Abstract
Purpose
Atopic dermatitis (AD) is a chronic inflammatory skin disease characterized by a wide spectrum of clinical phenotype. However, specific description of phenotypes of AD depending on the comorbidities in early childhood is lacking. This study aimed to investigate whether the AD phenotype in early childhood is related to childhood asthma and to elucidate the mechanisms involved.
Methods
Data on the first 3 years of life were collected prospectively from 1,699 children in the COhort for Childhood Origin of Asthma and allergic diseases (COCOA). We applied an unsupervised latent class analysis to the following five factors: food sensitization, inhalant sensitization, food allergy (FA), AD, and recurrent wheezing. The risks of developing FA, AD, allergic rhinitis (AR), and asthma in children aged 5–7 years were evaluated. Colonocyte transcriptome and ingenuity pathway analysis were performed.
Results
Four phenotypes were identified; no allergic diseases (78.4%), AD without sensitization (16.4%), FA with AD (2.9%), and AD with sensitization (7.8%). The FA with AD had the highest risk for FA, AR, and asthma and the highest cord blood immunoglobulin E (IgE) levels. In AD without sensitization and with sensitization, scoring of AD (SCORAD) in early childhood was higher than in FA with AD. Canonical pathway analysis with the colonocyte transcriptome revealed that the key pathway in FA with AD was ‘Wnt/β-catenin Signaling.’ The relative abundance of Wnt6 mRNA was positively correlated with food-specific IgE levels at 1 and 3 years.
Conclusions
When FA is present in various phenotypes of AD at early life, regardless of severity of eczema, it may be associated with gut Wnt signaling and later development of asthma.
Keywords: Asthma, dermatitis, atopic, phenotype, food hypersensitivity, latent class analysis, Wnt signaling pathway, transcriptome
INTRODUCTION
Atopic dermatitis (AD) is a complex disease with multiple causes and complex mechanistic pathways according to age of onset, severity, and comorbidity It has been well established that the atopic march only concerns from one-third to a half of children with AD.1 Early-onset, persistent AD, atopy, and family atopy are risk factors for childhood asthma or development of multiple comorbidities.2,3 In another high-risk cohort, most children with early-onset AD and no wheezing did not have an increased asthma risk.4 Therefore, the link between AD and asthma seems more complex than ever before. Moreover, although most of the atopic march studies have focused on T helper cell 2 responses,5 the exact mechanism of the atopic march has not been identified.6
Using various machine learning techniques, such as latent class analysis (LCA), several cohorts have deciphered various patterns of sensitization or AD from childhood to adolescence and their differential risks for various allergic diseases at later ages.7,8,9 Although food sensitization is frequent in children with early-onset AD, food allergy (FA) is not systematic. In the Observatory of Risks linked with Cutaneous Atopy (ORCA) study where all the children had early-onset (<12 months) moderate-to-severe AD, 57.5% had food sensitization at baseline, where FA was reported in only 11.7%.10 These results were very similar to those from the Danish Allergy Research Cohort (DARC) study.11 Therefore, it is necessary to analyze food sensitization and FA separately. However, no study with LCA analysis has focused on AD phenotypes, including FA, in early childhood.
We hypothesized that the risk of allergic diseases at a later age will vary according to the phenotype of AD in early childhood. We applied LCA to data from the COhort for Childhood Origin of Asthma and allergic diseases (COCOA) to examine the relationships of changes in AD, FA, wheeze, and sensitization from birth to age 3 with the development of allergic diseases in children aged 5–7 years. Furthermore, we investigated the early life colonocyte transcriptome in each phenotype, and ingenuity pathway analysis (IPA) was used to identify candidate pathways.
MATERIALS AND METHODS
Study design and participants
The methods of ‘COCOA’—a population-based, longitudinal birth cohort study in Korea—have previously been described.12 Of the 2,358 participants who provided consent, 583 were excluded due to their age being less than 3 years, and 76 were excluded due to a lack of data on a physician’s report at 1 and 3 years. The current LCA involved 1,699 children for whom complete data on specific immunoglobulin E (IgE) to milk and egg white at age 1 and the results of a skin prick test (SPT) at age 3 were available, and whose FA, AD, and recurrent wheezing status had been ascertained by clinical assessment by COCOA physicians at 6 months of age and annually thereafter. Allergic disease outcomes for 878 children were assessed by pediatric allergists in children aged 5, 6, and 7 years (Supplementary Table S1, Supplementary Fig. S1).
The ethics committees of each participating institution approved the current study protocol. This study was approved by the Institutional Review Boards of Asan Medical Center (IRB No. 2008-0616), Samsung Medical Center (IRB No. 2009-02-021), Severance Hospital (IRB No. 4-2008-0588), the CHA Medical Center (IRB No. 2010-010), and the Seoul National University Hospital (IRB No. H-1401-086-550).
