Abstract
Introduction
The aim of this study was to investigate endogenous retrovirus (ERV) expression and type I interferon (IFN) activation in human pleural mesothelioma (PM) and their association with clinical outcome.
Methods
The expression of ERV was determined from PM cohorts and mesothelial precursor RNA sequencing data. The expression of ERV was confirmed by quantitative polymerase chain reaction (qPCR). Methylation of genomic DNA was assessed by quantitative methylation-specific PCR. DNA demethylation was induced in cells by demethylating agent 5-Aza-2’-deoxycytidine (5-Aza-CdR) treatment. To block type I IFN signaling, the cells were treated with ruxolitinib or MAVS silencing. The expression of IFN-stimulated genes (ISGs) was determined by qPCR and Western blot. Circulating ERVs were detected by qPCR.
Results
Long terminal repeats (LTRs) represent the most abundant transposable elements up-regulated in PM. Within the LTR, ERVmap_1248 and LTR7Y, which are specifically enriched in PM, were further analyzed. The 5-Aza-CdR treatment increased the levels of ERVmap_1248 expression and induced ERVmap_1248 promoter demethylation in mesothelial cells. In addition, ERVmap_1248 promoter was more demethylated in the mesothelioma tissue compared with nontumor tissue. The 5-Aza-CdR treatment of the mesothelial cells also increased the levels of ISGs. Basal ISG expression was higher in the mesothelioma cells compared with the mesothelial cells, and it was significantly decreased by ruxolitinib treatment or MAVS silencing. Furthermore, ISG expression was higher in the tumor tissue with high expression levels of ERVmap_1248. High expression of ERVmap_1248 was associated with longer overall survival and BAP1 mutations. ERVmap_1248 and LTR7Y can be detected in the PM plasma.
Conclusions
We provide clues for patient stratification especially for immunotherapy where best clinical responses are associated with an activated basal immune response.
Keywords: Pleural mesothelioma, Endogenous retroviruses, Type I interferon, BRCA-associated protein 1
Introduction
Pleural mesothelioma (PM) is a rapidly fatal disease arising from the monolayer tissue lining the walls of the pleural cavity and the internal organs housed inside.1 Major drivers include CDKN2A/B, the more PM-specific BAP1, and NF2 mutations.1 Recent data suggest that subclonal NF2 mutations may occur later in mesothelioma development.2,3 Traditionally, the major histologic types of mesothelioma have been the main histologic indicators of prognosis. Indeed, patients with sarcomatoid and biphasic tumors have substantial worse overall survival (OS) compared with patients with epithelioid tumors.4 Recent studies based on multiomics approaches5, 6, 7, 8 have refined the classification into four groups or into gradients based on molecular profiles.
PM is mostly associated with previous exposure to asbestos fibers,1 and we have recently found that exposure to asbestos in mice leads to increased expression levels of endogenous retrovirus (ERV) sequences.9
ERVs are integrated retroviral elements that cover 8% of the human genome.10 They are part of the so-called transposable elements (TEs), which include retrotransposons using RNA as an intermediate that is reverse transcribed into DNA and integrated in the genome and DNA transposons directly excising themselves from one location before reinsertion (reviewed in Wells et al.11). ERV and long-interspersed nuclear elements (LINEs) are autonomous retroelements encoding required proteins for retrotransposition, whereas short-interspersed nuclear elements (SINEs) and SINE-VNTR-Alu (SVA) elements require the machinery from autonomous retrotransposons.
Many ERV sequences are expressed during embryo development and are subsequently epigenetically silenced.12 Nevertheless, certain ERV sequences are actively transcribed and are elevated in cancer.13 Most ERVs in the human genome are nonautonomous long terminal repeat (LTR) elements that are either solitary (solo) LTR or LTR flanking a small segment of internal ERV sequences and are short in length. They are likely to serve as genomic regulators and affect the transcription in cis.14 The autonomous LTRs, however, consist of LTRs that flank potential protein-coding sequences and are near full-length proviral sequences, which could encode disease-associated antigens or functional RNA that regulated gene expression in trans.14 In addition to the effects as transcription regulators, the expression of ERV has been recently explored for its property as inducers of viral mimicry response, especially in immunotherapy context, and we and others have observed that expression of interferon-induced genes is associated with clinical outcome in patients with mesothelioma.9,15
Studying ERV expression has not been regularly implemented in high-throughput studies because repetitive elements are not frequently investigated and the analysis of the few RNA sequencing (RNA-seq) data in mesothelioma has mostly focused on the investigations of known genes.5,7,16 Cancer-specific LTR retroelements are mostly cancer type specific,17 and mesothelioma is one of the cancer types with the highest number of expressed cancer-specific LTR retroelement (eighth of 31 cancer types)17; however, for the time being, ERV expression in mesothelioma has not been thoroughly explored.
In this study, we extended previous work on ERV expression in human cancers17 and mesothelioma experimental animal models,9 and we reveal the expression of ERVs in human mesothelioma which can be detected in the blood and is associated with type I interferon (IFN) signaling and better OS.
Materials and Methods
ERV Analysis
Mesothelioma RNA-seq reads included in the analysis were the following: The Cancer Genome Atlas (TCGA) Mesothelioma Cohort (n = 87) downloaded from the National Center for Biotechnology Information database of genotypes and phenotypes in 2019, under phs000178.v10.p8; the Pleural Mesothelioma cohort from the Bueno study (n = 211) downloaded from the European Genome-phenome Archive (EGA) in 2020, under EGAS00001001563 (EGAD00001001915 and EGAD00001001916); the genetically characterized pleural mesothelioma primary cultures (FunGeST, n = 64) provided by Didier Jean’s team in 2022 for which RNA-Seq was performed as described in Meiller et al.2; and the human embryonic stem cell-derived mesothelium (n = 10) downloaded from the National Center for Biotechnology Information Gene Expression Omnibus in 2020, under GSE113090 (GSM3096389–GSM3096398). The choice of using human embryonic stem cell-derived mesothelium as control was dictated by the fact that RNA-Seq data on normal mesothelial cells or pleura are not available and ERV expression is cancer specific. RNA-seq reads were preprocessed using fastp (0.20.0). For analysis of transposable element (TE) expression, TE transcripts18 was used to obtain TE counts, followed by differential expression analysis using DESeq2. TE loci were considered to be significantly differentially expressed when the adjusted p values were less than 0.01, and where the log2 of the fold change was more than 1 for up-regulated loci and less than −1 for down-regulated loci. Full-length ERV sequences were downloaded from the ERVmap.19 The ERV expression was quantified using feature Counts in the Bioconductor package Rsubreads, where reads mapped uniquely to ERVs were counted. Mutational status of BAP1, NF2, and CDKN2A was extracted from the TCGA7 data set.
PM Patients and Healthy Donors
Tumor tissue was collected from 155 patients with PM and nontumor tissue from six non-PM patients between 1999 and 201520 (Supplementary Table 1A). The study was approved by the Ethical Committee Zürich (KEK-ZH-2012-0094 and BASEC-No. 2020-02566), and either patients signed informed consent or waiver of consent was granted by the Ethical Committee (BASEC-No. 2020-02566). The study methodologies were conformed to the standards set by the Declaration of Helsinki. Tissue samples were processed immediately for total RNA extraction or frozen as previously described.20 To assess that extracted RNA corresponded to tumor tissue containing at least 50% tumor content, we evaluated the PM score, which is based on MSLN, CALB2, and PDPN expression levels as previously described.21
Plasma was collected from 76 patients with PM between 2005 and 2012 and 42 healthy donors between 2002 and 2017 (Supplementary Table 1B). The patients with PM were enrolled in the trial SAKK 17/04 (NCT00334594).22 The study was approved by the Ethics Committee of the Zurich University Hospital (KEK-StV-Nr. 24/05), and either patients signed informed consent or waiver of consent was granted by the Ethical Committee (KEK-StV-Nr. 24/05). The study methodologies conformed to the standards set by the Declaration of Helsinki.
