Abstract
Prostate segmentation in transrectal ultrasound (TRUS) image is an essential prerequisite for many prostate-related clinical procedures, which, however, is also a long-standing problem due to the challenges caused by the low image quality and shadow artifacts. In this paper, we propose a Shadow-consistent Semi-supervised Learning (SCO-SSL) method with two novel mechanisms, namely shadow augmentation (Shadow-AUG) and shadow dropout (Shadow-DROP), to tackle this challenging problem. Specifically, Shadow-AUG enriches training samples by adding simulated shadow artifacts to the images to make the network robust to the shadow patterns. Shadow-DROP enforces the segmentation network to infer the prostate boundary using the neighboring shadow-free pixels. Extensive experiments are conducted on two large clinical datasets (a public dataset containing 1,761 TRUS volumes and an in-house dataset containing 662 TRUS volumes). In the fully-supervised setting, a vanilla U-Net equipped with our Shadow-AUG&Shadow-DROP outperforms the state-of-the-arts with statistical significance. In the semi-supervised setting, even with only 20% labeled training data, our SCO-SSL method still achieves highly competitive performance, suggesting great clinical value in relieving the labor of data annotation. Source code is released at https://github.com/DIAL-RPI/SCO-SSL.
Keywords: Prostate segmentation, Semi-supervised learning, Fully convolutional network, Ultrasound image, Shadow artifact
I. Introduction
PROSTATE cancer is the most common type of cancer for men in the United States and the second leading cause of cancer mortality for this population [1]. Transrectal ultrasound (TRUS) imaging is widely used in prostate cancer diagnosis and treatment due to its great accessibility, low cost, and non-ionizing nature. Segmenting the whole prostate volume from TRUS acts as an essential prerequisite for a set of subsequent clinical procedures, such as image-guided biopsy, needle placement, and interventional therapy delivery. Manual segmentation not only consumes tremendous amount of time and labor but also varies significantly between different annotators. To this end, automatic prostate ultrasound segmentation is highly desired in practice with great clinical significance.
However, accurate prostate segmentation in TRUS images is a long-standing and challenging problem due to the following difficulties. First, TRUS images often suffer from low signal-to-noise ratio (SNR) and inhomogeneous intensity distribution, resulting in low-contrast and ambiguous boundaries of the prostate. Second, the large variability of the prostate shape and size across different patients increases the difficulty of segmentation. Last, but very importantly, the shadow artifacts caused by the ultrasonic absorption and reflection often lead to the missing prostate boundary in some local regions. All these facts complicate the prostate ultrasound segmentation and make it more challenging than other medical image segmentation tasks including the prostate segmentation in computed tomography (CT) [2]-[4] and magnetic resonance imaging (MRI) images [5], [6]. Fig. 1 shows four example TRUS images to illustrate the low image quality caused by the shadow artifacts.
Fig. 1:
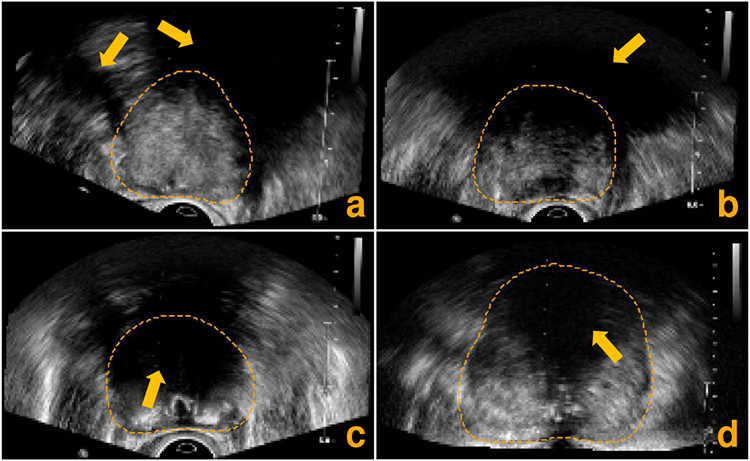
Axial slices extracted from four different 3D TRUS images illustrating the low image quality caused by the shadow artifacts (pointed out by the orange arrows). The orange dashed lines indicate the ground-truth prostate boundary.
Currently, the most popular methodology for prostate ultrasound segmentation is based on deep learning (DL), more specifically, the fully convolutional networks (FCNs) [7], [8]. The powerful representation ability of the self-learned hierarchical features makes FCN-based methods significantly outperform those conventional methods, whose performance largely relies on the hand-crafted image features. Some more advanced DL technologies, such as the long short-term memory [9], attention mechanism [10], [11], deep supervision [6], [12], and ensemble learning [13], further pushed the limits of segmentation performance.
Even though the FCN-based prostate segmentation methods have achieved impressive performance, significant challenges still remain. First, most of the existing methods count on the general DL techniques to facilitate the prostate segmentation but overlook the specificity of ultrasound images. Unlike other imaging modalities, ultrasound images often exhibit a unique kind of noise, the shadow artifacts as shown in Fig. 1. The information inside the shadow regions is largely lost and less reliable than that in the shadow-free regions. However, the existing methods tend to treat all the pixels equally no matter where they locate. Intuitively, treating the shadow pixels and the shadow-free pixels in ultrasound images is more desirable for accurate prostate segmentation. Second, the previous methods can only work in a fully-supervised learning manner, where the model performance highly depends on the size and quality of the labeled training data. However, large-scale and well-annotated TRUS images are very expensive to collect in clinic. In contrast, the raw TRUS images without any annotations are much easier to acquire. Leveraging the unlabeled TRUS images to facilitate the prostate segmentation on top of limited annotations is another problem yet to be solved in this domain.
In this paper, we propose a Shadow-consistent Semi-supervised Learning (SCO-SSL) method to address the above two issues in prostate ultrasound volume segmentation. The proposed method contains two novel mechanisms, namely shadow augmentation (Shadow-AUG) and shadow dropout (Shadow-DROP), which aim to encourage a segmentation network to extract discriminative features from the shadow-free regions at both image and feature levels, respectively. Specifically, the Shadow-AUG strategy enriches the training samples by adding the simulated shadow artifacts to the input ultrasound images to make the trained network robust to the potential shadow artifacts. The Shadow-DROP mechanism selectively erases a part of features extracted from the shadow regions and thus enforces the segmentation network to infer the prostate boundary using the neighboring shadow-free pixels for a reliable segmentation. Both Shadow-AUG and Shadow-DROP are independent of the network architectures and the training procedures. Thus, they can be easily incorporated into any FCN-based segmentation methods.
The main contributions of this work are four-fold, as summarized below.
We propose a novel data augmentation strategy, namely Shadow-AUG, for training prostate ultrasound image segmentation networks. By simulating the shadow artifacts in training samples, a deep segmentation network becomes more robust against such shadow artifacts and thus achieves higher segmentation accuracy.
We develop a Shadow-DROP mechanism to complement a deep network’s feature extraction ability, which encourages the trained network to infer the missing prostate boundaries using the neighboring shadow-free pixels by ignoring the features from a shadow region.
Both Shadow-AUG and Shadow-DROP can be easily incorporated into the consistency learning framework, reaching a novel SCO-SSL method for semi-supervised prostate ultrasound segmentation. To the best of our knowledge, this is the first attempt to leverage the power of semi-supervised learning to handle the challenging problem of prostate ultrasound segmentation.
We comprehensively evaluated the proposed SCO-SSL method on two large-scale clinical datasets, a public dataset with 1,761 TRUS volumes and an in-house dataset with 662 TRUS volumes. The experimental results show that, when trained with 100% labeled data, the proposed SCO-SSL method outperforms the state-of-the-art methods by statistically significant margins, especially inside the shadow regions. When trained with 20% labeled data, our method still yields competitive result in comparison to the state-of-the-art methods, which are trained using 100% labeled data.
II. Related works
A. Prostate ultrasound segmentation
Prostate ultrasound segmentation is a long-standing topic in medical image analysis. In the early stage of this domain, researchers designed various hand-crafted features to handle this challenging problem through statistical shape model based methods [14]-[16]. Conventional machine learning methods have also been explored [17]. The performance of these methods highly relies on the hand-crafted features, which tend to fail in segmenting low-contrast boundaries and regions affected by shadow artifacts.
In recent years, DL has become the dominant methodology in medical image segmentation. The powerful representation ability with self-learned hierarchical features makes the FCNs [7] significantly outperform the conventional methods. The state-of-the-art benchmark in medical image segmentation is U-Net [8], [18], which consists of an encoding path and a decoding path joint with several skip-connections in a U-shape architecture. With proper fine-tuning of a set of hyper-parameters, the U-Net may achieve superior performance on various medical image segmentation tasks, even surpassing some advanced networks that were specially designed for the specific tasks [18]. Specifically, on prostate ultrasound segmentation, Orlando et al. [19], [20] resampled series of 2D slices radially around the superior-inferior axis of the 3D TRUS image, and then utilized a standard U-Net to predict 2D contours on the extracted slices to handle the prostate appearance variation in transverse slices. The segmentation results are then combined to reconstruct the 3D prostate volume. Yang et al. [9] resampled the 2D TRUS image to a series of patches along the prostate boundary and exploited recurrent neural networks (RNNs) to infer the prostate shape sequentially, aiming to bridge the missing boundaries through long short-term memory (LSTM) networks. Wang et al. [10], [11] leveraged the attention mechanism to selectively extract multi-level features from TRUS images, facilitating the prostate segmentation by suppressing irrelevant background noise while enhancing prostate structural details. Lei et al. [12], [13] integrated multi-view ensemble learning and deep supervision strategies into 3D V-Nets [21] to refine the prostate segmentation with limited training data.
Although these methods have achieved promising performance on various TRUS image datasets, they have ignored the shadow artifacts and thus may fail to deal with the situation where severe shadow artifacts present. On the other hand, the training of these methods largely relies on the fully-annotated TRUS data, which is not always feasible to acquire in clinical practice. In contrast, our proposed SCO-SSL method provides special mechanisms (i.e., the Shadow-AUG and Shadow-DROP) to handle the complex imaging condition caused by the shadow artifacts. Furthermore, it supports the semi-supervised learning with limited annotated data and large portion of unlabeled data, which is more close to the real scenario in clinic.
