Abstract
Artificial molecular machines have found widespread applications ranging from fundamental studies to biomedicine. More recent advances in exploiting unique physical and chemical properties of DNA have led to the development of DNA-based artificial molecular machines. The unprecedented programmability of DNA provides a powerful means to design complex and sophisticated DNA-based molecular machines that can exert mechanical force or motion to realize complex tasks in a controllable, modular fashion. This Perspective highlights the potential and strategies to construct artificial molecular machines using double-stranded DNA, functional nucleic acids, and DNA frameworks, which enable improved control over reaction pathways and motion behaviors. We also outline the challenges and opportunities of using DNA-based molecular machines for biophysics, biosensing, and biocomputing.
Keywords: Artificial molecular machines, DNA nanotechnology, DNA framework, Functional nucleic acids, Nanomedicine
1. Introduction
Natural cellular machinery is essential for the function and processivity of a wide variety of biologic processes, ranging from adenosine triphosphate (ATP) synthase at the nanoscale to cell recognition at the microscale.1−7 In general, machinery is composed of a multitude of functional components working in tandem.8 These primitive biomolecules, in particular nucleic acids and proteins, exert mechanical force or motion in response to certain external stimuli (signal input). Inspired by these natural biomolecular machines, many researchers have repurposed artificial biomolecules to engineer artificial molecular machines that execute a list of functions and accomplish complex tasks in a controllable, modular manner.9−12 These studies lead to not only advancements in the creation of synthetic cells and lifelike entities but also insights into the structure and functionality of artificial molecular machines that have broad implications for a variety of fields ranging from fundamental biology to biomedical applications, including biomedical engineering, biosensing, and theragnostics.
Since the 1980s, a variety of chemicals and materials, including small organic molecules, nanoparticles, proteins, RNA, and DNA, have been utilized to construct diverse artificial molecular machines.13−16 As an exquisite illustration, Feringa and colleagues created organic molecule-based molecular machinery with a unidirectional rotation that performs complex tasks utilizing chemical fuels, light, or electrochemical react ions, as recognized by the 2016 Nobel Prize in Chemistry.17−20 Alternatively, DNA has also widened our vision and facilitated the development of artificial molecular machines.21−24 In particular, since Seeman’s pioneering work in the early 1980s, rapidly growing structural DNA nanotechnology has opened up new possibilities for the building of static and dynamic 2D and 3D DNA-based molecular machines (DMMs), that are built from exquisite architectures of DNA sequences and achieve mechanical operation by DNA in a dynamic, controllable, modular manner.25−27 Due to the flexible backbone of ssDNA and the rigid framework of double-helical DNA domains, the local stiffnesses of DNA nanostructures can be programmed and coupled to fit a wide range of structural requirements for each particular molecular machine design.28 Meanwhile, by exploring DNA’s highly predictable assembly qualities and the programmability of its hybridization processes, DNA offers a path for programming and controlling complicated motion or behavior at the nanoscale. In addition, the diverse kinds of DNA secondary structures found in nature, such as G-quadruplexes and i-motifs, offer an exquisite approach to controlling the functionalities and behaviors of DMMs.29,30 Thus, DNA rapidly evolved into this booming field of artificial molecular machine construction, closely resembling natural protein motors in terms of nanoscale size.31
With the development of DNA nanotechnology, initial research focused on creating and fabricating DMMs.32 To develop advanced applications, these DMMs must operate at the interface rather than in the liquid. The supporting interface, which is typically where biological systems or cascade-triggering events are integrated, would enable evaluating its functionality, such as direction control and programmability.33 Specifically, because unique DNA can be programmed in a variety of ways, localized motors on the surface are far more likely to be triggered by adjacent molecules or output signals, allowing for better control over reaction pathway direction. Moreover, once they are anchored to solid of soft surfaces, anchoring, addressability, and cooperative operation become critical, which provides a path to system complexity and considerable promise for the realization of functional DMMs.
In this Perspective, we focus on recent advances in DMMs. We first introduce the progress of DNA nanotechnology in the construction of DMMs and then describe methods for integrating DMMs at the interface as well as tactics for regulating motion and behavior. Following that, we provide a detailed review of recent reports on its biological application in biological rulers, biologic factories, biocomputing, and biosensing. Last, we discussed the future challenges and perspectives of this field.
2. DNA Nanotechnology-Enabled DMMs
2.1. DNA Nanotechnology
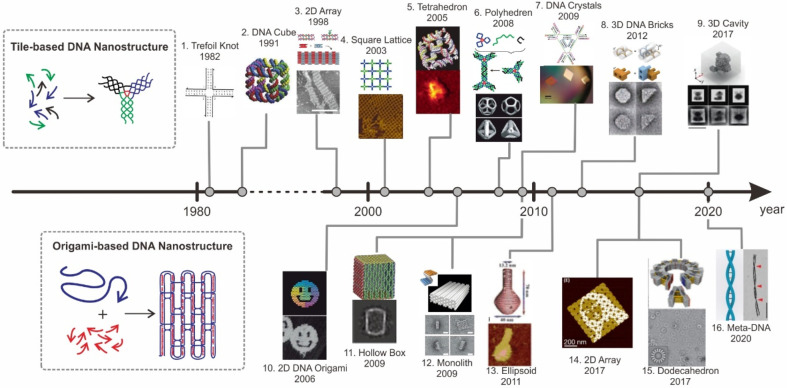
DNA is a lengthy natural biopolymer made up of four distinct bases: adenine (A), thymine (T), guanine (G), and cytosine (C), which constitute the basis for genetic information in living organisms.34−36 According to the Watson–Crick rule, the DNA strand hybridizes to its complementary sequences to form the B-form double helix structure, and the formed double helix structure has a diameter of 2 nm and a helix turn of 3.4 nm for every 10.5 base pairs. Seeman established in the 1980s that double-stranded DNA is also an ideal nanoscale building block via extremely sequence-specific hybridization.37−39 He suggested that arbitrary-shaped DNA nanostructures may be self-assembled by programming an immobile Holliday junction branching motif, and he succeeded in creating 2D and 3D DNA lattices beyond 1D linear double-strand DNA.40−43 Since then, advancements in structural DNA nanotechnology have enabled the fabrication of a range of intricate 2D networks and 3D DNA nanostructures, such as tetrahedrons and cubes, ushering in a new era of structural DNA nanotechnology (Figure 1).
Figure 1.
Timeline of tile-based and origami-based DNA nanostructures: (1) 2D DNA tiles including one-arm junction. Reproduced from ref (199). Copyright 2019, American Chemical Society. (2) DNA cube. Reproduced from ref (199). Copyright 2019, American Chemical Society. (3) Double-crossover (DX)-based alternating 2D array. Reproduced from ref (200). Copyright 2021, American Chemical Society. (4) 2D square lattice assembled with 4 × 4 DNA tiles. Reproduced from ref (199). Copyright 2019, American Chemical Society. (5) Tetrahedral DNA framework. Reproduced from ref (199). Copyright 2019, American Chemical Society. (6) Hierarchical polyhedral DNA framework. Reproduced from ref (199). Copyright 2019, American Chemical Society. (7) 3D DNA crystal self-assembled with a tensegrity DNA triangle motif. Reproduced from ref (199). Copyright 2019, American Chemical Society. (8) Tile DNA bricks-based 3D DNA framework. Reproduced from ref (199). Copyright 2019, American Chemical Society. (9) 3D DNA teddy bear. Reproduced from ref (201). Copyright 2021, American Chemical Society. (10) DNA-origami-based smiley face. Reproduced from ref (199). Copyright 2019, American Chemical Society. (11) 3D Hollow DNA box with a lid. Reproduced from ref (200). Copyright 2021, American Chemical Society. (12) 3D monolith with multiple pleated layers. Reproduced from ref (199). Copyright 2019, American Chemical Society. (13) 3D ellipsoid with complex curvature. Reproduced from ref (199). Copyright 2019, American Chemical Society. (14) Fractal assembly of DNA origami arrays with arbitrary patterns. Reproduced from ref (200). Copyright 2021, American Chemical Society. (15) Gigadalton-scale 3D dodecahedron. Reproduced from ref (200). Copyright 2021, American Chemical Society. (16) Double stranded meta-DNA. Reproduced with permission from ref (61). Copyright 2020, Nature Publishing Group.
Inspired by the sequence symmetry of the natural DNA-branched holiday junction, Seeman et al. created the first DNA nanostructure called double-crossover molecules (DX), laying the groundwork for DNA nanotechnology.37 The addition of single-stranded “sticky” ends to double-crossover molecules enables the creation of different tile-based DNA nanostructures, such as DNA cube, single-step formation of DNA tetrahedrons, hierarchical polyhedral DNA structures, and DNA branched junctions with 5-, 6-, 8-, and 12-arm double-helical arms surrounding a branch point.42−45 Following this remarkable development, a variety of DNA tiles were self-assembled into periodic 2D and 3D lattices with a proper sticky-ends design.40 Moreover, because the sizes of periodic 2D and 3D crystals are potentially infinite, rationally designed crystals with sizes of hundreds of micrometers were formed using man-made DNA tiles in 2009.46−48 Specifically, Seeman and co-workers created the first diffracting DNA crystal by employing a tensegrity rigid triangle motif with 3-fold rotational symmetry.
Furthermore, Yin et al. further introduced a chimeric conceptual form of DNA tile in 2008, dubbed single-stranded tile (SST), which is a 42-base-long single-stranded DNA.49 Rather than containing at least two or more DNA strands, the SST was composed of four concatenated sequences of nearly identical length. They were shown to assemble long tube circumferences by binding to their four local neighbors SST via their four concatenated motif. In 2012, they further demonstrated a conceptual extension of the SST, dubbed the ’DNA brick’, which is a 32-nt-long single-stranded DNA.50,51 By connecting to the four nearest neighbors in a square lattice arrangement of parallel helices, these DNA bricks were shown to form a variety of DNA nanostructures with 2D and 3D arbitrary shapes as well as crystal lattices with channels and pores.
In 2004, Paul Rothemund introduced a different approach, dubbed “scaffolded DNA origami”, which represents another major advance in structural DNA nanotechnology.52 In scaffolded DNA origami, a long single-strand DNA (ssDNA) oligomer was employed as a scaffold on which many DNA staple strands were stapled into desired forms in a one-pot method. This approach enables the construction of static DNA frameworks with predetermined sizes ranging from 50 to 500 nm with unprecedented precision and yield.53−57 Moreover, the local stiffness of a DNA origami nanostructure can be fine-tuned by programming its persistence length, allowing for the building of rigid and static designs. Furthermore, because these DNA staple strands are chemically generated with precise sequences, they can be changed and coupled to a variety of functional molecules, including enzymes, nanoparticles, and tiny compounds. Thus, the use of DNA origami nanostructures realizes the nanoscale control of molecular positioning and dynamic motion.
High-order hierarchical and large-sized DNA-framework constructions have become achievable by the stepwise assembly of DNA frameworks.46,50,51,58 One major approach to scaling up the DNA framework is the sticky end hybridization method, in which each DNA origami edge is extended with complementary ssDNA for more on-demand hybridization. Qian et al. used this process to generate larger 2D DNA origami 10 × 10 arrays with numerous square origami tiles, reaching 880 × 880 nm2. Following that, she then developed a “fractal assembly” with a three-stage self-assembly approach to produce a sophisticated DNA origami array with a maximum resolution of 8704 pixels and a size of 3.5 × 4.5 μm2.59 In addition, blunt end stacking was a sophisticated alternative method for scaling up DNA nanostructures. Similar to the key-and-lock concept, blunt end stacking is based on several DNA nanostructures docking in complementary shapes. As proven by Dietz et al., a microscale 3D rhombohedral nanostructure up to 2.5 μm in size has been successfully built.60 Furthermore, Our group introduced M-DNA, a six-helix DNA origami bundle as a basic building block, to assemble a scaling-up DNA framework with sizes up to sub-micrometers and micrometers.61 Research on the scaling up of DNA frameworks has enabled the development of resilient hierarchical DNA architectures.
2.2. DMM Construction
The booming advancements in structural DNA nanotechnology have attracted worldwide interest in harnessing DNA’s extraordinary architectural precision. Nature harnesses the modular framework to integrate numerous functions into natural molecular machines and perform complex tasks in tandem. This principle inspired the utilization of framework nucleic acids (FNA), including double-stranded DNA and static DNA frameworks, to build integrated and functional DMMs.62,63 By exploiting the programmability of structural DNA frameworks, the FNA enables the precise spatial arrangement of oligonucleotides in the modular DNA framework at the nanoscale. Moreover, owing to the advancements in the chemical synthesis of DNA, a variety of chemical groups, including fluorescent dyes, chemical-stimulus responsive chemicals, proteins, and nanoparticles, can be site-specifically modified at the ends of oligonucleotides.64−66 Furthermore, the intrinsic biocompatibility and biodegradability of FNA have made it a perfect biomaterial for programming DMM’s logical response to various chemical stimuli.67 Thus, the FNA features as an ideal molecular template for spatially organizing various functional molecules and materials in vitro and in vivo, presenting a promising strategy for developing DMM with integrated functions.
An interesting extension of this research is to transform FNA into a molecular machine.68,69 In 1999, Seeman and colleagues leveraged the sequence specificity of DNA and demonstrated the concept of DMMs.70 They developed dynamic DMMs capable of performing similar motions, as they demonstrated using the B-Z transition of DNA to switch the conformations of two stiff DNA “double-crossover” molecules. Hence, the incorporation of stimuli-responsive components into static DNA nanostructures may enable the generation of a range of sophisticated nanomechanical motions. Hereby, we focused on introducing several reconfigurable elements that have been incorporated into the DNA frameworks (Figure 2).
Figure 2.
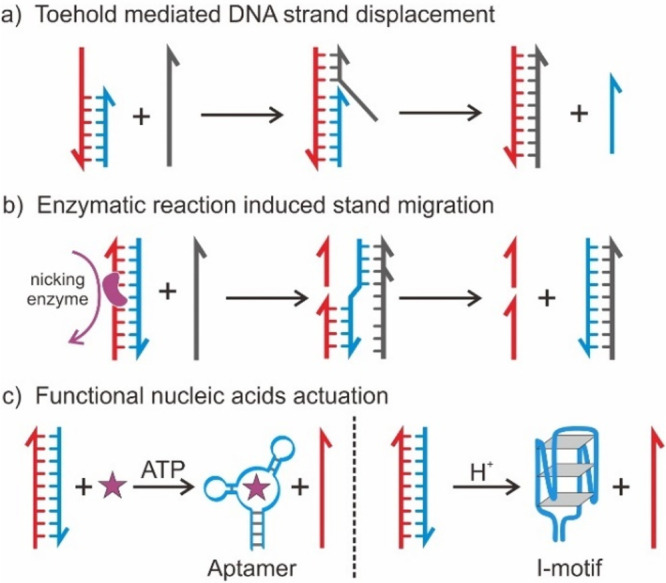
Reconfigurable elements and strategies for building DMMs. (a) Toehold mediated DNA strand displacement. (b) Degradation of DNA strands. (c) Special DNA motifs or target binding aptamers that undergo a conformational switch in response to environmental cues.
Much of the effort in engineering dynamic DMMs focused on integrating the Toehold mediated DNA strand displacement (TMSD) element into static DNA nanostructures, which was established by Yurke et al. in 2000.71 The TMSD typically begins with an ssDNA coupled to a partly dsDNA in its unhybridized domain. As a result of this process, the ssDNA undergoes branch migration to completely exchange the complementary ssDNA from the dsDNA.72 They used this technology to design a pair of DNA tweezers composed of two DNA helices and used an auxiliary strand of DNA to open and close them, resulting in the successful fabrication of a reversible DNA motor (Figure 3a).71 Recently, the TMSD approach has been used to reconfigure DNA origami nanostructures and lead to the creation of various complex and hieratical DMMs, such as nanoboxes, nanotweezers, and nanotubes. Moreover, by utilizing ssDNA as a fuel source, this strategy opened up new means for the building of processive DMMs with real-time coordinated motion. These accomplishments pave the way for the development of a library of programmable DMMs with modular and kinematic joints akin to those seen in macroscopic machines. Han et al., for example, used the TMSD approach to induce structural change in the DNA origami Möbius strip, a topological ribbon-like structure with a single side (Figure 3b).73 Using the TMSD approach, the DNA Möbius strip can generate about the double-sized topological supercoiled ring and catenane structures. Owing to its robustness in a wide range of temperatures and multiplexed nanosystems, the TMSD approach has been one of the most commonly used and established methods for controlling the motion of dynamic DNA nanostructures and DMMs.74
Figure 3.
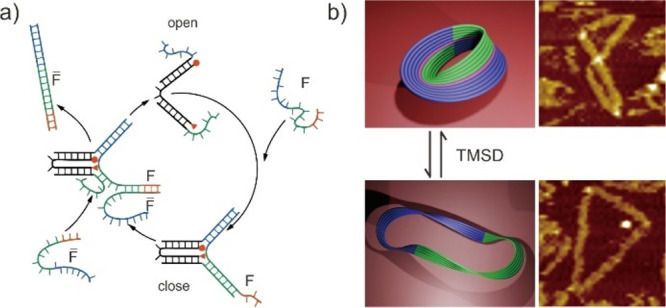
Toehold mediated DNA strand displacement (TMSD). (a) Schematic images of the reversible DNA tweezers by TMSD. Reproduced with permission from ref (71). Copyright 2000, Nature Publishing Group. (b) Schematic and AFM images of DNA origami Möbius strip. Reproduced with permission from ref (73). Copyright 2010, Nature Publishing Group.
