Abstract
Incorporating measures of taxonomic diversity into research and management plans has long been a tenet of conservation science. Increasingly, active conservation programs are turning toward multispecies landscape and regional conservation actions, and away from single species approaches. This is both a reflection of changing trends in conservation science and advances in foundational technologies, including genomics and geospatial science. Multispecies approaches may provide more fundamental insights into evolutionary processes and equip managers with a more holistic understanding of the landscapes under their jurisdiction. Central to this approach are data generation and analyses which embrace and reflect a broad range of taxonomic diversity. Here, we examine the family-level phylogenetic breadth of the California Conservation Genomics Project (CCGP) based on family-level phylogenetic diversity (PD), family-level phylogenetic distinctness, and family richness. We place this in the context of the diversity present in California and compare it to the 35-plus years of genetic research compiled in the CaliPopGen Database. We found that the family-level PD in the CCGP reflected that of California very well, slightly overrepresenting chordates and underrepresenting arthropods, and that 42% of CCGP PD represented new contributions to genetic data for the state. In one focused effort, the CCGP was able to achieve roughly half the family-level PD studied over the last several decades. To maximize studied PD, future work should focus on arthropods, a conclusion that likely reflects the overall lack of attention to this hyperdiverse clade.
Keywords: biodiversity, CCGP, conservation genetics, landscape genetics
Introduction
In a world of limited resources for biodiversity conservation, a key component of successful conservation management is the identification of meaningful and achievable priorities. Myriad approaches have been proposed, ranging from degree of threat and endemism (Myers et al. 2000), to “keystone,” “indicator,” or “charismatic” species (Simberloff 1998), to phylogenetic distinctness (Vane-Wright et al. 1991; Margules and Pressey 2000; Redding and Mooers 2006). As a result, particularly in the field of conservation genetics and genomics, the bulk of conservation management actions are carried out in single species frameworks. For example, in a recent compilation of population genetics research from across the state of California, the mean number of species per publication was 1.3 (range: 1 to 33, SD = 1.7, data derived from Beninde et al. 2022), underscoring the species-oriented approach of most genetic research over the last several decades. In addition to precluding work on broad-scale evolutionary processes for a wide range of taxonomic groups, single-species assessments may also concentrate research and management funds to well-known or well-studied species, diverting attention from lesser-known species (Martín-López et al. 2011).
Consequently, many active conservation programs are shifting their focus from species-based conservation priorities to comparative multispecies, or ecosystem-based frameworks (Simberloff 1998; Margules and Pressey 2000; Groves et al. 2002; Baldwin et al. 2018). For example, the state of California recently launched the 30 × 30 initiative to conserve 30% of the state’s land and coastal waters by the year 2030 in order to protect and restore biodiversity writ large, while also preparing to mitigate the effects of climate change (California Natural Resources Agency 2022). In so doing, the state will put into action many of the research-driven recommendations of the last 2 decades by identifying landscapes that maximize biodiversity value, as yet unprotected diversity, evolutionary processes, and the potential for adaptation to climate change (e.g. Hampe and Petit 2005; Thomassen et al. 2011; Stein et al. 2014; Hamilton et al. 2022).
The California Conservation Genomics Project (CCGP, www.ccgproject.org), embodies the type of conservation-oriented research program that will allow conservation managers to achieve the goals of multispecies landscape-scale conservation. The project will generate whole genome data for 253 taxa (231 plant and animal species) across the varied geography of California to identify regions of high and low genomic diversity, corridors of, and barriers to, gene-flow, and areas of risk and potential for adaptation to climate change (Shaffer et al. 2022). In addition to its wide taxonomic and geographic breadth, the CCGP is also the first project of this scale committed to bridging the so-called research-implementation gap (Knight et al. 2008), ensuring that its data and analytical products are easily accessible to managers inform conservation actions (Fiedler et al. 2022). At the core of the CCGP’s multispecies framework are data generated on a per-species basis, which are then aggregated and analyzed as a comprehensive, unitary dataset. This represents the best of both worlds, in that both individual species and a cross-section of the entire state’s biodiversity benefit from the project. Such a large-scale approach is made possible by innovations in DNA sequencing technologies, and associated bioinformatic and computational advances, as well as the commitment of the state of California to biodiversity conservation (“EO N-82-20” 2020; California Natural Resources Agency 2022).