Statistical Analysis
The figures represent the mean values from at least three independent experiments. Paired and unpaired t test, Mann-Whitney, chi-square test, Gehan-Breslow-Wilcoxon test, or Pearson correlation analysis was used and has been specified when used. Error bars indicate the SE of the mean. Statistical analysis was performed using Prism 8 (GraphPad 8.0.0).
All Other Methods
Detailed description of the methods used is available in the Supplementary Methods.
Results
LTRs Represent the Most Abundant TEs Up-Regulated or Down-Regulated in PM Patient Data Sets
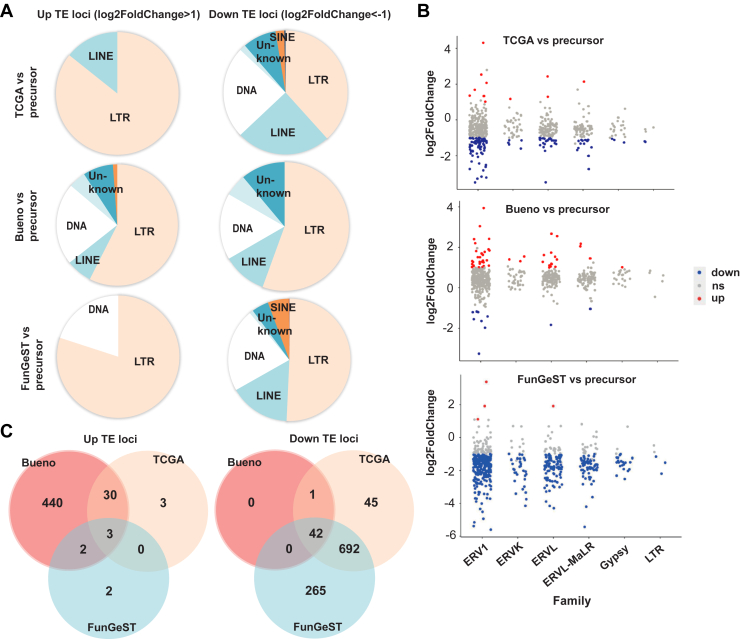
We used the RNA-seq data from TCGA,7 Bueno et al.’s5 tumor sample cohorts, or mesothelioma primary culture cell lines of FunGeST series6,23 and human embryonic stem cell-derived mesothelium24 as nontumor control, to determine which TE subfamilies are differentially expressed in PM tumors or primary cultures. We observed that LTRs represent the most abundant TEs up-regulated or down-regulated in PM patient data sets compared with mesothelial precursors. Furthermore, 86%, 57%, and 80% of the LTRs represent more than twofold up-regulated TEs and 38%, 56%, and 51% of the LTRs represent more than twofold down-regulated TEs in the TCGA, Bueno, and FunGeST data sets, respectively (Fig. 1A and Supplementary Table 3).
Figure 1.
LTRs represent the most abundant TE up-regulated or down-regulated in data sets of patients with PM. (A) Pie charts revealing up- versus down-regulated TE loci. (B) TE loci up- or down-regulated in TCGA, Bueno, or FunGeST versus mesothelial precursor. Results were filtered by padj less than 0.01 and abs (log2Fold changes) greater than 1. Results are revealed for loci within the stated LTR families. (C) Overlap of the significantly up-regulated genes in the three comparisons visualized as Venn diagram. Significance was defined by padj less than 0.01. LTR, long terminal repeat; ns, expression not significantly changed; PM, pleural mesothelioma; TCGA, The Cancer Genome Atlas; TE, transposable element.
The ERV1 family has the highest number of up-regulated or down-regulated loci in LTR (Fig. 1B). Within the ERV1 family, LTR48B, LTR7Y, and LTR6 were most often up-regulated in mesothelioma samples compared with mesothelial precursors (Fig. 1C, left panel). LTR7Y is an ERV specifically up-regulated in blastocyst stage of the human embryo.25 It is noteworthy that some LTR7Y up-regulated in blastocyst stage are located near genes such as WNT16 and FAM3C, which are enriched in PM translatome26 or NCF2 which is correlated to the so-called S-score, which defines the sarcomatoid component proportion of PM by transcriptome analysis8 (Supplementary Fig. 1). The ERV1 family constituted 43% of the most often down-regulated TE in mesothelioma samples compared with mesothelial precursors (Fig. 1C, right panel).
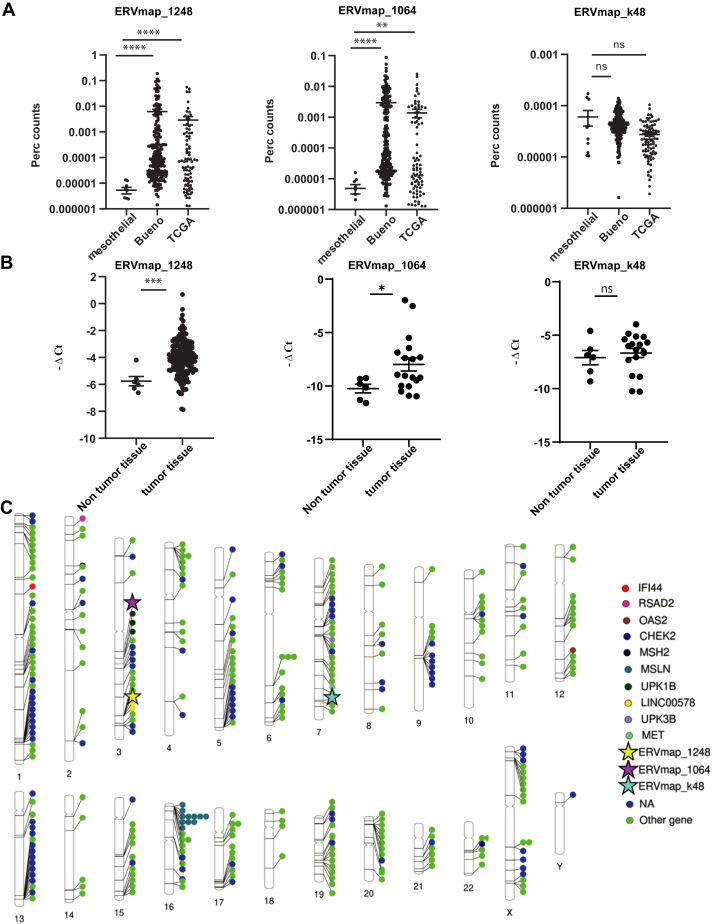
Next, we quantified locus-specific ERV expression comparing RNA-seq and reference ERV sequences using ERVmap,19 which uses stringent filtering criteria for RNA-seq reads that map to ERV loci and a 3220 ERV reference database of full-length proviral sequences.27
We were particularly interested to full-length proviral sequences because it has recently been suggested that viral mimicry primed cancers include elevated baseline expression of retrotransposons that form double-stranded RNA (dsRNA) and elevated levels of retrotransposon-derived antigenic peptide that forms tumor-associated antigens.28 We stratified the ERVmaps by their total counts and specific enrichment in tumor. Two representative ERVs—ERVmap_1248 (hg38, chr3: 177,657,043–177,671,140) and ERVmap_1064 (hg38, chr3: 112,413,019–112,423,381)—both belonging to ERVH group of the ERV1 family, were identified based on their enrichment in tumors. Another ERV sequence, ERVmap_k48 (also called HERV-K1529), where counts almost do not differ between PM cohorts and precursors, was also selected as control (Fig. 2A). Coverage of ERVmap_1248 locus was also enriched in FunGeST series, however to a lower extent (Supplementary Fig. 2A), likely due to the polyA RNA-seq protocol used which has been found to fail to detect several classes of repeat RNA.30 Of note, the patients expressing levels of ERVmap_1248 above the average were enriched for BAP1 mutations but not for NF2 or CDKN2A mutations (Supplementary Fig. 2B). We confirmed that the expressions of ERVmap_1248 and ERVmap_1064 but not of the control ERVmap_k48 were lower in normal compared with tumor tissue (Fig. 2B). The difference was maintained when females were excluded (Supplementary Fig. 2C). We next compared the chromosomal location of the two ERVmaps enriched in mesothelioma with the chromosomal location of ERVs specifically expressed in mesothelioma extracted from a pan-cancer analysis.17 Of note, 28 of 357 (8%) of those ERVs are located near genes relevant for mesothelioma, such as MSLN, MET, UPKB1, UPK3B, LINC00578, some interferon-stimulated genes (RSAD2, IFI44, OAS2), or DNA damage-related genes (CHEK2, MSH2) (Fig. 2C). MSLN locus on chromosome 16 is especially enriched in ERV expression in mesothelioma (11 ERVs). The ERV maps enriched in mesothelioma that we identified were located in ERV expression locations previously described17 (Fig. 2C).
