Abstract
Both preclinical and established rheumatoid arthritis (RA) patients display alterations in the gut microbiome. Prevotella spp. are preferentially enriched in a subset of RA patients. Here, we isolated a Prevotella strain, P. copri RA, from the feces of RA patients and showed that colonization of P. copri RA exacerbated arthritis in a collagen-induced arthritis (CIA) model. With the presence of P. copri RA colonization, a high-fiber diet exacerbated arthritis via microbial alterations and intestinal inflammation. Colonization of P. copri together with a high-fiber diet enabled the digestion of complex fiber, which led to the overproduction of organic acids, including fumarate, succinate and short-chain fatty acids. Succinate promoted proinflammatory responses in macrophages, and supplementation with succinate exacerbated arthritis in the CIA model. Our findings highlight the importance of dysbiosis when evaluating the effects of dietary interventions on RA pathogenesis and provide new insight into dietary interventions or microbiome modifications to improve RA management.
Keywords: Rheumatoid arthritis, Dysbiosis, Prevotella copri, High-fiber diet, Microbiota-derived metabolites
Subject terms: Autoimmunity, Mucosal immunology, Chronic inflammation
Background
Rheumatoid arthritis (RA) is a common systemic autoimmune disease caused by the interplay between genetic and environmental factors. Genetic studies on individuals with RA have identified multiple risk loci [1], and accumulating evidence has highlighted a critical role of environmental factors, including the microbiome and diet, in the pathogenesis of RA [2–4].
We have previously reported that alterations in the gut and oral microbiomes are associated with new-onset RA [5]. Compared to those of controls, the gut microbiomes of individuals with early onset or preclinical RA are particularly enriched in Prevotella copri [5–7]. Intestinal dysbiosis can alter mucosal immune responses, which may ultimately contribute to the development of inflammatory arthritis [8]. However, the role of Prevotella spp. in modulating the balance between health and disease is complex [9]. For instance, a high abundance of Prevotella is associated with dietary fiber-induced improvement in glucose metabolism by promoting glycogen storage [10], and human gut-derived P. histicola suppressed arthritis through induction of regulatory T cells (Tregs) [11]. In contrast, transplantation of feces enriched in P. copri from patients with RA has been found to promote a proinflammatory Th17 response in regional lymph nodes of arthritis-prone SKG mice [12]. Together, these data implicate a species- or strain-specific effect of Prevotella spp. on metabolic or immune modulation.
The diversity and prevalence of Prevotella species and strains in the human microbiome are affected by multiple factors, including lifestyle, diet and geography. In fact, Prevotella is usually enriched in non-Western populations, likely due to their relatively plant-rich diets [13]. Distinct Prevotella spp. bacteria outcompete commensal bacteria for polysaccharides by encoding specific carbohydrate-active enzymes (CAZymes), which allows them to reside in the human gut [14]. In addition to these differences at the species level, genomic analysis of over 1000 genomes of P. copri revealed substantial genetic diversity at the strain level, including genes encoding enzymes for utilization of diverse complex carbohydrates [15]. Indeed, different strains of P. copri differ in their abilities to breakdown plant polysaccharides and host-derived mucin [16, 17]. For instance, strains from non-Western subjects show a greater potential for complex fiber degradation [13].
The consumption of dietary fibers by gut microbes produces organic acids (OAs), in particular short-chain fatty acids (SCFAs). Fiber fermentation usually exerts beneficial effects on the host, i.e., promoting glucose homeostasis or facilitating an anti-inflammatory milieu in the gut. For instance, succinate from dietary fiber fermentation was identified as a substrate for intestinal gluconeogenesis, a process that maintains glucose homeostasis [18]. However, these fermentation products can also be detrimental in some situations. For instance, the fermentation of dietary soluble fibers by the gut microbiota induces cholestasis, hepatic inflammation and liver cancer in Toll-like receptor 5 (TLR5)-deficient mice [19]. Furthermore, in the context of intestinal inflammation, butyrate production following dietary fermentation has been shown to exacerbate inflammation by increasing NLRP3 activation [20]. Microbiota-derived succinate has been found to promote Clostridium difficile infection after antibiotic treatment or motility disturbance [21]. Succinate, which is produced during bacterial fermentation of dietary fiber, has also been reported in association with dysbiosis, as well as in patients with inflammatory bowel disease and in animal models of intestinal inflammation [22]. Thus, the effect of microbial fermentation of fiber on host health, mainly influenced by diet, can be both context dependent and species (strain) dependent.
RA patients and their physicians are often perplexed by what type of diet the patients should follow. A number of studies have implicated the impact of dietary intervention on disease development. The Mediterranean diet, characterized by a high fiber content, has been shown to reduce the risk of cardiovascular disease due to the anti-inflammatory properties of its contents [23]. As such, a few studies have set out to investigate the association between the Mediterranean diet and risk reduction of inflammatory diseases (i.e., RA) [24]. However, epidemiological studies have failed to show a reduction in RA risk or severity in patients on a Mediterranean diet [25, 26]. Intriguingly, the responses to a Mediterranean diet varied substantially among patients [24–26]. It remains unclear whether specific factors (i.e., the gut microbiome) may contribute to this variability. Notably, a fiber-enriched diet produced divergent immune responses in individuals with different baseline microbiota [27]. There are a few studies suggesting that a patient’s baseline microbiota is related to response to treatment [5], but baseline microbiota can also be related to response to diet. Diet components may interact with microbes, but there are few, if any, studies evaluating the combined effects. Thus, there is an urgent need to investigate the combinatory effect of diet and microbes on the pathogenesis or treatment options of RA, as such findings will provide rationales for adjusting diet to modulate the gut microbiome to improve RA management. In this study, we, for the first time, provided evidence that the presence of dysbiosis may interfere with the dietary effect of a high fiber content, which in turn exacerbated RA. Our findings may shed light on the complex interplay between dysbiosis and diet in RA management.
Materials and methods
P. copri isolation and genome sequencing
P. copri was isolated from the feces of RA patients with early RA (<1-year disease duration). For the P. copri isolate, cells were grown anaerobically (70% N2, 20% CO2, and 10% H2) in PYG medium supplemented with hematin (25 mg/L) and vitamin K1 (10 mg/L) at 37 °C for 2 days. The strain was sequenced on the Illumina HiSeq 2500 2 × 100 bp platform. Short reads were assembled, and the resulting contigs were then aligned to the P. copri reference genome (DSM 18205) with Mauve software.
Mice and induction of arthritis
DBA/1J mice were purchased from the Jackson Laboratory. All mice were maintained in a specific-pathogen free (SPF) facility. All studies were approved by the Animal Ethics Committee of Peking Union Medical College Hospital. All experiments were conducted with sex- and age-matched mice. To induce collagen-induced arthritis (CIA) in mice, female DBA/1 J mice (7–8 weeks old) were injected with an emulsion of bovine CII collagen and complete Freund’s adjuvant (CFA, Chondrex) containing a final concentration of 0.5 mg/ml of M. tuberculosis by the subcutaneous route. A booster injection with an emulsion of collagen and incomplete Freund’s adjuvant (IFA, Chondrex) was administered on Day 21.
Clinical assessment of arthritis
Clinical scoring for arthritis was performed as described previously [28]. The severity of arthritic limbs was scored as follows: 0 = normal, 1 = mild swelling confined to the tarsals or ankle joint, 2 = mild swelling extending from the ankle to the tarsals, 3 = moderate swelling extending from the ankle to the metatarsal joints, and 4 = severe swelling encompassing the ankle and foot or ankylosis of the limb. Then, the clinical score for each mouse was the sum of the scores for each paw.
