Abstract
We produced isogenic Escherichia coli K-12 lysogens of seven different Shiga toxin 2 (Stx2)-encoding bacteriophages derived from clinical Shiga toxin-producing E. coli (STEC) isolates of serotypes O157:H7, O145, O111, and O83 to assess the variability among these phages and determine if there were phage-related differences in toxin production. Phage genomic restriction fragment length polymorphisms (RFLP) and superinfection resistance studies revealed significant differences among these phages and allowed the seven phages to be placed into five distinct groups. Experiments revealed striking differences in spontaneous phage and toxin production that were correlated with the groupings derived from the RFLP and resistance studies. These results suggest that the genotype of the Stx2 prophage can influence the level of phage release and toxin expression by host strains and thus may be relevant to STEC pathogenesis.
Escherichia coli O157:H7 and other Shiga toxin-producing E. coli (STEC) are emerging pathogens responsible for both outbreaks and sporadic cases of diarrhea (12). In addition to diarrhea, some patients develop hemorrhagic colitis, hemolytic uremic syndrome, or thrombotic thrombocytopenic purpura after exposure to STEC (9). These severe clinical consequences of STEC infection are believed to be caused by the activity of Shiga toxins 1 and 2 (Stx1 and Stx2), although Stx2 appears to be more closely associated with these sequelae (4, 17, 20). E. coli isolates of more than 60 serotypes have been found to produce Shiga toxins and to be associated with human disease (2). The genes encoding the Shiga toxin A and B subunits in several well-studied STEC isolates have been found to reside on lambdoid prophages (11, 16), and STEC isolates appear, as a rule, to be lysogens, converted to toxinogenicity by bacteriophages. The role of the biology of these phages in host strain virulence, beyond merely carrying the Shiga toxin genes to the STEC strains during evolution, has been relatively unexplored. Recently, however, it was demonstrated that a phage-encoded transcription factor is able to activate Shiga toxin expression, suggesting a mechanism whereby phage regulation might influence host strain virulence (15). Furthermore, intraintestinal transmission of these phages has been demonstrated to occur among E. coli strains (3), and infectious Stx2-encoding phages were recently detected in municipal sewage (14), emphasizing that Stx-encoding bacteriophages have retained the capacity to disseminate Shiga toxin genes among nontoxigenic E. coli and other bacterial species as well.
Stx2-encoding bacteriophages may thus play a more active role in STEC pathogenesis than simply acting as the carrier of toxin genes, and an assessment of the level of biological diversity among these phages could reveal variations in, for example, levels of toxin expression relevant to the virulence of their hosts. Significant genomic divergence among Stx1-encoding phages isolated from different E. coli serotypes has been demonstrated by analysis of phage genome lengths and restriction fragment length polymorphisms (RFLP) (25). Recent studies have revealed some variation within O157-derived Stx2-encoding phages (6, 22). In the present study, to assess the level of variation among Stx2-encoding phages derived from multiple STEC serotypes and its possible relevance to toxin regulation, we produced isogenic lysogens of seven different Stx2-encoding phages. These seven lysogens were used in RFLP and superinfection resistance studies, which allowed their phages to be placed into five distinct groups. Investigation of spontaneous phage release and toxin production from these isogenic lysogens revealed striking differences in both phage titer and toxin production that were consistent with the RFLP and resistance groupings. These experiments suggest that the genotypes of the Stx2 prophages can influence the level of toxin expression by host strains and thus may be relevant to STEC pathogenesis.
Construction of isogenic lysogens, RFLP analysis, and superinfection resistance analysis.
We set out to examine the diversity of Stx2-encoding phages derived from various clinical STEC isolates of multiple serotypes and to investigate the possible contribution of phage heterogeneity to the regulation of toxin expression. The places and dates of isolation of the Stx2-encoding clinical isolates used in this study are presented in Table 1. These clinical isolates were obtained from stool samples of patients with either bloody diarrhea or hemolytic uremic syndrome (1). In order to eliminate the contribution of additional prophages or host factors in the diverse STEC clinical isolates from which Stx2-encoding phages were isolated, we purified Stx2-encoding phages from single plaques and subsequently lysogenized an E. coli K-12 derivative, MC1000 (5), with the purified phage. We were successful in producing MC1000 lysogens of six new Stx2-encoding phages, as well as phages λ, 933W (the prototype Stx2-encoding phage), and H19B (the prototype Stx1-encoding phage.)