Figure 2.
PM-specific ERV. (A) Expression of two representative ERV sequences ERVmap_1248 and ERVmap_1064 increases in tumors but not the control ERVmap_k-48. (∗) p less than 0.05; (∗∗) p less than 0.01; (∗∗∗) p less than 0.001; (∗∗∗∗) p less than 0.0001; ns, not significant, Mann-Whitney test. (B) RT-qPCR validation of ERV expression in samples from human nontumor tissue and tumor tissue. (∗) p less than 0.05; (∗∗) p less than 0.01; (∗∗∗) p less than 0.001; ns, not significant, Mann-Whitney test. (C) Location of ERVmaps and other ERVs with enriched expression in PM. The ideogram was created using http://visualization.ritchielab.org/phenograms/plot. ERV, endogenous retrovirus; PM, pleural mesothelioma; RT-qPCR, quantitative reverse-transcriptase polymerase chain reaction.
ERVmap_1248 Can Be Induced by 5-Aza-2’-Deoxycytidine Treatment in Mesothelial Cells
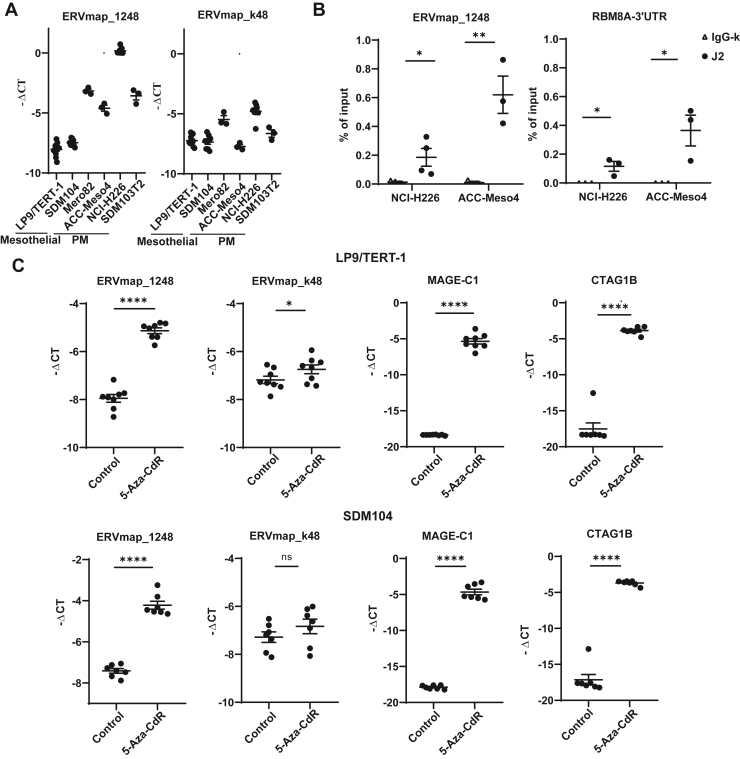
Among the selected ERVs, the most abundant one, ERVmap_1248, and the control ERV, ERVmap_k48, were further analyzed. Basal expression of ERVmap_1248 and ERVmap_k48 was investigated in human mesothelial cells LP9/TERT-1 and SDM104 and in human PM cells Mero82, ACC-Meso4, NCI-H226, and SDM103T2, used as surrogate for normal tissue versus mesothelioma, respectively. The expression of ERVmap_1248 is on average 16-fold more enriched in PM cells compared with ERVmap_k48, supporting the concept of ERVmap_1248 activation in mesothelioma (Fig. 3A). We had previously found that ERVs which are expressed in mouse mesothelioma cells form dsRNA9 and ERVmap_1248 is also predicted to form dsRNA (Supplementary Fig. 3). This was confirmed (Fig. 3B) by RNA pull-down experiments using J2 anti-dsRNA antibody. RNA Binding Motif 8A (RBM8A)-3’UTR which we have previously found to be a substrate for dsRNA editing31 was used as a positive control (Fig. 3B).
Figure 3.
ERVmap_1248 expression is lower in normal mesothelial cells compared with the PM cells, it can form dsRNA and its expression can be induced by 5-Aza-CdR in the mesothelial cells. (A) Basal expression of ERVmap_1248 and ERVmap_k48 in human PM cells and human mesothelial cells. (B) RT-qPCR analysis of ERVmap_1248 and RNA Binding Motif 8A (RBM8A)-3’UTR transcripts captured by J2 antibody in pulldown assay, (∗) p less than 0.05, (∗∗) p less than 0.01, two-tailed paired t tests. RBM8A-3’UTR is used as a positive control. (C) ERVmap_1248 expression increased significantly with 5-Aza-CdR treatment in LP9/TERT-1 and SDM104 mesothelial cells, whereas ERVmap_k48 expression increased by approximately 50% in LP9/TERT1 but not in SDM104 cells. (∗) p less than 0.05; (∗∗∗∗) p less than 0.0001; ns, not significant, two-tailed paired t test. MAGE-C1 and CTAG1B are used as positive controls (36). 5-Aza-CdR, 5-Aza-2’-deoxycytidine; dsRNA, double-stranded RNA; PM, pleural mesothelioma; RT-qPCR, quantitative reverse-transcriptase polymerase chain reaction.
