Highlights
-
•
CX-5461 may enhance immune checkpoint inhibitor (ICI) therapeutic efficacy.
-
•
CX-5461 triggers cGAS-STING-IFN pathway, eliciting STAT1-mediated PD-L1 expression.
-
•
CX-5461+ICI leads to improved immunotherapeutic effects in CRC.
-
•
They increase cytotoxic T-cell and reduce myeloid-derived suppressor cell numbers.
-
•
CX-5461 can turn cold CRC tumors into hot ones.
Keywords: CX-5461, G-quadruplex, Colorectal cancer, Immunotherapy, PD-L1
Abstract
Purpose
Immune checkpoint inhibitors (ICIs) alone or in combination with chemotherapy can improve the limited efficacy of colorectal cancer (CRC) immunotherapy. CX-5461 causes substantial DNA damage and genomic instability and can increase ICIs’ therapeutic efficacies through tumor microenvironment alteration.
Results
We analyzed whether CX-5461 enhances ICIs’ effects in CRC and discovered that CX-5461 causes severe DNA damage, including cytosolic dsDNA appearance, in various human and mouse CRC cells. Our bioinformatics analysis predicted CX-5461-based interferon (IFN) signaling pathway activation in these cells, which was verified by the finding that CX-5461 induces IFN-α and IFN-β secretion in these cells. Next, cGAMP, phospho-IRF3, CCL5, and CXCL10 levels exhibited significant posttreatment increases in CRC cells, indicating that CX-5461 activates the cGAS-STING-IFN pathway. CX-5461 also enhanced PD-L1 expression through STAT1 activation. CX-5461 alone inhibited tumor growth and prolonged survival in mice. CX-5461+anti-PD-1 or anti-PD-L1 alone exhibited synergistic growth-suppressive effects against CRC and breast cancer. CX-5461 alone or CX-5461+anti-PD-1 increased cytotoxic T-cell numbers and reduced myeloid-derived suppressor cell numbers in mouse spleens.
Conclusions
Therefore, clinically, CX-5461 combined with ICIs for CRC therapy warrants consideration because CX-5461 can turn cold tumors into hot ones.
Introduction
Colorectal cancer (CRC) is a malignancy occurring in the colon and rectum [1], the prognosis of which is positively correlated with disease stage at diagnosis. For example, the 5-year survival rates range from 14% to 91% depending on whether it is diagnosed in situ or after metastasis to other organs [2,3]. CRC treatment options include surgical local excision or resection of the colon with anastomosis, chemotherapy, radiation therapy, and targeted therapy [4]. Immunotherapy with different immune checkpoint inhibitors (ICIs) such as monoclonal antibodies against CTLA-4, PD-1, and programmed death ligand 1 (PD-L1) is a treatment option for rare CRC cases, where a tumor mutational burden (TMB) and microsatellite instability (MSI-H) levels are high [5], [6], [7]. André et al. reported that the single use of pembrolizumab, a PD-1 antibody, as a first-line therapy for MSI-H–mismatch repair deficiency (dMMR) metastatic CRC results in a significantly longer progression-free survival than does chemotherapy with or without targeted therapy [8]. However, only 10%–15% of patients with CRC harbor MSI-H, dMMR, or both [9,10]. Several studies have thus evaluated various ICIs in combination with other modalities including traditional 5-FU-based regimens, EGFR inhibitors, VEGF inhibitors, radiotherapy, and even vaccines in the setting of microsatellite stable (MSS) or mismatch repair protein (MMRp) CRC encompassing the majority of pathology noted in advanced disease on the basis of the hypothesis that a combination modality can evoke an immunogenic response enabling effective application of ICIs in patients with MSS or MMRp tumors [11]. As such, gemcitabine treatment in vitro has been reported to induce an upregulation of Wilms’ tumor 1 (WT1) mRNA, PD-L1, and calreticulin (an immunogenic cell death marker) in primary intrahepatic cholangiocarcinoma cells, cultured from a patient receiving gemcitabine followed by 5-FU, both combined with dendritic cells pulsed with WT1 peptides—suggesting that tumor immunogenicity can be altered by chemoimmunomodulating agents [12].
Pidnarulex (i.e., CX-5461), originally identified as an RNA polymerase I inhibitor capable of killing B-lymphoma cells selectively in vivo [13], causes G2-phase arrest and induces apoptosis in acute lymphoblastic leukemia cells by activating the canonical ATM/ATR pathway as well as by suppressing primary TERT-immortalized human foreskin fibroblast (BJ-T) proliferation by activating the noncanonical ATM/ATR pathway [14,15]. Notably, when combined with everolimus, CX-5461 can improve the survival of Eμ-Myc lymphoma-bearing mice considerably by inducing nucleolar stress and p53 pathway activation [16]. The significant survival advantage in both p53WT and p53null leukemic mice treated with CX-5461 is attributable to the activation of the checkpoint kinases 1/2, aberrant G2/M cell-cycle progression, and myeloid differentiation induction in leukemic blasts. CX-5461 can reduce both the leukemic granulocyte–macrophage progenitor and leukemia-initiating cell populations and suppress their clonogenic capacity [17]. CX-5461, used as a G-quadruplex stabilizer, causes severe death in homologous recombination (HR) and nonhomologous end joining (NHEJ)–deficient cancer cells by blocking replication forks and inducing ssDNA gaps or breaks [18]. CX-5461 also induces DNA damage checkpoint activation and G2-M arrest with increased levels of γH2AX staining in ovarian cancer cell lines as well as patient-derived xenografts, with an enhanced effect on chemoresistant cells [19]. A phase I dose-escalation study demonstrated that CX-5461 is safe a selective rDNA transcription inhibitor, exhibiting antitumor activity against advanced hematologic cancers [20]. By contrast with these findings, CX-5461 has also been demonstrated to exert its primary cytotoxic effect through topoisomerase II poisoning in various cancer cells [21,22]. Combination treatment with CX-5461 and talazoparib, a poly (ADP-ribose) polymerase (PARP) inhibitor, is effective against HR-proficient tumors unsuitable for monotherapy with PARP inhibitors [23]. A phase 1b clinical trial focused on determining the tolerable dose of CX-5461 for patients with various solid tumors with BRCA1/2, PALB2, or HRD mutations is underway (NCT04890613). Notably, ovarian cancer cell treatment with CX-5461 can cause cytosolic double-stranded (ds) DNA accumulation and type I interferon (IFN) production by activating the innate immune pathway cyclic GMP (cGMP)-AMP synthase (cGAS)-stimulator of interferon genes (STING)-TBK1-IRF3 [24]. In this study, we examined whether CX-5461 alone has direct cytotoxic effects on various human and mouse CRC cells and whether CX-5461 can induce type I IFN expression via the cGAS-STING-TBK1-IRF3 pathway. We determined whether the addition of CX-5461 enhances the therapeutic efficacy of two immune checkpoint inhibitors, namely anti-PD-1 and anti-PD-L1, in a Balb/c-CT26 murine CRC model.
Material and methods
Compound
CX-5461 [2-(4-Methyl-[1,4]diazepan-1-yl)-5-oxo-5H-7-thia-1,11b-diaza-benzo[c]fluorene-6-carboxylic acid (5-methyl-pyrazin-2-ylmethyl)-amide] was procured from Senhwa Biosciences. InVivoMAb antimouse PD-L1 (B7-H1) was purchased from BioCell (catalog# BE0101).
Cell culture
HT-29 and DLD-1 human colon adenocarcinoma cells and CT26 mouse colon and 4T1 mouse breast carcinoma cells were maintained in RPMI-1640 medium supplemented 10% fetal bovine serum (FBS), 100 U/mL penicillin, 100 µg/mL streptomycin, and 25 µg/mL amphotericin B (PSA, Biological Industries, USA) at 37°C in 5% CO2.
Flow cytometry
After treatment without or with CX-5461 for 24 h, the cells were detached and resuspended in RPMI medium containing the Alexa Fluor 647 Anti-Calreticulin (EPR3924, ab196159). After incubation at 4°C for 1 h, flow cytometry was applied to identify the subpopulations with surface translocation of calreticulin in each sample (FACSCanto, BD).
Comet assay
Cells (8 × 105) seeded in p60 dishes were treated with or without 10 µM CX-5461 for 24 h before being scraped, collected, and resuspended in phosphate-buffered saline (PBS) at a density of (1–3) × 105 cells/mL. In total, 50 µL of cell-containing LMAgarose was layered on a frosted glass slide; this was followed by electrophoresis at 21 V for 20 min. The cells on the slides were stained with Protech DNA dye for 1 h at a 1:10000 dilution and then observed under a fluorescent microscope.
