Abstract
Higher maternal pre-pregnancy body mass index (ppBMI) is associated with increased neonatal morbidity, as well as with pregnancy complications and metabolic outcomes in offspring later in life. The placenta is a key organ in fetal development and has been proposed to act as a mediator between the mother and different health outcomes in children. The overall aim of the present work is to investigate the association of ppBMI with epigenome-wide placental DNA methylation (DNAm) in 10 studies from the PACE consortium, amounting to 2631 mother-child pairs. We identify 27 CpG sites at which we observe placental DNAm variations of up to 2.0% per 10 ppBMI-unit. The CpGs that are differentially methylated in placenta do not overlap with CpGs identified in previous studies in cord blood DNAm related to ppBMI. Many of the identified CpGs are located in open sea regions, are often close to obesity-related genes such as GPX1 and LGR4 and altogether, are enriched in cancer and oxidative stress pathways. Our findings suggest that placental DNAm could be one of the mechanisms by which maternal obesity is associated with metabolic health outcomes in newborns and children, although further studies will be needed in order to corroborate these findings.
Subject terms: Epigenomics, Genetics research
A meta-analysis of pre-pregnancy maternal body mass index (ppBMI) and placental DNA methylation from 2631 mother-child pairs identifies 27 CpG sites associated with ppBMI, providing insight into how maternal obesity could be associated with metabolic health outcomes in offspring.
Introduction
Higher maternal pre-pregnancy body mass index (ppBMI) is associated with aberrant fetal growth1, macrosomia and increased neonatal morbidity and mortality2, and also with pregnancy complications such as pre-eclampsia, gestational diabetes, gestational hypertension, pre-term delivery and cesarean section3. It has been shown that maternal adipokine and insulin signaling in the placenta could contribute to regulate both the vascular development of this organ and the nutrient transport to the fetus, and therefore impact fetal development3. Additionally, it has also been observed that maternal ppBMI is associated with other offspring health outcomes in later life, including increased risk for obesity in children4. Observational studies have suggested links between maternal obesity and long-term risk of coronary heart disease, stroke, type 2 diabetes and asthma in offspring5. Very high maternal ppBMI has also been associated with poorer cognitive performance in children and greater risk of neurodevelopmental disorders6, while there is also preliminary evidence in favor of potential implications in immune and infectious disease-related outcomes3. These associations could be mediated by epigenetic changes, including DNA methylation (DNAm), but the implication of an early epigenetic reprogramming in utero deserves further research7.
A previous study carried out within the Pregnancy and Childhood Epigenetics (PACE) consortium8 has shown that maternal ppBMI is widely associated with differences in cord blood DNAm in the newborn9. However, the authors observed that many of the significant epigenetic effects were modest (<0.2% methylation per BMI unit) and they did not detect enrichment for any particular biological pathway, leaving open questions regarding potential intra-uterine mechanisms that could be affecting the epigenetic profile of the newborn9. In this context, while the epigenetic alterations in cord and peripheral blood have been thoroughly investigated9,10, the potential impact of maternal ppBMI in placental DNAm remains poorly explored. As far as we know, the most recent studies have performed methylation profiling with methylation arrays or reduced representation bisulfite sequencing in up to 300 term placentas of obese and non-obese mothers11,12. Although interesting, these studies have yielded a limited number of significant results, probably because of their relatively small sample size.
A recently published meta-analysis with 1700 placental samples by the PACE consortium13 has discovered a placental DNAm signature of maternal smoking during pregnancy that is quite different from what has been observed in cord blood14. Differentially methylated CpGs related to smoking in pregnancy fall within active regions of the placental epigenome, and nearby genes are involved in the response to environmental stressors, regulation of inflammatory activity, and growth factor signaling. The placenta is a transient organ at the maternal-fetal interface, with endocrine and substrate-transport functions, that is sensitive to pregnancy environmental influences – exogenous or endogenous. Maternal pre-pregnancy obesity is often characterized by an adverse metabolic milieu that may alter placental function by increasing oxidative stress, vascular endothelium thickening, and inflammatory lesions in placental tissues11. Altogether, these facts encourage the investigation of the placenta as a putative mediator of maternal obesity and health outcomes in the offspring, specifically through the modification of the placental DNAm landscape.
In this context, the overall aim of the current analyses was to investigate the association of maternal ppBMI with epigenome-wide placental DNAm in 10 studies from the PACE consortium amounting to 2631 mother-child pairs. We also conducted functional enrichment analyses and comparison of our results with maternal ppBMI-associated cord blood DNAm alterations previously reported by PACE.
Results
Study population
Eleven North-American, Australian, and European studies (N = 2631) contributed to the epigenome-wide association study (EWAS) to determine the associations of maternal ppBMI with placental DNAm (Table 1), including Asking Questions about Alcohol in pregnancy (AQUA),15 Early Autism Risk Longitudinal Investigation (EARLI)16, Study on the pre- and early postnatal determinants of child health and development (EDEN)17, Genetics of Glucose regulation in Gestation and Growth (Gen3G)18, Genetics, Early Life Environmental Exposures and Infant Development in Andalusia (GENEIDA)19, Harvard Epigenetic Birth cohort (HEBC)20, Environment and Childhood Project (INMA)21, The Intrauterine Sampling in Early Pregnancy Study (ITU)22, Markers of Autism Risk in Babies-Learning Early Signs (MARBLES)23, New Hampshire Birth Cohort Study (NHBCS)24, and Rhode Island Child Health Study (RICHS)25. MARBLES was excluded at a later stage as the sample size was too small and results were inconsistent with the other cohorts (Supplementary Fig. 1).
Table 1.
Effective sample size of the participating cohorts.
| Cohort | Country | N | Array |
|---|---|---|---|
| AQUA | Australia | 95 | 450 K |
| EARLI | USA | 54 | 450 K |
| EDEN | France | 664 | 450 K |
| HBEC | USA | 186 | 450 K |
| Gen3G | Canada | 448 | EPIC |
| GENEIDA | Spain | 103 | 450 K |
| INMA | Spain | 168 | 450 K |
| ITU | Finland | 352 | EPIC |
| NHBCS | USA | 311 | 450 K |
| RICHS | USA | 250 | 450 K |
| TOTAL | 2631 |
Maternal ppBMI (kg/m2) was generally self-reported. In those cases where ppBMI was not available, BMI in early pregnancy (1st trimester) was used. For simplicity, we will refer to both of them as maternal ppBMI. In all the analyses performed, we used ppBMI as a continuous variable. The cohort-specific average maternal ppBMI ranged from 22.9 in EDEN (France) to 27.6 in EARLI (USA) (standard deviation-SD = 1.60). In general, 548 (20.8%) and 369 (14.0%) mothers reported overweight (>25 BMI) and obesity (>30 BMI), respectively, while 115 (4.37%) appeared to be underweight (<18.5 BMI). Mean age of the mothers was 30.7 years (SD = 2.9). The distributions of other covariates by cohort are provided in Supplementary Data 1.