Next, we aimed at investigating whether promoter demethylation was a possible cause for increased ERVmap_1248 expression in PM as previously observed with murine mesothelioma ERV.9 Therefore, DNA demethylation was induced in human mesothelial cells LP9/TERT-1 and SDM104 by treatment with 5-Aza-2’-deoxycytidine (5-Aza-CdR), which is a DNA methyltransferase inhibitor. This treatment resulted in a significantly eightfold increase in ERVmap_1248 expression whereas the control ERV ERVmap_k48 increased by approximately 50% in LP9/TERT1 but not in SDM104 cells (Fig. 3C). The expression of two cancer-associated testis antigens CTAG1B and MAGE-C1 genes was used as positive controls as we previously described.32
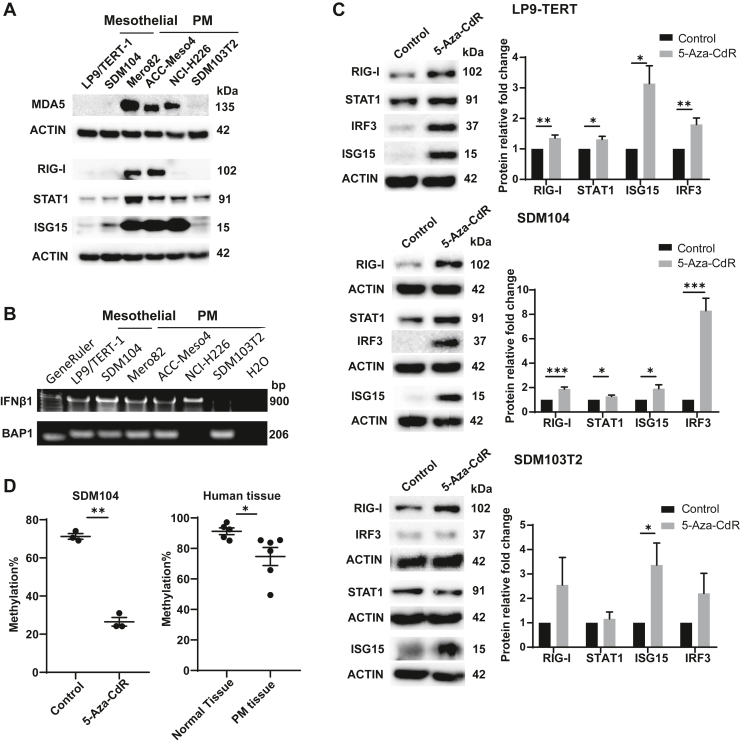
Basal Levels of ISG Expression Are Higher in PM Cells With Intact IFNB1 Coding Gene and 5-Aza-CdR Increases ISG Expression in Mesothelial Cells and It Is Associated With ERVmap_1248 Promoter Demethylation
To assess whether differential ERVmap_1248 expression between mesothelial and PM cells is associated with a differential type I IFN signaling activation as observed in the mouse model,9 we investigated the basal expression levels of various interferon-stimulated genes (ISGs).33 We observed that MDA5, RIG-I, STAT1, and ISG15 levels are higher in most PM cells when compared with mesothelial cells (Fig. 4A). Low levels of RIG-I in H226 cells are likely due to the mutation in RIG-I–encoding DDX58 gene resulting in G191_K193 duplication at the end of the second caspase activation and recruitment domain (http://www.cbioportal.org/). In addition, in PM cell line SDM103T2, ISG levels were at the same level as mesothelial cells (Fig. 4A). We have previously found that basal type I IFN signaling is present in cells with intact IFNB1 gene in PM.34 Accordingly, deficiency of IFNB1 gene was observed in SDM103T2 but not the other cell lines (Fig. 4B). A polymerase chain reaction (PCR) fragment covering exon 5 of BAP1 was used as control for genomic DNA input during PCR (Fig. 4B). BAP1 is known to be deleted in the NCI-H226 cells.35
Figure 4.
Basal ISG expression is higher in the PM cells with intact IFN coding genes and 5-Aza-CdR increased ISGs in mesothelial cells and is associated with promoter demethylation. (A) The expression of ISGs MDA5, RIG-I, STAT1, and ISG15 increased in the PM cells except SDM103T2 cells on protein level. (B) PCR-mediated detection of IFNB1 and BAP1 genes using genomic DNA from mesothelial and PM cells. The PCR fragment of BAP1 was used as DNA input control. (C) The expression of RIG-I, STAT1, IRF3, and ISG15 all increased on 5-Aza-CdR treatment in LP9/TERT-1 and SDM104 mesothelial cells but not all in SDM103T2 PM cells. WB quantification: n = 4–12. (∗) p less than 0.05; (∗∗) p less than 0.01; (∗∗∗) p less than 0.001, two-tailed paired t test. (D) The fraction of ERVmap_1248 promoter methylation decreased after 5-Aza-CdR treatment of SDM104 cells (left panel). The human PM tissue has less methylation percentage in ERVmap_1248 promoter compared with normal tissue. (∗) p less than 0.05; (∗∗) p less than 0.01, Mann-Whitney test. 5-Aza-CdR, 5-Aza-2’-deoxycytidine; IFN, interferon; PCR, polymerase chain reaction; PM, pleural mesothelioma.
We next assessed whether demethylation, which results in increased ERVmap_1248 expression in mesothelial cells, is also associated with increased ISG levels. As expected, we observed increased levels of RIG-I, STAT1, IRF3, and ISG15 in mesothelial cells LP9/TERT-1 and SDM104 on 5-Aza-CdR treatment. IFNB1-deficient PM cell line SDM103T2 was used as control. In this cell line, the increase of gene expression of CTAG1B and MAGE-C1 was observed as expected (Supplementary Fig. 4); however, we observed no significant increase of IFN-dependent STAT1, RIG-I, or IRF3. Only the expression of IFN-independent36 ISG15 was significantly increased (Fig. 4C).
To confirm that the increase in ERVmap_1248 expression observed upon treatment with 5-Aza-CdR is due to promoter demethylation, we first identified the target region based on the analysis of CpG islands (Supplementary Fig. 5) to design methylation-specific primer “M” and unmethylation-specific primer “U” for the region of the promoter of ERVmap_1248. These primers were used on sodium bisulfite-treated DNA, where all methyl-free cytosines are converted into uracils, whereas methylated cytosines remain unchanged allowing the use of quantitative methylation-specific PCR.
In the SDM104 cells, ERVmap_1248 promoter methylation decreased significantly upon 5-Aza-CdR treatment, and we also observed that the ERVmap_1248 promoter is significantly more demethylated in the tumor tissues compared with the nontumor tissues (Fig. 4D).
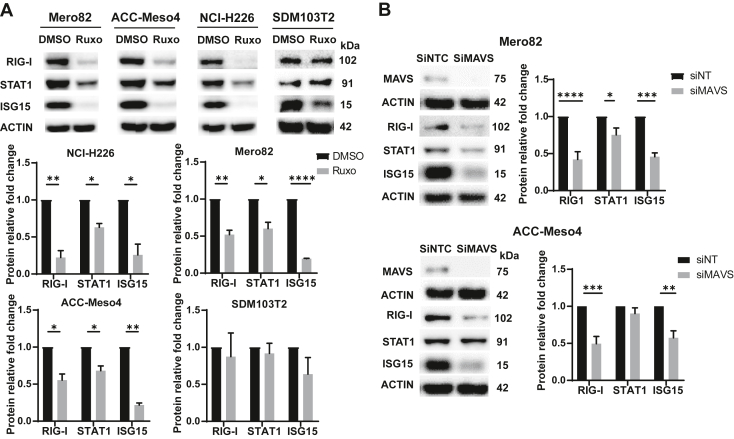
Expression of ISG Can Be Decreased in Mesothelioma Cells by Treatment With JAK Inhibitor Ruxolitinib and MAVS Silencing
To verify the activation of type I IFN signaling, PM cells were treated with ruxolitinib, a JAK1/2 inhibitor blocking the type I IFN signaling. This treatment resulted in decreased levels of ISG in the PM cell lines NCI-H226, Mero82, and ACC-Meso4 with an intact IFNB1 gene but not in IFNB1 gene-deficient cell line SDM103T2 (Fig. 5A and Supplementary Fig. 6). To confirm the involvement of dsRNA sensing, we silenced MAVS, which is downstream of the activation of dsRNA sensors RIG-I and MDA5. Silencing MAVS in the PM cells Mero82 and ACC-Meso4 resulted in a significant decrease of RIG-I, STAT1, and ISG15 (Fig. 5B).
Figure 5.
Basal levels of ISG can be decreased by treatment with JAK inhibitor ruxolitinib and MAVS silencing. (A) The expression of ISGs RIG-I, STAT1, and ISG15 decreased after ruxolitinib (Ruxo), a JAK1/2 inhibitor treatment in the PM cells except SDM103T2 cells. WB quantification: n = 3–4. (∗) p less than 0.05; (∗∗) p less than 0.01; (∗∗∗∗) p less than 0.0001, two-tailed paired t test. (B) Silencing MAVS decreased ISGs RIG-I, STAT1, and ISG15 levels in Mero82 and ACC-Meso4 PM cells. WB quantification: n = 3. (∗) p less than 0.05; (∗∗) p less than 0.01; (∗∗∗) p less than 0.001; (∗∗∗∗) p less than 0.0001, two-tailed paired t test. PM, pleural mesothelioma.