Here, we examine the extent to which the multispecies approach of the CCGP captures the family-level phylogenetic diversity (PD) present in the state of California using an explicitly phylogenetic, TimeTree-based approach. While PD is a metric more commonly applied at the species level, species-level datasets are incomplete for California. We therefore focus on family-level metrics of diversity. We calculated several measures of the fraction of the total family-level PD in California and compared the fraction of that diversity represented by the CCGP to that captured by 35 yr of genetics- and genomics-based research in the state collected in the CaliPopGen Database, a recent compilation of virtually all population genetic research in California from 1985 to 2020 (Beninde et al. 2022). These efforts allowed us to place the CCGP into an appropriate phylogenetic context for the state, identify taxonomic knowledge gaps in existing California biodiversity research, and provide recommendations for future efforts to increase conservation efficacy by maximizing phylogenetic coverage, across the state and beyond.
Methods
Plant and animal families of California
We used occurrence data from the Global Biodiversity Information Database (GBIF, www.gbif.org) to generate a comprehensive list of taxa that inhabit the terrestrial (including inland aquatic habitats) and coastal zones of California. To limit inferences to these regions of California, we first created a spatial mask by aggregating a polygon of the California state border (U.S. Geological Survey, National Geospatial Technical Operations Center 2022) with a polygon of the coastal zones of California (extending ~1,000 yards from the high tide line; Data Basin 2015; California Coastal Commission 2022) using the package raster v3.4-5 in the R Statistical Software v4.0.3 (R Core Team 2020). As the GBIF web search interface can only be spatially queried with rectangular extents, we specified the range geometry of the query as the extent of the aggregated polygon of the terrestrial and coastal zones created above (West: −124.5352, East: −114.1312, South: 32.5308, North: 42.00951). We further set parameters of the GBIF query to: “Coordinate uncertainly < 11600.0 m,” “Has geospatial issue = false,” “Occurrence status = present,” “Has coordinate = true.” As this rectangular spatial query includes occurrence records beyond the aggregated polygon of interest, we subsequently cropped all resulting GBIF records to the aggregated polygon using the raster package. Additionally, we removed records with greater than 10,000 m uncertainty and retained only those records from Kingdoms Animalia and Plantae with a family-level classification. We then tabulated the number of occurrences per unique species and retained only species with 10 or more occurrences.
We used GBIF’s Species Matching Tool (https://www.gbif.org/tools/species-lookup) to retrieve GBIF taxonomic classifications for all plant and animal species in CCGP species (N = 231) the CaliPopGen Database (N = 2,453).
Time-calibrated phylogeny
We first used TimeTree V5 (www.timetree.org, Kumar et al. 2022) to generate a time-calibrated phylogeny for all plant and animal species in California based on our curated list of taxa from GBIF. Because TimeTree will automatically substitute a related species if there are no data available for a specified taxon, we verified whether the families represented by the species in the resulting TimeTree phylogeny were still constrained to those known from California. If a TimeTree substitution resulted in a family in the tree that is not present in California, we removed it. We then pruned the TimeTree phylogeny to retain only 1 random species per family to represent the total familial diversity of taxa in California (“all-CA tree”). We generated 7 additional subtrees by pruning the all-CA tree using ape v5.5 (Paradis and Schliep 2019) which included representatives from: 1) all CCGP families (“CCGP”), 2) families in CCGP but not present in CaliPopGen (“CCGP.novel”), 3) all CaliPopGen families (“CaliPopGen”), 4) families in CaliPopGen, but not in CCGP (“CaliPopGen.novel”), and 3 “missing” subtrees comprised of species representative of families: 5) not present in CCGP (“missing.CCGP”), 6) not present in CaliPopGen (“missing.CaliPopGen”), and 7) not present in either CCGP or CaliPopGen (“missing.both”).
Diversity metrics
To compare the phylogenetic composition of the CCGP to that of all California and the CaliPopGen Database, we generated 3 metrics for each tree: Faith’s PD (Faith 1992), the sum of equal splits phylogenetic distinctness (ES, Redding and Mooers 2006), and family richness. PD is expressed as the sum of all branch lengths in each tree (Faith 1992), in our case measured as years. ES reflects a species’ evolutionary history and is calculated as the total branch length for each species where shared branches are equally split among descendant lineages (Redding and Mooers 2006; Vellend et al. 2011). We calculated ES values for each species in the all-CA tree and summed the ES across all tips (or subset of tips) to generate a single measure of family-level phylogenetic distinctness for each tree and subtree. PD and ES values were calculated using the R package picante v1.8.2 (Kembel et al. 2010). We calculated family richness of each tree as the total number of families present in each dataset (i.e. not based upon the families present in each tree, because some families are not represented in the trees due to incomplete phylogenetic data). We expressed the subtree family diversity as the proportion of the PD or ES out of the all-CA tree, and the proportion of family richness for each subset out of the total number of families in California based on the GBIF dataset.
