Abstract
Articular cartilage has a limited capacity to self-heal once damaged. Tissue-specific stem cells are a solution for cartilage regeneration; however, ex vivo expansion resulting in cell senescence remains a challenge as a large quantity of high-quality tissue-specific stem cells are needed for cartilage regeneration. Our previous report demonstrated that decellularized extracellular matrix (dECM) deposited by human synovium-derived stem cells (SDSCs), adipose-derived stem cells (ADSCs), urine-derived stem cells (UDSCs), or dermal fibroblasts (DFs) provided an ex vivo solution to rejuvenate human SDSCs in proliferation and chondrogenic potential, particularly for dECM deposited by UDSCs. To make the cell-derived dECM (C-dECM) approach applicable clinically, in this study, we evaluated ex vivo rejuvenation of rabbit infrapatellar fat pad-derived stem cells (IPFSCs), an easily accessible alternative for SDSCs, by the abovementioned C-dECMs, in vivo application for functional cartilage repair in a rabbit osteochondral defect model, and potential cellular and molecular mechanisms underlying this rejuvenation. We found that C-dECM rejuvenation promoted rabbit IPFSCs' cartilage engineering and functional regeneration in both ex vivo and in vivo models, particularly for the dECM deposited by UDSCs, which was further confirmed by proteomics data. RNA-Seq analysis indicated that both mesenchymal-epithelial transition (MET) and inflammation-mediated macrophage activation and polarization are potentially involved in the C-dECM-mediated promotion of IPFSCs’ chondrogenic capacity, which needs further investigation.
Keywords: Decellularized extracellular matrix, Tissue-specific stem cell, Urine-derived stem cell, Rejuvenation, Functional cartilage repair
Graphical abstract
Highlights
-
•
dECM expansion promoted IPFSCs' proliferation and chondrogenic potential.
-
•
UECM-preconditioned IPFSCs contributed to functional cartilage reconstruction.
-
•
The medial condyle displayed lower instantaneous modulus than the lateral condyle.
-
•
Macrophage transition is involved in dECM-mediated IPFSCs' rejuvenation.
-
•
dECM expansion modulates IPFSCs' MET/EMT signals.
1. Introduction
Once injured, articular cartilage does not have self-healing ability, often resulting in fibrocartilage and eventual osteoarthritis in the absence of treatment. Mesenchymal stem cells (MSCs) are a potential cell source for cartilage engineering and regeneration [1]. Given that MSCs located in different tissues have varied capacities to differentiate into a specific cell type [2], increasing evidence indicates that synovium-derived stem cells (SDSCs) are a tissue-specific stem cell for chondrogenic differentiation [3]. As a thin tissue (2–3 cell layers) lining the joints, however, a limited number of cells can be isolated from a synovial tissue biopsy through arthroscopy and harvested synovial tissue is often contaminated with sub-synovial connective tissue. Recent studies demonstrated that infrapatellar fat pad (IPFP)-derived stem cells (IPFSCs) can be used as an alternative stem cell source due to their strong chondrogenic potential and the spontaneous self-healing ability of IPFP [4,5].
Another challenge facing the successful application of stem cells is replicative senescence during ex vivo expansion due to the large number of stem cells needed for transplant [6]. By nature, MSCs are prone to instability because their fate is easily influenced by the surrounding environment [7,8]. Thanks to the wide use of decellularized extracellular matrix (dECM) in tissue engineering and regeneration [1,7], cell-derived dECM (C-dECM) has become an effective approach to rejuvenate MSCs by promoting proliferation and differentiation capacities [9,10] as well as preventing stem cell senescence [11,12] and inflammation [13,14] compared to tissue-derived dECM (T-dECM) [15]. Our recent report assessed human SDSCs' proliferation and chondrogenic capacity after expansion on dECMs from different human cell types, including SDSCs (SECM), adipose-derived stem cells (ADSCs) (AECM), urine-derived stem cells (UDSCs) (UECM), and dermal fibroblasts (DFs) (DECM), with tissue culture plastic (Plastic) as a substrate control [16]. Given that tissue-specific stem cells are significantly influenced by the tissue-specific matrix microenvironment they harbor [2], our initial expectation was that SDSCs grown on SECM could exhibit the greatest chondrogenic potential compared to the Plastic group (non-dECM control) and other dECM control groups including AECM (deposited by ADSCs with limited chondrogenic capacity) [4,5], UECM (deposited by UDSCs without chondrogenic capacity) [17], and DECM (deposited by DFs which are not stem cells). However, we found that human SDSCs exhibited the greatest proliferation and chondrogenic capacity when grown on UECM, followed by those grown on AECM and DECM, with those grown on SECM being the last; all C-dECM groups had superior performance compared to the Plastic group. In this study, we sought to determine whether this ex vivo finding applies to an in vivo cartilage repair model, whether this model works for rabbit IPFSCs, and the potential cellular and molecular mechanisms underlying C-dECM-mediated IPFSCs' rejuvenation. We hypothesized that rejuvenation on dECMs, particularly UECM, would dramatically enhance rabbit IPFSCs’ proliferation and chondrogenic capacity in both ex vivo and in vivo models.
2. Materials and methods
2.1. Preparation of C-dECMs
As described previously [16], human adult ADSCs (cat no. ASC-F-SL, from multiple donors, female, average 43 years old), DFs (cat no. DF-F, from multiple donors, female, average 42 years old), and SDSCs (4 donors, two male and two female, average 43 years old) were obtained from ZenBio Inc. (Research Triangle Park, NC). Human UDSCs (4 donors, male, 20–54 years old) were a generous gift from Dr. Yuanyuan Zhang (Wake Forest Institute for Regenerative Medicine). These cells were pooled to prepare C-dECMs, termed AECM, DECM, SECM, and UECM, respectively, following our protocol [18]. Briefly, Plastic flasks were pre-coated with 0.2% gelatin (cat. no. G9391, MilliporeSigma, St. Louis, MO) followed by treatment with 1% glutaraldehyde solution (cat. no. O2957, Fisher Scientific, Hampton, NH), and 1 M ethanolamine (cat. no. 02400, MilliporeSigma). Passage 5 ADSCs, SDSCs, UDSCs, and DFs were seeded at 6000 cells/cm2 on pre-coated Plastic. Confluent cells were treated with 250 μM l-ascorbic acid phosphate (Wako Chemicals USA Inc., Richmond, VA) for one week [19] in the growth medium [alpha MEM containing 10% fetal bovine serum (FBS) (cat. no. S11150, Bio-Techne, Minneapolis, MN), 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL fungizone]. Then, cells were removed by incubation at 37 °C for 10 min in lysis buffer containing 0.5% Triton X-100 and 20 mM ammonium hydroxide. The C-dECMs incubated in phosphate buffered solution (PBS)-diluted lysis buffer (1։1) at 4 °C overnight were rinsed with PBS followed by storing in PBS containing 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL fungizone at 4 °C until use.
2.2. Immunofluorescence staining of C-dECMs and expanded human cells
The C-dECMs and expanded human cells (to prepare C-dECMs) treated with 1% bovine serum albumin (BSA) (cat. no. BP1600, Fisher Scientific) for 1 h were incubated with primary antibodies against fibronectin [Clone 7.1; cat. no. HFN-3, Developmental Studies Hybridoma Bank (DSHB), Iowa City, IA], type IV collagen (cat. no. M3F7, DSHB), laminin (cat. no. Pa5-16287, Invitrogen, Waltham, MA), and perlecan (cat. no. sc-33707, Santa Cruz Biotechnology, Inc., Dallas, TX). Alexa Fluor Plus 555-conjugated anti-mouse secondary antibody (cat. no. A-32773, Invitrogen) was used to detect fibronectin and type IV collagen. Alexa Fluor 488-conjugated anti-rabbit (cat. no. A-11008, Invitrogen) and Alexa Fluor 488-conjugated anti-Rat (cat. no. A-11006, Invitrogen) were used as secondary antibodies to detect laminin and perlecan, respectively. Fluorescent intensity was visualized under a Zeiss Axiovert 40 CFL Inverted Microscope (Zeiss Oberkochen, Germany) using a 20 × objective lens.
2.3. Flow evaluation of cell proliferation and CD146 expression of rabbit IPFSCs
This experiment was approved by the Institutional Animal Care and Use Committee. IPFP tissue from four New Zealand White rabbits (male, 8 months old) (Envigo Global Services Inc., Denver, PA) were used to collect IPFSCs after a sequential digestion using 0.1% trypsin (Fisher Scientific) for 30 min and 0.1% collagenase P (Roche, Indianapolis, IN) for 2 h to release cells. The stemness of rabbit IPFSCs was characterized previously [5]. The pooled IPFSCs were cultured in growth medium at 37 °C in a humidified 5% CO2 incubator. The medium was changed every other day. Passage 5 rabbit IPFSCs expanded on Plastic and C-dECMs for five days were assessed for cell proliferation using Click-iT™ EdU (5-ethynyl-2′-deoxyuridine) Alexa Fluor™ 647 Flow Cytometry Assay Kit (cat. no. C10419, Invitrogen). Briefly, 5 × 105 expanded cells were treated with 10 μM EdU for 18 h followed by the staining procedure described in the manufacturer's protocol. Surface marker CD146 was also evaluated using flow cytometry. Briefly, 2 × 105 expanded cells were incubated in cold PBS containing 0.1% ChromPure Human IgG whole molecule (Jackson ImmunoResearch Laboratories, West Grove, PA) for 30 min followed by primary antibody CD146-PE (cat. no. 12-1469-42, eBioscience, Fisher Scientific) at 4 °C for 30 min. Fluorescence was assessed by a FACS Calibur (BD Biosciences, San Jose, CA) using the FCS Express software package (De Novo Software, Los Angeles, CA).
2.4. Preparation and evaluation of premature tissue constructs
To track the implanted tissue constructs, passage 2 rabbit IPFSCs were transduced with lentivirus carrying GFP (pRSC-SFFV-Luciferase-E2A-Puro-E2A-GFP-wpre) [20] in the presence of 4 μg/mL of protamine sulfate (MilliporeSigma). After an 8-day incubation on C-dECMs and Plastic, 0.2% collagenase type II was used to retrieve expanded cells by dissolving C-dECMs following our published protocol [18]. 3 × 106 transduced IPFSCs were seeded onto one polyglycolic acid (PLGA) mesh (5 mm diameter × 2 mm thickness; Synthecon, Houston, TX) in a spinner flask for three days according to our previous method [21], followed by a 21-day incubation in a chondrogenic medium containing high-glucose Dulbecco's modified Eagle's medium, 100 nM dexamethasone, 40 mg/mL proline, 0.1 mM l-ascorbic acid-2-phosphate, 100 U/mL penicillin, 100 μg/mL streptomycin, 0.25 μg/mL fungizone, and 1 × ITS™ Premix (Fisher Scientific) with the addition of 10 ng/mL transforming growth factor beta3 (TGFβ3, PeproTech, Rocky Hill, NJ). The diameter of tissue constructs was measured at days 0, 10, and 21 after chondrogenic induction. Expanded cells and Day 10 tissue constructs were evaluated using TaqMan real-time quantitative PCR (RT-qPCR) for chondrogenic marker genes including SOX9 (assay ID: Oc04096872_m1), COL1A1 (assay ID: Oc03396073_g1), COL2A1 (assay ID: Oc03396134_m1), COL10A1 (assay ID: Oc04097225_s1), and ACAN (assay ID: Oc06726465_m1); GAPDH (assay ID: Oc03823402_g1) was used as an endogenous control gene. qPCR was performed using Applied Biosystems™ 7500 Fast Real-Time PCR System (Applied Biosystems, Waltham, MA).