Bacterial strains and colonization of mice
P. copri RA was grown in PYG medium supplemented with hematin and vitamin K1. Bacteroides thetaiotaomicron ATCC 29148 was grown in TSB media with 1% glucose. DBA/1J mice were subjected to collagen-induced arthritis (CIA) and then treated with a cocktail of antibiotics containing metronidazole (1 g/L), neomycin (1 g/L), vancomycin (0.5 g/L), and ampicillin (1 g/L) in drinking water for 7 days (Day 0 to Day 6) after the second immunization. The mice were then provided with regular sterile water for 48 h (Day 7 to Day 8). A total of 108 CFU of P. copri or B. thetaiotamicron was administered to each group via oral gavage on Day 9 after the second immunization. Colonization of the microbiota was confirmed by quantitative PCR of specific bacterial genes. Genomic DNA was extracted from P. copri and diluted 10, 100, 1000 and 10,000 times to generate a standard curve for qPCR. The total bacterial DNA in feces was extracted, and total DNA concentrations were measured. The P. copri DNA concentration in each sample was determined using the standard curve. P. copri composition (%) = . Oligonucleotide sequences are given in Supplementary Table 1.
Histopathology
To assess tissue pathology, limbs and colons were collected and fixed in 4% paraformaldehyde. Histological sections were stained with hematoxylin and eosin (H&E) and scored in a blinded fashion by a pathologist. Histological sections of colons and joints were evaluated as described in Supplementary Table 2.
Cell culture and stimulation
The cell line HEK-blue-TLR4 (Invivogen) was cultured following the manufacturer’s instructions. These reporter HEK-Blue hTLR cells stably co-overexpress the human TLR and NF-κB/AP1-inducible SEAP (secreted embryonic alkaline phosphatase) genes. TLR4 activation was indicated by the absorbance of the SEAP reporter. For stimulation experiments, we determined the growth curves of the two bacterial strains. The two bacterial strains were grown to log phase in the same volume of medium and collected at OD600 = 0.6. The supernatants were collected and precipitated with trichloroacetic acid (TCA) acetone. The precipitants were resuspended in PBS and prepared for the following experiment. The bacterial cells were washed with PBS, ground with liquid nitrogen, and resuspended in PBS to obtain extracts. HEK-blue-TLR4 cells were stimulated with the bacterial cell extracts and supernatants of the strain P. copri RA and the control strain B. thetaiotamicron. LPS (10 ng/ml) from Escherichia coli (strain K12) was included as a positive control. Supernatants were collected and detected with the HEK-Blue detection kit (InvivoGen).
Diets and succinate treatments
Custom diets were based on modification to the AIN-93G control diet and were purchased from XieTong Pharmaceutical Bio-Engineering (China). The fiber-containing diet (FCD) was a diet enriched with 2.5% crude fiber and 7.5% fructooligosaccharide. The non-fiber-containing diet (NCD) was devoid of fiber. The composition of the two diets is provided in Supplementary Table 3. Mice were fed with these diets throughout the experiments after the induction of rheumatoid arthritis. For the sodium succinate treatment experiment, the mice were given sodium succinate (0.5%) at Day 9 after the second immunization until the end of the experiment.
Cytokine and immunoglobulin measurement
Serum cytokines were measured by enzyme-linked immunosorbent assay (ELISA) using Mouse IL-6 ELISA Sets (Raybiotech) according to the manufacturers’ protocols. Concentrations of anti‐Bovine type II collagen IgG were then measured using ELISA according to the manufacturers’ protocols. Total fecal IgA was measured using sandwich ELISA. For total IgA detection, fecal contents were resuspended in 1 ml of PBS and homogenized at 30 HZ for 30 s for 6 cycles and then centrifuged at 12,000 × g for 10 min. The supernatant was collected and assayed for free IgA by ELISA using the Mouse IgA Quantitation Set (Bethyl).
Cell isolation and flow cytometry analysis
MLN cells were isolated as previously described [29]. For Th17-cell subset quantification, isolated immune cells were stimulated with cell stimulation cocktail (BD) for 5 h. The cells were harvested and stained with CD4 (RM4-5, Invitrogen) and IL-17 antibodies (eBio1787, Invitrogen). Flow cytometry was conducted using a BD FACSCelesta Flow Cytometer. The analysis was performed using FlowJo software (Version 9).
SCFA quantification
SCFAs were analyzed as previously described [30] with modifications. Briefly, for each fecal sample, 20-mg aliquots were extracted with 0.2 ml of 50% H2SO4 and 0.3 ml of dichloroethane (containing 10 μg/ml acetic acid-d4), followed by vortex mixing for 1 min. The sample was centrifuged for 20 min at 12,000 rpm and 4 °C and kept at −20 °C for 30 min. The supernatant fluid was prepared for GC‒MS analysis. GC‒MS analysis was performed using an Agilent 8890 gas chromatograph system coupled with an Agilent 5977 mass spectrometer. SCFAs were separated on a DB-FFAP capillary column. A 1-μl aliquot of the analyte was injected in split mode (10:1). The initial temperature was kept at 45 °C for 2 min, then raised to 180 °C at a rate of 10 °C/min, then to 180 °C at a rate of 15 °C/min, then to 240 °C at a rate of 40 °C/min, and finally held at 240 °C for 3 min. The electronic energy was recorded at 70 eV. SCFA quantification was performed by calculating response factors for each SCFA relative to acetic acid-d4 using the injections of pure standards.
Fecal succinate and fumarate quantification
For the measurement of succinate and fumarate in the cecum of mice or the feces, the samples were quantified as described [31] with modifications. For each sample, 50-mg aliquots were extracted with 0.005 M NaOH containing 5 μg/mL acetic acid-d4 and derivatized with 1-propanol:pyridine (3:2, v/v) and propyl chloroformate (PCF). The propyl derivatives were extracted twice with hexane and were measured by an Agilent 8890 gas chromatograph system coupled with an Agilent 7010 mass spectrometer. The system utilized an Agilent DB-624 column (15 m × 250 μm × 1.4 μm). A 1-μl aliquot of the analyte was injected in split mode (10:1). The initial temperature was kept at 36 °C for 10 min, then raised to 180 °C at a rate of 5 °C/min, then to 240 °C at a rate of 40 °C/min, and finally held at 240 °C for 5 min. The energy was 70 eV in electron impact mode.
RNA sequencing and analysis
CIA mice, confirmed for the presence of P. copri by qPCR and treated with NCD or FCD diet, were used for transcriptome analysis. After sacrificing the mice, the colon of each mouse was collected and cooled with liquid nitrogen. mRNA was extracted and used for RNA-Seq library preparation following the instructions in the Illumina NEBNext Ultra RNA Library Prep Kit. Sequencing was run on an Illumina High Seq-2000. The resulting reads were aligned to the genome with HISAT2 v2.0.5, and gene counts were normalized using FPKM. Differential expression analysis was performed using the DESeq2 R package (1.16.1). Pathway enrichment analyses were performed using the clusterProfiler R package.
16 S rDNA gene sequencing and analysis
Total genomic DNA from samples was extracted using a QIAamp PowerFecal Pro DNA Kit for feces (Qiagen) according to the manufacturer’s instructions. 16 S rRNA genes of distinct regions (16 S V3-V4) were amplified using specific primers with barcodes. The mixed amplicons were then sequenced by Illumina PE250. Before 16 S rDNA data analysis, raw sequences were filtered, and paired-end reads were merged to tags by FLASH (V1.2.7). All tags were analyzed by scripts of QIIME 1.9.1, USEARCH v7.0.1090, and Vsearch 2.9.0. Specifically, the tags were clustered into operational taxonomic units (OTUs) with a 97% threshold by using UPARSE, and unique representative OTU sequences were obtained. OTU representative sequences were taxonomically classified using SILVA 132 and QIIME v1.9.1. The OTU table was used to calculate the beta diversities and to obtain taxonomic profiles. Subsequent statistical analysis was performed in R software.
CAZyme analysis in Prevotella genomes
Carbohydrate active enzymes (CAZymes) were identified using dbCAN2 [32] version v2.0.6. The results were filtered by the parameter (E-value < 1e-15, coverage > 0.35) based on the HMMER search. Predicted CAZymes were visualized in R version 4.0.2.