To assess how future work might contribute to the phylogenetic breadth of datasets for California, we quantified missing PD in several ways, focusing on phyla to provide a broad-scale perspective. We first examined which phyla were over- or underrepresented in the context of phylogenetic distinctness and family diversity within the CCGP dataset. For each phylum in each subset tree, we calculated the proportion of total ES for that phylum over the total ES for that tree. Of the taxa “missing” from CCGP or CaliPopGen (present in the all-CA dataset, and absent from CCGP or CaliPopGen, respectively), we calculated the total missing ES for both phyla and orders; and the fraction out of the total missing ES (summed across all phyla with missing data) for each phylum.
Results
Plant and animal families of California
In our initial GBIF search (GBIF.org 2022), we identified 6,255,475 California occurrence records across the tree of life and retained 4,361,218 occurrences after applying all filters. This yielded 12,951 species across 1,305 families of animals and plants in the state (Table 1). There were 231 unique plant and animal species included in the CCGP, representing 145 genera from 109 families (Table 2, Supplementary Table S1), and 411 unique plant and animal species representing 308 genera from 215 families in CaliPopGen (Supplementary Table S2).
Table 1.
Taxonomic diversity of California occurrence data for plants and animals derived from GBIF occurrence data, and the number retained at each level of classification in the final tree (before and after the “/,” respectively). We thinned the tree to 1 species (and therefore 1 genus) per family, and so constrained the number of genera and species in the tree. Values in parentheses for n.genera and n.species reflect the number of taxa for which TimeTree returned molecular time estimates before thinning to 1 representative species per family.
| Kingdom | n.phyla | n.orders | n.families | n.genera | n.species |
|---|---|---|---|---|---|
| Animalia | 14/9 | 206/159 | 1,030/595 | 3,429/(1,372) | 6,655/(2,033) |
| Plantae | 7/6 | 96/85 | 273/240 | 1,591/(1,230) | 6,296/(2,836) |
| Total | 21/15 | 302/244 | 1,305/835 | 5,020/(2,602) | 12,951/(4,869) |
Table 2.
Taxonomic diversity of species in the CCGP based on GBIF taxonomy, and the number retained at each level of classification in the final tree (before and after the “/,” respectively). We thinned the tree to 1 species (and therefore 1 genus) per family, and so constrained the number of genera and species in the tree. Values in parentheses for n.genera and n.species reflect the number of taxa for which TimeTree returned molecular time estimates before thinning to 1 taxon per family.
| Kingdom | n.phyla | n.orders | n.families | n.genera | n.species |
|---|---|---|---|---|---|
| Animalia | 5/5 | 47/42 | 81/74 | 108/(86) | 161/(95) |
| Plantae | 3/2 | 23/22 | 28/27 | 37/(35) | 70/(40) |
| Total | 8/7 | 70/64 | 109/101 | 145/(121) | 231/(135) |
Time-calibrated phylogeny
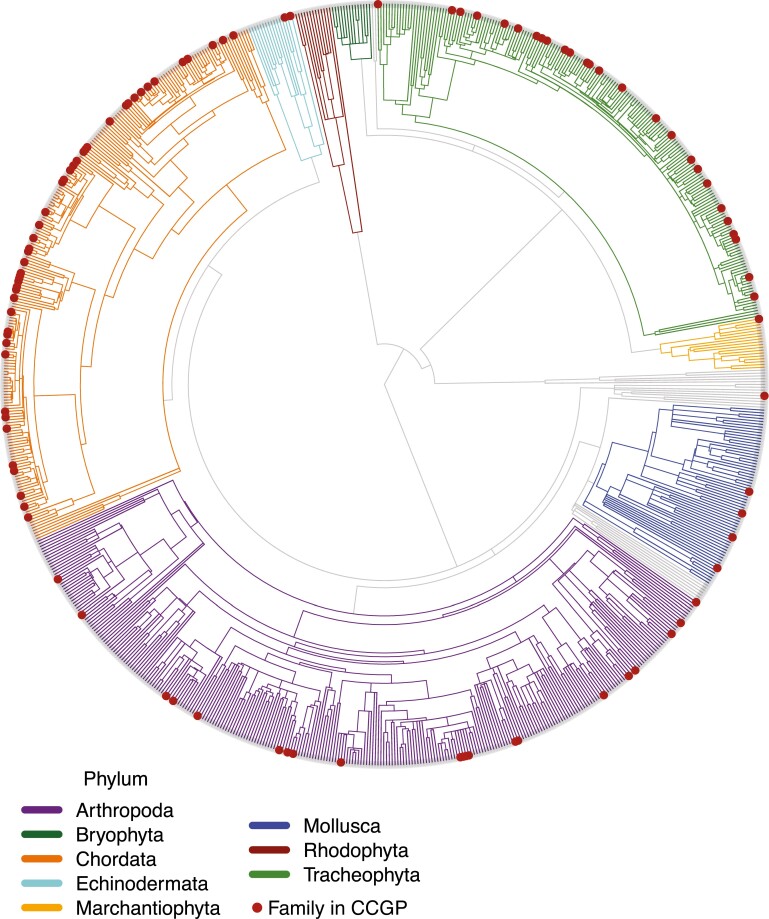
TimeTree returned a tree of 4,888 out of 12,951 species (38%) for the all-CA dataset. Missing species are those that did not have molecular clock estimates in the TimeTree database. We further removed 43 tips representing 23 non-California families. After pruning the phylogeny to retain only 1 random species per family, the tree included 835 families (out of the original 1,305 families based on the GBIF occurrence data), and these 835 families are shown as a TimeTree in Fig. 1. The CCGP tree included 101 out of 109 families (Table 2, Fig. 1), and the CaliPopGen tree included 172 of 215 families (Supplementary Table S2).
