Summary
Limb patterning by Sonic hedgehog (Shh), via either graded spatial or temporal signal integration, is a paradigm for “morphogen” function. Yet how Shh instructs distinct digit identities remains controversial. Here, we bypassed the Shh-requirement in cell survival during outgrowth and demonstrate that a transient, early Shh pulse is both necessary and sufficient for normal mouse limb development. Shh-response is only short-range and limited to the Shh-expressing zone during this time window. Shh patterns digits 1–3, anterior to this zone, by an indirect mechanism, rather than direct spatial or temporal signal integration. Using a genetic relay-signaling assay, we discovered Shh also specifies digit 1/thumb (thought to be exclusively Shh-independent) indirectly, implicating Shh in a unique regulatory hierarchy for digit 1 evolutionary adaptations, such as opposable thumbs. This study illuminates Shh as a trigger for an indirect downstream network that becomes rapidly self-sustaining, with mechanistic relevance for limb development, regeneration, and evolution.
Introduction
Shh is considered a prototypical vertebrate morphogen, with the limb and neural tube serving as a mainstay to elucidate its function. In contrast to neural tube, anterior-posterior (A-P) limb patterning yields different digit identities from a common set of cell types and tissues, with distinct skeletal morphologies that are not based on cell fate changes per se, but arise as an emergent property. Understanding how Shh specifies limb skeletal pattern is central to the problem of structural morphogenesis and informative to regenerative medicine. Shh is secreted by posterior limb bud mesoderm cells defined functionally as the “zone of polarizing activity” (ZPA). Shh/ZPA regulates the specification of distinct A-P digits 2–5 (d2-d5; index to pinky; reviewed by (Zhu and Mackem, 2017), excepting d1 (the thumb), which is thought to form Shh-independently (Chiang et al., 2001). Patterning and growth are coupled; Shh controls both digit type and number during its 2-day limb bud expression span (Figure 1A).
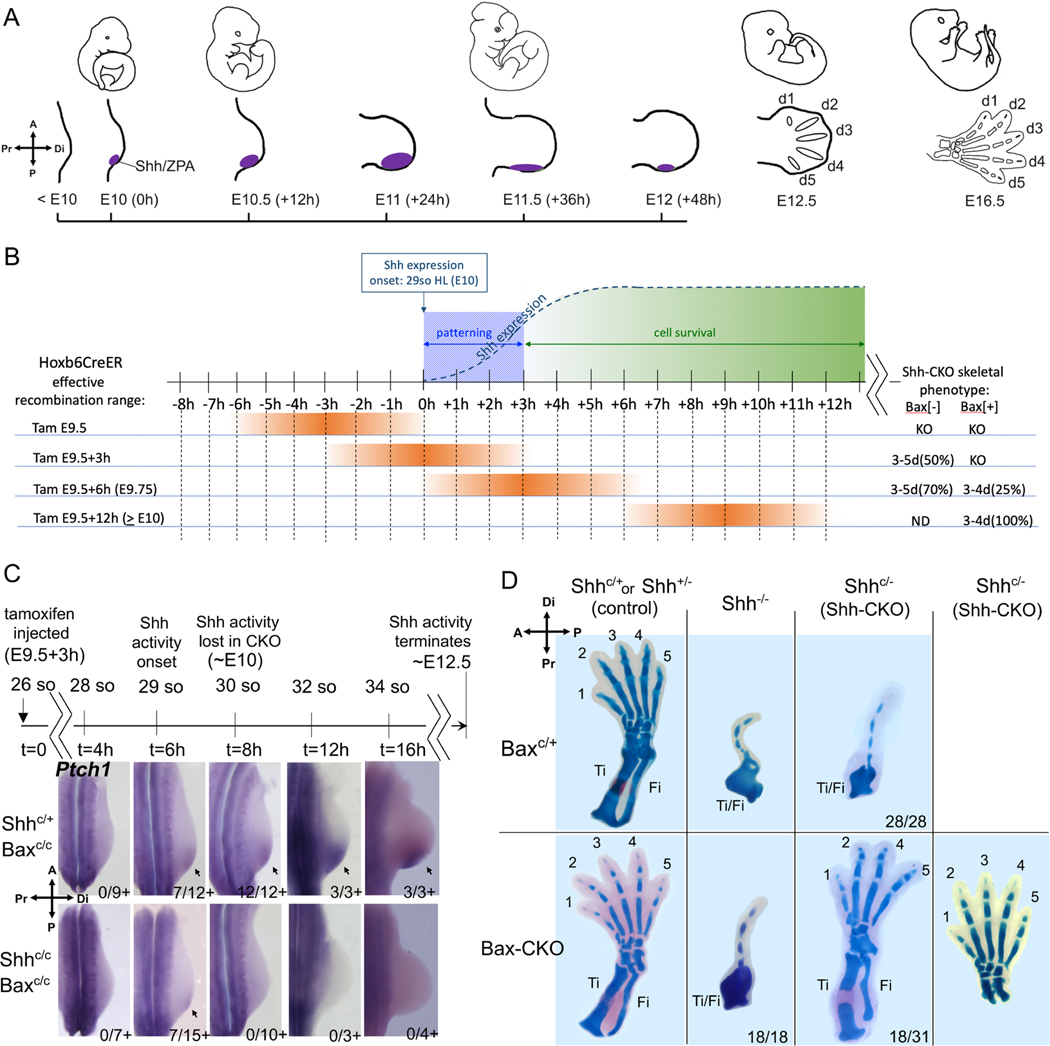
Figure 1. A transient Shh activity pulse suffices to specify all digits with enforced cell survival.
(A) Shh expression timeline in wildtype mouse hindlimb (Shh/ZPA, purple). Limb axis orientation indicated by compasses (upper left) in all figures. (B) Summary of recombination timelines (orange; see Methods for somite (so) staging/range) for tamoxifen (Tam)-induced Shh deletion at different times relative to Shh expression onset and subsequent digit outcomes (at right); KO, Shh null phenotype; ND, not done. At E9.5 Tam, deletion precedes Shh expression and all limbs have a Shh null phenotype. By E9.75 and later, deletion occurs after the Shh role in maintaining cell survival has commenced and both Bax[+] (apoptosis competent) and Bax[-] (cell survival enforced) embryos display some rescue of digit formation (see Table S1); >E10 data summarized from Zhu et al (2008). (C) Shh activity assayed by Ptch1 RNA (arrows) at times after Tam as indicated on timeline. Shh activity was first detected at 6hr (29 so) after injection in a subset of both control Shh+/C;Bax-CKO (7/12+), and Shh-CKO;Bax-CKO (7/15+) embryos, and became robust by 8hr (30 so) in control (12/12+), but was absent in all Shh-CKO;Bax-CKO embryos (0/10+) from t=8hrs (30 so) and later. (D) Skeletal staining (E16.5) after Bax/Bak and Shh removal by Tam (treated at E9.5+3h, as in C). In hindlimbs with Bax/Bak present (Baxc/+) all Shh-CKO embryos (28/28) have Shh null phenotype. In hindlimbs with Bax/Bak absent (Bax-CKO), about 50% of the Shh-CKO embryos (18/31) have 3–5 normal digits (5-digit phenotype shown in right-most panel) and normal zeugopod bones (tibia, Ti; fibula, Fi), but 100% Shh null embryos (Shh−/−; 18/18) with Bax/Bak removed still retain the null mutant phenotype. Related to Figure S1 and Table S1.
Yet, despite high functional conservation across tetrapods and intense investigation for over two decades, the mechanism by which Shh patterns digits remains controversial. Very disparate models have been proposed in different species with similar limb gene regulatory networks(Harfe et al., 2004; Towers et al., 2008; Yang et al., 1997; Zhu et al., 2008); reviewed by (Zhu and Mackem, 2017). Morphogen-based models of Shh function derive from chick studies showing that changes in concentration or duration can alter both digit identity and numbers, with posterior identities requiring higher concentrations or longer exposures(Scherz et al., 2007; Towers et al., 2008; Yang et al., 1997). Lineage tracing of ZPA-descendants in both chick and mouse have revealed that d4 and d5 arise from the descendants of Shh-expressing ZPA cells(Harfe et al., 2004; Towers et al., 2011), leading to the proposal that temporal integration of short-range Shh, rather than graded long-range signaling, specifies digit identities. Since non-ZPA cells are displaced further away from ZPA signals during outgrowth, the Shh-expressing d4,d5 progenitors receive the longest exposure(Harfe et al., 2004). But the normal coupling of Shh roles in digit identity (patterning) and number (growth) poses a challenge to tease apart the individual requirements for each. Comparing the effects of Shh inhibition with that of cell cycle blockade to reduce digit number in chick(Towers et al., 2008), led to the proposal that temporal signal integration acts to progressively “promote” digit territories to more posterior identity. Yet genetic lineage tracing indicated that Shh-expressing ZPA cells become refractory to Shh-response over time(Ahn and Joyner, 2004), seemingly at odds with temporal integration models to achieve posterior identity. Using a conditional Shh-mutant we previously found Shh was required for only a limited interval (∼8–10 hrs) to specify normal digits; later Shh removal caused digit loss in an order that reflected the normal temporal order in which digits arise, and not their AP position(Zhu et al., 2008). Accordingly, the remaining digits that formed were morphologically normal and not transformed to an anterior identity. These results suggested a biphasic model in which Shh is required early for patterning, but over an extended time to promote progenitor survival and expansion, potentially restricting Shh-morphogen action to a more limited time frame.
Here, we use a genetic strategy to uncouple the Shh requirement in digit patterning from expansion/cell survival and test how Shh acts to specify digit pattern. We demonstrate that a transient (∼2hr) burst of Shh activity, during which Shh-response is limited to d4-d5 progenitors, suffices to specify all digits normally, implying that non-ZPA derived digits are patterned by an indirect mechanism and that Shh acts neither as a spatial morphogen nor via temporal integration. A genetic assay to test for Shh-induced relay signals unexpectedly reveals that d1 specification is in fact Shh-dependent, requiring an indirect downstream relay signal and highlighting a role for Shh in the evolutionary emergence of a unique, polarized thumb.
Results
A transient Shh pulse restores normal limb development when cell survival is enforced.
To examine the early role of Shh selectively, we used a genetic strategy to uncouple the Shh requirement in digit patterning from that in outgrowth. We asked if enforcing cell survival can bypass late-phase Shh function and restore any digit formation. The pro-apoptotic Bax/Bak genes(Lindsten et al., 2000) were deleted in Shh conditional mutant limb buds exposed to a very transient, early Shh pulse (using ShhC/C;Hoxb6CreER, hereafter referred to as Shh-CKO, and BaxC/C;Bak−/− alleles, referred to as Bax-CKO; see Table 1 for list of all crosses and genotypes used). Our analysis focused on hindlimb, where Hoxb6CreER drives complete and robust recombination even prior to limb bud initiation (∼E9; see Figure 1A; (Nguyen et al., 2009). Since very early recombination will delete Shh prior to its expression onset, whereas deletion at too late a time will produce only mild digit loss phenotypes even despite reduced cell survival(Zhu et al., 2008), the proper tamoxifen timing to induce Cre and delete Shh is critical. Tamoxifen timing was optimized (to E9.5+3hrs; Figure 1B, Table S1) so that 100% of Shh-CKO sibling embryos that retained one wild-type Bax allele (cell survival not enforced) were invariably Shh germ-line null (Shh−/−) in hindlimb skeletal phenotype (28/28), providing a clear baseline to assess the effects of bypassing Shh late cell-survival function. This tamoxifen timing (at E9.5+3h) rescued digit formation in about 50% of embryos with enforced cell survival (Figure 1D; discussed below) and precedes the normal onset of Shh expression by about 9 hours(Lewis et al., 2001; Zhu et al., 2008). Earlier deletion (at E9.5) invariably produced a null phenotype irrespective of enforced cell survival (Table S1), consistent with previous work showing that E9.5 treatment deletes Shh efficiently before expression onset(Zhu et al., 2008). At later deletion times (by E9.5+6h), a partial rescue of digit formation occurred even in a portion of embryos without enforced cell survival (Bax[+], ∼25%, Table S1).
Table 1.