Statistical analysis
We analyzed the data with GraphPad Prism version 7 (San Diego, CA, USA). One-way ANOVA test, two-way ANOVA test, the Mann‒Whitney test, and t tests were used to compare experimental groups. All values are presented in the figures as the mean ± SEM, with *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, n.s. no significance.
Results
Colonization of CIA mice with the Prevotella copri strain isolated from RA exacerbated arthritis
We and other groups have previously shown an increased abundance of P. copri in patients with early RA [5, 6]. We isolated a Prevotella strain from the mixed feces of RA patients (P. copri RA, PC). The strain was sequenced, and high-quality reads were assembled. The resulting contigs were then aligned to the P. copri reference genome (DSM 18205) with Mauve software. There were rearrangements and inversions in the genome of PC (Supplementary Fig. 1), suggesting strain-level differences.
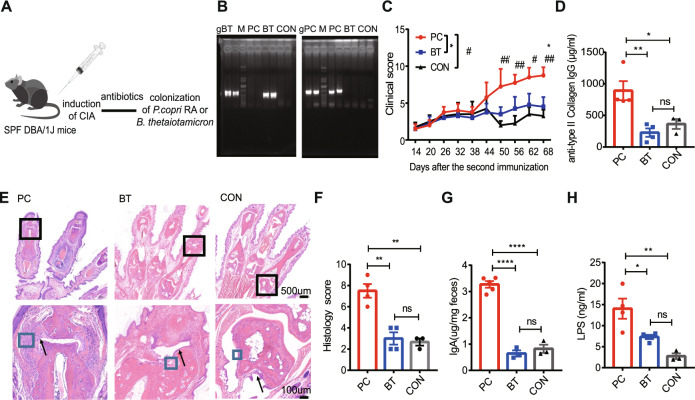
To determine whether the newly isolated P. copri RA contributes to RA pathogenesis, collagen-induced arthritis (CIA) was induced in DBA/1J mice, followed by colonization with PC after a course of antibiotic treatment (Fig. 1A). As a control, mice were inoculated with Bacteroides thetaiotaomicron (BT), a common host symbiotic bacterial strain in the intestine. We assessed the inflammatory profiles of the mice colonized with PC or BT, as well as the mice that received only antibiotic treatment (CON group). As expected, Prevotella spp. were detected in the feces of the PC group but not the BT and CON groups, as determined by gel electrophoresis analysis 3 weeks after inoculation (Fig. 1B). The PC group had higher clinical arthritis scores (Fig. 1C) and higher titers of anti-bovine type II collagen IgG than the BT and CON groups (Fig. 1D). The PC group developed more severe joint inflammation, as reflected by the proliferation of the synovial membrane, increased stromal cell density and infiltration of inflammatory cells (Fig. 1E, F).
Fig. 1.
P. copri RA colonization exacerbated arthritis. A Illustration of the colonization experiments with P. copri RA (PC) and B. thetaiotamicron (BT). B Confirmation of P. copri RA and B. thetaiotamicron colonization by agarose gel electrophoresis. gPC indicates P. copri genomic DNA (gPC), and B. thetaiotaomicron genomic DNA (gBT) was used as a positive control; PC, BT, and CON represent total bacterial DNA exacted from the feces of the corresponding group; M, DNA ladder. C Clinical scores of collagen-induced arthritis in the PC (n = 4), BT (n = 4) and CON (n = 3) groups. D Serum anti-bovine type II collagen IgG level. E Representative histology of joints. The black arrow indicates synovial lining-cell hyperplasia, and the blue box indicates stromal cells. F Histological scores of joints. G Levels of fecal IgA. H Serum LPS levels. Each symbol in (D, F, G, and H) represents an animal. Error bars indicate the SEM (C, D, F–H). P values were determined by two-way ANOVA (C), one-way ANOVA (D, F, G, H). *p < 0.05, **p < 0.01, ***p < 0.001, #p < 0.05, ##p < 0.01, ###p < 0.001
Given that gram-negative bacterial colonization may lead to LPS-induced inflammation and fecal IgA secretion, to better understand the mechanism by which PC may promote inflammation, we measured serum LPS levels and fecal IgA levels in the PC, BT and CON groups. We found that plasma LPS levels and fecal IgA levels were significantly higher in the PC group than in the BT and CON groups (Fig. 1G, H), suggesting that LPS may mediate, at least in part, the PC-induced inflammatory response. TLR4 is a major receptor for sensing microbial LPS and initiates the inflammatory response. To investigate the involvement of TLR signaling in PC-induced inflammation, we measured TLR4 activation in TLR4-transfected HEK-293 reporter cells treated with extracts and supernatant of PC or BT. The extracts from PC correspondingly displayed a stronger capacity to induce TLR4 signaling than the extracts from BT (Supplementary Fig. 2A). These findings were corroborated by the TLR4 activation reporting assay with supernatants from PC and BT cells (Supplementary Fig. 2B). These data demonstrate that PC is more capable of activating the TLR4 pathway and inducing inflammation.
PC exacerbated arthritis under a high-fiber diet
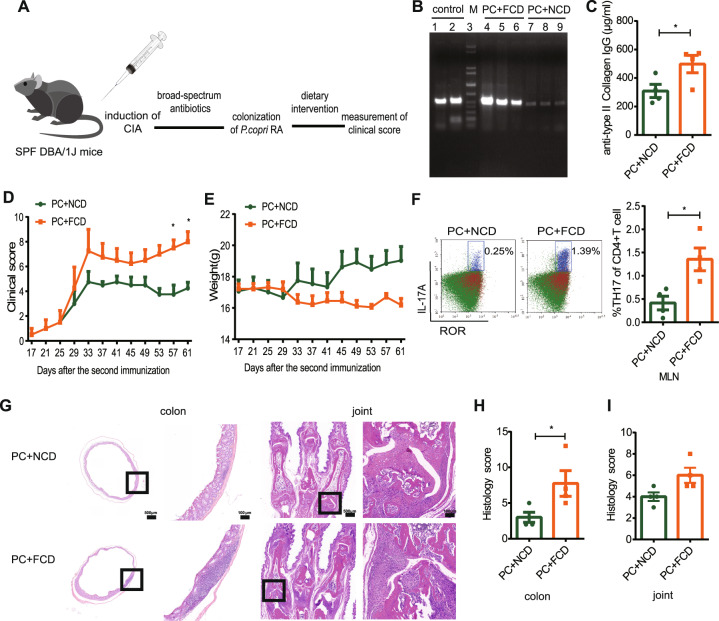
Since Prevotella spp. are highly capable of degrading cellulose and saccharides via hydrolysis and fermentation, resulting in the production of carboxylic acids [13], we next asked whether PC aggravates arthritis under a fiber-enriched diet. CIA mice were colonized by oral gavage with 1 × 108 CFUs of PC after a course of antibiotic treatment and then fed a fiber-containing diet (FCD) or a non-fiber-containing diet (NCD) at the same time point as oral gavage (Fig. 2A). PC colonization levels and the consequences of exposure to different dietary interventions were assessed.
Fig. 2.