Genome-wide DNAm meta-analyses
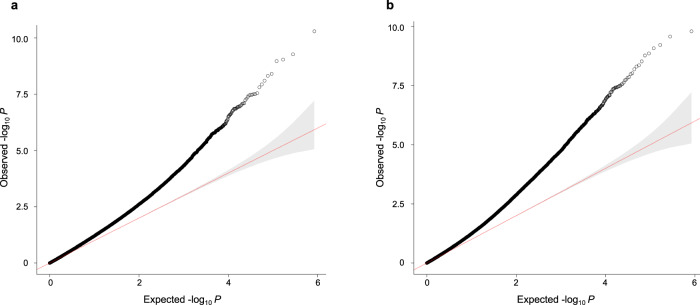
Each cohort analyst conducted two different EWAS, modeling DNAm beta-values at a maximum number of 419,460 CpG sites in relation to maternal ppBMI using robust linear regressions, with and without adjustment for putative cellular components. Cell composition was estimated using the reference-free deconvolution algorithm RefFreeCellMix26. The CpGs included in the analyses were among those shared by the two most common Illumina Infinium Beadchip arrays, 450 K and EPIC, since both arrays were used to assess the DNAm levels of the samples, as shown in Table 1. All models were adjusted for maternal age, parity, maternal education and maternal smoking. Genomic inflation factors from the cohort-specific models (ranging from λ = 0.692 to 1.472) and a summary of the results can be seen in Supplementary Data 2. Finally, after quality control of the results, we conducted an inverse variance-weighted fixed-effects meta-analysis using the software GWAMA27. The inflation factors from the meta-analyses (λ = 1.230 and 1.220 for the cell type-adjusted and -unadjusted models, respectively) (Fig. 1) revealed potential residual confounding and moderate inflation of test statistics.
Fig. 1. QQ-plots of the meta-analyses of the association between maternal ppBMI and placental DNAm.
Analyses were carried out a adjusting for putative cellular heterogeneity or b without adjusting for putative cellular heterogeneity (N = 2631 placental DNA samples). The red line represents the normal distribution and 95% confidence interval (gray shading).
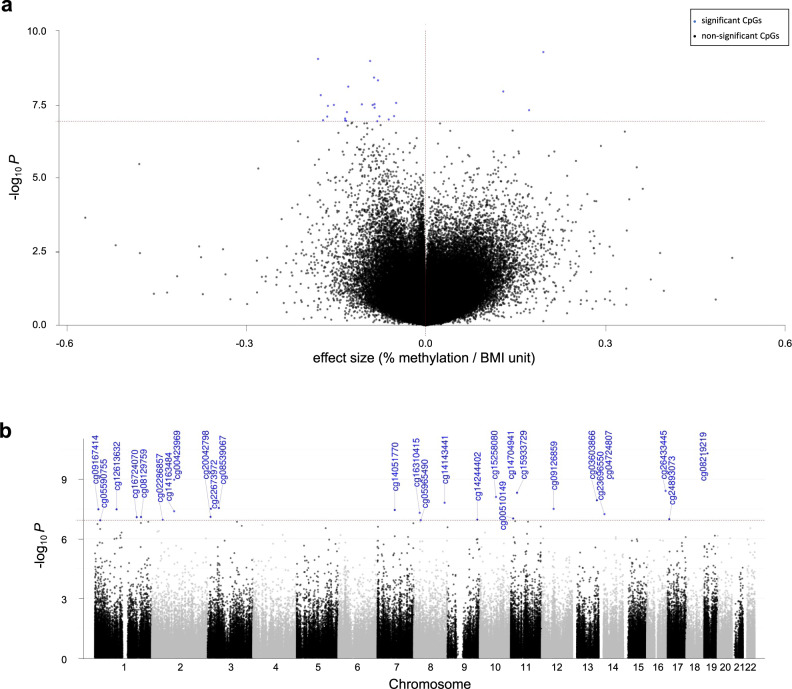
After applying the Bonferroni correction for multiple-testing (meta-analysis nominal p value <1.2e-07), we obtained 27 and 42 CpGs at which maternal ppBMI was significantly associated with placental DNAm in the models adjusted and unadjusted for cell type proportions, respectively. Full results for both models are provided in Supplementary Data 3 and 4, respectively. Higher maternal ppBMI was associated with lower placental DNAm in 24/27 differentially methylated CpGs identified in the cell type-adjusted model, while in the unadjusted model, 33/42 hits showed positive associations (higher maternal ppBMI associated with increased placental DNAm at the identified CpGs). However, beta-coefficients of CpGs that were differentially methylated in one model were positively correlated to the beta-coefficients of the same position in the other (Supplementary Fig. 2). Finally, the heterogeneity of associations across cohorts was lower for the model adjusted for cell type proportions compared to the unadjusted model (26/27 vs. 34/42 CpGs presented Cochran’s Q-test p-values > 0.01) and thus, we continued with the results from the fully adjusted model for all downstream analyses.
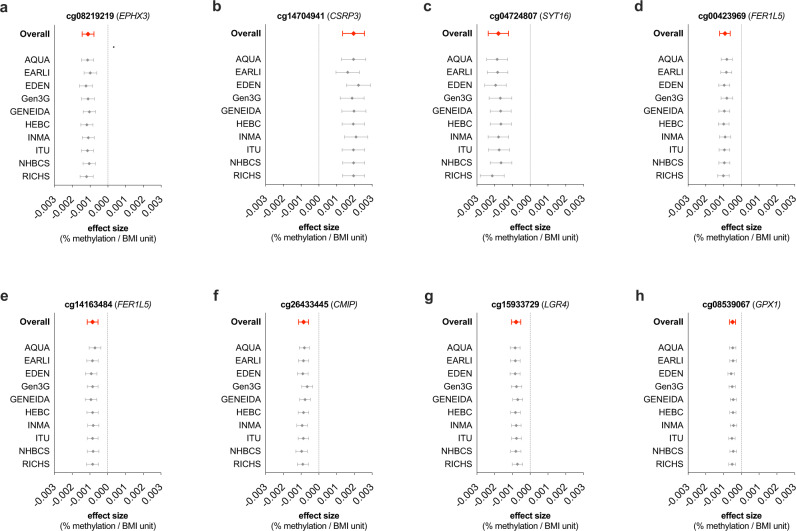
Among the 27 differentially methylated CpGs identified in our cell type-adjusted EWAS (Table 2, Fig. 2A, B), a few individual CpGs are worthy of mention. The most notable association was observed at cg08219219, located in the eighth exon of EPHX3, with the lowest p-value in the meta-analysis (Bonferroni-corrected, meta-analysis p-value = 2.12e-05) and a beta-coefficient of −1.12e-03, meaning that a 10-unit difference in maternal ppBMI is associated with a 1.1% lower DNAm at this specific CpG site. This association was consistent across all cohorts (Cochran’s Q-test p-value = 0.12) (Fig. 3a). The largest beta-coefficient was observed in cg14704941, in the first intron of CSRP3, with a positive beta-coefficient of 1.96e-03, corresponding to a 2% higher placental DNAm per 10-unit ppBMI (Bonferroni-corrected p-value = 2.24e-04 and Cochran’s Q-test p-value = 0.09) (Fig. 3b). In turn, the largest negative beta-coefficient was found in cg04724807 (more than 57 Kb upstream of SYT16) with 1.8% lower DNAm per 10-unit ppBMI (Bonferroni-corrected p-value = 3.83e-04 and Cochran’s Q-test p-value = 0.097) (Fig. 3c). The following CpGs reached the Bonferroni significance threshold and were not identified as highly heterogeneous across cohorts: cg00423969 and cg14163484, 1.5 kb upstream of the FER1L5 promoter, as well as cg26433445, cg15933729 and cg08539067, close to CMIP, LGR4 and GPX1, respectively (Fig. 3d–h). The remaining Bonferroni-significant hits and their heterogeneity across cohorts are shown in Supplementary Fig. 3.