TABLE 1.
Bacterial strains used in this study
| Strain | Descriptiona | Source or reference |
|---|---|---|
| MC1000 | E. coli K-12, F−galK16 galE15 relA1 rpsL150 spoT1 mcrB1 | 7 |
| C600 | E. coli K-12, F−thi-1 thr-1 leuB6 lacY1 tonA21 supE | 17 |
| C600(933W) | Lysogen of O157:H7-derived Stx2-encoding phage 933W | 31 |
| C600(λcI857) | Lysogen of phage λ with heat-labile repressor mutation cI857 | This study |
| C600(H19B) | Lysogen of Stx1-encoding lambdoid phage H19B | 18 |
| STEC clinical isolates | ||
| 933 | Oregon, 1982; serotype O157:H7 | 31 |
| STEC-10 | South Dakota, 1983; serotype O145:NM | 75-83(CDC)b |
| STEC-2 | Ohio, 1997; serotype O157:H7 | 9:100 (1) |
| STEC-3 | Michigan, 1997; serotype O157:H7 | 1:361 (1) |
| STEC-14 | Serotype O157:H7 | B94007314(CDC) |
| STEC-16 | Nebraska, 1985; serotype O111:NM | 3007-85(CDC) |
| STEC-19 | Ohio, 1990; serotype O83:H1 | 3119-90(CDC) |
| MC1000 lysogens | ||
| MC1000(933W) | Phage isolated from 933 (O157) | This study |
| MC1000(φ10) | Phage isolated from STEC-10 (O145) | This study |
| MC1000(φ2) | Phage isolated from STEC-2 (O157) | This study |
| MC1000(φ3) | Phage isolated from STEC-3 (O157) | This study |
| MC1000(φ14) | Phage isolated from STEC-14 (O157) | This study |
| MC1000(φ16) | Phage isolated from STEC-16 (O111) | This study |
| MC1000(φ19) | Phage isolated from STEC-19 (O83) | This study |
| MC1000(λcI857) | Lysogen of phage λ with heat-labile repressor mutation cI857 | This study |
| MC1000(H19B) | Lysogen of Stx1-encoding lambdoid phage H19B | This study |
For clinical isolates, date and location of isolation and serotype are given when known. For MC1000 lysogens, the strain from which each phage was isolated is given.
CDC indicates strains acquired from the Centers for Disease Control and Prevention.
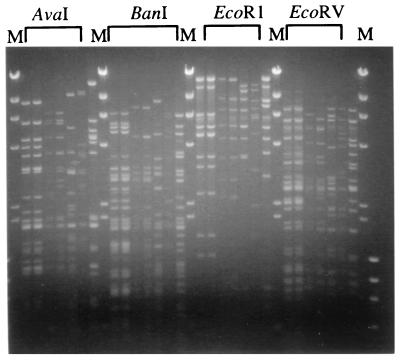
Phage genomic DNA was isolated from each of the seven Stx2-encoding phages (including 933W) by use of a commercial kit (Qiagen, Valencia, Calif.) RFLP among the seven phage genomes were detected by digestion with the enzymes AvaI, BanI, EcoRI, and EcoRV. The marked polymorphism in fragment lengths seen in Fig. 1 was used to assign the seven phages to five distinct RFLP groups. Lysogens of each phage were tested for resistance to superinfection by the other phages. This was done by spotting a sample of each of the seven phages on lawns of each lysogen and noting the presence or absence of lysis. Each lysogen was found to be resistant to infection by its own phage and susceptible to phages λ and H19B (Table 2). Furthermore, resistance was observed within but not among the five groups assigned by RFLP analysis. Thus, phages 933W and 10 shared a RFLP pattern and were mutually resistant to superinfection; phages 2 and 3 constituted a similar pair, while each of the remaining phages (phages 14, 16, and 19) constituted its own group. Our finding of five distinct groups among seven isolated phages implies that a high level of diversity exists among Stx2-encoding phages, especially considering the fact that our methods selected for phages sufficiently similar to lysogenize a common host. The relative conservation of the Shiga toxin gene sequences (as indicated by the fact that Stx2A sequences from all seven phages could be amplified with the same PCR primers) in the context of such diversity suggests that these genes were acquired relatively recently by this group of phages. This likely reflects significant ongoing recombination within the lambdoid phage gene pool (reviewed in reference 10).