Fig. 1.
Phylogeny of all California families, based on GBIF occurrence data. Taxa in the CCGP project (red circles at tips) encompass for 15.6% of PD, 8.9% of phylogenetic distinctness, and 12.1% of all families. Gray branches in the tree are from phyla with less than 10 families in the tree.
Diversity
Summaries of family-level PD and ES values summed for each subtree, and family richness are reported in Table 3, and total family richness and ES for phyla present and absent from CCGP and CaliPopGen are reported in Table 4. The phyla and orders with taxa not present in CCGP are reported in Supplementary Table S3 and are ranked by total ES for each phylum, then by the ES for each order. Higher values of ES and PD for any given tip or group represent relatively more PD or distinctness. The sum of the per-tip ES across all species in the all-CA tree was 146,631.8, and PD of the all-CA tree was 146,631.8 (the sum of the ES across all tips should equal the PD of the tree, Redding and Mooers 2006). Families present in CCGP comprised 8.9% of the total ES, 15.6% of the total PD, and 12.1% of the total family richness, while the CaliPopGen species comprised 18.2% of total ES, 25.9% of total PD, and 20.5% of family richness (Table 3). 78.1% of the total ES, 82.0% of the total PD, and 74.5% of family richness of the all-CA tree was missing from the combined CCGP and CaliPopGen subsets (Table 3). Among CCGP taxa, arthropods (Phylum: Arthropoda) were the group with both the highest total proportion of ES and the highest relative missingness (Table 4).
Table 3.
Family-level phylogenetic distinctness (ES) and diversity (PD) in the CCGP and the CaliPopGen Database for taxa for which there were molecular time estimates available in TimeTree.
| Subset | ES | Prop. ES | PD | Prop. PD | No. families | Prop. family richness |
|---|---|---|---|---|---|---|
| All-CA | 146,631.8 | 1.000 | 146,631.8 | 1.000 | 835 | 1.000 |
| CCGP | 13,061.21 | 0.089 | 22,829.67 | 0.156 | 101 | 0.121 |
| CCGP.novel | 5,550.66 | 0.038 | 12,386.20 | 0.084 | 42 | 0.050 |
| CaliPopGen | 26,794.87 | 0.182 | 38,032.76 | 0.259 | 171 | 0.205 |
| CaliPopGen.novel | 19,112.081 | 0.130 | 30,065.26 | 0.205 | 112 | 0.134 |
| missing.CCGP | 133,605.609 | 0.911 | 136,034.75 | 0.928 | 734 | 0.879 |
| missing.CaliPopGen | 120,044.188 | 0.819 | 124,803.59 | 0.851 | 664 | 0.795 |
| missing.both | 114,493.528 | 0.781 | 120,188.15 | 0.820 | 622 | 0.745 |
The total number of families in California was 1,305. The total ES and PD of the all-CA tree are both 146,631.8 (total ES = total PD in the all-CA tree). Proportions are expressed as the value of the subtree over that of the all-CA tree. In the table, ES = sum equal splits values among taxa in each subset; PD = the total family-level PD for each subset; No. families = number of families for each subset. CCGP.novel and CaliPopGen.novel include taxa exclusive to those groups. missing.CCGP, missing.CaliPopGen, and missing.both include taxa that are not found in those groups.
Table 4.