Western blotting
Cells were lysed at 4°C in RIPA buffer supplemented with protease and phosphatase inhibitors. Total lysates (containing 30 µg of protein) were separated through sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred onto PVDF membranes (Millipore, Bedford, MA, USA). After they had been blocked with 5% nonfat milk, the membranes were probed with specific antibodies [γH2AX, Millipore #05-636-I; p-IRF-3 (ser396), Invitrogen #70012; IRF-3, Cell Signaling #4302; STING, Cell Signaling #13647; NF-κB, Cell Signaling #8242; p-NF-κB (ser536), Cell Signaling #3033; Lamin A/C, Genetex #GTX101127; p-STAT1 (Try701), Cell Signaling #9167; STAT1, Santa Cruz #sc-417; PD-L1, Genetex #GTX104763, Abcam #ab269674; β-actin, Millipore #MAB1501] at 4°C overnight and then incubated with the horseradish peroxidase-conjugated secondary antibody for 1 h. All signals were visualized using the ECL-Plus detection kit (PerkinElmer Life Sciences, Boston, MA, USA).
Quantitative reverse transcription polymerase chain reaction
Reverse transcription was performed using 3 µg of total RNA with a reverse transcriptase by using a cDNA kit from Invitrogen, and real-time and quantitative polymerase chain reaction (PCR) was performed using primers designed for mRNA sequences to assess expression of MYC (forward: 5′-CCTGGTGCTCCATGAGGAGAC-3′ and reverse: 5′-CAGACTCTGACCTTTTGCCAGG-3′), CAV1 (forward: 5′-CCAAGGAGATCGACCTGGTCAA-3′ and reverse: 5′-GCCGTCAAAACTGTGTGTCCCT-3′), IFIT1 (forward: 5′-GCCTTGCTGAAGTGTGGAGGAA-3′ and reverse: 5′-ATCCAGGCGATAGGCAGAGATC-3′), IFIT2 (forward: 5′-GGAGCAGATTCTGAGGCTTTGC-3′ and reverse: 5′-GGATGAGGCTTCCAGACTCCAA-3′), IFIT3 (forward: 5′-CCTGGAATGCTTACGGCAAGCT-3′ and reverse: 5′-GAGCATCTGAGAGTCTGCCCAA-3′), PLAAT4 (forward: 5′-GCAGGAACTGTGAGCACTTTGTC-3′ and reverse: 5′-GCAACAACCAGGATTCCAAGCG-3′), CDKN1A (forward: 5′-AGGTGGACCTGGAGACTCTCAG-3′ and reverse: 5′-TCCTCTTGGAGAAGATCAGCCG-3′), PD-L1 (human) (forward: 5′-TGCCGACTACAAGCGAATTACTG-3′ and reverse: 5′-CTGCTTGTCCAGATGACTTCGG-3′) and PD-L1 (mouse) (forward: 5′-TGCGGACTACAAGCGAATCACG-3′ and reverse: 5′-CTCAGCTTCTGGATAACCCTCG-3′). Cycling conditions for real-time PCR included an initial denaturation cycle at 95°C for 15 min followed by 40 amplification cycles of 95°C for 15 s, 60°C for 60 s, and a final extension at 72°C for 30 sec followed by melting curve analysis with SYBR Green dye (Invitrogen).
Enzyme-linked immunosorbent assay
HT-29, DLD-1, and CT-26 cells were seeded in a 24-well plate in the absence or presence of 5 µM CX-5461. After incubation at 37°C for 0, 3, 6, 12, 18, and 24 h, the culture media were collected, and the concentrations of type I IFNs were determined using the following enzyme-linked immunosorbent assay (ELISA) kits: IFN-α (human), R&D # 41100-1; IFN-α (mouse), R&D # 42120-1; IFN-β (human), R&D # DIFNB0; IFN-β (mouse), R&D # MIFNB0. To analyze other cytokines, cells were seeded in the absence or presence of 1 and 5 µM CX-5461 for 48 h. Next, the culture media were collected, and the concentrations of TNF-α, IL-6, and CXCL10 were measured using the following ELISA kits: R&D #DTA00D, R&D #D6050, and R&D #DIP100, respectively. To measure cGAMP, the cells were collected and resuspended in RIPA buffer. After centrifugation, the cGAMP levels in the supernatant were determined using a 2′,3′-Cyclic GAMP ELISA Kit (ArborAssay #K067-H1). The results were calculated using an online tool provided from MyAssays (www.myassays.com/arbor-assays-detectx-2-3-cyclic-gamp-eia-kit.assay).
Cytosolic dsDNA detection
Immunofluorescent staining was performed to detect and analyze cytosolic dsDNA. HT-29, DLD-1, and CT-26 cells were seeded onto coverslips precoated with poly-D-lysine (Sigma-Aldrich, P6407) and then treated with or without 10 µM CX-5461 for 24 h. Next, the coverslips were fixed with 4% cold-paraformaldehyde, permeabilized using permeabilization buffer (0.1% Tween 20 and 0.01% Triton-X in PBS), and then blocked with PBS containing 1% BSA, 0.1% Tween 20, and 2.25% glycine. The coverslips were subsequently incubated with an anti-dsDNA (abcam, #ab27156, 1:1000) at 4°C overnight before being incubated with a rhodamine-conjugated rabbit secondary antibody (Jackson ImmunoResearch #211-295-109). Nuclei were then stained with DAPI. The coverslips were subsequently mounted with Flouromount-G (SouthernBiotech #0100-01) and then photographed using a charge-coupled device camera (CoolSNAP HQ2, Photometrics) mounted on an Olympus BX50 microscope by using Metaview.
Animal study
All animal experiments were performed in accordance with a protocol approved by the Academia Sinica Institutional Animal Care and Utilization Committee. To assess the growth-suppressive effects of CX-5461 alone, ICI alone, and their combinations against CRC, 1 × 106 CT-26 cells were resuspended in 100 µL of PBS and subcutaneously injected into male BALB/c mice (aged 6 weeks and weighing 20–25 g). When the tumors grew to 30 mm3, randomization was initiated for different treatments. The mice were equally divided among four groups—namely vehicle (twice per week, intraperitoneal injection), PD-L1 (200 µg twice per week, intraperitoneal injection), CX5461 (50 µg/kg twice per week, intraperitoneal injection), and CX5461+PD-L1. The volume of the tumors and body weight of the mice were measured weekly. Tumor masses were harvested after 5 weeks.
4T1 tumor cells were maintained in vitro in a monolayer culture in RPMI-1640 medium supplemented with 10% fetal bovine serum at 37°C under 5% CO2. Cells in their exponential growth phase were harvested and counted for tumor inoculation. Each mouse was inoculated in the right flank with 3 × 105 4T1 tumor cells in 0.1 mL of PBS for tumor development. The date of tumor cell inoculation was denoted as day 0. The randomization was initiated when the mean tumor size reached approximately 80–120 mm3. In total, 61 mice were enrolled in the efficacy study.
Microarray and in silico analyses
Microarray analysis was performed as described previously [25]. In brief, total RNAs were extracted from the HT-29 and DLD-1 cells after they were treated with low-dose (1 µM) or high-dose (10 µM) CX-5461 for 18 h and then reverse-transcribed into cDNAs. Affymetrix human U133 2.0 plus arrays were then performed, and the chips were scanned. Gene expression levels were normalized next and used to generate log2 values. The genes that exhibited a more than twofold change in expression were analyzed using Ingenuity Pathway Analysis (IPA; Qiagen, Valencia, CA, USA). We have uploaded the profile, GSE201899, to the GEO website (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE201899).
Statistical analysis
All data are presented as means ± standard deviations (SDs). Student's t test was applied to compare the differences between the experimental and control groups. A p value of <0.05, <0.01, and <0.005 indicated statistical significance.