P. copri RA-induced disease severity was promoted by a high-fiber diet. A Schematic of CIA mice with dietary intervention. B Confirmation of P. copri RA colonization by quantitative PCR. Control means P. copri genomic DNA; PC + FCD means DNA exacted from FCD group feces; PC + NCD means DNA exacted from NCD group feces. C Anti-bovine type II collagen IgG levels in the serum of PC + FCD (n = 4) or PC + NCD (n = 4) mice. D Clinical scores of collagen-induced arthritis. E Body weight of mice in the two groups. F Flow cytometric analysis of CD4+Th17+ cells in mesenteric lymph nodes. G Histological evaluation of the CIA colon and joint tissues. The scale bar is 100 μm. H The histology scores of the colon tissues. I The histology scores of the synovial tissues. Each symbol in (C, F, H, and I) represents an animal. P values were determined by two-way ANOVA (D, E), the Mann‒Whitney test (F, H, I), and Student’s t test (C). *p < 0.05, **p < 0.01, ****p < 0.0001. Abbreviations: PC P. copri RA, NCD non-fiber-containing diet, FCD fiber-containing diet
The FCD enhanced PC colonization compared to the NCD (Fig. 2B). Furthermore, FCD-fed mice displayed more severe disease characterized by elevated levels of anti-collagen antibodies, higher arthritis scores and greater body weight loss (Fig. 2C–E), as well as more Th17 cells in the mesenteric lymph nodes (MLNs) (Fig. 2F). Notably, FCD-fed mice developed more severe intestinal inflammation manifested as tissue erosion, more infiltration of inflammatory cells, and fewer intestinal crypts than NCD-fed mice (Fig. 2G, H). Histopathological analysis of joints revealed more severe joint damage in FCD-fed mice than in NCD-fed mice, as evidenced by the proliferation of synovial lining cells, pannus formation, and infiltration of inflammatory cells (Fig. 2G, I).
To determine whether PC may contribute to aggravating arthritis through the synergistic effects of diet with dysbiosis in inflammation, CIA mice were colonized by oral gavage with PC or BT and fed an FCD or NCD (Supplementary Fig. 3A). The consequences of PC or BT exposure to different dietary interventions were assessed. Prevotella spp. were detected in the feces of the PC but not the BT group, as demonstrated by gel electrophoresis analysis (Supplementary Fig. 3B–C). PC-colonized mice fed the FCD (PC + FCD group) displayed significantly higher arthritis scores and elevated levels of anti-collagen antibodies than PC-colonized mice fed the NCD (PC + NCD group). Moreover, the level of anti-collagen antibody in the PC + FCD group was markedly higher than that in the BT + FCD group (Supplementary Fig. 3D, F). The average body weight of the PC + FCD group was lower than that of the PC + NCD group, but there was no statistical significance (Supplementary Fig. 3E). The severity of intestinal inflammation in the PC + FCD group was slightly higher than that in the PC + NCD and BT + FCD groups, manifested as more infiltration of inflammatory cells and fewer intestinal crypts, but there were no significant differences (Supplementary Fig. 3G, H). The histology score of the PC + FCD group was significantly higher than that of the PC + NCD group, mainly manifested as the prominent proliferation of synovial lining cells, pannus formation, and massive infiltration of inflammatory cells. The synovial inflammation was slightly more severe in the PC + FCD group than in the BT + FCD group, but there were no significant differences (Supplementary Fig. 3I, J).
The fiber diet plus PC colonization induced inflammation at the transcriptional level
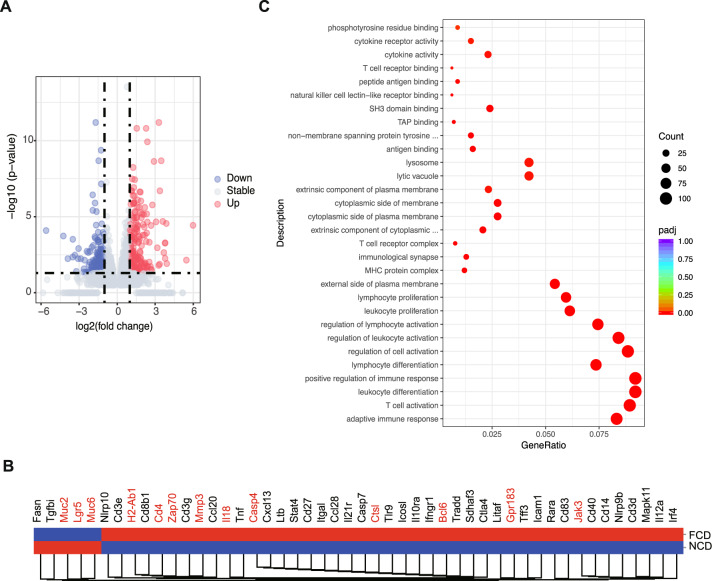
To understand how the FCD synergizing with PC colonization in CIA mice led to more severe inflammation, mRNA sequencing analysis was performed on freshly isolated colon tissues of FCD- and NCD-fed CIA mice colonized with PC. A total of 1444 genes were upregulated and 1487 genes were downregulated in the FCD group compared to the NCD group (Fig. 3A, Supplementary Table 4). Gene Ontology (GO) analysis revealed that the pathways of proinflammatory responses, adaptive immune dysregulation, T-cell activation, and leukocyte differentiation were upregulated in the FCD group (Fig. 3C). Specifically, we observed upregulation of genes involved in T-cell receptor (TCR) signaling, including Cd4 and Zap70, in the FCD group (Fig. 3B), indicating that the FCD may affect T-cell activation and proliferation. The FCD also resulted in the upregulation of Il18, Casp4, and Jak3, as well as Bcl6 and Gpr183, which encode a master regulator of GC B-cell differentiation and an orphan G protein-coupled receptor that is essential for the induction of early plasma blast responses, respectively [33] (Fig. 3B). In accordance with the decreased mucosal thickness (Fig. 4F), the gene expression levels of mucosal barrier markers (Muc2 and Muc6) and gastrointestinal stem cell markers (Lgr5) were significantly decreased in FCD mice. The expression levels of Mmp3 and Ctsl, both of which are implicated in joint destruction in RA [34], were also upregulated in FCD-treated mice (Fig. 3B). Collectively, these observations suggest that FCD intervention in the presence of PC colonization enhances inflammation through increased inflammatory cytokine production, adaptive immunity activation and gut barrier dysfunction.
Fig. 3.
The impact of dietary intervention on the colon transcriptome. A Volcano plots showing the expression of significantly downregulated (blue) and upregulated (red) genes in the FCD and NCD groups. Blue and red dots represent significant DEGs, with the lines denoting a cutoff P value of <0.05. B Heatmap showing the expression of genes involved in the colons of mice subjected to dietary intervention. The red square indicates upregulated genes, and the blue square indicates downregulated genes. For RNA-sequencing data, n = 4 per group. Key genes are highlighted in red. C Gene Ontology (GO) enrichment analysis among the significantly regulated transcripts from the colons of the FCD and NCD groups
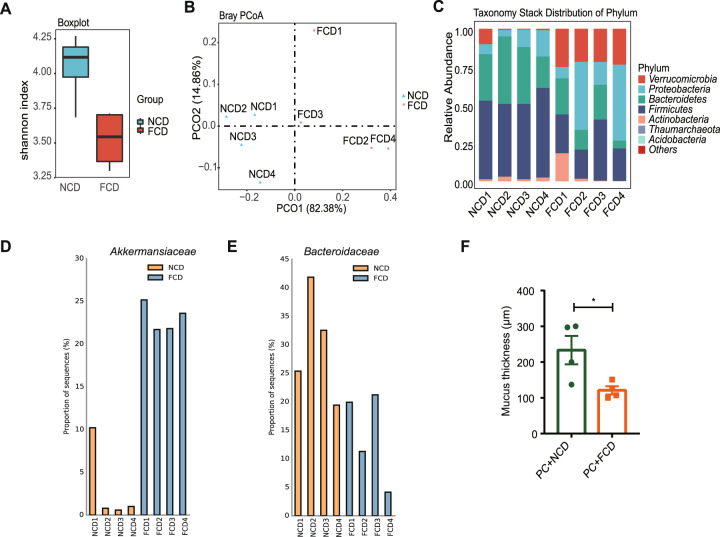
Fig. 4.