Definitions
A child is deemed to have atopy when at least one common allergen evokes a positive response (wheal diameter ≥ 3 mm), the mean wheal diameter to histamine exceeds 3 mm, and serum-specific IgE to milk or egg white at age 1 is > 0.35 kU/L. During the clinic visit at 6 months or 1, 2, and 3 years, children were assessed for atopic dermatitis (AD) and FA by interviewing their parents about symptoms in the past year and diagnosed by a pediatric allergist. The severity of AD using the scoring of AD (SCORAD) index was assessed annually. FA was diagnosed as the presence of definite allergic symptoms after ingestion of a specific causative food with a time interval of ≤4 hours from food ingestion to symptom awareness, followed by repeated allergic symptoms from the same definitive causative food or the complete avoidance of the definitive causative food after the development of allergy. Allergic symptoms included skin rashes/wheals or gastrointestinal/respiratory/cardiovascular symptoms. Recurrent wheezing was defined as the presence of at least two wheezing episodes during the first 3 years of life.
In 878 children aged 5, 6, and 7 years, COCOA children were defined as having asthma, allergic rhinitis (AR), AD, and FA with their respective diagnoses within the last 12 months and analyzed as clinical outcomes.
Total IgE and sensitization (specific IgE and SPT)
Total IgE levels of parents, cord blood, and children at age 1 and 3 were measured by using the UniCAP system (Pharmacia, Uppsala, Sweden). The SPTs were performed with 14 common inhalant allergens and 4 food allergens for children at 3 years of age.12 Levels of specific IgE to milk and egg white were measured in children at age 1. For details about measurement of IgE and SPTs were described in Supplementary Data S1.
Colonocyte transcriptome and pathway analysis
Transcriptome data on colonocytes were generated for 70 subjects (40 no-allergic disease, 15 AD without sensitization, 18 AD with sensitization, and 7 FA with AD). Fecal samples, collected from infants who did not take antibiotics at 6 months of age, were immediately frozen at −80°C before being processed for RNA extraction. Differentially expressed genes (DEGs) were identified based on a cutoff P value < 0.05 and an absolute fold change (FC) of 1.2. To identify canonical pathways for each group, especially FA with AD, we used the IPA software (Qiagen, Redwood City, CA, USA) with default parameters and selected Human Gene 2.0 ST Arrays as the platform (Supplementary Data S1).
Statistical analysis
To understand the overall patterns of allergic diseases in the data, we employed an exploratory, unsupervised LCA with no preconceived hypothesis. Ten factors involved between 1 and 3 years of age were evaluated to reveal distinct patterns of sensitization, FA, AD, and recurrent wheezing: food sensitization at 1 and 3 years; inhalant sensitization at 3 years; FA at 1, 2, and 3 years; AD at 1, 2, and 3 years; recurrent wheezing during the first 3 years of life (Supplementary Table S2, Supplementary Data S1). We evaluated the risk of demonstrating FA, AD, AR, and asthma at 5, 6, and 7 years for each of the identified LCA classes compared to the “no allergic disease” class. Odds ratios were obtained from multivariable logistic regression models with robust variance to account for the strong clustering effects of study centers.13 Correlations between the total IgE, specific IgE to milk, SCORAD at 12 months of age, and relative abundance of mRNA expression were analyzed using the Spearman correlation test. Stata 12 and LCA Stata plug-in were used for statistical analyses.
RESULTS
Study population and latent class characterized
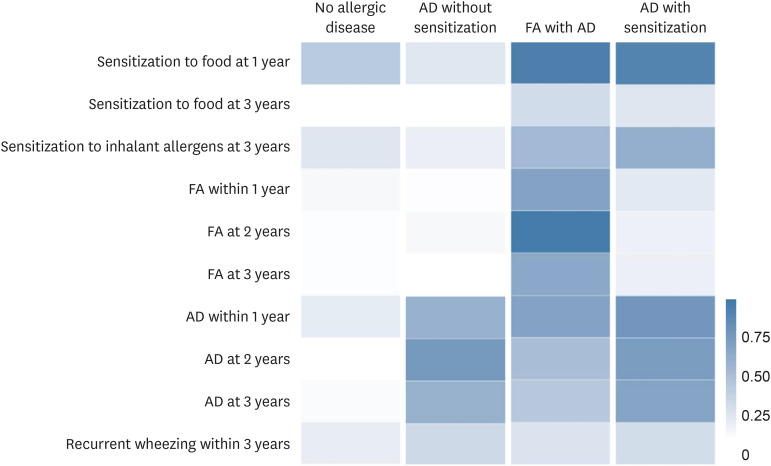
The LCA revealed solutions with 2 to 7 classes, with the best Akaike information criterion values for the 4-class solutions in this study (Supplementary Table S3). Item response probabilities for the 4 phenotypes of allergic disease based on the following 4 classes are shown in Fig. 1 and Supplementary Table S2.