Altogether, these data indicate that increased ISG expression is dependent on type I IFN receptor signaling.
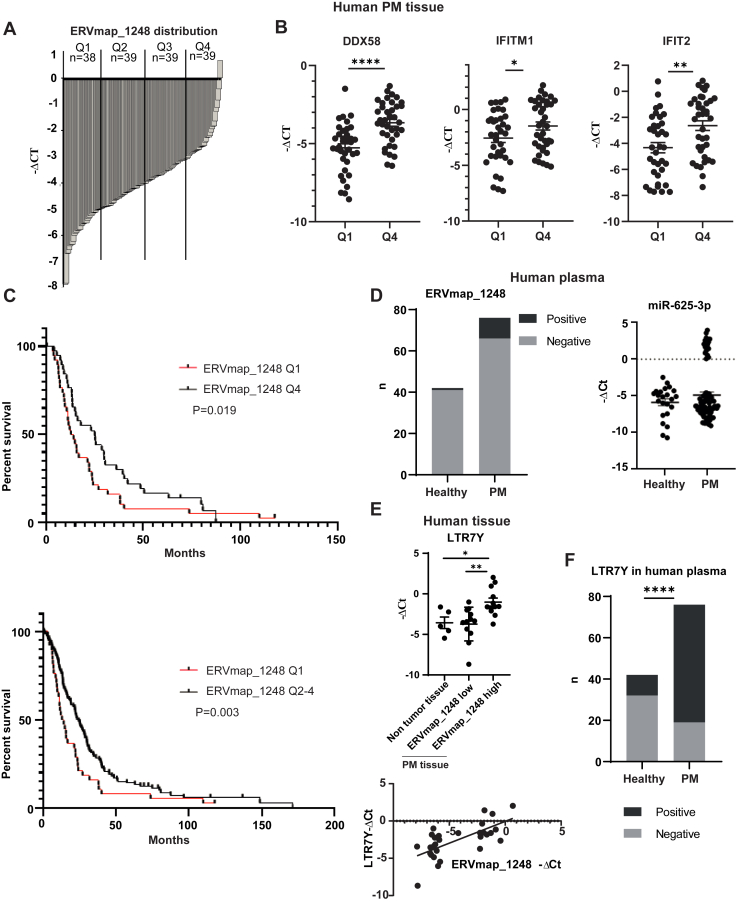
ISG Expression Is Higher in ERVmap_1248 High PM Tissues and Patients With Higher ERVmap1248 Expression Have Better Survival
Patients with PM were distributed into four groups (Q1–Q4) based on their ERVmap_1248 expression (Fig. 6A). Q1 has lowest and Q4 has highest ERVmap_1248 expression. The expression of ISGs, which we previously have associated with better OS,9 was higher in the tumor tissues with high expression levels of ERVmap_1248 (Fig. 6B). Furthermore, ERVmap_1248 expression was analyzed in relation to survival. OS was longer among patients with higher ERVmap_1248 expression (Q4 or Q2–4) compared with patients whose tumors had lower ERVmap_1248 expression (Q1) (Q4 versus Q1: median OS 25.16 versus 13.55 mo, hazard ratio = 1.57, p = 0.019; Q2–4 versus Q1: median OS 24.51 versus 13.55 mo, hazard ratio = 1.761, p = 0.003) (Fig. 6C). This observation was confirmed using the TCGA and Bueno data sets (Supplementary Fig. 7A) and is consistent with the TCGA cluster classification where the cluster with the best survival is related with type I IFN signaling and BAP1 mutation is associated with a longer overall patient survival rate.15 No significant association with histotype was observed (Supplementary Fig. 7B). To increase the clinical relevance of ERV-map1248 expression, we next assessed how its expression was associated with the four-gene (CD8A, STAT1, LAG3, and CD274) inflammatory signature score, which has recently been described to predict response to checkpoint inhibitors in the CheckMate743 trial.37 The four-gene signature score was significantly higher in Q4 compared with the Q1 group (Supplementary Fig. 7C).
Figure 6.
ERVmap_1248 high expression is associated with clinical outcome in PM. (A) ERVmap_1248 expression distribution in tumor tissues from patients with PM. (B) The expression of ISGs DDX58, IFITM1, and IFIT2 is higher in ERVmap_1248 high tumor tissues. (∗) p less than 0.05; (∗∗) p less than 0.01; (∗∗∗∗) p less than 0.0001, two-tailed unpaired t test. (C) Kaplan-Meier curves of overall survival according to ERVmap_1248 expression in patients with PM. Red and black curves represent lower (Q1) or higher ERVmap_1248 (Q4 or Q2–4) expression, respectively. Overall survival was calculated from date of diagnosis. Patients with PM from Q2 or Q2 to 4 have better survival rate than those from Q1. Gehan-Breslow-Wilcoxon test. (D) ERVmap_1248 detection in the plasma of patients with PM and healthy volunteers. Detection of miR-625-3p is used as comparison. (E) LTR7Y and ERVmap_1248 expressions are correlated. (∗) p less than 0.05; (∗∗) p less than 0.01, two-tailed unpaired t test. Correlation: R = 0.6798, p less than 0.0001, Pearson correlation analysis. (F) LTR7Y detection in the plasma of patients with PM and healthy volunteers. (∗∗∗∗) p less than 0.0001, chi-square test. PM, pleural mesothelioma; Q, quarter.
Finally, using plasma collected in the SAKK 17/04 (NCT00334594)22 trial, we determined that circulating ERVmap_1248 (Fig. 6D) can be detected in a small fraction (13%) of patients with PM, whereas only one normal control was positive of 42 normal plasma (2%). Relative levels could not be assessed due to difference in levels of the two normalizers used (Supplementary Fig. 8A). Circulating ERVmap_1248 was independent of the age of the patients (Supplementary Fig. 8B). The quality of RNA was assessed by quantification of relative levels of miR-625-3p where the levels of miR-16 used as normalizer were not different between healthy volunteers and patients with PM (Supplementary Fig. 8C). The bimodal detection of miR-625-3p previously described38 was observed only in the patients with PM (Fig. 6D).
We reasoned that the low sensitivity of ERV detection might be due to ERVmaps being full-length proviral sequences where the size is longer compared with solo-LTR. Therefore, we investigated whether LTR7Y expression could be used as circulating ERV instead of ERVmap_1248. We first determined that LTR7Y expression is enriched in the PM tissues with high ERVmap_1248 expression and that LTR7Y levels are significantly correlated with ERVmap1248 (Fig. 6E). LTR7Y was detected in 75% of the patients; however, it was less specific because it was observed in 24% of healthy controls (Fig. 6F). As observed for ERVmap_1248, circulating LTR7Y was independent of the age and sex (Supplementary Fig. 8D and E). The limitation of this investigation is that plasma samples did not match tumor samples.
Nevertheless, altogether, our data reveal that mesothelioma-specific ERV levels are associated with a better clinical outcome and can be detected in the blood.
Discussion
In this study, we provide evidence that ERV expression is associated with clinical outcome and viral mimicry response in human mesothelioma, consistent with our observation in a mouse model of mesothelioma development in mice exposed to asbestos.9
This is the first time that a specific ERV is associated with clinical outcome in human PM. A previous study17 had identified the location of mesothelioma-specific expressed ERVs based on the comparison of TCGA data with the average expression of ERV in GTEX data set. Interestingly, the location of some of these ERV is near genes relevant for mesothelioma, such as UPK3B and MSLN. For example, UPK3B is a marker for mesothelial cells39 and high levels of UPK3B expression are associated with better OS.5,6 In addition, UPK3B and MSLN are significantly positively correlated with mesothelioma E-score.8 In our study, we used three different data sets and validated the enrichment of specific ERVs, including in the blood from the patients with PM.