B. Ultrasound shadow segmentation
Acoustic shadow is a special artifact often encountered in ultrasound imaging. It can be useful for locating certain acoustic-reflecting/-absorbing objects [22]-[26], but can also hinder the ultrasound image analysis tasks such as segmentation [27]-[33]. In both scenarios, accurate segmentation of the shadow regions is favorable. Prior works in this domain range from the early-stage hand-crafted feature based methods [24], [27]-[30] to the more recent DL-based methods [25], [26], [31]-[33]. For example, Hellier et al. [27] used the geometrical and statistical features of the shadow distribution to detect the existence of such artifacts in ultrasound brain images. Basij et al. [28] designed a thresholding function to adaptively segment the shadow regions behind the calcification plaque in intra vascular ultrasound images. Karamalis et al. [29] utilized the random walker algorithm [34] to calculate a confidence map to measure the reliability of each ultrasound pixel in shadow conditions. Berton et al. [30] and Hacihaliloglu [24] sought to distinguish the shadow regions from shadow-free pixels by a set of hand-crafted features.
In the DL category, Meng et al. [31] proposed a weakly-supervised DL method for ultrasound shadow segmentation by training a classification network to tell whether the input image is shadow-free or not. The saliency map of this classifier was then used for shadow segmentation through a generative adversarial network (GAN) and GraphCut [35]. Meng et al. [32] further boosted the shadow segmentation accuracy by designing two co-trained FCNs equipped with attention mechanisms. Alsinan et al. [25] utilized a GAN to segment the shadow regions in ultrasound images. Wang et al. [26] proposed a multi-task network to separately estimate the coarse bone shadow enhancement and horizontal bone interval, both of which were then combined to generate the final shadow mask. Yasutomi et al. [33] trained an autoencoder with synthetic shadow masks to achieve the goal of semi-supervised shadow estimation.
The prior works on ultrasound shadow segmentation demonstrate the importance of shadow-robustness for ultrasound image analysis. It motivates us to design the Shadow-AUG and Shadow-DROP mechanisms in our prostate segmentation method.
C. Semi-supervised medical image segmentation
Semi-supervised learning is a special type of machine learning approach that falls into the category between fully-supervised learning and unsupervised learning. It combines the unlabeled data with a small portion of fully-annotated data during training, which aims to relieve the labor for labeling data while improve the model performance by leveraging the unlabeled data. The core assumption of semi-supervised learning is that the data points close to each other in the latent space should have similar or identical labels, which is often referred to as the smoothness assumption [36]. In other words, by conducting different transformations/augmentations on the same input image, the trained model can be regularized through consistency constraints on the output, no matter whether the input image is labeled or not. For example, Laine and Aila [36] proposed a self-ensembling framework to realize the semi-supervised learning, where the historic predictions of the trained model are averaged to generate the training target (or pseudo label) for the unlabeled data. As a further step of the self-ensembling learning, Tarvainen and Valpola [37] proposed to average the model parameters rather than the model predictions in the self-ensembling framework. The output of the resulting mean model (teacher) was used as the pseudo label to supervise the current trained model (student), leading to higher accuracy and more stable performance than the previous self-ensembling learning methods.
Since the raw medical images are much easier to collect than their annotations, semi-supervised learning methods are widely used in handling various of medical image segmentation tasks. Specifically, Yu et al. [38] proposed an uncertainty-aware self-ensembling model for semi-supervised segmentation of 3D left atrium, in which the teacher and student networks were interpreted as Bayesian networks through Monte Carlo dropout [39] to make the pseudo label aware of uncertainty. Li et al. [40], [41] applied different spatial augmentations on the same input image for the student and teacher models, aiming to build transformation-consistency during training. Xia et al. [42], [43] proposed a multi-view co-training method for semi-supervised 3D medical image segmentation. They trained three individual sub-networks using three orthogonal views (i.e., axial, coronal, and sagittal views) of the 3D images. The predictions of these three sub-networks were then combined to serve as pseudo labels for unsupervised training. In this paper, we propose a shadow-consistent semi-supervised learning method to improve prostate ultrasound segmentation by utilizing extra unlabeled images with shadow artifacts.
III. Method
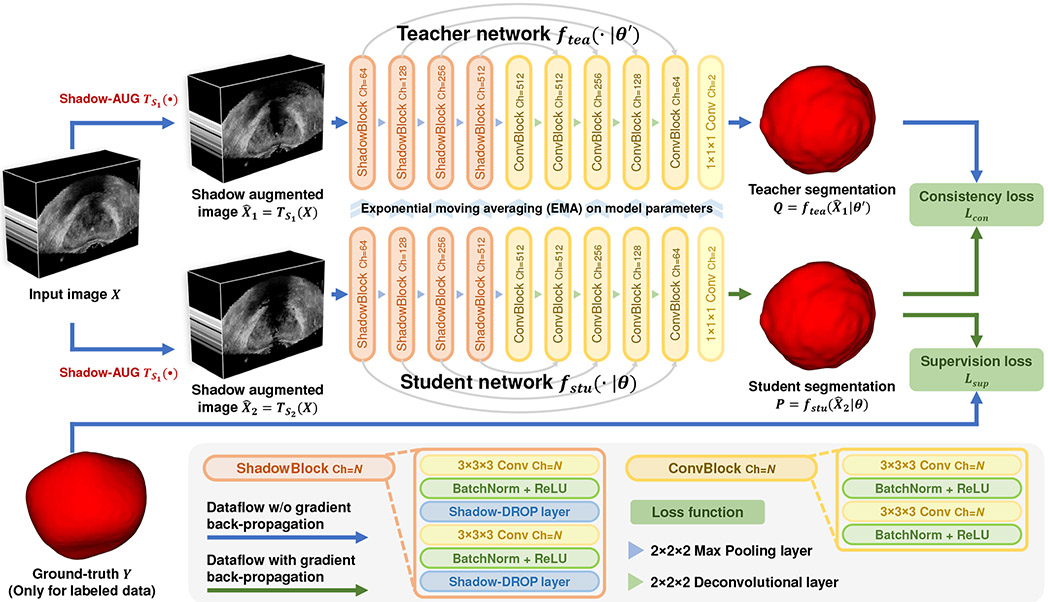
Fig. 2 gives an overview of the proposed SCO-SSL method for prostate ultrasound segmentation. The innovation of this SCO-SSL method lies in two components: a) the Shadow-AUG strategy applied to the input images and b) the Shadow-DROP layer acting on the intermediate feature maps, which will be introduced in Sections III-A and III-B, respectively. We then present the entire pipeline of the proposed SCO-SSL in Section III-C. The implementation details are provided in Section III-D.
Fig. 2:
Scheme of the proposed shadow-consistent semi-supervised learning (SCO-SSL) method for prostate segmentation in TRUS images.
A. Shadow augmentation strategy
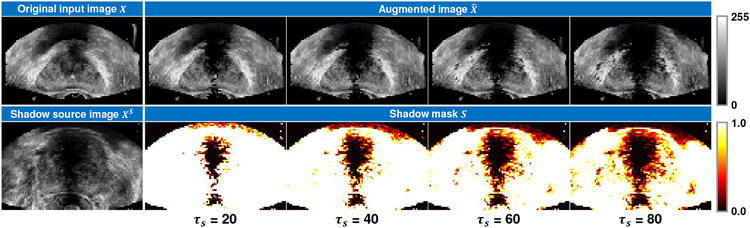
In order to make the segmentation network robust to the shadow artifacts appearing in the TRUS images, we propose the shadow augmentation strategy (Shadow-AUG) to simulate the shadow artifacts in the input images during model training. Fig. 3 gives a scheme of the proposed Shadow-AUG, whose core idea is to impose the shadow artifacts extracted from other TRUS images on the training image. Specifically, to generate realistic shadow artifacts for a given input TRUS image X, we randomly select another TRUS image Xs, namely shadow source image, from the training set and extract its shadow mask S using the following soft thresholding function:
| (1) |
where si, and denote the i-th voxel of the shadow mask S and the shadow source image Xs, respectively. τs is a hyper-parameter thresholding the range of the extracted shadow region. Fig. 4 shows example shadow images augmented using different values of τs. We use this soft thresholding function instead of a hard threshold here to avoid sharp boundaries in the shadow masks. After the extraction of the shadow mask S, the augmented image can be generated by masking S on the original input image X as follows:
| (2) |
where ⊗ denotes element-wise multiplication.
Fig. 3:
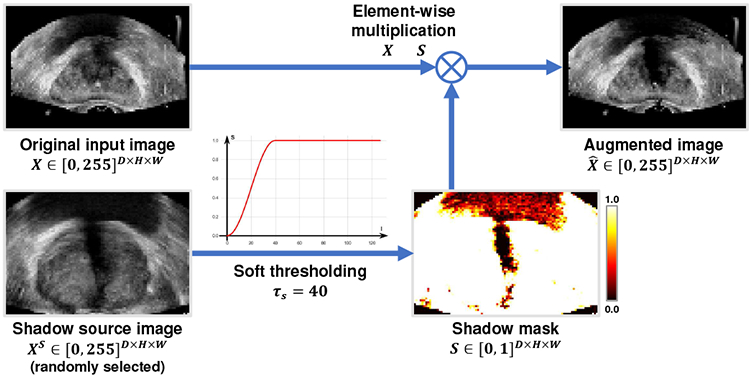
Illustration of shadow augmentation (Shadow-AUG) in 2D as an example. The actual Shadow-AUG is conducted in 3D on TRUS volumes.
Fig. 4:
Example shadow images augmented using different values of shadow threshold τs
Since the shadow source image Xs can be any TRUS image in the training set other than the input image X, the diversity of the augmented samples is extensively enriched, which largely alleviates the risk of over-fitting in training. On the other hand, since the shadow mask S is extracted from a real TRUS images Xs, the distribution of the simulated shadow artifacts may well approximate what exists in the real data. This is beneficial to the learning of discriminative and robust features for prostate segmentation in the presence of severe shadow artifacts.
