mukund K., Nayak P., Ashokkumar C., Rao S., Almeda J., Betancourt-Garcia M.M., Sindhi R., Subramaniam S. Immune response in severe and non-severe coronavirus disease 2019 (COVID-19) infection: a mechanistic landscape. Front. Immunol. 2021;12:738073. doi: 10.3389/fimmu.2021.738073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren X., Wen W., Fan X., Hou W., Su B., Cai P., Li J., Liu Y., Tang F., Zhang F., et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184:1895–1913.e19. doi: 10.1016/j.cell.2021.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y., Liu X., Xie L., Li J., Ye J., et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discovery. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho J., Moir S., Malaspina A., Howell M.L., Wang W., DiPoto A.C., O'Shea M.A., Roby G.A., Kwan R., Mican J.M., et al. Two overrepresented B cell populations in HIV-infected individuals undergo apoptosis by different mechanisms. Procd. Natl. Acad. Sci. USA. 2006;103:19436–19441. doi: 10.1073/pnas.0609515103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roul H., Mary-Krause M., Ghosn J., Delaugerre C., Pialoux G., Cuzin L., Launay O., Lacombe J.M., Menard A., De Truchis P., et al. CD4+ cell count recovery after combined antiretroviral therapy in the modern combined antiretroviral therapy era. Aids. 2018;32:2605–2614. doi: 10.1097/QAD.0000000000002010. [DOI] [PubMed] [Google Scholar]
- 24.Gandhi R.T., Spritzler J., Chan E., Asmuth D.M., Rodriguez B., Merigan T.C., Hirsch M.S., Shafer R.W., Robbins G.K., Pollard R.B., Team A. Effect of baseline- and treatment-related factors on immunologic recovery after initiation of antiretroviral therapy in HIV-1-positive subjects: results from ACTG 384. J. Acquir. Immune Defic. Syndr. 2006;42:426–434. doi: 10.1097/01.qai.0000226789.51992.3f. [DOI] [PubMed] [Google Scholar]
- 25.Rosado-Sanchez I., Herrero-Fernandez I., Alvarez-Rios A.I., Genebat M., Abad-Carrillo M.A., Ruiz-Mateos E., Pulido F., Gonzalez-Garcia J., Montero M., Bernal-Morell E., et al. A lower baseline CD4/CD8 T-cell ratio is independently associated with immunodiscordant response to antiretroviral therapy in HIV-infected subjects. Antimicrob. Agents Chemother. 2017;61:e00605–e00617. doi: 10.1128/AAC.00605-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorin V., Fernandez I., Masse-Ranson G., Bouvin-Pley M., Molinos-Albert L.M., Planchais C., Hieu T., Pehau-Arnaudet G., Hrebik D., Girelli-Zubani G., et al. Epitope convergence of broadly HIV-1 neutralizing IgA and IgG antibody lineages in a viremic controller. J. Exp. Med. 2022;219 doi: 10.1084/jem.20212045. e20212045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Titanji K., De Milito A., Cagigi A., Thorstensson R., Grutzmeier S., Atlas A., Hejdeman B., Kroon F.P., Lopalco L., Nilsson A., Chiodi F. Loss of memory B cells impairs maintenance of long-term serologic memory during HIV-1 infection. Blood. 2006;108:1580–1587. doi: 10.1182/blood-2005-11-013383. [DOI] [PubMed] [Google Scholar]
- 28.Kardava L., Moir S., Shah N., Wang W., Wilson R., Buckner C.M., Santich B.H., Kim L.J., Spurlin E.E., Nelson A.K., et al. Abnormal B cell memory subsets dominate HIV-specific responses in infected individuals. J. Clin. Investig. 2014;124:3252–3262. doi: 10.1172/JCI74351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salamango D.J., Harris R.S. Demystifying cell cycle arrest by HIV-1 Vif. Trends Microbiol. 2021;29:381–384. doi: 10.1016/j.tim.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang F., Bieniasz P.D. HIV-1 Vpr induces cell cycle arrest and enhances viral gene expression by depleting CCDC137. Elife. 2020;9:e55806. doi: 10.7554/eLife.55806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu X., Hill A., Packer J., Lin D., Ma Y.A., Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods. 2017;14:309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Single-cell RNA-seq data have been deposited at Sequence Read Archive (SRA) database in National Center for Biotechnology Information (NCBI) and are publicly available as of the date of publication. This paper does not report original code. The accession number for the scRNA-seq data reported in this paper is PRJNA783363 (BioProject).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
Codes of preprocessing, normalization, clustering and plotting of single-cell datasets are available on Zenodo: https://doi.org/10.5281/zenodo.7262476.