Dietary intervention affected the gut microbiota colonized with P. copri RA. A Shannon diversity index between the FCD and NCD groups. (p value = 0.0198). B Principal coordinate analysis (PCoA) of the microbial composition based on Bray Curtis. (p value = 0.031, permutational multivariate analysis of variance (PERMANOVA) by Adonis). C Taxonomy stack distribution of phyla. D Relative abundance of Akkermansiaceae. E Relative abundance of Bacteroidaceae. F Mucus layer thickness of the colon. Each symbol in (F) represents an animal
The fiber-containing diet altered the gut microbiota composition
Given that intestinal inflammation is closely associated with the perturbation of the intestinal microbiota, the effects of a high-fiber diet in the presence of PC colonization on the microbiota composition were investigated. Fecal samples from PC-colonized CIA mice receiving different dietary interventions were collected and subjected to 16 S rDNA sequencing. The gut microbial community structure was determined to identify potential pathobionts associated with RA pathogenesis. The FCD group exhibited decreased diversity, expressed in terms of the Shannon index, compared with that of the NCD group (Fig. 4A). Principal coordinate analysis (PCoA) of the microbial composition based on Bray–Curtis distances was also performed. The analysis showed clear distinctions between NCD and FCD samples (PERMANOVA, R2 = 0.64016, p value = 0.031) (Fig. 4B). Furthermore, the FCD and NCD groups displayed different abundances of the major bacterial phyla in the feces (Fig. 4C). Firmicutes and Bacteroidetes predominated in the NCD group but not in the FCD group. In contrast, Akkermansiaceae, a family within Verrucomicrobia, was the most enriched and overrepresented bacterial family in the FCD group (Fig. 4D). Our results were consistent with previous findings that the phylum Verrucomicrobia and genus Akkermansia were more abundant in RA patients than in controls [35]. A recent study indicated that a high abundance of mucus-degrading Akkermansiaceae is a potential risk factor leading to erosion of the colonic mucus layer in sugar-fed wild-type and Il10−/− mice, suggesting a role for these bacteria in the pathogenesis of sugar-induced inflammation [36]. In agreement with these findings, the FCD group displayed a reduced mucus layer (Fig. 4F), in accord with the increased levels of Akkermansiaceae. On the other hand, the abundance of Bacteroidaceae was markedly decreased in the FCD group compared to the NCD group (Fig. 4E), which is consistent with a previous study showing that the relative abundance of Bacteroides and Prevotella is inversely correlated in the intestine [17]. Taken together, our data indicate that FCD intervention combined with PC colonization led to intestinal dysbiosis characterized by an imbalance between Akkermansiaceae and Bacteroidaceae.
PC encodes distinct carbohydrate-active enzymes that might facilitate more efficient fermentation of complex fiber
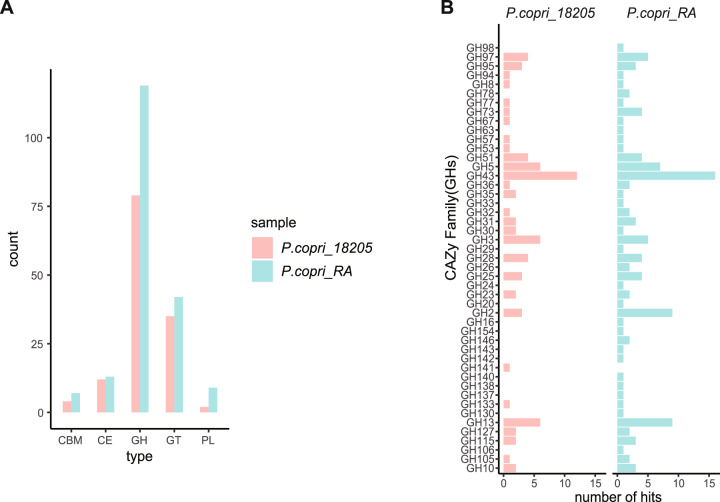
Given that the FCD differs from the NCD only in fiber content, we evaluated the carbohydrate degradation potential of PC. dbCAN2 (HMMER algorithm) [32] was utilized to predict the CAZymes in PC and the reference genome of P. copri DSM 18205. We found that PC encodes more CAZymes than P. copri 18205 (Fig. 5A). Specifically, 190 CAZyme-encoding genes were identified in the genome of PC, including 119 glycoside hydrolases (GHs), 42 glycosyltransferases (GTs), 13 carbohydrate esterases, 9 polysaccharide lyases (PLs) and 3 carbohydrate-binding modules (CBMs) (Supplementary Table 5). GH43, GH2 and GH13, which encode β-xylosidases, β-mannosidases and α-amylases, respectively, were the most abundant GH families in PC and P. copri 18205 (Fig. 5B). Comparative analysis of the GH families between the two strains revealed that PC contained a unique set of 17 GHs that were absent in P. copri 18205 (Fig. 5B). Of note, rhamnogalacturonan-II (RG-II), comprising endo-apiosidase (GH140), DHA hydrolase (GH143), β-l-arabinofuranosidases (GH142 and GH137) and α-galacturonidase (GH138), was identified only in PC (Fig. 5B) but not in P. copri 18205. This RG-II degradome was reported to be involved in the degradation of the most structurally complex glycan known [37]. Together, these data illustrate that PC encodes distinct glycan-degrading enzymes, which may give PC an advantage in the utilization and fermentation of complex fiber.
Fig. 5.
Unique carbohydrate-active enzyme (CAZyme) classes in the P. copri RA strain compared with P. copri 18205. A The distribution of CAZyme classes in P. copri 18205 and P. copri RA. B Presence or absence of glycoside hydrolases (GHs) in P. copri 18205 vs. P. copri RA. Differentially encoded GHs are listed. Abbreviations: CBM carbohydrate binding module, CE carbohydrate esterase, GH glycoside hydrolase, GT glycosyltransferase, PL polysaccharide lyase
The high-fiber diet skewed the production of inflammatory microbial metabolites
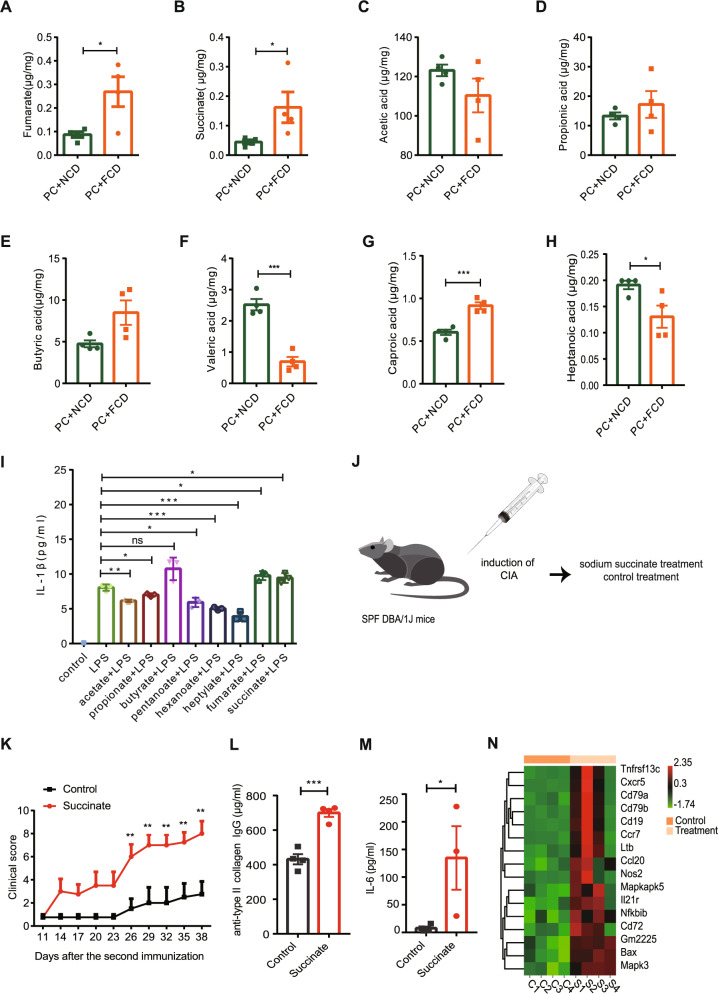
Previous studies have shown that a fiber-rich diet causes the accumulation of metabolites, such as carboxylic acid, in the cecum with concomitant changes in the gut microbiota composition [18]. To evaluate whether FCD aggravates arthritis by enhancing PC degradation of fiber into inflammatory metabolites, we applied a targeted metabolomics approach using GC‒MS analysis. We observed significantly higher levels of fumarate, succinate and caproic acid (Fig. 6A, B, G) and significantly lower levels of valeric acid and heptanoic acid (Fig. 6F, H) in the cecum in FCD-fed mice than in NCD-fed mice. No significant differences in the levels of acetic acid, propionic acid, or butyric acid were identified between the two groups (Fig. 6C–E).