Fig. 1. Probability of allergic diseases and allergic sensitization conditional on latent class memberships.
AD, atopic dermatitis; FA, food allergy.
• No allergic disease (78.4%)
• AD without sensitization (16.4%): Displayed very little sensitization to food and inhalant allergens and FA at 1 and 3 years; however, many had AD in early childhood (1 year: 54.1%, 2 years: 71.9%, 3 years: 54.9%).
• FA with AD (2.9%): Characterized by higher prevalence of FA during the first 3 years (1 year: 64.5%, 2 years: 94.8%, 3 years: 60.5%) compared to the other classes. As they aged, the prevalence of AD decreased significantly (1 year: 64.2%, 2 years: 44.6%, 3 years: 37.9%). Recurrent wheezing remained low (18.7%).
• AD with sensitization (7.8%): The 14.8% prevalence of FA at 1 year decreased to 9.4% and 9.8% at 2 and 3 years, despite persistent high prevalence of AD (1 year; 73.8%, 2 years: 69.2%, 3 years: 63.8%). Recurrent wheezing remained low (23.3%).
Risk factors for AD phenotypes
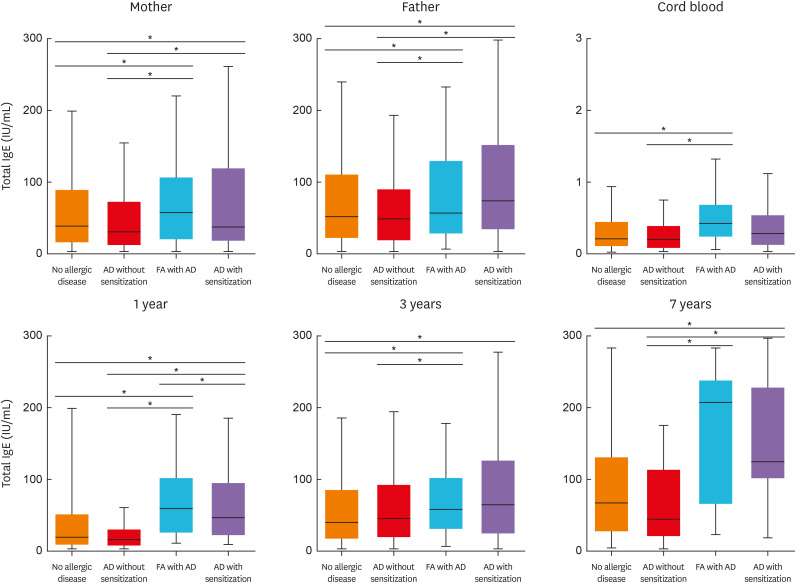
After adjustment for potential confounders in multinomial logistic regression, maternal FA and AR remained significant risk factors for FA with AD (Supplementary Table S4). Parental AD and atopy increased the risk of AD with sensitization. Cord blood total IgE levels were significantly higher in FA with AD than in the no-allergic disease and AD without sensitization (both P < 0.01, Fig. 2). Parental IgE and total IgE levels at 1 and 3 years were significantly higher in both FA with AD and AD with sensitization. In AD without sensitization and AD with sensitization, SCORAD in early childhood was higher than in FA with AD (Table).
Fig. 2. Comparison of parental and periodic IgE levels according to allergic disease phenotypes in early childhood (using analysis of variance test).
IgE, immunoglobulin E; AD, atopic dermatitis; FA, food allergy.
*P < 0.05.
Table. Comparison of SCORAD according to allergic disease phenotypes in early childhood.
| Variables | No-allergic disease | AD without sensitization | FA with AD | AD with sensitization | P value | ||||
|---|---|---|---|---|---|---|---|---|---|
| No. | Mean ± SD | No. | Mean ± SD | No. | Mean ± SD | No. | Mean ± SD | ||
| SCORAD at 1 year | 1,092 | 0.9 ± 3.76 | 122 | 6.0 ± 8.44 | 39 | 7.1 ± 9.75 | 103 | 9.7 ± 11.0 | 0.08 |
| SCORAD at 2 years | 1,051 | 0.5 ± 2.82 | 140 | 13.8 ± 11.2* | 38 | 5.7 ± 9.66 | 104 | 13.4 ± 11.9* | 0.01 |
| SCORAD at 3 years | 1,050 | 0.8 ± 3.81 | 125 | 10.8 ± 13.9* | 33 | 6.4 ± 9.75 | 96 | 13.1 ± 11.9* | < 0.01 |
P value: Least significant difference.
SCORAD, scoring of atopic dermatitis; AD, atopic dermatitis; FA, food allergy; SD, standard deviation.
*P < 0.05 compared with no-allergic disease.