B. Shadow dropout mechanism
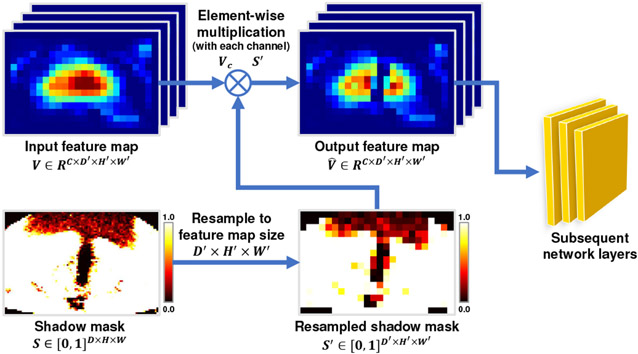
As the original image information in the shadow regions is largely eroded by the artifacts, the feature extracted from the shadow regions could be less reliable for accurate prostate segmentation. It is intuitive that a discriminative learning mechanism paying more attention to the shadow-free regions than the shadow regions could help to improve the robustness of the trained network. However, conventional convolutional neural networks tend to treat all the image voxels equally without telling whether they locate inside the shadow regions or not, which may hinder the learning of discriminative features for accurate prostate segmentation. To handle this issue, we design a novel Shadow-DROP layer to filter the features extracted from the shadow regions in the intermediate layers of the segmentation network. Fig. 5 shows the scheme of the proposed Shadow-DROP layer. Given an input feature map V with C channels, the proposed Shadow-DROP operates as:
| (3) |
where Vc and denote the c-th channel of the input feature map V and output feature map , respectively. S denotes the shadow mask, which is created in the same way as the one used in the aforementioned Shadow-AUG (see Eq. (1)&(2)). The function resample(S, Vc) resamples the shadow mask S to the same spatial size as the input feature Vc so that they can be merged through the element-wise multiplication operation ⊗.
Fig. 5:
Scheme of the shadow dropout (Shadow-DROP) layer. 2D feature map is used for illustration purpose only. The actual Shadow-DROP is conducted in 3D.
Essentially, the proposed Shadow-DROP layer works similarly as the standard dropout layer [44], [45], which randomly drops a part of the neural nodes out of the training by suppressing their output. This enforces the subsequent networks to leam generalized representations using the remaining neural nodes to meet the training objective. The major difference between the standard dropout layer and our proposed Shadow-DROP layer lies in the dropout mask. Our Shadow-DROP layer uses the shadow mask extracted from a real ultrasound image, where the spatial distribution of the dropped neural nodes submit to the prior knowledge of the shadow artifacts. Consequently, the trained networks learn to bridge the gap when dealing with the ultrasound images containing similar patterns of shadow artifacts. We will demonstrate this through an ablation study in Section IV-D.
C. Shadow-consistent semi-supervised learning framework
In order to utilize the unlabeled TRUS images to facilitate the prostate segmentation, we integrate the Shadow-AUG and Shadow-DROP mechanisms into the consistency learning framework [37], achieving the proposed SCO-SSL method for semi-supervised prostate ultrasound segmentation. As shown in Fig. 2, the SCO-SSL method consists of a student network fstu(·∣θ) and a teacher network ftea(·∣θ′), both of which have the same network architecture but different model parameters θ and θ′, respectively. Shadow-AUGs are applied on the input images. We exploit U-Net [8] as the backbone of the teacher/student networks, where the proposed Shadow-DROP layers are inserted to all the convolutional blocks in the encoding path (see “ShadowBlock” illustrated in the legend of Fig. 2).
Without loss of generality, we introduce the workflow of the proposed SCO-SSL method under the standard semi-supervised learning setting, where the training set contains Nl image-annotation pairs and Nu unlabeled images . Given an arbitrary image X from XL or XU, we first conduct two independent Shadow-AUGs on it to generate two different shadow augmented images and . These two augmented images are then fed to the student network and teacher network to generate the prostate segmentation and , respectively. According to the smoothness assumption in semi-supervised learning [36], the data point perturbing in image space should keep consistent in label space. In our problem, since the two augmented images and come from the same input image X, their segmentation masks should be the same through either the student network or the teacher network. Therefore, we can build a consistent constraint between the student prediction P and the teacher prediction Q by minimizing the following binary cross entropy (BCE) loss, namely consistency loss:
| (4) |
Note that, the consistency loss is applicable not only for the unlabeled images XU but also the labeled images XL since the smoothness assumption stands for both of them. For a labeled image XL, we also calculate the supervised loss on it using the corresponding ground-truth segmentation mask Y. In our method, we use Dice loss [21] as the supervised loss:
| (5) |
where pci and yci denote the i-th voxel in the c-th channel of the student prediction P and the ground-truth segmentation mask Y, respectively.
The student network parameters θ are updated through stochastic gradient descent (SGD) and back-propagation algorithm by minimizing the training objective ℒ = ℒsup + λ(T) · ℒcon, where λ(T) is a function of training epoch index T used to dynamically balance the supervised loss and the consistency loss. In our method, we adopt a Gaussian rampup function as λ(T), which is the same as other consistency learning methods [36], [38], [41]:
| (6) |
where λmax is the maximum weight for the consistency loss reached after Tmax training epochs. We empirically set λmax=0.1 and Tmax=200 epochs. The teacher network parameters θ′ are updated by calculating the exponential moving average (EMA) of the student model parameters θ:
| (7) |
where t indicates the index of training batches. Momentum term α controls the speed of teacher model updating, which is empirically set to 0.99. The student network parameters θ and teacher parameters θ′ are updated alternately during training. At inference stage, we adopt teacher network’s prediction as the final output since it is more stable and accurate than the student network’s prediction. Both the Shadow-AUG and the Shadow-DROP mechanisms merely work in the training stage, and thus they will bring no extra computational cost to the segmentation networks at inference time.
D. Implementation details
The proposed SCO-SSL method is implemented in 3D using PyTorch. Model parameters in the student network are initialized using Xavier algorithm [46] and optimized by SGD optimizer with a learning rate of 0.001 and momentum factor of 0.99. We train the model for 400 epochs and evaluate its performance on the validation set every epoch using Dice similarity coefficient (DSC) as the metric. The model achieving the highest DSC on the validation set is selected as the final model to be evaluated on the test set. The training batch size is set to 16 in fully-supervised setting and 36 in semi-supervised setting (12 labeled samples and 24 unlabeled samples). The input TRUS images are center-cropped and resampled to a uniform size of 96×64×96 with a spacing of 1.0×1.0×1.0mm3. The image intensities are normalized from [0.0, 255.0] to [0.0, 1.0]. Random translation ([−5,5]mm) and rotation ([−0.05,0.05]rad) are used to augment the training data. We keep the largest component in the predicted binary mask as the final segmentation of the prostate. Unless otherwise noted, all the competing methods and ablation models are trained and evaluated using the same configuration as our method. It is worth noting that, using the same training configuration for all the competing methods may not guarantee all of these methods to reach their full potential. However, this vanilla configuration can ensure all the performance disparities are caused by the method designs rather than the training strategies. For better reproducibility, the source code is released at https://github.com/DIAL-RPI/SCO-SSL.
IV. Experiments
A. Datasets and metrics
In this study, we conduct experiments on two large TRUS image datasets. One is a public dataset1 [47] shared by the Institute of Urologic Oncology, University of California-Los Angeles (UCLA) on the Cancer Imaging Archive (TCIA) platform [48]. The other one is an in-house dataset collected at the Nation Institutes of Health (NIH) from IRB-approved clinical trial. For brevity, we denote the two datasets as UCLA dataset and NIH dataset, respectively, in the following contents.
1). UCLA dataset:
The UCLA dataset contains 1,761 3D TRUS images collected from 1,150 patients2. All these image volumes are acquired by rotating a Hitachi Hi-Vision 5500 7.5 MHz end-fire probe or a Noblus C41V 2-10 MHz end-fire probe 200 degrees about its axis, and interpolating to resample the volume with isotropic resolution (spacing), which ranges from 0.21mm to 0.55mm. The image size varies from 342×226×342 to 452×290×452 (in voxel). Each TRUS volume has a prostate segmentation mask stored in the same size as the image.
We randomly divide the UCLA dataset into three parts with a proportion of 575(50%): 115(10%):460(40%) in terms of patients, resulting in a split of training/validation/test sets containing 895/169/697 TRUS image volumes, respectively. For fully-supervised learning, all the 575 patients (895 images) are used as the training samples. For semi-supervised learning, we randomly select 115 patients (20%, 194 images) in the training set as labeled samples and reserve the rest 460 patients (80%, 701 images) as unlabeled samples.
2). NIH dataset:
The NIH dataset contains 662 3D TRUS images collected from different patients. The image volumes are reconstructed from 2D TRUS frame sequences acquired by a Philips iU22 ultrasound scanner with C9-5 probes. All the images have isotropic voxel resolution (spacing) ranging from 0.40mm to 0.90mm. The image size varies from 173×113×122 to 218×184×337 (in voxel). 315 of the 662 TRUS images have a prostate segmentation mask manually annotated by two experienced physicians. The rest 347 TRUS images are unlabeled.
Considering the relatively smaller size of the NIH dataset compared with the UCLA dataset, we conducted a 3-fold cross validation on the NIH dataset to comprehensively evaluate method performance on it. Specifically, the 315 labeled samples are randomly divided into three folds with 105 samples in each fold. In each iteration of the cross validation, one fold (105 samples) is used for testing and the rest two folds (210 samples) are reserved for training and validation with a fixed split of 180 and 30 samples, respectively. The 347 unlabeled samples are only used for semi-supervised learning as the unlabeled training samples.
3). Metrics:
We use Dice similarity coefficient (DSC), average symmetric surface distance (ASD), and Hausdorff distance (HD) to quantitatively evaluate the model performance. We also calculate ASD-shadow to evaluate the segmentation performance inside the shadow regions, which is the ASD between the estimated boundary and ground-truth boundary inside the identified shadow regions. Paired t-tests on the above metrics are conducted to check the statistical significance between different results.
B. Comparison with fully-supervised methods
1). Benchmarking methods:
To justify the performance of our method, we compare it with five fully-supervised methods, including three state-of-the-art methods for general medical image segmentation and two recent methods dedicated to prostate ultrasound segmentation. Most of the competing methods are published in the past two years and thus represent the frontier performance in this domain.
V-Net [21] and U-Net [8]: Two popular FCNs with U-shape-like architectures that are widely used for various of medical image segmentation tasks.
nnU-Net [18]: The latest state-of-the-art method in many benchmark medical image segmentation tasks, which is actually a standard U-Net trained with specially tuned hyper-parameters. The value of the hyper-parameters are determined by a set of pre-defined guidelines regarding the dataset properties.
Radial-2.5D-UNet [20]: A 2.5D DL-based method dedicated for prostate ultrasound segmentation, where the TRUS volume is radially resampled to a set of slices around the superior-inferior axis and a 2D U-Net is trained to segment the prostate in the slices to reconstruct the 3D prostate volume.
DAF-Net [11]: A 3D DL-based method dedicated for prostate ultrasound segmentation, where a feature pyramid network combined with a special-designed attention module is trained through deep supervision to deal with the complex background condition in TRUS image.