Both selected solo-LTR and near full-length proviral sequences have elevated expression levels in mesothelioma. Three solo-LTR were most often enriched in the tumor tissues and mesothelioma primary cultures. Solo-LTR is known to act as an enhancer, and it is noteworthy that LTR7Y acts as stage-specific promoter in pluripotent epiblasts, one of three major cell types in the preimplantation blastocyst.25 LTR7Y expression is controlled by methylation40 and is frequently observed in enhancer regions in naive human embryonic stem cells,41 whereas it is not present in adult tissue.25 The observation that mesothelioma cells express pluripotent cell enhancers is consistent with our previous data where we had used a lentiviral fluorescence-based reporter construct sensing high SOX2 and OCT4 levels to identify and isolate a subpopulation of mesothelioma cells with cancer stem cell properties, characterized by chemoresistance and a higher tumor-initiating capacity in orthotopic xenograft and allograft mouse models.42 Future studies could take advantage of the new knowledge and use the recently described LTR7Y-driven reporter43 to further explore pluripotent mesothelioma cells.
Although not further investigated in this study, LTR7Y is in genomic regions enriched in retroposed genes, or genes linked to mesothelioma biology (e.g., S-score8), which is consistent with promoter or proximal enhancer effect of TE.12 Not much is known about the other two most often expressed solo-LTR, with the exception that a subset of LTR48B elements acquired enhancer activity in the pluripotent cells.44
Consistent with the enrichment in LTR7Y, the three ERVmap genes enriched in mesothelioma belong to the ERVH family, which is characterized by LTR7 promoter family and is expressed early in the embryo.45
In addition to embryogenesis, subsets of LTR7 and LTR7Y elements are known to be up-regulated in oncogenic states due to promoter demethylation.46
A correlation of the LTR7 transcriptional regulatory signals with human embryonic stem cell–specific expression of lncRNAs has been reported,47 including linc-ROR which is enriched in PM translatome.26 This is consistent with the observation that high level of transcription of several ERV loci promotes the expression of lncRNA,48 which seems important in controlling cell identity.49,50
Silencing of ERV in adult tissues occurs through binding of HERV-targeting KZFP, which recruit KAP1/TRIM28 co-repressor to induce heterochromatin formation.41 Therefore, the level of variation of HERVH-associated KZFP can potentially be the reason of differential expression of HERVH in PM. For example, the potential repressor ZNF534, which is particularly enriched in LTR7 and which is associated with pluripotency,45 is up-regulated in sarcomatoid compared with epithelioid mesothelioma.5
The ERVK family was less enriched in mesothelioma. ERVKs are the only ERVs that are human specific with intact open-reading frames, reported to generate viral-like proteins in teratocarcinoma cell lines and human blastocysts.51 ERVmap-k48 used as a control has a sequence of approximately 3900 bp encoding for Gag and is located near the housekeeping gene SSBP1, which has been hypothesized to drive its transcription and possibly explains the reason for stable levels in normal versus tumor tissue.29
KAP1/TRIM28 recruits chromatin modifiers including SETDB1, which is mutated in a subset of mesothelioma,5,7,16 thereby possibly also contributing to differential HERVH and HERVK expression.
According to the knowledge about epigenetic control of ERV expression,52 we observed that ERVmap_1248 expression increases on inhibition of methylation in normal mesothelial cells. Induction of the expression of ERV has been documented in studies supporting the use of viral mimicry in clinical trials, where effects of immune checkpoint inhibitor are tested in combination with demethylating agents.52,53 Basal ERV expression was correlated with low methylation pattern in a pan-cancer analysis.54 Changes in DNA methylation have been documented in human mesothelioma (reviewed in Vandenhoeck et al.55), and we recently discussed9 other factors such as control of DNA methylation that is dysregulated in mesothelioma besides KZFP.
Consistent with the association between expression of transposable elements and the viral mimicry response observed in cancer in general,28 and our own observation in a mesothelioma development model,9 we observed a basal activation of type I IFN signaling in tumors expressing high levels of ERVmap_1248. Of note, based on the mRNA expression profile, mesothelioma tumors have been clustered into four groups.5,7 Pathway-enriched analysis of genes expressed in the clusters revealed, among others, enrichment of reactome antiviral mechanism by ISG in one of the TCGA clusters, and this is confirmed in the epithelioid group of Bueno et al.5 Patients with this profile have a better clinical outcome7,9,15 and BAP1 mutations,15 consistent with our observations that tumors with high levels of ERVmap_1248 are associated with BAP1 mutations.
Mesothelioma cells were previously found to maintain the activation of the type I IFN signaling pathway.9,56 In addition, mesothelioma was described as being a cancer highly enriched for the 38-ISG signature, not always justified by the presence of immune cells in the microenvironment.57 We put forward the hypothesis that ERV expression reactivation in selected samples is a possible cause for type I IFN activation. These tumors are most likely associated with BAP1 but not CDKN2A/B mutations if we consider that IFNB1 and all the 15 other type I IFN genes are co-deleted in a large fraction of tumors bearing CDKN2A/B deletions.34
Activation of type I IFN was associated with response to immune checkpoint blockade in clear cell renal cancer.58 ERV expression is a predictor of patient response to immunotherapy in a urothelial cancer cohort,30 and, interestingly, in that study, it was a better predictor compared with type I IFN signature. Investigation of the expression of 6659 ERVs revealed that some ERVs are associated with both immune activation and checkpoint pathway up-regulation in clear cell renal cell carcinoma60 and expression levels of one of those ERVs predicted response to checkpoint blockade. In addition, high ERV expression was associated with better overall clinical outcome in a cohort of patients with melanoma whereas repression of ERV was observed in the cohort with worst outcome.61 Furthermore, ERVmap_2637 expression was higher in patients with melanoma with complete response to anti–programmed cell death protein 1 treatment62 and negatively correlated with KDM5B expression, which recruits SETDB1. Therefore, our observations are also important for mesothelioma therapy. Indeed, therapeutic approaches exploiting type I IFN pathway signaling have already been implemented in the clinic63 or proposed on the basis of preclinical studies.64,65 Future studies may investigate whether ERV expression could be a predictor of sensitivity to those therapeutic approaches and immune checkpoint inhibition, although it should be taken into account for therapies inducing type I IFN signaling that some mesothelioma have lost type I IFN genes34 and might therefore not be able to activate such signaling. ERV expression could be, for example, helpful to stratify patients with epithelioid histotype, which overall respond less well to immune checkpoint inhibition than those with sarcomatoid histotype.66
CRediT Authorship Contribution Statement
Suna Sun: Validation, Formal analysis, Investigation, Data curation, Writing—original draft, Writing—review and editing, and Visualization.
Weihong Qi: Methodology, Software, Validation, Formal analysis, Data curation, and Writing—review and editing.
Hubert Rehrauer: Conceptualization, Methodology, Software, Validation, and Writing—review and editing.
Manuel Ronner: Investigation and Writing—review and editing.
Ananya Hariharan: Investigation and Writing—review and editing.
Martin Wipplinger: Investigation and Writing—review and editing.
Clément Meiller: Formal analysis and Writing—review and editing.
Rolf Stahel: Resources and Writing—review and editing.
Martin Früh: Resources and Writing—review and editing.
Ferdinando Cerciello: Resources and Writing—review and editing.
Jean-François Fonteneau: Conceptualization, Methodology, and Writing—review and editing.
Didier Jean: Methodology and Writing—review and editing.
Emanuela Felley-Bosco: Conceptualization, Methodology, Formal analysis, Writing—original draft, Writing—review and editing, Supervision, and Funding acquisition.
