Abstract
Recent work examining nematode and tardigrade gut microbiomes has identified species-specific relationships between host and gut community composition. However, only a handful of species from either phylum have been examined. How microbiomes differ among species and what factors contribute to their assembly remains unexplored. Cyanobacterial mats within Antarctic Dry Valley streams host a simple and tractable natural ecosystem of identifiable microinvertebrates to address these questions. We sampled 2 types of coexisting mats (i.e., black and orange) across four spatially isolated streams, hand-picked single individuals of two nematode species (i.e., Eudorylaimus antarcticus and Plectus murrayi) and tardigrades, to examine their gut microbiomes using 16S and 18S rRNA metabarcoding. All gut microbiomes (bacterial and eukaryotic) were significantly less diverse than the mats they were isolated from. In contrast to mats, microinvertebrates’ guts were depleted of Cyanobacteria and differentially enriched in taxa of Bacteroidetes, Proteobacteria, and Fungi. Among factors investigated, gut microbiome composition was most influenced by host identity while environmental factors (e.g., mats and streams) were less important. The importance of host identity in predicting gut microbiome composition suggests functional value to the host, similar to other organisms with strong host selected microbiomes.
Subject terms: Biodiversity, Molecular ecology, Microbial communities
Introduction
Gut microbiomes of many animal species (e.g., aphids, cows, and humans) have been shown to play specific and vital roles in the physiology of their hosts (e.g., digestion of nutrients, protection against pathogens)1. In these animals, factors generated by host physical, chemical, or behavioral characteristics deterministically drive the assembly and composition of gut microbiomes in an eco-evolutionary pattern defined as phylosymbiosis2. However, hosts’ connection to their microbiomes can vary considerably. For example, gut microbiomes of some animals (e.g., dragonflies and lepidopteran larvae)3,4 are more reflective of microbial communities of the hosts’ environment and artificial removal of these microbiomes has no effect on host functioning5. Patterns and drivers of gut microbiome assembly can vary substantially, even among closely related host species5,6. For example, within the single family of Formicidae, canopy ants (i.e., Cephalotes) rely on gut microbiomes to cycle nitrogen and create essential amino acids7, but ground ants (i.e., Solenopsis and Pheidole) are devoid of any gut bacteria6 and instead associate with nitrogen rich plants not found in the canopy where the Cephalotes reside8. This suggests that gut microbiomes of some animals can depend solely on environmental factors and be independent of host identity and/or phylogeny, while other animals might be influenced by a combination of both9,10.
Despite our increased understanding of the assembly and function of gut microbiomes of many animals, the microbiomes of nematode and tardigrade species have only recently begun to be studied. Nematoda is an extremely diverse phylum of microscopic roundworms, with estimates of species richness ranging from 0.5 to 10 million, although < 30,000 have been currently described11. In addition to taxonomic diversity, nematodes present a wide diversity of life strategies (i.e., r-selected vs. K-selected) and feeding habits (e.g., bacterivores, omnivores, plant parasites, and predators)12. Tardigrada is the most closely related phylum to Nematoda13,14, and despite significant anatomical differences (e.g., segmented body and 8 legs), there is remarkable similarity in their digestive structures (i.e., a pharyngeal bulb and esophagus), average body size (< 1 mm), molting based growth, feeding habits (e.g., bacterivores, omnivores, and predators), and behavior (e.g., foraging and use of anhydrobiosis), as well as shared feeding niches15.
Due to the importance of nematodes in soil processes such as decomposition and nutrient cycling16 as well as in agriculture via plant and animal parasitism17, nematode gut microbiomes have received more attention than those of tardigrades. Initial studies using DNA clone libraries found that gut microbiomes of several bacterivorous nematode species were characterized by different levels of microbial diversity (e.g., higher in Acrobeles sp. than Prionchulus sp. and higher in Caenorhabditis elegans than Acrobeloides maximus)18,19. With metabarcoding methods, microbial communities of the model nematode C. elegans (a bacterivore) have been recently examined and described in more detail20–24. It is now well established that the gut microbiome of C. elegans is distinct from the microbiome of its natural habitat substrate (rotting fruit)25. Moreover, populations of C. elegans from widely distant geographic locations with different substrate microbial communities have remarkably similar gut microbiomes20,22. In addition, a comparison of the gut microbiome of C. elegans to that of its sister species, C. remanei, shows compositional differences despite similarity in both feeding habits and environmental niches20.
Although gut microbiomes of Caenorhabditis species appear to be driven by host identity, the exposure of C. elegans to different environmental conditions can affect the composition of the gut microbiome as well, but this remains not fully understood. For example, higher environmental temperatures can result in enrichment of Sphingobacterium within the soil substrate but depletion within C. elegans guts, while other bacterial taxa (e.g., Agrobacterium) can respond in exactly the opposite manner22 demonstrating the complex role of environmental factors in shaping the assembly of nematode gut microbiomes. While Tardigrada microbiomes have been studied far less than those of nematodes, they also appear to be host-specific and consistently distinct from their surrounding environment26–30. However, varying influence of host identity has been observed within the diverse phylum of Nematoda. For example, while two cryptic Litoditis marine species contained distinct microbiomes31, a more extensive survey of 33 marine genera found no importance of host identity or feeding habits, suggesting a more stochastic assembly32. Unfortunately, nematode and tardigrade gut microbiomes have been characterized for only a handful of species, leaving large gaps in our understanding of the factors driving the assembly of gut communities within the remaining majority of microinvertebrate species that are both phylogenetically diverse and vary greatly in their functional roles within ecosystems.
To begin assessing factors driving nematode and tardigrade microbiome assembly more systematically, we used the McMurdo Dry Valleys as a simplified model natural ecosystem. The McMurdo Dry Valleys are an ice-free region of Antarctica, characterized by unvegetated gravel-like soils, extremely high winds, and almost no precipitation. Temperatures within the soil can decline to − 59 °C33, restricting continuous biological activity for much of the year. However, during 8–12 weeks of the austral summer, temperatures sufficiently warm to melt adjacent glaciers, generating liquid water that flows down mountain slopes into the basin below34. Taylor Valley, the site of this study, contains a number of these seasonally active streams, flowing into a series of permanently ice-covered lakes (e.g., Lake Fryxell) (Fig. 1a). Within the streams reside different types of morphologically distinguishable cyanobacterial mats (e.g., black and orange) (Fig. 1b,c)35. Each type of mat is characterized by the dominance of different species of photosynthesizing Cyanobacteria (black by Nostoc and orange by Oscillatoriales)35 and supports a distinct but diverse microbial community directly adjacent to one another in a single stream36. In general, black mats establish at stream margins and orange mats establish within central flows. Both mat types provide a habitat for a finite and well characterized community of microinvertebrates consisting of only two nematode species (Plectus murrayi and Eudorylaimus antarcticus (currently undergoing taxonomic revaluation)), at least two tardigrade species (Acutuncus antarcticus and Milnesium sp.), and 3–5 rotifer species37–39. Due to its narrow funnel shaped unarmed stoma, P. murrayi is classified as a bacterial feeder while E. antarcticus contains a hypodermic needle-like odontostylet and is classified as an omnivore40. Although E. antarcticus has been thought to feed solely on algae41, a recent study in Taylor Valley’s Von Guerard stream using isotopic C and N ratios has indicated that while P. murrayi, tardigrades (e.g., A. antarcticus), and rotifers likely consume only microbes, E. antarcticus may rely on food sources consisting of the above mentioned microinvertebrates instead of or in addition to algae42. However, the exact food sources and gut microbiomes of these microinvertebrates have not been examined, therefore limiting a full understanding of their roles in the ecosystem.