Results
CX-5461 causes DNA damage and the appearance of cytosolic dsDNA in CRC cells
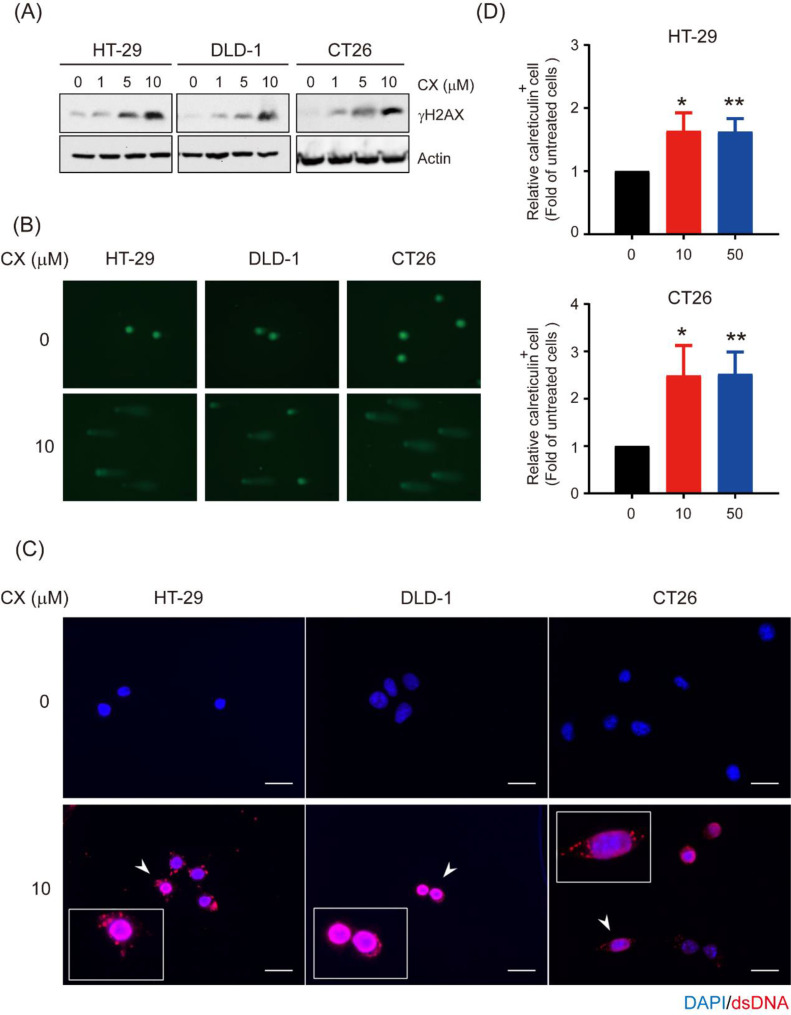
Because CX-5461 induces DNA damage in various cancer cells [24], we assessed whether this drug causes DNA damage in CRC cells by examining the levels of γH2AX, a hallmark of dsDNA breaks after CX-5461 treatment. A dose-dependent increase in γH2AX levels was observed in the human HT-29 and DLD-1 and in the mouse CT26 cells after they were treated with various concentrations of CX-5461 (Fig. 1A). We also performed the comet assay, frequently used to detect DNA damage at the single-cell level; the length of the comet tails increased considerably in all three cell lines after treatment with 10 µM CX-5461 for 24 h (Fig. 1B). We next determined whether this drug induced the appearance of cytosolic dsDNA, a key component of antitumor immune response [26], in these cells through immunofluorescence staining with an anti-dsDNA antibody. Perinuclear signals representing the dsDNA molecules were detected in three CRC cell lines only after treatment with CX-5461 (Fig. 1C). Because certain DNA-damaging drugs enhance the cross-presentation of tumor antigens by dendritic cells (DCs) characterized as immunogenic cell death (ICD) [27,28], we examined whether CX-5461 stimulates the release of some damage-associated molecular patterns. Flow cytometry demonstrated that CX-5461 treatment increased surface calreticulin levels in HT-29 and CT26 cells (Fig. 1D). Taken together, our data demonstrate that CX-5461 causes DNA damage, including the leakage of nuclear DNA into the cytosol and the cell surface translocation of calreticulin, a hallmark of ICD.
Fig. 1.
CX-5461 causes DNA damage in colorectal cancer cells. a Total lysates (30 μg) from HT-29, DLD-1, and CT26 cells were treated with 0, 1, 5, and 10 μM CX-5461 for 24 h and then subjected to Western blot analysis using anti-γH2AX as the probe. β-Actin was used the loading control. b Comet images of the HT-29, DLD-1, and CT26 cells treated with or without 10 μM CX-5461 for 24 h, stained with Protech DNA dye, and then observed under a fluorescent microscope (magnification: 200 ×). c Immunofluorescent staining of dsDNA (red) and nucleus (DPAI; blue) in the HT-29, DLD-1, and CT26 cells treated with or without 10 μM CX-5461 for 24 h. Photographs were taken using a fluorescent microscope (magnification: 200 ×). Scale bar=50μm. d Relative expression levels of calreticulin in each cell type after the administration of different CX-5461 dosages were measured through flow cytometry after the cells were incubated simultaneously with the APC-conjugated anticalreticulin. The quantitative results are presented as the mean ± SD from three independent experiments. *p < 0.05 and **p < 0.01, compared with untreated cells, Student's t test.
CX-5461 activates production of type I interferon and its signaling pathway
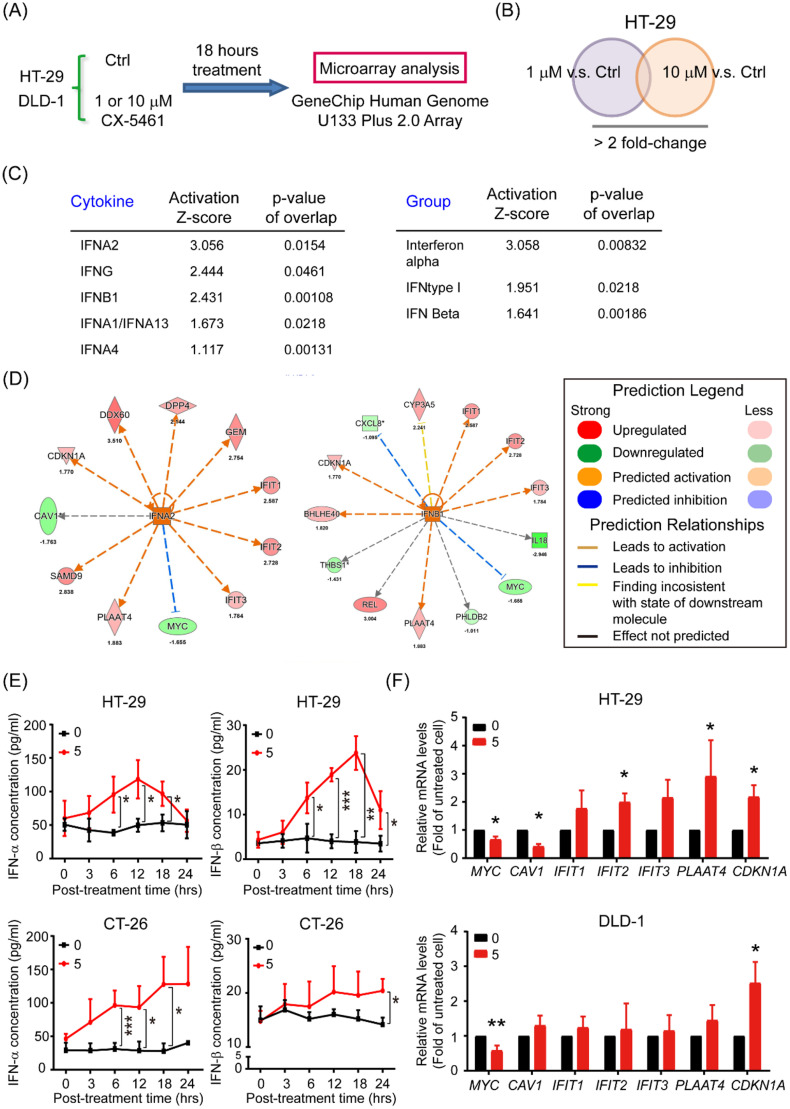
To investigate the potential signaling pathways involved in the effects of CX-5461, we performed a microarray analysis for HT-29 and DLD-1 cells to establish the transcriptomic datasets. After the treatment of cells with low (1 µM) and high (10 µM) doses of CX-5461 for 18 h, cDNA samples were hybridized on Human Genome U133 Plus 2.0 Array chips (Fig. 2A & supplementary table). After normalization, we selected probes with a >2.0-fold change in the 1 or 10 µM group compared with that in the controls for further prediction through IPA (Fig. 2B). Our core analysis indicated promising activation of type I or II interferons (IFNs) and the IFN signaling pathways (Fig. 2C). Notably, the genes encoding several common downstream mediators of IFNA2 and IFNB1, including MYC, CAV, IFIT1, IFIT2, IFIT3, PLAAT4, and CDKN1A (p21), were identified through core analysis (Fig. 2D); MYC downregulation and CDKN1A upregulation in CX-5461-treated HT-29 and DLD-1 cells were identified through RT-qPCR—consistent with the IPA prediction (Fig. 2E). To ensure that CX-5461 induces type I IFN production in CRC cells, we used ELISA to measure the amounts of IFN-α and IFN-β in HT-29 and CT26 cell culture media after they were treated with 5 µM CX-5461 for various periods. Even though the kinetics of IFN induction by this drug were dissimilar, particularly at later timepoints, in two cell lines, we detected significant increases in both IFN-α and IFN-β levels in these cells (Fig. 2F). These results demonstrate that the production of type I IFN and its downstream signaling pathway in both human and mouse CRC cells are activated by CX-5461 treatment.