List of genetic crosses, embryo numbers analyzed, and experimental outcomes related to each figure.
| Alleles crossed | shorthand notation | %rescue†/analysis | Tam Tx* | phenotype/analysis | Fig. |
|---|---|---|---|---|---|
|
| |||||
|
Shh+/C;Bax+/C;Bak−/−;Hoxb6CreER x ShhC/C;Bax C/C;Bak−/−;Hoxb6CreER |
Shh-CKO;Bax-CKO Shh-CKO;Bax+/c |
18/31 (Bax-KO); 0/28 (Bax-het) |
E9.5+3h | skeleton | 1 |
|
| |||||
|
Shh+/−;Bax+/C;Bak−/−;Hoxb6CreER x Shh+/−;Bax C/C;Bak−/− |
Shh−/−;Bax-CKO | 0/18 (Bax-KO) | E8.75–E9.5 | skeleton | 1 |
|
| |||||
|
Shh+/−;Bax
+/−;Bak−/− x Shh+/−;Bax +/−;Bak−/− |
Shh−/−;Bax−/− | 0/6 (Bax/Bak-KO) | NA | skeleton | 1 |
|
| |||||
|
Shh+/C;BaxC/C;Bak−/−;Hoxb6CreER x ShhC/C;Bax C/C;Bak−/−;Hoxb6CreER |
Shh-CKO;Bax-CKO | NA-see Figs. 1, S1, text | E9.5+3h | Shh activity duration |
1, S1 |
|
| |||||
|
Shh+/−;Bax+/C;Bak−/−;Hoxb6CreER x Shh−/C;Bax C/C;Bak−/−;Hoxb6CreER |
Shh-CKO;Bax-CKO Shh−/−;Bax-CKO |
NA-see Fig. S1, text | E9.5+3h | Cell survival | S1 |
|
| |||||
|
ShhCreER/+;RosaLacZ/LacZ x Gli1CreER/+;RosaLacZ/LacZ |
ShhCreER+ or Gli1CreER+ |
-51 embryos -60 embryos |
Varied-Fig 2 | Shh-expression, -response lineage | 2 |
|
| |||||
|
Shh+/−;BaxC/C;Bak−/−;Hoxb6CreER x Shh−/C;Bax C/C;Bak−/−;Hoxb6CreER |
Shh-CKO;Bax-CKO Shh−/−;Bax-CKO |
NA-see Figs. 3, S2 text | E9.5+3h | In situ HCR – Shh, Gli1 (response) |
3, S2 |
|
| |||||
|
Shh+/−;BaxC/C;Bak−/−;Hoxb6CreER x Shh−/C;Bax C/C;Bak−/−;Hoxb6CreER |
Shh-CKO;Bax-CKO Shh−/−;Bax-CKO |
NA-see Fig. 4, text | E9.5+3h | Hand2, Ptch1, Gli1 Gli3 western | 4 |
|
| |||||
|
Shh+/−;Gli3+/−;BaxC/C;Bak−/−;Hoxb6CreER x Shh+/−;Bax C/C;Bak−/−;Hoxb6CreER |
Shh−/−;Gli3+/−;BaxCKO | 10/10 (vs Shh−/−) | NA or E9.5+3h | skeleton - Gli3 dose modulation | 4 |
| Shh+/−;Gli3+/− x Shh+/− | Shh−/−;Gli3+/− | 8/8 (vs Shh−/−) | |||
| Shh+/−;Gli3+/− x ShhC/C;Hoxb6CreER | Shh-CKO;Gli3+/− | 12/12 (vs Shh-CKO) | |||
|
Shh+/−;BaxC/C;Bak−/−;Hoxb6CreER x Shh−/C;Bax C/C;Bak−/−;Hoxb6CreER |
Shh-CKO;Bax-CKO | 18/31 (vs Shh-CKO) | |||
|
| |||||
|
Shh+/−;BaxC/C;Bak−/−;Hoxb6CreER x Shh−/C;Bax C/C;Bak−/−;Hoxb6CreER |
Shh-CKO;Bax-CKO Shh−/−;Bax-CKO |
NA-see Fig. 5, text | E9.5+3h | Fgf8, Grem1, Hoxa13, Hoxd13 | 5 |
|
| |||||
|
Shh+/−;BaxC/C;Bak−/−;Hoxb6CreER x Shh−/C;Bax C/C;Bak−/−;Hoxb6CreER |
Shh-CKO;Bax-CKO Shh−/−;Bax-CKO |
NA-see Fig. S3, text | E9.5+3h | Jag1, Cyp26b1, Hoxd11, Hoxd13 | S3 |
|
| |||||
|
Shh+/−;BaxC/C;Bak−/−;Hoxb6CreER x Shh−/C;Bax C/C;Bak−/−;Hoxb6CreER |
Shh-CKO;Bax-CKO Shh−/−;Bax-CKO |
NA-see Fig. S4, text | E9.5+3h | Anterior pattern genes: Alx4, Irx3 | S4 |
|
| |||||
|
ShhCre/+;RosaLacZ/LacZ x Shh+/−;RosaSmoM2/SmoM2 |
ShhCre/−;Shh-SmoM2 | 21/32 (digit 1+) | NA | Skeleton; Hoxa13, Shh-lineage | 6 |
| ShhCre/+;RosaLacZ/LacZ x Shh+/− | ShhCre/− | 0/6 (digit 1+) | |||
|
| |||||
|
ShhCre/+;Bax+/−;Bak−/−;RosaLacZ/LacZ x Shh+/− ;Bax +/−;Bak−/−;RosaSmoM2/ SmoM2 |
ShhCre/−;Bax/Bak-KO; Shh-SmoM2 |
8/8 (only digit 1+) | NA | skeleton | 6 |
|
| |||||
|
ShhCre/+;RosaLacZ/LacZ x Shh+/−;RosaSmoM2/+ |
ShhCre/+;Shh-SmoM2 | 4/64 (ectopic ZPA, duplicated d1) | NA | skeleton Shh-lineage | S5 |
| ShhCre/−;Shh-SmoM2 ShhCre/− |
NA-see Fig. S5, text | Uncx4.1, Sox9 Ptch1, Gli1 | |||
|
| |||||
|
ShhCre/+;RosaLacZ/LacZ x Shh+/−;RosaSmoM2/+ ShhCre/+;RosaEYFP/EYFP x Shh+/−;RosaSmoM2/+ |
ShhCre/−;Shh-SmoM2 ShhCre/− |
−10 embryos − 6 embryos |
NA | Shh-lineage; cell survival | S5 |
skeletal numbers refer to embryo numbers (left+right hindlimb both rescued);
Tx, treatment time
To determine the total duration of Shh signaling activity after optimized tamoxifen treatment (at E9.5+3h), Shh response was assayed by detecting the direct Shh target RNAs (Gli1, Ptch1)(Litingtung et al., 2002; te Welscher et al., 2002b; Vokes et al., 2008) at one-somite intervals (2 hours per somite (Tam, 1981)) post-tamoxifen treatment (Figures 1C, S1A). Direct target detection is a highly sensitive and earlier read-out for Shh function than is the presence of Shh RNA itself(Lewis et al., 2001; Zhu et al., 2008), and also serves as a sensitive indicator of mosaic recombination at later stages, since residual Shh-expressing cells proliferate over time and would become readily apparent. Analysis of Shh-response at 1-somite intervals after tamoxifen detected a transient 2–3 hour window of Shh activity in about 50% of Shh-CKO embryos restricted to the 29 somite stage (7/15 Ptch1+, 4/10 Gli1+, Figures 1C, S1A). No activity was detected in the remaining 50%, presumably because recombination occurred prior to Shh expression onset in those embryos. Notably, transient Shh activity is also first detectable in controls at 29 somites (completely absent in all embryos at 28 somites; this study Figures 1C, 3, S1A, S2; in agreement with Zhu et al, 2008; Lex et al 2022). Within two hours later, at the 30 somite stage (+1 somite), when Shh activity (Ptch1, Gli1 expression) is detected in 100% of control (Shh+/−) sibling embryos, Shh activity has ceased in all Shh-CKO embryos (Figures 1C; S1A) and remains absent subsequently. Analysis of Shh-CKO;Bax-CKO embryos at multiple later stages also failed to detect the late emergence of any direct target Gli1 or Ptch1 expression (see below).
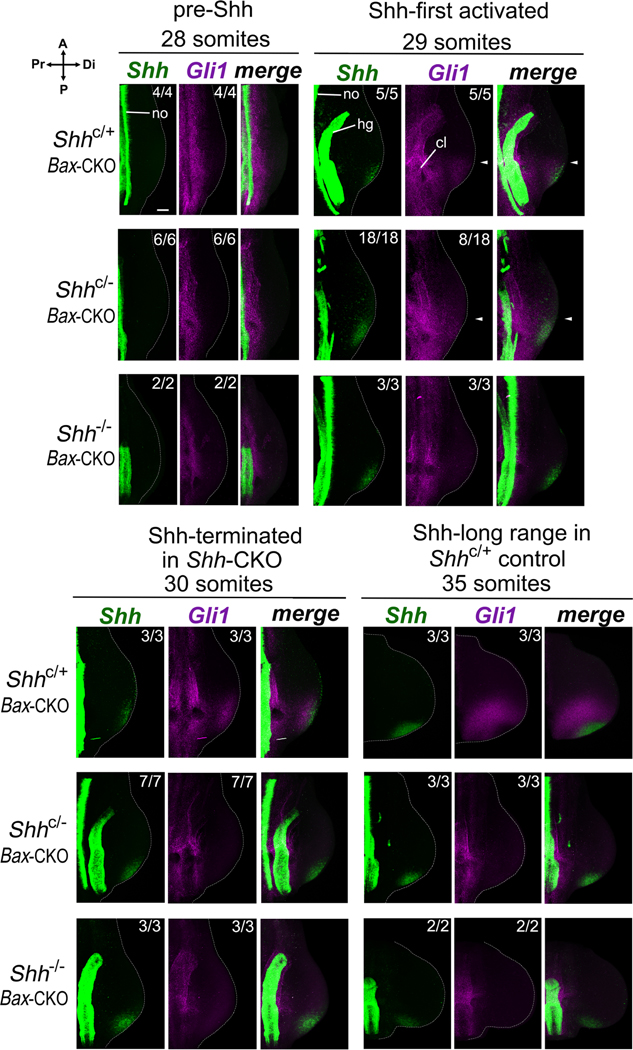
Figure 3. Only short-range Shh response is detected in Shh-CKO;Bax-CKO embryos during the transient Shh expression window.
Shh expression and activity was assayed by Shh (green) and Gli1 (purple) RNA in situ HCR(Choi et al., 2018) at somite stages indicated. After tamoxifen injection at E9.5+3h (as in Figure 1B,C), Shh response (Gli1) was detected at the 29 somite stage, shortly after Shh expression onset, in all control Shh+/−;Bax-CKO (5/5) and in a subset of Shh-CKO;Bax-CKO (8/18) embryos. By 30 somites (2h later) and at 35 somites (12h later), Shh response became robust in all controls but was absent in all Shh-CKO embryos. A-P expression extent of Gli1 in distal limb bud is very similar to that of Shh in both the control and the Shh-CKO embryos during the transient expression window (29 somites; merged panels, arrows). Note that non-functional (exon 2-deleted) Shh RNA remains detectable in both late-stage Shh-CKO and in Shh−/− null embryos (with absent Gli1 RNA signal). Numbers analyzed with result shown are indicated at bottom of each panel, with remainder negative for expression. no, notochord and hg, hindgut axial sources of Hh ligands and cl, cloacal-urogenital region, also responsive to local Shh and Ihh signaling(Haraguchi et al., 2007; Perriton et al., 2002). Scale bar = 100 μm (top left panel; all panels at same scale). Related to Figures 2 and S2.
Cell survival was completely restored in 100% of Shh mutant limb buds with Bax-CKO (20/20, Figure S1B), but notably Bax-CKO alone had no effects on limb skeletal patterning in Shh+/C;Bax-CKO sibling controls (Figure 1D). In Shh-CKO;Bax-CKO embryos, blocking cell death restored formation of from 3 to 5 digits with normal morphology in about 50% of embryos (18/31, Figure 1D); the remaining 50% retained the Shh−/− null mutant limb phenotype (13/31). The fraction of embryos that did not display any transient Shh activity early (Figures 1C, S1A; Table S1) correlated well with the subsequent frequency having a Shh null skeletal phenotype (Figure 1D, Table S1). In Shh-CKO;Bax-CKO embryos with rescued limbs (18/31), normal long bone morphology (tibia/fibula; zeugopod) was also restored (see also Figure 2C), and normal AP polarity was clearly evident in both the long bones and digits, including distinctive d1 and d5 identities. In contrast, all (100%) of sibling Shh-CKO embryos retaining one functional Bax allele (Shh-CKO;Bax+/C) had levels of apoptosis very similar to the Shh−/−;Bax+/C (Figure S1B) and later displayed a Shh null limb skeletal phenotype with a single dysmorphic digit and malformed zeugopod (28/28, Figure 1D).