FIG. 1.
RFLP in the Stx2-encoding genomes. Phage DNA was digested with the indicated restriction enzymes and electrophoresed in an agarose gel containing ethidium bromide. In the gel, the order of the digested phage DNA for each restriction enzyme is the same: φ933W, φ10, φ2, φ3, φ14, φ16, and φ19 (from left to right). M, HindIII-digested phage λ DNA and HaeIII-digested φX174 DNA markers.
TABLE 2.
Immunity or exclusion analysis of Stx2-encoding phages, phage λ, and phage H19B
| Recipient strain | Spot lysis with indicated phage
lysatea
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| φ933W | φ10 | φ2 | φ3 | φ14 | φ16 | φ19 | φλ | φH19B | |
| MC1000(933W) | − | − | + | + | + | + | + | + | + |
| MC1000(φ10) | − | − | + | + | + | + | + | + | + |
| MC1000(φ2) | + | + | − | − | + | + | + | + | + |
| MC1000(φ3) | + | + | − | − | + | + | + | + | + |
| MC1000(φ14) | + | + | + | + | − | + | + | + | + |
| MC1000(φ16) | + | + | + | + | + | − | + | + | + |
| MC1000(φ19) | + | + | + | + | + | + | − | + | + |
| MC1000(λcI857) | + | + | + | + | + | + | + | − | + |
| MC1000(H19B) | + | + | + | + | + | + | + | + | − |
Lawns of the indicated strains were tested for lysis by lysates containing the indicated phage. +, a spot of lysis on the lawn corresponding to the site of placement of a 10-μl drop of lysate; −, no lysis.
Differences between Stx2-encoding phages from serotype O157 STEC isolates have been reported (6, 22). Our results extend these findings to include Stx2-encoding phages derived from STEC of various serotypes, including O157, O145, O111, and O83. We found no evidence for a relationship between the RFLP and resistance properties of the phages and the serotype of the STEC strain from which they were isolated. Specifically, phages 933W and 10 were indistinguishable by RFLP and resistance analysis yet were isolated from strains of serotypes O157 and O145, respectively; conversely, phages 1, 3, and 14—all derived from O157 strains—were readily distinguished by RFLP and resistance studies. Among the various STEC serogroups, O157 strains have been shown to be closely related and are believed to share a common ancestry (24). It has been proposed that an early progenitor of the O157:H7 STEC complex was converted to toxinogenicity by an Stx2-encoding phage (23). Our findings of diverse Stx2 phages in different O157:H7 strains, taken together with the recent finding of two different O157-derived Stx2-encoding phages in Japan (22), suggests that Stx2 phage infection and lysogenic conversion of O157:H7 may have occurred more recently in STEC evolution than was previously appreciated and almost certainly rules out a single conversion event early in O157:H7 evolution.
Toxin and phage production by MC1000 lysogens and clinical isolates.
