Upon antigen stimulation, naïve T cells exit the quiescent state and undergo rapid clonal expansion and differentiation [1]. This cell fate transition requires accelerated protein synthesis, which is regulated at both the transcriptional and translational levels [2, 3]. Compared with well-documented transcriptional regulatory mechanisms, the translational regulation of T-cell activation has not been fully elucidated. A recent study by Liu et al. published in Nature Immunology suggested that the tRNA m1A58 modification is an important regulatory mechanism of T-cell activation [4].
During translation, tRNAs play important roles in decoding mRNA. In addition to their abundance, the modification and aminoacylation (charge) status of tRNAs is essential to ensure translation efficiency and fidelity [5, 6]. To date, more than 170 RNA modifications have been documented, and most of them have been identified in tRNAs [7]. On average, one tRNA molecule harbors 13 modifications that affect protein synthesis through several mechanisms, including tRNA stability, codon recognition, and aminoacylation capability [5]. Among these modifications, N1-methyladenosine (m1A), which is typically found at position 58 in cytoplasmic tRNA and is catalyzed by the TRMT6/61 A complex to enhance translation initiation and elongation, is among the most conserved [8]. A recent study reported that tRNA m1A58 methylation was required for liver tumorigenesis, but its physiological roles in T-cell activation remained to be investigated [9].
Upon T-cell activation, a large number of functional proteins are rapidly synthesized to drive T cells into mitosis and accelerate their proliferation [2]. To elucidate the translational control mechanisms, Liu et al. first performed tRNA sequencing to measure tRNA pool (abundance) changes at several timepoints after antigen stimulation. Interestingly, these tRNA transcripts are sequentially increased, and differential tRNA transcripts display distinct dynamic patterns. To further explore the molecular mechanisms, the mRNA expression levels at these timepoints were profiled. Notably, the number of transcripts involved in tRNA processing and tRNA modification are increased at an early stage. Since tRNA modifications are essential to ensure proper tRNA decoding and translation efficiency, the authors of the aforementioned study hypothesized that these modifications may play important regulatory roles during translation in T cells undergoing activation and proliferation. Therefore, they hypothesized that tRNA m1A58, one of the most abundant and prevalent tRNA modifications, may play a profound role in these T-cell processes.
To study the role of tRNA m1A58 methylation in T-cell activation, the authors first generated conditional Trmt61a-knockout mice. Trmt61a encodes the methyltransferase critical for the tRNA m1A58 modification. Upon Trmt61a depletion, the global m1A methylation level of activated CD4+ T cells decreased markedly. tRNA-m1A-seq further substantiated that the methylation levels of most tRNAs were reduced in Trmt61a-KO CD4+ T cells after antigen stimulation, although the magnitudes of the decrease in m1A methylation level differed among the modified tRNA transcripts. Furthermore, using an adoptive transfer model of colitis, the authors of this study revealed that the in vivo immune functions of CD4+ T cells were profoundly impaired after Trmt61a depletion. Potential causes of the attenuated immunologic function of Trmt61a-deficient CD4+ T cells were determined ex vivo; they included possible defective activation, compromised proliferation, aggravated apoptosis, and abnormal differentiation of CD4+ T cells. Using a CellTrace labeling assay combined with an in vitro T-cell differentiation system, the authors revealed that the proliferation and differentiation capacity of Trmt61a-deficient CD4+ T cells was impaired. Moreover, they confirmed that Trmt61a deficiency led to impaired proliferation and arrested cell cycle progression of CD4+ T cells in vivo. To determine whether the effects of Trmt61a on CD4+ T cells are dependent on tRNA m1A58 methylation, the authors introduced wild-type (WT) Trmt61a and m1A58 catalytically dead Trmt61a into Trmt61a-deficient CD4+ T cells. They found that the wild-type (WT) Trmt61a, but not m1A58 catalytically dead Trmt61a, restored the proliferation of these cells, as indicated by CellTrace labeling assay with the adoptive transfer model, confirming that the tRNA m1A58 modification is vital to restoring CD4+ T-cell immune function.
To explore the molecular mechanisms by which Trmt61a ensures normal T-cell homeostasis and proliferation, the TCR signaling pathway and IL-2 signaling pathway were first evaluated [10]. After in vitro stimulation, the activation of neither pathway was affected by Trmt61a deficiency. Furthermore, the authors found that the protein level of Myc, which can promote cell proliferation and is rapidly increased upon T-cell activation, was significantly increased in the early stage of CD4+ T-cell activation. Further investigations revealed that Trmt61a deficiency impaired the protein synthesis of Myc; however, it exerted no effect on its transcript level, indicating that Trmt61a regulates Myc expression at the translational level. Systematic analysis showed that the coding sequence of MYC is enriched with serine and leucine codons and that the m1A58 modification levels of the corresponding tRNAs were markedly decreased after Trmt61a deficiency. Thus, the authors hypothesized that the tRNA m1A58 modification modulates Myc protein expression in a codon-biased manner. Therefore, they constructed a mutant Myc sequence using the codon-switching method, in which the most abundant serine and leucine codons were replaced by respective synonymous codons, to render the corresponding tRNAs insensitive to TRMT61A deletion. Notably, the protein expression level of mutant Myc was not affected by TRMT61A deletion. Moreover, an in vivo hematopoietic stem cell (HSC) rescue assay showed that overexpressed MYC protein restored the proliferation of Trmt61a-KO CD4+ T cells. Collectively, the results of this study showed that the efficient translation of Myc was essential for CD4+ T-cell activation, which was precisely regulated by tRNA m1A58 modification (Fig. 1).