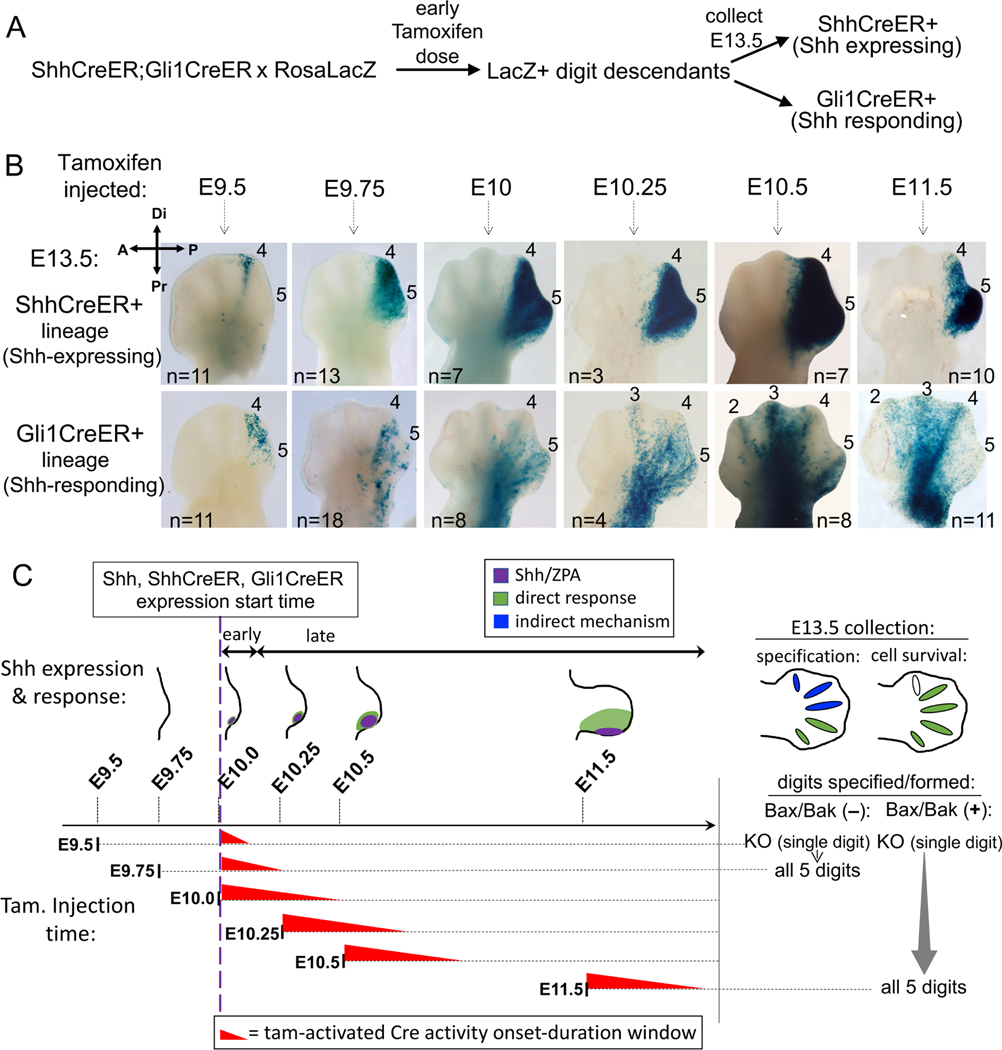
Figure 2. Lineage tracing of Shh response in normal limb buds at time of Shh onset.
(A) Diagram of strategy to compare Shh-expressing (ShhCreER) and Shh-responding (Gli1CreER) lineages in sibling embryos so that developmental ages and tamoxifen timing are identical. (B) Distribution of RosaLacZ reporter+ descendants (in E13.5 digit rays) after single-dose tamoxifen given at time indicated. After E9.5 or E9.75 tamoxifen (time as in Figure 1B), direct Shh-response is limited to digit 4/5 territory (ZPA domain; Shh-expressing descendants). Long-range response in digit 2–3 territory is evident by E10.5. n, numbers analyzed for each dosage time. (C) Summary of data in (B) showing overlap of tamoxifen (Tam) activity time window (red wedges); Tam duration estimated at ∼12hrs (from (Nakamura et al., 2006) and Figures 1B,C, S1A data). Shh expression(purple)/direct response(green) in schematics, and summary of digits specified and formed (to right), either with or without enforced cell survival, for same Shh activity time window (from Figure 1, Table S1) for each Tam dosage time. Early-responding cells (Tam at E9.5 or E9.75) contribute only to d4/5 (green, E13.5 hindlimb), indicating d1-d3 are specified by an indirect mechanism (blue). Later stage-responding cells (Tam after E10.25), when Shh acts to maintain cell survival, contribute to d2-d5 (green, E13.5 hindlimb).
These data indicate that a transient early Shh pulse (∼2–3h) suffices to specify digit progenitors, but not to maintain cell survival. We next asked if a short Shh pulse is even necessary when cell survival is enforced, using a non-conditional Shh null mutant completely devoid of Shh activity (Shh−/−;Bax-CKO). Cell death was completely blocked in Shh−/−;Bax-CKO limbs (9/9, Figure S1B), but enforced cell survival failed to rescue any digit or normal long bone formation (0/18, Figure 1D), even when a non-conditional germ-line Bax/Bak mutant (Shh−/−;Bax−/−;Bak−/−) was used to ensure complete Bax/Bak inactivation in the Shh−/− (0/6 skeletal rescue; Table 1). These results indicate that an early, transient Shh pulse (of 2–3hr) is both necessary and sufficient for normal digit and long bone formation, when the role of Shh in maintaining cell survival is bypassed by Bax/Bak removal. Consequently, later stage sustained Shh signaling acts mainly to support cell survival and limb bud expansion.
Rescued digits in the early Shh-CKO;Bax-CKO arise from cells that did not respond directly to Shh.
The short 2–3h Shh activity window required for normal limb development when cell survival is enforced is inconsistent with temporal Shh signal integration, but could be compatible with transient activity of a spatially graded morphogen. To assess if Shh acts as a long-range limb morphogen during this early time window in the normal limb bud, we used lineage tracing with Gli1CreER/+ (Ahn and Joyner, 2004) to track cells that had responded to Shh at early times immediately after Shh expression onset. We also compared Shh-response (Gli1CreER activity) with the spatial extent of the Shh-producing ZPA region at the same times (ShhCreER/+)(Harfe et al., 2004) as a guide in assessing the extent of long-range signaling. Lineage tracing crosses included both ShhCreER/+ and Gli1CreER/+ knock-in alleles to genetically mark cells (RosaLacZ reporter activation) in sibling embryos from the same litter that expressed either ShhCreER/+ or Gli1CreER/+ alone, ensuring that Shh production and Shh response were compared at identical embryonic ages and tamoxifen exposure times (Figure 2A). A single tamoxifen dose was given at closely spaced early times spanning Shh (and ShhCreER) expression onset (Figure 2B,C) and limb buds were collected at E13.5, after all digit rays have formed, so that the descendant (LacZ+) cell contributions to different digits can be easily scored (Figure 2A,B).
Genetic LacZ reporter marking revealed that Shh acts only very short-range at early times after expression onset. Notably, the ZPA is highly dynamic; the earliest Shh-expression and response begin in digit 4 territory and then extend to digit 5 progenitors. For tamoxifen treatment times prior to E10.25, Shh response (Gli1CreER activation) was confined to cells that later give rise to d4, d5 (Figure 2B). Long-range signaling was not detected until much later tamoxifen-induction times, initially extending toward d3 (E10.25) and later also including d2 territories (by E10.5, Figure 2B). Yet, tamoxifen treatment at a much earlier stage (E9.5+3h, Figure 1), provides a transient Shh pulse that is both necessary and sufficient to specify all 5 digits if cell survival is enforced. During this time interval, the lineage tracing shows that Shh acts only short-range. Shh response marked by Gli1CreER activity is limited to the Shh-producing ZPA region and directly impacts only d4, d5 progenitors. Consequently, other digit progenitors that do not respond directly to Shh during this time window must be specified by an indirect mechanism (Figure 2C-blue). Notably however, at later stages Shh does act as a long-range signal (by E10.5, Figures 2B and 2C-green), coinciding with the time frame when enforced cell survival (Bax/Bak loss) can replace Shh function.
Is the same time frame for short- and long-range Shh signaling also maintained in the Shh-CKO;Bax-CKO mutant? The need for a robust, early Cre driver to achieve efficient, rapid deletion in the Shh-CKO;Bax-CKO precludes genetic lineage analysis with Cre lines to track Shh response in the mutant context. Gli1LacZ is an alternative response reporter over short time frames (see below), but cannot be used to assess the digit contributions of early responding progenitors. Early-induced LacZ protein does not perdure over the 3-day time interval between transient Shh induction and digit ray formation (∼E10 to E13.5), and late Gli1LacZ induction by chondrogenesis-associated Ihh in digit rays further confounds interpretation(Witte et al., 2010). To determine if the extent of Shh response relative to early Shh expression is similar to wildtype in the Shh-CKO;Bax-CKO during the short Shh activity window after tamoxifen treatment (Figure 1), we used hybridization chain reaction (HCR)(Choi et al., 2018) for sensitive, high-resolution spatial detection of Shh RNA expression and response (Gli1 RNA expression) simultaneously, by multiplex labeling in the same limb bud to accurately compare spatial distributions. Note that the Shh probes used detect both the recombined (mutant) as well as functional Shh transcripts, and that midline hindgut Ihh signaling, preserved in the Shh−/− (Zhang et al., 2001), activates some proximal axial response in the urogenital-cloacal region(Haraguchi et al., 2007; Perriton et al., 2002) (see Shh and Gli1 RNAs in Figure 3 Shh−/− panels). Using HCR simultaneous detection, Gli1+ response is very similar in its A-P extent to Shh and is short range in both sibling controls and Shh-CKO;Bax-CKO mutants during the transient pulse when response is detected in the mutant limb buds (Figure 3, 29 somite panels, arrows). We also compared spatial A-P extents of Gli1 and Ptch1 RNAs by HCR to confirm that the extent coincided for both of the major direct Shh-response reporters (Figure S2). These analyses show that, as in the wildtype, Shh activity/response in the Shh-CKO;Bax-CKO mutant with rescued digit formation is limited to short-range pathway activation during the transient Shh pulse, and restricted to the territory that gives rise only to digit 4–5 (the Shh-expressing ZPA).
Digit Rescue in Shh-CKO;Bax-CKO is not a consequence of residual or re-activated Hh pathway function or pathway target de-repression.
There are several alternative possibilities to account for the observed rescue of normal limb development in Shh-CKO;Bax-CKO embryos that we excluded. First, a re-emergence of Hedgehog pathway activity, which might be the result of mosaic recombination of the Shh-floxed allele, de novo re-induction of a ZPA in a new cell population, or ectopic induction of an alternate Hh ligand. Each of these would result in, and be detected by, re-activation of direct Shh targets (Ptch1, Gli1 RNA) that provide a more sensitive readout of Hh pathway activity than measuring ligand expression. Monitoring of direct Shh target Ptch1 (0/7, 0/9) and Gli1 (0/7) RNAs at both early and late patterning stages, or inclusion of a Gli1LacZ/+ knock-in allele (0/8) to provide a highly sensitive Shh-response reporter with enzymatic amplification at early stages(Bai et al., 2002), all failed to detect any Hh pathway recovery in Shh-CKO;Bax-CKO embryos (Figure 4A). In particular, mosaic recombination of the Shh-CKO allele by the transient Cre activity would leave residual Shh-expressing cells that would proliferate and consequently be expected to increase the Ptch and Gli1 reporter signals over time.
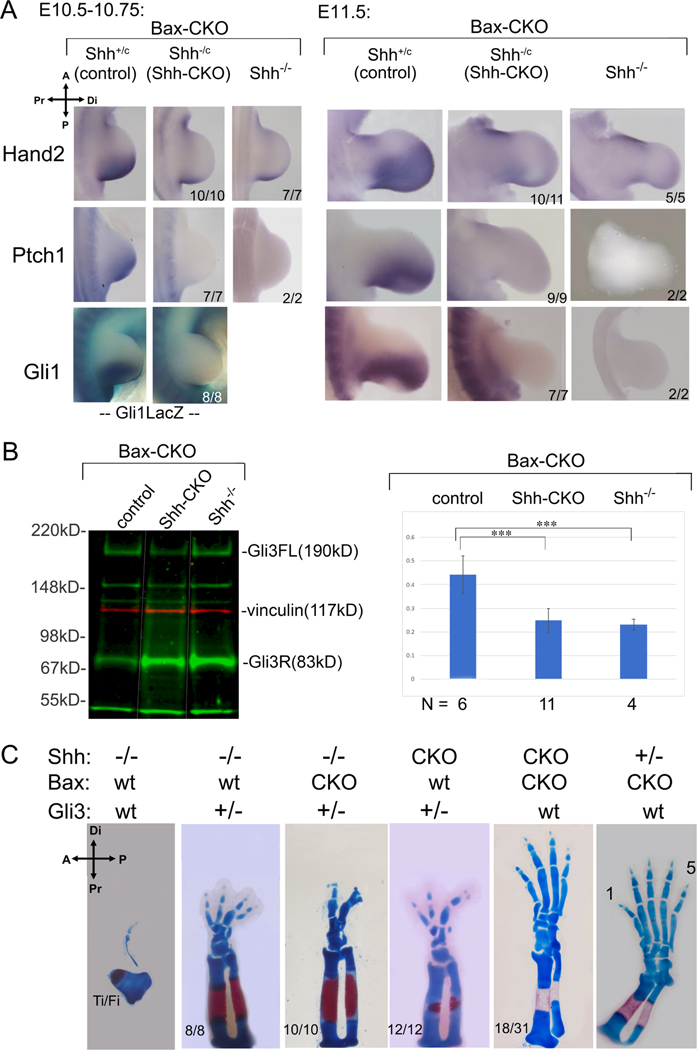
Figure 4. Digit rescue in Shh-CKO;Bax-CKO embryos is not due to persistent Shh pathway activity or re-activation.