Both U-Net and nnU-Net are implemented in 3D. The training configuration of nnUNet is tuned following the guidance provided in the original literature [18], which makes it different than the original UNet. Since the Radial-2.5D-UNet is a 2D network with a convergence behavior different from other 3D networks, it is trained using Adam optimizer (β1=0.9, β2=0.999) with a base learning rate of 1×10−4 for 200 epochs.
2). Intra-dataset comparison:
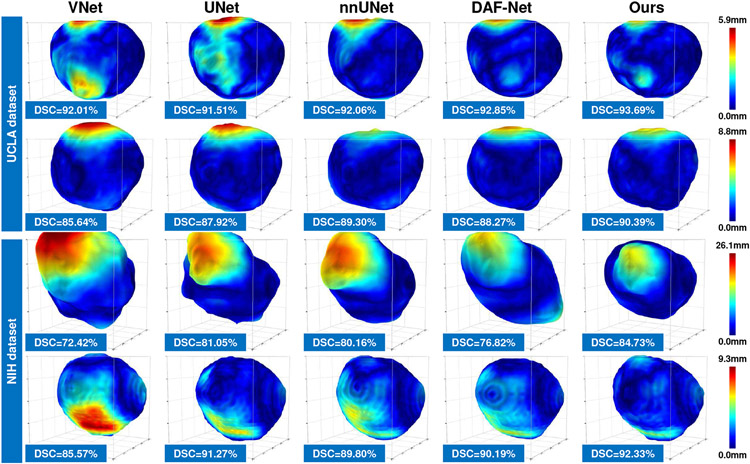
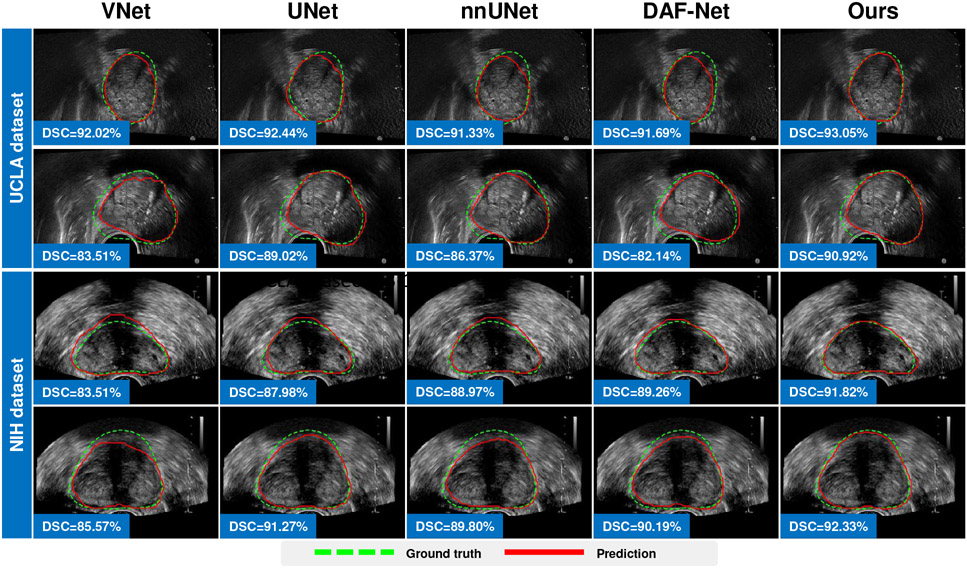
We first evaluate the model performance with the training and test data coming from the same dataset. In this case, the training data and test data share the same distribution. In this experiment, we evaluate the performance of our method working in two learning modes: 1) In fully-supervised mode, we train a 3D UNet equipped with our Shadow-AUG and Shadow-DROP mechanisms using all the labeled data in each dataset. 2) In semi-supervised mode, we train the proposed SCO-SSL method using a mixture of labeled and unlabeled data in each dataset. Table I lists the results of this intra-dataset comparison. It can be seen that, under the fully-supervised setting, our method (“SCO-SSL (full-supervised)” in Table I) generally achieves better performance than other competing methods on both datasets, and most of the improvement margins are statistically significant. Meanwhile, we observe that the segmentation accuracy in UCLA dataset is generally higher than that in NIH dataset. We attribute this performance gap to the data disparity between the two datasets. The NIH dataset contains fewer training samples but shows heavier shadow artifacts, both of which could degrade the performance of DL models. Notably, our method shows better performance inside the shadow regions in NIH dataset (see the column of “ASD-shadow” in Table I), which justifies the effectiveness of our Shadow-AUG and Shadow-DROP mechanisms. We also list the result of our SCO-SSL method trained under semi-supervised setting (“SCO-SSL (semi-supervised)” in Table I). On UCLA dataset, our SCO-SSL is trained with only 20% of image annotations while achieves comparable accuracy (DSC=91.60%) to that of the best competing method (nnUNet, DSC=92.17%) trained with 100% image annotations. On NIH dataset, by adding 347 extra unlabeled images to the training set, our SCO-SSL further improves the segmentation DSC from 89.85% to 90.12% with a significant margin (p<0.05). These results demonstrate the effectiveness of the semi-supervised learning framework. By incorporating unlabeled data into limited labeled data, the proposed SCO-SSL can present similar accuracy as the fully-supervised counterparts, which largely relieves the labor of data annotations in prostate ultrasound segmentation. On the other hand, by adding extra unlabeled data to regularize the model training, the accuracy of our SCO-SSL method can get further improved, which suggests an effective way to facilitate the DL-based prostate ultrasound segmentation with lower cost other than enlarging the data annotations. Fig. 6 and Fig. 7 visualize some results of this comparison.
TABLE I:
Intra-dataset evaluation of the fully-supervised methods. Bold values indicate the best results. Our method significantly outperformed most of the competing methods, except for those underlined entries (p>0.05).
| Methods | UCLA dataset | NIH dataset | ||||||
|---|---|---|---|---|---|---|---|---|
| DSC [%] |
ASD [mm] |
ASD-shadow [mm] |
HD [mm] |
DSC [%] |
ASD [mm] |
ASD-shadow [mm] |
HD [mm] |
|
| Radial-2.5D-UNet (2020) [20] | 88.56(2.78) | 1.46(0.41) | 1.81(0.72) | 7.21(2.16) | 86.13(5.49) | 1.80(1.07) | 2.33(1.78) | 8.38(4.08) |
| VNet (2016) [21] | 91.78(2.43) | 0.99(0.33) | 1.16(0.53) | 6.02(1.97) | 88.15(4.57) | 1.51(0.78) | 2.26(1.87) | 7.86(4.22) |
| UNet (2015) [8] | 91.96(2.38) | 0.98(0.34) | 1.16(0.54) | 6.22(2.33) | 89.28(4.55) | 1.34(0.68) | 1.92(1.28) | 7.36(3.65) |
| nnUNet (2021) [18] | 92.17(2.21) | 0.94(0.31) | 1.08 (0.50) | 5.79 (2.08) | 89.36(4.56) | 1.35(0.81) | 1.94 (1.53) | 7.09 (3.97) |
| DAF-Net (2019) [11] | 91.96(2.35) | 0.97(0.32) | 1.12(0.53) | 5.80 (1.95) | 89.07(4.15) | 1.39(0.74) | 2.02(1.56) | 6.92 (3.59) |
| SCO-SSL (semi-supervised) | 91.60(2.37) | 1.02(0.34) | 1.22(0.52) | 6.37(2.36) | 90.12 (3.61) | 1.23 (0.63) | 1.80 (1.18) | 6.65 (2.89) |
| SCO-SSL (full-supervised) | 92.25 (2.19) | 0.93 (0.29) | 1.10(0.46) | 5.89(1.93) | 89.85(3.30) | 1.26(0.58) | 1.84(1.13) | 6.88(3.00) |
Fig. 6:
Visualization of 3D distance error of fully-supervised prostate segmentation results of different methods. Each row illustrates one case.
Fig. 7:
Fully-supervised prostate segmentation results superimposed on 2D TRUS slices. Each row illustrates one case. Green dashed line and red solid line indicate the ground-truth boundary and predicted boundary, respectively.
3). Inter-dataset comparison:
We then evaluate the model performance with the training and test data coming from different datasets. The test data exhibit significant domain shifts from the training data. The purpose of this experiment is to evaluate the generalizability of our method. Same as the intra-dataset comparison, we evaluate the inter-dataset performance of our method under both fully-supervised and semi-supervised learning settings. All the models are tuned using the training/validation sets from one dataset and then evaluated using the test set from the other dataset. Table II lists the results of this inter-dataset comparison. It can be seen that, when compared with the intra-dataset results in Table I, all the competing methods suffer a large performance drop in the inter-dataset comparison. This performance degradation is caused by the large distribution gap between the training and test data, which are acquired using different types of ultrasound probes. On the other hand, we can see that both DAF-Net [11] and our method consistently show superior performance in this inter-dataset comparison. The outperformance margins over other competing methods are significantly larger than that in the intra-dataset comparison, demonstrating the good generalization ability of these two methods. Notably, our method achieves significantly better performance than DAF-Net (p<0.05) in the comparison where the models are trained on the smaller NIH training set (180 TRUS images) and tested on the larger UCLA test set (697 TRUS images). It is more challenging than the other comparison, where the models are trained on the larger UCLA training set (895 TRUS images) and tested on the smaller NIH test set (315 TRUS images).