Table 2.
Meta-analysis results from the model adjusted for maternal age, parity, maternal education, maternal smoking and putative cellular heterogeneity, showing the 27 Bonferroni significant CpGs associated with maternal ppBMI.
| CpG | Results from the meta-analysis | Meta-analysis heterogeneity | Location Annotations | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | 95% CI | P-value | Adj. P-value | Direction | HetISq | HetDf | HetPVal | Chr | Pos | Closest Gene | Distance to closest Gene | Relation to Island | |
| cg00423969 | −0.00092 | 0.00015 | −0.0012;−0.0006 | 1.07E-09 | 4.49E-04 | -----+---- | 0.474 | 10 | 0.047 | 2 | 97359879 | FER1L5 | 0 | Open Sea |
| cg00510149 | −0.00134 | 0.00025 | −0.0018;−0.0008 | 9.56E-08 | 4.01E-02 | --+------- | 0.531 | 10 | 0.023 | 11 | 10674966 | IRAG1 | 0 | Open Sea |
| cg02286857 | −0.00134 | 0.00025 | −0.0018;−0.0008 | 1.11E-07 | 4.66E-02 | ---------? | 0.602 | 9 | 0.010 | 2 | 47297177 | TTC7A | 0 | Open Sea |
| cg03603866 | 0.00129 | 0.00023 | 0.0008; 0.0017 | 1.16E-08 | 4.87E-03 | +-+-++?+++ | 0.167 | 9 | 0.294 | 13 | 107027625 | LINC00460 | 1284 | Open Sea |
| cg04724807 | −0.00179 | 0.00029 | −0.0024;−0.0012 | 9.12E-10 | 3.83E-04 | -+-------- | 0.391 | 10 | 0.097 | 14 | 62396305 | SYT16 | 57496 | Open Sea |
| cg05590755 | −0.00080 | 0.00015 | −0.0011;−0.0005 | 1.19E-07 | 4.99E-02 | --+------- | 0.439 | 10 | 0.066 | 1 | 23855149 | E2F2 | 0 | North Shore |
| cg05965490 | −0.00132 | 0.00025 | −0.0018;−0.0008 | 1.16E-07 | 4.87E-02 | --+------- | 0.398 | 10 | 0.092 | 8 | 30264627 | RBPMS | 0 | Open Sea |
| cg08129759 | −0.00077 | 0.00014 | −0.0010;−0.0005 | 8.09E-08 | 3.39E-02 | ---------- | 0.303 | 10 | 0.166 | 1 | 202091944 | GPR37L1 | 83 | Open Sea |
| cg08219219 | −0.00112 | 0.00017 | −0.0014;−0.0008 | 5.05E-11 | 2.12E-05 | ---------- | 0.361 | 10 | 0.119 | 19 | 15337971 | EPHX3 | 0 | South Shelf |
| cg08539067 | −0.00049 | 0.00009 | −0.0007;−0.0003 | 2.84E-08 | 1.19E-02 | --+----+-- | 0.491 | 10 | 0.039 | 3 | 49395985 | GPX1 | 193 | South Shore |
| cg09126859 | −0.00106 | 0.00019 | −0.0014;−0.0007 | 3.17E-08 | 1.33E-02 | --+------- | 0.415 | 10 | 0.081 | 12 | 52244063 | FIGNL2 | 18361 | South Shore |
| cg09167414 | −0.00152 | 0.00028 | −0.0021;−0.0010 | 3.31E-08 | 1.39E-02 | ---------- | 0.000 | 10 | 0.511 | 1 | 16076206 | TMEM82 | 1913 | South Shelf |
| cg12613632 | −0.00088 | 0.00016 | −0.0012;−0.0006 | 3.33E-08 | 1.40E-02 | ------+--- | 0.165 | 10 | 0.292 | 1 | 95385935 | CNN3 | 0 | Open Sea |
| cg14051770 | −0.00162 | 0.00029 | −0.0022;−0.0010 | 3.58E-08 | 1.50E-02 | ---------- | 0.000 | 10 | 0.833 | 7 | 76054572 | ZP3 | 0 | Open Sea |
| cg14143441 | −0.00174 | 0.00031 | −0.0023;−0.0011 | 1.55E-08 | 6.50E-03 | --+------- | 0.513 | 10 | 0.030 | 8 | 134387493 | NDRG1 | 77945 | Open Sea |
| cg14163484 | −0.00085 | 0.00015 | −0.0012;−0.0005 | 4.13E-08 | 1.73E-02 | ----?--?-- | 0.224 | 8 | 0.251 | 2 | 97359926 | FER1L5 | 0 | Open Sea |
| cg14244402 | −0.00170 | 0.00032 | −0.0023;−0.0011 | 1.10E-07 | 4.61E-02 | ---------- | 0.122 | 10 | 0.330 | 9 | 130681102 | ST6GALNAC4 | 1796 | South Shore |
| cg14704941 | 0.00196 | 0.00032 | 0.0013; 0.0026 | 5.33E-10 | 2.24E-04 | ++++++++?? | 0.432 | 8 | 0.090 | 11 | 19224659 | CSRP3 | 0 | Open Sea |
| cg15258080 | −0.00129 | 0.00022 | −0.0017;−0.0008 | 7.91E-09 | 3.32E-03 | ---------- | 0.419 | 10 | 0.078 | 10 | 71091204 | HK1 | 0 | Open Sea |
| cg15933729 | −0.00079 | 0.00014 | −0.0011;−0.0005 | 4.84E-09 | 2.03E-03 | -+-------- | 0.182 | 10 | 0.276 | 11 | 27504612 | LGR4 | 10277 | Open Sea |
| cg16310415 | 0.00172 | 0.00032 | 0.0011; 0.0023 | 4.99E-08 | 2.09E-02 | ++++-++-+? | 0.550 | 9 | 0.023 | 8 | 25898539 | PPP2R2A | 0 | North Shore |
| cg16724070 | −0.00163 | 0.00030 | −0.0022;−0.0010 | 8.22E-08 | 3.45E-02 | --+----+-? | 0.605 | 9 | 0.009 | 1 | 183133736 | LAMC1 | 19008 | Open Sea |
| cg20042798 | −0.00052 | 0.00010 | −0.0007;−0.0003 | 7.95E-08 | 3.33E-02 | ---------- | 0.000 | 10 | 0.827 | 3 | 13036713 | IQSEC1 | 0 | South Shore |
| cg22673972 | −0.00085 | 0.00015 | −0.0011;−0.0005 | 3.12E-08 | 1.31E-02 | ---------- | 0.000 | 10 | 0.949 | 3 | 14415346 | SLC6A6 | 28728 | Open Sea |
| cg23696550 | −0.00131 | 0.00024 | −0.0018;−0.0008 | 5.80E-08 | 2.43E-02 | -----+---- | 0.136 | 10 | 0.318 | 14 | 24732386 | TGM1 | 0 | Open Sea |
| cg24893073 | −0.00061 | 0.00012 | −0.0008;−0.0004 | 1.04E-07 | 4.36E-02 | ---------- | 0.000 | 10 | 0.845 | 17 | 7742126 | KDM6B | 1107 | N_Shelf |
| cg26433445 | −0.00086 | 0.00015 | −0.0011;−0.0006 | 3.92E-09 | 1.64E-03 | ---------- | 0.571 | 10 | 0.013 | 16 | 81764289 | CMIP | 18921 | Open Sea |
Beta: effect size, SE: Standard error, 95%CI: 95% confidence interval, Adjusted P-value: Bonferroni corrected p-value, HetISq: Heterogeneity test statistic I2, HetDf: Heterogeneity degrees of freedom, HetPVal: Heterogeneity test p-value. Direction: direction of the effect in each cohort, ordered in alphabetical order (+: positive; -: negative; and ?: CpG not present in the cohort).