(A) Shh pathway activity monitored by RNA (Hand2, Ptch1, Gli1) and by LacZ activity from a Gli1LacZ/+ knock-in allele after Shh removal by tamoxifen at E9.5+3h (as in Figure 1B). Mutant numbers analyzed with result shown are indicated in each panel. Hand2 at E11.5 was very slightly higher than Shh−/− in 1/11 Shh-CKO embryos. (B) Gli3 full-length (Gli3FL) and repressor (Gli3R) protein quantitation in E10.5 hindlimbs, after Bax/Bak and Shh removal as in Figure 1B,C. Blot shows representative example of Gli3 level (green) for different genotypes (noncontiguous lanes re-ordered from same blot, indicated by white lines). Molecular weight marker positions indicated to right; vinculin (red) loading control. Gli3 FL/R ratios (mean + SEM) shown at right in bar-graph with numbers analyzed (N) for each genotype. Gli3 FL/R is equivalently reduced in Shh-CKO;Bax-CKO (p=0.00002) and in Shh−/−;Bax-CKO (p=0.001) compared to control, and there is no significant difference in Gli3 FL/R between Shh-CKO;BaxCKO and Shh−/−;Bax-CKO (p=0.48). ***, p <0.001. (C) Effect of Gli3 dosage reduction (Gli3+/−) on Shh-CKO and on Shh−/− skeletal phenotypes (E16.5), compared to the effect of Bax/Bak removal (Bax-CKO).
Another major, important possibility to address, is potential loss of pathway repression by Gli3 repressor (Gli3R). Shh prevents processing of Gli2/Gli3 nuclear effectors from full-length activators (Gli2FL/Gli3FL; GliA) to truncated repressors (Gli2R/Gli3R) of Shh targets(Wang et al., 2000; Wang et al., 2010). Release from Gli3 repression arguably plays the main role in most Shh limb target regulation(Lewandowski et al., 2015; Litingtung et al., 2002; te Welscher et al., 2002b). Indeed, although Ptch1 and Gli1 are direct GliA targets, Gli1 is completely dispensible(Bai et al., 2002) and Ptch1 is not required for digit formation per se once Shh expression has begun(Butterfield et al., 2009). Consequently, key limb target activation may occur ligand-independently, without activating Gli1A-target reporters (Gli1, Ptch1), via either Gli3R removal or functional antagonism. We used several approaches to test for evidence of altered net Gli3R activity. First, Hand2, which induces Shh/ZPA by antagonizing Gli3(te Welscher et al., 2002a), is directly repressed by Gli3R(Vokes et al., 2008) and remained absent from rescued Shh-CKO;Bax-CKO in both early (10/10) and later (10/11) stage limb buds (Figure 4A). Secondly, Gli3R activity can be modulated at the protein level by altered processing or degradation(Wang et al., 2000; Wang et al., 2010). Although Bax/Bak removal in wildtype limb buds has no impact on skeletal patterning, an altered balance of Bcl2 family members has been reported to affect Gli-processing activity indirectly and could generate a Gli3R deficit(Wu et al., 2017). We examined Gli3 protein levels in early (E10.75) individual limb buds). Gli3FL/Gli3R ratios in Shh-CKO;Bax-CKO were unchanged from Shh−/−;Bax-CKO limb buds, and both were equally reduced compared to Shh-expressing controls (Figure 4B), indicating that rescue was not explained by reduced Gli3R protein. Thirdly, “effective” repressor activity of Gli3R may be altered by interacting protein partners without changing quantitative protein level(Chen et al., 2004; Galli et al., 2010). To exclude altered net Gli3R activity by any mechanism, we used a genetic test to compare the phenotypic effect of rescuing cell survival in Shh-CKO;Bax-CKO with that of intentional Gli3 dosage reduction. We compared Bax/Bak removal with Gli3 dosage (Gli3+/−) effects in both Shh-CKO and in Shh−/− limbs. Complete Gli3 loss alone rescues limb development in Shh−/− embryos, albeit with synpolydactyly and phalangeal morphology changes(Litingtung et al., 2002; te Welscher et al., 2002b). However, haploid Gli3 dosage (Shh−/−;Gli3+/−) has an intermediate effect on the Shh−/− null phenotype (improved zeugopod morphology, several small digit rudiments; Figure 4C, 8/8). In contrast to Shh−/−;Gli3+/−, no change in the Shh−/− null phenotype was observed when Bax/Bak was removed (Figure 1D; 18/18). Furthermore, removing Bax/Bak in the Shh−/−;Gli3+/− limb did not improve limb skeletal phenotype beyond the effect of Gli3 dosage reduction alone (Figure 4C, 10/10), suggesting that Bax/Bak removal did not impact the “effective” net Gli3R level significantly and that reduced Gli3 dosage, even together with enforced cell survival, in the Shh null does not mimic, or substitute for, the effect of transient Shh expression with enforced cell survival (Shh-CKO;Bax-CKO).
Whereas Bax/Bak removal had little effect on the Shh−/− null compared to Gli3 dosage reduction, the reverse holds for the Shh-CKO. Without enforced cell survival (Bax/Bak removal), the Shh-CKO;Gli3+/− was phenotypically identical to the Shh−/−;Gli3+/− (12/12; Figure 4C). In contrast, in Shh-CKO;Bax-CKO limbs with wildtype Gli3 dosage (Gli3+/+), both zeugopod and between 3–5 digits with normal morphologies and clear A-P polarity were restored (18/31; Figures 1D, 4C). Together, these results argue strongly against altered Gli3R activity per se as a mechanistic basis for the restoration of normally polarized limb development in the early Shh-CKO with enforced cell survival to bypass Shh late function.
Sustained expression of key outgrowth and patterning regulators in Shh-CKO;Bax-CKO with transient Shh activity and enforced cell survival.
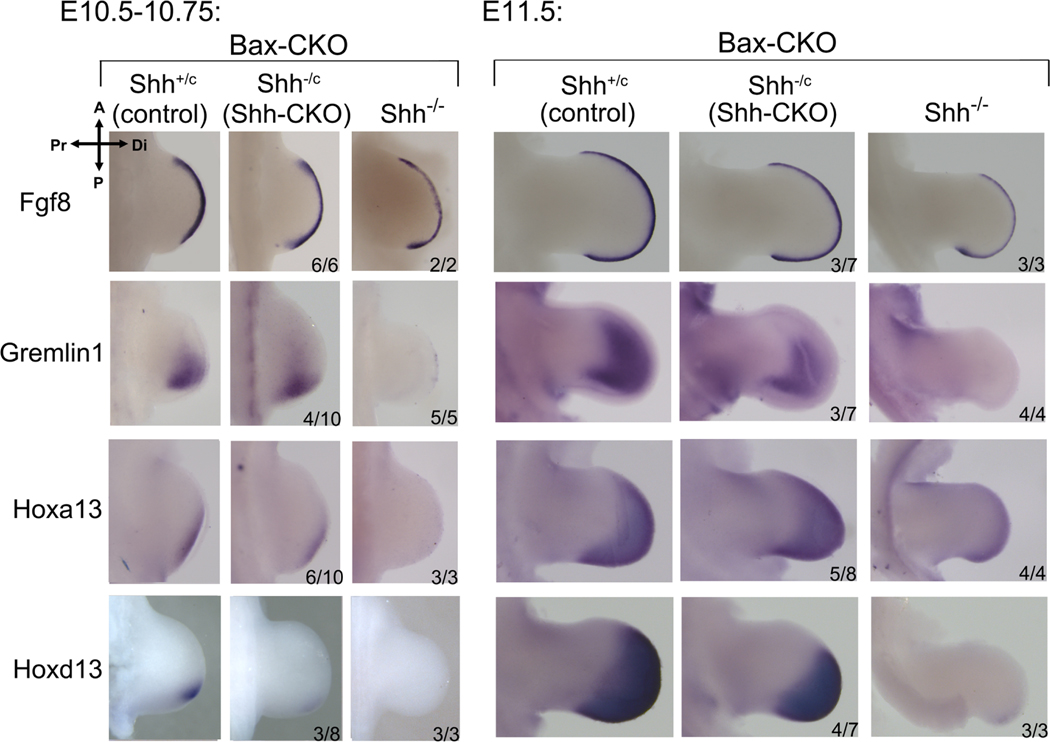
To assess whether downstream regulators of limb outgrowth and digit patterning are restored by transient Shh exposure, we examined expression of major direct and indirect downstream Shh targets that regulate limb bud outgrowth (AER/Fgf signaling(Mariani et al., 2008; Zuniga et al., 1999)) and digit patterning (5’Hox genes(Davis et al., 1995; Fromental-Ramain et al., 1996)). Unlike Hand2 and direct GliA-regulated targets (Ptch1, Gli1), expression of key outgrowth and patterning regulators is maintained in a subset of Shh-CKO;Bax-CKO embryos, roughly correlating with the observed 50% occurrence of an early, transient Shh activity pulse, and subsequent skeletal rescue (Figures 1, 3, 5, S1, S3). In the remainder, expression profiles in Shh-CKO;Bax-CKO resembled the null Shh−/−;Bax-CKO (Figures 5, S3).
Figure 5. Expression of key targets implicated in outgrowth and patterning is maintained in Shh-CKO;Bax-CKO embryos.
Expression of Shh target RNAs that regulate outgrowth (Fgf8, Grem1) and patterning (Hoxa13, Hoxd13) at early and later stages after Shh removal by tamoxifen at E9.5+3h (as in Figures 1B, 2A). Fgf8 expression is unaltered even in Shh−/− null hindlimb at E10.75(Chiang et al., 2001), but was reduced in Shh−/−;Bax-CKO by E11.5. Shh-CKO;Bax-CKO embryo numbers analyzed with result shown are indicated in each panel (expression maintained). In remainder, expression was unchanged from that seen in the null Shh−/−;Bax-CKO. Related to Figures S3, S4.
The direct Shh target Grem1 plays a key role in AER/Fgf8 maintenance(Zuniga et al., 1999) and consequently Fgf8 expression declines in null Shh−/− hindlimb after E10.5(Chiang et al., 2001). Shh−/−;Bax-CKO limb buds likewise lacked early and late Grem1 expression (absent in 5/5, 4/4) and by E11.5 Fgf8 expression was clearly reduced (3/3; Figure 5). In contrast, a subset of Shh-CKO;Bax-CKO embryos maintained early and late Grem1 expression (4/10 and 3/7), and preserved Fgf8 after E10.5 (3/7; Figure 5). The Fgf8-regulated target, Cyp26b1, required to clear retinoids for distal limb progression(Probst et al., 2011), was also maintained in Shh-CKO;Bax-CKO (4/6) but declined in Shh−/−;Bax-CKO by E11.5 (3/3, Figure S3).
Several 5’Hox genes regulate A-P patterning downstream of Shh. Hoxd13 and Hoxa13, critical for digit specification(Fromental-Ramain et al., 1996), are both expressed only at very late stages and at low levels in Shh−/−;Bax-CKO (3/3, 4/4) compared to controls (Figure 5). In contrast, low level distal Hoxa13 expression was already detected early in a subset of Shh-CKO,Bax-CKO hindlimb buds (6/10), and became robust at later stages (5/8). Hoxd13 was detected at a trace level early (3/8), but was clearly detectable at the onset of the second phase 5’Hoxd distal footplate expansion(Tarchini and Duboule, 2006) (4/7, E11.5), and well prior to the late condensation stage after all digit rays have formed, when both the Shh−/− null(Chiang et al., 2001) and Shh−/−;Bax-CKO re-express Hoxd13 (∼E12.5, Figure S3). Since 5’Hoxd genes act mainly during the distal expansion phase to determine digit identity by regulating late interdigit signaling centers(Huang et al., 2016), this sustained second phase Hoxd13 activation likely suffices for the morphogenesis of normal distinct digit types in Shh-CKO;Bax-CKO compared to Shh−/−;Bax-CKO (null) embryos. Hox11 paralogs play a key role in zeugopod patterning and growth(Davis et al., 1995), which is also highly perturbed in the Shh−/− but restored in Shh-CKO;Bax-CKO hindlimbs (tibia/fibula, Figures 1D, 4C). Early Hoxd11 expression was similar to controls even in Shh−/−;Bax-CKO (2/2), but became undetectable by the second distal expansion phase (2/2). In contrast, Hoxd11 was maintained at control levels in a subset of Shh-CKO;Bax-CKO limb buds at both early (5/5) and late (2/7) stages (Figure S3), consistent with zeugopod rescue frequency. Shh inhibition in short term mouse limb bud cultures(Lewandowski et al., 2015; Panman et al., 2006) also suggests that some direct Shh targets are maintained if Shh activity is curtailed after onset, as we have shown here (Grem1; Jag1 in Figures 5, S3). Yet in those studies other downstream targets, particularly 5’Hoxd genes, appeared to require sustained Shh activity for their continued expression. However, development does not progress normally in short-term ex vivo cultures, precluding analysis of the second phase 5’Hoxd expression (which is selectively restored in the Shh-CKO;Bax-CKO) in the cultured limb buds after early Shh inhibition.
We also examined the expression of anterior regulators that are repressed/antagonized by Shh activity in posterior limb bud. Unlike the posterior regulators, major anteriorly expressed regulators of patterning, Irx3(Li et al., 2014) and Alx4(te Welscher et al., 2002b), are only modestly altered in their early extent even in the Shh null (Shh−/−) mutant (Figure S4). However, the slightly extended posterior expression seen in Shh−/− was also observed in about 50% of the Shh-CKO;Bax-CKO embryos (Irx3 2/4; Alx4 5/8). Our results indicate that expression of targets important for both limb bud outgrowth and patterning are sustained by a transient pulse of Shh with enforced cell survival, providing a basis for the phenotypic rescue of limb development, but do not address the issue of how transient Shh activity leads to stable, graded A-P expression of certain Shh regulated targets in the limb.