Furthermore, enzymatic reactions, such as cleavage, degradation, and extension, are another versatile approach for manipulating reconfigurable components in DMMs. Rather than relying on strand displacement, this approach uses a variety of enzymes, including nuclease, DNAzyme, and polymerases, to trigger the release or capture of DNA oligomers, thereby driving DMMs to perform mechanic motion tasks.75 Because an enzymatic process is deemed autonomous and irreversible until no substrate is available, this technique is most frequently used to build a DNA walker, which typically comprises three essential complements: driving force, walking foot, and DNA track. Reif et al. reported the first enzymatic reaction-mediated DNA walker.76 Its walking strand, in particular, contains a short DNA fragment that connects to two distinct types of footholds that are attached at regular intervals along the DNA track. When the walking strand hybridizes with the foothold, a nick is formed, exposing the enzyme and initiating DNA cleavage. To ensure the DNA walker moved in a directed fashion, they employed two nicking enzymes, PflM I and BstAP I, and destroy the restriction sites of the DNA. Following this cycle, the walker proceeds along with the foothold until he reaches the track’s conclusion.
The concept of enzymatic reaction induced strand migration for DNA walkers was further developed by incorporating the DNAzyme system, which catalyzes the cleavage of a ribonucleotide phosphodiester link in ssDNA via a transesterification reaction.77 Due to DNAzyme’s superior performance with quicker strand displacement rates, the DNA walker displayed enhanced mobility but decreased attachment stability to shorter legs, driving the DNA walker moving along the DNA track. In 2005, Mao et al. demonstrated that the walking strand can be driven by DNAzyme cleavage of a specific RNA sequence (Figure 4a).78 Moreover, the motility and processivity of DNAzyme-based DNA walkers can be programmed via various parameters, including the length of the substrate-binding legs, the track type, and the DNAzyme structure. Willner et al. recently demonstrated the incorporation of a mixture of origami tiles into Zn2+- and/or Pb2+-dependent DNAzymes in order to program the operation of transformations in the origami nanocavities’ nanoholes (Figure 4b).79 Since DNAzyme is amenable to combinatorial selection, the incorporation of DNAzyme into the DNA motor not only broadens the range of work conditions available to the protein enzyme but also introduces a novel technique for DMMs with designable landscapes and programmable behavior.
Figure 4.
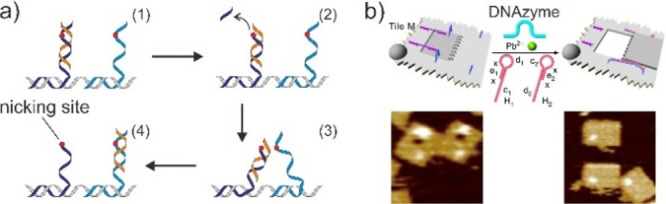
Enzymatic reaction induced strand migration. (a) DNAzyme powered the DNA walker along the track. Reproduced with permission from ref (78). Copyright 2005, John Wiley & Sons, Inc. (b) Schematic and AFM images of active generation of nanoholes in the origami scaffolds using DNAzymes. Reproduced with permission from ref (79). Copyright 2019, Nature Publishing Group.
In addition, functional nucleic acids, such as DNA triplex, aptamer, G-quadruplex, or i-motif, can be used as a reversible functional module or actuator in the construction of dynamic and sophisticated DMMs.80−83 The conformational transition of functional nucleic acids generates the mechanical force, enabling to drive the internal movement of the DMMs’ individual static nanostructures. Moreover, the incorporation of functional nucleic acids into dynamic DMMs enables unique controllability in response to a variety of ambient chemical stimuli, including pH, ATP, light, and Na+, as well as external physical cues, including light, thermal, electric, and magnetic fields. For example, Kuzuya et al. developed a DNA origami plier with shape transitions between closed and open states (Figure 5a).84 By incorporating different elements that bind together in the presence of the target into each of the levers, the DNA origami plier can be used as single-molecule beacons to visually detect a variety of biomolecules, including streptavidin, IgG, Ag+, and miRNA, via atomic force microscopy (AFM). Using this similar mechanism, we also designed a DNA origami nanocaliper as a dynamic shape-resolved nanodevice for pH sensing at the nanoscale (Figure 5b).85 By integrating pH-responsive Triplex DNA into the two arms of the origami nanocaliper, it can change its shape in response to changes in the local pH and act as a readout for transmission electron microscopy (TEM) imaging.
Figure 5.
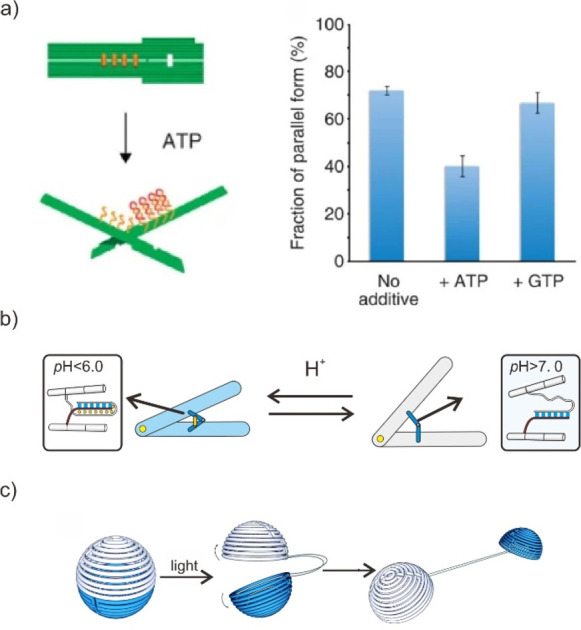
Functional nucleic acids actuation by chemical or physical cues. (a) Shape transitions of nanosized pinching DNA origami devices by a variety of chemical cues, such as ATP, Na+, Ag+, and miRNA. Reproduced with permission from ref (84). Copyright 2011, Nature Publishing Group. (b) DNA origami nanocaliper actuated through the incorporation of the pH-responsive i-motif sequence. Reproduced with permission from ref (85). Copyright 2022, Royal Society of Chemistry. (c) Schematic and TEM image of the light reconfigurable DNA origami sphere. Reproduced with permission from ref (87). Copyright 2015, Royal Society of Chemistry.
Furthermore, the light-induced conformational switch can also be employed to control the actuation of DMM. One major approach was to modify the DNA backbone with ortho-nitrobenzyl or azobenzene groups, which undergo isomerization when exposured to UV light.86 Particularly, the melting temperature of modified dsDNA were dominated by the photocleavage of ortho-nitrobenzyl or conformational switch of azobenzene-DNA, providing a means to regulate its association and dissociation with exposure to UV. For example, Han et al. demonstrated a structural reconfiguration of DNA origami sphere by modifying photocleavable ortho-nitrobenzyl between adjacent DNA helix (Figure 5c).87 The DNA origami sphere split into two hemispheres when exposed to light. Although light-based actuation is inefficient and slow, it has significant potential in biomedicine due to its noninvasive and conventional nature.
3. DMMs at the Interface
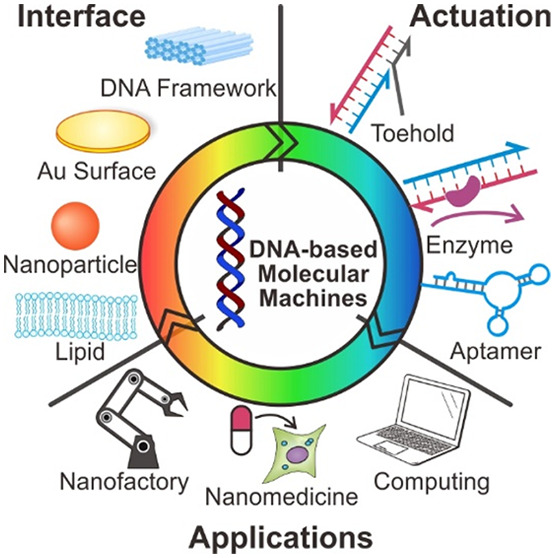
The incorporation of DMMs at the interface, rather than in the liquid, allows for the harnessing of nucleic acid structure and functioning. This approach further enables DMM to engineer the addressability and cooperative operation at the interface, enabling DMMs to achieve sophisticated tasks in a controllable, modular manner. The developments in solid-phase chemical synthesis of ssDNA have provided a convenient means for the synthesis of DNA with customized functionality.88−92 Specifically, diverse functional groups or domains can be modified at multiple sites of ssDNA, particularly the 5′- terminal and 3′- terminal of the ribose backbone. This customized DNA further enables the design of DMMs with unique functionality and additional properties, such as label attachment and surface anchoring. With these achievements in DNA modification and structural DNA technology, DMMs have been incorporated onto different solid and soft surfaces, which is a promising step toward realizing DMMs with controllable function (Figure 6).
Figure 6.
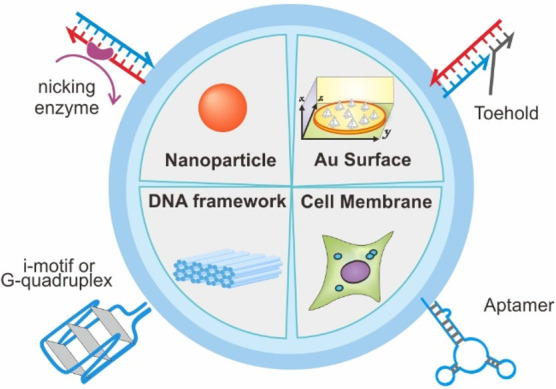
Substrates for DMM surface attachment. Harnessing the unique functionality and additional properties of DMM by immobilizing them at various interfaces.
3.1. Nanoparticle
The physicochemical features of nanoparticles include size-dependent optical and electrical properties.93−95 The anchoring of DMMs onto the surface of nanoparticles results in the creation of innovative hybrid systems, which allow to incorporate the advantageous properties of nanoparticles and DMMs. This endows DMMs with novel capabilities and functionalities, such as addressability and in vivo stability.96−99 One of the most impressive demonstrations of these DMMs on the gold nanoparticle (AuNP) surface is the AuNP-based core–satellite like nanosystem by Chan et al. (Figure 7).100 This nanosystem consists of a core nanoparticle surrounded by small satellites, and its conformation can be transformed in response to DNA via a toehold displacement mechanism, resulting alteration of its optical properties and biological interactions of the assembled nanosystem to the cell. Specifically, the DNA monolayer formed on the AuNP’s surface regulates the configuration of AuNP assemblies, enabling the assembly of shape-shifting DMMs with controlled biological functions.
Figure 7.
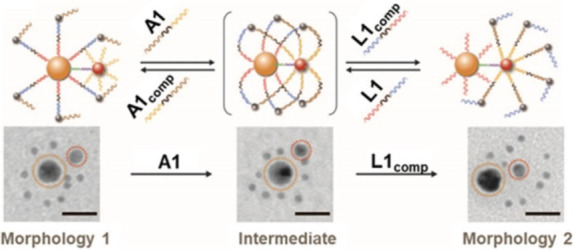
Harnessing of property of nanoparticles by modifying DMMs at their surface. Schematic illustration and corresponding TEM images of the shape change of nanoparticle assemblies mediated by DMM. Reproduced with permission from ref (100). Copyright 2016, The American Association for the Advancement of Science.
3.2. DNA Framework
For DMMs, nanoscale function control is confined to stabilizing various preprogrammed states, rather than operating the device directly and continuously with applied force, such as electronic and magnetic fields or directional flow. Direct real-time manipulation of DMMs with high spatial precision, subsecond response times, and customizable applied forces is a critical task for creating them. Apart from being used to build artificial molecular machines, DNA frameworks can also be employed to build tightly coupled DMMs. By precisely programming the DNA origami landscape and operation units of molecular machines, the incorporation of DMMs at the interface of DNA frameworks provides a promising means to programming its motion, in which the position or orientation of DMMs changes over time. The control of the motions enables a variety of functions, such as spatial manipulation of functional molecules, logic circuits, parallel processing, and signal transduction. Thus, the utilization of FNA permits the creation of highly ordered and tightly programmable surfaces with directional motion, and exceptional structural and functional programmability.
For the DMMs, translational motion and rotational motion represent the fundamental directional motion which lays the foundation for their different complex motions and behaviors.101,102 Translation, in which an object moves in a given direction, is the first motion achieved in the research of DMM. One of its remarkable examples is represented by the DNA walker, which was demonstrated by Reif et al. and Pierce and Shin in 2004.76,103 The DNA walker mostly consists of two components: the DNA walking strand and the DNA track. Typically, the walking strand continuously undergoes a nanoscale change of the position along the DNA track, similar to the DNA polymerase along with the DNA template. The continuous locomotion of the DNA walker is realized by transforming a variety of energy to disrupt the initial equilibrium of DNA interaction and progress down the DNA track. This walking mechanism endows DNA walkers with the fundamental properties of molecular machines, such as progressivity, processivity, and directionality.104,105 Since the pioneering work of Reif et al., most researchers designed bipedal walkers that used toehold-mediated strand displacement to remove the linker between the walker foot and track. To actuate the molecular machine, the second set of DNA linkers was added to the system and its hybridization resulted in the detachment of the foot and pairing to the next available foothold.
With the development of structural DNA nanotechnology, DNA origami offers opportunities to design and preprogram DNA walkers on the customized track with arbitrary shapes and structures. By exploiting the addressability of DNA nanostructures, different DNA walkers with autonomous motion over linear, 2D and 3D DNA ’tracks’ have been achieved. As a remarkable example, Yan et al. programmed the directional movement of DNA walkers by precisely defining the track on the DNA origami (Figure 8a).106 To ensure the walking strand anchoring on the surface track and moving route in the desired route, the walking strand was further designed with three DNA feet. Since each DNA foot moved independently along the DNA track, the cooperation of these three DNA feet would avoid the walking strand to be released from the origami DNA interface. Once actuated by the cleavage of foothold on the DNA track, the programmed translational motion is achieved by the recycled binding and cleaving of these legs. Meanwhile, Walter et al. optimized DNA walker with cartwheeling movements along the track on the DNA origami surface. This resulted in a significantly enhanced reaction speed of hybridization reactions, approximately 1 s–1 for a stepping rate constant, which is 10–100 times faster than that of previous DNA walkers (Figure 8b).107 This facilitated hybridization reaction further ensures the DNA walking strand moving along the DNA track.
Figure 8.
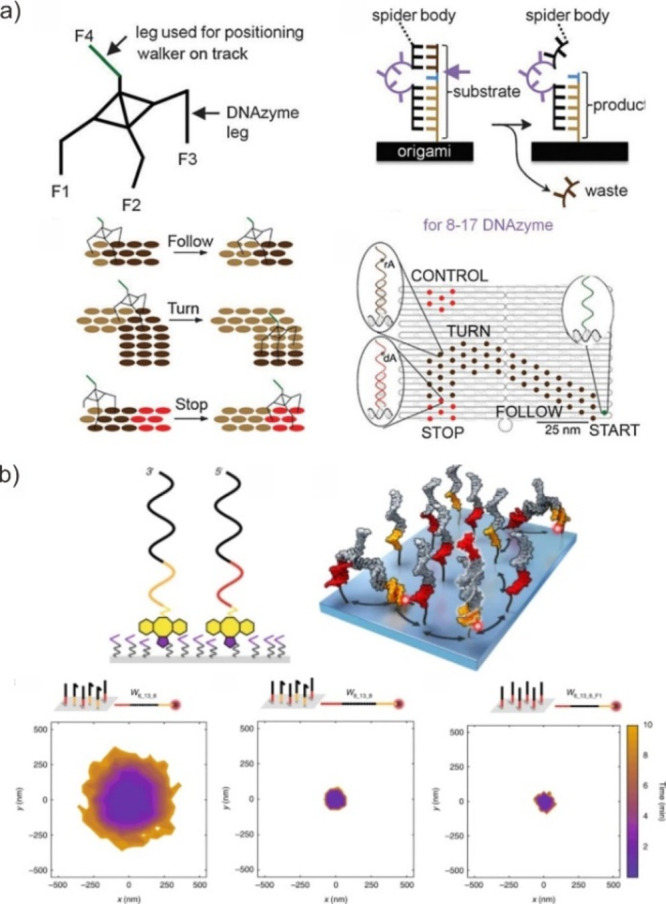
Translational movement at the DNA origami surface. (a) The walker directionally moves on a DNA origami track, hydrolyzing the complementary footholds. Reproduced with permission from ref (106). Copyright 2010, Nature Publishing Group. (b) Exploring the speed limit of toehold exchange on a DNA origami track. Reproduced with permission from ref (107). Copyright 2018, Nature Publishing Group.