CpGs that were not annotated with a gene name in the Illumina 450 K annotation file have been annotated with their closest gene.
Fig. 2. Association between maternal ppBMI and placental DNAm (N = 2631 placental DNA samples), after adjusting for maternal age, parity, maternal education, maternal smoking and putative cellular heterogeneity.
Association results are displayed as a volcano plot, where the X-axis shows the effect sizes (ranging between 0 and 1) in DNAm and b Manhattan plot, where the X-axis represents the genomic location of each CpG. In both panels blue dots indicate significantly associated CpGs (meta-analysis p-value < 1.2e-07).
Fig. 3. Forest plots of the leave-one-out analysis showing the fixed effects meta-analysis estimates of association between maternal ppBMI and placental DNAm.
Association of a cg08219219, b cg14704941, c cg04724807, d cg00423969, e cg14163484, f cg26433445, g cg15933729, h and cg08539067 with maternal ppBMI. In all panels, cohort names indicate the cohort excluded in each row, and error bars represent the 95% confidence interval of the effect size. Numerical source data for the figure are available in file Supplementary Data 9.
Gene-set and regulatory enrichment analyses
To gain insight into the biological processes that may be captured by placental DNAm associated with maternal ppBMI, we performed gene-set and regulatory enrichment analyses. To this end, first, we annotated CpGs to genes and regulatory elements as explained in the Material and Methods section. Then, we conducted gene-set enrichments for the 26 unique genes annotated to the 27 maternal ppBMI-sensitive CpGs with ConsensusPathDB28 using KEGG, Reactome, Wikipathways and Biocarta reference databases. Two gene-set pathways were significantly enriched (q value < 0.05), namely small cell lung cancer and oxidative stress-induced signaling pathway (Supplementary Data 5). This was also true when we reduced the background from default to only the genes that are represented in the Illumina 450 K array (~21,231). We also tested whether the genes annotated to maternal ppBMI-associated CpGs were enriched for regulatory regions of specific transcription factors (TFs). Most notably, our ppBMI-associated CpGs were enriched for genes regulated by ZNF217 (adjusted p value = 0.02).
We then examined whether the 27 maternal ppBMI-associated CpGs were enriched for CpG island locations, placenta-specific imprinting regions or parent-of-origin-specific germline differentially methylated regions29, regulatory features from the placenta-specific 15-chromatin state annotations from ROADMAP30,31, or placenta-specific partially methylated domains32 that contain placenta-specific repressed genes. We did not find any significant enrichment except for CpG island location and features: the maternal ppBMI-associated CpGs were depleted in CpG islands (χ2 = −2.927, p value = 8.4e-04) and highly enriched in open sea regions (χ2 = 2.742, p value = 1.3e-03) (Supplementary Fig. 4).
Proximity to genetic variants relevant for birth outcomes
We wanted to determine whether the maternal ppBMI-associated CpGs that we identified here were localized near genetic variants that have been associated with birth outcomes in previously published genome-wide association studies (GWAS). Thus, we investigated whether ppBMI-associated CpGs were within ± 0.5 Mb (1 Mb window) of single nucleotide polymorphisms (SNPs) that have been associated with birth weight (BW, N = 310), birth length (N = 5), head circumference (N = 3), gestational age (GA, N = 6) and BW + GA (N = 6)33–38. Of the total 330 birth outcome SNPs in autosomal chromosomes, 10 BW-associated SNPs were within 0.5 Mb of CpGs that were associated with maternal ppBMI. Therefore, more than a third of the 27 ppBMI-associated CpG sites were within 0.5 Mb of BW SNPs, including cg00423969 and cg14163484 (FER1L5), cg00510149 (IRAG1), cg02286857 (TTC7A), cg15258080 (HK1), cg22673972 (SLC6A6) and cg24893073 (KDM6B) (Supplementary Data 6).
Comparison with maternal ppBMI-associated CpGs in cord blood DNAm
We assessed whether the DNAm signatures of maternal ppBMI in the placenta were consistent with associations in cord blood previously reported by the PACE consortium9. We did not find any overlapping CpGs associated with maternal ppBMI between the two tissues. However, we reported three maternal ppBMI-associated CpGs in the placenta that were less than 0.5 Mb upstream or downstream from CpGs that had been associated with maternal ppBMI in cord blood: two out of the 3 loci identified showed consistent effect directions of the association with maternal ppBMI in both tissues (Supplementary Data 7).
Discussion
As far as we know, this is the largest EWAS meta-analysis conducted to date on placental DNAm. We have analyzed a total of 2631 mother-child pairs from 10 different PACE cohorts from Europe, America and Australia. We have identified 27 CpGs associated to maternal ppBMI, some of which showed up to 2% lower DNAm per 10-unit higher BMI. Although such a difference in BMI is unlikely in an individual woman in the context of pre-pregnancy interventions, we consider that it could represent the difference between women in the normal range of BMI and women with BMI in the obesity category.
The most significant association was observed for cg08219219, located in the eighth exon of EPHX3, for which a ppBMI difference of 10 units is associated with a 1.1% lower placental DNAm. It has been shown that soluble epoxide hydrolases such as EPHX3 have higher activity in obese mice39. Additionally, it has been suggested that this family of hydrolases could act as therapeutic targets for metabolic and cardiovascular abnormalities related to obesity40. We highlighted two other significant hits showing the largest positive and negative beta-coefficients. cg14704941, in the first intron of CSRP3, presented with 2% higher placental DNAm per 10-unit ppBMI. CRSP3 knockout mice develop dilated cardiomyopathy with hypertrophy and heart failure after birth41. The beta-coefficient of cg04724807, located upstream of SYT16, represented about 1.8% lower placental DNAm per 10-unit greater ppBMI. SYT16 is over-expressed in pancreatic islet cells upon high glucose challenge and is thought to play a role in insulin secretion42. As previously stated, maternal obesity has been described to be associated with obesity, diabetes and cardiometabolic conditions in offspring later in life4,5. The fact that our EWAS identified CpGs near these metabolically relevant genes highlights the plausibility that they may play a role in the link between maternal obesity and future health outcomes in children.
Among our significant signals, we also found two CpG sites, cg00423969 and cg14163484, 1.5 kb upstream of the FER1L5 promoter, presenting lower placental DNAm levels associated with higher maternal ppBMI. Remarkably, FER1L5 encodes a dysferlin- and myoferlin-related protein, which has been predicted to have a role in vesicle trafficking and muscle membrane fusion events43. Both vesicle trafficking and membrane fusion are crucial events in placental development, since they allow the formation of the syncytiotrophoblast, an uninterrupted and multi-nucleated mass that covers the placental villi and enables the interplay with the mother44. In addition, CMIP and GPX1, two of the genes annotated to maternal ppBMI-associated CpGs, may present relevant biological roles in pregnancy. For example, different CpGs surrounding CMIP have been associated with pre-eclampsia in a placental DNAm study45. GPX1 is an antioxidant gene and its mRNA levels are lower in the placenta of obese mothers compared to normal-weight mothers46. Finally, in the context of obesity, LGR4, another gene identified in the present study, bears an activating variant that contributes to abdominal visceral fat accumulation and therefore, to central obesity47, suggesting that both genetic and epigenetic regulation at this locus may have a role in obesity-related phenotypes.