Risks for allergic diseases at 5, 6, and 7 years
AD
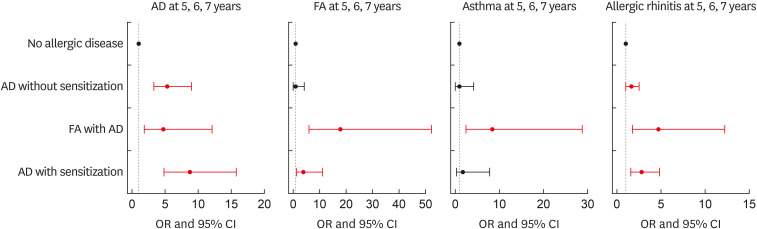
Compared to the no-allergic disease phenotype, children in AD with sensitization had the greatest risk of developing AD at 5–7 years, with an odds ratio (OR) of 8.70 (95% confidence interval [CI], 4.82–15.7; Fig. 3). Despite AD being much less common at 3 years of age, its risk was higher among children in the FA with AD as well as in AD without sensitization (adjusted OR [aOR], 4.72; 95% CI, 1.85–12.1 and aOR, 5.42; 95% CI, 3.28–8.96, respectively).
Fig. 3. Odds for AD, FA, AR, and asthma in preschool children (5, 6, and 7 years old) within each identified pattern.
AD, atopic dermatitis; FA, food allergy; AR, allergic rhinitis; OR, odds ratio; CI, confidence interval.
*Adjusted for sex, parental history of allergic diseases, maternal education, maternal age at delivery, second-hand smoking during pregnancy, and delivery mode.
FA
The risk of FA at 5–7 years was considerably elevated for those with FA with AD (aOR, 17.9; 95% CI, 6.08–52.7). Although the prevalence of FA decreased at ages 2 and 3 in AD with sensitization, the risk of FA increased at 5–7 years (aOR, 3.84; 95% CI, 1.31–11.3).
Asthma
FA with AD was the only phenotype to have increased odds of developing a childhood asthma (aOR, 8.40; 95% CI, 2.45–28.8). Children in AD with/without sensitization phenotypes were not at increased risk of developing asthma.
AR
Children in FA with AD were almost five times more likely to develop AR at age 5–7 years than those with no allergic disease (aOR, 4.72; 95% CI, 1.85–12.1) However, unlike asthma, the risk was also increased among children in AD with/without sensitization (aOR, 2.79; 95% CI, 1.61–4.85 and aOR, 1.66; 95% CI, 1.07–2.57, respectively).
IPA with colonocyte transcriptome
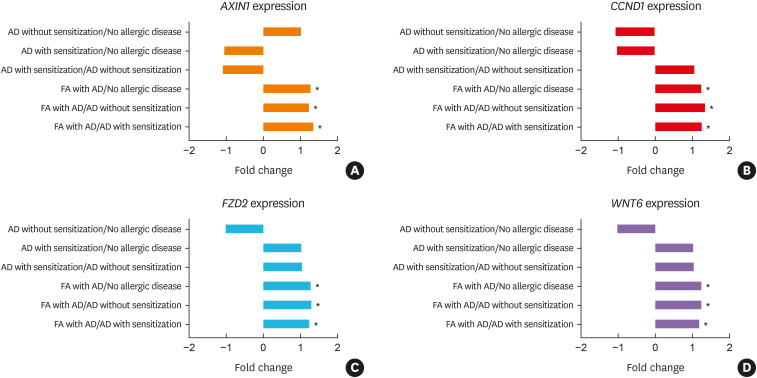
‘Colorectal Cancer Metastasis Signaling,’ ‘Wnt/β-catenin Signaling,’ and ‘Factors Promoting Cardiogenesis in Vertebrates’ showed predicted activation in all comparisons that included FA with AD phenotype (Supplementary Fig. S2).14,15,16,17 Activation of Axin 1 (AXIN1), Cyclin D1 (CCND1), Frizzled Class Receptor 2 (FZD2), and Wnt Family Member 6 (WNT6) was common among the genes of the three canonical pathways and was significantly higher in all comparisons that included FA with AD (Fig. 4, Supplementary Table S5). These four genes are all Wnt signaling pathway-related molecules. Next, we sought to identify up- and downstream molecules mediating ‘Wnt/β-catenin signaling’ in the transcriptome data sets using IPA software. As shown in Supplementary Fig. S3A, significant induction of the expression of their respective genes was observed. Canonical pathway analysis further revealed that the pathway depicted in ‘Role of Wnt/GSK-3β Signaling in the Pathogenesis of Influenza’18 exhibited high regulation of genes at the levels of both receptors and intracellular effectors in FA with AD compared to AD with sensitization (Supplementary Fig. S3B). In spite of the activation of WNT signaling, the antiviral response was reduced due to a decrease in interferon (IFN) levels.
Fig. 4. The expression FC of selected genes in each comparison from the microarray.