In addition, interlocked-molecule-based DMMs, such as catenanes and rotaxanes, also have attracted interest, due to their potential as molecular cargo shuttles to transport cargo along a predetermined track.108,109 For instance, Lin et al. engineered a DNA-origami-based rotaxanes with programmable transitional motion.110 Meanwhile, Simmel et al. created a large, rigid rotaxane using DNA origami, and a long-range transitional motion of up to 355 nm was realized.111 Therefore, not only do the nanostructure surfaces topologically arrange these supported DMMs in the desired pattern and route, but also they enable nucleotide-level manipulation of their structure and function, allowing for the performance of specific tasks at the nanoscale by these well-controlled DMMs.
In contrast to translational motion, rotational motion means circular movement around a fixed point. For DMMs, it generally operates by cycling through several multiple preprogrammed states in the response to energetic stimulation or the availability of a suitable fuel. To precisely control the specific configuration of DNA rotary machines, the pivot point for working components should be fixed on the anchoring surface. Dietz et al. constructed a fully synthetic DNA origami with different multilayers consisting of a rotor unit, which is powered by thermal fluctuation. The rotor has a hexagonal cross section and features a crank-lever-like axial protrusion and a body element that forms an axle bearing. The apparatus with greater structural complexity than previous mechanically interlocked objects reproduce some of the dynamic properties of the F1–ATP synthase motor and features a well-defined angular degree of freedom without restricting the range of rotation (Figure 9a).112 This system inspired new types of single-molecule assays by installing receptors in the bearing cavity and their ligands on the rotor due to the modularity. Moreover, Murata et al. further designed a thermal fluctuation powered DNA nanostructure with a rotor immobilized onto a substrate surface to evaluate its stepping operation (Figure 9b).113 Unlike the rotary motor presented by Dietz, this motor has a dynamic state (in which the rotor rotates randomly) and a static state (in which the rotor is stationary) at predetermined angles. Meanwhile, these states can be selected by a signal DNA strand given from the outside. These intrinsic DMMs demonstrate the importance of the surface to the controllability of the rotational motion.
Figure 9.
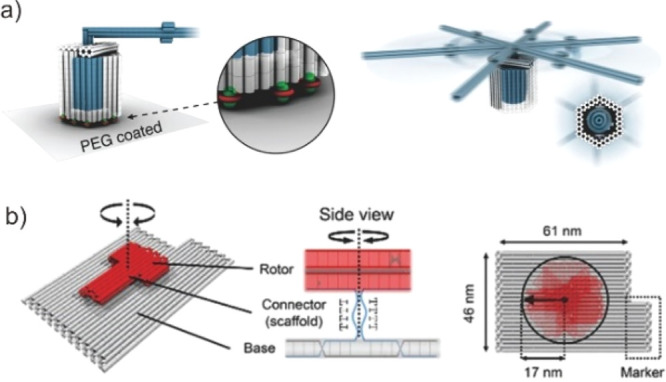
Rotational movement at the DNA origami surface in the absence of an energy source. Fully assembled trimeric rotor apparatus with closed brackets and a docked rotor. Reproduced with permission from ref (112). Copyright 2016, The American Association for the Advancement of Science. (b) Schematic illustration of the three parts. Colors: base (gray), rotor (red) and connector (blue). Reproduced with permission from ref (113). Copyright 2017, Royal Society of Chemistry.
In addition, the direct real-time manipulation of DNA nanodevices with exact spatial resolution, subsecond response times, and customizable applied pressures is a critical task for mechanical robot engineering. And this is a critical step toward enabling practical robotic systems at the molecular level using DMMs. Inspired by the design of macroscopic motors, starting with isolated joints for angular or linear motion, anchoring the pivot point for working components on the DNA nanostructure’s surface enables fine control of the DNA rotary motor’s particular configuration. Meanwhile, these rotaries can be manipulated using a variety of forces, including chemical forces associated with DNA displacement and electromagnetic forces.114,115 Simmel et al. used a computer-controlled electrical field to precisely switch the arm between arbitrary positions on the platform within milliseconds (Figure 10a).116 The DMM is at least 5 orders of magnitude faster than that previously reported for the fastest DNA motor systems and comparable to naturally occurring adenosine triphosphatase-driven biohybrids. Moreover, Castro et al. used magnetic force to control the specific motion of a rotary via a magnetic bead conjugated robotic arm by connecting a stiff microscale mechanical lever to the DNA nanostructure interface (Figure 10b).114 Its conformational change as well as the moment generated by the magnetic force can be visualized directly. Furthermore, Rant et al. demonstrated persistent orientation switching in a model DNA rotary that is end-tethered to a metal electrode and actuated by external voltages.117 They discovered that applying positive potentials to the surface facilitates selective immobilization of DNA nanostructures, allowing for the attachment of origamis in the presence of an abundance of staple strands from the reaction solution. These studies demonstrate that the DNA nanostructure surface paves the way for the development and characterization of a library of tunable DNA origami kinematic joints and their application in more complex controllable mechanisms resembling macroscopic machines.
Figure 10.
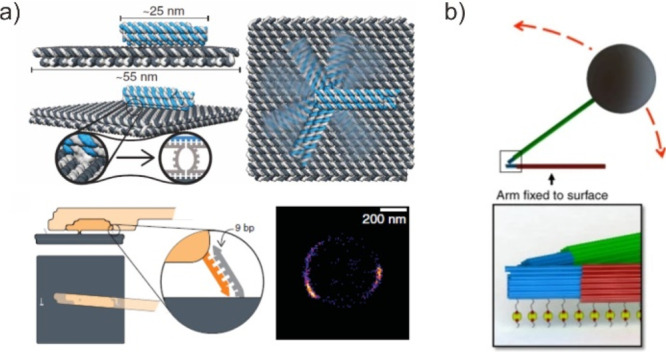
Harnessing the programmability of DMM at the surface of the DNA framework. (a) Sketch of the DNA origami structure inside (top left) and top (right) view. Reproduced with permission from ref (116). Copyright 2018, The American Association for the Advancement of Science. (b) Schematic of a nanorotor, in which two microlever arms are attached on both sides of the nanorotor arm. Reproduced with permission from ref (114). Copyright 2018, Nature Publishing Group.
3.3. Cell and Lipid Membrane
Micelles, vesicles, and membranes derived from lipids provide a unique surface for anchoring biomolecules, allowing functionally different compartments to be separated from one another. The lipid bilayer is characterized with a dynamic surface. The advantages of the lipid bilayer are its ease of preparation, its ability to build massive molecular assemblies (up to the micrometer scale), and the embedded components’ mobility. Hybrid systems composed of DNA nanostructures and lipids have been designed to exploit the physicochemical properties of membrane assemblies.62,118 Moreover, organizing DMMs within membranes reduces their dimensionality and enables them to interact more efficiently than when they are organized in three dimensions.119−123
To simulate intracellular communication and transport in cells, DMMs can be attached to lipid bilayer membranes via electrostatic attraction or by functionalization with membrane-interacting molecules, simulating membrane-associated proteins. In general, de novo construction of DNA nanostructures up to tens of nanometers in length is comparable to the natural proteins and polypeptides, such as tiny component sizes and short persistence. Existing DNA nanopores are mostly constructed using parallel-aligned DNA duplexes with optional curved sections serving as scaffolds. The scaffolds, in particular, contain lipid anchors such as cholesterol, porphyrin, or tocopherol, either on their membrane-spanning outer pore wall or in places next to the bilayer head groups, allowing for a secure attachment to the lip surface. A porphyrin-functionalized DNA-based hexagon was one of the first examples of a DNA nanostructure interacting with a lipid bilayer membrane. Simmel et al. used scaffolded DNA origami to produce the artificial membrane channel structure (Figure 11a).124 The bottom of the cap structure was functionalized with cholesterol to bind it to the membrane, allowing it to successfully penetrate lipid bilayer membranes during electrical recordings. In addition, Howorka et al. used DNA structure to develop an automictically determined molecular valve capable of regulating when and which cargo is carried across a bilayer (Figure 11b).125 The valve can bind a specific ligand and undergo a nanomechanical change in reaction, therefore opening the membrane-spanning channel. These DNA nanopores can differentiate with high selectivity the transport of small organic molecules that differ in their presentation of a positively or negatively charged group due to the narrow valve and DNA characteristic.126,127
Figure 11.
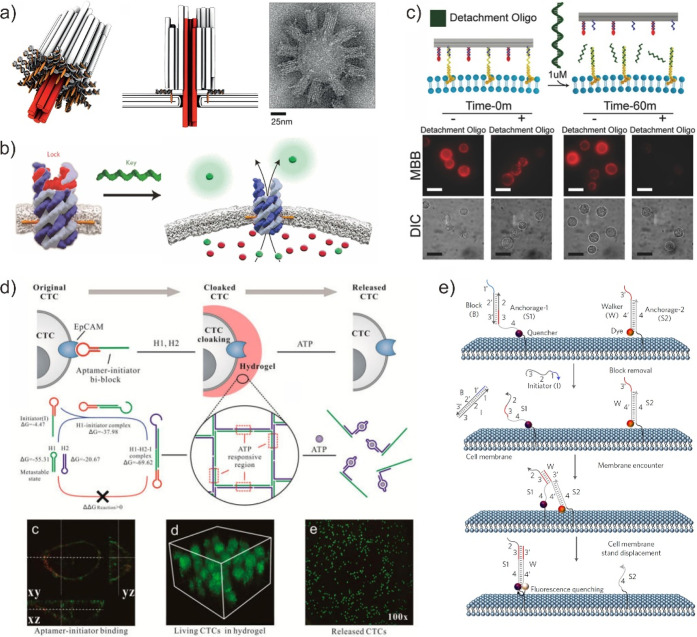
DMM-based synthetic molecular device and engineered cells. (a) DNA-based membrane channel. Reproduced with permission from ref (124). Copyright 2012, The American Association for the Advancement of Science. (b) A rationally designed DNA nanopore features a nanomechanical and sequence-specific gate to regulate transmembrane flux. Reproduced with permission from ref (125). Copyright 2016, Nature Publishing Group. (c) Detachment and hierarchical assembly of DMMs at the cell surface. Reproduced with permission from ref (128). Copyright 2017, John Wiley & Sons, Inc. (d) DNA gelation-based cloaking and decloaking of CTCs. Reproduced from ref (129). Copyright 2017, American Chemical Society. (e) Anchoring and operation scheme of DNA probe on the live cell membrane. Reproduced with permission from ref (130). Copyright 2017, Nature Publishing Group.
Another intriguing application for embedding DMMs on the cell surface is to engineer the function of the cell membrane. For example, Castro et al. used a DNA nanoplatform to build higher-order DNA assemblies on the cell surface and program cell–cell adhesion between homotypic and heterotypic cells via sequence-specific DNA hybridization (Figure 11c).128 This incorporation of DNA origami transforms the cell membrane into an engineered material capable of simulating, manipulating, and quantifying biophysical and biochemical functions found within plasma membranes of living cells. The robust membrane functionalization across epithelial, mesenchymal, and nonadherent immune cells further expands the enormous potential in biological engineering. For instance, our group demonstrated an aptamer-trigger clamped hybridization chain reaction (HCR)-based DMMs for in situ cloaking (or decloaking) of circulating tumor cells (CTCs) by forming porous DNA hydrogels (Figure 11d).129 DMMs containing aptamer-toehold biblocks specifically recognize epithelial cell adhesion molecule (EpCAM) on the CTC surface, initiating subsequent automated DNA polymerization (HCR) via toehold-initiated branch migration. DMMs also report biological events in living cells. For instance, Tan et al. demonstrated a novel DNA walker capable of transducing transient membrane encounter events into readable cumulative fluorescence signals (Figure 11e).130 The DNA walkers translocate from one anchor site to another via a toehold-mediated DNA strand displacement reaction. The DMMs revealed the rapid encounter events of membrane lipid domains and determined their rates and preferences during various live cell membrane signaling events.
3.4. Flat Surface
One of the primary goals of DMMs is to make them suitable for macroscopic applications. Although functional DMMs and a variety of other molecular machines have been developed recently and could provide a wealth of components for devices, the majority of these investigations have been conducted in solution. The difficult issue remains that when machines are fastened to a flat surface, their behavior may deviate dramatically from the behavior originally specified in the solution. For example, the motor may entirely cease to function. To overcome this issue, DNA nanostructures have been used to control the density of functional probe components fixed on the surface, hence enhancing molecular recognition capability at the surface.131−133 One of the most impressive demonstrations of a DNA switch on the macro surface is the DNA tetrahedral-based molecular pump (Figure 12a).134 This pump system was composed of a molecular scaffold, that could serve as the scaffold to ensure highly ordered orientation and spatial isolation of this nanomotor on a macroscopic gold surface, and a configurable motif within responsive edge. By incorporating a pH-sensitive i-motif sequence in the responsive edges, the proton-driven DNA tetrahedral machines can reversibly pump water and ferricyanide in response to environmental pH variation in a noncontinuous flow manner. In particular, it is probably even more intriguing to study water pumps of aquaporins, which are naturally existing proteins that channel water in and out of cells in an organized manner. Meanwhile, Liu et al. demonstrated a light-driven plasmonic DNA switch that can amplify the molecular motion of azobenzene through the host nanostructure and consequently translate it into reversible chiroptical function with large amplitude modulation.135
Figure 12.
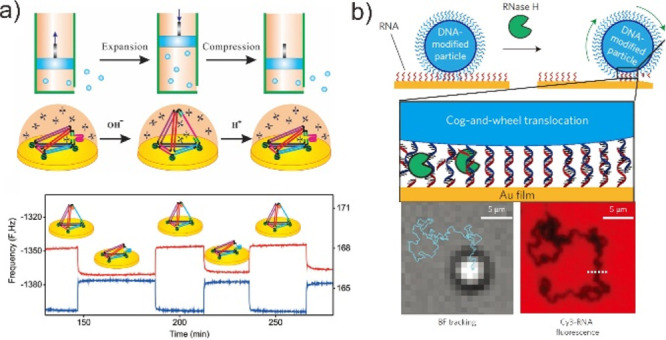
Incorporation of DMMs at the Au surface. (a) Scheme of the proton-driven, dynamic DNA switch with a DNA tetrahedral as a scaffold. This nanopump does not represent a continuous flow-type pump; instead it mimics a pump that can pump in and out via expansion and compression. Reproduced with permission from ref (134). Copyright 2016, John Wiley & Sons, Inc. (b) Approach used to generate RNA-fueled, enzyme-catalyzed autonomous DMMs. DNA-modified particles were hybridized into an RNA monolayer that presented a complementary strand. Particles were immobile until RNase H was added, which selectively hydrolyzes RNA duplexed to DNA. Reproduced with permission from ref (136). Copyright 2016, Nature Publishing Group.
Furthermore, by converting chemical energy into controlled motion, DNA walkers could be of use in applications on the macroscopic surface such as next-generation sensors, drug-delivery platforms, and biological computing. For example, Salaita et al. demonstrated a DNA walker made from DNA coated spherical particles that hybridize to a surface modified with complementary RNA (Figure 12b).136 The motion is achieved through the addition of RNase H, which selectively hydrolyzes the hybridized RNA. In particular, the DNA walker rolls rather than walks in a self-avoiding manner, and anisotropic particles can travel linearly without a track or external force. As the rolling mechanism of motion is highly sensitive to external stimuli, such as ATP and miRNA, the DNA walker represents an important step in bringing together the field of DNA-based sensing with the emerging area of DMMs, enabling one to engineer label free sensing assays. The demonstrated strategy exhibited excellent detection performancs for DNA analysis in a complex matrix such as human serum, which illuminated the practical application field of the sensing platform.
4. Applications of DMMs
Precision positioning of functional molecules is necessary for DMM to integrate and support multiple functions. By acting as a molecular landmark, the positional accuracy of DMM measurements at the nanoscale is a valuable tool for understanding biomolecular interaction at the interface. Moreover, the structural versatility and programmability of DNA nanostructures enable nanometer-precision positioning and coordination of the motions of diverse molecules at the interface, enabling the realization of advanced applications such as molecular synthesis, logic circuits, parallel processing, and signal transduction. DMMs could be used as imaging probes and drug carriers in living systems for nanomedicine, as one application. We have demonstrated extensive control over the motion of DMMs at the interface by constructing a biological network for biocomputing.
4.1. Biomolecular Ruler
To perform functions and regulate states, proteins in cells rely on their nanoscale distribution and rearrangement. Due to the precise positioning of multiple biological molecules, DMM has been used as a “molecular breadboard” to answer stoichiometric and quantitative questions about biomolecular interaction over the past decade.137−143 The DMM converts its structure or conformation into a biomolecular interaction or dissociation event, which is analyzed using force spectroscopy, by placing and arranging functional biomolecules in a predetermined pattern on addressable 2D and 3D DNA origami scaffolds. Probing protein assembly at the single molecular level is an exciting area of research. Dietz and Funke developed a V-shaped nanocaliper to study the nucleosome’s bimolecular force (Figure 13a).144 The nanocaliper was integrated with two nucleosomes in a defined relative orientation, and its hinge angle can be used as a force spectrometer in a manner analogous to force spectroscopy. Following that, the hinge angle of the nanocaliper was used as an easily visible output to amplify nanoscale molecular interaction and motion. Using nanometer-resolution TEM images, we determined the energy landscape underlying the frequency at which a particular hinge angle occurs, as well as nucleosome-nucleosome distances. Castro and colleagues took a similar approach in developing a DNA origami-based nanocapliper to study chromatin rearrangement in the presence of target DNA (Figure 13b).145 They established that the nanocaliper angle is an accurate measure of nucleosomal DNA’s end-to-end distance, allowing for the investigation of DNA protein interactions and chromatin conformational changes. Meanwhile, our group has integrated a pH-responsive Triplex DNA into this DMM to investigate pH variation on the nanoscale.85
Figure 13.