Shh is required indirectly to specify digit 1 (thumb).
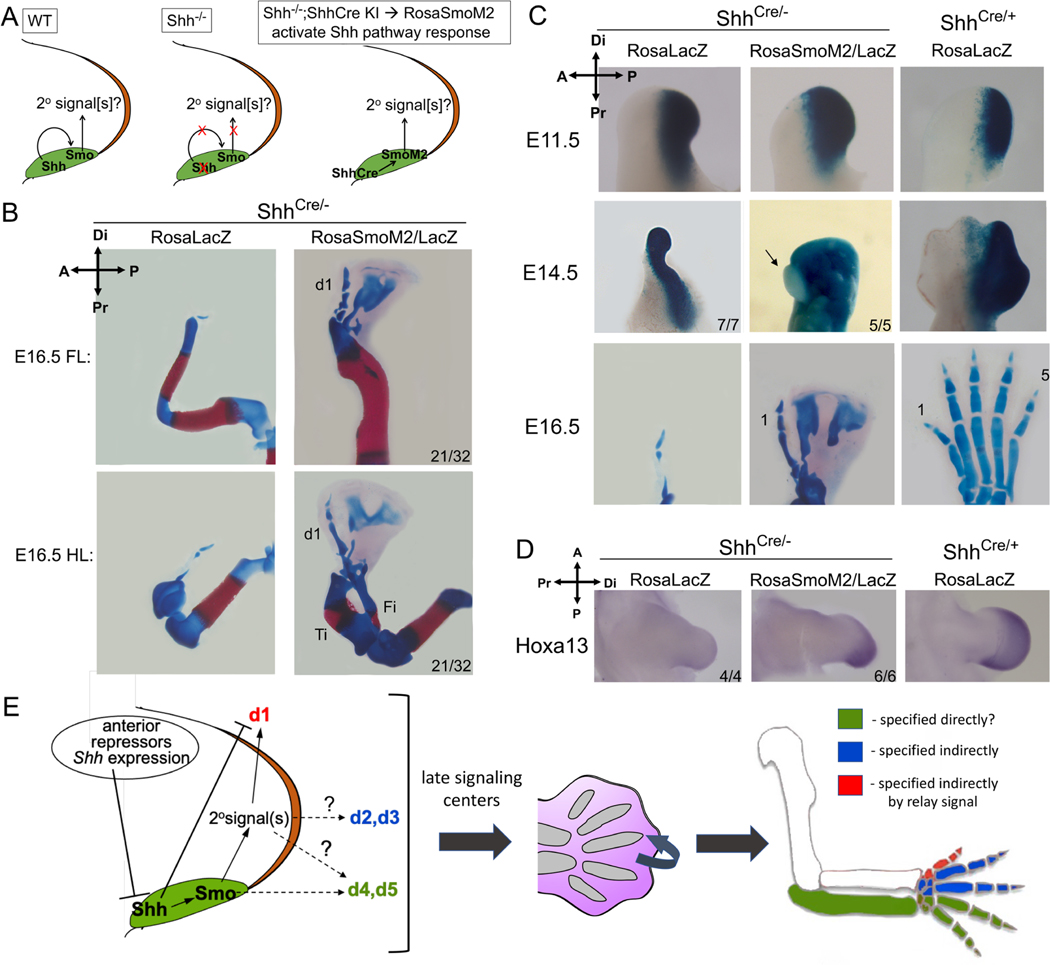
To test if Shh acts via a relay mechanism, we used a genetic strategy (Figure 6A) to activate Shh targets autonomously only in ZPA cells and ask if any non-ZPA-derived digits are rescued in the complete absence of Shh ligand (Shh null). Cell-autonomous pathway activation was achieved using a conditional transgene (RosaSmoM2), expressing a constitutively-active form of Smoothened (SmoM2)(Jeong et al., 2004), a membrane GPCR essential for transducing Shh(Kong et al., 2019). The ShhCre knock-in allele(Harfe et al., 2004) was used to restrict SmoM2 and Shh-target activation to ZPA cells (Shhcre;RosaSmoM2/+; referred to as Shh-SmoM2+), and evaluated in the Shh null background (Shhcre/-). Enforced Shh-response by SmoM2 in the ZPA also affects Ihh targets and chondrogenesis(Long et al., 2001), precluding morphologic evaluation of any ZPA-descendant digits (d4,d5 in wildtype), but differentiation of non-ZPA digits is unaffected (d1-d3, see Figure S5A). Unexpectedly, in both Shhcre/- fore- and hindlimbs, enforced, cell-autonomous pathway activation of Shhcre/-;Shh-SmoM2 in the ZPA rescued formation of a positionally and morphologically normal, biphalangeal digit 1 at high frequency (66%; 21/32 limbs, Figure 6B). Digit 1 specification is thought to be Hh-independent, based on the normal lack of any direct Shh-response in the d1 progenitor territory(Ahn and Joyner, 2004) (see also Figure 2B) and the persistence of a d1-like structure in Shh null hindlimbs(Chiang et al., 2001). However, our genetic lineage tracing reveals that the single dysmorphic digit in the Shh null mutant (Shhcre/-) is actually entirely descended from posterior ZPA (LacZ+) d4/d5 progenitor cells (Figures 6C, S5A), which lies outside of and posterior to the zone of broad anterior apoptosis present in Shh−/− limb buds (Figures S1B, S5D,E; see also (Chiang et al., 2001; Zhu et al., 2008)). In contrast to the posterior ZPA-origin of the single Shh−/− null hindlimb digit, our lineage analysis clearly demonstrates that the morphological “d1” rescued in the Shhcre/-;Shh-SmoM2 arises non-autonomously entirely from anterior (LacZ-negative) non-ZPA limb cells (5/5; Figure 6C) and condenses at the anterior limb margin where d1 normally forms (see Figure S5F). Digit 1 rescue was not the result of either cryptic Hh ligand or downstream pathway activation as seen in a small fraction of control Shh-SmoM2+ limb buds (Gli1, 0/8; Ptch1, 0/10; Figure S5A,C). Distal-anterior Hoxa13 expression, which is uniquely essential for d1 specification(Bastida et al., 2020; Fromental-Ramain et al., 1996), and is absent or greatly reduced in ShhCre/- null (0/4), was restored across the Shhcre/-;Shh-SmoM2+ distal limb bud (6/6; Fig. 6D), confirming d1 identity. Likewise, a late d1-specific marker (Uncx4.1) was also expressed in the rescued digit domain of Shhcre/-;Shh-SmoM2 (6/8), but not in ShhCre/- (0/4; Figure S5B). Together, these results strongly argue that a bona fide digit 1 is absent in the Shh null mutant but is restored in the Shhcre/-;Shh-SmoM2, indicating that d1 specification is indirectly Shh-dependent and requires a relay signal that is activated downstream of Shh-response in the ZPA.
Figure 6. Relay signals downstream of enforced Shh-response confined to ZPA restore normal digit 1 in Shh−/− null limb.
(A) diagram of Shh-response activation in ZPA of null limb bud (ShhCre/-) by SmoM2 (Shhcre/-;Shh-SmoM2+). Any effect on non-ZPA digits requires non-autonomous relay signal(s). (B) Skeletal stain showing normal digit 1 (d1) in Shhcre/-;Shh-SmoM2+ forelimbs and hindlimbs (21/32). Ti, tibia; Fi, fibula. (C) ZPA-lineage analysis and close-up of mutant footplate with activated SmoM2. The anterior-most digit in Shhcre/-;Shh-SmoM2+ remains devoid of LacZ+ cells (5/5, arrow), whereas residual digit in Shh−/− null arises entirely from LacZ+ ZPA-descended cells (7/7). (D) Hoxa13 RNA is restored in Shhcre/-;Shh-SmoM2+ (6/6) at E11.5, but is absent in Shh null (4/4; forelimb shown). (E) Model for digit specification by Shh via indirect mechanisms (see text discussion). Transient Shh specifies non-ZPA digits (d1-d3) by indirect mechanisms. Direct Shh signaling selectively inhibits(Li et al., 2014), but indirect relay signaling promotes, d1 specification, establishing a unique d1 (thumb) regulatory hierarchy. Relay signaling, initiated by early transient Shh, ultimately sets up late interdigit signaling centers to regulate final digit identity. Related to Figure S5.
Why was only d1, but not other non-ZPA digits (d2,3) restored by Shh-SmoM2? Shh cell-survival function remains absent and Shhcre/-;Shh-SmoM2+ limb buds display considerable anterior apoptosis (Figure S5D,E), but enforcing cell survival (with Bax−/−;Bak−/−, see Table 1) did not rescue any further digit formation besides d1 (8/8). We suspect that the failure to rescue d2–3 reflects a problem inherent in the timing of enforced SmoM2/Shh-response in ZPA cells (which is induced by ShhCre only after normal Shh onset). The delayed specification timing of d1 relative to other digits(Bastida et al., 2020) would be consistent with this selective d1 rescue.
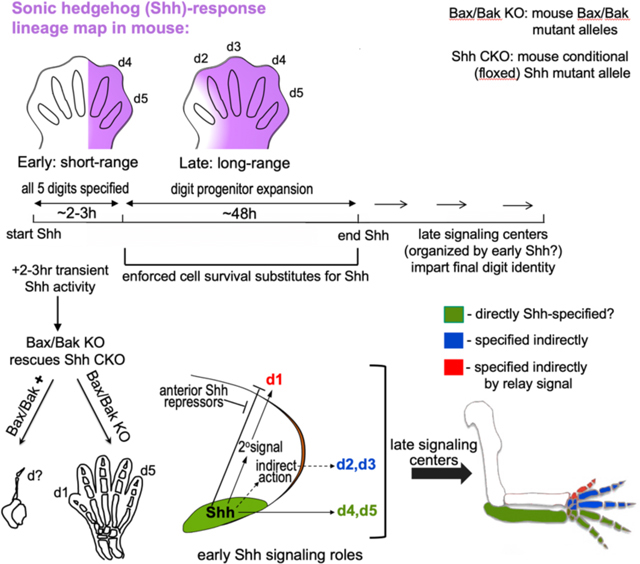
Discussion
Our genetic rescue reveals two distinct roles for Shh in the limb: a transient (2–3hr), early requirement that is critical to specify all digits, and a sustained requirement to promote cell survival. This transient Shh pulse is both necessary and sufficient for normal limb morphogenesis, if the role of Shh in maintaining cell survival is bypassed (by Bax/Bak removal). Yet genetic lineage tracing, together with HCR, shows that Shh-response is short-range and restricted to ZPA-derived digit progenitors (d4-d5) during the same time window of transient Shh activity that suffices to specify all digits. Both biochemical and genetic assays reveal that the Gli3R activity gradient is unperturbed by the short Shh activity window in the rescued Shh-CKO;Bax-CKO, consistent with the absence of any long-range Hh response. Together, these results indicate that Shh is not a limb morphogen but acts via an indirect mechanism to specify the non-ZPA digits (d1–3, Figure 6E). Shh may act locally to specify d4/5 directly, but our results don’t exclude an indirect contribution. Digit 1 specification, occurring relatively late, is further distinguished in being repressed by direct Shh response, yet indirectly Shh-dependent via a relay signal (discussed below). These different responses to Shh signaling define up to three distinct types of regulation leading to: specification of posterior ZPA-descended digits (4,5), anterior digits (2–3), and digit 1 (Figure 6E). Unexpectedly, long-range Shh signaling, which occurs only at later stages when the limb bud is expanding and well after specification is complete, plays a critical role in sustaining cell survival across the d2–5 territories. We previously showed the number of digits lost decreases progressively with later Shh deletion times, correlating with later onset of apoptosis ((Zhu et al., 2008); see also Table S1), but the entire late Shh requirement is completely bypassed by Bak/Bax removal in the early Shh-CKO.