FC, fold change; AD, atopic dermatitis; FA, food allergy; AXIN1, Axin 1; CCND1, Cyclin D1; FZD2, Frizzled Class Receptor 2; WNT6, Wnt Family Member 6.
*P < 0.05.
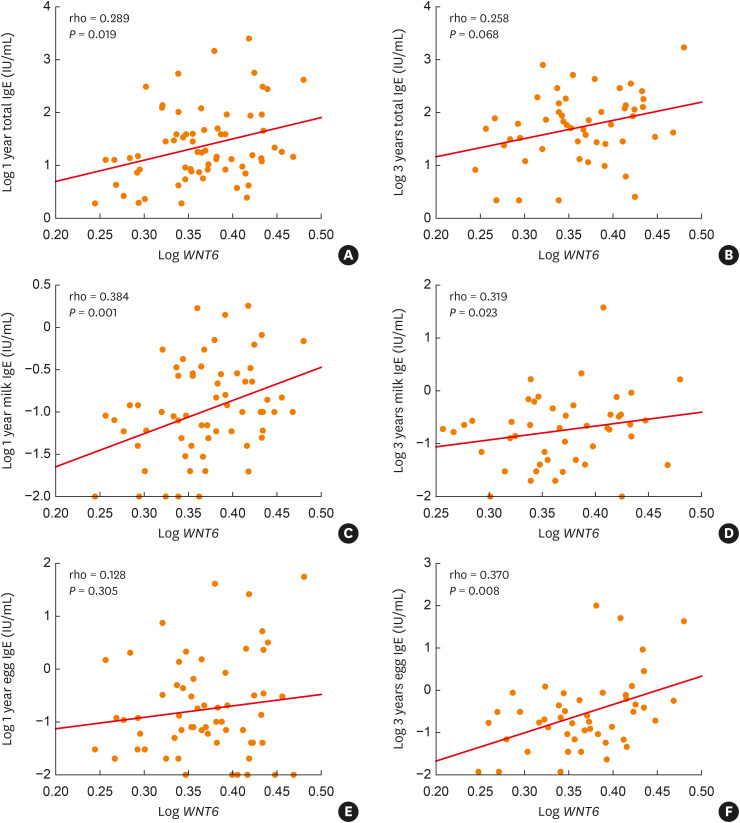
The relative abundance of Wnt6 mRNA levels was positively correlated with total IgE, specific IgE to milk at age 1, and specific IgE to milk and egg white at age 3 (P < 0.05, respectively; Fig. 5).
Fig. 5. Correlation between relative abundance of WNT6 mRNA levels and (A and B) total IgE at 1 and 3 years, (C and D) specific IgE to milk at 1 and 3 years, and (E and F) specific IgE to egg white at 1 and 3 years.
WNT6, Wnt Family Member 6; IgE, immunoglobulin E.
DISCUSSION
Children having FA with AD phenotype showed an increased risk of asthma at 5–7 years of age. The AD phenotype in early life is closely related to the development of asthma only in the cases of accompanying FA. IPA of the colonocyte transcriptome revealed that differentiation of FA with AD from the other 3 classes and from AD with sensitization was best described by the genes in ‘Wnt/β-catenin Signaling’ and ‘Role of Wnt/GSK-3β Signaling in the Pathogenesis of Influenza,’ respectively. When FA is present in various phenotypes of AD at early life, it is highly likely to progress to asthma later, and this mechanism might be regulated by Wnt signaling.
Although AD is a major risk factor for later respiratory allergic diseases, only a few children with early AD go on to develop asthma.10,19 The link between AD and respiratory allergic diseases is influenced by the age of AD onset and its severity. High-risk infants with early-onset persistent AD had a 3-fold higher risk of developing asthma and AR in later childhood than children with late-onset AD that began after 2 years of age.20 A 6-fold increase was reported in the risk of school-aged asthma in children with severe AD in the Multicenter Atopy Study (MAS) birth cohort, which is a high-risk cohort.4 However, it is important to note that this severe phenotype is infrequent, representing only 1% of the overall population.4,21 A characteristic of the allergic disease phenotype before 3 years of age in our study is that it is linked to early-onset AD. AD severity was less severe in FA with AD phenotype than in AD with sensitization and AD without sensitization phenotypes. However, the risk of respiratory allergic diseases at age 5–7 was increased in FA with AD phenotype, which mainly included mild AD. Therefore, other factors, in addition to severity, may contribute to the risk for subsequent respiratory allergic diseases. Unlike the MAS study, the severity of AD in this study does not seem to have any significant effect on subsequent asthma development, which may be due to the differences between the high-risk cohort and the general population-based cohort.