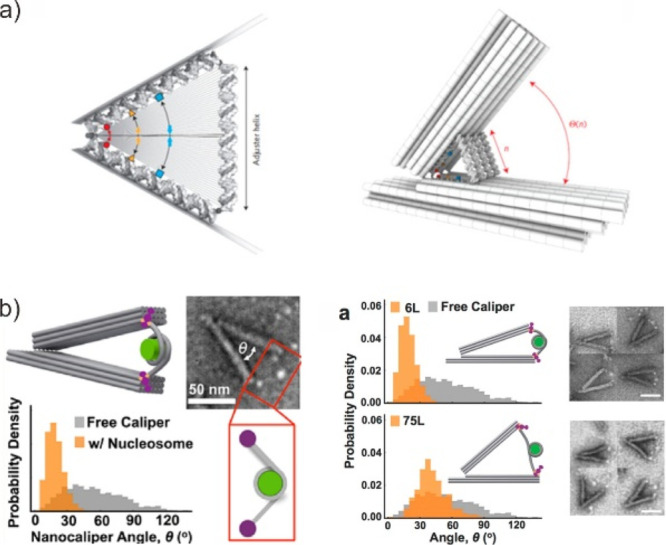
Probing biological questions with dynamic DMM. (a) DMM reveals the average distance between two molecules with atomic-scale precision by measuring the angle between the two structural units. Reproduced with permission from ref (144). Copyright 2016, Nature Publishing Group. (b) DNA origami nanocaplipers measure the transcription factor binding within the nucleosome. Reproduced from ref (145). Copyright 2016, American Chemical Society.
DMM has a high degree of structural rigidity and is easily visualized using direct imaging methods, such as AFM and TEM. It makes DMM uniquely suited for use in conjunction with high-resolution imaging techniques for understanding diverse subfields of biology at a single-molecule level.146,147 For instance, Zhuang et al. developed a fluorescent dye-labeled DNA origami rotor to quantify DNA rotation during various genome-processing reactions (Figure 14a).148 This DNA origami rotor featured four blades perpendicular to the rotation axis and a central dsDNA segment. The recBCD-induced unwinding of the DNA resulted in the spinning of the DNA origami rotor, which was tracked with millisecond time resolution at the single-molecule level. Moreover, using DNA origami as a “molecular landmark”, our group deduced the molecular mechanism by which protein nucleators mediate the formation of fibrils in cells (Figure 14b).149,150 We decorated the triangular DNA origami with CsgB, a protein that directs and accelerates the self-assembly of the CsgA monomer’s fibril subunit. We confirmed that CsgB accelerated fibril formation, but we also observed fibrils growing out from or toward the CsgB at the DNA origami using high-speed AFM. In a similar fashion, the distance-dependent binding of immunoglobulin Gs (IgGs) to epitopes was demonstrated by capturing its transient conformations on a triangular DNA origami with a programmed spatial distribution of epitopes (Figure 14c).151 Thus, DNA nanotechnology’s ability to amplify biomolecular motions sheds light on the molecular mechanism underlying nanoscale interaction.
Figure 14.
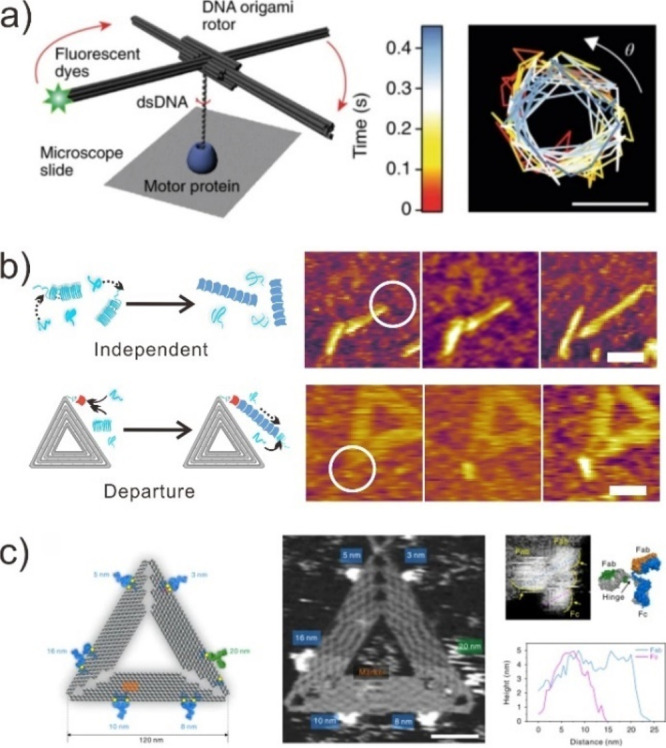
Single molecule chemical reactions are monitored by DMM. (a) Origami-rotor-based imaging and tracking (ORBIT) track the DNA rotations that result from unwinding by the RecBCD complex as well as from transcription by RNAP. Reproduced with permission from ref (148). Copyright 2019, Nature Publishing Group. (b) DNA origami-based DMM monitor the process of how protein nucleators mediate fibril formation. Reproduced with permission from ref (149). Copyright 2019, Nature Publishing Group. (c) DNA-origami-based DMM capture transient conformations of IgGs. Reproduced with permission from ref (151). Copyright 2020, Nature Publishing Group.
4.2. Biomolecular Factory
Combinatorial biosynthesis, such as mRNA-templated peptide synthesis during ribosomal translation, involves the use of protein or nucleic acid templates to direct reactions. This results in the selective generation of distinct multistep reaction products from a single set of substrates without the need for external intervention to modulate the effective molarities of substrates.152,153 However, the fabrication of nanoscale chemical species, such as nanoparticles and biomolecules, requires a continuous series of reactions. Specifically, due to thermodynamic and kinetic constraints, laboratory reactions for multistep synthetic products typically require multiple reactions using a variety of different substrates and reaction conditions. As they follow the track, DNA walkers change their physical location incrementally and autonomously over time. By simulating the biosynthesis process and protein, the programmable and controllable DNA nanostructure track can be used to template synthesize biomolecules.
The development of wholly synthetic DMMs with translational motion has been facilitated in particular by programmable DNA walkers with track lengths ranging from 20 nm to 1 μm, which provide an adequate track for multiple steps in synthesis. Moreover, the DNA motor can be programmed to transport a diverse range of chemical substrates, including RNA, aptamers, vaccines, and drugs. Thus, by combining the programming walking step with templated synthesis, these DMMs on the surface of the DNA nanostructures enable the walker’s arrival at each station to trigger a specific chemical reaction.154 Seeman et al., for example, demonstrate the use of a DNA walker to synthesize desired nanoscale chemical species of Au nanoparticles (Figure 15a).155 They connected individually selected nanoscale components in a stepwise and programmed manner on the track of a DNA origami surface. While the walker follows the track on the DNA nanostructure surface, the DNA motor encounters the three DNA devices sequentially and delivers its cargo to the desired location, promoting chemical reactions between attached moieties through proximity. Furthermore, Liu and He demonstrated an autonomous DNA walker capable of carrying out a series of amine acylation reactions while traversing a DNA track (Figure 15b).156 This system can synthesize multistep products at significantly higher yields than previous DNA-templated small molecule syntheses in a matter of hours.
Figure 15.
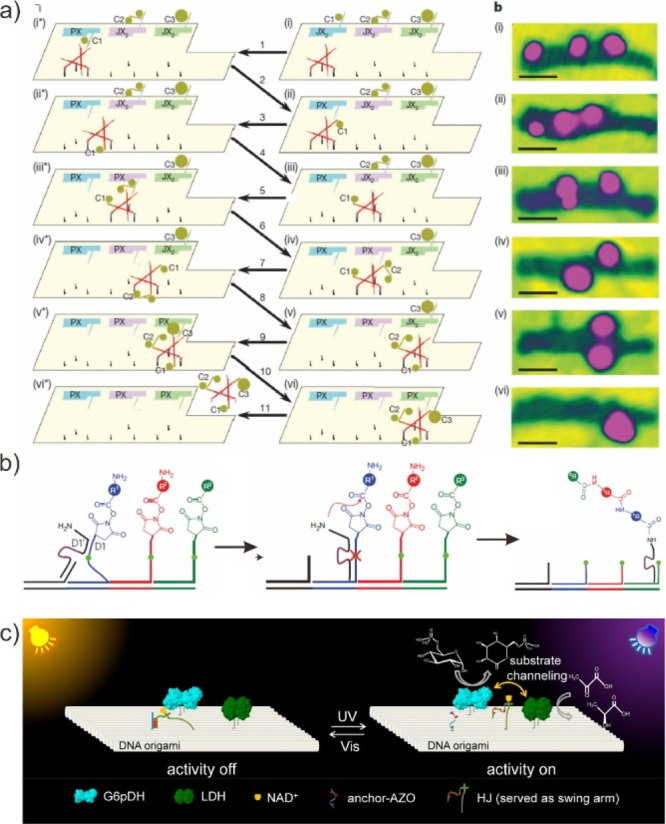
Autonomous synthesis with a DNA walker. (a) A DNA walker with “robotic arms” is capable of picking up different gold nanoparticles when traversing a programmed track with cargo-donating stations. AFM images show the change in nanoparticle arrangement at specific stages of the walker. Reproduced with permission from ref (155). Copyright 2010, Nature Publishing Group. (b) Sophisticated example of DNA-templated multistep synthesis programmed by a molecular walker. A DNAzyme-based walker modified with a primary amine is able to react with succinimidyl ester functionalized footholds to yield complex molecules. Reproduced with permission from ref (156). Copyright 2010, Nature Publishing Group. (c) Design and characterization of the enzyme pathway regulation system on DNA origami. Reproduced from ref (158). Copyright 2018, American Chemical Society.
It also has been demonstrated that DNA walkers regulate the function and performance of proteins and devices.157 Yang et al. and co-workers demonstrated a multienzyme synthetic system that enables precise and dynamic control of pathway activity (Figure 15c).158,159 They successfully and directly regulated the enzyme pathway with the aid of track by switching the active mediator between two enzyme pairs in a specific manner. Moreover, they demonstrated a novel photoresponsive DNA motor that converts energy light, a nonpolluting source of energy, to the motion of DNA through the incorporation of photoresponsive azobenzene molecules into DNA strands. The substrate channeling position and the interenzyme distance, which can be precisely controlled with light, dominated the performance of the enzyme cascade in this DNA walker.
4.3. Biocomputing
Unlike any other material system, DNA’s predictable and guided folding enables the manipulation of molecular-scale processes.160−163 Thus, DMMs can be combined with algorithms to create engineered and purposeful systems, such as computing networks, molecular sorting systems, and dynamic assembly lines that adapt to changing demands.164 However, solution-based DNA computing systems rely on sophisticated sequence design to ensure that hybridization reactions take place in the correct order. Specific sequence domains, in particular, are incompatible with reuse in other contexts, limiting the complexity and scalability of such systems. To overcome this constraint, several groups recently developed a DNA nanostructure equivalent of an electronics breadboard or scaffold. DMMs can be programmed and integrated into a variety of algorithms via a probe connected to the DNA nanostructure interface. Turberfield et al., for example, demonstrated the integration of long-range communication and information processing using a synthetic DNA-based system (Figure 16a).165 Moreover, Seelig et al. demonstrated that signal propagation transports across the logic gates and signal transmission lines on DNA origami.166 The path taken by a motor through a network of tracks with four possible routes can be programmed either externally or internally via instructions carried by the motor in this DNA walker.
Figure 16.
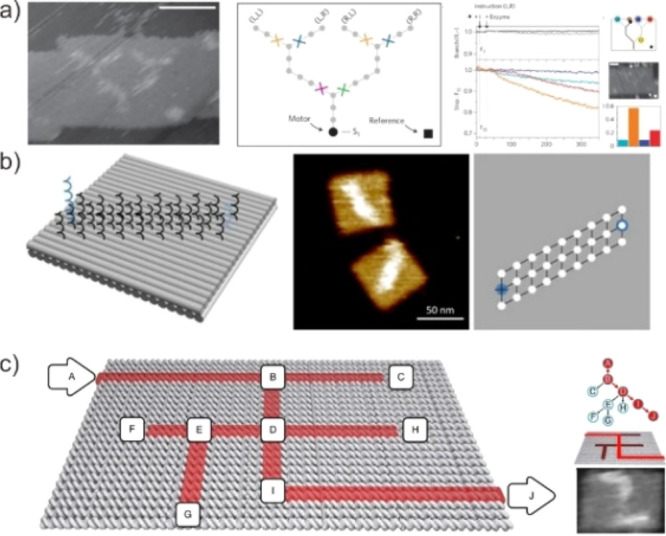
Biocomputing application of DMMs. (a) The DNA track network is assembled on a rectangular DNA origami substrate. Reproduced with permission from ref (165). Copyright 2012, Nature Publishing Group. (b) Random-walk building block and the cargo-sorting algorithm. Reproduced with permission from ref (167). Copyright 2017, The American Association for the Advancement of Science. (c) Maze design with ten vertices. Arrows indicate the entrance vertex A and the exit vertex J . Reproduced with permission from ref (168). Copyright 2019, Nature Publishing Group.
The integrated synthetic DNA walkers enable the development of molecular computing networks capable of sorting and processing cargo in accordance with the instructions carried by them. Qian et al. demonstrated this DNA walker using a computing network (Figure 16b).167 They developed a simple algorithm for sorting cargo in a DNA robot by adding a new building block for leaving pheromone-like signals along a path. DMMs, in particular, could be programmed to take the shortest path possible and efficiently transport cargo molecules. With more effort directed toward the development of modular and collective molecular robots, as well as simple and systematic approaches, molecular DNA walkers could eventually be easily programmed in the same way that macroscopic robots are, but for use in microscopic environments. Our group extended this work by developing a DNA motor system capable of making decisions and performing other types of information processing (Figure 16c).168 Specifically, this DNA walker employed the progression of hybridization chain reactions to perform a parallel depth-first search on a ten-vertex rooted tree defined on a 2D DNA nanostructure surface (HCR). Notably, the pathfinding mechanism of this DNA motor is based on a localized strand exchange cascade that initiates at a unique trigger site on the origami and proceeds automatically along paths defined by DNA hairpins containing a universal traversal sequence.
4.4. Biosensing
It has been demonstrated that incorporating stimulus-responsive functional molecules onto DMMs results in the formation of desirable DMMs capable of functioning in a living system.169−172 A specific stimulus can be applied as an input to the DMM to cause it to switch to a reconfiguration and function that have been predefined. These programmable dynamic DMMs can be used in biological solutions and living cells as sensors and nanomedicine. For example, Funck et al. developed a cross-shaped DNA origami containing chiral plasmonic signals for the detection of viral RNA (Figure 17a).173,174 The DNA origami was constructed using two DNA origami arms and a gold nanorod with a strong circular arrangement was attached to each arm. The addition of target nucleic acid sequences altered the configuration of DMM via TMSD, with modified gold nanorods arresting the DMM in the defined chiral state. Meanwhile, Douglas et al. developed a robotic boxlike DNA origami capable of transporting molecular payloads to the cell surface in response to a defined stimulus.175 The DNA origami is a hexagonal barrel formed by attaching two domains of single-stranded scaffold hinges at the rear. To secure the barrel noncovalently, an aptamer-based lock was integrated into the front of the stapes. When the aptamer-based locks were switched to an open state in response to specific antigens on the cell surface, the barrel would expose the inner payloads. Moreover, by tethering two gold nanorods (AuNRs) to each surface of two origami bundles with a reconfigurable DNA framework, Liu et al. created a reconfigurable 3D plasmonic rotary motor, which executes DNA-regulated conformational changes at the nanoscale (Figure 17b).176 Specifically, the addition of fuel DNA drives the metamolecules to distinct conformational states, resulting in transducing their conformational changes in situ into circular dichroism changes in the visible wavelength range simultaneously.
Figure 17.
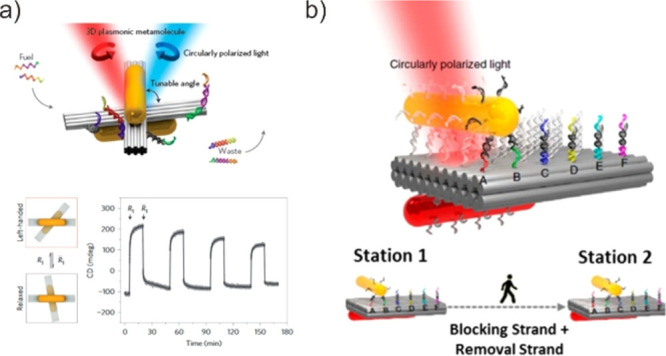
Biosensing application of DMMs. (a) Plasmonic nanostructures with dynamically controlled optical responses enabled by stimulus-driven DNA origami templates. Reproduced with permission from ref (173). Copyright 2014, Nature Publishing Group. (b) Plasmonic walker on DNA origami. Reproduced with permission from ref (176). Copyright 2015, Nature Publishing Group.