We considered the possibility that either persistent low-level, residual pathway activity, or a long-range response during the transient Shh expression pulse had escaped detection, but think this unlikely for the following reasons. 1) Low-level Shh activity persistence, owing to either incomplete Shh recombination or some compensatory feedback leading to ectopic Hh ligand activation should all result in the continued activation of direct targets Gli1 and Ptch1, which are detectable at much lower levels than is Shh RNA(Lewis et al., 2001; Lex et al., 2022; Zhu et al., 2008). Furthermore, Shh-expressing cells proliferate in the early limb bud to give rise to d4/d5, and activation of direct target reporters should be increasingly apparent over time (E10.5-11.5). Several methods (including reporter amplification steps; HCR, Gli1LacZ) failed to detect any Shh response in Shh-CKO;Bax-CKO limbs (Figures 1C, 3, 4A, S1), either by 3 hrs after Shh expression onset (28/28), at 12 hrs after onset (26/26), or at 36 hrs (16/16) nearing the end of normal Shh duration. 2) The skeletal phenotype of Bax+ Shh-CKO sibling embryos was uniformly identical to the Shh germline null (28/28, Figure 1D), which is incompatible with residual Shh signaling. Persistent, low level Shh (generated by deleting Shh with ShhCre) results in a very mild digit phenotype (16/16) and modest Ptch1 response(Scherz et al., 2007), despite only trace levels of Shh. 3) Long-range signaling effects on Gli3R activity (target de-repression) were not detected. Many Shh targets are regulated mainly by de-repression(Lewandowski et al., 2015; Lex et al., 2020), but long-range effects of Shh on Gli3R and target “de-repression” are not measured by GliA-dependent target reporters (Gli1, Ptch1). There is no single, uniform expression response for this target class, and consequently no good universal “reporters” to assay Shh-induced de-repression, because regulation of derepressed targets by other activating transcription factors varies highly from gene to gene(Lewandowski et al., 2015). But phenotypic comparison to the effects of Gli3 dosage reduction can serve as a gauge to assess perturbation of the anteriorly-biased Gli3R gradient caused by long-range signaling and target de-repression during transient Shh activity. We altered genetic Gli3 dosage to assess if Shh-CKO;Bax-CKO rescue might result from altered Gli3R activity. Our results (Figure 4C) indicate that Gli3 dosage reduction (Gli3+/−) does not phenocopy the digit rescue by enforced cell survival in the Shh-CKO, and has no impact on the Shh-CKO skeletal phenotype beyond the effect of Gli3+/− on the Shh−/− null mutant, arguing against long-range modulation of Gli3R by transient Shh signaling in the rescued Shh-CKO. Similarly, long range GliA response detection relies on two targets, Ptch1 and Gli1 (reported by HCR and by Gli1CreER). Given the limited repertoire of Hh-response reporters, we cannot definitively exclude a transient morphogen action of Shh at levels below their detection thresholds.
Digit identity is morphologic in nature, arising via distinct organizations of the same tissues and not based in cell fate changes, features suggesting progressive specification. Indeed, work in both chick and mouse indicates that late interdigit signaling centers impinge on digit tip progenitors to regulate phalanx numbers formed, determining final digit identities(Dahn and Fallon, 2000; Huang and Mackem, 2021; Huang et al., 2016; Suzuki et al., 2008; Witte et al., 2010). Shh may regulate digits specified at particular A-P limb positions via relay signals to establish such late signaling centers (Figure 6E). If Shh acts early as a trigger, it is not surprising that direct targets involved in signal transduction and response range modulation, such as Gli1 and Ptch1, become dispensable when cell survival is enforced. In contrast, downstream targets that regulate growth, such as Grem1, and later patterning, such as Hoxa13 and Hoxd13, appear to play key roles; their maintenance correlates with the limb rescue frequency by a transient Shh pulse.
Uncovering the mechanisms that sustain stable gene activation after only transient Shh exposure will be an important future focus to illuminate how Shh patterns the developing limb. Stable alterations in target gene expression following transient Shh exposure could involve several, non-mutually exclusive mechanisms including chromatin and/or DNA modifications and relay mechanisms incorporating lock-on circuitry(Alon, 2007). Recent work suggests that Shh targets are poised for expression as soon as Shh activity initiates and that Gli3-mediated repression commences only after this point(Lex et al., 2020; Lex et al., 2022). A transient burst of Shh activity could trigger a self-reinforcing bi-stable switch(Alon, 2007) if activating factors that reinforce target expression also block introduction of repressive marks by Gli3R post-Shh onset. One type of indirect mechanism would be via non-cell autonomous relay signals acting downstream of transient Shh activity to specify non-ZPA derived digits. Such relay signals can become rapidly self-sustaining via feedback loops, as occurs with Fgf10-Fgf8 signaling downstream of transient Tbx5 activity in the limb(Hasson et al., 2007). Other indirect mechanisms that remain less extensively explored, such as bioelectric or mechanical, are also possible(Harris, 2021; Piccolo et al., 2022).
One alternative to a relay mechanism for patterning digit territories not directly responsive to transient Shh signaling would be the maintenance of polarity, once initiated by transient Shh activity, at the level of antagonistic interactions between “posterior” (Shh target) and “anterior” (antagonizing) transcription factors, which are initially co-expressed in very early-stage limb bud mesenchymal cells(Osterwalder et al., 2014). However, such a scenario is difficult to reconcile with the finding that A-P polarized expression of some, but not other, targets is maintained in the rescued Shh-CKO;Bax-CKO limb buds and is accomplished in the absence of any Hand2 expression, which normally antagonizes anterior Gli3R to maintain posterior limb bud asymmetry downstream of Shh(te Welscher et al., 2002a). Furthermore, lineage tracing indicates that the response to “transient” Shh is entirely restricted to the posterior ZPA, which would generate clear-cut anterior and posterior limb bud compartments, rather than the graded A-P target gene expression seen for some target genes maintained in the rescued Shh-CKO;Bax-CKO (eg. Grem1, Hoxa13, Figure 5). Spatially graded target expression would require some type of non-autonomous effect, whether induced by Shh effectors (Gli2/3), or by other transcription factors acting downstream of short-range Shh response within the ZPA.
Whether Shh acts as a trigger, via an indirect mechanism, to specify digits has implications for both limb evolution and regeneration. Such an indirect mechanism is at odds with chick studies showing that Shh acts as a limb morphogen(Scherz et al., 2007; Towers et al., 2008; Yang et al., 1997). These studies rely on pharmacologic inhibition that may persist to later stages to affect Ihh in digit tip progenitors, leading to phalanx loss(Gao et al., 2009) that scores as digit identity changes attributed to Shh inhibition; reduced cell survival by Shh suppression may also impact late-stage digit morphogenesis. Other chick studies suggested involvement of downstream relay signals(Drossopoulou et al., 2000; Pickering et al., 2019; Yang et al., 1997) , but did not discriminate if Shh also acts as a morphogen. Whether apparent mouse-chick differences in Shh function reflect biologic or technical factors remains to be explored and will be important for understanding the degree of functional conservation in assessing evolutionary adaptation (for eg. proposed Shh role in thumb evolution, below). Our results indicating Shh acts mainly to promote cell survival after a transient requirement in specification also support the proposal that regulation of digit number and pattern by Shh can be uncoupled to facilitate adaptive evolution of digit number(Shapiro et al., 2003; Zhu et al., 2008). Shh acting as a short-range trigger in developing limb may also be more parsimonious with patterning events during limb regeneration(Nacu et al., 2016). Although Shh is implicated in AP patterning during axolotl limb regeneration, inductive interactions overall appear to be mainly short-range with little evidence for long-range morphogen function.
Our genetic relay signal assay, selectively enforcing Shh response solely in the ZPA of the Shh null (Shh−/−) mutant, unexpectedly revealed that d1 is indirectly Shh-dependent. The rescued digit arose entirely from non-ZPA anterior cells, and had all the features of bona fide digit 1 based on positional, morphologic, and gene expression criteria. Yet previous work has shown that direct Shh response selectively prevents formation of d1 territory, and indeed, a complex regulatory circuit that represses Shh anteriorly is required to specify d1(Li et al., 2014). Unlike other digits, the d1 territory also remains outside of the Shh signaling-response range (eg. Figure 2B) during the entire expansion phase when Shh maintains cell survival. Consequently d1 survival is sustained differently. Why impose such a complex regulatory hierarchy for d1 formation (see Figure 6E), involving repression by direct Shh signaling but requiring an indirect Shh-induced relay signal? We propose that the unique control and consequent delay in d1 specification and growth/expansion enabled its independent evolution by uncoupling its regulation and morphogenesis from that governing other digits. Such regulatory uncoupling would facilitate the evolution of an opposable thumb, as well as other grasping/clutching adaptations important for arboreal tetrapods, including birds and mammals.
Limitations of the Study.
Shh-response reporters showing that Shh acts only at short range and not beyond the ZPA in early limb bud may have failed to detect a very low level, transient longer-range activity. Restricted, short-range, early stage Shh activity and skeletal phenotypic outcome cannot be evaluated in the same embryo in Shh-CKO rescue experiments; the relationship remains correlative, albeit strongly so. Definitive demonstration that Shh acts indirectly, as a trigger rather than a morphogen, to pattern limb digits will require identifying the factors involved.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Susan Mackem (mackems@mail.nih.gov)
Materials availability
The Bax-deleted (Bax+/−) mouse line generated in this study can be obtained from the lead contact, Susan Mackem.
Data and Code Availability
Data
All microscopic and other imaging data reported in this paper will be shared by the lead contact upon request.
Code
No bioinformatic data or code were generated.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All animal studies were carried out according to the ethical guidelines of the Institutional Animal Care and Use Committee (IACUC) at NCI-Frederick under protocol #ASP-20-405. All mice were maintained in a specific pathogen-free facility with a 12-hr light cycle (6 am-6 pm) on a standard chow diet (LabDiet mouse breeder diet O/HS). Both male and female mice with mutant alleles displayed no sex differences in phenotypes, and embryos of both genders ranging from E9.5-17.5 were included in all analyses, but individual gender determination was not carried out. The Shh-floxed(Lewis et al., 2001), Shh null(Chiang et al., 1996), Bax-flox;Bak−/−(Takeuchi et al., 2005), Gli1LacZ/+(Bai et al., 2002), and Gli3 (Xt-J)(Buscher et al., 1998) mutant lines, and the Hoxb6CreER(Nguyen et al., 2009), ShhCre(Harfe et al., 2004), ShhCreER(Harfe et al., 2004), Gli1CreER(Ahn and Joyner, 2004), RosaSmoM2(Jeong et al., 2004), RosaLacZ(Soriano, 1999), and RosaEYFP(Srinivas et al., 2001) mouse lines were all described previously. All single and compound mouse lines were maintained on an outbred, mixed strain background (predominantly FVB/n and C57BL/6). A detailed summary of the crosses used to generate embryos for different experiments and outcomes is provided in Table 1. For timed matings, noon on the date of the vaginal plug was defined as E0.5. For phenotypic rescue with Hoxb6CreER, pregnant mice were injected intraperitoneally with a single dose of 3mg tamoxifen and 1mg progesterone (Nakamura et al., 2006) at E9.5+3hrs and embryos were collected at times indicated. Somite numbers were counted at time of collection for embryos that were harvested before E10.5. Sibling embryos were used in comparative analyses whenever possible. For lineage tracing with ShhCreER and Gli1CreER, a single dose of 0.5–1mg tamoxifen was injected at the times indicated.