TABLE II:
Inter-dataset evaluation of the fully-supervised methods. Bold values indicate the best result. Our method significantly outperformed most of the competing methods, except on those underlined entries (p>0.05).
| Methods | Train&val. on UCLA / test on NIH | Train&val. on NIH / test on UCLA | ||||||
|---|---|---|---|---|---|---|---|---|
| DSC [%] |
ASD [mm] |
ASD-shadow [mm] |
HD [mm] |
DSC [mm] |
ASD [mm] |
ASD-shadow [mm] |
HD [mm] |
|
| Radial-2.5D-UNet (2020) [20] | 63.10(16.41) | 4.14(1.68) | 5.21(2.13) | 14.59(4.67) | 75.41(8.04) | 3.56(1.10) | 4.16(1.89) | 17.67(5.18) |
| VNet (2016) [21] | 78.80 (10.55) | 3.09(1.75) | 4.99(3.49) | 16.88(7.47) | 54.86(13.00) | 9.21(3.45) | 12.01(5.03) | 35.01(9.25) |
| UNet (2015) [8] | 76.74(9.53) | 3.77(1.62) | 5.48(3.04) | 21.51(7.00) | 72.26(10.85) | 4.94(2.84) | 6.65(4.52) | 25.35(10.07) |
| nnUNet (2021) [18] | 74.42(13.27) | 4.42(2.72) | 7.80(5.70) | 22.27(11.09) | 74.47(11.74) | 4.52(2.72) | 6.60(4.70) | 23.47(9.67) |
| DAF-Net (2019) [11] | 79.97 (11.05) | 3.07(2.06) | 5.23(5.27) | 16.62(10.42) | 78.28 (10.57) | 3.56(2.73) | 4.76(4.65) | 19.37(10.65) |
| SCO-SSL (semi-supervised) | 75.58(11.14) | 4.15(2.17) | 5.26(3.65) | 23.03(8.96) | 80.93 (9.35) | 2.55 (2.06) | 3.23(2.53) | 13.30 (5.86) |
| SCO-SSL (full-supervised) | 79.51(10.06) | 2.71 (1.76) | 4.14 (2.48) | 12.97 (6.15) | 78.63(9.22) | 2.68(1.32) | 3.11 (1.89) | 13.89(4.44) |
C. Incorporation with semi-supervised frameworks
1). Benchmarking methods:
To demonstrate the flexibility of our SCO-SSL method, we incorporate it into five representative semi-supervised learning frameworks and evaluate the performance using both UCLA and NIH datasets. The selected semi-supervised baselines are:
Π-model and temporal ensembling model [36]: Two classical consistency learning methods for semi-supervised learning, where the trained network utilizes its own historic predictions to regularize the training on unlabeled data.
Mean-teacher [37]: A popular teacher-student model for semi-supervised learning, where the historic average of the trained model is used to generate the pseudo label to supervise the unlabeled data.
Uncertainty-aware mean-teacher (UA-MT) [38]: A variant of the mean-teacher [37] model designed for semi-supervised 3D left atrium segmentation in MRI images, which exploits Monte Carlo dropout [39] layers to estimate the uncertainty map of the pseudo label and guide the network to learn from more reliable targets.
Transformation-consistent mean-teacher (TC-MT) [41]: Another variant of the mean-teacher [37] model where the input of the student network and the teacher network are perturbed with different geometric transformations.
Since most of the existing semi-supervised segmentation methods can be seen as different variants of the five selected frameworks, the combination with these frameworks is sufficient to justify the flexibility of our SCO-SSL method. Considering different baselines may be implemented using different networks in their original literature, to eliminate the interference from network structures, we use a 3D version of U-Net [8] as the segmentation network for all the semi-supervised frameworks in this comparison.
2). Comparison results:
Table III lists the comparison result. It can be seen that, by incorporating our Shadow-AUG and Shadow-DROP mechanisms, the five semi-supervised methods show better performance than their baselines on both datasets. Most of the improvement margins are statistically significant (p<0.05). This overall performance gain demonstrates the effectiveness and flexibility of our shadow-consistent learning method when deployed in the mainstream semi-supervised frameworks. Among the five evaluated baselines, we find that our method achieves relatively better performance when combined with the mean-teacher [37] and UA-MT [38] frameworks. Since the UA-MT framework involves extra computations on uncertainty estimation, we finally build our SCO-SSL method upon the mean-teacher framework to balance the accuracy and efficiency.
TABLE III:
Comparison of different semi-supervised learning frameworks trained with/without our SCO-SSL method for prostate ultrasound segmentation. Bold values indicate the best performance. Our SCO-SSL method can significantly improve the performance of most of the semi-supervised learning frameworks, except those underlined entries (p>0.05).
| Methods | UCLA dataset labeled/unlabeled = 194/701 |
NIH dataset labeled/unlabeled = 180/347 |
||||||
|---|---|---|---|---|---|---|---|---|
| DSC [%] |
ASD [mm] |
ASD-shadow [mm] |
HD [mm] |
DSC [%] |
ASD [mm] |
ASD-shadow [mm] |
HD [mm] |
|
| Π-model (2017) [36] | 91.39 (2.54) | 1.04 (0.36) | 1.21 (0.59) | 6.28 (2.34) | 89.27 (5.18) | 1.35 (0.79) | 1.96 (1.44) | 7.33 (3.86) |
| + SCO-SSL | 91.47 (2.54) | 1.04 (0.34) | 1.21 (0.52) | 6.32(2.11) | 89.53 (5.94) | 1.35(1.39) | 1.96(1.96) | 7.40(3.98) |
| Temporal ensembling (2017) [36] | 91.11(2.70) | 1.09(0.38) | 1.29(0.64) | 6.95(2.77) | 88.72(4.28) | 1.44(0.78) | 2.15 (1.70) | 8.14(4.03) |
| + SCO-SSL | 91.61 (2.37) | 1.02 (0.34) | 1.23 (0.55) | 6.30 (2.28) | 89.63 (5.49) | 1.32 (1.03) | 1.96 (2.01) | 6.94 (3.58) |
| Mean-teacher (2017) [37] | 91.36(2.56) | 1.05(0.36) | 1.26(0.56) | 6.48(2.34) | 89.55(4.19) | 1.30(0.66) | 1.87(1.23) | 7.12(3.32) |
| + SCO-SSL | 91.60 (2.37) | 1.02 (0.34) | 1.22 (0.52) | 6.37 (2.36) | 90.12 (3.61) | 1.23 (0.63) | 1.80 (1.18) | 6.65 (2.89) |
| UA-MT (2019) [38] | 91.29(2.68) | 1.06(0.37) | 1.27(0.57) | 6.64(2.51) | 89.54(3.75) | 1.31(0.65) | 1.88(1.23) | 7.04(3.20) |
| + SCO-SSL | 91.80 (2.42) | 0.99 (0.32) | 1.17 (0.51) | 6.12 (2.08) | 90.07 (3.74) | 1.23 (0.61) | 1.78 (1.14) | 6.61 (2.87) |
| TC-MT (2020) [41] | 91.13(2.74) | 1.08(0.39) | 1.31(0.58) | 6.91(2.75) | 89.21(4.88) | 1.36(0.74) | 1.96(1.40) | 7.69(3.75) |
| + SCO-SSL | 91.48 (2.41) | 1.04 (0.35) | 1.25 (0.56) | 6.44 (2.45) | 89.94 (3.53) | 1.25 (0.62) | 1.85 (1.25) | 6.92 (2.96) |
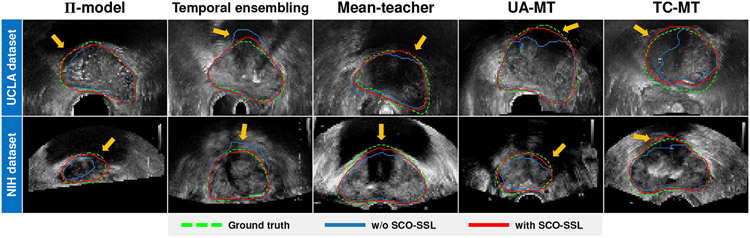
In Fig. 8, we visualize some results of the five evaluated semi-supervised learning frameworks when they are trained with/without our SCO-SSL method. It can be seen that, by incorporating our SCO-SSL, the semi-supervised learning methods achieve better performance in the shadow regions (pointed out by the orange arrows), where the corroded prostate boundaries can be merely inferred from the neighboring shadow-free boundaries.
Fig. 8:
Segmentation results of the semi-supervised learning methods trained with/without our SCO-SSL. Green dashed lines indicate the ground-truth segmentations. Red/blue solid lines indicate the segmentations by the semi-supervised learning methods equipped with/without our SCO-SSL, respectively. Orange arrows point to the shadow regions where SCO-SSL helps improve the segmentation.
D. Ablation study
1). Effectiveness of Shadow-AUG and Shadow-DROP:
We demonstrate the effectiveness of the two key components (i.e., Shadow-AUG and Shadow-DROP) of our SCO-SSL method through an ablation study on both the UCLA and NIH datasets. In this experiment, we adopt a 3D version of UNet [8] as the baseline model and evaluate its performance under a fully-supervised setting when combined with one or both of the two components. The experimental results are listed in Table IV. It can be seen that, by adding the Shadow-AUG and Shadow-DROP mechanisms to the baseline UNet model, the segmentation accuracy gradually get improved. This progressively increased accuracy suggests the effectiveness of the Shadow-AUG and Shadow-DROP mechanisms. Furthermore, the improvement margin caused by the Shadow-DROP is larger than that of the Shadow-AUG, indicating that applying the shadow masking in feature level (Shadow-DROP) is more effective than that in image level (Shadow-AUG) to facilitate the model training.
TABLE IV:
Ablation study of the Shadow-AUG and Shadow-DROP mechanisms. Bold values indicate the best result. Underlined values suggest results without statistical significance compared with the bottom row (p>0.05).
| UNet trained with | UCLA dataset | NIH dataset | |||||
|---|---|---|---|---|---|---|---|
| Shadow-AUG | Shadow-DROP | DSC[%] | ASD[mm] | HD[mm] | DSC[%] | ASD[mm] | HD[mm] |
| × | × | 91.96(2.38) | 0.98(0.34) | 6.22(2.33) | 89.28(4.55) | 1.34(0.68) | 7.36(3.65) |
| ✓ | × | 92.00(2.44) | 0.97(0.32) | 5.92 (2.02) | 89.55(3.72) | 1.31(0.65) | 6.96 (3.14) |
| × | ✓ | 92.11(2.74) | 0.95(0.36) | 6.11(2.35) | 89.79 (3.43) | 1.29 (0.65) | 7.42(3.49) |
| ✓ | ✓ | 92.25 (2.19) | 0.93 (0.29) | 5.89 (1.93) | 89.85 (3.30) | 1.26 (0.58) | 6.88 (3.00) |
2). Shadow-AUG using different shadow threshold τs:
In our Shadow-AUG strategy, the threshold τs in Eq. (1) controls the area of the shadow regions extracted from the source image. The optimal value of τs may vary from UCLA dataset to NIH dataset due to the different image properties. Therefore, we conduct an experiment to justify the choice of the shadow threshold τs on the two datasets. In Table V, we list the results of a fully-supervised 3D UNet trained with our Shadow-AUG and Shadow-DROP mechanisms when successively using τs=20/255, 40/255, 60/255, and 80/255 for shadow thresholding. It can be seen that, although the model accuracy varies across different τs, most of the differences are nonsignificant (p>0.05), suggesting that the shadow-consistent learning method is insensitive to the change of the shadow threshold τs in a wide range. We finally set τs to the value of 60/255 and 40/255 on UCLA dataset and NIH dataset, respectively, given the relatively smaller standard deviations.