In 2009, Krishnan et al. developed an i-motif DNA-based DMM that functions as a pH sensor for monitoring endosome maturation within living cells, representing the first instance of DMMs on the surface membrane being used in vivo.177 Due to the DMM’s biocompatibility and byproducts, they discovered that it had a negligible effect on processivity. They then extended the DMM to map enzymatic activity and membrane potential in intracellular organelles, demonstrating DMM’s universality in sensing and diagnostics in living systems.178−183
In addition, given the tremendous potential of functional AuNP-based, SNA (spherical nucleic acid) for imaging, diagnosis, and treatment of disease, the incorporation of DMM onto nanoparticles enables the development of smart DMM systems that can change their properties in response to biological stimuli.184,185 For example, the DNA monolayer formed on the surface of the AuNPs controls the configuration of AuNP assemblies, allowing to assemble shape-shifting molecular machines with controlled biological functions. More importantly, the combination of DMMs and nanoparticles enables endocytosis of the DNA motor-AuNP complex. This synergistic effect enables these DMMs to function within the living cell, enabling to detect living biomolecules rapidly and sensitively. For instance, Le et al. successfully constructed a DNAzyme motor that anchors to the surface of a nanoparticle in response to a specific intracellular target (Figure 18a).186 Moreover, Pei et al. further demonstrated that exonuclease III, like DNA, can also autonomously move on DNA tracks on nanoparticle surfaces (Figure 18b).187 Particularly, they further confirmed that the performance of the DNA walker is critically dependent on the DNA density and the track conformation by interrogating the morphological effect of the 3D track on the nuclease activity. Intracellular interaction between a target molecule and the motor system initiates the motor’s autonomous walking on the AuNP, allowing for amplified detection of the specific microRNA in individual cancer cells.
Figure 18.
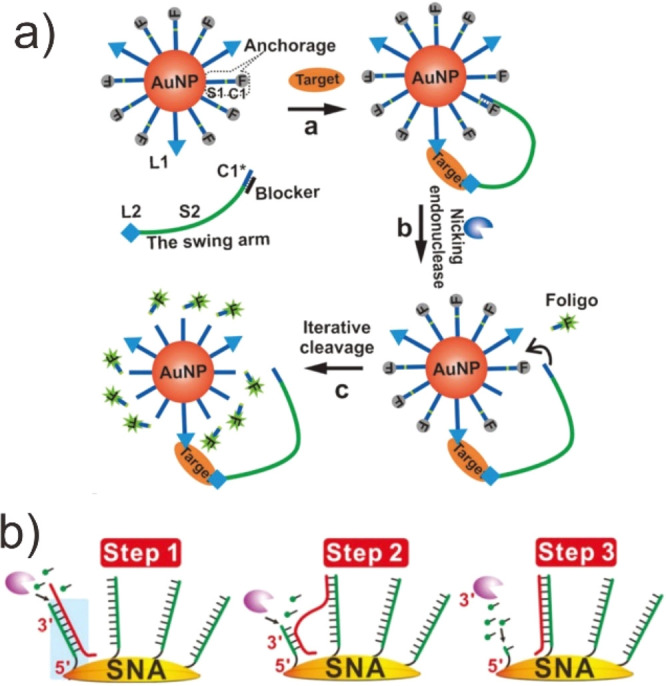
Imaging and diagnosis applications of DMMs. (a) Intracellular operation of a DNAzyme motor initiated by a specific miRNA. Reproduced with permission from ref (186). Copyright 2017, Nature Publishing Group. (b) Schematics of Exo III-powered stochastic DNA walkers that move on the SNA surfaces. Reproduced with permission from ref (187). Copyright 2017, John Wiley & Sons, Inc.
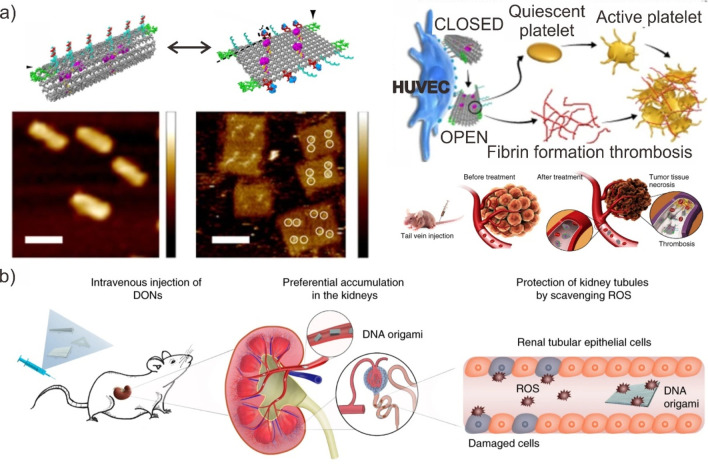
The integration of therapeutic components into the DMM enables the development of intelligent therapeutic carriers and sensors with enhanced in vivo reconfigurability. Ding et al. demonstrated a thrombin-activated DMM for cancer therapy via a vessel blockade (Figure 19a).188 In the DMM, thrombin was captured by extending strands from a specific location on a rectangular DNA origami sheet. The origami was then further assembled into a tubular shape using an aptamer that acts as a molecular lock for nucleolin, effectively encapsulating the thrombin within its inner cavity. When the DMM is delivered to cancer sites, the aptamer locks interact with the nucleolin expressed by tumor-associated endothelial cells, altering its configuration and thus inhibiting tubular origami. This rendered the encapsulated thrombin capable of inducing blood coagulation via thrombosis, resulting in tumor necrosis and inhibition of tumor growth due to a lack of oxygen and nutrients from blood vessels.
Figure 19.
Therapeutic applications of DMMs. (a) The nanorobot-Th binds to the vascular endothelium by recognizing nucleolin and opens to expose the encapsulated thrombin, which induces localized thromboses, tumor infarction, and cell necrosis. Reproduced with permission from ref (188). Copyright 2018, Nature Publishing Group. (b) DNA origami nanostructures can scavenge ROS and alleviate oxidative stress locally, protecting kidney structures and alleviating AKI. Reproduced with permission from ref (197). Copyright 2018, Nature Publishing Group.
They also used a similar method to construct a DMM for cancer vaccine therapy.189 The DMM used peptides as antigens, CpG DNA as a nucleic acid adjuvant, and triplex DNA as a molecular lock, rather than thrombin. When the mildly acidic intracellular environment of lysosomes in antigen-presenting cells is stimulated, the molecular locks of triplex DNA unfasten, exposing adjuvants and antigens for activation of a robust immune response. These DMMs served as a model for the development of intelligent nanoplatforms for biosensing and nanomedicine applications.190−194
Furthermore, the DMMs could be used as a novel therapeutic nanomedicine.195,196 Their therapeutic properties and biocompatibility, in particular, enable a programmable approach to medical therapies. For example, our group discovered that intact folding of DNA origami nanostructures larger than 100 nm resulted in their preferential accumulation in the kidneys of mice rather than their biodegradation via liver uptake (Figure 19b).197 We also demonstrated that these DNA frameworks have an active ROS scavenging effect in vivo, alleviating AKI-related clinical symptoms. Furthermore, Ge et al. modified a rectangular DNA origami nanostructure with a 3aC5a aptamer to create DMM for the exclusive retention of kidneys from oxidative stress-induced AKI.198 Since the 3aC5a aptamer inhibits C5a-mediated inflammation, the renally accumulating DMM may suppress inflammatory responses when the kidney retention time is prolonged (>12 h). As a result, DMMs are attractive candidates for the treatment of acute kidney injury associated with renal diseases.
5. Conclusions and Outlook
In summary, a variety of functional DNA molecules and nanostructures have been successfully used to construct complex and sophisticated DMMs.199−201 The advancement of DNA nanotechnology promotes the development of DNA walkers, motors, rotors, sensors, and robots. Moreover, the development of DMMs aids in the comprehension of physical theories of transport phenomena and aids in the clarification of the relationship between mechanical motions and chemical reactions. We have discussed recent advances in how integrating DMMs into various interfaces can help to empower their functions and applications in this Perspective. For nucleic acid chemistry, an increasing number of moieties and functional molecules have been developed, allowing for easy assembly and integration of diverse functions.77 The versatile intelligent DMMs were developed for a variety of biological applications, including biomolecular rulers, biosynthesis, biocomputing, and biosensing.202,203
One of the most significant challenges for artificial DMMs is to approach the complexity and intricacy of natural proteins. Since current methods focus on substituting allosteric and mechano-gating coupling into DNA nanostructures, engineering mechanochemical coupling requires a diverse set of fully predictable monomeric components.204 The highly precise discrete DNA nanostructures created by DNA oligomers or DNA origami provide an extremely useful method for guiding motion propagation. Nanometer-scale precision in molecule arrangement via DNA hybridization, in particular, enables the modification of DMMs with a variety of functions. Meanwhile, the inability to program DMM energy landscapes restricts the field of intelligent and efficient molecular machines. Later, as mass production of DNA and scaled-up DNA self-assembly technologies are developed, the cost and difficulty of building DMMs will decrease, thereby expanding the area of their biologic applications. Thus far, the development of integrating DMMs at the interface has paved the way for a sophisticated approach to the construction of artificial DMMs with complex structures and functions.
Acknowledgments
This work was financially supported by the National Science Foundation, National Key R&D Program of China (2021YFF1200300), NSFC (T2188102, 22025404, 22174092, 21904041), Natural Science Foundation of Shanghai (19JC1410300, 21ZR1439600), Innovative Research Team of High-Level Local Universities in Shanghai (SHSMU-ZLCX20212602), and the K. C. Wong Foundation at Shanghai Jiao Tong University.
The authors declare no competing financial interest.
References
- Guo H.; Bueler S. A.; Rubinstein J. L. Atomic Model for the Dimeric F-O Region of Mitochondrial Atp Synthase. Science 2017, 358, 936–940. 10.1126/science.aao4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q.; Li J. C.; Zhang M. J. Cargo Recognition and Cargo-Mediated Regulation of Unconventional Myosins. Acc. Chem. Res. 2014, 47, 3061–3070. 10.1021/ar500216z. [DOI] [PubMed] [Google Scholar]
- Mishra P.; Chan D. C. Mitochondrial Dynamics and Inheritance During Cell Division, Development and Disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 634–646. 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson S. L.; Redwine W. B.; Vale R. D.; Carter A. P. The Cytoplasmic Dynein Transport Machinery and Its Many Cargoes. Nat. Rev. Mol. Cell Biol. 2018, 19, 382–398. 10.1038/s41580-018-0004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A. P.; Luo M.; Zhou W. C.; Symersky J.; Bai D. Y.; Chambers M. G.; Faraldo-Gomez J. D.; Liao M. F.; Mueller D. M. High-Resolution Cryo-Em Analysis of the Yeast Atp Synthase in a Lipid Membrane. Science 2018, 360, 619–627. 10.1126/science.aas9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchihashi T.; Iino R.; Ando T.; Noji H. High-Speed Atomic Force Microscopy Reveals Rotary Catalysis of Rotorless F-1-Atpase. Science 2011, 333, 755–758. 10.1126/science.1205510. [DOI] [PubMed] [Google Scholar]
- Zagha E.; McCormick D. A. Neural Control of Brain State. Curr. Opin. Neurobiol. 2014, 29, 178–186. 10.1016/j.conb.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S.; Saijo S.; Suzuki K.; Mizutani K.; Kakinuma Y.; Ishizuka-Katsura Y.; Ohsawa N.; Terada T.; Shirouzu M.; Yokoyama S.; et al. Rotation Mechanism of Enterococcus Hirae V-1-Atpase Based on Asymmetric Crystal Structures. Nature 2013, 493, 703–707. 10.1038/nature11778. [DOI] [PubMed] [Google Scholar]
- Watson M. A.; Cockroft S. L. Man-Made Molecular Machines: Membrane Bound. Chem. Soc. Rev. 2016, 45, 6118–6129. 10.1039/C5CS00874C. [DOI] [PubMed] [Google Scholar]
- Abendroth J. M.; Bushuyev O. S.; Weiss P. S.; Barrett C. J. Controlling Motion at the Nanoscale: Rise of the Molecular Machines. ACS Nano 2015, 9, 7746–7768. 10.1021/acsnano.5b03367. [DOI] [PubMed] [Google Scholar]
- Astumian R. D. Design Principles for Brownian Molecular Machines: How to Swim in Molasses and Walk in a Hurricane. Phys. Chem. Chem. Phys. 2007, 9, 5067–5083. 10.1039/b708995c. [DOI] [PubMed] [Google Scholar]
- Krause S.; Feringa B. Towards Artificial Molecular Factories from Framework-Embedded Molecular Machines. Nat. Rev. Chem. 2020, 4, 550–562. 10.1038/s41570-020-0209-9. [DOI] [Google Scholar]
- van Leeuwen T.; Lubbe A. S.; Štacko P.; Wezenberg S. J.; Feringa B. L. Dynamic Control of Function by Light-Driven Molecular Motors. Nat. Rev. Chem. 2017, 1, 0096. 10.1038/s41570-017-0096. [DOI] [Google Scholar]
- Pan J.; Li F.; Cha T. G.; Chen H.; Choi J. H. Recent Progress on DNA Based Walkers. Curr. Opin. Biotechnol. 2015, 34, 56–64. 10.1016/j.copbio.2014.11.017. [DOI] [PubMed] [Google Scholar]
- Browne W. R.; Feringa B. L. Making Molecular Machines Work. Nat. Nanotechnol. 2006, 1, 25–35. 10.1038/nnano.2006.45. [DOI] [PubMed] [Google Scholar]
- Lancia F.; Ryabchun A.; Katsonis N. Life-Like Motion Driven by Artificial Molecular Machines. Nat. Rev. Chem. 2019, 3, 536–551. 10.1038/s41570-019-0122-2. [DOI] [Google Scholar]
- Sauvage J. P. From Chemical Topology to Molecular Machines (Nobel Lecture). Angew. Chem., Int. Edit 2017, 56, 11080–11093. 10.1002/anie.201702992. [DOI] [PubMed] [Google Scholar]
- Kassem S.; van Leeuwen T.; Lubbe A. S.; Wilson M. R.; Feringa B. L.; Leigh D. A. Artificial Molecular Motors. Chem. Soc. Rev. 2017, 46, 2592–2621. 10.1039/C7CS00245A. [DOI] [PubMed] [Google Scholar]
- Kocer A.; Walko M.; Meijberg W.; Feringa B. L. A Light-Actuated Nanovalve Derived from a Channel Protein. Science 2005, 309, 755–758. 10.1126/science.1114760. [DOI] [PubMed] [Google Scholar]
- van Delden R. A.; ter Wiel M. K. J.; Pollard M. M.; Vicario J.; Koumura N.; Feringa B. L. Unidirectional Molecular Motor on a Gold Surface. Nature 2005, 437, 1337–1340. 10.1038/nature04127. [DOI] [PubMed] [Google Scholar]
- Lubbe A. S.; Szymanski W.; Feringa B. L. Recent Developments in Reversible Photoregulation of Oligonucleotide Structure and Function. Chem. Soc. Rev. 2017, 46, 1052–1079. 10.1039/C6CS00461J. [DOI] [PubMed] [Google Scholar]
- Ramezani H.; Dietz H. Building Machines with DNA Molecules. Nat. Rev. Genet. 2020, 21, 5–26. 10.1038/s41576-019-0175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath J.; Turberfield A. J. DNA Nanomachines. Nat. Nanotechnol. 2007, 2, 275–284. 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- Budharaju H.; Zennifer A.; Sethuraman S.; Paul A.; Sundaramurthi D. Designer DNA Biomolecules as a Defined Biomaterial for 3d Bioprinting Applications. Mater. Horiz. 2022, 9, 1141–1166. 10.1039/D1MH01632F. [DOI] [PubMed] [Google Scholar]
- Rizzuto F. J.; Trinh T.; Sleiman H. F. Molecular Printing with DNA Nanotechnology. Chem. 2020, 6, 1560–1574. 10.1016/j.chempr.2020.06.012. [DOI] [Google Scholar]
- Seeman N. C.; Sleiman H. F. DNA Nanotechnology. Nat. Rev. Mater. 2018, 3, 17068. 10.1038/natrevmats.2017.68. [DOI] [Google Scholar]