Family richness and family-level phylogenetic distinctness of phyla in California and the CCGP.
| Phylum | No. families in CA | No. families in all-CA tree | All-CA: total ES | All-CA: group % of total ES | CCGP: total ES | CCGP: group % of total ES | Group % missing out of total missing |
|---|---|---|---|---|---|---|---|
| Kingdom Animalia | |||||||
| Arthropoda | 446 | 281 | 57,389.96 | 39.1% | 4,038.33 | 31.0% | 39.9% |
| Chordata | 238 | 216 | 21,694.03 | 14.8% | 4,003.52 | 30.7% | 13.2% |
| Mollusca | 248 | 65 | 18,241.78 | 12.4% | 1,324.53 | 10.2% | 12.7% |
| Echinodermata | 25 | 17 | 4,713.07 | 3.2% | 495.61 | 3.8% | 3.2% |
| Cnidaria | 24 | 8 | 4,199.38 | 2.9% | 545.69 | 4.2% | 2.7% |
| Annelida | 16 | 5 | 2,662.37 | 1.8% | — | 0.0% | 2.0% |
| Sipuncula | 1 | 1 | 665.50 | 0.5% | — | 0.0% | 0.5% |
| Brachiopoda | 5 | 1 | 583.82 | 0.4% | — | 0.0% | 0.4% |
| Phoronida | 1 | 1 | 583.82 | 0.4% | — | 0.0% | 0.4% |
| Bryozoa | 8 | 0 | — | — | — | — | — |
| Ctenophora | 1 | 0 | — | — | — | — | — |
| Nemertea | 3 | 0 | — | — | — | — | — |
| Platyhelminthes | 6 | 0 | — | — | — | — | — |
| Porifera | 8 | 0 | — | — | — | — | — |
| Kingdom Plantae | |||||||
| Tracheophyta | 190 | 188 | 19,861.00 | 13.5% | 2,428.40 | 18.6% | 13.0% |
| Rhodophyta | 28 | 13 | 5,363.44 | 3.7% | — | 0.0% | 4.0% |
| Marchantiophyta | 19 | 18 | 4,065.50 | 2.8% | 213.01 | 1.6% | 2.9% |
| Chlorophyta | 7 | 0 | 3,535.23 | 2.4% | — | 0.0% | 2.6% |
| Bryophyta | 28 | 14 | 2,315.75 | 1.6% | — | 0.0% | 1.7% |
| Anthocerotophyta | 2 | 2 | 780.04 | 0.5% | — | 0.0% | 0.6% |
| Charophyta | 1 | 5 | — | — | — | — | — |
The family richness of all plant and animal species in California (“No. families in CA”) is greater than that of the all-CA tree (“No. families in all-CA tree”) because some families identified in the GBIF occurrence data did not have molecular time estimates available in TimeTree. The “group % missing out of total missing” is the percentage of that group’s ES that is not covered by CCGP taxa out of the non-CCGP ES totaled across all groups.
Discussion
Advancing biodiversity conservation in the state of California
The CCGP has been successful in prioritizing research across a broad swath of the flora and fauna of California, including 12.1% of families, 15.6% of total PD, and 8.9% of total phylogenetic distinctness (Table 3). In comparison, the entire 35-yr history of population genetics research in California, summarized in the CaliPopGen Database (Beninde et al. 2022), covers 20.4%, 25.9%, and 18.2% of CA families, PD, and phylogenetic distinctness, respectively (Table 3), less than double the effect of the concerted effort on the part of the CCGP to prioritize diversity over the 5-yr time course of the project. Further, 42% of CCGP families (the total CCGP.novel ES out of the total ES for CCGP) have no representatives among published studies in the CaliPopGen Database and thereby contribute a substantial amount of phylogenetically novel genomic data for California.
While the CaliPopGen Database has considerably greater taxonomic coverage than CCGP, the historical data are overwhelmingly based upon microsatellites, AFLPs, mtDNA, and single or several gene nDNA studies, and are often limited to relatively small portions of each species’ geographic range (Beninde et al. in press). In contrast, the CCGP is generating whole genome sequencing data for individuals across the full extent of each species’ range in California (Shaffer et al. 2022), a dataset which delivers many benefits in applied conservation (reviewed extensively in e.g. Allendorf et al. 2010; Avise 2010; Benestan et al. 2016; Formenti et al. 2022). In the simplest case, genomic data can provide high accuracy estimates of genetic parameters relevant to conservation practitioners; in the most complex, it can help identify populations that might be most responsive to climate change. Taken together, the CCGP’s broad taxonomic approach coupled with genome-scale data should facilitate a much more complete, synthetic understanding of the state of biodiversity health across California, enabling conservation practitioners to move from data to action across a multispecies landscape framework.