Allergic sensitization is an important factor for determining the phenotype of allergic disease in early childhood with AD.9,22,23 Based on the Canadian Healthy Infant Longitudinal Development (CHILD) study, AD without concomitant allergic sensitization was not associated with an increased risk of asthma at age 3, whereas the combination of AD and allergic sensitization at age 1 was associated with an increased risk of asthma and FA at age 3.9 The MAS found that long-lasting food sensitization (i.e., food sensitization at age 1 and 2) was associated with an increased risk of asthma and AR compared to transient (i.e., food sensitization at only age 1 or 2 years) and never-sensitized children.24 A recent systematic review confirmed that early-life food sensitization leads to other allergic diseases.25 These studies concluded that early-life food sensitization should be used as a marker for developing subsequent allergic diseases that might benefit from preventive strategies. In our study, the allergic sensitization-related phenotypes were FA with AD and AD with sensitization. However, because FA with AD only increases the risk of subsequent asthma at age 7, it has been shown that FA is more important than food sensitization for developing asthma.
In the intestine, active Wnt signaling is essential for maintaining epithelial homeostasis, and pathway inhibition results in crypt loss and tissue degeneration.26 In inflammatory bowel disease, if and how Wnt/β-catenin signaling actually contributes to wound healing during colitis has yet to be formally established.27 Unlike other phenotypes of AD, Wnt/β-catenin signaling is activated in FA with AD phenotype, and the genes involved were AXIN1, CCND1, FZD2, and WNT6. We also found the Wnt6 mRNA level was correlated with specific IgE to foods in early childhood. Therefore, activation of Wnt signaling at early life is likely to be related to FA and is also shown to accelerate the proliferation of airway smooth muscle cells, which are involved in airway remodeling.28,29 Moreover, the genes depicted in the ‘Role of Wnt/GSK-3β Signaling in the Pathogenesis of Influenza’ pathway were activated in FA with AD, demonstrating that IFN reduction in influenza infection can reduce antiviral response compared to AD with sensitization. Therefore, the more frequent development of asthma in FA with AD phenotype suggests that the difference in lung development and antiviral response may be due to an increase in Wnt signaling. Further research is required to understand the functional mechanism.
This study has some limitations. First, the diagnosis of FA was based only on clinical symptoms and characteristics, and no provocation test was performed. However, the definition of FA used here was not only based on a physician’s diagnosis, but also on the presence of a definitive causative food, the period to the development of symptoms, and accompanying allergic symptoms as assessed by an allergy specialist.30 Second, as children in the COCOA study were only tested for 2 food allergens at age 1, milk and egg white, some of the children classified as sensitization-negative may have been sensitized to other food allergens that were not examined, such as wheat. However, hen’s egg (20.3%) was the most frequent cause of FA as the individual food item in young Korean children, followed by cow’s milk (13.2%).31 Third, the sample size used in transcriptome analysis was relatively small, so we need to be careful to generalize these results.
To the best of our knowledge, this is the first general population-based prospective birth cohort study that uniquely identifies AD phenotypes were analyzed with food sensitization and FA as different factors. For asthma, this study showed that FA should be considered a key factor in children with AD. In addition, our results show that the Wnt signaling pathway involved in the subsequent development of asthma may be differently regulated in the presence of FA, warranting further studies in the future.
ACKNOWLEDGMENTS
We thank all the children and their parents for active participation of COhort for Childhood Origin of Asthma and allergic diseases (COCOA) study. We also thank project manager Dr. Hea-Young Oh and all the field workers and laboratory personnel involved for their efforts.
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Science and ICT; MSIT) (NRF-2020R1A2C2012822) and by the Research Program funded by the Korea Disease Control and Prevention Agency (2008-E33030-00, 2009-E33033-00, 2011-E33021-00, 2012-E33012-00, 2013-E51003-00, 2014-E51004-00, 2014-E5 1004-01, 2014-E5 1004-02, 2017-E6 7002-00, 2017-E6 7002-01, and 2017-E6 7002-02).
Footnotes
Disclosure: There are no financial or other issues that might lead to conflict of interest.
SUPPLEMENTARY MATERIALS
Methods
Selection of study population
Probability of allergic diseases and allergic sensitization conditional on latent class memberships
Model fit criteria
Multinomial logistic regression to identify risk factors for phenotypes of allergic diseases, compared to baseline class of “no-allergic disease”
List of genes involved in the three canonical pathways
Flowchart of the study subjects.
Canonical pathways enriched in each comparison with a z-score. Enriched pathways (red) were activated by differentially expressed genes, whereas blue indicates inhibition of the enriched pathways.
The subcellular location of genes in the canonical pathway, (A) termed as “Wnt/β-catenin Signaling,” as determined via IPA analysis to compare transcriptomes in FA with AD/No allergic disease. (B) “Role of Wnt/GSK-3β Signaling in the Pathogenesis of Influenza” was identified as a unique canonical pathway to compare transcriptomes in FA with AD/AD with sensitization.