2.5. In vivo creation of osteochondral defects and tissue construct implantation
Adult NZW rabbits (n = 26, male, 3.20 ± 0.20 kg) (Envigo Global Services Inc.) with an average age of 9.1 months were used in this study. An intramuscular injection of midazolam (0.2–3.0 mg/kg) and ketamine (10–30 mg/kg) was given to induce anesthesia, which was maintained with inhalational isoflurane via nose cone. Rabbits were shaved from the hip to the hind legs and surgical site preparation was performed with povidone-iodine and alcohol. An anterior skin incision with subsequent medial parapatellar approach was utilized and the patella was dislocated laterally. A round 4.76 mm diameter × 2.00 mm depth osteochondral defect was created in the midline of the trochlear groove with the distal portion of the defect at the level of the extensor digitorum longus (EDL) insertion utilizing a custom designed hand drill [20]. The defect was then irrigated with 0.9% saline and implanted with premature tissue constructs (n = 8 knees for each C-dECM group), PLGA alone (n = 6 knees), or left untreated (Empty) (n = 6 knees). The patella was relocated taking care to elevate it to prevent chondral injury, and the capsule was repaired with 2-0 running vicryl suture. The superficial skin layer was then closed with a 4-0 running vicryl suture. The procedure was repeated on the contralateral knee. The rabbits were euthanized 26 weeks post operatively for mechanical and histological analyses.
2.6. Mechanical testing
Following exposure of the distal femur through the medial parapatellar approach, a sagittal saw (Stryker System 7, Kalamazoo, MI) was utilized to cut obliquely through the distal femur ensuring the cartilage defect was pointing directly upward for ex vivo mechanical cartilage testing. The excised specimens (n = 6 knees for each group) were stored at −80 °C until mechanical analysis [22,23]. A multiaxial mechanical tester (model Mach-1 v500css, Biomomentum, Canada) was configured with a calibrated 17 N multi-axis load cell and a 1 mm diameter spherical indenter. An automated displacement to each measurement position was accomplished through X and Y horizontal stages. Subsequently, the Z vertical stage was displaced at 0.1 mm/s velocity toward the specimen to determine the contact coordinates of the surface at measurement point and four neighboring positions. Using those contact coordinates, the surface angle was measured, and normal indentation to the articular cartilage was achieved by simultaneously moving the three stages at different speeds using the normal indentation function of the tester. The indentation was performed in stress relaxation with an amplitude of 0.1 mm at 0.1 mm/s velocity and relaxation of 180 s (for 3 positions per zone) and 5 s (for other positions). In order to obtain the cartilage thickness, the spherical indenter was then replaced by a needle probe. The probe was displaced toward the sample at 0.5 mm/s velocity and stopped when the load reached 1 N to ensure penetration in the subchondral bone. From the thickness curve, the cartilage surface and cartilage/subchondral bone interface positions were identified as the vertical thickness. It is necessary to multiply the vertical thickness by the cosine of the surface angle obtained in indentation. Once cartilage thickness was obtained, the stress relaxation curve was fit to an elastic model in indentation [24] to extract the instantaneous modulus at 20% strain, which characterized the dynamic (instantaneous) response of the tissue under load. Finally, the relaxation time was also extracted for positions that had a 180-s relaxation. From the load-time curves, peak load, equilibrium load, and the relaxation time (delay for the load to fall 37% of its peak value) were calculated.
2.7. Histological and immunostaining evaluation
Following excision, the patellar grooves of both knees (n = 8 knees for each C-dECM group, n = 6 knees for the Plastic group, and n = 6 knees for the Empty group) were photographed and fixed in 10% neutral buffered formalin at 4 °C for two days. Each specimen was incubated in 15% ethylenediaminetetraacetic acid (EDTA)/0.1% paraformaldehyde solution for decalcification. A 5-μm thick section of the repaired area was stained using Alcian blue (counterstained with fast red) for sGAG and hematoxylin-eosin staining (H&E) for identification of the intact tidemark line. For immunohistochemical analysis, 1% hydrogen peroxide (H2O2) in methanol was used to inactivate endogenous peroxidase activity. Sections were treated with 2 mg/mL hyaluronidase for 30 min followed by overnight incubation at 4 °C with monoclonal mouse antibodies against type I collagen (cat no. GTX26308, GeneTex, Irvine, CA), type II collagen (cat no. II-II6B3, DSHB), and type X collagen (cat no. GTX37732, GeneTex). Sections for GFP detection were treated with citrate unmasking solution for 20 min followed by overnight incubation at 4 °C with a monoclonal mouse antibody against GFP (4B10, Cell Signaling Technology, Danvers, MA). After washing with PBS, sections were incubated with secondary antibody for 30 min at room temperature. Immunostaining conducted with Vectastain® ABC reagent (Vector Laboratories, Burlingame, CA) was followed by 3,30-diaminobenzidine (DAB) staining and counterstaining was performed with hematoxylin (Vector Laboratories). Tissue sections were graded by three experts blinded to group assignment using a Modified O'Driscoll Scale (MODS) as previously described [20].
2.8. RNA-Seq evaluation
Total RNA was isolated from C-dECM- and Plastic-expanded cells (n = 3) and day 21 pellets (n = 3) using Trizol (Invitrogen) and purified using an RNeasy Mini Kit (Qiagen, Valencia, CA) following manufacturer's instructions. The WVU Core prepared the RNA-Seq library by using a KAPA mRNA HyperPrep Kit (KAPA Biosystems, Wilmington, MA). The Genomics Core of Marshall University sequenced the libraries by using HiSeq 2500. RNA-Seq data analysis was conducted as previously described [5,25]. Briefly, reads were aligned to the Oryctolagus cuniculus genome (oryCun2; rabbit) with subread-align version 2.0.1. Read counts were calculated by featureCounts from Rsubread version 2.0.1 against transcripts annotated from Ensembl (OryCun2.0.98) and summarized at the gene level. Gene expression level was measured by RPKM with an in-house script. Gene Set Enrichment Analysis against gene sets annotated by MSigDB was conducted by GSEA v4.2.3. RNA-Seq sequencing data were deposited to GEO: The following secure token has been created to allow review of record GSE207804 while it remains in private status: ojixwogotlethqn.
2.9. Proteomics evaluation
C-dECM- and Plastic-expanded cells (n = 3) and day 21 (D21) pellet samples (n = 3) were chemically and enzymatically digested prior to liquid chromatography tandem mass spectrometry (LC-MS/MS) as previously described [25,26] with the following exceptions. Digested samples were desalted and concentrated on a Thermo Scientific Pierce C18 tip and analyzed on a Thermo nanoEasy LC II coupled to a Q Exactive HF mass spectrometer (Thermo Fisher Scientific). Peptides were separated on an in-house constructed 20 cm C18 analytical column (100 μm ID) packed with 2.7 μm Phenomenex Cortecs C18 resin. Gradient conditions and MS acquisition parameters were described previously [27]. Raw files were directly loaded into the Proteome Discoverer 2.2 and searched against the human uniprotKB database (release date 2018.08) utilizing the Mascot search engine. Mass tolerances were set to ±10 ppm for MS parent ions and ±25 ppm for MS/MS fragment ions. Trypsin specificity was used allowing for one missed cleavage. Cys carbamidomethylation was set as a fixed modification and variable modifications included Met oxidation, Pro hydroxylation, protein N-terminal acetylation, and peptide N-terminal pyroglutamic acid formation. Search results were exported into an Excel spreadsheet for further processing with figures generated utilizing R. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier MassIVE MSV000089891.
2.10. Statistics analysis
The Mann-Whitney U test was used for comparisons between two conditions. All statistical analyses were performed with SPSS 20.0 statistical software (SPSS Inc., Chicago, IL). P values less than 0.05 were considered statistically significant.
3. Results
3.1. UDSC-derived dECM enhanced IPFSCs’ proliferation and chondrogenic potential
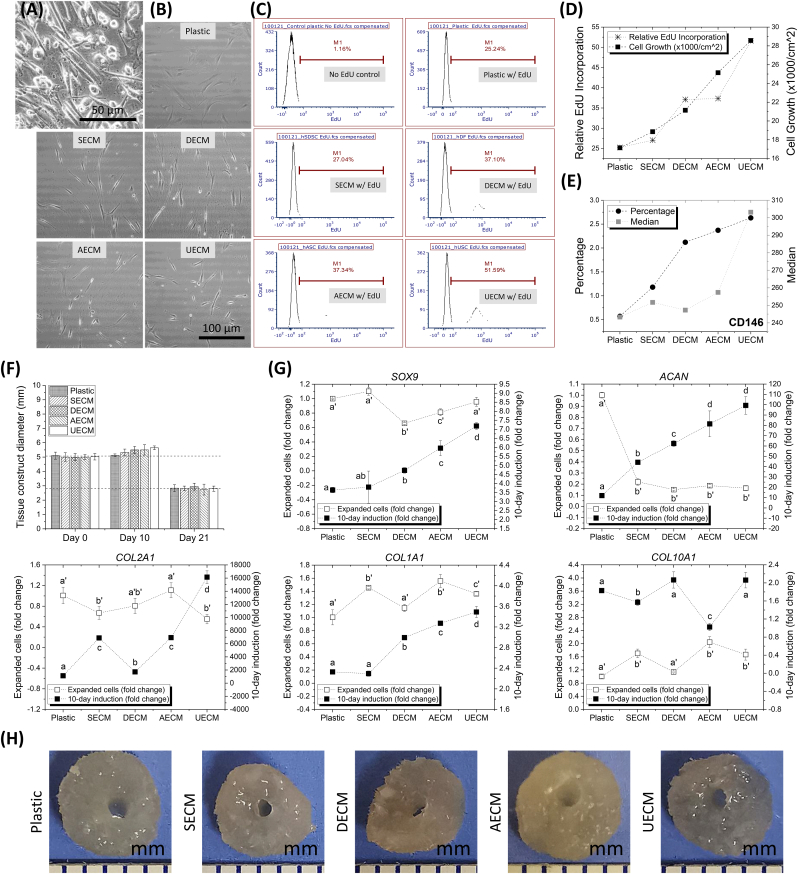
Unlike other human stem cells with a fibroblast-like shape, human UDSCs contained more cells with a cobblestone-like shape (Fig. 1A). Rabbit IPFSCs grown on Plastic appeared randomly distributed with an enlarged size; in contrast, C-dECM-expanded rabbit IPFSCs displayed tiny and glistening morphology and grew along matrix fibers (Fig. 1B). Flow cytometry was used to measure relative EdU incorporation of rabbit IPFSCs expanded on different substrates (Fig. 1C). We found that C-dECM-expanded cells exhibited increased DNA synthesis as compared to those grown on Plastic with the greatest increase in UECM-expanded cells, which is supported by cell growth data (Fig. 1D), indicating that expansion on the C-dECMs, particularly UECM, greatly enhanced rabbit IPFSCs’ proliferation ability. Cell proliferation data were also mirrored by expression of the MSC surface marker CD146 (Fig. 1E), a known marker for MSCs isolated from many adult and fetal organs [28].
Fig. 1.
Evaluation of rabbit IPFSCs' proliferation and chondrogenic capacity after expansion on dECMs deposited by human SDSCs (SECM), DFs (DECM), ADSCs (AECM), and UDSCs (UECM). The Plastic group serves as a control culture substrate. (A) Human UDSC cell morphology. (B) Rabbit IPFSCs' morphology after expansion on C-dECMs and Plastic. Cell proliferation was evaluated using relative EdU incorporation (C) and combination with cell growth (D), as well as CD146 surface marker expression (E) after expansion on C-dECMs and Plastic. (F) The size of tissue constructs (expanded cells + PLGA scaffolds) was measured at Days 0, 10, and 21 following chondrogenic induction. (G) Expanded cells (fold change adjusted by the data collected from the Plastic group) and Day 10 tissue constructs (fold change adjusted by the data collected from the corresponding cell samples) were used to evaluate chondrogenic marker gene expression including SOX9, COL2A1, ACAN, COL1A1, and COL10A1. Data are presented as average ± SD for n = 3. In each comparison, groups not connected by the same letter are significantly different (p < 0.05). (H) Representative tissue constructs were shown following 10-day chondrogenic induction.