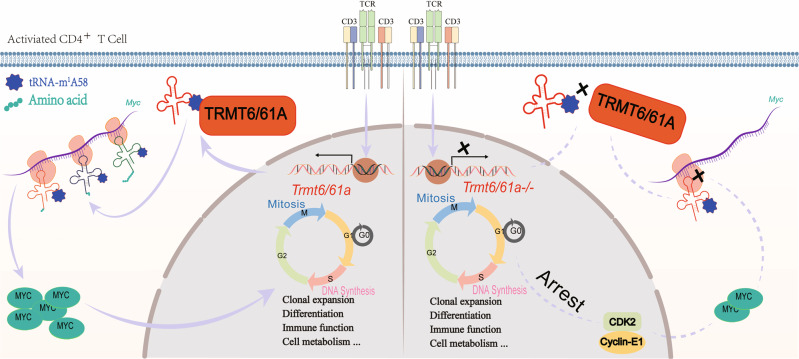
Fig. 1.
TRMT6/61A-mediated CD4+ T-cell activation through MYC synthesis. Upon CD4+ T-cell activation, Trmt61a is upregulated, leading to an increase tRNA-m1 A58 modification abundance, thus ensuring Myc translation, which is critical for mitosis and cell cycle progression. Myc also initiates the adaptive immune response, including clonal expansion, differentiation and effector functions. Specific deletion of Trmt61a in CD4+ T cells disrupted tRNA-m1 A58 modification and inhibited MYC translation, downregulating CDK2 and Cyclin-E1 and thus arresting the cell cycle.
To systematically study the regulatory roles of the tRNA m1A58 modification during CD4+ T-cell activation at the translational level, RiboTag-Seq was performed to identify the genes exhibiting decreased translation efficiency (TE) after TRMT61A deletion. An integrated analysis of RiboTag-Seq data and published T-cell proteomics data [2] revealed that TE-downregulated genes were significantly abundant among genes encoding functional proteins essential to early activation of CD4+ T cells. To elucidate the molecular mechanism of tRNA m1A58-mediated translational regulation, two properties of tRNAs involved in mRNA decoding were considered, including tRNA abundance and m1A58 methylation level changes. Considering the results of these analyses, the authors reported that TE-downregulated genes were enriched with codons encoding proteins with tRNAs that were upregulated early in translation. In addition, TRMT61A was associated with the preferential modification of these early-upregulated tRNAs, ensuring the normal decoding capability of these tRNAs.
In summary, this study proposes a new working model showing that tRNA abundance and modifications are orchestrated to maximize translation efficiency for the rapid synthesis of key functional proteins during CD4+ T-cell activation. In this process, the tRNA m1A58 modification is as an important epigenetic regulator for CD4+ T-cell activation, making it a potential therapeutic target for immunotherapy. In addition, this study also raises several open questions and suggests implications. For instance, in addition to Trmt61a-mediated tRNA m1A58 modification, other processes and modifications may be involved in this process. Moreover, the charge status of tRNAs can affect translation efficiency and can be rapidly changed, and the determination of whether a change in charge is involved in T-cell activation warrants further investigation.
Acknowledgements
We appreciate helpful discussions with Dr. Qin Tian and Dr. Yifei Wang.
Competing interests
The authors declare no competing interests.
References
- 1.Saravia J, Chapman NM, Chi H. Helper T cell differentiation. Cell Mol Immunol. 2019;16:634–43. doi: 10.1038/s41423-019-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan H, Yang K, Li Y, Shaw TI, Wang Y, Blanco DB, et al. Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity. 2017;46:488–503. doi: 10.1016/j.immuni.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548:338–42. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Zhou J, Li X, Zhang X, Shi J, Wang X, et al. tRNA-m1A modification promotes T cell expansion via efficient MYC protein synthesis. Nat Immunol. 2022;23:1433–44. doi: 10.1038/s41590-022-01301-3. [DOI] [PubMed] [Google Scholar]
- 5.Pan T. Modifications and functional genomics of human transfer RNA. Cell Res. 2018;28:395–404. doi: 10.1038/s41422-018-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat Rev Mol Cell Bio. 2021;22:375–92. doi: 10.1038/s41580-021-00342-0. [DOI] [PubMed] [Google Scholar]
- 7.Boccaletto P, Stefaniak F, Ray A, Cappannini A, Mukherjee S, Purta E, et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022;50:D231–D5. doi: 10.1093/nar/gkab1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell. 2016;167:1897. doi: 10.1016/j.cell.2016.11.045. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Wang J, Li X, Xiong X, Wang J, Zhou Z, et al. N(1)-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat Commun. 2021;12:6314. doi: 10.1038/s41467-021-26718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong Y, Walker SK, Pritykin Y, Leslie CS, Rudensky AY, Van der Veeken J. Hierarchical regulation of the resting and activated T cell epigenome by major transcription factor families. Nat Immunol. 2022;23:122. doi: 10.1038/s41590-021-01086-x. [DOI] [PMC free article] [PubMed] [Google Scholar]