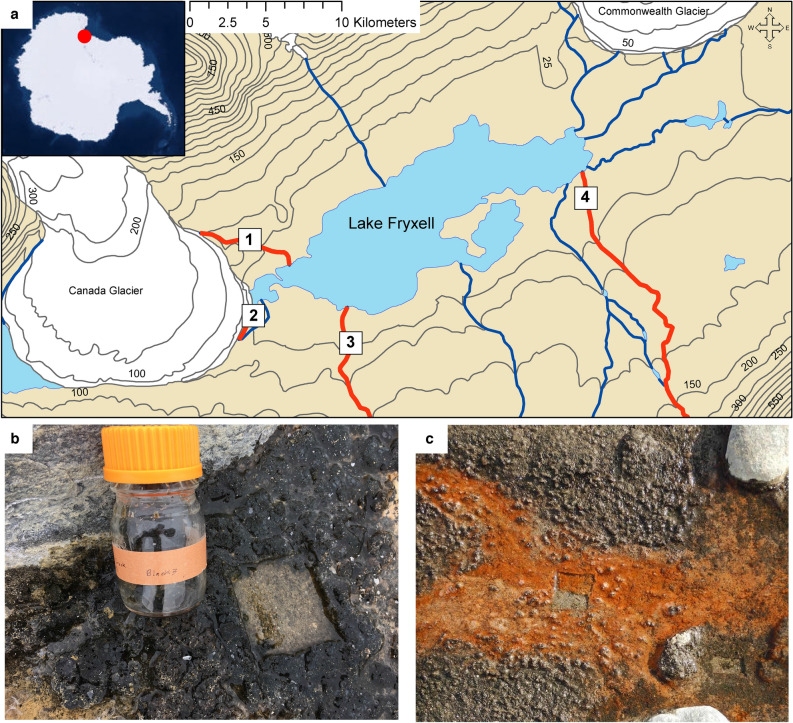
Figure 1.
Locations and types of mat samples used for this study. (a) Map of the Lake Fryxell Basin in Taylor Valley, Antarctica with exact sampling sites (white squares) along streams (red lines: 1. Canada Stream, 2. Bowles Creek, 3. Delta Stream, 4. Von Guerard Stream). Blue lines indicate other streams not included in this study. Types of mats included in this study: (b) black type and (c) orange type with orange type showing regrowth 12 months post sampling. Photo credit: Josh Darling and Mike Gooseff. Map was created with ArcGIS Desktop 10.8.2 (http://www.esri.com).
In this study, we characterized nematode and tardigrade gut microbiomes, as well as the potential factors (e.g., environment such as stream identity and mat type or host-specific factors such as host identity) that could play a role in structuring their assembly. Because this Antarctic ecosystem supports a remarkably simplified and morphologically tractable microinvertebrate community, patterns of gut microbiome assembly can be studied at the nematode species level under natural environmental conditions. In addition to describing the bacterial component of the gut microbiomes, we also examined host-associated microbial eukaryotes. We hypothesized that gut microbiomes of all microinvertebrates would be explained primarily by host identity rather than environmental factors and would therefore be consistent across black and orange mats, and all streams. Consequently, all host associated microbiomes (i.e., bacterial and eukaryotic) would be distinct from the microbial communities of the mats and from each other.
Results
Alpha diversity differences among communities
Nematode gut microbiomes were assigned into their respective species categories of E. antarcticus and P. murrayi based on 18S host data that was consistent with morphology (see Methods “Microinvertebrate haplotypes”). In contrast, due to recovery of three undiscernible 18S tardigrade haplotypes, the gut microbiomes were assigned to Tardigrada. Mat bacterial communities were significantly (Tukey's HSD, P < 2e−16) more diverse than communities of all microinvertebrate gut microbiomes for all four alpha diversity metrics tested (i.e., Richness, Shannon’s Index, Simpson’s Index, and Faith’s Phylogenic Diversity) (Fig. 2, Supplementary Table S1). In contrast to the significance of community type (i.e., mat, E. antarcticus, P. murrayi, Tardigrada) for all alpha diversity metrics (GLM, P < 0.001, χ2(3) > 58.21), there was no effect of mat type (P > 0.65, χ2(1) < 0.21) on bacterial alpha diversity, while stream was significant for Shannon’s and Simpson’s indices (P < 0.01, χ2(3) > 11.47) but not for Richness or Faith’s PD (P > 0.38, χ2(3) < 3.07) (Supplementary Table S1). The interaction between community type and stream was also significant (P < 0.001, χ2(9) > 23.59) for all metrics tested (Supplementary Table S1a). Gut microbiomes of E. antarcticus and P. murrayi were less diverse (Supplementary Table S1b) than those of Tardigrada for Shannon’s, Richness, and Faith’s PD (Tukey HSD, P < 0.05), but not for Simpson’s (Tukey HSD, P > 0.39). Although there was no difference in alpha diversity between nematode species gut microbiomes for Richness, Faith’s PD, and Simpson’s, the Shannon index indicated that gut communities of E. antarcticus were the least diverse (P < 0.05, Fig. 2).
Figure 2.
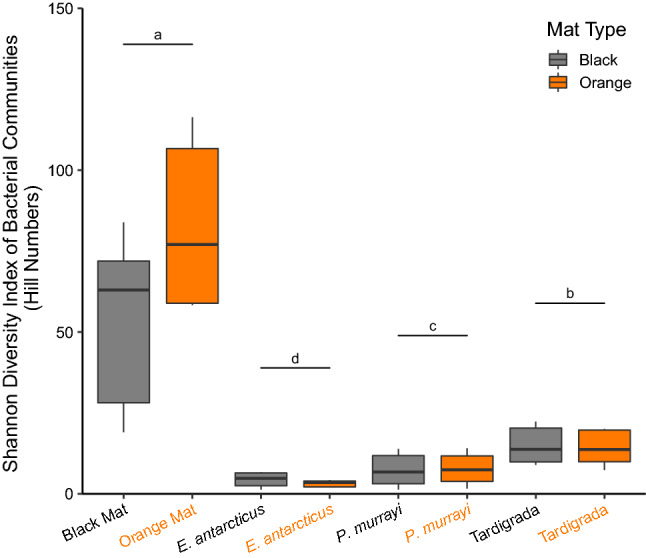
Bacterial ASV Shannon diversity (Hill Numbers) of microbial mats and microinvertebrate gut microbiomes from black and orange mats. Horizontal lines and letters indicate significant difference (P < 0.05) among the eight communities with GLM and then Tukey HSD.
Within microinvertebrates, most eukaryotic reads predictably assigned to the host (89.25% of E. antarcticus, 99.10% for P. murrayi, 99.45% of Tardigrada), however when removed, there was sufficient coverage and sequencing depth for further analysis of non-host eukaryotic communities. Mat eukaryotic communities were more diverse than all non-host eukaryotic gut communities for all metrics (Tukey HSD, Richness, Shannon’s, and Faith’s PD, P < 1.884e−05; Simpson’s P = 0.09) (Supplementary Table S2). Eukaryotic alpha diversity differed between mat types for only Shannon’s (GLM, P = 0.06, χ2(1) = 3.57) but not for Richness, Simpson’s, and Faith’s PD (P > 0.15, χ2(1) < 2.10). Among streams, Richness and Faith’s PD were significantly variable (P = 0.02, χ2(3) > 9.82, Supplementary Table S2a), with Canada Stream being more diverse (Tukey HSD, P < 0.05), but not for Shannon’s (P = 0.22) or Simpson’s indices (P = 0.12) (Supplementary Table S2). Similar to bacterial diversity, the Shannon index indicated that the non-host eukaryotic communities of E. antarcticus were the least diverse, followed by P. murrayi, and then Tardigrada as the most diverse (Tukey HSD, P < 0.05) (Table 2b). For Richness, all gut microbiomes were similarly less diverse than mats, while Faith’s PD showed an overlap of significance for E. antarcticus of the other two microinvertebrate communities although P. murrayi and Tardigrada did separate from each other (Tukey HSD, P < 0.05) (Supplementary Table S2).
Compositional differences among bacterial communities
Black and orange mats represented two dissimilar microbial communities. Examining only mats, although both mat type (PERMANOVA, P < 0.003, F(1) = 3.94) and stream (P < 0.003, F(3) = 4.04) significantly affected the composition of bacterial communities, the communities primarily clustered by mat type (Supplementary Fig. S1a), with only Canada Stream communities separating from those from the other streams (Supplementary Fig. S1b). Consequently, compared to mat type (10%), stream explained the most variation (32%) (Supplementary Table S3a) despite all other streams overlapping in NMDS space (Supplementary Fig. S1b). In contrast, the gut microbiomes of microinvertebrates did not cluster by mat type (Supplementary Fig. S2a) nor by stream, but instead by host identity (e.g., E. antarcticus, P. murrayi, Tardigrada) (Supplementary Fig. S2b). Although all investigated factors significantly affected gut microbiome compositions (PERMANOVA, P < 0.05, F(1–6) > 0.48), mat type and stream explained only 1% and 4% of the microbial community variation, respectively (Supplementary Table S4a). In contrast, host identity played the most dominant role in the assembly of gut microbiomes and explained 14% of total variation. However, most of the variation (72%) remained unexplained. At the genus level of taxonomic resolution, 75% of the taxa within Tardigrada, 78% within P. murrayi, and 87% within E. antarcticus were shared with mats. The remaining proportion of gut taxa was not found within any mats but was in low abundance across all samples. Black and orange mats shared 40% of ASVs, while 27% unique ASVs were assigned to black mats and 33% to orange mats. However, at the genus level, 71% of genera were observed in both mat types with 17% unique genera in black mats and 13% unique in orange mats.