- Endo M.; Sugiyama H. DNA Origami Nanomachines. Molecules 2018, 23, 1766. 10.3390/molecules23071766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J.; Karna D.; Mao H. DNA Origami Nano-Mechanics. Chem. Soc. Rev. 2021, 50, 11966–11978. 10.1039/D1CS00250C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X.; Li Q.; Zuo X.; Fan C. Catalytic Nucleic Acids for Bioanalysis. ACS Appl. Bio Mater. 2020, 3, 2674–2685. 10.1021/acsabm.9b00928. [DOI] [PubMed] [Google Scholar]
- He S.; Ge Z.; Zuo X.; Fan C.; Mao X. Dynamic Regulation of DNA Nanostructures by Noncanonical Nucleic Acids. NPG Asia Mater. 2021, 13, 42. 10.1038/s41427-021-00309-9. [DOI] [Google Scholar]
- Li Q.; Tong Z.; Cao Y.; Gu H. Dnas Catalyzing DNA Nanoconstruction. Chem. 2021, 7, 2556–2568. 10.1016/j.chempr.2021.08.008. [DOI] [Google Scholar]
- Fu J.; Oh S. W.; Monckton K.; Arbuckle-Keil G.; Ke Y.; Zhang T. Biomimetic Compartments Scaffolded by Nucleic Acid Nanostructures. Small 2019, 15, 1900256 10.1002/smll.201900256. [DOI] [PubMed] [Google Scholar]
- Wang W.; Yu S.; Huang S.; Bi S.; Han H.; Zhang J. R.; Lu Y.; Zhu J. J. Bioapplications of DNA Nanotechnology at the Solid-Liquid Interface. Chem. Soc. Rev. 2019, 48, 4892–4920. 10.1039/C8CS00402A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao R.; Zhao F.; Hamada S.; Yang P.; Xu H.; Luo D.; Yang D. DNA-Based Engineering System for Improving Human and Environmental Health: Identification, Detection, and Treatment. Nano Today 2020, 35, 100958. 10.1016/j.nantod.2020.100958. [DOI] [Google Scholar]
- Zhang L.; Ma X.; Wang G.; Liang X.; Mitomo H.; Pike A.; Houlton A.; Ijiro K. Non-Origami DNA for Functional Nanostructures: From Structural Control to Advanced Applications. Nano Today 2021, 39, 101154. 10.1016/j.nantod.2021.101154. [DOI] [Google Scholar]
- Seeman N. C. DNA in a Material World. Nature 2003, 421, 427–431. 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- Kallenbach N. R.; Ma R.; Seeman N. C. An Immobile Nucleic-Acid Junction Constructred from Oligonucleotides. Nature 1983, 305, 829–831. 10.1038/305829a0. [DOI] [Google Scholar]
- Evans C. G.; Winfree E. Physical Principles for DNA Tile Self-Assembly. Chem. Soc. Rev. 2017, 46, 3808–3829. 10.1039/C6CS00745G. [DOI] [PubMed] [Google Scholar]
- Heuer-Jungemann A.; Liedl T. From DNA Tiles to Functional DNA Materials. Trends Chem. 2019, 1, 799–814. 10.1016/j.trechm.2019.07.006. [DOI] [Google Scholar]
- Winfree E.; Liu F. R.; Wenzler L. A.; Seeman N. C. Design and Self-Assembly of Two-Dimensional DNA Crystals. Nature 1998, 394, 539–544. 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- Yan H.; Park S. H.; Finkelstein G.; Reif J. H.; LaBean T. H. DNA-Templated Self-Assembly of Protein Arrays and Highly Conductive Nanowires. Science 2003, 301, 1882–1884. 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- Goodman R. P.; Schaap I. A. T.; Tardin C. F.; Erben C. M.; Berry R. M.; Schmidt C. F.; Turberfield A. J. Rapid Chiral Assembly of Rigid DNA Building Blocks for Molecular Nanofabrication. Science 2005, 310, 1661–1665. 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- Chen J. H.; Seeman N. C. Synthesis from DNA of a Molecule with the Connectivity of a Cube. Nature 1991, 350, 631–633. 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- He Y.; Ye T.; Su M.; Zhang C.; Ribbe A. E.; Jiang W.; Mao C. Hierarchical Self-Assembly of DNA into Symmetric Supramolecular Polyhedra. Nature 2008, 452, 198–201. 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- Wang X.; Seeman N. C. Assembly and Characterization of 8-Arm and 12-Arm DNA Branched Junctions. J. Am. Chem. Soc. 2007, 129, 8169–8176. 10.1021/ja0693441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J. P.; Birktoft J. J.; Chen Y.; Wang T.; Sha R. J.; Constantinou P. E.; Ginell S. L.; Mao C. D.; Seeman N. C. From Molecular to Macroscopic Via the Rational Design of a Self-Assembled 3d DNA Crystal. Nature 2009, 461, 74–77. 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons C. R.; Zhang F.; Birktoft J. J.; Qi X. D.; Han D. R.; Liu Y.; Sha R. J.; Abdallah H. O.; Hernandez C.; Ohayon Y. P.; et al. Construction and Structure Determination of a Three-Dimensional DNA Crystal. J. Am. Chem. Soc. 2016, 138, 10047–10054. 10.1021/jacs.6b06508. [DOI] [PubMed] [Google Scholar]
- Wang T.; Sha R. J.; Birktoft J.; Zheng J. P.; Mao C. D.; Seeman N. C. A DNA Crystal Designed to Contain Two Molecules Per Asymmetric Unit. J. Am. Chem. Soc. 2010, 132, 15471–15473. 10.1021/ja104833t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P.; Hariadi R. F.; Sahu S.; Choi H. M. T.; Park S. H.; LaBean T. H.; Reif J. H. Programming DNA Tube Circumferences. Science 2008, 321, 824–826. 10.1126/science.1157312. [DOI] [PubMed] [Google Scholar]
- Ke Y.; Ong L. L.; Shih W. M.; Yin P. Three-Dimensional Structures Self-Assembled from DNA Bricks. Science 2012, 338, 1177–1183. 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong L. L.; Hanikel N.; Yaghi O. K.; Grun C.; Strauss M. T.; Bron P.; Lai-Kee-Him J.; Schueder F.; Wang B.; Wang P. F.; et al. Programmable Self-Assembly of Three-Dimensional Nanostructures from 10,000 Unique Components. Nature 2017, 552, 72–77. 10.1038/nature24648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothemund P. W. K. Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440, 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Han D. R.; Pal S.; Nangreave J.; Deng Z. T.; Liu Y.; Yan H. DNA Origami with Complex Curvatures in Three-Dimensional Space. Science 2011, 332, 342–346. 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; Dietz H.; Liedl T.; Hogberg B.; Graf F.; Shih W. M. Self-Assembly of DNA into Nanoscale Three-Dimensional Shapes. Nature 2009, 459, 414–418. 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E. S.; Dong M.; Nielsen M. M.; Jahn K.; Subramani R.; Mamdouh W.; Golas M. M.; Sander B.; Stark H.; Oliveira C. L. P.; et al. Self-Assembly of a Nanoscale DNA Box with a Controllable Lid. Nature 2009, 459, 73–77. 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- Han D. R.; Qi X. D.; Myhrvold C.; Wang B.; Dai M. J.; Jiang S. X.; Bates M.; Liu Y.; An B.; Zhang F.; et al. Single-Stranded DNA and Rna Origami. Science 2017, 358, 1402. 10.1126/science.aao2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.; Jiang S. X.; Wu S. Y.; Li Y. L.; Mao C. D.; Liu Y.; Yan H. Complex Wireframe DNA Origami Nanostructures with Multi-Arm Junction Vertices. Nat. Nanotechnol. 2015, 10, 779–784. 10.1038/nnano.2015.162. [DOI] [PubMed] [Google Scholar]
- Benson E.; Mohammed A.; Gardell J.; Masich S.; Czeizler E.; Orponen P.; Hogberg B. DNA Rendering of Polyhedral Meshes at the Nanoscale. Nature 2015, 523, 441–444. 10.1038/nature14586. [DOI] [PubMed] [Google Scholar]
- Tikhomirov G.; Petersen P.; Qian L. L. Fractal Assembly of Micrometre-Scale DNA Origami Arrays with Arbitrary Patterns. Nature 2017, 552, 67–71. 10.1038/nature24655. [DOI] [PubMed] [Google Scholar]
- Wagenbauer K. F.; Sigl C.; Dietz H. Gigadalton-Scale Shape-Programmable DNA Assemblies. Nature 2017, 552, 78–83. 10.1038/nature24651. [DOI] [PubMed] [Google Scholar]
- Yao G.; Zhang F.; Wang F.; Peng T.; Liu H.; Poppleton E.; Sulc P.; Jiang S.; Liu L.; Gong C.; et al. Meta-DNA Structures. Nat. Chem. 2020, 12, 1067–1075. 10.1038/s41557-020-0539-8. [DOI] [PubMed] [Google Scholar]
- Ge Z.; Gu H.; Li Q.; Fan C. Concept and Development of Framework Nucleic Acids. J. Am. Chem. Soc. 2018, 140, 17808–17819. 10.1021/jacs.8b10529. [DOI] [PubMed] [Google Scholar]
- Jones M. R.; Seeman N. C.; Mirkin C. A. Programmable Materials and the Nature of the DNA Bond. Science 2015, 347, 1260901. 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]
- Mao X. H.; Simon A. J.; Pei H.; Shi J. Y.; Li J.; Huang Q.; Plaxco K. W.; Fan C. H. Activity Modulation and Allosteric Control of a Scaffolded Dnazyme Using a Dynamic DNA Nanostructure. Chem. Sci. 2016, 7, 1200–1204. 10.1039/C5SC03705K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber R.; Do J.; Roller E. M.; Zhang T.; Schuller V. J.; Nickels P. C.; Feldmann J.; Liedl T. Hierarchical Assembly of Metal Nanoparticles, Quantum Dots and Organic Dyes Using DNA Origami Scaffolds. Nat. Nanotechnol. 2014, 9, 74–78. 10.1038/nnano.2013.253. [DOI] [PubMed] [Google Scholar]
- Zhao Z.; Fu J. L.; Dhakal S.; Johnson-Buck A.; Liu M. H.; Zhang T.; Woodbury N. W.; Liu Y.; Walter N. G.; Yan H. Nanocaged Enzymes with Enhanced Catalytic Activity and Increased Stability against Protease Digestion. Nat. Commun. 2016, 7, 10619. 10.1038/ncomms10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Fan C. H.; Pei H.; Shi J. Y.; Huang Q. Smart Drug Delivery Nanocarriers with Self-Assembled DNA Nanostructures. Adv. Mater. 2013, 25, 4386–4396. 10.1002/adma.201300875. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Pan V.; Li X.; Yang X.; Li H.; Wang P.; Ke Y. Dynamic DNA Structures. Small 2019, 15, 1900228 10.1002/smll.201900228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.; Li Z.; Wang P.; Meyer T.; Mao C.; Ke Y. Reconfiguration of DNA Molecular Arrays Driven by Information Relay. Science 2017, 357, 352. 10.1126/science.aan3377. [DOI] [PubMed] [Google Scholar]
- Mao C. D.; LaBean T. H.; Reif J. H.; Seeman N. C. Logical Computation Using Algorithmic Self-Assembly of DNA Triple-Crossover Molecules. Nature 2000, 407, 493–496. 10.1038/35035038. [DOI] [PubMed] [Google Scholar]
- Yurke B.; Turberfield A. J.; Mills A. P.; Simmel F. C.; Neumann J. L. A DNA-Fuelled Molecular Machine Made of DNA. Nature 2000, 406, 605–608. 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- Simmel F. C.; Yurke B.; Singh H. R. Principles and Applications of Nucleic Acid Strand Displacement Reactions. Chem. Rev. 2019, 119, 6326–6369. 10.1021/acs.chemrev.8b00580. [DOI] [PubMed] [Google Scholar]
- Han D. R.; Pal S.; Liu Y.; Yan H. Folding and Cutting DNA into Reconfigurable Topological Nanostructures. Nat. Nanotechnol. 2010, 5, 712–717. 10.1038/nnano.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P.; Choi H. M.; Calvert C. R.; Pierce N. A. Programming Biomolecular Self-Assembly Pathways. Nature 2008, 451, 318–322. 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Wang M. S.; Mao C. D. An Autonomous DNA Nanomotor Powered by a DNA Enzyme. Angew. Chem., Int. Edit 2004, 43, 3554–3557. 10.1002/anie.200453779. [DOI] [PubMed] [Google Scholar]
- Yin P.; Yan H.; Daniell X. G.; Turberfield A. J.; Reif J. H. A Unidirectional DNA Walker That Moves Autonomously Along a Track. Angew. Chem., Int. Edit 2004, 43, 4906–4911. 10.1002/anie.200460522. [DOI] [PubMed] [Google Scholar]
- Zhou W. H.; Saran R.; Liu J. W. Metal Sensing by DNA. Chem. Rev. 2017, 117, 8272–8325. 10.1021/acs.chemrev.7b00063. [DOI] [PubMed] [Google Scholar]
- Tian Y.; He Y.; Chen Y.; Yin P.; Mao C. D. A Dnazyme That Walks Processively and Autonomously Along a One-Dimensional Track. Angew. Chem., Int. Edit 2005, 44, 4355–4358. 10.1002/anie.200500703. [DOI] [PubMed] [Google Scholar]
- Wang J. B.; Yue L.; Li Z. Y.; Zhang J. J.; Tian H.; Willner I. Active Generation of Nanoholes in DNA Origami Scaffolds for Programmed Catalysis in Nanocavities. Nat. Commun. 2019, 10, 4963. 10.1038/s41467-019-12933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H.; Kim K. R.; Ahn D. R.; Lee J. E.; Yang E. G.; Kim S. Y. Reversible Regulation of Enzyme Activity by Ph-Responsive Encapsulation in DNA Nanocages. ACS Nano 2017, 11, 9352–9359. 10.1021/acsnano.7b04766. [DOI] [PubMed] [Google Scholar]
- Liu H.; Xu Y.; Li F.; Yang Y.; Wang W.; Song Y.; Liu D. Light-Driven Conformational Switch of I-Motif DNA. Angew. Chem., Int. Edit 2007, 46, 2515–2517. 10.1002/anie.200604589. [DOI] [PubMed] [Google Scholar]
- Liu X.; Lu C. H.; Willner I. Switchable Reconfiguration of Nucleic Acid Nanostructures by Stimuli-Responsive DNA Machines. Acc. Chem. Res. 2014, 47, 1673–1680. 10.1021/ar400316h. [DOI] [PubMed] [Google Scholar]
- Gerling T.; Wagenbauer K. F.; Neuner A. M.; Dietz H. Dynamic DNA Devices and Assemblies Formed by Shape-Complementary, Non-Base Pairing 3d Components. Science 2015, 347, 1446–1452. 10.1126/science.aaa5372. [DOI] [PubMed] [Google Scholar]
- Kuzuya A.; Sakai Y.; Yamazaki T.; Xu Y.; Komiyama M. Nanomechanical DNA Origami ’Single-Molecule Beacons’ Directly Imaged by Atomic Force Microscopy. Nat. Commun. 2011, 2, 196. 10.1038/ncomms1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y.; Pan L.; Guo R. Y.; Zhang Y. Y.; Li F.; Li M.; Li J.; Shi J. Y.; Qu F. L.; Zuo X. L.; et al. DNA Origami Nanocalipers for Ph Sensing at the Nanoscale. Chem. Commun. 2022, 58, 3673–3676. 10.1039/D1CC06701J. [DOI] [PubMed] [Google Scholar]
- Zhang J. J.; Wang J. X.; Tian H. Taking Orders from Light: Progress in Photochromic Bio-Materials. Mater. Horiz. 2014, 1, 169–184. 10.1039/C3MH00031A. [DOI] [Google Scholar]
- Kohman R. E.; Han X. Light Sensitization of DNA Nanostructures Via Incorporation of Photo- Cleavable Spacers. Chem. Commun. 2015, 51, 5747–5750. 10.1039/C5CC00082C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui L.; Zhang Q.; Deng W.; Liu H. DNA-Based Nanofabrication: Pathway to Applications in Surface Engineering. Small 2019, 15, 1805428 10.1002/smll.201805428. [DOI] [PubMed] [Google Scholar]
- Tjong V.; Tang L.; Zauscher S.; Chilkoti A. ″Smart″ DNA Interfaces. Chem. Soc. Rev. 2014, 43, 1612–1626. 10.1039/C3CS60331H. [DOI] [PubMed] [Google Scholar]
- Schneider A. K.; Niemeyer C. M. DNA Surface Technology: From Gene Sensors to Integrated Systems for Life and Materials Sciences. Angew. Chem., Int. Edit 2018, 57, 16959–16967. 10.1002/anie.201811713. [DOI] [PubMed] [Google Scholar]
- Zhang P.; Gao D.; An K.; Shen Q.; Wang C.; Zhang Y.; Pan X.; Chen X.; Lyv Y.; Cui C.; et al. A Programmable Polymer Library That Enables the Construction of Stimuli-Responsive Nanocarriers Containing Logic Gates. Nat. Chem. 2020, 12, 381–390. 10.1038/s41557-020-0426-3. [DOI] [PubMed] [Google Scholar]