Fig. 6.
High-fiber diet with P. copri RA colonization induced overproduction of inflammatory microbial metabolites and contributed to RA development. A, B The levels of fumarate and succinate in cecal samples. C–H Cecum SCFA levels. Error bars indicate the SEM (A–H). I The level of IL-1β in LPS-stimulated BMDMs pretreated with the designated acids. J Schematic of the sodium succinate treatment. K Clinical scores of collagen-induced arthritis in the succinate group (n = 4) and control group (n = 4). L Anti-bovine type II collagen IgG levels in serum. M Serum IL-6 levels. N Heatmap showing the expression of genes involved in the colon of mice subjected to sodium succinate treatment and control treatment. Each symbol in (A–H, L, and M) represents each animal. Error bars indicate the SEM. P values were determined by two-way ANOVA (K), and t test (A–I, L, M). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Dietary succinate aggravated arthritis
To assess how the abovementioned microbiota-derived metabolites affected immune responses, we isolated bone marrow-derived macrophages (BMDMs) from DBA/1J mice and tested IL-1β production by LPS-stimulated BMDMs treated with these metabolites. Succinate has been shown to be a proinflammatory mediator that promotes IL-1β production [38]. Consistent with these findings, we observed that succinate and fumarate enhanced LPS-mediated IL-1β production in BMDMs (Fig. 6I). In contrast, other SCFAs, including acetate, propionate, pentanoate, hexanoate and heptylate, suppressed LPS-induced IL-1β production in BMDMs (Fig. 6I). Together, these results suggest that in the presence of PC, the arthritis of FCD-fed mice was exacerbated by the production of more proinflammatory metabolites but fewer anti-inflammatory metabolites.
To determine whether succinate contributes to the development of RA, we fed CIA mice sodium succinate in drinking water (Fig. 6J). The succinate-fed group displayed a stronger proinflammatory response than the control, as determined by a higher clinical arthritis score (Fig. 6K) and elevated anti-collagen antibody and IL-6 levels (Fig. 6L, M). To identify the pathways involved in succinate supplementation, mRNA sequencing was performed. Intestinal transcriptomes revealed that succinate supplementation increased the expression of genes associated with a proinflammatory phenotype, such as membrane receptors (i.e., Ccr7), nitric oxide synthase 2 (Nos2), and chemokines (i.e., Ccl20) (Fig. 6N). In addition, succinate treatment affected the expression of genes involved in apoptosis, such as Bax and Mapk3 (Fig. 6N). Taken together, these data suggest that succinate plays a pathogenic role in the development of RA.
Discussion
Prevotella spp., in particular P. copri, are preferentially enriched in a subset of RA patients. When we colonized a P. copri strain isolated from the feces of RA patients (PC) to create a model of dysbiosis, we showed that an FCD promoted the development of RA through the skewed production of proinflammatory over anti-inflammatory metabolites by PC. The presence of PC enabled the digestion of complex fiber, which led to the production of proinflammatory metabolites, such as succinate, that contributed to RA pathogenesis. Our findings highlight that a high-fiber diet may synergize with dysbiosis in contributing to RA pathogenesis.
The presence and overexpansion of Prevotella spp. in the gut microbiome have been associated with detrimental outcomes on human health [6, 7, 10, 18]. Pianta et al. reported that a subgroup of RA patients displayed an aberrant immune response to P. copri, suggesting that P. copri is relevant to immune dysregulation in RA pathogenesis [39]. In this study, we demonstrated the pathogenic role of PC in a CIA model. We found that colonization with PC promoted the development of arthritis, characterized by elevated levels of anti-bovine type II collagen IgG and more severe joint inflammation, which were associated with higher levels of LPS. Furthermore, we showed that PC had a greater capacity to activate the TLR4 pathway, which may contribute to the enhanced immune response. Of interest, we recently reported that P. copri was also enriched in patients with ankylosing spondylitis (AS) [40], suggesting that different inflammatory arthritis diseases may share some common microbial signatures.
Dietary fiber intake has been considered to provide health benefits [41]. In this study, we tested the effect of a fiber diet on the development of RA. We introduced PC into the gut microbiota to mimic dysbiosis in RA patients. Intriguingly, we found that PC + FCD mice developed more severe clinical phenotypes than PC + NCD mice, with more severe intestinal inflammation and more pronounced intestinal microbiota perturbation. To further demonstrate whether PC contributes to the effect of diet, we also included BT-colonized mice receiving different diets. We found that the anti-type II collagen antibody in the serum of PC + FCD mice was markedly higher than that of BT + FCD mice, which confirmed that the synergistic effect of PC and FCD may aggravate arthritis. In the human colon, cellulose from dietary fiber is far more digestible than purified crystalline cellulose [42]. Cellulolytic bacteria (i.e., succinate-producing Prevotella spp.) are able to degrade polysaccharides to produce different breakdown products [43]. Different P. copri isolates can utilize distinct sets of polysaccharides from the diet due to their genomic diversity [17]. In this study, we found that PC, a strain isolated from RA patients, possessed some unique features, such as its expression of rhamnogalacturonan-II degradation-related enzymes, which allowed it to digest the most difficult-to-metabolize glycan in the human diet [37]. Thus, in the presence of an FCD, PC can metabolize more fiber into proinflammatory metabolites such as fumarate and succinate.
Nunns et al. showed that the levels of fumarate and succinate were significantly elevated in trauma patients who developed acute respiratory distress syndrome (ARDS) compared to those who did not, suggesting their involvement in inflammatory lung injury [44]. We demonstrated that succinate and fumarate synergized with LPS to promote BMDM production of IL-1β. We further showed that succinate supplementation exacerbated disease severity in an animal model of RA. In patients with RA, succinate is abundantly present in synovial fluids, and these fluids elicit IL-1β release from macrophages [45]. Thus, in the context of dysbiosis (i.e., overrepresentation of PC), an FCD can lead to more severe RA due to the excessive production of succinate and fumarate.
In the presence of PC, the FCD promoted microbial imbalances, characterized by an increased abundance of Akkermansiaceae and a decreased abundance of Bacteroidaceae. Of interest, our results recapitulated some of the features of human RA, as a recent study also demonstrated that RA patients had a higher abundance of Akkermansia and a lower abundance of Bacteroides [46]. Consistent with our findings, Lam et al. reported that a high-fiber diet favored colonization by Akkermansia, which led to the induction of type I interferon signaling and augmented dendritic cell counts [47]. The inflammatory role of Akkermansia was also confirmed by Fan et al., who showed that Akkermansia could induce an M1-like macrophage response [48]. In addition to stimulating the innate immune response, Akkermansia can induce the production of IgG1 antibodies and initiate antigen-specific T-cell responses in mice [49]. We also previously showed that the A. muciniphila abundance correlated with inflammation in systemic lupus erythematosus (SLE) patients [50]. Further mechanistic studies are warranted to determine the mechanisms by which Akkermansia contributes to RA pathogenesis. Taken together, these findings suggest that an FCD together with PC colonization promotes dysbiosis through the alteration of the microbiota composition, which has profound systemic effects and in turn promotes RA development.