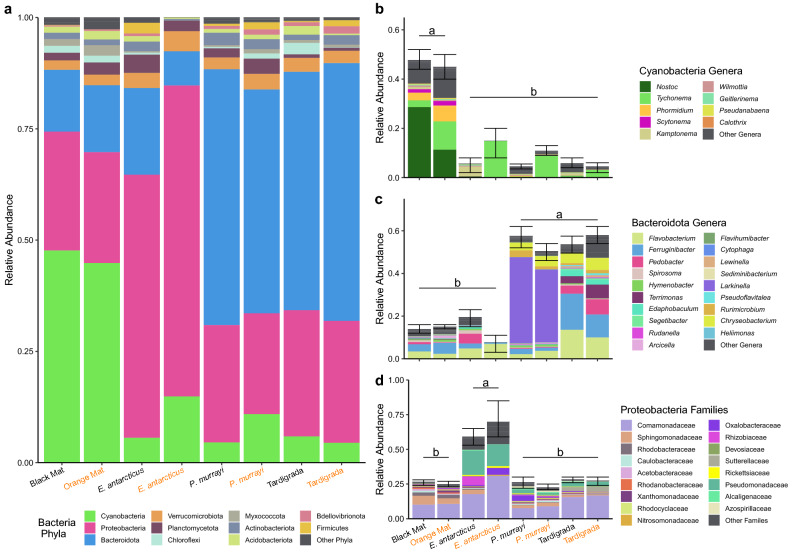
Cyanobacteria, Bacteroidota, and Proteobacteria were the most abundant phyla across all microbiomes comprising 86.40% of the total community composition (Fig. 3a, Supplementary Table S5a). Indicator species analysis confirmed Cyanobacteria, Bacteroidota, and Proteobacteria as significantly indicative phyla of the four microbial community types. Expectedly, Cyanobacteria was the most indicative phylum of the mat communities and although there were six other indicative phyla (Supplementary Table S6), their cumulative relative abundance was low (< 1.2%). Proteobacteria was the sole indicative phylum of the gut microbiomes of E. antarcticus. In contrast, Bacteroidota was the sole indicative phylum of the gut microbiomes of P. murrayi and was also indicative of Tardigrada. Although Patescibacteria was also indicative of Tardigrada, it comprised < 0.1% of all microbiomes and < 0.28% of Tardigrada gut microbiomes. A phylum of predatory bacterium, Bdellovibrionota, was enriched in all gut types (1.02%) vs. mats (0.2%). Due to both their high proportion within communities, and their significance as indicator species, taxa of the three most abundant phyla were selected for further analysis.
Figure 3.
Relative abundance of bacterial communities of mats and microinvertebrate gut microbiomes (a) bacterial phyla, (b) cyanobacterial genera, (c) bacteroidotal genera, and (d) proteobacterial families. Horizontal lines and letters indicate statistical differences (P < 0.05) at the phylum level among the eight communities tested with a GLMM and then Tukey HSD. Error bars represent SE of entire phylum.
There was no effect of mat type in respect to the relative abundance of the three dominant bacterial phyla, but there was a very strong effect of community type (GLMM, P < 1.39e−15, χ2(3) > 72.28, Supplementary Table S7a). Streams significantly affected the abundance of Cyanobacteria and Bacteroidota (P < 0.001, χ2(3) > 14.98), but not of Proteobacteria (P = 0.11, χ2(3) = 5.93). For Cyanobacteria, both mat types were dominated by a similar overall relative abundance (48% and 45% for black and orange, respectively, Fig. 3a, Supplementary Table S5a), but orange mats contained 70.8% more Phormidium, while black mats contained 87.2% more Nostoc (Fig. 3b, Supplementary Table S5b). In comparison to mats, all microinvertebrate gut microbiomes were significantly depleted of Cyanobacteria (Fig. 3b) and when compared to each other contained similar amounts of Cyanobacteria. The contribution of cyanobacterial Nostoc declined from an average 19.2% within both mat types to 0.2% within gut microbiomes. Although cyanobacterial Tychonema was an order of magnitude more abundant than Nostoc within gut microbiomes, its relative abundance was more than twice as abundant in all mats (7.5%) than in the guts (3.4%).
Compared to mats, gut microbiomes of P. murrayi and Tardigrada, but not of E. antarcticus, were similarly enriched in Bacteroidota (Tukey's HSD, P < 0.05) (Fig. 3c, Supplementary Table S5a). Although gut microbiomes of both P. murrayi and Tardigrada were characterized by similar relative abundance of Bacteroidota at the phylum level, at the genus level Tardigrada microbiome taxa were similar to the mats but at larger relative abundances. In contrast, gut microbiomes of P. murrayi were distinct from the mats and Tardigrada and significantly enriched by the single genus Larkinella making up 37% of its gut community compared to 0.03% of all other microbiome types (Fig. 3c, Supplementary Table S5b). In contrast to Tardigrada, P. murrayi, and mats that all contained similar proteobacterial communities, gut microbiomes of E. antarcticus were significantly enriched (Tukey HSD, P < 0.05) by Proteobacteria and that of the family Pseudomonadaceae in particular (Fig. 3d). Other Proteobacteria of interest within E. antarcticus included taxa in Rickettsiaceae, a family recognized for its intracellular symbionts. Wolbachia, a well-known intracellular bacterium was absent from any microbiome. Comamonadaceae, the most abundant family of Proteobacteria across all communities was almost entirely (97.91%) represented by Polaromonas.
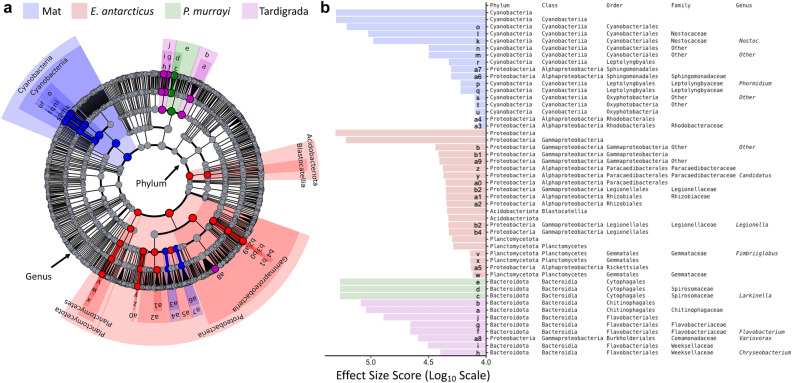
The linear discriminant analysis effect size algorithm (LEfSe) confirmed and further refined these compositional results. LEfSe identified a total of 49 distinctive taxa at different taxonomic ranks specifically affected by the four community types (i.e., mats, E. antarcticus, P. murrayi, Tardigrada) (LDA effect size = 4, P < 0.05) (Fig. 4), but no distinctive taxa when testing for stream (Canada, Bowles Creek, Delta, Von Guerard) or mat type (black or orange). Cyanobacteria (phylum) and Cyanobacteriia (class) were the two most relevant taxa for the mat communities, while Proteobacteria (phylum) and Gammaproteobacteria (subphylum) were the most relevant for gut communities of E. antarcticus (Fig. 4b). Acidobacteriota and Planctomycetota were two other indicative phyla of E. antarcticus but were lower in effect size score. In contrast, there were no characteristic phylum level taxa of P. murrayi nor Tardigrada gut microbiomes. Instead, the most significant taxa of P. murrayi gut microbiomes were Cytophagales (order), Spirosomaceae (family), and Larkinella (genus) (Fig. 4b), all highlighting taxonomic congruence as Cytophagales and Spirosomaceae are the order and family for Larkinella. The five most significant taxa within Tardigrada hosts were all Bacteroidia (class), and not a single taxon was significant above the order level (Fig. 4b). Highlighting the trophic level similarities, indicative taxa for P. murrayi and all but one taxon of Tardigrada were within the same class (i.e., Bacteroidia). However, all indicative taxa at the order, family, and genus level were host specific. Of indicative bacterial taxa for all groups, very little overlap of host types was observed among the entire bacterial tree (Fig. 4a), with only five of the 49 taxa of one host microbiome being nested within another.
Figure 4.