Remaining work to be done
Based only on georeferenced GBIF occurrence data, there are 12,951 species across 1,305 families of plants and animals present in California (Table 1). While this almost certainly underestimates total species richness, as many species are understudied and underreported to GBIF, if taken at face value the CCGP encompasses 9% to 16% of family-level PD in California, depending on the metric used. Combining all previous research in California plus the CCGP, 75% to 82% of family-level diversity lacks genetic or genomic study (“missing.both,” Table 3). Among the phyla present in the CCGP, the total family phylogenetic distinctness, expressed as the sum of ES across all tips in each phylum, is similar in rank order to that of all families across California (Table 4), reflecting the project’s commitment to phylogenetic representation. The proportions of each phylum in CCGP relative to all of California is similar, with a few notable exceptions. Some groups, particularly chordates, are relatively overrepresented (30.7% of CCGP total ES vs. 14.8% of California’s total ES), and vascular plants (Tracheophyta, 18.6% vs. 13.5%), while others (e.g. arthropods, mollusks) are underrepresented. Perhaps unsurprisingly because of their vast global diversity, arthropods comprise the largest portion of missing phylogenetic distinctness in the tree (39.9% of missing ES; 31.0% of CCGP ES vs. 39.1% of total ES, Table 4, Supplementary Table S3). Insects alone are responsible for pollinating 75% of flowering plant species (National Research Council 2007), and their numbers appear to be declining globally (Sánchez-Bayo and Wyckhuys 2019); insect conservation will be crucial to facilitating healthy ecosystems and food security, now and into the future. Ongoing efforts of other broad-scale projects such as the i5k initiative (Robinson et al. 2011) and USDA-ARS Ag100Pest Initiative (Childers et al. 2021) are attempting to fill this knowledge gap by delivering genome-scale data for thousands of arthropod species, although much of this effort is focused on reference genomes exclusively. Additionally, the disparity in ES and PD between over- and underrepresented groups is likely larger than we calculate here because our family-based approach underestimates ES and PD of more species-rich families relative to that of families with fewer species. Future contributions could focus on Coleopterans (beetles) and Hemipterans (true bugs), which together account for over 10% of unstudied PD in the state (Supplementary Table S3). Further, there are major taxonomic groups which remain unexplored relative to their total taxonomic diversity, including red and green algae (Rhodophyta and Chlorophyta, respectively), which play important functional roles as primary producers in aquatic ecosystems (Martin et al. 2013) and as ecosystem engineers (Pezzolesi et al. 2017).
We caution that our analysis of PD is based, by necessity, upon those species and families for which data to produce a time-calibrated phylogeny are available. Out of the 21 plant and animal phyla present in California, TimeTree was unable to assign any substitution for 6 (Animalia: Bryozoa, Ctenophora, Nemertea, Platyhelminthes, and Porifera; Plantae: Charophyta), and a total of 470 families (36%) were missing from the final phylogeny across all plants and animals (Tables 1 and 4). It is likely that some of these groups simply lack time-calibrated phylogenies and are therefore not represented in the tree despite ongoing genetics research. However, many of these species may be very poorly studied and therefore constitute novel research and conservation opportunities.
Conclusion
As genomic data become more widely accessible, researchers and conservation practitioners should shoulder the responsibility to quickly identify and fill gaps in our coverage of biodiversity. Here, we present several metrics that can be used to both assess the diversity that is already known to science and prioritize that which is absent from our datasets. The remarkable distribution of diversity that the CCGP has been able to include in a relatively short time speaks to the utility of concerted, multispecies-oriented project planning, which clearly can outpace single-species research. This approach has the potential to maximize the financial resources available for conservation, and minimize the research-implementation gap, resulting in greater conservation success and landscape-level recovery.
Supplementary Material
Acknowledgments
We thank Dr. Jack Craig for suggestions regarding methodological approaches, and Dr. Robert Cooper for insightful comments on early drafts of this manuscript. We also thank the external reviewer for their comments which helped to clarify parts of this work, as well as Journal of Heredity Associate Editor for their guidance.
Contributor Information
Erin Toffelmier, UCLA La Kretz Center for California Conservation Science, Institute of the Environment and Sustainability, University of California, Los Angeles, Los Angeles, CA, United States; Department of Ecology and Evolutionary Biology, University of California, Los Angeles, Los Angeles, CA, United States.
Joscha Beninde, UCLA La Kretz Center for California Conservation Science, Institute of the Environment and Sustainability, University of California, Los Angeles, Los Angeles, CA, United States; IUCN WCPA Connectivity Conservation Specialist Group, Gland, Switzerland.