Acknowledgments
This work was supported by the Swiss National Science Foundation, grant 320030_182690, Walter-Bruckerhoff Stiftung and Stiftung für Angewandte Krebsforchung. Dr. Sun is supported by China Scholarship Council. Dr. Jean is supported by Inserm, the Ligue Contre le Cancer (Ile de France committee), the Chancellerie des Universités de Paris (Legs POIX), and Cancéropôle Île-de-France (Emergence). Dr. Fonteneau is supported by La Ligue Régionale Grand Ouest contre le Cancer, ARSMESO44, Fondation ARC, Institut National du Cancer (INCA-PLBIO21-050), and LabEX IGO program.
The authors are grateful to SAKK 17/04 patients for donating their blood for translational investigation. We are grateful to Dr. Rheinwald for providing LP9/TERT-1 cells.
Footnotes
Disclosure: Dr. Früh reports receiving grants from Bristol-Myers Squibb and AstraZeneca and other support from AstraZeneca, Merck Sharp & Dohme, Roche, Bristol-Myers Squibb, Boehringer Ingelheim, Pfizer, and Takeda, outside of the submitted work. The remaining authors declare no conflicts of interest.
Note: To access the supplementary material accompanying this article, visit the online version of the JTO Clinical and Research Reports at www.jtocrr.org and at https://doi.org/10.1016/j.jtocrr.2022.100430.
Supplementary Data
References
- 1.Carbone M., Adusumilli P.S., Alexander H.R., Jr., et al. Mesothelioma: scientific clues for prevention, diagnosis, and therapy. CA Cancer J Clin. 2019;69:402–429. doi: 10.3322/caac.21572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meiller C., Montagne F., Hirsch T.Z., et al. Multi-site tumor sampling highlights molecular intra-tumor heterogeneity in malignant pleural mesothelioma. Genome Med. 2021;13:113. doi: 10.1186/s13073-021-00931-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang M., Luo J.L., Sun Q., et al. Clonal architecture in mesothelioma is prognostic and shapes the tumour microenvironment. Nat Commun. 2021;12:1751. doi: 10.1038/s41467-021-21798-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauter J.L., Dacic S., Galateau-Salle F., et al. The 2021 WHO classification of tumors of the pleura: advances since the 2015 classification. J Thorac Oncol. 2022;17:608–622. doi: 10.1016/j.jtho.2021.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Bueno R., Stawiski E.W., Goldstein L.D., et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48:407–416. doi: 10.1038/ng.3520. [DOI] [PubMed] [Google Scholar]
- 6.de Reynies A., Jaurand M.C., Renier A., et al. Molecular classification of malignant pleural mesothelioma: identification of a poor prognosis subgroup linked to the epithelial-to-mesenchymal transition. Clin Cancer Res. 2014;20:1323–1334. doi: 10.1158/1078-0432.CCR-13-2429. [DOI] [PubMed] [Google Scholar]
- 7.Hmeljak J., Sanchez-Vega F., Hoadley K.A., et al. Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov. 2018;8:1548–1565. doi: 10.1158/2159-8290.CD-18-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blum Y., Meiller C., Quetel L., et al. Dissecting heterogeneity in malignant pleural mesothelioma through histo-molecular gradients for clinical applications. Nat Commun. 2019;10:1333. doi: 10.1038/s41467-019-09307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun S., Frontini F., Qi W., et al. Endogenous retrovirus expression activates type-I interferon signaling in an experimental mouse model of mesothelioma development. Cancer Lett. 2021;507:26–38. doi: 10.1016/j.canlet.2021.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Hoyt S.J., Storer J.M., Hartley G.A., et al. From telomere to telomere: the transcriptional and epigenetic state of human repeat elements. Science. 2022;376 doi: 10.1126/science.abk3112. eabk3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wells J.N., Feschotte C. A field guide to eukaryotic transposable elements. Annu Rev Genet. 2020;54:539–561. doi: 10.1146/annurev-genet-040620-022145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedli M., Trono D. The developmental control of transposable elements and the evolution of higher species. Annu Rev Cell Dev Biol. 2015;31:429–451. doi: 10.1146/annurev-cellbio-100814-125514. [DOI] [PubMed] [Google Scholar]
- 13.Jansz N., Faulkner G.J. Endogenous retroviruses in the origins and treatment of cancer. Genome Biol. 2021;22:147. doi: 10.1186/s13059-021-02357-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fueyo R., Judd J., Feschotte C., Wysocka J. Roles of transposable elements in the regulation of mammalian transcription. Nat Rev Mol Cell Biol. 2022 doi: 10.1038/s41580-022-00457-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osmanbeyoglu H.U., Palmer G.E., Sagan C., et al. Isolated BAP1 loss in malignant pleural mesothelioma predicts immunogenicity with implications for immunotherapeutic response. bioRxiv. 2022 doi: 10.3390/cancers14225626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Creaney J., Patch A.M., Addala V., et al. Comprehensive genomic and tumour immune profiling reveals potential therapeutic targets in malignant pleural mesothelioma. Genome Med. 2022;14:58. doi: 10.1186/s13073-022-01060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Attig J., Young G.R., Hosie L., et al. LTR retroelement expansion of the human cancer transcriptome and immunopeptidome revealed by de novo transcript assembly. Genome Res. 2019;29:1578–1590. doi: 10.1101/gr.248922.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin Y., Tam O.H., Paniagua E., Hammell M. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics. 2015;31:3593–3599. doi: 10.1093/bioinformatics/btv422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tokuyama M., Kong Y., Song E., Jayewickreme T., Kang I., Iwasaki A. ERVmap analysis reveals genome-wide transcription of human endogenous retroviruses. Proc Natl Acad Sci U S A. 2018;115:12565–12572. doi: 10.1073/pnas.1814589115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oehl K., Kresoja-Rakic J., Opitz I., et al. Live-cell mesothelioma biobank to explore mechanisms of tumor progression. Front Oncol. 2018;8:40. doi: 10.3389/fonc.2018.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sidi R., Pasello G., Opitz I., et al. Induction of senescence markers after neo-adjuvant chemotherapy of malignant pleural mesothelioma and association with clinical outcome: an exploratory analysis. Eur J Cancer. 2011;47:326–332. doi: 10.1016/j.ejca.2010.09.044. [DOI] [PubMed] [Google Scholar]
- 22.Stahel R.A., Riesterer O., Xyrafas A., et al. Neoadjuvant chemotherapy and extrapleural pneumonectomy of malignant pleural mesothelioma with or without hemithoracic radiotherapy (SAKK 17/04): a randomised, international, multicentre phase 2 trial. Lancet Oncol. 2015;16:1651–1658. doi: 10.1016/S1470-2045(15)00208-9. [DOI] [PubMed] [Google Scholar]
- 23.Quetel L., Meiller C., Assié J.B., et al. Genetic alterations of malignant pleural mesothelioma: association with tumor heterogeneity and overall survival. Mol Oncol. 2020;14:1207–1223. doi: 10.1002/1878-0261.12651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colunga T., Hayworth M., Kress S., et al. Human pluripotent stem cell-derived multipotent vascular progenitors of the mesothelium lineage have utility in tissue engineering and repair. Cell Rep. 2019;26:2566–2579.e2510. doi: 10.1016/j.celrep.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goke J., Lu X., Chan Y.S., et al. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell. 2015;16:135–141. doi: 10.1016/j.stem.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Grosso S., Marini A., Gyuraszova K., et al. The pathogenesis of mesothelioma is driven by a dysregulated translatome. Nat Commun. 2021;12:4920. doi: 10.1038/s41467-021-25173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vargiu L., Rodriguez-Tome P., Sperber G.O., et al. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology. 2016;13:7. doi: 10.1186/s12977-015-0232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen R., Ishak C.A., De Carvalho D.D. Endogenous retroelements and the viral mimicry response in cancer therapy and cellular homeostasis. Cancer Discov. 2021;11:2707–2725. doi: 10.1158/2159-8290.CD-21-0506. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt K., Reichrath J., Roesch A., Meese E., Mayer J. Transcriptional profiling of human endogenous retrovirus group HERV-K(HML-2) loci in melanoma. Genome Biol Evol. 2013;5:307–328. doi: 10.1093/gbe/evt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solovyov A., Vabret N., Arora K.S., et al. Global cancer transcriptome quantifies repeat element polarization between immunotherapy responsive and T cell suppressive classes. Cell Rep. 2018;23:512–521. doi: 10.1016/j.celrep.2018.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abukar A., Wipplinger M., Hariharan A., et al. Double-stranded RNA structural elements holding the key to translational regulation in cancer: the case of editing in RNA-binding motif protein 8A. Cells. 2021;10 doi: 10.3390/cells10123543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kresoja-Rakic J., Kapaklikaya E., Ziltener G., et al. Identification of cis- and trans-acting elements regulating calretinin expression in mesothelioma cells. Oncotarget. 2016;7:21272–21286. doi: 10.18632/oncotarget.7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaunay T., Achard C., Boisgerault N., et al. Frequent homozygous deletions of type I interferon genes in pleural mesothelioma confer sensitivity to oncolytic measles virus. J Thorac Oncol. 2020;15:827–842. doi: 10.1016/j.jtho.2019.12.128. [DOI] [PubMed] [Google Scholar]