Fig. 2.
CX-5461 enhances interferon signaling in CRC cells. a, b Venn diagrams of the potential upstream regulators in HT-29 cells after treatment with low (1 μM) or high (10 μM) doses of CX-5461 and control group provided by IPA with a twofold change cutoff. c Ranking of the candidate upstream regulators of cytokines and group from CX-5461 treatment and vehicle control groups predicted using IPA. d Core analysis of the IFNA2 and IFNB1 signatures in CX-5461-treated and control HT-29 cells. e IFN-α and IFN-β levels in the supernatant of HT-29 and CT-26 cells collected from different posttreatment timepoints analyzed using ELISA. f Total RNAs (5 μg) isolated from HT-29 and DLD-1 cells treated with 5 μM CX-5461 for 24 h and then subjected to RT-qPCR analyses to determine the mRNA levels of the seven predicted putative downstream targets: MYC, CAV1, IFIT1, IFIT2, IFIT3, PLAAT4, and CDKN1A. Data are presented the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.005, compared with the respective untreated cells at 0 h, Student's t test.
cGAS-STING-IRF3 pathway in CRC cells is activated by CX-5461 treatment
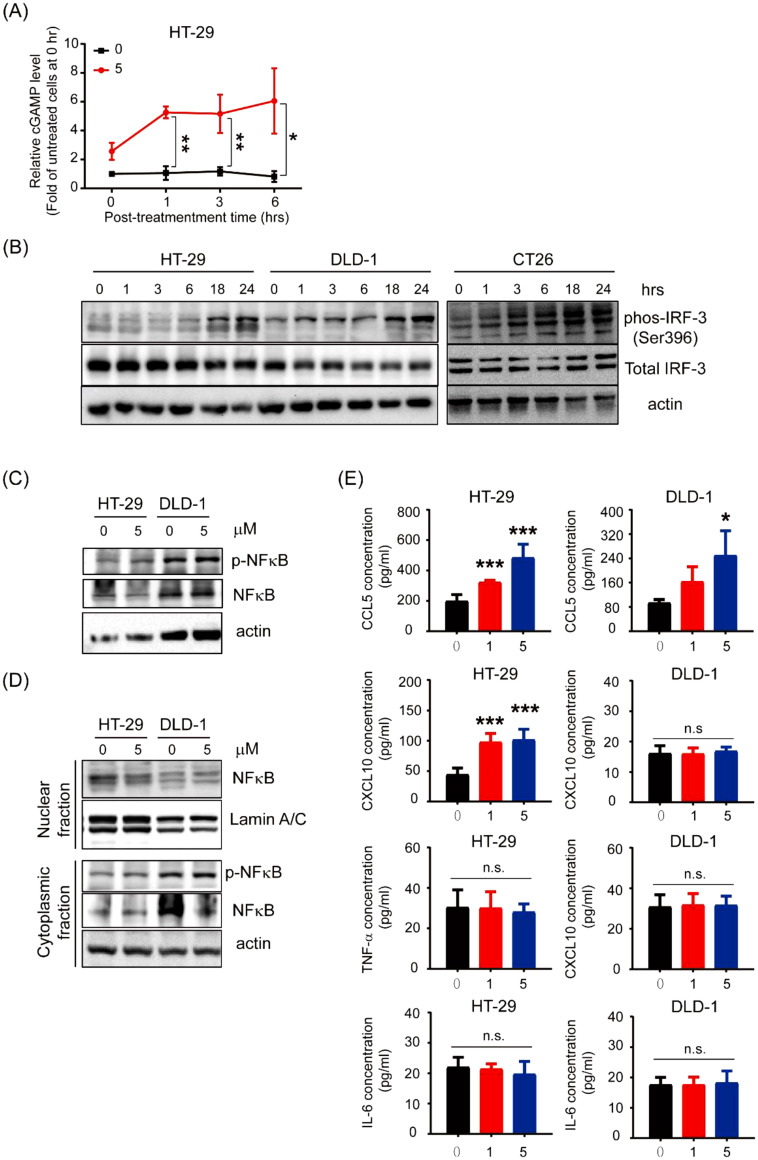
Numerous studies have reported that type I IFN production can be triggered by the cGAS-STING pathway, which is crucial in the immune response stimulated by cytosolic dsDNA [29], [30], [31]; therefore, we examined whether CX-5461 treatment induces cGAMP production. A significant increase in the cGAMP level in HT-29 cells was observed 1 to 6 h after drug treatment (Fig. 3A). Because IRF3 and NF-κB are the key transcription factors responsible for driving the expression of type I IFN and other proinflammatory cytokine genes, respectively, after STING activation [32], we examined whether CX-5461 treatment activates IRF3 in CRC cells. Significant increases in active IRF3 (phospho-Ser 396) levels in HT-29, DLD-1, and CT-26 cells were detected after treatment with 5 µM CX-5461 for ≥18 h (Fig. 3B). Active NF-κB levels in total lysates and nuclear NF-κB levels in HT-29 and DLD-1 cells treated with and without 5 µM CX-5461 for 24 h were analyzed. Neither p-NF-κB levels in total lysate nor those of nuclear NF-κB were affected after CX-5461 treatment (Fig. 3C & D). These results suggest that IRF3 but not NF-κB in CRC cells was activated by CX-5461. To confirm this speculation, ELISA was performed to analyze the levels of CCL5 and CXCL10—two chemokines whose genes are IRF3 downstream targets—as well as TNF-α and IL-6—two cytokines whose genes are NF-κB downstream targets—in HT-29 and DLD-1 cells after treatment with various CX-5461 concentrations. Although considerable increases in CCL5 levels were detected in both the cell lines, significant elevation in CXCL10 levels was noted in only HT-29 cells (Fig. 3E). By contrast, significant increases in TNF-α and IL-6 levels were observed in neither of the cell lines. Taken together, these data indicate that CX-5461 treatment induces IRF-3-mediated gene expression by activating the cGAS-STING pathway.
Fig. 3.
CX-5461 activates IRF-3, instead of NF-κB, through the cGAS-STING pathway. a Relative cGAMP levels in HT-29 cells treated with 5 μM CX-5461 at different posttreatment timepoints analyzed using ELISA. Data are present as the means ± SDs of three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.005, compared with the untreated cells at 0 h, Student's t test. b Total lysates (30 μg) were prepared from HT-29, DLD-1, and CT26 cells incubated with 5 μM CX-5461 for different durations and subjected to Western blot analyses using p-IRF3, IRF3, and STING antibodies as probes. β-Actin was used as the loading control. c Total lysates (30 μg) prepared from HT-29 and DLD-1 cells were subjected to Western blot analyses using primary antibodies against p-NF-κB (ser536) and total NF-κB. d Nuclear fractions (30 μg) prepared from HT-29 and DLD-1 cells were subjected to Western blot analyses to examine the nuclear levels of NF-κB. Cytoplasmic fractions were analyzed using p- NF-κB (ser536) and total NF-κB. Lamin A/C was used as the nuclear loading control. β-Actin was used as the cytoplasmic fractions loading control. e The expression levels of CCL5, CXCL10, TNF-α, and IL-6 in the supernatants of HT-29 and DLD-1 cells collected after exposure to different dosages were analyzed using ELISA. Data are present as the means ± SDs of three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.005, compared with the respective untreated cells, Student's t test.