After successful transduction with lentivirus carrying the green fluorescent protein (GFP) gene, 3 × 106 passage 5 rabbit IPFSCs expanded on Plastic and C-dECMs for five days were seeded on Poly Lactic-co-Glycolic Acid (PLGA) discs (5 mm diameter x 2 mm thickness) and grown in chondrogenic induction medium in spinner flasks for 21 days following our previous protocol [21]. Tissue construct size increased an average of 7.9% from Day 0 (5.02 ± 0.23 mm) to Day 10 (5.42 ± 0.28 mm) but dropped 56.2% from Day 0 to Day 21 (2.82 ± 0.24 mm) (Fig. 1F). Day 10 tissue constructs along with expanded cells were further assessed for chondrogenic gene expression using real-time qPCR (Fig. 1G). We found that, despite no increase in C-dECM-expanded cells compared with the Plastic group, SOX9 and COL2A1 expression levels significantly increased in tissue constructs after 10-day chondrogenic induction. Interestingly, in comparison with the Plastic group, C-dECM expansion dramatically decreased ACAN expression in expanded cells whereas 10-day chondrogenic induction groups were markedly increased, particularly for the tissue constructs from UECM-expanded cells. As an early marker of chondrogenesis [29], COL1A1 exhibited increased expression in tissue constructs from DECM-, AECM-, and UECM-expanded cells. We also found that, despite the increase of chondrogenic gene expression in the C-dECM groups under chondrogenic induction, hypertrophic gene COL10A1 did not show a corresponding increase. Therefore, day 10 tissue constructs (Fig. 1H) were chosen as premature cartilage constructs in the following implantation study in a rabbit model.
3.2. UDSC-derived dECM promoted IPFSCs’ functional cartilage reconstruction in a rabbit model
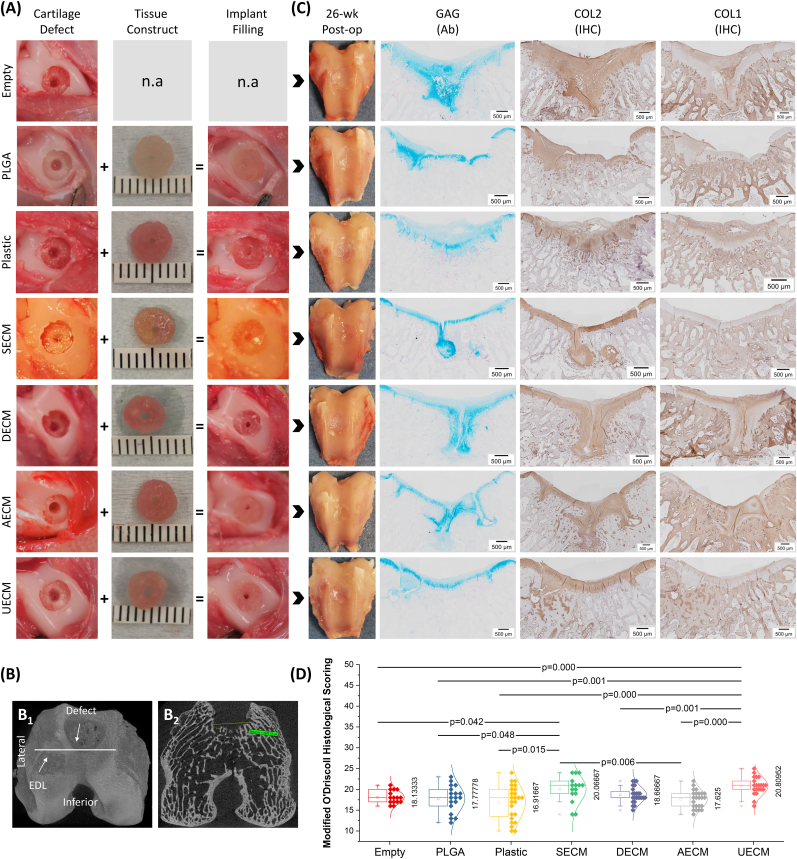
Day 10 premature tissue constructs (5.42 ± 0.28 mm) were pressed to fit in defects (5 mm diameter x 2 mm thickness) created in the trochlear groove (Fig. 2A); size and location of the defects were confirmed by micro-CT (Fig. 2B). At 26 weeks post operatively (Fig. 2C), there were no detectable signs of inflammation in the rabbit knee joints. Appearance of defects filled with newly formed tissue was improved in groups implanted with premature tissue constructs, particularly for the UECM and SECM groups, as compared to non-cell treated groups, evidenced by the intense staining of sulfated glycosaminoglycan (sGAG) and type II collagen in combination with decreased staining of type I collagen (Fig. 2C). This finding is supported by Modified O'Driscoll Histological (MODS) evaluation in which these two groups (UECM and SECM) were scored the highest compared to other groups (Fig. 2D).
Fig. 2.
Histological evaluation of cartilage resurfacing in a rabbit model. (A) Representative of experimental design: defect creation + tissue construct = implant filling. (B) μCT images of representative rabbit distal femur cartilage defect: (B1) 3D reconstruction of distal right femur demonstrating defect in trochlear groove with lower border at the level of the extensor digitorum longus (EDL) insertion and (B2) Axial μCT of right distal femur demonstrating width of the defect in the trochlear groove at time of harvest. (C) Appearance of cartilage resurfacing 26 weeks post operatively and histological assessment including Alcian blue (Ab) staining for sulfated GAG, and immunohistochemistry (IHC) for types I and II collagen from the Empty group (without implant), the PLGA group (with PLGA), the Plastic group (with Plastic-expanded cells), the SECM group (with SECM-expanded cells), the DECM group (with DECM-expanded cells), the AECM group (with AECM-expanded cells), and the UECM group (with UECM-expanded cells). (D) Modified O'Driscoll Histological Scoring of cartilage repair at 26 weeks post implant of the groups including EMPTY (n = 5), PLGA (n = 6), Plastic (n = 8), SECM (n = 5), DECM (n = 8), AECM (n = 8), and UECM (n = 7). Data are shown as average ± SD. p < 0.05 indicated statistically significant differences.
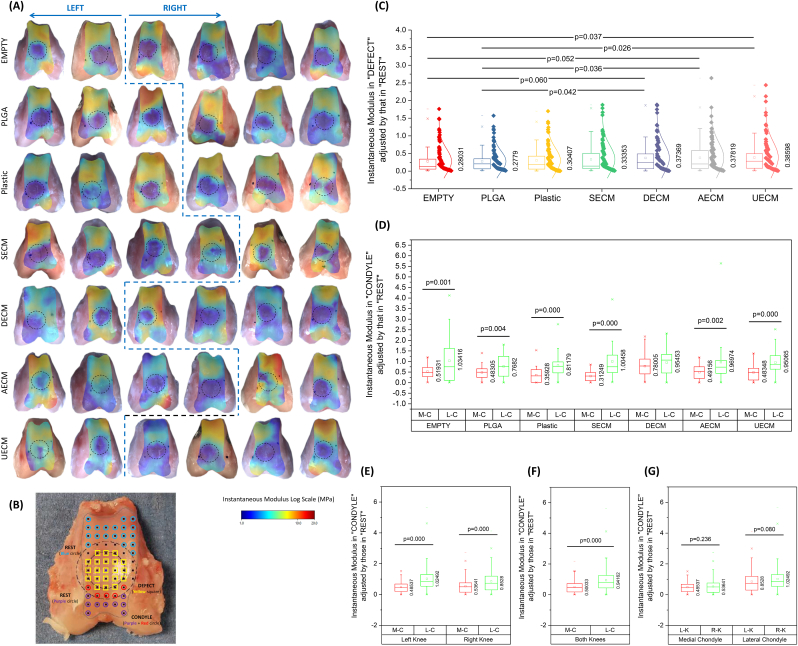
For mechanical evaluation, data from automated normal indentation mapping (Fig. 3A) showed superior instantaneous modulus in the “DEFECT” (adjusted by that in “REST”) (Fig. 3B) of the UECM, AECM, and DECM groups compared with control groups (PLGA alone and EMPTY) (Fig. 3C). Interestingly, we found that the instantaneous modulus in medial condyle cartilage was significantly lower than that of the corresponding lateral condyle cartilage except in the DECM group (Fig. 3D). This trend was still evident when data for either left or right knees were pooled (Fig. 3E). The instantaneous modulus of medial condyle cartilage from both knees was also significantly lower as compared to lateral condyle cartilage (Fig. 3F). The instantaneous modulus of medial condyle cartilage or lateral condyle cartilage between the left and right knees did not differ (Fig. 3G). Interestingly, the DECM group exhibited the least ratio of cartilage thickness in “DEFECT” (adjusted by that in “REST”) (Fig. S1A1/A2) while both UECM and AECM groups displayed the greatest ratio of relaxation time in “DEFECT” (adjusted by that in “REST”) (Fig. S1B1/B2).
Fig. 3.
Mechanical evaluation of cartilage resurfacing in a rabbit model. (A) Original measures of instantaneous modulus from six knees of both sides. (B) Design of three zones, including DEFECT zone in the solid circle with measured points – Yellow squares (TROCHLEA), REST zone of the trochlea located outside of the dashed circle with measured points – Blue circles and REST zone of the condyle located outside of the dashed circle with measured points – Purple and Red circles (very limited anterior region). (C) Functional evaluation of cartilage resurfacing using instantaneous modulus in “DEFECT” adjusted by that in “REST” of all groups: EMPTY (n = 124), PLGA (n = 119), Plastic (n = 121), SECM (n = 124), DECM (n = 117), AECM (n = 121), and UECM (n = 125). Instantaneous modulus in “CONDYLE” adjusted by that in “REST” was evaluated between medial condyle (M–C) and lateral condyle (L–C) in each group [EMPTY: M-C (n = 52) vs. L-C (n = 43), PLGA: M-C (n = 44) vs. L-C (n = 28), Plastic: M-C (n = 35) vs. L-C (n = 27), SECM: M-C (n = 50) vs. L-C (n = 33), DECM: M-C (n = 55) vs. L-C (n = 48), AECM: M-C (n = 54) vs. L-C (n = 47), and UECM: M-C (n = 48) vs. L-C (n = 40)] (D), or in either left knee [M-C (n = 157) vs. L-C (n = 124)] or right knees [M-C (n = 187) vs. L-C (n = 136)] (E), or in both knees [M-C (n = 338) vs. L-C (n = 266)] (F). Instantaneous modulus in “CONDYLE” adjusted by that in “REST” was also compared between left knee (L–K) and right knee (R–K) in either medial condyle [L-K (n = 157) vs. R-K (n = 187)] or lateral condyle [L-K (n = 136) vs. R-K (n = 124)] (G). Data are shown as average ± SD. p < 0.05 indicated statistically significant differences.
In all cell treated groups, we found that some areas in the interface between newly formed cartilage and subchondral bone stained positive for GFP, indicating that the cells and matrix in these brown stained areas were from originally implanted tissue constructs, given that all cells before implantation were transduced with lentivirus carrying GFP (Fig. 4). In addition, these GFP positive areas also immunostained positively for types I, II, and X collagen but not for sulfated GAGs stained by Alcian blue (Fig. 4), an indication of tissue remodeling of the implanted tissues. We also noticed that subchondral bone cysts existed in cell-implanted groups (Fig. 4), probably resulting from the degradation of the PLGA scaffold.
Fig. 4.
Location of implanted tissue constructs 26 weeks post implant. Immunohistochemistry (IHC) of GFP indicative of the leftover (*) tissue constructs with implanted cells and high-magnification of its inset, and IHC of types I, II, and X collagen, as well as Alcian blue (Ab) staining for sulfated GAG, and H&E staining for tissue matrix.