LEfSe (Linear Discriminant Analysis Effect Size) analysis displaying enrichment of most significant bacterial taxa at different levels of taxonomic resolution within mat, nematode (E. antarcticus and P. murrayi) and Tardigrada microbiomes. Results are visualized in a cladogram with (a) circles representing different ranks of taxonomic classification (phylum as the most inner and genus as most outer circle) and community specific significant clades (P < 0.05) colored in blue (mats), red (E. antarcticus), green (P. murrayi), purple (Tardigrada) determined with Kruskal–Wallis and Wilcoxon test. (b) LDA effect size scores of each significant clade showing its relative importance in comparison to other significant taxa. Clade identities are written on the cladogram itself or abbreviated and located adjacent to full strings at the base of the effect size scores.
Clearly distinct microbiomes within mats and microinvertebrates were also supported by preliminary predictions of their functional characteristics. Using PICRUSt2, functional profiles of mat communities (Supplementary Fig. S3) varied primarily by mat type (PERMANOVA, P = 0.04, F(1) = 2.76), and stream (P = 0.07, F(3) = 1.79) but stream explained more than twice the variation in the model (21.2% vs 10.9%). Similar to bacterial composition data, microinvertebrate gut microbiome functional profiles (Supplementary Fig. S4) statistically varied by both host identity (PERMANOVA, P < 0.01, F(2) = 21.10), mat type (P = 0.04, F(1) = 2.16), and stream (P < 0.01, F(3) = 8.60). However, host identity was the most important factor in the functional profile as it explained more variation (12.9%) than mat type (< 0.1%) or stream (7.2%) did.
Compositional differences among eukaryotic communities
Eukaryotic mat communities were dominated by tardigrades, rotifers, nematodes, and algae (Supplementary Fig. S5). Examining the eukaryotic composition of mats, there was a significant difference between mat types (PERMANOVA, P = 0.03, F(1) = 2.37) and among streams (P < 0.001, F(3) = 6.18), although stream explained a much larger amount of variation (40.7%) than mat type (5.2%) (Supplementary Table S3b). Canada Stream was particularly important for mat communities, making up half of the overall stream explained variation highlighting the uniqueness of mats from this location. Compositional differences of microinvertebrate non-host eukaryotic communities varied by microinvertebrate type. Although the effects of stream, mat type, and host type were all statistically significant to non-host eukaryotic gut communities (P < 0.001, F(1–3) > 1.78), similar to bacterial communities, host identity explained the most variation (3.7%), followed by stream (2.5%), and mat type (0.8%) (Supplementary Table S4b). Indicator species analysis identified Metazoa and Chloroplastida as associated with mat communities (P < 0.001, R2 = 0.69 and 0.43 respectively), while all three microinvertebrates shared the same importance of Fungi (P < 0.001, R2 = 0.69), at the kingdom level of taxonomic ranking.
Among microinvertebrate gut communities, Fungi comprised the largest (67.3%) component of each microbiome, followed by microbial eukaryotes (including the SAR supergroup), and metazoans (Supplementary Fig. S5). In comparison to mats, all microinvertebrate gut microbiomes were significantly (Tukey HSD, P < 3.54e−07) enriched in Fungi (3.3% vs. 48.9–78.5% respectively) (Supplementary Fig. S5b), resulting in a corresponding change in overall community composition. The relative abundance of Fungi in E. antarcticus was significantly lower (P < 0.01) than in P. murrayi and Tardigrada (48.9%, 76.7%, 78.5%, respectively) (Supplementary Fig. S5b). In contrast to mat types, streams affected the fungal abundance within guts (GLMM, P = 0.03, χ2(3) = 9.00, Supplementary Table S7). Compared to mats, all gut microbiomes were dominated by the subphylum Pezizomycotina and depleted of Blastocladiomycota (Tukey HSD, P < 0.05) (Supplementary Fig. S5b). Eurotiomycetes and Leotiomycetes were the most abundant fungal classes (15.7% and 9.5%), and all fungal communities of microinvertebrate guts contained a high diversity of taxa that were compositionally similar.
Metazoans significantly differed in relative abundance among community types (GLMM, P < 0.01, χ2(3) = 84.74), but there was no influence of mat type or stream (P > 0.47, χ2(1,3) < 2.51, Supplementary Table S7c). Non-host metazoan ASVs were significantly more common in the gut microbiome of E. antarcticus than P. antarcticus and Tardigrada (Tukey HSD, P < 0.001, Supplementary Fig. S5a). E. antarcticus eukaryotic communities were mostly of tardigrade (22.9%) and rotifer origin (5.2%) (Supplementary Fig. S5c). Interestingly, rotifer ASVs were detected in all microinvertebrate guts at similar relative abundances (Tukey HSD, P > 0.32). P. murrayi reads were detected in only a single specimen of E. antarcticus, and in seven out of 94 Tardigrada samples P. murrayi reads were in extremely low abundance (< 0.1%). No reads of E. antarcticus were found in any guts of P. murrayi nor those of tardigrades.
Microinvertebrate haplotypes
As expected, E. antarcticus and P. murrayi host 18S ASV data indicated a single species of each across all samples. However, host 18S ASV data indicated the potential presence of three molecular haplotypes of tardigrades all with equal assignments to known Dry Valley tardigrades (possibly Hypsibiidae Acutuncus antarcticus or Macrobiotidae Richtersius and Paramacrobiotus, or Milnesiidae Milnesium). None of the alpha diversity metrics tested nor the relative abundance of the three dominant bacterial phyla (i.e., Cyanobacteria, Proteobacteria, Bacteroidota) varied among the three haplotypes (GLM, GLMM, P > 0.43, χ2(2) < 4.21) (Supplementary Fig. S6). Although the overall bacterial community composition did statistically vary with stream and haplotype explaining 9.9% and 8.1% variation, respectively, they ordinated with complete overlap in NMDS space (Supplementary Fig. S7). Therefore, three possible host haplotypes were combined for this study due to undiscerned haplotype identity.
Discussion
Our study expands the phylogenetic coverage of nematode and tardigrade gut microbiomes as well as provides new insights into the significance of specific factors of their assembly. While many animals develop well-defined microbiomes, this pattern is not universal. Instead, microbial gut communities may be reflective of the surrounding environment, be random, or even absent5. Given that the number of described microbiomes of nematodes and tardigrades thus far is limited, which factors might drive their gut assembly have been hard to predict. Due to the importance of nematodes and tardigrades in ecosystem functioning16, it is critical to examine a wide range of hosts from different habitats.
As we hypothesized, microinvertebrate gut microbiomes were better explained by host identities than by environmental factors (e.g., mat type, stream). We observed that all gut microbiomes were less diverse and compositionally distinct from the microbial community of the environment they inhabited for both bacterial and eukaryotic components. At a coarse level of taxonomic resolution (phylum), indicator analysis and statistical tests of relative abundance suggested that microbiomes of bacterivorous P. murrayi and tardigrades were similar, and both were significantly distinct from the omnivorous E. antarcticus potentially indicating a role of feeding traits on gut microbiome assembly. However, at a finer level of taxonomic resolution, the LEfSe algorithm and relative abundance of orders, families, and genera showed that host identity was the most dominant factor in the assembly of nematode and tardigrade gut microbiomes. Although the potential role of environmental factors was also observed (e.g., mat type and stream), these factors explained less variation than the identity of all three microinvertebrates. These results are largely in line with the literature on C. elegans indicating the presence of a conserved gut microbiome that is relatively independent from their surrounding substrate25. Interestingly, previously described guts of C. elegans have been dominated by Proteobacteria25 rather than the Bacteroidota that dominated the guts of P. murrayi examined here. Although both nematode species are bacterivores, their different habitat preferences (rotting fruit vs. aquatic cyanobacterial mats) and perhaps phylogenetic placement (Rhabditida vs. Plectida) could be more important in the assembly of gut microbiomes than feeding traits. Nevertheless, Proteobacteria was still a major component of the P. murrayi microbiome. Proteobacteria was also the dominant gut component of the omnivorous E. antarcticus, similar to the sole other study of a terrestrial omnivorous nematode Dorylaimus stagnalis43, suggesting its possible importance to the entire nematode phylum. Tardigrade gut microbiomes were even more enriched in both Bacteroidota and Proteobacteria than in previous reports26,27 suggesting a functional role of these two bacterial phyla in nematode and tardigrade gut microbiomes. Flavobacterium and Ferruginibacter were the two most abundant genera of Bacteroidota across all microbiomes, as has been observed in other Antarctic tardigrades26 and specifically fully fed (i.e., not starved) Antarctic tardigrades30, suggesting this as a reoccurring food source. Interestingly, the LEfSe algorithm identified no phylum level significance for P. murrayi or tardigrades, but it did for E. antarcticus. This supports the idea of E. antarcticus as an omnivore not only for feeding on a wide range of food categories (i.e., fungi, bacteria, algae, micrometazoa), but also as a generalist feeding of taxa within each feeding category (i.e., many types of bacteria). However, individual E. antarcticus were less diverse than all other microinvertebrates, indicating that although a population of E. antarcticus can feed on a wide range of taxa, individuals do not.