References
- 1.van der Hulst AE, Klip H, Brand PL. Risk of developing asthma in young children with atopic eczema: a systematic review. J Allergy Clin Immunol. 2007;120:565–569. doi: 10.1016/j.jaci.2007.05.042. [DOI] [PubMed] [Google Scholar]
- 2.Singh AM, Evans MD, Gangnon R, Roberg KA, Tisler C, DaSilva D, et al. Expression patterns of atopic eczema and respiratory illnesses in a high-risk birth cohort. J Allergy Clin Immunol. 2010;125:491–493.e4. doi: 10.1016/j.jaci.2009.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacharier LB, Beigelman A, Calatroni A, Jackson DJ, Gergen PJ, O’Connor GT, et al. Longitudinal phenotypes of respiratory health in a high-risk urban birth cohort. Am J Respir Crit Care Med. 2019;199:71–82. doi: 10.1164/rccm.201801-0190OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Illi S, von Mutius E, Lau S, Nickel R, Grüber C, Niggemann B, et al. The natural course of atopic dermatitis from birth to age 7 years and the association with asthma. J Allergy Clin Immunol. 2004;113:925–931. doi: 10.1016/j.jaci.2004.01.778. [DOI] [PubMed] [Google Scholar]
- 5.Dharmage SC, Lowe AJ, Matheson MC, Burgess JA, Allen KJ, Abramson MJ. Atopic dermatitis and the atopic march revisited. Allergy. 2014;69:17–27. doi: 10.1111/all.12268. [DOI] [PubMed] [Google Scholar]
- 6.Tham EH, Leung DY. Mechanisms by which atopic dermatitis predisposes to food allergy and the atopic march. Allergy Asthma Immunol Res. 2019;11:4–15. doi: 10.4168/aair.2019.11.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee E, Lee SH, Kwon JW, Kim YH, Cho HJ, Yang SI, et al. Atopic dermatitis phenotype with early onset and high serum IL-13 is linked to the new development of bronchial hyperresponsiveness in school children. Allergy. 2016;71:692–700. doi: 10.1111/all.12844. [DOI] [PubMed] [Google Scholar]
- 8.Paternoster L, Savenije OE, Heron J, Evans DM, Vonk JM, Brunekreef B, et al. Identification of atopic dermatitis subgroups in children from 2 longitudinal birth cohorts. J Allergy Clin Immunol. 2018;141:964–971. doi: 10.1016/j.jaci.2017.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tran MM, Lefebvre DL, Dharma C, Dai D, Lou WYW, Subbarao P, et al. Predicting the atopic march: results from the Canadian Healthy Infant Longitudinal Development Study. J Allergy Clin Immunol. 2018;141:601–607.e8. doi: 10.1016/j.jaci.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 10.Amat F, Saint-Pierre P, Bourrat E, Nemni A, Couderc R, Boutmy-Deslandes E, et al. Early-onset atopic dermatitis in children: which are the phenotypes at risk of asthma? Results from the ORCA cohort. PLoS One. 2015;10:e0131369. doi: 10.1371/journal.pone.0131369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eller E, Kjaer HF, Høst A, Andersen KE, Bindslev-Jensen C. Food allergy and food sensitization in early childhood: results from the DARC cohort. Allergy. 2009;64:1023–1029. doi: 10.1111/j.1398-9995.2009.01952.x. [DOI] [PubMed] [Google Scholar]
- 12.Yang HJ, Lee SY, Suh DI, Shin YH, Kim BJ, Seo JH, et al. The Cohort for Childhood Origin of Asthma and allergic diseases (COCOA) study: design, rationale and methods. BMC Pulm Med. 2014;14:109. doi: 10.1186/1471-2466-14-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams RL. A note on robust variance estimation for cluster-correlated data. Biometrics. 2000;56:645–646. doi: 10.1111/j.0006-341x.2000.00645.x. [DOI] [PubMed] [Google Scholar]
- 14.Kanaan Z, Qadan M, Eichenberger MR, Galandiuk S. The actin-cytoskeleton pathway and its potential role in inflammatory bowel disease-associated human colorectal cancer. Genet Test Mol Biomarkers. 2010;14:347–353. doi: 10.1089/gtmb.2009.0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sánchez-Ovando S, Simpson JL, Barker D, Baines KJ, Wark PA. Transcriptomics of biopsies identifies novel genes and pathways linked to neutrophilic inflammation in severe asthma. Clin Exp Allergy. 2021;51:1279–1294. doi: 10.1111/cea.13986. [DOI] [PubMed] [Google Scholar]