3.3. Proteomics data confirmed C-dECM rejuvenation on IPFSCs’ chondrogenic capacity
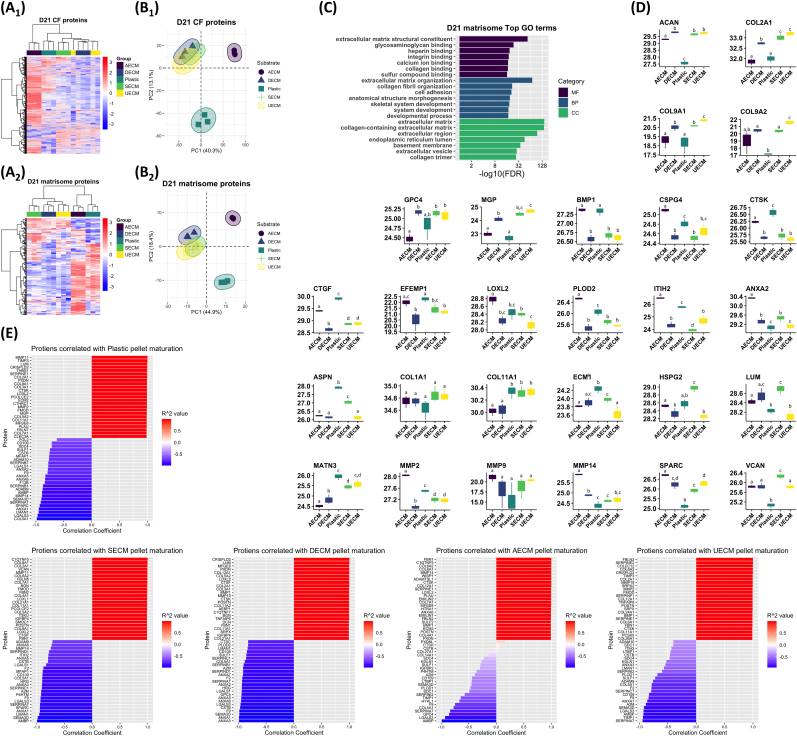
To gain deeper insight into cell differentiation and ECM production, the cell fraction (CF) proteome and matrisome (ECM and ECM-associated proteome) were analyzed independently. We first investigated changes to the C-dECM-expanded IPFSC proteome that occurred following 2D monolayer culture (Fig. S2). Hierarchical clustering of both the CF and matrisome proteomes revealed no distinct clustering among C-dECM substrates (Fig. S2A1/A2). Principal component analysis (PCA) showed significant overlap of all C-dECM groups in both CF and matrisome proteins (Fig. S2B1/B2). To generalize differences between IPFSCs cultured on C-dECM vs. Plastic, proteins that were significantly downregulated, upregulated, or that had no change in expression in C-dECM-expanded cells vs. Plastic-expanded cells were processed for gene ontology (GO) annotation. Of note, downregulated CF proteins were enriched for GO terms related to structural, cytoskeletal, and actin binding (Fig. S2C1). STRING networks for downregulated proteins included muscle protein, sarcoplasmic reticulum, and sarcomere organization (not shown). Upregulated CF proteins enriched for GO terms included small molecule binding as well as glycolytic and carbohydrate metabolism (Fig. S2C2). Likewise, upregulated STRING networks included carbon and pyruvate metabolism as well as glycolysis (not shown). Similar analyses on the 2D cultured cell matrisome indicated an upregulation in phospholipid binding activity (Fig. S2D1) while binding of many molecules remained unchanged (Fig. S2D2).
Next, to determine the influence of C-dECM substrates on IPFSC pellet chondrogenesis, we analyzed the day 21 (D21) pellet proteome (Fig. 5). Clustering of both CF and matrisome proteins by heatmap resulted in clear separation of C-dECM groups, with unique proteins upregulated within each group (Fig. 5A1/A2). PCA of both CF proteome and matrisome showed clear separation of the AECM and Plastic groups from the other substrate groups (Fig. 5B1/B2). To identify C-dECMs that produced superior cartilage ECM, we first performed GO annotation of matrisome proteins that were significantly different among all substrate groups, which resulted in the following GO terms: cartilage development and condensation, chondrocyte development and differentiation, and GAG biosynthetic process as well as endochondral bone growth, ossification, and osteoblast differentiation (Fig. 5C). We then looked at the expression of 27 ECM proteins that enriched for these terms (Fig. 5D). UECM, DECM, and SECM showed similar expression patterns of all 27 ECM proteins [i.e., all had upregulated COL2A1, downregulated bone morphogenetic protein 1 (BMP1), etc.], while Plastic and AECM showed similar expression patterns. The substrates UECM, DECM, and SECM resulted in the greatest D21 expression of canonical cartilage proteins (COL2A1, COL9A1, COL11A1) and Matrix Gla Protein (MGP), and generally low expression of BMP1 and proteases [matrix metalloproteinases (MMPs) and cathepsins] (Fig. 5D). Both Plastic and AECM had the opposite expression of these proteins. Analysis of other ECM proteins associated with cartilage formation including crosslinking enzymes (PLOD, LOXL, ITIH) and cartilage ECM stability proteins (ASPM, ANXA2, CTGF, ECM1) were mixed, but generally lower in the UECM, DECM, and SECM groups. A similar analysis of D21 CF proteins revealed no terms or pathways enriched for cartilage, chondrocytes, bone, or osteoblasts but rather enriched terms for basic cellular processes such as transcription and translation (not shown).
Fig. 5.
Proteomic results of chondrogenically induced IPFSCs following expansion on C-dECMs and Plastic. Hierarchical clustering (A1/A2) and principal component analysis (PCA) (B1/B2) of the cell fraction (CF) proteome (A1/B1) and matrisome (A2/B2). Enrichment analysis of matrisome proteins significantly different among C-dECM groups (C). Expression of proteins at day 21 (D21) pellet culture (D). Data are shown as average ± SD. In each comparison, groups not connected by the same letter are significantly different (p < 0.05). Correlation analyses of proteins with pellet maturation (D0-D10-D21) on different substrates (E).
To assess matrisome temporal patterns in pellets, we identified proteins positively or negatively correlated with time from D0 to D10 to D21 (Fig. 5E). Proteins associated with Plastic expansion include MMPs and cathepsin K (CTSK) as well as COL2A1, COL11A1, and MGP, while AECM expansion was associated with proteases MMP14, ADAMTS1, CTSK, and BMP1. Pellets from UECM-, DECM-, or SECM-expanded cells had high correlations with cartilage proteins (COL2A1, COL9, COL11, ACAN, VCAN), MMPs, and cathepsins (CTSK, CTSF). Enrichment analysis on proteins that increase with time on all C-dECMs produced GO terms for skeletal system development, and cartilage development and condensation.
3.4. RNA-Seq data proposed potential cellular and molecular pathways underlying C-dECM rejuvenation
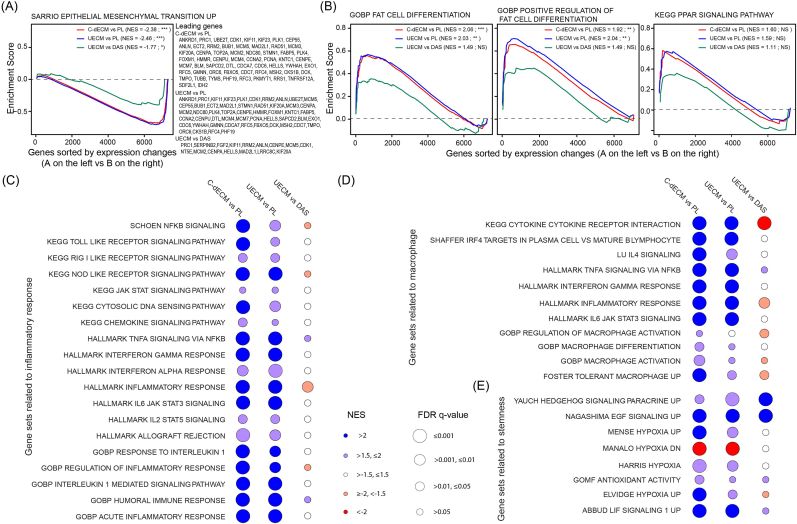
To determine cellular and molecular pathways in rabbit IPFSCs affected by expansion on varied C-dECMs compared to Plastic, gene set enrichment analysis (GSEA) for expression changes of expanded IPFSCs among different comparisons showed that: (1) the epithelial-mesenchymal transition (EMT) was significantly downregulated in C-dECM-expanded cells, particularly in the UECM group (Fig. 6A); (2) adipogenic differentiation was significantly upregulated in C-dECM-expanded cells as evidenced by gene ontology biological processes (GOBP) “fat cell differentiation” and “positive regulation of fat cell differentiation” and, to a lesser extent, in the KEGG PPAR (peroxisome proliferator-activated receptor) signaling pathway (Fig. 6B); (3) gene sets related to inflammatory response were significantly upregulated in C-dECM-expanded cells (Fig. 6C); (4) gene sets related to macrophages were significantly upregulated in C-dECM-expanded cells (Fig. 6D); and (5) gene sets related to stemness were significantly upregulated in C-dECM-expanded cells, especially for UECM vs. DAS in hedgehog and EGF (epidermal growth factor) signaling pathways (Fig. 6E).
Fig. 6.
Functional enrichment analysis by GSEA with RNA-Seq data of rabbit IPFSCs after expansion on C-dECMs (deposited by dermal fibroblasts, DECM; by synovium-derived stem cells, SECM; by adipose-derived stem cells, AECM; or by urine-derived stem cells, UECM) or tissue culture plastic (PL). (A) GSEA for expressed genes ranked by the fold change of expression in various comparisons (C-dECM as average of DECM, SECM, AECM, and UECM vs. PL as plastic control; UECM vs. PL; and UECM vs. DAS as average of DECM, AECM, and SECM) against the MSigDB gene sets “SARRIO epithelial mesenchymal transition up”. (B) GSEA for expressed genes ranked by the fold change of expression in varied comparisons (C-dECM vs. PL; UECM vs. PL; and UECM vs. DAS) against the MSigDB gene set “GOBP fat cell differentiation”, “GOBP positive regulation of fat cell differentiation”, and “KEGG PPAR signaling pathway”. NES: normalized enrichment score; ***: FDR q-value ≤ 0.001; **: FDR q-value ≤ 0.01 but >0.001; *: FDR q-value ≤ 0.05 but >0.01; NS: FDR q-value > 0.05. (C–E) Bubble plot visualization of NES (color) and FDR q-value (circle size) from GSEA for expressed genes ranked by fold change of expression in varied comparisons (C-dECM vs. PL, UECM vs. PL, and UECM vs. DAS) against gene sets related to inflammatory response (C), gene sets related to macrophage (D), and gene sets related to stemness (E).
In gene sets related to macrophages (Fig. 6D), we noticed that KEGG cytokine cytokine receptor interaction was significantly upregulated in both C-dECM vs. PL (NES = 2.41; FDR q = 0.000) and UECM vs. PL (NES = 2.19; FDR q = 0.000). Interestingly, expression of the core enrichment genes such as IL6, CCL2, IL1R1, IL1R2, HGF, VEGFA, and TGFBR2 were greatly increased in C-dECM-expanded cells (11.88-, 4.32-, 2.39-, 3.07-, 4.96-, 2.62-, and 1.62-fold; data not shown) and UECM-expanded cells (12.41-, 4.42-, 3.26-, 7.40-, 4.22-, 2.50-, and 1.70-fold; data not shown) compared to the Plastic group. In addition, UECM-expanded cells also exhibited a 1.49-fold increase in TGFB2 expression. Upregulation of these M2 macrophage related markers, KEGG toll like receptor signaling pathway (Fig. 6C), and LU IL4 signaling [30] (Fig. 6D) in C-dECM-expanded cells indicates that the C-dECM rejuvenation promotes macrophage switch from M1 to M2.