The abstraction of “host identity” likely involves a range of species-specific factors that together may influence the assembly of gut microbiomes. For example, although many nematodes and tardigrades are generally categorized as bacterivorous at the family or genus level, species-specific feeding habits44 could impose selective pressure on gut microbiome assembly. This could be explained by the fact that any potential component of gut microbiomes must first pass through the animal’s mouth likely acting as a selective barrier against certain bacterial taxa, with the size of an animal’s stoma imposing a layer of selection against bacteria too large to enter the mouth and digestive system. Easier intake of smaller bacteria by C. elegans45–47 would support this notion. Likewise, the depletion of large sized Nostoc (~ 4 μm diameter per bacterium48 and 2 cm–20 cm for colonies) from all microinvertebrate guts in our study could be explained by a small stoma size (~ 3 μm diameter for P. murrayi49), although it is unclear if E. antarcticus could pierce the larger cells with its odontostylet. Nematode esophageal morphology may also play a role in what can colonize the gut instead of simply passing through the nematode. For example, TEM imaging of C. elegans and P. pacificus cultured on E. coli showed broken vs. unbroken E. coli cells, respectively due to the presence vs. absence of an esophageal grinder50. Since P. murrayi contains an esophageal grinder, any bacteria able to colonize the gut must be small and/or resistant enough to maintain viability through this maceration step.
In addition to the filtering of communities resulting from morphological constraints, behavioral factors could also be important. For example, using food preference assays, it has been shown that C. elegans can discriminate between higher and lower quality (C:N ratios) bacteria46. This ability to discriminate among the quality of their bacterial food sources has been even observed for nematodes incapable of feeding on bacteria (e.g., plant parasitic nematodes)51, suggesting food quality sensing is an important factor to consider for all nematode microbiomes. However, it is important to remember that nematodes can be cultured on bacteria that do not colonize the gut. Other poorly understood behavioral factors may also play a role. For example, Plectus species from the Antarctic coastline showed preferences for feeding on proteobacterial Polaromonas52, the most abundant taxon of the family Comamonadaceae observed in all our microinvertebrates as well as mats. In addition, Polaromonas has been previously reported within Antarctic tardigrade microbiomes30, suggesting that Polaromonas may be a common food source for Antarctic microinvertebrates. Unfortunately, apart from a handful of feeding assay studies, very little is known about actual feeding preferences of most nematodes and tardigrades, hindering our understanding of their role in ecosystem functioning. Gut microbiome studies offer a novel mechanism to expediently expand this knowledge as has been done for other animals53.
Not everything that passes into the nematode or tardigrade gut can colonize the host and instead some bacteria may be nothing more than transient contents. Although it is methodologically hard to distinguish between the two, it has been shown that specimens of C. elegans with established diverse microbiomes in the natural environment maintain their gut microbiomes even when transferred to a culture of E. coli20, suggesting that once established, nematode gut microbiomes can be stable and resistant to change. A small proportion of gut microbiome taxa were intriguingly absent from mats (13% of genera within E. antarcticus, 22% of P. murrayi, 25% of tardigrades), although they were all in low abundance and of common phyla. However, it is possible that their lack of presence in mats was an artifact of the lower number of analyzed mat samples (24) than guts (251). Overall, all microinvertebrate species in this study displayed species-specific bacterial microbiomes with little variation across streams and mats suggesting that gut communities examined in this study were composed of actual resident microbes, but further study in this area is warranted.
Wolbachia, a common intracellular parasite sometimes reported in nematodes and tardigrades28 was not found within any microinvertebrate guts in this study. However, other intracellular bacteria within the sister family of Rickettsiaceae were detected, albeit at very low abundance. We also observed that Larkinella sp. was both the most abundant and the most indicative of any bacteria within P. murrayi guts. Not much is known about the function of this genus, but isolates grow into pink, horseshoe shaped cells54, opening up the possibility for future in vitro studies. Currently however, any possible symbiotic function, or reason Larkinella may be so enriched in P. murrayi guts is unknown.
All three microinvertebrates sampled in this study contained eukaryotic gut microbiomes primarily consisting of fungi. Fungal microbiomes (i.e., mycobiomes) have only been reported once for marine nematodes32 and once for tardigrades30, both suggesting well developed mycobiomes. Our data shows that the most abundant fungal ASVs of all hosts were represented by ascomycotous taxa such as Tetracladium furcatum and Pyronemataceae, both aquatic fungi known to feed on decomposing matter in streams. While the presence of fungi in the microbiome of an omnivorous nematode (E. antarcticus) is not surprising, it is interesting that the presumed bacterial-feeding P. murrayi contained higher levels of fungi than the omnivore. This could indicate that previously assigned nematode feeding categories may not be reflective of the exact nematode functional roles in the ecosystem, as P. murrayi appears to be intaking fungi even if it does not derive nutrition from it. Interestingly, fungal communities between all microinvertebrates guts were mostly similar, but distinct from previously recorded tardigrade mycobiomes from Italian glaciers30, suggesting a possible geographic role of community assembly. In our analysis of non-host eukaryotic gut contents, we found evidence to support the presumption of E. antarcticus as the sole omnivore/predator in this ecosystem. Its gut microbiome was characterized by the highest relative abundance of metazoan ASVs, mostly originating from tardigrades and rotifers. Although both P. murrayi and tardigrades did contain low relative abundance of other metazoans, especially rotifers, this is most likely a result of the filtering of free-floating DNA from broken cells rather than direct predation. Furthermore, there was no evidence of contamination in the negative controls. High numbers of P. murrayi reads observed within a single E. antarcticus gut could indicate that E. antarcticus may not consistently feed on other nematodes but retains the capability to do so.
Although host identity explained the largest proportion of microbiome variation in this study, most of the variation remained unexplained suggesting other factors may be at play including abiotic environmental factors, stochasticity, biogeography, or inter-microbiome interactions. Prior research of C. elegans has shown the presence of interbacterial competition within guts55, with greater substrate diversity resulting in less stochastic gut microbiomes56,57. Nevertheless, we show that the host identity (rather than feeding traits) relative to environmental factors measured is likely among the more important factors shaping the gut microbiomes of nematodes and tardigrades living in the Antarctic Dry Valleys streams. Although limited in coverage, the functional profiles referred from PICRUSt2 were in line with compositionally unique microbiomes suggesting their differential functional roles in this ecosystem. However, varying influence of host identity has been observed for Nematoda and so to confirm the role of host identity and any functional roles, additional studies using a wider range of species from a diverse range of geographic locations are needed.
Conclusions
Nematode and tardigrade gut microbiomes have only been described in a select few species, underrepresenting the wide phylogenetic and trophic diversity within these phyla. Of these, even fewer studies have been conducted to examine what drives the assembly of these microbial communities. Using cyanobacterial mats of Antarctic Dry Valleys streams as a natural model ecosystem, we were able to show that nematodes and tardigrades living in these mats have gut microbiomes that are both distinct from each other and from the surrounding microbial community they inhabit. Although at the bacterial phylum level, similar trophic level (bacterial-feeding) but phylogenetically distinct microinvertebrates (P. murrayi and tardigrades) had more similar microbiomes than the two phylogenetically closer (P. murrayi and E. antarcticus) but distinct trophic level (bacterivorous and omnivorous) nematodes, host identity was the most important factor of assembly at more precise levels of taxonomic classification. However, an examination of additional species representing the full range of trophic and phylogenetic diversity is required to fully support this idea.