- 16.Bakshi S, Zhang X, Godoy-Tundidor S, Cheng RY, Sartor MA, Medvedovic M, et al. Transcriptome analyses in normal prostate epithelial cells exposed to low-dose cadmium: oncogenic and immunomodulations involving the action of tumor necrosis factor. Environ Health Perspect. 2008;116:769–776. doi: 10.1289/ehp.11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;14:482–517. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marineau A, Khan KA, Servant MJ. Roles of GSK-3 and β-catenin in antiviral innate immune sensing of nucleic acids. Cells. 2020;9:897. doi: 10.3390/cells9040897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bantz SK, Zhu Z, Zheng T. The atopic march: progression from atopic dermatitis to allergic rhinitis and asthma. J Clin Cell Immunol. 2014;5:202. doi: 10.4172/2155-9899.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowe AJ, Angelica B, Su J, Lodge CJ, Hill DJ, Erbas B, et al. Age at onset and persistence of eczema are related to subsequent risk of asthma and hay fever from birth to 18 years of age. Pediatr Allergy Immunol. 2017;28:384–390. doi: 10.1111/pai.12714. [DOI] [PubMed] [Google Scholar]
- 21.Silverberg JI, Simpson EL. Association between severe eczema in children and multiple comorbid conditions and increased healthcare utilization. Pediatr Allergy Immunol. 2013;24:476–486. doi: 10.1111/pai.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurukulaaratchy RJ, Matthews S, Arshad SH. Defining childhood atopic phenotypes to investigate the association of atopic sensitization with allergic disease. Allergy. 2005;60:1280–1286. doi: 10.1111/j.1398-9995.2005.00890.x. [DOI] [PubMed] [Google Scholar]
- 23.Belgrave DC, Granell R, Simpson A, Guiver J, Bishop C, Buchan I, et al. Developmental profiles of eczema, wheeze, and rhinitis: two population-based birth cohort studies. PLoS Med. 2014;11:e1001748. doi: 10.1371/journal.pmed.1001748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulig M, Bergmann R, Tacke U, Wahn U, Guggenmoos-Holzmann I. Long-lasting sensitization to food during the first two years precedes allergic airway disease. The MAS Study Group, Germany. Pediatr Allergy Immunol. 1998;9:61–67. doi: 10.1111/j.1399-3038.1998.tb00305.x. [DOI] [PubMed] [Google Scholar]
- 25.Alduraywish SA, Lodge CJ, Campbell B, Allen KJ, Erbas B, Lowe AJ, et al. The march from early life food sensitization to allergic disease: a systematic review and meta-analyses of birth cohort studies. Allergy. 2016;71:77–89. doi: 10.1111/all.12784. [DOI] [PubMed] [Google Scholar]
- 26.Kuhnert F, Davis CR, Wang HT, Chu P, Lee M, Yuan J, et al. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A. 2004;101:266–271. doi: 10.1073/pnas.2536800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moparthi L, Koch S. Wnt signaling in intestinal inflammation. Differentiation. 2019;108:24–32. doi: 10.1016/j.diff.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Hussain M, Xu C, Lu M, Wu X, Tang L, Wu X. Wnt/β-catenin signaling links embryonic lung development and asthmatic airway remodeling. Biochim Biophys Acta BBAMol Basis Dis. 2017;1863:3226–3242. doi: 10.1016/j.bbadis.2017.08.031. [DOI] [PubMed] [Google Scholar]
- 29.Huo Y, Guan L, Xu J, Zhou L, Chen R. Tiotropium inhibits methacholine-induced extracellular matrix production via β-catenin signaling in human airway smooth muscle cells. Int J Chron Obstruct Pulmon Dis. 2018;13:1469–1481. doi: 10.2147/COPD.S158552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim YH, Kim KW, Lee SY, Koo KO, Kwon SO, Seo JH, et al. Maternal perinatal dietary patterns affect food allergy development in susceptible infants. J Allergy Clin Immunol Pract. 2019;7:2337–2347.e7. doi: 10.1016/j.jaip.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 31.Park M, Kim D, Ahn K, Kim J, Han Y. Prevalence of immediate-type food allergy in early childhood in Seoul. Allergy Asthma Immunol Res. 2014;6:131–136. doi: 10.4168/aair.2014.6.2.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods
Selection of study population
Probability of allergic diseases and allergic sensitization conditional on latent class memberships
Model fit criteria
Multinomial logistic regression to identify risk factors for phenotypes of allergic diseases, compared to baseline class of “no-allergic disease”
List of genes involved in the three canonical pathways
Flowchart of the study subjects.
Canonical pathways enriched in each comparison with a z-score. Enriched pathways (red) were activated by differentially expressed genes, whereas blue indicates inhibition of the enriched pathways.
The subcellular location of genes in the canonical pathway, (A) termed as “Wnt/β-catenin Signaling,” as determined via IPA analysis to compare transcriptomes in FA with AD/No allergic disease. (B) “Role of Wnt/GSK-3β Signaling in the Pathogenesis of Influenza” was identified as a unique canonical pathway to compare transcriptomes in FA with AD/AD with sensitization.