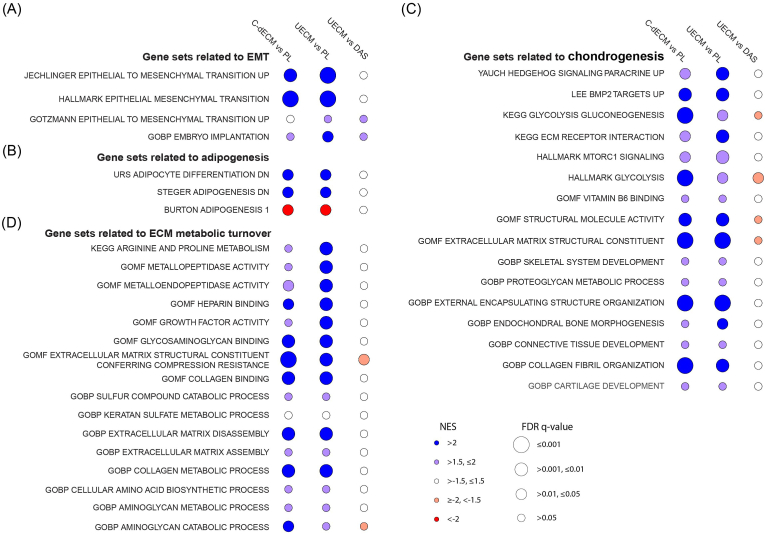
In contrast, C-dECM-expanded cells responded differently to chondrogenic induction compared to Plastic-expanded cells: GSEA with RNA-Seq data showed that C-dECM-expanded cells exhibited an upregulation of the EMT signaling pathway (Fig. 7A), a downregulation of adipogenic differentiation (Fig. 7B), an upregulation of chondrogenic differentiation (Fig. 7C), and an upregulation of metabolic turnover (Fig. 7D).
Fig. 7.
GSEA with RNA-Seq data of chondrogenically induced rabbit IPFSCs following expansion on C-dECMs (DECM, SECM, AECM, and UECM) or tissue culture plastic (PL). Bubble plot plots visualization of NES (color) and FDR q-value (circle size) from GSEA for expressed genes ranked by fold change of expression in varied comparisons (C-dECM as average of DECM, SECM, AECM, and UECM vs. PL as plastic control, UECM vs. PL, and UECM vs. DAS as average of DECM, AECM, and SECM) against gene sets related to the EMT signaling (A), gene sets related to adipogenesis (B), gene sets related to chondrogenesis (C), and gene sets related to metabolic turnover (D).
3.5. UECM showed unique expression and structure of basement membrane proteins compared to other C-dECMs
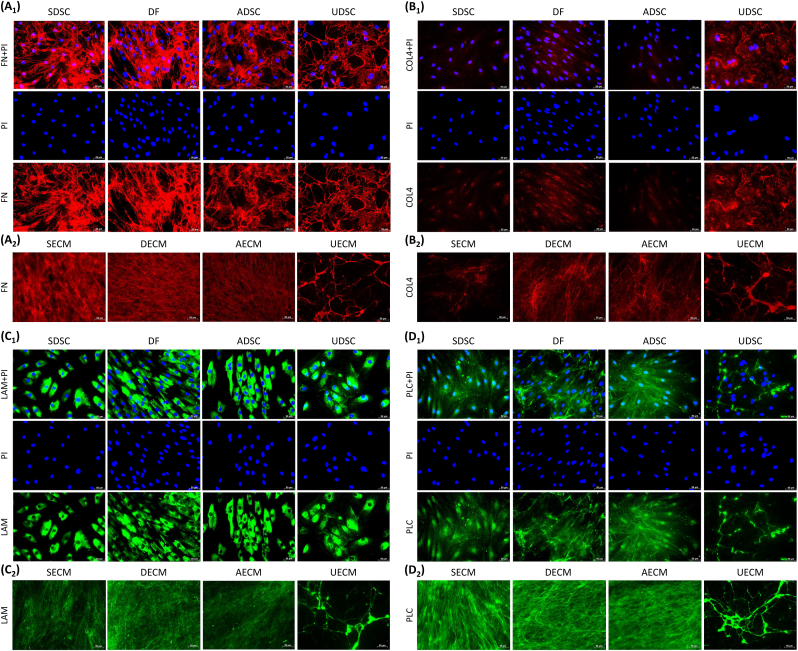
Upregulation of MET signaling in IPFSCs after expansion on C-dECMs indicated a potential contribution from the C-dECMs via basement membrane proteins [31,32]. Four C-dECMs and donated cells were assessed using immunostaining to detect three major basement membrane components (laminin, type IV collagen, and perlecan) and closely associated fibronectin [33] (Fig. 8). Staining for four matrix proteins demonstrated that fibronectin, COL4, laminin, and perlecan were present in all C-dECMs; expression of fibronectin was greatest and COL4 was least. UECM exhibited more intense staining in COL4 than all other C-dECMs. The polarization and fiber diameter of matrix proteins in SECM, DECM, and AECM differed from those in UECM with an isodirectional distribution of fine fibers vs. a net-like structure of thicker fibers in UECM. Prior to supplementation with ascorbic acid in cell expansion medium, fibronectin expression was detected in both cytoplasm and ECM. Interestingly, laminin was only detectable in cytoplasm whereas both perlecan and COL4 were localized in cytoplasm, the nucleus, and ECM.
Fig. 8.
Immunofluorescence of matrix proteins including fibronectin (FN, A1/A2), type IV collagen (COL4, B1/B2), laminin (LAM, C1/C2), and perlecan (PLC, D1/D2) in expanded human SDSCs, DFs, ADSCs, and UDSCs (A1-D1) and extracted SECM, DECM, AECM, and UECM (A2-D2).
4. Discussion
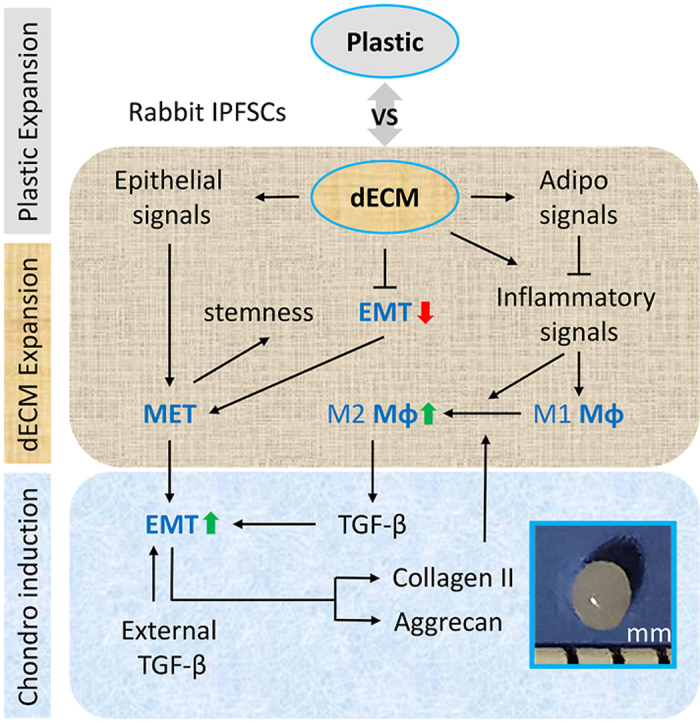
Our previous work indicated that human UDSCs themselves lack chondrogenic differentiation capacity but expansion on the dECM deposited by UDSCs could boost replicatively senescent human bone marrow stromal cell chondrogenesis [17]. Later, we found that dECM deposited by UDSCs exhibited the greatest rejuvenation effect on ex vivo human SDSCs in proliferation and chondrogenic capacities compared to dECMs deposited by human SDSCs, ADSCs, and DFs; notably, all C-dECM groups were superior to the Plastic group [16]. In this study, an in vivo rabbit model was used to verify whether UECM had any clinical potential in stem cell-based cartilage regeneration. Rabbit IPFSCs expanded on dECMs deposited by human SDSCs, ADSCs, DFs, and UDSCs or Plastic for one passage were used to grow 10-day premature tissue constructs for 26-week implantation to repair osteochondral defects in a rabbit model. We found that, in line with our previous observation [16], all C-dECM groups exhibited enhanced proliferation and chondrogenic potential, with UECM performing superior to all other groups. In vivo experiments demonstrated that UECM-expanded cells exhibited a robust resurfacing effect in both histological and mechanical assessments. We also found that the medial condyle displayed lower instantaneous modulus than the lateral condyle. Interestingly, GFP-expressing cells within the implanted tissues were detectable in the interface between newly formed cartilage tissue and subchondral bone. Mechanistically, RNA-Seq data indicated that inflammation-activated macrophage M1 to M2 transition as well as MET/EMT signaling had significant involvement in the C-dECM-mediated IPFSC rejuvenation and chondrogenic potential; the robust chondrogenic capacity exhibited by C-dECM-rejuvenated IPFSCs was further demonstrated by proteomics analysis.
Consistent with our previous report investigating the rejuvenation of human SDSCs [16], in this study, rabbit IPFSCs exhibited robust expansion and chondrogenic capacity after growth on C-dECMs, particularly for the UECM group, indicating that rejuvenation of tissue-specific stem cells is independent of stem cell type and species. Of note, different from other chondrogenic genes, ACAN was significantly downregulated in expanded IPFSCs from all C-dECM groups compared to the Plastic group. This change renders C-dECM-expanded cells with decreased chondrogenic gene expression but increased chondrogenic potential, which contributes to a significant increase of ACAN expression following subsequent chondrogenic induction, particularly for the UECM group. This rejuvenation effect in rabbit IPFSCs has also been observed in our previous reports, such as “change of GAG/DNA and ACAN mRNA in human SDSCs” [34] and “change of GAG/DNA in porcine SDSCs” [9]. For the first time, our rabbit study confirmed the greatest benefits of UECM rejuvenation in enhancing both histological and mechanical properties of newly formed cartilage tissue. Interestingly, the mechanical properties (instantaneous modulus) of newly formed cartilage tissue mirror the chondrogenic potential of corresponding ex vivo expansion groups, in order from greatest to least - UECM, AECM, DECM, SECM, and Plastic. Intriguingly, the AECM group exhibited the worst MODS score compared with other C-dECM groups, which is supported by the proteomics data indicating that the AECM had the lowest expression levels of chondrogenic markers such as aggrecan and types II and IX collagen in Day 21 pellets.
To track the implanted cells, immunostaining was used to evaluate the presence of GFP protein in lieu of direct fluorescence detection given that the GFP signal is susceptible to bleaching during the long-term process [35]. We found that cell-implanted groups had strong GFP signals limited to the interface between newly formed cartilage and subchondral bone. Given that cartilage hypertrophy adjacent to the defects is expected due to blood vessel invasion into the lesion site [36], it is not surprising to observe that these areas were positively stained for type X collagen. We also found these areas were positive for types I and II collagen and negative for GAG, indicative of fibrocartilage. Compared to hyaline cartilage, fibrocartilage typically degrades over time in response to repetitive load [37]. Our finding suggests that the functionality of implanted MSCs might be due to released trophic mediators instead of stem cell differentiation [38], which is in line with a previous report demonstrating that GFP-transfected autologous MSCs implanted in cartilage defects were detectable for four weeks following implantation even though no repair tissue was demonstrated in the histology [39]. The presence of subchondral bone cysts in cell-implanted groups indicates a prolonged degradation process and the accumulation of acid degradation products resulting from the implanted PLGA scaffold. Thus, a hydrogel with natural components like hyaluronic acid, chondroitin sulfate, and biocompatible synthetic biomaterials [40,41] will be considered in a future investigation.
Interestingly, we found that the medial femoral condyle exhibited significantly lower instantaneous modulus than the lateral femoral condyle. While there are no similar reports addressing the discrepancy between the medial and lateral femoral condyles in the repair of cartilage defects in animal models, this finding has been indicated in several clinical studies. For example, in 1000 knee arthroscopies, Hjelle and coworkers found 61% of patients with cartilage defects were mostly located in the medial femoral condyle (58%) as compared to 9% of cases in the lateral femoral condyle and 5–11% of cases in other tissues [42]. Similarly, after reviewing articular cartilage defects in 25,124 knee arthroscopies, Widuchowski et al. found that, with the exception of the patellar articular surface (36%), the medial femoral condyle (34%) was the most frequent site of cartilage lesions when compared to the lateral femoral condyle (9%) and other locations (6–8%) [43]. Adduction of the knee may be responsible for the uneven load distribution between the medial and lateral sides and eventually lead to the biomechanical abnormality of the medial femoral condyle in osteoarthritis [[44], [45], [46], [47]].