Methods
Sample collection and DNA processing
Cyanobacterial mats were collected from four seasonally active streams (1. Canada, 2. Bowles Creek, 3. Delta, 4. Von Guerard) in the Lake Fryxell Basin (77°36′21″S 163°07′32″E) in Taylor Valley, Antarctica (Fig. 1). Stream length varied from 0.49 km (Bowles Creek) to 5.89 km (Delta Stream) with an average length of 3.2 ± 2.3 km. Canada Stream and Bowles Creek both flow from the Canada Glacier located within the Asgard Range, while Delta Stream and Von Guerard Stream flow from glaciers located within the Kukri Hills. Within each stream, one representative circular plot (2 m radius) with both “black” and “orange” mat types was randomly selected upstream of any flow gauges. In January 2019, three replicates of each mat type were collected by cutting out one 7 cm × 7 cm piece with a sterile scalpel and placing each into sterile 100 mL glass bottles for a total of 24 mat samples (4 streams × 2 mat types × 3 replicates). Samples were frozen at − 20 °C, transported to the University of Florida, and stored at − 20 °C until processing. Mats were slowly defrosted to 4 °C (10 °C increase every 24 h) and examined in sterile Petri dishes under a dissecting microscope. Microinvertebrate individuals visually identified as Eudorylaimus antarcticus (n = 88) (currently undergoing taxonomic revaluation), Plectus murrayi (n = 176), and tardigrades (n = 94) were collected with a metal pick before being physically moved and agitated through 3 washes of cold sterile water. Previous studies have established similar processes to effectively remove externally adherent microorganisms22,32. Following washing, single microinvertebrate individuals were placed in separate 250 μL microcentrifuge tubes containing 25 μL of lysis buffer (800 μg/mL proteinase K, 0.2 M NaCl, 0.2 M Tris–HCL pH 8.0, 1% beta-mercaptoethanol)58. Tubes were inspected under a dissecting scope to confirm that only a single microinvertebrate was placed into the lysis buffer. Negative control tubes (n = 7) of lysis buffer, without any microinvertebrates, were prepared in the same manner. Total host and microbiome DNA was extracted by incubating tubes at 65 °C for 120 min to maximize activity of the proteinase K enzyme, followed by 100 °C for 10 min to inactivate it. Substrate mat DNA was extracted from 300 μL of mixed mat slurry using a DNeasy PowerSoil Kit (QIAGEN) according to manufacturer’s protocols. High-throughput metabarcoding was used to characterize bacterial and eukaryotic microbial communities present within microinvertebrate individuals and the mat substrate they were isolated from. Primers and PCR conditions from the Earth Microbiome Project were used to amplify 16S (515F/926R) and 18S (1391f./EukBr) rRNA gene markers59 (Supplemental Methods S1). PCR product was then visualized using gel electrophoresis to confirm successful amplification. Three PCR replicates (each amplified using 1μL of DNA template) with negative controls were pooled and sent to the Hubbard Center for Genome Studies, University of New Hampshire for the attachment of indexes (using Golay barcodes)59, library preparation, and paired-end sequencing on an Illumina HiSeq 2500 (2 × 250 bp) (Illumina Inc., CA, USA).
Bioinformatics and community processing
Following read demultiplexing by the sequencing facility, processing of reads was done using QIIME260. First, adapter and primer sequences were removed with cutadapt61 and quality filtering was performed in QIIME2 with the quality-filter plugin to trim reads by removing base pairs falling below an average quality score of 30. Reverse 16S reads were discarded due to an insufficient length after quality filtering and unpaired forward reads (196 bp) were denoised to create 100% similarity ASVs using the DADA2 pipeline62. Forward and reverse 18S reads were trimmed to 125 bp before joining within DADA2 to create 100% ASVs with an average length of 131 bp and maximum of 172 bp. For both pipelines, chimeras were removed using the built in DADA2 algorithms62. Taxonomy was assigned to each ASV using the assign_taxonomy.py script with BLAST in QIIME1.963, comparing against the SILVA v138 database for 16S and SILVA v111 for 18S, with both filtered to remove all reference sequences identified as “uncultured”64. Community filtering was conducted using the phyloseq package65. Non-bacterial sequences in the 16S ASV table and non-eukaryotic sequences in the 18S ASV table were removed. In addition, 18S sequences with hits to the specific microinvertebrate host were removed only from that host’s ASV table, as were 18S ASVs with poor assignments (i.e., below 90% of query coverage and 95% ID) removed from all hosts. Sequences identified in negative controls were subtracted from experimental samples. Due to the uncertain nature of the cyanobacterial clade of Phormidum/Phormidesmis66–68, reads assigned to Phormidesmis were assigned to Phormidium. Finally, samples with less than 100 total 16S reads and 100 18S reads were discarded based on species rarefaction curves reaching a horizontal asymptote. Furthermore, 9 additional samples of E. antarcticus were removed as extreme outliers of uncharacteristically low diversity. After processing and filtering, 4,233,899 bacterial reads and 1,040,761 eukaryotic reads were recovered across a total of 251 microinvertebrate (52 for E. antarcticus, 110 for P. murrayi, and 89 for tardigrades) and 24 mat samples. For simplicity we define and refer to a “microbiome” as the entire detected microbial community within a sample but recognize that further research may distinguish between resident and transient organisms within the gut.
Statistics and visualization
Statistics were performed in R Version 3.6.169 (http://www.r-project.org). Alpha diversity metrics (i.e., ASV Richness, Simpson’s, Shannon’s, and Faith’s Phylogenetic Diversity) were calculated with Hill Numbers using hill_taxa from the hillR package70. The effects of community type (mat, E. antarcticus, P. murrayi, Tardigrada), mat types (black, orange), streams (Canada, Bowles Creek, Delta, Von Guerard), and selected interactions on different measures of alpha diversity were tested using the same general linear model (GLM) with a standard normal distribution (AlphaDiversityMetric ~ Community*Mat*Stream). The significance of the tested variables in the models were evaluated by P and χ2 using a type II sum of squares ANOVA. All models were constructed for accuracy and selected based on examining residuals with the DHARMa package71. Tukey's HSD was used to compare significance between host identities of different communities (e.g., black mats compared to P. murrayi from orange mats, tardigrades from orange mats compared to tardigrades from black mats). Overall community compositional differences among mats as well as among microinvertebrate microbiomes were based on separate Bray Curtis dissimilarity matrices (one for only mats and one for only microinvertebrates) in order to more accurately evaluate the importance (R2) of examined factors to each group and were tested using permutational analysis of variance (PERMANOVA) with 9,999 permutations with the adonis function in vegan 2.5–772, and significance evaluated using P and F values. The PERMANOVA for mats was run using (DistanceMatrix ~ Mat*Stream), and for microinvertebrates with (DistanceMatrix ~ HostID*Mat*Stream). Ordinations were visualized using NMDS ordination plots made with the distance function within phyloseq65. Generalized linear mixed models (GLMMs) constructed with glmmTMB from the glmmTMB package were performed to test the effect of streams (Canada, Bowles Creek, Delta, Von Guerard), mat types (black, orange), community types (mat, E. antarcticus, P. murrayi, tardigrade), and their interactions on the relative abundances of selected bacterial phyla and genera, with all factors as fixed effects and stream as an additional random effect along with a beta distribution to account for overdispersion73 (RelativeAbundance ~ Community*Mat + Mat*Stream + Community*Stream + (1|Stream)). The significance of the tested variables in GLMM models were evaluated by P and χ2. To identify the most important taxa to microbial communities at the phylum level, indicator species analysis was conducted with the indicspecies package with 9,999 permutations74. In addition, linear discriminant analysis effect size (LEfSe) was performed down to the genus level75. The LEfSe algorithm identifies characteristic taxa that are both mathematically (Kruskal–Wallis sum-rank test P < 0.05) and biologically informative through the use of consistency assessments (unpaired Wilcoxon rank-sum tests P < 0.05) and effect size (LDA)75. LEfSe provides a taxonomically ranked examination of microbial communities that is not possible with other statistical tools, as for example LEfSe can highlight a family as indicative of a host microbiome, but not the genus within it or the order above it. For our analyses, we applied an LDA effect size score of 4 using the one-against-all method and otherwise default LEfSe parameters on the Huttenhower lab Galaxy server75 (https://huttenhower.sph.harvard.edu/). The LEfSe cladogram was exported from the Huttenhower lab Galaxy server and modified in Adobe illustrator for clarity. Initial prediction of the functional profile within microbial communities was performed using PICRUSt276 and tested using the PERMANOVA equations described above, but was not the focus of this study given the limitations of using amplicon data for predicting function77 and a limited database of Antarctic bacterial genomes. Sampling map was created using ArcGIS. All other figures were created in R using ggplot2.
Supplementary Information
Acknowledgements
The authors would like to thank Josh Darling and the McMurdo Dry Valleys LTER for the collection of samples used in this study. This work was funded by the NSF Office of Polar Programs Awards 1443578 and 1443373, and the University of Florida.