TABLE V:
Results of a fully-supervised 3D UNet trained with Shadow-AUG and Shadow-DROP mechanisms using different shadow threshold τs. Bold values indicate the best result. Underlined values suggest results without statistical significance compared with the best result (p>0.05).
| Models | UCLA dataset | NIH dataset | ||
|---|---|---|---|---|
| DSC[%] | ASD[mm] | DSC[%] | ASD[mm] | |
| τs = 20/255 | 92.21 (2.45) | 0.94 (0.32) | 89.68 (3.74) | 1.29(0.65) |
| τs = 40/255 | 92.17(2.34) | 0.94(0.32) | 89.85 (3.30) | 1.26 (0.58) |
| τs = 60/255 | 92.25 (2.19) | 0.93 (0.29) | 89.81 (3.86) | 1.27 (0.65) |
| τs = 80/255 | 92.25 (2.34) | 0.93 (0.31) | 89.84 (3.90) | 1.27 (0.60) |
3). Shadow-AUG/-DROP using hard thresholding function:
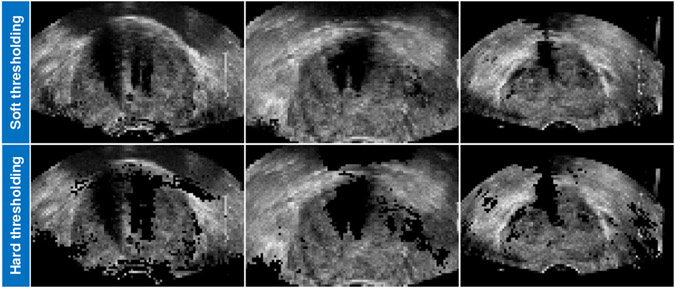
Our Shadow-AUG and Shadow-DROP mechanisms adopt a soft thresholding function in Eq. (1) to generate the shadow masks with continuous values between 0 and 1. We also try to use a simple hard thresholding function to generate binary shadow masks in the proposed method. In Table VI, we list the results of a fully-supervised 3D UNet trained with our Shadow-AUG and Shadow-DROP mechanisms when using hard/soft thresholding functions. It can be seen that the hard thresholding achieves slightly lower accuracy than the soft thresholding without statistical significance (p>0.05). However, when using the hard thresholding function, the simulated shadowed TRUS image exhibits abrupt pixel intensity change and looks less real as shown in Fig. 9. We thus chose to use the soft thresholding function in our method.
TABLE VI:
Results of a fully-supervised 3D UNet trained with Shadow-AUG and Shadow-DROP mechanisms using hard/soft shadow thresholding functions. Bold values indicate the best result. Underlined values suggest results without statistical significance compared with the bottom row (p>0.05).
| Models | UCLA dataset | NIH dataset | ||
|---|---|---|---|---|
| DSC[%] | ASD[mm] | DSC[%] | ASD[mm] | |
| Hard threshold | 92.24 (2.19) | 0.93 (0.30) | 89.82 (3.57) | 1.28 (0.63) |
| Soft threshold | 92.25 (2.19) | 0.93 (0.29) | 89.85 (3.30) | 1.26 (0.58) |
Fig. 9:
Contrast view of three Shadow-AUG images using soft thresholding function (top row) and hard thresholding function (bottom row).
4). Shadow-DROP at different stages of segmentation network:
In this experiment, we investigate the model performance when the Shadow-DROP layers are deployed at four different stages of the segmentation network, including 1) the encoder, 2) the bottle-neck, 3) the decoder, and 4) the whole network (excluding the last convolutional layer). We conduct the experiments on both the UCLA and NIH datasets. The segmentation network is a fully-supervised 3D UNet trained with our Shadow-AUG and Shadow-DROP mechanisms. The experimental results are listed in the odd rows of Table VII (start with “Shadow-DROP”). It can be seen that the model achieves the best performance when the Shadow-DROP layers are deployed at the encoder of the UNet. When we move the Shadow-DROP layers from the bottom layers (encoder) to the top layers (bottle-neck and decoder), the model accuracy gradually decreases. We attribute this result to the fact that the extracted features are more vulnerable to the neighboring shadow artifacts at the early stage of UNet, due to the relatively small size of the receptive field. These corroded features are harmful to the subsequent inference of prostate boundaries. Since the Shadow-DROP layer drops the feature associated with low-intensity pixels (shadow-like pixels), most of the corroded features can be filtered out if we deploy the Shadow-DROP at the early stage. As a consequence, the subsequent layers will suffer less interference and thus produce more discriminative features for better prostate segmentation. Based on these observations, we choose to deploy the Shadow-DROP layers to the encoder of the UNet in our method.
TABLE VII:
Results of a fully-supervised 3D UNet trained with our Shadow-AUG and Shadow-DROP mechanisms where the Shadow-DROP layers are deployed at different stages of the segmentation network. Bold values indicate the best result. Underlined values suggest results without statistical significance compared with the top row (p>0.05).
| Models | UCLA dataset | NIH dataset | ||||
|---|---|---|---|---|---|---|
| DSC[%] | ASD[mm] | HD[mm] | DSC[%] | ASD[mm] | HD[mm] | |
| Shadow-DROP at encoder | 92.25(2.19) | 0.93 (0.29) | 5.89(1.93) | 89.85 (3.30) | 1.26 (0.58) | 6.88 (3.00) |
| Standard dropout at encoder | 91.62(2.48) | 1.03(0.39) | 6.42(2.27) | 81.57(7.40) | 2.62(1.31) | 13.78(5.92) |
| Shadow-DROP at bottle-neck | 92.17(2.32) | 0.94(0.32) | 5.93 (1.97) | 89.61(3.56) | 1.30(0.63) | 7.20(3.37) |
| Standard dropout at bottle-neck | 92.32 (2.43) | 0.93(0.35) | 6.17(2.44) | 89.32(6.01) | 1.32(0.72) | 7.78(3.83) |
| Shadow-DROP at decoder | 92.01(2.41) | 0.97(0.34) | 6.14(2.22) | 89.36(4.23) | 1.34(0.70) | 7.72(3.85) |
| Standard dropout at decoder | 92.29 (2.40) | 0.93 (0.33) | 5.76 (2.08) | 89.21(6.00) | 1.41(1.60) | 7.63(5.02) |
| Shadow-DROP at all layers | 92.12(2.32) | 0.95(0.31) | 5.99(2.04) | 89.86 (3.40) | 1.29 (0.63) | 7.88(4.35) |
| Standard dropout at all layers | 91.48(2.44) | 1.05(0.38) | 6.48(2.33) | 82.75(6.73) | 2.27(1.22) | 11.82(4.94) |
We also compare our Shadow-DROP mechanism with the standard dropout mechanism [44]. Specifically, we replace all the Shadow-DROP layers in the above models with standard dropout layers and retrain them on the two datasets. The results of the standard dropout models are listed in the even rows of Table VII (start with “Standard dropout”). It can be seen that the standard dropout deployed at bottom layers (encoder) generally shows lower accuracy than that deployed at top layers (bottle-neck and decoder), which is opposite to the result of our Shadow-DROP. Since the low-level spatial feature extracted by the bottom layers contains more interference from the shadow artifacts in comparison with the high-level semantic feature extracted by the top layers, dropping out the low-level spatial features associated with low-intensity pixels can effectively suppress the interference caused by the shadow artifacts and thus facilitate the subsequent boundary inference. In contrast, the standard dropout layer drops the feature in a completely random way, either the shadow-associated features or the useful boundary features could be dropped if it is deployed at the bottom layers of the segmentation network. As a consequence, less useful features will remain for subsequent inference, which finally leads to performance degradation. This phenomenon is more pronounced when the images contain heavier shadow artifacts (which means more corroded information contained in the low-level spatial features). The significant performance degradation on NIH dataset (see “Standard dropout at encoder” on NIH dataset, Table VII) is an evidence for this explanation since the NIH dataset suffers worse image quality than the UCLA dataset.
5). Semi-supervised training with different ratio of labeled data:
The number of labeled training samples is a key factor affecting the performance of DL methods. In this experiment, we evaluate the performance of the competing methods in Sec. IV-B and our SCO-SSL method when they are successively trained with 10%, 20%, 40%, 60%, 80%, and 100% of the training samples. For our SCO-SSL method, we also take advantage of the rest part of the training samples as unlabeled samples through semi-supervised learning. Since the NIH dataset is in a relatively small size and partially labeled, we conduct this experiment using only the UCLA dataset. In Fig. 10, we plot the curves of testing DSC (y-axis) v.s. the ratio of labeled training samples (x-axis). It can be seen that, when we increase the labeled data ratio from 10% to 80%, the segmentation accuracy gradually gets improved with statistically significant margins. This observation is in line with our common sense that more labeled samples can bring stronger and more accurate regularizations to the model, and thus lead to better performance. However, when we further increase the labeled data ratio from 80% to 100%, the model accuracy does not get significant improvement. This can be attributed to the relatively large size of UCLA dataset where 80% of labeled samples is sufficient to represent the data distribution in the training set. On the other hand, our SCO-SSL method generally outperforms other competing methods throughout different ratios of labeled training data. The improvement margin gets larger when fewer labeled samples are involved during training. This result demonstrates the effectiveness of semi-supervised learning for prostate ultrasound segmentation, where the unlabeled images provide extra regularizations to the optimization and thus facilitate the model performance.
Fig. 10:
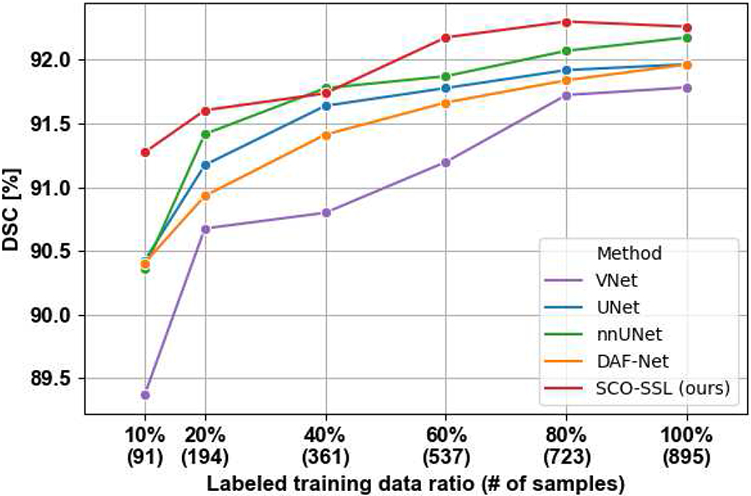
DSC curves of different prostate segmentation methods trained with different numbers of labeled samples. For brevity, we only show the curves of the best five methods.