To explore potential mechanisms underlying C-dECM rejuvenation, both RNA-Seq and proteomics were used to evaluate expanded cells and chondrogenically induced pellets. RNA-Seq data showed that C-dECM-expanded cells not only produced inflammatory factors such as interleukin 1 (IL1) and interferon gamma (IFNγ), but also exhibited increased expression of IL4 and IL6. Compared to IFNγ released by the classically-activated (M1) macrophage, IL6 produced by alternatively-activated (M2) macrophages that were polarized with IL4 tended toward an anti-inflammatory phenotype [48]. Four transcriptional factors, including PPAR gamma (PPARG), CCAAT/enhancer-binding protein beta (CEBPB), interferon regulatory factor 4 (IRF4), and the signal transducer and activator of transcription (STAT) family, were upregulated in C-dECM-expanded cells, indicating an active macrophage involvement in translating signals from the matrix microenvironment into a polarized phenotype, particularly for the dynamic transition from M1 to M2 macrophages [49,50]. This switch during inflammation facilitates the dual role of macrophages in first coordinating the onset of inflammation and afterward promoting healing and repair [51]. Given that the M2 macrophage promotes TGFβ-mediated chondrogenic differentiation [52] and type II collagen increases switching from M1 to M2 [53], positive feedback is indicated between C-dECM rejuvenation and chondrogenic differentiation through adaption of macrophage polarization to the local environment [54]. This finding was also supported by increased ECM dynamics in the pellets from C-dECM-expanded cells including, but not limited to, the strong enrichment for collagen-degradation enzymes (e.g., MMPs) and aminoglycan-degradation enzymes (e.g., hyaluronidase) as well as the enhanced integrin signaling, ribosome biogenesis, and amino acid biosynthesis, which benefits skeletal tissue regeneration [55,56]. In addition, we found that C-dECM-expanded cells exhibited increased expression of antioxidants and PPARG, in favor of maintenance of the mitochondrial membrane potential thereby avoiding senescence and apoptosis of the MSCs [57,58]. As opposed to oxidative phosphorylation needed in osteogenic and adipogenic differentiation, glycolysis, the major source of energy metabolism in chondrogenic differentiation [59], was enhanced in the pellets from C-dECM-expanded cells.
We also found that the EMT was significantly downregulated in IPFSCs following expansion on C-dECMs, particularly on UECM. This finding indicates that the MET is substantially involved in IPFSCs' rejuvenation, which is confirmed by activation of several critical downstream pathways, such as NF-κB, STAT3, MAPK, PI3K-AKT, and WNT [[60], [61], [62]]. Increasing evidence demonstrates that MET plays an essential role in early somatic cell reprogramming in mouse embryonic fibroblasts and human fibroblasts [63,64] following transduction of OSKM factors (OCT4, SOX2, KLF4, and MYC), in which SOX2, OCT4, and MYC suppress TGFβ-Snail signaling while KLF4 induces the epithelial program [65]. In the present study, prior to chondrogenic induction, C-dECM-expanded cells exhibited a significant downregulation of EMT and upregulation of epithelial signaling, along with an upregulation of stemness-associated genes and dynamic epigenetic changes, similar to the abovementioned somatic cell reprogramming. Upregulation of IL6 in C-dECM-expanded cells might contribute to the maintenance of MSCs' stemness [66] and promote chondrogenic capacity [67]. Following chondrogenic induction, RNA-Seq data from induced pellets suggested that prior expansion on C-dECMs later upregulated EMT and promoted IPFSCs’ chondrogenic differentiation in the pellets. Improved chondrogenic capacity might be associated with upregulation of TGFBR2 [68], BMP2 [69], hedgehog [70], vitamin B6 binding [71], and glycolysis [72], and downregulation of Wnt signals [73,74] in chondrogenically-induced C-dECM-expanded cells. These findings support the observation that MET signaling is a critical regulator of EMT and stem cell renewal in early embryonic development and adult stem cells [75].
Compared to type I EMT (embryogenesis and organ development) and type III EMT (cancer progression), type II EMT is involved in wound healing, tissue regeneration, and organ fibrosis, and has attracted more attention in the field of stem cells and tissue regeneration [76]. Mammary epithelial cells induced toward EMT by transduction of Twist, Snail, or TGFβ displayed multi-lineage differentiation potential similar to that of MSCs [77]; our results indicate that, for the first time, MET/EMT was actively involved in matrix microenvironment-mediated stem cell rejuvenation and chondrogenic differentiation. Proteomics data indicated that, compared to growth on Plastic, IPFSCs expanded on C-dECMs exhibited a downregulation of contractile fibers. Since there is an inverse relationship between proliferation and expression of contractile proteins [78], C-dECM expansion favors MSC proliferation. Furthermore, C-dECM-expanded IPFSCs exhibited increased levels of chondrogenic markers, with the exception of the AECM group. Of note, the UECM group yielded Day 21 pellets with superior expression of ACAN, COL2A1, COL9A1, COL9A2, and matrix Gla protein (MGP) and the least expression of cathepsin K (CTSK), lumican (LUM), and ECM1. Since MGP exhibits high affinity for hydroxyapatite crystals that are present in mineralized ECMs and inhibits calcification of the tissue where it is expressed [79,80], upregulation of MGP indicates less hypertrophic potential of UECM-expanded cells during chondrogenic induction. The downregulation of CTSK, LUM, and ECM1 in D21 pellets of the UECM group favors chondrogenesis due to their roles in promoting inflammation [81] and inhibiting chondrogenesis [82,83]. For instance, as a potent ECM-degrading protease, CTSK has specific cleavage sites toward type II collagen [84] and aggrecan [85]. Notably, an upregulation of CTSK expression has been observed in degraded cartilage caused by rheumatoid arthritis [86] and osteoarthritis [87], as well as dedifferentiation of chondrocytes during ex vivo expansion [83].
In contrast to the phenotypic transition in EMT involving epithelial cell disruption of the basement membrane [31], upregulation of MET in C-dECM-expanded IPFSCs indicates an assembly of the basement membrane in C-dECMs that facilitates IPFSCs' stemness. Our immunostaining data indicated that UECM exhibited a unique structure compared to other C-dECMs in major basement membrane proteins laminin, type IV collagen, and perlecan as well as fibronectin. It is well known that laminin isoforms crosslinking with type IV collagen provides basement membrane structure that allows for growth factors, such as fibroblast growth factor and TGFβ, to be tethered to the basement membrane via perlecan so that they can carry out their function. Notably, laminin promotes epithelial differentiation and MET [88] while fibrillar matrix components such as fibronectin and type I collagen promote mesenchymal differentiation and EMT [89]. Our previous report indicated that UECM exhibited advantageous major basement membrane proteins while other C-dECMs displayed dominant expression of fibrillary types I and III collagen; these findings might be responsible for the lowest stiffness of UECM and expanded cells compared to corresponding counterparts [16]. Given that ECM of higher stress within epithelial cells favors EMT while ECM of lower stress benefits the epithelial phenotype [90], in our study, UECM's soft matrix environment resulting from the dominant expression of basement membrane proteins and low expression of fibrillar matrix components might facilitate its promotion of MET in expanded IPFSCs. Expansion on UECM drives IPFSCs' rejuvenation to magnify subsequent chondrogenic induction driven by TGFβ, a so-called EMT process [91].
Funding
This work was supported by grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases, USA (Grant No. R01AR067747).
Data and materials availability
The raw/processed data required to reproduce these findings cannot be shared at this time as the data also form part of an ongoing study.
Ethics approval and consent to participate
I confirm that I have obtained all consents required by applicable law for the publication of any personal details or images of patients, research subjects or other individuals that are used in the materials submitted to KeAi. I have retained a written copy of all such consents and I agree to provide KeAi with copies of the consents or evidence that such consents have been obtained if requested by KeAi.
CRediT authorship contribution statement
Ming Pei: Conceptualization, Methodology, Visualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. Yixuan Amy Pei: Investigation, Visualization, Writing – review & editing. Sheng Zhou: Investigation, Visualization, Writing – review & editing. Elmira Mikaeiliagah: Investigation, Visualization, Writing – review & editing. Christopher Erickson: Methodology, Investigation, Visualization, Writing – review & editing. Benjamin Giertych: Investigation, Visualization, Writing – review & editing. Halima Akhter: Methodology, Investigation, Writing – review & editing. Lei Wang: Methodology, Investigation, Writing – review & editing. Amanda Stewart: Investigation, Writing – review & editing. Joshua Parenti: Investigation, Writing – review & editing. Bin Wang: Investigation, Writing – review & editing. Sijin Wen: Investigation, Supervision, Writing – review & editing. Sotcheadt Sim: Investigation, Visualization, Writing – review & editing. Eric Quenneville: Investigation, Visualization, Supervision, Writing – review & editing. Kirk C. Hansen: Investigation, Visualization, Writing – review & editing. Steven Frisch: Investigation, Writing – review & editing. Gangqing Hu: Methodology, Investigation, Visualization, Supervision, Writing – review & editing.
Declaration of competing interest
None.