CX-5461 induces PD-L1 expression in CRC cells through STAT1 activation
The IFN/JAK/STAT pathway plays critical roles in PD-L1 expression [33], where type I IFNs induce the JAK1/TYK2-mediated phosphorylation of STAT1 on the tyrosine residue 701 (Tyr701). This results in the formation of STAT1-STAT1 homodimers, which translocate to the nucleus and bind GAS (IFN-γ-activated site) elements to initiate the transcription of numerous genes, including PD-L1 [34], [35], [36]. We observed that CX-5461 treatment activates the expression of type I IFNs as well as their downstream signaling in CRC cells; therefore, we analyzed the status of STAT1 in these cells after drug treatment. Our results demonstrate that the levels of p-STAT1 (Tyr701) in three CRC lines were considerably elevated 32–48 h after drug treatment (Fig. 4A). Because IFN induces PD-L1 expression by activating STAT1 and STAT3 as well as their downstream target, IRF1, in melanoma cells [37], we next examined the protein levels of PD-L1 in HT-29 and DLD-1 cells after treatment with CX-5461 for 24 and 48 h. We detected a dose-dependent increase in the protein levels of PD-L1 in the cells treated with CX-5461 (Fig. 4B). Subsequently, we demonstrated that PD-L1 mRNA levels in the two human CRC cell lines and CT26 cells were significantly elevated after treatment with 5 µM CX-5461 for 48 h (Fig. 4C). Taken together, our data suggest that CX-5461 induces PD-L1 expression by activating the cGAS-STING-IFN-STAT1 pathway in CRC cells.
Fig. 4.
Interferon signaling pathway facilitates STAT1 phosphorylation and PD-L1 expression. a Total lysates (30 μg) prepared from HT-29, DLD-1, and CT26 cells incubated with 5 μM CX-5461 for different durations and subjected to Western blot analyses using primary antibodies against p-STAT1 (tyr701) and total STAT1. β-Actin was used as the loading control. b Total lysates (30 μg) prepared from HT-29 and DLD-1 cells treated with 0, 1, and 5 μM CX-5461 for 48 or 72 h were subjected to Western blot analysis using anti-PD-L1 as the probe. β-Actin was used as the loading control. c Total RNAs (5 μg) isolated from the HT-29, DLD-1, and CT-26 cells after treatment with or without 5 μM CX-5461 for 48 h were subjected to RT-qPCR to determine PD-L1 mRNA levels. Data are present as the means ± SDs of three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.005, compared with the respective untreated cells, Student's t test.
In vivo tumor growth suppressive effects of CX-5461 alone and its combination with an ICI on CT26 and 4T1 cells
To assess the in vivo antitumor effects of CX-5461 alone as well as its combination with ICI, BALB/c mice bearing syngeneic CT26 tumors were used as the models. When tumor volume was 30 mm3, mice were randomized into groups (n = 4) for different treatments (Fig. 5A). Our results indicate that the growth of CT26 (Fig. 5B) as well as their final weight (Fig. 5C) were considerably suppressed by the respective treatments of CX-5461 and an antimouse PD-L1 antibody (an ICI) alone. The highest therapeutic efficacy was detected in mice receiving CX-5461+anti-PD-L1. No significant change in body weight was observed in all the groups (Fig. 5D), suggesting that the dosage (50 µg/kg) of CX-5461 used here was well tolerated. To confirm these findings, a similar syngeneic tumor mouse model was applied with two main modifications: (1) higher dose of CX-5461 (62.5 mg/kg/week) administered intravenously and (2) anti-PD-L1 replaced by anti-mPD-1 (10 mg/kg, twice a week, intraperitoneal injection) for 4 weeks. The respective treatment with CX-5461 and anti-PD1 alone significantly prolonged mouse survival, and a much longer survival was observed in animals receiving the combination therapy (Fig. 5E). These data suggest that CX-5461 not only has a direct growth-suppressive effect on CRC but also can synergize with both anti-PD-1 and anti-PD-L1 antibodies against this malignancy.
Fig. 5.
CX-5461 may be a moderator of immunotherapy to suppress tumor growth in vivo. a Flow of the CT26 animal model treatment with CX-5461, anti-PD-L1, or their combination (n = 4 in each group). b–d Representative bar graph of tumor size b, photographs c, and body weight d of mice in the vehicle, anti-PD-L1, CX-5461, and anti-PD-L1+CX-5461 groups. e Kaplan–Meier plot of survival rates after different drug treatments. Data were analyzed using paired t test.
To assess whether CX-5461 alone can inhibit the in vivo growth of other cancer types as well as whether its combination with an ICI demonstrated synergism, BALB/c mice bearing syngeneic 4T1 breast tumors were selected as models. Tumor growth was inhibited by the treatments with CX-5461 alone, anti-PD-1 alone, and their combination (Fig. 6A). Consistent with our findings, the combination of CX-5461 and mPD-1 antibody exhibited stronger inhibition. In addition, no considerable body weight loss was observed in these animals, further suggesting the safety of a high dose (62.5 mg/kg) of CX-5461. To examine whether these treatments affected the composition of immune cells, which might be associated with their therapeutic effects in the major lymphoid organ, we first analyzed the T-cell composition in the spleens of mice after various treatments. Notably, the numbers of CD3+, CD4+, CD8+, activated CD8+, and even Treg cells considerably increased after treatments with CX-5461 alone and CX-5461+anti-PD-1. However, the effects of the combined treatment were similar to those of CX-5461 alone (Fig. 6C), suggesting that CX-5461 activates the cell-mediated immune system in tumor-bearing mice. By contrast, the populations of myeloid-derived suppressor cells (MDSCs) were significantly decreased by CX-5461 alone and CX-5461+anti-PD-1 (Fig. 6D).
Fig. 6.
CX-5461 enhances the immune system to improve tumor microenvironment in 4T1 mouse model. a, b Tumor volume a and body weight change b in 4T1 mouse model after treatment with vehicle, anti-PD-L1, CX-5461, and anti-PD-L1+CX-5461. c, d Bar graph displaying absolute count of different T-cell populations c and other immune-cell populations d in total live cells. PMN-MDSC: polymorphonuclear myeloid-derived suppressor cells; M-MDSC: monocytic myeloid-derived suppressor cells. Data are present as the means ± SDs of data from six independent mice. *p < 0.05, **p < 0.01, and ***p < 0.005 compared with the vehicle group, Student's t test.
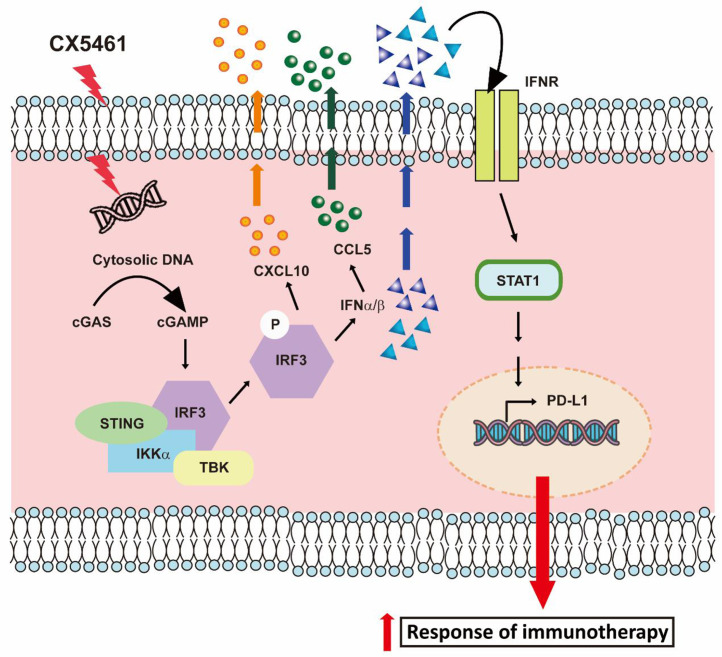
Taken together, our data demonstrate that CX-5461, by activating the cGAS-STING-IFN-STAT1-PD-L1 pathway, enhances the antitumor immune response in the tumor microenvironment, resulting in an improved efficacy of ICI therapy in CRC (Fig. 7).
Fig. 7.
Schematic representing how CX-5461 induces the activation of the cGAMP-STING-IFN pathway, leading to STAT1 phosphorylation and PD-L1 expression, and then facilitates immunotherapeutic responses in CRC.