- Zhu D.; Pei H.; Chao J.; Su S.; Aldalbahi A.; Rahaman M.; Wang L.; Wang L.; Huang W.; Fan C.; et al. Poly-Adenine-Based Programmable Engineering of Gold Nanoparticles for Highly Regulated Spherical Dnazymes. Nanoscale 2015, 7, 18671–18676. 10.1039/C5NR05366H. [DOI] [PubMed] [Google Scholar]
- Wu Y.; Ali M. R. K.; Chen K.; Fang N.; El-Sayed M. A. Gold Nanoparticles in Biological Optical Imaging. Nano Today 2019, 24, 120–140. 10.1016/j.nantod.2018.12.006. [DOI] [Google Scholar]
- Ma W.; Hao C. L.; Sun M. Z.; Xu L. G.; Xu C. L.; Kuang H. Tuning of Chiral Construction, Structural Diversity, Scale Transformation and Chiroptical Applications. Mater. Horiz. 2018, 5, 141–161. 10.1039/C7MH00966F. [DOI] [Google Scholar]
- Meng Y. C.; Chen Y.; Zhu J. J.; Qi Y.; Ding J. S.; Zhou W. H. Polarity Control of DNA Adsorption Enabling the Surface Functionalization of Cuo Nanozymes for Targeted Tumor Therapy. Mater. Horiz. 2021, 8, 972–986. 10.1039/D0MH01372B. [DOI] [PubMed] [Google Scholar]
- Rogers W. B.; Shih W. M.; Manoharan V. N. Using DNA to Program the Self-Assembly of Colloidal Nanoparticles and Microparticles. Nat. Rev. Mater. 2016, 1, 16008. 10.1038/natrevmats.2016.8. [DOI] [Google Scholar]
- Tian Y.; Wang T.; Liu W. Y.; Xin H. L.; Li H. L.; Ke Y. G.; Shih W. M.; Gang O. Prescribed Nanoparticle Cluster Architectures and Low-Dimensional Arrays Built Using Octahedral DNA Origami Frames. Nat. Nanotechnol. 2015, 10, 637–644. 10.1038/nnano.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. Y.; Lytton-Jean A. K. R.; Lee B.; Weigand S.; Schatz G. C.; Mirkin C. A. DNA-Programmable Nanoparticle Crystallization. Nature 2008, 451, 553–556. 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]
- Jung C.; Allen P. B.; Ellington A. D. A Stochastic DNA Walker That Traverses a Microparticle Surface. Nat. Nanotechnol. 2016, 11, 157–163. 10.1038/nnano.2015.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S.; Glancy D.; Chan W. C. W. DNA-Controlled Dynamic Colloidal Nanoparticle Systems for Mediating Cellular Interaction. Science 2016, 351, 841–845. 10.1126/science.aad4925. [DOI] [PubMed] [Google Scholar]
- Wang F.; Zhang X.; Liu X.; Fan C.; Li Q. Programming Motions of DNA Origami Nanomachines. Small 2019, 15, 1900013 10.1002/smll.201900013. [DOI] [PubMed] [Google Scholar]
- Wang Z. G.; Elbaz J.; Willner I. DNA Machines: Bipedal Walker and Stepper. Nano Lett. 2011, 11, 304–309. 10.1021/nl104088s. [DOI] [PubMed] [Google Scholar]
- Shin J. S.; Pierce N. A. A. Synthetic DNA Walker for Molecular Transport. J. Am. Chem. Soc. 2004, 126, 10834–10835. 10.1021/ja047543j. [DOI] [PubMed] [Google Scholar]
- Liber M.; Tomov T. E.; Tsukanov R.; Berger Y.; Nir E. A Bipedal DNA Motor That Travels Back and Forth between Two DNA Origami Tiles. Small 2015, 11, 568–575. 10.1002/smll.201402028. [DOI] [PubMed] [Google Scholar]
- Omabegho T.; Sha R.; Seeman N. C. A Bipedal DNA Brownian Motor with Coordinated Legs. Science 2009, 324, 67–71. 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund K.; Manzo A. J.; Dabby N.; Michelotti N.; Johnson-Buck A.; Nangreave J.; Taylor S.; Pei R.; Stojanovic M. N.; Walter N. G.; et al. Molecular Robots Guided by Prescriptive Landscapes. Nature 2010, 465, 206–210. 10.1038/nature09012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. M.; Johnson-Buck A.; Yang Y. R.; Shih W. M.; Yan H.; Walter N. G. Exploring the Speed Limit of Toehold Exchange with a Cartwheeling DNA Acrobat. Nat. Nanotechnol. 2018, 13, 723–729. 10.1038/s41565-018-0130-2. [DOI] [PubMed] [Google Scholar]
- Benson E.; Marzo R. C.; Bath J.; Turberfield A. J. A DNA Molecular Printer Capable of Programmable Positioning and Patterning in Two Dimensions. Sci. Robot. 2022, 7, eabn5459 10.1126/scirobotics.abn5459. [DOI] [PubMed] [Google Scholar]
- Stommer P.; Kiefer H.; Kopperger E.; Honemann M. N.; Kube M.; Simmel F. C.; Netz R. R.; Dietz H. A Synthetic Tubular Molecular Transport System. Nat. Commun. 2021, 12, 4393. 10.1038/s41467-021-24675-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. T.; Akhuetie-Oni B. O.; Zhang Z.; Lin C. X. DNA Origami Rotaxanes: Tailored Synthesis and Controlled Structure Switching. Angew. Chem., Int. Edit 2016, 55, 11412–11416. 10.1002/anie.201604621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- List J.; Falgenhauer E.; Kopperger E.; Pardatscher G.; Simmel F. C. Long-Range Movement of Large Mechanically Interlocked DNA Nanostructures. Nat. Commun. 2016, 7, 12414. 10.1038/ncomms12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketterer P.; Willner E. M.; Dietz H. Nanoscale Rotary Apparatus Formed from Tight-Fitting 3d DNA Components. Sci. Adv. 2016, 2, e1501209 10.1126/sciadv.1501209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaru T.; Suzuki Y.; Kawamata I.; Nomura S. M.; Murata S. Stepping Operation of a Rotary DNA Origami Device. Chem. Commun. 2017, 53, 7716–7719. 10.1039/C7CC03214E. [DOI] [PubMed] [Google Scholar]
- Lauback S.; Mattioli K. R.; Marras A. E.; Armstrong M.; Rudibaugh T. P.; Sooryakumar R.; Castro C. E. Real-Time Magnetic Actuation of DNA Nanodevices Via Modular Integration with Stiff Micro-Levers. Nat. Commun. 2018, 9, 1446. 10.1038/s41467-018-03601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marras A. E.; Zhou L.; Su H. J.; Castro C. E. Programmable Motion of DNA Origami Mechanisms. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 713–718. 10.1073/pnas.1408869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopperger E.; List J.; Madhira S.; Rothfischer F.; Lamb D. C.; Simmel F. C. A Self-Assembled Nanoscale Robotic Arm Controlled by Electric Fields. Science 2018, 359, 296–300. 10.1126/science.aao4284. [DOI] [PubMed] [Google Scholar]
- Kroener F.; Heerwig A.; Kaiser W.; Mertig M.; Rant U. Electrical Actuation of a DNA Origami Nanolever on an Electrode. J. Am. Chem. Soc. 2017, 139, 16510–16513. 10.1021/jacs.7b10862. [DOI] [PubMed] [Google Scholar]
- Langecker M.; Arnaut V.; List J.; Simmel F. C. DNA Nanostructures Interacting with Lipid Bilayer Membranes. Acc. Chem. Res. 2014, 47, 1807–1815. 10.1021/ar500051r. [DOI] [PubMed] [Google Scholar]
- Bae W.; Kocabey S.; Liedl T. DNA Nanostructures in Vitro, in Vivo and on Membranes. Nano Today 2019, 26, 98–107. 10.1016/j.nantod.2019.03.001. [DOI] [Google Scholar]
- Chiu Y. T. E.; Li H.; Choi C. H. J. Progress toward Understanding the Interactions between DNA Nanostructures and the Cell. Small 2019, 15, 1805416 10.1002/smll.201805416. [DOI] [PubMed] [Google Scholar]
- Mishra S.; Feng Y.; Endo M.; Sugiyama H. Advances in DNA Origami-Cell Interfaces. Chembiochem 2020, 21, 33–44. 10.1002/cbic.201900481. [DOI] [PubMed] [Google Scholar]
- Ge Z.; Liu J.; Guo L.; Yao G.; Li Q.; Wang L.; Li J.; Fan C. Programming Cell-Cell Communications with Engineered Cell Origami Clusters. J. Am. Chem. Soc. 2020, 142, 8800–8808. 10.1021/jacs.0c01580. [DOI] [PubMed] [Google Scholar]
- Liu J.; Li M.; Li F.; Ge Z.; Li Q.; Liu M.; Shi J.; Wang L.; Zuo X.; Fan C.; et al. Reconstructing Soma-Soma Synapse-Like Vesicular Exocytosis with DNA Origami. ACS Cent.Sci. 2021, 7, 1400–1407. 10.1021/acscentsci.1c00645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langecker M.; Arnaut V.; Martin T. G.; List J.; Renner S.; Mayer M.; Dietz H.; Simmel F. C. Synthetic Lipid Membrane Channels Formed by Designed DNA Nanostructures. Science 2012, 338, 932–936. 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J. R.; Seifert A.; Fertig N.; Howorka S. A Biomimetic DNA-Based Channel for the Ligand-Controlled Transport of Charged Molecular Cargo across a Biological Membrane. Nat. Nanotechnol. 2016, 11, 152–156. 10.1038/nnano.2015.279. [DOI] [PubMed] [Google Scholar]
- Bell N. A. W.; Engst C. R.; Ablay M.; Divitini G.; Ducati C.; Liedl T.; Keyser U. F. DNA Origami Nanopores. Nano Lett. 2012, 12, 512–517. 10.1021/nl204098n. [DOI] [PubMed] [Google Scholar]
- Howorka S. Building Membrane Nanopores. Nat. Nanotechnol. 2017, 12, 619–630. 10.1038/nnano.2017.99. [DOI] [PubMed] [Google Scholar]
- Akbari E.; Mollica M. Y.; Lucas C. R.; Bushman S. M.; Patton R. A.; Shahhosseini M.; Song J. W.; Castro C. E. Engineering Cell Surface Function with DNA Origami. Adv. Mater. 2017, 29, 1703632. 10.1002/adma.201703632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P.; Ye D. K.; Zuo X. L.; Li J.; Wang J. B.; Liu H. J.; Hwang M. T.; Chao J.; Su S.; Wang L. H.; et al. DNA Hydrogel with Aptamer-Toehold-Based Recognition, Cloaking, and Decloaking of Circulating Tumor Cells for Live Cell Analysis. Nano Lett. 2017, 17, 5193–5198. 10.1021/acs.nanolett.7b01006. [DOI] [PubMed] [Google Scholar]
- You M.; Lyu Y.; Han D.; Qiu L.; Liu Q.; Chen T.; Sam Wu C.; Peng L.; Zhang L.; Bao G.; et al. DNA Probes for Monitoring Dynamic and Transient Molecular Encounters on Live Cell Membranes. Nat. Nanotechnol. 2017, 12, 453–459. 10.1038/nnano.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Zhou S.; Li L.; Zhu J.-j. Nanomaterials-Based Sensitive Electrochemiluminescence Biosensing. Nano Today 2017, 12, 98–115. 10.1016/j.nantod.2016.12.013. [DOI] [Google Scholar]
- Wang L.; Wang X.; Wu Y.; Guo M.; Gu C.; Dai C.; Kong D.; Wang Y.; Zhang C.; Qu D.; et al. Rapid and Ultrasensitive Electromechanical Detection of Ions, Biomolecules and Sars-Cov-2 Rna in Unamplified Samples. Nat. Biomed. Eng. 2022, 6, 276–285. 10.1038/s41551-021-00833-7. [DOI] [PubMed] [Google Scholar]
- Song P.; Shen J.; Ye D.; Dong B.; Wang F.; Pei H.; Wang J.; Shi J.; Wang L.; Xue W.; et al. Programming Bulk Enzyme Heterojunctions for Biosensor Development with Tetrahedral DNA Framework. Nat. Commun. 2020, 11, 838. 10.1038/s41467-020-14664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D.; Pei H.; Yao G.; Wang L.; Su S.; Chao J.; Wang L.; Aldalbahi A.; Song S.; Shi J.; et al. A Surface-Confi Ned Proton-Driven DNA Pump Using a Dynamic 3d DNA Scaffold. Adv. Mater. 2016, 28, 6860–6865. 10.1002/adma.201506407. [DOI] [PubMed] [Google Scholar]
- Kuzyk A.; Yang Y.; Duan X.; Stoll S.; Govorov A. O.; Sugiyama H.; Endo M.; Liu N. A Light-Driven Three-Dimensional Plasmonic Nanosystem That Translates Molecular Motion into Reversible Chiroptical Function. Nat. Commun. 2016, 7, 10591. 10.1038/ncomms10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehl K.; Mugler A.; Vivek S.; Liu Y.; Zhang Y.; Fan M.; Weeks E. R.; Salaita K. High-Speed DNA-Based Rolling Motors Powered by Rnase H. Nat. Nanotechnol. 2016, 11, 184–190. 10.1038/nnano.2015.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A.; Hoffecker I. T.; Smyrlaki I.; Rosa J.; Grevys A.; Bratlie D.; Sandlie I.; Michaelsen T. E.; Andersen J. T.; Hogberg B. Binding to Nanopatterned Antigens Is Dominated by the Spatial Tolerance of Antibodies. Nat. Nanotechnol. 2019, 14, 184–190. 10.1038/s41565-018-0336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon P. S.; Ren S.; Kwon S. J.; Kizer M. E.; Kuo L.; Xie M.; Zhu D.; Zhou F.; Zhang F. M.; Kim D.; et al. Designer DNA Architecture Offers Precise and Multivalent Spatial Pattern-Recognition for Viral Sensing and Inhibition. Nat. Chem. 2020, 12, 26–35. 10.1038/s41557-019-0369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X.; Liu M.; Yan L.; Deng M.; Li F.; Li M.; Wang F.; Li J.; Wang L.; Tian Y.; et al. Programming Biomimetically Confined Aptamers with DNA Frameworks. ACS Nano 2020, 14, 8776–8783. 10.1021/acsnano.0c03362. [DOI] [PubMed] [Google Scholar]
- Willner E. M.; Kamada Y.; Suzuki Y.; Emura T.; Hidaka K.; Dietz H.; Sugiyama H.; Endo M. Single-Molecule Observation of the Photoregulated Conformational Dynamics of DNA Origami Nanoscissors. Angew. Chem., Int. Edit 2017, 56, 15324–15328. 10.1002/anie.201708722. [DOI] [PubMed] [Google Scholar]
- Bell N. A. W.; Keyser U. F. Digitally Encoded DNA Nanostructures for Multiplexed, Single-Molecule Protein Sensing with Nanopores. Nat. Nanotechnol. 2016, 11, 645–651. 10.1038/nnano.2016.50. [DOI] [PubMed] [Google Scholar]
- Mohapatra S.; Lin C. T.; Feng X. Y. A.; Basu A.; Ha T. Single-Molecule Analysis and Engineering of DNA Motors. Chem. Rev. 2020, 120, 36–78. 10.1021/acs.chemrev.9b00361. [DOI] [PubMed] [Google Scholar]
- Fu J. L.; Liu M. H.; Liu Y.; Woodbury N. W.; Yan H. Interenzyme Substrate Diffusion for an Enzyme Cascade Organized on Spatially Addressable DNA Nanostructures. J. Am. Chem. Soc. 2012, 134, 5516–5519. 10.1021/ja300897h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke J. J.; Dietz H. Placing Molecules with Bohr Radius Resolution Using DNA Origami. Nat. Nanotechnol. 2016, 11, 47–52. 10.1038/nnano.2015.240. [DOI] [PubMed] [Google Scholar]
- Le J. V.; Luo Y.; Darcy M. A.; Lucas C. R.; Goodwin M. F.; Poirier M. G.; Castro C. E. Probing Nucleosome Stability with a DNA Origami Nanocaliper. ACS Nano 2016, 10, 7073–7084. 10.1021/acsnano.6b03218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr N. D.; Goodman B. S.; Jungmann R.; Leschziner A. E.; Shih W. M.; Reck-Peterson S. L. Tug-of-War in Motor Protein Ensembles Revealed with a Programmable DNA Origami Scaffold. Science 2012, 338, 662–665. 10.1126/science.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Chao J.; Pan D.; Liu H.; Qiang Y.; Liu K.; Cui C.; Chen J.; Huang Q.; Hu J.; et al. DNA Origami-Based Shape Ids for Single-Molecule Nanomechanical Genotyping. Nat. Commun. 2017, 8, 14738. 10.1038/ncomms14738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri P.; Altheimer B. D.; Dai M. J.; Yin P.; Zhuang X. W. Rotation Tracking of Genome-Processing Enzymes Using DNA Origami Rotors. Nature 2019, 572, 136–140. 10.1038/s41586-019-1397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X.; Li K.; Liu M.; Wang X.; Zhao T.; An B.; Cui M.; Li Y.; Pu J.; Li J.; et al. Directing Curli Polymerization with DNA Origami Nucleators. Nat. Commun. 2019, 10, 1395. 10.1038/s41467-019-09369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K.; Li Y. F.; Wang X. Y.; Cui M. K.; An B. L.; Pu J. H.; Liu J. T.; Zhang B. Y.; Ma G. J.; Zhong C. Diatom-Inspired Multiscale Mineralization of Patterned Protein-Polysaccharide Complex Structures. Natl. Sci. Rev. 2021, 8, nwaa191. 10.1093/nsr/nwaa191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P.; Liu X.; Liu P.; Wang F.; Ariyama H.; Ando T.; Lin J.; Wang L.; Hu J.; Li B.; et al. Capturing Transient Antibody Conformations with DNA Origami Epitopes. Nat. Commun. 2020, 11, 3114. 10.1038/s41467-020-16949-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh G. C.; Burns J. R.; Howorka S. Comparing Proteins and Nucleic Acids for Next-Generation Biomolecular engineering. Nat. Rev. Chem. 2018, 2, 113–130. 10.1038/s41570-018-0015-9. [DOI] [Google Scholar]