H Bradley Shaffer, UCLA La Kretz Center for California Conservation Science, Institute of the Environment and Sustainability, University of California, Los Angeles, Los Angeles, CA, United States; Department of Ecology and Evolutionary Biology, University of California, Los Angeles, Los Angeles, CA, United States.
Funding
This work was funded by a grant provided to the University of California by the State of California, State Budget Act of 2019 [UC Award ID RSI-19-690224].
Data availability
The raw California occurrence dataset generated through GBIF.org is available through GBIF (https://doi.org/10.15468/dl.axfuhb).
References
- Allendorf FW, Hohenlohe P, Luikart G. Genomics and the future of conservation genetics. Nat Rev Genet. 2010;11(10):697–709. [DOI] [PubMed] [Google Scholar]
- Avise JC. Perspective: Conservation genetics enters the genomics era. Conserv Genet. 2010;11(2):665–669. [Google Scholar]
- Baldwin RF, Trombulak SC, Leonard PB, Noss RF, Hilty JA, Possingham HP, Scarlett L, Anderson MG. The future of landscape conservation. BioScience. 2018;68(2):60–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benestan LM, Ferchaud A, Hohenlohe PA, Garner B, Naylor GJP, Baums IB, Schwartz MK, Kelley JL, Luikart G. Conservation genomics of natural and managed populations: Building a conceptual and practical framework. Mol Ecol. 2016;25(13):2967–2977. [DOI] [PubMed] [Google Scholar]
- Beninde J, Toffelmier EM, Andreas A, Nishioka C, Slay M, Soto A, Bueno JP, Gonzalez G, Pham HV, Posta M, et al. CaliPopGen: a genetic and life history database for the fauna and flora of California. Sci Data. 2022;9(1):380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beninde J, Toffelmier E, Shaffer HB. A brief history of population genetic research in California and an evaluation of its utility for conservation decision-making. J Hered. 2022;113(6):604-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- California Coastal Commission. DS0990_20140326: coastal zone boundary [ds990] GIS dataset [Map]. 2022. https://map.dfg.ca.gov/metadata/ds0990.html. Accessed 01 July 2022.
- California Natural Resources Agency. Pathways to 30 × 30 California. 2022. https://canature.maps.arcgis.com/sharing/rest/content/items/8da9faef231c4e31b651ae6dff95254e/data. Accessed 01 July 2022.
- Childers AK, Geib SM, Sim SB, Poelchau MF, Coates BS, Simmonds TJ, Scully ED, Smith TPL, Childers CP, Corpuz RL, et al. The USDA-ARS Ag100Pest initiative: high-quality genome assemblies for agricultural pest arthropod research. Insects. 2021;12(7):626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Data Basin. California Coastal Zone Map [Map]. 2015. https://databasin.org/datasets/ece6ae2d026b43959cfa11cceb2c07ac/. Accessed 01 July 2022.
- EO N-82-20. State of California. 2020. https://www.library.ca.gov/wp-content/uploads/GovernmentPublications/executive-order-proclamation/40-N-82-20.pdf. Accessed 01 July 2022.
- Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61(1):1–10. [Google Scholar]
- Fiedler PL, Erickson B, Esgro M, Gold M, Hull JM, Norris J, Shapiro B, Westphal MF, Toffelmier E, Shaffer HB. Seizing the moment: the opportunity and relevance of the California Conservation Genomics Project to state and federal conservation policy. J Hered. 2022;113(6):589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formenti G, Theissinger K, Fernandes C, Bista I, Bombarely A, Bleidorn C, Ciofi C, Crottini A, Godoy JA, Höglund J, et al. The era of reference genomes in conservation genomics. Trends Ecol Evol. 2022;37(3):197–202. [DOI] [PubMed] [Google Scholar]
- GBIF.org. GBIF occurrence download. 2022. https://www.gbif.org/occurrence/download/0390831-210914110416597. Accessed 15 July 2022. [Google Scholar]
- Groves CR, Jensen DB, Valutis LL, Redford KH, Shaffer ML, Scott JM, Baumgartner JV, Higgins JV, Beck MW, Anderson MG. Planning for Biodiversity Conservation: putting Conservation Science into Practice: a seven-step framework for developing regional plans to conserve biological diversity, based upon principles of conservation biology and ecology, is being used extensively by the nature conservancy to identify priority areas for conservation. BioScience. 2002;52(6):499–512. [Google Scholar]