Author contributions
J.P.M. and D.L.P. conceived the study. D.L.P. and P.S. selected sampling sites. J.P.M. performed the lab work. J.P.M., E.G. and D.L.P. analyzed the data. K.G., E.G., and D.L.P. assisted with interpretation of the results. J.P.M. and D.L.P. wrote the manuscript and all authors edited and approved the manuscript. S.K.S. and D.L.P. provided funding.
Data availability
Raw reads are available at the NCBI Sequence Read Archive with the project ID PRJNA799934 (https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA799934). Documented code for the full bioinformatic pipeline, figure creation, and statistical analysis is available at http://www.wormsetal.com/antarcticstreamgutmicrobiomes and www.github.com/WormsEtAl/AntarcticStreamGutMicrobiomes.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-24206-5.
References
- 1.Gilbert SF, Sapp J, Tauber AI. A symbiotic view of life: We have never been individuals. Q. Rev. Biol. 2012;87:325–341. doi: 10.1086/668166. [DOI] [PubMed] [Google Scholar]
- 2.Brooks AW, et al. Phylosymbiosis: Relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 2016;14:e2000225. doi: 10.1371/journal.pbio.2000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammer TJ, et al. Caterpillars lack a resident gut microbiome. P. Natl. Acad. Sci. USA. 2017;114:9641–9646. doi: 10.1073/pnas.1707186114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deb R, Nair A, Agashe D. Host dietary specialization and neutral assembly shape gut bacterial communities of wild dragonflies. PeerJ. 2019;7:e8058. doi: 10.7717/peerj.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammer TJ, Sanders JG, Fierer N. Not all animals need a microbiome. FEMS Microbiol. Lett. 2019;366:fnz117. doi: 10.1093/femsle/fnz117. [DOI] [PubMed] [Google Scholar]
- 6.Sanders JG, et al. Dramatic differences in gut bacterial densities correlate with diet and habitat in rainforest ants. Integr. Comp. Biol. 2017;57:705–722. doi: 10.1093/icb/icx088. [DOI] [PubMed] [Google Scholar]
- 7.Hu Y, et al. Herbivorous turtle ants obtain essential nutrients from a conserved nitrogen-recycling gut microbiome. Nat. Commun. 2018;9:964. doi: 10.1038/s41467-018-03357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu DW, Davidson DW. Experimental Studies of Species-Specificity in Cecropia-Ant Relationships. Ecol. Monogr. 1997;67:273. doi: 10.2307/2963456. [DOI] [Google Scholar]
- 9.Zepeda-Mendoza ML, et al. Hologenomic adaptations underlying the evolution of sanguivory in the common vampire bat. Nat. Ecol. Evol. 2018;2:659–668. doi: 10.1038/s41559-018-0476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song SJ, et al. Comparative analyses of vertebrate gut microbiomes reveal convergence between birds and bats. MBio. 2020;11:e02901–e2919. doi: 10.1128/mBio.02901-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodda M. Phylum Nematoda: Trends in species descriptions, the documentation of diversity, systematics, and the species concept. Zootaxa. 2022;5114:290–317. doi: 10.11646/zootaxa.5114.1.2. [DOI] [PubMed] [Google Scholar]
- 12.Bongers T, Bongers M. Functional diversity of nematodes. Appl. Soil Ecol. 1998;10:239–251. doi: 10.1016/S0929-1393(98)00123-1. [DOI] [Google Scholar]
- 13.Dunn CW, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 14.Philippe H, Lartillot N, Brinkmann H. Multigene analyses of bilaterian animals corroborate the Monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia. Mol. Biol. Evol. 2005;22:1246–1253. doi: 10.1093/molbev/msi111. [DOI] [PubMed] [Google Scholar]
- 15.Schill RO. Water Bears: The Biology of Tardigrades. Springer International Publishing; 2018. [Google Scholar]
- 16.Neher DA. Role of nematodes in soil health and their use as indicators. J. Nematol. 2001;33:161–168. [PMC free article] [PubMed] [Google Scholar]
- 17.Nicol JM, et al. Current nematode threats to world agriculture. In: Jones J, Gheysen G, Fenoll C, et al., editors. Genomics and Molecular Genetics of Plant-Nematode Interactions. Springer; 2011. pp. 21–43. [Google Scholar]
- 18.Ladygina N, et al. Diversity of bacteria associated with grassland soil nematodes of different feeding groups: Bacteria associated with grassland soil nematodes. FEMS Microbiol. Ecol. 2009;69:53–61. doi: 10.1111/j.1574-6941.2009.00687.x. [DOI] [PubMed] [Google Scholar]
- 19.Baquiran J-P, et al. Culture-Independent Investigation of the Microbiome Associated with the Nematode Acrobeloides maximus. PLoS ONE. 2013;8:e67425. doi: 10.1371/journal.pone.0067425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dirksen P, et al. The native microbiome of the nematode Caenorhabditis elegans: Gateway to a new host-microbiome model. BMC Biol. 2016;14:38. doi: 10.1186/s12915-016-0258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berg M, Zhou XY, Shapira M. Host-specific functional significance of caenorhabditis gut commensals. Front. Microbiol. 2016;7:1622. doi: 10.3389/fmicb.2016.01622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berg M, et al. Assembly of the Caenorhabditis elegans gut microbiota from diverse soil microbial environments. ISME J. 2016;10:1998–2009. doi: 10.1038/ismej.2015.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samuel BS, et al. Caenorhabditis elegans responses to bacteria from its natural habitats. Proc. Natl. Acad. Sci. USA. 2016;113:E3941–E3949. doi: 10.1073/pnas.1607183113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmermann J, et al. The functional repertoire contained within the native microbiota of the model nematode Caenorhabditis elegans. ISME J. 2020;14:26–38. doi: 10.1038/s41396-019-0504-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F, et al. Caenorhabditis elegans as a model for microbiome research. Front. Microbiol. 2017;8:485. doi: 10.3389/fmicb.2017.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vecchi M, et al. The microbial community of tardigrades: environmental influence and species specificity of microbiome structure and composition. Microb. Ecol. 2018;76:467–481. doi: 10.1007/s00248-017-1134-4. [DOI] [PubMed] [Google Scholar]
- 27.Guidetti R, et al. Further insights in the Tardigrada microbiome: phylogenetic position and prevalence of infection of four new Alphaproteobacteria putative endosymbionts. Zool. J. Linn. Soc. 2019;188:925–937. doi: 10.1093/zoolinnean/zlz128. [DOI] [Google Scholar]
- 28.Kaczmarek Ł, et al. Integrative description of bisexual Paramacrobiotus experimentalis sp. Nov. (Macrobiotidae) from republic of Madagascar (Africa) with microbiome analysis. Mol. Phylogenet. Evol. 2020;145:106730. doi: 10.1016/j.ympev.2019.106730. [DOI] [PubMed] [Google Scholar]
- 29.Mioduchowska M, et al. Taxonomic classification of the bacterial endosymbiont Wolbachia based on next-generation sequencing: is there molecular evidence for its presence in tardigrades? Genome. 2021;64:951–958. doi: 10.1139/gen-2020-0036. [DOI] [PubMed] [Google Scholar]
- 30.Zawierucha K, et al. Trophic and symbiotic links between obligate-glacier water bears (Tardigrada) and cryoconite microorganisms. PLoS ONE. 2022;17:e0262039. doi: 10.1371/journal.pone.0262039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derycke S, et al. Coexisting cryptic species of the Litoditis marina complex (Nematoda) show differential resource use and have distinct microbiomes with high intraspecific variability. Mol. Ecol. 2016;25:2093–2110. doi: 10.1111/mec.13597. [DOI] [PubMed] [Google Scholar]
- 32.Schuelke T, Pereira TJ, Hardy SM, Bik HM. Nematode-associated microbial taxa do not correlate with host phylogeny, geographic region or feeding morphology in marine sediment habitats. Mol. Ecol. 2018;27:1930–1951. doi: 10.1111/mec.14539. [DOI] [PubMed] [Google Scholar]
- 33.Doran PT. Valley floor climate observations from the McMurdo dry valleys, Antarctica, 1986–2000. J. Geophys. Res. 2002;107:4772. doi: 10.1029/2001JD002045. [DOI] [Google Scholar]