A Mediterranean diet has been considered anti-inflammatory and to thus potentially exert a protective role in RA. However, prior work has suggested that a Mediterranean diet may be effective in only a subset of RA patients [51]. The mechanisms underlying the different effects of a Mediterranean diet on RA remain poorly understood but appear to be related to dysbiosis. Our results provide a potential explanation for the variation in response and suggest that specific gut microbiome signatures may be an important determinant of responses to different dietary interventions.
Our results suggested that dysbiosis mediates the effect of the FCD on RA development. Defining the mechanisms by which various dietary interventions influence RA in the context of existing dysbiosis is an important goal with clear clinical implications, especially in personalized medicine. Our study also provides an opportunity to explore potential biomarkers that could have predictive value in assessing the responses to a certain dietary intervention.
Supplementary information
Acknowledgements
We thank all donors who participated in this study.
Author contributions
Conception and design of the study: LJJ, ZHL, and XZ; implementation of in vivo and in vitro experiments: LJJ, MMS and SNY; analysis of 16 S rRNA sequencing data and the transcriptome data analysis: LJJ; statistical analysis: MMS and HZ; critical comments: YDL, YZZ, MW, TTW, XZ and ZHL; drafting of manuscript: LJJ and SNY; final approval of the version of the article to be published: all authors.
Funding
This work was supported by the National Natural Science Foundation of China (81788101, 82230060, 81630064, and 81701624), the CAMS Innovation Fund for Medical Sciences (CIFMS) (2021-I2M-1-017, 2021-I2M-1-047, 2021-I2M-1-040, and 2021-I2M-1-016), the Capital’s Funds for Health Improvement and Research (2020-2-4019) and the National Key Research and Development Program of China (Grant no. 2018YFE0207300).
Data availability
The genome sequences and RNA sequence data produced in this study are available at the Sequence Read Archive of the National Center for Biotechnology Information (the accession numbers for the sequences reported in this paper are NCBI SRA: SRR14576269).
Competing interests
The authors declare no competing interests.
Ethics approval
All studies reported here were approved by the Institutional Review Board and Animal Care and Use Committee of Peking Union Medical College Hospital (PUMCH).
Footnotes
These authors contributed equally: Lingjuan Jiang, Mengmeng Shang, Shengnan Yu.
Contributor Information
Zhihua Liu, Email: zhihualiu@mail.tsinghua.edu.cn.
Xuan Zhang, Email: zxpumch2003@sina.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-022-00934-6.
References
- 1.Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, Jia X, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–6. doi: 10.1038/ng.1076.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol. 2011;7:569–78. doi: 10.1038/nrrheum.2011.121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bang SY, Lee KH, Cho SK, Lee HS, Lee KW, Bae SC. Smoking increases rheumatoid arthritis susceptibility in individuals carrying the HLA-DRB1 shared epitope, regardless of rheumatoid factor or anti-cyclic citrullinated peptide antibody status. Arthritis Rheum. 2010;62:369–77. doi: 10.1002/art.27272.. [DOI] [PubMed] [Google Scholar]
- 4.Guerreiro CS, Calado A, Sousa J, Fonseca JE. Diet, microbiota, and gut permeability-the unknown triad in rheumatoid arthritis. Front Med (Lausanne) 2018;5:349. doi: 10.3389/fmed.2018.00349.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21:895–905. doi: 10.1038/nm.3914.. [DOI] [PubMed] [Google Scholar]
- 6.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2:e01202. doi: 10.7554/eLife.01202.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alpizar-Rodriguez D, Lesker TR, Gronow A, Gilbert B, Raemy E, Lamacchia C, et al. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann Rheum Dis. 2019;78:590–3. doi: 10.1136/annrheumdis-2018-214514.. [DOI] [PubMed] [Google Scholar]
- 8.Jubair WK, Hendrickson JD, Severs EL, Schulz HM, Adhikari S, Ir D, et al. Modulation of inflammatory arthritis in mice by gut microbiota through mucosal inflammation and autoantibody generation. Arthritis Rheumatol. 2018;70:1220–33. doi: 10.1002/art.40490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tett A, Pasolli E, Masetti G, Ercolini D, Segata N. Prevotella diversity, niches and interactions with the human host. Nat Rev Microbiol. 2021. 10.1038/s41579-021-00559-y. [DOI] [PMC free article] [PubMed]
- 10.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of prevotella. Cell Metab. 2015;22:971–82. doi: 10.1016/j.cmet.2015.10.001.. [DOI] [PubMed] [Google Scholar]
- 11.Marietta EV, Murray JA, Luckey DH, Jeraldo PR, Lamba A, Patel R, et al. Suppression of inflammatory arthritis by human gut-derived prevotella histicola in humanized mice. Arthritis Rheumatol. 2016;68:2878–88. doi: 10.1002/art.39785.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 2016;68:2646–61. doi: 10.1002/art.39783.. [DOI] [PubMed] [Google Scholar]
- 13.De Filippis F, Pasolli E, Tett A, Tarallo S, Naccarati A, De Angelis M, et al. Distinct genetic and functional traits of human intestinal prevotella copri strains are associated with different habitual diets. Cell Host Microbe. 2019;25:444–53 e443. doi: 10.1016/j.chom.2019.01.004.. [DOI] [PubMed] [Google Scholar]
- 14.Gálvez EJC, Iljazovic A, Amend L, Lesker TR, Renault T, Thiemann S, et al. Distinct polysaccharide utilization determines interspecies competition between intestinal prevotella spp. Cell Host Microbe. 2020;28:838–52 e836. doi: 10.1016/j.chom.2020.09.012.. [DOI] [PubMed] [Google Scholar]
- 15.Tett A, Huang KD, Asnicar F, Fehlner-Peach H, Pasolli E, Karcher N, et al. The Prevotella copri complex comprises four distinct clades underrepresented in Westernized populations. Cell Host Microbe. 2019;26:666–79 e667. doi: 10.1016/j.chom.2019.08.018.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright DP, Rosendale DI, Robertson AM. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol Lett. 2000;190:73–9. doi: 10.1111/j.1574-6968.2000.tb09265.x.. [DOI] [PubMed] [Google Scholar]
- 17.Fehlner-Peach H, Magnabosco C, Raghavan V, Scher JU, Tett A, Cox LM, et al. Distinct polysaccharide utilization profiles of human intestinal prevotella copri Isolates. Cell Host Microbe. 2019;26:680–90.e685. doi: 10.1016/j.chom.2019.10.013.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Vadder F, Kovatcheva-Datchary P, Zitoun C, Duchampt A, Bäckhed F, Mithieux G. Microbiota-produced succinate improves glucose homeostasis via intestinal gluconeogenesis. Cell Metab. 2016;24:151–7. doi: 10.1016/j.cmet.2016.06.013.. [DOI] [PubMed] [Google Scholar]
- 19.Singh V, Yeoh BS, Chassaing B, Xiao X, Saha P, Aguilera Olvera R, et al. Dysregulated microbial fermentation of soluble fiber induces cholestatic liver cancer. Cell. 2018;175:679–94 e622. doi: 10.1016/j.cell.2018.09.004.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh V, Yeoh BS, Walker RE, Xiao X, Saha P, Golonka RM, et al. Microbiota fermentation-NLRP3 axis shapes the impact of dietary fibres on intestinal inflammation. Gut. 2019;68:1801–1812. doi: 10.1136/gutjnl-2018-316250.. [DOI] [PubMed] [Google Scholar]