6). Choice of consistency loss function:
In the proposed SCO-SSL method, we use the BCE loss in Eq. (4) as the consistency loss to regularize the student network training on unlabeled data. This consistency loss can also be implemented with mean squared error (MSE) and Kullback-Leibler (KL) divergence, which are often used to minimize the disparity between two distributions. In Table VIII, we compare the performance of our SCO-SSL method when successively using MSE loss, KL loss, and BCE loss as the consistency loss for semi-supervised training. It shows that the three loss functions achieve similar performance on the two datasets. None of them is universally better than the other two loss functions.
TABLE VIII:
Results of our SCO-SSL method when using mean squared error (MSE) loss, Kullback-Leibler (KL) divergence loss, and binary cross entropy (BCE) loss as the consistency loss for semi-supervised learning.
| Losses | UCLA dataset | NIH dataset | ||
|---|---|---|---|---|
| DSC[%] | ASD[mm] | DSC[%] | ASD[mm] | |
| BCE | 91.60(2.37) | 1.02(0.34) | 90.12 (3.61) | 1.23 (0.63) |
| KL | 91.73(2.35) | 1.00(0.34) | 90.00(3.53) | 1.25(0.62) |
| MSE | 91.76 (2.35) | 1.00 (0.34) | 90.04(3.45) | 1.24(0.61) |
V. DISCUSSION
1). Clinical significance:
Prostate segmentation is in many ways a critical step of prostate cancer-related clinical workflows because improper identification of the prostate in image will result in inaccuracy of downstream processes. The clinical significance of our method comes from the following three aspects. 1) It achieves better performance inside shadow regions. The recent works using deep learning technology have substantially pushed the limit of prostate ultrasound segmentation to a very high level of accuracy (around 90% of DSC as shown in our experimental results in Table I). For most of the test samples, these methods can achieve good results. The remaining challenge is mainly about the shadow regions. According to our experimental results shown in Table I, our method exhibits lower boundary error inside the shadow regions in comparison to other methods (see the columns of “ASD-shadow [mm]” in Tables I&II). This improvement is clinically significant since the clinicians often need to draw much more attention and efforts to the shadow regions to make sure they are well and correctly processed. 2) Our method generalizes better on unseen datasets. According to our experimental results of the inter-dataset evaluation, our method shows superior performance when the test data comes from a completely unseen dataset. The outperformance margin over other competing methods is also significant. This is of great clinical value since the data from different clinical sites often suffer from significant quality disparities or domain shifts. 3) Our method requires fewer data annotations. Our method can work in the semi-supervised manner and achieve very competitive performance in comparison to the fully-supervised state-of-the-art methods (approximately 0.6% drop in DSC) while using only 20% data annotations. This can significantly lower the cost of data annotation in clinical practice.
2). Inter-/intra-observer variability:
Inter- and intra-observer variabilities are acknowledged as an inherent challenge in prostate ultrasound segmentation, especially inside the shadow regions, where the prostate boundaries are highly subject to the physicians’ experience and judgment. In this study, our goal is to develop a DL model that can approximate a specific expert physician who defined the ground-truth segmentations. To this end, any performance improvement of the DL model would be meaningful, since it can finally contribute to the efficiency and reproducibility of prostate segmentation. On the other hand, although our improvement over the competing methods is relatively small regarding the global performance, our method performed significantly better inside the shadow regions and on the unseen datasets, both of which are meaningful to the model deployment in clinical practice.
3). Accurate estimation of shadow regions:
In the proposed Shadow-AUG and Shadow-DROP mechanisms, we designed a thresholding function in Eq. (1) to extract the shadow masks from the ultrasound images. A number of non-shadow regions associated with low intensities (e.g., the region out of field-of-view and the bladder) could be included in these “over-segmented” shadow masks. However, our main purpose is to filter out the irrelevant regions that hardly contribute to the judgment of the prostate boundaries. Using these “over-segmented” shadow masks will not affect our target since the non-shadow regions associated with low intensities hardly contain useful information for prostate segmentation. On the other hand, it is certainly interesting if we can incorporate some advanced DL-based methods (such as [31]-[33]) into our framework to accurately estimate the shadow regions. We would like to set it as one of our future research aims.
4). Potential of shadow-consistent weakly-/self-supervised learning:
The Shadow-AUG and Shadow-DROP mechanisms manipulate input images and intermediate feature maps, respectively, without requiring the ground-truth segmentations. Hence it is possible to incorporate the proposed shadow-consistent learning method into other annotation-efficient DL paradigms such as weakly-supervised [49]-[51] and self-supervised learning [52]-[54]. For example, we can train a weakly-supervised UNet equipped with our Shadow-AUG and Shadow-DROP mechanisms by replacing the Dice loss with the bounding box tightness constraints [49], which do not require pixel-wise annotation and would thus further lower the cost of clinical data annotation for prostate segmentation.
5). Generalization to other types of ultrasound images:
In this study, the Shadow-AUG and Shadow-DROP mechanisms are designed to deal with the shadow artifacts encountered in the TRUS images. Since the shadow artifacts are not only seen in the TRUS images but also present in other ultrasound images, such as the breast ultrasound image [55], both Shadow-AUG and Shadow-DROP could be applied to the analysis tasks towards other types of ultrasound images. Investigating their performance on other types of ultrasound images may be explored in our future work.
VI. Conclusion
In this study, we propose a shadow-consistent semi-supervised learning (SCO-SSL) method to address the challenging problem of prostate ultrasound segmentation with large amount of unlabeled data on top of limited annotations. Two novel mechanisms, i.e., the Shadow-AUG and Shadow-DROP, are highlighted in the proposed SCO-SSL method to enhance the segmentation performance against shadow artifacts. We conduct extensive experiments on two large clinical datasets. The experimental results show that, in fully-supervised setting, our SCO-SSL outperforms the state-of-the-art methods on most of the metrics by statistically significant margins, demonstrating the superior performance of our design towards the challenging task of prostate ultrasound segmentation. In semi-supervised setting, even with 20% labeled training data, our SCO-SSL still achieves competitive results (approximately 0.6% lower DSC) in comparison to that of the state-of-the-art methods trained by fully annotated dataset, suggesting great clinical value in relieving the labor of data annotation for medical image analysis.
Acknowledgments
This work was partially supported by National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health (NIH) under awards R21EB028001 and R01EB027898, and through an NIH Bench-to-Bedside award made possible by the National Cancer Institute.
Footnotes
The original UCLA dataset contains 1,151 patients but one patient has no annotation on the TRUS image.
Contributor Information
Xuanang Xu, Department of Biomedical Engineering and the Center for Biotechnology and Interdisciplinary Studies at Rensselaer Polytechnic Institute, Troy, NY 12180 USA.
Thomas Sanford, State University of New York Upstate Medical University, Syracuse, NY 13210 USA.
Baris Turkbey, Molecular Imaging Program, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Sheng Xu, Center for Interventional Oncology, Radiology & Imaging Sciences at National Institutes of Health, Bethesda, MD 20892, USA.
Bradford J. Wood, Center for Interventional Oncology, Radiology & Imaging Sciences at National Institutes of Health, Bethesda, MD 20892, USA
Pingkun Yan, Department of Biomedical Engineering and the Center for Biotechnology and Interdisciplinary Studies at Rensselaer Polytechnic Institute, Troy, NY 12180 USA.