- 34.Fountain AG, et al. Physical controls on the taylor valley ecosystem, antarctica. Bioscience. 1999;49:961–972. doi: 10.1525/bisi.1999.49.12.961. [DOI] [Google Scholar]
- 35.McKnight D, et al. Dry Valley streams in antarctica: Ecosystems waiting for water. Bioscience. 1999;49:985–995. doi: 10.1525/bisi.1999.49.12.985. [DOI] [Google Scholar]
- 36.Van Horn DJ, et al. Patterns of bacterial biodiversity in the glacial meltwater streams of the McMurdo Dry Valleys, Antarctica. FEMS Microbiol. Ecol. 2016;92:fiw148. doi: 10.1093/femsec/fiw148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams BJ, et al. Diversity and distribution of Victoria Land biota. Soil Biol. Biochem. 2006;38:3003–3018. doi: 10.1016/j.soilbio.2006.04.030. [DOI] [Google Scholar]
- 38.Treonis AM, Wall DH, Virginia RA. Invertebrate biodiversity in Antarctic Dry Valley soils and sediments. Ecosystems. 1999;2:482–492. doi: 10.1007/s100219900096. [DOI] [Google Scholar]
- 39.Porazinska DL, Wall DH, Virginia RA. Population age structure of nematodes in the Antarctic Dry Valleys: perspectives on time, space, and habitat suitability. Arct. Antarct. Alp. Res. 2002;34:159–168. doi: 10.1080/15230430.2002.12003480. [DOI] [Google Scholar]
- 40.Wood FH. Nematode feeding relationships. Soil Biol. Biochem. 1973;5:593–601. doi: 10.1016/0038-0717(73)90049-7. [DOI] [Google Scholar]
- 41.Wall DH. Global change tipping points: Above- and below-ground biotic interactions in a low diversity ecosystem. Philos. Trans. R. Soc. B. 2007;362:2291–2306. doi: 10.1098/rstb.2006.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw EA, et al. Stable C and N isotope ratios reveal soil food web structure and identify the nematode Eudorylaimus antarcticus as an omnivore–predator in Taylor Valley, Antarctica. Polar Biol. 2018;41:1013–1018. doi: 10.1007/s00300-017-2243-8. [DOI] [Google Scholar]
- 43.Zheng F, et al. Mineral and organic fertilization alters the microbiome of a soil nematode Dorylaimus stagnalis and its resistome. Sci. Total Environ. 2019;680:70–78. doi: 10.1016/j.scitotenv.2019.04.384. [DOI] [PubMed] [Google Scholar]
- 44.Venette R, Ferris H. Influence of bacterial type and density on population growth of bacterial-feeding nematodes. Soil Biol. Biochem. 1998;30:949–960. doi: 10.1016/S0038-0717(97)00176-4. [DOI] [Google Scholar]
- 45.Fueser H, Rauchschwalbe M-T, Höss S, Traunspurger W. Food bacteria and synthetic microparticles of similar size influence pharyngeal pumping of Caenorhabditis elegans. Aquat. Toxicol. 2021;235:105827. doi: 10.1016/j.aquatox.2021.105827. [DOI] [PubMed] [Google Scholar]
- 46.Shtonda BB, Avery L. Dietary choice behavior in Caenorhabditis elegans. J. Exp. Biol. 2006;209:89–102. doi: 10.1242/jeb.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang-Yen C, Avery L, Samuel ADT. Two size-selective mechanisms specifically trap bacteria-sized food particles in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2009;106:20093–20096. doi: 10.1073/pnas.0904036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugiura K, Itoh S. Single-cell confocal spectrometry of a filamentous cyanobacterium Nostoc at room and cryogenic temperature. Diversity and Differentiation of pigment systems in 311 cells. Plant Cell Physiol. 2012;53:1492–1506. doi: 10.1093/pcp/pcs093. [DOI] [PubMed] [Google Scholar]
- 49.Maggenti AR. Revision of the genus Plectus (Nematoda: Plectidae) P. Helm. Soc. Wash. 1961;28:139–166. [Google Scholar]
- 50.Rae R, et al. Isolation of naturally associated bacteria of necromenic Pristionchus nematodes and fitness consequences. J. Exp. Biol. 2008;211:1927–1936. doi: 10.1242/jeb.014944. [DOI] [PubMed] [Google Scholar]
- 51.Tahseen Q, Clark IM. Attraction and preference of bacteriophagous and plant-parasitic nematodes towards different types of soil bacteria. J. Nat. Hist. 2014;48:1485–1502. doi: 10.1080/00222933.2013.873088. [DOI] [Google Scholar]
- 52.Newsham KK, Rolf J, Pearce DA, Strachan RJ. Differing preferences of Antarctic soil nematodes for microbial prey. Eur. J. Soil Biol. 2004;40:1–8. doi: 10.1016/j.ejsobi.2004.01.004. [DOI] [Google Scholar]
- 53.Kartzinel TR, et al. DNA metabarcoding illuminates dietary niche partitioning by African large herbivores. Proc. Natl. Acad. Sci. USA. 2015;112:8019–8024. doi: 10.1073/pnas.1503283112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vancanneyt M, et al. Larkinella insperata gen. nov., sp. nov., a bacterium of the phylum ‘Bacteroidetes’ isolated from water of a steam generator. Int. J. Syst. Evol. Microbiol. 2006;56:237–241. doi: 10.1099/ijs.0.63948-0. [DOI] [PubMed] [Google Scholar]
- 55.Ortiz A, Vega NM, Ratzke C, Gore J. Interspecies bacterial competition regulates community assembly in the C. elegans intestine. ISME J. 2021;15:2131–2145. doi: 10.1038/s41396-021-00910-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vega NM, Gore J. Stochastic assembly produces heterogeneous communities in the Caenorhabditis elegans intestine. PLoS Biol. 2017;15:e2000633. doi: 10.1371/journal.pbio.2000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.King KC, et al. Rapid evolution of microbe-mediated protection against pathogens in a worm host. ISME J. 2016;10:1915–1924. doi: 10.1038/ismej.2015.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bik HM, Thomas WK, Lunt DH, Lambshead PJD. Low endemism, continued deep-shallow interchanges, and evidence for cosmopolitan distributions in free-living marine nematodes (order Enoplida) BMC Evol. Biol. 2010;10:389. doi: 10.1186/1471-2148-10-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bolyen E, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 62.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raabová L, Kovacik L, Elster J, Strunecký O. Review of the genus Phormidesmis (Cyanobacteria) based on environmental, morphological, and molecular data with description of a new genus Leptodesmis. Phytotaxa. 2019;395:1–16. doi: 10.11646/phytotaxa.395.1.1. [DOI] [Google Scholar]
- 67.Komárek J, Kaštovský J, Ventura S, Turicchia S, Šmarda J. The cyanobacterial genus Phormidesmis. Algol. Stud. 2009;129:41–59. doi: 10.1127/1864-1318/2009/0129-0041. [DOI] [Google Scholar]
- 68.Thomazeau S, et al. The contribution of sub-saharan african strains to the phylogeny of cyanobacteria: focusing on the Nostocaceae (Nostocales, Cyanobacteria) J. Phycol. 2010;46:564–579. doi: 10.1111/j.1529-8817.2010.00836.x. [DOI] [Google Scholar]
- 69.R Core Team. R: A Language and Environment for Statistical Computing (2020).
- 70.Alberdi A, Gilbert MTP. A guide to the application of Hill numbers to DNA-based diversity analyses. Mol. Ecol. Resour. 2019;19:804–817. doi: 10.1111/1755-0998.13014. [DOI] [PubMed] [Google Scholar]
- 71.Hartig, F. DHARMa: Residual Diagnostics for HierARchical Models (2022)
- 72.Oksanen, J. et al. vegan: Community Ecology Package (2020).
- 73.Smithson M, Verkuilen J. A better lemon squeezer? Maximum-likelihood regression with beta-distributed dependent variables. Psychol. Methods. 2006;11:54–71. doi: 10.1037/1082-989X.11.1.54. [DOI] [PubMed] [Google Scholar]
- 74.Cáceres MD, Legendre P. Associations between species and groups of sites: indices and statistical inference. Ecology. 2009;90:3566–3574. doi: 10.1890/08-1823.1. [DOI] [PubMed] [Google Scholar]
- 75.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Douglas GM, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sevigny JL, et al. Marker genes as predictors of shared genomic function. BMC Genomics. 2019;20:268. doi: 10.1186/s12864-019-5641-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads are available at the NCBI Sequence Read Archive with the project ID PRJNA799934 (https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA799934). Documented code for the full bioinformatic pipeline, figure creation, and statistical analysis is available at http://www.wormsetal.com/antarcticstreamgutmicrobiomes and www.github.com/WormsEtAl/AntarcticStreamGutMicrobiomes.