- Hamilton H, Smyth RL, Young BE, Howard TG, Tracey C, Breyer S, Cameron DR, Chazal A, Conley AK, Frye C, et al. Increasing taxonomic diversity and spatial resolution clarifies opportunities for protecting US imperiled species. Ecol Appl. 2022;32(3):e2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe A, Petit RJ. Conserving biodiversity under climate change: the rear edge matters. Ecol Lett. 2005;8(5):461–467. [DOI] [PubMed] [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26(11):1463–1464. [DOI] [PubMed] [Google Scholar]
- Knight AT, Cowling RM, Rouget M, Balmford A, Lombard AT, Campbell BM. Knowing but not doing: selecting priority conservation areas and the research-implementation gap. Conserv Biol. 2008;22(3):610–617. [DOI] [PubMed] [Google Scholar]
- Kumar S, Suleski M, Craig JM, Kasprowicz AE, Sanderford M, Li M, Stecher G, Hedges SB. TimeTree 5: an expanded resource for species divergence times. Mol Biol Evol. 2022;39(8):msac174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margules CR, Pressey RL. Systematic conservation planning. Nature. 2000;405(6783):243–253. [DOI] [PubMed] [Google Scholar]
- Martin S, Charnoz A, Gattuso J-P. Photosynthesis, respiration and calcification in the Mediterranean crustose coralline alga Lithophyllum cabiochae (Corallinales, Rhodophyta). Eur J Phycol. 2013;48(2):163–172. [Google Scholar]
- Martín-López B, González JA, Montes C. The pitfall-trap of species conservation priority setting. Biodivers Conserv. 2011;20(3):663–682. [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, Da Fonseca GA, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403(6772):853–858. [DOI] [PubMed] [Google Scholar]
- National Research Council. Status of pollinators in North America. Washington (DC): National Academies Press; 2007. [Google Scholar]
- Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35(3):526–528. [DOI] [PubMed] [Google Scholar]
- Pezzolesi L, Falace A, Kaleb S, Hernandez-Kantun JJ, Cerrano C, Rindi F. Genetic and morphological variation in an ecosystem engineer, Lithophyllum byssoides (Corallinales, Rhodophyta). J Phycol. 2017;53(1):146–160. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing; 2020. https://www.R-project.org/. Accessed 01 July 2022. [Google Scholar]
- Redding DW, Mooers A. Incorporating evolutionary measures into conservation prioritization. Conserv Biol. 2006;20(6):1670–1678. [DOI] [PubMed] [Google Scholar]
- Robinson GE, Hackett KJ, Purcell-Miramontes M, Brown SJ, Evans JD, Goldsmith MR, Lawson D, Okamuro J, Robertson HM, Schneider DJ. Creating a buzz about insect genomes. Science. 2011;331(6023):1386. [DOI] [PubMed] [Google Scholar]
- Sánchez-Bayo F, Wyckhuys KAG. Worldwide decline of the entomofauna: a review of its drivers. Biol Conserv. 2019;232:8–27. [Google Scholar]
- Shaffer HB, Toffelmier E, Corbett-Detig RB, Escalona M, Erickson B, Fiedler P, Gold M, Harrigan RJ, Hodges S, Luckau TK, et al. Landscape genomics to enable conservation actions: the California Conservation Genomics Project. J Hered. 2022;113(.6 ):577–588. [DOI] [PubMed] [Google Scholar]
- Simberloff D. Flagships, umbrellas, and keystones: is single-species management passé in the landscape era? Biol Conserv. 1998;83(3):247–257. [Google Scholar]
- Stein BA, Glick P, Edelson N, Staudt A. (eds). Climate-smart conservation: putting adaption principles into practice. Washington (DC): National Wildlife Federation; 2014. [Google Scholar]
- Thomassen HA, Fuller T, Buermann W, Milá B, Kieswetter CM, Jarrín-V P, Cameron SE, Mason E, Schweizer R, Schlunegger J, et al. Mapping evolutionary process: a multi-taxa approach to conservation prioritization. Evol Appl. 2011;4(2):397–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Geological Survey, National Geospatial Technical Operations Center. USGS National Boundary Dataset (NBD) in California 20220624 State or Territory Shapefile [Map]. 2022. https://www.sciencebase.gov/catalog/item/59fa9f5ae4b0531197affb17. Accessed 01 July 2022.
- Vane-Wright RI, Humphries CJ, Williams PH. What to protect? Systematics and the agony of choice. Biol Conserv. 1991;55(3):235–254. [Google Scholar]
- Vellend M, Cornwell WK, Magnuson-Ford K, Mooers AØ. Measuring phylogenetic biodiversity. In: Magurran AE, McGill BJ, editors. Biological diversity: frontiers in measurement and assessment. Oxford (UK): Oxford University Press; 2011. p. 194–207. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw California occurrence dataset generated through GBIF.org is available through GBIF (https://doi.org/10.15468/dl.axfuhb).