Bax-deleted (Bax+/−) mice were generated by crossing Bax-flox males with Prrx1Cre(Logan et al., 2002) females to produce germ-line recombination and offspring were out-crossed to remove Prrx1Cre and then crossed with the lines listed in Table 1. The Bax-deleted allele genotype was detected using PCR (see Key Resources Table for primers).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| goat anti-Gli3 antibody | R&D Systems | Cat#AF3690; RRID: AB_2232499 |
| rabbit anti-Gli3 antibody | Chen et al., 2004 | N/A |
| mouse anti-Vinculin antibody | Sigma-Aldrich | Cat#V9264; RRID: AB_10603627 |
| IRDye 800CW goat anti-rabbit secondary antibody | LI-COR | Cat#926-32211; RRID: AB_621843 |
| IRDye 800CW donkey anti-goat secondary antibody | LI-COR | Cat#926-32214; RRID: AB_621846 |
| IRDye 680RD donkey anti-mouse secondary antibody | LI-COR | Cat#926-68072; RRID: AB_10953628 |
| Anti-Digoxigenin-AP antibody | Sigma-Aldrich | Cat#11093274910; RRID: AB_2734716 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma-Aldrich | Cat#T5648; CAS: 10540-29-1 |
| Progesterone | West-Ward Pharmaceuticals | Cat#NDC 0143-9725-01; CAS: 57-83-0 |
| Lysotracker Red | ThermoFisher Scientific | Cat#L7528 |
| Alcian Blue 8 GX | Sigma-Aldrich | Cat#A-5268; CAS: 33864-99-2 |
| Alizarin Red S | Sigma-Aldrich | Cat#A5533; CAS: 130-22-3 |
| Ethanol | Sigma-Aldrich | Cat# 107017; CAS: 64-17-5 |
| Acetone | Sigma-Aldrich | Cat# 179124; CAS: 67-64-1 |
| Methanol | Sigma-Aldrich | Cat#322415; CAS: 67-56-1 |
| Paraformaldehyde | Sigma-Aldrich | Cat#441244; CAS: 30525-89-4 |
| Glutaraldehyde | Polyscience | Cat#00216; CAS: 111-30-8 |
| Glycerol | Sigma-Aldrich | Cat#G7757; CAS: 56-81-5 |
| Formamide | Sigma-Aldrich | Cat#F7503; CAS: 75-12-7 |
| X-Gal | GoldBio | Cat#X4281C; CAS: 7240-90-6 |
| Potassium ferrocyanide | Sigma-Aldrich | Cat#P3289; CAS: 14459-95-1 |
| Potassium ferricyanide | Sigma-Aldrich | Cat#702587; CAS: 13746-66-2 |
| BM-Purple | Sigma-Aldrich | Cat#11442074001 |
| NBT | Sigma-Aldrich | Cat#11383213001; CAS: 298-83-9 |
| BCIP | Sigma-Aldrich | Cat#11383221001; CAS: 6578-06-9 |
| Benzyl alcohol | Sigma-Aldrich | Cat#108006; CAS: 100-51-6 |
| Benzyl benzoate | Sigma-Aldrich | Cat#B6630; CAS: 120-51-4 |
| NuPAGE LDS Sample Buffer (4X) | ThermoFisher Scientific | Cat#NP0007 |
| cOmplete, Mini Protease Inhibitor Cocktail | Sigma-Aldrich | Cat# 11836153001 |
| Critical Commercial Assays | ||
| in situ Hybridization Chain Reaction v3.0 | Molecular Instruments (Choi et al., 2018) | N/A |
| NuPAGE 3 to 8%, Tris-Acetate protein gels | ThermoFisher Scientific | Cat# EA0378BOX |
| Experimental Models: Organisms/Strains | ||
| Mouse: Shhflox/flox | The Jackson Laboratory | Cat#004293; RRID:IMSR_JAX:004293 |
| Mouse: ShhCre/+ | The Jackson Laboratory | Cat#005622; RRID:IMSR_JAX:005622 |
| Mouse: ShhCreER/+ | The Jackson Laboratory | Cat#005623; RRID:IMSR_JAX:005623 |
| Mouse: Gli1CreER/+ | The Jackson Laboratory | Cat#007913; RRID:IMSR_JAX:007913 |
| Mouse: Baxflox/flox;Bak−/− | The Jackson Laboratory | Cat#006329; RRID:IMSR_JAX:006329 |
| Mouse: Prrx-Cre tg | The Jackson Laboratory | Cat#005584; RRID:IMSR_JAX:005584 |
| Mouse: Rosa-EYFP tg | The Jackson Laboratory | Cat#006148; RRID:IMSR_JAX:006148 |
| Mouse: Rosa-SmoM2 | The Jackson Laboratory | Cat#005130; RRID:IMSR_JAX:005130 |
| Mouse: Shh+/− | Chiang et al., 1996 | N/A |
| Mouse: Gli3XtJ/+ | Buscher et al., 1998 | N/A |
| Mouse: Gli1LacZ/+ | Bai et al., 2002 | N/A |
| Mouse: Rosa-LacZ | Soriano, 1999 | N/A |
| Mouse: Hoxb6-CreER tg | Nguyen et al., 2009 | N/A |
| Mouse: Bax+/− | This study – see STAR method; Table 1 | N/A |
| Oligonucleotides | ||
| Genotyping primers for Bax-deleted allele | This study; 300bp PCR product | F:
5′-GAACCCTAGGACCCCTCCG-3′ R: 5′-CAACTCCTACCGCAAGTCCTGG-3′ |
| Genotyping primers for other mouse lines | Integrated DNA Technologies | Sequence information available from the Jackson Lab website and original published papers |
| Recombinant DNA | ||
| pBluescript-mGli1 | Gift from Dr. A. Joyner; Dev. Biol. Program, Sloan Kettering Institute, NY, NY |
N/A |
| pGEMT-mPtch1 | Gift from Dr. H. Arnheiter; NINDS, NIH, Bethesda, MD |
N/A |
| pBluescript-mHand2 | Gift from Dr. M. Ros; IBBTEC, Univ. Cantabria, Santander, Spain |
N/A |
| pBluescript-mFgf8 | Gift from Dr. G. Martin; UCSF, San Francisco, CA |
N/A |
| pBluescript-mGremlin1 | Gift from Dr. Y. Yang; Dept. Dev. Biol., Harvard Sch. Dental Medicine, Boston, MA |
N/A |
| mCyp26b1 ORF clone | Origene Technologies | Cat#MR222344 |
| pGEMT-mHoxd13 | Gift from Dr. D. Duboule; Dept. Genetics and Evolution, Univ. Geneva, Switzerland |
N/A |
| pGEMT-mHoxd11 | Gift from Dr. D. Duboule;as above | N/A |
| pXCMI-mHoxa13 | Gift from Dr. S. Stadler; Div. Skeletal Biology, Shriners Hospital for Children, Portland,OR |
N/A |
| pBluescript-mJagged1 | Gift from Dr. P. Koopman; Inst. Mol. Bioscience, Univ. Queensland, Brisbane, Australia |
N/A |
| pBluescript-mIrx3 | Gift from Dr. Kmita; Genetics and Dev. Res. Unit, Institut de Recherches Cliniques de Montréal, Québec, Canada |
N/A |
| pBluescript-mAlx4 | Gift from Dr. M. Ros; as above | N/A |
| pBluescript-mUncx4.1 | Gift from Dr. B. Herrmann; Max Planck Inst. for Molecular Genetics, Dept. Dev. Genetics, Berlin, Germany |
N/A |
| pBluescript-mSox9 | Gift from Dr. R.R. Behringer; Dept. Mol. Genetics, UT-MDACC, Houston, TX |
N/A |
| Software and Algorithms | ||
| LI-COR Odyssey Image Studio v5.2 | LI-COR | RRID: SCR_014579 |
| Imaris v9.1 | Bitplane/Oxford Instruments | RRID: SCR_007370 |
| Microsoft excel ver 16.59 | Microsoft | N/A |
METHOD DETAILS
Determination of embryo ages for Shh-response onset and loss in Shh-CKOs
To determine embryo ages for Shh-response onset and loss in Shh-CKOs (Figures 1, 3, S1), somite numbers were counted at the times of embryo collection. At this stage, somites form at a predictable rate of 1 somite every 2 hours(Tam, 1981) and our results in Figures 1, 3, S1, S2. Somite numbers present at time of tamoxifen injection were calculated retrospectively (Figure 1B), based on the actual somite number present when the embryos were collected several hours later for analysis. Somite number of embryos at a given collection time showed some variation (+/− 1 somite) and this was reflected in the time range of CreER-mediated recombination shown in Figure 1B. Somite-matched controls and Shh-CKOs came from the same litters to ensure the same tamoxifen treatment conditions. Importantly, the first appearance of Shh activity/response (Ptch1 and Gli1 RNA) in hindlimb buds reproducibly occurred at 29 somites, consistent with both our previous detailed time-course analysis and with other reports(Lewis et al., 2001; Lex et al., 2022; Zhu et al., 2008). Shh inactivation in Shh-CKOs, determined by the absence of Ptch1 and Gli1 expression compared to control siblings, was consistently 100% by 30 somites (within 2 hrs of first detection).
Whole mount in situ hybridization
Hybridizations were carried out following a previously described detailed protocol(Wilkinson, 1992). Embryos were fixed in 4% w/v paraformaldehyde in PBS overnight at 4°C, washed in PBS, gradually changed to absolute methanol and bleached in 5:1 methanol/30% hydrogen peroxide for 2 hrs at room temperature, and stored in methanol at −20°C until hybridization. Embryos with different genotypes were treated together, in one tube, with 20ug/ml proteinase K in PBS for 8–16 mins based on the embryo age (this step was omitted for Fgf8 probe). Gene-specific, digoxigenin-UTP labeled probes were synthesized from cDNA templates and incubated with embryos in hybridization buffer with 50% v/v formamide overnight at 70°C. The embryos were then washed with a series of buffers and incubated in alkaline-phosphatase-conjugated antidigoxigenin antibody overnight at 4°C. After washing with 0.1% v/v Tween in Tris buffered saline, embryos were incubated in BM purple or in 350 ug/ml NBT and 175 ug/ml BCIP in Tris buffered saline (pH 9.5) to detect hybridized RNA.
Hybridization chain reaction (HCR) in situ analysis
Mouse embryos were fixed, bleached, and stored the same way as described in the whole mount in situ hybridization above. Whole mount in situ hybridization of limb buds at ages indicated was performed using recommended conditions and solutions for third generation hybridization chain reaction (HCR) probes(Choi et al., 2018) designed by Molecular Instruments (Los Angeles, CA) and analyzed by confocal microscopy. Maximum projection images of fluorescence intensity from confocal image stacks spanning the entire dorsoventral limb bud thickness were generated to compare the A-P extent of Shh, Gli1 and Ptch1 RNA signals. For both Figures 3 and S2, all image panels are shown at the same scale, with scale bar of 100μm.
Skeletal staining
For skeletal staining(Kessel and Gruss, 1991), embryos were collected at E15.5-E17.5, eviscerated and fixed in absolute ethanol overnight, followed by absolute acetone overnight, and by staining in 150 ug/ml alcian blue and 50 ug/ml alizarin red in 95% v/v ethanol in H2O overnight. After clearing in 1% w/v KOH in H2O for several hours followed by 1% KOH w/v in 20% v/v glycerol in H2O, embryos were stored and imaged in 50% v/v glycerol in H2O.
Lysotracker staining
Embryos were collected in PBS and immediately incubated in 5uM lysotracker red in PBS with calcium and magnesium for 30 mins at 37°C. Embryos were then washed in PBS and fixed in 4% w/v paraformaldehyde in PBS overnight at 4°C. Embryos were washed in PBS, transferred to absolute methanol in graded steps and cleared in 1:2 v/v benzyl alcohol/benzyl benzoate (BABB) solution to visualize staining.
Western blot and quantification analysis
For western blot analysis, one pair of hindlimb buds from individual E10.75 embryos were dissected in PBS, lysed and sonicated in 1x NuPAGE LDS sample buffer with 1% w/v SDS and proteinase inhibitors. Reducing agent was added and samples were heated to 95 °C for 10 mins before loading. Two hindlimb buds (from 1 embryo) were loaded per lane, and electrophoresed in NuPAGE 3–8% Tris-Acetate protein gels. Proteins transferred to nitrocellulose membranes were probed with either affinity-purified polyclonal rabbit anti-Gli3(Chen et al., 2004) or goat polyclonal anti-Gli3 (1:1000, R&D, AF3690) and mouse anti-vinculin (1:1000, Sigma, V9264) and visualized with fluorescent secondary antibodies (1:10,000, LI-COR IRDye 800CW, 926-32211, anti-rabbit green; 926-32214, anti-goat green; and with #680RD, #926-68072, anti-mouse red) using LI-COR Odyssey CLx. Band intensities were quantified with Image Studio software v5.2. Number of independent samples analyzed for each genotype is listed in Fig. 4. For statistical analysis of western data (Gli3 FL/R), standard 2-sided t-test was used to calculate p values. Levels of alpha in t-tests <0.05 are considered significant.
Beta-galactosidase (LacZ) staining
Embryos were fixed in 2% w/v paraformaldehyde with 0.2% v/v glutaraldehyde for 1h at 4°C, washed in PBS with 0.1% v/v Tween (PBT) and stained with XGal (1mg/ml) in PBT and 2mM MgCl2, 5mM Ferro-CN, 5mM Ferri-CN, at 37 °C for several hours.
QUANTIFICATION AND STATISTICAL ANALYSIS
For immunoblots, Image Studio software v5.2 in the LI-COR Odyssey CLx system was used to quantify band fluorescence signals. Statistical significance of differences was determined using the standard 2-sided t-test. Numbers of biological replicates, means, SEM, and p values for immunoblots are reported in the main text and/or figure legends. Levels of alpha in t-tests <0.05 are considered significant.
Supplementary Material
Acknowledgements:
We thank Cliff Tabin and Marian Ros for stimulating discussions and critical reading of the manuscript and Sohyun Ahn, Chin Chiang, Alex Joyner, Andy McMahon, Cliff Tabin, Heiner Westphal and Yingzi Yang for providing mouse lines.
Funding:
This research was supported by the CCR, NCI (to SM, Intramural Research Program).
Footnotes
Competing interests: None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data and materials availability:
All data is available in manuscript and supplementary materials.