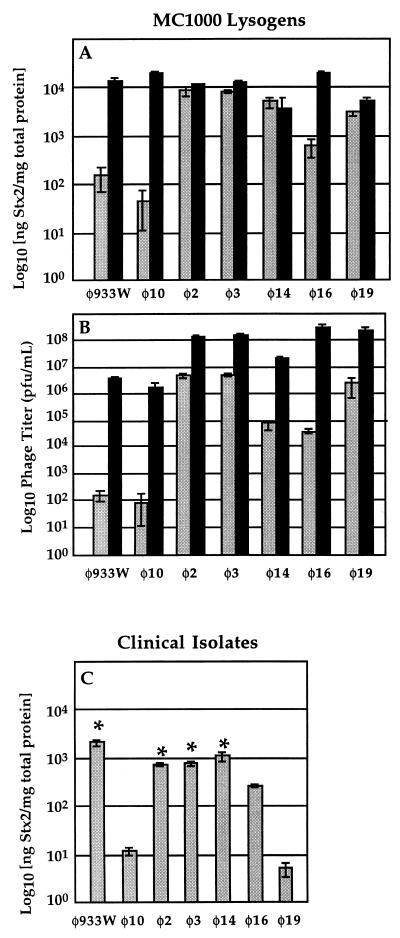
The significant genomic variation revealed by RFLP analysis suggested the possibility that differences in toxin production among the lysogens might also exist. Therefore, we examined amounts of spontaneous phage and toxin produced by the MC1000 lysogens. Phage titers were obtained by infecting lawns of MC1000, and total Stx2 from sonicated cultures was measured by a previously described enzyme-linked immunosorbent assay (7). Figure 2A and B presents the striking differences observed among the seven MC1000 strains in both phage and toxin production measured during mid-logarithmic growth phase. Large differences were observed among but not within the groups defined by the RFLP and resistance studies. Furthermore, some correlation between the amounts of toxin and phage produced is evident from the observation that the order of the seven lysogens ranked by phage titer is nearly identical to that ranked by toxin production. When normalized to total culture protein concentration, differences in toxin production by these isogenic strains varied by as much as 200-fold at 3 h after subculture (Fig. 2A) and exceeded 800-fold after overnight growth (data not shown). Differences in phage titer reached 105-fold at 3 h (Fig. 2B). Since our MC1000 lysogens differ only in their Stx2 prophage genotype, our finding of significant variation in toxin production indicates that phage genotype has an unmistakable influence on toxin expression by the lysogens. To the list of phenotypic variations among Shiga toxin-encoding bacteriophages, then, we add the potentially clinically germane characteristic of toxin expression level in a common host strain.
FIG. 2.
(A) Toxin production in the presence (black bars) or absence (gray bars) of mitomycin C (0.5 μg/ml) by MC1000 lysogens of the phages indicated. (B) Phage production in the presence (black bars) or absence (gray bars) of mitomycin C by MC1000 lysogens of the phages indicated. (C) Toxin production in the absence of mitomycin C by the clinical isolates from which the indicated phages were obtained. ∗, O157:H7 clinical isolates. Mean values are shown along with standard deviations derived from three independent trials.
In order to determine whether the variation we observed in toxin production among the MC1000 lysogens also exists among the clinical STEC isolates from which the Stx2 phages were isolated, we measured the Stx2 concentration in cultures of each clinical isolate grown overnight, as shown in Fig. 2C. Corrected for total protein concentration, uninduced Stx2 production varied by up to 480-fold. No relationship was found between toxin production by a given clinical isolate and toxin production by the MC1000 lysogen of the phage derived from that clinical isolate. For instance, Fig. 2C shows a large difference in toxin production by the clinical hosts of phages 933W and 10, despite the fact that MC1000 lysogens of these phages make similar amounts of toxin. A relationship between serotype and toxin production is, however, suggested by the result that the four O157:H7 strains produced more toxin than the non-O157 strains (Fig. 2C). Unlike the single plaque-purified MC1000 lysogens, the clinical isolates are generally lysogens of multiple prophages. Our inability to distinguish plaques from different phages made an assessment of specific phage release from clinical isolates impractical.