Regarding enrichment analyses, one of the most interesting findings is that several significant CpGs are located close to cancer-related genes. It has been recently described that the placenta is organized as a big mass of tumoral clones, with rapid cell divisions that enable selection for good cells that will eventually form the baby. Additionally, cancer and the syncytiotrophoblast of the placenta are both invasive tissues with many biological parallelisms48,49. Indeed, it is not surprising that factors that are relevant to the placenta, such as maternal obesity, could affect genes that are relevant to cancer. The other pathway that was enriched for altered genes is oxidative stress. It is well known that excessive fat mass accumulation is linked to oxidative stress. Moreover, peroxisomal fatty acid oxidation seems to be enhanced in the placenta of obese women, while mitochondrial activity is impaired, with a greater lipid storage and an altered transfer of lipids to the fetus50. Altogether, there is growing evidence suggesting that obesity-induced oxidative stress is a central factor involved in the risk for adverse outcomes in pregnancy51,52.
Another interesting finding that deserves further investigation is the observation that differentially methylated CpGs are enriched for ZNF217 binding sites. This TF is epigenetically altered in placental cells under hypoxia53, and it has been suggested that maternal obesity during pregnancy causes placental hypoxia54. However, whether this TF can drive the methylation machinery to selected regions of the genome and cause epigenetic changes has not yet been explored. Similarly, the overlap between our CpGs and BW-associated regions suggests that both fetal genetic and placental epigenetic factors may contribute to the regulation of fetal growth, but this requires further research.
Our study has notable strengths but also several limitations. As previously mentioned, we have been able to coordinate a large number of cohorts and thus, to obtain an important sample size. Additionally, we have the experience of previous works, in which robust pipelines had already been implemented for EWAS, and we have run the quality control and meta-analysis in two independent institutions. Finally, none of the maternal ppBMI-associated CpGs from the current study are among the problematic probes with absolute methylation differences greater than 10% between Illumina 450 K and EPIC arrays that we identified in a previous study55.
Regarding limitations, we did not have access to individual data addressing whether each ppBMI measurement was self-reported or taken at the end of the first trimester of pregnancy. Therefore, we cannot use this variable as a covariate nor compare between measurement types. We are very aware that self-reported ppBMI may not be the most accurate measurement for our variable of interest, as may also be the case of the measurement of BMI at the end of the first trimester. However, self-reporting of ppBMI has been shown to be reliable and highly correlated to measured BMI at 12 weeks of gestation (r = 0.96; p-value <0.0001)56. Second, the unavailability of genotype data in some of the participating cohorts did not allow to add genotype principal components to our models and there might be residual confounding by population structure that we did not account for. Third, most of the cohorts were composed by a majority of individuals from European descent, which limits generalizability of our findings to other populations.
On the other hand, we are aware that RefFreeCellMix, the R package employed for adjustment of cell mixtures, is a principal component analysis-type correction method, and therefore presents the risk of over-correcting the results, especially in dense signal scenarios like the Illumina Beadchips, due to the capture of the signal by some of the top components of the estimation57. This, together with the fact that Bonferroni-correction is very strict and that our approach does not take into account the correlation between nearby CpGs, may have let some discoveries out of the focus. However, we have preferred to be strict and report only the most robust results.
In summary, here we present the largest EWAS of maternal ppBMI in association with placental DNAm performed to date. We identify 27 CpG sites at which we observe placental DNAm variations of 0.5–2.0% by 10-unit maternal ppBMI difference. Additionally, our DNAm findings seem to be placenta-specific, showing minimal overlap with a previous meta-analysis in cord-blood DNAm in relation to maternal ppBMI. The differentially methylated CpGs are mainly located in open sea regions, with a complete depletion from CpG islands, and enriched in cancer and oxidative stress- related pathways. These observations, together with the fact that maternal ppBMI is associated with placental DNAm at CpGs located close to obesity-related genes, leads us to hypothesize that placental DNAm could be one of the mechanisms by which maternal obesity is associated with aberrant fetal growth and maybe, other metabolic health outcomes in offspring later in life. However, we cannot rule out that the changes observed could be markers of exposure to high ppBMI and therefore, our findings will need to be supplemented by functional studies or causal inference analyses to better understand if they truly have a role in pregnancy complications or long-term metabolic outcomes.
Methods
Participating cohorts
Cohorts that are members of the PACE consortium, had existing DNAm data from placental tissue obtained with the Illumina 450 K or EPIC BeadChips, and had maternal BMI information prior to the beginning of pregnancy were invited to participate in the present study. The ten cohorts that contributed to the meta-analysis were AQUA, EARLI, EDEN, Gen3G, GENEIDA, HEBC, INMA, ITU, NHBCS and RICHS. All cohorts obtained ethics approval and informed consent from participants prior to data collection through their Institutional Ethics Boards. Exclusion criteria for this study were: non-singleton births, pre-eclampsia, and DNAm data not derived from the fetal facing side of the placenta. All participants included in this meta-analysis were of European ancestry. Detailed methods for each cohort are provided in Supplementary Note 1.
DNAm data quality control and normalization
All DNAm data processing and analyses were conducted in R 3.3.258, with the exception of the meta-analyses, which were performed with the GWAMA software (https://genomics.ut.ee/en/tools/gwama)27. DNAm from the fetal-facing side of the placenta was assessed with the Illumina 450 K or EPIC arrays. See Supplementary Note 1 for extra details on placenta collection, DNA extraction and DNAm acquisition in each cohort. In general, samples were randomized across the different arrays to avoid group differences derived from batch effects. Quality control of DNAm was standardized across all cohorts. Low-quality samples (showing a shifted beta-value distribution) were filtered out and probes with detection p-values >0.01 were excluded (for cohort-specific probe lists see Supplementary Data 8). DNAm beta-values were normalized with functional normalization59 and beta-mixture quantile normalization (BMIQ)60 was applied to correct for probe type bias. Cohorts examined their data for batch effects by depicting box-plots that divided the samples into different groups according to suspicious variables, and applied ComBat when applicable; all but one cohort (GENEIDA) identified batch effects and used ComBat to remove this source of variation. Probes that were exclusive for the EPIC array, hybridized to the X/Y chromosomes, cross-hybridizing probes and probes with SNPs at the CpG site, extension site, or within 10 bp of the extension site with an average minor allele frequency > 0.01 were filtered out61. Overall, 419,460 probes were available in the ten participating cohorts to assess placental DNAm. Methylation beta-values were modeled in robust linear regressions considering maternal ppBMI as a continuous variable. Finally, DNAm extreme outliers (<25th percentile - 3*IQR or >75th percentile + 3*IQR across all the samples) were trimmed.