- Wilhelm D.; Bruck J.; Qian L. L. Probabilistic Switching Circuits in DNA. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 903–908. 10.1073/pnas.1715926115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra R.; Sharma J.; Liu Y.; Yan H. Addressable Molecular Tweezers for DNA-Templated Coupling Reactions. Nano Lett. 2006, 6, 978–983. 10.1021/nl060212f. [DOI] [PubMed] [Google Scholar]
- Gu H.; Chao J.; Xiao S.-J.; Seeman N. C. A Proximity-Based Programmable DNA Nanoscale Assembly Line. Nature 2010, 465, 202–205. 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.; Liu D. R. Autonomous Multistep Organic Synthesis in a Single Isothermal Solution Mediated by a DNA Walker. Nat. Nanotechnol. 2010, 5, 778–782. 10.1038/nnano.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. N.; Chao J.; Liu H. J.; Wang F.; Su S.; Liu B.; Zhang L.; Shi J. Y.; Wang L. H.; Huang W.; et al. Transfer of Two-Dimensional Oligonucleotide Patterns onto Stereocontrolled Plasmonic Nanostructures through DNA-Origami-Based Nanoimprinting Lithography. Angew. Chem., Int. Edit 2016, 55, 8036–8040. 10.1002/anie.201512022. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Ke G.; Ma Y.; Zhu Z.; Liu M.; Liu Y.; Yan H.; Yang C. J. A Synthetic Light-Driven Substrate Channeling System for Precise Regulation of Enzyme Cascade Activity Based on DNA Origami. J. Am. Chem. Soc. 2018, 140, 8990–8996. 10.1021/jacs.8b05429. [DOI] [PubMed] [Google Scholar]
- Ke G. L.; Liu M. H.; Jiang S. X.; Qi X. D.; Yang Y. R.; Wootten S.; Zhang F.; Zhu Z.; Liu Y.; Yang C. J.; et al. Directional Regulation of Enzyme Pathways through the Control of Substrate Channeling on a DNA Origami Scaffold. Angew. Chem., Int. Edit 2016, 55, 7483–7486. 10.1002/anie.201603183. [DOI] [PubMed] [Google Scholar]
- Li J.; Green A. A.; Yan H.; Fan C. Engineering Nucleic Acid Structures for Programmable Molecular Circuitry and Intracellular Biocomputation. Nat. Chem. 2017, 9, 1056–1067. 10.1038/nchem.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.; Wu C.; You M.; Zhang T.; Wan S.; Chen T.; Qiu L.; Zheng Z.; Liang H.; Tan W. A Cascade Reaction Network Mimicking the Basic Functional Steps of Adaptive Immune Response. Nat. Chem. 2015, 7, 835. 10.1038/nchem.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R.; Xu L.; Wang H.; Lyu Y.; Wang D.; Bi C.; Cui C.; Fan C.; Liu Q.; Zhang X.; et al. DNA-Based Artificial Molecular Signaling System That Mimics Basic Elements of Reception and Response. Nat. Commun. 2020, 11, 978. 10.1038/s41467-020-14739-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D. Q.; Wang E. K.; Dong S. J. Upconversion-Chameleon-Driven DNA Computing: The DNA-Unlocked Inner-Filter-Effect (Du-Ife) for Operating a Multicolor Upconversion Luminescent DNA Logic Library and Its Biosensing Application. Mater. Horiz. 2019, 6, 375–384. 10.1039/C8MH01151F. [DOI] [Google Scholar]
- Wang F.; Lv H.; Li Q.; Li J.; Zhang X.; Shi J.; Wang L.; Fan C. Implementing Digital Computing with DNA-Based Switching Circuits. Nat. Commun. 2020, 11, 121. 10.1038/s41467-019-13980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham S. F.; Bath J.; Katsuda Y.; Endo M.; Hidaka K.; Sugiyama H.; Turberfield A. J. A DNA-Based Molecular Motor That Can Navigate a Network of Tracks. Nat. Nanotechnol. 2012, 7, 169–173. 10.1038/nnano.2011.253. [DOI] [PubMed] [Google Scholar]
- Chatterjee G.; Dalchau N.; Muscat R. A.; Phillips A.; Seelig G. A Spatially Localized Architecture for Fast and Modular DNA Computing. Nat. Nanotechnol. 2017, 12, 920–927. 10.1038/nnano.2017.127. [DOI] [PubMed] [Google Scholar]
- Thubagere A. J.; Li W.; Johnson R. F.; Chen Z.; Doroudi S.; Lee Y. L.; Izatt G.; Wittman S.; Srinivas N.; Woods D.; et al. A Cargo-Sorting DNA Robot. Science 2017, 357, 1095. 10.1126/science.aan6558. [DOI] [PubMed] [Google Scholar]
- Chao J.; Wang J.; Wang F.; Ouyang X.; Kopperger E.; Liu H.; Li Q.; Shi J.; Wang L.; Hu J.; et al. Solving Mazes with Single-Molecule DNA Navigators. Nat. Mater. 2019, 18, 273–279. 10.1038/s41563-018-0205-3. [DOI] [PubMed] [Google Scholar]
- Chen J.; Luo Z.; Sun C.; Huang Z.; Zhou C.; Yin S.; Duan Y.; Li Y. Research Progress of DNA Walker and Its Recent Applications in Biosensor. TrAC, Trends Anal. Chem. 2019, 120, 115626. 10.1016/j.trac.2019.115626. [DOI] [Google Scholar]
- Li J.; Dai J.; Jiang S.; Xie M.; Zhai T.; Guo L.; Cao S.; Xing S.; Qu Z.; Zhao Y.; et al. Encoding Quantized Fluorescence States with Fractal DNA Frameworks. Nat. Commun. 2020, 11, 2185. 10.1038/s41467-020-16112-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. X.; Zuo X. L.; Li Q.; Chen F.; Chen Y. R.; Deng J. Q.; Han D.; Hao C. L.; Huang F. J.; Huang Y. Y.; et al. Nucleic Acids Analysis. Sci. China: Chem. 2021, 64, 171–203. 10.1007/s11426-020-9864-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong H. T.; Zhang J. C.; Li J.; Wang J.; Shin H. J.; Tai R. Z.; Yan Q. L.; Xia K.; Hu J.; Wang L. H.; et al. Genetically Encoded X-Ray Cellular Imaging for Nanoscale Protein Localization. Natl. Sci. Rev. 2020, 7, 1218–1227. 10.1093/nsr/nwaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyk A.; Schreiber R.; Zhang H.; Govorov A. O.; Liedl T.; Liu N. Reconfigurable 3d Plasmonic Metamolecules. Nat. Mater. 2014, 13, 862–866. 10.1038/nmat4031. [DOI] [PubMed] [Google Scholar]
- Funck T.; Nicoli F.; Kuzyk A.; Liedl T. Sensing Picomolar Concentrations of Rna Using Switchable Plasmonic Chirality. Angew. Chem., Int. Edit 2018, 57, 13495–13498. 10.1002/anie.201807029. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; Bachelet I.; Church G. M. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335, 831–834. 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- Zhou C.; Duan X. Y.; Liu N. A Plasmonic Nanorod That Walks on DNA Origami. Nat. Commun. 2015, 6, 8102. 10.1038/ncomms9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi S.; M. G. S.; Goswami D.; Gupta G. D.; Mayor S.; Krishnan Y. A DNA Nanomachine That Maps Spatial and Temporal Ph Changes inside Living Cells. Nat. Nanotechnol. 2009, 4, 325–330. 10.1038/nnano.2009.83. [DOI] [PubMed] [Google Scholar]
- Modi S.; Nizak C.; Surana S.; Halder S.; Krishnan Y. Two DNA Nanomachines Map Ph Changes Along Intersecting Endocytic Pathways inside the Same Cell. Nat. Nanotechnol. 2013, 8, 459–467. 10.1038/nnano.2013.92. [DOI] [PubMed] [Google Scholar]
- Bhatia D.; Surana S.; Chakraborty S.; Koushika S. P.; Krishnan Y. A Synthetic Icosahedral DNA-Based Host-Cargo Complex for Functional in Vivo Imaging. Nat. Commun. 2011, 2, 339. 10.1038/ncomms1337. [DOI] [PubMed] [Google Scholar]
- Saminathan A.; Devany J.; Veetil A. T.; Suresh B.; Pillai K. S.; Schwake M.; Krishnan Y. A DNA-Based Voltmeter for Organelles. Nat. Nanotechnol. 2021, 16, 96–103. 10.1038/s41565-020-00784-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan K.; Veetil A. T.; Chakraborty K.; Krishnan Y. DNA Nanodevices Map Enzymatic Activity in Organelles. Nat. Nanotechnol. 2019, 14, 252–259. 10.1038/s41565-019-0365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash V.; Tsekouras K.; Venkatachalapathy M.; Heinicke L.; Presse S.; Walter N. G.; Krishnan Y. Quantitative Mapping of Endosomal DNA Processing by Single Molecule Counting. Angew. Chem., Int. Edit 2019, 58, 3073–3076. 10.1002/anie.201811746. [DOI] [PubMed] [Google Scholar]
- Saha S.; Prakash V.; Halder S.; Chakraborty K.; Krishnan Y. A Ph-Independent DNA Nanodevice for Quantifying Chloride Transport in Organelles of Living Cells. Nat. Nanotechnol. 2015, 10, 645–651. 10.1038/nnano.2015.130. [DOI] [PubMed] [Google Scholar]
- Ran X.; Wang Z. Z.; Pu F.; Ju E. G.; Ren J. S.; Qu X. G. Nucleic Acid-Driven Aggregation-Induced Emission of Au Nanoclusters for Visualizing Telomerase Activity in Living Cells and in Vivo. Mater. Horiz. 2021, 8, 1769–1775. 10.1039/D0MH01875A. [DOI] [PubMed] [Google Scholar]
- Li Y. Q.; Liu J. W. Nanozyme’s Catching Up: Activity, Specificity, Reaction Conditions and Reaction Types. Mater. Horiz. 2021, 8, 336–350. 10.1039/D0MH01393E. [DOI] [PubMed] [Google Scholar]
- Peng H.; Li X. F.; Zhang H.; Le X. C. A Microrna-Initiated Dnazyme Motor Operating in Living Cells. Nat. Commun. 2017, 8, 14378. 10.1038/ncomms14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X.; Zhu D.; Yao G.; Su S.; Chao J.; Liu H.; Zuo X.; Wang L.; Shi J.; Wang L.; et al. An Exonuclease Iii-Powered, on-Particle Stochastic DNA Walker. Angew. Chem., Int. Edit 2017, 56, 1855–1858. 10.1002/anie.201611777. [DOI] [PubMed] [Google Scholar]
- Li S.; Jiang Q.; Liu S.; Zhang Y.; Tian Y.; Song C.; Wang J.; Zou Y.; Anderson G. J.; Han J.-Y.; et al. A DNA Nanorobot Functions as a Cancer Therapeutic in Response to a Molecular Trigger in Vivo. Nat. Biotechnol. 2018, 36, 258–264. 10.1038/nbt.4071. [DOI] [PubMed] [Google Scholar]
- Liu S.; Jiang Q.; Zhao X.; Zhao R.; Wang Y.; Wang Y.; Liu J.; Shang Y.; Zhao S.; Wu T.; et al. A DNA Nanodevice-Based Vaccine for Cancer Immunotherapy. Nat. Mater. 2021, 20, 421–430. 10.1038/s41563-020-0793-6. [DOI] [PubMed] [Google Scholar]
- Seitz I.; Shaukat A.; Nurmi K.; Ijas H.; Hirvonen J.; Santos H. A.; Kostiainen M. A.; Linko V. Prospective Cancer Therapies Using Stimuli-Responsive DNA Nanostructures. Macromol. Biosci. 2021, 21, 2100272 10.1002/mabi.202100272. [DOI] [PubMed] [Google Scholar]
- Pitikultham P.; Wang Z.; Wang Y.; Shang Y.; Jiang Q.; Ding B. Stimuli-Responsive DNA Origami Nanodevices and Their Biological Applications. ChemMedChem 2022, 17, e202100635 10.1002/cmdc.202100635. [DOI] [PubMed] [Google Scholar]
- Keller A.; Linko V. Challenges and Perspectives of DNA Nanostructures in Biomedicine. Angew. Chem., Int. Edit 2020, 59, 15818–15833. 10.1002/anie.201916390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. B.; Song L. L.; Liu S. L.; Zhao S.; Jiang Q.; Ding B. Q. Atailored DNA Nanoplatform for Synergistic Rnai-/Chemotherapy of Multidrug-Resistant Tumors. Angew. Chem., Int. Edit 2018, 57, 15486–15490. 10.1002/anie.201809452. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Song L.; Liu Q.; Tian R.; Shang Y.; Liu F.; Liu S.; Zhao S.; Han Z.; Sun J.; et al. A Tubular DNA Nanodevice as a Sirna/Chemo-Drug Co-Delivery Vehicle for Combined Cancer Therapy. Angew. Chem., Int. Edit 2021, 60, 2594–2598. 10.1002/anie.202009842. [DOI] [PubMed] [Google Scholar]
- Wang J.; Li Y.; Nie G. Multifunctional Biomolecule Nanostructures for Cancer Therapy. Nat. Rev. Mater. 2021, 6, 766–783. 10.1038/s41578-021-00315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. G.; Blum N. T.; Lin J.; Shi J. J.; Zhang C.; Huang P. Chemotherapeutic Drug-DNA Hybrid Nanostructures for Anti-Tumor Therapy. Mater. Horiz. 2021, 8, 78–101. 10.1039/D0MH00715C. [DOI] [PubMed] [Google Scholar]
- Jiang D.; Ge Z.; Im H.-J.; England C. G.; Ni D.; Hou J.; Zhang L.; Kutyreff C. J.; Yan Y.; Liu Y.; et al. DNA Origami Nanostructures Can Exhibit Preferential Renal Uptake and Alleviate Acute Kidney Injury. Nat. Biomed. Eng. 2018, 2, 865–877. 10.1038/s41551-018-0317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q.; Ding F.; Zhang S. Y.; Li Q.; Liu X. G.; Song H. Y.; Zuo X. L.; Fan C. H.; Mou S.; Ge Z. L. Sequential Therapy of Acute Kidney Injury with a DNA Nanodevice. Nano Lett. 2021, 21, 4394–4402. 10.1021/acs.nanolett.1c01044. [DOI] [PubMed] [Google Scholar]
- Hu Q. Q.; Li H.; Wang L. H.; Gu H. Z.; Fan C. H. DNA Nanotechnology-Enabled Drug Delivery Systems. Chem. Rev. 2019, 119, 6459–6506. 10.1021/acs.chemrev.7b00663. [DOI] [PubMed] [Google Scholar]
- Krissanaprasit A.; Key C. M.; Pontula S.; LaBean T. H. Self-Assembling Nucleic Acid Nanostructures Functionalized with Aptamers. Chem. Rev. 2021, 121, 13797–13868. 10.1021/acs.chemrev.0c01332. [DOI] [PubMed] [Google Scholar]
- Li M.; Yin F. F.; Song L.; Mao X. H.; Li F.; Fan C. H.; Zuo X. L.; Xia Q. Nucleic Acid Tests for Clinical Translation. Chem. Rev. 2021, 121, 10469–10558. 10.1021/acs.chemrev.1c00241. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Y.; Li M.; Li Z. H.; Li Q.; Aldalbahi A.; Shi J. Y.; Wang L. H.; Fan C. H.; Zuo X. L. Recognizing Single Phospholipid Vesicle Collisions on Carbon Fiber Nanoelectrode. Sci. China: Chem. 2017, 60, 1474–1480. 10.1007/s11426-017-9036-0. [DOI] [Google Scholar]
- Li Y.; Lu H.; Qu Z. B.; Li M. Q.; Zheng H. R.; Gu P. L.; Shi J. Y.; Li J.; Li Q.; Wang L. H.; et al. Phase Transferring Luminescent Gold Nanoclusters Via Single-Stranded DNA. Sci. China: Chem. 2022, 65, 1212–1220. 10.1007/s11426-022-1238-2. [DOI] [Google Scholar]
- Yang F.; Zuo X. L.; Fan C. H.; Zhang X. E. Biomacromolecular Nanostructures-Based Interfacial Engineering: From Precise Assembly to Precision Biosensing. Natl. Sci. Rev. 2018, 5, 740–755. 10.1093/nsr/nwx134. [DOI] [Google Scholar]