Discussion
Surgery remains the primary option for patients with CRC; however, in patients with unresectable CRC, chemotherapy, radiation, targeted therapy, or a combination thereof may be administered before or after resection [38,39]. For instance, conventional chemotherapeutic drugs such as 5-fluorouracil (5-FU) with folic acid (leucovorin) and oxaliplatin (FOLFOX) or irinotecan (FOLFIRI) or capecitabine plus oxaliplatin (XELOX) could be combined with monoclonal antibodies against VEGF (bevacizumab) or EGFR (cetuximab or panitumumab) as first-line therapies for patients with unresectable metastatic CRC [40], [41], [42], [43], [44]. However, these regimens have several side effects and limitations, which must be resolved during the treatment course.
Immune checkpoint inhibition using antibodies against CTLA-4 or the PD-1/PD-L1 axis may be effective in the treatment of various malignancies. However, regardless of the cancer type, the response rates for the single treatment with these ICIs are relatively low except in patients with specific biomarkers such as MSI-H, TMB, and PD-L1 tumor proportion score [45,46]. Similarly, a clinical study revealed that high somatic mutation loads were associated with prolonged progression-free survival in both CRC and non-CRC patients receiving pembrolizumab (an anti-PD1 antibody) [47]. In addition, the results of a clinical trial suggested that nivolumab (an anti-PD1 antibody) plus low-dose ipilimumab (an anti-CTLA-4 antibody) is a promising treatment option for patients with dMMR/MSI-H Mcrc [48]. However, only approximately 5% of patients with mCRC have MSI-H or dMMR tumors [9]; hence, increasing the response rate of the remaining major CRC patients to immunotherapy is necessary.
We examined the feasibility of combining various ICIs with CX-5461, a novel anticancer drug with G-quadruplex stabilizing activity, because it can block replication forks and induce ssDNA gaps or breaks, which can be repaired by the Breast Cancer gene (BRCA) and nonhomologous end joining (NHEJ) pathways [18], in CRC therapy. We noted that CX-5461 treatment induced significant increases in several critical DNA damage markers including γH2AX and comet tail length as well as cytosolic dsDNA appearance in both human and mouse CRC cells (Fig. 1). In silico analysis of the microarray data then predicted the activation of IFNs and their downstream target genes in CRC cells treated with CX-5461 (Fig. 2A-D), which was confirmed by ELISA and RT-qPCR analyses, respectively (Fig. 2E & F). Taken together, these findings suggest that CX-5461 treatment activates the cGAS-STING pathway in CRC cells; this postulation is supported by the increases in cGAMP and active IRF3 levels in various CRC cells after drug treatment (Fig. 3A & B). By contrast, our results (Fig. 3C-E) do not support the activation of the NF-κB pathway by CX-5461 in CRC cells. Regarding the downstream mediator of type 1 IFN, we noted the activation of STAT1 and PD-L1, one of its well-known target genes, (Fig. 4). Because PD-L1 expression is one of three predictive biomarkers approved by the US Food and Drug Administration for anti-PD-(L)1 therapies in patients with advanced NSCLC, we performed animal experiments to examine the antitumor effects of CX-5461 alone, anti-PD-1 or PD-L1 antibody alone, and the combinations of CX-5461 and either ICI. Notably, the suppressive effects of CX-5461 alone on tumor grown from both CT26 and 4T1 cells were even more substantial than those of either antibody alone. The highest antitumor efficacies were detected in mice receiving the combination therapies (Figs. 5 & 6), which might be attributable to the robust increase in PD-L1 levels in tumor cells after CX-5461 treatment (Fig. 4). To explain the highly effective in vivo antitumor effect of CX-5461 alone, we analyzed the numbers of various immune cells in the spleen of mice under different treatments. Our results demonstrate that CX-5461 alone as well as its combination with anti-PD1 antibody but not the antibody alone significantly increased the numbers of various T-cell types (CD3+, CD4+, regular and activated CD8+, as well as regulatory T cells) in the spleen of mice. In addition, considerable decreases in the numbers of MDSCs were observed in the spleen of mice receiving CX-5461 alone and CX-5461+anti-PD1 (Fig. 6C & D). Taken together, our results demonstrate that CX-5461 can kill CRC (and other tumor) cells directly through various mechanisms as well as indirectly by activating the cGAS-STING-IFN pathway and then triggering STAT1-mediated PD-L1 expression; this may enhance the therapeutic efficacies of available ICIs. Hence, this study provides strong evidence supporting the inclusion of CX-5461 in cancer immunotherapy.
Declarations
The authors have declared no conflict of interest.
Ethical Approval and Consent to participate
All animal experiments were performed in accordance with a protocol approved by the Academia Sinica Institutional Animal Care and Utilization Committee.
Human Ethics
None
Consent for publication
Not applicable
Availability of supporting data
The datasets generated and analyzed during the current study are not publicly available but are available from the corresponding author upon reasonable request.
Funding
This work was supported by Taiwan Cancer Clinic Foundation and Melissa Lee Cancer Foundation. We like to thank Academia Sinica for their supports. Additional support was provided by the Taipei Veterans General Hospital (V109C-028 to M. Chen), the Ministry of Science and Technology (MOST 109-2314-B-075-082-MY3 to MH. Chen; MOST 109-2320-B-010-023 to Y. Su) and Taipei, Taichung, Kaohsiung Veterans General Hospital, Tri-Service General Hospital, Academia Sinica Joint Research Program [AS-VTA-108-07] to M.H.C and M.H. Additinal support was provided by the Yen Tjing Ling medical foundation (CI-112-7 to N.J Chiang).
Authors' contributions
Conception and design: Shin-Yi Chung, Yu-Chan Chang, Yeu Su, Ming-Huang Chen
Development of methodology: Shin-Yi Chung, Yu-Chan Chang, Yeu Su, Ming-Huang Chen
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Shin-Yi Chung, Yu-Chan Chang, Michael Hsiao, John Soong, Yeu Su, Ming-Huang Chen
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Shin-Yi Chung, Yu-Chan Chang, Dennis Shin-Shian Hsu, Ya-Chi Hong, Meng-Lun Lu, John Soong
Writing, review, and/or revision of the manuscript: Shin-Yi Chung, Yu-Chan Chang, Dennis Shin-Shian Hsu, Yi-Ping Hung, Nai-Jung Chiang, Chun-Nan Yeh, Michael Hsiao, John Soong, Yeu Su, Ming-Huang Chen
Administrative, technical, or material support (i.e., reporting or organizing data, constricting databases): Michael Hsiao, John Soong, Yeu Su, Ming-Huang Chen
Study supervision: Yu-Chan Chang, Yeu Su, Ming-Huang Chen
Authors' information
Authors and Affiliations
Taipei Veterans General Hospital, Taiwan
Shin-Yi Chung, Ya-Chi Hung, Meng-Lun Lu, Yi-Ping Hung, Nai-Jung Chiang, Ming-Huang Chen
National Yang Ming Chiao Tung University, Taiwan
Yu-Chan Chang, Yeu Su
Asclepiumm Taiwan Co., Ltd, Taiwan
Dennis Shin-Shian Hsu
Chang Gung Memorial Hospital, Chang Gung University, Taiwan
Chun-Nan Yeh
Genomics Research Center, Academia Sinica, Taiwan
Michael Hsiao
Senhwa Biosciences, Inc., Taiwan
John Soong
Declaration of Competing Interest
The authors have no competing interests to declare that are relevant to the content of this article.
Acknowledgements
We thank Professor Michael Hsiao for supporting the microarray and in silico analysis. We also thank Senhwa biosciences, Inc. for providing the CX-5461 compound for the work. This manuscript was edited by Wallace Academic Editing.
Contributor Information
Yeu Su, Email: yeusu@nycu.edu.tw.
Ming-Huang Chen, Email: mhchen9@vghtpe.gov.tw.