Estimates of putative cellular heterogeneity
Putative cellular heterogeneity was estimated from DNAm data using a reference-free cell-mixture deconvolution method (RefFreeCellMix)26. The number of components varied between cohorts and ranged from 2 to 6, maybe because different sampling protocols result in differential heterogeneity across cohorts, or since the approach is data driven, those components could be capturing other major sources of variation in the array data, such as residual technical artifacts. Models for differential DNAm were corrected for the number of surrogate variables minus one to reduce multi-collinearity.
Genome-wide differential DNAm analyses
Within each cohort, robust linear regression from the MASS package62 in R was run to account for potential heteroskedasticity and to test the associations between normalized placental DNAm beta-values at each CpG and maternal ppBMI. Models were adjusted for maternal age, parity, maternal education and maternal smoking during pregnancy. Cohorts ran models both with and without adjustment for RefFreeCellMix cell type proportions. Covariate data are described in more detail in Supplementary Note 1.
Meta-analyses
We performed inverse variance-weighted fixed effects meta-analyses using GWAMA27. The meta-analysis was performed independently by two groups to ensure consistent results and identical results were reproduced. We used the Bonferroni adjustment to correct for multiple testing. Secondary analyses were only performed on CpGs that passed the Bonferroni correction, particularly in the RefFreeCellMix-adjusted model. It is worth mentioning that both the meta-analysis and shadow meta-analysis teams performed a general quality control separately prior to the meta-analysis itself, showing consistent results. In summary, we checked that conflictive probes had been removed and drew the cohort-specific qq-plots, as well as the correlation between sample sizes and significant hits across cohorts. Finally, we checked the genomic inflation of the whole meta-analysis and plotted forest plots of the significant hits after leaving one cohort out at a time, to see whether any of the cohorts was guiding the associations. The MARBLES cohort was excluded due to its small sample size and to the fact that forest plots showed inconsistencies compared to the rest of the cohorts (Supplementary Fig. 1).
Functional and regulatory enrichment analyses
We annotated CpGs to their closest genes and to CpG islands with annotations from the Illumina Human Methylation 450 K annotation file, and with several regulatory features using publicly available data: placental 15-chromatin states30 released from the ROADMAP Epigenomics Mapping Consortium31 (ChromHMM v1.10), placental germline differentially methylated regions29 and placental partially methylated domains32.
Over-representation analyses for gene-sets or pathways were performed at the gene level with ConsensusPathDB28 using KEGG, Reactome, Wikipathways and Biocarta as reference databases. ConsensusPathDB performs a hypergeometric test, with a default background equal to the number of ConsensusPathDB entities that are annotated with an ID of the type the user has provided, and participate in at least one pathway. Finally, the program corrects multiple-testing with FDR. Enrichment for TFs was assessed at the gene level with EnrichR using ENCODE and ChEA consensus TFs from ChIP-X database. EnrichR results were ranked using the combined score (p-value computed using Fisher’s exact test combined with the z-score of the deviation from the expected rank)63.
Overlap of ppBMI-sensitive CpG sites and birth outcome SNPs
We assessed the genomic proximity between CpGs identified by our maternal ppBMI placental DNAm EWAS (Bonferroni significant in the cell-type adjusted model) and SNPs previously associated with BW, birth length, head circumference and GA30–35. Briefly, we verified the genomic proximity between SNPs from the largest GWAS performed to date on the above-mentioned birth outcomes and our identified CpGs by using the Genomic Ranges package64 in R, within 1 Mb windows (±0.5 Mb) surrounding each of the 367 autosomal SNPs.
Comparison of ppBMI-associated CpGs in placenta and in cord blood
We examined whether maternal ppBMI-associated CpGs in placenta were the same as those previously reported in cord blood9. As no overlap was found between the hits that passed the Bonferroni correction in each study, we searched for CpGs from the cord blood study present 0.5 Mb upstream or downstream of each of the maternal ppBMI-associated CpGs in placenta (1 Mb windows), by using the GenomicRanges R package, with the aim of finding genomic regions where DNAm was related to ppBMI in the two different tissues.
Statistics and reproducibility
DNAm data from up to 419,460 CpG obtained with the Illumina 450 K or EPIC BeadChips in 2631 placental samples of the fetal side were normalized with functional normalization and BMIQ, and corrected for batch effects with ComBat if applicable. Afterwards, DNAm was correlated with maternal ppBMI in each of the participating cohorts using robust linear regressions with the MASS package in R 3.3.2. The code to perform the full analysis and the details are publicly available as described in the Code Availability section. The cohort-specific results were then meta-analyzed using the inverse variance-weighted fixed effects method in the GWAMA software, simultaneously in two independent laboratories. Results were fully consistent. Statistical significance was set at a meta-analysis nominal p-value = 1.2e-07, after Bonferroni correction for multiple-testing. Only CpGs below this threshold were taken into account in the downstream analyses. Generally, the statistical methods used in each of the downstream analyses were the ones suggested by the developers of each of the analytic tool implemented. More details in each of the analytical steps included in this study can be obtained in the specific section in Methods, Supplementary Note 1 and the code, as described in the Code Availability section.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We would like to thank the Pregnancy and Childhood Epigenetics (PACE) consortium, as well as all the families that participated in these studies for their generous contribution. This work was partially funded by GVSAN2018111086 from the Basque Department of Health and PI18/01142 from ISCIII - Spanish Ministry of Science and Innovation - cofounded by the ERDF “A way to make Europe” to JRB and LSM, respectively; and by the Joint Programming Initiative – A Healthy Diet for a Healthy Life (JPI HDHL) (NutriPROGRAM). ACP was supported by grant GVSAN2019111085 from the Basque Department of Health to NFJ. Detailed acknowledgements and funding for each participating cohort are described in Supplementary Note 1.
Author contributions
N.F.-J., M.B., M.F.H. & J.R.B. conceived and designed the study. Study-specific analyses were completed by J. Loke. (AQUA), K.M.B. & J.F.D. (EARLI & MARBLES), A.F & B.H. (EDEN), R.F. (Gen3G), P.C.-S. & J.M.-M. (GENEIDA), A.M.B. (HEBC), N.F.-J. (INMA), T.K. (ITU), F.-Y.T. (NHBCS) and C.L. (RICHS). N.F.-J. & R.F. meta-analyzed the results. N.F-J. & A.C.-P. performed the follow-up analyses. N.F.- J., R.F., A.C.-P., M.F.H. & J.R.B. interpreted the results. N.F.-J., R.F., A.C.-P., M.F.H. & J.R.B. wrote the first draft of the manuscript. M.F.H. & J.R.B. contributed equally to guiding the direction of the project. All authors (N.F.-J., R.F., A.C.-P., J.L., P.P., T.K., F.-Y.T, C.L., A.M.B., M. Lozano., J-M.-M., Y.J.L., K.M.B., Y.Z., A.F., S.S., T.M.E., J.C., K.B.M., T.B., P.C.-S., J.H., M.D.F., J.M.L., J.T., D.C., M.F.F., A.G.-M., J.M.C., B.G.-A., R.J.S., J.F.D., E.M., M. Lacasaña., M.V., C.J.M., M.R.K., K.R., L.B., B.H., L.S.M., M.B., M.F.H. & J.R.B.) made substantial contributions to the acquisition, analysis, or interpretation of data, and read and critically revised the manuscript.
Peer review
Peer review information
Communications Biology thanks Tina Bianco-Miotto, Chiea Chuen Khor, Markos Tesfaye and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Eirini Trompouki, Anam Akhtar, Christina Karlsson Rosenthal and George Inglis. Peer reviewer reports are available.