Mitomycin C was added to cultures of each MC1000 lysogen to study the effects of exogenous induction on phage release and toxin production. After 3 h (by which time lysis was visually apparent in each mitomycin-treated culture), we determined the phage titer and Stx2 concentration in each lysate. As shown in Fig. 2, mitomycin C induction substantially reduced the differences in the phage and toxin production described above among the cultures of uninduced lysogens. Thus, the Stx2 concentration, when corrected for total protein concentration, varied by no more than 6-fold (Fig. 2A), while variation in the phage titer was reduced to less than 200-fold (Fig. 2B). Among the seven lysogens, the augmentation of toxin and phage production following mitomycin C treatment was most pronounced in those strains which had the lowest titers of toxin and phage in the absence of mitomycin C treatment (Fig. 2). Mitomycin C induction of clinical isolates was not carried out because of the presence of multiple prophage repression systems in each strain. One possible explanation for our findings of variation in phage and toxin production is that the different phages, upon lysogenization of the MC1000 host, could in some way damage the capacity of the host to generate phage or toxin. The equilibration of phage and toxin production by mitomycin C illustrates that the various strains retain a comparable capacity for phage and toxin release and suggests that in certain strains phage and toxin expression are held in check by some factor that is removed upon treatment with mitomycin C.
The genotypic variation among the bacteriophages which accounts for the diverse levels of toxin production by the uninduced MC1000 lysogens is not known. While a variety of mechanisms for phage control of toxin production are conceivable and more than one may be important, the most parsimonious explanation involves differences in prophage repression. The λ prophage encodes a repressor, cI, which is inactivated by RecA-dependent autocleavage as part of the SOS response. This cleavage is the critical molecular event associated with the switch from the lysogenic to the lytic phase of the temperate phage life cycle (19). Previous work has established that SOS-inducing agents, such as mitomycin C, increase both phage titer and toxin production from particular Shiga toxin phage lysogens (13). Furthermore, the Shiga toxin genes in 933W and H19B have been located in the late region of each prophage (15, 18). Following induction and repressor cleavage in lambdoid prophages, phage-encoded Q antiterminators (reviewed in reference 8) act at DNA qut sites located between promoters and terminators to modify passing RNA polymerase complexes in a way that renders them insusceptible to termination. In this way, the late genes, held quiescent in lysogenic phase by the presence of a strong stem-loop terminator, tR′, just downstream of the late promoter, can be expressed following the lytic switch when the Q antiterminator becomes available. Neely and Friedman (15) showed that overexpression of the Q antiterminator of 933W enhances toxin expression from its prophage in the K-12 derivative C600, as well as expression of downstream lysis genes. This suggests that lysis genes and the toxin subunit genes are cotranscribed from the late phage promoter pR′, a possibility that is particularly attractive since no specific Stx2 secretory mechanism has been described, and that phage-mediated lysis may be a mechanism for toxin release. This body of evidence suggests that the regulation of toxin production is dependent on phage development, i.e., that toxin production is part of the coordinated expression of late genes at the appropriate time in the phage lytic cycle following induction. This represents a departure from the conventional view that Stx2-converting phages strictly serve as vectors for horizontal transfer of toxin genes among STEC strains (21).
Our observations add to this model in two ways. First, we observed the trend among seven Stx2-encoding phages that phage particle production and toxin production vary together, which strengthens the idea that they are coordinately regulated. Second, although we have not strictly shown that our phages exhibit immunity in the lambdoid sense (i.e., immunity based upon common repressors rather than, for example, surface exclusion), our finding that various phage groups are susceptible to superinfection by each other indicates that they differ in their repressor regions. Thus, in cases where significant differences in phage and toxin production exist between two lysogens, we know that, at the least, they have different prophage repression systems. Conversely, in MC1000 lysogen pairs that appear to be homoimmune (i.e., to share functionally identical repressors), we observed no significant difference in toxin production. It is possible that some Stx2-encoding phages have weaker repressors than others, weaker in this context meaning more susceptible to RecA-mediated cleavage in MC1000. We speculate that such repressor differences are the prophage genotypes most likely responsible for the observed differences in toxin and phage production in the MC1000 background.
Other host- or bacteriophage-encoded factors besides or in addition to relative repressor strength may be influencing toxin and phage production as well. For instance, different phage integration sites could at least partially explain the differences in toxin expression by the MC1000 lysogens. Since most lambdoid phages integrate site specifically via phage-encoded recombinases, this again would be an example of a phage genotype directing the level of toxin expression. Alternatively, differences in expression and activity of lysis genes could influence the stability of lysogeny. Despite these other possibilities, the observation that variation among the MC1000 lysogens was substantially blunted by mitomycin C—an agent known to bring about RecA-mediated repressor cleavage—strongly implies that repressor differences explain toxin and phage production differences.