References
- 1.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi: 10.1016/S0140-6736(13)61649-9. Apr 26Epub 2013 Nov 11. PMID: 24225001. [DOI] [PubMed] [Google Scholar]
- 2.Recio-Boiles A, Kashyap S, Tsoris A, et al. StatPearls [Internet]. Treasure Island (FL) StatPearls Publishing; 2022. Rectal Cancer. [Updated 2021 Dec 15]https://www.ncbi.nlm.nih.gov/books/NBK493202/ Jan-. Available from. [Google Scholar]
- 3.Fietkau R, Zettl H, Klöcking S, Kundt G. Incidence, therapy and prognosis of colorectal cancer in different age groups. A population-based cohort study of the Rostock Cancer Registry. Strahlenther Onkol. 2004;180(8):478–487. doi: 10.1007/s00066-004-1260-z. AugPMID: 15292968. [DOI] [PubMed] [Google Scholar]
- 4.Labianca R, Beretta GD, Kildani B, Milesi L, Merlin F, Mosconi S, Pessi MA, Prochilo T, Quadri A, Gatta G, de Braud F. Wils J. Colon cancer. Crit Rev Oncol Hematol. 2010;74(2):106–133. doi: 10.1016/j.critrevonc.2010.01.010. May. [DOI] [PubMed] [Google Scholar]
- 5.Zeng Z, Yang B, Liao Z. Biomarkers in Immunotherapy-Based Precision Treatments of Digestive System Tumors. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.650481. Mar 11PMID: 33777812; PMCID: PMC7991593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strickler JH, Hanks BA, Khasraw M. Tumor Mutational Burden as a Predictor of Immunotherapy Response: Is More Always Better? Clin Cancer Res. 2021;27(5):1236–1241. doi: 10.1158/1078-0432.CCR-20-3054. Mar 1Epub 2020 Nov 16. PMID: 33199494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, Diaz LA., Jr. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16(6):361–375. doi: 10.1038/s41575-019-0126-x. JunPMID: 30886395; PMCID: PMC7295073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.André T, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, Smith D, Garcia-Carbonero R, Benavides M, Gibbs P, de la Fouchardiere C, Rivera F, Elez E, Bendell J, Le DT, Yoshino T, Van Cutsem E, Yang P, Farooqui MZH, Marinello P, Diaz LA., Jr KEYNOTE-177 Investigators. Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N Engl J Med. 2020;383(23):2207–2218. doi: 10.1056/NEJMoa2017699. Dec 3PMID: 33264544. [DOI] [PubMed] [Google Scholar]
- 9.De' Angelis GL, Bottarelli L, Azzoni C, De' Angelis N, Leandro G, Di Mario F, Gaiani F, Negri F. Microsatellite instability in colorectal cancer. Acta Biomed. 2018;89(9-S):97–101. doi: 10.23750/abm.v89i9-S.7960. Dec 17PMID: 30561401; PMCID: PMC6502181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webber EM, Kauffman TL, O'Connor E, Goddard KA. Systematic review of the predictive effect of MSI status in colorectal cancer patients undergoing 5FU-based chemotherapy. BMC Cancer. 2015;15:156. doi: 10.1186/s12885-015-1093-4. Mar 21PMID: 25884995; PMCID: PMC4376504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golshani G, Zhang Y. Advances in immunotherapy for colorectal cancer: a review. Therap Adv Gastroenterol. 2020;13 doi: 10.1177/1756284820917527. Jun 1PMID: 32536977; PMCID: PMC7268115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koido S., Kan S., Yoshida K., Yoshizaki S., Takakura K., Namiki Y., Tsukinaga S., Odahara S., Kajihara M., Okamoto M., Ito M., Yusa S., Gong J., Sugiyama H., Ohkusa T., Homma S., Tajiri H. Immunogenic modulation of cholangiocarcinoma cells by chemoimmunotherapy. Anticancer research. 2014;34(11):6353–6361. [PubMed] [Google Scholar]
- 13.Bywater M.J., Poortinga G., Sanij E., Hein N., Peck A., Cullinane C., Wall M., Cluse L., Drygin D., Anderes K., Huser N., Proffitt C., Bliesath J., Haddach M., Schwaebe M.K., Ryckman D.M., Rice W.G., Schmitt C., Lowe S.W., Johnstone R.W., Hannan R.D. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer cell. 2012;22(1):51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Negi S.S., Brown P. rRNA synthesis inhibitor, CX-5461, activates ATM/ATR pathway in acute lymphoblastic leukemia, arrests cells in G2 phase and induces apoptosis. Oncotarget. 2015;6(20):18094–18104. doi: 10.18632/oncotarget.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quin J., Chan K.T., Devlin J.R., Cameron D.P., Diesch J., Cullinane C., Ahern J., Khot A., Hein N., George A.J., Hannan K.M., Poortinga G., Sheppard K.E., Khanna K.K., Johnstone R.W., Drygin D., McArthur G.A., Pearson R.B., Sanij E., Hannan R.D. Inhibition of RNA polymerase I transcription initiation by CX-5461 activates non-canonical ATM/ATR signaling. Oncotarget. 2016;7(31):49800–49818. doi: 10.18632/oncotarget.10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devlin J.R., Hannan K.M., Hein N., Cullinane C., Kusnadi E., Ng P.Y., George A.J., Shortt J., Bywater M.J., Poortinga G., Sanij E., Kang J., Drygin D., O'Brien S., Johnstone R.W., McArthur G.A., Hannan R.D., Pearson R.B. Combination Therapy Targeting Ribosome Biogenesis and mRNA Translation Synergistically Extends Survival in MYC-Driven Lymphoma. Cancer discovery. 2016;6(1):59–70. doi: 10.1158/2159-8290.CD-14-0673. [DOI] [PubMed] [Google Scholar]
- 17.Hein N., Cameron D.P., Hannan K.M., Nguyen N.N., Fong C.Y., Sornkom J., Wall M., Pavy M., Cullinane C., Diesch J., Devlin J.R., George A.J., Sanij E., Quin J., Poortinga G., Verbrugge I., Baker A., Drygin D., Harrison S.J., Rozario J.D., Hannan R.D. Inhibition of Pol I transcription treats murine and human AML by targeting the leukemia-initiating cell population. Blood. 2017;129(21):2882–2895. doi: 10.1182/blood-2016-05-718171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, Di Antonio M, McKinney S, Mathew V, Ho B, O'Neil NJ, Santos ND, Silvester J, Wei V, Garcia J, Kabeer F, Lai D, Soriano P, Banáth J, Chiu DS, Yap D, Le DD, Ye FB, Zhang A, Thu K, Soong J, Lin SC, Tsai AH, Osako T, Algara T, Saunders DN, Wong J, Xian J, Bally MB, Brenton JD, Brown GW, Shah SP, Cescon D, Mak TW, Caldas C, Stirling PC, Hieter P, Balasubramanian S, Aparicio S. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat Commun. 2017;8:14432. doi: 10.1038/ncomms14432. Feb 17PMID: 28211448; PMCID: PMC5321743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornelison R., Dobbin Z.C., Katre A.A., Jeong D.H., Zhang Y., Chen D., Petrova Y., Llaneza D.C., Steg A.D., Parsons L., Schneider D.A., Landen C.N. Targeting RNA-Polymerase I in Both Chemosensitive and Chemoresistant Populations in Epithelial Ovarian Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2017;23(21):6529–6540. doi: 10.1158/1078-0432.CCR-17-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosiol N, Juranek S, Brossart P, Heine A, Paeschke K. G-quadruplexes: a promising target for cancer therapy. Mol Cancer. 2021;20(1):40. doi: 10.1186/s12943-021-01328-4. Feb 25PMID: 33632214; PMCID: PMC7905668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruno P.M., Lu M., Dennis K.A., Inam H., Moore C.J., Sheehe J., Elledge S.J., Hemann M.T., Pritchard J.R. The primary mechanism of cytotoxicity of the chemotherapeutic agent CX-5461 is topoisomerase II poisoning. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(8):4053–4060. doi: 10.1073/pnas.1921649117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan M., Wright W.C., Chapple R.H., Zubair A., Sandhu M., Batchelder J.E., Huddle B.C., Low J., Blankenship K.B., Wang Y., Gordon B., Archer P., Brady S.W., Natarajan S., Posgai M.J., Schuetz J., Miller D., Kalathur R., Chen S., Connelly J.P., Geeleher P. The chemotherapeutic CX-5461 primarily targets TOP2B and exhibits selective activity in high-risk neuroblastoma. Nature communications. 2021;12(1):6468. doi: 10.1038/s41467-021-26640-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence M.G., Porter L.H., Choo N., Pook D., Grummet J.P., Pezaro C.J., Sandhu S., Ramm S., Luu J., Bakshi A., Goode D.L., Sanij E., Pearson R.B., Hannan R.D., Simpson K.J., Taylor R.A., Risbridger G.P., Furic L. CX-5461 Sensitizes DNA Damage Repair-proficient Castrate-resistant Prostate Cancer to PARP Inhibition. Molecular cancer therapeutics. 2021;20(11):2140–2150. doi: 10.1158/1535-7163.MCT-20-0932. [DOI] [PubMed] [Google Scholar]