References
- [1].Siegel RL, Miller KD, and Jemal A, “Cancer statistics, 2020,” CA: A Cancer J. Clinicians, vol. 70, no. 1, pp. 7–30, 2020. [DOI] [PubMed] [Google Scholar]
- [2].Wang S, He K, Nie D, Zhou S, Gao Y, and Shen D, “CT male pelvic organ segmentation using fully convolutional networks with boundary sensitive representation,” Med. Image Anal, vol. 54, pp. 168–178, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Xu X et al. , “Asymmetrical multi-task attention u-net for the segmentation of prostate bed in CT image,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent. Springer, 2020, pp. 470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Xu X, Lian C, Wang S, Zhu T, Chen RC, Wang AZ, Royce TJ, Yap P-T, Shen D, and Lian J, “Asymmetric multi-task attention network for prostate bed segmentation in computed tomography images,” Medical Image Analysis, p. 102116, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nie D, Wang L, Gao Y, Lian J, and Shen D, “Strainet: Spatially varying stochastic residual adversarial networks for MRI pelvic organ segmentation,” IEEE Trans. Neural Netw. and Learn. Syst, vol. 30, no. 5, pp. 1552–1564, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhu Q, Du B, and Yan P, “Boundary-weighted domain adaptive neural network for prostate MR image segmentation,” IEEE Trans. Med. Imag, vol. 39, no. 3, pp. 753–763, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Long J, Shelhamer E, and Darrell T, “Fully convolutional networks for semantic segmentation,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2015, pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- [8].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent. Springer, 2015, pp. 234–241. [Google Scholar]
- [9].Yang X et al. , “Fine-grained recurrent neural networks for automatic prostate segmentation in ultrasound images,” in Proc. AAAI Conf. Artif. Intell, vol. 31, no. 1, 2017. [Google Scholar]
- [10].Wang Y et al. , “Deep attentional features for prostate segmentation in ultrasound,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent. Springer, 2018, pp. 523–530. [Google Scholar]
- [11].—, “Deep attentive features for prostate segmentation in 3D transrectal ultrasound,” IEEE Trans. Med. Imag, vol. 38, no. 12, pp. 2768–2778, 2019. [DOI] [PubMed] [Google Scholar]
- [12].Lei Y et al. , “Ultrasound prostate segmentation based on 3D V-Net with deep supervision,” in Med. Imag. 2019: Ultrason. Imag. and Tomography, vol. 10955. Int. Soc. Opt. Photon., 2019, p. 109550V. [Google Scholar]
- [13].—, “Ultrasound prostate segmentation based on multidirectional deeply supervised V-Net,” Med. Phys, vol. 46, no. 7, pp. 3194–3206, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shen D, Zhan Y, and Davatzikos C, “Segmentation of prostate boundaries from ultrasound images using statistical shape model,” IEEE Trans. Med. Imag, vol. 22, no. 4, pp. 539–551, 2003. [DOI] [PubMed] [Google Scholar]
- [15].Yan P, Xu S, Turkbey B, and Kruecker J, “Discrete deformable model guided by partial active shape model for TRUS image segmentation,” IEEE Trans. Biomed. Eng, vol. 57, no. 5, pp. 1158–1166, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].—, “Adaptively learning local shape statistics for prostate segmentation in ultrasound,” IEEE Trans. Biomed. Eng, vol. 58, no. 3, pp. 633–641, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ghose S et al. , “A supervised learning framework of statistical shape and probability priors for automatic prostate segmentation in ultrasound images,” Med. Image Anal, vol. 17, no. 6, pp. 587–600, 2013. [DOI] [PubMed] [Google Scholar]
- [18].Isensee F, Jaeger PF, Kohl SA, Petersen J, and Maier-Hein KH, “nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation,” Nature Methods, vol. 18, no. 2, pp. 203–211, 2021. [DOI] [PubMed] [Google Scholar]
- [19].Orlando N, Gillies DJ, Gyacskov I, and Fenster A, “Deep learning-based automatic prostate segmentation in 3D transrectal ultrasound images from multiple acquisition geometries and systems,” in Proc. Med. Imag.: Image-Guided Procedures, Robot. Interventions, Model, vol. 11315. Int. Soc. Opt. Photon., 2020, p. 113152I. [Google Scholar]
- [20].Orlando N, Gillies DJ, Gyacskov I, Romagnoli C, DSouza D, and Fenster A, “Automatic prostate segmentation using deep learning on clinically diverse 3D transrectal ultrasound images,” Med. Phys, vol. 47, no. 6, pp. 2413–2426, 2020. [DOI] [PubMed] [Google Scholar]
- [21].Milletari F, Navab N, and Ahmadi S-A, “V-net: Fully convolutional neural networks for volumetric medical image segmentation,” in Proc. Fourth Int. Conf. 3D Vision (3DV). IEEE, 2016, pp. 565–571. [Google Scholar]
- [22].Dunmire B, Harper JD, Cunitz BW, Lee FC, Hsi R, Liu Z, Bailey MR, and Sorensen MD, “Use of the acoustic shadow width to determine kidney stone size with ultrasound,” The Journal of urology, vol. 195, no. 1, pp. 171–177, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dai JC, Dunmire B, Sternberg KM, Liu Z, Larson T, Thiel J, Chang HC, Harper JD, Bailey MR, and Sorensen MD, “Retrospective comparison of measured stone size and posterior acoustic shadow width in clinical ultrasound images,” World journal of urology, vol. 36, no. 5, pp. 727–732, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hacihaliloglu I, “Enhancement of bone shadow region using local phase-based ultrasound transmission maps,” International Journal of Computer Assisted Radiology and Surgery, vol. 12, no. 6, pp. 951–960, 2017. [DOI] [PubMed] [Google Scholar]
- [25].Alsinan AZ, Patel VM, and Hacihaliloglu I, “Bone shadow segmentation from ultrasound data for orthopedic surgery using gan,” International Journal of Computer Assisted Radiology and Surgery, vol. 15, no. 9, pp. 1477–1485, 2020. [DOI] [PubMed] [Google Scholar]
- [26].Wang P, Vives M, Patel VM, and Hacihaliloglu I, “Robust bone shadow segmentation from 2d ultrasound through task decomposition,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent. Springer, 2020, pp. 805–814. [Google Scholar]
- [27].Hellier P, Coupé P, Morandi X, and Collins DL, “An automatic geometrical and statistical method to detect acoustic shadows in intraoperative ultrasound brain images,” Med. Image Anal, vol. 14, no. 2, pp. 195–204, 2010. [DOI] [PubMed] [Google Scholar]
- [28].Basij M, Moallem P, Yazdchi M, and Mohammadi S, “Automatic shadow detection in intra vascular ultrasound images using adaptive thresholding,” in 2012 IEEE International Conference on Systems, Man, and Cybernetics (SMC). IEEE, 2012, pp. 2173–2177. [Google Scholar]
- [29].Karamalis A, Wein W, Klein T, and Navab N, “Ultrasound confidence maps using random walks,” Med. Image Anal, vol. 16, no. 6, pp. 1101–1112, 2012. [DOI] [PubMed] [Google Scholar]
- [30].Berton F, Cheriet F, Miron M-C, and Laporte C, “Segmentation of the spinous process and its acoustic shadow in vertebral ultrasound images,” Comput. in Biology and Medicine, vol. 72, pp. 201–211, 2016. [DOI] [PubMed] [Google Scholar]
- [31].Meng Q, Baumgartner C, Sinclair M, Housden J, Rajchl M, Gomez A, Hou B, Toussaint N, Zimmer V, Tan J et al. , “Automatic shadow detection in 2d ultrasound images,” in Data Driven Treatment Response Assessment and Preterm, Perinatal, and Paediatric Image Analysis. Springer, 2018, pp. 66–75. [Google Scholar]
- [32].Meng Q, Sinclair M, Zimmer V, Hou B, Rajchl M, Toussaint N, Oktay O, Schlemper J, Gomez A, Housden J et al. , “Weakly supervised estimation of shadow confidence maps in fetal ultrasound imaging,” IEEE Trans. Med. Imag, vol. 38, no. 12, pp. 2755–2767, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yasutomi S, Arakaki T, Matsuoka R, Sakai A, Komatsu R, Shozu K, Dozen A, Machino H, Asada K, Kaneko S et al. , “Shadow estimation for ultrasound images using auto-encoding structures and synthetic shadows,” Applied Sciences, vol. 11, no. 3, p. 1127, 2021. [Google Scholar]
- [34].Grady L, “Random walks for image segmentation,” IEEE Trans. Pattern Anal. and Mach. Intell, vol. 28, no. 11, pp. 1768–1783, 2006. [DOI] [PubMed] [Google Scholar]
- [35].Boykov Y and Kolmogorov V, “An experimental comparison of mincut/max-flow algorithms for energy minimization in vision,” IEEE Trans. Pattern Anal. and Mach. Intell, vol. 26, no. 9, pp. 1124–1137, 2004. [DOI] [PubMed] [Google Scholar]
- [36].Laine S and Aila T, “Temporal ensembling for semi-supervised learning,” arXiv preprint arXiv:1610.02242, 2016. [Google Scholar]
- [37].Tarvainen A and Valpola H, “Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results,” arXiv preprint arXiv:1703.01780, 2017. [Google Scholar]
- [38].Yu L, Wang S, Li X, Fu C-W, and Heng P-A, “Uncertainty-aware self-ensembling model for semi-supervised 3D left atrium segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent. Springer, 2019, pp. 605–613. [Google Scholar]
- [39].Kendall A and Gal Y, “What uncertainties do we need in bayesian deep learning for computer vision?” arXiv preprint arXiv:1703.04977, 2017. [Google Scholar]
- [40].Li X, Yu L, Chen H, Fu C-W, and Heng P-A, “Semi-supervised skin lesion segmentation via transformation consistent self-ensembling model,” arXiv preprint arXiv:1808.03887, 2018. [Google Scholar]
- [41].Li X, Yu L, Chen H, Fu C-W, Xing L, and Heng P-A, “Transformation-consistent self-ensembling model for semisupervised medical image segmentation,” IEEE Trans. Neural Netw. and Learn. Syst, 2020. [DOI] [PubMed] [Google Scholar]
- [42].Xia Y et al. , “3D semi-supervised learning with uncertainty-aware multi-view co-training,” in Proc. IEEE Winter Conf. Appl. Comput. Vis., 2020, pp. 3646–3655. [Google Scholar]
- [43].—, “Uncertainty-aware multi-view co-training for semi-supervised medical image segmentation and domain adaptation,” Med. Image Anal, vol. 65, p. 101766, 2020. [DOI] [PubMed] [Google Scholar]
- [44].Hinton GE, Srivastava N, Krizhevsky A, Sutskever I, and Salakhutdinov RR, “Improving neural networks by preventing co-adaptation of feature detectors,” arXiv preprint arXiv:1207.0580, 2012. [Google Scholar]
- [45].Srivastava N, Hinton G, Krizhevsky A, Sutskever I, and Salakhutdinov R, “Dropout: a simple way to prevent neural networks from overfitting,” J. Mach. Learn. Res, vol. 15, no. 1, pp. 1929–1958, 2014. [Google Scholar]
- [46].Glorot X and Bengio Y, “Understanding the difficulty of training deep feedforward neural networks,” in Proc. Thirteenth Int. Conf. Artif. Intell. and Statist. JMLR Workshop and Conference Proceedings, 2010, pp. 249–256. [Google Scholar]
- [47].Sonn GA et al. , “Targeted biopsy in the detection of prostate cancer using an office based magnetic resonance ultrasound fusion device,” J. Urology, vol. 189, no. 1, pp. 86–92, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Clark K et al. , “The cancer imaging archive (TCIA): maintaining and operating a public information repository,” J. Digit. Imag, vol. 26, no. 6, pp. 1045–1057, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wang J and Xia B, “Bounding box tightness prior for weakly supervised image segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent. Springer, 2021, pp. 526–536. [Google Scholar]
- [50].Zhang D, Zeng W, Yao J, and Han J, “Weakly supervised object detection using proposal-and semantic-level relationships,” IEEE Trans. Pattern Anal. and Mach. Intell, 2020. [DOI] [PubMed] [Google Scholar]
- [51].Zhang D, Han J, Cheng G, and Yang M-H, “Weakly supervised object localization and detection: A survey,” IEEE Trans. Pattern Anal. and Mach. Intell, 2021. [DOI] [PubMed] [Google Scholar]
- [52].Chen T, Kornblith S, Norouzi M, and Hinton G, “A simple framework for contrastive learning of visual representations,” in Int. Conf. Mach. Learn. PMLR, 2020, pp. 1597–1607. [Google Scholar]
- [53].Grill J-B, Strub F, Altché F, Tallec C, Richemond PH, Buchatskaya E, Doersch C, Pires BA, Guo ZD, Azar MG et al. , “Bootstrap your own latent: A new approach to self-supervised learning,” arXiv preprint arXiv:2006.07733, 2020. [Google Scholar]
- [54].Jing L and Tian Y, “Self-supervised visual feature learning with deep neural networks: A survey,” IEEE Trans. Pattern Anal. and Mach. Intell, 2020. [DOI] [PubMed] [Google Scholar]
- [55].Zhou Y, Chen H, Li Y, Liu Q, Xu X, Wang S, Yap P-T, and Shen D, “Multi-task learning for segmentation and classification of tumors in 3d automated breast ultrasound images,” Medical Image Analysis, vol. 70, p. 101918, 2021. [DOI] [PubMed] [Google Scholar]