No relationship was found between the amount of toxin produced by the MC1000 lysogens and that produced by the clinical isolates from which each MC1000 lysogen's phage was derived. In most instances, the uninduced clinical isolates produced much less toxin than their uninduced MC1000 counterparts at comparable points in mid-logarithmic phase (data not shown), making it necessary to measure toxin release by the clinical isolates after overnight growth (Fig. 2C) instead of after 3 h of growth, as in the MC1000 strains (Fig. 2A). This may reflect the evolution of the prophage relationship with its natural host, in which selection would presumably favor repressor systems which act to more stably maintain lysogeny, with less lytic switching and therefore less toxin production. Further complicating matters in the clinical isolates is the fact that STEC isolates generally contain numerous prophages. Frequently, Stx1- and Stx2-encoding prophages are found in the same strain, along with non-Shiga toxin-encoding prophages, and in fact at least one of our clinical isolates contains two heteroimmune Stx2-encoding phages. With each additional prophage repression system present, the control of Stx2 production may move further away from the lytic switch of the encoding prophage and more toward a complex matrix of interactions between phage repressors and host factors like RecA. Clinical intervention to prevent the lytic switch by prophages in STEC strains would represent a novel strategy—based upon the underlying phage biology—to avoid the clinical sequelae of STEC infection.
Acknowledgments
We are grateful to B. Davis, F. Boyd, and A. Kane for critical reading of this manuscript and A. Kane and the NEMC GRASP Digestive Disease Center for preparing the microbiologic media for our studies.
This work was supported by grants AI-42347 to M.K.W., AI-39067 to D.W.K.A. and P30DK-34928 for the NEMC GRASP Digestive Center. M.K.W. is a Pew Scholar in the Biomedical Sciences and a Tupper Research Fellow. P.L.W. is supported by a Harvard Medical School Student Research Fellowship and a Howard Hughes Medical Institute Research Training Fellowship for Medical Students.
REFERENCES
- 1.Acheson D W, Frankson K, Willis D. Abstracts of the 98th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1998. Multicenter prevalence study of Shiga toxin-producing Escherichia coli, abstr. C-205; p. 165. [Google Scholar]
- 2.Acheson D W K, Keusch G T K. Which Shiga-toxin producing types of E. coli are important? ASM News. 1996;62:302–306. [Google Scholar]
- 3.Acheson D W K, Reidl J, Zhang X, Keusch G T, Mekalanos J J, Waldor M K. In vivo transduction with Shiga toxin 1-encoding phage. Infect Immun. 1998;66:4496–4498. doi: 10.1128/iai.66.9.4496-4498.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boerlin P, McEwen S A, Boerlin-Petzold F, Wilson J B, Johnson R P, Gyles C L. Associations between virulence factors of Shiga toxin-producing Escherichia coliand disease in humans. J Clin Microbiol. 1999;37:497–503. doi: 10.1128/jcm.37.3.497-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 6.Datz M, Janetzki-Mittmann C, Franke S, Gunzer F, Schmidt H, Karch H. Analysis of the enterohemorrhagic Escherichia coliO157 DNA region containing lambdoid phage gene p and Shiga-like toxin structural genes. Appl Environ Microbiol. 1996;62:791–797. doi: 10.1128/aem.62.3.791-797.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donohue-Rolfe A, Acheson D W K, Kane A K, Keusch G T. Purification of Shiga-like toxins I and II by receptor analog affinity chromatography with immobilized P1 glycoprotein and production of cross-reactive monoclonal antibodies. Infect Immun. 1989;57:3888–3893. doi: 10.1128/iai.57.12.3888-3893.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman D I, Court D L. Transcription antitermination: the lambda paradigm updated. Mol Microbiol. 1995;18:191–200. doi: 10.1111/j.1365-2958.1995.mmi_18020191.x. [DOI] [PubMed] [Google Scholar]