Data availability
The full genome-wide results of the meta-analysis are presented in Supplementary Data 3 and 4 and have been uploaded to the EWAS Catalogue, available in Zenodo with identifier 10.5281/zenodo.731453465. The individual level data used are not publicly available for several reasons. First, participants were not explicitly informed about this in the informed consent. Second, there are some studies that suggest that DNA methylation data has enough information to identify participants. Third, each PACE cohort follows different internal regulations in regards to public access of the data. Individual level data can still be shared with external researchers after signature of a data transfer agreement (DTA) with each of the participant cohorts, listed in Supplementary Note 1. More information is available in the PACE consortium website (https://www.niehs.nih.gov/research/atniehs/labs/epi/pi/genetics/pace/index.cfm). Source data underlying Fig. 3 is presented in Supplementary Data 9.
Code availability
Scripts to reproduce the analysis have been deposited in a public GitHub repository and are available in Zenodo with identifier 10.5281/zenodo.731396666.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Nora Fernandez-Jimenez, Ruby Fore, Ariadna Cilleros-Portet.
These authors jointly supervised this work: Marie-France Hivert, Jose Ramon Bilbao.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-04267-y.
References
- 1.Maffeis C, Morandi A. Effect of Maternal Obesity on Foetal Growth and Metabolic Health of the Offspring. Obes. Facts. 2017;10:112–117. doi: 10.1159/000456668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marchi J, et al. Risks associated with obesity in pregnancy, for the mother and baby: a systematic review of reviews. Obes. Rev. 2015;16:621–638. doi: 10.1111/obr.12288. [DOI] [PubMed] [Google Scholar]
- 3.Howell KR, Powell TL. Effects of maternal obesity on placental function and fetal development. Reproduction. 2017;153:R97–R108. doi: 10.1530/REP-16-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mamun AA, Mannan M, Doi SA. Gestational weight gain in relation to offspring obesity over the life course: a systematic review and bias-adjusted meta-analysis. Obes. Rev. 2014;15:338–347. doi: 10.1111/obr.12132. [DOI] [PubMed] [Google Scholar]
- 5.Godfrey KM, et al. Influence of maternal obesity on the long-term health of offspring. Lancet Diabetes Endocrinol. 2017;5:53–64. doi: 10.1016/S2213-8587(16)30107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivera HM, Christiansen KJ, Sullivan EL. The role of maternal obesity in the risk of neuropsychiatric disorders. Front. Neurosci. 2015;9:194. doi: 10.3389/fnins.2015.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desai M, Jellyman JK, Ross MG. Epigenomics, gestational programming and risk of metabolic syndrome. Int. J. Obes. (Lond.) 2015;39:633–641. doi: 10.1038/ijo.2015.13. [DOI] [PubMed] [Google Scholar]
- 8.Felix JF, et al. Cohort profile: Pregnancy and childhood epigenetics (PACE) consortium. Int. J. Epidemiol. 2018;47:22–23u. doi: 10.1093/ije/dyx190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharp GC, et al. Maternal BMI at the start of pregnancy and offspring epigenome-wide DNA methylation: findings from the pregnancy and childhood epigenetics (PACE) consortium. Hum. Mol. Genet. 2017;26:4067–4085. doi: 10.1093/hmg/ddx290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharp GC, et al. Maternal pre-pregnancy BMI and gestational weight gain, offspring DNA methylation and later offspring adiposity: findings from the Avon Longitudinal Study of Parents and Children. Int. J. Epidemiol. 2015;44:1288–1304. doi: 10.1093/ije/dyv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shrestha D, et al. Placental DNA methylation changes associated with maternal prepregnancy BMI and gestational weight gain. Int. J. Obes. (Lond.). 2020;44:1406–1416. doi: 10.1038/s41366-020-0546-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thakali KM, Zhong Y, Cleves M, Andres A, Shankar K. Associations between maternal body mass index and diet composition with placental DNA methylation at term. Placenta. 2020;93:74–82. doi: 10.1016/j.placenta.2020.02.018. [DOI] [PubMed] [Google Scholar]
- 13.Everson TM, et al. Placental DNA methylation signatures of maternal smoking during pregnancy and potential impacts on fetal growth. Nat. Commun. 2021;12:5095. doi: 10.1038/s41467-021-24558-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joubert BR, et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. Am. J. Hum. Genet. 2016;98:680–696. doi: 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muggli E, et al. Study protocol: Asking QUestions about Alcohol in pregnancy (AQUA): A longitudinal cohort study of fetal effects of low to moderate alcohol exposure. BMC Pregnancy Childbirth. 2014;14:302. doi: 10.1186/1471-2393-14-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newschaffer CJ, et al. Infant siblings and the investigation of autism risk factors. J. Neurodev. Disord. 2012;4:7. doi: 10.1186/1866-1955-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heude B, et al. Cohort Profile: The EDEN mother-child cohort on the prenatal and early postnatal determinants of child health and development. Int. J. Epidemiol. 2016;45:353–363. doi: 10.1093/ije/dyv151. [DOI] [PubMed] [Google Scholar]
- 18.Guillemette L, et al. Genetics of Glucose regulation in Gestation and Growth (Gen3G): a prospective prebirth cohort of mother–child pairs in Sherbrooke, Canada. BMJ Open. 2016;6:e010031. doi: 10.1136/bmjopen-2015-010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aguilar-Lacasaña S, et al. Methylenetetrahydrofolate Reductase (MTHFR) Gene Polymorphism and Infant’s Anthropometry at Birth. Nutrients. 2021;13:831. doi: 10.3390/nu13030831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michels KB, Harris HR, Barault L. Birthweight, maternal weight trajectories and global DNA methylation of LINE-1 repetitive elements. PLoS One. 2011;6:e25254. doi: 10.1371/journal.pone.0025254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guxens M, et al. Cohort Profile: The INMA—INfancia y Medio Ambiente—(Environment and Childhood) Proj. Int. J. Epidemiol. 2012;41:930–940. doi: 10.1093/ije/dyr054. [DOI] [PubMed] [Google Scholar]
- 22.Dieckmann L, et al. Characteristics of epigenetic aging across gestational and perinatal tissues. Clin. Epigenetics. 2021;13:97. doi: 10.1186/s13148-021-01080-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hertz-Picciotto I, et al. A Prospective Study of Environmental Exposures and Early Biomarkers in Autism Spectrum Disorder: Design, Protocols, and Preliminary Data from the MARBLES Study. Environ. Health Perspect. 2018;126:117004. doi: 10.1289/EHP535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert-diamond D, Emond JA, Baker ER, Korrick SA, Karagas MR. Relation between in Utero Arsenic Exposure and Birth Outcomes in a Cohort of Mothers and Their Newborns from New Hampshire. Environ. Health Perspect. 2016;124:1299–1307. doi: 10.1289/ehp.1510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Appleton AA, et al. Prenatal Programming of Infant Neurobehaviour in a Healthy Population. Paediatr. Perinat. Epidemiol. 2016;30:367–375. doi: 10.1111/ppe.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houseman EA, et al. Reference-free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinforma. 2016;17:259. doi: 10.1186/s12859-016-1140-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinforma. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamburov A, et al. ConsensusPathDB: Toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:712–717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamada H, et al. Allele-Specific Methylome and Transcriptome Analysis Reveals Widespread Imprinting in the Human Placenta. Am. J. Hum. Genet. 2016;99:1045–1058. doi: 10.1016/j.ajhg.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roadmap Epigenomics Consortium. et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–329. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroeder DI, et al. The human placenta methylome. Proc. Natl Acad. Sci. 2013;110:6037–6042. doi: 10.1073/pnas.1215145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horikoshi M, et al. Genome-wide associations for birth weight and correlations with adult disease. Nature. 2016;538:248–252. doi: 10.1038/nature19806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beaumont RN, et al. Genome-wide association study of offspring birth weight in 86-577 women identifies five novel loci and highlights maternal genetic effects that are independent of fetal genetics. Hum. Mol. Genet. 2018;27:742–775. doi: 10.1093/hmg/ddx429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Valk RJP, et al. A novel common variant in DCST2 is associated with length in early life and height in adulthood. Hum. Mol. Genet. 2015;24:1155–1168. doi: 10.1093/hmg/ddu510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taal H, et al. Common variants at 12q15 and 12q24 are associated with infant head circumference. Nat. Genet. 2012;44:532–538. doi: 10.1038/ng.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang G, et al. Genetic Associations with Gestational Duration and Spontaneous Preterm Birth. N. Engl. J. Med. 2017;377:1156–1167. doi: 10.1056/NEJMoa1612665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warrington NM, et al. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat. Genet. 2019;51:804–814. doi: 10.1038/s41588-019-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Decker M, et al. EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides. J. Lipid Res. 2012;53:2038–2045. doi: 10.1194/jlr.M024448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Taeye BM, et al. Expression and regulation of soluble epoxide hydrolase in adipose tissue. Obesity. 2010;18:489–498. doi: 10.1038/oby.2009.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun L, et al. CRISPR/Cas9 mediated establishment of a human CSRP3 compound heterozygous knockout hESC line to model cardiomyopathy and heart failure. Stem Cell. Res. 2020;49:102077. doi: 10.1016/j.scr.2020.102077. [DOI] [PubMed] [Google Scholar]
- 42.Hall E, et al. The effects of high glucose exposure on global gene expression and DNA methylation in human pancreatic islets. Mol. Cell. Endocrinol. 2018;472:57–67. doi: 10.1016/j.mce.2017.11.019. [DOI] [PubMed] [Google Scholar]
- 43.Usha Kalyani R, et al. Fer1L5, a Dysferlin Homologue Present in Vesicles and Involved in C2C12 Myoblast Fusion and Membrane Repair. Biology. 2020;9:386. doi: 10.3390/biology9110386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knöfler M, et al. Human placenta and trophoblast development: key molecular mechanisms and model systems. Cell. Mol. Life Sci. 2019;76:3479–3496. doi: 10.1007/s00018-019-03104-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Almomani SN, et al. Identification and validation of DNA methylation changes in pre-eclampsia. Placenta. 2021;110:16–23. doi: 10.1016/j.placenta.2021.05.005. [DOI] [PubMed] [Google Scholar]
- 46.Diceglie C, et al. Placental Antioxidant Defenses and Autophagy-Related Genes in Maternal Obesity and Gestational Diabetes Mellitus. Nutrients. 2021;13:1303. doi: 10.3390/nu13041303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zou Y, et al. Association of a gain-of-function variant in LGR4 with central obesity. Obes. (Silver Spring) 2017;25:252–260. doi: 10.1002/oby.21704. [DOI] [PubMed] [Google Scholar]
- 48.Coorens THH, et al. Inherent mosaicism and extensive mutation of human placentas. Nature. 2021;592:80–85. doi: 10.1038/s41586-021-03345-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costanzo V, Bardelli A, Siena S, Abrignani S. Exploring the links between cancer and placenta development. Open Biol. 2018;8:180081. doi: 10.1098/rsob.180081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calabuig-Navarro V, et al. Effect of Maternal Obesity on Placental Lipid Metabolism. Endocrinology. 2017;158:2543–2555. doi: 10.1210/en.2017-00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alcala M, et al. Antioxidants and Oxidative Stress: Focus in Obese Pregnancies. Front. Physiol. 2018;9:1569. doi: 10.3389/fphys.2018.01569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malti N, et al. Oxidative stress and maternal obesity: feto-placental unit interaction. Placenta. 2014;35:411–416. doi: 10.1016/j.placenta.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 53.Yuen RK, Chen B, Blair JD, Robinson WP, Nelson DM. Hypoxia alters the epigenetic profile in cultured human placental trophoblasts. Epigenetics. 2013;8:192–202. doi: 10.4161/epi.23400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernandez-Twinn DS, et al. Exercise rescues obese mothers’ insulin sensitivity, placental hypoxia and male offspring insulin sensitivity. Sci. Rep. 2017;7:44650. doi: 10.1038/srep44650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernandez-Jimenez N, et al. Comparison of Illumina 450 K and EPIC arrays in placental DNA methylation. Epigenetics. 2019;14:1177–1182. doi: 10.1080/15592294.2019.1634975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Casas M, et al. Maternal pre-pregnancy overweight and obesity, and child neuropsychological development: two Southern European birth cohort studies. Int. J. Epidemiol. 2013;42:506–517. doi: 10.1093/ije/dyt002. [DOI] [PubMed] [Google Scholar]
- 57.Chen J, et al. Fast and robust adjustment of cell mixtures in epigenome-wide association studies with SmartSVA. BMC Genomics. 2017;18:413. doi: 10.1186/s12864-017-3808-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/ (2021).
- 59.Fortin J-P, et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15:503. doi: 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teschendorff AE, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen J, et al. CpGFilter: model-based CpG probe filtering with replicates for epigenome-wide association studies. Bioinformatics. 2016;32:469–471. doi: 10.1093/bioinformatics/btv577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S. (Springer, 2002).
- 63.Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics14, (2013). [DOI] [PMC free article] [PubMed]
- 64.Lawrence M, et al. Software for Computing and Annotating Genomic Ranges. PLOS Computational Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fernandez-Jimenez, N. et al. A meta-analysis of pre-pregnancy maternal body mass index and placental DNA methylation identifies 27 CpG sites with implications for mother-child health. [Data set]. Zenodo. 10.5281/zenodo.7314534 (2022). [DOI] [PMC free article] [PubMed]
- 66.Cilleros-Portet A. ariadnacilleros/EWAS-maternal-BMI-vs-placental-methylation: v1.0 (v1.0) Zenodo. 2022 doi: 10.5281/zenodo.7313966. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The full genome-wide results of the meta-analysis are presented in Supplementary Data 3 and 4 and have been uploaded to the EWAS Catalogue, available in Zenodo with identifier 10.5281/zenodo.731453465. The individual level data used are not publicly available for several reasons. First, participants were not explicitly informed about this in the informed consent. Second, there are some studies that suggest that DNA methylation data has enough information to identify participants. Third, each PACE cohort follows different internal regulations in regards to public access of the data. Individual level data can still be shared with external researchers after signature of a data transfer agreement (DTA) with each of the participant cohorts, listed in Supplementary Note 1. More information is available in the PACE consortium website (https://www.niehs.nih.gov/research/atniehs/labs/epi/pi/genetics/pace/index.cfm). Source data underlying Fig. 3 is presented in Supplementary Data 9.
Scripts to reproduce the analysis have been deposited in a public GitHub repository and are available in Zenodo with identifier 10.5281/zenodo.731396666.