- 35.Jensen D.E., Proctor M., Marquis S.T., et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 ring finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 36.Grandvaux N., Servant M.J., tenOever B., et al. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters S., Scherpereel A., Cornelissen R., et al. First-line nivolumab plus ipilimumab versus chemotherapy in patients with unresectable malignant pleural mesothelioma: 3-year outcomes from CheckMate 743. Ann Oncol. 2022;33:488–499. doi: 10.1016/j.annonc.2022.01.074. [DOI] [PubMed] [Google Scholar]
- 38.Kresoja-Rakic J., Szpechcinski A., Kirschner M.B., et al. Vol. 5. Noncoding RNA; 2019. miR-625-3p and lncRNA GAS5. (Liquid Biopsies for Predicting the Outcome of Malignant Pleural Mesothelioma Patients Treated With Neo-adjuvant Chemotherapy and Surgery). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanamori-Katayama M., Kaiho A., Ishizu Y., et al. LRRN4 and UPK3B are markers of primary mesothelial cells. PLOS ONE. 2011;6 doi: 10.1371/journal.pone.0025391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Theunissen T.W., Friedli M., He Y., et al. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19:502–515. doi: 10.1016/j.stem.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pontis J., Planet E., Offner S., et al. Hominoid-specific transposable elements and KZFPs facilitate human embryonic genome activation and control transcription in naive human ESCs. Cell Stem Cell. 2019;24:724–735. doi: 10.1016/j.stem.2019.03.012. e725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blum W., Pecze L., Felley-Bosco E., Wu L., de Perrot M., Schwaller B. Stem cell factor-based identification and functional properties of in vitro-selected subpopulations of malignant mesothelioma cells. Stem Cell Rep. 2017;8:1005–1017. doi: 10.1016/j.stemcr.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szczerbinska I., Gonzales K.A.U., Cukuroglu E., et al. A chemically defined feeder-free system for the establishment and maintenance of the human naive pluripotent state. Stem Cell Rep. 2019;13:612–626. doi: 10.1016/j.stemcr.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casanova M., Moscatelli M., Chauviere L.E., et al. A primate-specific retroviral enhancer wires the XACT lncRNA into the core pluripotency network in humans. Nat Commun. 2019;10:5652. doi: 10.1038/s41467-019-13551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter T.A., Singh M., Dumbovic G., Chobirko J.D., Rinn J.L., Feschotte C. Mosaic cis-regulatory evolution drives transcriptional partitioning of HERVH endogenous retrovirus in the human embryo. eLife. 2022:11. doi: 10.7554/eLife.76257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kong Y., Rose C.M., Cass A.A., et al. Transposable element expression in tumors is associated with immune infiltration and increased antigenicity. Nat Commun. 2019;10:5228. doi: 10.1038/s41467-019-13035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelley D., Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012;13:R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kapusta A., Kronenberg Z., Lynch V.J., et al. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLOS Genet. 2013;9 doi: 10.1371/journal.pgen.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu X., Sachs F., Ramsay L., et al. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol. 2014;21:423–425. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- 50.Wang J., Xie G., Singh M., et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516:405–409. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- 51.Grow E.J., Flynn R.A., Chavez S.L., et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiappinelli K.B., Strissel P.L., Desrichard A., et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roulois D., Loo Yau H., Singhania R., et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith C.C., Beckermann K.E., Bortone D.S., et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J Clin Invest. 2018;128:4804–4820. doi: 10.1172/JCI121476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vandenhoeck J., van Meerbeeck J.P., Fransen E., et al. DNA methylation as a diagnostic biomarker for malignant mesothelioma: a systematic review and meta-analysis. J Thorac Oncol. 2021;16:1461–1478. doi: 10.1016/j.jtho.2021.05.015. [DOI] [PubMed] [Google Scholar]
- 56.Chernova T., Sun X.M., Powley I.R., et al. Molecular profiling reveals primary mesothelioma cell lines recapitulate human disease. Cell Death Differ. 2016;23:1152–1164. doi: 10.1038/cdd.2015.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu H., Golji J., Brodeur L.K., et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat Med. 2019;25:95–102. doi: 10.1038/s41591-018-0302-5. [DOI] [PubMed] [Google Scholar]
- 58.de Cubas A.A., Dunker W., Zaninovich A., et al. DNA hypomethylation promotes transposable element expression and activation of immune signaling in renal cell cancer. JCI Insight. 2020;5 doi: 10.1172/jci.insight.137569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mayer J., Blomberg J., Seal R.L. A revised nomenclature for transcribed human endogenous retroviral loci. Mob DNA. 2011;2:7. doi: 10.1186/1759-8753-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Panda A., de Cubas A.A., Stein M., et al. Endogenous retrovirus expression is associated with response to immune checkpoint blockade in clear cell renal cell carcinoma. JCI Insight. 2018;3 doi: 10.1172/jci.insight.121522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Badal B., Solovyov A., Di Cecilia S., et al. Transcriptional dissection of melanoma identifies a high-risk subtype underlying TP53 family genes and epigenome deregulation. JCI Insight. 2017;2 doi: 10.1172/jci.insight.92102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang S.M., Cai W.L., Liu X., et al. KDM5B promotes immune evasion by recruiting SETDB1 to silence retroelements. Nature. 2021;598:682–687. doi: 10.1038/s41586-021-03994-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sterman D.H., Recio A., Haas A.R., et al. A phase I trial of repeated intrapleural adenoviral-mediated interferon-beta gene transfer for mesothelioma and metastatic pleural effusions. Mol Ther. 2010;18:852–860. doi: 10.1038/mt.2009.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vanbervliet-Defrance B., Delaunay T., Daunizeau T., et al. Cisplatin unleashes toll-like receptor 3-mediated apoptosis through the downregulation of c-FLIP in malignant mesothelioma. Cancer Lett. 2020;472:29–39. doi: 10.1016/j.canlet.2019.12.016. [DOI] [PubMed] [Google Scholar]
- 65.Achard C., Boisgerault N., Delaunay T., et al. Sensitivity of human pleural mesothelioma to oncolytic measles virus depends on defects of the type I interferon response. Oncotarget. 2015;6:44892–44904. doi: 10.18632/oncotarget.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baas P., Scherpereel A., Nowak A.K., et al. First-line nivolumab plus ipilimumab in unresectable malignant pleural mesothelioma (CheckMate 743): a multicentre, randomised, open-label, phase 3 trial. Lancet. 2021;397:375–386. doi: 10.1016/S0140-6736(20)32714-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.