Acknowledgments
We thank Suzanne Danley for editing the manuscript.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2022.11.012.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Toh W.S., Foldager C.B., Pei M., Hui J.H. Advances in mesenchymal stem cell-based strategies for cartilage repair and regeneration. Stem Cell Rev. Rep. 2014;10(5):686–696. doi: 10.1007/s12015-014-9526-z. [DOI] [PubMed] [Google Scholar]
- 2.Pizzute T., Lynch K., Pei M. Impact of tissue-specific stem cells on lineage specific differentiation: a focus on musculoskeletal system. Stem Cell Rev. Rep. 2015;11(1):119–132. doi: 10.1007/s12015-014-9546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones B., Pei M. Synovium-derived stem cells: a tissue-specific stem cell for cartilage tissue engineering and regeneration, Tissue Eng. Part B Rev. 2012;18(4):301–311. doi: 10.1089/ten.TEB.2012.0002. [DOI] [PubMed] [Google Scholar]
- 4.Sun Y., Chen S., Pei M. Comparative advantages of infrapatellar fat pad: an emerging stem cell source for regeneration medicine. Rheumatology. 2018;57(12):2072–2086. doi: 10.1093/rheumatology/kex487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang T., Hill R.C., Dzieciatkowska M., Zhu L., Infante A.M., Hu G., Hansen K.C., Pei M. Site-Dependent lineage preference of adipose stem cells. Front. Cell Dev. Biol. 2020;8:237. doi: 10.3389/fcell.2020.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J.T., Pei M. Cell senescence: a challenge in cartilage engineering and regeneration, Tissue Eng. Part B Rev. 2012;18(4):270–287. doi: 10.1089/ten.TEB.2011.0583. [DOI] [PubMed] [Google Scholar]
- 7.Pei M. Environmental preconditioning rejuvenates adult stem cells' proliferation and chondrogenic potential. Biomaterials. 2017;117:10–23. doi: 10.1016/j.biomaterials.2016.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sisakhtnezhad S., Alimoradi E., Akrami H. External factors influencing mesenchymal stem cell fate in vitro. Eur. J. Cell Biol. 2017;96(1):13–33. doi: 10.1016/j.ejcb.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 9.He F., Chen X., Pei M. Reconstruction of an in vitro tissue-specific microenvironment to rejuvenate synovium-derived stem cells for cartilage tissue engineering. Tissue Eng. 2009;15(12):3809–3821. doi: 10.1089/ten.TEA.2009.0188. [DOI] [PubMed] [Google Scholar]
- 10.Pei M., Li J.T., Shoukry M., Zhang Y. A review of decellularized stem cell matrix: a novel cell expansion system for cartilage tissue engineering. Eur. Cell. Mater. 2011;22:333–343. doi: 10.22203/ecm.v022a25. [DOI] [PubMed] [Google Scholar]
- 11.Li J., He F., Pei M. Creation of an in vitro microenvironment to enhance human fetal synovium-derived stem cell chondrogenesis. Cell Tissue Res. 2011;345(3):357–365. doi: 10.1007/s00441-011-1212-8. [DOI] [PubMed] [Google Scholar]
- 12.Pei M., He F. Extracellular matrix deposited by synovium-derived stem cells delays replicative senescent chondrocyte dedifferentiation and enhances redifferentiation. J. Cell. Physiol. 2012;227(5):2163–2174. doi: 10.1002/jcp.22950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X., Zhou L., Chen X., Liu T., Pan G., Cui W., Li M., Luo Z.P., Pei M., Yang H., Gong Y., He F. Culturing on decellularized extracellular matrix enhances antioxidant properties of human umbilical cord-derived mesenchymal stem cells. Mater. Sci. Eng. C Mater. Biol. Appl. 2016;61:437–448. doi: 10.1016/j.msec.2015.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y., Pizzute T., Li J., He F., Pei M. sb203580 preconditioning recharges matrix-expanded human adult stem cells for chondrogenesis in an inflammatory environment - a feasible approach for autologous stem cell based osteoarthritic cartilage repair. Biomaterials. 2015;64:88–97. doi: 10.1016/j.biomaterials.2015.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y., Yan L., Chen S., Pei M. Functionality of decellularized matrix in cartilage regeneration: a comparison of tissue versus cell sources. Acta Biomater. 2018;74:56–73. doi: 10.1016/j.actbio.2018.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J.T., Narayanan K., Zhang Y., Hill R.C., He F., Hansen K.C., Pei M. Role of lineage-specific matrix in stem cell chondrogenesis. Biomaterials. 2020;231 doi: 10.1016/j.biomaterials.2019.119681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pei M., Li J., Zhang Y., Liu G., Wei L., Zhang Y. Expansion on a matrix deposited by nonchondrogenic urine stem cells strengthens the chondrogenic capacity of repeated-passage bone marrow stromal cells. Cell Tissue Res. 2014;356(2):391–403. doi: 10.1007/s00441-014-1801-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J.T., Pei M. A protocol to prepare decellularized stem cell matrix for rejuvenation of cell expansion and cartilage regeneration. Methods Mol. Biol. 2018;1577:147–154. doi: 10.1007/7651_2017_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pizzute T., Zhang Y., He F., Pei M. Ascorbate-dependent impact on cell-derived matrix in modulation of stiffness and rejuvenation of infrapatellar fat derived stem cells toward chondrogenesis. Biomed. Mater. 2016;11(4) doi: 10.1088/1748-6041/11/4/045009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Z., Zhou S., Vaida J., Gao G., Stewart A., Parenti J., Yan L., Pei M. Unfavorable contribution of a tissue-engineering cartilage graft to osteochondral defect repair in young rabbits. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.595518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pei M., Solchaga L.A., Seidel J., Zeng L., Vunjak-Novakovic G., Caplan A.I., Freed L.E. Bioreactors mediate the effectiveness of tissue engineering scaffolds. Faseb. J. 2002;16(12):1691–1694. doi: 10.1096/fj.02-0083fje. [DOI] [PubMed] [Google Scholar]
- 22.Changoor A., Fereydoonzad L., Yaroshinsky A., Buschmann M.D. Effects of refrigeration and freezing on the electromechanical and biomechanical properties of articular cartilage. J. Biomech. Eng. 2010;132(6) doi: 10.1115/1.4000991. [DOI] [PubMed] [Google Scholar]
- 23.Kiefer G.N., Sundby K., McAllister D., Shrive N.G., Frank C.B., Lam T., Schachar N.S. The effect of cryopreservation on the biomechanical behavior of bovine articular cartilage. J. Orthop. Res. 1989;7(4):494–501. doi: 10.1002/jor.1100070406. [DOI] [PubMed] [Google Scholar]
- 24.Hayes W.C., Keer L.M., Herrmann G., Mockros L.F. A mathematical analysis for indentation tests of articular cartilage. J. Biomech. 1972;5(5):541–551. doi: 10.1016/0021-9290(72)90010-3. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Hu G., Hill R.C., Dzieciatkowska M., Hansen K.C., Zhang X.B., Yan Z., Pei M. Matrix reverses immortalization-mediated stem cell fate determination. Biomaterials. 2021;265 doi: 10.1016/j.biomaterials.2020.120387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J.T., Hansen K.C., Zhang Y., Dong C., Dinu C.Z., Dzieciatkowska M., Pei M. Rejuvenation of chondrogenic potential in a young stem cell microenvironment. Biomaterials. 2014;35(2):642–653. doi: 10.1016/j.biomaterials.2013.09.099. [DOI] [PubMed] [Google Scholar]
- 27.Reisz J.A., Nemkov T., Dzieciatkowska M., Culp-Hill R., Stefanoni D., Hill R.C., Yoshida T., Dunham A., Kanias T., Dumont L.J., Busch M., Eisenmesser E.Z., Zimring J.C., Hansen K.C., D'Alessandro A. Methylation of protein aspartates and deamidated asparagines as a function of blood bank storage and oxidative stress in human red blood cells. Transfusion. 2018;58(12):2978–2991. doi: 10.1111/trf.14936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Covas D.T., Panepucci R.A., Fontes A.M., Silva W.A., Jr., Orellana M.D., Freitas M.C., Neder L., Santos A.R., Peres L.C., Jamur M.C., Zago M.A. Multipotent mesenchymal stromal cells obtained from diverse human tissues share functional properties and gene-expression profile with CD146+ perivascular cells and fibroblasts. Exp. Hematol. 2008;36(5):642–654. doi: 10.1016/j.exphem.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 29.Kosher R.A., Kulyk W.M., Gay S.W. Collagen gene expression during limb cartilage differentiation. J. Cell Biol. 1986;102(4):1151–1156. doi: 10.1083/jcb.102.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rőszer T. Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediat. Inflamm. 2015;2015 doi: 10.1155/2015/816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horejs C.M. Basement membrane fragments in the context of the epithelial-to-mesenchymal transition. Eur. J. Cell Biol. 2016;95(11):427–440. doi: 10.1016/j.ejcb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Pitsidianaki I., Morgan J., Adams J., Campbell K. Mesenchymal-to-epithelial transitions require tissue-specific interactions with distinct laminins. J. Cell Biol. 2021;220(8) doi: 10.1083/jcb.202010154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J., Doyle A.D., Shinsato Y., Wang S., Bodendorfer M.A., Zheng M., Yamada K.M. Basement membrane regulates fibronectin organization using sliding focal adhesions driven by a contractile winch. Dev. Cell. 2020;52(5):631–646. doi: 10.1016/j.devcel.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Li J., Davis M.E., Pei M. Delineation of in vitro chondrogenesis of human synovial stem cells following preconditioning using decellularized matrix. Acta Biomater. 2015;20:39–50. doi: 10.1016/j.actbio.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu X., Zhu J., Li X., Zhang X., Meng Q., Yuan L., Zhang J., Fu X., Duan X., Chen H., Ao Y. Dextran-coated fluorapatite crystals doped with Yb3+/Ho3+ for labeling and tracking chondrogenic differentiation of bone marrow mesenchymal stem cells in vitro and in vivo. Biomaterials. 2015;52:441–451. doi: 10.1016/j.biomaterials.2015.02.050. [DOI] [PubMed] [Google Scholar]
- 36.Pei Y.A., Chen S., Pei M. The essential anti-angiogenic strategies in cartilage engineering and osteoarthritic cartilage repair, Cell. Mol. Life Sci. 2022;79(1):71. doi: 10.1007/s00018-021-04105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armiento A.R., Alini M., Stoddart M.J. Articular fibrocartilage - why does hyaline cartilage fail to repair? Adv. Drug Deliv. Rev. 2019;146:289–305. doi: 10.1016/j.addr.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 38.Caplan A.I., Dennis J.E. Mesenchymal stem cells as trophic mediators. J. Cell. Biochem. 2006;98(5):1076–1084. doi: 10.1002/jcb.20886. [DOI] [PubMed] [Google Scholar]
- 39.Hindle P., Baily J., Khan N., Biant L.C., Simpson A.H., Péault B. Perivascular mesenchymal stem cells in sheep: characterization and autologous transplantation in a model of articular cartilage repair. Stem Cell. Dev. 2016;25(21):1659–1669. doi: 10.1089/scd.2016.0165. [DOI] [PubMed] [Google Scholar]
- 40.Li J., Huang Y., Song J., Li X., Zhang X., Zhou Z., Chen D., Ma P.X., Peng W., Wang W., Zhou G. Cartilage regeneration using arthroscopic flushing fluid-derived mesenchymal stem cells encapsulated in a one-step rapid cross-linked hydrogel. Acta Biomater. 2018;79:202–215. doi: 10.1016/j.actbio.2018.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X., Xu Q., Johnson M., Wang X., Lyu J., Li Y., McMahon S., Greiser U.A.S., Wang W. A chondroitin sulfate based injectable hydrogel for delivery of stem cells in cartilage regeneration. Biomater. Sci. 2021;9(11):4139–4148. doi: 10.1039/d1bm00482d. [DOI] [PubMed] [Google Scholar]
- 42.Hjelle K., Solheim E., Strand T., Muri R., Brittberg M. Articular cartilage defects in 1,000 knee arthroscopies. Arthroscopy. 2002;18(7):730–734. doi: 10.1053/jars.2002.32839. [DOI] [PubMed] [Google Scholar]
- 43.Widuchowski W., Widuchowski J., Trzaska T. Articular cartilage defects: study of 25,124 knee arthroscopies. Knee. 2007;14(3):177–182. doi: 10.1016/j.knee.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Andriacchi T.P. Dynamics of knee malalignment, Orthop. Clin. North Am. 1994;25(3):395–403. [PubMed] [Google Scholar]
- 45.Levinger P., Menz H.B., Morrow A.D., Bartlett J.R., Feller J.A., Bergman N.R. Relationship between foot function and medial knee joint loading in people with medial compartment knee osteoarthritis. J. Foot Ankle Res. 2013;6(1):33. doi: 10.1186/1757-1146-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyazaki T., Wada M., Kawahara H., Sato M., Baba H., Shimada S. Dynamic load at baseline can predict radiographic disease progression in medial compartment knee osteoarthritis. Ann. Rheum. Dis. 2002;61(7):617–622. doi: 10.1136/ard.61.7.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schipplein O.D., Andriacchi T.P. Interaction between active and passive knee stabilizers during level walking. J. Orthop. Res. 1991;9(1):113–119. doi: 10.1002/jor.1100090114. [DOI] [PubMed] [Google Scholar]