- 24.Cornelison R., Biswas K., Llaneza D.C., Harris A.R., Sosale N.G., Lazzara M.J., Landen C.N. CX-5461 Treatment Leads to Cytosolic DNA-Mediated STING Activation in Ovarian Cancer. Cancers. 2021;13(20):5056. doi: 10.3390/cancers13205056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang Y.C., Yang Y.F., Chiou J., Tsai H.F., Fang C.Y., Yang C.J., Chen C.L., Hsiao M. Nonenzymatic function of Aldolase A downregulates miR-145 to promote the Oct4/DUSP4/TRAF4 axis and the acquisition of lung cancer stemness. Cell death & disease. 2020;11(3):195. doi: 10.1038/s41419-020-2387-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spada S, Yamazaki T. Vanpouille-Box C. Detection and quantification of cytosolic DNA. Methods Enzymol. 2019;629:17–33. doi: 10.1016/bs.mie.2019.07.042. Epub 2019 Aug 9. PMID: 31727239. [DOI] [PubMed] [Google Scholar]
- 27.Paludan SR, Reinert LS, Hornung V. DNA-stimulated cell death: implications for host defence, inflammatory diseases and cancer. Nat Rev Immunol. 2019;19(3):141–153. doi: 10.1038/s41577-018-0117-0. MarPMID: 30644449; PMCID: PMC7311199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roh JS, Sohn DH. Damage-Associated Molecular Patterns in Inflammatory Diseases. Immune Netw. 2018;18(4):e27. doi: 10.4110/in.2018.18.e27. Aug 13PMID: 30181915; PMCID: PMC6117512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcus A, Mao AJ, Lensink-Vasan M, Wang L, Vance RE, Raulet DH. Tumor-Derived cGAMP Triggers a STING-Mediated Interferon Response in Non-tumor Cells to Activate the NK Cell Response. Immunity. 2018;49(4):754–763. doi: 10.1016/j.immuni.2018.09.016. Oct 16e4PMID: 30332631; PMCID: PMC6488306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24(1):93–103. doi: 10.1016/j.immuni.2005.12.003. JanPMID: 16413926. [DOI] [PubMed] [Google Scholar]
- 31.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, Sato S, Yamamoto M, Uematsu S, Kawai T, Takeuchi O, Akira S. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7(1):40–48. doi: 10.1038/ni1282. JanEpub 2005 Nov 13. Erratum in: Nat Immunol. 2006 Apr;7(4):427. PMID: 16286919. [DOI] [PubMed] [Google Scholar]
- 32.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17(10):1142–1149. doi: 10.1038/ni.3558. Sep 20PMID: 27648547. [DOI] [PubMed] [Google Scholar]
- 33.Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31(4):539–550. doi: 10.1016/j.immuni.2009.09.002. Oct 16PMID: 19833085; PMCID: PMC2774226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadzak I, Schiff M, Gattermeier I, Glinitzer R, Sauer I, Saalmüller A, Yang E, Schaljo B, Kovarik P. Recruitment of Stat1 to chromatin is required for interferon-induced serine phosphorylation of Stat1 transactivation domain. Proc Natl Acad Sci U S A. 2008;105(26):8944–8949. doi: 10.1073/pnas.0801794105. Jul 1Epub 2008 Jun 23. PMID: 18574148; PMCID: PMC2435588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheon H, Stark GR. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci U S A. 2009;106(23):9373–9378. doi: 10.1073/pnas.0903487106. Jun 9Epub 2009 May 28. PMID: 19478064; PMCID: PMC2688000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pilz A, Ramsauer K, Heidari H, Leitges M, Kovarik P, Decker T. Phosphorylation of the Stat1 transactivating domain is required for the response to type I interferons. EMBO Rep. 2003;4(4):368–373. doi: 10.1038/sj.embor.embor802. AprEpub 2003 Mar 21. PMID: 12671680; PMCID: PMC1319158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X, Parisi G, Saus CP, Torrejon DY, Graeber TG, Comin-Anduix B, Hu-Lieskovan S, Damoiseaux R, Lo RS, Ribas A. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2019;29(11):3766. doi: 10.1016/j.celrep.2019.11.113. Erratum for: Cell Rep. 2017 May 9;19(6):1189-1201. PMID: 31825850. [DOI] [PubMed] [Google Scholar]
- 38.Matsuda T., Yamashita K., Hasegawa H., Oshikiri T., Hosono M., Higashino N., Yamamoto M., Matsuda Y., Kanaji S., Nakamura T., Suzuki S., Sumi Y., Kakeji Y. Recent updates in the surgical treatment of colorectal cancer. Annals of gastroenterological surgery. 2018;2(2):129–136. doi: 10.1002/ags3.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costas-Chavarri A., Nandakumar G., Temin S., Lopes G., Cervantes A., Cruz Correa M., Engineer R., Hamashima C., Ho G.F., Huitzil F.D., Malekzadeh Moghani M., Sharara A.I., Stern M.C., Teh C., Vázquez Manjarrez S.E., Verjee A., Yantiss R., Shah M.A. Treatment of Patients With Early-Stage Colorectal Cancer: ASCO Resource-Stratified Guideline. Journal of global oncology. 2019;5:1–19. doi: 10.1200/JGO.18.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Douillard J.Y., Oliner K.S., Siena S., Tabernero J., Burkes R., Barugel M., Humblet Y., Bodoky G., Cunningham D., Jassem J., Rivera F., Kocákova I., Ruff P., Błasińska-Morawiec M., Šmakal M., Canon J.L., Rother M., Williams R., Rong A., Wiezorek J., Patterson S.D. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. The New England journal of medicine. 2013;369(11):1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 41.Lo Nigro C., Ricci V., Vivenza D., Granetto C., Fabozzi T., Miraglio E., Merlano M.C. Prognostic and predictive biomarkers in metastatic colorectal cancer anti-EGFR therapy. World journal of gastroenterology. 2016;22(30):6944–6954. doi: 10.3748/wjg.v22.i30.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siena S., Falcone A., Ychou M., Humblet Y., Bouché O., Mineur L., Barone C., Adenis A., Tabernero J., Yoshino T., Lenz H.J., Goldberg R.M., Sargent D.J., Cihon F., Cupit L., Wagner A., Laurent D. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled. phase 3 trial. Lancet (London, England) 2013;381(9863):303–312. doi: 10.1016/S0140-6736(12)61900-X. [DOI] [PubMed] [Google Scholar]
- 43.Venook A.P., Niedzwiecki D., Lenz H.J., Innocenti F., Fruth B., Meyerhardt J.A., Schrag D., Greene C., O'Neil B.H., Atkins J.N., Berry S., Polite B.N., O'Reilly E.M., Goldberg R.M., Hochster H.S., Schilsky R.L., Bertagnolli M.M., El-Khoueiry A.B., Watson P., Benson A.B., Blanke C. Effect of First-Line Chemotherapy Combined With Cetuximab or Bevacizumab on Overall Survival in Patients With KRAS Wild-Type Advanced or Metastatic Colorectal Cancer: A Randomized Clinical Trial. JAMA. 2017;317(23):2392–2401. doi: 10.1001/jama.2017.7105. 3rd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ottaiano A., Capozzi M., Tafuto S., De Stefano A., De Divitiis C., Romano C., Avallone A., Nasti G. Folfiri-Aflibercept vs. Folfiri-Bevacizumab as Second Line Treatment of RAS Mutated Metastatic Colorectal Cancer in Real Practice. Frontiers in oncology. 2019;9:766. doi: 10.3389/fonc.2019.00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi M., Jiao D., Xu H., Liu Q., Zhao W., Han X., Wu K. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Molecular cancer. 2018;17(1):129. doi: 10.1186/s12943-018-0864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai R., Lv Z., Xu D., Cui J. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomarker research. 2020;8:34. doi: 10.1186/s40364-020-00209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Overman M.J., Lonardi S., Wong K., Lenz H.J., Gelsomino F., Aglietta M., Morse M.A., Van Cutsem E., McDermott R., Hill A., Sawyer M.B., Hendlisz A., Neyns B., Svrcek M., Moss R.A., Ledeine J.M., Cao Z.A., Kamble S., Kopetz S., André T. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2018;36(8):773–779. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 48.Le D.T., Uram J.N., Wang H., Bartlett B.R., Kemberling H., Eyring A.D., Skora A.D., Luber B.S., Azad N.S., Laheru D., Biedrzycki B., Donehower R.C., Zaheer A., Fisher G.A., Crocenzi T.S., Lee J.J., Duffy S.M., Goldberg R.M., de la Chapelle A., Koshiji M., Diaz L.A. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. The New England journal of medicine, 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are not publicly available but are available from the corresponding author upon reasonable request.