- 9.Griffin P M. Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 739–762. [Google Scholar]
- 10.Hendrix R W, Smith M C M, Burns R N, Ford M E, Hatfull G F. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang H, Friesen J, Bruton J L. Characterization of a bacteriophage that carries the genes for production of Shiga-like toxin 1 in Escherichia coli. J Bacteriol. 1987;169:4308–4312. doi: 10.1128/jb.169.9.4308-4312.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaper J B, O'Brien A D. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C: ASM Press; 1998. [Google Scholar]
- 13.Muhldorfer I, Hacker J, Keusch G T, Acheson D W, Tshape H, Kane A V, Ritter A, Olschlager T, Donohue-Rolfe A. Regulation of Shiga-like toxin II operon in Escherichia coli. Infect Immun. 1996;64:495–501. doi: 10.1128/iai.64.2.495-502.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muniesa M, Jofre J. Abundance in sewage of bacteriophages that infect Escherichia coliO157:H7 and that carry the Shiga toxin 2 gene. Appl Environ Microbiol. 1998;64:2443–2448. doi: 10.1128/aem.64.7.2443-2448.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neely M N, Friedman D I. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol Microbiol. 1998;28:1255–1268. doi: 10.1046/j.1365-2958.1998.00890.x. [DOI] [PubMed] [Google Scholar]
- 16.O'Brien A D, Newland J W, Miller S F, Holmes R K, Williams Smith H, Formal S B. Shiga-like toxin-converting phages from Escherichia colistrains that cause hemorrhagic colitis or infantile diarrhea. Science. 1984;226:694–696. doi: 10.1126/science.6387911. [DOI] [PubMed] [Google Scholar]
- 17.Ostroff S M, Tarr P I, Neill M A, Lewis J H, Hargrett-Bean N, Kobayashi J M. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coliO157:H7 infections. J Infect Dis. 1989;160:994–998. doi: 10.1093/infdis/160.6.994. [DOI] [PubMed] [Google Scholar]
- 18.Plunkett G, Rose D J, Durfee T J, Blattner F R. Sequence of Shiga toxin 2 phage 933W from Escherichia coliO157:H7: Shiga toxin as phage late-gene product. J Bacteriol. 1999;181:1767–1778. doi: 10.1128/jb.181.6.1767-1778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ptashne M. A genetic switch: phage λ and higher organisms. Cambridge, Mass: Cell Press; 1992. [Google Scholar]
- 20.Wadolkowski E A, Sung L M, Burris J A, Samuel J E, O'Brien A D. Acute renal tubular necrosis and death of mice orally infected with Escherichia colistrains that produce Shiga-like toxin type II. Infect Immun. 1990;58:3959–3965. doi: 10.1128/iai.58.12.3959-3965.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waldor M K. Bacteriophage biology and bacterial virulence. Trends Microbiol. 1998;6:295–297. doi: 10.1016/s0966-842x(98)01320-1. [DOI] [PubMed] [Google Scholar]
- 22.Watarai M, Sato T, Kobayashi M, Shimizu T, Yamasaki S, Tobe T, Sasakawa C, Takeda Y. Identification and characterization of a newly isolated Shiga toxin 2-converting phage from Shiga toxin-producing Escherichia coli. Infect Immun. 1998;66:4100–4107. doi: 10.1128/iai.66.9.4100-4107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whittam T. Evolution of Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. In: Kaper J B, O'Brien A D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C: ASM press; 1998. pp. 195–209. [Google Scholar]
- 24.Whittam T S, Wachsmuth I K, Wilson R A. Genetic evidence of clonal descent of Escherichia coliO157:H7 associated with hemorrhagic colitis and hemolytic uremic syndrome. J Infect Dis. 1988;157:1124–1133. doi: 10.1093/infdis/157.6.1124. [DOI] [PubMed] [Google Scholar]
- 25.Willshaw G A, Smith H R, Scotland S M, Field A M, Rowe B. Heterogeneity of Escherichia coliphages encoding vero cytotoxins: comparison of cloned sequences determining VT1 and VT2 and development of specific gene probes. J Gen Microbiol. 1987;133:1309–1317. doi: 10.1099/00221287-133-5-1309. [DOI] [PubMed] [Google Scholar]