- 48.Casella G., Garzetti L., Gatta A.T., Finardi A., Maiorino C., Ruffini F., Martino G., Muzio L., Furlan R. IL4 induces IL6-producing M2 macrophages associated to inhibition of neuroinflammation in vitro and in vivo. J. Neuroinflammation. 2016;13(1):139. doi: 10.1186/s12974-016-0596-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biswas S.K., Lopez C.E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 2009;30(10):475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 50.Porta C., Rimoldi M., Raes G., Brys L., Ghezzi P., Liberto D.D., Dieli F., Ghisletti S., Natoli G., Baetselier P.D., Mantovani A., Sica A. Tolerance and M2 (alternative) macrophage polarization are related processes orchestrated by p50 nuclear factor kappaB. Proc. Natl. Acad. Sci. U.S.A. 2009;106(35):14978–14983. doi: 10.1073/pnas.0809784106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawrence T., Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol. 2011;11(11):750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 52.Fernandes T.L., Gomoll A.H., Lattermann C., Hernandez A.J., Bueno D.F., Amano M.T. Macrophage: a potential target on cartilage regeneration. Front. Immunol. 2020;11:111. doi: 10.3389/fimmu.2020.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dai M., Sui B., Xue Y., Liu X., Sun J. Cartilage repair in degenerative osteoarthritis mediated by squid type II collagen via immunomodulating activation of M2 macrophages, inhibiting apoptosis and hypertrophy of chondrocytes. Biomaterials. 2018;180:91–103. doi: 10.1016/j.biomaterials.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 54.Viola A., Munari F., Sánchez R.R., Scolaro T., Castegna A. The metabolic signature of macrophage responses. Front. Immunol. 2019;10:1462. doi: 10.3389/fimmu.2019.01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bastow E.R., Byers S., Golub S.B., Clarkin C.E., Pitsillides A.A., Fosang A.J. Hyaluronan synthesis and degradation in cartilage and bone, Cell. Mol. Life Sci. 2008;65(3):395–413. doi: 10.1007/s00018-007-7360-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hall B.K., Miyake T. All for one and one for all: condensations and the initiation of skeletal development. Bioessays. 2000;22(2):138–147. doi: 10.1002/(SICI)1521-1878(200002)22:2<138::AID-BIES5>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 57.Hu Y., Huang L., Shen M., Liu Y., Liu G., Wu Y., Ding F., Ma K., Wang W., Zhang Y., Shao Z., Cai X., Xiong L. Pioglitazone protects compression-mediated apoptosis in nucleus pulposus mesenchymal stem cells by suppressing oxidative stress. Oxid. Med. Cell. Longev. 2019;2019 doi: 10.1155/2019/4764071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L., Guo X., Guo X., Zhang X., Ren J. Decitabine promotes apoptosis in mesenchymal stromal cells isolated from patients with myelodysplastic syndromes by inducing reactive oxygen species generation. Eur. J. Pharmacol. 2019;863 doi: 10.1016/j.ejphar.2019.172676. [DOI] [PubMed] [Google Scholar]
- 59.Pattappa G., Heywood H.K., de Bruijn J.D., Lee D.A. The metabolism of human mesenchymal stem cells during proliferation and differentiation. J. Cell. Physiol. 2011;226(10):2562–2570. doi: 10.1002/jcp.22605. [DOI] [PubMed] [Google Scholar]
- 60.Müller M., Morotti A., Ponzetto C. Activation of NF-kappaB is essential for hepatocyte growth factor-mediated proliferation and tubulogenesis. Mol. Cell Biol. 2002;22(4):1060–1072. doi: 10.1128/MCB.22.4.1060-1072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Syed Z.A., Yin W., Hughes K., Gill J.N., Shi R., Clifford J.L. HGF/c-met/Stat3 signaling during skin tumor cell invasion: indications for a positive feedback loop. BMC Cancer. 2011;11:180. doi: 10.1186/1471-2407-11-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vermeulen L., De Sousa E Melo F., van der Heijden M., Cameron K., de Jong J.H., Borovski T., Tuynman J.B., Todaro M., Merz C., Rodermond H., Sprick M.R., Kemper K., Richel D.J., Stassi G., Medema J.P. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 2010;12(5):468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 63.Høffding M.K., Hyttel P. Ultrastructural visualization of the Mesenchymal-to-Epithelial Transition during reprogramming of human fibroblasts to induced pluripotent stem cells. Stem Cell Res. 2015;14(1):39–53. doi: 10.1016/j.scr.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 64.Subramanyam D., Lamouille S., Judson R.L., Liu J.Y., Bucay N., Derynck R., Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 2011;29(5):443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q., Qin B., Xu J., Li W., Yang J., Gan Y., Qin D., Feng S., Song H., Yang D., Zhang B., Zeng L., Lai L., Esteban M.A., Pei D. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7(1):51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 66.Pricola K.L., Kuhn N.Z., Haleem-Smith H., Song Y., Tuan R.S. Interleukin-6 maintains bone marrow-derived mesenchymal stem cell stemness by an ERK1/2-dependent mechanism. J. Cell. Biochem. 2009;108(3):577–588. doi: 10.1002/jcb.22289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kondo M., Yamaoka K., Sakata K., Sonomoto K., Lin L., Nakano K., Tanaka Y. Contribution of the interleukin-6/STAT-3 signaling pathway to chondrogenic differentiation of human mesenchymal stem cells. Arthritis Rheumatol. 2015;67(5):1250–1260. doi: 10.1002/art.39036. [DOI] [PubMed] [Google Scholar]
- 68.Spagnoli A., O'Rear L., Chandler R.L., Granero-Molto F., Mortlock D.P., Gorska A.E., Weis J.A., Longobardi L., Chytil A., Shimer K., Moses H.L. TGF-beta signaling is essential for joint morphogenesis. J. Cell Biol. 2007;177(6):1105–1117. doi: 10.1083/jcb.200611031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shu B., Zhang M., Xie R., Wang M., Jin H., Hou W., Tang D., Harris S.E., Mishina Y., O'Keefe R.J., Hilton M.J., Wang Y., Chen D. BMP2, but not BMP4, is crucial for chondrocyte proliferation and maturation during endochondral bone development. J. Cell Sci. 2011;124(Pt 20):3428–3440. doi: 10.1242/jcs.083659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iwamoto M., Enomoto-Iwamoto M., Kurisu K. Actions of hedgehog proteins on skeletal cells, Crit. Rev. Oral. Biol. Med. 1999;10(4):477–486. doi: 10.1177/10454411990100040401. [DOI] [PubMed] [Google Scholar]
- 71.Deiana M., Malerba G., Dalle Carbonare L., Cheri S., Patuzzo C., Tsenov G., Moron Dalla Tor L., Mori A., Saviola G., Zipeto D., Schena F., Mottes M., Valenti M.T. Physical activity prevents cartilage degradation: a metabolomics study pinpoints the involvement of vitamin B6. Cells. 2019;8(11):1374. doi: 10.3390/cells8111374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong Y., Caplan A.I., Welter J.F., Baskaran H. Glucose availability affects extracellular matrix synthesis during chondrogenesis in vitro. Tissue Eng. 2021;27(19–20):1321–1332. doi: 10.1089/ten.tea.2020.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fang M., Alfieri C.M., Hulin A., Conway S.J., Yutzey K.E. Loss of β-catenin promotes chondrogenic differentiation of aortic valve interstitial cells. Arterioscler. Thromb. Vasc. Biol. 2014;34(12):2601–2608. doi: 10.1161/ATVBAHA.114.304579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goodnough L.H., Chang A.T., Treloar C., Yang J., Scacheri P.C., Atit R.P. Twist1 mediates repression of chondrogenesis by β-catenin to promote cranial bone progenitor specification. Development. 2012;139(23):4428–4438. doi: 10.1242/dev.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jeon H.M., Lee J. MET: roles in epithelial-mesenchymal transition and cancer stemness. Ann. Transl. Med. 2017;5(1):5. doi: 10.21037/atm.2016.12.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marconi G.D., Fonticoli L., Rajan T.S., Pierdomenico S.D., Trubiani O., Pizzicannella J., Diomede F. Epithelial-mesenchymal transition (EMT): the type-2 EMT in wound healing, tissue regeneration and organ fibrosis. Cells. 2021;10(7):1587. doi: 10.3390/cells10071587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Battula V.L., Evans K.W., Hollier B.G., Shi Y., Marini F.C., Ayyanan A., Wang R.Y., Brisken C., Guerra R., Andreeff M., Mani S.A. Epithelial-mesenchymal transition-derived cells exhibit multilineage differentiation potential similar to mesenchymal stem cells. Stem Cell. 2010;28(8):1435–1445. doi: 10.1002/stem.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fager G., Hansson G.K., Gown A.M., Larson D.M., Skalli O., Bondjers G. Human arterial smooth muscle cells in culture: inverse relationship between proliferation and expression of contractile proteins. Vitro Cell Dev. Biol. 1989;25(6):511–520. doi: 10.1007/BF02623563. [DOI] [PubMed] [Google Scholar]
- 79.Roy M.E., Nishimoto S.K. Matrix Gla protein binding to hydroxyapatite is dependent on the ionic environment: calcium enhances binding affinity but phosphate and magnesium decrease affinity. Bone. 2002;31(2):296–302. doi: 10.1016/s8756-3282(02)00821-9. [DOI] [PubMed] [Google Scholar]
- 80.Yagami K., Suh J.Y., Enomoto-Iwamoto M., Koyama E., Abrams W.R., Shapiro I.M., Pacifici M., Iwamoto M. Matrix GLA protein is a developmental regulator of chondrocyte mineralization and, when constitutively expressed, blocks endochondral and intramembranous ossification in the limb. J. Cell Biol. 1999;147(5):1097–1108. doi: 10.1083/jcb.147.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barreto G., Senturk B., Colombo L., Brück O., Neidenbach P., Salzmann G., Zenobi-Wong M., Rottmar M. Lumican is upregulated in osteoarthritis and contributes to TLR4-induced pro-inflammatory activation of cartilage degradation and macrophage polarization. Osteoarthritis Cartilage. 2020;28(1):92–101. doi: 10.1016/j.joca.2019.10.011. [DOI] [PubMed] [Google Scholar]
- 82.Kong L., Zhao Y.P., Tian Q.Y., Feng J.Q., Kobayashi T., Merregaert J., Liu C.J. Extracellular matrix protein 1, a direct targeting molecule of parathyroid hormone-related peptide, negatively regulates chondrogenesis and endochondral ossification via associating with progranulin growth factor. Faseb. J. 2016;30(8):2741–2754. doi: 10.1096/fj.201600261R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y., Li J., Zhu J., Zhou G., Zhang W.J., Cao Y., Liu W. Enhanced cartilage formation by inhibiting cathepsin K expression in chondrocytes expanded in vitro. Biomaterials. 2012;33(30):7394–7404. doi: 10.1016/j.biomaterials.2012.06.070. [DOI] [PubMed] [Google Scholar]
- 84.Kafienah W., Bromme D., Buttle D.J., Croucher L.J., Hollander A.P. Human cathepsin K cleaves native type I and II collagens at the N-terminal end of the triple helix. Biochem. J. 1998;331(Pt 3):727–732. doi: 10.1042/bj3310727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hou W.S., Li Z., Buttner F.H., Bartnik E., Bromme D. Cleavage site specificity of cathepsin K toward cartilage proteoglycans and protease complex formation. Biol. Chem. 2003;384(6):891–897. doi: 10.1515/BC.2003.100. [DOI] [PubMed] [Google Scholar]
- 86.Skoumal M., Haberhauer G., Kolarz G., Hawa G., Woloszczuk W., Klingler A. Serum cathepsin K levels of patients with longstanding rheumatoid arthritis: correlation with radiological destruction. Arthritis Res. Ther. 2005;7(1):R65–R70. doi: 10.1186/ar1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morko J.P., Soderstrom M., Saamanen A.M., Salminen H.J., Vuorio E.I. Up regulation of cathepsin K expression in articular chondrocytes in a transgenic mouse model for osteoarthritis. Ann. Rheum. Dis. 2004;63(6):649–655. doi: 10.1136/ard.2002.004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramirez N.E., Zhang Z., Madamanchi A., Boyd K.L., O'Rear L.D., Nashabi A., Li Z., Dupont W.D., Zijlstra A., Zutter M.M. The α₂β₁ integrin is a metastasis suppressor in mouse models and human cancer. J. Clin. Invest. 2011;121(1):226–237. doi: 10.1172/JCI42328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scott L.E., Weinberg S.H., Lemmon C.A. Mechanochemical signaling of the extracellular matrix in epithelial-mesenchymal transition. Front. Cell Dev. Biol. 2019;7:135. doi: 10.3389/fcell.2019.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gomez E.W., Chen Q.K., Gjorevski N., Nelson C.M. Tissue geometry patterns epithelial-mesenchymal transition via intercellular mechanotransduction. J. Cell. Biochem. 2010;110(1):44–51. doi: 10.1002/jcb.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kalluri R., Neilson E.G. Epithelial-mesenchymal transition and its implications for fibrosis. J. Clin. Invest. 2003;112(12):1776–1784. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.