- 21.Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 2014;16:770–7. doi: 10.1016/j.chom.2014.11.003.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Connors J, Dawe N & Van Limbergen J. The role of succinate in the regulation of intestinal inflammation. Nutrients. 2018;11. 10.3390/nu11010025. [DOI] [PMC free article] [PubMed]
- 23.Battino M, Forbes-Hernández TY, Gasparrini M, Afrin S, Cianciosi D, Zhang J, et al. Relevance of functional foods in the Mediterranean diet: the role of olive oil, berries and honey in the prevention of cancer and cardiovascular diseases. Crit Rev Food Sci Nutr. 2019;59:893–920. doi: 10.1080/10408398.2018.1526165.. [DOI] [PubMed] [Google Scholar]
- 24.Petersson S, Philippou E, Rodomar C, Nikiphorou E. The Mediterranean diet, fish oil supplements and Rheumatoid arthritis outcomes: evidence from clinical trials. Autoimmun Rev. 2018;17:1105–14. doi: 10.1016/j.autrev.2018.06.007.. [DOI] [PubMed] [Google Scholar]
- 25.Hu Y, Costenbader KH, Gao X, Hu FB, Karlson EW, Lu B. Mediterranean diet and incidence of rheumatoid arthritis in women. Arthritis Care Res (Hoboken) 2015;67:597–606. doi: 10.1002/acr.22481.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingegnoli F, Schioppo T, Scotti I, Ubiali T, De Lucia O, Murgo A, et al. Adherence to Mediterranean diet and patient perception of rheumatoid arthritis. Complement Ther Med. 2020;52:102519. doi: 10.1016/j.ctim.2020.102519.. [DOI] [PubMed] [Google Scholar]
- 27.Wastyk HC, Fragiadakis GK, Perelman D, Dahan D, Merrill BD, Yu FB, et al. Gut-microbiota-targeted diets modulate human immune status. Cell. 2021;184:4137–53 e4114. doi: 10.1016/j.cell.2021.06.019.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brand DD, Latham KA, Rosloniec EF. Collagen-induced arthritis. Nat Protoc. 2007;2:1269–75. doi: 10.1038/nprot.2007.173.. [DOI] [PubMed] [Google Scholar]
- 29.Qiu Z, Sheridan BS. Isolating lymphocytes from the mouse small intestinal immune system. J Vis Exp. 2018. 10.3791/57281. [DOI] [PMC free article] [PubMed]
- 30.Wang Y, Li N, Yang JJ, Zhao DM, Chen B, Zhang GQ, et al. Probiotics and fructo-oligosaccharide intervention modulate the microbiota-gut brain axis to improve autism spectrum reducing also the hyper-serotonergic state and the dopamine metabolism disorder. Pharm Res. 2020;157:104784. doi: 10.1016/j.phrs.2020.104784.. [DOI] [PubMed] [Google Scholar]
- 31.Zheng X, Qiu Y, Zhong W, Baxter S, Su M, Li Q, et al. A targeted metabolomic protocol for short-chain fatty acids and branched-chain amino acids. Metabolomics. 2013;9:818–27. doi: 10.1007/s11306-013-0500-6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–101. doi: 10.1093/nar/gky418.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gatto D, Paus D, Basten A, Mackay CR, Brink R. Guidance of B cells by the orphan G protein-coupled receptor EBI2 shapes humoral immune responses. Immunity. 2009;31:259–69. doi: 10.1016/j.immuni.2009.06.016.. [DOI] [PubMed] [Google Scholar]
- 34.Ikeda Y, Ikata T, Mishiro T, Nakano S, Ikebe M, Yasuoka S. Cathepsins B and L in synovial fluids from patients with rheumatoid arthritis and the effect of cathepsin B on the activation of pro-urokinase. J Med Invest. 2000;47:61–75. [PubMed] [Google Scholar]
- 35.Chiang H-I, Li J-R, Liu C-C, Liu P-Y, Chen H-H, Chen Y-M, et al. An association of gut microbiota with different phenotypes in chinese patients with rheumatoid arthritis. J Clin Med. 2019;8. 10.3390/jcm8111770. [DOI] [PMC free article] [PubMed]
- 36.Khan S, Waliullah S, Godfrey V, Khan MAW, Ramachandran RA, Cantarel BL, et al. Dietary simple sugars alter microbial ecology in the gut and promote colitis in mice. Sci Transl Med. 2020;12. 10.1126/scitranslmed.aay6218. [DOI] [PubMed]
- 37.Ndeh D, Rogowski A, Cartmell A, Luis AS, Baslé A, Gray J, et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature. 2017;544:65–70. doi: 10.1038/nature21725.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, et al. Succinate is an inflammatory signal that induces IL-1 beta through HIF-1 alpha. Nature. 2013;496:238-+. doi: 10.1038/nature11986.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pianta A, Arvikar S, Strle K, Drouin EE, Wang Q, Costello CE, et al. Evidence of the immune relevance of prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol. 2017;69:964–75. doi: 10.1002/art.40003.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou C, Zhao H, Xiao XY, Chen BD, Guo RJ, Wang Q, et al. Metagenomic profiling of the pro-inflammatory gut microbiota in ankylosing spondylitis. J Autoimmun. 2020;107:102360. doi: 10.1016/j.jaut.2019.102360.. [DOI] [PubMed] [Google Scholar]
- 41.Anderson JW, Baird P, Davis RH, Jr, Ferreri S, Knudtson M, Koraym A, et al. Health benefits of dietary fiber. Nutr Rev. 2009;67:188–205. doi: 10.1111/j.1753-4887.2009.00189.x.. [DOI] [PubMed] [Google Scholar]
- 42.Slavin JL, Brauer PM, Marlett JA. Neutral detergent fiber, hemicellulose and cellulose digestibility in human subjects. J Nutr. 1981;111:287–97. doi: 10.1093/jn/111.2.287.. [DOI] [PubMed] [Google Scholar]
- 43.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–31. doi: 10.1038/nrmicro1817.. [DOI] [PubMed] [Google Scholar]
- 44.Nunns GR, Vigneshwar N, Kelher MR, Stettler GR, Gera L, Reisz JA, et al. Succinate activation of SUCNR1 predisposes severely injured patients to neutrophil-mediated ARDS. Ann Surg. 2020. 10.1097/SLA.0000000000004644. [DOI] [PMC free article] [PubMed]
- 45.Littlewood-Evans A, Sarret S, Apfel V, Loesle P, Dawson J, Zhang J, et al. GPR91 senses extracellular succinate released from inflammatory macrophages and exacerbates rheumatoid arthritis. J Exp Med. 2016;213:1655–62. doi: 10.1084/jem.20160061.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Ma C, Liu L, He J, Zhu C, Zheng F, et al. Analysis of gut microbiota and metabolites in patients with rheumatoid arthritis and identification of potential biomarkers. Aging (Albany NY). 2021;13. 10.18632/aging.203641. [DOI] [PMC free article] [PubMed]
- 47.Lam KC, Araya RE, Huang A, Chen Q, Di Modica M, Rodrigues RR, et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell. 2021;184:5338–56.e5321. doi: 10.1016/j.cell.2021.09.019.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan L, Xu C, Ge Q, Lin Y, Wong CC, Qi Y, et al. A. muciniphila suppresses colorectal tumorigenesis by inducing TLR2/NLRP3-mediated M1-Like TAMs. Cancer Immunol Res. 2021;9:1111–24. doi: 10.1158/2326-6066.CIR-20-1019.. [DOI] [PubMed] [Google Scholar]
- 49.Ansaldo E, Slayden LC, Ching KL, Koch MA, Wolf NK, Plichta DR, et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science. 2019;364:1179–84. doi: 10.1126/science.aaw7479.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen BD, Jia XM, Xu JY, Zhao LD, Ji JY, Wu BX, et al. An autoimmunogenic and proinflammatory profile defined by the gut microbiota of patients with untreated systemic lupus erythematosus. Arthritis Rheumatol. 2021;73:232–43. doi: 10.1002/art.41511.. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen Y, Salliot C, Gelot A, Gambaretti J, Mariette X, Boutron-Ruault MC, et al. Mediterranean diet and risk of rheumatoid arthritis: findings from the French E3N-EPIC cohort study. Arthritis Rheumatol. 2021;73:69–77. doi: 10.1002/art.41487.. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences and RNA sequence data produced in this study are available at the Sequence Read Archive of the National Center for Biotechnology Information (the accession numbers for the sequences reported in this paper are NCBI SRA: SRR14576269).