References
- Ahn S, and Joyner AL. (2004). Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell 118, 505–516. 10.1016/j.cell.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Alon U. (2007). An introduction to systems biology : design principles of biological circuits (Chapman & Hall/CRC; ). [Google Scholar]
- Bai CB, Auerbach W, Lee JS, Stephen D, and Joyner AL. (2002). Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129, 4753–4761. [DOI] [PubMed] [Google Scholar]
- Bastida MF, Perez-Gomez R, Trofka A, Zhu J, Rada-Iglesias A, Sheth R, Stadler HS, Mackem S, and Ros MA. (2020). The formation of the thumb requires direct modulation of Gli3 transcription by Hoxa13. Proc Natl Acad Sci U S A 117, 1090–1096. 10.1073/pnas.1919470117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscher D, Grotewold L, and Ruther U. (1998). The XtJ allele generates a Gli3 fusion transcript. Mamm Genome 9, 676–678. 10.1007/s003359900845. [DOI] [PubMed] [Google Scholar]
- Butterfield NC, Metzis V, McGlinn E, Bruce SJ, Wainwright BJ, and Wicking C. (2009). Patched 1 is a crucial determinant of asymmetry and digit number in the vertebrate limb. Development 136, 3515–3524. 10.1242/dev.037507. [DOI] [PubMed] [Google Scholar]
- Chen Y, Knezevic V, Ervin V, Hutson R, Ward Y, and Mackem S. (2004). Direct interaction with Hoxd proteins reverses Gli3-repressor function to promote digit formation downstream of Shh. Development 131, 2339–2347. 10.1242/dev.01115. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Harris MP, Simandl BK, Li Y, Beachy PA, and Fallon JF. (2001). Manifestation of the limb prepattern: limb development in the absence of sonic hedgehog function. Dev Biol 236, 421–435. 10.1006/dbio.2001.0346. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, and Beachy PA. (1996). Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383, 407–413. 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, and Pierce NA. (2018). Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145. 10.1242/dev.165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahn RD, and Fallon JF. (2000). Interdigital regulation of digit identity and homeotic transformation by modulated BMP signaling. Science 289, 438–441. 10.1126/science.289.5478.438. [DOI] [PubMed] [Google Scholar]
- Davis AP, Witte DP, Hsieh-Li HM, Potter SS, and Capecchi MR. (1995). Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature 375, 791–795. 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- Drossopoulou G, Lewis KE, Sanz-Ezquerro JJ, Nikbakht N, McMahon AP, Hofmann C, and Tickle C. (2000). A model for anteroposterior patterning of the vertebrate limb based on sequential long- and short-range Shh signalling and Bmp signalling. Development 127, 1337–1348. [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain C, Warot X, Messadecq N, LeMeur M, Dolle P, and Chambon P. (1996). Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development 122, 2997–3011. [DOI] [PubMed] [Google Scholar]
- Galli A, Robay D, Osterwalder M, Bao X, Benazet JD, Tariq M, Paro R, Mackem S, and Zeller R. (2010). Distinct roles of Hand2 in initiating polarity and posterior Shh expression during the onset of mouse limb bud development. PLoS Genet 6, e1000901. 10.1371/journal.pgen.1000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Hu J, Stricker S, Cheung M, Ma G, Law KF, Witte F, Briscoe J, Mundlos S, He L, et al. (2009). A mutation in Ihh that causes digit abnormalities alters its signalling capacity and range. Nature 458, 1196–1200. 10.1038/nature07862. [DOI] [PubMed] [Google Scholar]
- Haraguchi R, Motoyama J, Sasaki H, Satoh Y, Miyagawa S, Nakagata N, Moon A, and Yamada G. (2007). Molecular analysis of coordinated bladder and urogenital organ formation by Hedgehog signaling. Development 134, 525–533. 10.1242/dev.02736. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian F, McMahon AP, and Tabin CJ. (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517528. Doi 10.1016/J.Cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Harris MP. (2021). Bioelectric signaling as a unique regulator of development and regeneration. Development 148. 10.1242/dev.180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson P, Del Buono J, and Logan MP. (2007). Tbx5 is dispensable for forelimb outgrowth. Development 134, 85–92. 10.1242/dev.02622. [DOI] [PubMed] [Google Scholar]
- Huang BL, and Mackem S. (2021). Rethinking positional information and digit identity: The role of late interdigit signaling. Dev Dyn. 10.1002/dvdy.440. [DOI] [PubMed] [Google Scholar]
- Huang BL, Trofka A, Furusawa A, Norrie JL, Rabinowitz AH, Vokes SA, Mark Taketo M, Zakany J, and Mackem S. (2016). An interdigit signalling centre instructs coordinate phalanx-joint formation governed by 5’Hoxd-Gli3 antagonism. Nat Commun 7, 12903. 10.1038/ncomms12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Mao J, Tenzen T, Kottmann AH, and McMahon AP. (2004). Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev 18, 937–951. 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel M, and Gruss P. (1991). Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell 67, 89–104. 10.1016/0092-8674(91)90574i. [DOI] [PubMed] [Google Scholar]
- Kong JH, Siebold C, and Rohatgi R. (2019). Biochemical mechanisms of vertebrate hedgehog signaling. Development 146. 10.1242/dev.166892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski JP, Du F, Zhang S, Powell MB, Falkenstein KN, Ji H, and Vokes SA. (2015). Spatiotemporal regulation of GLI target genes in the mammalian limb bud. Dev Biol 406, 92–103. 10.1016/j.ydbio.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PM, Dunn MP, McMahon JA, Logan M, Martin JF, St-Jacques B, and McMahon AP. (2001). Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell 105, 599–612. 10.1016/s0092-8674(01)00369-5. [DOI] [PubMed] [Google Scholar]
- Lex RK, Ji Z, Falkenstein KN, Zhou W, Henry JL, Ji H, and Vokes SA. (2020). GLI transcriptional repression regulates tissue-specific enhancer activity in response to Hedgehog signaling. Elife 9, e50670. 10.7554/eLife.50670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lex RK, Zhou W, Ji Z, Falkenstein KN, Schuler KE, Windsor KE, Kim JD, Ji H, and Vokes SA. (2022). GLI transcriptional repression is inert prior to Hedgehog pathway activation. Nat Commun 13, 808. 10.1038/s41467-022-28485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DY, Sakuma R, Vakili NA, Mo R, Puviindran V, Deimling S, Zhang XY, Hopyan S, and Hui CC. (2014). Formation of Proximal and Anterior Limb Skeleton Requires Early Function of Irx3 and Irx5 and Is Negatively Regulated by Shh Signaling. Developmental Cell 29, 233–240. 10.1016/j.devcel.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Lindsten T, Ross AJ, King A, Zong WX, Rathmell JC, Shiels HA, Ulrich E, Waymire KG, Mahar P, Frauwirth K, et al. (2000). The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol Cell 6, 1389–1399. 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litingtung Y, Dahn RD, Li Y, Fallon JF, and Chiang C. (2002). Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature 418, 979–983. 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- Logan M, Martin JF, Nagy A, Lobe C, Olson EN, and Tabin CJ. (2002). Expression of Cre Recombinase in the developing mouse limb bud driven by a Prx1 enhancer. Genesis 33, 77–80. 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- Long F, Zhang XM, Karp S, Yang Y, and McMahon AP. (2001). Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development 128, 5099–5108. [DOI] [PubMed] [Google Scholar]
- Mariani FV, Ahn CP, and Martin GR. (2008). Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature 453, 401–405. 10.1038/nature06876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacu E, Gromberg E, Oliveira CR, Drechsel D, and Tanaka EM. (2016). FGF8 and SHH substitute for anterior-posterior tissue interactions to induce limb regeneration. Nature 533, 407–410. 10.1038/nature17972. [DOI] [PubMed] [Google Scholar]
- Nakamura E, Nguyen MT, and Mackem S. (2006). Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev Dyn 235, 2603–2612. 10.1002/dvdy.20892. [DOI] [PubMed] [Google Scholar]
- Nguyen MT, Zhu J, Nakamura E, Bao X, and Mackem S. (2009). Tamoxifen-dependent, inducible Hoxb6CreERT recombinase function in lateral plate and limb mesoderm, CNS isthmic organizer, posterior trunk neural crest, hindgut, and tailbud. Dev Dyn 238, 467–474. 10.1002/dvdy.21846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M, Speziale D, Shoukry M, Mohan R, Ivanek R, Kohler M, Beisel C, Wen X, Scales SJ, Christoffels VM, et al. (2014). HAND2 targets define a network of transcriptional regulators that compartmentalize the early limb bud mesenchyme. Dev Cell 31, 345–357. 10.1016/j.devcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panman L, Galli A, Lagarde N, Michos O, Soete G, Zuniga A, and Zeller R. (2006). Differential regulation of gene expression in the digit forming area of the mouse limb bud by SHH and gremlin 1/FGF-mediated epithelial-mesenchymal signalling. Development 133, 3419–3428. 10.1242/dev.02529. [DOI] [PubMed] [Google Scholar]
- Perriton CL, Powles N, Chiang C, Maconochie MK, and Cohn MJ. (2002). Sonic hedgehog signaling from the urethral epithelium controls external genital development. Dev Biol 247, 26–46. 10.1006/dbio.2002.0668 [DOI] [PubMed] [Google Scholar]
- Piccolo S, Sladitschek-Martens HL, and Cordenonsi M. (2022). Mechanosignaling in vertebrate development. Dev Biol 488, 54–67. 10.1016/j.ydbio.2022.05.005. [DOI] [PubMed] [Google Scholar]
- Pickering J, Chinnaiya K, and Towers M. (2019). An autoregulatory cell cycle timer integrates growth and specification in chick wing digit development. Elife 8, e47625. 10.7554/eLife.47625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst S, Kraemer C, Demougin P, Sheth R, Martin GR, Shiratori H, Hamada H, Iber D, Zeller R, and Zuniga A. (2011). SHH propagates distal limb bud development by enhancing CYP26B1-mediated retinoic acid clearance via AER-FGF signalling. Development 138, 1913–1923. 10.1242/dev.063966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz PJ, McGlinn E, Nissim S, and Tabin CJ. (2007). Extended exposure to Sonic hedgehog is required for patterning the posterior digits of the vertebrate limb. Dev Biol 308, 343–354. 10.1016/j.ydbio.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, Hanken J, and Rosenthal N. (2003). Developmental basis of evolutionary digit loss in the Australian lizard Hemiergis. J Exp Zool B Mol Dev Evol 297, 48–56. 10.1002/jez.b.19. [DOI] [PubMed] [Google Scholar]
- Soriano P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21, 70–71. Doi 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, and Costantini F. (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC developmental biology 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Hasso SM, and Fallon JF. (2008). Unique SMAD1/5/8 activity at the phalanx-forming region determines digit identity. Proc Natl Acad Sci U S A 105, 4185–4190. 10.1073/pnas.0707899105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Fisher J, Suh H, Harada H, Malynn BA, and Korsmeyer SJ. (2005). Essential role of BAX,BAK in B cell homeostasis and prevention of autoimmune disease. Proc Natl Acad Sci U S A 102, 11272–11277. 10.1073/pnas.0504783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PP. (1981). The control of somitogenesis in mouse embryos. J Embryol Exp Morphol 65 Suppl, 103–128. [PubMed] [Google Scholar]
- Tarchini B, and Duboule D. (2006). Control of Hoxd genes’ collinearity during early limb development. Dev Cell 10, 93–103. 10.1016/j.devcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- te Welscher P, Fernandez-Teran M, Ros MA, and Zeller R. (2002a). Mutual genetic antagonism involving GLI3 and dHAND prepatterns the vertebrate limb bud mesenchyme prior to SHH signaling. Genes Dev 16, 421–426. 10.1101/gad.219202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Welscher P, Zuniga A, Kuijper S, Drenth T, Goedemans HJ, Meijlink F, and Zeller R. (2002b). Progression of vertebrate limb development through SHH-mediated counteraction of GLI3. Science 298, 827–830. 10.1126/science.1075620. [DOI] [PubMed] [Google Scholar]
- Towers M, Mahood R, Yin Y, and Tickle C. (2008). Integration of growth and specification in chick wing digit-patterning. Nature 452, 882–886. 10.1038/nature06718. [DOI] [PubMed] [Google Scholar]
- Towers M, Signolet J, Sherman A, Sang H, and Tickle C. (2011). Insights into bird wing evolution and digit specification from polarizing region fate maps. Nat Commun 2, 426. 10.1038/ncomms1437. [DOI] [PubMed] [Google Scholar]
- Vokes SA, Ji H, Wong WH, and McMahon AP. (2008). A genome-scale analysis of the cisregulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev 22, 2651–2663. 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Fallon JF, and Beachy PA. (2000). Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100, 423–434. 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Wang C, Pan Y, and Wang B. (2010). Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development 137, 2001–2009. 10.1242/dev.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson D. (1992). Whole mount in situ hybridization of vertebrate embryos. In In Situ Hybridization: A practical Approach, Wilkinson DG, ed. (Oxford: IRL Press; ), pp. 75–83. [Google Scholar]
- Witte F, Chan D, Economides AN, Mundlos S, and Stricker S. (2010). Receptor tyrosine kinase-like orphan receptor 2 (ROR2) and Indian hedgehog regulate digit outgrowth mediated by the phalanx-forming region. Proc Natl Acad Sci U S A 107, 14211–14216. 10.1073/pnas.1009314107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhang LS, Toombs J, Kuo YC, Piazza JT, Tuladhar R, Barrett Q, Fan CW, Zhang X, Walensky LD, et al. (2017). Extra-mitochondrial prosurvival BCL-2 proteins regulate gene transcription by inhibiting the SUFU tumour suppressor. Nat Cell Biol 19, 1226–1236. 10.1038/ncb3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Drossopoulou G, Chuang PT, Duprez D, Marti E, Bumcrot D, Vargesson N, Clarke J, Niswander L, McMahon A, and Tickle C. (1997). Relationship between dose, distance and time in Sonic Hedgehog-mediated regulation of anteroposterior polarity in the chick limb. Development 124, 4393–4404. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Ramalho-Santos M, and McMahon AP. (2001). Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R symmetry by the mouse node. Cell 106, 781–792. [PubMed] [Google Scholar]
- Zhu J, and Mackem S. (2017). John Saunders’ ZPA, Sonic hedgehog and digit identity - How does it really all work? Dev Biol 429, 391–400. 10.1016/j.ydbio.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Nakamura E, Nguyen MT, Bao X, Akiyama H, and Mackem S. (2008). Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev Cell 14, 624–632. 10.1016/j.devcel.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga A, Haramis AP, McMahon AP, and Zeller R. (1999). Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature 401, 598–602. 10.1038/44157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data
All microscopic and other imaging data reported in this paper will be shared by the lead contact upon request.
Code
No bioinformatic data or code were generated.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
All data is available in manuscript and supplementary materials.